For close to 100 years, Chlamydomonas reinhardtii has been used as a reference organism for exploring cilia and organelle function, photosynthesis, and metabolism.
Abstract
The unicellular alga Chlamydomonas reinhardtii is a classical reference organism for studying photosynthesis, chloroplast biology, cell cycle control, and cilia structure and function. It is also an emerging model for studying sensory cilia, the production of high-value bioproducts, and in situ structural determination. Much of the early appeal of Chlamydomonas was rooted in its promise as a genetic system, but like other classic model organisms, this rise to prominence predated the discovery of the structure of DNA, whole-genome sequences, and molecular techniques for gene manipulation. The haploid genome of C. reinhardtii facilitates genetic analyses and offers many of the advantages of microbial systems applied to a photosynthetic organism. C. reinhardtii has contributed to our understanding of chloroplast-based photosynthesis and cilia biology. Despite pervasive transgene silencing, technological advances have allowed researchers to address outstanding lines of inquiry in algal research. The most thoroughly studied unicellular alga, C. reinhardtii, is the current standard for algal research, and although genome editing is still far from efficient and routine, it nevertheless serves as a template for other algae. We present a historical retrospective of the rise of C. reinhardtii to illuminate its past and present. We also present resources for current and future scientists who may wish to expand their studies to the realm of microalgae.
WHY ALGAE (IN GENERAL) AND CHLAMYDOMONAS (SPECIFICALLY)?
Algae represent a very large and diverse polyphyletic group of photosynthetic eukaryotes (Blaby-Haas and Merchant, 2019). They occupy all possible ecological niches on the planet, and therefore constitute a potential reservoir of untapped functional capabilities for adaptation to the environment. Algae are primary producers that contribute ∼50% of total carbon fixation worldwide (Field et al., 1998), which makes their study fundamental to our understanding of global primary production and carbon flux. They also offer a low-cost option for the large-scale production of high-value molecules, since algae only require water, salts, air, and light. Unicellular algae such as the ciliated green alga Chlamydomonas reinhardtii offer high signal-to-noise during experiments due to the ease of growth in controlled medium and environments (temperature and light regimes) and the homogenous nature of the cultures, and they grow much more rapidly than classic plant models.
With its haploid genome, C. reinhardtii is well suited for classical genetics, as loss-of-function mutations are immediately expressed and more likely lead to observable phenotypes compared with diploid organisms. C. reinhardtii not only responds to light, but it also anticipates environmental transitions like dawn and dusk under the supervision of a circadian system, which coordinates cell division, photosynthesis, and cilia biogenesis, representing three fundamental research topics (Noordally and Millar, 2015). Other topics of research using C. reinhardtii include the carbon-concentrating mechanism (CCM) and in situ structure determination of protein complexes by electron cryotomography. Responses to excess light and the dissipation of light energy to prevent cellular damage are another research avenue that has benefitted enormously from the analysis of C. reinhardtii. Additional research questions being addressed by the study of C. reinhardtii focus on metabolism (starch, nitrogen, amino acids, sulfur, phosphorus, and metal micronutrients), biosynthetic pathways (glycerolipids, chlorophyll, hemes, and carotenoids), anoxia, thioredoxins, chaperones and proteases, and the control of chloroplast gene expression (Harris, 2008). Inside its 2000 pages and three volumes, The Chlamydomonas Sourcebook encompasses major research topics, history, and methodology (Harris, 2008). We will feature selected examples below where C. reinhardtii has played a key role before examining the events that promoted this alga to the forefront.
SELECTED RESEARCH HIGHLIGHTS
Cell Cycle and Cell Division
The eukaryotic cell cycle machinery was largely deciphered in budding yeast (Saccharomyces cerevisiae) using synchronized cultures. Despite conservation of the underlying proteins (for example, cyclins, cyclin-dependent kinases, and the anaphase-promoting complex), budding yeast is evolutionarily distant from algae and plants. C. reinhardtii makes an excellent photosynthetic system whose progression through the cell cycle can be synchronized by daily light-dark cycles, as is common for many algae: cells enter G1 during the day, followed by the S and M phases around dusk. This separation of phase along the diurnal cycle provides a predictable temporal cascade that starts with cell growth (fueled by photosynthesis), followed by the commitment to divide, resorption of cilia, doubling of DNA and histone contents, mitosis, and the growth of new cilia. With the help of robotics and semiautomated imaging, it is possible to identify temperature-sensitive mutants affected in some aspect of cell cycle progression, and whole-genome sequencing can be used to pinpoint the causal mutation. So far, ∼150 mutants have been identified with a defect in division or exit from G1 phase (Tulin and Cross, 2014; Breker et al., 2018). These mutants define genes with important roles in the cell cycle that may be applicable to plants as well, as ∼75% of these genes have clear homologs in Arabidopsis (Arabidopsis thaliana). A clear challenge is to understand the role of each cell cycle gene, but looking at their expression windows over the diurnal cycle should help narrow down the options (Zones et al., 2015; Strenkert et al., 2019).
Chloroplast Biogenesis and Photosynthesis
C. reinhardtii is a facultative autotroph that can handle mutations in the photosynthetic apparatus if grown in the presence of a reduced carbon source such as acetate, offering a number of advantages when studying chloroplast biogenesis and function. Mutants with defects in the light reactions can be specifically enriched by the addition of the bactericidal agent metronidazole. Also known as Flagyl, this compound is reduced to its toxic form by ferredoxin; only cells that cannot reduce ferredoxin survive in its presence (Schmidt et al., 1977). Through classic genetic screens, acetate-requiring (ac) photosynthetic mutants were isolated based on the requirement for acetate as a reduced carbon source (hence the name acetate requiring) or increased chlorophyll fluorescence, a consequence of disrupted electron transport (Levine, 1960a; Bennoun and Levine, 1967; Bennoun et al., 1980). Chlorophyll fluorescence reflects the progression of electron flow through the electron transport chain: its blockage increases fluorescence, as light energy can no longer be used for photochemistry. The same fluorescence assay provides a powerful means to determine allelism and genomic rescue when identifying the underlying causal loci. In land plants, mutants in the homologous genes also display high chlorophyll fluorescence but are generally seedling-lethal and must be maintained as a segregating stock (Maiwald et al., 2003). Homozygous mutant plants can, however, survive on medium supplemented with sucrose, which is taken up by dedicated transporters (missing in Chlamydomonas).
Early evidence for nucleus- and plastid-encoded components of the photosynthetic apparatus came from radiolabeling C. reinhardtii cells blocked selectively in cytosolic or plastid translation (Chua and Gillham, 1977; Delepelaire, 1983; Merchant and Selman, 1984). Pulse-chase labeling of cells with 35S in the presence or absence of a plastid translation inhibitor also revealed that the small subunit of Rubisco (translated in the cytoplasm) is rapidly degraded when the large subunit is missing (Schmidt and Mishkind, 1983), mirroring observations collected on photosystem II (PSII) proteins in the absence of plastid-encoded D1 and D2 proteins (Erickson et al., 1986; Jensen et al., 1986). The control of epistasy of synthesis (CES) model describes how the translation potential of individual components of a photosynthetic protein complex depends on the presence of the other constituents (Choquet and Wollman, 2002).
The genetic dissection of photosynthesis owes much to the collection of ac mutants from Levine, followed by new mutants from career-long screens from the Rochaix and Wollman laboratories. ac mutants were used to define nuclear loci that control chloroplast transcription, RNA stability, and translation. Analysis of some mutants provided experimental validation for trans-splicing of the plastid-encoded psaA gene (Kück et al., 1987; Choquet et al., 1988) , which is transcribed as three separate RNAs that are later spliced together via the concerted action of at least 14 nucleus-encoded factors. Coordination between nucleus- and plastid-encoded subunits of photosynthetic complexes and enzymes is critical for proper chloroplast function. For example, the nucleus-encoded NAC2 protein is an RNA binding protein with tetratricopeptide-like repeats that stabilizes psbD RNA (Boudreau et al., 2000). Such RNA binding proteins exhibit sequence specificity for a given chloroplast RNA and are encoded by large gene families in land plants that have not yet expanded in algae (Jalal et al., 2015).
Photosynthesis and State Transitions
Analyses of other mutants have led to the genetic dissection of state transitions. At their core, light-dependent reactions drive electron flow from water to NADP+ via PSII and PSI while generating a proton gradient via proton transfers at multiple steps. Under conditions referred to as state 1, each photosystem receives photons from its cognate chlorophyll binding antenna proteins: Light Harvesting Complex I (LHCI) or LHCII for PSI and PSII, respectively. When PSII becomes too excited, reduced plastoquinone accumulates, resulting in the phosphorylation of LHCII proteins and their subsequent dissociation from PSII and association with PSI: this is known as state 2. Healthy cells continuously shuttle LHCII proteins between PSII and PSI complexes as a function of light quality and quantity. State transitions balance the distribution of excitation energy, allowing the modulation of NADPH and ATP production.
Although state transitions were first discovered in other organisms, the biochemical underpinnings of this process were elucidated in C. reinhardtii, wherein as much as 80% of LHCII dissociates from PSII, in contrast to ∼20% in land plants (Delosme et al., 1996). State transitions require cytochrome (Cyt) b6f to sense the redox status of the plastoquinone pool, as a Cyt b6f mutant is locked in state 1 and primarily accumulates unphosphorylated LHCII proteins (Wollman and Lemaire, 1988). The transition to state 2 involves the (reversible) phosphorylation of LHCII proteins (Wollman and Delepelaire, 1984). Mutants lacking state transitions display identical chlorophyll fluorescence whether the plastoquinone pool is highly oxidized (state 1) or reduced (state 2). In the stt7 mutant (for state-transition deficient), LHCII proteins are only found in their nonphosphorylated form. The stt7 mutant can still grow in the absence of acetate, albeit at a slower rate, indicating that state transitions contribute to fitness (Fleischmann et al., 1999). STT7 encodes the kinase responsible for LHCII phosphorylation (Depège et al., 2003). The interaction of Cyt b6f with STT7 is promoted by reduced plastoquinone and results in the autophosphorylation and activation of the kinase. STT7 is then released from Cyt b6f and diffuses to PSII within the thylakoid membrane to phosphorylate associated LHCII proteins, thereby initiating state 2 (Dumas et al., 2017). The STT7 homolog in Arabidopsis, STN7, also catalyzes LHCII phosphorylation in response to the redox status of plastoquinone, and the stn7 mutant shows growth defects under a fluctuating light regime, which more accurately mimics real-life growth conditions (Bellafiore et al., 2005). Therefore, the dissection of a process in C. reinhardtii is applicable to plants. This process may allow an increase in photosynthetic yields by better recycling excess light photon energy away from dissipation.
Photosynthesis and the CCM
Oxygen and carbon dioxide compete for the active site of Rubisco, causing a decrease in photosynthetic efficiency. Rubisco evolved under low-oxygen conditions, but oxygenic photosynthesis altered the surrounding environment. Biological innovations increased local relative CO2 concentrations to enhance photosynthetic performance. These innovations include carboxysomes in cyanobacteria, C4 metabolism in land plants, and the CCM in C. reinhardtii and other algae. A functional CCM complex starts with the uptake of bicarbonate from the environment, its delivery to the pyrenoid (a site of concentrated Rubisco), and its conversion to CO2 by carbonic anhydrases. The zinc-finger transcription factor CIA5/CCM1 was identified from genetic screens for mutants lacking the pyrenoid and unable to adapt to low CO2 levels. CIA5/CCM1 orchestrates most transcriptional responses to variations in CO2 levels (Fang et al., 2012). The putative membrane-bound transporters responsible for inorganic carbon uptake, the carbonic anhydrases, and the transcriptional network controlled by inorganic carbon availability were identified through comparative transcriptome analysis of cells grown under low and high CO2 concentrations and through genetic screens (Jungnick et al., 2014; Memmola et al., 2014; Wang et al., 2015b). In addition, the biochemical identification of proteins in purified pyrenoid components was validated genetically (Mackinder et al., 2016; Zhan et al., 2018). Most CCM components were tentatively arranged along the bicarbonate chemical gradient created within the cell (from the cell wall to the pyrenoid matrix) based on their localization (Mackinder et al., 2017). Surprisingly, proteins can localize to distinct layers and puncta within the pyrenoid, in addition to the typical matrix localization of Rubisco and the Rubisco binding Essential Pyrenoid Component 1 (EPYC1). Time-course observations of the pyrenoid during cell division solved its mode of inheritance: most daughter cells acquire their pyrenoid by elongation and fission of the mother cell’s pyrenoid (Freeman Rosenzweig et al., 2017). The direct observation of the Rubisco small subunit, RBCS1, fused to YFP also resolved the organization of the pyrenoid matrix as a liquid whose contents can mix internally, unlike the more rigid cyanobacterial carboxysomes (Freeman Rosenzweig et al., 2017). The introduction of the Rubisco linker protein EPYC1 into plants (or a version that can interact with plant Rubisco) may be an important step toward the engineering of plants to form pyrenoids to enhance photosynthetic output.
Channelrhodopsins and Optogenetics
For a unicellular organism, C. reinhardtii possesses a surprisingly large number of putative photoreceptors, including 12 rhodopsins that bind retinal (Greiner et al., 2017). As suggested by their names, channelrhodopsins (ChRs) behave as light-activated ion channels to conduct protons and sodium cations across the membrane, normally to elicit cilia movement. Chlamydomonas ChRs decrease the local membrane potential in Chlamydomonas and after transient or stable transformation of the genetically encoded photoreceptor in cell cultures, nematodes, fruit flies (Drosophila melanogaster), and mammals via a method known as optogenetics (Deisseroth and Hegemann, 2017). The chromophore for ChRs, all-trans-retinal, is widespread from algae to mammals, although the feeding of retinal analogs can shift wavelength sensitivity (Foster et al., 1984).
When expressed in neurons and activated by light, ChRs elicit neuronal firing only in the cell types and within the membrane domain where they are expressed. Genetic engineering applied to ChRs created variants in the duration of the channel’s open state, which drive longer neuronal activation upon stimulation by blue light and can be closed on command using green or yellow light pulses. Genome mining added to the optogenetics molecular toolkit, yielding rhodopsins that respond maximally to other wavelengths or cause hyperpolarization of the plasma membrane, resulting in the inhibition of neuronal impulses. Together with the technical innovations that supported the precise delivery of light pulses to single cells, optogenetics has opened up new avenues in the neurosciences by disturbing neural excitation and observing behavioral consequences, therefore going much beyond the simple recording of neuronal activity (Zhang et al., 2007; Rost et al., 2017).
Cilia Biogenesis and Function
C. reinhardtii uses cilia for phototaxis to optimize light exposure during photosynthesis and for cell-cell recognition during mating. It became clear during early mutant screens that cilia are dispensable in this organism in a laboratory setting. The same cannot be said for most eukaryotic systems that have retained cilia; mutations in human ciliary proteins lead to diseases (referred to as ciliopathies) that include male infertility, kidney and eye malfunction, polydactyly, and brain disorders. Fungi (including budding yeast) and seed plants have lost genes for cilia, but the overall structure of cilia is well conserved between C. reinhardtii and other ciliated eukaryotes.
The biogenesis of C. reinhardtii cilia has been dissected genetically, biochemically, and microscopically over the past several decades. Transmission electron micrography analysis of wild-type and mutant cilia helped identify affected structures associated with various motility defects in mutants (Goodenough and Heuser, 1982, 1985a). A comparison of the protein complement within the cilia core (the axoneme) between wild type and various paralyzed flagella (pf) mutants can provide clues about function. Perhaps unique to C. reinhardtii is the analysis of dikaryons: newly fused gametes exist for several hours as tetraflagellate diploids whose cytoplasm is derived from two genotypes, allowing the study of rescue or complementation of mutant ciliary phenotypes (Dutcher, 2014). Intact cilia detach from cells following a pH shock or mechanical shearing, lending themselves to biochemical analysis: the cilium proteome is composed of over 1000 proteins, as identified by mass spectrometry (Pazour et al., 2005). Based on genome sequences, proteins found in ciliates (animals, C. reinhardtii, Caenorhabditis elegans) but missing in fungi and plants were compiled in the “CiliaCut” list, consisting of ∼200 proteins (Li et al., 2004; Merchant et al., 2007), over half of which were detected in the cilium proteome. One major task will be to associate each protein with a function in cilia biogenesis and control, which will be aided by the availability of large insertion mutant libraries (Li et al., 2016; Cheng et al., 2017b).
Cilia grow from their tips, but their structural constituents are synthesized in the cytosol and must therefore be transported to the growing tip. Using differential interference contrast microscopy, researchers in Rosenbaum’s laboratory noticed particles moving up and down the length of cilia between the membrane and doublet microtubules, which were most readily visualized in mutants with paralyzed cilia (Kozminski et al., 1993). This intraflagellar transport (IFT) system was first described and characterized in C. reinhardtii (Piperno and Mead, 1997). Concurrently, technical advances in microscopy allowed the direct visualization of nonmotile (or primary) cilia in mammals, and a mouse (Mus musculus) model was developed for polycystic kidney disease using an insertional mutant (Moyer et al., 1994; Satir, 2017). The gene disrupted in the mouse model has one C. reinhardtii homolog, encoding a protein belonging to the IFT proteome: IFT88 (Pazour et al., 2000). ift88 mutants in C. reinhardtii and mouse have short or no flagella. The major polycystin proteins PC1 and PC2 were later found to localize to the primary cilia membranes of kidney cells after being transported there along the IFT system (Pazour et al., 2002).
In Situ Electron Cryotomography
By necessity, researchers tend to focus on a handful of genes to uncover the mechanisms controlling their expression and the functions of their products, with the ultimate goal of placing the encoded proteins into the (much larger) context of a living cell. C. reinhardtii cells are composed of several compartments, each with a specialized function that is reflected in its structure. While microscopic observations of fixed cells have revealed much, sample preparation may cause artifacts and distort the underlying structures. Protocols based on living cells face their own challenges and are limited to locating single molecules within the cell at the expense of cellular structures. Quick-freeze, deep-etch techniques afford a glimpse of the substructure of the axoneme but remain limited to the exposed surface (Goodenough and Heuser, 1982, 1985a). Recent advances in in situ electron cryotomography and computer-assisted 3D reconstruction bring the promise of imaging organellar structures and protein aggregates. In the early days, this technique was hampered by the dense cytoplasm of yeast cells, but it worked very well in C. reinhardtii, especially for the mat3 mutant, which is defective in the algal homolog of the RETINOBLASTOMA gene (Umen and Goodenough, 2001a). Smaller cells are indeed advantageous during vitrification (Albert et al., 2017). Flash-frozen cells still need to be reduced to sections of 300 nm or less for optimal resolution, a process currently attained via focused ion beam-mediated thinning of vitrified cells (Schaffer et al., 2017). In recent years, Engel and coworkers have successfully visualized the chloroplast and pyrenoid, Golgi apparatus, COPI-coated vesicles, proteasome, and nuclear pore complex of Chlamydomonas (Engel et al., 2015; Albert et al., 2017; Bykov et al., 2017; Freeman Rosenzweig et al., 2017). These recent advances highlight the potential for visualizing organellar networks in unmatched detail, as well as protein complexes of moderate size in their natural state and place. For instance, thylakoid membranes are connected to the pyrenoid via tubules embedded into the starch sheath surrounding the pyrenoid and may therefore provide an access route to deliver CO2 to the Rubisco enzymes concentrated inside the pyrenoid (Engel et al., 2015). These tubules also contain minitubules that are only wide enough for unidirectional trafficking, for example to deliver ATP to Rubisco activase and to export sugars from the pyrenoid after carbon fixation. Within the pyrenoid matrix, densely-packed Rubisco complexes were initially thought to assemble into hexameric ring-like structures (Mackinder et al., 2016). However, more recent higher-resolution imaging resolved Rubisco distribution within the matrix as liquid-like rose (Freeman Rosenzweig et al., 2017). .
