Whole-genome sequencing (WGS) is a highly sensitive method for identifying genetic relatedness and transmission of Clostridioides difficile strains. Previous studies suggest that as few as 3 core genome single-nucleotide variants (SNVs) discriminate between genetically distinct isolates. Because a single C. difficile colony is selected from culture for WGS, significant within-host genetic diversity could preclude identification of transmission events.
KEYWORDS: Clostridium difficile, genomics, pediatrics, transmission, whole-genome sequencing
ABSTRACT
Whole-genome sequencing (WGS) is a highly sensitive method for identifying genetic relatedness and transmission of Clostridioides difficile strains. Previous studies suggest that as few as 3 core genome single-nucleotide variants (SNVs) discriminate between genetically distinct isolates. Because a single C. difficile colony is selected from culture for WGS, significant within-host genetic diversity could preclude identification of transmission events. To evaluate the likelihood of missed transmission events using WGS of single colonies from culture, we examined within-host genetic diversity among C. difficile isolates collected from children. We performed WGS using an Illumina MiSeq instrument on 8 C. difficile colonies randomly selected from each culture performed on stool collected from 10 children (8 children diagnosed with C. difficile infection and 2 children with asymptomatic carriage); 77/80 (96%) isolate sequences were successfully assembled. Among 8/10 (80%) children, all isolates were the same sequence type (ST). The other 2 children each had mixed infection with two STs, although one ST predominated. Among 9/10 (90%) children, isotypic isolates differed by ≤2 SNVs; an isotypic isolate in the remaining child differed by 3 to SNVs relative to the other isolates from that child. Overall, among the 77 isolates collected from 10 stool cultures, 74/77 (96%) were clonal (i.e., same ST and ≤2 core genome SNVs) to other isolates in stool culture. In summary, we identified rare C. difficile within-host genetic diversity in children, suggesting that WGS of a single colony from stool is likely to appropriately characterize isolate clonality and putative transmission events in the majority of cases.
INTRODUCTION
Clostridioides (Clostridium) difficile is the most common health care-associated pathogen in the United States, causing approximately 500,000 C. difficile infections (CDI) and 30,000 deaths annually among U.S. adults (1). Recent investigation of C. difficile transmission using whole-genome sequencing (WGS) has demonstrated that acquisition from other hospitalized patients with CDI is uncommon among both adults (2, 3) and children (4); other common reservoirs of infection are likely but remain unidentified. WGS is the preferred method for tracking C. difficile because of its exceptional sensitivity in discriminating strain genetic relatedness and identifying putative transmission events (2). Prior work, based on analysis of C. difficile within-host microevolution in adults with CDI relapses, suggests that as few as 3 core genome single-nucleotide variants (SNVs) discriminate between genetically distinct isolates collected within 4 months of each other (2). This microevolutionary analysis also provided an estimate of the expected within-host genetic diversity (i.e., number of SNVs) of isotypic strains isolated from the same stool (2).
Using multilocus sequence typing (MLST), which is significantly less discriminatory than WGS, multiple distinct C. difficile sequence types (ST) are found in stool from the same CDI in 7% of adult patients, suggesting that mixed infection occurs (5). These findings have not yet been confirmed in children. Although the expected number of SNVs among isotypic strains from the same CDI has been estimated based on measurements of C. difficile within-host microevolutionary rate in adults with CDI relapses (2), these estimates have not yet been confirmed. WGS is typically performed on a single isolate randomly selected from stool culture. Thus, the presence of significant within-host C. difficile genetic diversity could preclude identification of a putative transmission event if the transmitted strain is not the one selected for WGS. To evaluate the likelihood of missed C. difficile transmission events in children using WGS of single colonies from stool culture, we examined the within-host genetic diversity among C. difficile isolates collected from children.
MATERIALS AND METHODS
Patients and setting.
