Abstract
Background.
Benign giant cell tumor of bone (GCT) is a primary skeletal neoplasm with an unpredictable pattern of biologic aggressiveness and cytogenetic findings characterized by telomeric associations and telomeric reduction. The role of maintaining telomeric integrity is performed by telomerase. To determine if telomerase activity is present, cell extracts from fibroblasts and tumor cells from five patients with GCT were analyzed and compared with HeLa (a positive control cell line).
Methods.
Telomerase activity was detected by visualizing the extension of radioactive telomeric repeats on DNA sequencing gels. Telomere reduction was assessed using southern blot analyses of the restriction enzyme Hinf I digested DNA with a radio-labeled telomere probe.
Results.
Telomerase or telomerase-like activity was detected in the cell extracts from HeLa and tumor cells. However, GCT telomerase activity varied and was less than that observed in HeLa, but no activity was detected from fibroblasts. In addition, telomere reduction was seen in DNA isolated from both HeLa and GCT but not in fibroblasts or age-matched controls.
Conclusion.
Telomere reduction and telomerase activity may be oncogenic sustaining events required to maintain the transformed phenotype seen in GCT.
Keywords: telomerase, telomere reduction, telomeric associations, giant cell tumor of bone, HeLa
Introduction
Giant cell tumor of bone (GCT) is a benign, primary skeletal neoplasm and has an unpredictable pattern of biologic aggressiveness that may manifest with benign pulmonary metastases in about 2% of patients.1–3 It typically occurs in the epiphyses around major joints in young adults. GCT represents an enigma because it is a neoplasm behaving intermediately between a true benign and overtly malignant tumor. Cytogenetic analyses of GCTs are characterized by telomeric associations (tas) or nonrandom end-to-end fusion of chromosomes in the majority of patient.4–13 Aberrant telomeres are involved in tas. The telomere is functionally defined as that region of DNA at the end of a linear chromosome that is required for replication and stability of the chromosome. The demonstration of tas in GCT supports the hypothesis of chromosomal instability in tumorigenesis.
Telomeric DNA has a special structure so that binding to the ends of DNA from other chromosomes is avoided, thus preventing tas and other spurious recombinations.14 Terminal repeat arrays of base pairs characterized by clusters of G residues in the 3′ strand (TTAGGG)n are consistently identified in telomeres. Repeat base pairs isolated from human telomeres are evolutionarily conserved throughout nature.15,16 Molecular analysis has demonstrated that the length of the telomere in GCT is reduced relative to each individual’s nonneoplastic control fibroblasts.17 Interestingly, telomeric reduction is also observed during the in vitro senescence of human fibroblasts,18 in vivo aging,17,19 and in several human solid tumors (e.g., colon cancer20 and intracranial tumors).21.
In humans, 5 to 16 kbp of identical TTAGGG repeats are found at the ends of all chromosomes forming telemores. Telomeric integrity is required to protect termini from recombination. The role of maintaining telomere integrity is performed by a specific telomere terminal transferase (telomerase), a ribonucleoprotein enzyme. This enzyme catalyzes de novo telomere reconstitution by the addition of TTAGGG repeats that can be quantitated in vitro.14,22,23 The TTAGGG repeat addition lengthens and stabilizes the telomere. Telomerase counteracts molecular senescence and is most active in germ cells that have significantly longer telomeres than found in somatic cells.15,20 In addition, telomerase is inactive in somatic cells where telomere shortening naturally occurs. Telomerase activity has been detected in HeLa cells,22 an immortalized tumor-derived cell line, and ovarian carcinoma24 (late stage tumor cells fractionated from nontumor cells obtained during paracentesis of ascitic fluid), but telomerase activity has never been reported from freshly harvested solid tumor specimens.
We hypothesize that the abnormal presence of telomerase activity must exist in GCT for this tumor to maintain and propagate its transformed neoplastic phenotype and that variable levels of telomerase activity and telomere reduction found in different GCTs may correlate with the biologic aggressiveness of the particular tumor. Hence, we report our cytogenetic and molecular results using a telomerase assay in five GCT patients.
