Abstract
Fusion of secretory granules and synaptic vesicles with the plasma membrane is driven by SNARE protein interactions. Intensive investigations in vitro, which include x-ray crystallography, cryoelectron microscopy, and NMR analyses by numerous groups, have elucidated structures relevant to the function of these proteins. Although function depends on the proteins being membrane bound, for experimental reasons, most of the studies have used cytosolic domains, as exemplified by the groundbreaking studies that elucidated the structure of a tetrapeptide helical bundle formed by interaction of the cytosolic domains of syntaxin1A, SNAP25 (two peptides) and synaptobrevin 2. Because the cytosolic fragments were unfettered by membrane attachments, it is likely that the tetrapeptide helical bundle reflects the lowest energy state, such as that found in the “cis” interactions of the SNARE motifs after fusion when they co-localize in the plasma membrane. Much more difficult to study and still poorly understood are critical “trans” interactions between the synaptic vesicle SNARE protein synaptobrevin 2 and the plasma membrane syntaxin1A/SNAP25 complex that initiate the fusion event. In a series of articles from the laboratory of Lukas Tamm, the spontaneous orientation of the SNARE motif of membrane-bound, full-length syntaxin1A with respect to the membrane hosting syntaxin’s transmembrane domain was investigated with nanometer precision under a variety of conditions, including those that model aspects of the “trans” configuration. The studies rely on fluorescence interference-contrast microscopy, a technique that utilizes the pattern of constructive and destructive interference arising from incoming and reflected excitation and emission light at the surface of a silicon chip that has been layered with oxidized silicon of varying depths. This Perspective discusses their findings, including the unexpected influence of the degree of lipid unsaturation on the orientation of the SNARE complex.
Main Text
Fusion of secretory granules and synaptic vesicles with the plasma membrane is driven by SNARE protein interactions. Intensive investigations in vitro, which include x-ray crystallography, cryoelectron microscopy, and NMR analyses by numerous groups, have elucidated structures relevant to the function of these proteins. Although function depends on the proteins being membrane-bound, for experimental reasons, most of the studies have used cytosolic domains as exemplified by the groundbreaking study elucidating the structure of a tetrapeptide helical bundle formed by the interaction of the cytosolic domains of neuronal syntaxin1A, SNAP25 (two peptides) and synaptobrevin 2 (1), hereafter termed “the complex.” Because the cytosolic fragments were unfettered by any membrane attachments, it is generally accepted that the complex reflects the lowest energy state, such as that found in the “cis” interactions of the SNARE motifs after fusion when they co-localize in the plasma membrane. Indeed, a more recent x-ray crystallography study using detergent-solubilized syntaxin1A and synaptobrevin 2 with their transmembrane domains intact identified the same tetrapeptide helical structure (2). The study also found that syntaxin1A and synaptobrevin interactions in the “cis” configuration extends through the linker regions that join the SNARE motifs to the transmembrane domains, suggesting that the interactions of the SNARE motifs could exert forces on the bilayer membrane throughout the critical transition from trans to cis orientations addressed below. This possibility is consistent with studies that demonstrated the importance of the linker region of syntaxin1A (3) and the lumenal (4) and transmembrane domains (5) of synaptobrevin 2 in exocytosis.
Much more difficult to study and still poorly understood are critical “trans” interactions between the synaptic vesicle SNARE protein synaptobrevin 2 and the plasma membrane syntaxin1A/SNAP25 SNARE complex that initiate the fusion event. The interaction of synaptobrevin 2 with the acceptor complex (syntaxin1A and SNAP25) is thought to begin with interaction of the N-terminal regions of the SNARE domains (6, 7, 8) with subsequent “zippering” toward the C-termini of the SNAREs. Zippering could provide energy to do the work of bringing membranes closer (9), presumably until the force of hydration repulsion of the phospholipid headgroups is balanced by the force of the Van der Waals attraction at a trans lipid headgroup separation of ∼2 nm (10). However, it is unclear whether zippering of the domains immediately adjacent to the membrane is necessary because the zippering force of the membrane-proximal domains is modest (11, 12).
