Cell enlargement and nuclear genome replication in the absence of mitosis involve the transcriptional repression of Anaphase Promoting Complex/Cyclosome activator genes by a Mediator complex subunit.
Abstract
Endoreduplication, the replication of the nuclear genome in the absence of mitosis, is often associated with cell growth and differentiation in plants and animals, but the molecular mechanisms underlying endoreduplication in plants have not been fully elucidated. Here, we show that the Mediator complex subunit MED16 acts as a negative regulator of endoreduplication to influence cell growth in Arabidopsis (Arabidopsis thaliana). The med16 mutant exhibits larger and more numerous cells than the wild type, resulting in enlarged organs. The large cells in med16 are associated with high DNA ploidy levels. MED16 associates with the promoters of the Anaphase Promoting Complex/Cyclosome activators CELL CYCLE SWITCH52 A1 (CCS52A1) and CCS52A2 (encoding important factors for endoreduplication and cell growth) and represses their expression. MED16 interacts physically with the transcriptional repressor DEL1 to repress the expression of CCS52A2. Genetic analysis suggested that MED16 is partially dependent on CCS52A1/A2 to control endoreduplication and cell growth. Our results indicate that the transcriptional repression of CCS52A1/A2 by MED16 regulates endoreduplication and cell growth in Arabidopsis.
INTRODUCTION
Plant organ growth and development begins with an initial proliferative phase, which results in an increase in cell number, followed by the cell expansion phase in which cell size increases. The transition from cell proliferation to cell expansion is often correlated with a switch from the mitotic cell cycle to the endoreduplication cycle, during which DNA rereplication is stimulated and mitosis is completely repressed, resulting in cells with higher ploidy levels (Sugimoto-Shirasu and Roberts, 2003; Breuer et al., 2010, 2014). Endoreduplication is a common feature among animals and plants and is frequently correlated with large cells. In Drosophila melanogaster, large cells with high DNA ploidy levels in the salivary glands are readily observed (Lilly and Duronio, 2005). Large cells in Arabidopsis (Arabidopsis thaliana) leaves are positively correlated with high DNA ploidy levels (Inzé and De Veylder, 2006; Gegas et al., 2014). In plants, endoreduplication is primarily controlled by the Anaphase Promoting Complex/Cyclosome (APC/C). The CELL CYCLE SWITCH52 proteins (CCS52A1/A2), two activators of the APC/C complex, are crucial for endoreduplication and influence cell growth in Arabidopsis. Leaves of the ccs52a1 and ccs52a2 mutants have smaller cells coupled with lower ploidy levels compared with the wild type (Larson-Rabin et al., 2009; Breuer et al., 2012; Liu et al., 2012; Baloban et al., 2013). The transcriptional repressor DEL1 (DP-E2F-LIKE1/E2Fe) specifically associates with the promoter of CCS52A2 and represses its expression (Lammens et al., 2008).
Transcriptional regulation is crucial for plant growth and development. The Mediator complex, an evolutionarily conserved transcriptional cofactor, mediates various signaling pathways from transcription factors to the RNA polymerase II machinery, thereby influencing gene expression (Kim et al., 1994; Koleske and Young, 1994). Several Mediator complex subunits influence various aspects of organ growth and development in Arabidopsis. For example, mutations in the Mediator complex subunit MED14 cause various defects in growth and development (Autran et al., 2002). Mutations in MED25 result in large organs with larger and slightly more cells than the wild type by influencing the expression of several expansin genes (Xu and Li, 2011). However, little is known about how Mediator complex subunits cooperate with transcription factors to regulate the expression of endoreduplication and cell growth-related genes in plants.
MED16 regulates flowering time, freezing tolerance, disease resistance, and iron homeostasis (Knight et al., 1999, 2008, 2009; Wathugala et al., 2012; Zhang et al., 2012, 2013, 2014; Hemsley et al., 2014; Yang et al., 2014b), but how MED16 influences endoreduplication and cell growth is currently unclear. Here, we show that MED16 functions as a negative regulator of endoreduplication and cell growth. MED16 associates with the promoters of CCS52A1 and CCS52A2 and represses their expression. MED16 physically interacts with the transcriptional repressor DEL1 to repress the expression of CCS52A2. Thus, the transcriptional repression of CCS52A1/A2 by MED16 controls endoreduplication and cell growth in Arabidopsis.
RESULTS
Identification of an Enhancer of da1-1
The Arabidopsis da1-1 mutant (DA means “large” in Chinese) forms large organs due to enhanced cell proliferation (Li et al., 2008; Dong et al., 2017). DA1 encodes a ubiquitin receptor with peptidase activity. To investigate the genetic mechanisms of DA1 action and identify other plant growth and developmental regulators, we searched for modifiers of da1-1 using ethyl methanesulfonate mutagenesis. One enhancer of da1-1, eod9-1, obviously increased organ growth in da1-1 (Figures 1A to 1G). The eod9-1 da1-1 double mutants exhibited much larger leaves than da1-1 (Figures 1A, 1B, and 1E). The eod9-1 da1-1 double mutants also formed larger flowers with larger petals and sepals than da1-1 (Figures 1C, 1D, 1F, and 1G). These results indicate that the eod9-1 mutation enhances the organ growth phenotypes of da1-1.
Figure 1.
eod9-1 Enhances the Phenotypes of da1-1.
(A) to (D) Thirty-two-day-old plants (A), the sixth leaves (B), sepals (C), and flowers (D) of Col-0, da1-1, and da1-1 eod9-1 (from left to right).
(E) Leaf area (LA), leaf length (LL), and leaf width (LW) of the sixth leaves of Col-0, da1-1, and da1-1 eod9-1 plants (n = 12).
(F) Sepal area (SA), sepal length (SL), and sepal width (SW) of Col-0, da1-1, and da1-1 eod9-1 plants (n = 70).
(G) Petal area (PA), petal length (PL), and petal width (PW) of Col-0, da1-1, and da1-1 eod9-1 plants (n = 80).
Error bars represent se. Different letters above the columns indicate significant differences among different groups, P < 0.05. Bars = 4 cm (A), 1 cm (B), and 1 mm ([C] and [D]).
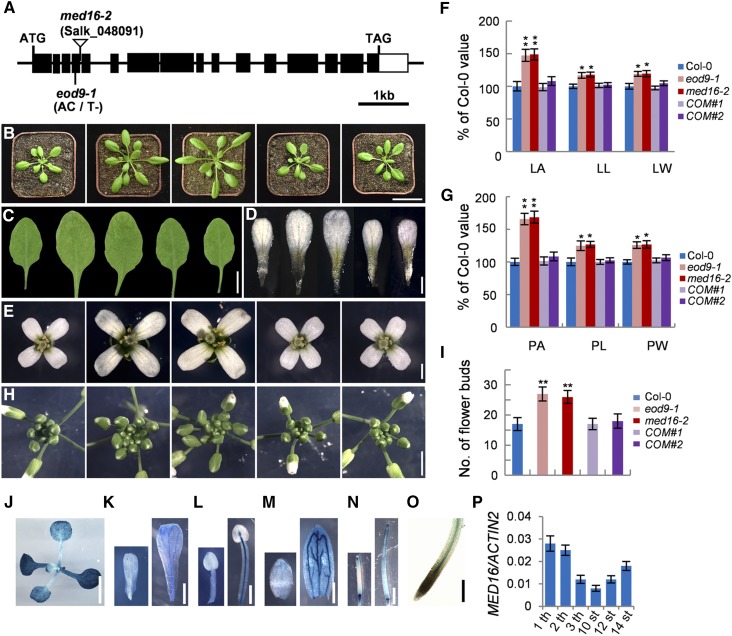
EOD9 Encodes MED16
To identify the eod9-1 mutation, we crossed eod9-1 da1-1Col-0 and da1-1Ler to obtain an F2 segregation population. The eod9-1 mutation was mapped to a genomic region between markers P3 and P6 on chromosome 4 (Supplemental Figure 1A). DNA sequencing revealed that eod9-1 has an A-to-T transition and a 1-bp deletion in the fourth exon of MED16, resulting in a frameshift that produces a truncated protein (Figure 2A; Supplemental Figure 1B). We developed the dCAPS1 marker based on these mutations in eod9-1, finding that it cosegregated with the eod9-1 phenotypes (Supplemental Figures 1A and 1C), suggesting that MED16 is the candidate gene.
Figure 2.
Identification and Molecular Characterization of the EOD9 Gene.
(A) MED16/EOD9 gene structure, showing the mutation site of eod9-1 and the T-DNA insertion site of med16-2. Black boxes represent exons, lines represent introns, and the white box represents the 3′ untranslated region.
(B) and (C) Plants before bolting (B) and the sixth leaves (C) of Col-0, eod9-1, med16-2, gMED16;eod9-1 #1 (COM#1), and gMED16;eod9-1 #2 (COM#2) (from left to right). The gMED16;eod9-1 plants were generated by transforming eod9-1 plants with a genomic fragment (gMED16) containing the 2080-bp promoter and the MED16 (At4g04920) gene.
(D) and (E) Petals (D) and flowers (E) of Col-0, eod9-1, med16-2, COM#1, and COM#2 plants (from left to right).