Materials and Methods
Tumor samples (1cm3) were obtained after informed consent intraoperatively from five new previously unreported patients with histologically confirmed GCT (Table 1). The tumor specimens used in our analysis were carefully analyzed and only tumor from the core or middle of the tumor specimens were collected. Thus, the likelihood was increased that the tumor specimen used in our analysis did not contain nonneoplastic cells. Fibroblast cultures were concomitantly established from the GCT patients. HeLa cells were obtained commercially (National Institute of General Medical Sciences, Human Genetic Mutant Cell Repository, Camden, NJ). Routine cytogenetic studies were performed on 20 tumor cells from each GCT patient following methods described elsewhere.6,8,17
Table 1.
Clinical, Cytogenetic, and Molecular Data for Giant Cell Tumor Patients
| Patient* | Sex | Age (yr) |
Location and surgical procedure |
Average telomere length (kb) |
Telomerase activity [% activity compared with positive control (HeLa cells)] |
Telomeric associations |
Local recurrence of tumor (months followed) |
|
|---|---|---|---|---|---|---|---|---|
| GCT | Control | |||||||
| 1 | F | 43 | Tibia curettage | 14.0 | 14.0 | 5.8 | Yes | Yes (5) |
| 2 | F | 33 | Femur curettage | 13.5 | 26.0 | 1.5 | Yes | No (13) |
| 3 | F | 20 | Sacrum resection (wide) | 11.5 | 14.3 | 4.4 | NA | No (12) |
| 4 | F | 23 | Tibia curettage | 11.0 | 9.8 | 0.7 | Yes | No (12) |
| 5 | M | 33 | Tibia curettage | 10.5 | 19.0 | 3.4 | No | No (12) |
NA: not available.
Correlates with lanes in Figure 1.
Genomic DNA was isolated and digested with the restriction enzyme, Hinf I, which does not digest the telomere DNA segment (TTAGGG)n. A minigel electrophoresis was performed to check for completeness of digestion. Quantitative southern hybridization was performed to assess telomere length. Five micrograms of digested genomic DNA was electrophoresed for 20 hours at 22 volts using a 0.8% agarose gel. The fragments were transferred to a nylon membrane and hybridized with a radiolabeled probe (TTAGGG)50, following protocols previously described.17 After exposure to X-ray film, the autoradiographs were analyzed with a densitometer to quantitate the hybridization signal and to characterize the length of the telomere.
Telomerase or telomerase-like activity (TA) was assayed after a modification of the protocols described by Counter et al.22 and Morin.23 Briefly, an aliquot of cytoplasmic S-100 cell extract from approximately 5×106 cells was diluted with an equal volume of reaction mixture [(2mM dATP, 2mM dTTP, 1mM MgCl2, 1uM (TTAGGG)3 primer, 3.1 uM [α−32P] dGTP, 1mM spermidine, 5mM B-mercaptoethanol, 50mM potassium acetate and 50mM Tris-acetate (pH 8.5)]. The reaction was incubated for 60 minutes at 30°C, stopped by the addition of Tris-EDTA (pH 7.5) containing 0.1 mg/ml RNase A and followed by incubation for 15 minutes at 37°C. As a control, RNase A was added to parallel reactions to determine if TA was inhibited (telomerase is a ribonucleoprotein inactivated with RNase). The proteins were digested with 0.3mg/ml proteinase K in Tris-HC1 (pH 7.5) in 0.5% sodium dodecyl sulfate for 15 minutes at 37°C, followed by extraction with phenol and then 2.5M ammonium acetate was added with 4μg of carrier tRNA. The DNA was precipitated with ethanol at −20°C. DNA pellets were resuspended in a for-mamide loading dye, boiled, chilled on ice and loaded onto a (8% polyacrylamide-7 M urea) sequencing gel then run at 2000V for 2.5 hours in 1 × Tris, boric acid, ethylene diaminetetraacetic acid buffer. Dried gels were exposed to Amersham film (Hyperfilm-MP, Arlington Heights, IL) using an enhancing screen at −70°C for approximately 1 to 7 days. TA was determined by visualizing the size of the telomeric fragments assembled in a ladder fashion from the cytoplasmic extracts by using the telomerase reaction described above and by the telomeric DNA signal intensity determined by densitometry from the autoradiograph. Telomerase inactivity was determined if no signal was detected on the autoradiograph indicating little or no assembling of the telomeric repeats. TA was noted in the positive control after long term exposure (greater than 7 days).
