Vaginitis is often diagnosed by microscopy and limited to testing for bacterial vaginosis (BV), vulvovaginal candidiasis, and trichomoniasis. Approximately 10% of vaginal swabs are negative but designated “altered flora” by BV Nugent score, leaving clinicians unsure how to treat patients. Accurate and comprehensive vaginitis diagnostics are needed to direct treatment and reduce risks of recurrent or more severe infections.
KEYWORDS: 16s rRNA gene, Nugent score, aerobic vaginitis, bacterial vaginosis, dysbiosis, microbiome, microscopy, molecular diagnostics, qPCR
ABSTRACT
Vaginitis is often diagnosed by microscopy and limited to testing for bacterial vaginosis (BV), vulvovaginal candidiasis, and trichomoniasis. Approximately 10% of vaginal swabs are negative but designated “altered flora” by BV Nugent score, leaving clinicians unsure how to treat patients. Accurate and comprehensive vaginitis diagnostics are needed to direct treatment and reduce risks of recurrent or more severe infections. Vaginal swabs were collected from 93 women (mean age, 23.53 years; range, 18 to 42 years) in a cross-sectional study. Microscopy results for BV and Candida were compared to those from two molecular approaches: (i) a comprehensive quantitative PCR (qPCR) assay, including testing for aerobic vaginitis (AV), Candida, sexually transmitted infections (STI), and BV (Applied Biosystems) with an accompanying BV interpretive algorithm (Coriell Life Sciences), and (ii) microbiome profiling of the 16S rRNA gene (Illumina). Microscopy plus BV Nugent score had 76% overall agreement with the qPCR plus BV interpretive algorithm method (24 positive, 47 negative). OF the nine samples designated altered flora by Nugent, five were categorized BV positive and four were BV negative by the qPCR method. Although BV negative, 3/4 of the latter samples had positive AV targets with one also was STI positive. Microscopic identification of Candida versus that by qPCR had 94% agreement (9 positive, 78 negative). The comprehensive qPCR assay revealed alternative etiologies summarized as 38% BV, 10% AV, 5% Candida, 2% STI, 10% mixed infection (positive targets in multiple panels), and 35% negative for all targets. 16S microbiome analysis confirmed the bacterial qPCR results and identified differentiating patterns between AV, BV, and Lactobacillus-dominated vaginal microbiomes.
INTRODUCTION
The healthy vaginal microbiome is generally dominated by Lactobacillus species such as L. crispatus and L. jensenii, which have protective effects through competitive adherence to vaginal epithelial cells and produce antimicrobials such as hydrogen peroxide, lactic acid, and bacteriocins (1). Factors such as age, ethnicity, and geography have been associated with differences in vaginal microbiome patterns in previous studies, prompting continued research to understand what constitutes a “healthy” vaginal microbiome (2–4). Abnormal vaginal discharge and vaginitis symptoms are experienced by millions of women globally (5, 6) and have a broad range in severity and associated risk factors depending on etiology (7). The most common diagnoses for tested patients are bacterial vaginosis (BV; 40% to 50%), vulvovaginal candidiasis (20% to 25%), and trichomoniasis (15% to 20%); noninfectious causes such as irritants or allergic reactions account for 5% to 10% of cases (8).
BV is a polymicrobial anaerobic dysbiosis characterized by a decrease in Lactobacillus spp. overcome by a heterogenous profile of mixed anaerobes such as Gardnerella, Atopobium, Prevotella, and Megasphaera species (9). Symptoms associated with BV include vaginal itching and production of whitish-gray discharge with an unpleasant odor, but women can be asymptomatic (10). BV has been associated with an increased risk of developing more serious health complications such as upper urogenital tract disorders (11), sexually transmitted infections (12), and preterm delivery (13). Aerobic vaginitis (AV) is a different vaginal dysbiotic condition which also involves decreased Lactobacillus spp.; however, AV includes inflammation and increased presence of aerobic intestinal-associated bacteria such as Escherichia coli and group B Streptococcus (reviewed in reference 14).
Diagnosis of AV and BV has historically relied on the use of clinical assessment and microscopic scoring. AV is diagnosed by phase-contrast microscopic examination of a wet-mount preparation to enumerate Lactobacillus, toxic leucocytes, and immature epithelial cells (14). To diagnose BV, at least three of the following Amsel criteria require positivity: pH >4.5, gray discharge, positive amine test, and the presence of epithelial “clue” cells (15). Nugent scoring has been the gold standard for laboratory diagnosis of BV, whereby a Gram-stained vaginal smear is microscopically examined and scored based on morphotype enumeration of Gram-positive Lactobacillus versus Gram-negative bacteria (16, 17). Microscopy, however, is labor intensive and allows subjective uncertainty, as some BV-associated bacteria have Gram-variable morphotypes, making them difficult to score accurately. The Nugent score designates a sample negative (score of 0 to 3), altered/indeterminate (score of 4 to 6), or positive for BV (score of 7 to 10). Antibiotic treatment is recommended for BV; however, studies have shown recurrence rates to be very high (20% to 67%) due to biofilm formation of the abnormal anaerobic flora that create clue cells (18). An altered Nugent result indicates some degree of vaginal dysbiosis; however, this may not transition to disease. In some cases, altered vaginal flora may represent “partial BV,” or an early transition toward BV but it may also indicate the development of other vaginal dysbiotic conditions such as AV that would require a different clinical treatment (14, 19). An altered Nugent score leaves approximately 10% of women without a clear treatment plan (20, 21). Likewise, a symptomatic woman with a negative BV result by Nugent may require follow-up laboratory tests to determine a diagnosis and appropriate treatment.
The first of three major goals of this pilot study was to compare the current microscopy with Nugent scoring against a targeted quantitative PCR (qPCR) panel (Applied Biosystems) with a BV diagnostic interpretive algorithm (Coriell Life Sciences) to diagnose BV. The second was to identify alternative diagnoses for cases designated altered or negative by Nugent score using a comprehensive qPCR panel of pathogens associated with AV, sexually transmitted infections (STI), and candidiasis (yeast). The third goal was to utilize high-throughput sequencing (HTS) to compare the bacterial microbiome variation in samples categorized by the qPCR results. To the best of our knowledge, this is the first study to not only compare BV diagnostics but also include alternative infectious etiologies of vaginitis (AV, yeast, and STI) using qPCR and additionally use HTS 16S profiling to further investigate variability patterns in the vaginal microflora.
MATERIALS AND METHODS
Ethics.
This study was conducted following review and approval from the Conjoint Health Research Ethics Board at the University of Calgary (REB14-0764).
Recruitment and sample collection.
Enrollment of women attending a clinic for an annual physical or presenting with vaginitis symptoms occurred at either the sexual reproductive health (SRH) or the sexually transmitted infection (STI) clinics in Calgary, Canada, over a 3-month period. The inclusion criteria were women of reproductive age that had regular menstrual cycles, were not pregnant, were not using contraceptives that are delivered directly to the vaginal mucosa, and had not been on antibiotics in the preceding 30 days. Samples were included in the study if the microscopic score was complete, extracted DNA produced high-quality qPCR and 16S rRNA gene sequence data, and they were accompanied by a completed clinical history from the physician. Ninety-three high-quality patient samples met this criteria and were included in the study. Due to the anonymity policies of the STI clinic, we were unable to obtain patient information about ethnicity or additional risk factors (i.e., number of sexual partners) for this study.
