Abstract
Background
The aim of this study was to clarify the biological function of microRNA-449b-5p in the progression of osteosarcoma (OS) and to define the underlying mechanism.
Material/Methods
Relative levels of microRNA-449b-5p in OS tissues and cell lines was determined by quantitative real-time polymerase chain reaction (qRT-PCR). The correlation between microRNA-449b-5p level and pathological characteristics of OS patients was analyzed by chi-square test. Kaplan-Meier analysis was used for survival analysis of OS patients based on their expression level of microRNA-449b-5p. Regulatory effects of microRNA-449b-5p on cellular behaviors of OS cells were evaluated by cell counting kit-8 (CCK-8) and Transwell assay. The binding relationship between microRNA-449b-5p and c-Met was verified through dual-luciferase reporter gene assay, and their interaction in OS progression was further examined through a series of rescue experiments.
Results
MicroRNA-449b-5p was expressed at low levels in OS. Lower levels of microRNA-449b-5p were seen in OS tissues with worse tumor grade or histological differentiation. OS patients with low levels of microRNA-449b-5p had worse overall survival relative to those with high level of microRNA-449b-5p. Overexpression of microRNA-449b-5p markedly attenuated proliferative, migratory, and invasive abilities of OS cells. C-Met is the downstream target of microRNA-449b-5p, and its level was inhibited in OS cells overexpressing microRNA-449b-5p. Importantly, c-Met partially rescued the inhibitory effects of microRNA-449b-5p on behavior of OS cells.
Conclusions
MicroRNA-449b-5p is downregulated in OS, which alleviates the malignant progression of OS by targeting c-Met.
MeSH Keywords: Cell Proliferation, MicroRNAs, Osteosarcoma
Background
Osteosarcoma (OS) is a highly malignant primary bone tumor that mainly involves the long bones. Its accounts for 2.4% of all adolescent malignancies [1,2]. Biological characteristics of OS include rapid growth, strong invasiveness, and high rate of distant metastasis. The prognosis of OS patients is poor, with high mortality and disability rates [1]. It is reported that the 5-year survival of OS is as low as to 30–75%. About 80% of OS patients develop metastases in the lungs or other organs [3,4]. Although great strides have been made in surgical resection, tumor prosthesis replacement, chemotherapy, and radiotherapy, clinical outcomes of OS patients are unsatisfactory [5,6]. Therefore, it is of great importance to discover the pathogenic mechanism of OS.
MicroRNAs (miRNAs) are a class of non-coding, small RNAs that bind to the 3′UTR of their target mRNAs. They are capable of degrading target mRNAs or inhibiting their translation process [7]. miRNAs participate in diverse biological processes [8,9]. Dysregulated miRNAs are reported to be closely related to tumor progression [10–14]. Hence, tumor-related miRNAs could be utilized as diagnostic and therapeutic targets for tumors. Several OS-related miRNAs have been identified. Zhu et al. [15] found that miR-183 downregulates the expression of Ezrin to inhibit the invasive and migratory capacities of OS cells. Novello et al. [16] found that miRNA-34a suppresses the metastasis of OS through directly targeting p53 and c-Met. Cao et al. [17] reported that miR-802 is upregulated in OS and serves as an oncogene to promote cell proliferation by regulating p27. MicroRNA-449b-5p is involved in many types of tumors [18–20]. However, the specific role of microRNA-449b-5p in OS is unclear.
To provide a novel target for clinical treatment of OS, we assessed microRNA-449b-5p levels in OS tissues and cell lines and investigated the potential role of microRNA-449b-5p in the progression of OS.
Material and Methods
OS patients
Tumor tissues and paracancerous tissues were surgically resected from 60 OS patients treated in Weifang People’s Hospital from December 2016 to February 2018. They did not receive preoperative anti-tumor therapy and were pathologically diagnosed. All subjects volunteered to participate in the study and signed written informed consent. This study was approved by the Ethics Committee of Weifang People’s Hospital (22 Nov 2016).
