Summary
Endogenous rhythmic behaviors are evolutionarily conserved and essential for life. In mammalian and invertebrate models, well characterized neuronal circuits and evolutionarily conserved mechanisms regulate circadian behavior and sleep [1–4]. In Drosophila, neuronal populations located in multiple brain regions mediate arousal, sleep drive and homeostasis (reviewed in [3,5–7]). Similar to mammals [8], there is also evidence that fly glial cells modulate the neuronal circuits controlling rhythmic behaviors including sleep [1]. Here, we describe a novel gene (CG14141; aka Nkt) that is required for normal sleep. NKT is a 162-amino acid protein with a single IgC2 immunoglobulin (Ig) domain and a high quality signal peptide [9], and we show evidence that it is secreted, similar to its C. elegans ortholog (OIG-4; [10]). We demonstrate that Nkt null flies or those with selective knockdown in either neurons or glia have decreased and fragmented night sleep, indicative of a non-redundant requirement in both cell types. We show that Nkt is required in fly astrocytes and in a specific set of wake-promoting neurons – the mushroom body (MB) α’β’ cells that link sleep to memory consolidation [11]. Importantly, Nkt gene expression is required in the adult nervous system for normal sleep, consistent with a physiological rather than developmental function for the Ig-domain protein.
Results
Knockdown of CG14141 leads to high nocturnal activity and altered sleep amount.
We identified CG14141 in an RNAi-based screen for genes that show enriched expression in astrocytes and regulate locomotor activity, circadian behavior or seizure-like behavior [12]. Flies with a pan-glial (repo-GAL4-dependent) knockdown of CG14141 have high levels of locomotor activity selectively in the night (Figure S1A). Similarly, Nkt knockdown in fly astrocytes, but not cortex or ensheathing glia, led to high levels of locomotor activity (Table S1). Notwithstanding the behavioral phenotype, there are no obvious effects of the knockdown on development, viability or gross nervous system morphology (data not shown). In addition, CG14141 has not been associated with phenotypes in other RNAi-based screens according to the Genome RNAi database [13]. The CG14141 gene was named Noktochor (Nkt), meaning “nocturnal” in Bengali, based on the high night-activity phenotype. Although there are no predicted off-target effects of the Nkt RNAi (Nkt.IR; listed in Key Resources), we nonetheless verified that Nkt.IR expression targets the Nkt gene. We measured RNA levels in head tissues of tub-GAL4>Nkt.IR flies that are expected to have a ubiquitous knockdown of the gene; those flies exhibited high nocturnal activity similar to flies with pan-glial knockdown (Figure S1B). In addition to Nkt, we assayed the Plod gene, because Nkt is nested within an intron of that gene (Flybase [14]). Suggesting RNAi specificity, quantitative real-time PCR revealed a significant reduction in Nkt mRNA abundance (>90%), relative to controls, with no effect on Plod expression (Figure S1C).
KEY RESOURCE TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Software | ||
| DAM acquisition software | Trikinetics, Inc. | http://www.trikinetics.com |
| Flytoolbox software | Joel Levine, U. Toronto, Canada | [52] |
| Sleep analysis software | Paul Shaw, Wash. U., St. Louis, MO) | [51] |
| MatLab v. 2011b | The MathWorks, Natick, MA, USA | https://www.mathworks.com/products/matlab.html |
| Other | ||
| DAM activity monitors | Trikinetics, Inc. | http://www.trikinetics.com |
| Nkt cDNA clone RH33338 | Drosophila Genome Resource Center (DGRC) | https://dgrc.bio.indiana.edu/Home |
| fosmid clone fTRG3207692103829951_C07 | Source Bioscience, Nottingham, UK | https://www.sourcebioscience.com/ |
| Anti-REPO and anti-ELAV antibodies | Developmental Studies Hybridoma Bank | http://dshb.biology.uiowa.edu |
| Anti-GFP antibody | ThermoFisher, Inc. | https://www.thermofisher.com/us/en/home.html |
The high nocturnal activity phenotype of Nkt knockdown flies suggests a sleep deficit. To explore this possibility, we assayed sleep in flies with glial Nkt knockdown and controls using standard procedures (see Methods). Day sleep was normal in knockdown animals and controls (Figure 1A, C), similar to the activity phenotype, but night sleep was reduced on average by ~3 h (Figure 1A, D). As a consequence, total daily sleep was also decreased (Figure S2A). Indicative of sleep fragmentation, night sleep bout duration was decreased several fold (Figure S2C) and night sleep bout number was increased with glial Nkt knockdown (Figure S2D). Typical of many sleep mutants, latency – the time until the first sleep bout after lights off – was increased with Nkt knockdown (Figure S2B). However, the waking activity index – expressed as activity events per min – was in the normal range in knockdown flies, indicating wild-type locomotor health (U-Nkt.IR control, 4.0; repo-GAL4>U-Nkt.IR, 3.5; w1118, 3.5).
Figure 1. Nkt deficits lead to reduced night sleep with no effect on day sleep.
