Chloroplast outer membrane proteins with β-barrel-shaped transmembrane domains are sorted to the chloroplasts by amino-terminal transit peptides and/or intrinsic targeting information, but in both cases the proteins use the general import apparatus that also serves proteins destined for the interior compartments of the chloroplast.
Abstract
Chloroplasts evolved from a cyanobacterial endosymbiont that resided within a eukaryotic cell. Due to their prokaryotic heritage, chloroplast outer membranes contain transmembrane β-barrel proteins. While most chloroplast proteins use N-terminal transit peptides to enter the chloroplasts through the translocons at the outer and inner chloroplast envelope membranes (TOC/TIC), only one β-barrel protein, Toc75, has been shown to use this pathway. The route other β-barrel proteins use has remained unresolved. Here we use in vitro pea (Pisum sativum) chloroplast import assays and transient expression in Nicotiana benthamiana to address this. We show that a paralog of Toc75, outer envelope protein 80 kD (OEP80), also uses a transit peptide but has a distinct envelope sorting signal. Our results additionally indicate that β-barrels that do not use transit peptides also enter the chloroplast using components of the general import pathway.
INTRODUCTION
Chloroplasts arose from an ancient endosymbiotic relationship between a species of cyanobacteria and a eukaryotic cell. These two organisms evolved together and diversified to become glaucophytes, red algae, green algae, and land plants. As the cyanobacterial endosymbiont became integrated with the host cell, a protein import machinery spanning its double membrane envelope evolved. This import apparatus allowed the targeting of new proteins to the endosymbiont as well as the migration of most cyanobacterial genes to the nucleus. In modern plant cells, the import apparatus consists of translocons at the outer and inner envelope membranes of the chloroplast (TOC/TIC). The TOC complex contains two GTPase receptors, Toc34 and Toc159, as well as Toc75, which is thought to form the pore (Kessler et al., 1994; Schnell et al., 1994). The exact protein composition of the TIC is a matter of debate (Bölter, 2018; Nakai, 2018). Proteins destined for the interior compartments of the chloroplast, including the inner membrane, stroma, and thylakoids, are directed to the protein import apparatus by N-terminal transit peptides (Lee and Hwang, 2018). These transit peptides are removed after import by the stromal processing peptidase (SPP; Robinson and Ellis, 1984; Richter and Lamppa, 1998).
Proteins of both prokaryotic and eukaryotic origin make up the chloroplast import apparatus (Shi and Theg, 2013). Notably Toc75 evolved from a cyanobacterial protein and is part of a large prokaryotic protein family called “outer membrane protein 85 kD (Omp85) homologs” (Bölter et al., 1998; Reumann and Keegstra, 1999; Inoue and Potter, 2004). In bacteria, Omp85 homologs function in protein export and assembly of outer membrane (OM) transmembrane β-barrel proteins (Voulhoux et al., 2003; Selkrig et al., 2012). The latter function is essential for diderm bacteria envelope biogenesis and is thought to be the function of the cyanobacterial Omp85 from which Toc75 evolved. Like the OM of bacteria, the chloroplast OM contains transmembrane β-barrel proteins. These are hypothesized to be inserted into the OM by a paralog of Toc75, OEP80 (Eckart et al., 2002; Inoue and Potter, 2004; Schleiff and Soll, 2005; Day et al., 2014). According to this hypothesis, the ancestral Omp85 homolog was duplicated either before or after gaining a function in chloroplast import, with Toc75 specializing in protein import and OEP80 retaining the ancestral function. Due to Toc75’s evolutionary origin, understanding how chloroplast OM β-barrels are sorted in modern plants can help us understand the mechanism by which the chloroplast protein import apparatus evolved.
β-barrels are translated on eukaryotic ribosomes outside the chloroplast, as are most chloroplast proteins. Due to their unique position at the interface between the chloroplast and the cytosol where proteins are translated, it seems possible that the OM β-barrels could be inserted directly without first entering the chloroplast through the TOC complex. The first chloroplast OM β-barrel whose targeting was investigated was Toc75. The import of Toc75 shares many similarities with the import of other chloroplast proteins. These include reliance on an N-terminal transit peptide and use of the general import pathway (Tranel et al., 1995; Tranel and Keegstra, 1996). Unlike proteins targeted to the interior compartments of the chloroplast, Toc75 uses an additional targeting peptide directly following the transit peptide. This peptide contains a polyglycine stretch that is required for sorting to the envelope (Inoue and Keegstra, 2003; Endow et al., 2016). It is unclear how much of the mature region of Toc75 passes the OM before insertion. Other OM β-barrels, including OEP24, OEP37, OEP21, and OEP40, were later shown to not use a cleavable transit peptide (Pohlmeyer et al., 1998; Bölter et al., 1999; Hofmann and Theg, 2005; Harsman et al., 2016). We previously showed that the OEP80 ortholog from Arabidopsis (Arabidopsis thaliana) was processed to its mature size during in vitro import, suggesting it may use a transit peptide (Day et al., 2014). Unlike Toc75, the N terminus of OEP80 does not contain a polyglycine stretch, so it is not clear how it is sorted to the envelope.
Because of our limited understanding of the sorting of OM β-barrels, we cannot rule out the possibility that they are directly inserted into OM without first passing through the TOC machinery. Perhaps they are directly sorted to an OEP80-containing complex from the cytosol. In bacteria, OM β-barrels are translated in the interior, then exported through the Sec machinery. They approach the OM from the periplasm, which is homologous to the chloroplast intermembrane space (IMS), then are folded and inserted by an Omp85 homolog (Voulhoux et al., 2003). If chloroplast β-barrels are inserted from the cytosol, OEP80 would have needed to evolve the capacity to accept substrates from the opposite side of the membrane. A study by Sommer et al. (2011) proposed that such an adaptation was accomplished by a change in orientation of the chloroplast Omp85 homologs. This possibility was especially interesting because it suggested a compelling scenario for the evolution of the TOC complex. Omp85 homologs share a conserved domain layout, including a variable number of polypeptide transport-associated (POTRA) domains and a C-terminal transmembrane β-barrel. The POTRA domains are thought to act as receptors for incoming substrates (Sánchez-Pulido et al., 2003). In bacteria, these are located in the periplasm and accept substrates recently released from the Sec translocase. Sommer et al. (2011) proposed that, during evolution, the POTRA domains of the endosymbiont’s Omp85 homolog shifted to face the cytosol. After this shift, the homolog could accept β-barrel substrates translated on eukaryotic ribosomes. Later, the Omp85 homolog could have evolved the capacity to accept new substrates that would pass completely through the OM rather that insert into it, thus leading to the evolution of the Toc75.
Although this hypothesis provides a molecular mechanism for the evolution of chloroplast protein import (that is, with the POTRA domains facing the cytosol), later studies established that the orientation of Toc75 is conserved with bacterial Omp85 homologs (Chen et al., 2016; Paila et al., 2016). Sommer et al. (2011) used the same experimental system to show that OEP80 faces the cytosol, so this finding was also called into question. Assuming β-barrel precursors must be inserted from the same side of the membrane as the Omp85’s POTRAs, then they must first pass through the chloroplast OM. On the other hand, if Omp85s can assemble β-barrels from either side of the membrane, chloroplast OM proteins could be directly inserted from the cytosol. Although it has been shown that Toc75 is at least partially imported through the import apparatus, it is not known if OEP80 and OM β-barrels that do not use transit peptides enter the chloroplast through the TOC complex.
Results in this study from in vitro chloroplast import assays and transient expression in Nicotiana benthamiana demonstrate that the N terminus of OEP80 behaves like a canonical transit peptide. Unlike Toc75, OEP80’s N terminus is not responsible for envelope sorting. Instead, the C-terminal transmembrane is required to prevent full translocation into the stroma. We also show that the reason OEP80 requires a transit peptide, while most other OM β-barrels do not, is due to its IMS-localized POTRA domains. Lastly, results from import competition assays and pulldowns after transient expression suggest that both OM β-barrels with or without transit peptides use components of the general import apparatus.
