Cryptococcus gattii, one of the etiological agents of cryptococcosis, can be distinguished from its sister species Cryptococcus neoformans by growth on d-amino acids. C. gattii MYO5 affected the growth of C. gattii on d-amino acids. The myo5Δ cells accumulated high levels of various substrates from outside the cells, and excessively accumulated d-amino acids appeared to have caused toxicity in the myo5Δ cells. We provide evidence on the alteration of membrane properties in the myo5Δ mutants. Additionally, alteration in the myo5Δ membrane permeability causing higher substrate accumulation is associated with the changes in the sterol distribution. Furthermore, myosin-I in three other yeasts also manifested a similar role in substrate accumulation. Thus, while fungal myosin-I may function as a classical myosin-I, it has hitherto unknown additional roles in regulating membrane permeability. Since deletion of fungal myosin-I causes significantly elevated susceptibility to multiple antifungal drugs, it could serve as an effective target for augmentation of fungal therapy.
KEYWORDS: Cryptococcus, actin patches, endocytosis, membrane permeability, myosin-I
ABSTRACT
We found a novel role of Myo5, a type I myosin (myosin-I), and its fortuitous association with d-amino acid utilization in Cryptococcus gattii. Myo5 colocalized with actin cortical patches and was required for endocytosis. Interestingly, the myo5Δ mutant accumulated high levels of d-proline and d-alanine which caused toxicity in C. gattii cells. The myo5Δ mutant also accumulated a large set of substrates, such as membrane-permeant as well as non-membrane-permeant dyes, l-proline, l-alanine, and flucytosine intracellularly. Furthermore, the efflux rate of fluorescein was significantly increased in the myo5Δ mutant. Importantly, the endocytic defect of the myo5Δ mutant did not affect the localization of the proline permease and flucytosine transporter. These data indicate that the substrate accumulation phenotype is not solely due to a defect in endocytosis, but the membrane properties may have been altered in the myo5Δ mutant. Consistent with this, the sterol staining pattern of the myo5Δ mutant was different from that of the wild type, and the mutant was hypersensitive to amphotericin B. It appears that the changes in sterol distribution may have caused altered membrane permeability in the myo5Δ mutant, allowing increased accumulation of substrate. Moreover, myosin-I mutants generated in several other yeast species displayed a similar substrate accumulation phenotype. Thus, fungal type I myosin appears to play an important role in regulating membrane permeability. Although the substrate accumulation phenotype was detected in strains with mutations in the genes involved in actin nucleation, the phenotype was not shared in all endocytic mutants, indicating a complicated relationship between substrate accumulation and endocytosis.
INTRODUCTION
Type I myosin (myosin-I) belongs to a large superfamily of actin-dependent molecular motors (1) and plays important roles in diverse cellular processes such as polarized morphogenesis, cell migration, and endocytosis (for reviews, see references 2, to ,7). Myosin-I is composed of a globular head or motor domain and neck and tail of various lengths. The head or motor domain binds to actin in an ATP-dependent manner and generates force by ATP hydrolysis. The N-terminal head region is connected to the neck which consists of an IQ motif that provides binding sites for myosin light chains. The carboxyl-terminal tail contains TH1 and TH2 (tail homology 1 and 2) and SH3 domains which are thought to specify the role of myosin-I in protein-protein interactions, membrane binding, and intracellular signaling. Myosin-I has been extensively studied in several yeast species. For instance, MYO3 and MYO5 are the two myosin-I-encoding genes in Saccharomyces cerevisiae, and both Myo5 and Myo3 are colocalized with actin cortical patches (8). Deletion of either MYO3 or MYO5 is tolerated by cells, but a deletion of both genes causes defects in endocytosis as well as in growth (9, 10). Myo5 of Candida albicans is also colocalized with actin patches at the bud and hyphal tips, and the myo5 mutant is impaired in polarized growth, hyphal formation, and endocytosis (11, 12). Also, disruption of myo1 in Schizosaccharomyces pombe causes growth retardation and aberrance in cell morphology associated with an abnormal distribution of F-actin patches (13, 14).
Cryptococcus gattii and Cryptococcus neoformans are two closely related basidiomycetous yeasts that cause cryptococcosis in humans and animals (15). The ability to utilize d-proline or d-alanine as the sole source of nitrogen can be used to differentiate C. gattii isolates from C. neoformans isolates (16, 17). d-Amino acids have been found in the mammalian forebrain and various tissues (18–20). In addition, plants synthesize d-amino acid derivatives, and some d-amino acids, such as d-alanine, are widely found in higher plants (21, 22). Since the major ecological niche of C. gattii is plants (15), d-amino acid utilization may bear significance in pathobiology as well as ecology of the species. During the screening of a library constructed by T-DNA insertion in C. gattii strain R-265, we isolated a clone that failed to grow robustly on d-proline as the sole nitrogen source (23). The clone was identified to have an insertion in the gene encoding the putative type I myosin (CNBG_4524). Although myosin-I is involved in diverse cellular processes, there is no report of myosin-I function related to d-amino acid utilization. This observation prompted us to explore the function of the C. gattii myosin-I gene, which we designated MYO5. Myo5 was found to colocalize with actin cortical patches and was required for endocytosis. We further established the possible cause of how the myo5 mutant failed to grow when d-proline was the sole source of nitrogen. Unexpectedly, deletion of MYO5 caused higher accumulation of d-proline, which is toxic and thereby restricted the growth of the myo5Δ mutant on d-proline. We hypothesize and provide evidence that the properties of the cell membrane in the myo5 mutant have been altered to cause higher accumulation of foreign substrates inside the cells. Furthermore, we demonstrated that similar phenotypes are also conserved in C. neoformans and two other well-studied yeast species unrelated to Cryptococcus.
RESULTS
MYO5 deletion in C. gattii causes growth defects on d-amino acids.
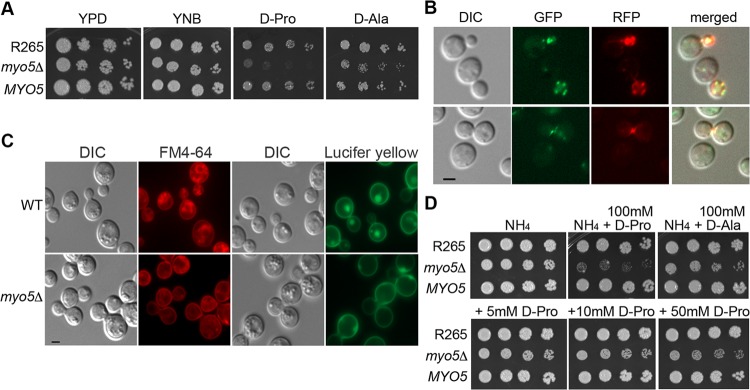
To further characterize the role of MYO5 in the utilization of d-amino acids, we deleted the MYO5 gene in the C. gattii strain R265. The myo5Δ mutant was viable, and its growth was only slightly reduced on YEPD and YNB media (Fig. 1A). The myo5Δ mutant, however, grew poorly on media containing d-proline, and growth was also clearly reduced on media with d-alanine as the sole nitrogen source (Fig. 1A). Reconstitution of the myo5Δ mutant with the wild-type MYO5 complemented the growth defects on these media, indicating that MYO5 plays a hitherto unidentified role in d-amino acid utilization.
FIG 1.
Identification of MYO5 function. (A) MYO5 is important for d-proline utilization. Cells of the wild type, the myo5Δ mutant, and the myo5Δ complemented strain (MYO5) were serially diluted and spotted on the indicated media. Plates were incubated at 30°C for 3 days and photographed. (B) Myo5 is colocalized with actin cortical patches. Myo5 was tagged with green fluorescent protein (GFP) (green color). Actin was visualized with Lifeact-RFP (red color). Fluorescent and differential interference contrast (DIC) images were taken from log-phase cells. (C) MYO5 is involved in endocytosis. Cells were stained with FM4-64 to determine membrane internalization (DIC and FM4-64 columns) or stained with Lucifer yellow to determine fluid phase endocytosis (DIC and Lucifer yellow columns). For FM4-64 staining, cells were stained with 16 μM FM4-64 on ice for 15 min, washed, and incubated in YEPD for 80 min before imaging. Bar = 2 μm. WT, wild type. (D) d-Amino acids are toxic to the myo5Δ mutant. Cells were serially diluted and spotted on media containing 10 mM ammonium sulfate (NH4) with or without supplementation with 100 mM d-proline or d-alanine (top row). Cells were also spotted on NH4 media supplemented with 5, 10, or 50 mM d-proline (bottom row). Plates were incubated at 30°C for 3 days and photographed.
