Abstract
Background
Light is a key environmental factor in regulation of anthocyanin biosynthesis. Through a large number of bagging screenings, we obtained non-photosensitive eggplants that still have decent amount of anthocyanin synthesized after bagging. In the present study, transcriptome was made to explore the molecular mechanism of dark-regulated anthocyanin synthesis in non-photosensitive eggplant.
Results
The transcriptome of the pericarp at 0 h, 0.5 h, 4 h, and 8 h after bag removal were sequenced and analyzed. Comparison of the sequencing data with those of photosensitive eggplant for the same time period showed that anthocyanin synthesis genes had different expression trends. Based on the expression trends of the structural genes, it was discovered that 22 transcription factors and 4 light signal transduction elements may be involved in the anthocyanin synthesis in two types of eggplants. Through transcription factor target gene prediction and yeast one-hybrid assay, SmBIM1, SmAP2, SmHD, SmMYB94, SmMYB19, SmTT8, SmYABBY, SmTTG2, and SmMYC2 were identified to be directly or indirectly bound to the promoter of the structural gene SmCHS. These results indicate that the identified 9 genes participated in the anthocyanin synthesis in eggplant peel and formed a network of interactions among themselves.
Conclusions
Based on the comparative transcription, the identified 22 transcription factors and 4 light signal transduction elements may act as the key factors in dark regulated anthocyanin synthesis in non-photosensitive eggplant. The results provided a step stone for further analysis of the molecular mechanism of dark-regulated anthocyanin synthesis in non-photosensitive eggplant.
Electronic supplementary material
The online version of this article (10.1186/s12864-019-6023-4) contains supplementary material, which is available to authorized users.
Keywords: Eggplant (Solanum melongena L.), Non-photosensitive, Dark-regulated, Transcriptome, Anthocyanin biosynthesis
Background
Anthocyanins are a class of secondary metabolites that contribute to the red, blue, and purple colors in a range of flowers and fruits [1]. Anthocyanins play an important role not only in plant physiology, pollinators and visual appeal of seed communicators, but also in human health, such as preventing cardiovascular disease, controlling obesity and reducing diabetes [2, 3]. Anthocyanin biosynthesis in plants is affected by many factors, including light, temperature, sugar and hormones, with light being the most important [4–7].
Light signaling plays a pivotal role in the control of plant metabolism, growth and development. Light signals are perceived by photoreceptors including phytochrome (PHYA-PHYE), cryptochromes (CRYs), phototropins (PHOTs) and UV resistance locus 8 (UVR8, [8–12]). CRYs are blue light receptor proteins of which some functions have been reported, such as the blue light-dependent hypocotyl inhibition, floral initiation in Arabidopsis, and anthocyanin accumulation in eggplant [13–15]. Studies showed that BIC (blue-light inhibitor of cryptochromes) binds to CRY2 to suppress the blue light–dependent dimerization, photobody formation, phosphorylation, and degradation, resulting in the reduction of physiological activities of CRY2 and thus the inhibition of photomorphogenesis [16]. Phototropins are blue light receptors that control many responses to optimize photosynthesis efficiency. It was shown that FaPHOT2 played a role in sensing blue light and mediating anthocyanin biosynthesis in strawberry fruits [17]. The phytochrome family of photoreceptors (phyA–phyE) monitor the environment for informational light signals and induce plant growth and developmental responses appropriate to the prevailing conditions. Arabidopsis phytochrome regulates the accumulation of anthocyanins under nitrogen, phosphorus, and cold induction conditions [18, 19]. UVR8 was identified originally as a UV-resistance gene shown to contribute to the UV-B-induced flavonoid accumulation and UV-B protection. In Arabidopsis uvr8 mutant, CHS expression and flavonoid content were reduced significantly [19, 20].
After the photoreceptors perceive light signal, they will further regulate the expression of transcription factors (TFs), such as MYBs, bHLHs, WD40, thereby activating or inhibiting the expression of structural genes and ultimately affecting anthocyanin biosynthesis. COP1 is an E3 ubiquitin ligase that promotes the ubiquitination and degradation of many genes that are related to light signal transmission [13, 21–25]. In addition, COP1 can interact with suppressor of PHYA (SPA) to inhibit photomorphogenesis, while spa mutants exhibited characteristics similar to those of cop1 mutants under dark conditions in Arabidopsis [26–28]. The TFs that control anthocyanin biosynthesis include those of the v-myb avian myeloblastosis viral oncogene homolog (MYB), the basic helix-loop-helix (bHLH), and the Trp-Asp forty amino acid repeat (WD40) proteins [29, 30]. MYB TFs, especially the R2R3-MYB class, play a key role in anthocyanin biosynthesis. The functions of R2R3 MYB TFs in anthocyanin accumulation had been well studied, such as AtMYB75 in Arabidopsis, MdMYB1 in apple, NnMYB5 in N. nucifera, PyMYB10 in Pyrus pyrifolia, etc. [31–34]. The bHLHs family can be divided into 26 subgroups, of which subgroup IIIf is associated with flavonoids [35]. In Arabidopsis, AtTT8, AtGL3 and AtEGL3 have been characterized to positively regulate anthocyanin synthesis [36–39]. In addition, the bHLHs were found to regulate anthocyanin synthesis in other plant species, such as MdbHLH3 in apple, LcbHLH in Litchi chinensis, MtbHLH (MtTT8) in Medicago truncatula, and VvbHLH (VvMYC1) in grape [29, 40–43]. Other transcription factors, including Elongated Hypocotyl 5 (HY5), TCP, and squamosa promoter-binding protein-like 9 (SPL9), have also been shown to be associated with anthocyanin biosynthesis [44–47].
Anthocyanin biosynthesis is regulated by transcription factors that induce the expression of structural genes coded for enzymes in the biosynthesis pathway [48, 49]. Anthocyanins are synthesized from phenylalanine. The synthesis was catalyzed first by phenylalanine ammonia lyase (PAL) and 4-coumarate-CoA ligase (4CL), and then by early flavonoid biosynthetic genes (EBGs) and late biosynthetic genes (LBGs) [50]. The EBGs encode enzymes such as chalcone synthase (CHS), chalcone isomerase (CHI), and flavanone 3-hydroxylase (F3H). The LBGs include dihydroflavonol reductase (DFR), anthocyanidin synthase (ANS), flavonoid 5-Oglucosyltranferase (5GT) and flavonoid 3-Oglucosyltranferase (3GT), which regulate the production of anthocyanins [50].
Eggplant (Solanum melongena L.) is a horticultural crop of high economic value and has more than one thousand years of plantation history in China. Greenhouse cultivation and low light conditions, however, often lead to poor coloration of eggplant and reduce its quality. In this study, we obtained non-photosensitive eggplant through a large number of bagging screenings that still synthesized a significant amount of anthocyanin after bagging. Twelve RNA samples of non-photosensitive eggplant before and after light induction were sequenced with the Illumina Hiseq platform. Through comparative analysis of transcriptome data of both photosensitive and non-photosensitive eggplant samples at the same time points, we aimed to find key genes that took part in dark-regulated anthocyanin biosynthesis in non-photosensitive eggplant.
Results
Dark treatment and determination of anthocyanin content
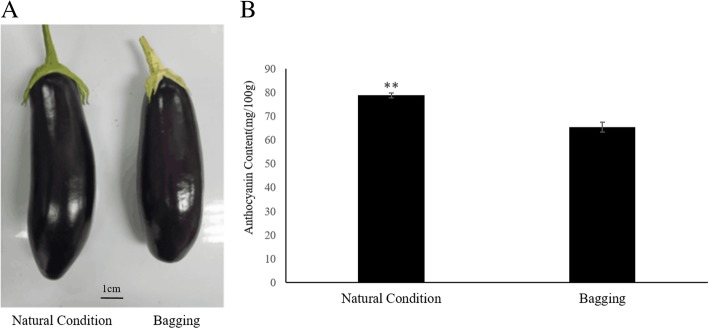
Our previous study showed that the peel of photosensitive eggplant was white when it was bagged, while the anthocyanin content of peel gradually increased after bag removal [15]. Non-photosensitive eggplant, on the other hand, can synthesize anthocyanins under dark conditions. After the non-photosensitive materials were bagged for 24 days, the color of their pericarp was almost the same as that of the un-bagged eggplant (Fig. 1a). The anthocyanin contents of the un-bagged and the bagged eggplants were 78.7 mg/100 g and 65.4 mg/100 g, respectively. Variance analysis of the anthocyanin content showed that darkness also affected the anthocyanin biosynthesis in non-photosensitive eggplant; however, the eggplant was evenly colored and the fruit attributes were not affected (P value = 0.0012) (Fig. 1b).
Fig. 1.