Cell culture and transfection
Osteoblasts (hFOB1.19) and OS cell lines (Saos2, HOS, U2OS, and MG63) were provided by Cell Bank (Shanghai, China). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, Rockville, MD, USA) containing 10% fetal bovine serum (FBS) (Gibco, Rockville, MD, USA), 100 μg/mL penicillin, and 0.1 mg/mL streptomycin, in a 37°C, 5% CO2 incubator.
Cells were inoculated in a 6-well plate and cultured until 80% confluence. Transfection of 50 nmol/L microRNA-449b-5p mimics or NC was conducted using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA).
Cell counting kit-8 (CCK-8)
Cells were inoculated in a 96-well plate (2×103 cells/well). Absorbance (A) at 450 nm was recorded at predetermined time points using the CCK-8 kit (Dojindo Laboratories, Kumamoto, Japan) to produce a viability curve.
Quantitative real-time polymerase chain reaction (qRT-PCR)
TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was applied to extract cellular RNAs, and they were reversely transcribed into complementary deoxyribose nucleic acid (cDNA). Subsequently, PCR was conducted using the SYBR Green method. Primer sequences were:
MicroRNA-449b-5p: F: 5′-GGGAGGCAGTGTATTGTTA-3′,
R: 5′-CAGTGCGTGTCGTGGAGT-3′;
C-Met: F: 3′-GAGGCAGTGCAGCATGTAGT-5′,
R: 3′-GATGATTCCCTCGGTCAGAA-5′.
Transwell assay
Diluted Matrigel was used to pre-coat the Transwell chamber overnight at 4°C. Cell density was adjusted to 2×105/mL in serum-free medium. We added 500 μL of medium containing 10% FBS into the basolateral chamber of the 24-well plate. We seeded 200 μL of cell suspension into the apical chamber, and 24 h later, cells were fixed in methanol for 30 min and stained with 0.1% crystal violet for another 30 min. Under an inverted microscope, invasive cells were photographed. Migration assay was conducted following the same procedures except for Matrigel pre-coating (BD Biosciences, Franklin Lakes, NJ, USA).
Western blot analysis
Using radioimmunoprecipitation assay (RIPA), cellular protein was isolated and its concentration was measured by bicinchoninic acid (BCA) method (Pierce, Rockford, IL, USA). Electrophoresis was performed and protein was transferred on a polyvinylidene fluoride (PVDF) membrane (Millipore, Billerica, MA, USA). After 2-h non-specific antigen blockage in PBS containing 5% skim milk, membranes were reacted with primary and secondary antibodies. Electrochemiluminescence (ECL) was conducted to expose bands, followed by grey-value analysis using Image J Software (NIH, Bethesda, MD, USA).
Dual-luciferase reporter gene assay
Cells seeded into a 24-well plate with 2×105 cells per well co-transfected with WT c-Met 3′UTR/MUT c-Met 3′UTR and microRNA-449b-5p mimic/NC. Cells were lysed 48 h later to assess relative luciferase activity (Promega, Madison, WI, USA).
Statistical analysis
Statistical Product and Service Solutions (SPSS) 17.0 software (SPSS, Inc., Chicago, IL, USA) was used for data analyses. Data are expressed as mean ± standard deviation. Differences between 2 groups were analyzed by the t test. The chi-square test was performed to assess the correlation between microRNA-449b-5p levels and pathological indexes of OS patients. Survival analysis was carried out using Kaplan-Meier method. P<0.05 was set as the level of statistical significance.
