Abstract
Background
Myocardial infarction (MI) is a life-threatening disease, often leading to heart failure. Defining therapeutic targets at an early time point is important to prevent heart failure.
Methods
MicroRNA screening was performed at early time points after MI using paired samples isolated from the infarcted and remote myocardium of pigs. We also examined the microRNA expression in plasma of MI patients and pigs. For mechanistic studies, AAV9-mediated microRNA knockdown and overexpression were administrated in mice undergoing MI.
Findings
MicroRNAs let-7a and let-7f were significantly downregulated in the infarct area within 24 h post-MI in pigs. We also observed a reduction of let-7a and let-7f in plasma of MI patients and pigs. Inhibition of let-7 exacerbated cardiomyocyte apoptosis, induced a cardiac hypertrophic phenotype, and resulted in worsened left ventricular ejection fraction. In contrast, ectopic let-7 overexpression significantly reduced those phenotypes and improved heart function. We then identified TGFBR3 as a target of let-7, and found that induction of Tgfbr3 in cardiomyocytes caused apoptosis, likely through p38 MAPK activation. Finally, we showed that the plasma TGFBR3 level was elevated after MI in plasma of MI patients and pigs.
Interpretation
Together, we conclude that the let-7-Tgfbr3-p38 MAPK signalling plays an important role in cardiomyocyte apoptosis after MI. Furthermore, microRNA let-7 and Tgfbr3 may serve as therapeutic targets and biomarkers for myocardial damage.
Fund
Ministry of Science and Technology, National Health Research Institutes, Academia Sinica Program for Translational Innovation of Biopharmaceutical Development-Technology Supporting Platform Axis, Thematic Research Program and the Summit Research Program, Taiwan.
Keywords: MicroRNA let-7, TGFBR3, Gene therapy, Biomarkers
Research in Context section.
Evidence before this study
MicroRNAs have been studied using rodent disease models. Given the large burden of coronary artery disease and limited success of pharmacological therapies to blunt myocardial infarct size and remodelling, we considered whether there is a research gap between rodent animal studies and clinics. Conducting the current studies using pigs is optimal for several reasons. In particular, there are close similarities in MI pathophysiology between pigs and humans; comparative whole genome sequencing studies have shown that compared to mice, genomic microRNA sequences in pigs are more closely evolutionarily conserved with humans.
Added value of this study
We identified microRNAs let-7a and let-7f as novel factors that are significantly downregulated in the infarcted myocardium in pigs within 24 h post-MI. The plasma levels of let-7a and let-7f were also decreased in pigs and in ST elevation MI patients. AAV9 mediated let-7 inhibition or overexpression suggested that let-7a and let-7f are cardiac protective factors after injury. In addition, we proposed that Let-7-TGFBR3-p38 MAPK signalling regulates cardiomyocyte apoptosis. Moreover, plasma TGFBR3 in STEMI patients also responds to cardiac injury in pigs and humans.
Implications of all the available evidence
This study provides translational potential for prevention of heart failure after infarction. For example, administration of let-7a and let-7f or TGFBR3 antagonist may protect STEMI patients from deterioration. Furthermore, the plasma levels of microRNA let-7a, let-7f and TGFBR3 may serve as diagnosis and prognosis biomarkers for patients with MI.
Alt-text: Unlabelled Box
1. Introduction
Cardiomyocyte apoptosis is considered a significant event during the development of cardiomyopathy. Because the mammalian heart has minimal regenerative capacity [1,2], protection of cardiomyocytes from injury is extremely important for preventing progression to heart failure. Rodent animal models of acute myocardial infarction (MI) are well established for the study of the pathogenesis from MI to heart failure. Several critical genes, microRNAs and long-noncoding RNAs have been identified from the rodent MI model. However, little is known about how these studies relate to clinical MI or whether these candidates can be utilized for clinical applications. Considering these issues, we chose pig as our experimental animal. Pigs are similar to humans in physiology and pathophysiology after MI [3], and comparative whole genome sequencing studies have shown that compared to mice, genomic sequences, including microRNAs, in pigs are closely evolutionarily conserved with humans [4]. MicroRNAs are considered good targets for gene therapy because they are small enough to be synthesized and modified. In this study, we identified microRNAs whose expression responds to MI at the acute phase and explored their underlying mechanisms to further evaluate their potential for gene therapy.
The microRNA let-7 family was one of the earliest microRNA families discovered [5]. It is evolutionarily conserved, and is required for development in most organisms [5]. It has been widely studied in cancer [6], pluripotency [7] and diabetes [8]. A recent study has suggested that the let-7 family is the most up-regulated microRNA (miRNA) family member in matured cardiomyocytes derived from human embryonic stem cells (hESC-CMs) during one year maturation [9]. Overexpression of let-7 family members in hESC-CMs enhances maturation mediated by metabolic switch [9]. However, the role of let-7 in the context of MI is very controversial. For example, Seeger et al. reported that let-7 is upregulated after infarction and inhibition of let-7 protects heart function through epithelial–mesenchymal transition of epicardial cells [10]. Conversely, Sun et al. demonstrated that another let-7 member, miR-98, is downregulated after MI, and administration of miR-98 agomir protects cardiomyocytes from apoptosis through the Fas-Caspase3 pathway [11]. These opposite results may be due to the technical variations in these small animal studies, such as the delivery method in both studies being based on local injection of chemically modified nucleotides, which requires a high level of expertise and is not applicable to the clinic. To restart the investigation, a cardiac tropism virus, adeno-associated virus 9 (AAV9), was used in our study to deliver let-7 inhibitor and expression vector by intravenous (i.v.) injection. Gene delivery by AAVs is less invasive and has been used in clinics [12].
Transforming growth factor beta receptor III (TGFBR3), also named Betaglycan, is a coreceptor of the TGF-β superfamily. It can bind TGFb1, TGFb2, TGFb3 [13], BMPs [14], inhibin [15] and bFGF [16] to modulate signalling. TGFBR3 is important for cardiac development, especially endocardial or pericardial cell migration and proliferation. Early in 1999, Brown et al. demonstrated that TGFBR3 is required for endocardial cell epithelial–mesenchymal transition in the heart responding to TGFb2 [17]. Knockout of TGFBR3 in mice led to embryonic lethality at embryonic day 13.5 with heart and liver defects [18]. In addition to its role in signalling, TGFBR3 can also undergo ectodomain shedding, releasing a soluble form into the extracellular space [19]. A ligand independent role of TGFBR3 has also been proposed to inhibit myogenesis and promote fibrosis, through p38 MAPK signalling, and the cytoplasmic domain of TGFBR3 is required [20]. However, its role in cardiovascular disease is unclear.
In this study, we carefully examined the expression of microRNAs during the acute phase of MI in the infarct and the remote regions of pig hearts. Blood samples from ST elevation myocardial infarction (STEMI) patients were collected to evaluate the association between the animal studies and the clinical situation. We also re-investigated the role of microRNA let-7 in a mouse MI model using AAV9 with i.v. injection and identified the important downstream effects of let-7, including the role of its target gene TGFBR3 in cardiomyocyte apoptosis.
2. Materials and methods
Detailed methods are available in the Supplemental Materials.
2.1. LAD ligation in a porcine model
MI was induced by permanent ligation of the left anterior descending (LAD) artery in ~22 kg Lanyu minipigs. The ligation site and subsequent infarct area are indicated in Fig. S1A. Success of MI surgery was confirmed by the presence of ST-segment elevation in ECG recordings (Fig. S1B).
2.2. LAD ligation and echocardiography
Ten-week-old mice were anesthetized by isofluorane (Halocarbon). The left anterior descending coronary artery was visualized and occluded. Heart function was evaluated with echocardiography, when heart rates were 400–500 bpm, LV end-systolic and end-diastolic diameter were measured using M-mode in 3 short-axis views, at papillary muscle, between papillary muscle and apex and apex, and used to calculate ejection fraction. The results for each mouse were the average from 3 views. All experiments involving animals were conducted in accordance with the Guide for the Use and Care of Laboratory Animals. All animal protocols were approved by the Institutional Animal Care and Use Committee of Academia Sinica (IACUC No. 106083 and 11–09-211).
2.3. Total RNA and plasma miRNA extraction
Total RNA was extracted from tissues or cells using Trizol reagent (Invitrogen). To detect plasma miRNAs, MirVana PARIS Kit (Ambion) and miRNeasy Serum/Plasma Kit (Qiagen) were used following the manufacturer's protocol. In the MirVana method, small RNAs were extracted from 200 μl of plasma with five femto-moles of synthetic microRNA, cel-mir-39, as an internal control for technical variation. In the miRNeasy method, 100 μl of plasma were used with 40 million copies of cel-mir-39 as control.