Myo5 functions as classical type I myosin.
It has been shown that myosin-I is located at cortical actin patches in several fungi (8, 12). To determine whether Myo5 is associated with actin, we tagged Myo5 with green fluorescent protein, mNG (24), at its C terminus. Homologous integration of Myo5-mNG into the myo5Δ locus complemented the myo5Δ mutant’s growth defect in d-proline, suggesting that tagging of Myo5 with mNG did not interfere with its function. To determine whether Myo5 is localized to cortical actin patches, we transformed the Lifeact-RFP plasmid to the Myo5-mNG-tagged strain. Lifeact has a 17-amino-acid peptide sequence of Abp140 from S. cerevisiae which is a versatile marker for localization of actin (25), and Lifeact-RFP has stained typical actin patches and cables in C. neoformans (26). We found that Myo5-mNG colocalized with Lifeact-RFP at cortical actin patches (Fig. 1B, top row) and occasionally at the isthmus between the mother and daughter cells (Fig. 1B, bottom row).
To determine whether the cryptococcal Myo5 functions as a classical myosin-I, we investigated the effect of MYO5 deletion on endocytosis. First, FM4-64, a styryl dye, was used to determine the behavior of membrane internalization. When the R265 cells were stained with FM4-64, bright ring-like structures inside cells depicting the endosomes were observed (Fig. 1C, DIC and FM4-64 columns). However, myo5Δ cells stained with FM4-64 lacked bright ring-like structure inside the cell, and most of the FM4-64 staining was on the cell membrane. Second, analysis of fluid phase endocytosis with Lucifer yellow revealed that the dye was internalized and trafficked to vacuoles in the wild type but not in the myo5Δ mutant (Fig. 1C, DIC and Lucifer yellow columns). These results suggested that the myo5Δ mutant was defective in endocytosis. Thus, cryptococcal Myo5 behaves as a classical myosin-I in relation to endocytosis, as is the case in several other fungi.
MYO5 is involved in substrate accumulation.
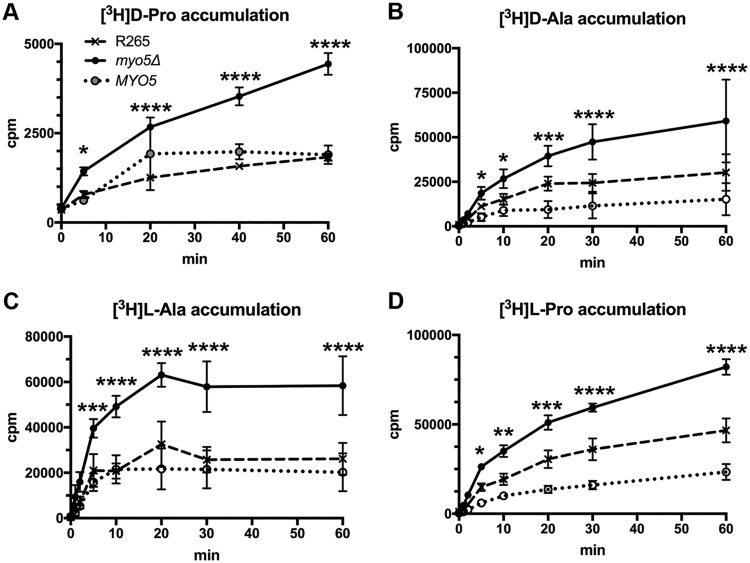
We showed that C. gattii Myo5 behaves as a typical type I myosin. However, the involvement of myosin-I in the utilization of d-amino acids as the sole nitrogen source remained obscure. d-Amino acids have been reported to be toxic to animals and plants (27–29). Although the growth of the wild type was not affected on defined media containing 10 mM ammonium sulfate supplemented with 100 mM d-proline or 100 mM l-proline, the myo5Δ mutant grew poorly on media supplemented with 100 mM d-proline, and its growth was slightly reduced on media supplemented with 100 mM d-alanine (Fig. 1D, top). In addition, the lowest concentration of the supplemented d-proline to cause visible growth reduction in the myo5Δ mutant was 5 mM (Fig. 1D, bottom). Together, these data suggested that the growth defect of the myo5Δ mutant may not be due to a loss of the d-amino acid utilization pathway but may instead be due to toxicity of excess d-amino acids. To test this hypothesis, we measured the accumulation of [3H]d-proline and [3H]d-alanine in the cells. Figure 2A and B show that the accumulation of tritium-labeled amino acids is significantly higher in the myo5Δ mutant than in the control strains. These results are consistent with the hypothesis that the myo5Δ cells accumulate d-amino acids from the culture media which causes growth retardation due to toxicity of excess d-amino acids. To our surprise, however, when we measured the uptake of [3H]l-proline or l-alanine as a control, the accumulation of tritium-labeled amino acids was also significantly higher in the myo5Δ mutant than in the control strains (Fig. 2C and D). This observation prompted us to speculate that deletion of MYO5 may have caused changes in membrane permeability and affected the cell’s ability to take up or transport substrates from outside the cells.
FIG 2.
The myo5Δ mutant accumulates high levels of amino acids. Log-phase cells of the wild type, myo5Δ mutant, and the myo5Δ complemented strain (MYO5) were incubated with the indicated 3H-labeled amino acids, and the time course of intracellular accumulation of each amino acid was determined. The experiments were repeated three times, and the error bars represent the standard deviations. Two-way ANOVA with Dunnett’s multiple-comparison test was used to compare the differences at the indicated time points between the corresponding R265 strain and myo5Δ mutant (*, P < 0.0332; **, P < 0.0021; ***, P < 0.0002; ****, P < 0.0001).
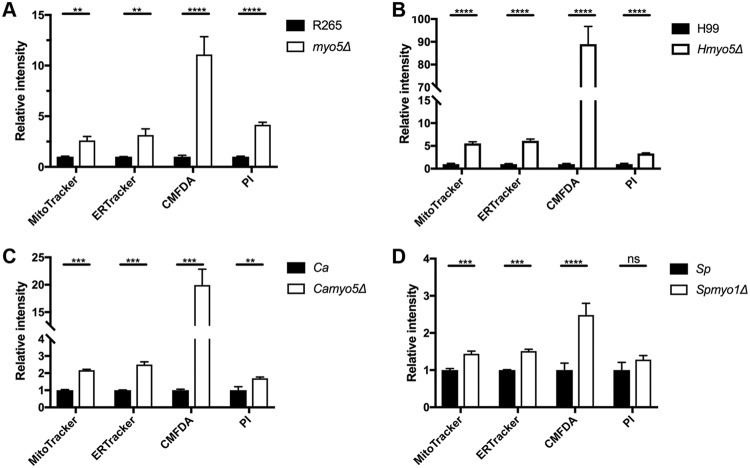
To examine the possible importance of Myo5 in membrane permeability, we compared the staining patterns of three different membrane-permeant dyes. ER-Tracker Red stains the endoplasmic reticulum. CellTracker Green CMFDA is a colorless and nonfluorescent dye until its acetate groups are cleaved by intracellular esterases. MitoTracker Red CMXRos contains a mildly thiol-reactive chloromethyl moiety that labels mitochondria. The localization patterns of these dyes were according to the property of individual dyes as expected and were similar between the cells of the myo5Δ mutant and the wild type (Fig. 3). However, the signal intensity of each dye was considerably higher in the myo5Δ cells than in the wild type. These data indicate that deletion of MYO5 affects neither the specificity nor distribution of the membrane-permeant dyes but allows higher accumulation of the fluorescence dyes within the cell. Quantitation of the fluorescence intensity by flow cytometry confirmed that the fluorescence intensity of all three membrane-permeant dyes was higher in the myo5Δ cells than in the wild type (Fig. 4A). To further characterize the importance of MYO5 in membrane permeability, we compared the staining patterns of a membrane-nonpermeant dye, propidium iodide (PI). PI is a positively charged membrane-impermeable molecule that is generally excluded from viable cells. The PI staining patterns were similar between the myo5Δ mutant and the wild type, but the myo5Δ mutant clearly accumulated more PI than the wild type did (Fig. 3D and Fig. 4A). Together, these results support the possible alteration of the membrane permeability in the myo5Δ mutant.