Results
Downregulated microRNA-449b-5p in OS
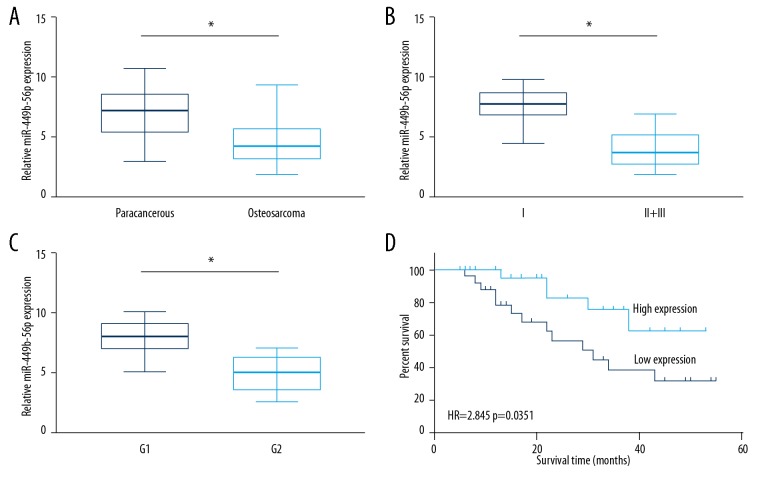
Compared with paracancerous tissues, microRNA-449b-5p was downregulated in OS tissues (Figure 1A). Furthermore, microRNA-449b-5p levels remained lower in OS patients in stage II+III relative to those in stage I (Figure 1B). Based on the histological differentiation stage, OS patients in G1 presented higher levels of microRNA-449b-5p relative to those in G2 (Figure 1C). Compared with OS patients with high levels of microRNA-449b-5p, those had low levels of microRNA-449b-5p had worse prognosis (Figure 1D). Through analyzing the follow-up data of OS patients, microRNA-449b-5p level was identified to be correlated to tumor grade and histological differentiation of OS patients (Table 1). These results indicate that microRNA-449b-5p may be a prognostic hallmark of OS.
Figure 1.
Downregulated miR-449b-5p in OS. (A) Relative levels of miR-449b-5p in OS tissues and paracancerous tissues. (B) Relative levels of miR-449b-5p in OS patients in stage II+III and stage I. (C) Relative levels of miR-449b-5p in OS patients in G1 and G2. (D) Kaplan-Meier curves showing overall survival of OS patients with high and low levels of miR-449b-5p.
Table 1.
Correlation between miR-449b-5p level and pathological indexes of osteosarcoma patients (n=60).
| Clinicopathologic features | Number of cases | miR-449b-5p expression | P-value | |
|---|---|---|---|---|
| Low (n=30) | High (n=30) | |||
| Age (years) | 0.795 | |||
| ≤24 | 27 | 14 | 13 | |
| >24 | 33 | 16 | 17 | |
| Gender | 0.297 | |||
| Male | 26 | 11 | 15 | |
| Female | 34 | 19 | 15 | |
| Tumor size | 0.432 | |||
| ≤5 CM | 25 | 11 | 14 | |
| >5 CM | 35 | 19 | 16 | |
| Clinical stage | 0.028* | |||
| I | 20 | 6 | 14 | |
| II+III | 40 | 24 | 16 | |
| Histological differentiation | 0.003* | |||
| G1 | 23 | 6 | 17 | |
| G2 | 37 | 24 | 13 | |
Overexpression of microRNA-449b-5p suppressed the ability of OS cells to proliferate, migrate, and invade
