Abstract
Clinical and pathological stage are defining parameters in oncology, which direct a patient’s treatment options and prognosis. Pathology reports contain a wealth of staging information that is not stored in structured form in most electronic health records (EHRs). Therefore, we evaluated three supervised marchine learning methods (Support Vector Machine, Decision Trees, Gradient Boosting) to classify free-text pathology reports for prostate cancer into T, N and M stage groups.
Keywords: Prostate Cancer, TNM Staging, Natural Language Processing, Pathology Reports
Introduction
Prostate cancer is the commonest non-cutaneous malignancies in men, with over 260,000 new cases annually in the United States.[1] The staging of these newly diagnosed cancer patients is one of the most important factors in determining treatment options and predicting patient survival. [3] Free-text pathology reports contain a wealth of staging information that is not captured in structured form in most electronic health records (EHRs). The ability to automatically extract stage from pathology reports would facilitate the creation of research cohorts from the EHR (e.g. pragmatic trials), provide a framework for quality assurance over time (e.g. assess bone scan adherence), and assist with harmonizing data across sites (e.g. evaulate population-level trends).
Natural language processing (NLP) has emerged as a promising tool for extracting stage from clinical texts. There have been various attempts to apply NLP to automatically extract stage from progress clnical notes and pathology reports across a range of tumor types including lung, breast, colorectal and prostate [4–9]. The majority of these studies have used a rule-based approach, relying on regular expressions associated with stage descriptions or smart text forms. However, rule-based approaches often have limited generalizability between tumor types and across institutions. Therefore, in this study, we aimed to evaluate the performance of different machine learning approaches for extracting staging information from pathology reports in prostate cancer using a more generalizable machine learning approach. This may help to inform the strategy of automated stage extraction from unstructured clinical text.
Methods
The Stanford prostate cancer research database was used for analysis, which is described in detail elsewhere [10]. We identified a cohort of prostate cancer subjects with at least one pathology report. This study was made possible due to linkage of the EHR with an institutional cancer registry, which contained ground-truth stage labels manually abstracted from the clinical notes. Stage annotations were defined at the time of diagnosis using the T, N, M classification (i.e. each document had a separate T, N and M annotation).
We included only reports within one year of the diagnosis date. As we are a teritary cancer center, one year post-diagnosis was used to ensure patients on active surveillance seeking secondary opinions were includedIn the case where multiple reports appeared within one year of diagnosis, we treated each report as a separate training sample. In an effort to simplify the classification task, stage labels from the cancer registry were clustered intro groups under the guidance of clinical advisors (e.g. 7 separate T stage labels were grouped into 3). The cohort contained only tumors of T stage 2 and above, as lower-stage tumors were not biopsied.
The pipeline was built with Python (version 3.6) using the Natural Language Toolkit (NLTK) for preprocessing, and scikit-learn for feature extraction and classification. Each report was put through a pre-processing pipeline consisting of stemming, lemmatizing, stop-word and punctuation removal. Subsequently, term frequency-inverse document frequency (TF-iDF) scores were generated for each term-document pair [11].
A bag-of-words representation for each document was generated, with word weighting by TF-iDF scores. A vocabulary was constructed using the entire document corpus. This vocabulary was used to generate document-level word vectors. Neural embeddings were not used because of the limited size of the corpus, and the fact that pre-trained embeddings such as GloVe (GLobal Vectors for Word Representation) were not well suited to the vocabulary of pathology reports. The NLP pipeline is illustrated in Figure 1.
Figure 1:
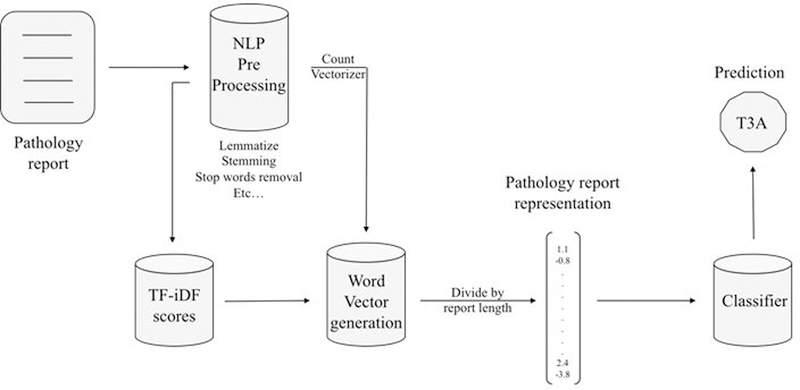
Architecture of the NLP pipeline for classifying pathology reports into T, N, M stage categories.
For each of T, N and M stage classifications, we used the document-level vector representations to train a classifier against the ground-truth pooled stage labels from the cancer registry. We used an 80/10/10% split for training/validation/test sets. The following classifiers were trialed: support vector machines (SVM), random forest (RF), and extreme gradient boosting (XGB). With F1-score as our target metric, we used random hyperparameter search to tune our classifiers.