2.4. Preparation of adeno-associated virus 9
Let-7 Tough Decoy RNA sequence was sub-cloned from MISSION microRNA Inhibitors (Sigma) and let-7a/f overexpressing sequence was sub-cloned from pLKO_TRC014-let-7 into pAAV-U6-GFP plasmid (Vigene Biosciences). Adeno-associated viral vectors were produced according to a previously published protocol with minor modification [21]. Three plasmids: pHelper, pXX9 and expression plasmid (pAAV-U6-GFP, pAAV-U6-TuD or pAAV-U6-let-7) were co-transfected into 293 T cells using calcium phosphate. Viral particles were purified by two rounds of CsCl2 gradient centrifugation and titered by real-time PCR using AAV-EGFP primers (Table S3).
2.5. Statistics
Data are presented as means ± SEM with numbers (N) indicated in the bars or Figure legends. Differences between two groups were analysed by unpaired or paired Student's t-test. Differences between multiple groups were analysed by one-way ANOVA with post-hoc Tukey HSD Test. Differences between control group and other groups were analysed by one-way ANOVA with Dunnett's test. P values of <0.05 are indicated by one asterisk (*), P values of less or equal than 0.01 are indicated by with two asterisks (**) and P values of less or equal than 0.001 are indicated by three asterisks (***).
2.6. Study approval
IRB approval (AS-IRB02–06163) was granted for plasma samples obtained from human samples.
3. Results
3.1. Dynamics of let-7a and let-7f expression over 24 hours following acute myocardial infarction in pigs and humans
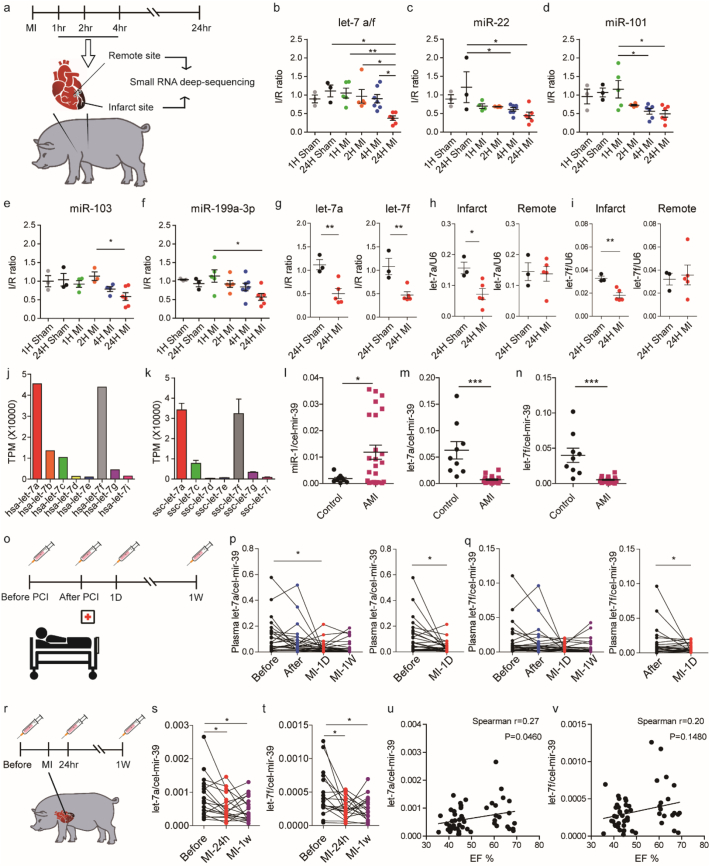
To characterize the changes in microRNA expression following acute MI in pigs, myocardial samples were collected from the infarct and remote areas of pigs that were sacrificed at 1, 2, or 4 h post-MI for small RNA deep-sequencing (NGS). Remote myocardial samples were additionally utilized as internal controls to account for intra-pig variation in baseline miRNA expression (Fig. 1A). TUNEL staining was also performed in porcine hearts at 24 h post-MI (Fig. S1C) to verify that appropriate sampling of infarct and remote areas had been performed.
Fig. 1.
Expression of micro RNA let-7a and let-7f is decreased after acute myocardial infarction in experimental pigs and human patients.
(A) Experimental design. Pigs were subjected to LAD ligation and sacrificed after 1, 2 or 4 h. Next-generation sequencing was performed on RNA extracts from both infarct and remote tissues. Tissue was also collected from sham animals and a 24-h post-MI group. (B—F) Stem-loop qPCR validation of miRNA expression in sham and infarcted hearts at different time points. I/R ratio indicates the ratio of miRNA expression level in the infarct compared to the remote area, which was used as an internal control for each individual pig. (G) TaqMan qPCR validation of let-7a and let-7f expression in sham and infarcted hearts. (H—I) U6-normalized let-7a and let-7f expression levels in the infarct and remote area at 24 h post MI. (J) Abundance of let-7 family members in human hearts based on NGS data from Meunier, J. et al., 2013 [25]. (K) Abundance of let-7 family members in pig hearts based on next-generation sequencing data derived from the remote area of three individual pigs. TPM: transcripts per million. (L) Human plasma miR-1 expression levels in healthy subjects (n = 9) and acute MI patients (n = 24). (M-N) Human plasma let-7a and let-7f expression levels in healthy subjects (n = 9) and acute MI patients (n = 24). Samples were collected by National Taiwan University Hospital (NTUH) when patients arrived in the emergency room. Plasma microRNAs were extracted by the MirVana method with 5 f-moles of cel-mir-39 as a spike-in control. (O) Experimental design. Human plasma were collected at different time points: before percutaneous coronary intervention (PCI), after PCI, 1 day after MI and one week after MI. (P-Q) Human plasma let-7a and let-7f expression levels at several time points: before therapeutic intervention (Before), after therapeutic intervention (After), 1 day and 1 week post-MI (MI-1D and MI-1 W) (n = 28). Samples were collected by Far Eastern Memorial Hospital (FEMH), Taipei, and extracted by the Qiagen method with 40 million copies of cel-mir-39 RNA used as a spike-in control. Differences between the before group and the MI-1D group were analysed by paired Student's t-test (*, P < .05). (R) Experimental design. Pig plasma samples were collected from pigs subjected to the MI model at different time points: before MI (Before), one day and one week post-MI (MI-24 h and MI-1w). (S-T) Pig plasma let-7a and let-7f expression levels at baseline, 1 day and 7 days post-MI. Plasma microRNAs were extracted by the MirVana method with 5 f-moles of cel-mir-39 as a spike-in control (n = 18). (U—V) Correlation of plasma let-7a/f expression and ejection fraction in pigs. Data are presented as means ± SEM. Differences between multiple groups (B—F) were analysed by one-way ANOVA with post-hoc Tukey HSD Test. (*, P < .05; **, P < .001). Differences between two groups (G-I, L-N) were analysed by unpaired Student's t-test. (*, P < .05; **, P < .001; ***, P < .0001). Differences between control group and other groups (P-Q, S-T) were analysed by one-way ANOVA with Dunnett's test. (*, P < .05; **, P < .001).
Heart enriched microRNAs were selected as shown in online Table I for further candidate selection. Based on changes in microRNAs at 4 time points, 1, 2, 4 and 24 h post MI, in the infarct relative to the remote area (I/R ratio), microRNAs let-7a, let-7f, miR-22, miR-101, miR-103 and miR-199a-3p were selected as candidates for further validation using stem-loop qPCR analysis. The entire panel of microRNAs showed significant downregulation at 24 h post-MI (Fig. 1B-1F). Next, we performed the same model in mice 24 h after MI. Interestingly, only let-7a/f and miR-101 demonstrated the same patterns in downregulation after MI, with the other candidates showing no change in expression level (Fig. S1E—I). This suggested that microRNAs let-7a/f and miR-101 are conserved in the regulatory response to acute MI in pigs and mice.
Given the conservation of let-7a and let-7f expression across mice and pigs, we selected these microRNAs amongst the candidates as our focus for further studies. To this end, we used specific TaqMan probes to specifically distinguish between the expression levels of let-7a and let-7f, and confirmed that both showed significant downregulation in the infarct at 24 h post-MI (Fig. 1G). In mice, such downregulation persisted after 1 and 2 days post-MI and recovered 6 days post-MI (Fig. S1J—K). Specifically, we observed that let-7a was downregulated in the peri-infarct cardiomyocytes (Fig. S1L). This result contrasts with Seeger et al. who reported that, in mice, let-7a is upregulated in the myocardial border zone over the course of the first week after MI [10]. To verify whether the apparent decrease in let-7a and let-7f I/R ratio in pigs was actually due to an upregulation in the remote area affecting the I/R ratio calculation, the expression levels of let-7a and let-7f relative to U6 in 24-h infarct and remote samples were further compared. Decreased I/R ratio was confirmed to be due to significant downregulation of let-7a and let-7f in the infarct area, with the remote area showing no significant changes in the expression level (Fig. 1H and I). Thus, our results are more in-line with other studies which demonstrated that a downregulation of let-7 occurred in early MI [11,22,23].