Children older than 12 months of age who were either diagnosed with CDI (diarrhea and positive stool tcdB [toxin B gene] PCR) or identified as asymptomatic C. difficile carriers (positive stool tcdB PCR but without diarrhea) at the Ann & Robert H. Lurie Children’s Hospital of Chicago were enrolled into unrelated studies. From these cohorts, we randomly selected stools from 10 children (8 children with CDI and 2 children with asymptomatic C. difficile carriage) for the present study. We randomly selected stools with heavy growth of C. difficile by culture to permit selection of several distinct C. difficile colonies for further studies (see below). The Lurie Children’s Hospital Institutional Review Board approved this study.
C. difficile isolation and toxin testing.
Stools that tested positive in the clinical microbiology laboratory by the Xpert C. difficile tcdB PCR (Cepheid, Sunnyvale, CA) were aliquoted and stored at −80°C for further testing. Stools from the 10 study subjects were cultured anaerobically onto taurocholate-cycloserine-cefoxitin-fructose agar (TCCFA) in an anaerobic chamber and incubated at 37°C for 48 h, as previously described (6). Eight distinct C. difficile colonies, identified based on typical morphology, were randomly selected from each of the 10 TCCFA plates and subcultured onto blood agar plates. Stool underwent testing for C. difficile toxin using the Quik Chek Complete (QCC; Techlab, Blacksburg, VA) and the Bartels Clostridium difficile cytotoxicity assay (Trinity Biotech, Buffalo, NY), both per the manufacturer’s instructions. All stools were tested for C. difficile toxin after a single freeze-thaw cycle, with the exception of that from patient 8; this stool underwent toxin testing after multiple freeze-thaw cycles.
Whole-genome sequencing and bioinformatics.
Genomic DNA was extracted from C. difficile isolates using the BiOstic bacteremia DNA isolation kit (Qiagen, Inc., Germantown, MD). Paired-end sequencing libraries were prepared using the Nextera XT DNA library prep kit (Illumina, San Diego, CA), and WGS was performed using the Illumina MiSeq platform to produce 300-bp paired-end reads, as previously described (4), De novo genome assembly was performed using SPAdes (v3.9.1; http://cab.spbu.ru/software/spades/) (7). In silico multilocus sequence typing (MLST) (8) was performed using PubMLST (https://pubmlst.org/cdifficile/), which permitted isolate sequence type (ST) and clade assignment.
Within-host C. difficile isolate genetic diversity was assessed by comparing the STs of the isolates from each patient. Among strains identified as the same ST within each patient (i.e., isotypic isolates), core genome relatedness was determined by performing pairwise comparisons of SNVs as adapted from methods described by Eyre et al. (2) and that we have previously applied (4). Illumina reads were trimmed and filtered for low-quality bases and adapter sequences using Trimmomatic v0.36 (9). Reads were then aligned to the chromosomal sequence of the clade-specific reference strain using Stampy (10) (v1.0.29) with an expected substitution rate setting of 0.01. The reference strain used for each isolate alignment was based on the major C. difficile clade assignment as determined by MLST, as follows: clade 1, reference sequence 630 (GenBank accession number AM180355.1); clade 2, reference sequence R20291 (GenBank accession number FN545816.1). SNVs relative to the reference were called using the mpileup function of samtools (v0.1.19-44428cd) with the following options: -E (recalculate extended BAQ), -M 0 (cap mapping quality at 0), -Q 25 (skip bases with BAQ less than 25), -q 30 (skip alignments with mapQ less than 30), -m 2 (minimum gapped reads for indel candidates of 2), -D (output per-sample DP in BCF), -S (output per-sample strand bias P value in BCF), and -g (generate BCF output). SNVs were filtered if they failed to meet one or more of the following criteria: minimum SNV quality score of 200; minimum read consensus of 75%; minimum of 5 reads covering the SNV position; maximum of × the median read depth of the total alignment; minimum of 1 read in either direction covering the SNV position; homozygous under the diploid model; and not within a repetitive region as determined by BLAST alignment of fragments of the clade-specific reference strain sequence against itself. For each strain, the clade-specific reference strain sequence was used as the base sequence. Any positions with SNVs that passed the above filters were changed to the SNV base. Any positions with SNVs that did not pass the above filters were changed to a missing base character. Any non-SNV position with coverage of fewer than 5 reads was changed to a missing base character. After filtering, positions with a base in less than 100% of all genomes in the clade were excluded (i.e., included only the core genome). Isolates were considered to meet the clonality definition if they were the same ST and differed by ≤2 core genome SNVs from each other, as previously described (2).