Two nylon-flocked Copan ESwabs were used to collect midvaginal samples that were immediately placed into Amie’s transport medium; the first was used for microscopic examination and the second swab was used for molecular DNA analysis (qPCR assays and 16S rRNA gene sequencing). An additional vaginal swab was collected from all patients using the APTIMA vaginal swab, which was tested with a Trichomonas vaginalis nucleic acid amplification test (NAAT) using the APTIMA T. vaginalis assay. However, Neisseria gonorrhoeae and Chlamydia trachomatis NAATs on the Panther platform (Hologic, San Diego, CA) were only performed if ordered by the physician according to the Alberta treatment guidelines for STI (22).
Sample storage and processing.
All completed study kits were transported within 4 to 6 h postcollection to the centralized clinical microbiology laboratory. A vaginal smear was immediately made from one Copan ESwab, Gram-stained, read, and reported the same day by a trained laboratory technologist using Nugent’s scoring criteria (23). The Gram-stained smear was also assessed for the presence of yeast and reported as indicative of vaginal candidiasis. The second Copan ESwab was immediately stored at −80°C to be batch processed for molecular testing. Total DNA was isolated using the QIAamp DNA Mini kit (Qiagen, Hilden, Germany). Briefly, the samples were centrifuged (13,000 × g for 5 min), and the bacterial pellets were pretreated in an enzymatic lysis buffer containing lysozyme (20 mg/ml; Sigma-Aldrich, Dorset, UK) and incubated for 30 min at 37°C; proteinase K and buffer AL were then added and the mixture was further incubated for 30 min at 56°C. The QIAamp spin column protocol was followed for the remaining steps according to the manufacturer’s instructions, and the DNA was eluted in 100 μl molecular-grade water. All APTIMA vaginal swabs were tested within 24 h of collection for the presence of T. vaginalis and other STI pathogens (N. gonorrhoeae and C. trachomatis as clinically directed) using the respective NAAT assays on the Panther platform.
Laboratory methods.
(i) Microscopic examination. The current standard protocol at Calgary Laboratory Services (CLS) to diagnose BV and yeast requires microscopic identification and enumeration of Gram-stained vaginal smears. Briefly, the number of Gram-positive Lactobacillus spp. is counted against that of the Gram-variable coccobacilli, Gardnerella/Bacteroides, and Gram-negative bacilli to derive a Nugent score for each case (16, 23). A Nugent score of 0 to 3 is considered BV negative, 4 to 6 represents altered vaginal flora, and 7 to 10 is BV positive. The presence or absence of yeast on the slide is also identified and reported.
(ii) Molecular assays. The purified DNA samples were tested using a commercial qPCR assay (TaqMan OpenArray vaginal microbiota comprehensive plate; Applied Biosystems, part of Thermo Fisher Scientific, Waltham, MA) according to the manufacturer’s specifications. This qPCR assay used a targeted approach to detect 14 unique bacteria for bacterial vaginosis (BV; 4 commensal lactobacilli and 10 pathobionts), 4 targets for aerobic vaginitis (AV), 7 for Candida spp. or yeast (YST), and 7 specific for sexually transmitted infections (STI) analyzed with QuantStudio software (Thermo Fisher Scientific). The BV panel consists of both pathobiont and commensal targets unlike the remaining panels with only pathogen/pathobiont targets. To interpret the BV panel qPCR results, the data were run through the recently validated CLS2.0q algorithm (Coriell Life Sciences, Philadelphia, PA). The CLS2.0q algorithm is a quantitative molecular diagnostic based on the relative abundances of fourteen bacterial species. Ten of these are considered to be pathobionts or pathogenic and four are commensal Lactobacillus species thought to confer beneficial effects on the overall balance and stability of the vaginal microbiome. The approach involves a linear transformation that yields a set of weighted species-specific values which are then combined to produce a score for each patient. In previous evaluations, it has shown an overall sensitivity of 93% and a specificity of 90% compared to gold-standard testing (24).
Purified DNA from the vaginal swabs was also tested by next-generation sequencing (NGS) using high-throughput 16S rRNA gene amplicon profiling on the MiSeq instrument (Illumina, San Diego, CA) at the Nicole Perkins Microbial Community Laboratory sequencing facility at the University of Calgary. The V3 variable region was amplified using modified 341F and 518R primers developed previously (25). PCRs were performed in triplicates, with each 50-μl reaction mixture containing 10 mM each deoxynucleotide triphosphate (dNTP), 1.5 mM MgCl2, 25 pmol of each primer, and 0.5 U Taq polymerase. PCR was completed in the S1000 thermocycler (Bio-Rad, Mississauga, ON) with an initial 2-min denaturation at 94°C followed by 30 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 30 s before a final extension step at 72°C for 10 min. The amplicons were pooled and then quantified with the Bioanalyzer high-sensitivity DNA chip (Agilent, Mississauga, ON) before sequencing on the MiSeq using a reagent kit v3 according to the manufacturer’s instructions (Illumina). The sequencing run included a no-template negative sample as an internal control for contamination.
Bioinformatics analysis.
The 16S rRNA gene data were assessed with FastQC 0.11.7 (www.bioinformatics.babraham.ac.uk/projects/fastqc/) and MultiQC 1.0 (26) before primers and host contaminant reads were removed with BBMap (https://sourceforge.net/projects/bbmap/). The reads were further processed using the DADA2 package 1.9.0 (27) in R 3.4.4 (28). Default settings in DADA2 were used unless otherwise specified. The fastqFilter function was used to trim and filter the reads with a maximum expected error of 1. Error rates of the filtered dereplicated reads were estimated using 100,000 sequences and used to create amplicon sequence variants (ASV) for each sample. The reverse reads were not used in the analysis due to low-quality data, likely due to a power outage during sequencing. In DADA2, chimeras were removed using BimeraDenovo and taxonomy assigned using the assignTaxonomy function. Two reference databases were used for training the naive Bayesian classifier for comparison: a specialized vaginal microbiome reference database (29) and the RDP training set 16 (30). Both reference databases were formatted using TaxMan (31) and utilized for taxonomy assignment. A maximum likelihood phylogenetic tree was constructed using the R package phangorn (32).
Statistical analysis was also performed in R, with phyloseq 1.25.2 (33) and vegan 2.5-2 (34). Diversity measures were calculated, grouping samples by qPCR panel results: negative, BV, or AV. Samples that were negative for BV and AV but were positive for STI and/or YST targets were classified as negative based on α-diversity comparisons by an analysis of variance (ANOVA) (see Fig. S1 in the supplemental material). The α-diversity calculations were done in vegan, including richness, a count of the features/ASVs in each sample, and the Simpson diversity index, a measure of evenness within the community.