(F) Leaf area (LA), leaf length (LL), and leaf width (LW) of the sixth leaves of Col-0, eod9-1, med16-2, COM#1, and COM#2 plants (n = 12).
(G) Petal area (PA), petal length (PL), and petal width (PW) of Col-0, eod9-1, med16-2, COM#1, and COM#2 plants (n = 80).
(H) and (I) Inflorescences (H) and number of flower buds (I) of Col-0, eod9-1, med16-2, COM#1, and COM#2 plants (n = 6).
(J) to (O) The expression patterns of MED16 in 9-d-old seedlings (J), developing petals (K), stamens (L), sepals (M), siliques (N), and root tips (O) of MED16pro:GUS plants.
(P) The expression levels of MED16 in the first leaf (1 th), second leaf (2 th), and third leaf (3 th) from 9-d-old Col-0 seedlings and 10th stage petals (10 st), 12th stage petals (12 st), and 14th stage petals (14 st) from Col-0 flowers (n = 3).
Error bars represent se. Asterisks indicate significant differences from Col-0: * P < 0.05 and ** P < 0.01. Bars = 4 cm (B), 1 cm (C), 0.25 cm ([J] and [N]), 3 mm (H), 1 mm ([D], [E], [K], and [M]), 0.5 mm (L), and 100 μm (O).
We isolated the eod9-1 single mutant from an eod9-1 da1-1/Col-0 F2 population and backcrossed it three times into the wild-type Col-0 background. The eod9-1 single mutant produced obviously longer and wider leaves and petals than the wild type (Figures 2B and 2E to 2G). The inflorescences of eod9-1 were also markedly larger than those of the wild type, resulting in more floral buds (Figures 2H and 2I). We obtained the T-DNA insertion homozygous mutant, med16-2 (SALK_048091; Knight et al., 2009; Zhang et al., 2014). We identified the T-DNA insertion in the fourth intron of MED16 by PCR analysis using T-DNA-specific and flanking primers and by sequencing the PCR products (Supplemental Data Set 1; Supplemental Figures 2A and 2B). The expression of MED16 was barely detected in med16-2 (Supplemental Figure 2C), indicating that med16-2 is a loss-of-function allele. Similar to eod9-1, med16-2 formed larger leaves, flowers, and inflorescences and more floral buds than the wild type (Figures 2B to 2I; Supplemental Figure 3), further suggesting that MED16 is the EOD9 gene. The identity of the EOD9 gene was confirmed by transforming eod9-1 plants with a genomic fragment (gMED16) containing a 2080-bp promoter and the MED16 (At4g04920) gene. The phenotypes of eod9-1 were rescued in gMED16;eod9-1 transgenic plants (Figures 2B to 2I), demonstrating that EOD9 encodes the MED16 gene product. MED16 affects stress responses, flowering time, and iron homeostasis in Arabidopsis (Knight et al., 1999, 2008, 2009; Wathugala et al., 2012; Zhang et al., 2012, 2013, 2014; Hemsley et al., 2014; Yang et al., 2014b), but how it affects organ growth and development has been unclear.
To examine the tissue-specific expression patterns of MED16, we generated MED16pro:GUS transgenic Arabidopsis plants containing a 2080-bp region of the promoter of MED16. In seedlings, the leaves and roots showed GUS activity (Figures 2J and 2O; Supplemental Figure 4). In flowers, MED16 expression was detected in petals, stamens, sepals, and siliques (Figures 2K to 2N). These expression patterns of MED16 are consistent with its roles in controlling leaf and flower size. Interestingly, we detected higher expression of MED16 in relatively old organs versus younger ones (Figures 2J to 2N and 2P). GUS activity was detected throughout the leaves of MED16pro:GUS, although GUS activity gradually decreased from the tip to the basal region of the leaf at different time points of GUS staining (Supplemental Figure 4B), indicating that MED16 is expressed during cell proliferation and cell expansion. In addition, GUS in MED16pro:GUS plants was expressed in the cell proliferative region, which was marked by GUS activity in the CYCLINB1;1pro:CDB-GUS marker line (harboring a cell cycle reporter construct) after 4 h of staining (Supplemental Figure 4A). To examine the subcellular localization of MED16, we expressed a MED16-green fluorescent protein (GFP) fusion protein driven by the 35S promoter. GFP fluorescence was observed exclusively in the nuclei of petal cells from 35S:MED16-GFP transgenic plants (Supplemental Figures 5A to 5D), which is in agreement with previous findings (Knight et al., 2009; Yang et al., 2014b; Zhang et al., 2014).
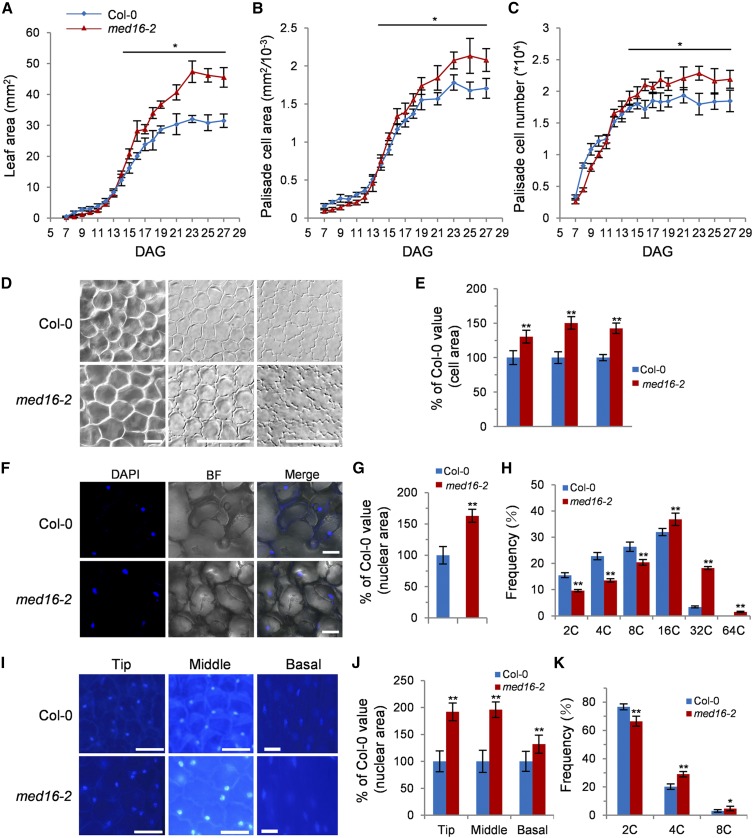
MED16 Is Required for Normal Endoreduplication and Cell Growth
To further explore the functions of MED16, we traced leaf development over time by harvesting the first pair of leaves from med16-2 and wild-type plants to measure the leaf area and quantify cell area and cell number. At 14 DAG (days after germination), the area of med16-2 leaves was significantly larger than that of wild-type leaves, which resulted from more and larger cells (Figures 3A to 3C), indicating that MED16 is involved in regulating both cell proliferation and cell expansion during leaf development.
Figure 3.
MED16 Is Involved in Regulating Cell Proliferation, Cell Growth, and Endoreduplication.
(A) to (C) Leaf area (A), palisade cell area (B), and palisade cell number (C) of the first pair of leaves in Col-0 and med16-2 from 5 to 25 DAG (n = 4).
(D) The palisade cells in the sixth leaves and adaxial and abaxial epidermis cells in petals of Col-0 and med16-2 (from left to right).
(E) The area of the palisade cells in the sixth leaves and adaxial and abaxial epidermis cells in petals of Col-0 and med16-2 (n = 300).
(F) DAPI staining of the sixth leaves of Col-0 and med16-2.
(G) Nuclear area of the sixth leaves of Col-0 and med16-2 (n = 40).
(H) Distribution of nuclear ploidy in the sixth leaves of Col-0 and med16-2 (n = 3).
(I) DAPI staining of the tip, middle, and basal regions of Col-0 and med16-2 petals.
(J) The nuclear area in the tip, middle, and basal regions of petals from Col-0 and med16-2 (n = 40).
(K) Distribution of nuclear ploidy in Col-0 and med16-2 petals (n = 3).
Error bars represent se. Asterisks indicate significant differences from Col-0: * P < 0.05 and ** P < 0.01. Bars = 50 μm (D) and 10 μm ([F] and [I]).
In accordance with the first pair of leaves, the med16-2 mutant had significantly larger cells in the sixth leaves and petals than the wild type (Figures 3D and 3E). As cell size is frequently correlated with the size of the nucleus and DNA ploidy levels in plants (Joubès and Chevalier, 2000; Sugimoto-Shirasu and Roberts, 2003), we measured the nuclei of cells in the sixth leaves and petals of the wild type and med16-2. As shown in Figures 3F, 3G, 3I, and 3J, the nuclear area of cells in med16-2 was obviously larger than that in the wild type. To investigate whether the enlargement of cells and nuclei in med16-2 leaves and petals is associated with an increase in DNA ploidy levels, we performed flow cytometry of the nuclei from the sixth leaves and petals of the wild type and med16-2. There were substantially more 16C and 32C nuclei in med16-2 leaves than in wild-type leaves. 64C nuclei were observed in med16-2 leaves but not in wild-type leaves. By contrast, the population of 2C, 4C, and 8C nuclei in med16-2 leaves was lower than that in wild-type leaves (Figure 3H; Supplemental Figures 6A and 6B). Similarly, higher DNA ploidy was observed in the nuclei of med16-2 petals compared with wild-type petals (Figure 3K; Supplemental Figures 6C and 6D). These results indicate that MED16 regulates endoreduplication and cell growth.