FIG 3.
The myo5Δ mutant accumulates high levels of membrane-permeant and -nonpermeant dyes as determined by microscopy. (A to D) Cells were incubated with the indicated fluorescent dyes and observed with a microscope. (A) ER-Tracker Red. (B) CellTracker Green CMFDA. (C) MitoTracker Red CMXRos. (D) Propidium iodide (PI). Fluorescent and DIC images were photographed. The staining patterns of each dye were similar between the wild type (WT) and the myo5Δ mutant, but the fluorescent intensity was greater in the myo5Δ mutant than in the wild type. Bars = 5 μm.
FIG 4.
Myosin-I mutants of different fungal species accumulate high levels of membrane-permeant or -nonpermeant dyes. Cells of indicated strains were incubated with each membrane-permeant or -nonpermeant dye, and accumulation of each dye was assessed by flow cytometry. The experiments were repeated three times, and the error bars represent the standard deviations. Ca, C. albicans; Sp, S. pombe. Values that are significantly different are indicated by bars and asterisks as follows: **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001. Values that are not significantly different (P > 0.05) are indicated (ns).
Since the deletion of MYO5 affected substrate accumulation, it was probable that the myo5Δ mutant could also accumulate larger amounts of antifungal drugs and cause hypersensitivity to the drugs. We determined the susceptibility of the myo5Δ mutant to several antifungal drugs by using E-tests, including flucytosine, amphotericin B, and three triazoles, fluconazole, itraconazole, and voriconazole. It was clear that the myo5Δ mutant had lower MICs and was hypersensitive toward all antifungal drugs compared to strain R265 (Table 1). These observations suggested that the myo5Δ mutant accumulates higher levels of the drugs, leading to its hypersensitivity. To validate such a possibility, we quantified the time course accumulation of [3H]flucytosine in the wild type and the myo5Δ mutant. Compared to the wild type, myo5Δ cells accumulated substantially more [3H]flucytosine (Fig. 5A), attesting that the high accumulation of flucytosine results in its hypersensitivity to flucytosine in the myo5Δ mutant.
TABLE 1.
MICs of antifungal drugs
| Strain or species |
MIC (μg/ml)a
|
||||
|---|---|---|---|---|---|
| 5-FC | AmB | Flc | Itr | Vor | |
| R265 | 0.19 | 2 | 16 | 0.5 | 0.19 |
| myo5Δ | 0.094 | 0.75 | 3 | 0.19 | 0.047 |
| vrp1Δ | 0.047 | 1.0 | 8 | 0.38 | 0.094 |
| arp2Δ | 0.064 | 1.0 | 0.032 | 0.002 | 0.003 |
| arp3Δ | 0.016 | 0.5 | 0.75 | 0.016 | 0.008 |
| C. albicans | 0.094 | 2 | 1b | 0.016b | 0.023 |
| Camyo5Δ | 0.016 | 1.0 | 0.125b | 0.008b | 0.008 |
| S. pombe | Resistant | 0.75 | 32 | 0.094 | 0.25 |
| Spmyo1Δ | Resistant | 0.5 | 16 | 0.032 | 0.047 |
| H99 | 0.19 | 2 | 32 | 1.0 | 0.094 |
| Hmyo5Δ | 0.016 | 0.75 | 16 | 0.5 | 0.047 |
| las17Δ | 0.032 | ||||
| sla1Δ | 0.125 | ||||
| KN99 | 0.125 | ||||
| abp1Δ | 0.125 | ||||
| crn1Δ | 0.125 | ||||
| B3501 | 0.047 | ||||
| abp1Δ | 0.047 | ||||
| sla2Δ | 0.047 | ||||
Drug abbreviations: 5-FC, 5-fluorocytosine; AmB, amphotericin B; Flc, fluconazole; Itr, itraconazole; Vor, voriconazole.
Test was done on YNB medium.
FIG 5.
Myosin-I mutants of different fungal species accumulate higher levels of flucytosine. Cells of the indicated strains were incubated with [3H]labeled flucytosine, and the time course of intracellular accumulation of 3H was determined. The experiments were repeated three times, and the error bars represent the standard deviations. Ca, C. albicans; Sp, S. pombe. Two-way ANOVA with Dunnett’s multiple-comparison test was used to compare the differences at the indicated time point between the corresponding R265 strain and the myo5Δ mutant (ns, not significant; *, P < 0.0332; **, P < 0.0021; ***, P < 0.0002; ****, P < 0.0001).
Deletion of MYO5 does not affect the distribution of permeases.
It is known that amino acids and flucytosine generally enter cells through transporters/permeases. For instance, several amino acid permeases can regulate the uptake of l-amino acids in C. neoformans (30, 31), and transport of cytosine/flucytosine requires the function of transporters in Candida species and S. cerevisiae (32, 33). Since permeases are normally removed from the cell surface by endocytosis, the endocytosis defect in the myo5Δ mutant might result in higher levels of permeases on the cell surface, an alternative explanation for increased accumulation of substrates in the mutants. To address this possibility, we attempted to tag the permeases and studied their localization. In order to tag the appropriate permeases, we first identified the permease for flucytosine by deleting the gene encoding the putative purine-cytosine permease (CNBG_3227) in strain R265 which shares high similarity to the S. cerevisiae Fcy2. When the FCY2 gene was deleted, the resulting strains were resistant to flucytosine (Fig. 6A), suggesting that the R265 FCY2 functions as the purine-cytosine permease, as is the case in S. cerevisiae. To identify the permease(s) for proline, we first deleted the two putative proline permease genes (CNBG_4785 and CNAG_00597) which show high similarity to the proline permease genes identified in C. neoformans (30). However, deletion of both genes individually or in combination did not affect the proline utilization in R265. As an alternative approach to identify permease genes, we made use of a preliminary RNASeq data set for WM276, another genome-sequenced strain of the C. gattii species complex. C. gattii strain WM276 was grown in media containing ammonium sulfate or proline as the sole nitrogen source for the RNAseq experiment (data not shown [to be presented in a separate study]). We identified two putative amino acid transporter genes CGB_J2080C and CGB_F2450W, whose transcripts were highly induced in the cells grown in proline (67.3- and 61.6-fold increase, respectively). We then deleted the putative orthologs of these two genes in strain R265, CNBG_6051 and CNBG_1852, respectively, and found that only CNBG_1852, which we designated AAP9, was required for R265 to grow in d- or l-proline as the sole nitrogen source, indicating that Aap9 is the permease for d- and l-proline in R265 (Fig. 6B). We tagged Fcy2 and Aap9 with the green fluorescent protein (GFP) at its native locus, respectively, and confirmed that GFP tagging of either protein did not affect the function of the gene in flucytosine resistance and d- and l-proline utilization (data not shown). Next, we examined the localization of Fcy2 and Aap9 in strain R265 and the myo5Δ mutant and found that the fluorescent intensity of both proteins was indistinguishable between the myo5Δ mutant and R265 (Fig. 6C and D). These results indicate that a defect in myo5Δ does not affect the distribution of both permeases, and as such, the observed phenotypes in the accumulation of d-/l-proline and flucytosine are not due to the endocytic defect of the myo5Δ mutant in removing permeases from the cell surface.
FIG 6.
(A) FCY2 is required for resistance to flucytosine. Cells of the indicated strains were spotted on YNB medium with or without 0.5 μg/ml of flucytosine. (B) AAP9 is required for the growth in media containing d-/l-proline as the sole nitrogen source. Cells of the indicated strains were spotted on media containing 10 mM ammonium sulfate, l-proline, or d-proline. Plates were incubated at 30°C for 3 days and photographed. (C and D) Deletion of MYO5 does not affect the distribution of transporters for flucytosine or proline. The Fcy2 and Aap9 proteins were separately tagged with green fluorescent protein in the myo5Δ mutant and the wild type, respectively. Strains were grown in YPD (C) or YNB supplemented with 10 mM Asn as the sole nitrogen source (D). Fluorescent and DIC images were taken from log-phase cells. Bar = 2 μm.
Deletion of MYO5 affects the rate of accumulation and efflux.