Phenotype and Anthocyanin Content of Non-photosensitive Eggplant under Natural Condition and Bagging. (a) Image of non-photosensitive eggplants (b) The anthocyanin content. The error bar indicates the standard deviation, based on three biological replicates. Asterisks indicate significant differences, according to Student’s t-test (**, P < 0.01)
De novo assembly and analyses of RNA-Seq data
In our previous study, RNA-Seq was performed for photosensitive eggplant collected at four time points (0, 0.5, 4 and 8 h after bag removal), and 869 genes were found to be involved in the light-induced anthocyanin biosynthesis [51]. In order to compare the similarities and differences of anthocyanin synthesis in different photosensitive eggplants and to understand the dark-regulated mechanism of anthocyanin synthesis in non-photosensitive eggplant, the same four time points after bag removal were chosen to analyze the transcription of non-photosensitive eggplant. RNA-Seq was performed for three biological replicates at each time point. On average, about 4.45 Gb bases were generated from each sample by Illumina Hiseq platform. After mapping sequenced reads to reference genome and reconstruct transcripts, a total of 27,889 novel transcripts were obtained from all samples. Of these, 14,595 are previously unknown splicing event for known genes, and 1034 are novel coding transcripts without any known features. The remaining 12,260 are long noncoding RNAs. After novel transcript detection, the novel coding transcripts were merged with reference transcript to get the complete reference. The clean reads were then mapped to the reference using Bowtie. The gene expression summary is shown in Additional file 1. To test if the samples chosen were reliable, the correlation value between each two samples was calculated based on FPKM. As shown in Fig. 2, the three replicates at four time points showed a good correlation.
Fig. 2.
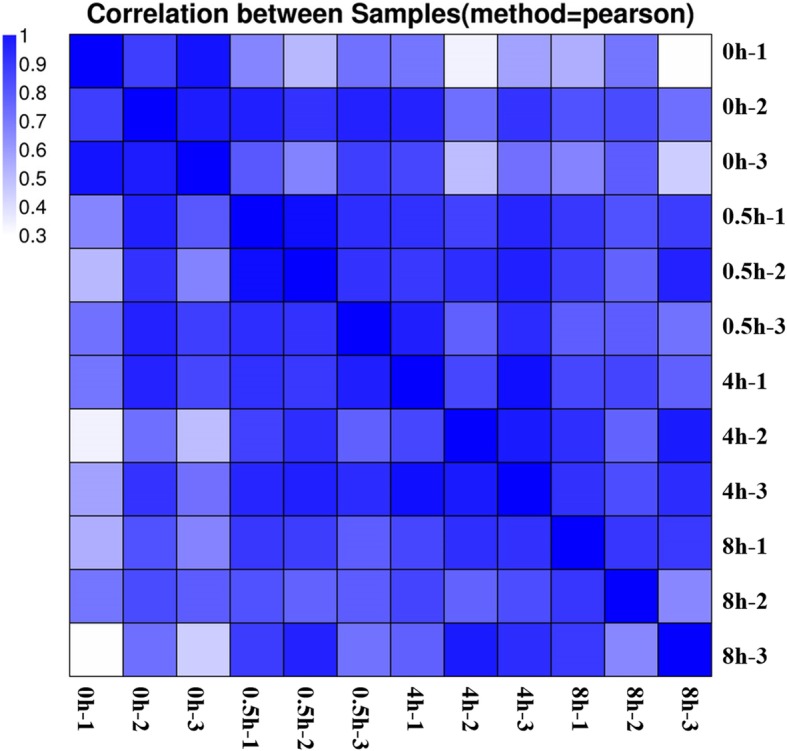
Correlation coefficients for every two samples. Heat map color represents the correlation coefficient; the darker the color, the higher the correlation
Analysis of the differentially expressed genes (DEGs)
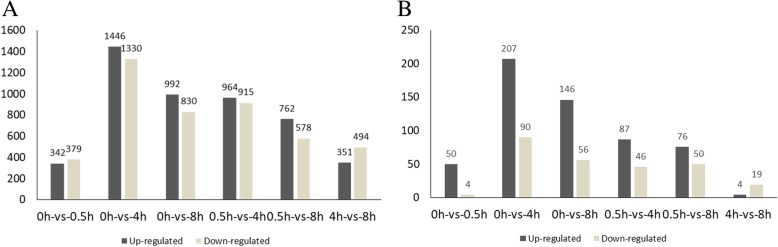
Three thousand eight hundred forty-one DEGs were identified by comparing FPKM values between different libraries under the thresholds of false discovery rate ≤ 0.001, absolute Log 2 Ratio value ≥1, and divergence probability ≥0.8 (Additional file 2). In non-photosensitive eggplant, 721 DEGs were found at 0.5 h vs 0 h (342 up-regulated, 379 down-regulated), 1879 DEGs at 4 h vs 0.5 h (964 up-regulated, 915 down-regulated), and 845 DEGs at 8 h vs 4 h (351 up-regulated, 494 down-regulated) (Fig. 3a). In photosensitive eggplant, 843 DEGs were found at 0.5 h vs 0 h (703 up-regulated, 140 down-regulated), 865 DEGs at 4 h vs 0.5 h (385 up-regulated, 480 down-regulated), and 223 DEGs at 8 h vs 4 h (65 up-regulated, 158 down-regulated) [51]. By taking the intersection of DEGs of the two materials, there were only 50 co-up-regulated and 4 co-down-regulated DEGs at 0.5 h vs 0 h, 87 co-up-regulated and 46 co-down-regulated DEGs at 4 h vs 0.5 h, and 4 co-up-regulated and 19 co-down-regulated DEGs at 8 h vs 4 h (Fig. 3b). The results indicate that there is much difference between photosensitive and non-photosensitive eggplants in terms of light response patterns and anthocyanin synthesis mechanisms.
Fig. 3.
Statistics of DEGs After Opening the Bags. a Statistics of up-regulated genes and down-regulated genes at 0 h, 0.5 h, 4 h and 8 h in non-photosensitive eggplant. b Statistics of co-up-regulated genes and co-down-regulated genes at 0 h, 0.5 h, 4 h and 8 h in both photosensitive and non-photosensitive eggplant
qRT-PCR validation of differentially expressed genes
In order to validate the accuracy of RNA-seq data, SmMYB1(Sme2.5_05099.1_g00002.1), SmMYB73(Sme2.5_24183.1_g00002.1), SmTT8(Sme2.5_00592.1_g00005.1), SmDFR (Sme2.5_01401.1_g00004.1), SmHY5(Sme2.5_03211.1_g00004.1), SmWD40(Sme2.5_05196.1_g00001.1), SmF3H(Sme2.5_00015.1_g00020.1), SmCHI (Sme2.5_01193.1_g00009.1), SmbHLH (Sme2.5_01795.1_g00002.1) and SmWD40(Sme2.5_04405.1_g00003.1) related to anthocyanin biosynthesis were selected and subjected to qRT-PCR analysis. The results showed that qRT-PCR highly repeated RNA-seq data and thus verified the accuracy of RNA-seq data (R = 0.78–0.99) (Additional file 3).
GO analysis of the DEGs
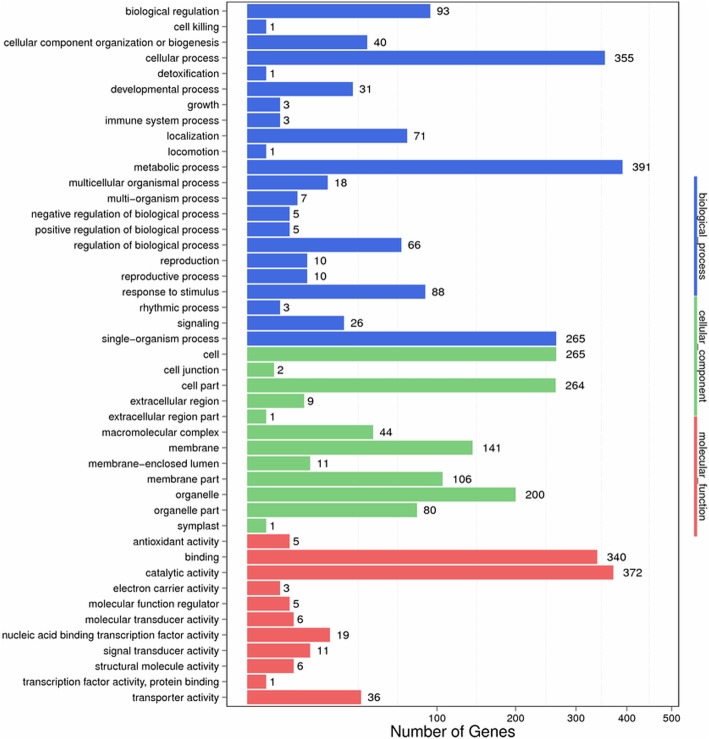
GO is a useful program for annotation and functional categorization of genes [52]. The GO assignment was used here to classify the functions of the DEGs in eggplant peel. A total of 3841 DEGs were assigned to different GO ontologies based on their sequence similarity to the genes with previously known functions; specifically, 804, 1493, and 1124 DEGs were assigned to the molecular function, the biological process, and the cellular component categories, respectively (Fig. 4). Same as the analysis result for the DEGs in photosensitive eggplant [51], the largest two subcategories in each of the ‘biological process’, the ‘cellular component’, and the ‘molecular function’ categories were ‘metabolic process’ and ‘cellular process’, the ‘cell’ and ‘cell part’, and the ‘catalytic activity’ and ‘binding activity’, respectively.
Fig. 4.