Previously published databases have shown that in human hearts, the most abundant let-7 members are let-7a and let-7f [24] (Fig. 1J). Our NGS data also showed that let-7a and let-7f were the most abundant let-7 members in pig heart tissues (Fig. 1K), whilst mice show an abundance of let-7a and let-7b, let-7c and let-7f [24] (Fig. S1D), suggesting that the basal expression of microRNAs in pigs is closer to that in humans.
Let-7a has previously been reported to be detectable within extracellular vesicles in human plasma [25]. To investigate whether these findings would be clinically relevant, we evaluated the expression of plasma let-7a and let-7f in patients with ST elevation myocardial infarction (STEMI). Samples were collected independently from two hospitals. In samples from National Taiwan University Hospital, the plasma was collected when patients arrived in the emergency room. MiR-1, which was reported to be upregulated upon MI [26], was used as a positive control. Accordingly, we observed an increase in plasma miR-1 in our patient samples when compared to the healthy subjects (Fig. 1L). In these samples, we observed that the plasma levels of let-7a and let-7f were significantly lower in the plasma of MI patients (Fig. 1M and N). In order to trace the dynamic changes of plasma let-7a and let-7f in MI patients, another study was initiated at Far Eastern Memorial Hospital. The plasma was collected at different time points: before percutaneous coronary intervention (PCI), after PCI, 1 day after MI and one week after MI (Fig. 1O). During this time course, we noticed that the plasma let-7a was significantly downregulated one day after MI (Fig. 1P and Q). Meanwhile, we tried to compare the concentration of cardiac troponin T, a conventional cardiac injury biomarker, with the expression of plasma let-7 and found there was no correlation. Cardiac troponin T expression rose immediately after PCI (Fig. S1M), but the expression of plasma let-7 remained the same before and after PCI (Fig. S1N—O). This suggests that their regulatory mechanism may be independent. In order to eliminate the influence of therapeutic treatment and the variety of individuals, plasma samples were also collected from pigs subjected to the MI model at different time points: before MI, one day and one week post-MI (Fig. 1R). Similar to the findings in humans, we observed that both let-7a and let-7f levels were significantly decreased at both one day and one week post-MI (Fig. 1S and T). Interestingly, both let-7a and let-7f expression levels showed a trend towards correlation with left ventricular function as indicated by echocardiography-derived ejection fraction (Fig. 1U and V). These results indicate that tissue injury after MI can lead to changes in the expression levels of circulating let-7a and let-7f, and that these changes are conserved across mice, pigs and humans.
To gain mechanistic insight into the downregulation of let-7a/f upon MI, primary let-7a/f transcript, pre-let-7a-1, pre-let-7a-2, pre-let-7f-2 and LIN28A expression levels were also evaluated. LIN28A is an RNA binding protein which was shown to repress let-7 maturation during development [7] and disease [27], and is involved in several biological processes [8,28]. Interestingly, we found that the primary let-7a and let-7f cluster transcript was not decreased in the infarct area (Fig. S2A). Instead, pre-let7a-1, pre-let-7a-2 and pre-let-7f-2 were all upregulated (Fig. S2B-D). The data indicated that the biogenesis of let-7 was impaired. Although LIN28A mRNA expression was increased (Fig. S2E), there was no significant difference in the protein level between the infarct and remote areas (Fig. S2F-G), suggesting that let-7a/f downregulation in the infarct area could be due to a LIN28A-independent repression of transcript maturation rather than a reduction of let-7a/f transcription. Another potential regulator of let-7 maturation is the adenosine deaminase ADAR1, by adenosine-to-inosine RNA editing [29]. Its expression was also examined in our pig MI model. Although there was no significant difference at its mRNA and protein expression level after infarction (Fig. S2H-J), we noticed an alteration in post-translational modification (PTM) of ADAR1, a putative sumoylation band, which may inhibit the activity of ADAR1 [30]. The PTM of ADAR1 was diminished in the infarct area (Fig. S2I), which means the enzyme activity of ADAR1 might be higher in the infarct area and then inhibit the maturation of let-7.
3.2. Inhibition of let-7 in vivo leads to exacerbation of cardiac dysfunction after myocardial infarction in mice
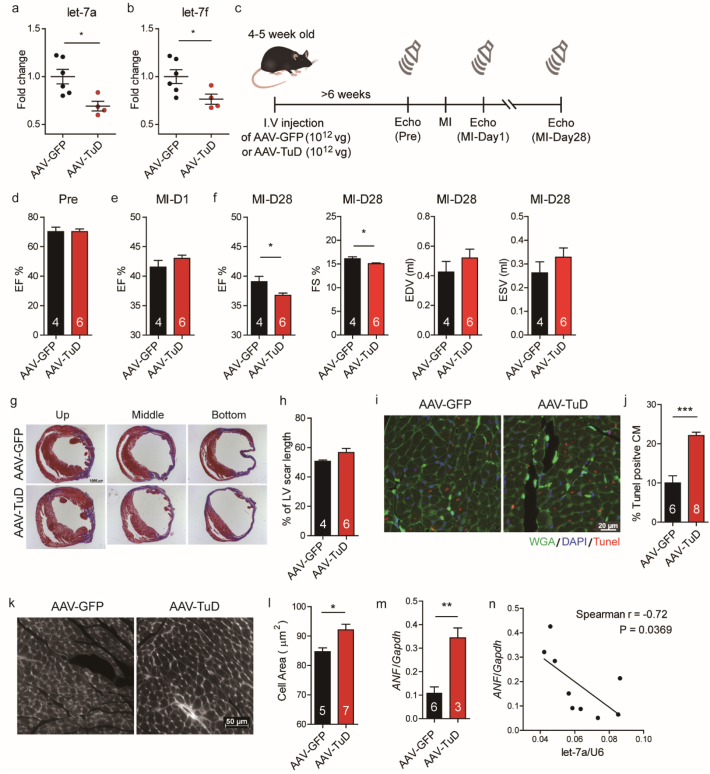
As mentioned above, the function of let-7 is unclear; therefore, we decided to re-evaluate its role in MI. We performed an in vivo loss-of-function study using Tough Decoy (TuD) RNA - an artificial sequence containing two miRNA recognition sites designed to suppress microRNA expression [31]. It was previously shown by Xie et al. that adeno-associated virus (AAV)-mediated TuD expression leads to superior long-term microRNA inhibition efficiency in mice [32]. In addition, AAV9 is myocardium tropic when systemically delivered into mice [33]. Thus, AAV9 viruses carrying let-7 targeting TuD RNA (AAV-TuD) were generated.
To ensure the expression of transduced plasmids, mice were intravenously injected with AAV-TuD or AAV-GFP for six weeks before induction of MI, and the knockdown efficiency of cardiac let-7a and let-7f levels were confirmed (Fig. 2A and B). We subsequently examined the impact of let-7a and let-7f knockdown on MI-induced cardiac dysfunction. Serial echocardiography was performed before MI, one day and one month post-MI (Fig. 2C). We found that at one month post-MI, AAV-TuD-infected mice displayed exacerbated cardiac dysfunction compared to the AAV-GFP control group (Fig. 2F). Although there was a trend towards increased LV scar length in the AAV-TuD group, this did not reach statistical significance (Fig. 2G and H). Further, TUNEL staining showed a two-fold increase in the number of apoptotic cardiomyocytes in the border zone of AAV-TuD-infected hearts (Fig. 2I and J). Unexpectedly, we found that AAV-TuD-infected cardiomyocytes already began to display increased cell size at one day post-MI in the remote area (Fig. 2K and L). We further checked atrial natriuretic factor (ANF) expression, a marker of hypertrophy, and observed a higher expression level of ANF in the AAV-TuD mice compared to the AAV-GFP mice (Fig. 2M). Interestingly, ANF expression was inversely correlated with that of let-7a (Fig. 2N). To understand the role of let-7 in cardiac fibrosis during cardiac remodelling, fibrosis-related genes were analysed post-MI day 1, day 6, and day 28. The result showed the fibrosis-related genes were increased 6 days post infarction, but there was no significant difference between AAV-GFP mice and AAV-TuD mice (Fig. S3). These results indicate that let-7a/f inhibition leads to exacerbation of cardiac dysfunction after MI, likely through increased apoptosis and cell death upon infarction. This was also associated with evidence of early onset cardiomyocyte hypertrophy. Taken together, these results suggest that let-7a/f could have cardioprotective potential following injury.
Fig. 2.
AAV9-mediated let-7 knockdown increases cardiomyocyte apoptosis and reduces cardiac function after experimental myocardial infarction in mice.