Within-host accessory genome relatedness was also assessed among isotypic isolates within each patient using bioinformatics resources as previously described (11). The core genome was determined for each set of isotypic isolates from each patient using Spine (v0.3.2; http://vfsmspineagent.fsm.northwestern.edu/cgi-bin/spine.cgi) (12). Using AGEnt (v0.3.1; http://vfsmspineagent.fsm.northwestern.edu/cgi-bin/agent.cgi), accessory genomic elements were designated as those sequences in each isolate that were not identified as core genome by Spine (12). The distributions of individual accessory genomic elements among each set of isotypic isolates were determined using ClustAGE (v0.8.1; http://vfsmspineagent.fsm.northwestern.edu/cgi-bin/clustage.cgi) (13). Accessory genomic element distributions among isotypic isolates from the same patient were manually verified by alignment of sequencing reads to the element sequences using Stampy (v1.0.29) (10) and by visualization using Tablet (v1.17.08.17) (14).
Data availability.
This whole-genome shotgun project (BioProject PRJNA518899) has been deposited at DDBJ/ENA/GenBank under the accession numbers SEOS00000000 to SERQ00000000.
RESULTS
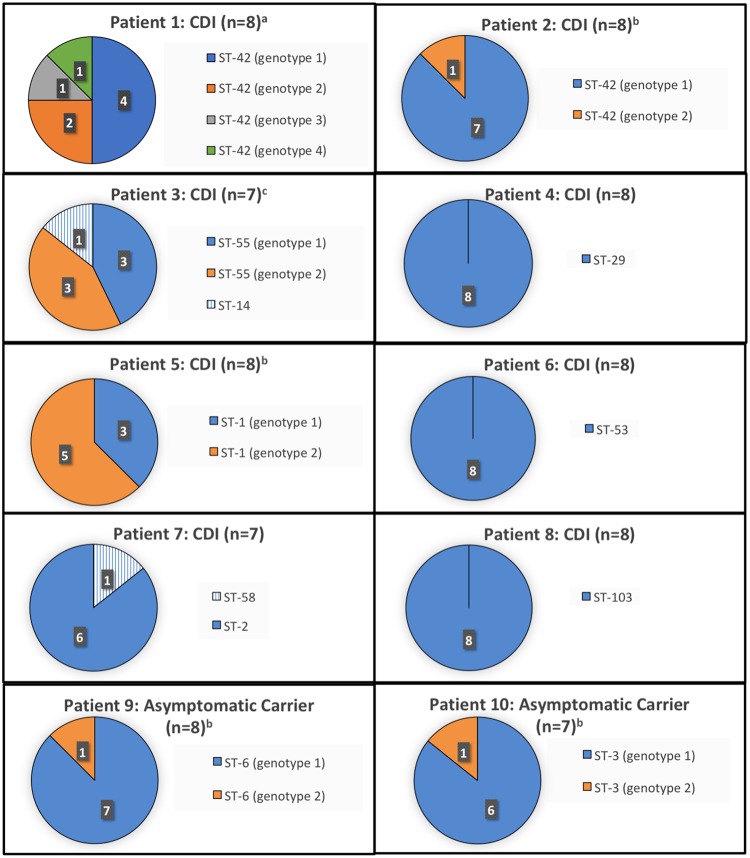
In total, 80 C. difficile isolates underwent WGS (8 isolates per child), and 77 C. difficile isolate sequences (8 from 7 children each and 7 from 3 children each) were successfully assembled. Characteristics of the 10 children selected for this study are listed in Table 1. Figure 1 illustrates the STs of each C. difficile isolate and the genotypic distribution (i.e., number of SNVs) of isotypic isolates. Among 8/10 (80%) children (6 children with CDI and 2 children with asymptomatic C. difficile carriage), all isolates were the same ST. The other 2 children with CDI (patients 3 and 7) each had mixed infection with two STs, although one ST predominated in each child. Among 9/10 (90%) children, isotypic isolates differed by ≤2 SNVs; an isotypic isolate in the remaining child (patient 1, genotype 4) differed by 2 to 4 SNVs relative to the other isolates from the same child. In the isolate with up to 4 SNVs difference relative to other isotypic isolates, 3 were nonsynonymous SNVs in protein coding regions, and the other SNV was in a non-protein coding region. The number of