Results
This study cohort included 4,470 prostate cancer subjects with at least one pathology report, yielding a total of 13,595 unique reports. Table 1 shows the results of each classifier for the T, N, M classification tasks. The optimal F1-score achieved was 0.80 on pooled T stage (3 labels), 0.71 on unpooled T stage (7 labels), 0.98 on N stage and 0.99 on M stage.
Table 1:
Evaluation Results
| Classifier | Model | Precision | Recall | F-Score |
|---|---|---|---|---|
| T (3 labels) | SVM | 0.77 | 0.79 | 0.77 |
| Decision Trees | 0.76 | 0.75 | 0.76 | |
| Gradient Boosting | 0.80 | 0.81 | 0.80 | |
| T (7 labels) | SVM | 0.61 | 0.64 | 0.61 |
| Decision Trees | 0.63 | 0.62 | 0.62 | |
| Gradient Boosting | 0.71 | 0.71 | 0.71 | |
| N (2 labels) | SVM | 0.98 | 0.98 | 0.97 |
| Gradient Boosting | 0.99 | 0.98 | 0.98 | |
| M (2 labels) | SVM | 0. 99 | 0. 99 | 0. 99 |
| Gradient Boosting | 0.99 | 0. 99 | 0. 99 | |
This study is limited in analyzing pathology reports from a single institution, albeit one with a very diverse clinician and patient population over an extended timeframe. Further work is warranted to apply the pipeline to pathology reports from other sites in order to validate the putative generalizability of this machine learning approach relative to rule-based methods. In addition, the classification tasks were affected by the class imbalances in the dataset, especially between prostate M0 and M1, and breast M0 and M1. We have also made assumptions that a pathology report within one year of diagnosis date reflects the stage at the time of diagnosis - it is conceivable that the stage listed by the registry is not accurate at the time of the report.
Conclusions
Our NLP pipeline is able to efficiently classify pathology reports into T, N, and M stage categories, with strongest performance for N and M stage. This may be a more scalable method than rule-based systems for extracting staging data from unstructured text, which has implications for auto-populating registries and identifying observational research cohorts from EHRs.
Acknowledgements
Research reported in this publication was supported by the Stanford AstraZeneca Research grant and National Cancer Institute of the National Institutes of Health under Award Number R01CA183962. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- [1].Siegel RL, Miller KD, Jemal A, Cancer statistics, 2018, CA Cancer J Clin 68(1) (2018) 7–30. [DOI] [PubMed] [Google Scholar]
- [2].Siegel RL, Miller KD, Jemal A, Cancer statistics, 2015, CA Cancer J Clin 65(1) (2015) 5–29. [DOI] [PubMed] [Google Scholar]
- [3].Buyyounouski MK, Choyke PL, McKenney JK, Sartor O, Sandler HM, Amin MB, Kattan MW, Lin DW, Prostate cancer - major changes in the American Joint Committee on Cancer eighth edition cancer staging manual, CA Cancer J Clin 67(3) (2017) 245–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Evans TL, Gabriel PE, Shulman LN, Cancer Staging in Electronic Health Records: Strategies to Improve Documentation of These Critical Data, J Oncol Pract 12(2) (2016) 137–9. [DOI] [PubMed] [Google Scholar]
- [5].McCowan I, Moore D, Fry MJ, Classification of cancer stage from free-text histology reports, Conf Proc IEEE Eng Med Biol Soc 1 (2006) 5153–6. [DOI] [PubMed] [Google Scholar]
- [6].McCowan IA, Moore DC, Nguyen AN, Bowman RV, Clarke BE, Duhig EE, Fry MJ, Collection of cancer stage data by classifying free-text medical reports, J Am Med Inform Assoc 14(6) (2007) 736–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Nguyen A, Moore D, McCowan I, Courage MJ, Multi-class classification of cancer stages from free-text histology reports using support vector machines, Conf Proc IEEE Eng Med Biol Soc 2007 (2007) 5140–3. [DOI] [PubMed] [Google Scholar]
- [8].Nguyen AN, Lawley MJ, Hansen DP, Bowman RV, Clarke BE, Duhig EE, Colquist S, Symbolic rule-based classification of lung cancer stages from free-text pathology reports, J Am Med Inform Assoc 17(4) (2010) 440–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Warner JL, Levy MA, Neuss MN, ReCAP: Feasibility and Accuracy of Extracting Cancer Stage Information From Narrative Electronic Health Record Data, J Oncol Pract 12(2) (2016) 157–8; e169–7. [DOI] [PubMed] [Google Scholar]
- [10].Seneviratne MG, Seto T, Blayney DW, Brooks JD, Hernandez-Boussard T, Architecture and Implementation of a Clinical Research Data Warehouse for Prostate Cancer, EGEMS (Wash DC) 6(1) (2018) 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ramos J, Using tf-idf to determine word relevance in document queries, Proceedings of the first instructional conference on machine learning, 2003. [Google Scholar]