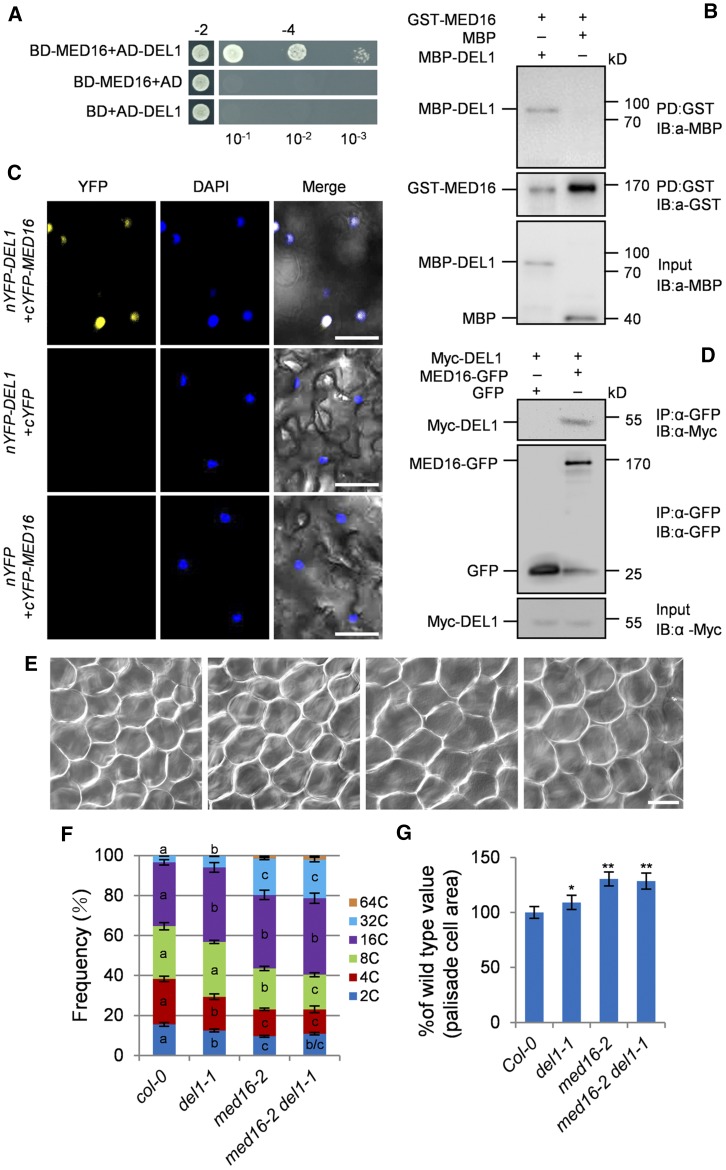
MED16 Physically Associates with the Transcriptional Repressor DEL1 in Arabidopsis
To explore the roles of MED16 in endoreduplication and cell growth, we performed yeast two-hybrid screening to identify MED16-interacting proteins. Several candidate proteins were identified in this screen (Supplemental Table). One of these proteins was the transcriptional repressor, DEL1. This protein controls endoreduplication by binding to the promoter of CCS52A2 and repressing its expression (Lammens et al., 2008), implying that DEL1 is a good candidate for a MED16-interacting protein. We further confirmed that MED16 interacted with the full-length DEL1 in yeast cells (Figure 4A). We then used a pull-down assay to verify the interaction between MED16 and DEL1. As shown in Figure 4B, glutathione S-transferase (GST)-MED16 physically interacted with MBP-DEL1 in vitro.
Figure 4.
MED16 Interacts Physically and Genetically with DEL1 to Control Cell Size and Endoreduplication.
(A) MED16 interacts with DEL1 in yeast cells. The BD-MED16 and AD-DEL1 constructs were cotransformed into Y2H Gold yeast cells. Cotransformed yeast cells were selected on medium −2 (SD/-Leu/-Trp). The interaction was tested on medium −4 (SD/-Ade/-His/-Leu/-Trp) with different dilution series (10−1, 10−2, and 10−3). BD-MED16/AD and BD/AD-DEL1 were used as the negative controls. BD and AD represent the pGBKT7 and pGADT7 vectors, respectively.
(B) Pull-down assay showing the interaction between MED16 and DEL1 in vitro. All of the proteins were expressed in E. coli BL21 (DE3). MBP or MBP-DEL1 was incubated with GST-MED16 and pulled down by GST-Trap-A agarose beads. The interactions were detected by immunoblotting with anti-GST or anti-MBP antibody, respectively.
(C) Bimolecular fluorescence complementation assays showing that MED16 interacts with DEL1 in N. benthamiana. cYFP-MED16 and nYFP-DEL1 were coexpressed in N. benthamiana leaves. YFP fluorescence was observed with a laser-scanning confocal microscope. Blue dots represent nuclei after DAPI staining. Bars = 50 μm.
(D) Coimmunoprecipitation analysis showing the interaction between MED16 and DEL1 in Arabidopsis. Total protein extracts of 35S:Myc-DEL1;35S:GFP and 35S:Myc-DEL1;35S:MED16-GFP transgenic plants were incubated with GFP-Trap-A agarose beads, and precipitates were detected by immunoblotting with anti-GFP or anti-Myc antibody, respectively.
(E) The palisade cells in the sixth leaves of Col-0, del1-1, med16-2, and med16-2 del1-1 (from left to right). Bar = 50 μm.
(F) Distribution of nuclear ploidy in the sixth leaves of Col-0, del1-1, med16-2, and med16-2 del1-1 (n = 3).
(G) Relative palisade cell area of the sixth leaves of Col-0, del1-1, med16-2, and med16-2 del1-1 (n = 300).
Error bars represent se. Different letters in columns of the same color indicate significant differences among different groups: P < 0.05. Asterisks indicate significant differences from Col-0: * P < 0.05 and ** P < 0.01.
We confirmed the interaction between MED16 and DEL1 in planta by performing bimolecular fluorescence complementation assays. We transiently coexpressed C-terminal yellow fluorescent protein (cYFP)-MED16 with N-terminal (n)YFP-DEL1 in wild tobacco (Nicotiana benthamiana) leaves. As shown in Figure 4C, when we coexpressed cYFP-MED16 with nYFP-DEL1, strong YFP fluorescence was observed in the nuclei of epidermal cells. To investigate whether MED16 associates with DEL1 in Arabidopsis, we generated a 35S:Myc-DEL1 transgenic Arabidopsis line and crossed it with 35S:MED16-GFP or 35S:GFP transgenic plants to obtain 35S:Myc-DEL1;35S:MED16-GFP and 35S:Myc-DEL1;35S:GFP plants, respectively. Coimmunoprecipitation analysis revealed that DEL1 physically associated with MED16 but not with the GFP control (Figure 4D). These results indicate that MED16 and DEL1 can form a protein complex in Arabidopsis.
MED16 Acts in a Common Genetic Pathway with DEL1 to Control Endoreduplication and Cell Growth
DEL1 represses CCS52A2 expression, thereby regulating endoreduplication in Arabidopsis (Lammens et al., 2008). Since MED16 interacts with DEL1, we asked whether MED16 and DEL1 act in a common pathway to control endoreduplication. To test this, we crossed med16-2 with del1-1 to generate the med16-2 del1-1 double mutant. We performed a flow cytometry assay with the nuclei of the sixth leaves of various plants. The del1-1 mutant had higher ploidy levels than the wild type (Figure 4F; Supplemental Figure 7), which is consistent with previous reports (Vlieghe et al., 2005; Lammens et al., 2008; Heyman et al., 2017). Similarly, med16-2 showed increased ploidy levels compared with the wild type. med16-2 del1-1 leaves contained similar levels of 64C, 32C, and 16C cells to med16-2 single mutant leaves (Figure 4F; Supplemental Figure 7). We then examined the sizes of palisade cells in wild-type, med16-2, del1-1, and med16-2 del1-1 leaves. Palisade cells were significantly larger in del1-1 leaves than in wild-type leaves (Figures 4E and 4G). med16-2 leaves also contained larger palisade cells than wild-type leaves (Figures 4E and 4G). The size of palisade cells in med16-2 del1-1 leaves was comparable to that of med16-2 leaves (Figures 4E and 4G). These results suggest that MED16 and DEL1 act in a common genetic pathway to control endoreduplication and cell growth.