While a protein complex is tentatively identified based on its 3D shape, its true identity requires the confirmation that the corresponding C. reinhardtii genes exist. To bridge the gap between the observation and identification of proteins, it should be possible to add a tag to a protein of interest that is small enough to maintain the overall shape and structures within the cell but bulky enough to be observed in tomograms.
HISTORY OF C. REINHARDTII
Early Observations
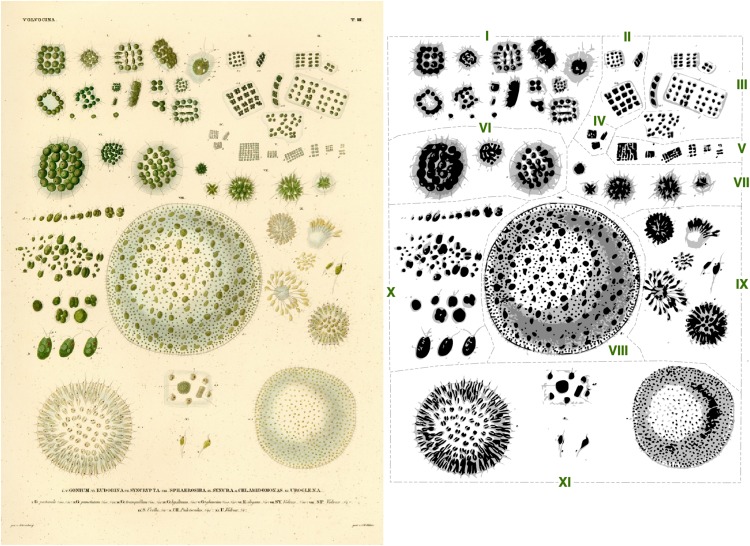
Where did this unicellular alga come from and how did it emerge as a reference organism? The Chlamydomonas genus was first mentioned in the 1830s, when the German naturalist Christian Ehrenberg drew Chlamydomonas and Volvox (Ehrenberg, 1838). Naturalists described the world around them, and Ehrenberg was truly prolific (Jahn, 1998). His Chlamidomonas [sic] drawings are rather underwhelming and do not foretell its scientific future (Figure 1). Ehrenberg’s slides and samples reside at the Museum für Naturkunde in Berlin (Lazarus and Jahn, 1998).
Figure 1.
Taxonomic Basis of Chlamydomonas and Volvox.
Ehrenberg’s drawings of Chlamydomonas and Volvox cells, published in 1838. Cells that belong to the same species are indicated by Roman numerals in the right panel. Reproduced with permission, Museum für Naturkunde, Berlin. I, Gonium pectorale; II, Gonium punctatum; III, Gonium tranquillum; IV, Gonium hyalinum; V, Gonium glaucum; VI, Eudorina elegans; VII, Syncrypta volvox; VIII, Sphaerosira volvox; IX, Synura uvella; X, Chlamidomonas pulvisculus; XI, Uroglena volvox. The species was identified as Chlamidomonas pulvisculus but renamed Chlamydomonas reinhardtii in 1888.
Early observations of unicellular algae illustrated two key properties: sexual reproduction and phototaxis. The basis of sexual reproduction was a major topic of inquiry and curiosity, and unicellular algae were a prime research subject, as all steps of the sexual cycle can be readily observed by examining isolates from soil or freshwater ponds under a microscope (Johnson, 1914). The observation of aggregation and fusion of gametes under a microscope offered early evidence that Chlamydomonas cells are present as two mating types. Likewise, Faminzin (1866) described phototaxis in Chlamydomonas and Euglena. The alga was renamed from Chlamydomonas pulvisculus (the species drawn by Ehrenberg) to C. reinhardtii by the French botanist Pierre Dangeard in honor of the Ukranian botanist Ludwig Reinhardt in 1888, and the new species name caught on.
This propelled Chlamydomonas to become a promising system to study sexual reproduction and phototaxis, possessing much of the convenience normally associated with microbes but with the added benefit of sexual dimorphism (Johnson, 1914). A report of phenotypic segregation in crosses between two Chlamydomonas species (Pascher, 1918) empirically confirmed Mendel’s segregation rules, and it introduced tetrad analysis as a powerful tool for the genetic dissection of segregation.
Early Genetics in Chlamydomonas
Chlamydomonas research first entered the spotlight in the 1930s with Franz Moewus’ biochemical genetics studies. Moewus proclaimed to have identified several sexual hormones related to carotenoids in the Chlamydomonas strain Chlamydomonas eugametos that activated male or female gametes selectively. He also reported the isolation and genetic mapping of mutants along their biosynthetic pathway. He described the analysis of 200,000 zygotes for 10 phenotypes scored over a 10-year period, which represents an enormous amount of work for anyone (Gowans, 1976). Unfortunately, most of his results could not be reproduced, even by his own hands, casting doubt on his scientific claims.
The influence of Franz Moewus on modern Chlamydomonas research is, nevertheless, undeniable (Sapp, 1987). First, Moewus demonstrated that Chlamydomonas mutants could be isolated. His results, however implausible, remarkably supported the correct view that genes are located on chromosomes and that genetic mutations could result in the loss of enzymatic activity (the one gene-one enzyme concept). Second, as discussed below, these results motivated Gilbert Morgan Smith to isolate (and later share) C. reinhardtii strains in order to repeat Moewus’ experiments.
The Modern C. reinhardtii Era and Gilbert Smith
Smith almost dropped out of high school and only discovered a keen interest in biology and botany when he was sent to a remedial academy. There, he gained enough confidence to enroll in college (Wiggins, 1962). During his Ph.D. work, he developed techniques to isolate pure cultures from field samples. He eventually moved to Stanford to become a Botany Professor until he retired in 1950. Smith was keen to repeat some of Moewus’ experiments about Chlamydomonas, which he even made the focus of his address as the retiring President of the Botanical Society of America (Smith, 1946). Smith spent a year cultivating and separating mixed cultures into pure isolates. Of the over 700 soil samples collected all over the United States, he isolated several Chlamydomonas colonies. He specifically isolated spores rather than haploid vegetative cells, thereby producing the mating pairs required for genetic analysis. One of the 15 species Smith collected is the modern C. reinhardtii strain now in use worldwide.
Smith was not the only biologist isolating sexually competent algae, but he was lucky. On the East Coast, Luigi Provasoli identified a pair of Chlamydomonas moewusii strains (Provasoli et al., 1951) and shared them with Ralph Lewin, who was himself interested in recombination and sexual inheritance. Lewin isolated the first mutants with paralyzed flagella (Lewin, 1954) but quickly discovered that C. moewusii was not suited for mutants with photosynthetic defects, being an obligate photoautotroph (Lewin, 1950). Luckily for the field, C. reinhardtii is a facultative photoautotroph, can survive without photosynthesis if provided a reduced carbon source such as acetate, and remains green in the dark. Photosynthetic mutants emerged from work by Paul Levine using the Smith strains (Levine, 1960a).
Sharing Is Caring
Smith was indeed generous and shared his pure algal cultures. (Moewus was not, perhaps for the better, although his strains did end up in various hands over the years and are now available in algal repositories as C. eugametos Moewus.) Smith sent mating pairs to a number of eager colleagues (Ruth Sager at Rockefeller University; Bill Ebersold, David Regnery, Bill Eversole, and Edward Tatum, all at Stanford; John N. Hartshorne at the University of Manchester). Smith and Regnery determined that the inheritance of sexuality in C. reinhardtii was exclusively heterothallic (Smith and Regnery, 1950). At first, these experiments were limited to the segregation of the mating locus, but soon Smith, Tatum, Ebersold, and Eversole accumulated mutants to serve as phenotypically tractable markers.
Mutants in nuclear genes segregated in the 2:2 ratio typical for a haploid genome, but some mutants showed a 4:0 segregation ratio, providing evidence for non-Mendelian segregation. This piqued the interest of Sager, who isolated mutants conferring resistance to streptomycin whose causal locus mapped to the chloroplast genome (Sager, 1954; Ohta and Sager, 1975). She also discovered nutritional control of sexuality in Chlamydomonas: vegetative cells starved for nitrogen (and of opposite mating types) will readily fuse to form zygotes that, after a few days in the dark, will undergo meiosis and germinate when plated onto nitrogen-replete medium in the light (Sager and Granick, 1954).
Around the same time, Paul Levine, a Drosophila geneticist wishing to study recombination, was convinced by Sager and Lewin to switch to C. reinhardtii (Goodenough, 2015). Levine set up over 250 crosses between 45 mutants gathered by Smith, Tatum, Eversole, and Ebersold (Levine and Ebersold, 1960) and published the first algal genetic maps (Ebersold and Levine, 1959; Ebersold et al., 1962).
The isolation of many mutants between 1950 and 1960 laid the foundation for cilia biology and photosynthesis research. Analysis of pf mutants with paralyzed flagella demonstrated that cilia are dispensable for viability in laboratory settings and offered the promise of genetic analysis. Similarly, the ac mutants consolidated C. reinhardtii’s position as a system of choice for photosynthetic research decades before Arabidopsis research took off. Levine posited that ac mutants were defective in some step of the photosynthetic process because they could not fix radioactive CO2, the prototypical mutant being ac-21 (Levine, 1960b). This strain carries a mutation in the chloroplast Rieske iron-sulfur protein PETC. Other genes identified from the ac screen include those encoding plastocyanin (ac-208), factors involved in the biosynthesis of cytochromes b (ac-141) and f (ac-206) from the cytochrome b6f complex, and factors involved in the biosynthesis of the PSII D2 protein (ac-115; Harris, 2008). All mutants were isolated in the strain CC-124 and CC-125 backgrounds (originally named 137c until they were deposited at the Chlamydomonas Resource Center; Table 1), one of the mating pairs from Smith.
Table 1. Community Resources and Biological Repositories for Chlamydomonas Research.
Overcoming Limitations to Transformation and Transgene Expression
While the 1960s and 1970s were a fertile period for C. reinhardtii research, the 1980s witnessed the introduction of recombinant DNA, allowing the cloning and propagation of genes in bacteria, as well as DNA sequencing, paving the way for the molecular characterization of classical mutants. The functional rescue of a mutant via the introduction of an intact copy of the presumptive causal gene demands transformation protocols, which were developed fairly early on for yeast (Hinnen et al., 1978) and Drosophila (Rubin and Spradling, 1982) but initially seemed out of reach for the Chlamydomonas community, possibly due to poor penetrance of selection markers. The unusually high GC content of the Chlamydomonas genome (Chiang and Sueoka, 1967a) was thought to contribute to the poor expression of some selectable markers (Rochaix and van Dillewijn, 1982; Cerutti et al., 1997), but the expression of foreign genes with matching GC content fared just as poorly (Stevens et al., 1996), and the number of confirmed transformants was always low (Debuchy et al., 1989; Kindle et al., 1989; Diener et al., 1990; Mayfield and Kindle, 1990).
A major breakthrough in Chlamydomonas nuclear transformation came about in 1990, with the development of a modified yeast transformation protocol based on agitation of cells in the presence of glass beads coated with DNA (Kindle, 1990). Previous transformation methods had necessitated the use of biolistic gene guns to cross the cell wall and deliver DNA to the nucleus. A critical innovation was the weakening of cell wall strength to facilitate the entry of DNA into the cell, either genetically or following treatment with autolysin, a hydrolase secreted by gametes during mating that digests the cell wall. Electroporation was later applied to any strain with efficiencies equal to or higher than the glass bead method (Shimogawara et al., 1998). Organellar transformation was achieved with biolistics (Boynton et al., 1988; Remacle et al., 2006) and was instrumental in the dissection of plastid and mitochondrial gene function; in this case, transgenes are not silenced and are expressed at high levels.
Sequencing the algal genome in the 2000s (Merchant et al., 2007) introduced the community to whole-genome transcriptome profiling, an approach made even more powerful by the microbe-like nature of C. reinhardtii: unicellular, fast-growing, and very amenable to quick changes in growth medium composition or growth conditions.
Such is the history of the rise of C. reinhardtii as a model for green unicellular algae.
CHARACTERISTICS
Morphology
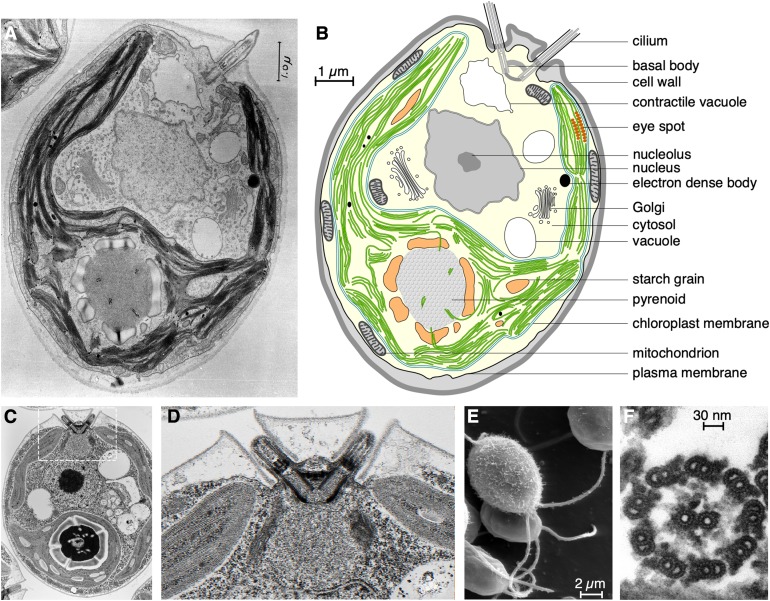
Since the 1800s, algal taxonomy has been based on distinguishing cellular features. C. reinhardtii is unicellular, oval-shaped, and measures ∼5 × 10 µm. The cell is surrounded by a cell wall composed of glycoproteins and carbohydrates, arranged in seven layers, as seen in quick-freeze, deep-etched samples (Goodenough and Heuser, 1985b). During the course of sexual reproduction, gametes shed their cell wall before fusion. Many laboratory strains carry cell wall (cw) mutations and lack cell walls but are fully viable, although they are perhaps more sensitive to some stressors and may be more difficult to cross due to their shorter cilia. Transmission electron microscopy best reveals the underlying cellular anatomy, as shown in Figures 2A and 2C and in the cartoon in Figure 2B. The cell is characterized by a pair of cilia of equal length located at one pole (Figures 2D and 2E). Chlamydomonas cilia (previously called flagella) are prototypes of motile animal cilia, and their axonemes show the same 9+2 symmetry consisting of nine microtubule pairs surrounding the central pair microtubules (Figure 2F). Close to the same pole, the cell accumulates vacuoles of various sizes, named contractile vacuoles, that control intracellular tonicity by expelling excess water that comes in through a passive gradient (as most Chlamydomonas algae are freshwater algae). These vacuoles are only dynamic under hypo-osmosis: small vacuoles fuse into a larger vacuole, which eventually fuses with the plasma membrane and releases its contents outside the cell (Luykx et al., 1997; Xu et al., 2016). The main osmolyte is thought to be potassium, while water molecules are brought into the contractile vacuole via aquaporin channels (Xu et al., 2016).
Figure 2.
Chlamydomonas Anatomy.
(A) Transmission electron micrograph (TEM) of a cell. Originally published by Ohad et al. (Ohad et al., 1967) and made available on the Cell Image Library website ( CIL:37252, C reinhardtii. CIL. Data set: https://doi.org/doi:10.7295/W9CIL37252).
(B) Drawing of a C. reinhardtii cell based on the TEM image in (A).
(C) and (D) Another TEM of a C. reinhardtii cell, showing a complete basal body (white box, enlarged in [D]), nucleus, and pyrenoid. Image courtesy of Dr. William Dentler (University of Kansas).
(E) Scanning electron micrograph of C. reinhardtii cells. Note the two cilia per cell.
(F) TEM of a cross section through isolated axonemes.
The images in (E) and (F) were generated at the Dartmouth College Rippel Electron Microscope Facility by Louisa Howard, Elizabeth Smith, and Erin Dymek (Smith and Lefebvre, 1996).
A single cup-shaped chloroplast occupies over half the cell volume (Sager and Palade, 1957). Starch granules are found between thylakoid membrane stacks and surrounding a spherical structure located at the pole opposite the cilia known as the pyrenoid. This structure is the site of CO2 fixation, a process facilitated by high local concentrations of the enzymes Rubisco, Rubisco activase, and EPYC1 (Mackinder et al., 2016; Freeman Rosenzweig et al., 2017). Finally, the eyespot is a specialized domain of the chloroplast membranes containing carotenoid granules. The eyespot reflects light back onto ChR-type photoreceptors located in the overlaying plasma membrane to control light-mediated flagellar movement (Eitzinger et al., 2015; Engel et al., 2015). The chloroplast envelope protein EYE2 connects pigment granules inside plastids to the cytoskeleton outside the organelle. In addition, EYE2 contributes to the positioning of the single eyespot to one side of the chloroplast so that cells can detect the direction and intensity of incoming light for phototactic movement (Boyd et al., 2011).
Vegetative Life Cycle
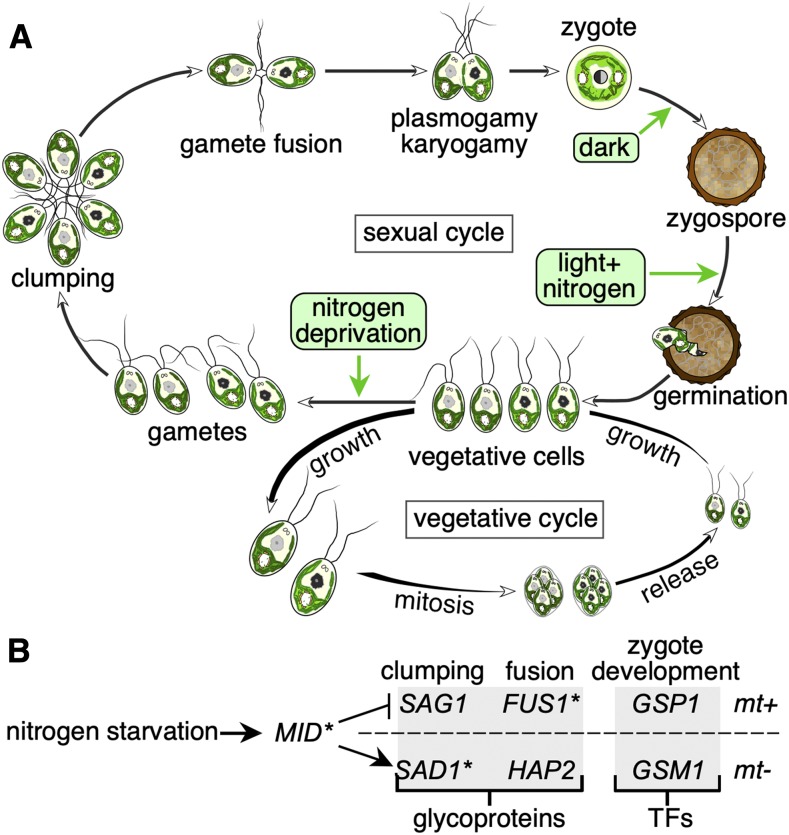
When resources are not limiting, Chlamydomonas multiplies asexually by simple mitotic division. Each daughter cell will carry the identical genetic information encoded by the nuclear, mitochondrial, and chloroplast genomes of its parent (Figure 3). The time to divide is dictated by two check-points during the G1 phase of the cell cycle and is based on cell size. The first check-point, termed “commitment,” is equivalent to “start” in budding yeast and marks the irreversible decision to divide at least once (Cross and Umen, 2015). The second check-point measures the volume of the mother cell and fixes the number of successive rounds of S phase and mitosis to produce daughter cells of equal size (Umen and Goodenough, 2001a). Therefore, like many chlorophyte algae, C. reinhardtii undergoes multiple fission divisions during mitosis, unlike budding yeast, where one daughter buds off the mother. Cells grown in constant light can divide every 8 to 10 h, but the cell cycle can be lengthened to 24 h by imposing daily light-dark periods. These conditions provide very good synchronization of cell populations and allow the biochemical, physiological, and molecular characterization of events leading to cell division (Chiang and Sueoka, 1967a, 1967b; Voigt and Münzner, 1987; Tulin and Cross, 2014; Zones et al., 2015). Cilia resorb before cell division and provide a visual marker for cell-cycle progression. After division(s), the daughter cells remain enclosed within the mother cell wall until they grow new cilia, which then release the hatching enzyme sporangin responsible for the degradation of the mother cell wall (Kubo et al., 2009). Therefore, under diurnal conditions, cells lack cilia at night but synthesize them anew before the next dawn (Figure 3A; Cross and Umen, 2015; Zones et al., 2015; Strenkert et al., 2019). Algae such as C. reinhardtii possess a circadian clock that shares some similarity with known Arabidopsis clock components, which generate oscillations with strong amplitudes and help synchronize cellular metabolism and cell division to imposed diurnal cycles (Matsuo et al., 2008; Noordally and Millar, 2015; Müller et al., 2017). Biosynthetic pathways for classical phytohormones are present in most algae, but the genes encoding their associated receptors are mostly missing from algal genomes (Lu and Xu, 2015; Wang et al., 2015a). The addition of individual phytohormones can affect growth rates, but the underlying molecular mechanisms are currently unclear. Perhaps hormones found in land plants have been coopted from existing biosynthetic pathways during evolution.
