Abstract
We describe clonal shifts in vanA Enterococcus faecium isolates from clinical samples obtained from patients in Denmark from 2015 to the first quarter (Q1) of 2019. During Q1 2019, the vancomycin-variable enterococci (VVE) ST1421-CT1134 vanA E. faecium became the most dominant vanA E. faecium clone and has spread to all five regions in Denmark. Among 174 E. faecium isolates with vanA, vanB or vanA/vanB genes in Q1 2019, 44% belonged to this type.
Keywords: VRE, VVE, Enterococci, vanA, vanB, MLST
We describe the clonal shift for vanA Enterococcus faecium during the last 4 years and the national spread of a vancomycin-variable vanA E. faecium ST1421-CT1134 clone in Denmark. The aim is to highlight the importance of using molecular methods for detecting vancomycin-variable enterococci (VVE), and to alert other countries about this emerging nosocomial clone.
Vancomycin-variable enterococci
Vancomycin-variable enterococci (VVE) are E. faecium harboring the vanA gene complex, but being phenotypically vancomycin susceptible [1,2]. VVE can only be detected by molecular methods and cannot be cultured on selective vancomycin-containing media. Different clones of VVE have caused nosocomial outbreaks and development of vancomycin-resistant revertant mutants in vitro and in vivo has been described [1,3-5]. This makes the detection of VVE highly important in clinical samples in order to assure relevant antibiotic treatment and in screening samples to avoid nosocomial spread. In 2015 and 2016, sporadic VVE with different genetic background were detected in the Capital Region of Denmark, in connection with vancomycin-resistant enterococci (VRE) outbreaks (data not shown). In 2016, a VVE clone belonging to ST1421-CT1134, which displays variable vancomycin susceptibility (minimum inhibitory concentration (MIC) 1 to ≥ 256 mg/ml) was detected in screening samples from a hospital in the Capital Region [5]. One strain, Efm-V1511, belonging to this clone was characterised by Hansen et al. [5]. Efm-V1511 had a 49.6 Kp plasmid, which carried the Tn1546 (vanA transposon). Tn1546 was truncated in vanX by a 252 bp 3' deletion explaining the vancomycin susceptibility of Efm-V1511. In ST1421-CT1134 isolates resistant to vancomycin, resistance could be attributed to changes in ddl disrupting gene function sometimes accompanied by changes in vanS, increased pHVH-V1511 copy number or the existence of an additional vanA-containing plasmid encoding a functional vanX [5].
National surveillance of vancomycin-resistant and vancomycin-variable enterococci
We have previously described the surveillance of vancomycin-resistant enterococci (VRE) in clinical isolates in Denmark from 2005 to 2015 [6]. In the present study, we follow up and describe the data from isolates obtained from 2016 through the first quarter (Q1) of 2019. Since 2005, VRE isolates from clinical samples, e.g. urine, blood and tissue, as opposed to screening (faecal) isolates have been voluntarily submitted to Statens Serum Institut (SSI) from Danish Departments of Clinical Microbiology (DCM) for species identification, genotyping and surveillance (Figure 1) [7]. Only one isolate per patient per 12 months was included. All VRE isolates (699 E. faecium and 30 E. faecalis) were tested for the presence of vancomycin resistance genes vanA and vanB by PCR from 2005 through 2014.
Figure 1.
The five healthcare regions and the 10 Departments of Clinical Microbiology, Denmark, 2019
DCM: Department of Clinical Microbiology; ND: not detected.
Modified from DANMAP 2017 [7].
From 2015 through Q1 2019, all clinical VRE/VVE isolates (n = 1,935) underwent whole-genome sequencing (WGS) as previously described [6]. From the WGS data, multilocus sequence type (MLST), and van genes were extracted in silico. The isolates were further subtyped in SeqSphere + (Ridom GmbH, Münster, Germany (http://www.ridom.de/seqsphere/)) using the cgMLST scheme by de Been et al. [8] for E. faecium.
VVE diagnostic algorithms have differed substantially over time and between the five Danish regions. In 2017, testing of phenotypically vancomycin-susceptible E. faecium isolates from blood cultures for the presence of vanA/vanB genes by PCR was introduced in the DCMs in the Capital Region. During 2018, this was expanded to testing of all clinical E. faecium isolates. During 2018, molecular testing by PCR of E. faecium from all clinical samples was also implemented in one of the four DCMs in the Region of Southern Denmark. Furthermore, E. faecium isolates from blood cultures were tested by PCR for vanA/vanB genes in another DCM in the Region of Southern Denmark and in the DCM in the Central Denmark Region in 2018. In Q1 2019, diagnostic algorithms to detect VVE have expanded. Most of the DCMs across Denmark test at least all blood culture E. faecium isolates for the presence of vanA genes using PCR.