Reliability of the assay was assessed by using more than one aliquot from the same cell extraction (e.g., 12.5μg, 25μg and 50μg of HeLa extract) and the signal intensity recorded for each reaction on the same auto-radiograph. A linear response was found for densito-metric determinations of aliquots from the same cell extract per specimen (i.e., a 5μg HeLa cell extract aliquot showed approximately twice the DNA intensity signal compared with a 25μg aliquot from the same autoradiograph). In addition, an average densitometric value was calculated after several recordings per reaction or lane from the autoradiograph. Generally less than a 10% variation was found based on multiple densitometer recordings of the same telomerase reaction.
Cell viability was assessed with trypan-blue staining (viable cells do not stain blue whereas nonviable cells do stain). Cells were suspended with an equal volume of trypan-blue (0.4%) in phosphate buffered saline and allowed to stain for 2–3 minutes at room temperature. One-tenth ml of cell suspension was transferred to a hemocytometer counting grid for analysis following established guidelines.25
Protein concentrations were determined by the Pierce BCA protein assay (Pierce, Rockford, IL), using standardized bovine serum albumin26 and hemoglobin was determined by absorbance spectrophotometry at a wavelength of 415 nm. The hemoglobin concentration was then subtracted from the total protein concentration from each of the cytoplasmic extracts. By so doing, the protein concentration (excluding hemoglobin) was standardized per aliquot and used in the telomerase assay.
Results
The clinical, cytogenetic, and molecular data of the patients with GCT are summarized in Table 1. Fourteen tas were observed in three of the four GCT patients demonstrating tas and studied cytogenetically. One GCT patient (No. 2) showed a chromosome ring 14 in two tumor cells. No other chromosomal abnormalities were detected.
Tumor cells from GCT patients demonstrated telomere reduction in three of five patients when compared with internal controls (e.g., fibroblasts) (Figs. 1 and 2) although their telomere reduction was not statistically significant (P > 0.05 for two-tailed matched Student’s t test). However, significant telomere reduction was observed previously in a larger number of GCT patients when compared with age-matched controls.17
Figure 1.
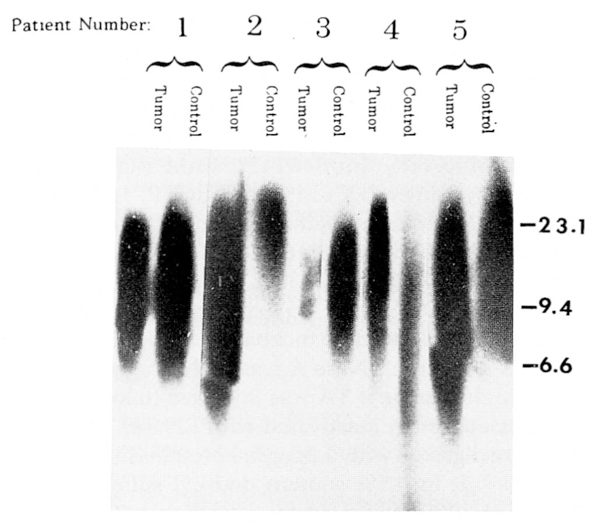
Autoradiograph of radio-labeled telomere probe (TTAGGG)50 to Hinf I digested genomic DNA (5μg per lane) from five patients with GCT and their controls. DNA from smaller telomere regions migrate farther by electrophoresis and thus show a longer signal length. Peak migration distance was determined by densitometry and mean kilobase size of the telomeric fragments determined by comparison with the migration distance of fragments of a known marker (lambda DNA digested with Hind III). The approximate locations of the high weight marker fragments (in kilobases) are shown on the vertical axis for the known marker. The patient numbers correspond to those seen in Table 1.