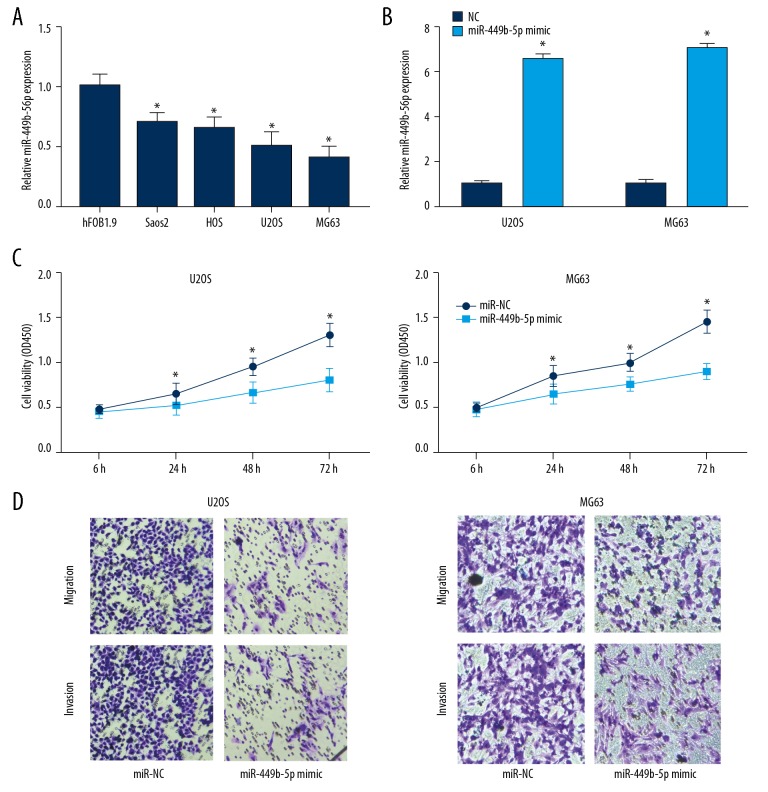
Identical to the expression pattern of microRNA-449b-5p in OS tissues, it was expressed at lower levels in OS cell lines than in osteoblasts (Figure 2A). U2OS and MG63 cells with a relatively low expression of microRNA-449b-5p were selected for the following experiments. The transfection efficacy of microRNA-449b-5p mimic was verified in OS cells (Figure 2B). CCK-8 assay revealed inhibited viability in OS cells overexpressing microRNA-449b-5p (Figure 2C). Moreover, migratory and invasive abilities were attenuated after transfection of microRNA-449b-5p mimic in U2OS and MG63 cells (Figure 2D).
Figure 2.
Overexpression of miR-449b-5p suppressed ability of OS cells to proliferate, migrate, and invade. (A) Relative levels of miR-449b-5p in osteoblasts and OS cell lines. (B) Transfection efficacy of miR-449b-5p mimic in U2OS and MG63 cells. (C) CCK-8 assay showed viability in U2OS and MG63 cells transfected with NC or miR-449b-5p mimic. (D) Transwell assay showing migration and invasion abilities in U2OS and MG63 cells transfected with NC or miR-449b-5p mimic.
C-Met is the downstream target of microRNA-449b-5p
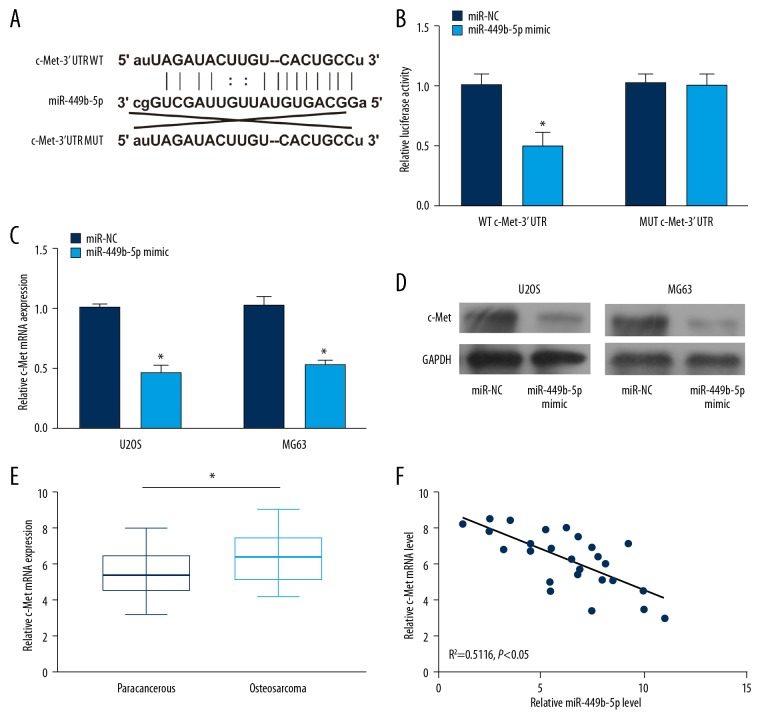
Through TargetScan prediction, potential binding sequences of c-Met to microRNA-449b-5p were identified (Figure 3A). Luciferase activity was markedly decreased in cells co-transfected with microRNA-449b-5p mimic and WT c-Met 3′UTR, suggesting the binding relationship between microRNA-449b-5p and c-Met (Figure 3B). After transfection of microRNA-449b-5p mimic in U2OS and MG63 cells, the protein level of c-Met was markedly downregulated (Figure 3C, 3D). A negative correlation was observed between expression levels of microRNA-449b-5p and c-Met in OS, further confirming that microRNA-449b-5p can bind to c-Met and negatively regulate its level. By detecting c-Met levels in OS tissues, we found they were higher in OS tissues than in paracancerous tissues (Figure 3E, 3F).