RESULTS
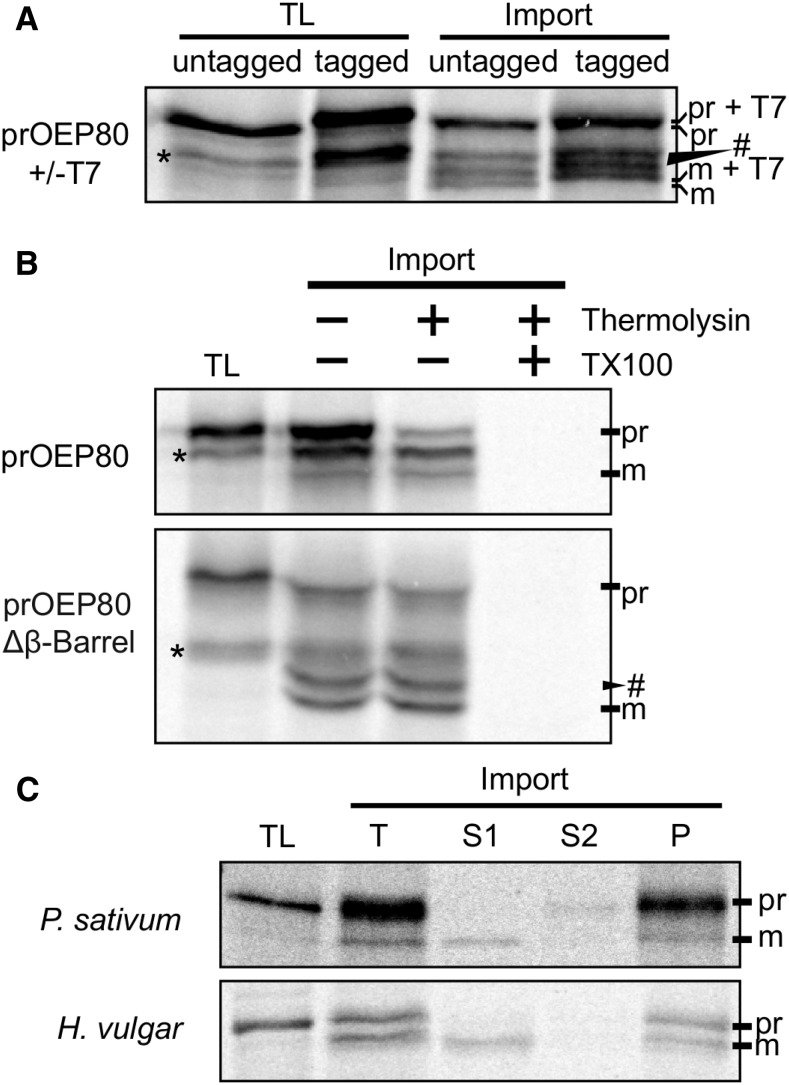
The N Terminus of OEP80 Is Removed After Import
Results from our previous work showed that the OEP80 ortholog from Arabidopsis (AtOEP80) was processed to its mature size during in vitro import assays (Day et al., 2014). This suggested that, like Toc75, it may use N-terminal targeting information. To test if the processing of AtOEP80 occurred at the N terminus, we added a C-terminal T7 tag to the precursor and then checked whether the tag remained after processing (Figure 1). Figure 1A shows that the processed, tagged protein retained the slight increase in size over the untagged version, meaning the processing must occur at the N terminus. As previously reported, three processed bands appear after import. The smallest of these bands comigrates with endogenous AtOEP80 (Day et al., 2014). The signal from the tagged form of the protein appears more intense than the untagged likely due to the three additional Met residues in the T7 sequence. To confirm that the processing occurs at the N terminus, we performed in vitro import of a construct lacking the C-terminal transmembrane β-barrel (Figure 1B). This construct was imported and processed, demonstrating that the processing occurs at the N terminus and that the transmembrane β-barrel is not required for import. Lastly, import of OEP80 orthologs from Pisum sativum (pea) and Hordeum vulgare (barley) into isolated pea chloroplasts showed that they are also processed (Figure 1C). This indicates that the presence of N-terminal targeting information is not unique to Arabidopsis OEP80. Indeed, the program ChloroP (DTU Bioinformatics; http://www.cbs.dtu.dk/services/ChloroP/) predicts the presence of transit peptides on many OEP80 orthologs throughout Viridiplantae (Supplemental Figure 1).
Figure 1.
OEP80 Is Processed at the N Terminus.
(A) In vitro-translated, radiolabeled AtOEP80 ± a C-terminal T7 tag were incubated with isolated chloroplasts (import). After 20 min the chloroplasts were reisolated and run on SDS-PAGE next to 10% of the translation product input (TL). Radioactive bands were detected with autoradiography.
(B) In vitro-translated, radiolabeled AtOEP80 ± the C-terminal transmembrane β-barrel were incubated with isolated chloroplasts. After 20 min, the chloroplasts were reisolated, then treated with or without thermolysin or solubilized with 1% (v/v) Triton X100 (TX100) and treated with thermolysin. The protease was stopped with 10-mM EDTA. Intact chloroplasts were reisolated. Proteins in the solubilized sample were precipitated in 80% (v/v) acetone. Samples were run on SDS-PAGE and visualized as in (A). In (A) and (B), the asterisks (*) near the TL lane indicate bands resulting from translation starting at the second and/or third methionines in the coding sequence. The pound sign (#) indicates additional processed bands, other than the mature-sized one.
(C) In vitro-translated, radiolabeled OEP80 orthologs from pea (P. sativum) and barley (H. vulgare) were incubated with isolated chloroplasts. After 20 min, the chloroplasts were reisolated and half were lysed in 10-mM HEPES at pH 8.0 and 10-mM MgCl2. The soluble (S1) and membrane fractions were separated by centrifugation. The membranes were washed with 0.1-M sodium carbonate. The wash (S2) and pellet (P) were separated by centrifugation. Samples were run on SDS-PAGE and visualized as in (A). pr, precursor; m, mature.
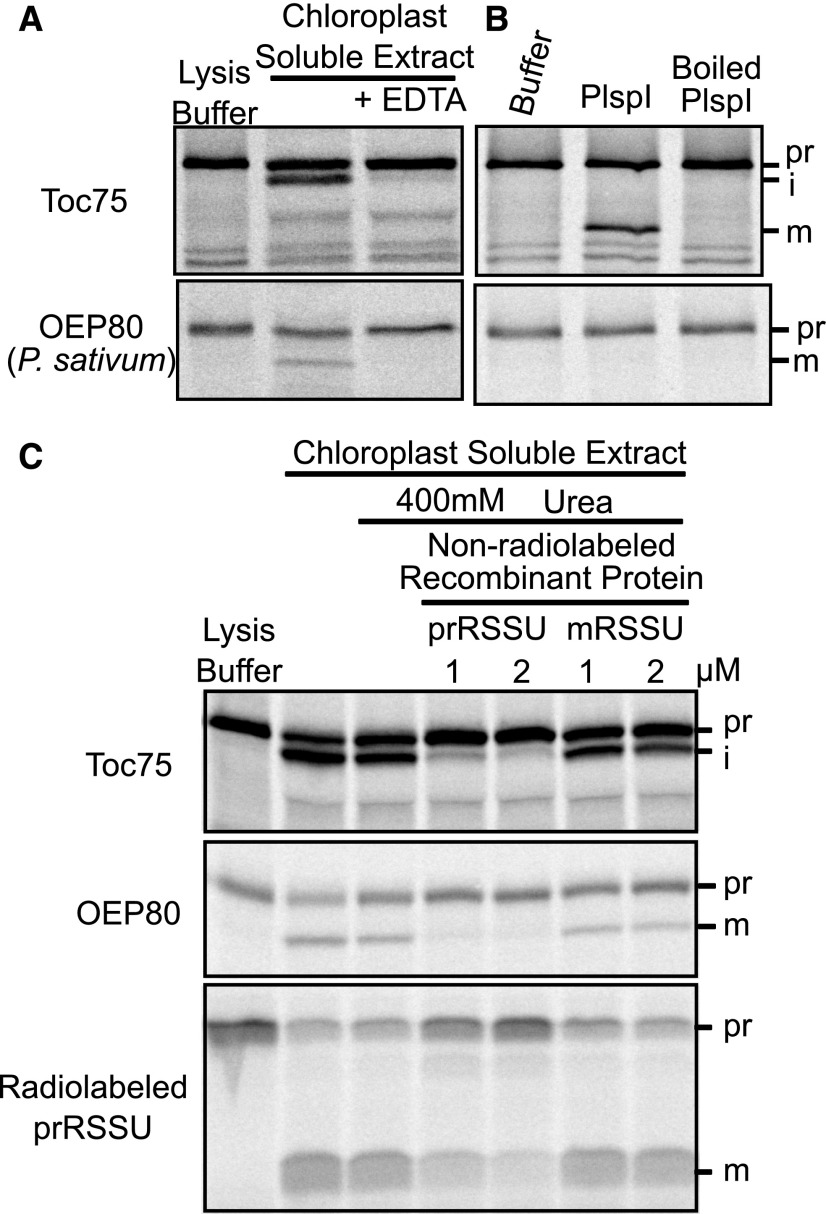
OEP80 Is Processed by the Stromal Processing Peptidase
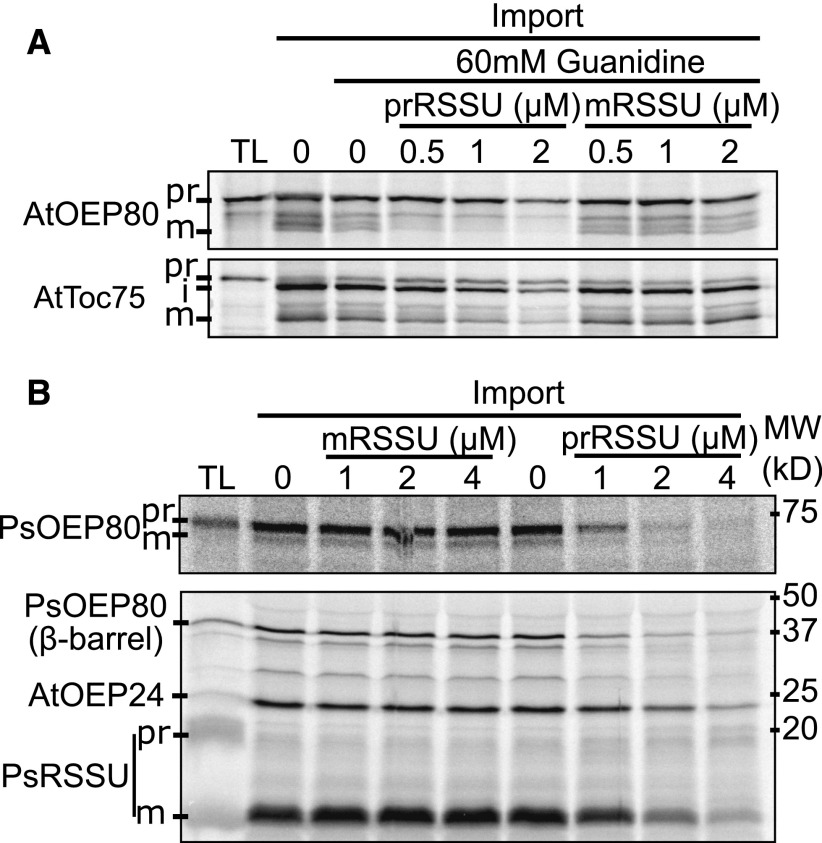
Early studies demonstrated that transit peptides from several chloroplast proteins can be removed during incubation with the soluble fraction from chloroplasts due to the presence of the SPP (Robinson and Ellis, 1984; Tranel and Keegstra, 1996; Richter and Lamppa, 1998). This was the case for OEP80 from pea (PsOEP80; Figure 2), but not AtOEP80 (Supplemental Figure 2). EDTA inhibited this processing (Figure 2A), supporting the hypothesis that PsOEP80 is processed by the SPP, which requires a zinc ion (VanderVere et al., 1995). During import, Toc75 is sequentially processed by the SPP, then plastidic type I signal peptidase1 (Plsp1; Tranel and Keegstra, 1996; Inoue et al., 2005). Recombinant Plsp1 was capable of processing Toc75 to its mature size but did not affect PsOEP80 (Figure 2B), suggesting PsOEP80 is only processed by the SPP. To further test that the SPP is responsible, we performed processing competition assays with recombinant precursor of the small subunit of RUBISCO (prRSSU) or with the mature small subunit of RUBISCO (mRSSU). Inclusion of prRSSU, but not of mRSSU, prevented the processing of PsOEP80, showing that they are processed by the same peptidase (Figure 2C). Results from an additional in vitro processing competition assay are included in Supplemental Figure 3. These results indicate that unlike Toc75, a portion of the mature PsOEP80 must reach the stroma. It is not clear why the Arabidopsis OEP80 was not processed by the soluble extract. Perhaps it is processed by a different protease or does not adopt the proper conformation while soluble. Notably, in another study Tic22 was also not processed by a chloroplast soluble extract (Vojta et al., 2007).