GO classification of DEGs
Cluster analysis of expression patterns in the DEGs
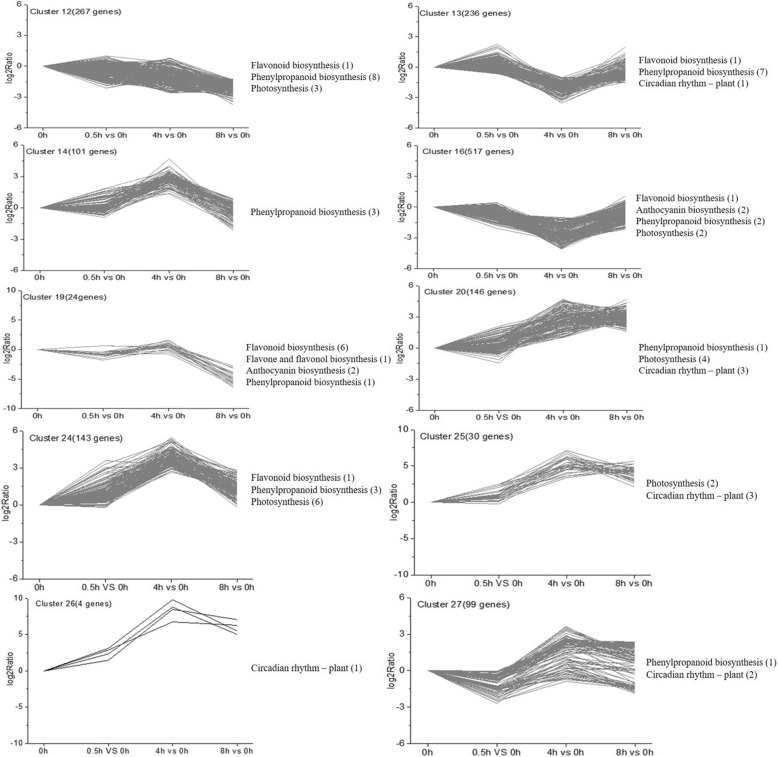
Three thousand eight hundred forty-one DEGs were subjected to complete-linkage hierarchical clustering using a Euclidean distance metric and divided into 27 clusters (Additional file 4). Genes associated with the anthocyanin biosynthesis were enriched in cluster 12, 13, 14, 16, 19, 20, 24, 25, 26 and 27 (Fig. 5). The expression levels of genes in cluster 12 were down-regulated during 0 h–8 h. The expression levels of genes in clusters 13 and 16 changed slightly at 0.5 h, but were down-regulated at 4 h, and then up-regulated at 8 h. The expression levels of genes in clusters 14, 19, 20, 24, 25, 26, 27 also changed very little at 0.5 h but peaked at 4 h, and were down-regulated at 8 h. Through cluster analysis, it was found that most DEGs related to anthocyanin biosynthesis were down-regulated during the period while in photosensitive eggplant, most such DEGs were up-regulated [51]. This result further illustrates that the regulatory modes for non-photosensitive and photosensitive eggplants were different.
Fig. 5.
Cluster analysis and KEGG pathway enrichment analysis of DEGs. The x-axis showed the time point comparison. The y-axis shows the relative log2 (ratio) of each comparison. The anthocyanin-related KEGG pathways are listed to the right of each cluster
The structural anthocyanin biosynthesis genes
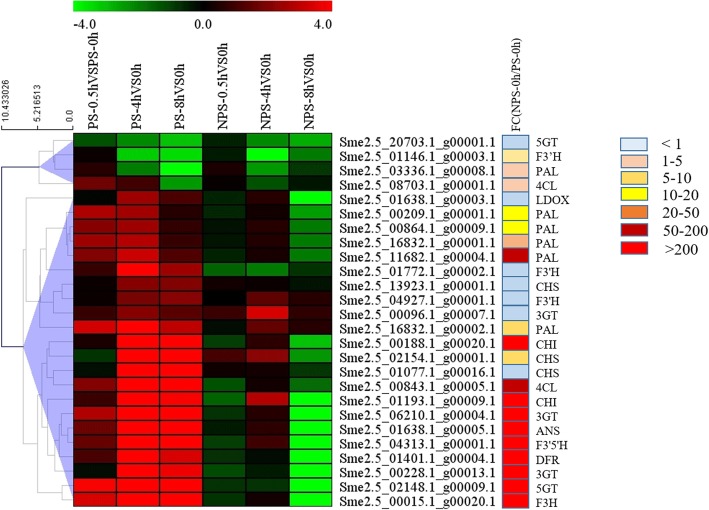
From the cluster analysis, it was found that the structural anthocyanin biosynthesis genes had different expression patterns for photosensitive and non-photosensitive eggplants. In order to find the key genes for the synthesis of anthocyanin in non-photosensitive eggplants under dark conditions, structural anthocyanin biosynthesis genes with significant changes after opening the bags in photosensitive and non-photosensitive eggplants were merged. As show in Fig. 6, 26 structural anthocyanin biosynthesis genes were divided into two clusters. The genes in cluster 1 were down-regulated after bag removal in both photosensitive and non-photosensitive eggplants, and their expression levels at 0 h were similar to each other, suggesting that these genes did not participate in the anthocyanin synthesis in non-photosensitive eggplant under dark conditions. The genes in cluster 2, most of which had high expression levels in dark conditions in non-photosensitive eggplant, were up-regulated after bag removal in photosensitive eggplant but were down-regulated or unchanged in non-photosensitive eggplant (Fig. 6). The expression levels of PAL (Sme2.5_11682.1_g00004.1) and 4CL (Sme2.5_00843.1_g00005.1) in non-photosensitive eggplant were more than 50 times that in photosensitive eggplant. The expression level of CHI (Sme2.5_01193.1_g00009.1, Sme2.5_00188.1_g00020.1), 3GT (Sme2.5_00228.1_g00013.1, Sme2.5_06210.1_g00004.1), ANS (Sme2.5_01638.1_g00005.1), DFR (Sme2.5_01401.1_g00004.1), 5GT (Sme2.5_02148.1_g00009.1), F3H (Sme2.5_00015.1_g00020.1) and F3’5’H (Sme2.5_04313.1_g00001.1) in non-photosensitive eggplant were more than 200 times that in photosensitive eggplant (Fig. 6). Therefore, it is believed that these genes were involved in the dark-regulated anthocyanin synthesis in non-photosensitive eggplant.
Fig. 6.
Expression patterns of the structural anthocyanin biosynthesis genes. The color scale represents the Log2 (fold-change to the 0 h time point). Genes that are up-regulated appear in red, and those that are down-regulated appear in green. FC (NPS-0 h / PS-0 h) represents the fold change in gene expression of non-photosensitive and photosensitive eggplants under dark conditions. PS: photosensitive, NPS: non-photosensitive
Light signal perception and transduction
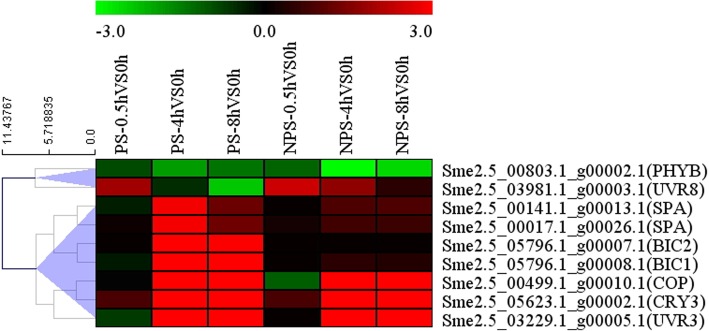
In order to clarify the difference between the two eggplants in expression patterns of light signal related genes, the DEGs from photosensitive and non-photosensitive eggplants were combined for comparison. As show in Fig. 7, 9 DEGs were divided into two clusters. In cluster 1, PHYB (Sme2.5_00803.1_g00002.1) had the same expression pattern in both photosensitive and non-photosensitive eggplants, but UVR8 (Sme2.5_03981.1_g00003.1) showed different expression trends in photosensitive and non-photosensitive eggplants after 0.5 h exposure to light. In cluster 2, COP, UVR3 and CRY3 were up-regulated in both photosensitive and non-photosensitive eggplants after 0.5 h exposure to light; two SPAs, BIC1 and BIC2, were up-regulated significantly in photosensitive eggplant, but the up-regulation was insignificant in non-photosensitive eggplant. Thus, we believe that UVR8, SPAs, BIC1 and BIC2 may be involved in dark-regulated anthocyanin biosynthesis in non-photosensitive eggplant.
Fig. 7.