Figure 3.
C-Met was the target gene of miR-449b-5p. (A) Potential binding sequences between miR-449b-5p and c-Met. (B) Luciferase activity in cells co-transfected with WT c-Met 3′UTR/MUT c-Met 3′UTR and miR-449b-5p mimic/NC. (C) mRNA levels of c-Met in U2OS and MG63 cells transfected with NC or miR-449b-5p mimic. (D) Protein levels of c-Met in U2OS and MG63 cells transfected with NC or miR-449b-5p mimic. (E) Relative levels of c-Met in OS tissues and paracancerous tissues. (F) Negative correlation between c-Met and miR-449b-5p in OS tissues.
By targeting c-Met, microRNA-449b-5p suppressed the ability of OS cells to proliferate, migrate, and invade
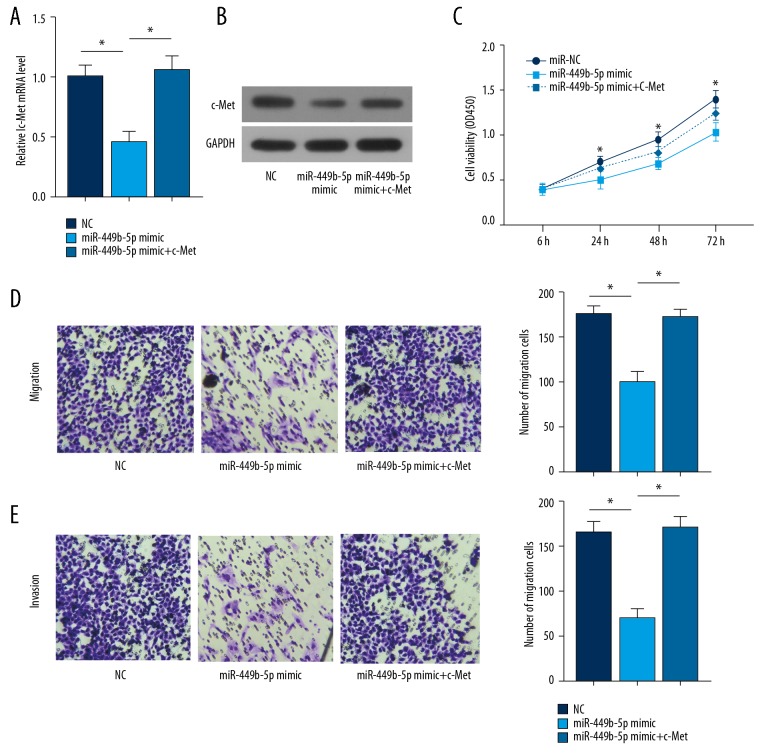
To clarify the involvement of microRNA-449b-5p/c-Met in the progression of OS, rescue experiments were performed. Transfection of microRNA-449b-5p mimic in OS cells markedly downregulated c-Met levels, which was reversed by overexpression of c-Met (Figure 4A, 4B). Overexpression of microRNA-449b-5p inhibited the viability of OS cells, but was reversed by transfection of pcDNA-c-Met (Figure 4C). Similarly, the inhibited migratory and invasive abilities of OS cells overexpressing microRNA-449b-5p were reversed by overexpression of c-Met (Figure 4D). These results suggest that microRNA-449b-5p suppresses the cellular behaviors of OS cells by downregulating c-Met.