Figure 2.
OEP80 Is Processed by the SPP.
(A) In vitro-translated, radiolabeled precursors were incubated in lysis buffer or chloroplast soluble extract with or without 50-mM EDTA. Samples were run on SDS-PAGE and radioactive bands were detected with autoradiography.
(B) The same precursors were incubated with recombinant Plsp1, Boiled Plsp1, or empty buffer. Results were visualized as in (A).
(C) Precursors were incubated in chloroplast soluble extract with or without urea-solubilized recombinant precursor (prRSSU) or mRSSU. Results were visualized as in (A). i, intermediate.
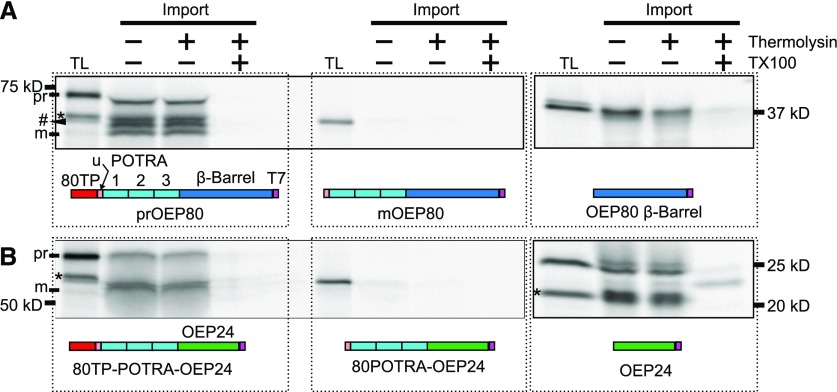
OEP80 Requires an N-Terminal Transit Peptide for Import of IMS-Localized Domains
Like many chloroplast proteins, AtOEP80 is predicted to contain a transit peptide by the program ChloroP (Emanuelsson et al., 1999). We asked if the predicted transit peptide, hereafter referred to as “80TP,” is required for chloroplast import. ChloroP predicts that the first 93 amino acids of the AtOEP80 constitute the transit peptide (Supplemental Figure 1; Emanuelsson et al., 1999). We performed an in vitro import reaction with a construct lacking amino acids 2 to 93 (mOEP80; Figure 3). Note that the AtOEP80 constructs labeled as prOEP80 in figures showing results from in vitro import assays start at the second Met of the predicted AtOEP80 sequence, amino acid 53, because the region before this was not required for targeting and function in vivo (Hsu et al., 2012), and this was the construct used in our previous work (Day et al., 2014). mOEP80 was not imported into isolated chloroplasts, indicating that the 80TP is required for chloroplast targeting (Figure 3A). Importantly, the size of the engineered mOEP80 is slightly larger than the processed OEP80, meaning the real cut site is a bit downstream of the 93rd amino acid. Interestingly, if the POTRA domains were removed in addition to 80TP leaving only the transmembrane domain, chloroplast import was restored (Figure 3A). Like other OM β-barrels, it appears that the AtOEP80 β-barrel has intrinsic chloroplast targeting information. This leads to a compelling answer to the question of why Toc75 and OEP80 require N-terminal targeting information whereas other OM β-barrels do not—80TP seems to be required to facilitate the import of the POTRA domains. To further support this hypothesis, we added AtOEP80’s POTRA domains to another OM β-barrel, OEP24b, which is encoded by the A. thaliana gene At5g42960. OEP24 is imported into isolated chloroplasts without processing (Figure 3B, right). The band in the translation product and import lanes at 20 kD is a result of translation from the third Met. When the POTRA domains from AtOEP80 are added to OEP24, import is abolished (Figure 3B, center). OEP24 has intrinsic targeting information; however, this signal is not sufficient to accommodate additional protein sections. Addition of 80TP before the POTRAs on this chimeric construct restored import (Figure 3B, left).
Figure 3.
The N Terminus of OEP80 Is Necessary for the Import of the IMS-Localized POTRA Domains But Not the Transmembrane Domain.
(A) In vitro-translated, radiolabeled sections of AtOEP80 (illustrated below the gels) were incubated with isolated chloroplasts (import). Chloroplasts were reisolated, then treated with or without thermolysin or solubilized with 1% (v/v) Triton X100 (TX100) and treated with thermolysin. The protease was stopped with 10-mM EDTA, and intact chloroplasts were reisolated. Proteins in the solubilized sample were precipitated in 80% (v/v) acetone. Samples were separated by SDS-PAGE next to 10% of the input translation product (TL). Radioactive bands were detected with autoradiography. 80TP, transit peptide of OEP80; u, uncharacterized region between the transit peptide and the first POTRA. The three POTRA domains are labeled as “1,” “2,” and “3.” The pound sign (#) indicates additional processed bands, other than the mature-sized one.