Expression patterns of the photoreceptors and genes related to light signal transduction. The color scale represents the Log2 (fold-change to the 0 h time point). Genes that are up-regulated appear in red, and those that are down-regulated appear in green
Transcription factors related to dark-regulated anthocyanin synthesis
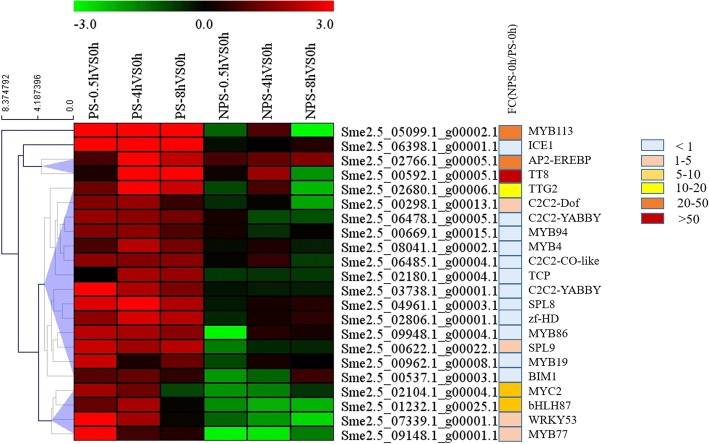
Analysis of the structural and light signal sensing and transduction genes related to anthocyanin biosynthesis showed that those likely involved in dark regulation of anthocyanin synthesis in non-photosensitive eggplant were up-regulated in photosensitive eggplant after bag removal (Fig. 6). However, in non-photosensitive eggplant, the structural genes were down-regulated at 0.5 h and 8 h. Although up-regulated at 4 h, only a few genes’ expressions reached the significant level; the light signal sensing and transduction genes either had the same expression level or had an up-regulated expression trend, but did not reach the significant level at 4 h and 8 h. Since genes with the same expression trend usually have the same function, TFs related to anthocyanin synthesis can be differentiated based on the expression trends of anthocyanin structural genes and light signal sensing and transduction genes. We obtained 338 TFs that changed significantly after removing the bags in photosensitive or non-photosensitive eggplants by merging (Additional file 5); then, we identified 22 TFs that were up-regulated in photosensitive eggplant, but down-regulated or had no significant change in non-photosensitive eggplant (Fig. 8). In the 22 TF genes, MYB113, TT8, TTG2 had high expression levels at 0 h in non-photosensitive eggplant. These three TF genes were found to participate in eggplant anthocyanin synthesis in our previous study [15, 53]; AP2-EREBP, MYC2, MYB77, bHLH87 and WRKY53 also had high expression level at 0 h in non-photosensitive eggplant, implying their importance to anthocyanin synthesis; Other 14 TF genes were expressed similarly at 0 h in photosensitive and non-photosensitive eggplants. Since one of these TF genes, SmMYB86, was considered as a negative regulator of anthocyanin synthesis in our previous studies [51], it is reasonable to believe that other TF genes of the same type may also have participated in dark-regulated anthocyanin synthesis in non-photosensitive eggplant.
Fig. 8.
Expression patterns of all the TFs related to dark regulated anthocyanin synthesis. The color scale represents the Log2 (fold-change to the 0 h time point). Genes that are up-regulated appear in red, and those that are down-regulated appear in green. FC (NPS-0 h / PS-0 h) represents the fold change in gene expression of non-photosensitive and photosensitive eggplants under dark conditions
Interaction network construction and target gene prediction of the TFs related to dark regulated anthocyanin synthesis
To verify whether the TFs identified in the analysis results were involved in anthocyanin synthesis, we analyzed the 50–1500 bp upstream of the initiation codon of the anthocyanin structural genes and TFs using the I-sanger cloud platform (https://www.i-sanger.com/) for regions that the transcription factors may bind. It was found that these TFs could not only regulate structural genes, but also regulate each other (Additional file 6). Possible interactions among these TFs were also predicted through a protein interaction site (http://netbio.sjtu.edu.cn/arappinet/). As shown in Fig. 9, there were interaction relationships between some of the TFs, and the interaction scores were greater than 0.5 (Additional file 7). These findings suggest that these transcription factors may have the same function and are involved in the dark-regulated anthocyanin biosynthesis in non-photosensitive eggplant.
Fig. 9.
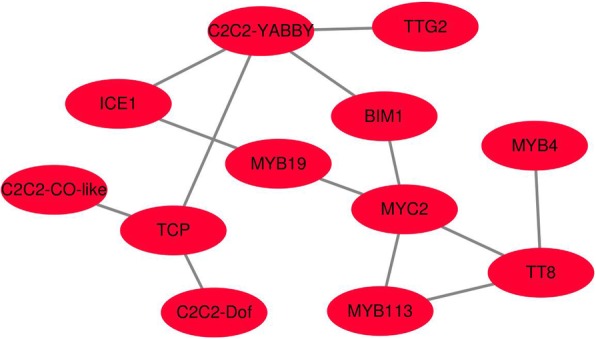
Interaction network of TFs related to dark regulated anthocyanin synthesis
Experimental verification of regulatory relationship between dark regulated anthocyanin synthesis related TFs and target genes
In order to confirm the predicted regulatory relationship between TFs and target genes, the promoters of the non-photosensitive eggplant anthocyanin synthetic structural genes and the coding region of transcription factors were cloned according to target gene prediction results (Additional file 6). Through yeast one-hybrid assay, 8 TFs were identified to be direct participants in anthocyanin synthesis, of which SmMYB94, SmBIM1, SmTT8, SmMYC2, SmTTG2, SmAP2, and SmMYB19 can bind to the promoter of SmCHS (Sme2.5_13923.1_g00001.1), SmHD can bind to the promoter of SmCHS (Sme2.5_13923.1_g00001.1), SmANS (Sme2.5_01638.1_g00005.1) and Sm3GT (Sme2.5_00228.1_g00013.1), and SmYABBY can bind to the promoter of SmTT8 (Sme2.5_00592.1_g00005.1) (Fig. 10). Since previous study showed that overexpression of SmCHS and SmANS can promote anthocyanin synthesis [54]. Thus, TFs that bind to their promoters are likely involved in anthocyanin synthesis.
Fig. 10.
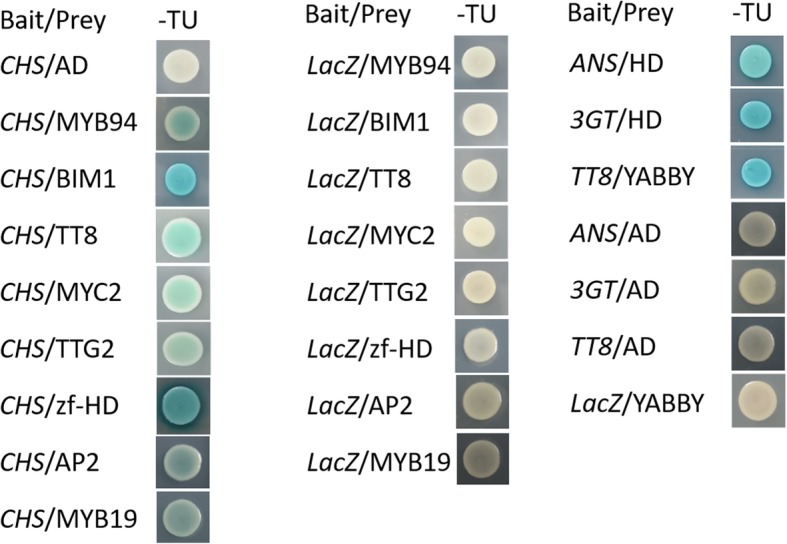
Interactions/bindings between TFs and the promoters of anthocyanin biosynthesis genes, revealed by yeast one-hybrid assays. Indicated combinations of constructs were co-expressed and grown on synthetic dropout –Trp-Ura (−TU). Blue precipitates represented corresponding ß- galactosidase activities
Discussion
Our previous study shows that photosensitive eggplant did not synthesize anthocyanins under dark conditions, and anthocyanin synthesis genes and regulatory factors were not expressed; however, the anthocyanin synthesis genes and regulatory factors were significantly induced at 0.5 h, 4 h and 8 h after exposure to light, and anthocyanins were synthesized within a few days [15]. In order to find the key regulators of synthetic anthocyanins in dark conditions to solve the problem of poor coloration of eggplant peel under low light conditions, transcriptome data of photosensitive and non-photosensitive eggplant peel at 0 h, 0.5 h, 4 h, 8 h after illumination were compared and analyzed in this study.
Cluster analysis showed that most of the genes in the phenylpropanoid biosynthesis, flavonoid biosynthesis and anthocyanin biosynthesis pathways were down-regulated in non-photosensitive eggplant after opening bags (Fig. 6). On the other hand, they were up-regulated in photosensitive eggplant [51]. This difference in gene regulation suggests that the mechanisms for anthocyanin biosynthesis in photosensitive and non-photosensitive eggplants were not the same. Through transcriptome comparison, we found that PAL (Sme2.5_11682.1_g00004.1), 4CL (Sme2.5_00843.1_g00005.1), CHI (Sme2.5_01193.1_g00009.1, Sme2.5_00188.1_g00020.1), 3GT (Sme2.5_00228.1_g00013.1, Sme2.5_06210.1_g00004.1), ANS (Sme2.5_01638.1_g00005.1), DFR (Sme2.5_01401.1_g00004.1), 5GT (Sme2.5_02148.1_g00009.1), F3H (Sme2.5_00015.1_g00020.1) and F3’5’H (Sme2.5_04313.1_g00001.1) were up-regulated in photosensitive eggplant, but down-regulated in non-photosensitive eggplant after bags removal, and they have a high expression level in dark conditions in non-photosensitive eggplant (Fig. 6). Many studies demonstrated that overexpression or antisense of these structural genes resulted in changes in anthocyanin accumulation. Compared to untransformed lines, overexpression of CHS gene from Malus crabapple in tobacco resulted in anthocyanin accumulation [55]. In Malus domestica, chs knockout lines had no detectable anthocyanins and had radically reduced concentrations of flavonoids [56]. Overexpression of the CHI gene of petunia in tomato (Solanum lycopersicum) resulted in flavonol accumulation in fruit peel [57]. F3H-silenced strawberry fruit displayed reduced anthocyanins and flavonol content [58]. Overexpressing PtrDFR1 in Chinese white poplar (P. tomentosa Carr.) increased the accumulation of anthocyanins and condensed tannins [59]. Silencing DFR expression in sweet potato (Dioscorea esculenta) significantly decreased anthocyanin content in storage roots and stems [60]. Overexpression of CHS, CHI, F3H and DFR of Solanum melongena in Arabidopsis thaliana successfully increased the pigmentation in the stems and siliques [54]. The anthocyanin content was significantly lower in the Pe3GT-suppressed Phalaenopsis flowers [61]. Taken as a whole, both the anthocyanin accumulation and pigmentation phenotype were in agreement with the above results.