(A-B) Myocardial expression levels of let-7a and let-7f 6 weeks after injection of AAV9 viruses carrying either GFP (AAV-GFP) or TuD (AAV-TuD) expressing plasmids. (C) Experimental schematic. Mice were injected with AAV-GFP or AAV-TuD 6 weeks prior to induction of MI. Serial echocardiography was performed to evaluate heart function before surgery, 1 day and 28 days after MI. (D) Ejection fraction (EF) in AAV-TuD and AAV-GFP infected mice before MI. (E) Ejection fraction in AAV-TuD and AAV-GFP infected mice one day after MI. (F) Ejection fraction, fraction shortening (FS), end diastolic volume (EDV) and end systolic volume (ESV) in AAV-TuD and AAV-GFP infected mice one month after MI. (G) Representative trichrome staining of LV sections 28 days post-MI. (H) Quantification of LV scar length 28 days post-MI. (I) TUNEL staining of mouse peri-infarct tissue 1 day post MI. (J) Quantification of TUNEL positive cells in AAV-TuD and AAV-GFP hearts 1 day post-MI. (K) WGA staining for cell size quantification. (L) Quantification of cell size. (M) ANF expression level in let-7 inhibition hearts. (N) Correlation of ANF and let-7a expression level in mouse hearts. Data are presented as means ± SEM. Differences between groups were analysed by unpaired Student's t-test (*, P < .05; **, P < .01; ***, P < .001).
3.3. AAV9-mediated let-7 overexpression in vivo preserves heart function in myocardial infarction
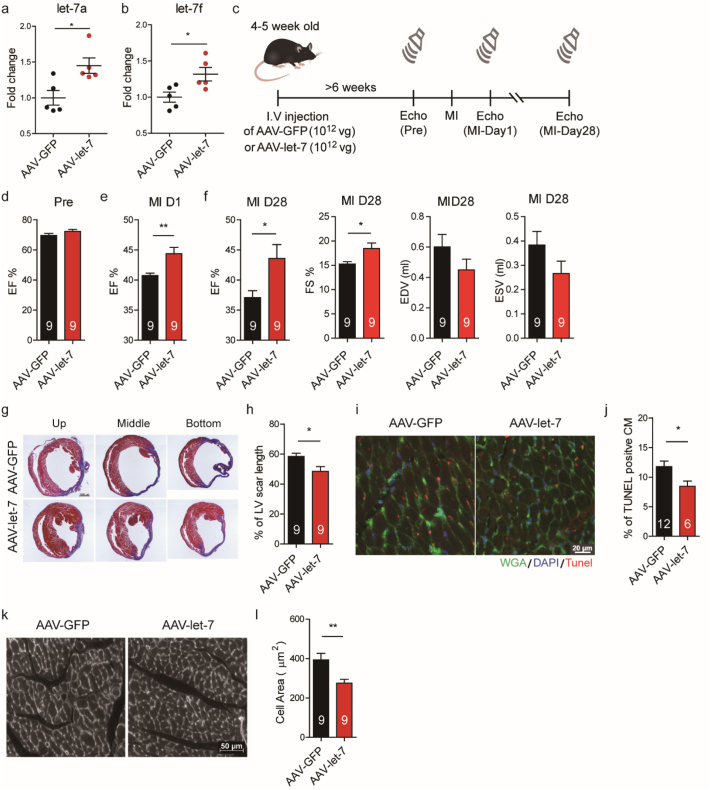
Having observed that let-7 inhibition impaired cardiac function by increasing cell apoptosis and cardiac hypertrophy, we tested whether let-7 overexpression could be applied to protect heart function. To this end, let-7 overexpression was achieved using AAV9 viruses carrying a microRNA cluster let-7a and let-7f expressing plasmid (AAV-let-7). Mice were intravenously injected with AAV-let-7 six weeks before MI induction and confirmed to demonstrate significant cardiac let-7a and let-7f upregulation compared to control (Fig. 3A and B).
Fig. 3.
AAV9-mediated let 7 overexpression reduces cardiomyocyte apoptosis and improves cardiac function after experimental myocardial infarction in mice.
(A-B) Let-7a/f overexpression in mouse hearts after infection with either AAV-let-7a/f or AAV-GFP. (C) Experimental schematic. Mice were intravenously injected with AAV9 viruses carrying let-7 expressing plasmid for at least six weeks before LAD ligation. Heart function was evaluated by echocardiography before surgery, one day and four weeks after MI. (D) Ejection fraction in AAV-let-7 and AAV-GFP infected mice before MI. (E) Ejection fraction in AAV-let-7 and AAV-GFP infected mice one day after MI. (F) Ejection fraction, fraction shortening, end diastolic volume and end systolic volume in AAV-let-7 and AAV-GFP infected mice one month after MI. (G) Trichrome staining of LV sections 28 days post-MI. (H) Quantification of scar length 28 days post-MI. (I-J) Reduction of cardiomyocyte apoptosis in the border zone following let-7a/f overexpression based on TUNEL staining. (K-L) Attenuation of cardiomyocyte hypertrophy in the remote area 28 days post-MI following let-7a/f overexpression based on WGA staining. Data are presented as means ± SEM. Differences between groups were analysed by unpaired t-test (*, P < .05; **, P < .01).
Serial echocardiography was performed to evaluate the heart function after MI (Fig. 3C). Surprisingly, the results revealed that mice receiving AAV-let-7 showed a significant increase in cardiac function at one month post-MI (Fig. 3F), and such protection appeared as early as one day post-MI (Fig. 3E). In addition, there was a significant reduction in the scar length (Fig. 3G and H), and a reduction in degree of cardiomyocyte hypertrophy in the remote area (Fig. 3K and L). TUNEL staining performed one day post-MI revealed a significant reduction in apoptotic cells in the border zone of mice receiving AAV-let-7 (Fig. 3I and J).
Taken together, these results suggest that let-7a/f is a cardioprotective factor and that AAV-mediated let-7 delivery can confer therapeutic benefit. This is likely due to a reduction of cardiomyocyte apoptosis in the acute phase of MI, leading to subsequent preservation of myocardial integrity and cardiac function going into the chronic phase.
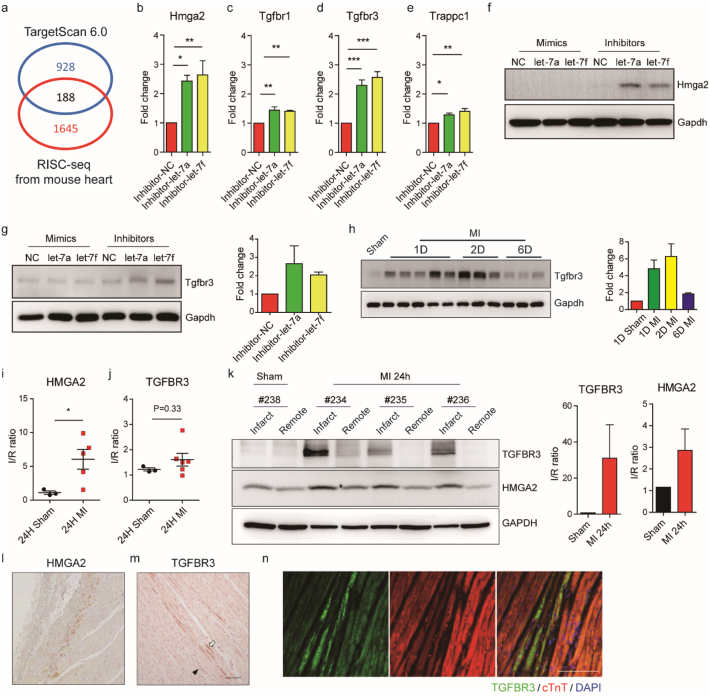
3.4. Hmga2, Tgfbr1, Tgfbr3 and Trappc1 are potential targets of let-7 in Cardiomyocytes
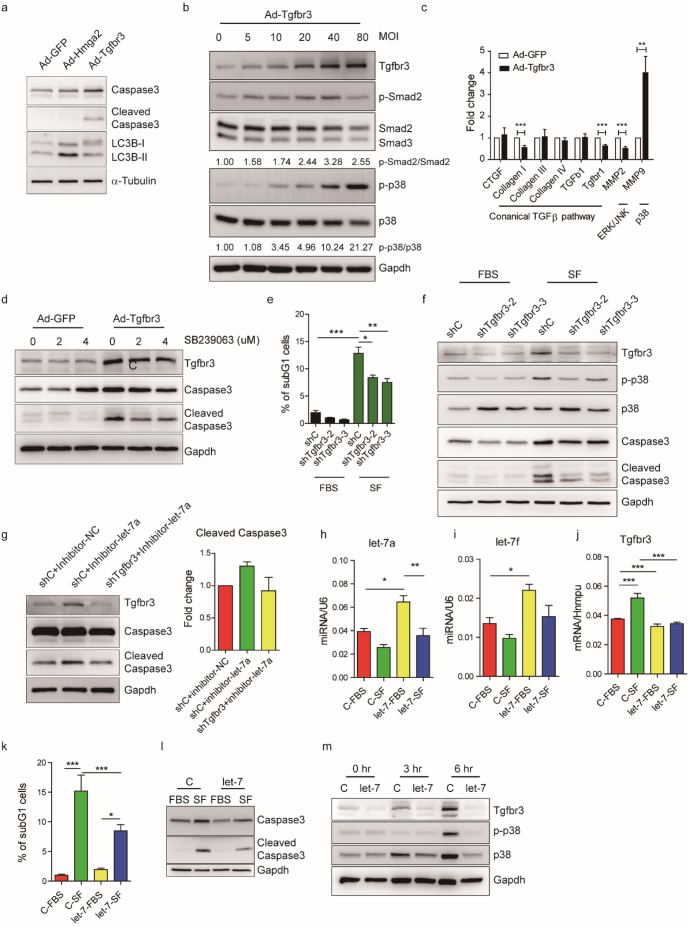
To identify the downstream targets of let-7a and let-7f, an existing heart RISC-seq database [34] was used in combination with TargetScan software [35]. We selected 29 candidates from 188 potential targets (Fig. 4A) with Hmga2 as a positive control [36,37]. They were subsequently validated in rat neonatal cardiomyocytes to examine response to let-7a and let-7f inhibition (Fig. S4). Following transfection of let-7a and let-7f inhibitors, we observed significant upregulation amongst several of the candidate genes, including Hmga2, Tgfbr1, Tgfbr3 and Trappc1 (Fig. 3B-3E and Fig. S4). Based on the degree of fold-change in gene expression levels, we selected Hmga2 and Tgfbr3 to confirm the induction of protein expression following let-7a and let-7f inhibition (Fig. 4F and G).