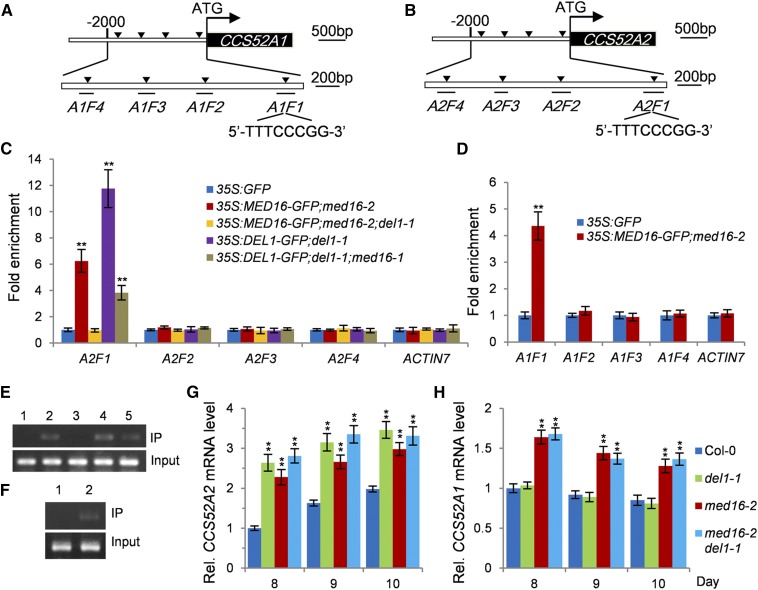
MED16 Associates with the Promoters of CCS52A1/A2 and Represses Their Expression
The transcriptional repressor, DEL1, regulates endoreduplication by specifically binding to the typical E2F cis-acting element in the promoter of CCS52A2 and repressing its expression (Lammens et al., 2008). MED16 physically and genetically interacts with DEL1 and regulates endoreduplication and cell growth. We therefore asked whether MED16 associates with the promoter of CCS52A2 via DEL1 and regulates its expression. To test this, we conducted chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR) analysis using 35S:GFP and 35S:MED16-GFP;med16-2 plants. As shown in Figures 5B, 5C, and 5E, the A2F1 fragment in the promoter of CCS52A2 containing a typical E2F cis-acting element (5′-TTTCCCGG-3′) was strongly enriched compared with the other fragments (A2F2–A2F4) without the typical E2F cis-acting element and the negative control (a fragment of the ACTIN7 promoter), indicating that MED16 associates with the promoter of CCS52A2 in vivo. We then asked whether MED16 associates with the promoter of CCS52A2 through DEL1. To address this, we generated 35S:DEL1-GFP;del1-1 and 35S:MED16-GFP;med16-2;del1-1 plants. As shown in Figures 5C and 5E, MED16 did not associate with the A2F1 fragment in the promoter of CCS52A2 in 35S:MED16-GFP;med16-2;del1-1 plants, revealing that the association of MED16 with the CCS52A2 promoter depends on the transcriptional repressor, DEL1. We then asked whether MED16 influences the association of DEL1 with the CCS52A2 promoter. DEL1 associated with the A2F1 fragment in the CCS52A2 promoter in 35S:DEL1-GFP;del1-1, which is consistent with a previous study (Lammens et al., 2008). By contrast, the enrichment of the A2F1 fragment in the promoter of CCS52A2 in 35S:DEL1-GFP;del1-1;med16-2 plants was significantly reduced compared with that in 35S:DEL1-GFP;del1-1 plants (Figure 5C), indicating that MED16 also influences the association of DEL1 with the CCS52A2 promoter.
Figure 5.
MED16 Associates with the Promoters of CCS52A1/A2 and Represses Their Expression.
(A) Schematic diagram of the CCS52A1 promoter containing a typical E2F binding box (5′-TTTCCCGG-3′) in the A1F1 fragment. A1F1 to A1F4 represent the DNA fragments used for ChIP-qPCR analysis.
(B) Schematic diagram of the CCS52A2 promoter containing a typical E2F binding box (5′-TTTCCCGG-3′) in the A2F1 fragment. A2F1 to A2F4 represent the DNA fragments used for ChIP-qPCR analysis.
(C) ChIP-qPCR assays showing that MED16 associates with the promoter of CCS52A2 in planta. Chromatin from 35S:GFP, 35S:MED16-GFP;med16-2, 35S:MED16-GFP;med16-2;del1-1, 35S:DEL1-GFP;del1-1, and 35S:DEL1-GFP;del1-1;med16-2 seedlings at 16 DAG was incubated with ChIP anti-GFP antibody and coprecipitated by ChIP protein A+G magnetic beads. The enrichment of the fragments was determined by qPCR. The ACTIN7 promoter was used as a negative control. Error bars represent se. Asterisks indicate significant differences from Col-0: ** P < 0.01 (n = 3).
(D) ChIP-qPCR assays showing that MED16 associates with the promoter of CCS52A1 in planta. Chromatin from 35S:GFP and 35S:MED16-GFP;med16-2 seedlings at 16 DAG was incubated with ChIP anti-GFP antibody and coprecipitated by ChIP protein A+G magnetic beads. The enrichment of the fragments was determined by qPCR. The ACTIN7 promoter was used as a negative control. Error bars represent se. Asterisks indicate significant differences compared with the 35S:GFP control: ** P < 0.01 (n = 3).
(E) The A2F1 fragment products of the CCS52A2 promoter from ChIP-qPCR analysis examined by agarose gel electrophoresis. Chromatin prior to immunoprecipitation from 35S:GFP, 35S:MED16-GFP;med16-2, 35S:MED16-GFP;med16-2;del1-1, 35S:DEL1-GFP;del1-1, and 35S:DEL1-GFP;del1-1;med16-2 seedlings was used as input. Lanes 1 to 5 represent the A2F1 fragment products of 35S:GFP, 35S:MED16-GFP;med16-2, 35S:MED16-GFP;med16-2;del1-1, 35S:DEL1-GFP;del1-1, and 35S:DEL1-GFP;del1-1;med16-2, respectively.
(F) The A1F1 fragment products of the CCS52A1 promoter from ChIP-qPCR analysis examined by agarose gel electrophoresis. Chromatin prior to immunoprecipitation from 35S:GFP and 35S:MED16-GFP;med16-2 seedlings was used as input. Lanes 1 and 2 represent the A2F1 fragment products of 35S:GFP and 35S:MED16-GFP;med16-2, respectively.
(G) and (H) Relative expression levels of CCS52A2 (G) and CCS52A1 (H) in the first pair of leaves from 8- to 10-d-old Col-0, med16-2, del1-1, and med16-2 del1-1 seedlings detected by qPCR. Error bars represent se. Asterisks indicate significant differences from Col-0: ** P < 0.01 (n = 3).
Considering that the promoters of both CCS52A1 and CCS52A2 contain a typical E2F cis-acting element (Figures 5A and 5B; Supplemental Figure 8), we asked whether MED16 also associates with the CCS52A1 promoter. We performed ChIP-qPCR assays using 35S:GFP and 35S:MED16-GFP;med16-2 plants. As shown in Figures 5D and 5F, the A1F1 fragment in the CCS52A1 promoter (containing the typical E2F cis-acting element) was significantly enriched compared with the other fragments in the CCS52A1 promoter (A1F2–A1F4) and the negative control (a fragment of the ACTIN7 promoter), indicating that MED16 also associates with the CCS52A1 promoter in Arabidopsis.
Considering that MED16 associates with the promoters of both CCS52A1 and CCS52A2, which regulate endoreduplication and cell growth (Fülöp et al., 2005; Larson-Rabin et al., 2009; Vanstraelen et al., 2009; Breuer et al., 2012; Baloban et al., 2013), we asked whether MED16 affects the expression of CCS52A1 and CCS52A2 in Arabidopsis. We examined the expression levels of CCS52A1/A2 in the first pair of leaves from 8- to 10-d-old wild-type, del1-1, med16-2, and med16-2 del1-1 seedlings by qPCR. As shown in Figures 5G and 5H, CCS52A2 expression gradually increased, and CCS52A1 expression gradually decreased, in 8- to 10-d-old wild-type seedlings, which is consistent with previous results (Lammens et al., 2008). The expression levels of CCS52A1 and CCS52A2 were significantly higher in med16-2 compared with the wild type. The del1-1 mutation increased the expression of CCS52A2 but did not affect the expression of CCS52A1, which is consistent with previous findings (Lammens et al., 2008; Heyman et al., 2017). Interestingly, the expression level of CCS52A2 in the med16-2 del1-1 double mutant was similar to that in the del1-1 single mutant, suggesting that MED16 relies on DEL1 to repress the expression of CCS52A2. Together, these results demonstrate that MED16 associates with the promoters of CCS52A1/A2 and represses their expression.