Figure 2.
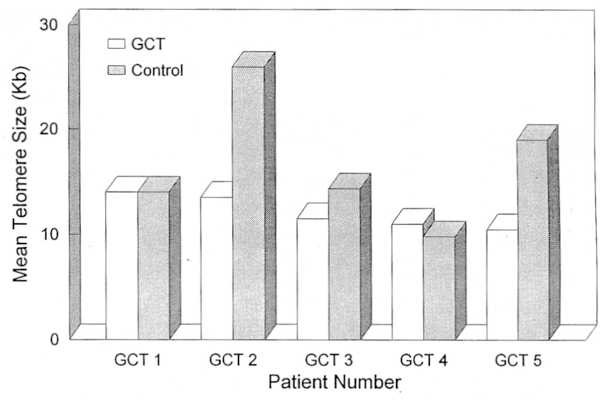
Graph showing telomere reduction in three of five GCT patients. Paired samples of GCT and control DNA for each patient are shown. Peak migration distance is converted to size in kilobases by comparison to known marker (lambda DNA digested with Hind III). Numbers on horizontal axis represents lanes as seen in Figure 1.
Telomerase assays were undertaken from cytoplasmic S100 extracts of tumor cells from five GCT patients as well as from HeLa cells and control fibroblasts. TA was detected in GCT and HeLa cells but not in fibroblasts (used as negative control) (Fig. 3). The TA was determined by visualizing the extension of radioactive telomeric repeats on the autoradiograph and recorded by densitometry. It was inhibited by pretreatment of the cytoplasmic S100 cell extract with proteinase K and RNase A. Quantitative TA was variable among the GCT patients studied (Fig. 4). GCT TA averaged approximately 3% (range 0.7–5.8%) for the five GCT patients compared with the TA found in HeLa cells (used as positive control) (Table 1, Figs. 3, 4, and 5).
Figure 3.
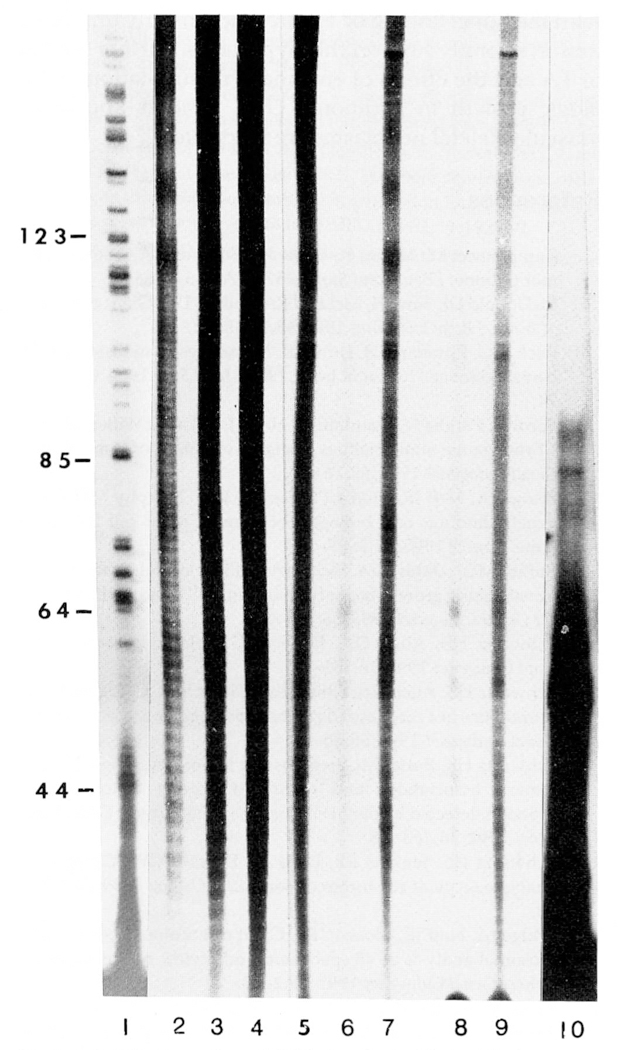
DNA sequencing gel (8%) showing evidence of telomerase or telomerase-like activity from cell extracts of tumor cells from HeLa and GCT patients. Lane 1 = base pair sizes from the nucleotide C-lane of a standard DNA sequencing reaction from intron III and exon III segments of the growth hormone gene (courtesy of J. Cogan). DNA fragment sizes can be determined by calculating the difference between the known base pair markers represented by numbers on the vertical axis. Lane 2, HeLa (12.5μg); Lane 3, HeLa (25μg); Lane 4, HeLa (50μg); Lanes 5–9, patients with GCT 1–5, respectively, (250μg each); and Lane 10, primer (TTAGGG)3. Telomerase activity was detected in the cytoplasmic extracts of HeLa cells and GCT.