Principal-coordinate analysis (PCoA) was used to plot the ordination of ASVs in a reduced space with the Bray-Curtis and unweighted UniFrac distance methods using capscale in vegan to visualize the β-diversity between the biome groups. The spread of the groups was plotted using the vegan ordellipse function, which represents the 95% confidence interval of the standard error of the comparison groups. The distance measures used were Bray-Curtis to quantify compositional differences between the groups using ASV abundance counts and unweighted UniFrac, which considers the phylogenetic differences between branch lengths. Sources of β-diversity variation were compared using a permutational multivariate analysis of variance (PERMANOVA) with vegan’s adonis function with the distance matrices described above. Pairwise PERMANOVA calculations were done with the pairwise.adonis function (35). Second, the multivariate homogeneity of dispersion between the groups was measured with the vegan betadisper function, examining the distances between each point and the group centroids before reducing the original distances (Bray Curtis, unweighted UniFrac) to principal coordinates and then comparing them by ANOVA. The phylogenetic composition within each sample was characterized by calculating abundance-weighted nearest relative index (NRI) and nearest taxa index (NTI) to determine if the bacterial microbial communities were driven more by competition (stochastic) or by environmental pressure (deterministic) factors using the Picante R package (36, 37).
Sparse partial least-squares discriminant analysis (sPLS-DA) was performed to identify the most differentiating features (in this case, this is based on abundance at the genus level) to classify samples into the 3 biome groups: BV, AV, or negative (38). The mixOmics R package was used for the sPLS-DA analysis according to methods outlined elsewhere (39, 40). Briefly, the procedure constructs artificial latent components of the predicted data set [genera table denoted as X(N × P) collated at genus level] and the response variable (denoted as Y with categorical information of samples, e.g., BV, AV, or negative) by factorizing these matrices into scores and loading vectors in a new space, such that the covariance between the scores of these two matrices in this space is maximized under two constraints: (i) ensuring the loading vector to have unit magnitude (requirement of the procedure) and (ii) ensuring that for the features that do not vary between the categories, the corresponding loading vector coefficients go to zero. This is conducted by using the sparsity negative parameter, which can be adjusted to enforce shrinkage of loading vector coefficients. According to the recommendations given in the mixOmics package (http://www.mixomics.org), before applying the procedure splsda, we prefiltered 1% of the lowest abundant genera and then perform total sum scaling followed by centralized log ratio (TSS+CLR) normalization. To predict the number of latent components (associated loading vectors) and the number of discriminants, the perf.plsda and tune.splsda functions were used, respectively. In the latter case, we fine-tuned the model using leave-one-out cross-validation by splitting the data into training and testing sets and then finding the classification error rates using overall error rates.
Accession number(s).
Sequences were submitted to the Short Read Archive under accession number PRJNA513873.
RESULTS
Clinical summary.
There were 93 vaginal swabs with associated clinical information included in the study. The average age of the participants was 23 years (range, 18 to 42 years). Table 1 summarizes the patient characteristics. Fifty-seven women were symptomatic when they presented at the clinic, with abnormal discharge being the most common symptom (41%). In the symptomatic group, 15/57 (26%) reported at least one previous BV diagnosis compared to only 6/36 (17%) in the asymptomatic group.
TABLE 1.
Clinical summary for the 93 patients included in this studya
| Patient state at time of clinic visit | Value |
||
|---|---|---|---|
| Symptomatic | Asymptomatic | Total | |
| No. of patients | 57 | 36 | 93 |
| Average age (years [range]) | 23.9 (18–42) | 21.2 (18–34) | 23.53 (18–42) |
| No. (%) previous BV | 15 (26) | 6 (17) | 21 (23) |
| Symptoms (n [%]) | |||
| Abnormal discharge | 38 (67) | 38 (41) | |
| Pelvic pain | 7 (12) | 7 (8) | |
| Dyspareunia | 6 (11) | 6 (6) | |
| Dysuria | 2 (4) | 2 (2) | |
| Fever | 0 | 0 | |
| Vulvovaginal pruritus | 10 (18) | 10 (11) | |
| Contraceptive use (n [%]) | |||
| Oral | 20 (35) | 20 (56) | 40 (43) |
| IUD | 9 (16) | 1 (3) | 10 (11) |
| Condom | 0 | 1 (3) | 1 (1) |
| Patch | 0 | 1 (3) | 1 (1) |
| None | 25 (44) | 13 (36) | 38 (41) |
| Not reported | 2 (4) | 1 (3) | 3 (3) |
Summarized results of self-reported clinical history with clinical assessment at time of enrollment.
Self-reported contraceptive use was also recorded, with oral contraceptives (43%), no contraceptive use (41%), intrauterine devices (11%), and one patient each reporting condom or patch use. There were no significant differences in contraceptive use between symptomatic and asymptomatic groups (P > 0.5 by Fisher’s exact test).
Performance of current diagnostic methods compared to that of the qPCR assay.
Microscopic investigation of Gram-stained slides followed by Nugent scoring to determine BV status found 59 negative samples (Nugent 1 to 3), 25 positive for BV (Nugent 7 to 10), and 9 with “altered flora” (Nugent 4 to 6). Yeast consistent with Candida spp. was identified microscopically in 13 samples, of which 11 were BV negative and only 2 BV positive by Nugent score.
The qPCR results are summarized in relation to the microscopy results with Nugent BV status in Table 2. BV panel results are presented as the algorithm designated, but all other panels are presented as positive if any targets within the panel met the confidence criteria for positivity. There were 35 (38%) samples positive for BV only, 9 (10%) positive for AV targets only, 5 (5%) for Candida spp. only (herein abbreviated as YST), and 2 (2%) for STI only. There were 33 (35%) samples negative for all targets, while nine (10%) had mixed panel positivity by qPCR.
TABLE 2.
Summary of microscopy and qPCR results
| Nugent resulta | No. positive for yeasta | qPCR panel results |
Total no. of samples | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CLS2.0q BVb | No. of negative samples | No. of samples positive for: |
||||||||||
| BV only | AV only | YST | STI | BV+AV+STI | BV+YST | AV+STI | AV+YST | |||||
| Negative | 8 | Negative | 32 | 6 | 5 | 2 | 2 | 47 | ||||
| Negative | 3 | Positive | 8 | 1 | 3 | 12 | ||||||
| Positive | Negative | 1 | 1 | |||||||||
| Positive | 2 | Positive | 22 | 1 | 1 | 24 | ||||||
| Altered | Negative | 1 | 2 | 1 | 4 | |||||||
| Altered | Positive | 5 | 5 | |||||||||
Microscopy results.
qPCR results were categorized as positive/negative for BV by the CLS2.0q interpretive algorithm.
Comparing definitive BV results (altered flora samples excluded), the qPCR sensitivity was 96%, the specificity was 80%, and the negative and positive predictive values were 98% and 67%, respectively, compared to the gold standard microscopy plus Nugent score.