MED16 Acts through CCS52A1/A2 to Control Endoreduplication and Cell Growth
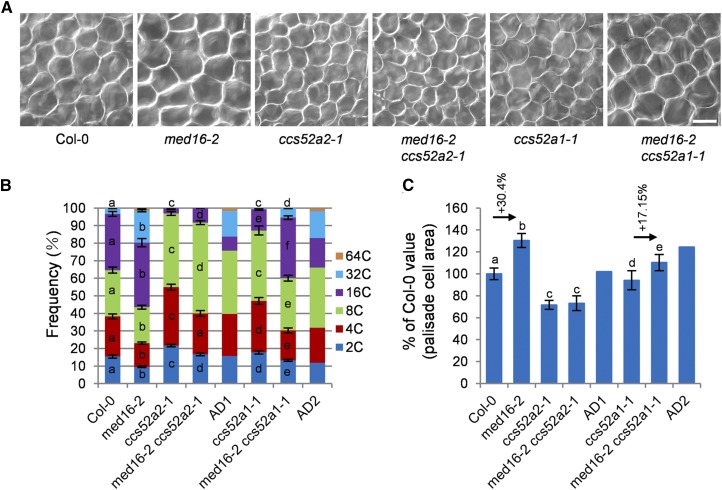
CCS52A1/A2, two activators of the APC/C, affect endoreduplication and cell growth (Vanstraelen et al., 2009; Liu et al., 2012; Baloban et al., 2013). MED16 associates with the CCS52A1 and CCS52A2 promoters and represses their expression. We therefore asked whether MED16 and CCS52A1/A2 might function in a common genetic pathway to control endoreduplication and cell growth. We crossed med16-2 with ccs52a2-1 (SALK_001978) or ccs52a1-1 (SALK_082656) to generate the med16-2 ccs52a2-1 and med16-2 ccs52a1-1 double mutants, respectively. The ccs52a2-1 plants were smaller than the wild type (Supplemental Figure 9), which is consistent with previous results (Baloban et al., 2013). med16-2 ccs52a2-1 double mutant plants showed similar morphology to ccs52a2-1 plants (Figures 6A to 6C; Supplemental Figure 9). We performed a flow cytometry assay of the nuclei of the sixth leaves. As shown in Figure 6B and Supplemental Figure 10, the ccs52a2-1 mutant contained more 2C, 4C, and 8C nuclei, fewer 16C nuclei, and no 32C nuclei compared with the wild type, indicating that the ccs52a2-1 mutant has reduced ploidy levels in leaves, which is consistent with previous findings (Liu et al., 2012; Baloban et al., 2013). The ccs52a2-1 mutation mostly but not entirely suppressed the high-ploidy phenotype of med16-2 (Figure 6B; Supplemental Figure 10). We then examined the size of palisade cells in wild-type, med16-2, ccs52a2-1, and med16-2 ccs52a2-1 leaves. As shown in Figures 6A and 6C, the size of palisade cells in med16-2 ccs52a2-1 leaves was similar to that in ccs52a2-1 leaves. These results indicate that ccs52a2-1 is epistatic to med16-2 with respect to cell size and mostly but not entirely suppresses the endoreduplication phenotype of med16-2.
Figure 6.
MED16 Genetically Interacts with CCS52A1/A2 to Control Endoreduplication and Cell Growth.
(A) Palisade cells in the sixth leaves of Col-0, med16-2, ccs52a2-1, med16-2 ccs52a2-1, ccs52a1-1, and med16-2 ccs52a1-1 (from left to right). Bar = 50 μm.
(B) Distribution of nuclear ploidy in the sixth leaves of Col-0, med16-2, ccs52a2-1, med16-2 ccs52a2-1, ccs52a1-1, and med16-2 ccs52a1-1 (n = 3). AD1 indicates the expected med16-2 ccs52a2-1 value if med16-2 and ccs52a2-1 have additive effects. AD2 indicates the expected med16-2 ccs52a1-1 value if med16-2 and ccs52a1-1 have additive effects. Different letters in columns of the same color indicate significant differences among different groups: P < 0.05. Error bars represent se.
(C) Palisade cell area in the sixth leaves of Col-0, med16-2, ccs52a2-1, med16-2 ccs52a2-1, ccs52a1-1, and med16-2 ccs52a1-1 (n = 300). AD1 indicates the expected med16-2 ccs52a2-1 value if med16-2 and ccs52a2-1 have additive effects. AD2 indicates the expected med16-2 ccs52a1-1 value if med16-2 and ccs52a1-1 have additive effects. Different letters above the columns indicate significant differences among different groups: P < 0.05. Error bars represent se.
We then investigated the genetic interaction between MED16 and CCS52A1. The ccs52a1-1 mutant plants appeared to be slightly smaller than the wild type (Supplemental Figure 9), which is consistent with a previous study (Baloban et al., 2013). Compared with med16-2 plants, med16-2 ccs52a1-1 plants were smaller (Supplemental Figure 9). We conducted flow cytometry assays of the nuclei of the sixth leaves. The ccs52a1-1 mutant showed reduced ploidy levels in leaves compared with the wild type (Figure 6B; Supplemental Figure 10), which is consistent with previous findings (Larson-Rabin et al., 2009; Baloban et al., 2013). The ccs52a1-1 mutation strongly suppressed the high-ploidy phenotype of med16-2 (Figure 6B; Supplemental Figure 10). The average DNA ploidy level of med16-2 increased by 57.8% compared with that of wild-type plants. By contrast, the average DNA ploidy level of med16-2 ccs52a1-1 only increased by 24.14% compared with ccs52a1-1 (Supplemental Figure 10B). These results indicate that the ccs52a1-1 mutation strongly suppresses the DNA ploidy level of med16-2 and that MED16 acts partially through CCS52A1 to control endoreduplication. Finally, we examined the size of palisade cells in wild-type, med16-2, ccs52a1-1, and med16-2 ccs52a1-1 leaves. The palisade cells of med16-2 were 30.4% larger than those of wild-type plants, but the palisade cells of med16-2 ccs52a1-1 were only 17.15% larger than those of ccs52a1-1 (Figure 6C), indicating that MED16 functions partially through CCS52A1 to control cell growth.
DISCUSSION
Endoreduplication is often associated with cell growth and differentiation in plants and animals, but the mechanisms underlying plant endoreduplication have not been fully elucidated. In this study, we demonstrated that the Mediator subunit MED16 associates with the promoters of CCS52A1/A2 and represses their expression, thereby regulating endoreduplication and cell growth in Arabidopsis. Our results support the notion that the transcriptional repression of CCS52A1/A2 by MED16 regulates endoreduplication and cell growth in Arabidopsis.
The Mediator complex transduces information from transcription factors to RNA polymerase II, thereby influencing transcription (Malik and Roeder, 2005). The Mediator complex subunits function in various processes in Arabidopsis, such as cold responses, embryo patterning, defensive responses, and flowering time (Autran et al., 2002; Dhawan et al., 2009; Kidd et al., 2009; Gillmor et al., 2010). Here, we identified the eod9/med16 mutant as an enhancer of da1-1. The genetic data indicate that EOD9/MED16 acts independently of DA1 to control organ growth (Supplemental Figure 11). MED16 regulates multiple biological processes, such as cold responses, plant basal defense and abiotic stress responses, and iron homeostasis (Knight et al., 1999, 2008, 2009; Wathugala et al., 2012; Zhang et al., 2012, 2013, 2014; Hemsley et al., 2014; Yang et al., 2014b), but how MED16 affects endoreduplication and cell growth has been unclear. Here, we showed that MED16 negatively regulates endoreduplication, cell growth, as well as cell proliferation (Figures 3A to 3C). We previously demonstrated that MED25/EOD8 controls cell expansion and cell proliferation in Arabidopsis (Xu and Li, 2011). MED25 influences cell expansion by repressing the expression of several expansin genes but does not affect endoreduplication (Xu and Li, 2011). These findings suggest that MED25 and MED16 use different mechanisms to regulate cell expansion. Consistent with this notion, our genetic analysis suggested that MED16 and MED25 function independently to control organ growth (Supplemental Figure 12). In addition, MED14/SWP promotes cell proliferation and limits cell growth (Autran et al., 2002). MED14 interacts with the transcription corepressor LEUNIG to regulate gene expression (Gonzalez et al., 2007). Thus, it is possible that different modular Mediator complexes coexist in the nucleus, which might mediate the transduction of various signals from distinct transcription factors to regulate gene expression.