Fig. 4.
HMGA2 and TGFBR3 are let-7 targeted genes and are highly expressed in cardiomyocytes after acute infarction.
(A) Overview of target gene identification. A heart RISC-seq database [34] and TargetScan 6.0 prediction [35] were used to identify candidate genes from a common pool of 188. (B-E) Gene expression of Hmga2, Tgfbr1, Tgfbr3 and Trappc1 after transfection of let-7a and let-7f inhibitors in rat neonatal cardiomyocytes. Histograms and error bars indicate mean and SEM, respectively, from four independent experiments. Differences between the control group and other groups were analysed by one-way ANOVA with Dunnett's test. (*, P < .05; **, P < .001; **, P < .0001). (F) Western blot of Hmga2 in rat neonatal cardiomyocytes transfected with let-7a and let-7f mimics and inhibitors. (G) Western blot of Tgfbr3 in rat neonatal cardiomyocytes transfected with let-7a and let-7f mimics and inhibitors. Quantification of western blot is shown in the histogram. Histograms and error bars indicate mean and SEM, respectively, from two independent experiments. (H) Characterization of Tgfbr3 expression in sham and 1, 2 and 6 day post-MI in mice. Quantification of western blot is shown in the histogram. Histograms and error bars indicate mean and SEM, respectively. (I-J) HMGA2 and TGFBR3 RNA expression levels in pig hearts 1 day post-MI. Data are presented as means ± SEM. Differences between groups were analysed by unpaired Student's t-test (*, P < .05). (K) Western blot of TGFBR3 and HMGA2 in pig infarct and remote tissues 1 day post-MI. Quantification of western blot is shown in the histogram. Histograms and error bars indicate mean and SEM, respectively. (L) Immunochemistry staining of HMGA2 in pig infarct tissue 1 day post-MI. (M) Immunochemistry staining of TGFBR3 in pig infarct tissue 1 day post-MI. TGFBR3 positive cells were indicated by white arrows. Hematoxylin was used to stain nuclei. Clustered nuclei are indicated by black arrow heads. Scale bar: 100 μm. (N) Immunofluorescence staining of TGFBR3 and cardiac troponin T in pig infarct tissue 1 day post-MI. DAPI was used to stain cell nuclei. Scale bar: 100 μm.
Hmga2 has previously been implicated in cardiogenesis in Xenopus, and the expression of Hmga2 in a mouse model of MI has been reported to be upregulated one day after MI [38]. Tgfbr3, also named Betaglycan, is a co-receptor of the TGF-β superfamily, and has been reported to have multiple functions in a variety of cancers [39,40]. Recently, it has been confirmed to be a direct target of let-7 through luciferase assay [41]. To detect mouse endogenous Tgfbr3, we generated anti-mouse Tgfbr3 monoclonal antibodies. Using a homemade antibody for western blotting, we noticed that endogenous Tgfbr3 expression was induced in the acute phase of MI and then subsequently downregulated (Fig. 4H). Over the same period of time, we observed an inverse trend in let-7a and let-7f expression relative to that of Tgfbr3 (Fig. S1J and S1K). This contrast suggests an antagonistic relationship between the two factors. We subsequently examined HMGA2 and TGFBR3 expression in pig samples taken 24 h post-MI. The protein and the mRNA of HMGA2 was upregulated at 24 h post-MI (Fig. 4I and K). Although there was no significant change in mRNA, TGFBR3 protein was notably upregulated in the pig infarct (Fig. 4J and K). MicroRNAs regulate gene expression by translational suppression and mRNA degradation [42]. In the case of TGFBR3 protein expression, post-transcriptional regulation, such as translation, may be more preferable in vivo.
We further performed immunohistochemistry to visualize the expression of HMGA2 and TGFBR3 in the pig infarct (Fig. 4L and M). Interestingly, the expression of HMGA2 was localized to a boundary-like strip between the necrotic and intact myocardium (Fig. 4L). TGFBR3 was found in cardiomyocytes surrounded by immune infiltrate based on proximity to patches of clustered nuclei (Fig. 4M and N). Taken together, these results suggest that HMGA2 and TGFBR3, two potential target genes of let-7 in the myocardium, are expressed upon infarction, and may play roles in the progression of ischemic injury.
3.5. Tgfbr3 activates p38 MAPK and enhances Cardiomyocyte apoptosis
To further investigate let-7 downstream signalling, we infected rat neonatal cardiomyocytes with Hmga2 and Tgfbr3 overexpressing adenoviruses. Markers for apoptosis and autophagy were examined because these two modalities of cell death are commonly observed during early MI. Interestingly, overexpression of Hmga2 induced autophagy, based on observation of LC3B cleavage, whereas overexpression of Tgfbr3 induced apoptosis, based on observation of caspase-3 cleavage (Fig. 5A). Further investigation revealed that Hmga2 induction of autophagy was associated with downregulation of Ppargc1a/b expression (Fig. S5A) and increased AMPK signalling (Fig. S5B). These results indicated that Hmga2 may regulate metabolism and autophagy in response to cardiac injury.
Fig. 5.
MicroRNA let-7-Tgfbr3-p38 MAPK signalling regulates cardiomyocyte apoptosis.
(A) Western blots of cleaved LC3B (marker of autophagy) and caspase 3 (marker of apoptosis) in rat neonatal cardiomyocytes 1 day after adenovirus-mediated Hmga2 and Tgfbr3 expression. (B) Dose dependent activation of Smad2 and p38 MAPK signalling following Tgfbr3 expression. MOI: multiplicity of infection. The relative ratios of phospho-Smad2 to Smad2 and phospho-p38 to p38 are indicated by the numbers at the bottoms of the lanes. (C) Induction of p38 downstream target gene MMP9 signalling, instead of canonical TGFβ signalling, following overexpression of Tgfbr3 in rat neonatal cardiomyocytes. The histograms and error bars indicate mean and SEM, respectively, from six independent experiments. Differences between groups were analysed by unpaired Student's t-test (**, P < .01; ***, P < .001). (D) Partial rescue of Tgfbr3 induced apoptosis in rat neonatal cardiomyocytes upon treatment with p38 MAPK inhibitor, SB239063. (E) Quantification of rescue of apoptosis by flow cytometry. Percentage of subG1 cells from three independent experiments is shown. (F) Western blot of Tgfbr3, p38 activity and caspase 3 indicating rescue of apoptosis following knockdown of Tgfbr3 in serum-depleted (SF) H9C2 cells. (G) Rescue of let-7 inhibition-enhanced apoptosis following shRNA knockdown of Tgfbr3 in serum-depleted H9C2 cells. Quantification of cleaved caspase 3 is shown in the histogram from two independent experiments. (H—I) Validation of lentiviral-mediated let-7a/f overexpression. Histograms and error bars indicate mean and SEM, respectively, from three independent experiments. (J) Tgfbr3 RNA expression level upon serum depletion (SF) and let-7 overexpression from four independent experiments. (K) Quantification of let-7a/f overexpression rescue of serum depletion (SF)-induced apoptosis by flow cytometry from five independent experiments. (L) Reduction of caspase 3 cleavage upon let-7 overexpression following serum depletion. (M) Effect of let-7a/f overexpression on Tgfbr3 and p38 activity at 0, 3 and 6 h serum depletion time points. Histograms and error bars indicate mean and SEM, respectively. Differences between multiple groups (E, H, I—K) were analysed by one-way ANOVA with post-hoc Tukey HSD Test. (*, P < .05; **, P < .001; ***, P < .0001).