Figure 4.
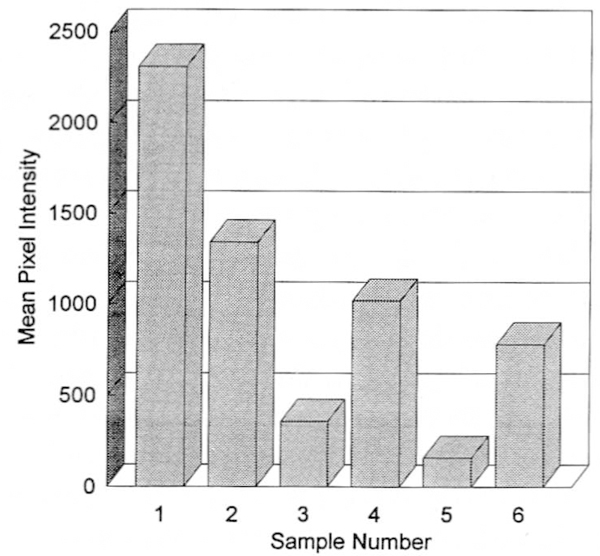
Graph showing telomeric DNA signal intensity (in pixels) for a specific area (3mm width × 300mm length) for each lane of tumor cells from five patients with GCT and HeLa by analyzing autoradiographs exposed for 18 hours from the telomerase activity assay. Sample number 1, HeLa; sample numbers 2–6, GCT patients 1–5, respectively, (25 μg of HeLa cell extract and 250μg of GCT cell extract were used per patient per analysis).
Figure 5.
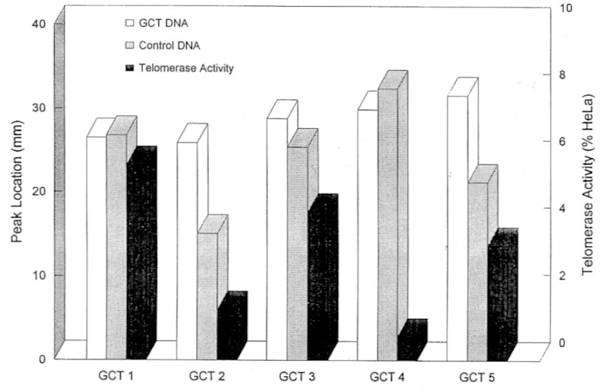
Graph showing peak migration distance of telomeric DNA using southern hybridization with GCT and control DNA and the corresponding GCT telomerase activity as a percentage of HeLa telomerase activity. Increased peak migration distance reflects shortening of the telomere. GCT patient (No. 4) did not show telomeric reduction but did demonstrate the lowest telomerase activity of the five patients with GCT.