Structural genes of anthocyanin biosynthesis are simultaneously regulated by the activated TFs. In this study, through comparative transcriptiome we also found 22 TFs that have the same expression pattern as the structural genes (Fig. 8). The 22 TFs can be divided into 8 categories which contained six MYBs, five bHLHs, two WRKYs, four C2C2s, two SPLs, HD, TCP, and AP2. Among these TFs, three MYBs (MYB113, MYB94 and MYB19), three bHLHs (TT8, MYC2 and BIM1), WRKY (TTG2), HD, and AP2 can bind directly to the promoter of CHS (Sme2.5_13923.1_g00001.1), and one C2C2 (YABBY) can bind to the promoter of TT8 (Fig. 10). Although other transcription factors can not bind directly to the promoters of the structural genes, they were predicted to be involved in anthocyanin synthesis by binding to the promoter of TFs which can bind directly to the promoter of anthocyanin structural genes (Additional file 6). Studies also showed that MYB113 (MYB1 or MYB75) [30, 33, 62, 63], TT8 [40, 64, 65] and TTG2 (WRKY44, [66, 67]) were involved in anthocyanin synthesis in many plant species. In addition, other TFs such as SPL [47], TCP (TCP3 and TCP15, [45, 46], AP2 [44], WRKY (WRKY41 and WRKY11, [68, 69]) and MYC (MYC1, [29]) were also characterized to be involved in anthocyanin synthesis. At present, there is no report on the involvement of homeodomain-like protein (HD) and C2C2 type zinc finger protein in the synthesis of anthocyanins and flavonoids. In this study, MYB113, TT8, TTG2, MYC2 and AP2 have a high expression level in dark condition in non-photosensitive eggplant, and the expression levels of MYB94, MYB19, BIM1 and HD were not much different between the two eggplants in dark condition, but they have different expression patterns in the two materials. Since these TFs bind directly to the promoter of CHS (Sme2.5_13923.1_g00001.1), they were thought to have participated in anthocyanin biosynthesis in eggplant.
While light is one of the key environmental factors for the anthocyanin synthesis in eggplant, this study found that anthocyanins can still proceed without light. Transcriptome comparison analysis showed that UVR8 has different expression pattern among the two types of eggplants (Fig. 7). It has also been shown that UVR8 can promote anthocyanin synthesis in radish sprouts and Arabidopsis, but the synthesis was dependent of light [70, 71]. It was reported that AtUVR8 can interact directly with AtBIM1 and AtWRKY36 to participate in hypocotyl elongation [72, 73]. We also confirmed in this study that SmBIM1 and SmWRKY53 can directly bind to the promoter of SmCHS (Sme2.5_13923.1_g00001.1). Based on these results, we hypothesize that SmUVR8 can directly interact with SmWRKY53 and SmBIM1 to regulate eggplant anthocyanin synthesis. COP1, an E3 ubiquitin ligase, can form a complex with SPA to regulate anthocyanin synthesis of Arabidopsis; cop1 and spa mutants can produce anthocyanins also in the dark [26]. Jiang et al. reported that SmCOP1 (Sme2.5_00128.1_g00013.1) can interact with SmHY5 and SmMYB113 to regulate anthocyanin biosynthesis and restore the phenotype of the Arabidopsis cop mutant [15]. In this study, we found two SPAs and COP1 (Sme2.5_00499.1_g00010.1) which had different expression patterns in the two materials and thus might have participated in the dark-regulated anthocyanin synthesis in non-photosensitive eggplant (Fig. 7). Cryptochromes are blue-light receptors that regulate plant development and the circadian clock. CRY2 undergoes blue light–dependent homodimerization to become physiologically active. The binding of BICs to CRY2 inhibits the blue light–dependent dimerization, thereby regulating cell photosensitivity [16, 74]. In this study, we found that BIC and BIC2 were up-regulated in photosensitive eggplant after bag removal; yet, there was essentially no expression of these genes in non-photosensitive eggplant, leading us to suspect that the non-photosensitive eggplant peel cells might have lost photosensitivity in some cases.
Conclusions
In this study, comparative transcriptome was applied to elucidate the underlying molecular mechanism for dark-regulated anthocyanin biosynthesis in non-photosensitive eggplant. The analyses revealed that the structural genes and the TFs involved in anthocyanin synthesis were down-regulated after bag removal, while most of them have high expression levels in dark conditions in non-photosensitive eggplant. Through transcription factor target gene prediction and yeast one-hybrid verification, we believe that SmBIM1, SmHD, SmMYB94, SmMYB19, SmTT8, SmYABBY, SmTTG2, SmAP2 and SmMYC2 may be involved in dark-regulated anthocyanin synthesis in non-photosensitive eggplant. Our results shaded some lights on the understanding of molecular mechanism of dark-regulated anthocyanin synthesis in non-photosensitive eggplant.
Methods
Plant materials
The non-photosensitive eggplant material ‘145’, obtained through a large number of bagging screening of fruits with different eggplant genotypes, was provided by the Shanghai Academy of Agricultural Sciences, Shanghai, China. ‘145’ was grown in the greenhouse of Shanghai Jiao Tong University, Shanghai, China, under natural light conditions. For fruit bagging treatment, sepals were covered with double-layer Kraft paper bags after full bloom. After the bagging of the materials for 2 weeks, the bags were opened to collect materials at 8:00 am. Three materials were selected as biological replicates, with each individual material being sampled from every bagged plant at 0 h, 0.5 h, 4 h, and 8 h after the bags were removed. All peel samples were frozen immediately with liquid nitrogen and kept at − 80 °C.
Anthocyanin content determination
Anthocyanin content was measured with the pH differential spectrophotometry method [75]. 0.5 g materials were used to extract anthocyanin with 0.01% HCl in 0.5 ml methanol at 4 °C for 12 h. The extracts were centrifuged at 8500 g for 20 min. The supernatant was transferred to a 10 ml tube and the sediments were further processed with 5 ml extraction solution at 4 °C for 6 h. The extracts from the sediments were also centrifuged at 8500 g for 20 min. Spectrophotometrical absorbance was measured at 510 nm and at 700 nm in buffers at pH 1.0 and pH 4.5. The anthocyanin content was calculated according to the following formula: TA = A × MW × 5 × 100 × V/ε, where TA stands for total anthocyanin content (mg/100 g), V is the final volume (ml), A equals to [A510 nm (pH 1.0) − A700 nm (pH 1.0)] − [A510 nm (pH 4.5) − A700 nm (pH 4.5)], ε is the molar absorptivity which is 26,900, MW is the molecular weight with a value of 449.2, and A510 and A700 represent the absorbance values at 510 and 700 nm, respectively [76]. Three measurements were performed for each sample replicate.
RNA extraction, cDNA library construction, and RNA-Seq
Total RNAs were extracted from pericarp using the RNAiso plus kit (TaKaRa, Otsu, Shiga, Japan) and following the manufacturer’s instructions therein. RNase-free DNase (Takara, Japan) was used to remove the potential genomic DNAs in extracted RNAs. After total RNAs were extracted and treated with DNase I, Oligo (dT) was used to isolate mRNA. The mRNAs were fragmented through mixing with the fragmentation buffer. cDNA was then synthesized using the mRNA fragments as templates. Short fragments were purified and resolved with EB buffer for end reparation and single nucleotide A (adenine) addition. The short fragments were then connected with adapters. Suitable fragments were selected for the PCR amplification. The quality and quantity of RNAs were determined by Agilent 2100 Bioanalyzer and ABI StepOne Plus Real-Time PC System. Finally, the library was sequenced using Illumina HiSeq™ 2000.
Sequence assembly and gene annotation
Low-quality, adaptor-pollute, and high content of unknown base (N) reads were removed by QC before downstream analyses. After reads filtering, the clean reads were mapped to the reference genome by HISAT. Eggplant reference genome database (http://vegmarks.nivot.affrc.go.jp/VegMarks/jsp/index.jsp) was used as the reference. After the novel transcripts were obtained, their coding transcripts were merged with reference transcripts to get the complete reference which was used for gene expression analysis.
Identification and annotation of DEGs
After the setup of the complete reference, the clean reads were mapped to the reference library using Bowtie2; then gene expression level for each sample was calculated by RSEM. DEGs were detected by DEseq2 and fold change ≥ 2.00 and adjusted p-value ≤ 0.05 were taken as conditions. Gene Ontology (GO) classification and functional enrichment were performed by WEGO software (http://wego.genomics.org.cn/cgi-bin/wego/index.pl). KEGG pathway classification and functional enrichment for DEGs were performed by the database-Kyoto Encyclopedia of Genes and Genomes. Cluster analysis of expression patterns were analyzed by MeV (4.9).
