Abstract
Background
Animal trypanosomosis is endemic in Nigeria, while the human disease caused by Trypanosoma brucei gambiense is rarely reported nowadays after efforts to bring it under control in the 20th century. The University of Nigeria Veterinary Teaching Hospital (UNVTH) is a reference centre located within the Nsukka area and serves Enugu and neighboring states, Benue, Kogi, Anambra and Delta. Among dogs presented to the UNVTH with canine trypanosomosis, T. brucei is frequently reported as the causative agent. However, this is by morphological identification under the microscope, which does not allow distinction of human-infective (T. b. gambiense) and non-human-infective (T. b. brucei) subspecies. Here, we used subspecies-specific PCR tests to distinguish T. b. gambiense and T. b. brucei.
Methods
Blood samples were collected on FTA cards from 19 dogs presenting with clinical signs of trypanosomosis at the UNVTH from January 2017 to December 2018. All dogs had a patent parasitaemia. DNA was extracted from the FTA cards using Chelex 100 resin and used as template for PCR.
Results
All infections were initially identified as belonging to subgenus Trypanozoon using a generic PCR test based on the internal transcribed spacer 1 (ITS1) of the ribosomal RNA locus and a PCR test specific for the 177 bp satellite DNA of subgenus Trypanozoon. None of the samples were positive using a specific PCR test for T. evansi Type A kinetoplast DNA minicircles. Further PCR tests specific for T. b. gambiense based on the TgsGP and AnTat 11.17 genes revealed that two of the dogs harboured T. b. gambiense. In addition to trypanosomes of subgenus Trypanozoon, T. congolense savannah was identified in one dog using a species-specific PCR test for this taxon.
Conclusions
Nineteen dogs presenting with canine African trypanosomosis at UNVTH were infected with trypanosomes of the T. brucei group and in two cases the trypanosomes were further identified to subspecies T. b. gambiense using specific PCR tests. Thus T. b. gambiense is one of the parasites responsible for canine African trypanosomosis in the Nsukka area of Nigeria and represents a serious danger to human health.
Keywords: Canine trypanosomosis, Trypanosoma brucei gambiense, Trypanosoma brucei brucei, Trypanosoma congolense, Nsukka, Nigeria, Corneal opacity, TGSGP, AnTat 11.17
Background
Human African trypanosomosis (HAT), or sleeping sickness, is caused by protozoan parasites belonging to the Trypanosoma brucei complex in sub-Saharan Africa. The subspecies Trypanosoma brucei gambiense is the causative agent of the chronic form of the disease found in Central and West Africa, while T. b. rhodesiense is the agent of the virulent form in eastern and southern Africa. Trypanosoma b. brucei infects only domestic and wild animals [1]. Trypanosoma b. gambiense is divided into two sub-types or groups: the majority of isolates from human patients across the endemic region present a homogenous genetic composition, are avirulent in nature and belong to Group 1 T. b. gambiense, while a small minority identified predominantly in Côte d’Ivoire and Burkina Faso are genetically heterogeneous, show high virulence in experimental animals and belong to Group 2 [2–4].
Gambiense HAT caused by Group 1 T. b. gambiense (Tbg1) is considered to be an anthroponotic disease and consequently control programmes are generally aimed at stopping transmission by treating human cases and eliminating the tsetse vector [5]. However, animal reservoirs may be responsible for the endemic nature of HAT and its resurgence in the historic foci of West and Central Africa [5, 6]. Tbg1 has been isolated from pigs in Cameroon and Ivory Coast [2, 6–8], in sheep and goats in Cameroon, Equatorial Guinea and Congo [6, 9–11] and in pigs and a dog in Liberia [12]. Despite the identification of Tbg1 in various animals, there is an argument concerning their potential as animal reservoirs in sustaining Tbg1 transmission, based on the fact that these animals may not hold the disease for a long time. For example, dogs are considered to be sentinels for trypanosome infection rather than reservoir hosts, because dogs are very susceptible to trypanosome infection (T. brucei subspecies, T. evansi, T. congolense) and succumb rapidly, with death occurring within a few weeks without treatment [13]. In Kenya, outbreaks of T. b. rhodesiense in humans have been associated with outbreaks of blindness (corneal opacity) in dogs [14].