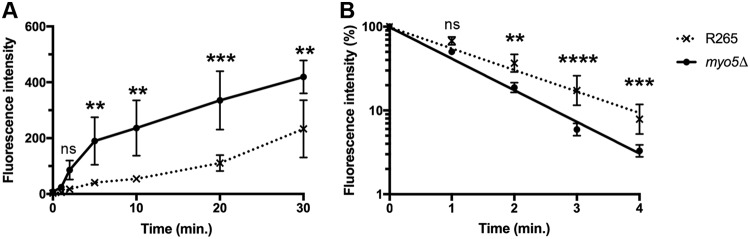
Since accumulation of substrates inside the cells depends on both the rate of influx as well as efflux of the substrates, we compared the efflux rates between the wild type and the myo5Δ mutant. Fluorescein diacetate (FDA) has been used for studying the properties of membrane permeability by measuring the kinetics of intracellular accumulation and efflux of fluorescein (34, 35). We first evaluated the time course accumulation of fluorescein in both strain R265 and the myo5Δ mutant. As shown in Fig. 7A, the myo5Δ mutant accumulates more fluorescein and faster than R265 does. After cells were loaded with a similar quantity of fluorescein, the cells were washed to remove residual FDA, and the efflux rate of fluorescein was determined. It was clear that the fluorescein efflux rate was higher in the myo5Δ mutant than in R265 (Fig. 7B). Thus, deletion of MYO5 affects both the accumulation and efflux of fluorescein, indicating that the membrane in the myo5Δ mutant is more permeable than the membrane in the wild type.
FIG 7.
Fluorescein accumulation and efflux rate are higher in the myo5Δ mutant. (A) Time course of intracellular fluorescein accumulation. Cells were incubated with 10 μM FDA, and cellular fluorescent intensity was determined at the indicated time by flow cytometry. (B) Kinetics of fluorescein efflux. Cells were loaded with similar amounts of fluorescein, and the efflux of fluorescein was determined by measuring the intracellular fluorescent intensity by flow cytometry. The experiments were repeated three times, and the error bars represent standard deviations. Two-way ANOVA with Dunnett’s multiple-comparison test was used to compare the differences at the indicated time point between the corresponding R265 strain and myo5Δ mutant (ns, not significant; **, P < 0.0021; ***, P < 0.0002; ****, P < 0.0001).
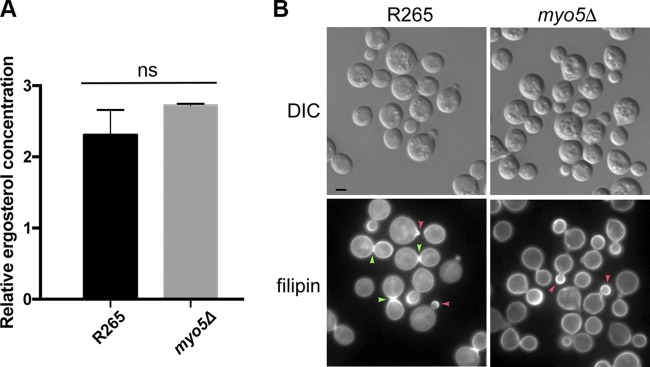
The sterol distribution is altered in the myo5Δ mutant.
It is known that myosin-I can bind to membrane lipids (36), and myosin-I mutant displays an abnormal sterol distribution in the plasma membrane of fission yeast (13). It is possible that Myo5 may control membrane sterol distribution, which in turn regulates the membrane property, since it is known that the sterol contents can influence membrane fluidity and permeability (for a review, see reference 37). We used LCMS to measure the major sterol contents but found no difference in total concentration of ergosterol between the wild type and the myo5Δ mutant (Fig. 8A). To determine whether the distribution of sterol is different in the membrane, log-phase-grown cells of the wild type and myo5Δ were stained with filipin. Filipin is a naturally fluorescent sterol-binding antibiotic (38) and known to stain the areas of polarized growth as well as mating projections in S. pombe and the areas of mating projections in S. cerevisiae (39, 40). In the wild-type R265, filipin staining was primarily concentrated at the tip of the emerging bud (red arrowhead) and at the isthmus between the mother and matured daughter cells (green arrowheads) (Fig. 8B, bottom left panel), which was similar to the patterns observed in C. neoformans (41). We noted that the intensely stained isthmus was predominantly found in the mother-daughter pair where the size of daughter cells has reached close to 70% of the size of the mother cells (green arrowheads). This indicates that sterols were enriched at the isthmus during the later stages of cell replication before the release of daughter cells. In the myo5Δ mutant, filipin stained wider areas of emerging bud surface (red arrowheads) than in the wild type. In addition, intensely stained isthmus was rarely observed, and filipin staining was more evenly distributed at the cell periphery of the mother and daughter cells in the myo5Δ mutant. Since filipin binds sterol, it is likely that the distribution of sterol is altered in the myo5Δ cells and that the changes in sterol distribution are discernible in a developmental stage-dependent manner.
FIG 8.
Ergosterol distribution appears to be altered in the myo5Δ mutant. (A) Ergosterol content is similar between the wild type and myo5Δ mutant. Ergosterol was extracted from cells and quantitated by LCMS. The experiments were repeated three times, and the error bars represent the standard deviations. (B) Filipin staining patterns are different between the wild type and the myo5Δ mutant. Log-phase grown cells were stained with 5 μg/ml filipin. The fluorescent and DIC images were photographed. Red arrowhead, emerging bud; green arrowhead, isthmus. Bar = 2 μm.
MYO5 is involved in substrate accumulation in a closely related species.
To determine whether the observed myo5Δ phenotypes were C. gattii specific, we deleted MYO5 in a closely related species, C. neoformans serotype A strain H99. Since C. neoformans cannot use d-proline as the sole nitrogen source, the d-proline growth phenotype of the H99 myo5Δ (Hmyo5Δ) mutant was not determined. However, as was the case with the R265 myo5Δ mutant, accumulation of all the membrane-permeant dyes and nonpermeant dye was markedly increased in the Hmyo5Δ mutant than in strain H99 (Fig. 4B). Similarly, the Hmyo5Δ mutant was more susceptible to all the antifungal drugs tested (Table 1) and accumulated more [3H]flucytosine than the H99 strain (Fig. 5B). Therefore, MYO5 of both C. neoformans and C. gattii plays important roles for substrate accumulation.
Myosin-I is also involved in substrate accumulation in other yeast species.
It was of interest to determine whether myosin-I was involved in the substrate accumulation in other species of pathogenic and nonpathogenic yeast phylogenetically distanced from Cryptococcus. We found that the myo5Δ mutant of C. albicans behaves like the myo5Δ mutant of Cryptococcus species; the accumulation of all the membrane-permeant dyes and nonpermeant dye was higher in the C. albicans myo5Δ mutant than in the wild type (Fig. 4C), and the C. albicans myo5Δ mutant was hypersensitive to five antifungal drugs (Table 1). Interestingly, the levels of [3H]flucytosine accumulated in the wild-type C. albicans was more than 10-fold higher than those in the Cryptococcus species, and the C. albicans myo5Δ mutant accumulated even more [3H]flucytosine than the C. albicans wild type did (Fig. 5C). However, the differences in accumulation between the mutant and the wild type were smaller in C. albicans than in Cryptococcus species. The myosin-I of S. pombe has been designated Myo1p. We found that the S. pombe myo1Δ mutant accumulated more membrane-permeant dyes than the wild type did, but no significant difference was found between the wild type and myo1Δ in the accumulation of PI, a membrane-nonpermeant dye (Fig. 4D). Interestingly, the wild type and the myo1Δ mutant of S. pombe were intrinsically resistant to flucytosine, and both strains accumulated low levels of [3H]flucytosine (Table 1 and Fig. 5D). However, the S. pombe myo1Δ mutant was hypersensitive to the other four antifungal drugs (Table 1). Thus, myosin-I of C. albicans plays a clear role in substrate accumulation, but the role of S. pombe myosin-I for substrate accumulation was not as versatile.