Figure 4.
miR-449b-5p suppressed the ability of OS cells to proliferate, migrate, and invade by targeting c-Met. U2OS cells were transfected with NC, miR-449b-5p mimic, or miR-449b-5p mimic + pcDNA-c-Met. (A) Relative mRNA levels of c-Met in each group. (B) Relative protein levels of c-Met in each group. (C) CCK-8 assay showing the viability in each group. (D) Transwell assay showing migration and invasion ability in each group.
Discussion
OS is a malignant tumor that has the potential of unlimited proliferation and evading growth suppressors [21]. OS is prone to metastasis at an early stage and is highly invasive. Prevention and control of proliferation and metastasis of OS are important aspects of OS research. This study identified that microRNA-449b-5p was downregulated in OS. Low levels of microRNA-449b-5p were closely related to advanced tumor stage, histological differentiation, and poor overall survival. Our data suggest the potential of microRNA-449b-5p as a therapeutic target for OS.
miRNAs are single-stranded, small-molecule RNAs that accounts for about 1% of the human genome. miRNAs are highly conserved and participate in diverse cellular activities [22,23]. The regulatory effects of miRNAs on tumors, serving as oncogenes or tumor suppressors, have been well documented [24]. A growing number of studies have demonstrated the crucial involvement of miRNAs in the progression of OS. Xie et al. [25] found that miR-876-5p can inhibit proliferation, migration, and invasion abilities of OS cells by targeting c-Met. Fan et al. [26] found that miRNA-145 downregulates VEGF levels, thereby inhibiting the local invasion and metastasis of OS cells. MicroRNA-449b-5p is reported to exert a carcinogenic role in multiple types of tumors [18]. In this study, overexpression of microRNA-449b-5p in U2OS and MG63 cells attenuated abilities of cells to proliferate, migrate, and invade.
C-Met belongs to the RTKs family, which is mainly expressed in epithelial cells. Serving as an oncogene, c-Met activates multiple pathways and amplifies the transduction cascade to further mediate tumor cell behaviors [27]. C-Met is also considered as predictive marker of prognosis of tumor patients [28–30]. In this study, c-Met was upregulated in OS tissues and cell lines. Through TargetScan prediction and dual-luciferase reporter gene verification, c-Met was identified to be the direct target of microRNA-449b-5p. C-Met was negatively correlated with microRNA-449b-5p in OS. Previous studies have shown that c-Met regulates the progression of OS by mediating multiple miRNAs [31–33]. In this study, overexpression of c-Met reversed the inhibitory effects of microRNA-449b-5p on behaviors of OS cells. We found that microRNA-449b-5p suppressed proliferative, migratory, and invasive abilities of OS cells by targeting c-Met.
Conclusions
MicroRNA-449b-5p is downregulated in OS, which attenuates the malignant progression of OS by targeting c-Met.
Footnotes
Source of support: The National Natural Science Foundation of China (Grant Nos. 81802054 and 81541093), the Natural Science Foundation of Shandong Province, China (Grant Nos. ZR2015HL075), and A Project of Shandong Province Higher Educational Science and Technology Program (J17KA252)
Conflict of interest
None.