Figure 3.
Chlamydomonas Life Cycles.
(A) The sexual and vegetative cycles of C. reinhardtii. See text for details. Adapted from a figure originally drawn by Karen VanWinkle-Swift and published in The Chlamydomonas Sourcebook, Volume 1 (Harris, 2008).
(B) Genes involved in gamete differentiation, agglutination, fusion, and zygote development as a function of mating type. SAD1, SAG1, FUS1, and HAP2 encode glycoproteins critical for gamete mating type recognition, while GSP1 and GSM1 encode transcription factors (TFs) important for zygote development. MT-linked genes are indicated by asterisks. Adapted from Joo et al. (2017).
Reproductive Cycle
Under more adverse conditions of nutrient scarcity, vegetative cells differentiate into gametes of one of two mating types, mt+ and mt−, as determined genetically by the MT locus. Nitrogen deprivation induces gamete differentiation in the laboratory (Sager and Granick, 1954). Cells on solid medium may already be nitrogen-limited if grown for more than 5 d and are therefore sexually competent cells. Exposure to light, perceived by the blue light photoreceptor phototropin (Huang and Beck, 2003), commits nitrogen-starved cells to the gametic fate, but this step is reversible by incubation in darkness. C. reinhardtii is isogamous, as both gametes are identical in terms of size and morphology.
Gametic cilia are decorated by hydroxyproline-rich glycoproteins called agglutinins used for cell-cell recognition. Each mating type expresses a distinct agglutinin: SEXUAL ADHESION1 (SAD1) in mt− gametes and SEXUAL AGGLUTINATION1 (SAG1) in mt+ gametes (Ferris et al., 2005). SAD1 and SAG1 share the same secondary structure but only weak sequence similarity (Ferris et al., 2005). Only SAD1 maps to the mating locus. The proper expression of agglutinins is dictated by the transcription factor MINUS DOMINANCE (MID; Ferris and Goodenough, 1997; Lin and Goodenough, 2007). MID is found within the mt− mating locus, is expressed upon nitrogen starvation, and is necessary and sufficient to specify the mt− fate by contributing to SAD1 induction in mt− gametes while preventing SAG1 expression (Lin and Goodenough, 2007). The mt+ mating locus encodes a third agglutinin critical for cell fusion: FUS1. As illustrated in Figure 3, mt+ and mt− gametes first clump in oriented groups. Shortly thereafter, pairs of gametes of opposite mating types break off from the clump and fuse to form a diploid zygote. sad1 and sag1 mutants cannot form clumps, but fus1 mutants can (Ferris et al., 1996, 2005).
Following agglutination and pairing, cells release an enzyme (autolysin) that digests the cell wall (Matsuda et al., 1978). Specialized mating structures between the cilia are activated to connect the two gametes: a fertilization tubule for the mt+ gamete and a domed structure for the mt− gamete. In a parallel to ciliary recognition during agglutination, proteins decorate these mating structures and take on the appearance of a fringe in transmission electron micrographs (Goodenough et al., 1982). The glycoprotein FUS1 is a fringe component from mt+ gametes (Ferris et al., 1996). The mt− counterpart to FUS1 is HAPLESS2 (HAP2; Liu et al., 2008). fus1 and hap2 gametes cannot fuse to form a diploid zygote (Goodenough et al., 1978; Liu et al., 2008). Compatible mating structures fuse, followed by the shortening of the fertilization tube prior to cell fusion. The early zygote has four cilia that are resorbed after 1 to 2 h.
Agglutination is accompanied by a transient increase in intracellular cAMP levels. Treating cells with dibutyryl-cAMP (a more soluble form of cAMP) and with papaverine or isobutylmethylxanthine (inhibitors of phosphodiesterases) initiates the formation of the fertilization tubule and dome, triggers the release of the gametic cell wall, and leads to gametic fusion into a zygote. In fact, dibutyryl-cAMP and isobutylmethylxanthine can rescue agglutination-defective and paralyzed flagella gametes and allow progression through the sexual cycle (Pasquale and Goodenough, 1987; Dutcher, 1995).
Zygote development is initiated by the dimerization of the homeodomain transcription factors GAMETE-SPECIFIC PLUS1 and MINUS1, which are contributed by the mt+ and mt− gametes, respectively (Lee et al., 2008). GSP1 and GSM1 are cytosolic proteins in gametes, but they translocate to the nucleus upon fusion and trigger the zygote developmental program; the regulatory cascade of the algal sexual cycle is summarized in Figure 3B. Diploid zygotes plated on solid growth medium lacking nitrogen quickly secrete a cell wall that acts as a chemical and physical protectant barrier (Figure 3). Cell wall formation starts soon after gamete fusion from hydroxyproline-rich glycoproteins (Suzuki et al., 2000) and later becomes more desiccation-resistant via the activity of the polyketide synthase PKS1 (Heimerl et al., 2018).
Based on electron microscopy observations, the two nuclei are the first to fuse following gamete fusion, followed by the two chloroplasts 1 h later (Blank et al., 1978). Inheritance of the chloroplast genome in C. reinhardtii is uniparental as a consequence of the complete digestion of the plastid genome originating from the mt− parent, although parts of it can escape degradation (and recombine with the mt+ plastid DNA after fusion) ∼10% of the time (Gillham, 1969). This genome elimination is performed within 24 h after mating (before chloroplast fusion) by the nonspecific DNA nuclease MDN (for Mt+-specific DNase; Nishimura et al., 2002). MDN is thought to reside in the cytosol of mt+ gametes before relocating to the mt− chloroplast upon zygote formation, where high Ca2+ concentrations activate its enzymatic activity. It remains unclear how the mt+ plastid genome escapes degradation and whether plastid DNA methylation plays a role in this process (Lopez et al., 2015). Another nonexclusive scenario calls upon the differential replication rates of the chloroplast genomes derived from the mt+ and mt− gametes during zygote germination (Umen and Goodenough, 2001b). The mitochondrial genome is itself inherited from the mt− parent during sexual reproduction (Beckers et al., 1991; Nakamura, 2010).
The isolation and manipulation of zygotes are facilitated by plating mating mixtures on 4% agar plates, as zygotes adhere strongly to the surface of this medium. Zygotes are later induced to germinate (undergo meiosis) via transfer onto rich medium in the light, which is perceived by the blue light photoreceptors cryptochrome and phototropin (Huang and Beck, 2003; Müller et al., 2017; Zou et al., 2017). Chlamydomonas can be used for tetrad analysis via the dissection of the four germinated meiotic products, as they carry one or the other mating type, which segregate in a 2:2 ratio. The germinated products globally carry a mosaic of both parental nuclear genomes that have been shuffled during meiosis (Liu et al., 2018).
Systematics of the Chlamydomonas Family
Traditional taxonomic keys used to attribute algae to a given genus rely on the presence/absence and position of key cellular features described above (Ettl, 1976; Harris, 2008). Green algae containing a cell wall, two cilia of equal length near each other at one cellular pole, and a single cup-shaped chloroplast with a single pyrenoid at the pole opposite the cilia belong to the genus Chlamydomonas Ehrenberg, including C. reinhardtii and over 1000 other green algae. Based solely on morphological characteristics observed by microscopy, the Chlorophyceae class of green algae can be subdivided into 10 subgenera or sections, but several of the deciding criteria can also be found in algae that belong to other taxa. In addition, many early descriptions of algae were performed “in the wild” under natural conditions, which often meant small sample size and heterogeneity. Based on analysis of rRNA and internal transcribed spacer sequences, the Chlorophyceae are now considered to be made up of 18 monophyletic lineages or clades. Representative species within these clades include well-studied algae: Dunaliella, Polytoma, Chloromonas, and Scenedesmus (Pröschold et al., 2001; Harris, 2008). These molecular criteria represent a departure from the taxonomic keys, as Chloromonas and Polytoma lack pyrenoids.
As in animals, members of the same algal species must be interfertile and form diploid zygotes that can later germinate, but interfertility has not been systematically tested. Approximately 300 Chlamydomonas strains are available in algae repositories, providing an opportunity to confirm relationships based on phylogenetic analyses and reproductive compatibility.
Modern C. reinhardtii is now the archetype for the Reinhardtinia clade, to which the multicellular green alga Volvox carteri also belongs (Pröschold et al., 2001). The genomes of C. reinhardtii and V. carteri share extensive synteny; with a sequence similarity of ∼74% for mutual best BLAST hits, the two algae are as closely related to each other as humans are to chickens or Arabidopsis is to poplar (Populus spp; Prochnik et al., 2010). Volvox forms colonies of only two cell types: small vegetative cells (∼2000) and large germ cells (∼16), encapsulated within a spheroid body. Comparative analysis of the Volvox and Chlamydomonas genomes may provide clues about the evolution of multicellularity (Umen and Olson, 2012).
Most laboratory strains are derived from the isolates provided by Smith to Sager, Ebersold, and Hartshorne. The Ebersold lineage cannot utilize nitrate as its nitrogen source due to two unlinked mutations in the nitrate reductase gene NIT1 and the transcription factor gene NIT2, whose expression is repressed by ammonium (Gallaher et al., 2015). It is not clear when this lineage acquired these mutations, although it probably happened shortly after the strains made their way to Ebersold, since all mutants isolated in this background are unable to grow on nitrate.
Ecology
The Chlamydomonas genus is found worldwide, although the presence or absence of an isolate may be more reflective of the distribution of phycologists than of the genus itself (Harris, 2008). The first C. reinhardtii isolates were collected in Europe, with more recent isolates coming from eastern North America (Massachusetts, Minnesota, North Carolina, Pennsylvania, Florida, and Quebec) (Harris, 2008; Flowers et al., 2015) and Japan (Nakada et al., 2014). All isolates are interfertile and are therefore considered to belong to the C. reinhardtii clade (Harris, 2008; Nakada et al., 2014).
Chlamydomonas species have been collected from a variety of habitats with a vast range in humidity, temperature, and salinity conditions. Species have been found on damp soil surfaces and in fresh and marine water. Chlamydomonas species are fairly tolerant to pollution and can also be found in sewage wastewater (Prescott, 1968). Algae can be picked up by strong winds and spread over land; viable algae (including Chlamydomonas spp) can be found in the air at high altitude (Brown et al., 1964).
Cobalamin (vitamin B12) is an important cofactor for the Met synthase METH. An alternative Met synthase, encoded by METE (Helliwell et al., 2011), is present in half the eukaryotic unicellular algae, including C. reinhardtii. Only Gonium pectoral and Volvox species lack a functional METE gene (Helliwell et al., 2011) and thus depend on exogenous vitamin B12 scavenged from the environment or obtained via the establishment of beneficial interactions with bacteria. The secretion of potential carbon sources for nearby respiring prokaryotes in the form of organic compounds such as acetate, ethanol, and formate by C. reinhardtii at night (Yang et al., 2014; van Lis et al., 2017) also speaks to such an interaction.
Whether in a mutualistic interaction or alone, Chlamydomonas isolates originating from soil or water samples contain a mixture of cells at any stage along the algal vegetative or sexual cycle (illustrated in Figure 3). Pure C. reinhardtii cultures of vegetative haploid cells can be obtained by streaking, serial dilutions, or single-cell isolation with antibiotics to kill off bacteria. Phototaxis can be used to separate algae from bacteria when soil samples are resuspended in water (Gross et al., 1988), as can chlorophyll fluorescence, coupled with fluorescence-activated cell sorting. As a final purification step, single cells may be sprayed onto agar plates and green colonies selected (Wiedeman et al., 1964). Cultures can be maintained on solid agar plates or slants or in liquid cultures with or without bubbling (Figure 4A).
Figure 4.
Chlamydomonas Culture and Transformation.
(A) Chlamydomonas growing on agar plates, slants, and liquid medium.
(B) Example of negative phototaxis. The white arrow represents directed strong white light. After 30 min, most cells have swum away.
(C) Transformation of C. reinhardtii by electroporation or glass beads. Cultures are grown to a cell density of 1 to 2 × 106 cells/mL and concentrated by light centrifugation before agitation with glass beads or electroporation. After recovery in fresh medium for 8 to 16 h, the cells are plated onto selective plates, and colonies derived from individual transformants develop within 7 to 10 d.
GENOMICS
The Chlamydomonas Genome: Sequencing-Aided Genealogy
Until recently, Smith’s paired strains were presumed to be isogenic outside of the mating locus. The genealogy of all Chlamydomonas strains was tentatively retraced based on whole-genome sequencing of 39 independent laboratory strains and 12 field isolates (Flowers et al., 2015; Gallaher et al., 2015). Field strains exhibited sequence diversity of ∼2.8% between themselves and 2.4% when examined against laboratory strains. A comparison of laboratory strains yielded a number of surprises: (1) laboratory strains are not identical and can diverge by as many as 500,000 single-nucleotide polymorphisms (SNPs), or 0.4% of the genome; this number is lower than the ∼2.8% diversity seen between field strains because (2) sequence divergence is concentrated inside polymorphic blocks that display ∼2% sequence diversity; (3) these blocks are not random and can be shared by many strains; (4) these blocks come only in two flavors: haplotype 1 from the sequenced reference strain CC-503 and a polymorphic haplotype 2. Haplotype 2 only covers ∼25% of the total genome. Laboratory strains can thus be thought of as limited recombinant inbred lines. Since CC-503 is mt+, strains of the opposite mating type are polymorphic at the beginning of chromosome 6, where the MT locus resides.
How did this happen? It is clear that the strains shared by Smith are not, in fact, equivalent. He sent strains as compatible mating pairs, but no pairs are isogenic, and different pairs were sent to different laboratories. The history of all lineages is likely further muddied by backcrosses since the original gifts. One Ebersold strain gave rise to CC-503, whose genome was sequenced as the reference. The apparent lack of polymorphisms in CC-503 is only an illusion, since polymorphic means “different from CC-503,” just like polymorphic in Arabidopsis accessions really means “different from the reference accession Col-0.” Nonetheless, CC-503 surely carries blocks derived from both its parents and may represent one of the four original zoospores.
One take-home message is “know thy strain.” Laboratory strains can behave rather differently under various conditions (Gallaher et al., 2015). Haplotype 2 carries ∼65,000 nonsynonymous codon changes as well as ∼600 insertions/deletions predicted to have major effects. There is therefore ample room for loss-of-function alleles within haplotype 2, as revealed under the appropriate growth conditions (Flowers et al., 2015; Gallaher et al., 2015).
The Chlamydomonas Nuclear Genomic Space
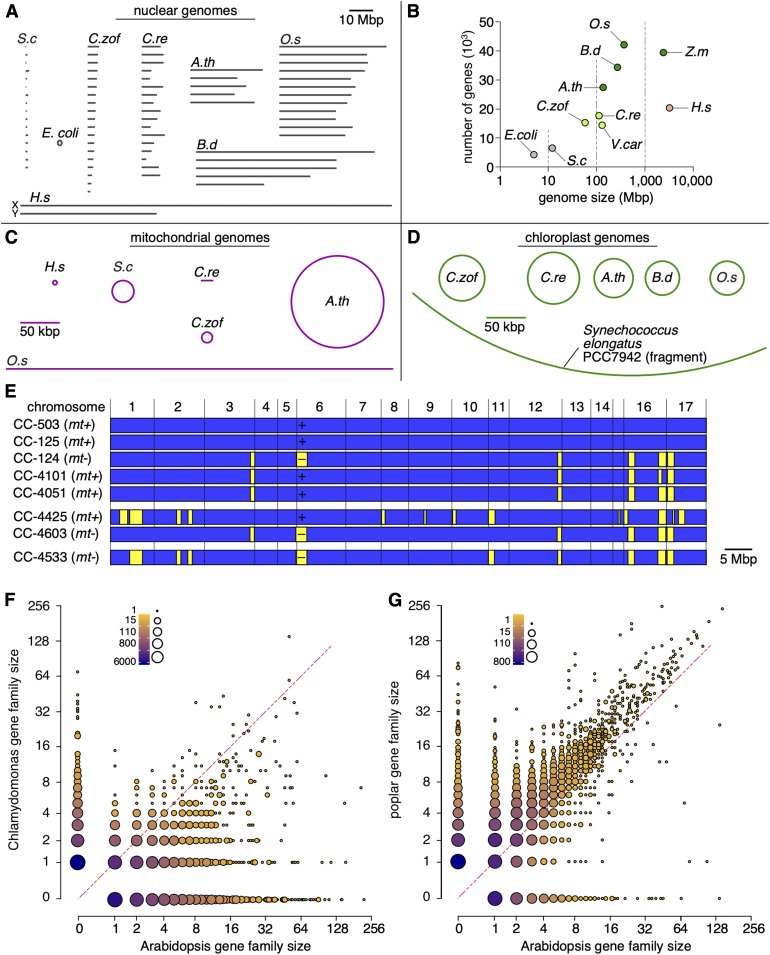
Regardless of strain history, the nuclear genome of C. reinhardtii is ∼111 Mb and is organized into 17 chromosomes (Figure 5A). These numbers are comparable to those of other algae such as V. carteri (Prochnik et al., 2010) and Chromochloris zofingiensis (Roth et al., 2017), although the C. reinhardtii genome is two to three times larger than those of other green algae such as Chlorella spp. The size of the C. reinhardtii genome sits between the small genomes of bacteria (Escherichia coli) and unicellular nonphotosynthetic eukaryotes (yeast) and the larger genomes of plant and animal reference organisms (Arabidopsis, Brachypodium, rice [Oryza sativa], maize [Zea mays], and Homo sapiens; Figure 5B).
Figure 5.
Genome Sizes and Chromosome Numbers in Bacteria, Green Algae, Plants, Yeast, and Humans.
(A) Nuclear genomes. All chromosomes were drawn to scale and ordered by their assigned number. Chromosomes in C. zofingiensis were sorted by length (Roth et al., 2017). The human X and Y chromosomes were drawn for reference.
(B) Genome size versus gene space, showing the number of predicted protein-coding loci as a function of nuclear genome size.
(C) The mitochondrial genome is a linear molecule (with inverted repeats at each end for replication) in C. reinhardtii, C. zofingiensis, and rice.