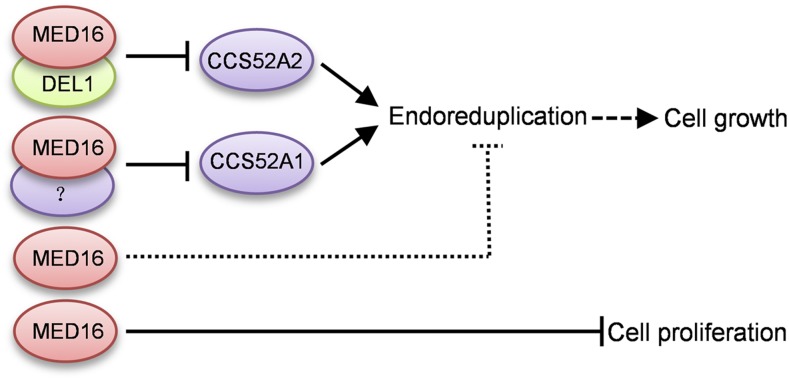
Mediator complex subunits can act as transcriptional coactivators or corepressors, depending on whether they associate with transcription factors that activate or repress gene transcription. For example, Arabidopsis MED18 interacts with the transcription factor YIN YANG1 to bind to the promoter regions of disease-related genes and represses their expression (Meng, 2015). MED14 interacts with the transcription corepressor LEUNIG to regulate gene expression (Gonzalez et al., 2007). By contrast, MED18 also associates with the transcription factor ABI4 (ABSCISIC ACID INSENSITIVE4) to activate ABI5 expression (Meng, 2015). In the current study, we determined that MED16 physically interacts with the transcriptional repressor DEL1 in vitro and in vivo (Figures 4A to 4D). DEL1 specifically binds to the promoter of CCS52A2 and represses its expression (Lammens et al., 2008). MED16 associates with the CCS52A1/A2 promoters and represses their expression (Figures 5C to 5H). DEL1 is required for the association of MED16 with the CCS52A2 promoter. Meanwhile, MED16 influences the association of DEL1 with the CCS52A2 promoter (Figures 5C and 5E). Therefore, MED16 and DEL1 interact with each other to repress CCS52A2 expression. Consistent with previous reports (Lammens et al., 2008; Heyman et al., 2017), CCS52A1 expression levels were similar in del1-1 and Col-0 plants (Figure 5H), indicating that DEL1 is not involved in the transcriptional regulation of CCS52A1. DEL1 binds to the promoter of CCS52A2 but not CCS52A1 (Lammens et al., 2008). Perhaps MED16 interacts with other (currently unknown) transcription factors to repress CCS52A1 expression (Figure 7).
Figure 7.
Working Model of the Role of MED16 in Controlling Cell Growth and Proliferation.
MED16 regulates endoreduplication by associating with the promoters of CCS52A1 and CCS52A2 and repressing their expression. MED16 interacts with DEL1 to repress CCS52A2 expression by binding to its promoter. MED16 also associates with the promoter of CCS52A1 and represses its expression in conjunction with an unknown transcription factor. In addition, MED16 limits cell proliferation via an unknown mechanism. The dashed line with arrow represents partially dependent relationships. The dotted line represents an unknown mechanism.
The med16 mutant showed increased cell size and cell number (Figures 3A to 3E). The del1 mutants have large cells and reduced cell number (Heyman et al., 2017). Therefore, perhaps MED16 acts independently of DEL1 to control cell proliferation (Figure 7). We compared expression patterns of MED16 and DEL1 during leaf development in MED16pro:GUS and DEL1pro:GUS lines (Supplemental Figure 13A). The expression levels of MED16 in the first pair of leaves in 5- and 6-d-old seedlings gradually decreased from the tip to the basal region of the leaf. By contrast, the expression levels of DEL1 in the first pair of leaves in 5- and 6-d-old seedlings gradually increased from the tip to the basal region. Although MED16 and DEL1 were highly expressed in the tip region and the basal region of the first pair of leaves, respectively, their regions of expression partially overlapped (Supplemental Figure 13A). In addition, we cut the first pair of leaves from the 8- to 10-d-old Col-0 and med16-2 seedlings into top and basal halves and investigated the expression of MED16, DEL1, CCS52A1, and CCS52A2 on agarose gels by RT-PCR. As shown in Supplemental Figure 13B, MED16, DEL1, CCS52A1, and CCS52A2 expression was detected in both the top and basal regions in the first pair of leaves from 8- to 10-d-old Col-0 seedlings, suggesting that MED16, DEL1, CCS52A1, and CCS52A2 have at least partially overlapping regions of expression during leaf development. The expression patterns of CCS52A1 and CCS52A2 are consistent with the GUS activity detected in CCS52A1pro:GUS and CCS52A2pro:GUS plants (Liu et al., 2012; Baloban et al., 2013). DEL1 expression gradually decreased from 8 to 10 d in both top and basal regions, while the expression of CCS52A2 in both the top and basal regions gradually increased, which is consistent with a previous study (Lammens et al., 2008). CCS52A1 and CCS52A2 were expressed at higher levels in med16-2 than in Col-0, which is consistent with the results of qPCR (Figures 5G and 5H). MED16 expression in both the top and basal regions of leaves gradually increased in 8- to 10-d-old Col-0 leaves. Considering that MED16 regulates multiple physiological processes, such as stress responses, flowering time, iron homeostasis, and so on (Knight et al., 1999, 2008, 2009; Wathugala et al., 2012; Zhang et al., 2012, 2013, 2014; Hemsley et al., 2014; Yang et al., 2014b), it seems reasonable that MED16 and DEL1 have partially overlapping expression regions in developing leaves and have different expression levels. Similarly, different proteins in the same complex or pathway have been shown to have partially overlapping expression patterns in several studies (Cortellino et al., 2009; Fernández-Calvo et al., 2011).
CCS52A2 and its homolog, CCS52A1, regulate endoreduplication and cell growth in Arabidopsis (Fülöp et al., 2005; Larson-Rabin et al., 2009; Vanstraelen et al., 2009; Breuer et al., 2012; Liu et al., 2012; Baloban et al., 2013). CCS52A1 and CCS52A2 promote endoreduplication and arrest cell division. Both loss of function and overexpression of CCS52A2 result in reduced cell number in leaves (Lammens et al., 2008; Larson-Rabin et al., 2009; Baloban et al., 2013). By contrast, med16-2 produced more cells in leaves than the wild type. Therefore, perhaps MED16 acts independently of CCS52A1/A2 to control cell proliferation (Figure 7). Genetic analysis showed that ccs52a2-1 is epistatic to med16-2 with regard to cell size (Figures 6A and 6C). The ccs52a2-1 mutation mostly but not entirely suppressed the endoreduplication phenotype of med16-2 (Figure 6B). The ccs52a1-1 mutation strongly but not entirely suppressed the endoreduplication and cell size phenotypes of med16-2 (Figures 6A to 6C), suggesting that MED16 acts partially through CCS52A1 to regulate endoreduplication and cell size. The ccs52a1 or ccs52a2 mutation did not entirely suppress the endoreduplication phenotype of med16-2, suggesting that MED16 also regulates endoreduplication through other (as yet unknown) pathways (Figure 7). Thus, these findings support the notion that the transcriptional repression of CCS52A1/A2 by MED16 is crucial for endoreduplication and cell growth in Arabidopsis.
METHODS
Plant Materials and Growth Conditions
The mutants used in this study were in the Arabidopsis (Arabidopsis thaliana) Col-0 ecotype background, apart from da1-1Ler in the Landsberg erecta background. The da1-1 eod9-1 double mutant was obtained from an M2 population of da1-1 treated with ethyl methanesulfonate. The eod9-1 single mutant was isolated from an eod9-1 da1-1/Col-0 F2 population and backcrossed three times into wild-type Col-0. The T-DNA insertion lines med16-2 (SALK_048091), ccs52a1-1 (SALK_082656), ccs52a2-1 (SALK_001978), del1-1 (SALK_105648), and med25-2 (SALK_080230) were obtained from the ABRC or NASC Arabidopsis stock center. These mutants were verified by PCR analysis using T-DNA-specific and flanking primers and sequencing of the PCR products (Supplemental Data Set). The seeds were surface sterilized with 75% (v/v) ethanol for 3 min and 10% (v/v) bleach for 15 min, washed three times with sterile water, and plated on Murashige and Skoog medium. The plates were stored in the dark at 4°C for 4 d before they were transferred to the light. Plants were grown at 22°C under a 16-h-light (28 W/6500 K)/8-h-dark cycle.
Map-Based Cloning
To map the eod9-1 mutation, the segregation F2 population of a cross between eod9-1 da1-1Col-0 and da1-1Ler was used. The eod9-1 mutation was mapped to an interval between markers P3 and P6 on chromosome 4 using specific DNA markers T19B17 and F1K3 (Supplemental Figure 1A). The candidate gene was further verified by DNA sequencing.
Plant Transformation and Transgenic Plant Screening
Transgenic Arabidopsis plants were obtained by Agrobacterium tumefaciens-mediated transformation (Zhang et al., 2006). The developing Arabidopsis inflorescences were dipped into a solution containing 0.05% (v/v) Silwet L-77, 5% (m/v) sucrose, 0.22% (m/v) Murashige and Skoog medium, 0.02% (m/v) MES (pH 5.7), and A. tumefaciens cells carrying the chosen vectors for a few seconds. T1 seeds were plated on selective media to screen transgenic Arabidopsis plants.
Morphological and Cellular Analyses
The areas of leaves and petals were measured with ImageJ software after photographing. For cell size, leaves and petals were cleared in solution (10 mL of glycerol, 80 g of chloral hydrate, and 30 mL of water) and photographed under a differential interference contrast microscope (Leica, DM2500). Cell size was then measured with ImageJ software.
GUS Staining
A 2080-bp promoter region of MED16 and a 2141-bp promoter region of DEL1 were cloned into the attR1/attR2 sites of pMDC164 vector to generate the MED16pro:GUS and DEL1pro:GUS constructs, respectively. MED16pro:GUS and DEL1pro:GUS plants were obtained by A. tumefaciens-mediated transformation. Samples were stained with X-gluc buffer [100 mM NaPO4, pH 7, 750 µg mL−1 X-gluc, 3 mM K3Fe(CN)6, 10 mM EDTA, and 0.1% (v/v) Nonidet P-40] at 37°C for 4 to 9 h after 1 h of vacuum infiltration. The samples were cleared with 75% (v/v) ethanol before photographing.