(B) In vitro-translated, radiolabeled OEP24 and OEP80-OEP24 chimeras were incubated with isolated chloroplasts, then treated as in (A). pr, precursor; m, mature.
In both (A) and (B), the asterisks (*) near the TL lane indicates bands resulting from translation starting at the second and/or third methionines in the coding sequence.
Envelope Sorting of OEP80
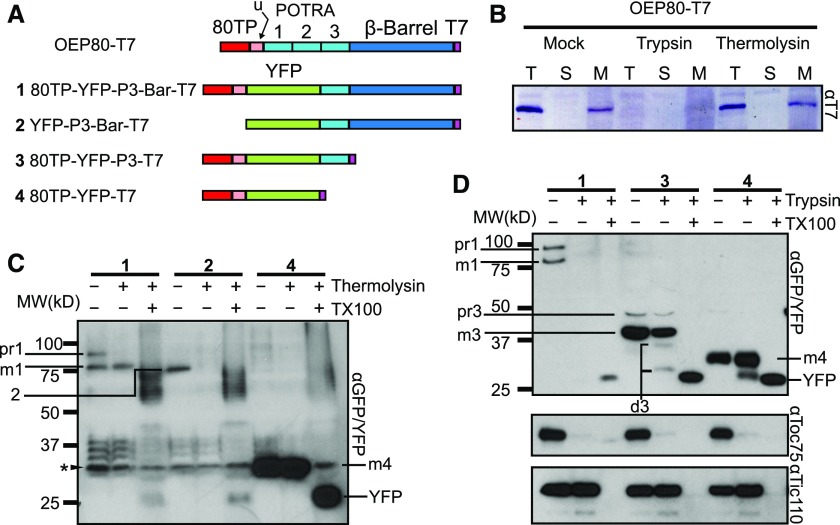
At this point, our results show that OEP80 uses a cleavable transit peptide for chloroplast targeting. Because it lacks a polyglycine stretch like Toc75, it is not clear how OEP80 is sorted to the envelope. Due to its OM localization, endogenous OEP80 is found in a membrane fraction and is digested during trypsin treatment of intact chloroplasts (Hsu et al., 2012). Trypsin is a protease that is believed to have access to proteins in both the chloroplast OM and IMS (Cline et al., 1984; Kouranov et al., 1998). Our previous results using in vitro import assays showed that the majority of processed AtOEP80 was in the soluble fraction and protected from trypsin (Day et al., 2014). This indicates that in vitro import assays do not reliably report the sorting of OEP80 to the envelope. To address this, we employed transient expression in N. benthamiana (Figure 4). Transiently expressed T7 tagged AtOEP80 displayed properties in common with endogenous OEP80, including protection from thermolysin but not trypsin, and recovery in the membrane upon chloroplast fractionation (Figure 4B).
Figure 4.
The N Terminus of OEP80 Is Sufficient for Targeting to the Chloroplast But the C Terminus is Required for Envelope Sorting.
(A) Diagram of constructs tested. 80TP, transit peptide of OEP80; u, uncharacterized region between the transit peptide and first POTRA. The three POTRA domains are labeled as “1,” “2,” and “3.”
(B) Chloroplasts were isolated from plants transiently expressing a T7-tagged version of OEP80. Chloroplasts were treated with no protease, trypsin, or thermolysin. Proteases were quenched, then the chloroplasts were reisolated. Chloroplasts were fractionated into soluble (S) and membrane (M). T, Total chloroplasts. Results were visualized by immunoblot with an anti-T7 antibody.
(C) Chloroplasts were isolated from plants transiently expressing YFP-tagged sections of OEP80. The chloroplasts were treated with or without thermolysin or solubilized with 1% (v/v) Triton X100 (TX100) and treated with thermolysin. After 30 min, the protease was quenched, and intact chloroplasts were reisolated. Proteins in the solubilized sample were precipitated in 80% (v/v) acetone. Samples were visualized by immunoblotting with anti-GFP antibodies. A nonspecific band recognized by the GFP antibody is indicated with an asterisks (*).
(D) The same experiment was performed as in (C), but with trypsin rather than thermolysin. Immunoblots of endogenous Toc75 and Tic110 are included as trypsin-sensitive and -insensitive controls, respectively.
In (C) and (D), the identities of bands are indicated by their number in (A) as well as “pr” to indicate the precursor form, “m” to indicate the processed form, or “d” to indicate degradation products. Free YFP that is the result of degradation of the OEP80 sections is indicated as “YFP.”
To determine the sections of AtOEP80 necessary for envelope sorting, we transiently expressed constructs tagged with the citrine variant of yellow fluorescent protein (YFP; Griesbeck et al., 2001) and then analyzed their localization in isolated chloroplasts using protease treatments. A construct with YFP replacing the first and second POTRA domains was protected from thermolysin but degraded by trypsin, suggesting that it was properly sorted to the envelope (Figures 4C and 4D, construct 1). Interestingly, a band corresponding in size to the unprocessed chimeric protein also appeared. This was degraded by thermolysin, indicating that it could represent an early binding intermediate. When 80TP was removed, the resulting protein associated with the chloroplasts but was degraded by thermolysin (Figure 4C, construct 2), demonstrating that the YFP moiety was not taken across the OM. A construct that did not contain the transmembrane β-barrel was predominantly resistant to trypsin, indicating its stromal localization (Figure 4D, construct 3). Another construct lacking any part of OEP80 other than the N-terminal targeting information was mostly resistant to both proteases, indicating that it had passed into the stroma (Figures 4C and 4D, construct 4). Faint degradation products did appear after trypsin treatment for both C-terminal truncation constructs, suggesting that a small amount of these proteins might be sorted to the envelope or stalled in the translocon. These faint bands might also arise due to incomplete quenching of the trypsin after the treatment. These results suggest that the N terminus of AtOEP80 is sufficient for chloroplast localization, but not envelope sorting. Instead, some portion of the third POTRA and/or the transmembrane region is responsible for envelope sorting.
Import of Chloroplast OM β-Barrels Competes with Import of RSSU
Hitherto, the only chloroplast OM β-barrel known to use the general import pathway was Toc75 (Tranel et al., 1995). If OEP80 uses a canonical transit peptide, it should enter the chloroplast through the general import apparatus. To test this, we performed import competition assays with recombinant precursor to prRSSU (Figure 5). Import of both the Arabidopsis and pea orthologs of OEP80 was reduced by prRSSU, but not the mature form (mRSSU), indicating that they share the same translocon (Figures 5A and 5B, top). We also asked whether β-barrels that do not use transit peptides enter through the general import apparatus. To answer this, we included OEP24 and a synthetic truncated form of PsOEP80 β-barrel in these experiments. These also competed with prRSSU (Figure 5B, bottom). Together, our results suggest that chloroplast OM β-barrels engage at least the TOC complex regardless of the presence of a transit peptide. Additional import competition assays are included in Supplemental Figure 3.
Figure 5.
Import OM β-Barrels With or Without Cleavable Targeting Information Competes with Import of a Stromal Protein.
(A) In vitro-translated, radiolabeled precursors were incubated with isolated pea chloroplasts in the presence or absence of guanidine-solubilized recombinant prRSSU or mRSSU. After 20 min, chloroplasts were reisolated through 40% (v/v) Percoll.
(B) In vitro-translated, radiolabeled precursors were incubated with isolated chloroplasts in the presence or absence of recombinant prRSSU or mRSSU. The radiolabeled PsOEP80 barrel, AtOEP24, and prRSSU were imported into the same chloroplasts. After 20 min, chloroplasts were reisolated through 40% (v/v) Percoll, treated with thermolysin, then reisolated again.
In both (A) and (B), samples were separated by SDS-PAGE next to 10% (AtOEP80, AtToc75, PsOEP80) or 3.3% (PsOEP80 barrel, AtOEP24, and PsprRSSU) of the input translation product (TL). Radioactive bands were detected with autoradiography. pr, precursor; i, intermediate; m, mature.
Transiently Expressed Chloroplast OM Proteins Interact with TOC Components
Other studies identified components of the chloroplast import assay by stalling substrates in the import apparatus during in vitro import assays (Kessler et al., 1994; Schnell et al., 1994). A similar approach was used to show that chloroplast OM α-helix transmembrane proteins use Toc75 (Tu et al., 2004). Production of import-competent recombinant protein can be difficult, so we used an alternative approach. Recent studies have shown that under some conditions, protein import into organelles can stall in vivo. Lee et al. (2018) showed that mutation of key Pro residues in the transit peptide of a thylakoid protein led to accumulation at the chloroplast OM during transient expression in Arabidopsis protoplasts. Also, Weidberg and Amon (2018) demonstrated that overexpression of mitochondria inner membrane proteins that use a stop transfer signal in yeast led to accumulation of other mitochondrial proteins in the import apparatus.