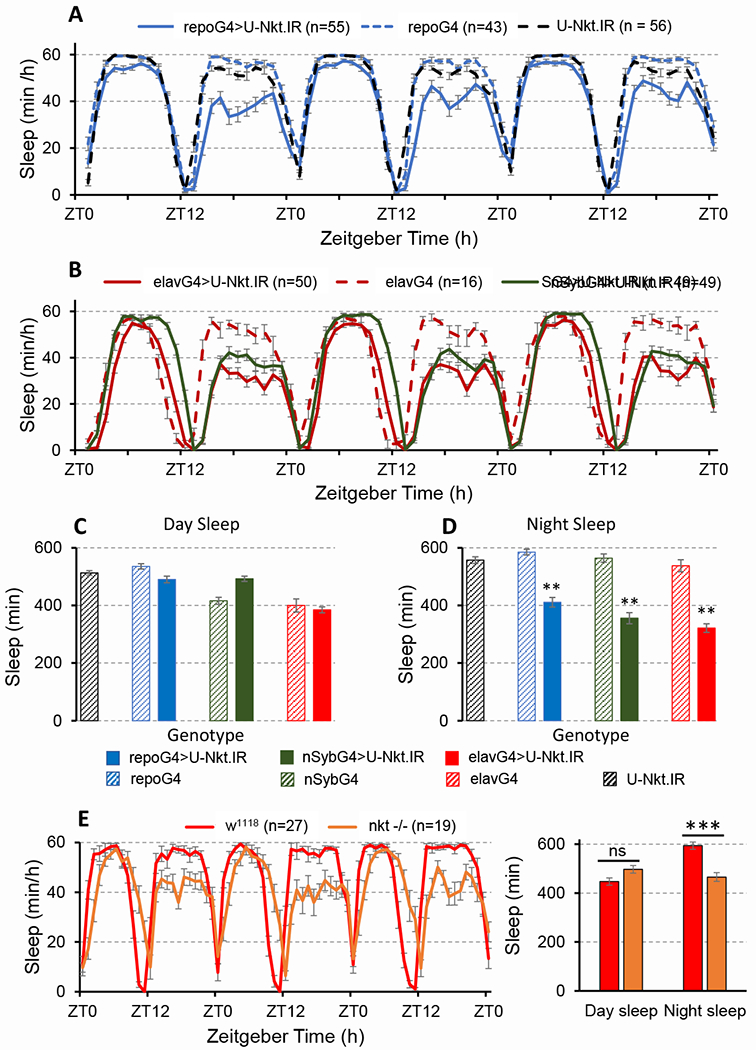
A-B) Population sleep profiles (Day 2 -Day 4) for flies in LD 12:12 with either pan-glial (A) or pan-neuronal (B) Nkt knockdown. RG4, repo-GAL4; eG4, elav-GAL4; SG4, NSyb-GAL4. C) Average day sleep in the different genotypes. D) average night sleep. Histograms represent averages for 3 days of LD data. E, F) Sleep phenotype of the Nkt L2-11 null mutant. The mutation was backcrossed into a w1118 background, the control shown in this panel. In this figure and all others, error bars are SEM. **, p<0.01 or ***, p<0.001 relative to U-Nkt.IR or GAL4 controls. See also Figures S1, S2, S3 and S4 and Table S1.
We identified Nkt as a gene showing enriched expression in fly astrocytes [12]. Therefore, we examined sleep in flies with a knockdown of the gene in astrocytes, using two different GAL4 drivers: eaat1-GAL4 [15] and GMR86E01-GAL4 which was recently described as a reference line for fly astrocytes [16]. Both drivers have a broad astrocyte pattern of expression. We observed reduced sleep with Nkt knockdown using either GAL4 strain, although GMR86E01-GAL4 was associated with a strong sleep phenotype similar to that observed with repo-GAL4. As with pan-glial knockdown, there was no effect on day sleep (Figure S2G and Mendeley Data, http://dx.doi.org/10.17632/d6jx953yjy.1).
Nkt is expressed in neurons [17] but has highly enriched astrocyte expression, and we imagined the gene might have an astrocyte-specific requirement in behavior. Surprisingly, elav-GAL4-driven pan-neuronal expression of Nkt.IR was also associated with high nocturnal activity, relative to genetic background controls, as was knockdown using nSyb-GAL4, another pan-neuronal driver (Figure S1A). A high activity phenotype was observed in constant darkness (DD) with pan-neuronal or pan-glial Nkt knockdown, but activity nonetheless exhibited circadian changes (data not shown). Given the activity phenotype with neuronal knockdown, we examined sleep in flies of similar genotype. Pan-neuronal knockdown with either elav-GAL4 or nSyb-GAL4 resulted in reduced night sleep relative to controls (Figure 1B, D) with no effect on day sleep (Figure 1A, C). Sleep was also fragmented similar to that seen with glial knockdown of the gene (Figure S2C, D) and latency was increased (Figure S2B). However, the waking activity index was in the normal range (3.5 for both nSyb>U-Nkt.IR and w1118 flies). Thus, NKT is required in both astrocytes and neurons.
Elav-GAL4-dependent knockdown of Nkt results in strong night sleep loss. To ask whether Nkt is required for sleep homeostasis, we sleep deprived neuronal knockdown and control flies. Although baseline night sleep was decreased in elavGAL4>Nkt.IR flies, sleep loss and gain after 12 hours of deprivation were similar in experimental and control populations (Figure S3). Similar results were obtained in a replicate experiment and with the use of a different pan-neuronal GAL4 for knockdown (nSybGAL4). We conclude that Nkt regulates night sleep amount and consolidation but not homeostasis.
Sleep deficits are caused by decreased Nkt gene function.
Since only a single RNAi transgene exists for Nkt, we wished to be certain that sleep deficits observed in knockdown animals were not a consequence of ‘off-target’ effects of the Nkt RNAi. We explored this issue in 2 ways. First, we showed that Nkt knockdown phenotypes can be partially rescued by co-expression of Nkt+ with Nkt RNAi in the same cell type (either glia or neurons; Figure S4; see figure legend for details). Overexpression of Nkt+ in a wild-type background had no effect on day or night sleep (data not shown), with the exception of Dv_Nkt which slightly increased sleep relative to the GAL4 control; the biological significance of this small effect is unclear. Lack of effect for overexpression is consistent with tight physiological regulation of NKT amount, similar to what has been observed for overexpression of Pigment Dispersing Factor (PDF), a clock neuropeptide [18]. Second, we generated null alleles of the gene using CRISPR/Cas9 methods (see Methods). Null mutants of Nkt had a sleep phenotype similar to that observed with knockdown of the gene in either glia or neurons, confirming a role in sleep regulation (Figures 1E and S2E, F). Of interest, the null animals had a slightly extended daytime ‘siesta’, perhaps indicative of compensatory sleep due to the night sleep deficit. In the null background, expression of Nkt+ in glia or neurons alone did not rescue the sleep phenotype, demonstrating an independent function in the two cell types. Conversely, an Nkt+ genomic transgene expressing a tagged version of NKT, under control of the endogenous promoter (see Methods) did rescue (all results in Mendeley Data, http://dx.doi.org/10.17632/7nsphs4fjm.2). Altogether, these results demonstrate that Nkt deficits cause the sleep phenotypes.
We wondered whether the night-sleep phenotype was a consequence of an altered light response, and therefore examined the phenotype in constant light and constant darkness. As expected, sleep became arrhythmic in LL due to loss of circadian control; however, it was still reduced in the Nkt null compared to control w1118 flies (data not shown). Because the curves were flat in LL, it was not possible to specify selective effects on day or night sleep. In DD, sleep was reduced in the mutant but became unimodal in both genotypes (data not shown); thus, night and day sleep bouts were not easily identified. We conclude that the effect on sleep is not simply due to an altered response to light.
Conditional, adult knockdown of Nkt also results in sleep and activity phenotypes.