Discussion
Healthy somatic cells in adults do not demonstrate TA.22,23 However, we have demonstrated TA in cytoplasmic extracts from GCT cells although lower than that seen in immortalized cells (e.g., HeLa cells). No TA was detected in fibroblasts (used as negative control) established from the GCT patients. The presence of activity in HeLa cells (abnormal malignant cells), absence of activity in fibroblasts (healthy somatic cells) and inhibition of activity by RNase and proteinase K (data not shown) reinforces the validity of our assay.
GCT TA as measured by signal intensity ranged from 0.7 to 5.8% of the activity detected in HeLa cells for the five GCT patients. This telomerase variation among GCT patients may illustrate cell to cell difference within the same GCT. Whether this variation suggests cell survival by selection or can be translated into biologic aggressiveness among the GCT tumors is not known. Interestingly, the GCT patient with the lowest TA (0.7%) did not show telomere reduction or metastasis and had longer average telomere size (Table 1, Fig. 5). The GCT patients with telomeric reductions had higher TA than the patient with slightly longer telomeres in the GCT cells but well below that observed in HeLa cells. The clinical and cytogenetic characteristics of our five previously unreported GCT patients did not differ significantly from GCT patients described earlier.4,5,10,17 Factors that may contribute to variation in TA may include quantity of telomerase recovered from cytoplasmic extract; number of viable cells for analysis; and amount of cytoplasmic extract used per lane in the assay. To control for these variables the number of viable cells was determined with trypan-blue staining and the quantity of cytoplasmic extract used per lane was the same for each GCT patient (250 μg). This quantity was ten times the amount used per lane for the HeLa cells to compare signal intensities for the same telomerase assay. Cytoplasmic nonhemoglobin protein concentration per extract aliquot was also similar as determined by standardized protein assay.
To date, GCT is the only human neoplasm for which tas, telomeric reduction, and TA have been demonstrated. These data suggest a fundamental role for a dysfunctional telomere in GCT oncogenesis. Our data show that TA can be detected in (i.e., in fresh tumor specimens early phases of tumorigenesis and not from cells or cell lines derived from tumor of advanced stage that may have been treated with radiation/chemotherapy) and is present in tumors of low grade malignant potential.
It is postulated that telomeric reduction and TA are oncogenic sustaining events required to maintain the transformed phenotype. The independent observation of TA identified from another solid neoplasm, along with the novel finding of TA presence in early untreated tumor, strongly support this hypothesis. Further studies of TA and the effects of enzymatic manipulation on biologic growth in additional GCT patients and other musculoskeletal neoplasms are warranted.
Acknowledgments
This research was partially funded by Orthopaedic Research and Education Foundation, Grant 93 005 (H.S.S.).
The authors thank Janie Falkenberg for expert preparation of this manuscript.
References
- 1.Campanacci M, Baldini N, Boriani S, Sudanesa A. Giant cell tumor of bone. J Bone Joint Surg 1987;69A:106–14. [PubMed] [Google Scholar]
- 2.McDonald DJ, Sim FH, McLeod RA, Dahlin DC. Giant cell tumor of bone. J Bone Joint Surg 1986;68A:235–42. [PubMed] [Google Scholar]
- 3.Rock MG, Pritchard DJ, Unni KK. Metastases from histologically benign giant cell tumor of bone. J Bone Joint Surg 1984; 66A:269–74. [PubMed] [Google Scholar]
- 4.Bardi G, Pandis N, Mandahl N, Heim J, Sfikas K, Willen H, et al. Chromosome abnormalities in giant cell tumor of bone. Cancer Genet Cytogenet 1991;57:161–7. [DOI] [PubMed] [Google Scholar]