References
- 1.Chen Z, Zhao G, Zhang Y, et al. miR-199b-5p promotes malignant progression of osteosarcoma by regulating HER2. J BUON. 2018;23:1816–24. [PubMed] [Google Scholar]
- 2.Zhang ZF, Li GR, Cao CN, et al. MicroRNA-1294 targets HOXA9 and has a tumor suppressive role in osteosarcoma. Eur Rev Med Pharmacol Sci. 2018;22:8582–88. doi: 10.26355/eurrev_201812_16621. [DOI] [PubMed] [Google Scholar]
- 3.Li W, Zhang S. Survival of patients with primary osteosarcoma and lung metastases. J BUON. 2018;23:1500–4. [PubMed] [Google Scholar]
- 4.Serlo J, Tarkkanen M, Sampo M, et al. Incidence, treatment and survival of paediatric patients with bone sarcomas in Finland from 1991 to 2005. Acta Paediatr. 2015;104:738–45. doi: 10.1111/apa.12986. [DOI] [PubMed] [Google Scholar]
- 5.Friebele JC, Peck J, Pan X, et al. Osteosarcoma: A meta-analysis and review of the literature. Am J Orthop (Belle Mead NJ) 2015;44:547–53. [PubMed] [Google Scholar]
- 6.He X, Gao Z, Xu H, et al. A meta-analysis of randomized control trials of surgical methods with osteosarcoma outcomes. J Orthop Surg Res. 2017;12:5. doi: 10.1186/s13018-016-0500-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lampe L, Levashina EA. The role of microRNAs in Anopheles biology-an emerging research field. Parasite Immunol. 2017;39(2) doi: 10.1111/pim.12405. [DOI] [PubMed] [Google Scholar]
- 8.Eckstein F. Small non-coding RNAs as magic bullets. Trends Biochem Sci. 2005;30:445–52. doi: 10.1016/j.tibs.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 9.Wu W, Sun M, Zou GM, Chen J. MicroRNA and cancer: Current status and prospective. Int J Cancer. 2007;120:953–60. doi: 10.1002/ijc.22454. [DOI] [PubMed] [Google Scholar]
- 10.Ueda T, Volinia S, Okumura H, et al. Relation between microRNA expression and progression and prognosis of gastric cancer: A microRNA expression analysis. Lancet Oncol. 2010;11:136–46. doi: 10.1016/S1470-2045(09)70343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chistiakov DA, Chekhonin VP. Contribution of microRNAs to radio- and chemoresistance of brain tumors and their therapeutic potential. Eur J Pharmacol. 2012;684:8–18. doi: 10.1016/j.ejphar.2012.03.031. [DOI] [PubMed] [Google Scholar]
- 12.Chen CZ. MicroRNAs as oncogenes and tumor suppressors. N Engl J Med. 2005;353:1768–71. doi: 10.1056/NEJMp058190. [DOI] [PubMed] [Google Scholar]
- 13.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baumhoer D, Zillmer S, Unger K, et al. MicroRNA profiling with correlation to gene expression revealed the oncogenic miR-17-92 cluster to be up-regulated in osteosarcoma. Cancer Genet. 2012;205:212–19. doi: 10.1016/j.cancergen.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 15.Zhu J, Feng Y, Ke Z, et al. Down-regulation of miR-183 promotes migration and invasion of osteosarcoma by targeting Ezrin. Am J Pathol. 2012;180:2440–51. doi: 10.1016/j.ajpath.2012.02.023. [DOI] [PubMed] [Google Scholar]
- 16.Novello C, Pazzaglia L, Conti A, et al. p53-dependent activation of microRNA-34a in response to etoposide-induced DNA damage in osteosarcoma cell lines not impaired by dominant negative p53 expression. Plos One. 2014;9:e114757. doi: 10.1371/journal.pone.0114757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao ZQ, Shen Z, Huang WY. MicroRNA-802 promotes osteosarcoma cell proliferation by targeting p27. Asian Pac J Cancer Prev. 2013;14:7081–84. doi: 10.7314/apjcp.2013.14.12.7081. [DOI] [PubMed] [Google Scholar]