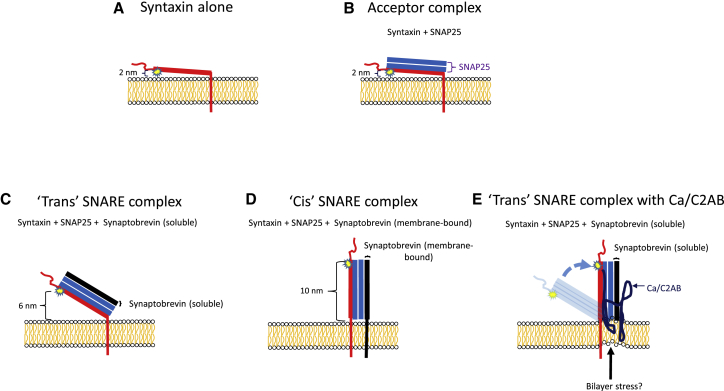
In a series of articles from the laboratory of Lukas Tamm, the spontaneous orientation of the SNARE motif of membrane-bound syntaxin1A was investigated. The studies rely on fluorescence interference-contrast microscopy (13), a technique that utilizes the pattern of constructive and destructive interference arising from incoming and reflected excitation and emission light at the surface of a silicon chip that has been layered with silicon oxide films of varying depths. The varying intensity serves as a ruler for the distance from the silicon-silicon oxide interface of a fluorophore that is excited by the light. Fluorescence intensities are averaged from ∼500 sets of patterned pixelated depths on the silicon chip. The technique allows measurement of the average distance with nanometer precision of a fluorophore from a supported bilayer that has been deposited on the chip. Alexa Fluor 546 was covalently linked to amino acid 192 of syntaxin1A at the N-terminus of the SNARE motif. The technique demonstrated that the fluorescent dye-labeled SNARE motif of membrane-bound syntaxin1A in the absence of other SNARE proteins lies mostly parallel to the bilayer at a distance of 1–2 nm (14) (Fig. 1 A). The N-terminus of the SNARE motif of syntaxin1a in the “acceptor complex,” formed in bilayers reconstituted with syntaxin1a and membrane-bound SNAP25 (dodecylated), was at a similar distance from the bilayer (Fig. 1 B; (15)).
Figure 1.
The cartoons are modeled after the figures in Kiessling et al. (15). Membrane-bound syntaxin1A was labeled with Alexa Fluor 546 at amino acid 192, located on the N-terminus of its SNARE motif (thick red line). The N-terminal Habc domain of syntaxin is not shown, although it was present in most of the experiments. The two SNARE motifs of SNAP25 are depicted as two parallel blue lines without showing the linker region. Membrane-bound synaptobrevin 2 (black line) was used to model the cis SNARE complex (D). Synaptobrevin 2 without the transmembrane domain was used to model the trans-SNARE complex without (C) and with (E) the C2AB domain of synaptotagmin1. The possibility that C2AB confers stress to the bilayer when the SNAREs are in a trans configuration is indicated in (E). The indicated distances correspond to the distances of the fluorophore from the supported bilayer. They are approximate and depend on experimental conditions including lipids in the supported bilayer. The 2-nm distance in (A) is from (14). The distances in (B–E) were taken from (15). The distance for the acceptor complex in (B) was determined with full-length syntaxin1A and dodecylated (membrane-bound) SNAP25. The distances in (C–E) were obtained with cysteine-free SNAP25 (without acyl fatty acids). Similar results were obtained for the cis-SNARE complex depicted in (D) when the complex was reconstituted with full length syntaxin1A, dodecylated (membrane-bound) SNAP25, and full-length (membrane-bound) synaptobrevin 2 (V. Kiessling, personal communication). To see this figure in color, go online.
The ”cis,” “trans,” and Ca2+/C2AB-activated “trans” SNARE complexes were modeled in supported bilayers using the same Alexa Fluor 546-labeled syntaxin1A (15). The “cis” complex was formed in the supported bilayer with membrane-bound synaptobrevin 2, membrane-bound syntaxin1A, and soluble SNAP25. Consistent with previous structural studies (see (1, 2)), the N-terminal end of the SNARE motif of syntaxin1A in complex with the other SNAREs was ∼10 nm from the bilayer (Fig. 1 D), further away than in the preformed acceptor complex without synaptobrevin 2. A similar result was obtained when membrane-bound (dodecylated) SNAP25 rather than soluble SNAP25 was used (V. Kiessling, personal communication). The “trans” SNARE complex that is presumed to form before fusion was modeled by using the cytosolic domain of synaptobrevin 2 (without the transmembrane domain). The important characteristic of this configuration is that the SNARE motif of synaptobrevin 2 is not constrained by being in the same bilayer as the acceptor complex, thereby differentiating it from the cis SNARE complex. The N-terminus of syntaxin1A was at an intermediate distance from the bilayer (∼6 nm, Fig. 1 C) compared to the acceptor complex alone and the cis complex. It should be noted that this configuration lacks an important characteristic of the actual trans-SNARE complex, being dually constrained by both fusing membranes. The Ca2+ trigger for exocytosis was modeled by the addition of Ca2+ and the C2AB domain of synaptotagmin. Remarkably, the distance of the “trans” domain SNARE complex increased to 10 nm (similar to that of the “cis” complex, Fig. 1 E). There was little effect of Ca2+/C2AB on the “acceptor” complex or the “cis” complex. This specificity supports the contention that the experiments model the physiologically relevant SNARE complexes. It is not known whether this change in distance of the “trans” domain SNARE complex is simply due to the steric hindrance caused by protein adhesion onto syntaxin1A or a bona fide conformational change in syntaxin1A at its membrane attachment region. Whatever the correct interpretation, the experiments are consistent with the hypothesis that the Ca2+-bound C2AB forces the orientation of the trans-SNARE complex to that resembling the fused (“cis”) state.