Real-time qPCR analysis
Real-time quantitative reverse transcription-PCR was used to estimate the accuracy of RNA-Seq. Total RNA was treated with DNase to remove trace of DNA. 1 μg RNA was used for synthesis of cDNA with the PrimeScript RT Master Mix Perfect Real Time Kit (Takara). Quantitative real-time RT-PCR (qPCR) was performed on FTC-3000 real-time PCR System (Funglyn Biotech, Canada) using the following program: 95 °C for 30 s, followed by 40 cycles of 95 °C for 20 s, 58 °C for 30 s, and 72 °C for 30 s, which was consistent with the manufacturer’s instructions of SYBR Premix Ex Taq II Kit (Takara). The Actin gene (GU984779.1) was used as an internal reference, and relative expression was calculated using the 2ΔCt method. Each qPCR analysis was performed in triplicate. The relative expression levels of the amplified products were analyzed using the comparative CT method based on CT values. All primers used in this study are listed in Additional file 8.
Yeast one-hybrid assay
The procedure for the yeast one-hybrid assay was described in a previous report [34]. 50–1500 bp upstream of the initiation codon of the anthocyanin structural genes was amplified from non-photosensitive eggplant and fused to the vector LacZ. The coding regions of the transcription factors were amplified from non-photosensitive eggplant and fused to the vector PB4 2 AD. The constructs were co-transformed into the yeast strain EGY48 in the indicated combinations. After 3 days of transformation, 6 yeast clones grown on the medium-synthesized sputum-Trp-Ura (SD-TU) were selected and cultured in liquid SD-TU medium for 24 h, and the culture was inoculated to contain 80 mg/ L X-gal in SD-TU medium.
Additional files
Table S2. Gene Expression Summary. (DOCX 15 kb)
Table S3. Differentially expressed genes (DEGs) in non-photosensitive eggplant. (XLSX 239 kb)
Figure S1. qRT-PCR Analysis of DEGs after Opening the Bags. (DOCX 522 kb)
Figure S2. Cluster analysis and KEGG pathway enrichment analysis of DEGs. (DOCX 1176 kb)
Table S4. Differentially expressed TF genes in non-photosensitive and photosensitive eggplant. (XLSX 45 kb)
Table S5. Transcription factors target gene prediction results. (DOCX 35 kb)
Table S6. The interaction score of transcription factors in Fig. 9. (DOCX 13 kb)
Table S1. List of all the primer sequences used in this study. (DOCX 15 kb)
Acknowledgements
The project was supported by the National Natural Science Foundation of China (31471870 and 31872944).
Ethics approval consent to participate
Not applicable, as this study did not involve human or animal subjects, and the seeds of eggplant were stored in School of Agriculture and Biology, Shanghai Jiao Tong University.
Abbreviations
- 4CL
4-coumarate-CoA ligase
- ANS
Anthocyanin synthase
- AT
Acylation
- CHI
Chalcone isomerase
- CHS
Chalcone synthase
- COL
CONSTANS-like
- COP1
Constitutive Photomorphogenic 1
- CRY3
Cryptochrome 3
- DEGs
Differentially expressed genes
- DFR
Dihydroflavonol reductase
- F3H
Flavanone 3-hydroxyl enzyme
- FDR
False discovery rate
- GO
Gene ontology
- GT
Glycosylation
- HFR
Hypocotyl in Far-Red 1
- HY5
Hypocotyl 5
- HYH
Hypocotyl homolog
- KEGG
Kyoto encyclopedia of genes and genomes
- LDOX
Leucoanthocyanidin dioxygenase
- MT
Methylation
- NPS
non-photosensitive
- Nr
NCBI non-redundant database
- PAL
Phenylalanine ammonia lyase
- PHOTs
Phototropins
- PHYs
Phytochromes
- PS
photosensitive
- QC
Quality control
- SPL9
Squamosa promoter binding protein-like 9
- SRA
Sequence read archive
- UVR8
UV Resistance Locus 8
Authors’ contributions
H-YC, YL, Y-JH, HC and LZ planned and designed the research. Y-JH, HC and LZ performed experiments. Y-JH analyzed data and wrote the manuscript. All authors read and approved the manuscript.
Funding
The financial of this research was supported by National Natural Science Foundation of China (31471870 and 31872944).
Availability of data and materials
The Illumina RNA-seq data generated from non-photosensitive eggplant are available in the NCBI SRA with accessions number SRA613313, and the RNA-seq data of photosensitive eggplant are available in the NCBI SRA with accessions numbers SRR5650714, SRR5651526, SRR5658205 and SRR5658226 [51].
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yongjun He, Email: hyj016150910024@sjtu.edu.cn.
Hang Chen, Email: tuer10086@alumni.sjtu.edu.cn.
Lu Zhou, Email: zhoulu@sjtu.edu.cn.
Yang Liu, Email: liuyangtl@sjtu.edu.cn.
Huoying Chen, Email: chhy@sjtu.edu.cn.
References
- 1.Takos AM, Jaffé FW, Jacob SR, Bogs J, Robinson SP, Walker AR. Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 2006;142:1216–1232. doi: 10.1104/pp.106.088104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.He J, Giusti MM. Anthocyanins: natural colorants with health-promoting properties. Annu Rev Food Sci Technol. 2010;1:163–187. doi: 10.1146/annurev.food.080708.100754. [DOI] [PubMed] [Google Scholar]
- 3.Stintzing FC, Carle R. Functional properties of anthocyanins and betalains in plants, food, and in human nutrition. Trends Food Sci Technol. 2004;15:19–38. doi: 10.1016/j.tifs.2003.07.004. [DOI] [Google Scholar]
- 4.Mori K, Saito H, Goto-Yamamoto N, Kitayama M, Kobayashi S, Sugaya S, et al. Effects of abscisic acid treatment and night temperatures on anthocyanin composition in pinot noir grapes. Vitis - J Grapevine Res. 2005;44:161–165. [Google Scholar]
- 5.Yamane T, Seok TJ, Goto-Yamamoto N, Koshita Y, Kobayashi S. Effects of temperature on anthocyanin biosynthesis in grape berry skins. Am J Enol Vitic. 2006;57:54–59. [Google Scholar]
- 6.Kami Chitose, Lorrain Séverine, Hornitschek Patricia, Fankhauser Christian. Current Topics in Developmental Biology. 2010. Light-Regulated Plant Growth and Development; pp. 29–66. [DOI] [PubMed] [Google Scholar]
- 7.Christie PJ, Alfenito MR, Walbot V. Impact of low-temperature stress on general phenylpropanoid and anthocyanin pathways: enhancement of transcript abundance and anthocyanin pigmentation in maize seedlings. Planta. 1994;194:541–549. doi: 10.1007/BF00714468. [DOI] [Google Scholar]
- 8.Yin R, Skvortsova MY, Loubéry S, Ulm R. COP1 is required for UV-B–induced nuclear accumulation of the UVR8 photoreceptor. Proc Natl Acad Sci. 2016;113:E4415–E4422. doi: 10.1073/pnas.1607074113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharrock RA, Clack T. Patterns of expression and normalized levels of the five Arabidopsis Phytochromes. Plant Physiol. 2002;130:442–456. doi: 10.1104/pp.005389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rizzini L., Favory J.-J., Cloix C., Faggionato D., O'Hara A., Kaiserli E., Baumeister R., Schafer E., Nagy F., Jenkins G. I., Ulm R. Perception of UV-B by the Arabidopsis UVR8 Protein. Science. 2011;332(6025):103–106. doi: 10.1126/science.1200660. [DOI] [PubMed] [Google Scholar]
- 11.Möglich A, Yang X, Ayers RA, Moffat K. Structure and function of plant photoreceptors. Annu Rev Plant Biol. 2010;61:21–47. doi: 10.1146/annurev-arplant-042809-112259. [DOI] [PubMed] [Google Scholar]
- 12.Liu B, Liu H, Zhong D, Lin C. Searching for a photocycle of the cryptochrome photoreceptors. Curr Opin Plant Biol. 2010;13:578–586. doi: 10.1016/j.pbi.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zuo Z, Liu H, Liu B, Liu X, Lin C. Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in arabidopsis. Curr Biol. 2011;21:841–847. doi: 10.1016/j.cub.2011.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris SR, Henbest KB, Maeda K, Pannell JR, Timmel CR, Hore PJ, et al. Effect of magnetic fields on cryptochrome-dependent responses in Arabidopsis thaliana. J R Soc Interface. 2009;6:1193–1205. doi: 10.1098/rsif.2008.0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang M, Ren L, Lian H, Liu Y, Chen H. Novel insight into the mechanism underlying light-controlled anthocyanin accumulation in eggplant (Solanum melongena L.) Plant Sci. 2016;249:46–58. doi: 10.1016/j.plantsci.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 16.Wang Q, Zuo Z, Wang X, Gu L, Yoshizumi T, Yang Z, et al. Photoactivation and inactivation of Arabidopsis cryptochrome 2. Science (80- ). 2016;354:343–347. [DOI] [PMC free article] [PubMed]
- 17.Kadomura-Ishikawa Y, Miyawaki K, Noji S, Takahashi A. Phototropin 2 is involved in blue light-induced anthocyanin accumulation in Fragaria x ananassa fruits. J Plant Res. 2013;126:847–857. doi: 10.1007/s10265-013-0582-2. [DOI] [PubMed] [Google Scholar]