(D) The plastid genome is a circular molecule in green algae and plants. A portion of the single chromosome from Synechococcus elongatus PCC7942 is shown as a reference for the genome size of the cyanobacterium ancestor that gave rise to modern-day chloroplasts. A.th, Arabidopsis thaliana; B.d, Brachypodium distachyon; C.re, Chlamydomonas reinhardtii; C.zof, Chromochloris zofingiensis; H.s, Homo sapiens; O.s, Oryza sativa; S.c, Saccharomyces cerevisiae; V.car, Volvox carteri; Z.m, Zea mays.
(E) Similarities among the genomes of selected strains, represented within the context of the full genome. Haplotype 1 blocks are shown in blue, and haplotype 2 blocks are shown in yellow.
(F) and (G) Comparison of gene family sizes between Arabidopsis and Chlamydomonas (F) and poplar (G). The number of genes associated with each gene family was extracted from Guo (2013).
The quality and completion of a genome assembly can be measured based on the number of scaffolds and the sizes of the remaining gaps. The latest C. reinhardtii genome assembly release covers ∼99.5% of the genome in only 30 scaffolds (17 chromosome-length scaffolds [1.9–9.7 Mb] and 13 smaller scaffolds ranging from 52 to 272 kb). Some gaps remain along the genome, but they only account for ∼3.6% of the genomic sequence, or half that of the previous 2009 release.
Wild isolates have proven to be very useful for generating genetic maps and mapping mutants (Gross et al., 1988; Kathir et al., 2003; Rymarquis et al., 2005; Liu et al., 2018). During reanalysis of published recombination data, we found that some markers are misplaced. For example, the phytoene synthase gene PSY is assigned to chromosome 2 in the latest genome release but maps to chromosome 11 based on our segregation and historical data (Kathir et al., 2003; McCarthy et al., 2004). The misallocation of the PSY locus to chromosome 2 happened between 2009 and 2012. The chromosomal locations of several additional markers are not supported by segregation data, suggesting that the physical map can be improved by incorporating recombination data.
The physical locations of centromeres are still unclear. In Arabidopsis and rice, centromeric regions contain few genes and are rich in transposons and repetitive sequences that are heavily methylated on cytosine residues (Zhang et al., 2006; Feng et al., 2010). Gene and transposable element densities are relatively uniform in C. reinhardtii: the reference genome contains ∼850 transposable elements, while Arabidopsis boasts over 31,000 transposable elements heavily concentrated at and around centromeres (Zhang et al., 2006; Feng et al., 2010). Cytosine (CG) methylation of genomic DNA is much less prevalent in C. reinhardtii (∼0.5 versus ∼22% for Arabidopsis). However, CG methylation is clearly concentrated in discrete genomic regions (Lopez et al., 2015): for 15 of the 17 chromosomes, high CG methylation was detected over one to three genomic regions, often flanking known sequence gaps, themselves indicative of repetitive sequences that are notoriously difficult to assemble. The centromeres of the green alga C. zofingiensis were identified by mapping repetitive sequences (Roth et al., 2017). It is therefore reasonable to associate these regions of high CG methylation with C. reinhardtii centromeres.
Genome Annotation and Gene Prediction
To facilitate genome annotation, over 200,000 ESTs were generated from cells grown under various conditions and sequenced at both ends (Grossman et al., 2003; Shrager et al., 2003; Asamizu et al., 2004). ESTs were generated from two polymorphic Chlamydomonas strains to facilitate the design of molecular markers to anchor physical and genetic maps and for map-based cloning of mutated genes (Kathir et al., 2003; Rymarquis et al., 2005). The Chlamydomonas community also undertook a major manual annotation effort and provided functions and symbols for almost 3000 genes (Merchant et al., 2007). Some genes received names based on motifs present in their encoded proteins: for example, Ser-Thr kinases were given the generic name STK. Therefore, gene names do not always equate with functional information. Gene names anchored in functional data can supersede historical names (from the original mutant): ac-208 is now known as plastocyanin (PCY1; Quinn et al., 1993). More recently, gene model predictions have been enriched and corrected with the help of RNA sequencing (RNA-seq) data sets, improved gene prediction algorithms, and sequence similarity and synteny with Volvox. Details on the applied methods were described recently (Blaby et al., 2014). The latest genome annotation lists 17,741 loci, with the potential to encode 19,526 proteins (due to alternative splicing). This list excludes microRNAs (miRNA), long noncoding RNAs, and other small RNAs but does include all tRNAs.
Inspired by the Arabidopsis genetic nomenclature, the Chlamydomonas community uses a unique and permanent locus identifier that indicates the chromosome a gene maps to as well as a unique genomic “zipcode” along that chromosome. For instance, plastocyanin has the locus identifier Cre03.g182551: it is located on chromosome 3, between genes Cre03.g182550 and Cre03.g182560. Most often, the zipcode of two neighboring genes increases by a count of 50, which leaves more than enough room for new genes in the intervening space. Note that, contrary to Arabidopsis, where the zipcode resets for each chromosome, Chlamydomonas zipcodes keep increasing, such that the first gene on the last chromosome (chromosome 17) is Cre17.g692564 (and not Cre17.g000050 if following Arabidopsis rules).
Most C. reinhardtii genes contain at least one intron and are composed of an average of seven exons of 200 to 240 bp, which are comparable to exon lengths in V. carteri but ∼50% shorter than those in Arabidopsis. Introns are over twice as long in C. reinhardtii as in Arabidopsis and almost three times as long in Volvox (Merchant et al., 2007; Prochnik et al., 2010; Kianianmomeni et al., 2014; Cheng et al., 2017a). In addition, both Chlamydomonas and Volvox transcripts are distinguished by long 3′ untranslated regions (UTRs) relative to land plants, with a mean length of ∼800 bp, compared with ∼220 bp in Arabidopsis (Shen et al., 2008). The same trend is also observed for the 5′ UTRs, although not as drastically: ∼280 bp in Chlamydomonas and 131 bp in Arabidopsis (Kim et al., 2007). UTR sizes scale with gene length.
The wealth of sequence information (RNA-seq and ESTs) can be mined for consensus motifs for polyadenylation signals. For 65% of human and 60% of fruit fly genes, mRNAs present the consensus sequences AAUAAA or AUUAAA within the last 50 nucleotides before the start of the poly(A) signal (Graber et al., 1999). However, the same motifs are only detected in ∼10% of yeast and Arabidopsis coding sequences (Graber et al., 1999; Loke et al., 2005), and no other consensus sequence has been identified. Chlamydomonas appears to favor the polyadenylation signal UGUAA, which is found in 50% of all genes (Wodniok et al., 2007). The motif UGUAA was also found in the handful of characterized Volvox genes (Dietmaier et al., 1995).
Chlamydomonas gene families are generally accepted to be much smaller than those of land plants, whose genomes have undergone multiple rounds of whole-genome duplications and rearrangements. Over 85% of gene families in C. reinhardtii are composed of single genes and represent almost 60% of the gene complement. Gene families of three genes or more account for only 6.5% of all gene families but contribute 28% of all genes; the situation is completely reversed in Arabidopsis (Arabidopsis Genome Initiative, 2000; Merchant et al., 2007; Guo, 2013). This picture is, however, incomplete: only one-third of Arabidopsis gene families have a match in Chlamydomonas (Figure 5E), whereas 7 out of 10 Arabidopsis gene families have a counterpart in poplar (Figure 5F). Of the gene families with a match in both organisms, the size does tend to be smaller in C. reinhardtii.
The Other Chlamydomonas Gene Space: The Mitochondrial and Chloroplast Genomes
The C. reinhardtii mitochondrial genome is a 15.8-kb linear molecule that encodes 8 proteins, 7 of which are either subunits of the respiratory chain or ATP synthase, as well as 3 tRNAs and 15 rRNA fragments (Boer et al., 1985; Gallaher et al., 2018). This small size came as a surprise, since mitochondrial genomes in land plants are 10 to 150 times larger (Figure 5C). Each cell contains ∼130 copies of the mitochondrial genome. This organelle relies on importing missing tRNAs from the cytosol to sustain its translation needs (Salinas et al., 2012). Mitochondrial genes are expressed as two divergent and multicistronic RNAs that are subsequently processed into individual transcripts by endonucleolytic cleavage. RNA degradation in organelles is initiated by the addition of a poly(A) tail, but analysis of RNA-seq data sets revealed the preponderance of C-rich, polynucleotide tails for all protein-coding RNAs (Salinas-Giegé et al., 2017; Gallaher et al., 2018). Poly(C) tail length appears to be quite variable and may represent a previously undescribed early step in RNA maturation restricted to members of the Chlorophyceae lineage and absent in land plants (Salinas-Giegé et al., 2017).
The first cloned and sequenced C. reinhardtii chloroplast gene was rbcL, encoding the large subunit of Rubisco (Gelvin et al., 1977; Dron et al., 1982). The full chloroplast genome was assembled from National Center for Biotechnology Information (NCBI) sequences and targeted subcloning to fill in gaps into a circular molecule of 205 kb with two inverted repeats (each 21.2 kb) (Maul et al., 2002). This structure is similar to the structures of chloroplast genomes in C. zofingiensis, Arabidopsis, Brachypodium, and rice (Figure 5D). The C. reinhardtii plastid genome distinguishes itself by a high repeat content, which accounts for over 20% of the organellar genome and is limited to intergenic regions. The chloroplast genome is almost identical across all laboratory strains, which is in agreement with (1) uniparental chloroplast inheritance and (2) their common origin (Gallaher et al., 2018). The chloroplast genome sequence was recently improved by incorporating data initially filtered out while assembling the nuclear genomes of laboratory strains (Flowers et al., 2015; Gallaher et al., 2015), which corrected the orientation of some genomic fragments, in addition to a few polymorphisms present in earlier releases. This new genome release confirms the absence of RNA editing in the chloroplast transcriptome.
The chloroplast genome has a low GC content of 34.6%, with each cell containing ∼80 copies (Gelvin et al., 1977; Gallaher et al., 2018), and is organized in large protein-DNA complexes called nucleoids. Plastid nucleoids are associated with DNA replication and repair, transcription, RNA processing, and translation (Krupinska et al., 2013). Unlike land plants, plastid gene transcription is performed solely by a plastid-encoded RNA polymerase (PEP); a single nucleus-encoded sigma factor participates in PEP-mediated transcription (Surzycki and Shellenbarger, 1976). The plastid genome contains 99 genes, including a full set of 30 tRNA genes, 5 rRNA genes, 17 ribosomal protein genes, 32 genes involved in photosynthesis (including the large subunit of Rubisco), and 5 genes for PEP subunits. One plastid gene contains cis-spliced introns (psbA), while another is present as three distinct loci that are independently transcribed (psaA) and posttranscriptionally joined in a process known as trans-splicing. The number of intron-containing plastid genes increased during the evolution of land plants, from 2 in C. reinhardtii to 20 in Arabidopsis (Bonen and Vogel, 2001).
RESOURCES AND DATABASES
The cumulative information collected on the nuclear genome described above (sequences, annotations, and gene identifiers) is accessible via the Joint Genome Institute’s platform Phytozome and the European Nucleotide Archive (Table 1). The six most recent genome releases, including genome assemblies and annotation files, can be downloaded from the Genome Portal at the Joint Genome Institute upon the creation of a (free) account. Also available for download are polymorphisms inferred from the resequencing of laboratory strains (Gallaher et al., 2015). The short reads for 12 field isolates (including S1 D2, used to generate roughly half of the sequenced ESTs deposited at NCBI) and 8 laboratory strains were uploaded at NCBI (Flowers et al., 2015).
The nuclear genome can be browsed on Phytozome, with optional tracks for Vista plots (for sequence conservation between Chlamydomonas, Volvox, Dunaliella, Arabidopsis, and rice), repeat elements, and remaining gaps in the current genome assembly. The positions of mapped reads from a subset of 13 RNA-seq experiments (out of the 46 published as of 2019) can be visualized at Phytozome and the UCSC’s browser at UCLA (Table 1) and used as empirical support for gene annotation and gene models. We hope to launch a new visualization database with all RNA-seq data sets in the future, with an updated browser. Only 5% to 10% of genes have some functional annotation information. Metabolic pathways can be perused on the Kyoto Encyclopedia of Genes and Genomes (KEGG), while Gene Ontology enrichment can be tested at the Protein ANalysis THrough Evolutionary Relationships (PANTHER) Classification System website (Table 1). In both cases, annotations are based on the initial genome release from 2007 and use either the original gene identifier numbers (KEGG, with links to DNA and protein sequences, which need to be checked against the latest genome release for the Cre number) or corresponding Uniprot IDs (PANTHER).
The distribution of biological material is greatly aided by the availability of a dedicated stock center; for the Chlamydomonas community, this role is filled by the Chlamydomonas Resource Center (CRC). Strains from the core collection have the designation “CC” preceding a unique number. CRC maintains stocks for almost 3200 strains: laboratory strains and wild C. reinhardtii isolates, C. reinhardtii relatives, transgenic strains, and mutants, including original extant mutants isolated in the 1950s. CRC also holds ∼400 plasmids, the indexed BAC genomic library produced from the reference strain CC-503 for genome sequencing, and several lambda phage cDNA libraries.
Several collections have been added to the CRC portfolio. The Jonikas lab recently deposited 185 strains expressing Venus YFP and/or mCherry fusions with a number of target proteins, many of them involved in the CCM, as well as cellular localization markers for microscopy and cloning vectors to fuse proteins of interest to the fluorescent proteins (Table 1; Mackinder et al., 2017). In addition, large-scale insertion mutant collections have been deposited. Insertion sites for all CLiP (Chlamydomonas Library Project) mutants (Li et al., 2016) are indicated on the Chlamydomonas Phytozome web portal. This National Science Foundation-funded project generated ∼54,000 mutants by random mutagenic insertion in the CC-4533 strain selected for good recovery from cryogenic storage, transformation rate by electroporation, and absence of clumping when grown in liquid cultures. As with other large-scale projects, ∼80% of mutants carry an insertion; the presence of the insertion should be confirmed by PCR. This mutant collection joins a previous set of ∼49,000 insertion mutants generated by Niyogi in the CC-4051 strain (Dent et al., 2015), ∼450 of which are available at CRC (Table 1). Finally, the CRC also houses a collection of 383 temperature-sensitive mutants generated by Fred Cross by UV mutagenesis of strain CC-124. The putative causal mutations are known in over half the collection, leaving another ∼160 mutations in unassigned loci (Tulin and Cross, 2014). One last mutant collection, not distributed by CRC but available, was generated in strain CC-4101 (T222+) and assembled in France by Wollman and colleagues and consists of ∼500 photosynthetic mutants (Table 1).
Luckily for the community, the strains used for these large-scale mutant collections are closely related. CC-4533 (mt−) was derived from a cross between CC-4425 (mt+) and CC-4603 (mt−), itself essentially identical to CC-124 (mt−). CC-4051 (mt+) is isogenic to CC-4603 outside of the mating locus on chromosome 6 and is also very similar to CC-4101, except for a block at the end of chromosome 16 (Figure 5E; Gallaher et al., 2015). As a reminder, the historical ac and pf mutants were generated in CC-124. All but CC-4533 are thus closely related and only differ by ∼20,000 SNPs, not counting those linked to the mating locus; CC-4533 is separated by an additional 100,000 SNPs due to haplotype 2 blocks on chromosomes 1, 2, and 11. Each haplotype block can be genotyped using allele-specific primers and selected when building double mutants (Gallaher et al., 2015).
CRC handles the distribution and long-term maintenance of the CLiP collection. In the past 2 years, CRC has distributed ∼3,100 CLiP mutants (Matt Laudon, personal communication). Periodic passage of the collection has been accompanied by very minimal colony loss. In addition, cryopreservation is possible, although it is somewhat challenging and requires storage under liquid nitrogen, since frozen cells die within 24 h when stored at −80°C (Crutchfield et al., 1999). In both cases (cryopreservation and maintenance), fragile mutants may not recover past the initial isolation, first passage, or thawing.
In addition to CRC, the University of Texas at Austin in the United States and the Culture Collection of Algae at the University of Göttingen in Germany maintain stocks for almost 2600 and 2300 algal strains, respectively, including Chlamydomonas strains (177 in Austin and 320 in Göttingen; Table 1). Additional algal repositories are listed on the CRC website.
AVAILABLE TOOLS AND TECHNOLOGIES
Armed with the biology and genomics of the organism, we can now design some experiments. Many tested protocols are available online: for example, the 2006 EMBO practical course collection of protocols covers culture handling, crosses, transformation, and biochemical characterization of proteins. CRC has also compiled protocols and resources as well as unpublished “wisdom” from researchers (Table 1). We recommend our readers familiarize themselves with these detailed protocols before starting experiments. Below, we highlight important technical aspects of C. reinhardtii research and some intrinsic caveats and work-arounds.
Classical Genetics and Mutagens
C. reinhardtii performs admirably well for classical forward genetics: mutations can be induced in each of the three genomes by chemical, radiation, or insertional mutagenesis using a selection cassette (such as a linearized plasmid or PCR product that includes a selection marker) (Jinkerson and Jonikas, 2015). Insertional mutants within coding sequences tend to be null alleles. This is problematic for studying essential genes, in which case temperature-sensitive or weak alleles produced with chemical or radiation mutagens may be more appropriate. However, weak alleles can be present in insertional collections if the cassette integrated in noncoding regions. One drawback of a haploid genome is the inability to distinguish between loss- and gain-of-function mutations. However, dominance can be tested in diploid vegetative cells (discussed below).
Crosses: Double Mutants, Dominance, and Allelism Tests
Mutants may be combined, or sorted into complementation groups, by crossing. This assumes that mutants are available in both mating types, either by performing the screen in two isogenic strains differing at the mating locus or by crossing mutants to an isogenic strain of opposite mating type and selecting appropriate genetic combinations in the progeny. As detailed earlier, gamete formation is induced by nitrogen depletion, preferably on plates, before mixing gametes in distilled water. The mating reaction is plated and kept in the dark for 4 to 7 d to prevent premature germination. Zygotes adhere strongly and partially embed themselves into the agar surface, and are resistant to chloroform fumes, whereas unmated haploid cells can be washed off, scraped off with a razor blade, or killed by chloroform fumes.
Mapping a mutation by genotyping large numbers of progeny is possible but somewhat limited by the number of tetrads one is willing and able to dissect. At a smaller scale, for example allelism tests, two mutants are crossed and multiple tetrads are phenotyped. If all progeny display only the mutant phenotype, the two mutants are allelic (or tightly linked, depending on how many tetrads are analyzed). A second technique is based on noncomplementation in vegetative diploids, which form during mating reactions at low rates. To facilitate the selection of vegetative diploids, arg2 and arg7 are crossed into the mutants of interest; both mutations map to ARG7 but result in intragenic complementation and therefore confer Arg autotrophy in diploids (Debuchy et al., 1989). The same strategy can be undertaken for dominance tests, using a wild-type strain as one of the mating partners.
In yeast, mating type is controlled by the MAT locus and switches back and forth when the allele present at MAT is replaced by the opposite mating type from a duplicated but silenced copy found on the same chromosome by homologous recombination (HR; Haber, 2012). Each Chlamydomonas cell harbors only one copy of the MT locus and cannot simply switch. Nevertheless, gametes induced from an mt+ strain transformed with the MID gene differentiate as mt− and are competent to mate with mt+ gametes, although with reduced efficiency (Ferris and Goodenough, 1997). While this remains to be empirically tested, it may therefore be possible to transform mt+ mutants by introducing MID by transformation and using the transformants for crosses with other mt+ mutants for allelism tests or for generating double mutants.