Tgfbr3 mediates many different signalling cascades. Thus, to distinguish whether Tgbr3 signalling in cardiomyocytes follows canonical or noncanonical TGF-β signalling, a dose-dependent Tgfbr3 overexpression experiment was performed. Neonatal cardiomyocytes were infected with Tgfbr3-overexpressing adenoviruses at different infection units (MOI). We observed an obvious dose-dependent activation of non-canonical Tgfbr3 signalling in the form of p38 MAPK activation, and a slight dose-dependent activation of Smad2 up to 40 MOI (Fig. 5B). Additional examination of several TGF-β downstream targets was performed in cells infected with 20 MOI, which showed an obvious induction of MMP9 expression - a p38 downstream target [43,44] (Fig. 5C). Activation of p38 MAPK has been shown to play a role in cardiomyocyte apoptosis and cardiac remodelling after MI [45]. Treatment of 20 MOI Tgfbr3-overexpressing cardiomyocytes with a p38 MAPK inhibitor, SB239063, partially rescued apoptosis (Fig. 5D). On the other hand, treatment with canonical TGF-β signalling inhibitor, ALK5 inhibitor II, did not influence apoptosis despite an obvious reduction in Smad2 activation (Fig. S5C). Interestingly, Tgfbr3 overexpression appeared to be associated with downregulation of Tgfbr1 mRNA expression (Fig. 5C), whereas inhibition of Tgfbr1 by ALK5i enhanced Tgfbr3 expression (Fig. S5C), suggesting that a potential negative co-regulation may exist between Tgfbr1 and Tgfbr3. In addition, we found our homemade antibodies were able to serve as Tgfbr3 agonists, since they activated p38 MAPK (Fig. S5D) and increased caspase 3 cleavage (Fig. S5E) in HL-1 cells, similar to the effects by Tgfbr3 overexpression in rat neonatal cardiomyocytes. In vivo, mice that received the Tgfbr3 agonist (R3–9) by i.v. injection one day post-MI showed worse cardiac function by a decrease of the LV ejection fraction and increases of the LV end-diastolic volume (EDV) and end-systolic volume (ESV) (Fig. S5I-3 K) one month after MI. The one-month-post-MI survival rate of R3–9 treated mice was 50% (4 out of 8 mice with LAD ligation), which was lower than that of mice treated with control IgM, ~83% (5 out of 6 mice with LAD ligation). Together these findings suggest that Tgfbr3 activation induces cardiomyocyte apoptosis and impairs cardiac function after MI through non-canonical TGF-β/p38 MAPK signalling.
To elaborate the results of our Tgfbr3 overexpression experiments, we next explored whether Tgfbr3 knockdown could protect cardiomyocytes from apoptosis. Preliminary screening was performed in several in vitro models of various cardiac stresses associated with acute MI such as ATP-release, ROS, hypoxia, TNFα, and serum depletion in H9C2 or rat neonatal cardiomyocytes. We also investigated a GSK3 inhibition and Wnt3a activation model to mimic early events in infarction [46,47]. Amongst these, we found that serum depletion in H9C2 cells demonstrated the greatest suitability. We thus generated two Tgfbr3 knockdown H9C2 cell lines using shRNAs and subjected them to serum depletion to induce apoptosis. Based on the percentage of subG1 cells, serum depletion-induced apoptosis was significantly reduced in these knockdown cells (Fig. 5E). The knockdown efficiency was confirmed under both normal and serum depleted conditions and a significant reduction in p38 MAPK and caspase-3 activation under serum depletion was also found in Tgfbr3 knockdown cells (Fig. 5F). In addition, let-7 inhibition in H9C2 cells enhanced serum depletion-induced apoptosis and Tgfbr3 knockdown rescued this enhancement (Fig. 5G). We therefore suggest that the let-7-Tgfbr3 axis is involved in cardiomyocyte apoptosis, primarily through p38 MAPK activation. As we found that inhibition of Tgfbr3 expression under serum deprivation stress could reduce apoptosis, we hypothesized that a cardioprotective effect could also be achieved by let-7 overexpression. Thus, a let-7 overexpressing H9C2 cell line was generated and confirmed by qPCR (Fig. 5H and I). Ectopic let-7 overexpression reduced serum depletion-induced apoptosis, as measured by flow cytometric quantification of the subG1 population (Fig. 5K) and expression of cleaved caspase 3 (Fig. 5L). Induction of Tgfbr3 signalling in response to serum depletion was also suppressed on both the mRNA and protein levels upon let-7 overexpression (Fig. 5J and M). Whilst p38 MAPK signalling was activated at 6 h after serum depletion in the control group, overexpression of let-7 delayed the activation of p38 MAPK (Fig. 5M). These results indicate that the cardioprotective effect of let-7 may be attributed to downregulation of Tgfbr3 and its subsequent activation of p38 MAPK.
3.6. Acute MI is associated with changes in circulating TGFBR3 levels
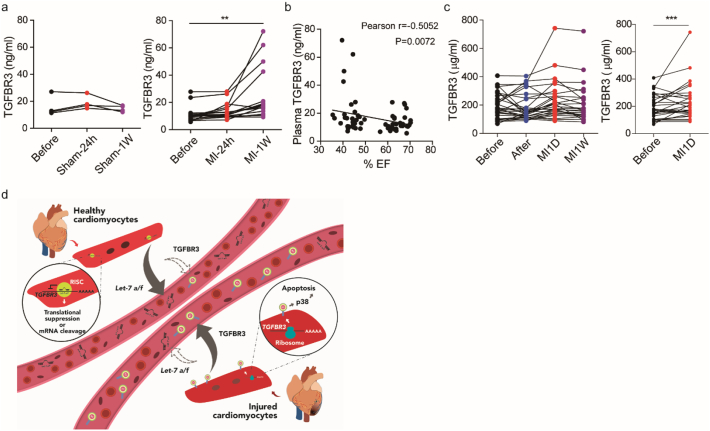
TGFBR3 is a membrane protein that can be cleaved and released into the extracellular matrix [48,49]. As such, we hypothesized that TGFBR3 is upregulated in the infarct because the de-repression of let-7 may also lead to an increase in the plasma TGFBR3 level. In pig plasma, we observed a trend in upregulation of plasma TGFBR3 level after MI, which reached significance at one week post-MI, and such upregulation was not observed in the sham control (Fig. 6A). Furthermore, the plasma concentration of TGFBR3 was inversely correlated with left ventricular function based on ejection fraction (Fig. 6B). In humans, we also observed a similar trend to that found in pigs, i in STEMI patients, the concentration of plasma TGFBR3 increased after MI. When comparing its plasma concentration between the pre-therapeutic intervention group and the 1 day post MI group, there was a significant increase (Fig. 6C). These results demonstrate the novel potential of TGFBR3 as a biomarker reflecting cardiac injury after MI. Nevertheless, whether it also reflects the severity of injury or predicts the outcome of heart failure requires further research.
Fig. 6.
Plasma level of TGFBR3 is increased after acute myocardial infarction in pigs and in human patients.
(A) Pig plasma TGFBR3 expression levels at baseline, 1 day and 7 days post-sham and MI (n = 16). One-way ANOVA was performed with Dunnett's test. (**, P < .01) (B) Correlation of plasma TGFBR3 expression and ejection fraction in pigs. (C) Human plasma TGFBR3 expression levels at several time points: before therapeutic intervention (Before), after therapeutic intervention (After), 1 day and 1 week post-MI (MI-1D and MI-1 W). Differences between the before group and the MI-1D group were analysed by paired Student's t-test (*, P < .05). (D) Graphical summary. MicroRNA let-7a and let-7f are abundant in healthy cardiomyocytes to suppress the expression of pro-apoptosis factor TGFBR3. Meanwhile, microRNA let-7a and let-7f are enriched in the plasma and the concentration of plasma TGFBR3 is low (left). Once infarction occurs, the expression of let-7a and let-7f are decreased and TGFBR3 is de-repressed. The expression of TGFBR3 initiates p38 MAPK signalling and promotes myocyte apoptosis. Meanwhile, plasma let-7a and let-7f respond to the injury by reducing the amount of their expression. The concentration of plasma TGFBR3 is increased as well. MicroRNA let-7-TGFBR3-p38 MAPK regulates cardiomyocyte apoptosis and the plasma microRNAs and TGFBR3 reflect the events of injury.