Enterococcus faecium and Enterococcus faecalis isolates from clinical samples carrying vanA and vanB genes
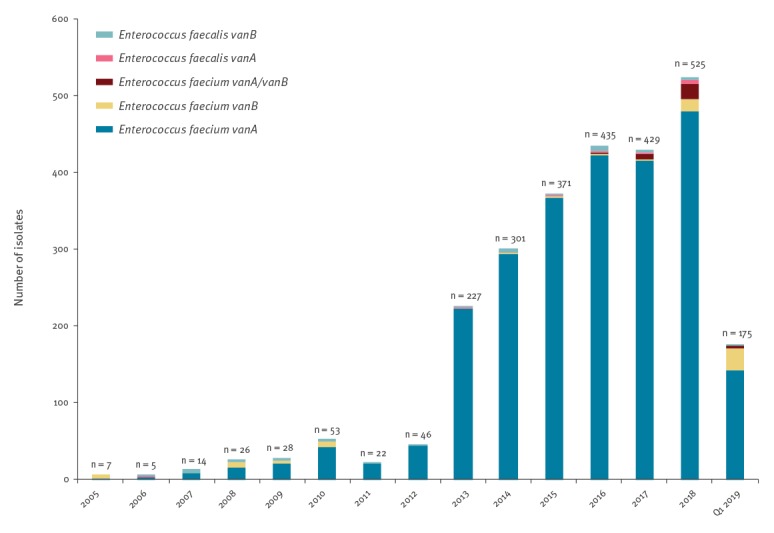
From 2005 to Q1 2019, 2,503 vanA E. faecium, 74 vanB E. faecium, 32 vanA/vanB E. faecium, 12 vanA E. faecalis, and 43 vanB E. faecalis from clinical samples were submitted to SSI (Figure 2).
Figure 2.
Vancomycin-resistant and vancomycin-variable Enterococcus faecium and vancomycin-resistant E. faecalis isolates from clinical samples carrying van genes, Denmark, 2005–Q1 2019 (n = 2,664)
Q1: first quarter.
Emergence and disappearance of major Enterococcus faecium clones
Of the 1,935 VRE/VVE isolates obtained from 2015 through Q1 2019, 1,910 were E. faecium and 25 E. faecalis (Figure 2).
The E. faecium isolates belonged to 29 sequence types (STs). ST80 (22%), ST203 (65%) and ST1421 (9%) were most prevalent. Typing by cgMLST revealed 156 different complex types (CTs).
The 13 most common types of vanA, vanB and vanA/vanB E. faecium from 2015 to Q1 2019 are shown in Table 1. From 2015 to 2019, three types were dominating: ST80-CT14 vanA E. faecium, ST203-CT859 vanA E. faecium and ST1421-CT1134 vanA E. faecium (Table 1).
Table 1. Description of the most common types of vanA and/or vanB Enterococcus faecium by MLST and cgMLST, Denmark, 2015–Q1 2019 (n = 1,910).
| Types | 2015 (n = 369) |
2016 (n = 427) |
2017 (n = 425) |
2018 (n = 515) |
Q1 2019 (n = 174) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | n | % | n | % | |
| ST80-CT14 vanA | 81 | 22 | 38 | 9 | 15 | 4 | 1 | < 1 | ND | ND |
| ST80-CT24 vanA | 23 | 6 | 19 | 4 | 11 | 3 | 2 | < 1 | 4 | 2 |
| ST80-CT860 vanA | 7 | 2 | 11 | 3 | ND | ND | ND | ND | ND | ND |
| ST80-CT866 vanA | 14 | 4 | 10 | 2 | 7 | 2 | ND | ND | ND | ND |
| ST80-CT991 vanA | ND | ND | 11 | 3 | 9 | 2 | 6 | 1 | ND | ND |
| ST80-CT1160 vanA | ND | ND | ND | ND | 7 | 2 | 10 | 2 | ND | ND |
| ST80-CT1064 vanA/vanB | ND | ND | 2 | < 1 | 8 | 2 | 23 | 5 | 4 | 2 |
| ST80-CT1729 vanA | ND | ND | ND | ND | ND | ND | 22 | 4 | 2 | 1 |
| ST117-CT873 vanA | 5 | 1 | 12 | 3 | ND | ND | ND | ND | ND | ND |
| ST117-CT1180 vanA | ND | ND | ND | ND | 9 | 2 | 30 | 6 | 7 | 4 |
| ST117-CT36 vanB | ND | ND | ND | ND | ND | ND | 2 | < 1 | 16 | 9 |
| ST203-CT859 (subtypes CT1051 and CT1507) vanA | 188 | 51 | 271 | 64 | 265 | 63 | 161 | 31 | 20 | 12 |
| ST1421-CT1134 vanA | ND | ND | 2 | < 1 | 13 | 3 | 176 | 34 | 77 | 44 |
| Other types | 51 | 14 | 51 | 12 | 81 | 19 | 82 | 16 | 44 | 25 |
CT: cluster type (cgMLST); MLST: multilocus sequence typing; ND: not detected; ST: sequence type (MLST); Q1: first quarter.