SNV differences among the 4 genotypes identified in stool collected from patient 1 is illustrated in Table 2. After manual visualization of SNV positions among isotypic isolates from patient 1, genetic recombination was determined to be an unlikely explanation of the identified genetic diversity (i.e., the 4 SNVs were scattered throughout the genome [adjacent SNVs separated by 3,000 and 395,000 bp], suggesting independent serial acquisition of SNVs rather than genetic recombination). Overall, among the 77 isolates collected from 10 stool cultures, 74/77 (96%) were clonal (i.e., same ST and ≤2 core genome SNVs) to other isolates in stool culture. No differences in accessory genomic elements were identified among isotypic isolates from the same patient.
TABLE 1.
Clinical characteristics of study subjects
| Patient no. (category) | Age (years) | No./total (%) of clonal isolatesc | Toxin EIA/CCNA result/PCR CTa | Comorbidity | Antibiotic(s) received during previous 30 daysb |
|---|---|---|---|---|---|
| 1 (CDI) | 12 | 7/8 (88%) | Pos/Pos/21.6 | Malignancy | TMP-SMX, metronidazole |
| 2 (CDI) | 1 | 8/8 (100%) | Pos/Pos/22.4 | Respiratory failure | Amoxicillin, ampicillin, ceftazidime, ceftriaxone, vancomycin |
| 3 (CDI) | 18 | 6/7 (86%) | Neg/Neg/24.2 | Solid organ transplant | Azithromycin, TMP-SMX |
| 4 (CDI) | 4 | 8/8 (100%) | Pos/Pos/22.3 | Lymphatic malformation | Ceftriaxone |
| 5 (CDI) | 13 | 8/8 (100%) | Neg/Neg/24.8 | None | None |
| 6 (CDI) | 17 | 8/8 (100%) | Pos/Pos/22.0 | Recurrent bacterial infections | None |
| 7 (CDI) | 10 | 6/7 (86%) | Pos/Pos/21.8 | Malignancy | Cefepime, TMP-SMX, vancomycin |
| 8 (CDI) | 3 | 8/8 (100%) | Pos/Pos/20.1 | Inflammatory bowel disease | None |
| 9 (asymptomatic carrier) | 3 | 8/8 (100%) | Neg/Pos/31.6 | Malignancy | TMP-SMX |
| 10 (asymptomatic carrier) | 6 | 7/7 (100%) | Pos/Pos/26.9 | Cystic fibrosis | Ceftazidime, tobramycin, TMP-SMX |
EIA, enzyme immunoassay; CCNA, cell culture cytotoxicity neutralization assay; CT, threshold cycle (inverse measure of C. difficile stool burden); Pos, positive; Neg, negative.
TMP-SMX, trimethoprim-sulfamethoxazole.
Clonal isolates are those that are the same sequence type (ST) and differ by no more than 2 core genome SNVs from the other stool culture isolates based on existing clonality definitions.