- 18.Jiang J, Yang X, He X, et al. MicroRNA-449b-5p suppresses the growth and invasion of breast cancer cells via inhibiting CREPT-mediated Wnt/beta-catenin signaling. Chem Biol Interact. 2019;302:74–82. doi: 10.1016/j.cbi.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 19.Zhao Z, Ma X, Shelton SD, et al. A combined gene expression and functional study reveals the crosstalk between N-Myc and differentiation-inducing microRNAs in neuroblastoma cells. Oncotarget. 2016;7:79372–87. doi: 10.18632/oncotarget.12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhen L, Yun-Hui L, Hong-Yu D, et al. Long noncoding RNA NEAT1 promotes glioma pathogenesis by regulating miR-449b-5p/c-Met axis. Tumour Biol. 2016;37:673–83. doi: 10.1007/s13277-015-3843-y. [DOI] [PubMed] [Google Scholar]
- 21.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 22.Cui N, Hao G, Zhao Z, et al. MicroRNA-224 regulates self-renewal of mouse spermatogonial stem cells via targeting DMRT1. J Cell Mol Med. 2016;20:1503–12. doi: 10.1111/jcmm.12838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He Y, Li HB, Li X, et al. miR-124 promotes the growth of retinal ganglion cells derived from muller cells. Cell Physiol Biochem. 2018;45:973–83. doi: 10.1159/000487292. [DOI] [PubMed] [Google Scholar]
- 24.Rupaimoole R, Calin GA, Lopez-Berestein G, Sood AK. miRNA deregulation in cancer cells and the tumor microenvironment. Cancer Discov. 2016;6:235–46. doi: 10.1158/2159-8290.CD-15-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie W, Xiao J, Wang T, et al. MicroRNA-876-5p inhibits cell proliferation, migration and invasion by targeting c-Met in osteosarcoma. J Cell Mol Med. 2019;23(5):3293–301. doi: 10.1111/jcmm.14217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fan L, Wu Q, Xing X, et al. MicroRNA-145 targets vascular endothelial growth factor and inhibits invasion and metastasis of osteosarcoma cells. Acta Biochim Biophys Sin (Shanghai) 2012;44:407–14. doi: 10.1093/abbs/gms019. [DOI] [PubMed] [Google Scholar]
- 27.Rothenberger NJ, Stabile LP. Hepatocyte growth factor/c-Met signaling in head and neck cancer and implications for treatment. Cancers (Basel) 2017;9(4) doi: 10.3390/cancers9040039. pii: E39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Samson P, Lockhart AC. Biologic therapy in esophageal and gastric malignancies: Current therapies and future directions. J Gastrointest Oncol. 2017;8:418–29. doi: 10.21037/jgo.2016.11.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu CW, Wang WX, Wu MJ, et al. Comparison of the c-MET gene amplification between primary tumor and metastatic lymph nodes in non-small cell lung cancer. Thorac Cancer. 2017;8:417–22. doi: 10.1111/1759-7714.12455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Refaat T, Donnelly ED, Sachdev S, et al. c-Met overexpression in cervical cancer, a prognostic factor and a potential molecular therapeutic target. Am J Clin Oncol. 2017;40:590–97. doi: 10.1097/COC.0000000000000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X, Sun X, Wu J, Li Z. MicroRNA-613 suppresses proliferation, migration and invasion of osteosarcoma by targeting c-MET. Am J Cancer Res. 2016;6:2869–79. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Niu G, Li B, Sun J, Sun L. miR-454 is down-regulated in osteosarcomas and suppresses cell proliferation and invasion by directly targeting c-Met. Cell Prolif. 2015;48:348–55. doi: 10.1111/cpr.12187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan K, Gao J, Yang T, et al. MicroRNA-34a inhibits the proliferation and metastasis of osteosarcoma cells both in vitro and in vivo. PLoS One. 2012;7:e33778. doi: 10.1371/journal.pone.0033778. [DOI] [PMC free article] [PubMed] [Google Scholar]