- 18.Oh Sookyung, Warnasooriya Sankalpi N, Montgomery Beronda L. Mesophyll-localized phytochromes gate stress- and light-inducible anthocyanin accumulation inArabidopsis thaliana. Plant Signaling & Behavior. 2014;9(3):e28013. doi: 10.4161/psb.28013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rockwell NC, Su Y-S, Lagarias JC. PHYTOCHROME STRUCTURE and signaling mechanisms. Annu Rev Plant Biol. 2006;57:837–858. doi: 10.1146/annurev.arplant.56.032604.144208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kliebenstein DJ, Lim JE, Landry LG, Last RL. Arabidopsis UVR8 regulates ultraviolet-B signal transduction and tolerance and contains sequence similarity to human regulator of chromatin condensation 1. Plant Physiol. 2002;130:234–243. doi: 10.1104/pp.005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jang I-C, Henriques R, Seo HS, Nagatani A, Chua N-H. Arabidopsis PHYTOCHROME INTERACTING FACTOR proteins promote Phytochrome B Polyubiquitination by COP1 E3 ligase in the nucleus. Plant Cell. 2010;22:2370–2383. doi: 10.1105/tpc.109.072520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lian HL, He SB, Zhang YC, Zhu DM, Zhang JY, Jia KP, et al. Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev. 2011;25:1023–1028. doi: 10.1101/gad.2025111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osterlund MT, Ang LH, Deng XW. The role of COP1 in repression of Arabidopsis photomorphogenic development. Trends Cell Biol. 1999;9:113–118. doi: 10.1016/S0962-8924(99)01499-3. [DOI] [PubMed] [Google Scholar]
- 24.Osterlund MT, Hardtke CS, Ning W, Deng XW. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature. 2000;405:462–466. doi: 10.1038/35013076. [DOI] [PubMed] [Google Scholar]
- 25.Seo HS, Watanabe E, Tokutomi S, Nagatani A, Chua NH. Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome a signaling. Genes Dev. 2004;18:617–622. doi: 10.1101/gad.1187804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maier A, Schrader A, Kokkelink L, Falke C, Welter B, Iniesto E, et al. Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in Arabidopsis. Plant J. 2013;74:638–651. doi: 10.1111/tpj.12153. [DOI] [PubMed] [Google Scholar]
- 27.Hoecker Ute, Xu Yong, Quail Peter H. SPA1: A New Genetic Locus Involved in Phytochrome A–Specific Signal Transduction. The Plant Cell. 1998;10(1):19–33. doi: 10.1105/tpc.10.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laubinger S, Fittinghoff K, Hoecker U. The SPA quartet: a family of WD-repeat proteins with a central role in suppression of photomorphogenesis in arabidopsis. Plant Cell. 2004;16:2293–2306. doi: 10.1105/tpc.104.024216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hichri I, Heppel SC, Pillet J, Léon C, Czemmel S, Delrot S, et al. The basic helix-loop-helix transcription factor MYC1 is involved in the regulation of the flavonoid biosynthesis pathway in grapevine. Mol Plant. 2010;3:509–523. doi: 10.1093/mp/ssp118. [DOI] [PubMed] [Google Scholar]
- 30.Gonzalez A, Zhao M, Leavitt JM, Lloyd AM. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008;53:814–827. doi: 10.1111/j.1365-313X.2007.03373.x. [DOI] [PubMed] [Google Scholar]
- 31.Sun S-S, Gugger PF, Wang Q-F, Chen J-M. Identification of a R2R3-MYB gene regulating anthocyanin biosynthesis and relationships between its variation and flower color difference in lotus ( Nelumbo Adans.) Peer J. 2016;4:e2369. doi: 10.7717/peerj.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feng S, Wang Y, Yang S, Xu Y, Chen X. Anthocyanin biosynthesis in pears is regulated by a R2R3-MYB transcription factor PyMYB10. Planta. 2010;232:245–255. doi: 10.1007/s00425-010-1170-5. [DOI] [PubMed] [Google Scholar]
- 33.Zuluaga DL, Gonzali S, Loreti E, Pucciariello C, Degl’Innocenti E, Guidi L, et al. Arabidopsis thaliana MYB75/PAP1 transcription factor induces anthocyanin production in transgenic tomato plants. Funct Plant Biol. 2008;35:606–618. doi: 10.1071/FP08021. [DOI] [PubMed] [Google Scholar]
- 34.Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 2007;49:414–427. doi: 10.1111/j.1365-313X.2006.02964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pires N, Dolan L. Origin and diversification of basic-helix-loop-helix proteins in plants. Mol Biol Evol. 2010;27:862–874. doi: 10.1093/molbev/msp288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feyissa DN, Løvdal T, Olsen KM, Slimestad R, Lillo C. The endogenous GL3, but not EGL3, gene is necessary for anthocyanin accumulation as induced by nitrogen depletion in arabidopsis rosette stage leaves. Planta. 2009;230:747–754. doi: 10.1007/s00425-009-0978-3. [DOI] [PubMed] [Google Scholar]
- 37.Nesi N. The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell. 2000;12:1863–1878. doi: 10.1105/tpc.12.10.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cominelli E, Gusmaroli G, Allegra D, Galbiati M, Wade HK, Jenkins GI, et al. Expression analysis of anthocyanin regulatory genes in response to different light qualities in Arabidopsis thaliana. J Plant Physiol. 2008;165:886–894. doi: 10.1016/j.jplph.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 39.Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 2004;39:366–380. doi: 10.1111/j.1365-313X.2004.02138.x. [DOI] [PubMed] [Google Scholar]
- 40.Li P, Chen B, Zhang G, Chen L, Dong Q, Wen J, et al. Regulation of anthocyanin and proanthocyanidin biosynthesis by Medicago truncatula bHLH transcription factor MtTT8. New Phytol. 2016;210:905–921. doi: 10.1111/nph.13816. [DOI] [PubMed] [Google Scholar]
- 41.Bin XX, Li S, Zhang RF, Zhao J, Chen YC, Zhao Q, et al. The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ. 2012;35:1884–1897. doi: 10.1111/j.1365-3040.2012.02523.x. [DOI] [PubMed] [Google Scholar]
- 42.Qiu Z, Wang X, Gao J, Guo Y, Huang Z, Du Y. The tomato Hoffman’s anthocyaninless gene encodes a bHLH transcription factor involved in anthocyanin biosynthesis that is developmentally regulated and induced by low temperatures. PLoS One. 2016;11:1–22. doi: 10.1371/journal.pone.0151067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lai B, Du L, Liu R, Hu B, Su W, Qin Y, et al. Two LcbHLH transcription factors Interacting with LcMYB1 in regulating late structural genes of anthocyanin biosynthesis in Nicotiana and Litchi chinensis during anthocyanin accumulation. Front Plant Sci. 2016;7:1–15. doi: 10.3389/fpls.2016.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen Y, Wu P, Zhao Q, Tang Y, Chen Y, Li M, et al. Overexpression of a phosphate starvation response AP2/ERF gene from physic nut in Arabidopsis alters root morphological traits and phosphate starvation-induced anthocyanin accumulation. Front Plant Sci. 2018;9. [DOI] [PMC free article] [PubMed]
- 45.Li S, Zachgo S. TCP3 interacts with R2R3-MYB proteins, promotes flavonoid biosynthesis and negatively regulates the auxin response in Arabidopsis thaliana. Plant J. 2013;76:901–913. doi: 10.1111/tpj.12348. [DOI] [PubMed] [Google Scholar]
- 46.Viola IL, Camoirano A, Gonzalez DH. Redox-dependent modulation of anthocyanin biosynthesis by the TCP transcription factor TCP15 during exposure to high light intensity conditions in Arabidopsis. Plant Physiol. 2016;170:74–85. doi: 10.1104/pp.15.01016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gou J-Y, Felippes FF, Liu C-J, Weigel D, Wang J-W. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell. 2011;23:1512–1522. doi: 10.1105/tpc.111.084525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petroni K, Tonelli C. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 2011;181:219–229. doi: 10.1016/j.plantsci.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 49.Broun Pierre. Transcriptional control of flavonoid biosynthesis: a complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Current Opinion in Plant Biology. 2005;8(3):272–279. doi: 10.1016/j.pbi.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 50.Huang ZA, Zhao T, Wang N, Zheng S Song. Ectopic expression of Lc differentially regulated anthocyanin biosynthesis in the floral parts of tobacco (Nicotiana tobacum L.) plants. Bot Stud. 2016;57. [DOI] [PMC free article] [PubMed]
- 51.Li J, He YJ, Zhou L, Liu Y, Jiang M, Ren L, et al. Correction: Transcriptome profiling of genes related to light-induced anthocyanin biosynthesis in eggplant (Solanum melongena L.) before purple color becomes evident. BMC Genomics. 2018;19. [DOI] [PMC free article] [PubMed]
- 52.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li J, Ren L, Gao Z, Jiang M, Liu Y, Zhou L, et al. Combined transcriptomic and proteomic analysis constructs a new model for light-induced anthocyanin biosynthesis in eggplant (Solanum melongena L.) Plant Cell Environ. 2017;40:3069–3087. doi: 10.1111/pce.13074. [DOI] [PubMed] [Google Scholar]
- 54.Jiang MM, Liu Y, Ren L, Lian HL, Chen HY. Molecular cloning and characterization of anthocyanin biosynthesis genes in eggplant (Solanum melongena L.). Acta Physiol Plant. 2016;38.