There were 12 discordant samples which were negative by Nugent score but BV positive by qPCR. Further investigation of these 12 results revealed that 8 were positive for BV pathobionts that are Gram variable (i.e., Gardnerella vaginalis, Atopobium vaginae) or unseen by Gram stain (i.e., Mycoplasma hominis, Ureaplasma urealyticum). The remaining 4 samples were mixed, being positive for BV and AV, YST, and/or STI targets. Only 1 sample was positive by Nugent but negative by qPCR, for which the molecular assay identified AV-associated pathogens, suggesting an alternative diagnosis. Thus, we conclude that the low positive predictive value (PPV) value is due to the higher resolution by qPCR to identify targets at the species level which are ambiguous or unable to be seen by microscopy.
Nine samples were categorized as altered flora by Nugent (score 4 to 6) but designated positive or negative by qPCR assay. Of these samples, 4 were negative for BV by qPCR but contained positive targets in the AV, YST, and/or STI panels, confirming the observation of altered flora, but the higher resolution method suggested a non-BV diagnosis. The remaining 5 “altered” samples were classified as BV positive by qPCR.
Currently, STI pathogens are tested using a nucleic acid amplification test (NAAT; as described in Materials and Methods). The NAATs found 1 sample positive for Trichomonas vaginalis, 3 for Chlamydia trachomatis, and 1 positive sample for Neisseria gonorrhoeae. The qPCR STI panel results agreed with 2/3 C. trachomatis samples and the N. gonorrhoeae sample; however, Herpes simplex virus 2 (HSV2) was the only pathogen detected in the T. vaginalis-positive sample by NAAT, and HSV1 was found in an additional sample. HSV is not included in the current STI testing at our laboratory.
Microbiome analysis.
Amplicon sequencing of the 16S rRNA gene V3 variable region generated a total of 13,712,478 reads from 93 samples (mean, 147,446 reads per sample; range, 34,584 to 259,831). Only sequences assigned to the kingdom Bacteria were included, with the conservative approach of including only amplicon sequence variants (ASV) present in at least 2% of samples (adapted from reference 41). After filtering, the final high-confidence data set consisted of 1,364 ASVs with a total of 13,447,475 reads (mean, 144,596 per sample; range, 32,500 to 259,701).
Samples were divided into groups based on the qPCR results and α-diversity calculated for both richness and evenness (Simpson diversity index) before comparison by ANOVA (see Fig. S1 in the supplemental material). Due to small group sizes and α-diversity pairwise comparisons, samples containing STI, YST, or mixed panel targets were grouped based on the closest mean by Simpson diversity index (pairwise comparisons, P > 0.05). Therefore, downstream analyses were conducted with all samples grouped into 3 biome categories: AV, BV, or negative. As the 16S rRNA gene primers do not identify yeast, we cannot infer any changes beyond the bacterial microflora.
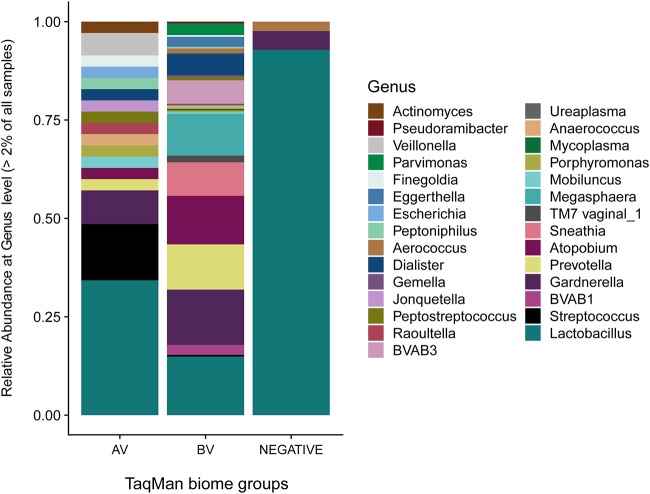
Figure 1 illustrates the relative abundance of 29 unique taxa at the genus level identified in >2% of all samples by biome group (AV, BV, or negative). Our findings corroborate those from previous studies where 16S rRNA gene profiling showed Lactobacillus dominance in the negative group samples (relative abundance, 92%) which decreased in the AV and BV dysbiotic groups (42–44). Lactobacillus still held the highest relative abundance within the AV group (34%), followed by Streptococcus and Gardnerella (14% and 8%, respectively). In the BV group, the relative abundance of Lactobacillus was equivalent to that of Gardnerella (14% for both), followed by other genera: Atopobium (12%), Prevotella (11%), and Megasphaera (11%). Detailed relative abundance data identified in Fig. 1 are listed in Table S1.
FIG 1.
Relative abundances of all genera present in the total data set >2% using the 16S rRNA gene V3 region amplicon data sequenced on the MiSeq platform (Illumina). Samples are grouped by biome as determined by the qPCR results and preliminary comparison of microbiome diversity (see Fig. S1 in the supplemental material).
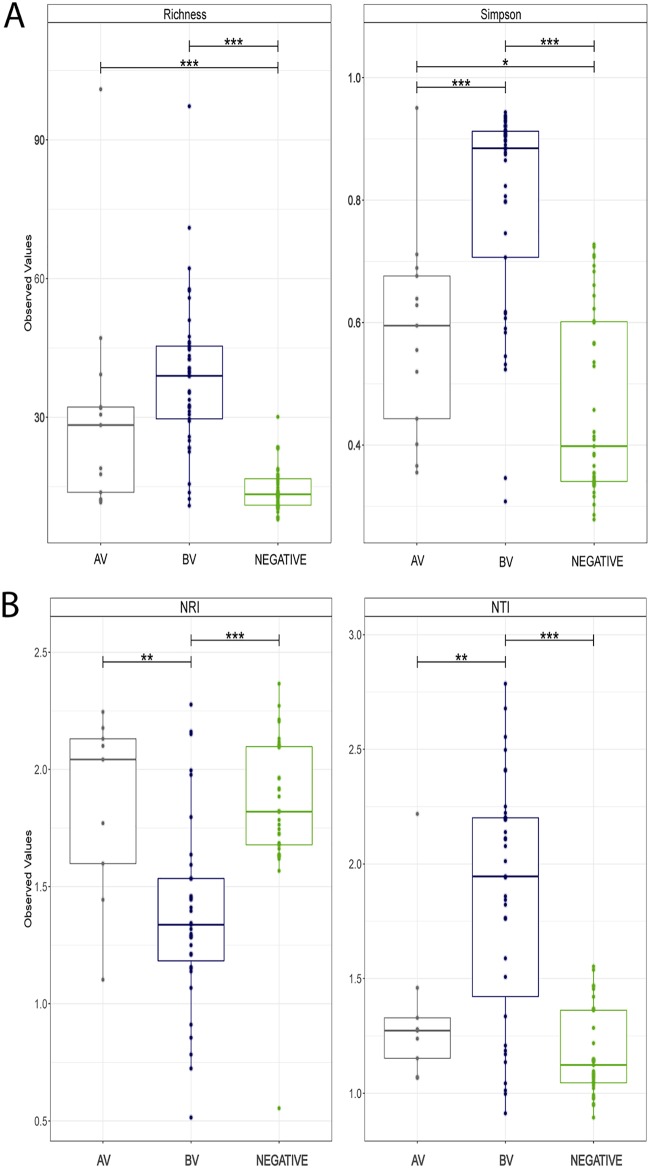
We then estimated the α-diversity between biome groups AV, BV, and negative (Fig. 2A). With respect to richness, the AV and BV groups were both significantly higher than the negative group (P < 0.001), owing to the increased number of species within the dysbiotic states. All pairwise comparisons were significant by the Simpson diversity index (P < 0.05), suggesting not only variability between species abundance but also differences in evenness across the biome groups (Fig. 2A). Estimation of β-diversity by PCoA was also completed and distances compared by PERMANOVA (see Fig. S2). The results suggested that the biome groups were significant sources of variation by both Bray-Curtis (unadjusted R2 = 0.1978, P < 0.001) and unweighted UniFrac (unadjusted R2 = 0.1216, P < 0.001). Homogeneity of dispersion using the same distance matrices also suggested significant differences between the dysbiotic groups (AV and BV) compared to the negative group (P < 0.05). Wide dispersion and overlap of samples within the biome groups was observed, suggesting that greater sample numbers and categorical parameters are needed to fully understand and differentiate the dysbiotic states from the “healthy” microbiome.