RT-PCR and qPCR Analyses
Total RNA was isolated from the first pair of leaves from Arabidopsis seedlings with an RNAprep pure kit (Tiangen). RT-PCR was performed with an RT-PCR kit (Tiangen). The qPCR analysis was performed with SYBR Green І (Roche) using a Mastercycler RealPlex2 (Eppendorf) under the following conditions: denaturation for 2 min at 95°C, 40 cycles of 10 s at 95°C for denaturation, 10 s at 55°C for annealing, and 30 s at 68°C for extension. ACTIN2 was used as the internal control.
ChIP-qPCR Analysis
Chromatin affinity purification was conducted as described previously (Yamaguchi et al., 2014). Fourteen-day-old Arabidopsis seedlings were used in this study. Chromatin from seedlings was incubated with ChIP anti-GFP antibody (1:3000; Invitrogen, A-6455) and coprecipitated by ChIP protein A+G magnetic beads (Magna, 16-663). The enrichment of the fragments was tested by qPCR. The ACTIN7 promoter was used as a negative control.
Yeast Two-Hybrid Assays
The coding sequence (CDS) of MED16 was cloned into the SalI site of pGBKT7 vector (Clontech) to generate the pGBKT7-MED16 construct. An Arabidopsis cDNA library (Clontech) was cloned into the EcoRI site of pGADT7 vector (Clontech). The prey and bait constructs were cotransformed into Y2H Gold yeast cells (Clontech) and selected on SD medium −3 (SD/-His/-Leu/-Trp). The CDSs of positive clones were isolated by PCR and further identified by DNA sequencing. The interactions were further verified on SD medium −4 (SD/-Trp/-His/-Leu/-Ade) with different dilution series (10−1, 10−2, and 10−3). The pGBKT7-MED16/pGADT7 and pGBKT7/pGADT7-DEL1 combinations were used as negative controls.
Pull-Down Assays
Pull-down assays were performed as previously described (Xia et al., 2013). The CDSs of MED16 and DEL1 were cloned into the EcoRI site of pGEX4T1-GST vector and the BamHI site of pMALC2-MBP vector, respectively. Proteins were expressed in Escherichia coli BL21 (DE3) cells via induction with 0.5 mM IPTG at 28°C for 4 h and extracted via ultrasonication in solution (50 mM HEPES, 150 mM NaCl, 1 mM EDTA, 1.5 mM MgCl2, 10% [v/v] glycerol, 1% [v/v] Triton X-100, and 1 mM PMSF, pH 7.5). MBP or MBP-DEL1 was incubated with GST-MED16 and coprecipitated by GST-Trap-A agarose beads (New England Biolabs). The interactions were checked by immunoblotting with anti-GST (1:5000; Abmart, M20007) or anti-MBP (1:5000; Abmart, T40007) antibodies.
Coimmunoprecipitation
The CDSs of MED16 and DEL1 were cloned into the attR1/attR2 sites of pK7FWG2-GFP vector and the KpnI site of pCambia1300-221-Myc vector to generate the 35S:MED16-GFP and 35S:Myc-DEL1 constructs, respectively. 35S:MED16-GFP and 35S:Myc-DEL1 transgenic Arabidopsis plants were obtained by A. tumefaciens-mediated transformation. 35S:Myc-DEL1;35S:MED16-GFP and 35S:Myc-DEL1;35S:GFP plants were generated by crossing 35S:Myc-DEL1 with 35S:MED16-GFP or 35S:GFP, respectively. Coimmunoprecipitation was performed as described previously (Yang et al., 2014a). After grinding, total protein was obtained from the samples in isolation buffer (50 mM Tris-HCl, 20% [v/v] glycerol, 150 mM NaCl, 2% [v/v] Triton X-100, 1 mM EDTA, and 1× protease inhibitor cocktail, pH 7.5) and incubated with GFP-Trap agarose beads (Chromotek). Immunoblotting was used to detect the coprecipitated proteins with anti-Myc (1:10,000; Abmart, M20002) or anti-GFP (1:5000; Abmart, M20004) antibodies.
Bimolecular Fluorescence Complementation Assays
The CDSs of MED16 and DEL1 were cloned into the XbaI and SalI sites of pGWB414-cYFP and pGWB414-nYFP vectors to generate the cYFP-MED16 and nYFP-DEL1 constructs, respectively. Combinations of cYFP-MED16/nYFP-DEL1, cYFP-MED16/nYFP, and cYFP/nYFP-DEL1 were coinfiltrated into Nicotiana benthamiana leaves by A. tumefaciens-mediated transformation. Forty-eight hours later, YFP fluorescent signals in N. benthamiana leaf cells were observed under an LSM710 confocal laser-scanning microscope (Zeiss).
Flow Cytometry
The sixth leaves and petals of plants were dipped in cold nuclear extraction buffer (20 mM MOPS, pH 5.8, 30 mM sodium citrate, 45 mM MgCl2, and 0.1% [v/v] Triton X-100) and chopped with a razor blade. Nuclei were obtained through a 300 mesh nylon filter. The nuclei were stained with 2 μg mL−1 4′,6-diamidino-2-phenylindole (DAPI) and analyzed with a BD FACSCalibur flow cytometer. A total of 10,000 nuclei were analyzed per experiment.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL library under the following accession numbers: MED16 (AT4G25540), DEL1 (AT3G48160), CCS52A1 (AT4G22910), and CCS52A2 (AT4G11920).
Supplemental Data
Supplemental Figure 1. Identification of the EOD9 gene.
Supplemental Figure 2. Identification of med16-2 mutants.
Supplemental Figure 3. med16-2 produces large leaves.
Supplemental Figure 4. The expression patterns of MED16.
Supplemental Figure 5. Subcellular localization of MED16.
Supplemental Figure 6. MED16 regulates nuclear ploidy levels.
Supplemental Figure 7. MED16 acts in a common pathway with DEL1 to control endoreduplication.
Supplemental Figure 8. Alignment of A1F1 of the CCS52A1 promoter with A2F1 of the CCS52A2 promoter.
Supplemental Figure 9. Genetic analysis between MED16 and CCS52A1/A2.
Supplemental Figure 10. MED16 acts through CCS52A1/A2 to control endoreduplication.
Supplemental Figure 11. Genetics analysis between DA1 and EOD9/MED16.
Supplemental Figure 12. Genetics analysis between MED16 and MED25.
Supplemental Figure 13. Expression of MED16, DEL1, CCS52A1, and CCS52A2.
Supplemental Table. List of MED16-interacting proteins identified by yeast two-hybrid screening.
Supplemental Data Set. List of primers used in this study.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Michael Lenhard for the CYCLINB1;1pro:CDB-GUS line and the Arabidopsis stock centers ABRC and NASC for the ccs51a1-1, ccs52a2-1, and del1-1 mutants. This work was supported by grants from the National Natural Science Foundation of China (31425004, 31872663, 91735302, and 91535203) and the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB27010102).
AUTHOR CONTRIBUTIONS
Z.L., G.C., F.G., R.X., N.L., and Y.Z. performed experiments. Z.L., G.C., and Y.L. analyzed data. Z.L., G.C., and Y.L. wrote the article.