4. Discussion
In this study, we identified the let-7/Tgfbr3/p38 axis as an important mediator of cardiomyocyte apoptosis after MI. In particular, we found that the onset of ischemia leads to rapid downregulation of let-7a and let-7f expression in the infarct. This leads to subsequent Tgfbr3 signalling followed by p38-induced cardiomyocyte apoptosis (Fig. 6D). We showed that compensating for these changes via AAV-mediated delivery of let-7 in a mouse model of MI significantly reduced cardiomyocyte apoptosis and improved cardiac function. This suggests that let-7 could potentially be utilized as a candidate for gene therapy after MI. We also found that changes of circulating let-7a and let-7f as well as TGFBR3 concentrations in the plasma were associated with cardiac injury both in pigs and humans with MI (Fig. 6D), suggesting that let-7a, let-7f and TGFBR3 have potential for use as diagnostic and prognostic biomarkers post-MI.
The downregulation of microRNA let-7 in the myocardium may be due to several reasons, including the change in microRNAs in specific cell types and the change in cell population after injury because of immune infiltration, cell death and fibroblast proliferation. Here, we demonstrated that microRNA let-7 was dysregulated in the peri-infarct cardiomyocytes although it does not rule out other possibilities that may cause the overall changes in microRNA let-7 in the myocardium. The source of circulating let-7a/f and TGFBR3 are even more elusive as microRNA let-7 is ubiquitous in all organs and almost all cell types, and TGFBR3 may be enriched in other tissues [50]. Here, we showed that the changes in plasma microRNA let-7 and TGFBR3 may be due to the change in microRNA let-7 and TGFBR3 in the myocardium. However, other possibilities may also exist. For example, after MI, the activity of metalloproteinases (MMPs) increased, and membrane bound TGFBR3 could be cleaved by MMPs [48]. The increase in plasma TGFBR3 may be the result of the combination of increased MMP activity and TGFBR3 expression itself. Notably, TargetScan prediction showed that both MiR-101 and MiR-103 may also target Tgfbr3, which suggests Tgfbr3 may be regulated by multiple microRNAs under injury.
In light of our results, further work in a porcine model of MI would validate the therapeutic efficacy of delivering let-7 as a cardioprotective gene therapy. An additional cardiac specific Tgfbr3 knock-out model would be instrumental in further investigating the let-7/Tgfbr3/p38 axis, as well as to identify other possible pathways. Furthermore, the potential of circulating let-7a/f and TGFBR3 as biomarkers should also be further investigated.
Data sharing
The data, analytical methods, and study materials for the purposes of reproducing the results or replicating procedures can be made available on request to the corresponding author who manages the information.
Funding sources
This work was supported by the Ministry of Science and Technology, Taiwan (MOST 106-2811-B-001-036, 1062319-B-001-003 and 107-2314-B-004), the National Health Research Institutes grant EX106-10512SI and the Academia Sinica Program for Translational Innovation of Biopharmaceutical Development-Technology Supporting Platform Axis (AS-KPQ-106-TSPA), the Thematic Research Program (AS-107-TP-B12) and the Summit Research Program (AS-SUMMIT-108).
Declaration of Competing Interest
P.C. Hsieh and C.Y. Chen are co-inventors of two pending patent applications for the use of let-7 and Tgfbr3 for treatment and diagnosis of myocardial infarction. P.C. Hsieh received research support from AstraZeneca, Takeda, and Gilead. The other authors declare that they have no competing interests.
Author contributions
C.Y.C.: conception and design, data collection, analysis and interpretation, manuscript writing and final approval of manuscript; O.K·C and L.W.L.: collection of data and data analysis; Y.C·C, M.R.W, M.H·C, T.J.T, Y.W·W, L.C.L, Y.L.C.: data collection; S.C.L. and W.C.L.: data analysis; C.Y·Y: manuscript writing; T.A.H. and T.J.K.: critically revised the paper; P.C.H.H.: conception and design and final approval of manuscript.
Acknowledgements
We gratefully acknowledge R.H. Lin and B.L. Lin for their assistance in animal surgery and echocardiography. We thank the core facilities at the Institute of Biomedical Sciences, Academia Sinica for equipment and technical support, and Professor Shu Chien (Department of Bioengineering, University of California at San Diego, San Diego, CA 92093, USA) for helpful discussion.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.08.001.
Appendix A. Supplementary data
Supplementary material
References
- 1.Hsieh P.C., Segers V.F., Davis M.E., MacGillivray C., Gannon J., Molkentin J.D. Evidence from a genetic fate-mapping study that stem cells refresh adult mammalian cardiomyocytes after injury. Nat Med. 2007;13:970–974. doi: 10.1038/nm1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Senyo S.E., Steinhauser M.L., Pizzimenti C.L., Yang V.K., Cai L., Wang M. Mammalian heart renewal by pre-existing cardiomyocytes. Nature. 2013;493:433–436. doi: 10.1038/nature11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Munz M.R., Faria M.A., Monteiro J.R., Aguas A.P., Amorim M.J. Surgical porcine myocardial infarction model through permanent coronary occlusion. Comp Med. 2011;61:445–452. [PMC free article] [PubMed] [Google Scholar]
- 4.Wernersson R., Schierup M.H., Jorgensen F.G., Gorodkin J., Panitz F., Staerfeldt H.H. Pigs in sequence space: a 0.66X coverage pig genome survey based on shotgun sequencing. BMC Genomics. 2005;6:70. doi: 10.1186/1471-2164-6-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reinhart B.J., Slack F.J., Basson M., Pasquinelli A.E., Bettinger J.C., Rougvie A.E. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 6.Johnson S.M., Grosshans H., Shingara J., Byrom M., Jarvis R., Cheng A. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 7.Melton C., Judson R.L., Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu H., Shyh-Chang N., Segre A.V., Shinoda G., Shah S.P., Einhorn W.S. The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147:81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuppusamy K.T., Jones D.C., Sperber H., Madan A., Fischer K.A., Rodriguez M.L. Let-7 family of microRNA is required for maturation and adult-like metabolism in stem cell-derived cardiomyocytes. Proc Natl Acad Sci U S A. 2015;112:E2785–E2794. doi: 10.1073/pnas.1424042112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seeger T., Xu Q.F., Muhly-Reinholz M., Fischer A., Kremp E.M., Zeiher A.M. Inhibition of let-7 augments the recruitment of epicardial cells and improves cardiac function after myocardial infarction. J Mol Cell Cardiol. 2016;94:145–152. doi: 10.1016/j.yjmcc.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 11.Sun C., Liu H., Guo J., Yu Y., Yang D., He F. MicroRNA-98 negatively regulates myocardial infarction-induced apoptosis by down-regulating Fas and caspase-3. Sci Rep. 2017;7:7460. doi: 10.1038/s41598-017-07578-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scott L.J. Alipogene tiparvovec: a review of its use in adults with familial lipoprotein lipase deficiency. Drugs. 2015;75:175–182. doi: 10.1007/s40265-014-0339-9. [DOI] [PubMed] [Google Scholar]
- 13.Wang X.F., Lin H.Y., Ng-Eaton E., Downward J., Lodish H.F., Weinberg R.A. Expression cloning and characterization of the TGF-beta type III receptor. Cell. 1991;67:797–805. doi: 10.1016/0092-8674(91)90074-9. [DOI] [PubMed] [Google Scholar]
- 14.Kirkbride K.C., Townsend T.A., Bruinsma M.W., Barnett J.V., Blobe G.C. Bone morphogenetic proteins signal through the transforming growth factor-beta type III receptor. J Biol Chem. 2008;283:7628–7637. doi: 10.1074/jbc.M704883200. [DOI] [PubMed] [Google Scholar]
- 15.Lewis K.A., Gray P.C., Blount A.L., MacConell L.A., Wiater E., Bilezikjian L.M. Betaglycan binds inhibin and can mediate functional antagonism of activin signalling. Nature. 2000;404:411–414. doi: 10.1038/35006129. [DOI] [PubMed] [Google Scholar]
- 16.Andres J.L., DeFalcis D., Noda M., Massague J. Binding of two growth factor families to separate domains of the proteoglycan betaglycan. J Biol Chem. 1992;267:5927–5930. [PubMed] [Google Scholar]