FIG 2.
Diversity and community composition analyses of the qPCR-determined biome groups using the 16S rRNA gene data generated with HTS. (A) α-diversity measurements of both richness and evenness of the bacterial microflora within each biome group. (B) Estimations of phylogenetic clustering and environmental pressure on the bacterial communities within each biome group using abundance-weighted nearest relative index (NRI) and nearest taxa index (NTI). Statistical comparisons were performed by ANOVA.*, P < 0.1; **, P < 0.05; ***, P < 0.001.
Further investigation of the phylogenetic structure within the biome groups using NRI (net relatedness index) and NTI (nearest taxon index) is presented in Fig. 2B. The NRI plot shows that the AV and negative groups are more phylogenetically clustered than the BV group (by ANOVA: P < 0.01 and 0.001, respectively), suggesting environmental pressures influence the community compositions more than within the BV group. NTI values of samples within the BV group were significantly higher than in the AV and negative groups (P < 0.001); however, the values in AV samples were also higher than in the negative group (P < 0.05). NTI differences are likely due to clustering of the AV-associated Gram-negative aerobes and the BV-associated Gram-negative anaerobic species compared to the Lactobacillus-dominated communities in the negative group samples.
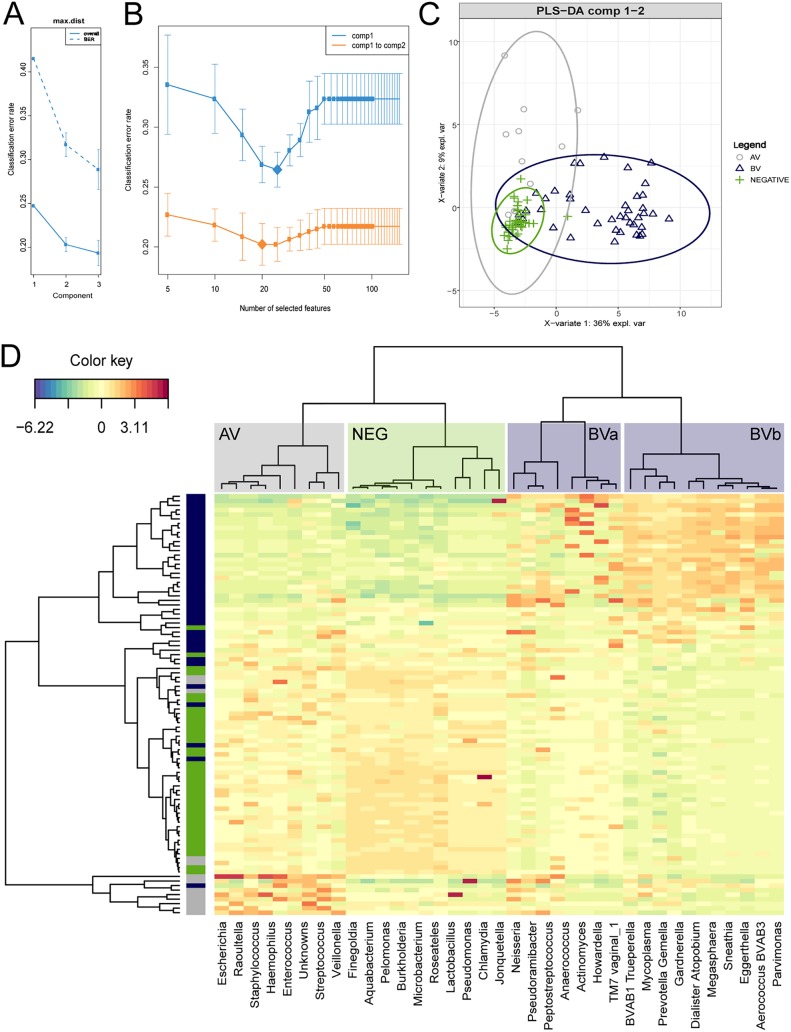
Figure 3 illustrates further analysis using sPLS-DA which identified 42 genera that were differentially abundant between the biome groups. Figure 3A and B shows the classification error rate reductions with optimization of tuning components and numbers of features (genera) for modeling. Figure 3C shows the first 2 components of the PLS-DA with 95% confidence ellipses around each biome group. Figure 3D is a clustered heat map illustrating the discriminatory genera with the agglomerative clustering shown as dendrograms and the abundance values displayed as the normalized log ratio-transformed values (39, 45). Labeled clusters across the top of the heat map show the AV-associated bacteria include genera such as Escherichia, Staphylococcus, Enterococcus, Streptococcus, Haemophilus, Raoultella, Veillonella, and Finegoldia. The next block is consistent with species dominant in the negative group samples, such as Lactobacillus, Pelomonas, Burkholderia, and Microbacterium. Some samples have high Chlamydia and Neisseria abundance, which can be members of the normal vaginal flora; however, the qPCR data confirmed these samples are positive for the species associated with STI, C. trachomatis or N. gonorrhoeae. The BVa block appears more variably present in BV samples, but the species are also in higher abundance within many of the AV samples (Peptostreptococcus, Anaerococcus, Actinomyces, and Howardella). The BVb block consists of bacteria associated with BV, such as BVAB 1 and 3, Prevotella, Gardnerella, Atopobium, Megasphaera, and Sneathia. Importantly, there are highly abundant bacteria found in BV samples which are not included in the qPCR assay (i.e., Gemella, Dialister, and Eggerthella). The most discriminative features (genera) that characterize each biome group are shown in Fig. S3. The coefficients from the loading vector on the first and second components are displayed as relative proportions between the biome groups (Fig. S3, left and right, respectively).
FIG 3.