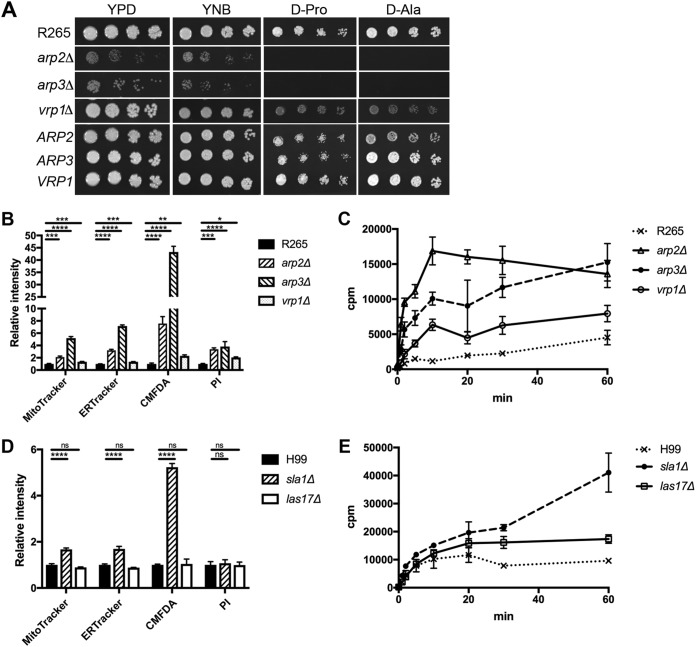
ARP2, ARP3, and VRP1 are involved in substrate accumulation.
To determine whether other genes involved in the endocytosis process also controlled substrate accumulation, we deleted ARP2, ARP3, and VRP1 separately in strain R265. In budding and fission yeasts, myosin-I interacts with Vrp1/WIP, which in turn binds G-actin (9, 42) and delivers actin monomers to the Arp2/3 complex (for a review, see reference 43). The genome of strain R265 contains highly conserved ARP2 and ARP3 and also contains a CNBG_5343 gene, which we designated VRP1. VRP1 encodes a protein with a predicted WH2 motif at its N terminus and shares low similarity with Vrp1 of S. cerevisiae. Figure 9A shows that deletion of ARP2 or ARP3 in strain R265 showed a severe growth defect in d-proline and d-alanine utilization. The growth of the vrp1Δ mutant on d-proline and d-alanine was slightly reduced, and the growth defect was not as severe as in the myo5Δ mutant. Furthermore, complementation of each mutant with its wild-type gene restored the growth defect of each mutant (Fig. 9A). The arp2Δ, arp3Δ, and vrp1Δ mutants were hypersensitive toward antifungal drugs compared to the wild type (Table 1). Similarly, the accumulation of membrane-permeant dyes and nonpermeant dye was significantly higher in the arp2Δ, arp3Δ, and vrp1Δ mutants than in the wild type, although the amount of accumulation in the vrp1Δ mutant was not as dramatically increased as in the myo5Δ, arp2Δ, and arp3Δ mutants (Fig. 9B). In addition, the arp2Δ, arp3Δ, and vrp1Δ mutants accumulated higher levels of [3H]flucytosine than the wild-type strain did (Fig. 9C). These results suggest that the three genes involved in actin nucleation also play a role in substrate accumulation.
FIG 9.
Several genes involved in endocytosis play a role in membrane permeability. (A) ARP2, ARP3, and VRP1 are important for d-proline utilization. Cells of the indicated mutants and the corresponding complemented strains (ARP2, ARP3, and VRP1) were serially diluted and spotted on the indicated media. Plates were incubated at 30°C for 3 days and photographed. (B and C) Deletion of ARP2, ARP3, and VRP1 causes accumulation of different fluorescent dyes (B) or [3H]flucytosine (C). (D and E) Not every endocytic mutant accumulates different fluorescent dyes (D) or [3H]flucytosine (E). (ns, not significant [P > 0.05]; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001).
Not all the endocytic mutants display the phenotype of substrate accumulation.
To investigate whether other known components of endocytosis were involved in substrate accumulation, we took advantage of the large collection of deletion constructs available in the H99 background (Fungal Genetics Stock Center; provided by J. Lodge in 2007) and its derivative strain, KN99α (Fungal Genetics Stock Center; provided by H. Madhani in 2015). We examined the flucytosine sensitivity of the abp1Δ and crn1Δ mutants derived from strain KN99α as well as the sla1Δ and las17Δ mutants derived from strainH99. In addition, the abp1Δ and sla2Δ mutants were also available in the background of a C. neoformans serotype D strain, B-3501 (26). All the selected genes have been shown to be involved in endocytosis in other organisms (43). Out of the six tested strains, only the las17Δ and sla1Δ mutants displayed reduced resistance toward flucytosine compared to the respective wild-type strains (Table 1). We chose the las17Δ and sla1Δ mutants to determine the accumulation of fluorescent dyes and [3H]flucytosine. The sla1Δ mutant, but not the las17Δ mutant, showed higher accumulation of the membrane-permeant dyes than strain H99 did (Fig. 9D). In contrast, the levels of accumulated membrane-nonpermeant dye, PI, were similar between las17Δ mutant, sla1Δ mutant, and the wild type. However, the accumulation of [3H]flucytosine was higher in both the las17Δ and sla1Δ mutants than in H99 (Fig. 9E). Therefore, not all the endocytic mutants displayed the phenotype of substrate accumulation in C. neoformans.
DISCUSSION
Myosin-I is a molecular motor and plays various cellular roles related to membrane dynamics and trafficking. In this study, we demonstrate a hitherto unknown role of fungal myosin-I in controlling membrane permeability. We showed that the C. gattii myo5Δ mutant accumulates three types of molecules that can enter cells by different means: membrane-permeant molecules, membrane-nonpermeant molecules, and transporter/permease-mediated molecules. It is possible that the increased accumulation of the membrane-permeant molecules can be attributed to disrupted membrane recycling due to endocytosis defect rather than increased membrane permeability. However, microscopic examination showed no accumulation of fluorescence dyes on the cell surface, and the location of each fluorescent dye within the cells was similar for the myo5Δ mutant and the wild type, except for much higher fluorescent intensities in the myo5Δ mutant. Additionally, there was no noticeable deficiency in removing permeases/transporters from the cell surface in the myo5Δ mutant, indicating that the substrate accumulation phenotype was not likely due to a defect in permease recycling by endocytosis. In addition, the kinetics of fluorescein accumulation and efflux rates were higher in the myo5Δ mutant. These results indicate that the myo5Δ mutant in C. gattii has altered membrane permeability. Furthermore, we show that involvement of myosin-I in substrate accumulation is common among four different fungal species tested, although it was less clear in S. pombe. Together, these data indicate that fungal myosin-I plays a role in controlling membrane permeability, which has not been reported in other eukaryotic organisms.
The importance of myosin-1 for endocytosis has been well established. There are more than 50 different proteins involved in endocytosis (43–45) which form functional modules in different stages of endocytosis and are recruited to the endocytic site with an organized spatial and temporal order (44, 46, 47). In the current study, we have selected many genes known to be associated with the process of endocytosis in other organisms, including the genes involved in the formation stages of the coat complex (sla1Δ and sla2Δ), WASP/Myo module (las17Δ, vrp1Δ, and myo5Δ), and actin network (arp2Δ, arp3Δ, abp1Δ, and crn1Δ). Interestingly, mutants from each stage of endocytosis either displayed or did not display the substrate accumulation phenotype, and the levels of accumulated substrates were not the same among all those that displayed the phenotype. Thus, our results not only indicate that several genes functioning in endocytosis play a role in controlling substrate accumulation, but also exhibit the complexity of the relationship between membrane permeability and endocytosis. It is not clear whether severities of endocytic defects vary among these mutants or what the nature of the relationship between endocytosis and substrate accumulation phenotype is. Future studies in quantification of endocytic defects and the extent of miscellaneous substance accumulation in different endocytic mutants may provide insights into their biological connections with altered membrane permeability.