Fusion requires major and complex lipid rearrangements (16). How do the lipids influence SNARE interactions? Kiessling et al. (15) investigated the lipid dependencies of the tilt of the membrane-bound syntaxin1A SNARE motif. Not surprisingly, the experiments revealed the importance of negatively charged phospholipids, phosphatidylserine, and PI-4,5-P2, which enhance the interaction of synaptotagmin1 with membranes (17). Unexpectedly, the studies identified an important role for the balance of unsaturated and saturated fatty acid acyl chains of the phospholipids. The Ca2+/C2AB-dependent structural changes were optimal when the phospholipid acyl chains were mixed: 1-palmitoyl-2-oleoyl rather than just dipalmitoyl (saturated acyl chains, 16:0) or dioleoyl (single double-bond acyl chains, 18:1) phospholipids. In fact, the SNARE arrangement that modeled the “trans” complex remained parallel to bilayers that contained only dipalmitoyl phospholipids in the presence of Ca2+/C2AB. Lipid unsaturation increases acyl chain splaying in the bilayer and alters numerous physical parameters, including membrane thickness, spontaneous monolayer curvature, and compressibility. It is impossible at this time to tell which of these factors contribute to the changes in distance measured in these experiments.
The role of bilayer lipids in the orientation of the SNARE complex is consistent with other studies, suggesting more than a passive role of lipids in exocytotic fusion. Exocytosis of sea urchin egg dense core cortical granules show inhibition by lipids promoting positive monolayer spontaneous curvature (18). Curvature altering lipids alter the life times of the early fusion pore in chromaffin cells (19). A direct role for lipids in SNARE function has also been suggested in recent study concerning the function of SNAP25 in exocytosis (20).
A role for lipids in allowing “trans” SNARE interactions is also suggested based upon geometric considerations. A perfectly spherical 50-nm diameter synaptic vesicle or a 300-nm diameter secretory granule adherent to a planar plasma membrane attains a separation distance of greater than 2 nm (comparable to the tetrahelical SNARE diameter) only at distances greater than 10 nm or 24 nm, respectively, from the point of contact. Thus, there would have to be a deformation at the point of contact of the vesicle/granule membrane (outward dimpling) or of the plasma membrane (inward dimpling (21)) to accommodate zippering of the tetrahelical bundle at the point of contact.
Kiessling et al. (15) also provide evidence that the structural changes that they observed are relevant to secretory granule fusion. The authors used a previously described system in which fusion of secretory granules isolated from PC12 cells is reconstituted with supported bilayers (22). The granules were isolated from cells in which synaptotagmin was knocked down. The probability of fusion of the secretory granules with supported bilayers harboring acceptor SNARE complexes that were used in the structural studies was correlated with the altered tilt of the SNARE complex.
In summary, recent articles from the Tamm laboratory measured for the first time the orientation of the SNARE motif of syntaxin1A relative to a bilayer membrane hosting its transmembrane domain during different states of polypeptide interactions relevant to membrane fusion. The nanometer resolution required the averaging of many optical measurements, thereby preventing a fine-grained and time-dependent analysis of the configurations. Thus, changes in the average distance of the SNARE motif under the various experimental conditions may not reflect discrete distances but rather the time average of different orientations whose frequency depends upon the details of the complex. Nevertheless, the work is an example of the creative efforts of numerous laboratories that are leading us to the (still distant) holy grail of understanding in real time the detailed molecular interactions and structural changes of proteins and lipids that occur as an individual fusion event unfolds.
Acknowledgments
This work was supported by National Institutes of Health grant R01-GM110289 to R.W.H. and in part by the Division of Intramural Research of the NICHD to J.Z.
Editor: Brian Salzberg.