FIG 1.
Characterization of C. difficile sequence types (ST) and discrete genotypes among isotypic isolates collected from stool of children with C. difficile infection (CDI) and asymptomatic C. difficile carriage. a, 1 to 4 SNVs among isotypic strains; b, 1 SNV among isotypic strains; c, 2 SNVs among isotypic strains.
TABLE 2.
Pairwise comparisons of numbers of single nucleotide variants among the four distinct genotypes isolated from patient 1
| Genotypea | Genotype (no. of SNVs) |
|||
|---|---|---|---|---|
| GT1 | GT2 | GT3 | GT4 | |
| GT1 (n = 4) | ||||
| GT2 (n = 2) | 1 | |||
| GT3 (n = 1) | 1 | 2 | ||
| GT4 (n = 1) | 3 | 2 | 4 | |
GT, genotype; n, number of isolates of each specific genotype.
DISCUSSION
After performance of WGS of multiple distinct C. difficile isolates from stool culture of children with CDI or asymptomatic C. difficile carriage, we identified very little within-host genetic diversity. Specifically, among the 77 isolates collected from 10 stool cultures, 96% were clonal (i.e., same ST and ≤2 core genome SNVs) to the other isolates in stool culture. Thus, our data support the position that existing C. difficile clonality definitions (≤2 core genome SNVs), based on estimates of within-host genetic diversity from measurements of C. difficile microevolution in adults with CDI relapses (2), will be minimally biased from random selection of a single colony from stool culture for WGS. With recent studies suggesting that WGS is the optimal method for identifying putative transmission events with exquisite sensitivity (2–4), our data suggest that using WGS of single colonies to identify putative C. difficile transmission events will appropriately characterize the vast majority of events using existing clonality definitions.
A strength of this study is the use of rigorous bioinformatics analyses adapted from methods described by Eyre et al. (2) and that we have previously applied to investigation of C. difficile transmission in children (4), Our study is limited by a relatively small sample size, and this prevents an extensive assessment for the host and pathogen factors that contribute to rare cases of within-host diversity. Children with evidence of within-host diversity did not have broader antibiotic exposure than those without within-host diversity. While the three children with within-host diversity were all immunocompromised, there are also other children with malignancy in this study who did not demonstrate within-host diversity. Despite the small sample size, the number of SNVs among isotypic strains in stool that we directly measured in the present study are similar to that estimated through mathematical modeling in a previous large cohort study of adults with recurrent CDI (2). It is possible that we would have detected more frequent within-host genetic diversity if we had sampled more than 8 colonies per culture. In a previous study in adults (15), 36 colonies per culture were isolated from adults with CDI, and half of the cultures had more than 1 (median of 2) unique ribotype; the presence of multiple ribotypes at the time of initial CDI was associated with subsequent CDI recurrence.
In summary, we identified rare within-host genetic diversity of C. difficile in stool of children with CDI or asymptomatic C. difficile carriage. Among the minority of children in our cohort in whom some within-host genetic diversity was identified, only a single isolate was either a different ST or an isotypic strain that differed by >2 SNVs from other isolates in culture. Although our data suggest that WGS of a single colony from stool will appropriately characterize isolate clonality and putative transmission events in the majority of cases, it is possible that genome-based tracking could occasionally underestimate transmission events when only a single isolate is selected for WGS. Future investigation includes better assessment of host and pathogen factors that may contribute to rare instances of C. difficile within-host genetic diversity.