Complementation by Transformation of Genomic Libraries
Causal genes may be identified by genomic rescue, whereby random genomic fragments subcloned into cosmids or BACs are introduced into the mutant. The Arg auxotrophy mutant arg7 is crossed with the mutant of interest; the resulting double mutant is then cotransformed with the ARG7 locus and pools of cosmid or BAC DNA clones. Alternatively, dominant selectable markers that confer resistance to antibiotics or herbicides may be used in place of ARG7-based auxotrophic complementation (Purton and Rochaix, 1994; Zhang et al., 1994). This method is best suited for mutants amenable to plate screening techniques, such as chlorophyll fluorescence or autotrophy rescue. Only 1% to ∼20% of transformants typically exhibit full rescue due to a combination of low expression of transgenes and diluted individual clone representation within DNA pools. These frequencies are obviously higher than those that might be expected of spontaneous second-site suppressors, but they may startle researchers new to C. reinhardtii. As in microbes, mutational reversion does occur at variable frequencies. Intragenic reversion can provide support for the identification of the causal gene, while extragenic suppressors will complicate matters and potentially lead to heterogeneous stocks.
Transformation Methods and Selectable Markers
Stable transformation methods for C. reinhardtii were developed in the 1980s and 1990s. Recombinant DNA is introduced into cells by vortexing in the presence of glass beads coated with DNA, by electroporation, or by bombardment of tungsten or gold particles coated with DNA (Figure 4C; Kindle et al., 1989; Diener et al., 1990; Kindle, 1990; Mayfield and Kindle, 1990; Shimogawara et al., 1998; Yamano et al., 2013). The glass bead method works best for strains without a cell wall, which otherwise limits the amount of DNA reaching the nucleus. The cell wall may be removed by autolysin digestion (Neupert et al., 2012). Strains with a cell wall can be transformed at higher frequencies by electroporation. A recent study using the NEPA21 electroporator from NepaGene reported transformation efficiencies at least 10 times higher than those obtained using the Gene-Pulser from Bio-Rad (Yamano et al., 2013). Biolistics-mediated transformation requires specialized equipment and is generally low-efficiency but is the prime method for manipulating the chloroplast (Boynton et al., 1988) and mitochondrial (Remacle et al., 2006) genomes. Chlamydomonas is the only system where all three genomes can be genetically modified. In fact, the Chlamydomonas chloroplast genome was the first chloroplast genome to be manipulated by transgenesis (Boynton et al., 1988).
Selectable markers that confer antibiotic or herbicide resistance or complement auxotrophic growth facilitate the selection of stable integration of recombinant DNA into the genome(s) (Tables 2–4). Popular markers for nuclear transformation confer growth on medium lacking Arg (ARG7; see above) or ammonium (NIT1) or resistance via antibiotic detoxification: paromomycin (aphVIII), spectinomycin (aadA), and zeocin (ble). A gene encoding a mutant form of ribosomal protein S14, known as cry1-1, provides a dominant resistance marker against the drug emetine (Tables 2–4). The effective dosages of zeocin and emetine that kill all nontransformants should be tested empirically due to strain-by-strain variation. The selection of chloroplast transformants is possible using aadA (Goldschmidt-Clermont, 1991) or a mutant allele of the 16S-23S plastid rRNA that prevents antibiotic binding (Newman et al., 1990). The selection of mitochondrial transformants relies on the rescue of respiration capacity in respiration-deficient mutants by reintroducing a fragment of the mitochondrial genome (Remacle et al., 2006). Organellar transformation is a very rare event but is mitigated by the absence of transgene silencing.
Table 2. Selectable Markers in Use for Chlamydomonas Transformation.
| Marker Name | Selection Type | Marker Origin | Organelle | Reference |
|---|---|---|---|---|
| NIT1a | Auxotrophy rescue (nitrate use) | Chlamydomonas | Nucleus | (Kindle et al., 1989) |
| ARG4a | Auxotrophy rescue (Arg) | Budding yeast | Nucleus | (Rochaix et al., 1984) |
| ARG7a | Auxotrophy rescue (Arg) | Chlamydomonas | Nucleus | (Debuchy et al., 1989) |
| ARG9a | Auxotrophy rescue (Arg) | Chlamydomonas | Nucleus | (Remacle et al., 2009) |
| ARG9a | Auxotrophy rescue (Arg) | Arabidopsis | Chloroplast | (Remacle et al., 2009) |
| NIC7a | Auxotrophy rescue (nicotinamide) | Chlamydomonas | Nucleus | (Ferris, 1995) |
| THI10a | Auxotrophy rescue (thiamine) | Chlamydomonas | Nucleus | (Ferris, 1995) |
| OEE1a,b | Auxotrophy rescue (acetate requiring) | Chlamydomonas | Nucleus | (Mayfield and Kindle, 1990) |
| psbAa,b | Herbicide resistance (DCMU) | Chlamydomonas | Nucleus | (Erickson et al., 1984) |
| PDS1b | Herbicide resistance (norflurazon) | Chlamydomonas | Nucleus | (Brueggeman et al., 2014) |
| PPOb | Herbicide resistance (oxyfluorfen) | Chlamydomonas | Nucleus | (Brueggeman et al., 2014) |
| GAT | Herbicide resistance (glyphosate) | Agrobacterium spp | Nucleus | (Brueggeman et al., 2014) |
| SUL | Herbicide resistance (sulfadiazine) | E. coli | Nucleus | (Tabatabaei et al., 2019) |
| CRY1-1c | Drug resistance (emetine) | Chlamydomonas | Nucleus | (Nelson et al., 1994) |
| aphVIII | Antibiotic resistance (paromomycin) | Streptomyces rimosus | Nucleus | (Sizova et al., 2001) |
| aphA-6 | Antibiotic resistance (kanamycin) | Acetinobacter | Chloroplast | (Bateman and Purton, 2000) |
| aadA | Antibiotic resistance (spectinomycin) | E. coli | Nucleus, chloroplast | (Goldschmidt-Clermont, 1991) |
| blec | Antibiotic resistance (bleomycin, zeocin) | Streptoalloteichus hindustanus | Nucleus | (Stevens et al., 1996) |
| aph7′′ | Antibiotic resistance (hygromycin) | Streptoalloteichus hindustanus | Nucleus | (Berthold et al., 2002) |
| NPTII | Antibiotic resistance (kanamycin) | E. coli Tn5 transposon | Nucleus | (Barahimipour et al., 2016) |
| er, spr, sr (16S, 23S cp rRNA)d | Antibiotic resistance (spectinomycin, erythromycin) | Chlamydomonas | Chloroplast | (Newman et al., 1990) |
| cob-nd4-nd5 | Auxotrophy rescue (respiration) | Chlamydomonas | Mitochondrion | (Remacle et al., 2006) |
| cobmud2 | Drug resistance (myxothiazol) | Chlamydomonas | Mitochondrion | (Remacle et al., 2006) |
Require the mutation to be present in the relevant background for auxotrophy rescue.
Herbicide resistance is conferred by a point mutation in the presumed target of the herbicide, but the mutants have no phenotype on their own.
Can potentially damage DNA.
Antibiotics target the plastid ribosomes, and the resistance-conferring mutations are in the plastid 16S or 23S rRNAs.
Table 4. Selectable Promoters in Use in Chlamydomonas Transformation.
| Promoter | Expression Type | Origin | Reference |
|---|---|---|---|
| RBCS2 | Constitutive | Chlamydomonas | (Stevens et al., 1996) |
| PsaD | Constitutive | Chlamydomonas | (Fischer and Rochaix, 2001) |
| HSP70A | Constitutive, heat-inducible | Chlamydomonas | (Schroda et al., 2000) |
| RBCS2:HSP70A | Constitutive, heat-inducible | Chlamydomonas | (Schroda et al., 2000) |
| NIT1 | Ammonium-inducible, nitrogen-repressible | Chlamydomonas | (Ohresser et al., 1997) |
| CYC6 | Nickel-inducible, copper-repressible | Chlamydomonas | (Pérez-Martín et al., 2015) |
| THI4a | Thiamine-repressible | Chlamydomonas | (Croft et al., 2007) |
| METE | Vitamin B12-repressible | Chlamydomonas | (Helliwell et al., 2014) |
| 35S | Constitutive | CaMV | (Díaz-Santos et al., 2013) |
| Nopaline synthase (NOS) | Constitutive | Agrobacterium | (Díaz-Santos et al., 2013) |
THI4 is a riboswitch found in the THI4 transcript that provides thiamine-repressible control of gene expression when fused to a promoter.
Table 3. Selectable Reporters in Use in Chlamydomonas Transformation.
| Reporter | Reporter Origin | Reference |
|---|---|---|
| ARSa | Chlamydomonas | (Davies et al., 1992) |
| luciferase | Gaussia princeps, Renilla reniformis, firefly | (Matsuo et al., 2006; Shao and Bock, 2008) |
| GFP, BFP, CFP, YFP | Aequorea victoriab | (Rasala et al., 2013) |
| mCherry | Sea anemone | (Rasala et al., 2013) |
| mRuby | Sea anemone | (Rasala et al., 2013) |
| tdTomato | Sea anemone | (Rasala et al., 2013) |
| aequorin | Aequorea victoria | (Aiyar et al., 2017) |
ARS can be used to dissect promoter elements, when cloning them upstream of the β-tubulin minimal promoter.
GFP was cloned from the jellyfish, but the other colored variants were engineered in the laboratory by mutagenesis.
Multiple selection markers can be used sequentially, but this is not routinely performed, possibly due to the likelihood of transgene silencing upon serial transformation. Gene synthesis is strongly recommended for all selectable markers that are not derived from C. reinhardtii to match codon usage and promote high-level expression and translation of the transgene (Barahimipour et al., 2015).
Adequate transgene expression has indeed long been an issue when generating nuclear transformants in C. reinhardtii, but a number of strong promoters and regulatory sequences derived from C. reinhardtii genes have been adopted. Strong constitutive promoters include PSAD, RBCS2, and a chimeric RBCS2 promoter combined with regulatory elements from the HSP70A promoter (Table 4). Inducible promoters may prevent silencing. The 5′ UTR of the THIAMINE4 (THI4) gene contains a riboswitch. In the presence of thiamine, the 5′ UTR undergoes alternative splicing and includes a premature stop codon in the resulting transcript, effectively providing vitamin B1-dependent repression of gene expression (Croft et al., 2007). The nitrate-repressible, ammonium-inducible NIT1 promoter offers tight control for transgene expression, but it only works in strains that have not lost the ability to grow on nitrate, as have most laboratory strains (Schmollinger et al., 2010). The CYC6 promoter confers copper-dependent repression and nickel-dependent induction of gene expression. While copper deficiency is not associated with visible phenotypes, nickel may cause an upregulation of autophagy (Pérez-Martín et al., 2015). The applicability of a steroid-inducible system (Aoyama and Chua, 1997) has yet to be demonstrated. Adding introns to transgenes increases their expression, for instance RBCS2 intron 1. This intron is included in many selection marker cassettes, as one (aphVIII) or two (ble) copies, and is often included in the chimeric RBCS2/HSP70A promoter mentioned earlier. The construction of shuttle vectors is based on decades of observations associated with efficient transgene expression (Tables 2 and 4). Most recently, a series of constructs based on Golden Gate technology was developed that allows combinatorial cloning of regulatory and selection elements (promoter, 5′ UTR, intron, 3′ UTR, and selection marker; Crozet et al., 2018) and decreases the cloning bottleneck. This study also illustrates the importance of the choice of promoter and untranslated regulatory sequences for expression potential. It should be noted that the lengths of common 5′ UTRs have not been optimized and are rather short, which may hinder ribosome landing onto mRNAs.
Reverse Genetics Approaches for the Nuclear Genome
The wealth of RNA-seq-generated expression data sets for C. reinhardtii covers many growth conditions and can be mined to generate hypotheses that can be tested by reverse genetic approaches. Insertional mutant libraries (see above) present one route; targeted knockdowns or gene inactivation offer another route.
C. reinhardtii possesses most proteins necessary for the biogenesis of short RNAs, small interfering RNAs, and miRNAs. C. reinhardtii miRNAs can induce mRNA cleavage and/or translational repression depending on the miRNA and target. Cleavage by the Argonaute complex is not very accurate, and mutants that lack all small RNAs and miRNAs are completely viable with no clear phenotypes (Voshall et al., 2017; Yamasaki and Cerutti, 2017). Most of the miRNAs initially described do not survive stringent reevaluation for annotation as true miRNAs (Tarver et al., 2012). Of the six remaining robust miRNAs, the highly expressed Cre-miR1157 miRNA has provided the backbone for knocking down candidate genes by modifying the miRNA target site to match a gene of interest following plant miRNA target selection rules (Schwab et al., 2005). Success rates are variable, partially due to transgene silencing. This can be minimized with the use of an inducible promoter such as NIT1 but is restricted to NIT+ strains (Schmollinger et al., 2010) or by selecting for luciferase activity derived from a bicistronic message combining Gaussia princeps luciferase and the artificial microRNA (amiRNA; Hu et al., 2014). In addition, amiRNA design may be hindered by the high GC content of the genome, which places a strong burden on the nucleotide composition of putative target sites: natural plant amiRNAs indeed tend to have an adenine at the presumptive cleavage site (Schwab et al., 2005). RNA interference via expression of a double-stranded RNA hairpin construct can be effective and offers the advantage of enabling signal amplification over the entire coding sequence (Rohr et al., 2004). Small RNA-seq also revealed putative trans-acting small interfering RNAs that may be produced by phased cleavage downstream of a miRNA target site (Zhao et al., 2007). In Arabidopsis, the 22-nucleotide miR173 sequence is sufficient to trigger the formation of trans-acting small interfering RNAs from a gene fragment cloned downstream (Felippes et al., 2012) but is dependent on the activity of the RNA-dependent RNA polymerase RDR6, which C. reinhardtii apparently lacks.
Genome editing by zinc-finger nucleases (ZFNs; Sizova et al., 2013; Greiner et al., 2017) and transcription activator-like effectors (TALEs) has been reported in C. reinhardtii but is not routine. ZFNs are modified versions of an endonuclease that consists of a nonspecific DNA cleavage domain and zinc-finger DNA binding domains. Each zinc finger targets three nucleotides, so they are stacked to attain desired specificity. Target sequences have been matched with potential ZFNs for eight organisms, including C. reinhardtii (Reyon et al., 2011). Sequence specificity should be tested in vitro against the intended target prior to transformation. TALEs from Xanthomonas offer another avenue for genome editing. Each module recognizes 1 bp and can be combined to achieve sequence specificity. The C terminus of a TALE acts as a transcriptional activator. Constitutive expression of a gene of interest can therefore be achieved by targeting its promoter with a suitably designed TALE, but the C terminus can also be substituted for an endonuclease domain to generate breaks in the DNA (Gao et al., 2015).
Gene inactivation and genomic manipulation by HR were reported in C. reinhardtii as early as 1993. Using this technique, a selection cassette is flanked by 200 bp or more of genomic sequence from a gene of interest. HR borrows the endogenous DNA repair machinery to replace the gene with the selection cassette. The efficiency is poor, with 1000 random insertion events for each true HR event (Sodeinde and Kindle, 1993; Gumpel et al., 1994; Nelson and Lefebvre, 1995). The establishment of HR would partially alleviate technical difficulties associated with the expression of engineered nucleases such as ZFNs (see above) and Cas9 (see below) but will require higher efficiency. Complementation of auxotrophic mutants by HR-mediated replacement of the inactive allele with a functional copy may provide a positive selection method that does not rely on the random insertion of a selection cassette (Gumpel et al., 1994; Mages et al., 2007). In Neurospora crassa and other fungi, the detection of HR was greatly improved by performing all manipulations in a background that lacked the nonhomologous end-joining protein KU70 or KU80 (Colot et al., 2006). The C. reinhardtii genome contains a single copy of each KU gene, and insertion mutants are available.
Cas9/CRISPR technology has quickly been adapted for use in many reference organisms since the discovery of this bacterial immune system, including C. reinhardtii (Baek et al., 2016; Ferenczi et al., 2017; Greiner et al., 2017). Target specificity is achieved with an RNA molecule, the single guide RNA (sgRNA), which binds to and activates the Cas9 nuclease. While several groups have published successful genome editing with Cas9 (and related nucleases such as Cpf1) and sgRNAs where Cas9/Cpf1 is either transiently expressed in cells or delivered preloaded with the sgRNA by electroporation, the efficiency remains low to very low. A robust selection method for edited cells is clearly needed; thus far, the Cas9 and sgRNA constructs have been cotransformed with an antibiotic selection cassette, or two sgRNAs have been cotransformed (one for the gene of interest and one targeting FKBP12, whose loss of function leads to rapamycin resistance).
Genetic Manipulation of Organellar Genomes
HR is highly efficient for organellar genomes, using homologous sequences as short as 25 bp (Esland et al., 2018). Transgenes may be targeted to “neutral sites” that do not affect the expression of nearby genes or to endogenous organellar genes to replace them by versions carrying point mutations or deletions to study gene function. The design of transformation constructs should be given considerable thought to ensure successful transgene expression. Several rounds of selection might be needed to reach homoplasmy (when all organellar genome copies carry the transgene). Lingering heteroplasmy may indicate that the gene under study is essential; a conditional system will then be more appropriate.
Rochaix’s laboratory devised such a system involving the nucleus-encoded, chloroplast-localized protein NAC2, which binds to and stabilizes the psbD RNA via its 5′ UTR. The conditional repression system exploits and diverts NAC2-dependent stabilization of psbD RNA with two modifications. First, the expression of NAC2 is driven by the METE promoter, which is turned off by vitamin B12 (Helliwell et al., 2014), and the construct is introduced into a nac2 mutant. Second, the 5′ UTR of the selected plastid gene is replaced by that of psbD, thereby becoming NAC2-dependent. The 5′ UTR of the psbD gene itself is replaced by the petA 5′ UTR to maintain an otherwise functional chloroplast (Ramundo and Rochaix, 2015). The role of a plastid gene can be uncovered by the addition of vitamin B12, resulting in the repression of NAC2 and the destabilization of the chimeric plastid transcript carrying the psbD 5′ UTR.
(Heterologous) Protein Expression
The production of metabolites or peptides of high pharmaceutical or nutritional value would be ideal in large-scale bioreactors if not for common nuclear transgene silencing in C. reinhardtii. Matching codon usage and GC content to the Chlamydomonas nuclear genome increases protein accumulation dramatically (Barahimipour et al., 2015, 2016), as does the use of strains selected for high-level transgene expression. The UVM11 strain was identified by UV mutagenesis-induced derepression of a transgenic cry1-1 gene, which confers resistance to emetine. This strain does not exhibit silencing even after long-term cultivation (Neupert et al., 2009). Identifying the causal mutation will pave the way for conducting parallel loss- and gain-of-function studies in the same background, assuming that a CLiP insertion allele exists.
Transgenes inserted into the chloroplast genome by HR are not silenced and offer a platform for the production of recombinant proteins at high titer (Dyo and Purton, 2018). Increasing the number of cloning vectors and selectable markers specifically designed for chloroplast genome transformation would broaden the range of heterologous protein expression within the organelle (Esland et al., 2018).
Metabolic Studies in a Green Microbe
One great advantage of C. reinhardtii is the ease with which growth medium composition can be modulated to study responses to nutrient deficiency or to label proteins with stable or radioactive tracer isotopes. Cells can be grown in replete medium and then transferred to a medium lacking an element such as sulfur, nitrogen, or phosphorus. As these elements are fundamental building blocks for proteins, their absence will severely limit cell growth and metabolism within 24 h (Saroussi et al., 2017). Cellular and physiological responses to various essential metal concentrations can also be investigated; iron deficiency is the easiest, as the chloroplast constitutes the major iron sink in the cell and requires a high iron quota to sustain photosynthesis. Deficiencies in other metals (zinc, copper, and manganese) are more difficult to observe and require the use of ultra-pure water and acid-washed glassware to remove any trace contamination from glass surfaces (Blaby-Haas and Merchant, 2017). Trace element solutions can be purchased from CRC (therefore alleviating the significant effort of preparing them); alternatively, cells grow very well when using a revised mineral nutrient supplement that more than covers cellular requirements and is easy to prepare (Kropat et al., 2011).