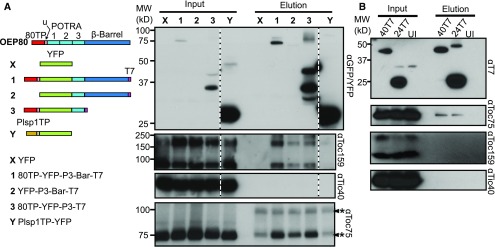
Our experiments using transient overexpression in N. benthamiana suggested that a similar phenomenon takes place for chloroplast OM β-barrels (Figure 6). We transiently overexpressed various portions of AtOEP80 fused to YFP (Figure 6A). As controls, we used YFP without a chloroplast targeting signal and YFP with the transit peptide from the thylakoid and inner membrane protein Plsp1. When the YFP-tagged proteins were immunoprecipitated from solubilized chloroplasts with GFP affinity beads, the constructs that contained portions of AtOEP80 co-eluted with Toc159, whereas the others did not (Figure 6A). Toc75 also appeared to co-elute; however, the antibody also detected faint bands in the controls that came from the affinity beads. This may be because the GFP binding protein was purified in a similar way as the original antigen used to generate the Toc75 antibody. Interestingly, constructs lacking one of either the transit peptide or the β-barrel pulled down TOC components, suggesting that either are sufficient for interaction with the import apparatus. The YFP construct containing the transit peptide of Plsp1 did not pull down components of the TOC, even though it presumably uses the import apparatus. This is likely due to the higher import efficiency resulting in very little of it stalling in the translocon.
Figure 6.
Transiently Expressed Sections of OEP80 and OEP24 Pull Down TOC Components.
(A) Diagram of constructs used (left). Chloroplasts from plants transiently expressing YFP-tagged constructs were solubilized with 1% (w/v) dodecyl-maltoside (input) and mixed with GFP affinity beads. Beads were washed, then bound proteins eluted. The inputs and eluates were subjected to immunoblotting with antibodies against GFP, Toc159, Tic40, and Toc75. The blot for GFP, Toc159, and Tic40 are from one membrane each; however, some regions were removed for clarity (indicated by dotted white and black lines). The asterisks indicate bands from the GFP affinity beads recognized by the Toc75 antibody. One of these comigrated with Toc75, but eluates 1, 2, and 3 showed a higher signal in this region. 80TP, transit peptide of OEP80; Plsp1TP, transit peptide of Plsp1; u, uncharacterized region between the transit peptide and the first POTRA. The three POTRA domains are labeled as 1, 2, and 3.
(B) Chloroplasts from uninfiltrated (UI) plants or plants transiently expressing T7-tagged OEP24 (24T7) or OEP40 (40T7) were solubilized with 1% (w/v) dodecyl-maltoside (input) and mixed with T7 affinity beads. The inputs and eluates were subjected to immunoblotting with antibodies against T7, Toc75, Toc159, and Tic40.
Overexpressed, T7-tagged Arabidopsis OEP24 and OEP40 also pulled down Toc75 (Figure 6B). Toc75 was not pulled down when chloroplasts from uninfiltrated leaves were used (Figure 6B). Unlike the tagged OEP80 fragments, OEP24-T7 and OEP40-T7 did not pull down Toc159. It is not clear why this is the case, but it may be that they only use Toc75, as has been proposed for OM α-helical proteins (Tu et al., 2004). Neither sets of tagged OM β-barrels pulled down Tic40 (Figures 6A and 6B). This suggests that either these proteins do not use the TIC, Tic40 is not part of the TIC, or under the conditions used the substrate did not remain bound to the TIC complex. In aggregate, these results indicate that chloroplast OM β-barrels, including those that do not use a transit peptide, enter the chloroplast through the TOC complex.
DISCUSSION
Mitochondria and chloroplast OMs contain proteins with both α-helical and β-barrel transmembrane domains. The vast majority of these differ from internal organellar proteins in that they do not require cleavable N-terminal targeting information; only the chloroplast β-barrel protein Toc75 was shown to be an exception (Tranel et al., 1995; Tranel and Keegstra, 1996). Exactly why it was an exception was not clear because its paralog, OEP80, was originally thought to not use a transit peptide (Inoue and Potter, 2004). Our more recent results suggested that OEP80 does indeed use a transit peptide (Figures 1 and 2; Day et al., 2014). This led us to hypothesize that the IMS-localized POTRA domains found in both Toc75 and OEP80 may be the reason they require transit peptides. Our results shown in Figure 3 support this hypothesis. Also, even when the first two POTRA domains are replaced with YFP, the protein could not pass the OM without the transit peptide (Figure 4C). This suggests that any large IMS-localized domain probably would require a transit peptide for proper targeting. On the other hand, chloroplast OM β-barrels that primarily consist of a transmembrane domain, including OEP24 and OEP40, do not require cleavable transit peptides. Instead, they must rely on intrinsic information within the mature protein. The exact location and nature of this intrinsic information is not clear. A recent study by Klinger et al. (2019) implicated the penultimate β-strand in distinguishing targeting between the chloroplast and mitochondria. This may represent the chloroplast import signal. Because removal of the POTRA domains from OEP80 allows import of the β-barrel without the aid of a transit peptide, full-length OEP80 may contain both types of targeting information.
Even though OEP80 is an outer envelope protein, the fact that it is related to Toc75 and shares a similar domain layout makes it seems unremarkable that it also uses a transit peptide. However, the N terminus of OEP80 lacks the polyglycine stretch required for envelope sorting of Toc75. Some OEP80 orthologs have short regions of glycines, but many do not (Supplemental Figure 1). Considering that even small changes to the Toc75 polyglycine region leads to mistargeting (Baldwin and Inoue, 2006), it seems unlikely that OEP80’s N terminus would be sufficient for envelope sorting. Indeed, our results show that OEP80’s N terminus takes YFP completely into the stroma and that proper envelope sorting requires the β-barrel at the C terminus (Figure 4D). It remains to be shown how the β-barrel leads to envelope sorting. Perhaps import of OEP80 is coupled with insertion of the transmembrane domain and the tight association with the OM prevents full translocation through the TIC. On the other hand, the nascent OEP80 may be passed to an OEP80-containing assembly complex, which binds it tightly enough to prevent translocation. If either of these are correct, it seems curious that Toc75 could not use a similar sorting mechanism. Further studies are needed to discover the reason for their distinct envelope-sorting mechanisms.
Similar to BamA in bacteria, ancestors of Toc75 and OEP80 likely functioned in β-barrel assembly and insertion. During the early stages of chloroplast evolution, this ancestral Omp85 homolog was selected to act as a general protein import pore. The molecular reason this protein was selected remains unknown. One hypothesis that has been proposed is that transmembrane β-barrels were the first proteins to be targeted to the endosymbiont from the host cytosol (Gross and Bhattacharya, 2009). In this scenario, the ancestral Omp85 was capable of inserting substrates from the exterior of the endosymbiont cell. The ancestral Omp85 then gained the ability to accept other substrates and transfer them completely through the OM. At some point this protein diverged into Toc75, which specialized in non-β-barrels, and OEP80, which specialized in β-barrels. If this is the case, an OEP80-containing complex should be capable of directly inserting β-barrels from the cytosol. Our results showing that β-barrels use the TOC complex indicate that they are not directly inserted from the cytosol by an OEP80-containing complex. Instead, they favor the previously proposed model that predicts β-barrels traverse the OM before being inserted from the IMS side of the membrane (Lee et al., 2014; Richardson et al., 2014; Day and Theg, 2018). This model is comparable to β-barrel targeting to the mitochondrial OM where insertion by the Omp85 containing sorting and assembly machinery is coupled to import through the TOM machinery (Qiu et al., 2013). Furthermore, our results indicate that both Toc75 and OEP80 require transit peptides for chloroplast sorting. This makes it unlikely that the functional copies of their ancestral genes could have migrated to the host nucleus before the evolution of a protein import apparatus that could handle transit peptides.
In summary, we demonstrate that the N terminus of OEP80 behaves like a canonical transit peptide. It is removed by the SPP (Figure 2). It is necessary for the import of the IMS-localized POTRA domains, but an OEP80 construct consisting only of the β-barrel did not require the transit peptide for chloroplast targeting. This suggests that, like other β-barrels, its transmembrane domain contains intrinsic targeting information (Figure 3). Lastly, The N terminus of OEP80 is sufficient for the import of heterologous passenger proteins into the stroma, but the C terminus prevents OEP80 from completely crossing the envelope membranes (Figure 4). Our results also show that OEP80 and β-barrels without transit peptides use the TOC complex (Figures 5 and 6). Further studies are needed to determine how chloroplast β-barrels are inserted into the OM, and what other proteins are involved.