Nkt shows enriched expression in adult astrocytes [12] and is reported to have nervous system-specific expression in adult animals (FlyAtlas [19]). To ask if NKT was required in the adult brain, we performed temperature-dependent, adult-specific knockdowns using the TARGET (UAS/GAL4/tub-GAL80ts) system [20] along with the nSyb-GAL4 or repo-GAL4 drivers. Within one day of the 30°C treatment, night sleep of knockdown flies was decreased to ~2/3 that of controls (Figure 2A, B), with no effect on day sleep parameters (Figure 2). There was a small but significant decrease in night sleep at 23°C (Figure 2A), but this is probably due to incomplete inhibition with Gal80ts at that temperature [20]. Conditional glial knockdown of Nkt using repo-GAL4 also resulted in a selective decrease of night sleep (Figure 2E, F), but only at 30°C with no effect on day sleep. Of note, night sleep was decreased in these experiments, but unlike constitutive knockdown it was not severely fragmented in either case (compare Figure 2C–D and G–H to Figure S2). This may result from less efficacious knockdown of Nkt in conditional experiments. Not surprisingly, conditional adult knockdown in either glia or neurons caused increased night activity, and this effect was completely reversible (all results in Mendeley Data, http://dx.doi.org/10.17632/tpt29br8cj.2). Together with the effects on sleep, these results indicate a physiological function for NKT protein in the adult brain.
Figure 2. Conditional pan-neuronal or pan-glial knockdown of Nkt in adult flies results in decreased sleep.
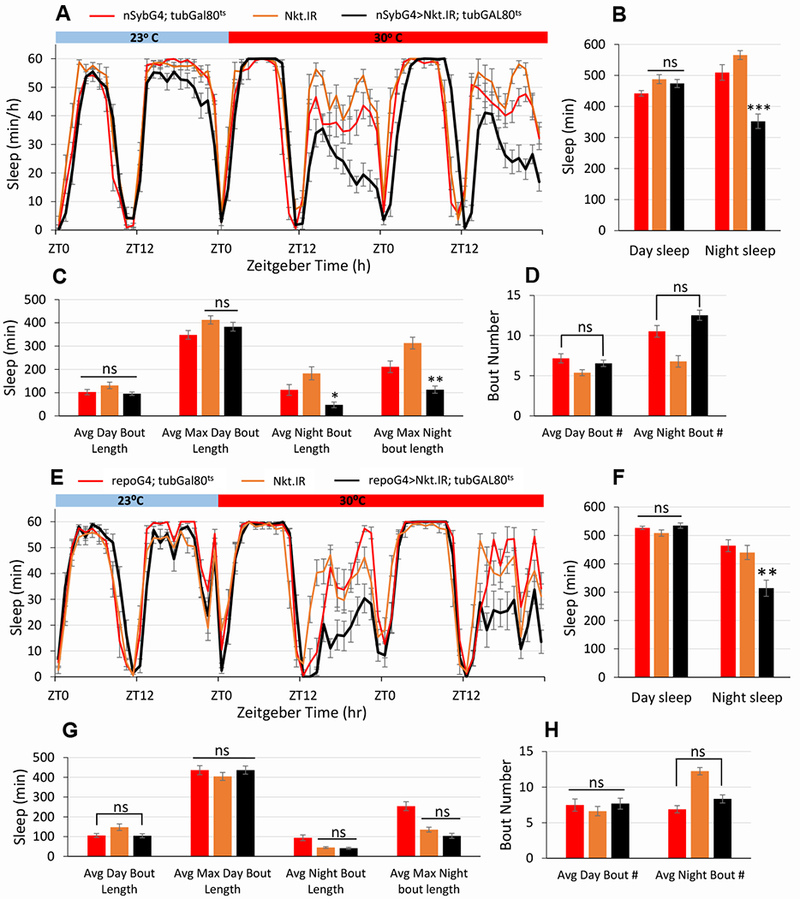
Experimental (nSybG4>Nkt.IR; tubGAL80ts or repoG4>Nkt.IR; tubGAL80ts) and control flies were reared at 23°C and LD 12:12 and then maintained in the same conditions as adults for 4 days. A 30°C step-up in temperature was used to inactivate GAL80ts and turn on GAL4 activity. Although only 2 days are shown in panels A and E, sleep was significantly decreased on each of the 4 days at 30°C in experimental flies compared to controls. A) Four-day sleep curves for nSybG4>Nkt.IR; tubGAL80ts and controls. B) Average day and night sleep amounts for experimental and control populations. C) Day and night sleep average and maximum bout lengths. D) Average bout numbers for day and night sleep. n = 12-15 flies for all genotypes. E) Four-day sleep curves for repoG4>Nkt.IR; tubGAL80ts and controls. F) Average day and night sleep amounts for experimental and control populations. G) Day and night sleep average and maximum bout lengths. H) Average bout numbers for day and night sleep. For both neuronal and glial knockdown, similar results were obtained in two independent biological replicates. n = 11-16 flies for all genotypes. ***, p < 0.001, **, p < 0.01 and *, p < 0.05 compared to controls. See also Figures S1, S2 and S4.
Nkt mRNA does not show a circadian rhythm in abundance.
As sleep is regulated, in part, by circadian processes, we considered whether Nkt RNA is under circadian control. In unpublished studies, we have examined genome-wide mRNA abundance profiles using total RNA from fly head tissues. This analysis demonstrated that Nkt mRNA does not exhibit circadian rhythms in abundance when assayed over a period of 2 days in constant darkness (Figure S1D). Thus, if NKT is under clock regulation, this likely occurs via post-transcriptional mechanisms (translation, degradation or secretion of NKT). Alternatively, Nkt expression may respond to sleep pressure to control night sleep although the gene does not appear to regulate homeostasis (Figure S3).
NKT protein localizes to neural processes and behaves as a secreted protein.
NKT protein has a high quality signal peptide, predictive of secretion, based on analysis by SignalP [21]. According to TMHMM [22] and PredGPI [23], it has neither a transmembrane domain nor a predicted GPI anchor, consistent with secretion into the extracellular space. Indeed, the C. elegans ortholog (OIG-4) is known to be a secreted protein [10], and this suggests that NKT may be secreted as well. The protein is highly conserved among invertebrate species, and in insects it is remarkably similar (70-90% identical) in species ranging from Dipterans to honeybees, ants and butterflies. Although there is not an obvious mammalian NKT ortholog, the Ig domain of NKT is similar to that of many Ig-containing mammalian proteins with neural functions including several short forms of Neuregulin 2.