Sparse partial least-squares discriminant analysis (sPLS-DA) was performed to identify the most differentiating genera for each biome group. (A and B) Classification error rates across the components and the numbers of optimal features (genera) included in the model, chosen by the lowest error rates. (C) The first 2 components of the PLS-DA with 95% confidence ellipses around each biome group. (D) Clustered heat map illustrating the discriminating genera with the agglomerative clustering shown as dendrograms. Abundance values are displayed as normalized log ratio-transformed values.
DISCUSSION
To the best of our knowledge, this is the largest comprehensive study on diagnostic methods for infectious vaginitis. Our study compared the microscopic results for BV and Candida with an extensive qPCR commercial assay that included panels for BV, AV, Candida, and STI pathogens. We also conducted 16S rRNA gene profiling to explore differentiating patterns in the bacterial microflora categorized by the qPCR results. Our data showed that the comprehensive qPCR assay was able to confirm or resolve BV status with higher resolution than the microscopy plus Nugent scoring and also provide alternative etiologies.
Vaginitis is a common concern for many women, which has led to a growing body of literature on diagnostic methods (20, 44, 46–48), epidemiology, and treatment of BV and other causes of vaginitis (7, 49, 50). Similarly to the present work, the evidence consistently supports the benefits of molecular methods to diagnose BV over traditional microscopic methods. Shipitsyna et al. (47) published a foundational study using HTS and qPCR to determine quantitative thresholds for BV prediction as defined by Amsel criteria. Our study agrees with the authors’ recommendation to use quantitative relative abundance (Lactobacillus spp. versus BV-associated bacteria) as the best determination of BV status, although interestingly, their study did not report identification of any AV-associated bacteria such as Escherichia or Streptococcus spp. It would be valuable to the field of women’s reproductive health to investigate comprehensive vaginal infectious etiologies in a larger population to understand the global prevalence of AV.
The diversity and phylogenetic structure analysis demonstrated significant differences between the three biome groups: BV, AV, and negative (Fig. 2). A recent paper by Cartwright et al. (20) recommended an optimal cutoff using the Simpson diversity index (SDI = 0.82) for identifying BV positivity. Data in the present study would support a high SDI cutoff to differentiate BV status (mean SDI = 0.88), but due to overlap in sample distributions, we are not confidently able to recommend an α-diversity threshold for diagnostic purposes at this time. Further work on multivariate modeling of microbiome data accompanied by complete clinical information is needed to robustly differentiate between dysbiotic conditions and provide women with an accurate diagnosis.
There were experimental limitations in our study; however, we do not believe they altered the trends observed. First, we suspect a power outage occurred during the Illumina run, leaving the reverse reads short and of very poor quality, unlike the forward reads which were produced to the full length and quality expected; therefore, the analysis was completed as single end. Recently, Korpela et al. (51) conducted a comparative analysis of different read lengths on single/paired-end data with the conclusion that single-end 150-bp data are favorable for microbiome analyses, similar to those performed in this study. Second, DNA extraction was completed with an enzymatic lysis protocol instead of bead beating as commonly reported; however, a recent comparison of lysis methods found that enzymatic lysis alone does not statistically affect the diversity measures for vaginal microbiome analysis (52). Third, due to the anonymity policies of the clinics participating in the study, we were unable to collect important patient information to make robust conclusions regarding the association of ethnicity or hormone-producing contraceptive methods and the vaginal microbiome. Previous studies have shown vaginal microbiome compositions differ by ethnicity, with some patterns suggestive of BV (2, 4, 53); unfortunately, we were unable to assess if there was any correlation between BV status and ethnicity in this pilot study. Contraceptive use is another factor shown to influence BV prevalence and microbiome changes (52, 54); however, we did not observe statistically significant differences in BV proportion or community diversity metrics (data not shown) between women taking oral contraceptives and those using other methods or no contraception. We did not have information on the oral contraceptive brand or whether the intrauterine device (IUD) was hormone producing, which may influence the vaginal flora and could be important details for future research. There were 14 symptomatic women with negative results for all qPCR targets and by microscopy, of which 13/14 used oral contraceptives or had an IUD (6 and 7 women, respectively). We aim to explore the correlation of vaginitis symptoms and diagnoses with ethnicity and hormone-producing contraceptive methods in the larger follow-up study by our research group.
In conclusion, our study confirmed that molecular methods are more accurate and objective for BV diagnosis than microscopy, as qPCR was able to provide higher resolution with species-level data. Additionally, the BV interpretive algorithm was able to resolve the BV status for samples designated altered flora by Nugent (24). The comprehensive qPCR assay included possibilities for alternative diagnoses, revealing a higher prevalence of AV pathogens than expected. AV is likely underreported at present, because reliable tests have not been commercially available and wet mounts are not routinely performed (at least in North American practice). This is clinically important, as BV treatments are not effective for AV, and as such, we recommend including AV in the diagnostic workup for patients seeking medical care for vaginitis symptoms (14, 19). The broad-range 16S rRNA gene analysis provided a relatively unbiased characterization of the bacterial community compositions of the AV, BV, and negative biome groups which will contribute to our understanding of the vaginal microbiome and toward the use of HTS for diagnosing dysbiotic conditions. Future work by our group aims to further contribute to understanding variations in “normal” or “healthy” vaginal microbiomes, especially with respect to adverse pregnancy outcomes and decision making for treatment (11, 55–57). This study is important for women’s health, as it furthers our understanding of the vaginal microbiome in the context of dysbiotic conditions and highlights the need for more accurate and comprehensive diagnostics.
Supplementary Material
ACKNOWLEDGMENTS
We thank Calgary Laboratory Services Microbiology Department, Karen Poon at the Nicole Perkins Microbial Community Laboratory for the high-throughput sequencing, the nurses and clinic managers at the STI and SRH clinics for their invaluable roles in patient enrollment and data collection, and Matthew Workentine and Tannistha Nandi for their analytical advice and review of the manuscript.
Umer Zeeshan Imaj is funded by a NERC Independent Research Fellowship (NE/L011956/1) and a Lord Kelvin Adam Smith Leadership Fellowship (University of Glasgow).
E.D. is an employee of Thermo Fisher Scientific, and J.P.J. and J.A.S. are Coriell Life Sciences employees. E.D. oversaw the running of DNA samples on the qPCR platform in the Thermo Fisher laboratory. The qPCR presence/absence results after confidence filtering were shared for further analysis. J.P.J. and J.A.S. developed the BV diagnostic algorithm and provided insight regarding result interpretation of discordant results. All samples were coded with no additional information shared. All authors reviewed and approved the manuscript.
The qPCR assay run on the study samples was funded by Thermo Fisher Scientific. The samples were sent to the company with no information other than a study identification number, and so the commercial company did not have access to any other test results or patient information. No other monetary gains were had by any parties of the project.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.00300-19.