Large numbers of cells can easily be collected and assayed for transcriptomics, proteomics, and metabolomics studies. While yeast two-hybrid screens are technically possible and have been performed with C. reinhardtii genes (Lee et al., 2008), this technique suffers from the high GC content of the cDNA library and the ensuing low expression levels of baits and preys. The exploration of protein complexes and organellar proteomes has greatly benefited from proteomics approaches and was bolstered following the completion of the genome. The flux of carbon, nitrogen, and sulfur within the cell may be followed by pulse-labeling with stable isotopes.
CONCLUSION AND OUTLOOK
Chlamydomonas has had a long, rich history as a model organism for studying photosynthesis and cilia biology. More recently, studies of Chlamydomonas have branched out into light responses, metabolism, and the cell cycle, in large part due to its microbe-like nature. This alga is a choice model for cell biology and microscopy, as well as for defining growth conditions to study the effect of nutrient deficiencies or to bend cellular metabolism to suit our needs. Foundational work on chloroplast and cilia biogenesis should be quantitatively revisited at a global scale using isotope labeling. Certainly, genome sequencing, deep transcriptomics, and proteomics studies have reenergized the community. The state of completion of the Chlamydomonas genome (sequence, assembly, and annotation) currently stands as a standard to which other algae should aspire. Chlamydomonas now finds itself at another turning point in its long history: large-scale mutant collections bring hypothesis-driven science and reverse genetics within grasp, while we wait for the optimization of genome-editing methods.
Many classical mutants remain uncharacterized (at the molecular level), while large insertional mutant collections are being screened for phenotypes of interest, with thousands of algal genes not annotated or associated with a clear function. In the near future, Chlamydomonas studies will involve the integration of both forward and reverse genetics approaches within the technical challenges still facing the organism. The upcoming bounty of algal genomes (Cheng et al., 2018) may advocate for a focus on conserved algal genes; longer term, they may offer a new algal model with easier genome-editing possibilities and minimal transgene silencing. As the most thoroughly characterized unicellular alga, C. reinhardtii can transcend its impact in algal biology by becoming a model for future microalgae and biotechnology for decades to come.
Accession Numbers
Sequence data from this article can be found in the Phytozome and TAIR databases under the accession numbers listed in the Supplemental Data Set.
Supplemental Data
Supplemental Data Set. Accession numbers of the major genes mentioned in this article.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank four anonymous reviewers as well as Jean-David Rochaix, Alison Smith, Ursula Goodenough, Ralph Bock, and Sean Gallaher for comments and insightful discussions on earlier versions of this article. We apologize to those whose work could not be mentioned due to word count limits. We thank the reviewers for their comments on an earlier version of this article. Some work in the Merchant lab on the Chlamydomonas genome was supported by a cooperative agreement with the U.S. Department of Energy Office of Science, Office of Biological and Environmental Research program, under award DE-FC02-02ER63421.
AUTHOR CONTRIBUTIONS
P.A.S. prepared all figures and wrote the article, with supervision from S.S.M.
Footnotes
Articles can be viewed without a subscription.
References
- Aiyar P., Schaeme D., García-Altares M., Carrasco Flores D., Dathe H., Hertweck C., Sasso S., Mittag M. (2017). Antagonistic bacteria disrupt calcium homeostasis and immobilize algal cells. Nat. Commun. 8: 1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert S., Schaffer M., Beck F., Mosalaganti S., Asano S., Thomas H.F., Plitzko J.M., Beck M., Baumeister W., Engel B.D. (2017). Proteasomes tether to two distinct sites at the nuclear pore complex. Proc. Natl. Acad. Sci. USA 114: 13726–13731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama T., Chua N.H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11: 605–612. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815. [DOI] [PubMed] [Google Scholar]
- Asamizu E., Nakamura Y., Miura K., Fukuzawa H., Fujiwara S., Hirono M., Iwamoto K., Matsuda Y., Minagawa J., Shimogawara K., Takahashi Y., Tabata S. (2004). Establishment of publicly available cDNA material and information resource of Chlamydomonas reinhardtii (Chlorophyta) to facilitate gene function analysis. Phycologia 43: 722–726. [Google Scholar]
- Baek K., Kim D.H., Jeong J., Sim S.J., Melis A., Kim J.S., Jin E., Bae S. (2016). DNA-free two-gene knockout in Chlamydomonas reinhardtii via CRISPR-Cas9 ribonucleoproteins. Sci. Rep. 6: 30620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barahimipour R., Strenkert D., Neupert J., Schroda M., Merchant S.S., Bock R. (2015). Dissecting the contributions of GC content and codon usage to gene expression in the model alga Chlamydomonas reinhardtii. Plant J. 84: 704–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barahimipour R., Neupert J., Bock R. (2016). Efficient expression of nuclear transgenes in the green alga Chlamydomonas: Synthesis of an HIV antigen and development of a new selectable marker. Plant Mol. Biol. 90: 403–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman J.M., Purton S. (2000). Tools for chloroplast transformation in Chlamydomonas: Expression vectors and a new dominant selectable marker. Mol. Gen. Genet. 263: 404–410. [DOI] [PubMed] [Google Scholar]
- Beckers M.C., Munaut C., Minet A., Matagne R.F. (1991). The fate of mitochondrial DNAs of mt+ and mt− origin in gametes and zygotes of Chlamydomonas. Curr. Genet. 20: 239–243. [DOI] [PubMed] [Google Scholar]
- Bellafiore S., Barneche F., Peltier G., Rochaix J.D. (2005). State transitions and light adaptation require chloroplast thylakoid protein kinase STN7. Nature 433: 892–895. [DOI] [PubMed] [Google Scholar]
- Bennoun P., Levine R.P. (1967). Detecting mutants that have impaired photosynthesis by their increased level of fluorescence. Plant Physiol. 42: 1284–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennoun P., Masson A., Delosme M. (1980). A method for complementation analysis of nuclear and chloroplast mutants of photosynthesis in Chlamydomonas. Genetics 95: 39–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthold P., Schmitt R., Mages W. (2002). An engineered Streptomyces hygroscopicus aph 7" gene mediates dominant resistance against hygromycin B in Chlamydomonas reinhardtii. Protist 153: 401–412. [DOI] [PubMed] [Google Scholar]
- Blaby I.K., et al. (2014). The Chlamydomonas genome project: A decade on. Trends Plant Sci. 19: 672–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaby-Haas C.E., Merchant S.S. (2017). Regulating cellular trace metal economy in algae. Curr. Opin. Plant Biol. 39: 88–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaby-Haas C.E., Merchant S.S. (2019). Comparative and functional algal genomics. Annu. Rev. Plant Biol. 70: 605–638. [DOI] [PubMed] [Google Scholar]
- Blank R., Grobe B., Arnold C.G. (1978). Time-sequence of nuclear and chloroplast fusions in the zygote of Chlamydomonas reinhardii. Planta 138: 63–64. [DOI] [PubMed] [Google Scholar]
- Boer P.H., Bonen L., Lee R.W., Gray M.W. (1985). Genes for respiratory chain proteins and ribosomal RNAs are present on a 16-kilobase-pair DNA species from Chlamydomonas reinhardtii mitochondria. Proc. Natl. Acad. Sci. USA 82: 3340–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonen L., Vogel J. (2001). The ins and outs of group II introns. Trends Genet. 17: 322–331. [DOI] [PubMed] [Google Scholar]
- Boudreau E., Nickelsen J., Lemaire S.D., Ossenbühl F., Rochaix J.D. (2000). The Nac2 gene of Chlamydomonas encodes a chloroplast TPR-like protein involved in psbD mRNA stability. EMBO J. 19: 3366–3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd J.S., Mittelmeier T.M., Dieckmann C.L. (2011). New insights into eyespot placement and assembly in Chlamydomonas. Bioarchitecture 1: 196–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boynton J.E., Gillham N.W., Harris E.H., Hosler J.P., Johnson A.M., Jones A.R., Randolph-Anderson B.L., Robertson D., Klein T.M., Shark K.B., Sanford J.C. (1988). Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science 240: 1534–1538. [DOI] [PubMed] [Google Scholar]
- Breker M., Lieberman K., Cross F.R. (2018). Comprehensive discovery of cell-cycle-essential pathways in Chlamydomonas reinhardtii. Plant Cell 30: 1178–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown R.M. Jr., Larson D.A., Bold H.C. (1964). Airborne algae: Their abundance and heterogeneity. Science 143: 583–585. [DOI] [PubMed] [Google Scholar]
- Brueggeman A.J., Kuehler D., Weeks D.P. (2014). Evaluation of three herbicide resistance genes for use in genetic transformations and for potential crop protection in algae production. Plant Biotechnol. J. 12: 894–902. [DOI] [PubMed] [Google Scholar]
- Bykov Y.S., Schaffer M., Dodonova S.O., Albert S., Plitzko J.M., Baumeister W., Engel B.D., Briggs J.A. (2017). The structure of the COPI coat determined within the cell. eLife 6: e32493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti H., Johnson A.M., Gillham N.W., Boynton J.E. (1997). A eubacterial gene conferring spectinomycin resistance on Chlamydomonas reinhardtii: Integration into the nuclear genome and gene expression. Genetics 145: 97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S., et al. (2018). 10KP: A phylodiverse genome sequencing plan. Gigascience 7: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C.Y., Krishnakumar V., Chan A.P., Thibaud-Nissen F., Schobel S., Town C.D. (2017a). Araport11: A complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 89: 789–804. [DOI] [PubMed] [Google Scholar]
- Cheng X., Liu G., Ke W., Zhao L., Lv B., Ma X., Xu N., Xia X., Deng X., Zheng C., Huang K. (2017b). Building a multipurpose insertional mutant library for forward and reverse genetics in Chlamydomonas. Plant Methods 13: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang K.S., Sueoka N. (1967a). Replication of chloroplast DNA in Chlamydomonas reinhardi during vegetative cell cycle: Its mode and regulation. Proc. Natl. Acad. Sci. USA 57: 1506–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang K.S., Sueoka N. (1967b). Replication of chromosomal and cytoplasmic DNA during mitosis and meiosis in the eucaryote Chlamydomonas reinhardi. J. Cell. Physiol. 70 (suppl.): 89–112. [DOI] [PubMed] [Google Scholar]
- Choquet Y., Wollman F.A. (2002). Translational regulations as specific traits of chloroplast gene expression. FEBS Lett. 529: 39–42. [DOI] [PubMed] [Google Scholar]
- Choquet Y., Goldschmidt-Clermont M., Girard-Bascou J., Kück U., Bennoun P., Rochaix J.D. (1988). Mutant phenotypes support a trans-splicing mechanism for the expression of the tripartite psaA gene in the C. reinhardtii chloroplast. Cell 52: 903–913. [DOI] [PubMed] [Google Scholar]
- Chua N.H., Gillham N.W. (1977). The sites of synthesis of the principal thylakoid membrane polypeptides in Chlamydomonas reinhardtii. J. Cell Biol. 74: 441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colot H.V., Park G., Turner G.E., Ringelberg C., Crew C.M., Litvinkova L., Weiss R.L., Borkovich K.A., Dunlap J.C. (2006). A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA 103: 10352–10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft M.T., Moulin M., Webb M.E., Smith A.G. (2007). Thiamine biosynthesis in algae is regulated by riboswitches. Proc. Natl. Acad. Sci. USA 104: 20770–20775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross F.R., Umen J.G. (2015). The Chlamydomonas cell cycle. Plant J. 82: 370–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozet P., et al. (2018). Birth of a photosynthetic chassis: A MoClo toolkit enabling synthetic biology in the microalga Chlamydomonas reinhardtii. ACS Synth. Biol. 7: 2074–2086. [DOI] [PubMed] [Google Scholar]
- Crutchfield A.L.M., Diller K.R., Brand J.J. (1999). Cryopreservation of Chlamydomonas reinhardtii (Chlorophyta). Eur. J. Phycol. 34: 43–52. [Google Scholar]
- Davies J.P., Weeks D.P., Grossman A.R. (1992). Expression of the arylsulfatase gene from the beta 2-tubulin promoter in Chlamydomonas reinhardtii. Nucleic Acids Res. 20: 2959–2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debuchy R., Purton S., Rochaix J.D. (1989). The argininosuccinate lyase gene of Chlamydomonas reinhardtii: An important tool for nuclear transformation and for correlating the genetic and molecular maps of the ARG7 locus. EMBO J. 8: 2803–2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisseroth K., Hegemann P. (2017). The form and function of channelrhodopsin. Science 357: 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delepelaire P. (1983). Characterization of additional thylakoid membrane polypeptides synthesized inside the chloroplast in Chlamydomonas reinhardtii. Photobiochemistry and Photobiophysics 6: 279–291. [Google Scholar]
- Delosme R., Olive J., Wollman F.A. (1996). Changes in light energy distribution upon state transitions: An in vivo photoacoustic study of the wild type and photosynthesis mutants from Chlamydomonas reinhardtii. Biochim. Biophys. Acta 1273: 150–158. [Google Scholar]
- Dent R.M., Sharifi M.N., Malnoë A., Haglund C., Calderon R.H., Wakao S., Niyogi K.K. (2015). Large-scale insertional mutagenesis of Chlamydomonas supports phylogenomic functional prediction of photosynthetic genes and analysis of classical acetate-requiring mutants. Plant J. 82: 337–351. [DOI] [PubMed] [Google Scholar]
- Depège N., Bellafiore S., Rochaix J.D. (2003). Role of chloroplast protein kinase Stt7 in LHCII phosphorylation and state transition in Chlamydomonas. Science 299: 1572–1575. [DOI] [PubMed] [Google Scholar]
- Díaz-Santos E., de la Vega M., Vila M., Vigara J., León R. (2013). Efficiency of different heterologous promoters in the unicellular microalga Chlamydomonas reinhardtii. Biotechnol. Prog. 29: 319–328. [DOI] [PubMed] [Google Scholar]
- Diener D.R., Curry A.M., Johnson K.A., Williams B.D., Lefebvre P.A., Kindle K.L., Rosenbaum J.L. (1990). Rescue of a paralyzed-flagella mutant of Chlamydomonas by transformation. Proc. Natl. Acad. Sci. USA 87: 5739–5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietmaier W., Fabry S., Huber H., Schmitt R. (1995). Analysis of a family of ypt genes and their products from Chlamydomonas reinhardtii. Gene 158: 41–50. [DOI] [PubMed] [Google Scholar]
- Dron M., Rahire M., Rochaix J.D. (1982). Sequence of the chloroplast DNA region of Chlamydomonas reinhardii containing the gene of the large subunit of ribulose bisphosphate carboxylase and parts of its flanking genes. J. Mol. Biol. 162: 775–793. [DOI] [PubMed] [Google Scholar]
- Dumas L., Zito F., Blangy S., Auroy P., Johnson X., Peltier G., Alric J. (2017). A stromal region of cytochrome b6f subunit IV is involved in the activation of the Stt7 kinase in Chlamydomonas. Proc. Natl. Acad. Sci. USA 114: 12063–12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher S.K. (1995). Mating and tetrad analysis in Chlamydomonas reinhardtii. Methods Cell Biol. 47: 531–540. [DOI] [PubMed] [Google Scholar]
- Dutcher S.K. (2014). The awesome power of dikaryons for studying flagella and basal bodies in Chlamydomonas reinhardtii. Cytoskeleton (Hoboken) 71: 79–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyo Y.M., Purton S. (2018). The algal chloroplast as a synthetic biology platform for production of therapeutic proteins. Microbiology 164: 113–121. [DOI] [PubMed] [Google Scholar]
- Ebersold W.T., Levine R.P. (1959). A genetic analysis of linkage group I of Chlamydomonas reinhardi. Z. Vererbungsl. 90: 74–82. [DOI] [PubMed] [Google Scholar]
- Ebersold W.T., Levine R.P., Levine E.E., Olmsted M.A. (1962). Linkage maps in Chlamydomonas reinhardi. Genetics 47: 531–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenberg C.G. (1838). Die Infusionsthierchen als Vollkommene Organismen: Ein Blick in das Tiefere Organische Leben der Natur. (Leipzig, Germany: L. Voss; ). [Google Scholar]
- Eitzinger N., Wagner V., Weisheit W., Geimer S., Boness D., Kreimer G., Mittag M. (2015). Proteomic analysis of a fraction with intact eyespots of Chlamydomonas reinhardtii and assignment of protein methylation. Front. Plant Sci. 6: 1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel B.D., Schaffer M., Kuhn Cuellar L., Villa E., Plitzko J.M., Baumeister W. (2015). Native architecture of the Chlamydomonas chloroplast revealed by in situ cryo-electron tomography. eLife 4: e04889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson J.M., Rahire M., Bennoun P., Delepelaire P., Diner B., Rochaix J.D. (1984). Herbicide resistance in Chlamydomonas reinhardtii results from a mutation in the chloroplast gene for the 32-kilodalton protein of photosystem II. Proc. Natl. Acad. Sci. USA 81: 3617–3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson J.M., Rahire M., Malnoë P., Girard-Bascou J., Pierre Y., Bennoun P., Rochaix J.D. (1986). Lack of the D2 protein in a Chlamydomonas reinhardtii psbD mutant affects photosystem II stability and D1 expression. EMBO J. 5: 1745–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esland L., Larrea-Alvarez M., Purton S. (2018). Selectable markers and reporter genes for engineering the chloroplast of Chlamydomonas reinhardtii. Biology (Basel) 7: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettl C.G. (1976). Die Gattung Chlamydomonas Ehrenberg. (Vaduz, Liechtenstein: J. Cramer; ). [Google Scholar]
- Faminzin A. (1866). Die Wirkung des Lichtes auf die Bewegung der Chlamidomonas pulvisculus Ehr., Euglena viridis Ehr. und Oscillatoria insignis Tw. Mélanges Biologiques tirés du Bulletin de l’Académie Imperial des Sciences de St-Petersbourg 6: 73–93. [Google Scholar]
- Fang W., Si Y., Douglass S., Casero D., Merchant S.S., Pellegrini M., Ladunga I., Liu P., Spalding M.H. (2012). Transcriptome-wide changes in Chlamydomonas reinhardtii gene expression regulated by carbon dioxide and the CO2-concentrating mechanism regulator CIA5/CCM1. Plant Cell 24: 1876–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felippes F.F., Wang J.W., Weigel D. (2012). MIGS: MiRNA-induced gene silencing. Plant J. 70: 541–547. [DOI] [PubMed] [Google Scholar]
- Feng S., et al. (2010). Conservation and divergence of methylation patterning in plants and animals. Proc. Natl. Acad. Sci. USA 107: 8689–8694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenczi A., Pyott D.E., Xipnitou A., Molnar A. (2017). Efficient targeted DNA editing and replacement in Chlamydomonas reinhardtii using Cpf1 ribonucleoproteins and single-stranded DNA. Proc. Natl. Acad. Sci. USA 114: 13567–13572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris P.J. (1995). Localization of the nic-7, ac-29 and thi-10 genes within the mating-type locus of Chlamydomonas reinhardtii. Genetics 141: 543–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris P.J., Goodenough U.W. (1997). Mating type in Chlamydomonas is specified by mid, the minus-dominance gene. Genetics 146: 859–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris P.J., Woessner J.P., Goodenough U.W. (1996). A sex recognition glycoprotein is encoded by the plus mating-type gene fus1 of Chlamydomonas reinhardtii. Mol. Biol. Cell 7: 1235–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris P.J., Waffenschmidt S., Umen J.G., Lin H., Lee J.H., Ishida K., Kubo T., Lau J., Goodenough U.W. (2005). Plus and minus sexual agglutinins from Chlamydomonas reinhardtii. Plant Cell 17: 597–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field C.B., Behrenfeld M.J., Randerson J.T., Falkowski P. (1998). Primary production of the biosphere: Integrating terrestrial and oceanic components. Science 281: 237–240. [DOI] [PubMed] [Google Scholar]