To determine the endogenous expression pattern of NKT, transgenic flies were generated carrying insertions of an ~30-kb fosmid clone in which the C-terminal NKT coding region is tagged, in frame, with sequences encoding multiple epitopes including superfolder GFP (sGFP; Figure 3A; [24]). As already mentioned, this genomic construct can rescue the sleep behavior of an Nkt null mutant. Flies carrying one or two copies of the transgene expressed NKT:sGFP in glial cells and neurons in many regions of the fly brain. Punctate signal for NKT:sGFP was detected in processes of Repo-positive glial cells within the optic lobe (Figure 3B) and other brain regions. It was also broadly localized within Elav-positive neurons and within neuropil (Figure 3C), similar to that observed for overexpression of an NKT:GFP fusion.
Figure 3. NKT:sGFP is trafficked to glial and neuronal processes and it behaves as a secreted protein.
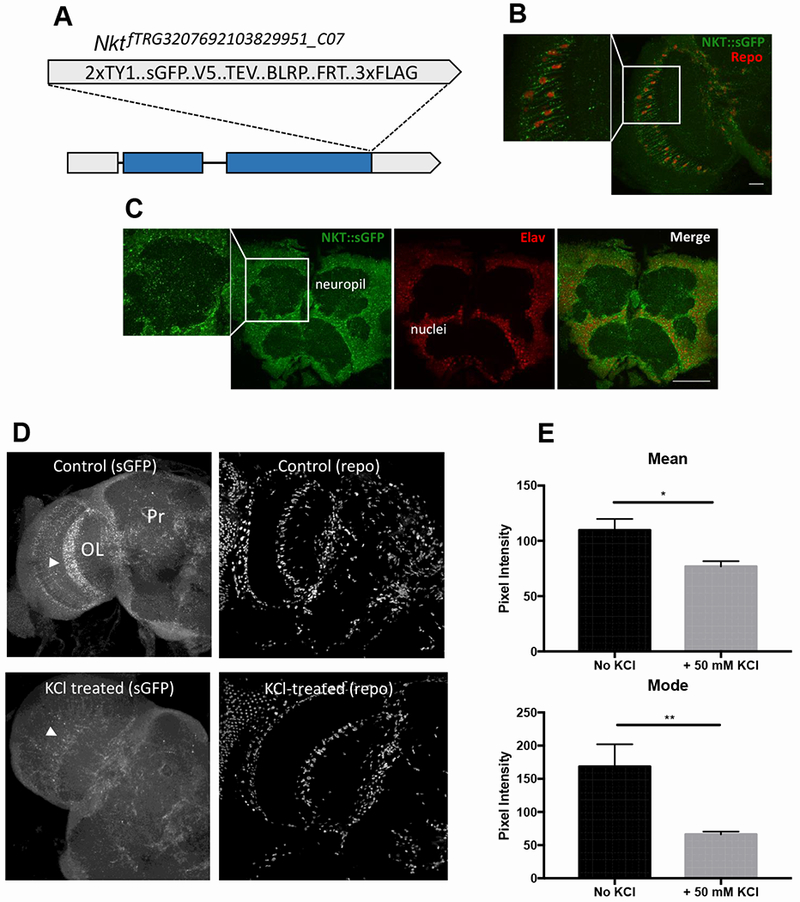
A) Schematic representation of endogenously tagged Nkt. B) NKT::sGFP expression in optic lobe glial cells of transgenic flies expressing the tagged protein from the endogenous promoter. Repo signal shows the positions of glial cell nuclei. C) Endogenous NKT in adult brain cells as assessed by NKT::sGFP localization. Elav signal shows the positions of neuronal nuclei. D) NKT:sGFP signal within astrocytes of the medulla optic lobe (arrowheads) is decreased in 90 sec by 50 mM KCl treatment. Repo antibody signal is not altered by KCl treatment. E) Mean and modal sGFP pixel intensities (arbitrary units) with and without KCl treatment. n=8 brains for both control and KCl-treated samples. Scale bars are 20 μm for B and 50 μm for C. *, p<0.05; **, p<0.01.
To test whether endogenous NKT was secreted, we asked whether brief KCl treatment of brains – expected to cause release of neuropeptides – could decrease the NKT:sGFP signal within processes of glial astrocytes. We chose to examine easily identified astrocytes of the fly optic medulla (Figure 3B) in these experiments. As shown in Figure 3D–E, a 90-sec KCl treatment of brains resulted in substantially reduced signal within these cells. In contrast, repo antibody signal was not noticeably altered by the KCl treatment (Figure 3D), indicating that the reduced sGFP signal does not result from effects of KCl treatment but is likely due to release of NKT. These results suggest that NKT is a secreted protein, similar to its C. elegans ortholog.
Nkt expression is required in α’ β’ mushroom body neurons for normal night sleep.
Astrocytes are found in all regions of the fly brain and there are few characterized GAL4 drivers that distinguish spatially distinct subtypes. Thus, we focused on the neuronal requirement for Nkt as there are GAL4 drivers that distinguish the different sleep-regulating neuronal populations. Multiple regions of the fly brain, including the mushroom body (MB), participate in the regulation of sleep. In a set of experiments designed to localize circuits requiring Nkt for nocturnal sleep regulation, we included mb247-GAL80 [25]) in the background of flies with a pan-neuronal knockdown of the gene; this GAL80 is predicted to inhibit GAL4 activity in the MB. If Nkt is required in the MB, then expression of mb247-GAL80 should ameliorate the Nkt knockdown phenotype by blocking GAL4 activity and U-Nkt.IR expression. As shown in Figure 4A, mb247-GAL80 blocked MB expression of GFP in nSyb-GAL4>Nkt::GFP flies, whereas GFP expression was not affected in the FB and Ellipsoid body. More important, expression of mb247-GAL80 in flies with a pan-neuronal Nkt knockdown was sufficient to significantly rescue the night sleep defect (Figure 4B). To follow up on this finding, we utilized a panel of MB GAL4 drivers to verify the requirement for Nkt in this brain region. In confirmation, expression of U-Nkt.IR under control of 2 different pan-MB drivers (238-GAL4 and OK107-GAL4) resulted in nocturnal sleep loss (Figure 4C and data not shown).
Figure 4. Nkt is required in mushroom body (MB) α’β’ neurons for normal sleep.