METHODS
Plasmids
The untagged Arabidopsis (Arabidopsis thaliana) OEP80 construct used for import experiments shown in Figures 1A and 5A and the Toc75 construct in Figures 2 and 5A were the same as those used in Day et al. (2014). OEP24 used in Figure 5B was derived from the coding sequence of At5g42960 in the pGEM‐T Easy vector (Promega) under the SP6 promotor. The barley OEP80 was clone AK355572 at https://barleyflc.dna.affrc.go.jp/bexdb/index.html (Sato et al., 2009; Matsumoto et al., 2011). The TPPlsp1-YFP construct was the same as described in Endow and Inoue (2013).
PsOEP80, PsOEP80Δaa1−318 (PsOEP80 β-barrel in Figure 5B), and the T7-tagged Arabidopsis OEP80-T7 (Figures 4A and 4B), OEP80Δaa1−52-T7 (prOEP80 in Figures 1B and 3A), OEP80Δaa2−93-T7 (mOEP80 in Figure 3A), OEP80Δaa2−386-T7 (OEP80 β-barrel in Figure 3A), OEP24-T7, and OEP40-T7 were cloned using the Gateway Cloning system (Thermo Fisher Scientific). The coding sequences were amplified using PCR, and the nucleotide sequences encoding the T7 tag and gateway Attb sites were added via primer overhangs. The templates for the PsOEP80 construct were Pisum sativum cDNA. The sequence cloned corresponds to contig p. sativum_CSFL_reftransV2_0048650 at www.coolseasonfoodlegume.org (Humann et al., 2017). The template for the Arabidopsis OEP80-T7 was the OEP80 plasmid described in Day et al. (2014). Plasmids containing these full-length constructs served as templates for their truncated forms. The template for OEP24-T7 was the OEP24 plasmid described in the previous paragraph. The template for OEP40-T7 was A. thaliana genomic DNA, because the gene, At3g57990, does not contain introns. The PCR fragments were cloned into pDONR207 using BP Clonase II enzyme mix (cat. no. 11,789,020; Invitrogen).
Coding sequences for the fusion protein constructs including AtOEP80Δaa1-52; Δaa400−732-OEP24-T7 (80TP-POTRA-OEP24 in Figure 3), AtOEP80Δaa2-93;Δaa400−732-OEP24-T7 (80POTRA-OEP24 in Figure 3), AtOEP80 aa1-158(GGGS)-Citrine-(GGSGGGSG)-OEP80 aa313-732-T7 (80TP-YFP-P3-Bar-T7 in Figures 4 and 6), and Citrine-(SGGGSG)-OEP80 aa313-732-T7 (YFP-P3-Bar-T7 in Figures 4 and 6), were produced using sections of the constructs listed in the previous paragraph and the long flanking overhang PCR method described in Endow and Inoue (2013). These constructs were also initially placed on pDONR207.
The constructs with C-terminal deletions, including AtOEP80Δaa1-52;Δaa 401−732-T7 (prOEP80Δβ-barrel in Figure 1B), 80TP-YFP-P3-T7 (Figures 4 and 6), and 80TP-YFP-T7 (Figure 4), were created by PCR amplification of entire plasmids OEP80Δaa1−52 and 80TP-YFP-P3-Bar-T7 on pDonr207 with primers flanking the region to be deleted. This method is commonly referred to as “round-the-horn” PCR (Moore and Prevelige, 2002; Moore, 2018). For prOEP80Δβ-barrel and 80TP-YFP-P3-Bar-T7, amino acids 401–732 from the Arabidopsis OEP80 protein sequence were deleted and a His codon was added between the OEP80 sequence and the T7 tag sequence, creating a NdeI cut site. For 80TP-YFP-T7, an additional sequence encoding the amino acid sequence “GGSRITK” was added using primer overhangs.
All constructs on pDONR207 were cloned into destination vectors using LR Clonase II enzyme mix (cat. no. 11,791,100; Invitrogen). The destination vector used for in vitro import and processing assays was pIVTGW-SP6, described in Endow et al. (2015). The destination vector used for transient expression was pMDC32 (Curtis and Grossniklaus, 2003).
In Vitro Import and Processing Assays
In vitro import assays and post-import thermolysin treatments were performed as in Day et al. (2014). Loading of all import reactions was normalized based on chlorophyll content. For the import competition assay shown in Figure 5A, recombinant prRSSU and mRSSU were produced as described in Perry and Keegstra (1994). The guanidine-solubilized recombinant protein was added to import reactions 5 min before the radiolabeled precursors. For the competition assay shown in Figure 5B, C-terminally hexa-His–tagged RSSU constructs were expressed in bacteria and purified with Ni-NTA Agarose (Qiagen) under denaturing conditions following the manufacturer’s protocol. The urea-denatured purified protein was buffer-exchanged to 50-mM HEPES-KOH at pH 8.0 with Ultra-15 10K centrifugal filters (cat. no. UFC901024; Millipore). The concentration of the recombinant protein was measured by Coomassie staining of SDS-PAGE and comparison to bovine serum albumin standards. The recombinant protein was added to import reactions 5 min before the addition of the radiolabeled precursors.
Plsp1 processing assays were performed as described in Midorikawa et al. (2014). For chloroplast soluble extract processing assays, isolated pea chloroplasts were lysed in lysis buffer (10 mM of HEPES-KOH at pH 8.0 and 10 mM of MgCl2) at a concentration of 0.25 mg of chlorophyll/mL lysis buffer. Membranes were pelleted at 16,200g for 10 min at 4°C. The supernatant was concentrated four times in an Amicon Ultra-4 30K centrifugal filter (cat. no. UFC803008; Millipore). The concentrated soluble extract was incubated with translation product at 7 μL:1 μL for 2 h at room temperature. For processing competition assays, recombinant prRSSU and mRSSU were produced as in Perry and Keegstra (1994), however, inclusion bodies were solubilized in a buffer containing 8 M of urea, 10 mM of Tris, and 100 mM of sodium phosphate at pH 8.0 rather than a guanidine-containing buffer.
Transient Expression in Nicotiana benthamiana
Five-mL seed cultures of Agrobacterium tumefaciens GV3101 were grown overnight in Luria-Bertani medium with 50 μg/mL of kanamycin, 25 μg/mL of gentamycin, and 34 μg/mL of rifampicin at 28°C. The agrobacteria contained the indicated constructs on pMDC32 (Curtis and Grossniklaus, 2003) or the suppressor of silencing, tomato bushy stunt virus p19 (Voinnet et al., 2003). Thirty-five mL Luria-Bertani medium with the same antibiotics was added to the seed culture and grown for 3 h to 4 h. Bacteria were pelleted and resuspended to an optical density600 of 0.5 in induction buffer containing 10 mM of MES-KOH at pH 5.6, 1 mM of MgCl2, 0.2% (w/v) Glu, and 0.3 mM of acetosyringone. Bacteria were incubated in this buffer for 2 h at 28°C with gentle shaking. Bacteria were pelleted and resuspended in injection buffer containing 5% (w/v) Suc and 0.3 mM of acetosyringone. Agrobacteria containing test constructs and p19 were mixed at a 4:1 ratio. The resulting solutions were injected into 4 to 8 leaves of 4- to 6-week–old N. benthamiana. Plants remained in a growth chamber for 3–4 d before analysis. Plants were grown before and after infiltration in a model no. CMP 5090 with Metal Halide lighting set (Conviron) to level 2, a 12-h light/dark cycle, 22°C, and 52% relative humidity.
Nicotiana Chloroplast Isolation and Protease Treatment
Infiltrated plants were placed in darkness the evening before chloroplast isolation to prevent overaccumulation of starch. Infiltrated leaves were homogenized in a small kitchen blender with 40 mL of grinding buffer containing 50 mM of HEPES-KOH at pH 8.0, 330 mM of sorbitol, 2 mM of EDTA, and 0.5% (w/v) BSA. This mixture was passed through cheese cloth, then centrifuged for 5 min at 4,000 rpm at 4°C in a model no. HB6 rotor (Beckman). The resulting pellet was resuspended in grinding buffer and loaded over a Percoll step gradient. The gradient was centrifuged for 10 min at 7,000 rpm at 4°C and intact chloroplasts were collected from the interface of 40% (v/v) and 80% (v/v) Percoll solutions. Chloroplasts were washed and resuspended in import buffer.