How does a type I fungal myosin, a molecular motor, control substrate accumulation? It is possible that the putative motor function and/or lipid interacting function of type I myosin together with the components involved in endocytosis process may modulate the functions of transporters and regulate the properties of membrane permeability. It has been shown that type I myosin can bind to membrane lipids (36). Furthermore, the S. pombe myosin-I, myo1p, is required for proper organization of the large sterol-rich membrane domains (13). It has been proposed that the basic TH1 domain of myo1p may bind to phospholipids that directly affect the local composition of the membrane, or alternatively, myo1p may contribute to membrane tension and could affect membrane organization (48). Since it is known that sterols can influence membrane fluidity and permeability (for a review, see reference 37), it is probable that the changes in sterol organization have contributed to the observed alterations in membrane characteristics. By filipin staining, we showed that deletion of MYO5 perturbs organization of the large sterol-rich domains. Filipin is a traditional sterol marker (38), and it has been used to stain the large sterol-rich domains in several fungal species (41, 49). The likelihood of altered sterol organization in the myosin-I mutants was further supported by their hypersensitivity to amphotericin B (Table 1). Amphotericin B is known to target ergosterol to form aggregates and increase the permeability of the cell membrane (50, 51) or to extract ergosterol from lipid bilayers and kill fungal cells (52). However, the detailed mechanisms of interaction between myosin-I and sterol/lipids for the regulation of membrane properties remain to be elucidated. It is also currently unknown whether the contents or distribution of membrane lipids have been modified in the myo5Δ mutant. Future studies focused on lipidomics may shed light on the possible relationship between type I myosin and the structures, functions, and dynamics of cellular lipids. Finally, since myosin-I mutants were hypersensitive to several antifungal drugs and the carboxyl-terminal tail of fungal myosin-I is different from the tail of human myosin-I (see Fig. S1 in the supplemental material), it is possible that combination therapy with drugs targeting the fungal myosin or components of endocytosis process may offer new treatments for fungal infections.
Amino acid alignment of myosin-I. Amino acid sequences of myosin-I from strain R265, Candida albicans (Ca), Saccharomyces cerevisiae (Sc), Schizosaccharomyces pombe (Sp), and human myosin-Ie and myosin-If (NP_004989.2 and NP_036467.2, respectively) were compared by CLUSTER W alignment program. Download FIG S1, PDF file, 0.1 MB (147.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
MATERIALS AND METHODS
Strains and culture conditions.
Strains relevant to the study are listed in Table S1 in the supplemental material. Standard culture conditions and microbiological methods were used as described previously (23). Spot assays were performed on yeast extract-peptone-dextrose (YEPD) or yeast nitrogen base (YNB) without amino acids and without ammonium sulfate (YNB-A-N; 1.7g/liter) with 2% glucose supplemented with 10 mM indicated nitrogen sources.
List of strains relevant to this study. Download Table S1, DOCX file, 0.01 MB (13.7KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Gene manipulation.
The entire protein coding sequence of genes of interest were deleted by homologous recombination as described previously (23). Homologous integration was confirmed by PCR and Southern blot analysis (data not shown). Deletants were complemented by homologous integration using biolistic transformation. The MYO5, FCY2, and AAP9 genes were tagged with mNG (24) at its native locus to visualize its localization.
Microscopy.
A Zeiss Axiovert fluorescence microscope equipped with an AxioCam MRm digital camera was used to take differential interference contrast (DIC) microscopy and fluorescent images. The exposure times and other microscope settings were held constant between compared images. An endocytosis assay of FM4-64 and Lucifer yellow was performed as described previously (53). Filipin staining was carried out as described previously (54) at a final concentration of 5 μg/ml for 2 min at room temperature.
Amino acids and flucytosine accumulation assay.
For amino acid uptake assays, cells were grown to log phase in YNB medium containing 10 mM asparagine as the sole nitrogen source. Cells were harvested, washed with cold YNB-A-N without glucose, and adjusted to 10 optical density units/ml. Each 100 μl of cells was mixed with 100 μl of YNB-A-N with glucose supplemented with 0.5 mM concentration of assayed amino acid and 0.5 μCi of 3H-labeled assayed amino acid. The 3H-labeled amino acid used in the experiment was d-[3,4-3H]proline (40 Ci/mmol; 1 mCi/ml) (Moravek Inc.), l-[3,4-3H]proline (60 Ci/mmol; 1 mCi/ml) (Moravek Inc.), d-[2,3-3H]-alanine (60 Ci/mmol; 1 mCi/ml) (Moravek Inc.), and l-[2,3-3H]alanine (55 Ci/mmol; 1 mCi/ml) (Moravek Inc.). Cells were incubated at 30°C in a water bath shaker, and 50-μl aliquots of the sample were removed at 0, 1, 2, 5, 10, 20, 30, and 60 min and added to 2 ml ice-cold YNB-A-N supplemented with 50 mM concentration of the assayed amino acid. Cells were immediately filtered with 24-mm Whatman GF/C filter and washed with cold YNB-A-N and assayed for radioactivity. The flucytosine accumulation assay was performed as described previously with modifications (55). The cells were grown and harvested as described above. Each 100 μl of cells was mixed with 100 μl of 2× Asn media with glucose supplemented with 200 μM flucytosine and 0.8 μCi of [3H]flucytosine (4.0 Ci/mmol; 1 mCi/ml) (Moravek Inc.). Cells were incubated at 30°C in a water bath shaker, and 50-μl aliquots of sample were removed at 0, 1, 2, 5, 10, 20, 30, and 60 min and added to 2 ml of cold media containing 20 mM flucytosine. Cells were immediately filtered with 24-mm Whatman GF/C filter, washed with cold YNB-A-N, and assayed for radioactivity.
Uptake of fluorescent dyes.
Each strain was grown to the log phase in 10 mM Asn media and adjusted to an optical density at 600 nm (OD600) of 0.3. Cells were incubated at 30°C for 30 min in the presence of 0.1 μM Mito Tracker Red CMXRos, 5 μM CMFDA, or 1 μM ER-Tracker Red. For propidium iodide staining, cells were washed with phosphate-buffered saline (PBS) and treated with 10 μg/ml of dye for 10 min at room temperature (RT). The accumulation of each dye was immediately assessed by flow cytometry or by observation with a microscope. For each sample, 30,000 events were analyzed using Flow Jo software. FDA accumulation and efflux rate measurement were performed as described previously with modifications (34). For FDA accumulation, log-phase-grown cells were washed in ice-cold 10 mM potassium phosphate (pH 6.0) and adjusted to an OD600 of 0.8. FDA was added to 10 μM and incubated at 30°C. At the indicated time, 100 μl of cells was taken out and transferred to 400 μl of ice-cold 10 mM potassium phosphate (pH 6.0) and analyzed immediately by flow cytometry. For fluorescein efflux, a similar amount of fluorescein was loaded to each strain by controlling the time of incubation with FDA as described above. The cells were washed in ice-cold 10 mM potassium phosphate (pH 6.0) and resuspended in 1 ml of ice-cold potassium phosphate (pH 6.0). Cells were removed from ice and incubated at room temperature. At the indicated time, 100 μl of cells was taken out and transferred to 400 μl of ice-cold 10 mM potassium phosphate (pH 6.0) and analyzed immediately by flow cytometry. Efflux rate constants were calculated from the slopes of the fitted lines obtained by regression analysis.
Drug susceptibility test.
MICs of different drugs, flucytosine (5-FC), amphotericin B (AmB), fluconazole (Flc), itraconazole (Itr), and voriconazole (Vor), were determined by plating 2 × 106 cells on YEPD or YNB agar plates followed by application of Epsilometer test strips (Etest strips; AB Biodisk). The plates were incubated at 30°C, and photographs were taken after 72 h.
Sterol analysis.
Log-phase-grown cells were used for sterol extraction as described previously with modification (56). The extracted sterols were analyzed and characterized by liquid chromatography/mass spectrometry (LCMS). LCMS analysis was performed with a Waters Acquity I-class UPLC (Waters Corp., Milford, MA) and a Thermo Scientific Q-Exactive mass spectrometer (high resolution – accurate mass) with a HESI-II electrospray source in positive ion mode and at a resolution of 35,000. Samples were kept at 5°C in the temperature-controlled autosampler. The separation was performed with a sample volume injection of 1 μl on a 1.6-μm Cortecs C8 column (2.1 by 100 mm) (Waters Corp., Milford, MA) maintained at 40°C. Each sample was analyzed three times with separate injections. The isocratic chromatography used solvent A (2% H2O with 0.1% formic acid) and solvent B (98% methanol [MeOH] with 0.1% formic acid) at a flow rate of 250 μl/min. The mass spectrometer method used a full scan function from m/z 300 to 450 without a lock mass. The compounds were identified by a comparison with the standards (cholesterol, ergosterol, and lanosterol) on the basis of their retention time and high-resolution accurate-mass (HR-AM) mass spectrometry, or tentatively identified (squalene, ergosta-7-enol, eburicol, 4-alpha-methylfecosterol) by HR-AM mass spectrometry.