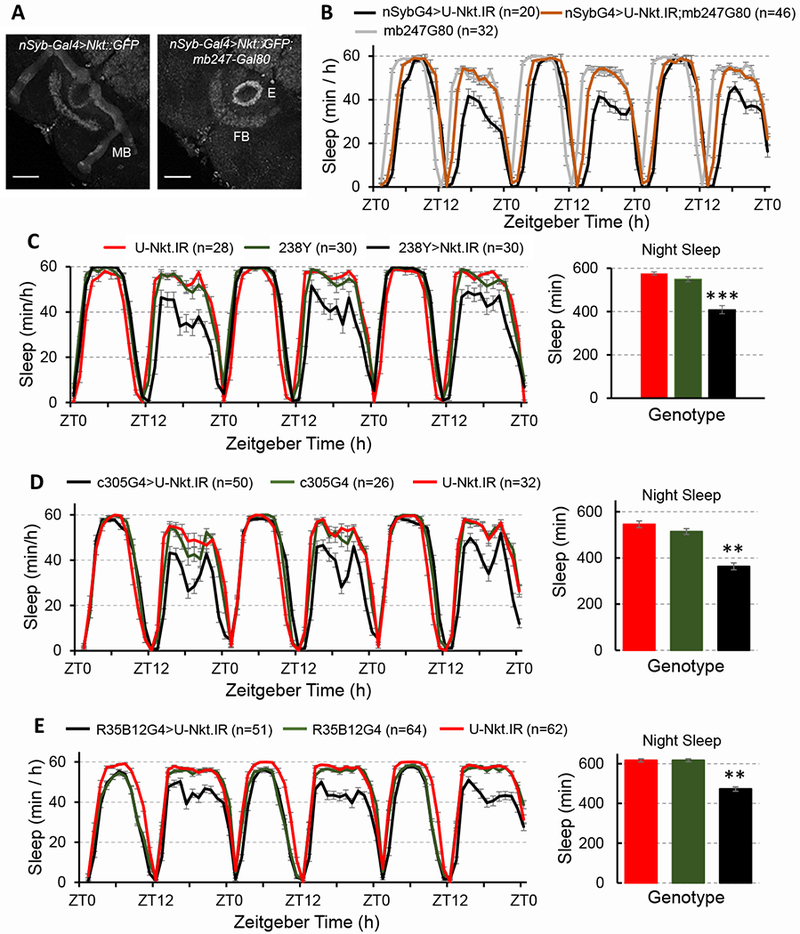
A) mb247-Gal80 blocks Gal4 activity in the MB but not the ellipsoid body (E) or fan-shaped (FB) body neurons. Signals were enhanced in Photoshop for the right-hand image to document lack of GFP expression in the MB. B) Night sleep phenotype of syb-Gal4 (SG4)>Nkt.IR; mb247-Gal80 (mb-G80) flies and controls. Night sleep for the nSybG4>U-Nkt.IR; mb247G80 population (a 3-day average) is significantly different from that of the nSybG4>U-Nkt.IR knockdown control; p<0.01. C) Flies with 238Y-GAL4-driven knockdown have reduced night sleep and sleep bout fragmentation. This panel shows one of 3 biological replicates with similar results. Knockdown of Nkt with a different pan-MB driver (OK107-GAL4) also reduced night sleep. D, E) Nkt knockdown in the α’β’ lobes of the MB using either R35B12-GAL4 (C, C1) or c305a-GAL4 (D, D1) causes a reduction in night sleep with no effect on day sleep. For C, D and E, histograms represent averages for 3 days of LD data. **, p<0.01; ***, p<0.001 relative to controls.
We asked whether specific cells of the MB require Nkt. It has been reported, for example, that the α’β’ MB neurons [26] promote wakefulness and the integration of sleep with memory consolidation [11], the latter another function of the MB [27]. Thus, we asked if Nkt if required within this subset of MB neurons, using two different GAL4 drivers that express in the α’β’ cells (c305a-GAL4 and R35B12-GAL4 [25,28]). Indeed, we found that knockdown of Nkt with either driver was associated with decreased night sleep but normal day sleep (Figure 4D, E), similar to the phenotype observed in flies with a pan-MB or pan-neuronal Nkt deficit. These results indicate a specific requirement for Nkt in important sleep-regulating MB neurons.
Discussion
Our study has identified a novel astrocyte-enriched Ig-domain protein that regulates night sleep. Conditional knockdown experiments indicate that Nkt functions in the adult brain for the physiological regulation of sleep. The results demonstrate localization of NKT in processes of neurons and astrocytes and suggest that the protein is secreted from these cells, similar to the C. elegans ortholog OIG-4 [10]. Although secreted neuronal peptides, including PDF, are known to be important regulators of circadian behavior and sleep [29–37], very few secreted glial peptides regulating sleep have been reported. However, it has been reported that glia-neuron signaling via Notch-Delta interactions regulate sleep homeostasis [38], and a recent paper describes a function for fly astrocyte TNFα (Eiger) and its receptor (Wengen) in this process [39]. More generally, glia-neuron interactions are known to be important for sleep regulation in mammals and Drosophila [38,40]. Mammalian astrocytes modulate sleep homeostasis by an action of adenosine on A1 neuronal receptors (reviewed in [8]). In Drosophila, several glial classes have been implicated in sleep regulation. Perturbations of endocytosis in fly surface glia, for example, enhance sleep and lead to resistance to sleep deprivation [41]. It has also recently been reported that ensheathing glia and a taurine transporter (Eaat2) localized in these cells promote sleep during the daytime [42], the opposite of NKT.
NKT is required in both astrocytes and specific cells of the mushroom body (MB), of interest as the MB has been implicated in sleep regulation [43,44]. The MB is innervated by multiple populations of neurons – including the inhibitory Dorsal Paired Medial (DPM) and the Anterior Paired Lateral (APL) cells [11,45,46] – and the DPMs can promote sleep by inhibiting the wake-promoting MB α’β’ neurons [11]. As NKT is required in the α’β’ neurons, we suggest it is secreted from those cells to enhance inhibitory inputs, thus promoting sleep. There are two possible mechanisms by which NKT secretion might regulate synaptic inhibition: (1) by activation of sleep-promoting DPM or APL neurons through a retrograde action of the secreted protein, or (2) through an autocrine action of NKT on the α’β’ neurons, themselves, to regulate the cellular response to neurotransmitters. An intriguing possibility is that NKT acts to regulate neurotransmitter receptor amounts or clustering on the α’β’ neurons, similar to the autocrine action of C. elegans OIG-4 on muscle acetylcholine receptors [10].