For thermolysin (cat. no. P1512, Sigma-Aldrich) treatments, both chlorophyll and protease were brought to 0.25 mg/mL in import buffer containing 2.5 mM of CaCl2. The mixtures were incubated on ice for 30 min with mixing at 15 min, after which thermolysin was inhibited with EDTA at a final concentration of 10 mM. Intact chloroplasts were reisolated through 1 mL of 40% (v/v) Percoll in import buffer with 10 mM of EDTA. For trypsin treatments, chlorophyll was brought to 0.333 mg/mL and trypsin (cat. no. T1426; Sigma-Aldrich) to 0.2 mg/mL in import buffer. The suspensions were incubated on ice for 15 min. Chloroplasts that had settled at the bottom of the tube were resuspended by gentle pipetting then incubated for an additional 15 min on ice. Trypsin was inhibited by the addition of soybean trypsin inhibitor (cat no. T9003; Sigma-Aldrich) to a final concentration of 1 mg/mL. Intact chloroplasts were reisolated through 40% (v/v) Percoll in import buffer containing 0.1 mg/mL of trypsin inhibitor. The pellet was washed two times with import buffer containing 0.1 mg/mL of trypsin inhibitor. The proteins from chloroplasts solubilized by 1% (v/v) Triton X-100 were precipitated with 80% (v/v) cold acetone.
Immunoprecipitations from Solubilized Chloroplasts
GFP affinity beads were prepared as described in Katoh et al. (2015). Briefly, a fusion protein containing glutathione s-transferase and the GFP binding nanobody was produced in Escherichia coli and bound to glutathione sepharose. Chloroplasts were brought to 0.5 mg/mL in solubilization buffer containing 25 mM of HEPES-NaOH at pH 7.5, 50 mM of NaCl, 2 mM of EDTA, 2 mM of EGTA, 1 mM of phenylmethylsulfonyl fluoride, 1% (v/v) plant protease inhibitor cocktail (cat. no. P9599; Sigma-Aldrich), and 1% (w/v) n-Dodecyl β-d-maltoside (DDM). Insoluble material was pelleted by centrifugation at 18,000g for 30 min at 4°C. The solubilized chloroplast solution was mixed with buffer-equilibrated GFP affinity beads at a ratio of 1 mL of chloroplast solution to 40 μL of bed-volume beads. The mixture was rotated end-over-end for 4 h at 4°C. The beads were pelleted and washed three times with solubilization buffer, and then two times with 50 mM of Tris-HCl at pH 8.0 with 0.5% (w/v) DDM. The bound proteins were eluted in 50 mM of Tris-HCl at pH 8.0 with 0.5% (w/v) DDM and 10 mM of reduced glutathione.
Chloroplasts used for T7 protein purification were solubilized as described in the previous paragraph, except the buffer contained 125 mM of NaCl rather than 50 mM, as well as 9 mM of KCl. Solubilized chloroplast solution was mixed with T7 affinity agarose (cat. no. 69,026; Millipore) at a ratio of 1.5-mL to 100-μL bed volume beads. The mixture was rotated end-over-end for 3 h at 4°C. The beads were pelleted and washed five times with solubilization buffer. Bound proteins were eluted with 100 mM of Gly at pH 2.2 and 0.5% (w/v) DDM.
Antibodies
Antibodies against Toc75, Toc159, and Tic110 were used as described in Hsu et al. (2012). Tic40 antisera was diluted 1:6,000 in blocking buffer (Tris-buffered saline with 0.5% [v/v] TWEEN 20 and 5% [w/v] milk protein). The T7 antibody (cat. no. AB3790; Millipore) was diluted 1:3,000 in blocking buffer. Citrine-tagged constructs were detected using a polyclonal GFP antiserum (cat. no. sc-8334; Santa Cruz Biotechnology) diluted 1:5,000 in blocking buffer. Secondary antibodies were goat anti-rabbit–conjugated to horseradish peroxidase (cat. no. 074-1506; Kirkegaard and Perry Laboratories) and goat anti-rabbit–conjugated to alkaline phosphatase (cat. no. ADI-SAB-301-J; Enzo).
Accession Numbers
Protein sequences for the proteins analyzed in this article can be found in the GenBank/EMBL data libraries under accession numbers AtOEP80 (Q9C5J8.1), AtToc75 (Q9STE8.1) AtOEP24 (Q8H0Y1.1), AtOEP40 (NP_191358.1), AtPlsp1 (ANM63871.1), and HvOEP80 (BAJ86791.1). The full protein sequence of PsOEP80 is not available on GenBank, but the transcript sequence can be found at the Cool Season Food Legume Crop Database (Humann et al., 2017), https://www.coolseasonfoodlegume.org/, as the sole coding sequence on contig p. sativum_CSFL_reftransV2_0048650.
Supplemental Data
Supplemental Figure 1. Many OEP80 orthologs are predicted to contain transit peptides.
Supplemental Figure 2. Processing of the A. thaliana OEP80 by Isolated chloroplast soluble extract is not detectible.
Supplemental Figure 3. Additional competition assays.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
This work was supported by the U.S. National Science Foundation (grant 1050602 to K.I.), the Division of Chemical Sciences, Geosciences, and Biosciences, 408 Office of Basic Energy Sciences of the U.S. Department of Energy (grant DE-SC0017035 to S.M.T.), University of California Davis Department of Plant Sciences Graduate Student Research Assistantship (to P.M.D.), and a Henry A. Jastro Graduate Research Award (to P.M.D.). We thank Kenneth Keegstra for the Toc75, Tic110, and Tic40 antibodies, and Masato Nakai for the Toc159 antibody.
AUTHOR CONTRIBUTIONS
P.M.D., K.I., and S.M.T. designed the experiments; P.M.D. performed the experiments; P.M.D. and S.M.T. wrote the article.
References
- Baldwin A.J., Inoue K. (2006). The most C-terminal tri-glycine segment within the polyglycine stretch of the pea Toc75 transit peptide plays a critical role for targeting the protein to the chloroplast outer envelope membrane. FEBS J. 273: 1547–1555. [DOI] [PubMed] [Google Scholar]
- Bölter B. (2018). En route into chloroplasts: Preproteins’ way home. Photosynth. Res. 138: 263–275. [DOI] [PubMed] [Google Scholar]
- Bölter B., Soll J., Schulz A., Hinnah S., Wagner R. (1998). Origin of a chloroplast protein importer. Proc. Natl. Acad. Sci. USA 95: 15831–15836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bölter B., Soll J., Hill K., Hemmler R., Wagner R. (1999). A rectifying ATP-regulated solute channel in the chloroplastic outer envelope from pea. EMBO J. 18: 5505–5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.L., Chen L.J., Li H.M. (2016). Polypeptide transport-associated domains of the Toc75 channel protein are located in the intermembrane space of chloroplasts. Plant Physiol. 172: 235–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline K., Werner-Washburne M., Andrews J., Keegstra K. (1984). Thermolysin is a suitable protease for probing the surface of intact pea chloroplasts. Plant Physiol. 75: 675–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day P.M., Theg S.M. (2018). Evolution of protein transport to the chloroplast envelope membranes. Photosynth. Res. 138: 315–326. [DOI] [PubMed] [Google Scholar]
- Day P.M., Potter D., Inoue K. (2014). Evolution and targeting of Omp85 homologs in the chloroplast outer envelope membrane. Front. Plant Sci. 5: 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckart K., Eichacker L., Sohrt K., Schleiff E., Heins L., Soll J. (2002). A Toc75-like protein import channel is abundant in chloroplasts. EMBO Rep. 3: 557–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen H., von Heijne G. (1999). ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8: 978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endow J.K., Inoue K. (2013). Stable complex formation of thylakoidal processing peptidase and PGRL1. FEBS Lett. 587: 2226–2231. [DOI] [PubMed] [Google Scholar]
- Endow J.K., Singhal R., Fernandez D.E., Inoue K. (2015). Chaperone-assisted post-translational transport of plastidic type I signal peptidase 1. J. Biol. Chem. 290: 28778–28791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endow J.K., Rocha A.G., Baldwin A.J., Roston R.L., Yamaguchi T., Kamikubo H., Inoue K. (2016). Polyglycine acts as a rejection signal for protein transport at the chloroplast envelope. PLoS One 11: e0167802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griesbeck O., Baird G.S., Campbell R.E., Zacharias D.A., Tsien R.Y. (2001). Reducing the environmental sensitivity of yellow fluorescent protein. Mechanism and applications. J. Biol. Chem. 276: 29188–29194. [DOI] [PubMed] [Google Scholar]
- Gross J., Bhattacharya D. (2009). Mitochondrial and plastid evolution in eukaryotes: An outsiders’ perspective. Nat. Rev. Genet. 10: 495–505. [DOI] [PubMed] [Google Scholar]