In the Nsukka area of Nigeria, T. brucei is highly prevalent in dogs [15] and also in pigs [16, 17], West African dwarf sheep and goats [18]. Tsetse flies (Glossina tachinoides) are abundant in the Nsukka area [19] and are found infected with trypanosomes (T. brucei and T. congolense) [19]. However, such reports relied on morphological identification by microscopy, which does not allow the distinction of different species and subspecies within subgenus Trypanozoon. Importantly, morphological identification fails to discriminate between human-infective and non-human-infective trypanosomes. The human serum resistance test, as originally devised by Rickman & Robson [20], was used to identify potentially human infective trypanosomes in one trade pig in Nsukka Area of Enugu State [21]. However, there has never been any report of human trypanosomosis in the Nsukka area of Enugu State, and HAT is not among the diseases commonly screened for by hospitals in Nigeria, even in areas where the tsetse vectors abound and trypanosomosis is reported in animals. In 2016, a case of HAT caused by Tbg1 was reported in a 58-year-old Nigerian woman visiting UK, who lived near Warri in Delta State, Nigeria [22]; according to the authors, no cases of HAT had been reported from Nigeria since 2012.
This study of dogs presenting with clinical signs of trypanosomosis at the UNVTH was conducted to determine which trypanosome species cause canine trypanosomosis in the Nsukka area of Nigeria and whether any dogs harbor the human-infective trypanosome, Tbg1.
Methods
Study population
Nsukka is located at 6°52ʹ–6°58ʹN, 7°20ʹ–7°27ʹE, covers an area of 1810 km2 and has a population of 309,633 [23]. The climatic conditions are characterized by high temperatures, averaging 27–28 °C. There are two seasons: the wet season extends from April to October, whilst the dry season extends from November to March. The annual rainfall range is 1680–1700 mm [24].
Blood samples were collected from 19 dogs presented to UNVTH for veterinary attention between 23rd January 2017 and 8th December 2018. On examination these dogs showed clinical signs of canine trypanosomosis including corneal opacity and enlarged lymph nodes, and were screened for trypanosomes by microscopy of wet blood smears. Demographic data, signalment (age, sex, breed and season) and clinical signs were recorded for each dog (Table 1). Blood samples from parasitologically-positive dogs were spotted on Whatman FTA cards, which were air-dried and stored in a cool, dry place until DNA extraction.
Table 1.
Summary of patient data for 19 cases of Canine African Trypanosomosis presented at UNVTH
| Case | Sex | Breed | Approx. age | LGA | Lymph node enlargement | Corneal opacity | Parasitaemia | PCV (%) | Outcome |
|---|---|---|---|---|---|---|---|---|---|
| 1 | M | Rottweiler | 5 years | Nsukka | + | + | +++ | 22 | Death |
| 2 | F | Mongrel | 2 years | Nsukka | + | + | ++ | 19 | nk |
| 3 | – | – | – | – | − | − | ++ | nd | nk |
| 4 | F | Mongrel | 2 years | Nsukka | + | + | +++ | 18 | Death |
| 5 | F | – | 4 years | Igbo-Eze North | + | + | +++ | 31 | Death |
| 6 | F | Rottweiler | 7 years | Nsukka | + | + | +++ | nd | Death |
| 7 | M | Mastiff | 2 years | Nsukka | + | - | ++ | 28 | Death |
| 8 | M | Rottweiler | – | Nsukka | + | - | +++ | nd | Death |
| 9 | F | – | 5 months | Udenu | + | + | +++ | nd | nk |
| 10 | F | Mastiff | 8 months | Nsukka | + | + | ++ | 12 | Death |
| 11 | F | Mongrel | 2.5 years | Nsukka | + | + | ++ | nd | Recovery |
| 12 | – | – | – | Nsukka | + | + | +++ | nd | nk |
| 13 | F | Caucasian | 2 years | Udenu | + | + | +++ | 26 | Death |
| 14 | M | – | 9 months | Nsukka | + | + | +++ | nd | Death |
| 15 | M | – | 2 years | Nsukka | + | + | + | 12 | nk |
| 16 | F | Caucasian | 6 months | Nsukka | + | + | +++ | 19 | Death |
| 17 | M | Mongrel | 2 years | Igbo-Eze South | − | − | +++ | nd | nk |
| 18 | F | Mongrel | 1.5 years | Nsukka | + | + | +++ | 20 | Recovery |
| 19 | M | Mongrel | 1.5 years | Nsukka | + | + | +++ | nd | nk |
Abbreviations: F, female; M, male; LGA, local government area; nd, not done; nk, not known
Key: +, <1 trypanosome/field; ++, 1–5 trypanosomes/field, +++, >5 trypanosomes/field
DNA extraction
DNA was extracted from the FTA cards using Chelex 100 resin using a method adapted from [25]. Briefly, five 2-mm discs were removed from the center of each blood spot using a Harris Uni-Core disposable punch and washed twice in 1 ml of sterile distilled water for 10 min at room temperature with occasional vortexing. Samples were then centrifuged for 3 min at maximum speed (14,500× rpm) in a microcentrifuge and the water was removed. Two hundred microliters of a 5% w/v suspension of Chelex 100 resin in sterile water was added and samples were incubated at 56 °C for 20 min with vortexing every 10 min, followed by incubation at 95 °C for 10 min. The samples were vortexed, centrifuged as before for 5 min and 150 µl of the supernatant was then transferred to a clean microcentrifuge tube, being careful to avoid carrying over any Chelex 100 resin. DNA extracts were stored frozen at − 20 °C until use.