ACKNOWLEDGMENTS
We acknowledge the NUSeq Core at Northwestern University Feinberg School of Medicine for their assistance with performance of whole-genome sequencing. This work was supported by grants from the National Institute of Allergy and Infectious Diseases at the National Institutes of Health [K23AI123525 to L.K.K.], and the American Cancer Society [MRSG-13-220-01 to E.A.O.]. Research reported in this publication was supported, in part, by the National Institutes of Health’s National Center for Advancing Translational Sciences, Grant Number UL1TR001422. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, Farley MM, Holzbauer SM, Meek JI, Phipps EC, Wilson LE, Winston LG, Cohen JA, Limbago BM, Fridkin SK, Gerding DN, McDonald LC. 2015. Burden of Clostridium difficile infection in the United States. N Engl J Med 372:825–834. doi: 10.1056/NEJMoa1408913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eyre DW, Cule ML, Wilson DJ, Griffiths D, Vaughan A, O'Connor L, Ip CL, Golubchik T, Batty EM, Finney JM, Wyllie DH, Didelot X, Piazza P, Bowden R, Dingle KE, Harding RM, Crook DW, Wilcox MH, Peto TE, Walker AS. 2013. Diverse sources of C. difficile infection identified on whole-genome sequencing. N Engl J Med 369:1195–1205. doi: 10.1056/NEJMoa1216064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mawer DPC, Eyre DW, Griffiths D, Fawley WN, Martin JSH, Quan TP, Peto TEA, Crook DW, Walker AS, Wilcox MH. 2017. Contribution to Clostridium difficile transmission of symptomatic patients with toxigenic strains who are fecal toxin negative. Clin Infect Dis 64:1163–1170. doi: 10.1093/cid/cix079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kociolek LK, Gerding DN, Espinosa RO, Patel SJ, Shulman ST, Ozer EA. 2018. Clostridium difficile whole genome sequencing reveals limited transmission among symptomatic children: A single center analysis. Clin Infect Dis 67:229–234. doi: 10.1093/cid/ciy060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eyre DW, Walker AS, Griffiths D, Wilcox MH, Wyllie DH, Dingle KE, Crook DW, Peto TE. 2012. Clostridium difficile mixed infection and reinfection. J Clin Microbiol 50:142–144. doi: 10.1128/JCM.05177-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kociolek LK, Patel SJ, Shulman ST, Gerding D. 2015. Molecular epidemiology of Clostridium difficile infections in children: a retrospective cohort study. Infect Control Hosp Epidemiol 36:445–451. doi: 10.1017/ice.2014.89. [DOI] [PubMed] [Google Scholar]
- 7.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jolley KA, Maiden MC. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lunter G, Goodson M. 2011. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res 21:936–939. doi: 10.1101/gr.111120.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kociolek LK, Gerding DN, Hecht DW, Ozer EA. 2018. Comparative genomics analysis of Clostridium difficile epidemic strain DH/NAP11/106. Microbes Infect 20:245–253. doi: 10.1016/j.micinf.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ozer EA, Allen JP, Hauser AR. 2014. Characterization of the core and accessory genomes of Pseudomonas aeruginosa using bioinformatic tools Spine and AGEnt. BMC Genomics 15:737–754. doi: 10.1186/1471-2164-15-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ozer EA. 2018. ClustAGE: a tool for clustering and distribution analysis of bacterial accessory genomic elements. BMC Bioinformatics 19:150. doi: 10.1186/s12859-018-2154-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milne I, Stephen G, Bayer M, Cock PJ, Pritchard L, Cardle L, Shaw PD, Marshall D. 2013. Using Tablet for visual exploration of second-generation sequencing data. Brief Bioinform 14:193–202. doi: 10.1093/bib/bbs012. [DOI] [PubMed] [Google Scholar]
- 15.Seekatz AM, Wolfrum E, DeWald CM, Putler RKB, Vendrov KC, Rao K, Young VB. 2018. Presence of multiple Clostridium difficile strains at primary infection is associated with development of recurrent disease. Anaerobe 53:74–81. doi: 10.1016/j.anaerobe.2018.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This whole-genome shotgun project (BioProject PRJNA518899) has been deposited at DDBJ/ENA/GenBank under the accession numbers SEOS00000000 to SERQ00000000.