- 17.Brown C.B., Boyer A.S., Runyan R.B., Barnett J.V. Requirement of type III TGF-beta receptor for endocardial cell transformation in the heart. Science. 1999;283:2080–2082. doi: 10.1126/science.283.5410.2080. [DOI] [PubMed] [Google Scholar]
- 18.Stenvers K.L., Tursky M.L., Harder K.W., Kountouri N., Amatayakul-Chantler S., Grail D. Heart and liver defects and reduced transforming growth factor beta2 sensitivity in transforming growth factor beta type III receptor-deficient embryos. Mol Cell Biol. 2003;23:4371–4385. doi: 10.1128/MCB.23.12.4371-4385.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopez-Casillas F., Payne H.M., Andres J.L., Massague J. Betaglycan can act as a dual modulator of TGF-beta access to signaling receptors: mapping of ligand binding and GAG attachment sites. J Cell Biol. 1994;124:557–568. doi: 10.1083/jcb.124.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Santander C., Brandan E. Betaglycan induces TGF-beta signaling in a ligand-independent manner, through activation of the p38 pathway. Cell Signal. 2006;18:1482–1491. doi: 10.1016/j.cellsig.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 21.Chen C.C., Sun C.P., Ma H.I., Fang C.C., Wu P.Y., Xiao X. Comparative study of anti-hepatitis B virus RNA interference by double-stranded adeno-associated virus serotypes 7, 8, and 9. Mol Ther. 2009;17:352–359. doi: 10.1038/mt.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joshi S., Wei J., Bishopric N.H. A cardiac myocyte-restricted Lin28/let-7 regulatory axis promotes hypoxia-mediated apoptosis by inducing the AKT signaling suppressor PIK3IP1. Biochim Biophys Acta. 2016;1862:240–251. doi: 10.1016/j.bbadis.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tolonen A.M., Magga J., Szabo Z., Viitala P., Gao E., Moilanen A.M. Inhibition of Let-7 microRNA attenuates myocardial remodeling and improves cardiac function postinfarction in mice. Pharmacol Res Perspect. 2014;2 doi: 10.1002/prp2.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meunier J., Lemoine F., Soumillon M., Liechti A., Weier M., Guschanski K. Birth and expression evolution of mammalian microRNA genes. Genome Res. 2013;23:34–45. doi: 10.1101/gr.140269.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arroyo J.D., Chevillet J.R., Kroh E.M., Ruf I.K., Pritchard C.C., Gibson D.F. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang R., Niu H., Ban T., Xu L., Li Y., Wang N. Elevated plasma microRNA-1 predicts heart failure after acute myocardial infarction. Int J Cardiol. 2013;166:259–260. doi: 10.1016/j.ijcard.2012.09.108. [DOI] [PubMed] [Google Scholar]
- 27.Viswanathan S.R., Powers J.T., Einhorn W., Hoshida Y., Ng T.L., Toffanin S. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet. 2009;41:843–848. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iliopoulos D., Hirsch H.A., Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zipeto M.A., Court A.C., Sadarangani A., Delos Santos N.P., Balaian L., Chun H.J. ADAR1 activation drives Leukemia stem cell self-renewal by impairing Let-7 biogenesis. Cell Stem Cell. 2016;19:177–191. doi: 10.1016/j.stem.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Desterro J.M., Keegan L.P., Jaffray E., Hay R.T., O'Connell M.A., Carmo-Fonseca M. SUMO-1 modification alters ADAR1 editing activity. Mol Biol Cell. 2005;16:5115–5126. doi: 10.1091/mbc.E05-06-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haraguchi T., Ozaki Y., Iba H. Vectors expressing efficient RNA decoys achieve the long-term suppression of specific microRNA activity in mammalian cells. Nucleic Acids Res. 2009;37:e43. doi: 10.1093/nar/gkp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie J., Ameres S.L., Friedline R., Hung J.H., Zhang Y., Xie Q. Long-term, efficient inhibition of microRNA function in mice using rAAV vectors. Nat Methods. 2012;9:403–409. doi: 10.1038/nmeth.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inagaki K., Fuess S., Storm T.A., Gibson G.A., McTiernan C.F., Kay M.A. Robust systemic transduction with AAV9 vectors in mice: efficient global cardiac gene transfer superior to that of AAV8. Mol Ther. 2006;14:45–53. doi: 10.1016/j.ymthe.2006.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matkovich S.J., Van Booven D.J., Eschenbacher W.H., Dorn G.W., 2nd RISC RNA sequencing for context-specific identification of in vivo microRNA targets. Circ Res. 2011;108:18–26. doi: 10.1161/CIRCRESAHA.110.233528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shin C., Nam J.W., Farh K.K., Chiang H.R., Shkumatava A., Bartel D.P. Expanding the microRNA targeting code: functional sites with centered pairing. Mol Cell. 2010;38:789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mayr C., Hemann M.T., Bartel D.P. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Copley M.R., Babovic S., Benz C., Knapp D.J., Beer P.A., Kent D.G. The Lin28b-let-7-Hmga2 axis determines the higher self-renewal potential of fetal haematopoietic stem cells. Nat Cell Biol. 2013;15:916–925. doi: 10.1038/ncb2783. [DOI] [PubMed] [Google Scholar]
- 38.Monzen K., Ito Y., Naito A.T., Kasai H., Hiroi Y., Hayashi D. A crucial role of a high mobility group protein HMGA2 in cardiogenesis. Nat Cell Biol. 2008;10:567–574. doi: 10.1038/ncb1719. [DOI] [PubMed] [Google Scholar]
- 39.Knelson E.H., Gaviglio A.L., Tewari A.K., Armstrong M.B., Mythreye K., Blobe G.C. Type III TGF-beta receptor promotes FGF2-mediated neuronal differentiation in neuroblastoma. J Clin Invest. 2013;123:4786–4798. doi: 10.1172/JCI69657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dong M., How T., Kirkbride K.C., Gordon K.J., Lee J.D., Hempel N. The type III TGF-beta receptor suppresses breast cancer progression. J Clin Invest. 2007;117:206–217. doi: 10.1172/JCI29293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang S., Zhou H., Wu D., Ni H., Chen Z., Chen C. MicroRNA let-7a regulates angiogenesis by targeting TGFBR3 mRNA. J Cell Mol Med. 2019;23:556–567. doi: 10.1111/jcmm.13960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huntzinger E., Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 43.Ringshausen I., Dechow T., Schneller F., Weick K., Oelsner M., Peschel C. Constitutive activation of the MAPkinase p38 is critical for MMP-9 production and survival of B-CLL cells on bone marrow stromal cells. Leukemia. 2004;18:1964–1970. doi: 10.1038/sj.leu.2403544. [DOI] [PubMed] [Google Scholar]
- 44.Simon C., Simon M., Vucelic G., Hicks M.J., Plinkert P.K., Koitschev A. The p38 SAPK pathway regulates the expression of the MMP-9 collagenase via AP-1-dependent promoter activation. Exp Cell Res. 2001;271:344–355. doi: 10.1006/excr.2001.5374. [DOI] [PubMed] [Google Scholar]
- 45.Ren J., Zhang S., Kovacs A., Wang Y., Muslin A.J. Role of p38alpha MAPK in cardiac apoptosis and remodeling after myocardial infarction. J Mol Cell Cardiol. 2005;38:617–623. doi: 10.1016/j.yjmcc.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 46.Duan J., Gherghe C., Liu D., Hamlett E., Srikantha L., Rodgers L. Wnt1/betacatenin injury response activates the epicardium and cardiac fibroblasts to promote cardiac repair. EMBO J. 2012;31:429–442. doi: 10.1038/emboj.2011.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cai W.Y., Wei T.Z., Luo Q.C., Wu Q.W., Liu Q.F., Yang M. The Wnt-beta-catenin pathway represses let-7 microRNA expression through transactivation of Lin28 to augment breast cancer stem cell expansion. J Cell Sci. 2013;126:2877–2889. doi: 10.1242/jcs.123810. [DOI] [PubMed] [Google Scholar]
- 48.Velasco-Loyden G., Arribas J., Lopez-Casillas F. The shedding of betaglycan is regulated by pervanadate and mediated by membrane type matrix metalloprotease-1. J Biol Chem. 2004;279:7721–7733. doi: 10.1074/jbc.M306499200. [DOI] [PubMed] [Google Scholar]
- 49.Mendoza V., Vilchis-Landeros M.M., Mendoza-Hernandez G., Huang T., Villarreal M.M., Hinck A.P. Betaglycan has two independent domains required for high affinity TGF-beta binding: proteolytic cleavage separates the domains and inactivates the neutralizing activity of the soluble receptor. Biochemistry. 2009;48:11755–11765. doi: 10.1021/bi901528w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Uhlen M., Fagerberg L., Hallstrom B.M., Lindskog C., Oksvold P., Mardinoglu A. Proteomics. Tissue-based map of the human proteome. Science. 2015;347 doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material