Molecular identification by PCR
All PCRs were performed using DreamTaq polymerase (Thermo Fisher Scientific, UK) in 25 µl reaction volumes containing 5 µl of the template DNA and 0.4 µM primers (Table 2). Cycling conditions for ITS1 PCR were as specified by Adams et al. [26]; for other PCRs, cycling conditions were 95 °C for 3 min followed by 30 cycles of 95 °C for 45 s, × °C for 45 s and 72 °C for 45 s (where the annealing temperature × °C is specified in Table 2), ending with an extension reaction at 72 °C for 5 min. Positive and negative controls were included in each set of reactions: purified DNA of T. b. brucei, T. b. gambiense Group 1, T. evansi or T. congolense savannah, and water as negative control. Amplified products were resolved by electrophoresis through 1.7 % agarose gels and visualized by staining with ethidium bromide.
Table 2.
PCR for detection of African trypanosomes
| Target taxon/gene | Primer name | Primer sequence (5’–3’) | Annealing temperature (°C) | Amplicon size (bp) | Reference |
|---|---|---|---|---|---|
| Trypanosoma ITS1 | TRYP3 | TGCAATTATTGGTCGCGC | 54 | Various sizes according to species | [26] |
| TRYP4 | CTTTGCTGCGTTCTT | ||||
| Subgenus Trypanozoon 177-bp satellite repeat | TBR1 | GAATATTAAACAATGCGCAG | 60 | 164 (monomer) | [27, 32] |
| TBR2 | CCATTTATTAGCTTTGTTGC | ||||
| T. congolense savannah 350-bp satellite repeat | TCS1 | CGAGAACGGGCACTTTGCGA | 60 | 316 (monomer) | [27] |
| TCS2 | GGACAAACAAATCCCGCACA | ||||
| T. evansi Type A kDNA minicircle | EVA1 | ACATATCAACAACGACAAAG | 60 | 139 | [33] |
| EVA2 | CCCTAGTATCTCCAATGAAT | ||||
| T. b. gambiense Group 1 TgsGP gene | TgsGP-F | GCTGCTGTGTTCGGAGAGC | 50 | 308 | [28] |
| TgsGP-R | GCCATCGTGCTTGCCGCTC | ||||
| T. b. gambiense Group 1 AnTat 11.17 VSG gene | AnTA-outer | CACAGACGACAGAAGCGATA | 50 | 653 | [29] |
| AnTB-outer | GAAAGTGGGAGTTGTTGCTC | ||||
| AnTC-inner | GCCTTCGAAGACACAAGCAG | 50 | 381 | ||
| AnTD-inner | XCGTCGTGCTGAAGTCTCCTG |
Results
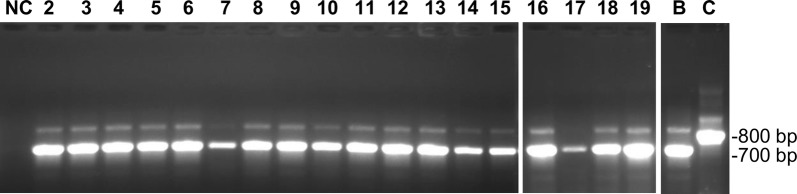
Trypanosomes in the blood samples from the 19 dogs were initially identified by a generic PCR test based on the size of the ITS1 amplicon [26]; all 19 samples produced an amplicon of ~ 700 bp, consistent with identification as subgenus Trypanozoon (Fig. 1, Table 3). This result was confirmed using primers specific for the 177-bp satellite repeat of subgenus Trypanozoon (TBR1 and 2; Table 3).