REFERENCES
- 1.Boris S, Barbés C. 2000. Role played by lactobacilli in controlling the population of vaginal pathogens. Microbes Infect 2:543–546. doi: 10.1016/S1286-4579(00)00313-0. [DOI] [PubMed] [Google Scholar]
- 2.Hickey RJ, Zhou X, Pierson JD, Ravel J, Forney LJ. 2012. Understanding vaginal microbiome complexity from an ecological perspective. Transl Res 160:267–282. doi: 10.1016/j.trsl.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gupta VK, Paul S, Dutta C. 2017. Geography, ethnicity or subsistence-specific variations in human microbiome composition and diversity. Front Microbiol 8:1162. doi: 10.3389/fmicb.2017.01162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SSK, McCulle SL, Karlebach S, Gorle R, Russell J, Tacket CO, Brotman RM, Davis CC, Ault K, Peralta L, Forney LJ. 2011. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A 108 Suppl:4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koumans EH, Sternberg M, Bruce C, McQuillan G, Kendrick J, Sutton M, Markowitz LE. 2007. The prevalence of bacterial vaginosis in the United States, 2001–2004; associations with symptoms, sexual behaviors, and reproductive health. Sex Transm Dis 34:864–869. doi: 10.1097/OLQ.0b013e318074e565. [DOI] [PubMed] [Google Scholar]
- 6.Wang H, Huang Z, Wu Z, Qi X, Lin D. 2017. An epidemiological study on vaginitis in 6,150 women of reproductive age in Shanghai. New Microbiol 40:113–118. [PubMed] [Google Scholar]
- 7.Bitew A, Abebaw Y, Bekele D, Mihret A. 2017. Prevalence of bacterial vaginosis and associated risk factors among women complaining of genital tract infection. Int J Microbiol 2017:4919404. doi: 10.1155/2017/4919404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paladine HL, Desai UA. 2018. Vaginitis: diagnosis and treatment. Am Fam Physician 97:321–329. [PubMed] [Google Scholar]
- 9.van de Wijgert J, Jespers V. 2017. The global health impact of vaginal dysbiosis. Res Microbiol 168:859–864. doi: 10.1016/j.resmic.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 10.Sobel JD. 2000. Bacterial vaginosis. Annu Rev Med 51:349–356. doi: 10.1146/annurev.med.51.1.349. [DOI] [PubMed] [Google Scholar]
- 11.Taylor BD, Darville T, Haggerty CL. 2013. Does bacterial vaginosis cause pelvic inflammatory disease? Sex Transm Dis 40:117–122. doi: 10.1097/OLQ.0b013e31827c5a5b. [DOI] [PubMed] [Google Scholar]
- 12.Bautista CT, Wurapa E, Sateren WB, Morris S, Hollingsworth B, Sanchez JL. 2016. Bacterial vaginosis: a synthesis of the literature on etiology, prevalence, risk factors, and relationship with chlamydia and gonorrhea infections. Mil Med Res 3:4. doi: 10.1186/s40779-016-0074-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leitich H, Bodner-Adler B, Brunbauer M, Kaider A, Egarter C, Husslein P. 2003. Bacterial vaginosis as a risk factor for preterm delivery: a meta-analysis. Am J Obstet Gynecol 189:139–147. doi: 10.1067/mob.2003.339. [DOI] [PubMed] [Google Scholar]
- 14.Ruban K, Grinceviciene S, Donders GGG, Vieira-Baptista P, Bellen G. 2017. Aerobic vaginitis: no longer a stranger. Res Microbiol 168:845–858. doi: 10.1016/j.resmic.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 15.Amsel R, Totten PA, Spiegel CA, Chen KC, Eschenbach D, Holmes KK. 1983. Nonspecific vaginitis. Diagnostic criteria and microbial and epidemiologic associations. Am J Med 74:14–22. doi: 10.1016/0002-9343(83)91112-9. [DOI] [PubMed] [Google Scholar]
- 16.Nugent RP, Krohn MA, Hillier SL, Zariffard MR, Cohen MH, Spear GT. 1991. Reliability of diagnosing bacterial vaginosis is improved by a standardized method of Gram stain interpretation. J Clin Microbiol 29:297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhat G, Kotigadde SS, Shenoy S. 2011. Comparison of the methods of diagnosis of bacterial vaginosis. J Clin Diagnostic Res 5:498–501. [Google Scholar]
- 18.Machado D, Castro J, Palmeira-de-Oliveira A, Martinez-de-Oliveira J, Cerca N. 2016. Bacterial vaginosis biofilms: challenges to current therapies and emerging solutions. Front Microbiol 6:1528. doi: 10.3389/fmicb.2015.01528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaambo E, Africa C, Chambuso R, Passmore JS. 2018. Vaginal microbiomes associated with aerobic vaginitis and bacterial vaginosis. Front Public Heal 6:78. doi: 10.3389/fpubh.2018.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cartwright CP, Pherson AJ, Harris AB, Clancey MS, Nye MB. 2018. Multicenter study establishing the clinical validity of a nucleic-acid amplification-based assay for the diagnosis of bacterial vaginosis. Diagn Microbiol Infect Dis 92:173–178. doi: 10.1016/j.diagmicrobio.2018.05.022. [DOI] [PubMed] [Google Scholar]
- 21.Van De Wijgert J, Borgdorff H, Verhelst R, Crucitti T, Francis S, Verstraelen H, Jespers V. 2014. The vaginal microbiota: what have we learned after a decade of molecular characterization? PLoS One 9:e105998. doi: 10.1371/journal.pone.0105998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.The Government of Alberta. 2018. Alberta treatment guidelines for sexually transmitted infections (STI) in adolescents and adults. Government of Alberta, Edmonton, Canada. [Google Scholar]
- 23.Church D, Miller B. 2011. Alberta guideline for laboratory processing and interpretation of vaginal specimens for bacterial vaginosis. Alberta Health Services, Edmonton, Canada. [Google Scholar]
- 24.Jarvis JP, Rains D, Kradel SJ, Elliott J, Diamond EE, Avaniss-Aghajani E, Yasharpour F, Shaman JA. 2018. Diagnosing bacterial vaginosis with a novel, clinically-actionable molecular diagnostic tool. J Appl Microb Res 1:1–8. [Google Scholar]
- 25.Bartram AK, Lynch MDJ, Stearns JC, Moreno-Hagelsieb G, Neufeld JD. 2011. Generation of multimillion-sequence 16S rRNA gene libraries from complex microbial communities by assembling paired-end Illumina reads. Appl Environ Microbiol 77:3846–3852. doi: 10.1128/AEM.02772-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ewels P, Magnusson M, Lundin S, Käller M. 2016. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.R Core Team. 2017. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 29.Srinivasan S, Hoffman NG, Morgan MT, Matsen FA, Fiedler TL, Hall RW, Ross FJ, McCoy CO, Bumgarner R, Marrazzo JM, Fredricks DN. 2012. Bacterial communities in women with bacterial vaginosis: high resolution phylogenetic analyses reveal relationships of microbiota to clinical criteria. PLoS One 7:e37818. doi: 10.1371/journal.pone.0037818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brandt BW, Bonder MJ, Huse SM, Zaura E. 2012. TaxMan: a server to trim rRNA reference databases and inspect taxonomic coverage. Nucleic Acids Res 40:W82–W87. doi: 10.1093/nar/gks418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schliep KP. 2011. phangorn: phylogenetic analysis in R. Bioinformatics 27:592–593. doi: 10.1093/bioinformatics/btq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McMurdie PJ, Holmes S. 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos PM, Stevens MHH, Szoecs E, Wagner H. 2018. vegan: Community Ecology Package. https://CRAN.R-project.org/package=vegan.