- 55.Tai Deqiang, Tian Ji, Zhang Jie, Song Tingting, Yao Yuncong. A Malus Crabapple Chalcone Synthase Gene, McCHS, Regulates Red Petal Color and Flavonoid Biosynthesis. PLoS ONE. 2014;9(10):e110570. doi: 10.1371/journal.pone.0110570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dare AP, Tomes S, Jones M, McGhie TK, Stevenson DE, Johnson RA, et al. Phenotypic changes associated with RNA interference silencing of chalcone synthase in apple (Malus × domestica) Plant J. 2013;74:398–410. doi: 10.1111/tpj.12140. [DOI] [PubMed] [Google Scholar]
- 57.Muir SR, Collins GJ, Robinson S, Hughes S, Bovy A, Ric De Vos CH, et al. Overexpression of petunia chalcone isomerase in tomato results in fruit containing increased levels of flavonols. Nat Biotechnol. 2001;19:470–474. doi: 10.1038/88150. [DOI] [PubMed] [Google Scholar]
- 58.Jiang F, Wang JY, Jia HF, Jia WS, Wang HQ, Xiao M. RNAi-mediated silencing of the flavanone 3-hydroxylase gene and its effect on flavonoid biosynthesis in strawberry fruit. J Plant Growth Regul. 2013;32:182–190. doi: 10.1007/s00344-012-9289-1. [DOI] [Google Scholar]
- 59.Huang Yan, Gou Jiqing, Jia Zhichun, Yang Li, Sun Yimin, Xiao Xunyan, Song Feng, Luo Keming. Molecular Cloning and Characterization of Two Genes Encoding Dihydroflavonol-4-Reductase from Populus trichocarpa. PLoS ONE. 2012;7(2):e30364. doi: 10.1371/journal.pone.0030364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Hongxia, Fan Weijuan, Li Hong, Yang Jun, Huang Jirong, Zhang Peng. Functional Characterization of Dihydroflavonol-4-Reductase in Anthocyanin Biosynthesis of Purple Sweet Potato Underlies the Direct Evidence of Anthocyanins Function against Abiotic Stresses. PLoS ONE. 2013;8(11):e78484. doi: 10.1371/journal.pone.0078484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen WH, Hsu CY, Cheng HY, Chang H, Chen HH, Ger MJ. Downregulation of putative UDP-glucose: flavonoid 3-O-glucosyltransferase gene alters flower coloring in Phalaenopsis. Plant Cell Rep. 2011;30:1007–1017. doi: 10.1007/s00299-011-1006-1. [DOI] [PubMed] [Google Scholar]
- 62.An J-P, Wang X-F, Li Y-Y, Song L-Q, Zhao L-L, You C-X, et al. EIN3-LIKE1, MYB1, and ETHYLENE RESPONSE FACTOR3 act in a regulatory loop that synergistically modulates Ethylene biosynthesis and anthocyanin accumulation. Plant Physiol. 2018;178:808–823. doi: 10.1104/pp.18.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou LL, Zeng HN, Shi MZ, Xie DY. Development of tobacco callus cultures over expressing Arabidopsis PAP1/MYB75 transcription factor and characterization of anthocyanin biosynthesis. Planta. 2008;229:37–51. doi: 10.1007/s00425-008-0809-y. [DOI] [PubMed] [Google Scholar]
- 64.Sompornpailin K, Kanthang S. Tobacco randomly inserted TT8 differenly enhance light signals and flavonoid accumulation. Pakistan J Bot. 2015;47:1303–1309. [Google Scholar]
- 65.Lim S-H, Kim D-H, Kim JK, Lee J-Y, Ha S-H. A radish basic helix-loop-helix transcription factor, RsTT8 acts a positive regulator for anthocyanin biosynthesis. Front Plant Sci. 2017;8. [DOI] [PMC free article] [PubMed]
- 66.Gonzalez A, Brown M, Hatlestad G, Akhavan N, Smith T, Hembd A, et al. TTG2 controls the developmental regulation of seed coat tannins in Arabidopsis by regulating vacuolar transport steps in the proanthocyanidin pathway. Dev Biol. 2016;419:54–63. doi: 10.1016/j.ydbio.2016.03.031. [DOI] [PubMed] [Google Scholar]
- 67.Li P, Chen X, Sun F, Dong H. Tobacco TTG2 and ARF8 function concomitantly to control flower colouring by regulating anthocyanin synthesis genes. Plant Biol. 2017;19:525–532. doi: 10.1111/plb.12560. [DOI] [PubMed] [Google Scholar]
- 68.Duan S, Wang J, Gao C, Jin C, Li D, Peng D, et al. Functional characterization of a heterologously expressed Brassica napus WRKY41-1 transcription factor in regulating anthocyanin biosynthesis in Arabidopsis thaliana. Plant Sci. 2018;268:47–53. doi: 10.1016/j.plantsci.2017.12.010. [DOI] [PubMed] [Google Scholar]
- 69.Wang N, Liu W, Zhang T, Jiang S, Xu H, Wang Y, et al. Transcriptomic analysis of red-fleshed apples reveals the novel role of MdWRKY11 in flavonoid and anthocyanin biosynthesis. J Agric Food Chem. 2018;66:7076–7086. doi: 10.1021/acs.jafc.8b01273. [DOI] [PubMed] [Google Scholar]
- 70.Mao Ke, Wang Lina, Li Yuan-Yuan, Wu Rongling. Molecular Cloning and Functional Analysis of UV RESISTANCE LOCUS 8 (PeUVR8) from Populus euphratica. PLOS ONE. 2015;10(7):e0132390. doi: 10.1371/journal.pone.0132390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu Q, Su N, Zhang X, Liu Y, Cui J, Liang Y. Hydrogen peroxide, nitric oxide and UV RESISTANCE LOCUS8 interact to mediate UV-B-induced anthocyanin biosynthesis in radish sprouts. Sci Rep. 2016;6. [DOI] [PMC free article] [PubMed]
- 72.Yang Y, Liang T, Zhang L, Shao K, Gu X, Shang R, et al. UVR8 interacts with WRKY36 to regulate HY5 transcription and hypocotyl elongation in Arabidopsis. Nat Plants. 2018;4:98–107. doi: 10.1038/s41477-017-0099-0. [DOI] [PubMed] [Google Scholar]
- 73.Liang T, Mei S, Shi C, Yang Y, Peng Y, Ma L, et al. UVR8 Interacts with BES1 and BIM1 to Regulate Transcription and Photomorphogenesis in Arabidopsis. Dev Cell. 2018;44:512–523.e5. doi: 10.1016/j.devcel.2017.12.028. [DOI] [PubMed] [Google Scholar]
- 74.Wang X, Wang Q, Han Y-J, Liu Q, Gu L, Yang Z, et al. A CRY-BIC negative feedback circuitry regulating blue light sensitivity of Arabidopsis. Plant J. 2017;:1–11. [DOI] [PMC free article] [PubMed]
- 75.Niu SS, Xu CJ, Zhang WS, Zhang B, Li X, Lin-Wang K, et al. Coordinated regulation of anthocyanin biosynthesis in Chinese bayberry (Myrica rubra) fruit by a R2R3 MYB transcription factor. Planta. 2010;231:887–899. doi: 10.1007/s00425-009-1095-z. [DOI] [PubMed] [Google Scholar]
- 76.Romero I, Teresa Sanchez-Ballesta M, Maldonado R, Isabel Escribano M, Merodio C. Anthocyanin, antioxidant activity and stress-induced gene expression in high CO<inf>2</inf>−treated table grapes stored at low temperature. J Plant Physiol. 2008;165:522–530. doi: 10.1016/j.jplph.2006.12.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S2. Gene Expression Summary. (DOCX 15 kb)
Table S3. Differentially expressed genes (DEGs) in non-photosensitive eggplant. (XLSX 239 kb)
Figure S1. qRT-PCR Analysis of DEGs after Opening the Bags. (DOCX 522 kb)
Figure S2. Cluster analysis and KEGG pathway enrichment analysis of DEGs. (DOCX 1176 kb)
Table S4. Differentially expressed TF genes in non-photosensitive and photosensitive eggplant. (XLSX 45 kb)
Table S5. Transcription factors target gene prediction results. (DOCX 35 kb)
Table S6. The interaction score of transcription factors in Fig. 9. (DOCX 13 kb)
Table S1. List of all the primer sequences used in this study. (DOCX 15 kb)
Data Availability Statement
The Illumina RNA-seq data generated from non-photosensitive eggplant are available in the NCBI SRA with accessions number SRA613313, and the RNA-seq data of photosensitive eggplant are available in the NCBI SRA with accessions numbers SRR5650714, SRR5651526, SRR5658205 and SRR5658226 [51].