Fig. 1.
ITS1 PCR. Lane NC: water negative control; Lanes 2–19: blood samples from dogs (Table 1); Lane B: T. b. brucei J10; Lane C: T. congolense savannah WG81
Table 3.
PCR results for 19 blood samples from dogs with canine trypanosomosis
| Case | ITS1 (size in bp) | Tz satellite repeat | Tcs satellite repeat | Tev kDNA minicircle | Tbg1 TgsGP | Tbg1 AnTat 11.17 | PCR ID |
|---|---|---|---|---|---|---|---|
| 1 | 700 | + | − | − | − | − | Tbb |
| 2 | 700 | + | − | − | + | − | Tbg1 |
| 3 | 700 | + | − | − | − | − | Tbb |
| 4 | 700 | + | − | − | − | − | Tbb |
| 5 | 700 | + | − | − | − | − | Tbb |
| 6 | 700 | + | − | − | − | − | Tbb |
| 7 | 700 | + | − | − | − | − | Tbb |
| 8 | 700 | + | − | − | − | − | Tbb |
| 9 | 700 | + | − | − | − | − | Tbb |
| 10 | 700 | + | − | − | − | − | Tbb |
| 11 | 700 | + | − | − | − | − | Tbb |
| 12 | 700 | + | − | − | − | − | Tbb |
| 13 | 700 | + | + | − | − | − | Tbb, Tcs |
| 14 | 700 | + | − | − | − | − | Tbb |
| 15 | 700 | + | − | − | − | − | Tbb |
| 16 | 700 | + | − | − | − | − | Tbb |
| 17 | 700 | + | − | − | − | − | Tbb |
| 18 | 700 | + | − | − | − | − | Tbb |
| 19 | 700 | + | − | − | + | + | Tbg1 |
Abbreviations: ITS1, internal transcribed spacer; Tz, subgenus Trypanozoon; Tcs, Trypanosoma congolense savannah; Tev, T. evansi; Tbg1, T. brucei gambiense Group 1
Key: +, amplicon of expected size present, −, no amplicon present
No samples were identified as T. evansi using primers specific for the T. evansi Type A kinetoplast DNA minicircle (EVA1 and 2; Table 3). We conclude that all 19 dogs were infected with T. brucei and had therefore probably been infected by tsetse bite.
As T. congolense had been identified in previous cases of canine trypanosomosis examined [15] (P. U. Umeakuana, unpublished), the 19 blood samples were also analysed by PCR specific for T. congolense savannah using primers targeted to the ~ 350-bp satellite DNA repeat [27]. One sample was positive (Fig. 2). As this sample had already been shown to be positive for T. brucei spp., this dog had a mixed infection. However, the expected ITS1 amplicon of ~ 800 bp for T. congolense savannah was not apparent (Fig. 1); we presume this is because trypanosomes of subgenus Trypanozoon were more numerous and/or the smaller 700-bp amplicon was preferentially amplified in the PCR reaction.
Fig. 2.