- Fischer N., Rochaix J.D. (2001). The flanking regions of PsaD drive efficient gene expression in the nucleus of the green alga Chlamydomonas reinhardtii. Mol. Genet. Genomics 265: 888–894. [DOI] [PubMed] [Google Scholar]
- Fleischmann M.M., Ravanel S., Delosme R., Olive J., Zito F., Wollman F.A., Rochaix J.D. (1999). Isolation and characterization of photoautotrophic mutants of Chlamydomonas reinhardtii deficient in state transition. J. Biol. Chem. 274: 30987–30994. [DOI] [PubMed] [Google Scholar]
- Flowers J.M., et al. (2015). Whole-genome resequencing reveals extensive natural variation in the model green alga Chlamydomonas reinhardtii. Plant Cell 27: 2353–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster K.W., Saranak J., Patel N., Zarilli G., Okabe M., Kline T., Nakanishi K. (1984). A rhodopsin is the functional photoreceptor for phototaxis in the unicellular eukaryote Chlamydomonas. Nature 311: 756–759. [DOI] [PubMed] [Google Scholar]
- Freeman Rosenzweig E.S., et al. (2017). The eukaryotic CO2-concentrating organelle is liquid-like and exhibits dynamic reorganization. Cell 171: 148–162.e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallaher S.D., Fitz-Gibbon S.T., Glaesener A.G., Pellegrini M., Merchant S.S. (2015). Chlamydomonas genome resource for laboratory strains reveals a mosaic of sequence variation, identifies true strain histories, and enables strain-specific studies. Plant Cell 27: 2335–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallaher S.D., Fitz-Gibbon S.T., Strenkert D., Purvine S.O., Pellegrini M., Merchant S.S. (2018). High-throughput sequencing of the chloroplast and mitochondrion of Chlamydomonas reinhardtii to generate improved de novo assemblies, analyze expression patterns and transcript speciation, and evaluate diversity among laboratory strains and wild isolates. Plant J. 93: 545–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Wang Y., Fei X., Wright D.A., Spalding M.H. (2015). Expression activation and functional analysis of HLA3, a putative inorganic carbon transporter in Chlamydomonas reinhardtii. Plant J. 82: 1–11. [DOI] [PubMed] [Google Scholar]
- Gelvin S., Heizmann P., Howell S.H. (1977). Identification and cloning of the chloroplast gene coding for the large subunit of ribulose-1,5-bisphosphate carboxylase from Chlamydomonas reinhardi. Proc. Natl. Acad. Sci. USA 74: 3193–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillham N.W. (1969). Uniparental inheritance in Chlamydomonas reinhardi. Am. Nat. 103: 355. [Google Scholar]
- Goldschmidt-Clermont M. (1991). Transgenic expression of aminoglycoside adenine transferase in the chloroplast: A selectable marker of site-directed transformation of Chlamydomonas. Nucleic Acids Res. 19: 4083–4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough U. (2015). Historical perspective on Chlamydomonas as a model for basic research: 1950-1970. Plant J. 82: 365–369. [DOI] [PubMed] [Google Scholar]
- Goodenough U.W., Heuser J.E. (1982). Substructure of the outer dynein arm. J. Cell Biol. 95: 798–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough U.W., Heuser J.E. (1985a). Substructure of inner dynein arms, radial spokes, and the central pair/projection complex of cilia and flagella. J. Cell Biol. 100: 2008–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough U.W., Heuser J.E. (1985b). The Chlamydomonas cell wall and its constituent glycoproteins analyzed by the quick-freeze, deep-etch technique. J. Cell Biol. 101: 1550–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough U.W., Hwang C., Warren A.J. (1978). Sex-limited expression of gene loci controlling flagellar membrane agglutination in the Chlamydomonas mating reaction. Genetics 89: 235–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough U.W., Detmers P.A., Hwang C. (1982). Activation for cell fusion in Chlamydomonas: Analysis of wild-type gametes and nonfusing mutants. J. Cell Biol. 92: 378–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowans C.S. (1976). Publications by Franz Moewus on the Genetics of Algae. (Berkeley: University of California Press; ). [Google Scholar]
- Graber J.H., Cantor C.R., Mohr S.C., Smith T.F. (1999). In silico detection of control signals: mRNA 3′-end-processing sequences in diverse species. Proc. Natl. Acad. Sci. USA 96: 14055–14060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner A., Kelterborn S., Evers H., Kreimer G., Sizova I., Hegemann P. (2017). Targeting of photoreceptor genes in Chlamydomonas reinhardtii via zinc-finger nucleases and CRISPR/Cas9. Plant Cell 29: 2498–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross C.H., Ranum L.P., Lefebvre P.A. (1988). Extensive restriction fragment length polymorphisms in a new isolate of Chlamydomonas reinhardtii. Curr. Genet. 13: 503–508. [DOI] [PubMed] [Google Scholar]
- Grossman A.R., Harris E.E., Hauser C., Lefebvre P.A., Martinez D., Rokhsar D., Shrager J., Silflow C.D., Stern D., Vallon O., Zhang Z. (2003). Chlamydomonas reinhardtii at the crossroads of genomics. Eukaryot. Cell 2: 1137–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumpel N.J., Rochaix J.D., Purton S. (1994). Studies on homologous recombination in the green alga Chlamydomonas reinhardtii. Curr. Genet. 26: 438–442. [DOI] [PubMed] [Google Scholar]
- Guo Y.L. (2013). Gene family evolution in green plants with emphasis on the origination and evolution of Arabidopsis thaliana genes. Plant J. 73: 941–951. [DOI] [PubMed] [Google Scholar]
- Haber J.E. (2012). Mating-type genes and MAT switching in Saccharomyces cerevisiae. Genetics 191: 33–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris E.C. (2008). The Chlamydomonas Sourcebook: Introduction into Chlamydomonas and Its Laboratory Use. (Oxford, UK: Elsevier Academic Press; ). [Google Scholar]
- Heimerl N., Hommel E., Westermann M., Meichsner D., Lohr M., Hertweck C., Grossman A.R., Mittag M., Sasso S. (2018). A giant type I polyketide synthase participates in zygospore maturation in Chlamydomonas reinhardtii. Plant J. 95: 268–281. [DOI] [PubMed] [Google Scholar]
- Helliwell K.E., Wheeler G.L., Leptos K.C., Goldstein R.E., Smith A.G. (2011). Insights into the evolution of vitamin B12 auxotrophy from sequenced algal genomes. Mol. Biol. Evol. 28: 2921–2933. [DOI] [PubMed] [Google Scholar]
- Helliwell K.E., Scaife M.A., Sasso S., Araujo A.P., Purton S., Smith A.G. (2014). Unraveling vitamin B12-responsive gene regulation in algae. Plant Physiol. 165: 388–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnen A., Hicks J.B., Fink G.R. (1978). Transformation of yeast. Proc. Natl. Acad. Sci. USA 75: 1929–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Deng X., Shao N., Wang G., Huang K. (2014). Rapid construction and screening of artificial microRNA systems in Chlamydomonas reinhardtii. Plant J. 79: 1052–1064. [DOI] [PubMed] [Google Scholar]
- Huang K., Beck C.F. (2003). Phototropin is the blue-light receptor that controls multiple steps in the sexual life cycle of the green alga Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 100: 6269–6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn R. (1998). C. G. Ehrenberg: The man and his contribution to botanical science. In Christian Gottfried Ehrenberg (1795–1876): The Man and His Legacy, Williams D.M. and Huxley R., eds (London, UK: Special Publication of The Linnean Society, Academic Press; ), pp. 15–29. [Google Scholar]
- Jalal A., Schwarz C., Schmitz-Linneweber C., Vallon O., Nickelsen J., Bohne A.V. (2015). A small multifunctional pentatricopeptide repeat protein in the chloroplast of Chlamydomonas reinhardtii. Mol. Plant 8: 412–426. [DOI] [PubMed] [Google Scholar]
- Jensen K.H., Herrin D.L., Plumley F.G., Schmidt G.W. (1986). Biogenesis of photosystem II complexes: Transcriptional, translational, and posttranslational regulation. J. Cell Biol. 103: 1315–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinkerson R.E., Jonikas M.C. (2015). Molecular techniques to interrogate and edit the Chlamydomonas nuclear genome. Plant J. 82: 393–412. [DOI] [PubMed] [Google Scholar]
- Johnson D.S. (1914). The history of the discovery of sexuality in plants. Annual Report Smithsonian Institution 69: 383–406. [Google Scholar]
- Joo S., Nishimura Y., Cronmiller E., Hong R.H., Kariyawasam T., Wang M.H., Shao N.C., El Akkad S.E., Suzuki T., Higashiyama T., Jin E., Lee J.H. (2017). Gene regulatory networks for the haploid-to-diploid transition of Chlamydomonas reinhardtii. Plant Physiol. 175: 314–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungnick N., Ma Y., Mukherjee B., Cronan J.C., Speed D.J., Laborde S.M., Longstreth D.J., Moroney J.V. (2014). The carbon concentrating mechanism in Chlamydomonas reinhardtii: Finding the missing pieces. Photosynth. Res. 121: 159–173. [DOI] [PubMed] [Google Scholar]
- Kathir P., LaVoie M., Brazelton W.J., Haas N.A., Lefebvre P.A., Silflow C.D. (2003). Molecular map of the Chlamydomonas reinhardtii nuclear genome. Eukaryot. Cell 2: 362–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kianianmomeni A., Ong C.S., Rätsch G., Hallmann A. (2014). Genome-wide analysis of alternative splicing in Volvox carteri. BMC Genomics 15: 1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B.H., Cai X., Vaughn J.N., von Arnim A.G. (2007). On the functions of the h subunit of eukaryotic initiation factor 3 in late stages of translation initiation. Genome Biol. 8: R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle K.L. (1990). High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 87: 1228–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle K.L., Schnell R.A., Fernández E., Lefebvre P.A. (1989). Stable nuclear transformation of Chlamydomonas using the Chlamydomonas gene for nitrate reductase. J. Cell Biol. 109: 2589–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski K.G., Johnson K.A., Forscher P., Rosenbaum J.L. (1993). A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc. Natl. Acad. Sci. USA 90: 5519–5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropat J., Hong-Hermesdorf A., Casero D., Ent P., Castruita M., Pellegrini M., Merchant S.S., Malasarn D. (2011). A revised mineral nutrient supplement increases biomass and growth rate in Chlamydomonas reinhardtii. Plant J. 66: 770–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupinska K., Melonek J., Krause K. (2013). New insights into plastid nucleoid structure and functionality. Planta 237: 653–664. [DOI] [PubMed] [Google Scholar]
- Kubo T., Kaida S., Abe J., Saito T., Fukuzawa H., Matsuda Y. (2009). The Chlamydomonas hatching enzyme, sporangin, is expressed in specific phases of the cell cycle and is localized to the flagella of daughter cells within the sporangial cell wall. Plant Cell Physiol. 50: 572–583. [DOI] [PubMed] [Google Scholar]
- Kück U., Choquet Y., Schneider M., Dron M., Bennoun P. (1987). Structural and transcription analysis of two homologous genes for the P700 chlorophyll a-apoproteins in Chlamydomonas reinhardii: Evidence for in vivo trans-splicing. EMBO J. 6: 2185–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarus D., Jahn R. (1998). Using the Ehrenberg Collection. Diatom Res. 13: 273–291. [Google Scholar]
- Lee J.H., Lin H., Joo S., Goodenough U. (2008). Early sexual origins of homeoprotein heterodimerization and evolution of the plant KNOX/BELL family. Cell 133: 829–840. [DOI] [PubMed] [Google Scholar]
- Levine R.P. (1960a). Genetic control of photosynthesis in Chlamydomonas reinhardi. Proc. Natl. Acad. Sci. USA 46: 972–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine R.P. (1960b). A screening technique for photosynthetic mutants in unicellular algae. Nature 188: 339–340. [DOI] [PubMed] [Google Scholar]
- Levine R.P., Ebersold W.T. (1960). The genetics and cytology of Chlamydomonas. Annu. Rev. Microbiol. 14: 197–216. [DOI] [PubMed] [Google Scholar]
- Lewin J.C. (1950). Obligate autotrophy in Chlamydomonas moewusii Gerloff. Science 112: 652–653. [DOI] [PubMed] [Google Scholar]
- Lewin R.A. (1954). Mutants of Chlamydomonas moewusii with impaired motility. J. Gen. Microbiol. 11: 358–363. [DOI] [PubMed] [Google Scholar]
- Li J.B., et al. (2004). Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell 117: 541–552. [DOI] [PubMed] [Google Scholar]
- Li X., Zhang R., Patena W., Gang S.S., Blum S.R., Ivanova N., Yue R., Robertson J.M., Lefebvre P.A., Fitz-Gibbon S.T., Grossman A.R., Jonikas M.C. (2016). An indexed, mapped mutant library enables reverse genetics studies of biological processes in Chlamydomonas reinhardtii. Plant Cell 28: 367–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Goodenough U.W. (2007). Gametogenesis in the Chlamydomonas reinhardtii minus mating type is controlled by two genes, MID and MTD1. Genetics 176: 913–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Huang J., Sun X., Li J., Hu Y., Yu L., Liti G., Tian D., Hurst L.D., Yang S. (2018). Tetrad analysis in plants and fungi finds large differences in gene conversion rates but no GC bias. Nat. Ecol. Evol. 2: 164–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Tewari R., Ning J., Blagborough A.M., Garbom S., Pei J., Grishin N.V., Steele R.E., Sinden R.E., Snell W.J., Billker O. (2008). The conserved plant sterility gene HAP2 functions after attachment of fusogenic membranes in Chlamydomonas and Plasmodium gametes. Genes Dev. 22: 1051–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loke J.C., Stahlberg E.A., Strenski D.G., Haas B.J., Wood P.C., Li Q.Q. (2005). Compilation of mRNA polyadenylation signals in Arabidopsis revealed a new signal element and potential secondary structures. Plant Physiol. 138: 1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez D., Hamaji T., Kropat J., De Hoff P., Morselli M., Rubbi L., Fitz-Gibbon S., Gallaher S.D., Merchant S.S., Umen J., Pellegrini M. (2015). Dynamic changes in the transcriptome and methylome of Chlamydomonas reinhardtii throughout its life cycle. Plant Physiol. 169: 2730–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Xu J. (2015). Phytohormones in microalgae: A new opportunity for microalgal biotechnology? Trends Plant Sci. 20: 273–282. [DOI] [PubMed] [Google Scholar]
- Luykx P., Hoppenrath M., Robinson D.G. (1997). Structure and behavior of contractile vacuoles in Chlamydomonas reinhardtii. Protoplasma 198: 73–84. [Google Scholar]
- Mackinder L.C., et al. (2016). A repeat protein links Rubisco to form the eukaryotic carbon-concentrating organelle. Proc. Natl. Acad. Sci. USA 113: 5958–5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackinder L.C.M., Chen C., Leib R.D., Patena W., Blum S.R., Rodman M., Ramundo S., Adams C.M., Jonikas M.C. (2017). A spatial interactome reveals the protein organization of the algal CO2-concentrating mechanism. Cell 171: 133–147.e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mages W., Heinrich O., Treuner G., Vlcek D., Daubnerova I., Slaninova M. (2007). Complementation of the Chlamydomonas reinhardtii arg7-8 (arg2) point mutation by recombination with a truncated nonfunctional ARG7 gene. Protist 158: 435–446. [DOI] [PubMed] [Google Scholar]
- Maiwald D., Dietzmann A., Jahns P., Pesaresi P., Joliot P., Joliot A., Levin J.Z., Salamini F., Leister D. (2003). Knock-out of the genes coding for the Rieske protein and the ATP-synthase delta-subunit of Arabidopsis: Effects on photosynthesis, thylakoid protein composition, and nuclear chloroplast gene expression. Plant Physiol. 133: 191–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda Y., Tamaki S.-I., Tsubo Y. (1978). Mating type specific induction of cell wall lytic factor by agglutination of gametes in Chlamydomonas reinhardtii. Plant Cell Physiol. 19: 1253–1261. [Google Scholar]
- Matsuo T., Onai K., Okamoto K., Minagawa J., Ishiura M. (2006). Real-time monitoring of chloroplast gene expression by a luciferase reporter: evidence for nuclear regulation of chloroplast circadian period. Mol. Cell. Biol. 26: 863–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo T., Okamoto K., Onai K., Niwa Y., Shimogawara K., Ishiura M. (2008). A systematic forward genetic analysis identified components of the Chlamydomonas circadian system. Genes Dev. 22: 918–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul J.E., Lilly J.W., Cui L., dePamphilis C.W., Miller W., Harris E.H., Stern D.B. (2002). The Chlamydomonas reinhardtii plastid chromosome: Islands of genes in a sea of repeats. Plant Cell 14: 2659–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield S.P., Kindle K.L. (1990). Stable nuclear transformation of Chlamydomonas reinhardtii by using a C. reinhardtii gene as the selectable marker. Proc. Natl. Acad. Sci. USA 87: 2087–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy S.S., Kobayashi M.C., Niyogi K.K. (2004). White mutants of Chlamydomonas reinhardtii are defective in phytoene synthase. Genetics 168: 1249–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memmola F., Mukherjee B., Moroney J.V., Giordano M. (2014). Carbon allocation and element composition in four Chlamydomonas mutants defective in genes related to the CO2 concentrating mechanism. Photosynth. Res. 121: 201–211. [DOI] [PubMed] [Google Scholar]
- Merchant S.S., et al. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant S., Selman B.R. (1984). Synthesis and turnover of the chloroplast coupling factor 1 in Chlamydomonas reinhardi. Plant Physiol. 75: 781–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer J.H., Lee-Tischler M.J., Kwon H.Y., Schrick J.J., Avner E.D., Sweeney W.E., Godfrey V.L., Cacheiro N.L., Wilkinson J.E., Woychik R.P. (1994). Candidate gene associated with a mutation causing recessive polycystic kidney disease in mice. Science 264: 1329–1333. [DOI] [PubMed] [Google Scholar]
- Müller N., Wenzel S., Zou Y., Künzel S., Sasso S., Weiß D., Prager K., Grossman A., Kottke T., Mittag M. (2017). A plant cryptochrome controls key features of the Chlamydomonas circadian clock and its life cycle. Plant Physiol. 174: 185–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada T., Tsuchida Y., Arakawa K., Ito T., Tomita M. (2014). Hybridization between Japanese and North American Chlamydomonas reinhardtii (Volvocales, Chlorophyceae). Phycol Res 62: 232–236. [Google Scholar]
- Nakamura S. (2010). Paternal inheritance of mitochondria in Chlamydomonas. J. Plant Res. 123: 163–170. [DOI] [PubMed] [Google Scholar]