Statistical analysis.
All experiments were performed in triplicate, and statistics and graphs were obtained using Graph Pad Prism 7. Student’s t test and analysis of variance (ANOVA) followed by multiple-comparison test were used to determine whether there are significant differences between the indicated samples.
ACKNOWLEDGMENTS
We thank P. J. Espenshade and M. Whiteway for providing the S. pombe and C. albicans strains, respectively. Deletion strains of KN99 and H99 were obtained from the Fungal Genetics Stock Center (Manhattan, Kansas, USA). We also thank Shannon Moyer and Michael Davis for critical reading of the manuscript.
This work was supported by the Division of Intramural Research (DIR), NIAID, NIH.
Footnotes
Citation Khanal Lamichhane A, Garraffo HM, Cai H, Walter PJ, Kwon-Chung KJ, Chang YC. 2019. A novel role of fungal type I myosin in regulating membrane properties and its association with d-amino acid utilization in Cryptococcus gattii. mBio 10:e01867-19. https://doi.org/10.1128/mBio.01867-19.
Contributor Information
James W. Kronstad, University of British Columbia.
J. Andrew Alspaugh, Duke University Medical Center.
Scott Filler, Los Angeles Biomedical Research Institute at Harbor-UCLA Medical Center.
REFERENCES
- 1.Sellers JR. 2000. Myosins: a diverse superfamily. Biochim Biophys Acta 1496:3–22. doi: 10.1016/s0167-4889(00)00005-7. [DOI] [PubMed] [Google Scholar]
- 2.Coluccio LM. 2008. Myosin I, p 95–124. In Coluccio LM. (ed), Myosins: a superfamily of molecular motors. Springer, Dordrecht, The Netherlands. doi: 10.1007/978-1-4020-6519-4. [DOI] [Google Scholar]
- 3.Masters TA, Kendrick-Jones J, Buss F. 2017. Myosins: domain organisation, motor properties, physiological roles and cellular functions. Handb Exp Pharmacol 235:77–122. doi: 10.1007/164_2016_29. [DOI] [PubMed] [Google Scholar]
- 4.McIntosh BB, Ostap EM. 2016. Myosin-I molecular motors at a glance. J Cell Sci 129:2689–2695. doi: 10.1242/jcs.186403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim SV, Flavell RA. 2008. Myosin I: from yeast to human. Cell Mol Life Sci 65:2128–2137. doi: 10.1007/s00018-008-7435-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mermall V, Post PL, Mooseker MS. 1998. Unconventional myosins in cell movement, membrane traffic, and signal transduction. Science 279:527–533. doi: 10.1126/science.279.5350.527. [DOI] [PubMed] [Google Scholar]
- 7.Giblin J, Fernandez-Golbano IM, Idrissi FZ, Geli MI. 2011. Function and regulation of Saccharomyces cerevisiae myosins-I in endocytic budding. Biochem Soc Trans 39:1185–1190. doi: 10.1042/BST0391185. [DOI] [PubMed] [Google Scholar]
- 8.Goodson HV, Anderson BL, Warrick HM, Pon LA, Spudich JA. 1996. Synthetic lethality screen identifies a novel yeast myosin I gene (MYO5): myosin I proteins are required for polarization of the actin cytoskeleton. J Cell Biol 133:1277–1291. doi: 10.1083/jcb.133.6.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun Y, Martin AC, Drubin DG. 2006. Endocytic internalization in budding yeast requires coordinated actin nucleation and myosin motor activity. Dev Cell 11:33–46. doi: 10.1016/j.devcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 10.Lewellyn EB, Pedersen RT, Hong J, Lu R, Morrison HM, Drubin DG. 2015. An engineered minimal WASP-myosin fusion protein reveals essential functions for endocytosis. Dev Cell 35:281–294. doi: 10.1016/j.devcel.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kachurina N, Turcotte B, Whiteway M. 2012. Motor protein Myo5p is required to maintain the regulatory circuit controlling WOR1 expression in Candida albicans. Eukaryot Cell 11:626–637. doi: 10.1128/EC.00021-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oberholzer U, Iouk TL, Thomas DY, Whiteway M. 2004. Functional characterization of myosin I tail regions in Candida albicans. Eukaryot Cell 3:1272–1286. doi: 10.1128/EC.3.5.1272-1286.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takeda T, Chang F. 2005. Role of fission yeast myosin I in organization of sterol-rich membrane domains. Curr Biol 15:1331–1336. doi: 10.1016/j.cub.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 14.Toya M, Motegi F, Nakano K, Mabuchi I, Yamamoto M. 2001. Identification and functional analysis of the gene for type I myosin in fission yeast. Genes Cells 6:187–199. doi: 10.1046/j.1365-2443.2001.00414.x. [DOI] [PubMed] [Google Scholar]
- 15.Kwon-Chung KJ, Bennett JE. 1992. Cryptococcosis In Medical mycology. Lea & Febiger, Philadelphia, PA. [Google Scholar]
- 16.Ngamskulrungroj P, Chang Y, Roh J, Kwon-Chung KJ. 2012. Differences in nitrogen metabolism between Cryptococcus neoformans and C. gattii, the two etiologic agents of cryptococcosis. PLoS One 7:e34258. doi: 10.1371/journal.pone.0034258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dufait R, Velho R, De Vroey C. 1987. Rapid identification of the two varieties of Cryptococcus neoformans by d-proline assimilation. Mykosen 30:483. [PubMed] [Google Scholar]
- 18.Hashimoto A, Nishikawa T, Oka T, Takahashi K, Hayashi T. 1992. Determination of free amino acid enantiomers in rat brain and serum by high-performance liquid chromatography after derivatization with N-tert.-butyloxycarbonyl-l-cysteine and o-phthaldialdehyde. J Chromatogr B 582:41–48. doi: 10.1016/0378-4347(92)80300-f. [DOI] [PubMed] [Google Scholar]
- 19.Schell MJ, Molliver ME, Snyder SH. 1995. d-Serine, an endogenous synaptic modulator: localization to astrocytes and glutamate-stimulated release. Proc Natl Acad Sci U S A 92:3948–3952. doi: 10.1073/pnas.92.9.3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D’Aniello A, D’Onofrio G, Pischetola M, D’Aniello G, Vetere A, Petrucelli L, Fisher GH. 1993. Biological role of d-amino acid oxidase and d-aspartate oxidase. Effects of d-amino acids. J Biol Chem 268:26941–26949. [PubMed] [Google Scholar]
- 21.Robinson T. 1976. d-Amino acids in higher plants. Life Sci 19:1097–1102. doi: 10.1016/0024-3205(76)90244-7. [DOI] [PubMed] [Google Scholar]
- 22.Friedman M. 1999. Chemistry, nutrition, and microbiology of d-amino acids. J Agric Food Chem 47:3457–3479. doi: 10.1021/jf990080u. [DOI] [PubMed] [Google Scholar]
- 23.Chang YC, Khanal Lamichhane A, Bradley J, Rodgers L, Ngamskulrungroj P, Kwon-Chung KJ. 2015. Differences between Cryptococcus neoformans and Cryptococcus gattii in the molecular mechanisms governing utilization of d-amino acids as the sole nitrogen source. PLoS One 10:e0131865. doi: 10.1371/journal.pone.0131865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shaner NC, Lambert GG, Chammas A, Ni Y, Cranfill PJ, Baird MA, Sell BR, Allen JR, Day RN, Israelsson M, Davidson MW, Wang J. 2013. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat Methods 10:407–409. doi: 10.1038/nmeth.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, Sixt M, Wedlich-Soldner R. 2008. Lifeact: a versatile marker to visualize F-actin. Nat Methods 5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang YC, Lamichhane AK, Kwon-Chung KJ. 2012. Role of actin-bundling protein SacVI in growth of Cryptococcus neoformans at low oxygen concentration. Eukaryot Cell 11:943–951. doi: 10.1128/EC.00120-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soutourina J, Plateau P, Blanquet S. 2000. Metabolism of d-aminoacyl-tRNAs in Escherichia coli and Saccharomyces cerevisiae cells. J Biol Chem 275:32535–32542. doi: 10.1074/jbc.M005166200. [DOI] [PubMed] [Google Scholar]