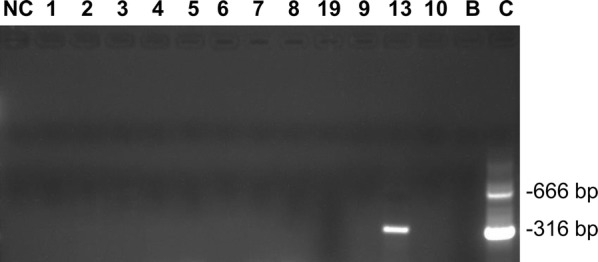
PCR specific for Trypanosoma congolense savannah. Lane NC, water negative control; Lanes 1–10, 13, 19: blood samples from dogs (Table 1, selected samples); Lane B: T. b. brucei J10; Lane C: T. congolense savannah WG81
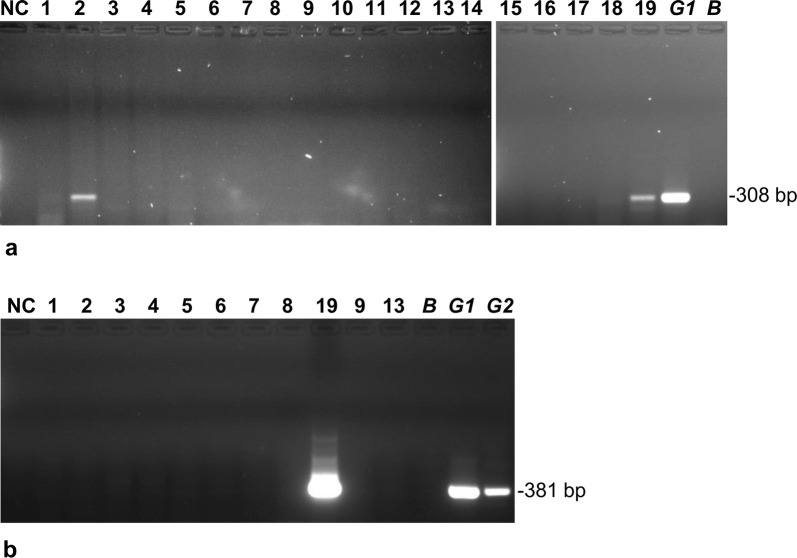
To test whether any of the dogs were infected with the human pathogen Tbg1, two subspecies-specific PCRs were carried out using primers specific for the TgsGP gene [28] and the AnTat 11.17 variant surface glycoprotein (VSG) gene, using a nested PCR [29]. Two of the 19 samples were positive for TgsGP and one was also confirmed to have the Tbg1-specific VSG gene AnTat 11.17 (Fig. 3, Table 3). As the presence of the TgsGP gene is an unequivocal marker for Tbg1 [28, 30], we conclude that two of the 19 dogs were infected with Tbg1. This may have been as the sole infection or mixed with T. b. brucei. The additional positive result for one sample with AnTat 11.17 supports the identification of Tbg1. However, loss of VSG genes from the repertoire is not uncommon, so the absence of this gene in the other sample does not detract from its identification as Tbg1; indeed absence of AnTat 11.17 in Tbg1 has been reported previously [31].
Fig. 3.
Tbg1 specific PCR. a PCR amplification of the TgsGP gene; dog blood samples 2 and 19 are positive. b Nested PCR of the AnTat11.17 VSG gene; sample 19 is positive. Lane NC: water negative control; Lanes 1–19: blood samples from dogs (Table 1); Lane B: T. b. brucei J10; Lane G1: Tbg1 Bida 3; Lane G2: Tbg1 NW2
Discussion
All 19 dogs sampled in this study from the Nsukka area of Nigeria had canine trypanosomosis caused by trypanosomes of the T. brucei group, and in one case also T. congolense savannah. These dogs typically showed corneal opacity and were reported to have become blind by their owners. Several of the dogs were in extremely poor condition and died despite treatment with Diminazene aceturate. Most of the dogs had fever with temperatures of 40–42 °C and showed high parasitaemia with low PCV values. Anorexia, inappetence, unilateral and bilateral enlargement of superficial lymph nodes (popliteal, prescapular and submandibular lymph nodes) were common observations in the infected dogs. Other clinical aberrations observed were pale mucous membranes and evidence of loss of skin turgor.
Two of the dogs were shown to be infected with the human pathogen T. b. gambiense Group 1 (Tbg1) by subspecies-specific PCR tests. To the knowledge of the authors, no cases of HAT have been identified in the Nsukka area for the past 50 years, but the identification of two dogs harbouring the causative organism is worrying. Previously, a human serum resistant trypanosome was isolated from a trade pig in the Nsukka area [21]. Thus it is possible that HAT is endemic in the Nsukka area, but that sporadic cases of HAT have been misdiagnosed and gone unreported. Alternatively, the parasite may have been imported into the area through the movement of infected tsetse flies and/or animals. The Nsukka area is in Enugu State and shares a border with Benue State, in which one of the oldest HAT foci in Nigeria, i.e. Gboko, is located. Gboko neighbors the HAT endemic focus of Fontem in the Republic of Cameroon, which could make trans-boundary movement a possibility. The recently reported case from Nigeria [22] was from Warri in Delta State, which is approximately 225 km from Nsukka.