- Nelson J.A., Lefebvre P.A. (1995). Targeted disruption of the NIT8 gene in Chlamydomonas reinhardtii. Mol. Cell. Biol. 15: 5762–5769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson J.A., Savereide P.B., Lefebvre P.A. (1994). The CRY1 gene in Chlamydomonas reinhardtii: Structure and use as a dominant selectable marker for nuclear transformation. Mol. Cell. Biol. 14: 4011–4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert J., Karcher D., Bock R. (2009). Generation of Chlamydomonas strains that efficiently express nuclear transgenes. Plant J. 57: 1140–1150. [DOI] [PubMed] [Google Scholar]
- Neupert J., Shao N., Lu Y., Bock R. (2012). Genetic transformation of the model green alga Chlamydomonas reinhardtii. Methods Mol. Biol. 847: 35–47. [DOI] [PubMed] [Google Scholar]
- Newman S.M., Boynton J.E., Gillham N.W., Randolph-Anderson B.L., Johnson A.M., Harris E.H. (1990). Transformation of chloroplast ribosomal RNA genes in Chlamydomonas: Molecular and genetic characterization of integration events. Genetics 126: 875–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y., Misumi O., Kato K., Inada N., Higashiyama T., Momoyama Y., Kuroiwa T. (2002). An mt(+) gamete-specific nuclease that targets mt(−) chloroplasts during sexual reproduction in C. reinhardtii. Genes Dev. 16: 1116–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordally Z.B., Millar A.J. (2015). Clocks in algae. Biochemistry 54: 171–183. [DOI] [PubMed] [Google Scholar]
- Ohad I., Siekevitz P., Palade G.E. (1967). Biogenesis of chloroplast membranes. I. Plastid dedifferentiation in a dark-grown algal mutant (Chlamydomonas reinhardi). J. Cell Biol. 35: 521–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohresser M., Matagne R.F., Loppes R. (1997). Expression of the arylsulphatase reporter gene under the control of the nit1 promoter in Chlamydomonas reinhardtii. Curr. Genet. 31: 264–271. [DOI] [PubMed] [Google Scholar]
- Ohta N., Sager R. (1975). Identification of a chloroplast ribosomal protein altered by a chloroplast mutation in Chlamydomonas. J. Biol. Chem. 250: 3655–3659. [PubMed] [Google Scholar]
- Pascher A. (1918). Über die Beziehung der Reduktionsteilung sur Mendelschen Spaltung. Ber. Dtsch. Bot. Ges. 36: 163–168. [Google Scholar]
- Pasquale S.M., Goodenough U.W. (1987). Cyclic AMP functions as a primary sexual signal in gametes of Chlamydomonas reinhardtii. J. Cell Biol. 105: 2279–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G.J., Dickert B.L., Vucica Y., Seeley E.S., Rosenbaum J.L., Witman G.B., Cole D.G. (2000). Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J. Cell Biol. 151: 709–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G.J., San Agustin J.T., Follit J.A., Rosenbaum J.L., Witman G.B. (2002). Polycystin-2 localizes to kidney cilia and the ciliary level is elevated in orpk mice with polycystic kidney disease. Curr. Biol. 12: R378–R380. [DOI] [PubMed] [Google Scholar]
- Pazour G.J., Agrin N., Leszyk J., Witman G.B. (2005). Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 170: 103–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Martín M., Blaby-Haas C.E., Pérez-Pérez M.E., Andrés-Garrido A., Blaby I.K., Merchant S.S., Crespo J.L. (2015). Activation of autophagy by metals in Chlamydomonas reinhardtii. Eukaryot. Cell 14: 964–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G., Mead K. (1997). Transport of a novel complex in the cytoplasmic matrix of Chlamydomonas flagella. Proc. Natl. Acad. Sci. USA 94: 4457–4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott G.W. (1968). The Algae: A Review. (Boston: Houghton Mifflin; ). [Google Scholar]
- Prochnik S.E., et al. (2010). Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science 329: 223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pröschold T., Marin B., Schlösser U.G., Melkonian M. (2001). Molecular phylogeny and taxonomic revision of Chlamydomonas (Chlorophyta). I. Emendation of Chlamydomonas Ehrenberg and Chloromonas Gobi, and description of Oogamochlamys gen. nov. and Lobochlamys gen. nov. Protist 152: 265–300. [DOI] [PubMed] [Google Scholar]
- Provasoli L., Hutner S.H., Pintner I.J. (1951). Destruction of chloroplasts by streptomycin. Cold Spring Harb. Symp. Quant. Biol. 16: 113–120. [DOI] [PubMed] [Google Scholar]
- Purton S., Rochaix J.D. (1994). Complementation of a Chlamydomonas reinhardtii mutant using a genomic cosmid library. Plant Mol. Biol. 24: 533–537. [DOI] [PubMed] [Google Scholar]
- Quinn J., Li H.H., Singer J., Morimoto B., Mets L., Kindle K., Merchant S. (1993). The plastocyanin-deficient phenotype of Chlamydomonas reinhardtii Ac-208 results from a frame-shift mutation in the nuclear gene encoding preapoplastocyanin. J. Biol. Chem. 268: 7832–7841. [PubMed] [Google Scholar]
- Ramundo S., Rochaix J.D. (2015). Controlling expression of genes in the unicellular alga Chlamydomonas reinhardtii with a vitamin-repressible riboswitch. Methods Enzymol. 550: 267–281. [DOI] [PubMed] [Google Scholar]
- Rasala B.A., Barrera D.J., Ng J., Plucinak T.M., Rosenberg J.N., Weeks D.P., Oyler G.A., Peterson T.C., Haerizadeh F., Mayfield S.P. (2013). Expanding the spectral palette of fluorescent proteins for the green microalga Chlamydomonas reinhardtii. Plant J. 74: 545–556. [DOI] [PubMed] [Google Scholar]
- Remacle C., Cardol P., Coosemans N., Gaisne M., Bonnefoy N. (2006). High-efficiency biolistic transformation of Chlamydomonas mitochondria can be used to insert mutations in complex I genes. Proc. Natl. Acad. Sci. USA 103: 4771–4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remacle C., Cline S., Boutaffala L., Gabilly S., Larosa V., Barbieri M.R., Coosemans N., Hamel P.P. (2009). The ARG9 gene encodes the plastid-resident N-acetyl ornithine aminotransferase in the green alga Chlamydomonas reinhardtii. Eukaryot. Cell 8: 1460–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyon D., Kirkpatrick J.R., Sander J.D., Zhang F., Voytas D.F., Joung J.K., Dobbs D., Coffman C.R. (2011). ZFNGenome: A comprehensive resource for locating zinc finger nuclease target sites in model organisms. BMC Genomics 12: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix J.D., van Dillewijn J. (1982). Transformation of the green alga Chlamydomonas reinhardii with yeast DNA. Nature 296: 70–72. [DOI] [PubMed] [Google Scholar]
- Rochaix J.D., van Dillewijn J., Rahire M. (1984). Construction and characterization of autonomously replicating plasmids in the green unicellular alga Chlamydomonas reinhardii. Cell 36: 925–931. [DOI] [PubMed] [Google Scholar]
- Rohr J., Sarkar N., Balenger S., Jeong B.R., Cerutti H. (2004). Tandem inverted repeat system for selection of effective transgenic RNAi strains in Chlamydomonas. Plant J. 40: 611–621. [DOI] [PubMed] [Google Scholar]
- Rost B.R., Schneider-Warme F., Schmitz D., Hegemann P. (2017). Optogenetic tools for subcellular applications in neuroscience. Neuron 96: 572–603. [DOI] [PubMed] [Google Scholar]
- Roth M.S., Cokus S.J., Gallaher S.D., Walter A., Lopez D., Erickson E., Endelman B., Westcott D., Larabell C.A., Merchant S.S., Pellegrini M., Niyogi K.K. (2017). Chromosome-level genome assembly and transcriptome of the green alga Chromochloris zofingiensis illuminates astaxanthin production. Proc. Natl. Acad. Sci. USA 114: E4296–E4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G.M., Spradling A.C. (1982). Genetic transformation of Drosophila with transposable element vectors. Science 218: 348–353. [DOI] [PubMed] [Google Scholar]
- Rymarquis L.A., Handley J.M., Thomas M., Stern D.B. (2005). Beyond complementation: Map-based cloning in Chlamydomonas reinhardtii. Plant Physiol. 137: 557–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager R. (1954). Mendelian and non-Mendelian inheritance of streptomycin resistance in Chlamydomonas reinhardi. Proc. Natl. Acad. Sci. USA 40: 356–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager R., Granick S. (1954). Nutritional control of sexuality in Chlamydomonas reinhardi. J. Gen. Physiol. 37: 729–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager R., Palade G.E. (1957). Structure and development of the chloroplast in Chlamydomonas. I. The normal green cell. J. Biophys. Biochem. Cytol. 3: 463–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas T., Duby F., Larosa V., Coosemans N., Bonnefoy N., Motte P., Maréchal-Drouard L., Remacle C. (2012). Co-evolution of mitochondrial tRNA import and codon usage determines translational efficiency in the green alga Chlamydomonas. PLoS Genet. 8: e1002946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas-Giegé T., Cavaiuolo M., Cognat V., Ubrig E., Remacle C., Duchêne A.M., Vallon O., Maréchal-Drouard L. (2017). Polycytidylation of mitochondrial mRNAs in Chlamydomonas reinhardtii. Nucleic Acids Res. 45: 12963–12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapp J. (1987). What counts as evidence or who was Franz Moewus and why was everybody saying such terrible things about him? Hist. Philos. Life Sci. 9: 277–308. [PubMed] [Google Scholar]
- Saroussi S., Sanz-Luque E., Kim R.G., Grossman A.R. (2017). Nutrient scavenging and energy management: Acclimation responses in nitrogen and sulfur deprived Chlamydomonas. Curr. Opin. Plant Biol. 39: 114–122. [DOI] [PubMed] [Google Scholar]
- Satir P. (2017). Cilia: Before and after. Cilia 6: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer M., Mahamid J., Engel B.D., Laugks T., Baumeister W., Plitzko J.M. (2017). Optimized cryo-focused ion beam sample preparation aimed at in situ structural studies of membrane proteins. J. Struct. Biol. 197: 73–82. [DOI] [PubMed] [Google Scholar]
- Schmidt G.W., Mishkind M.L. (1983). Rapid degradation of unassembled ribulose 1,5-bisphosphate carboxylase small subunits in chloroplasts. Proc. Natl. Acad. Sci. USA 80: 2632–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt G.W., Matlin K.S., Chua N.H. (1977). A rapid procedure for selective enrichment of photosynthetic electron transport mutants. Proc. Natl. Acad. Sci. USA 74: 610–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmollinger S., Strenkert D., Schroda M. (2010). An inducible artificial microRNA system for Chlamydomonas reinhardtii confirms a key role for heat shock factor 1 in regulating thermotolerance. Curr. Genet. 56: 383–389. [DOI] [PubMed] [Google Scholar]
- Schroda M., Blöcker D., Beck C.F. (2000). The HSP70A promoter as a tool for the improved expression of transgenes in Chlamydomonas. Plant J. 21: 121–131. [DOI] [PubMed] [Google Scholar]
- Schwab R., Palatnik J.F., Riester M., Schommer C., Schmid M., Weigel D. (2005). Specific effects of microRNAs on the plant transcriptome. Dev. Cell 8: 517–527. [DOI] [PubMed] [Google Scholar]
- Shao N., Bock R. (2008). A codon-optimized luciferase from Gaussia princeps facilitates the in vivo monitoring of gene expression in the model alga Chlamydomonas reinhardtii. Curr. Genet. 53: 381–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Ji G., Haas B.J., Wu X., Zheng J., Reese G.J., Li Q.Q. (2008). Genome level analysis of rice mRNA 3′-end processing signals and alternative polyadenylation. Nucleic Acids Res. 36: 3150–3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimogawara K., Fujiwara S., Grossman A., Usuda H. (1998). High-efficiency transformation of Chlamydomonas reinhardtii by electroporation. Genetics 148: 1821–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrager J., Hauser C., Chang C.W., Harris E.H., Davies J., McDermott J., Tamse R., Zhang Z., Grossman A.R. (2003). Chlamydomonas reinhardtii genome project: A guide to the generation and use of the cDNA information. Plant Physiol. 131: 401–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizova I., Fuhrmann M., Hegemann P. (2001). A Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii. Gene 277: 221–229. [DOI] [PubMed] [Google Scholar]
- Sizova I., Greiner A., Awasthi M., Kateriya S., Hegemann P. (2013). Nuclear gene targeting in Chlamydomonas using engineered zinc-finger nucleases. Plant J. 73: 873–882. [DOI] [PubMed] [Google Scholar]
- Smith G.M. (1946). The nature of sexuality in Chlamydomonas. Am. J. Bot. 33: 625–630. [PubMed] [Google Scholar]
- Smith E.F., Lefebvre P.A. (1996). PF16 encodes a protein with armadillo repeats and localizes to a single microtubule of the central apparatus in Chlamydomonas flagella. J. Cell Biol. 132: 359–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G.M., Regnery D.C. (1950). Inheritance of sexuality in Chlamydomonas reinhardi. Proc. Natl. Acad. Sci. USA 36: 246–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodeinde O.A., Kindle K.L. (1993). Homologous recombination in the nuclear genome of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 90: 9199–9203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens D.R., Rochaix J.D., Purton S. (1996). The bacterial phleomycin resistance gene ble as a dominant selectable marker in Chlamydomonas. Mol. Gen. Genet. 251: 23–30. [DOI] [PubMed] [Google Scholar]
- Strenkert D., Schmollinger S., Gallaher S.D., Salomé P.A., Purvine S.O., Nicora C.D., Mettler-Altmann T., Soubeyrand E., Weber A.P.M., Lipton M.S., Basset G.J., Merchant S.S. (2019). Multiomics resolution of molecular events during a day in the life of Chlamydomonas. Proc. Natl. Acad. Sci. USA 116: 2374–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surzycki S.J., Shellenbarger D.L. (1976). Purification and characterization of a putative sigma factor from Chalamydomonas reinhardi. Proc. Natl. Acad. Sci. USA 73: 3961–3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki L., Woessner J.P., Uchida H., Kuroiwa H., Yuasa Y., Waffenschmidt S., Goodenough U.W., Kuroiwa T. (2000). A zygote-specific protein with hydroxyproline-rich glycoprotein domains and lectin-like domains involved in the assembly of the cell wall of Chlamydomonas reinhardtii (Chlorophyta). J. Phycol. 36: 571–583. [DOI] [PubMed] [Google Scholar]
- Tabatabaei I., Dal Bosco C., Bednarska M., Ruf S., Meurer J., Bock R. (2019). A highly efficient sulfadiazine selection system for the generation of transgenic plants and algae. Plant Biotechnol. J. 17: 638–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarver J.E., Donoghue P.C., Peterson K.J. (2012). Do miRNAs have a deep evolutionary history? BioEssays 34: 857–866. [DOI] [PubMed] [Google Scholar]
- Tulin F., Cross F.R. (2014). A microbial avenue to cell cycle control in the plant superkingdom. Plant Cell 26: 4019–4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umen J.G., Goodenough U.W. (2001a). Control of cell division by a retinoblastoma protein homolog in Chlamydomonas. Genes Dev. 15: 1652–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umen J.G., Goodenough U.W. (2001b). Chloroplast DNA methylation and inheritance in Chlamydomonas. Genes Dev. 15: 2585–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umen J.G., Olson B.J. (2012). Genomics of volvocine algae. Adv. Bot. Res. 64: 185–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Lis R., Popek M., Couté Y., Kosta A., Drapier D., Nitschke W., Atteia A. (2017). Concerted up-regulation of aldehyde/alcohol dehydrogenase (ADHE) and starch in Chlamydomonas reinhardtii increases survival under dark anoxia. J. Biol. Chem. 292: 2395–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt J., Münzner P. (1987). The Chlamydomonas cell cycle is regulated by a light/dark-responsive cell-cycle switch. Planta 172: 463–472. [DOI] [PubMed] [Google Scholar]
- Voshall A., Kim E.J., Ma X., Yamasaki T., Moriyama E.N., Cerutti H. (2017). miRNAs in the alga Chlamydomonas reinhardtii are not phylogenetically conserved and play a limited role in responses to nutrient deprivation. Sci. Rep. 7: 5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Liu Y., Li S.S., Han G.Z. (2015a). Insights into the origin and evolution of the plant hormone signaling machinery. Plant Physiol. 167: 872–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Stessman D.J., Spalding M.H. (2015b). The CO2 concentrating mechanism and photosynthetic carbon assimilation in limiting CO2: How Chlamydomonas works against the gradient. Plant J. 82: 429–448. [DOI] [PubMed] [Google Scholar]
- Wiedeman V.E., Walne P.L., Trainor F.R. (1964). A new technique for obtaining axenic cultures of algae. Can. J. Bot. 42: 958–959. [Google Scholar]
- Wiggins I.L. (1962). Gilbert Morgan Smith, a Biographical Memoir. (Washington, D.C.: National Academy of Sciences; ). [Google Scholar]
- Wodniok S., Simon A., Glöckner G., Becker B. (2007). Gain and loss of polyadenylation signals during evolution of green algae. BMC Evol. Biol. 7: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollman F.A., Delepelaire P. (1984). Correlation between changes in light energy distribution and changes in thylakoid membrane polypeptide phosphorylation in Chlamydomonas reinhardtii. J. Cell Biol. 98: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollman F.A., Lemaire C. (1988). Studies on kinase-controlled state transitions in photosystem II and b6 jf mutants from Chlamydomonas reinhardtii which lack quinone-binding proteins. Biochim. Biophys. Acta 933: 85–94. [Google Scholar]
- Xu F., Wu X., Jiang L.H., Zhao H., Pan J. (2016). An organelle K+ channel is required for osmoregulation in Chlamydomonas reinhardtii. J. Cell Sci. 129: 3008–3014. [DOI] [PubMed] [Google Scholar]
- Yamano T., Iguchi H., Fukuzawa H. (2013). Rapid transformation of Chlamydomonas reinhardtii without cell-wall removal. J. Biosci. Bioeng. 115: 691–694. [DOI] [PubMed] [Google Scholar]
- Yamasaki T., Cerutti H. (2017). Cooperative processing of primary miRNAs by DUS16 and DCL3 in the unicellular green alga Chlamydomonas reinhardtii. Commun. Integr. Biol. 10: e1280208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., et al. (2014). Alternative acetate production pathways in Chlamydomonas reinhardtii during dark anoxia and the dominant role of chloroplasts in fermentative acetate production. Plant Cell 26: 4499–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan Y., et al. (2018). Pyrenoid functions revealed by proteomics in Chlamydomonas reinhardtii. PLoS One 13: e0185039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Wang L.P., Brauner M., Liewald J.F., Kay K., Watzke N., Wood P.G., Bamberg E., Nagel G., Gottschalk A., Deisseroth K. (2007). Multimodal fast optical interrogation of neural circuitry. Nature 446: 633–639. [DOI] [PubMed] [Google Scholar]
- Zhang H., Herman P.L., Weeks D.P. (1994). Gene isolation through genomic complementation using an indexed library of Chlamydomonas reinhardtii DNA. Plant Mol. Biol. 24: 663–672. [DOI] [PubMed] [Google Scholar]
- Zhang X., Yazaki J., Sundaresan A., Cokus S., Chan S.W., Chen H., Henderson I.R., Shinn P., Pellegrini M., Jacobsen S.E., Ecker J.R. (2006). Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126: 1189–1201. [DOI] [PubMed] [Google Scholar]
- Zhao T., Li G., Mi S., Li S., Hannon G.J., Wang X.J., Qi Y. (2007). A complex system of small RNAs in the unicellular green alga Chlamydomonas reinhardtii. Genes Dev. 21: 1190–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zones J.M., Blaby I.K., Merchant S.S., Umen J.G. (2015). High-resolution profiling of a synchronized diurnal transcriptome from Chlamydomonas reinhardtii reveals continuous cell and metabolic differentiation. Plant Cell 27: 2743–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y., Wenzel S., Müller N., Prager K., Jung E.M., Kothe E., Kottke T., Mittag M. (2017). An animal-like cryptochrome controls the Chlamydomonas sexual cycle. Plant Physiol. 174: 1334–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]