- 28.Ohnishi E, Macleod H, Horowitz NH. 1962. Mutants of Neurospora deficient in d-amino acid oxidase. J Biol Chem 237:138–142. [PubMed] [Google Scholar]
- 29.Champney WS, Jensen RA. 1970. Molecular events in the growth inhibition of Bacillus subtilis by d-tyrosine. J Bacteriol 104:107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martho KF, de Melo AT, Takahashi JP, Guerra JM, Santos DC, Purisco SU, Melhem MS, Fazioli RD, Phanord C, Sartorelli P, Vallim MA, Pascon RC. 2016. Amino acid permeases and virulence in Cryptococcus neoformans. PLoS One 11:e0163919. doi: 10.1371/journal.pone.0163919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saier MH., Jr. 2000. Families of transmembrane transporters selective for amino acids and their derivatives. Microbiology 146:1775–1795. doi: 10.1099/00221287-146-8-1775. [DOI] [PubMed] [Google Scholar]
- 32.Fasoli MO, Kerridge D. 1990. Uptake of pyrimidines and their derivatives into Candida glabrata and Candida albicans. J Gen Microbiol 136:1475–1481. doi: 10.1099/00221287-136-8-1475. [DOI] [PubMed] [Google Scholar]
- 33.Weber E, Rodriguez C, Chevallier MR, Jund R. 1990. The purine-cytosine permease gene of Saccharomyces cerevisiae: primary structure and deduced protein sequence of the FCY2 gene product. Mol Microbiol 4:585–596. doi: 10.1111/j.1365-2958.1990.tb00627.x. [DOI] [PubMed] [Google Scholar]
- 34.Prosperi E, Croce AC, Bottiroli G, Supino R. 1986. Flow cytometric analysis of membrane permeability properties influencing intracellular accumulation and efflux of fluorescein. Cytometry 7:70–75. doi: 10.1002/cyto.990070110. [DOI] [PubMed] [Google Scholar]
- 35.Sengbusch GV, Couwenbergs C, Kuhner J, Muller U. 1976. Fluorogenic substrate turnover in single living cells. Histochem J 8:341–350. doi: 10.1007/BF01003822. [DOI] [PubMed] [Google Scholar]
- 36.Adams RJ, Pollard TD. 1989. Binding of myosin I to membrane lipids. Nature 340:565–568. doi: 10.1038/340565a0. [DOI] [PubMed] [Google Scholar]
- 37.Dufourc EJ. 2008. Sterols and membrane dynamics. J Chem Biol 1:63–77. doi: 10.1007/s12154-008-0010-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Drabikowski W, Lagwińska E, Sarzala MG. 1973. Filipin as a fluorescent probe for the location of cholesterol in the membranes of fragmented sarcoplasmic reticulum. Biochim Biophys Acta 291:61–70. doi: 10.1016/0005-2736(73)90060-6. [DOI] [PubMed] [Google Scholar]
- 39.Wachtler V, Rajagopalan S, Balasubramanian MK. 2003. Sterol-rich plasma membrane domains in the fission yeast Schizosaccharomyces pombe. J Cell Sci 116:867–874. doi: 10.1242/jcs.00299. [DOI] [PubMed] [Google Scholar]
- 40.Bagnat M, Simons K. 2002. Cell surface polarization during yeast mating. Proc Natl Acad Sci U S A 99:14183–14188. doi: 10.1073/pnas.172517799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nichols CB, Fraser JA, Heitman J. 2004. PAK kinases Ste20 and Pak1 govern cell polarity at different stages of mating in Cryptococcus neoformans. Mol Biol Cell 15:4476–4489. doi: 10.1091/mbc.e04-05-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sirotkin V, Beltzner CC, Marchand JB, Pollard TD. 2005. Interactions of WASp, myosin-I, and verprolin with Arp2/3 complex during actin patch assembly in fission yeast. J Cell Biol 170:637–648. doi: 10.1083/jcb.200502053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goode BL, Eskin JA, Wendland B. 2015. Actin and endocytosis in budding yeast. Genetics 199:315–358. doi: 10.1534/genetics.112.145540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weinberg J, Drubin DG. 2012. Clathrin-mediated endocytosis in budding yeast. Trends Cell Biol 22:1–13. doi: 10.1016/j.tcb.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaksonen M, Roux A. 2018. Mechanisms of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol 19:313–326. doi: 10.1038/nrm.2017.132. [DOI] [PubMed] [Google Scholar]
- 46.Mund M, van der Beek JA, Deschamps J, Dmitrieff S, Hoess P, Monster JL, Picco A, Nedelec F, Kaksonen M, Ries J. 2018. Systematic nanoscale analysis of endocytosis links efficient vesicle formation to patterned actin nucleation. Cell 174:884–896.e17. doi: 10.1016/j.cell.2018.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kukulski W, Schorb M, Kaksonen M, Briggs JA. 2012. Plasma membrane reshaping during endocytosis is revealed by time-resolved electron tomography. Cell 150:508–520. doi: 10.1016/j.cell.2012.05.046. [DOI] [PubMed] [Google Scholar]
- 48.Dai J, Ting-Beall HP, Hochmuth RM, Sheetz MP, Titus MA. 1999. Myosin I contributes to the generation of resting cortical tension. Biophys J 77:1168–1176. doi: 10.1016/s0006-3495(99)76968-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alvarez FJ, Douglas LM, Konopka JB. 2007. Sterol-rich plasma membrane domains in fungi. Eukaryot Cell 6:755–763. doi: 10.1128/EC.00008-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bolard J. 1986. How do the polyene macrolide antibiotics affect the cellular membrane properties? Biochim Biophys Acta 864:257–304. doi: 10.1016/0304-4157(86)90002-x. [DOI] [PubMed] [Google Scholar]
- 51.Baginski M, Czub J. 2009. Amphotericin B and its new derivatives - mode of action. Curr Drug Metab 10:459–469. doi: 10.2174/138920009788898019. [DOI] [PubMed] [Google Scholar]
- 52.Anderson TM, Clay MC, Cioffi AG, Diaz KA, Hisao GS, Tuttle MD, Nieuwkoop AJ, Comellas G, Maryum N, Wang S, Uno BE, Wildeman EL, Gonen T, Rienstra CM, Burke MD. 2014. Amphotericin forms an extramembranous and fungicidal sterol sponge. Nat Chem Biol 10:400–406. doi: 10.1038/nchembio.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baggett JJ, Shaw JD, Sciambi CJ, Watson HA, Wendland B. 2003. Fluorescent labeling of yeast. Curr Protoc Cell Biol Chapter 4:Unit 4.13. doi: 10.1002/0471143030.cb0413s20. [DOI] [PubMed] [Google Scholar]
- 54.Takeda T, Kawate T, Chang F. 2004. Organization of a sterol-rich membrane domain by cdc15p during cytokinesis in fission yeast. Nat Cell Biol 6:1142–1144. doi: 10.1038/ncb1189. [DOI] [PubMed] [Google Scholar]
- 55.Costa C, Ponte A, Pais P, Santos R, Cavalheiro M, Yaguchi T, Chibana H, Teixeira MC. 2015. New mechanisms of flucytosine resistance in C. glabrata unveiled by a chemogenomics analysis in S. cerevisiae. PLoS One 10:e0135110. doi: 10.1371/journal.pone.0135110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chang YC, Khanal Lamichhane A, Garraffo HM, Walter PJ, Leerkes M, Kwon-Chung KJ. 2014. Molecular mechanisms of hypoxic responses via unique roles of Ras1, Cdc24 and Ptp3 in a human fungal pathogen Cryptococcus neoformans. PLoS Genet 10:e1004292. doi: 10.1371/journal.pgen.1004292. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Amino acid alignment of myosin-I. Amino acid sequences of myosin-I from strain R265, Candida albicans (Ca), Saccharomyces cerevisiae (Sc), Schizosaccharomyces pombe (Sp), and human myosin-Ie and myosin-If (NP_004989.2 and NP_036467.2, respectively) were compared by CLUSTER W alignment program. Download FIG S1, PDF file, 0.1 MB (147.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
List of strains relevant to this study. Download Table S1, DOCX file, 0.01 MB (13.7KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.