- 5.Bridge JA, Neff JR, Bhatia PS, Sanger WG, Murphy MD. Cytogenetic findings and biologic behavior of giant cell tumors of bone. Cancer 1990;65:2697–703. [DOI] [PubMed] [Google Scholar]
- 6.Butler MG, Dahir GA, Schwartz HS. Molecular analysis of transforming growth factor beta in giant cell tumor of bone. Cancer Genet Cytogenet 1993; 66:108–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwartz HS, Allen GA, Butler MG. Telomeric associations. Appl Cytogenet 1990;16:133–7. [PMC free article] [PubMed] [Google Scholar]
- 8.Schwartz HS, Allen GA, Chudoba I, Butler MG. Cytogenetic abnormalities in a rare case of giant cell osteogenic sarcoma. Cancer Genet Cytogenet 1992;58:60–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwartz HS, Butler MG, Jenkins RB, Miller DA, Moses HL. Telomeric associations and consistent growth factor overexpression detected in giant cell tumor of bone. Cancer Genet Cytogenet 1991; 56,263–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwartz HS, Jenkins RB, Dahl RJ, Dewald GW. Cytogenetic analyses on giant cell tumor of bone. Clin Orthop 1989; 240:250–60. [PubMed] [Google Scholar]
- 11.Bridge JA, Neff JR, Mouron BS. Giant cell tumor of bone. Chromosomal analysis of 48 specimens and review of the literature. Cancer Genet Cytogenet 1992;58:2–13. [DOI] [PubMed] [Google Scholar]
- 12.Noguera R, Llombart-Bofch A, Lopez-Gines C, Carda C, Fernandez Cl. Giant cell tumor of bone, stage II, displaying translocation t(12; 19) (q13;q13). Virchows Arch A Pathol Anat Histopathol 1989; 415:377–82. [DOI] [PubMed] [Google Scholar]
- 13.Su CC, Tzeng CC, Chow NH, Lin CJ. Tissue culture, DNA flow cytometric and cytogenetic studies of a giant cell tumor of bone. Chin Med J 1991;48:419–26. [PubMed] [Google Scholar]
- 14.Blackburn EH. Telomeres and their synthesis. Science 1990;249: 489–90. [DOI] [PubMed] [Google Scholar]
- 15.Allshire RC, Dempster M, Hastie ND. Human telomeres contain at least three types of G-rich repeats distributed non-randomly. Nucleic Acids Res 1989;17:4611–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, et al. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosome. Proc Natl Acad Sci USA 1988; 85:6622–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwartz HS, Dahir GA, Butler MG. Telomere reduction in giant cell tumor of bone and with aging. Cancer Genet Cutosenet 1993;71:132–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harley CB, Futcher AB, Greider CW. Telomeres shorten during aging of human fibroblasts. Nature 1990;345:458–60. [DOI] [PubMed] [Google Scholar]
- 19.Lindsey J, McGill NI, Lindsey LA, Green DK, Cooke HJ. In vivo loss of telomeric repeats with age in humans. Mutat Res 1991;256:45–8. [DOI] [PubMed] [Google Scholar]
- 20.Hastie ND, Dempster M, Dunlop MG, Thompson AM, Green DK, Allshire RC. Telomere reduction in human colorectal carcinoma and with aging. Nature 1990;346:866–8. [DOI] [PubMed] [Google Scholar]
- 21.Nürnberg P, Thiel G, Weber F, Epplen JT. Changes of telomere lengths in human intracranial tumors. Hum Genet 1993;91:190–2. [DOI] [PubMed] [Google Scholar]
- 22.Counter CM, Avilion AA, Lefeuvre CE, Stewart NG, Greider CW, Harley CB, and Bacchetti S. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J 1992; 11:1921–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morin GB. The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell 1989;59:521–9. [DOI] [PubMed] [Google Scholar]
- 24.Counter CM, Hirte HW, Bacchetti S, Harley CB. Telomerase activity in human ovarian carcinoma. Proc Natl Acad Sci USA 1994;91:2900–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kearney JF, Radbruch A, Liesebang B, Rajewsky K. Current protocols in molecular biology. J Immunol 1979; 123:1458–550. [Google Scholar]
- 26.Smith PK, Krohn RI, Hermarson GT, Mallia AK, Gartner FH, Provenzano MD, et al. Measurement of protein using bicinchoninic acid. Anal Biochem 1985; 150: 76–85. [DOI] [PubMed] [Google Scholar]