The epidemiological implications of our finding are controversial. Dogs have been adjudged to be sentinels of infection rather than reservoir hosts, because of their susceptibility to infection and the short course of disease, which is two to four weeks without treatment [13]. On the other hand, these are pet dogs harboring a dangerous human pathogen and living in close proximity to their owners and families. In addition, there is the possibility that other animals such as cattle, sheep, goats and pigs also have cryptic infection with the human pathogen. Thus, there is a need for systematic screening of livestock as well as dogs in this area to determine the level of prevalence of Tbg1. Importantly, human health practitioners in the area also need to be aware of the possibility of HAT in patients reporting with fever and/or other signs of trypanosome infection such as enlarged lymph glands and neurological problems.
Conclusions
Nineteen dogs presenting with canine African trypanosomosis at UNVTH were infected with trypanosomes of the T. brucei group and in two cases the trypanosomes were further identified to subspecies T. b. gambiense using specific PCR tests. Thus T. b. gambiense is one of the parasites responsible for canine African trypanosomosis in the Nsukka area of Nigeria and represents a serious danger to human health.
Acknowledgements
The authors are grateful to Professor J. I. Ihedioha, the Director University of Nigeria Veterinary Teaching Hospital, Nsukka (UNVTH) for giving initial approval for this study to be done in the UNVTH. We are also grateful to subsequent directors Professor D. N. Onah and Professor S. O. Udegbunam for giving their approval for the study to be continued during their tenures. We are grateful to Mrs Ezinne Kalu and all the other staff of UNVTH for collaborating and assisting in collecting blood samples from the dogs.
Abbreviations
- ITS1
internal transcribed spacer
- PCR
polymerase chain reaction
- Tbg1
T. brucei gambiense Group 1
- Tcs
Trypanosoma congolense savannah
- Tev
T. evansi
- Tz
subgenus Trypanozoon
Authors’ contributions
PUU, RCE and BME designed the study. PUU carried out the clinical work at UNVTH and PCR analysis in Bristol assisted by WG. PUU and WG drafted the manuscript. All authors read and approved the final manuscript.
Funding
This work was funded by a Global Challenges Research Fund Ad Hoc grant provided by the UK BBSRC (Biotechnology and Biological Sciences Research Council), administered by the University of Bristol, Grant Number BB/GCRF-IAA/03.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Ethics approval and consent to participate
This study was approved by the Director UNVTH and Department of Veterinary Medicine, Faculty of Veterinary Medicine, University of Nigeria, Nsukka and observed all the guidelines governing the use of animals in research.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Paschal Ugochukwu Umeakuana, Email: paschal.umeakuana@uniabuja.edu.ng.
Wendy Gibson, Email: w.gibson@bristol.ac.uk.
Romanus Chukwuduruo Ezeokonkwo, Email: romanus.ezeokonkwo@unn.edu.ng.
Boniface Maduka Anene, Email: boniface.anene@unn.edu.ng.
References
- 1.Hoare CA. The trypanosomes of mammals. Oxford: Blackwell Scientific Publications; 1972. [Google Scholar]
- 2.Mehlitz D, Zillmann U, Scott CM, Godfrey DG. Epidemiological studies on the animal reservoir of gambiense sleeping sickness. III. Characterisation of Trypanozoon stocks by isoenzymes and sensitivity to human serum. Tropenmed Parasitol. 1982;33:113–118. [PubMed] [Google Scholar]
- 3.Gibson WC. Will the real Trypanosoma brucei gambiense please stand up? Parasitol Today. 1986;2:255–257. doi: 10.1016/0169-4758(86)90011-6. [DOI] [PubMed] [Google Scholar]