References
- Autran D., Jonak C., Belcram K., Beemster G.T.S., Kronenberger J., Grandjean O., Inzé D., Traas J. (2002). Cell numbers and leaf development in Arabidopsis: A functional analysis of the STRUWWELPETER gene. EMBO J. 21: 6036–6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baloban M., Vanstraelen M., Tarayre S., Reuzeau C., Cultrone A., Mergaert P., Kondorosi E. (2013). Complementary and dose-dependent action of AtCCS52A isoforms in endoreduplication and plant size control. New Phytol. 198: 1049–1059. [DOI] [PubMed] [Google Scholar]
- Breuer C., Ishida T., Sugimoto K. (2010). Developmental control of endocycles and cell growth in plants. Curr. Opin. Plant Biol. 13: 654–660. [DOI] [PubMed] [Google Scholar]
- Breuer C., Morohashi K., Kawamura A., Takahashi N., Ishida T., Umeda M., Grotewold E., Sugimoto K. (2012). Transcriptional repression of the APC/C activator CCS52A1 promotes active termination of cell growth. EMBO J. 31: 4488–4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer C., Braidwood L., Sugimoto K. (2014). Endocycling in the path of plant development. Curr. Opin. Plant Biol. 17: 78–85. [DOI] [PubMed] [Google Scholar]
- Cortellino S., Wang C., Wang B., Bassi M.R., Caretti E., Champeval D., Calmont A., Jarnik M., Burch J., Zaret K.S., Larue L., Bellacosa A. (2009). Defective ciliogenesis, embryonic lethality and severe impairment of the Sonic Hedgehog pathway caused by inactivation of the mouse complex A intraflagellar transport gene Ift122/Wdr10, partially overlapping with the DNA repair gene Med1/Mbd4. Dev. Biol. 325: 225–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhawan R., Luo H., Foerster A.M., Abuqamar S., Du H.N., Briggs S.D., Mittelsten Scheid O., Mengiste T. (2009). HISTONE MONOUBIQUITINATION1 interacts with a subunit of the mediator complex and regulates defense against necrotrophic fungal pathogens in Arabidopsis. Plant Cell 21: 1000–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H., et al. (2017). Ubiquitylation activates a peptidase that promotes cleavage and destabilization of its activating E3 ligases and diverse growth regulatory proteins to limit cell proliferation in Arabidopsis. Genes Dev. 31: 197–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Calvo P., et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fülöp K., Tarayre S., Kelemen Z., Horváth G., Kevei Z., Nikovics K., Bakó L., Brown S., Kondorosi A., Kondorosi E. (2005). Arabidopsis anaphase-promoting complexes: Multiple activators and wide range of substrates might keep APC perpetually busy. Cell Cycle 4: 1084–1092. [PubMed] [Google Scholar]
- Gegas V.C., Wargent J.J., Pesquet E., Granqvist E., Paul N.D., Doonan J.H. (2014). Endopolyploidy as a potential alternative adaptive strategy for Arabidopsis leaf size variation in response to UV-B. J. Exp. Bot. 65: 2757–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillmor C.S., Park M.Y., Smith M.R., Pepitone R., Kerstetter R.A., Poethig R.S. (2010). The MED12-MED13 module of Mediator regulates the timing of embryo patterning in Arabidopsis. Development 137: 113–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez D., Bowen A.J., Carroll T.S., Conlan R.S. (2007). The transcription corepressor LEUNIG interacts with the histone deacetylase HDA19 and mediator components MED14 (SWP) and CDK8 (HEN3) to repress transcription. Mol. Cell. Biol. 27: 5306–5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemsley P.A., Hurst C.H., Kaliyadasa E., Lamb R., Knight M.R., De Cothi E.A., Steele J.F., Knight H. (2014). The Arabidopsis mediator complex subunits MED16, MED14, and MED2 regulate mediator and RNA polymerase II recruitment to CBF-responsive cold-regulated genes. Plant Cell 26: 465–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyman J., Polyn S., Eekhout T., De Veylder L. (2017). Tissue-specific control of the endocycle by the Anaphase Promoting Complex/Cyclosome inhibitors UVI4 and DEL1. Plant Physiol. 175: 303–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inzé D., De Veylder L. (2006). Cell cycle regulation in plant development. Annu. Rev. Genet. 40: 77–105. [DOI] [PubMed] [Google Scholar]
- Joubès J., Chevalier C. (2000). Endoreduplication in higher plants. Plant Mol. Biol. 43: 735–745. [DOI] [PubMed] [Google Scholar]
- Kidd B.N., Edgar C.I., Kumar K.K., Aitken E.A., Schenk P.M., Manners J.M., Kazan K. (2009). The mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis. Plant Cell 21: 2237–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.J., Björklund S., Li Y., Sayre M.H., Kornberg R.D. (1994). A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 77: 599–608. [DOI] [PubMed] [Google Scholar]
- Knight H., Veale E.L., Warren G.J., Knight M.R. (1999). The sfr6 mutation in Arabidopsis suppresses low-temperature induction of genes dependent on the CRT/DRE sequence motif. Plant Cell 11: 875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight H., Thomson A.J., McWatters H.G. (2008). Sensitive to freezing6 integrates cellular and environmental inputs to the plant circadian clock. Plant Physiol. 148: 293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight H., Mugford S.G., Ulker B., Gao D., Thorlby G., Knight M.R. (2009). Identification of SFR6, a key component in cold acclimation acting post-translationally on CBF function. Plant J. 58: 97–108. [DOI] [PubMed] [Google Scholar]
- Koleske A.J., Young R.A. (1994). An RNA polymerase II holoenzyme responsive to activators. Nature 368: 466–469. [DOI] [PubMed] [Google Scholar]
- Lammens T., Boudolf V., Kheibarshekan L., Zalmas L.P., Gaamouche T., Maes S., Vanstraelen M., Kondorosi E., La Thangue N.B., Govaerts W., Inzé D., De Veylder L. (2008). Atypical E2F activity restrains APC/CCCS52A2 function obligatory for endocycle onset. Proc. Natl. Acad. Sci. USA 105: 14721–14726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson-Rabin Z., Li Z., Masson P.H., Day C.D. (2009). FZR2/CCS52A1 expression is a determinant of endoreduplication and cell expansion in Arabidopsis. Plant Physiol. 149: 874–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zheng L., Corke F., Smith C., Bevan M.W. (2008). Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Genes Dev. 22: 1331–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly M.A., Duronio R.J. (2005). New insights into cell cycle control from the Drosophila endocycle. Oncogene 24: 2765–2775. [DOI] [PubMed] [Google Scholar]
- Liu Y., Ye W., Li B., Zhou X., Cui Y., Running M.P., Liu K. (2012). CCS52A2/FZR1, a cell cycle regulator, is an essential factor for shoot apical meristem maintenance in Arabidopsis thaliana. BMC Plant Biol. 12: 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S., Roeder R.G. (2005). Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem. Sci. 30: 256–263. [DOI] [PubMed] [Google Scholar]
- Meng L.S. (2015). Transcription coactivator Arabidopsis ANGUSTIFOLIA3 modulates anthocyanin accumulation and light-induced root elongation through transrepression of Constitutive Photomorphogenic1. Plant Cell Environ. 38: 838–851. [DOI] [PubMed] [Google Scholar]
- Sugimoto-Shirasu K., Roberts K. (2003). “Big it up”: Endoreduplication and cell-size control in plants. Curr. Opin. Plant Biol. 6: 544–553. [DOI] [PubMed] [Google Scholar]
- Vanstraelen M., Baloban M., Da Ines O., Cultrone A., Lammens T., Boudolf V., Brown S.C., De Veylder L., Mergaert P., Kondorosi E. (2009). APC/C-CCS52A complexes control meristem maintenance in the Arabidopsis root. Proc. Natl. Acad. Sci. USA 106: 11806–11811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlieghe K., Boudolf V., Beemster G.T., Maes S., Magyar Z., Atanassova A., de Almeida Engler J., De Groodt R., Inzé D., De Veylder L. (2005). The DP-E2F-like gene DEL1 controls the endocycle in Arabidopsis thaliana. Curr. Biol. 15: 59–63. [DOI] [PubMed] [Google Scholar]
- Wathugala D.L., Hemsley P.A., Moffat C.S., Cremelie P., Knight M.R., Knight H. (2012). The Mediator subunit SFR6/MED16 controls defence gene expression mediated by salicylic acid and jasmonate responsive pathways. New Phytol. 195: 217–230. [DOI] [PubMed] [Google Scholar]
- Xia T., Li N., Dumenil J., Li J., Kamenski A., Bevan M.W., Gao F., Li Y. (2013). The ubiquitin receptor DA1 interacts with the E3 ubiquitin ligase DA2 to regulate seed and organ size in Arabidopsis. Plant Cell 25: 3347–3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R., Li Y. (2011). Control of final organ size by Mediator complex subunit 25 in Arabidopsis thaliana. Development 138: 4545–4554. [DOI] [PubMed] [Google Scholar]
- Yamaguchi N., Winter C.M., Wu M.F., Kwon C.S., William D.A., Wagner D. (2014). PROTOCOLS: Chromatin immunoprecipitation from Arabidopsis tissues. The Arabidopsis Book 12: e0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.W., Fu J.X., Li J., Cheng X.L., Li F., Dong J.F., Liu Z.L., Zhuang C.X. (2014a). A novel co-immunoprecipitation protocol based on protoplast transient gene expression for studying protein-protein interactions in rice. Plant Mol. Biol. Rep. 32: 153–161. [Google Scholar]
- Yang Y., Ou B., Zhang J., Si W., Gu H., Qin G., Qu L.J. (2014b). The Arabidopsis Mediator subunit MED16 regulates iron homeostasis by associating with EIN3/EIL1 through subunit MED25. Plant J. 77: 838–851. [DOI] [PubMed] [Google Scholar]
- Zhang X., Henriques R., Lin S.S., Niu Q.W., Chua N.H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1: 641–646. [DOI] [PubMed] [Google Scholar]
- Zhang X., Wang C., Zhang Y., Sun Y., Mou Z. (2012). The Arabidopsis mediator complex subunit16 positively regulates salicylate-mediated systemic acquired resistance and jasmonate/ethylene-induced defense pathways. Plant Cell 24: 4294–4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Yao J., Zhang Y., Sun Y., Mou Z. (2013). The Arabidopsis Mediator complex subunits MED14/SWP and MED16/SFR6/IEN1 differentially regulate defense gene expression in plant immune responses. Plant J. 75: 484–497. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Wu H., Wang N., Fan H., Chen C., Cui Y., Liu H., Ling H.Q. (2014). Mediator subunit 16 functions in the regulation of iron uptake gene expression in Arabidopsis. New Phytol. 203: 770–783. [DOI] [PubMed] [Google Scholar]