- 35.Arbizu M. 2017. pairwiseAdonis: pairwise multilevel comparison using adonis. R package version 0.0.1 R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 36.Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, Blomberg SP, Webb CO. 2010. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26:1463–1464. doi: 10.1093/bioinformatics/btq166. [DOI] [PubMed] [Google Scholar]
- 37.Swenson NG. 2009. Phylogenetic resolution and quantifying the phylogenetic diversity and dispersion of communities. PLoS One 4:e4390. doi: 10.1371/journal.pone.0004390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lê Cao KA, Boitard S, Besse P. 2011. Sparse PLS discriminant analysis: biologically relevant feature selection and graphical displays for multiclass problems. BMC Bioinformatics 12:253. doi: 10.1186/1471-2105-12-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rohart F, Gautier B, Singh A, Lê Cao KA. 2017. mixOmics: an R package for ‘omics feature selection and multiple data integration. PLoS Comput Biol 13:e1005752. doi: 10.1371/journal.pcbi.1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kostrytsia A, Papirio S, Morrison L, Ijaz UZ, Collins G, Lens PNL, Esposito G. 2018. Biokinetics of microbial consortia using biogenic sulfur as a novel electron donor for sustainable denitrification. Bioresour Technol 270:359–367. doi: 10.1016/j.biortech.2018.09.044. [DOI] [PubMed] [Google Scholar]
- 41.Callahan BJ, Sankaran K, Fukuyama JA, McMurdie PJ, Holmes SP. 2016. Bioconductor workflow for microbiome data analysis: from raw reads to community analyses. F1000Res 5:1492. doi: 10.12688/f1000research.8986.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang B, Fettweis JM, Brooks JP, Jefferson KK, Buck GA. 2014. The changing landscape of the vaginal microbiome. Clin Lab Med 34:747–761. doi: 10.1016/j.cll.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Donders GG. 2007. Definition and classification of abnormal vaginal flora. Best Pract Res Clin Obstet Gynaecol 21:355–373. doi: 10.1016/j.bpobgyn.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 44.Jespers V, Menten J, Smet H, Poradosú S, Abdellati S, Verhelst R, Hardy L, Buvé A, Crucitti T. 2012. Quantification of bacterial species of the vaginal microbiome in different groups of women, using nucleic acid amplification tests. BMC Microbiol 12:83. doi: 10.1186/1471-2180-12-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koren O, Knights D, Gonzalez A, Waldron L, Segata N, Knight R, Huttenhower C, Ley RE. 2013. A guide to enterotypes across the human body: meta-analysis of microbial community structures in human microbiome datasets. PLoS Comput Biol 9:e1002863. doi: 10.1371/journal.pcbi.1002863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van der Veer C, van Houdt R, van Dam A, de Vries H, Bruisten S. 2018. Accuracy of a commercial multiplex PCR for the diagnosis of bacterial vaginosis. J Med Microbiol 67:1265–1270. doi: 10.1099/jmm.0.000792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shipitsyna E, Roos A, Datcu R, Hallén A, Fredlund H, Jensen JS, Engstrand L, Unemo M. 2013. Composition of the vaginal microbiota in women of reproductive age–sensitive and specific molecular diagnosis of bacterial vaginosis is possible? PLoS One 8:e60670. doi: 10.1371/journal.pone.0060670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nyirjesy P, Kodsi S, Gaydos CA, Schwebke JR, Cooper CK, Paradis S. 2018. Diagnostic performance of a molecular test versus clinician assessment of vaginitis. J Clin Microbiol 56:e00252-18. doi: 10.1128/JCM.00252-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Schalkwyk J, Yudin MH, Yudin MH, Allen V, Bouchard C, Boucher M, Boucoiran I, Caddy S, Castillo E, Kennedy VL, Money DM, Murphy K, Ogilvie G, Paquet C, van Schalkwyk J. 2015. Vulvovaginitis: screening for and management of trichomoniasis, vulvovaginal candidiasis, and bacterial vaginosis. J Obstet Gynaecol Can 37:266–274. doi: 10.1016/S1701-2163(15)30316-9. [DOI] [PubMed] [Google Scholar]
- 50.Nasioudis D, Linhares IM, Ledger WJ, Witkin SS. 2017. Bacterial vaginosis: a critical analysis of current knowledge. BJOG 124:61–69. doi: 10.1111/1471-0528.14209. [DOI] [PubMed] [Google Scholar]
- 51.Korpela K, Zijlmans MAC, Kuitunen M, Kukkonen K, Savilahti E, Salonen A, de Weerth C, de Vos WM. 2017. Childhood BMI in relation to microbiota in infancy and lifetime antibiotic use. Microbiome 5:26. doi: 10.1186/s40168-017-0245-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Achilles SL, Austin MN, Meyn LA, Mhlanga F, Chirenje ZM, Hillier SL. 2018. Impact of contraceptive initiation on vaginal microbiota. Am J Obstet Gynecol 218:622.e1. doi: 10.1016/j.ajog.2018.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fettweis JM, Paul Brooks J, Serrano MG, Sheth NU, Girerd PH, Edwards DJ, Strauss JF, Jefferson KK, Buck GA. 2014. Differences in vaginal microbiome in African American women versus women of European ancestry. Microbiology 160:2272–2282. doi: 10.1099/mic.0.081034-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fosch SE, Ficoseco CA, Marchesi A, Cocucci S, Nader-Macias MEF, Perazzi BE. 2018. Contraception: influence on vaginal microbiota and identification of vaginal lactobacilli using MALDI-TOF MS and 16S rDNA sequencing. Open Microbiol J 12:218–229. doi: 10.2174/1874285801812010218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brown RG, Marchesi JR, Lee YS, Smith A, Lehne B, Kindinger LM, Terzidou V, Holmes E, Nicholson JK, Bennett PR, MacIntyre DA. 2018. Vaginal dysbiosis increases risk of preterm fetal membrane rupture, neonatal sepsis and is exacerbated by erythromycin. BMC Med 16:9. doi: 10.1186/s12916-017-0999-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brotman RM, Latey Bradford L, Conrad M, Gajer P, Ault K, Peralta L, Forney LJ, Carlton JM, Abdo Z, Ravel J. 2012. Association between Trichomonas vaginalis and vaginal bacterial community composition among reproductive-age women. Sex Transm Dis 39:807–812. doi: 10.1097/OLQ.0b013e3182631c79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hyman RW, Fukushima M, Jiang H, Fung E, Rand L, Johnson B, Vo KC, Caughey AB, Hilton JF, Davis RW, Giudice LC. 2014. Diversity of the vaginal microbiome correlates with preterm birth. Reprod Sci 21:32–40. doi: 10.1177/1933719113488838. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.