In 2015, 22% of the E. faecium isolates belonged to ST80-CT14 vanA E. faecium. The type decreased during 2016.
ST203-CT859 vanA E. faecium isolates were first detected during the end of 2014 [6]. It emerged very fast and was the most prevalent vanA E. faecium type (together with its subtypes CT1051 and CT1507) during 2015 to 2017, but decreased in 2018 (Table1). In Q1 2019 only 12% of the VRE/VVE E. faecium isolates belonged to ST203-CT859.
In 2017, 3% of the E. faecium isolates belonged to the VVE clone, ST1421-CT1134 vanA E. faecium. This type was only detected from clinical samples from the Capital Region. In 2018, 34% of the E. faecium isolates belonged to ST1421-CT1134 and were detected in the Capital Region, the Region Zealand and from one DCM in the Region of Southern Denmark (Table 1, Table 2). During Q1 2019, ST1421-CT1134 vanA E. faecium was the most prevalent type (44%) (Table 1). It was detected in all five regions of Denmark, 50 isolates from the Capital Region, one isolate from Region Zealand, 23 isolates from the Region of Southern Denmark, two isolates from Central Denmark Region and one isolate from the North Denmark Region (Table 2). Furthermore, ST1421-CT1134 vanA E. faecium spread to the Faroe Islands during 2018 and 2019 (data not shown).
Table 2. Regional occurrence of ST1421-CT1134 vanA E. faecium, Denmark, 2016–Q1 2019 (n = 268).
| Region | 2016 (n = 2) |
2017 (n = 13) |
2018 (n = 176) |
Q1 2019 (n = 77) |
|---|---|---|---|---|
| Capital Region of Denmark | 2 | 9 | 158 | 50 |
| Region Zealand | ND | 3 | 9 | 1 |
| Region of Southern Denmark | ND | ND | 9 | 23 |
| Central Denmark Region | ND | ND | ND | 2 |
| North Denmark Region | ND | 1 | ND | 1 |
Q1: first quarter.
Discussion and conclusion
During 2005 to Q1 2019, most of the Danish clinical VRE isolates have been vanA E. faecium isolates. This study shows that predominating clones shifted over time and, importantly, the emergence of a vancomycin-variable clone, ST1421-CT1134 vanA E. faecium, that has spread to all the five Danish regions in 2019.
Although the E. faecium isolates belonged to 156 CTs, three types (ST80-CT14 vanA E. faecium, ST203-CT859 vanA E. faecium, ST1421-CT1134 vanA E. faecium) have dominated during the last 4 years.
ST80-CT14 vanA E. faecium was highly prevalent in the Capital Region during 2012 to 2015 [9]. The vanA E. faecium constituting Group2_ST80 in the paper by Pinholt et al. [9] belonged to ST80-CT14 (data not shown). On a national level, the numbers of ST80-CT14 vanA E. faecium decreased during 2016 to 2018, and this clone was not detected during Q1 2019.
ST203-CT859 vanA E. faecium emerged during 2015 through 2017 and nearly disappeared 2019. This clone has spread to Sweden, the Faroe Islands and Greenland [6,7].
Because of differences in diagnostic algorithms, there is a detection bias of VVE. It seems very likely that ST1421-CT1134 vanA E. faecium have been under-reported in some regions at least during some periods. Thus, the rising incidence could partly be explained by increasing molecular testing of vancomycin susceptible isolates. However, a sharply increasing incidence has also been seen in DCM with extensive testing for VVE.
The origin of ST80-CT14 vanA E. faecium and ST203-CT859 vanA E. faecium are still unknown. vanA E. faecium isolates belonging to ST1421-CT1134 have also been reported from Australia, but these isolates have not been VVE [10]. Why these three clones were so successful is unknown.
The spread of the VVE clone, ST1421-CT1134 vanA E. faecium, in Denmark is of concern, especially since VVE diagnostic is challenging. Because of this, the clone is likely to be underdiagnosed, which facilitates further spread. Since cross-border spread has been described for VRE, countries with patients transferred from Denmark should be aware of the vancomycin-variable ST1421-CT1134 vanA E. faecium clone.
Acknowledgements
We thank Karin Sixhøj Pedersen and Frank Hansen for their excellent technical assistance.
Conflict of interest: None declared.