STAR*METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, F. Rob Jackson (rob.jackson@tufts.edu). Transgenic Drosophila strains generated in this study will be made available for use upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Drosophila melanogaster males were employed in all studies.
METHODS DETAILS
Fly Stocks and maintenance:
A standard cornmeal-agar medium, with added wheatgerm, was used for crosses and maintenance of Drosophila cultures. All crosses or stocks were reared at 25°C in a light/dark cycle consisting of 12 hours of light and 12 hours of dark (LD 12:12) unless otherwise stated. The sources of Drosophila strains are indicated in the Key Resource Table.
DNA Cloning and production of transgenic flies:
The UAS-Nkt::GFP plasmid construct was generated using a full-length cDNA clone for Nkt (RH33338). A 530-bp cDNA encoding the entire Nkt gene was PCR amplified using a forward primer containing an EcoRI site and a reverse primer containing an XhoI site (Table S2). The amplified fragment was cloned into the pUAST transformation vector. Sequence encoding Emerald-GFP(EMD) was obtained by KpnI digestion from a pUAST vector containing an ANF-EMD fusion [47] and ligated into the pUAST-Nkt vector to generate pUAS-Nkt-GFP (EMD is hereafter referred to as GFP). D. virilis Nkt cDNA and all other synthetic DNAs were obtained from GenScript, Inc. The D. virilis ortholog encodes a protein that is >90% similar to D. melanogaster NKT, but the coding sequence is not predicted to be targeted by D. melanogaster U-Nkt.IR (RNAi) expression. NotI and BamHI sites were included at the 5’ and 3’ ends, respectively, to facilitate cloning into pUAST. Fosmid clone clone fTRG3207692103829951_C07 of the TransgeneOme collection was employed to generate transgenic flies expressing NKT::sGFP under control of the endogenous promoter. This clone contains >10-kb of upstream and downstream sequences surrounding the Nkt gene. Generation of these fosmid genomic clones has previously been described in detail [24].
Production of CRISPR Nkt alleles:
Nkt knockout lines were engineered using a CRISPR strategy with eye color selection markers [48]. A pCFD5 construct [49] containing two sgRNAs against Nkt and one sgRNA against w (U6:3-t:: gRNA-nkt x 2:w) was inserted into attP40 embryos. The sequences of sgRNAs are shown in Table S2. Males from two resulting lines (L1 and L2) were mated with y1, M{vas-cas9}ZH-2A (Bloomington 66554) virgin females. Possible transgenic CRISPR Nkt knockout lines were created by backcrossing individual male progeny with red and white mottled eyes suggestive of leaky Cas9-mediated w interference. These lines were denoted with the original pCFD5-containing line and individual clone number. For example, L2-11, the strain shown in Figure 1, represents clonal line 11 from the L2 cross). Each line was backcrossed to remove balancer markers and create homozygous Nkt knockout lines. Homozygous lines were screened for Nkt using PCR genotyping and were confirmed via sequencing (primer sequences shown in Table S2). All mutants lack the NKT Ig domain and are predicted to produce amino-terminal truncated NKT proteins of 19-44 amino acids.
Quantitative PCR:
RNA was isolated from whole head tissues using the TRIzol method, and cDNA was generated using a kit from Invitrogen. Primers shown in Table S2.
Immunohistochemistry and imaging:
For most immunochemical experiments, adult fly brains were dissected in ice cold 1X phosphate buffered saline (PBS) and incubated in 4% Paraformaldehyde (PFA; EMS Cat. # 15712-S) for 30 minutes on ice. Brains were then washed in 1X PBST (0.3% TritonX-100), blocked with 4% normal goat serum (NGS in 1x PBST) and incubated overnight with primary antibody at 4 °C. Primary antibodies used were anti-REPO (mouse, 1:500 dilution) and anti-ELAV (mouse, 1:100 dilution). Fluorophore-conjugated secondary antibodies (Alexa fluor 555) were used at 1:1000 dilution. For visualizing sGFP, flies were fixed for 30 minutes in 4% PFA prior to dissection in PBST. After dissection, brains were fixed in 4% PFA for 20 minutes and blocked with 5% NGS for 3 hours at room temperature before primary antibody incubation. Anti-GFP (Life Technologies) was applied at 1:1000 for 2 days at 4°C. After washing, brains were incubated with anti-rabbit Alexa Fluor 488 at 1:500 for 1 day at 4°C. Brains were washed in PBST following antibody application and mounted in Vectashield. Two υm optical sections were acquired using Leica TCS SP2 or Nikon A1R confocal microscopes.
To visualize NKT-sGFP signal in KCl-treated brains, flies were anesthetized on ice before dissection in PBS. Immediately after dissection, treated brains were transferred, one at a time, to a 50 mM KCl solution diluted in PBS for 90 seconds. Thereafter, they were fixed as described above. For controls, brains were fixed immediately after dissection. All brains were then blocked as described above and then incubated in primary antibodies (Anti-GFP, 1:500 and Anti-REPO, 1:500) for 1 day at 4°C. After washing in PBST, brains were incubated with anti-rabbit Alexa Fluor 488 and anti-mouse Alex Fluor 647, both at 1:500, for 1 day at 4°C. Brains were then washed and mounted and images acquired as described above. A region of interest (ROI) was drawn around astrocytes to quantify sGFP signal. Pixel intensity was quantified using Fiji/ImageJ.
QUANTIFICATION AND STATISTICAL ANALYSIS
Behavior Assays:
Locomotor activity was assayed using monitors and software from Trikinetics, Inc. Activity was collected in 1-min bins from 3-5 day-old males. For sleep data analysis, activity data was collected 24 h after CO2 administration. For conditional knockdown assays, flies expressing tub-GAL80ts were reared at 23°C. Two to three-day old flies were loaded in activity tubes and entrained to LD 12:12 at the same temperature and then exposed to higher temperature (30°C) to inactivate temperature sensitive GAL80ts and induce dsRNA (RNAi) expression. Sleep deprivation experiments were performed using an automated SNAP device [50]. Analyses of sleep and activity were performed using an Excel-based package obtained from Paul Shaw [51] and a MatLab-based package (Fly Toolbox) from Joel Levine [52].