- 4.Truc P, Formenty P, Diallo PB, Komoinoka C, Lauginie F. Confirmation of two distinct classes of zymodemes of Trypanosoma brucei infecting man and wild mammals in Côte d’Ivoire: suspected difference in pathogenicity. Ann Trop Med Parasitol. 1997;91:951–956. doi: 10.1080/00034983.1997.11813224. [DOI] [PubMed] [Google Scholar]
- 5.Büscher P, Bart JM, Boelaert M, Bucheton B, Cecchi G, Chitnis N, et al. Do cryptic reservoirs threaten Gambiense-Sleeping Sickness elimination? Trends Parasitol. 2018;34:197–207. doi: 10.1016/j.pt.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Njiokou F, Nimpaye H, Simo G, Njitchouang GR, Asonganyi T, Cuny G, et al. Domestic animals as potential reservoir hosts of Trypanosoma brucei gambiense in sleeping sickness foci in Cameroon. Parasite. 2010;17:61–66. doi: 10.1051/parasite/2010171061. [DOI] [PubMed] [Google Scholar]
- 7.Simo G, Asonganyi T, Nkinin SW, Njiokou F, Herder S. High prevalence of Trypanosoma brucei gambiense group 1 in pigs from the Fontem sleeping sickness focus in Cameroon. Vet Parasitol. 2006;139:57–66. doi: 10.1016/j.vetpar.2006.02.026. [DOI] [PubMed] [Google Scholar]
- 8.Nkinin SW, Njiokou F, Penchenier L, Grebaut P, Simo G, Herder S. Characterization of Trypanosoma brucei s.l. subspecies by isoenzymes in domestic pigs from the Fontem sleeping sickness focus of Cameroon. Acta Trop. 2002;81:225–232. doi: 10.1016/S0001-706X(01)00213-3. [DOI] [PubMed] [Google Scholar]
- 9.Scott CM, Frezil J-L, Toudic A, Godfrey DG. The sheep as a potential reservoir of human trypanosomiasis in the Republic of the Congo. Trans R Soc Trop Med Hyg. 1983;77:397–401. doi: 10.1016/0035-9203(83)90172-4. [DOI] [PubMed] [Google Scholar]
- 10.Noireau F, Paindavoine P, Lemesre JL, Toudic A, Pays E, Gouteux JP, et al. The epidemiological importance of the animal reservoir of Trypanosoma brucei gambiense in the Congo: characterisation of the T. brucei complex. Tropenmed Parasitol. 1989;40:9–11. [PubMed] [Google Scholar]
- 11.Cordon-Obras C, Berzosa P, Ndong-Mabale N, Bobuakasi L, Buatiche JN, Ndongo-Asumu P, et al. Trypanosoma brucei gambiense in domestic livestock of Kogo and Mbini foci (Equatorial Guinea) Trop Med Int Health. 2009;14:535–541. doi: 10.1111/j.1365-3156.2009.02271.x. [DOI] [PubMed] [Google Scholar]
- 12.Gibson WC, Mehlitz D, Lanham SM, Godfrey DG. The identification of Trypanosoma brucei gambiense in Liberian pigs and dogs by isoenzymes and by resistance to human plasma. Tropenmed Parasitol. 1978;29:335–345. [PubMed] [Google Scholar]
- 13.Greene CE. Infectious diseases of the dog and cat. 3. St Louis: Elsevier; 2006. [Google Scholar]
- 14.Matete G. Occurrence, clinical manifestation and the epidemiological implications of naturally occurring canine trypanosomosis in western Kenya. Ond J Vet Res. 2003;70:317–323. doi: 10.4102/ojvr.v70i4.296. [DOI] [PubMed] [Google Scholar]
- 15.Umeakuana PU, Mohammed BR, Anene BM. Canine trypanosomosis in the University of Nigeria Veterinary Teaching Hospital (UNVTH), Enugu State, Nigeria, sub-Saharan Africa. J Vet Adv. 2016;6:1350–1356. doi: 10.5455/jva.19691231040001. [DOI] [Google Scholar]
- 16.Anene BM, Ifebigh AO, Igwilo IA, Umeakuana PU. Prevalence and haemato-biochemical parameters of trypanosome-infected pigs at Nsukka, Nigeria. Comp Clin Pathol. 2011;20:15–18. doi: 10.1007/s00580-009-0944-2. [DOI] [Google Scholar]
- 17.Onah DN. Porcine trypanosomiasis in Nigeria: infection in local and exotic pigs in Nsukka area of Anambra State. Trop Anim Health Prod. 1991;23:141–146. doi: 10.1007/BF02356992. [DOI] [PubMed] [Google Scholar]