References
- 1.Sutton R.B., Fasshauer D., Brunger A.T. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 2.Stein A., Weber G., Jahn R. Helical extension of the neuronal SNARE complex into the membrane. Nature. 2009;460:525–528. doi: 10.1038/nature08156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lam A.D., Tryoen-Toth P., Stuenkel E.L. SNARE-catalyzed fusion events are regulated by syntaxin1A-lipid interactions. Mol. Biol. Cell. 2008;19:485–497. doi: 10.1091/mbc.E07-02-0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ngatchou A.N., Kisler K., Lindau M. Role of the synaptobrevin C terminus in fusion pore formation. Proc. Natl. Acad. Sci. USA. 2010;107:18463–18468. doi: 10.1073/pnas.1006727107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang C.W., Chiang C.W., Jackson M.B. Lipid-anchored synaptobrevin provides little or no support for exocytosis or liposome fusion. J. Biol. Chem. 2016;291:2848–2857. doi: 10.1074/jbc.M115.701169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu T., Rammner B., Jahn R. Inhibition of SNARE complex assembly differentially affects kinetic components of exocytosis. Cell. 1999;99:713–722. doi: 10.1016/s0092-8674(00)81669-4. [DOI] [PubMed] [Google Scholar]
- 7.Chen Y.A., Scales S.J., Scheller R.H. Sequential SNARE assembly underlies priming and triggering of exocytosis. Neuron. 2001;30:161–170. doi: 10.1016/s0896-6273(01)00270-7. [DOI] [PubMed] [Google Scholar]
- 8.Melia T.J., Weber T., Rothman J.E. Regulation of membrane fusion by the membrane-proximal coil of the t-SNARE during zippering of SNAREpins. J. Cell Biol. 2002;158:929–940. doi: 10.1083/jcb.200112081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharma S., Lindau M. Molecular mechanism of fusion pore formation driven by the neuronal SNARE complex. Proc. Natl. Acad. Sci. USA. 2018;115:12751–12756. doi: 10.1073/pnas.1816495115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zimmerberg J. Membrane biophysics. Curr. Biol. 2006;16:R272–R276. doi: 10.1016/j.cub.2006.03.050. [DOI] [PubMed] [Google Scholar]
- 11.Gao Y., Zorman S., Zhang Y. Single reconstituted neuronal SNARE complexes zipper in three distinct stages. Science. 2012;337:1340–1343. doi: 10.1126/science.1224492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mostafavi H., Thiyagarajan S., O’Shaughnessy B. Entropic forces drive self-organization and membrane fusion by SNARE proteins. Proc. Natl. Acad. Sci. USA. 2017;114:5455–5460. doi: 10.1073/pnas.1611506114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kiessling V., Tamm L.K. Measuring distances in supported bilayers by fluorescence interference-contrast microscopy: polymer supports and SNARE proteins. Biophys. J. 2003;84:408–418. doi: 10.1016/S0006-3495(03)74861-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang B., Kiessling V., Tamm L.K. Prefusion structure of syntaxin-1A suggests pathway for folding into neuronal trans-SNARE complex fusion intermediate. Proc. Natl. Acad. Sci. USA. 2013;110:19384–19389. doi: 10.1073/pnas.1314699110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kiessling V., Kreutzberger A.J.B., Tamm L.K. A molecular mechanism for calcium-mediated synaptotagmin-triggered exocytosis. Nat. Struct. Mol. Biol. 2018;25:911–917. doi: 10.1038/s41594-018-0130-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chernomordik L.V., Zimmerberg J., Kozlov M.M. Membranes of the world unite! J. Cell Biol. 2006;175:201–207. doi: 10.1083/jcb.200607083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bai J., Tucker W.C., Chapman E.R. PIP2 increases the speed of response of synaptotagmin and steers its membrane-penetration activity toward the plasma membrane. Nat. Struct. Mol. Biol. 2004;11:36–44. doi: 10.1038/nsmb709. [DOI] [PubMed] [Google Scholar]
- 18.Chernomordik L.V., Vogel S.S., Zimmerberg J. Lysolipids reversibly inhibit Ca(2+)-, GTP- and pH-dependent fusion of biological membranes. FEBS Lett. 1993;318:71–76. doi: 10.1016/0014-5793(93)81330-3. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Z., Jackson M.B. Membrane bending energy and fusion pore kinetics in Ca(2+)-triggered exocytosis. Biophys. J. 2010;98:2524–2534. doi: 10.1016/j.bpj.2010.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shaaban A., Dhara M., Mohrmann R. The SNAP-25 linker supports fusion intermediates by local lipid interactions. eLife. 2019;8:e41720. doi: 10.7554/eLife.41720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ornberg R.L., Reese T.S. Beginning of exocytosis captured by rapid-freezing of Limulus amebocytes. J. Cell Biol. 1981;90:40–54. doi: 10.1083/jcb.90.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kreutzberger A.J.B., Kiessling V., Tamm L.K. Reconstitution of calcium-mediated exocytosis of dense-core vesicles. Sci. Adv. 2017;3:e1603208. doi: 10.1126/sciadv.1603208. [DOI] [PMC free article] [PubMed] [Google Scholar]