Statistics:
The D’Agostino & Pearson normality test was used to assess normality in data sets. A one-way Tukey-Kramer Multiple Comparisons test was used to assess statistical significance if data were normally distributed. If the absence of normality, we used the Kruskal-Wallis nonparametric ANOVA with Dunn’s Multiple Comparison test. For experiments with two genotypes, a two-tailed Student’s t-test was used. Significance was determined as p<0.05.
DATA AND CODE AVAILABILITY
The datasets supporting the current study have not been deposited in a public repository due to the number of raw data files resulting from the analysis of behavior, but are available from the corresponding author on request. The studies did not generate new code.
Supplementary Material
Highlights.
A novel, secreted immunoglobulin-domain protein (NKT) regulates fly sleep.
Decreased NKT function selectively decreases fly night sleep.
NKT is required in both astrocytes and wake-promoting neurons for normal sleep.
It is likely that NKT mediates intercellular signaling in the adult brain.
Acknowledgements
We thank the Bloomington Stock and Vienna Drosophila Resource Centers for fly stocks, the Drosophila Genomic Resource Center for plasmid clones, the Developmental Studies Hybridoma Bank for antibodies, FlyBase for access to genetic and genomic information, and Jason Gerstner and Paul Shaw for providing sleep analysis programs. We thank Sumedha Sahay and Jessica Fok for help with characterization of an Nkt null allele and sleep deprivation experiments and Ivanna Joseph for help with genetic rescue experiments and sleep analysis. We are indebted to Alder M. Yu (University of Wisconsin - La Crosse) for providing Nkt RNA abundance data. We are grateful to Alenka Lovy and the CNR Imaging Core for advice and expertise in confocal microscopy. This work was supported by NIH R01 NS065900, NIH R01 MH099554, NIH R21 NS096991 and P30 NS047243 to FRJ. LBC was supported, in part, by NIH postdoctoral training grant K12GM074869.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
REFERENCES
- 1.Jackson FR, Ng FS, Sengupta S, You S, and Huang Y (2015). Glial cell regulation of rhythmic behavior. Methods Enzymol. 552: 45–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Helfrich-Forster C, Nitabach MN, and Holmes TC (2011). Insect circadian clock outputs. Essays Biochem. 49: 87–101. [DOI] [PubMed] [Google Scholar]
- 3.Tomita J, Ban G, and Kume K (2017). Genes and neural circuits for sleep of the fruit fly. Neurosci. Res [DOI] [PubMed] [Google Scholar]
- 4.Herzog ED, Hermanstyne T, Smyllie NJ, and Hastings MH (2017). Regulating the Suprachiasmatic Nucleus (SCN) Circadian Clockwork: Interplay between Cell-Autonomous and Circuit-Level Mechanisms. Cold Spring Harb. Perspect. Biol. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donlea JM (2017). Neuronal and molecular mechanisms of sleep homeostasis. Curr. Opin. Insect Sci. 24: 51–57. [DOI] [PubMed] [Google Scholar]
- 6.Dubowy C and Sehgal A (2017). Circadian Rhythms and Sleep in Drosophila melanogaster. Genetics 205: 1373–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allada R, Cirelli C, and Sehgal A (2017). Molecular Mechanisms of Sleep Homeostasis in Flies and Mammals. Cold Spring Harb. Perspect. Biol. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haydon PG (2017). Astrocytes and the modulation of sleep. Curr. Opin. Neurobiol 44: 28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vogel C, Teichmann SA, and Chothia C (2003). The immunoglobulin superfamily in Drosophila melanogaster and Caenorhabditis elegans and the evolution of complexity. Development 130: 6317–6328. [DOI] [PubMed] [Google Scholar]
- 10.Rapti G, Richmond J, and Bessereau JL (2011). A single immunoglobulin-domain protein required for clustering acetylcholine receptors in C. elegans. EMBO J. 30: 706–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haynes PR, Christmann BL, and Griffith LC (2015). A single pair of neurons links sleep to memory consolidation in Drosophila melanogaster. Elife. 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ng FS, Sengupta S, Huang Y, Yu AM, You S, Roberts MA, Iyer LK, Yang Y, and Jackson FR (2016). TRAP-seq Profiling and RNAi-Based Genetic Screens Identify Conserved Glial Genes Required for Adult Drosophila Behavior. Front Mol. Neurosci. 9: 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmidt EE, Pelz O, Buhlmann S, Kerr G, Horn T, and Boutros M (2013). GenomeRNAi: a database for cell-based and in vivo RNAi phenotypes. Nucleic Acids Res D1021–D1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Attrill H, Falls K, Goodman JL, Millburn GH, Antonazzo G, Rey AJ, and Marygold SJ (2016). FlyBase: establishing a Gene Group resource for Drosophila melanogaster. Nucleic Acids Res 44: D786–D792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rival T, Soustelle L, Cattaert D, Strambi C, Iche M, and Birman S (2006). Physiological requirement for the glutamate transporter dEAAT1 at the adult Drosophila neuromuscular junction. J. Neurobiol 66: 1061–1074. [DOI] [PubMed] [Google Scholar]
- 16.Kremer MC, Jung C, Batelli S, Rubin GM, and Gaul U (2017). The glia of the adult Drosophila nervous system. Glia 65: 606–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomas A, Lee PJ, Dalton JE, Nomie KJ, Stoica L, Costa-Mattioli M, Chang P, Nuzhdin S, Arbeitman MN, and Dierick HA (2012). A versatile method for cell-specific profiling of translated mRNAs in Drosophila. PLoS. One. 7: e40276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helfrich-Förster C, Tauber M, Park JH, Muhlig-Versen M, Schneuwly S, and Hofbauer A (2000). Ectopic expression of the neuropeptide pigment-dispersing factor alters behavioral rhythms in Drosophila melanogaster. J. Neurosci 20: 3339–3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chintapalli VR, Wang J, and Dow JA (2007). Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet 39: 715–720. [DOI] [PubMed] [Google Scholar]
- 20.McGuire SE, Mao Z, and Davis RL (2004). Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Sci. STKE. 2004: l6. [DOI] [PubMed] [Google Scholar]
- 21.Emanuelsson O, Brunak S, von HG, and Nielsen H (2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc 2: 953–971. [DOI] [PubMed] [Google Scholar]
- 22.Moller S, Croning MD, and Apweiler R (2001). Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics. 17: 646–653. [DOI] [PubMed] [Google Scholar]