- 18.Fakae BB, Chiejina SN. The prevalence of concurrent trypanosome and gastrointestinal nematode infections in West African dwarf sheep and goats in Nsukka area of eastern Nigeria. Vet Parasitol. 1993;49:313–318. doi: 10.1016/0304-4017(93)90129-B. [DOI] [PubMed] [Google Scholar]
- 19.Mmadubunyi LC. Trypanosome infection in Glossina spp. inhabiting peridomestic agroecosystem in Nsukka area, Anambra State, Nigeria. J Trop Med Parasitol. 1987;81:319–329. doi: 10.1080/00034983.1987.11812126. [DOI] [PubMed] [Google Scholar]
- 20.Rickman LR, Robson J. The testing of proven Trypanosoma brucei and T. rhodesiense strains by the blood incubation infectivity test. Bull WHO. 1970;42:911–916. [PMC free article] [PubMed] [Google Scholar]
- 21.Onah DN, Ebenebe OO. Isolation of a human serum-resistant Trypanosoma brucei from a naturally infected pig in the Nsukka area of Enugu State. Nig Vet J. 2003;24:37–45. [Google Scholar]
- 22.Luintel A, Lowe P, Cooper A, MacLeod A, Büscher P, Brooks T, et al. Case of Nigeria-acquired human african trypanosomiasis in United Kingdom, 2016. Emerg Infect Dis. 2017;23:1225–1227. doi: 10.3201/eid2307.170695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abonyi FO, Omeh CVO, Machebe NS. Neonatal mortality of pigs in Nsukka, Southeast Nigeria. Afr J Biotechnol. 2016;11:13228–13234. [Google Scholar]
- 24.Ozor N, Ozioko R, Acheampong E. Rural-urban interdependence in food systems in Nsukka Local Government Area of Enugu State, Nigeria. J Agric Ext. 2015;19:157–183. [Google Scholar]
- 25.GE-Healthcare. Reliable extraction of DNA from Whatman™ FTA™ cards. Application Note 28-9822-22 AA. 2010. https://us.vwr.com/assetsvc/asset/en_US/id/16147319/contents. Accessed 26 Feb 2019.
- 26.Adams ER, Malele II, Msangi AR, Gibson WC. Trypanosome identification in wild tsetse populations in Tanzania using generic primers to amplify the ribosomal RNA ITS-1 region. Acta Trop. 2006;100:103–109. doi: 10.1016/j.actatropica.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 27.Masiga DK, Smyth AJ, Hayes PJ, Bromidge TJ, Gibson WC. Sensitive detection of trypanosomes in tsetse flies by DNA amplification. Int J Parasitol. 1992;22:909–918. doi: 10.1016/0020-7519(92)90047-O. [DOI] [PubMed] [Google Scholar]
- 28.Radwanska M, Claes F, Magez S, Magnus E, Perez-Morga D, Pays E, et al. Novel primer sequences for polymerase chain reaction-based detection of Trypanosoma brucei gambiense. Am J Trop Med Hyg. 2002;67:289–295. doi: 10.4269/ajtmh.2002.67.289. [DOI] [PubMed] [Google Scholar]
- 29.Bromidge T, Gibson W, Hudson KM, Dukes P. Identification of Trypanosoma brucei gambiense by PCR amplification of variant surface glycoprotein genes. Acta Trop. 1993;53:107–119. doi: 10.1016/0001-706X(93)90023-5. [DOI] [PubMed] [Google Scholar]
- 30.Uzureau P, Uzureau S, Lecordier L, Fontaine F, Tebabi P, Homble F, et al. Mechanism of Trypanosoma brucei gambiense resistance to human serum. Nature. 2013;501:430–434. doi: 10.1038/nature12516. [DOI] [PubMed] [Google Scholar]
- 31.Enyaru JC, Allingham R, Bromidge T, Kanmogne GD, Carasco JF. The isolation and genetic heterogeneity of Trypanosoma brucei gambiense from north-west Uganda. Acta Trop. 1993;54:31–39. doi: 10.1016/0001-706X(93)90066-K. [DOI] [PubMed] [Google Scholar]
- 32.Moser DR, Cook GA, Ochs DE, Bailey CP, McKane MR, Donelson JE. Detection of Trypanosoma congolense and T. brucei subspecies by DNA amplification using the polymerase chain reaction. Parasitology. 1989;99:57–66. doi: 10.1017/S0031182000061023. [DOI] [PubMed] [Google Scholar]
- 33.Masiga DK. The development and application of a polymerase chain reaction methodology for the identification of African trypanosomes. PhD Thesis, University of Bristol, UK; 1994.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.