Authors’ contributions: Mette Pinholt, Louise Roer, Hülya Kaya, Peder Worning, Sanne Nygaard, Marianne Engell Clausen, Karen Leth Nielsen, Jurgita Samulioniené, Mona Kjærsgaard, Claus Østergaard, Ulrik S Justesen, John Coia, Turid Snekloth Søndergaard, Shahin Gaini, Kristian Schønning, Henrik Westh, Henrik Hasman and Barbara Holzknecht contributed to the revision of the manuscript and approved the final version. Louise Roer, Hülya Kaya, Anette M Hammerum and Henrik Hasman did the molecular analysis. Mette Pinholt, Peder Worning, Kristian Schønning and Henrik Westh shared WGS data for many of the VRE/VVE isolates from DCM Hvidovre. Ulrik S Justesen, Mette Pinholt, Marianne Engell Clausen, Karen Leth Nielsen, Sanne Nygaard, Michael Kemp, Jurgita Samulioniené, Mona Kjærsgaard, Claus Østergaard, John Coia, Turid Snekloth Søndergaard, Kristian Schønning, Henrik Westh and Barbara Holzknecht detected VRE/VVE at the DCMs in Denmark. Shahin Gaini shared isolates and data on the VRE/VVE from the Faroe Islands.
Anette M Hammerum and Barbara Holzknecht drafted the manuscript. Anette M Hammerum incorporated comments, additions and feedback throughout the revision.
References
- 1. Sivertsen A, Pedersen T, Larssen KW, Bergh K, Rønning TG, Radtke A, et al. A Silenced vanA Gene Cluster on a Transferable Plasmid Caused an Outbreak of Vancomycin-Variable Enterococci. Antimicrob Agents Chemother. 2016;60(7):4119-27. 10.1128/AAC.00286-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kohler P, Eshaghi A, Kim HC, Plevneshi A, Green K, Willey BM, et al. Toronto Invasive Bacterial Diseases Network (TIBDN) Prevalence of vancomycin-variable Enterococcus faecium (VVE) among vanA-positive sterile site isolates and patient factors associated with VVE bacteremia. PLoS One. 2018;13(3):e0193926. 10.1371/journal.pone.0193926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Szakacs TA, Kalan L, McConnell MJ, Eshaghi A, Shahinas D, McGeer A, et al. Outbreak of vancomycin-susceptible Enterococcus faecium containing the wild-type vanA gene. J Clin Microbiol. 2014;52(5):1682-6. 10.1128/JCM.03563-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Coburn B, Low DE, Patel SN, Poutanen SM, Shahinas D, Eshaghi A, et al. Vancomycin-variable Enterococcus faecium: in vivo emergence of vancomycin resistance in a vancomycin-susceptible isolate. J Clin Microbiol. 2014;52(5):1766-7. 10.1128/JCM.03579-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hansen TA, Pedersen MS, Nielsen LG, Ma CMG, Søes LM, Worning P, et al. Emergence of a vancomycin-variable Enterococcus faecium ST1421 strain containing a deletion in vanX. J Antimicrob Chemother. 2018;73(11):2936-40. 10.1093/jac/dky308 [DOI] [PubMed] [Google Scholar]
- 6. Hammerum AM, Baig S, Kamel Y, Roer L, Pinholt M, Gumpert H, et al. Emergence of vanA Enterococcus faecium in Denmark, 2005-15. J Antimicrob Chemother. 2017;72(8):2184-90. 10.1093/jac/dkx138 [DOI] [PubMed] [Google Scholar]
- 7.The Danish Integrated Antimicrobial Resistance Monitoring and Research Programme (DANMAP). DANMAP 2017 - Use of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from food animals, food and humans in Denmark. Copenhagen: DANMAP; 2017. Available from: https://www.danmap.org/-/media/arkiv/projekt-sites/danmap/danmap-reports/danmap-2017/danmap2017.pdf?la=en.
- 8. de Been M, Pinholt M, Top J, Bletz S, Mellmann A, van Schaik W, et al. Core Genome Multilocus Sequence Typing Scheme for High- Resolution Typing of Enterococcus faecium. J Clin Microbiol. 2015;53(12):3788-97. 10.1128/JCM.01946-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pinholt M, Bayliss SC, Gumpert H, Worning P, Jensen VVS, Pedersen M, et al. WGS of 1058 Enterococcus faecium from Copenhagen, Denmark, reveals rapid clonal expansion of vancomycin-resistant clone ST80 combined with widespread dissemination of a vanA-containing plasmid and acquisition of a heterogeneous accessory genome. J Antimicrob Chemother. 2019;74(7):1776-85. 10.1093/jac/dkz118 [DOI] [PubMed] [Google Scholar]
- 10. van Hal SJ, Beukers AG, Timms VJ, Ellem JA, Taylor P, Maley MW, et al. Relentless spread and adaptation of non-typeable vanA vancomycin-resistant Enterococcus faecium: a genome-wide investigation. J Antimicrob Chemother. 2018;73(6):1487-91. 10.1093/jac/dky074 [DOI] [PubMed] [Google Scholar]