- 23.Pierleoni A, Martelli PL, and Casadio R (2008). PredGPI: a GPI-anchor predictor. BMC. Bioinformatics. 9: 392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sarov M, Barz C, Jambor H, Hein MY, Schmied C, Suchold D, Stender B, Janosch S, VV KJ, Krishnan RT et al. (2016). A genome-wide resource for the analysis of protein localisation in Drosophila. Elife. 5: e12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krashes MJ, Keene AC, Leung B, Armstrong JD, and Waddell S (2007). Sequential use of mushroom body neuron subsets during drosophila odor memory processing. Neuron 53: 103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanaka NK, Tanimoto H, and Ito K (2008). Neuronal assemblies of the Drosophila mushroom body. J. Comp Neurol. 508: 711–755. [DOI] [PubMed] [Google Scholar]
- 27.Roman G and Davis RL (2001). Molecular biology and anatomy of Drosophila olfactory associative learning. BioEssays 23: 571–581. [DOI] [PubMed] [Google Scholar]
- 28.Jenett A, Rubin GM, Ngo TT, Shepherd D, Murphy C, Dionne H, Pfeiffer BD, Cavallaro A, Hall D, Jeter J et al. (2012). A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2: 991–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kunst M, Hughes ME, Raccuglia D, Felix M, Li M, Barnett G, Duah J, and Nitabach MN (2014). Calcitonin gene-related peptide neurons mediate sleep-specific circadian output in Drosophila. Curr. Biol 24: 2652–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parisky KM, Agosto J, Pulver SR, Shang Y, Kuklin E, Hodge JJ, Kang K, Liu X, Garrity PA, Rosbash M et al. (2008). PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit. Neuron 60: 672–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chung BY, Kilman VL, Keath JR, Pitman JL, and Allada R (2009). The GABA(A) receptor RDL acts in peptidergic PDF neurons to promote sleep in Drosophila. Curr. Biol 19: 386–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheeba V, Fogle KJ, Kaneko M, Rashid S, Chou YT, Sharma VK, and Holmes TC (2008). Large ventral lateral neurons modulate arousal and sleep in Drosophila. Curr. Biol 18: 1537–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shang Y, Donelson NC, Vecsey CG, Guo F, Rosbash M, and Griffith LC (2013). Short neuropeptide F is a sleep-promoting inhibitory modulator. Neuron 80: 171–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He C, Yang Y, Zhang M, Price JL, and Zhao Z (2013). Regulation of sleep by neuropeptide Y-like system in Drosophila melanogaster. PLoS. One. 8: e74237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Potdar S and Sheeba V (2018). Wakefulness Is Promoted during Day Time by PDFR Signalling to Dopaminergic Neurons in Drosophila melanogaster. eNeuro. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oh Y, Yoon SE, Zhang Q, Chae HS, Daubnerova I, Shafer OT, Choe J, and Kim YJ (2014). A homeostatic sleep-stabilizing pathway in Drosophila composed of the sex peptide receptor and its ligand, the myoinhibitory peptide. PLoS. Biol 12: e1001974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toda H, Williams JA, Gulledge M, and Sehgal A (2019). A sleep-inducing gene, nemuri, links sleep and immune function in Drosophila. Science 363: 509–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seugnet L, Suzuki Y, Merlin G, Gottschalk L, Duntley SP, and Shaw PJ (2011). Notch signaling modulates sleep homeostasis and learning after sleep deprivation in Drosophila. Curr. Biol 21: 835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vanderheyden WM, Goodman AG, Taylor RH, Frank MG, Van Dongen HPA, and Gerstner JR (2018). Astrocyte expression of the Drosophila TNF-alpha homologue, Eiger, regulates sleep in flies. PLoS. Genet 14: e1007724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen WF, Maguire S, Sowcik M, Luo W, Koh K, and Sehgal A (2014). A neuron-glia interaction involving GABA transaminase contributes to sleep loss in sleepless mutants. Mol. Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Artiushin G, Zhang SL, Tricoire H, and Sehgal A (2018). Endocytosis at the Drosophila blood-brain barrier as a function for sleep. Elife. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stahl BA, Peco E, Davla S, Murakami K, Caicedo Moreno NA, van Meyel DJ, and Keene AC (2018). The Taurine Transporter Eaat2 Functions in Ensheathing Glia to Modulate Sleep and Metabolic Rate. Curr. Biol 28: 3700–3708. [DOI] [PubMed] [Google Scholar]
- 43.Joiner WJ, Crocker A, White BH, and Sehgal A (2006). Sleep in Drosophila is regulated by adult mushroom bodies. Nature 441: 757–760. [DOI] [PubMed] [Google Scholar]
- 44.Pitman JL, McGill JJ, Keegan KP, and Allada R (2006). A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature 441: 753–756. [DOI] [PubMed] [Google Scholar]
- 45.Pitman JL, Huetteroth W, Burke CJ, Krashes MJ, Lai SL, Lee T, and Waddell S (2011). A pair of inhibitory neurons are required to sustain labile memory in the Drosophila mushroom body. Curr. Biol 21: 855–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu X and Davis RL (2009). The GABAergic anterior paired lateral neuron suppresses and is suppressed by olfactory learning. Nat. Neurosci 12: 53–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rao S, Lang C, Levitan ES, and Deitcher DL (2001). Visualization of neuropeptide expression, transport, and exocytosis in Drosophila melanogaster. J. Neurobiol 49: 159–172. [DOI] [PubMed] [Google Scholar]
- 48.Ge DT, Tipping C, Brodsky MH, and Zamore PD (2016). Rapid Screening for CRISPR-Directed Editing of the Drosophila Genome Using white Coconversion. G3. (Bethesda. ) 6: 3197–3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Port F and Bullock SL (2016). Augmenting CRISPR applications in Drosophila with tRNA-flanked sgRNAs. Nat. Methods 13: 852–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shaw PJ, Tononi G, Greenspan RJ, and Robinson DF (2002). Stress response genes protect against lethal effects of sleep deprivation in Drosophila. Nature 417: 287–91. [DOI] [PubMed] [Google Scholar]
- 51.Andretic R and Shaw PJ (2005). Essentials of sleep recordings in Drosophila: moving beyond sleep time. Methods Enzymol 393: 759–72. [DOI] [PubMed] [Google Scholar]
- 52.Levine JD, Funes P, Dowse HB, and Hall JC (2002). Signal analysis of behavioral and molecular cycles. BMC Neurosci. 3 (1): 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting the current study have not been deposited in a public repository due to the number of raw data files resulting from the analysis of behavior, but are available from the corresponding author on request. The studies did not generate new code.