- Harsman A., Schock A., Hemmis B., Wahl V., Jeshen I., Bartsch P., Schlereth A., Pertl-Obermeyer H., Goetze T.A., Soll J., Philippar K., Wagner R. (2016). OEP40, a regulated glucose-permeable β-barrel solute channel in the chloroplast outer envelope membrane. J. Biol. Chem. 291: 17848–17860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann N.R., Theg S.M. (2005). Chloroplast outer membrane protein targeting and insertion. Trends Plant Sci. 10: 450–457. [DOI] [PubMed] [Google Scholar]
- Hsu S.C., Nafati M., Inoue K. (2012). OEP80, an essential protein paralogous to the chloroplast protein translocation channel Toc75, exists as a 70-kD protein in the Arabidopsis thaliana chloroplast outer envelope. Plant Mol. Biol. 78: 147–158. [DOI] [PubMed] [Google Scholar]
- Humann J.L., et al. (2017). Cool Season Food Legume Genome Database: A resource for pea, lentil, faba bean and chickpea genetics, genomics and breeding. Proceedings of the International Plant and Animal Genome Conference: January 2017, San Diego, CA. [Google Scholar]
- Inoue K., Keegstra K. (2003). A polyglycine stretch is necessary for proper targeting of the protein translocation channel precursor to the outer envelope membrane of chloroplasts. Plant J. 34: 661–669. [DOI] [PubMed] [Google Scholar]
- Inoue K., Potter D. (2004). The chloroplastic protein translocation channel Toc75 and its paralog OEP80 represent two distinct protein families and are targeted to the chloroplastic outer envelope by different mechanisms. Plant J. 39: 354–365. [DOI] [PubMed] [Google Scholar]
- Inoue K., Baldwin A.J., Shipman R.L., Matsui K., Theg S.M., Ohme-Takagi M. (2005). Complete maturation of the plastid protein translocation channel requires a type I signal peptidase. J. Cell Biol. 171: 425–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh Y., Nozaki S., Hartanto D., Miyano R., Nakayama K. (2015). Architectures of multisubunit complexes revealed by a visible immunoprecipitation assay using fluorescent fusion proteins. J. Cell Sci. 128: 2351–2362. [DOI] [PubMed] [Google Scholar]
- Kessler F., Blobel G., Patel H.A., Schnell D.J. (1994). Identification of two GTP-binding proteins in the chloroplast protein import machinery. Science 266: 1035–1039. [DOI] [PubMed] [Google Scholar]
- Klinger A., Gosch V., Bodensohn U., Ladig R., Schleiff E. (2019). The signal distinguishing between targeting of outer membrane β-barrel protein to plastids and mitochondria in plants. Biochim Biophys Acta Mol Cell Res 1866: 663–672. [DOI] [PubMed] [Google Scholar]
- Kouranov A., Chen X., Fuks B., Schnell D.J. (1998). Tic20 and Tic22 are new components of the protein import apparatus at the chloroplast inner envelope membrane. J. Cell Biol. 143: 991–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.W., Hwang I. (2018). Evolution and design principles of the diverse chloroplast transit peptides. Mol. Cells 41: 161–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.W., Yoo Y.J., Razzak M.A., Hwang I. (2018). Prolines in transit peptides are crucial for efficient preprotein translocation into chloroplasts. Plant Physiol. 176: 663–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Kim D.H., Hwang I. (2014). Specific targeting of proteins to outer envelope membranes of endosymbiotic organelles, chloroplasts, and mitochondria. Front. Plant Sci. 5: 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto T., et al. (2011). Comprehensive sequence analysis of 24,783 barley full-length cDNAs derived from 12 clone libraries. Plant Physiol. 156: 20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midorikawa T., Endow J.K., Dufour J., Zhu J., Inoue K. (2014). Plastidic type I signal peptidase 1 is a redox-dependent thylakoidal processing peptidase. Plant J. 80: 592–603. [DOI] [PubMed] [Google Scholar]
- Moore S. (2018). Round-the-horn site-directed mutagenesis (OpenWetWare). https://openwetware.org/wiki/'Round-the-horn_site-directed_mutagenesis
- Moore S.D., Prevelige P.E. Jr (2002). A P22 scaffold protein mutation increases the robustness of head assembly in the presence of excess portal protein. J. Virol. 76: 10245–10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai M. (2018). New perspectives on chloroplast protein import. Plant Cell Physiol. 59: 1111–1119. [DOI] [PubMed] [Google Scholar]
- Paila Y.D., Richardson L.G., Inoue H., Parks E.S., McMahon J., Inoue K., Schnell D.J. (2016). Multi-functional roles for the polypeptide transport associated domains of Toc75 in chloroplast protein import. eLife 5: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry S.E., Keegstra K. (1994). Envelope membrane proteins that interact with chloroplastic precursor proteins. Plant Cell 6: 93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlmeyer K., Soll J., Grimm R., Hill K., Wagner R. (1998). A high-conductance solute channel in the chloroplastic outer envelope from Pea. Plant Cell 10: 1207–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J., et al. (2013). Coupling of mitochondrial import and export translocases by receptor-mediated supercomplex formation. Cell 154: 596–608. [DOI] [PubMed] [Google Scholar]
- Reumann S., Keegstra K. (1999). The endosymbiotic origin of the protein import machinery of chloroplastic envelope membranes. Trends Plant Sci. 4: 302–307. [DOI] [PubMed] [Google Scholar]
- Richardson L.G., Paila Y.D., Siman S.R., Chen Y., Smith M.D., Schnell D.J. (2014). Targeting and assembly of components of the TOC protein import complex at the chloroplast outer envelope membrane. Front. Plant Sci. 5: 269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter S., Lamppa G.K. (1998). A chloroplast processing enzyme functions as the general stromal processing peptidase. Proc. Natl. Acad. Sci. USA 95: 7463–7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson C., Ellis R.J. (1984). Transport of proteins into chloroplasts. Partial purification of a chloroplast protease involved in the processing of important precursor polypeptides. Eur. J. Biochem. 142: 337–342. [DOI] [PubMed] [Google Scholar]
- Sánchez-Pulido L., Devos D., Genevrois S., Vicente M., Valencia A. (2003). POTRA: A conserved domain in the FtsQ family and a class of beta-barrel outer membrane proteins. Trends Biochem. Sci. 28: 523–526. [DOI] [PubMed] [Google Scholar]
- Sato K., Shin-I T., Seki M., Shinozaki K., Yoshida H., Takeda K., Yamazaki Y., Conte M., Kohara Y. (2009). Development of 5006 full-length CDNAs in barley: A tool for accessing cereal genomics resources. DNA Res. 16: 81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiff E., Soll J. (2005). Membrane protein insertion: Mixing eukaryotic and prokaryotic concepts. EMBO Rep. 6: 1023–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell D.J., Kessler F., Blobel G. (1994). Isolation of components of the chloroplast protein import machinery. Science 266: 1007–1012. [DOI] [PubMed] [Google Scholar]
- Selkrig J., et al. (2012). Discovery of an archetypal protein transport system in bacterial outer membranes. Nat Struct Mol Biol 19: 506–510. [DOI] [PubMed] [Google Scholar]
- Shi L.X., Theg S.M. (2013). The chloroplast protein import system: From algae to trees. Biochim. Biophys. Acta 1833: 314–331. [DOI] [PubMed] [Google Scholar]
- Sommer M.S., Daum B., Gross L.E., Weis B.L., Mirus O., Abram L., Maier U.G., Kühlbrandt W., Schleiff E. (2011). Chloroplast Omp85 proteins change orientation during evolution. Proc. Natl. Acad. Sci. USA 108: 13841–13846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranel P.J., Keegstra K. (1996). A novel, bipartite transit peptide targets OEP75 to the outer membrane of the chloroplastic envelope. Plant Cell 8: 2093–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranel P.J., Froehlich J., Goyal A., Keegstra K. (1995). A component of the chloroplastic protein import apparatus is targeted to the outer envelope membrane via a novel pathway. EMBO J. 14: 2436–2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu S.L., Chen L.J., Smith M.D., Su Y.S., Schnell D.J., Li H.M. (2004). Import pathways of chloroplast interior proteins and the outer-membrane protein OEP14 converge at Toc75. Plant Cell 16: 2078–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanderVere P.S., Bennett T.M., Oblong J.E., Lamppa G.K. (1995). A chloroplast processing enzyme involved in precursor maturation shares a zinc-binding motif with a recently recognized family of metalloendopeptidases. Proc. Natl. Acad. Sci. USA 92: 7177–7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33: 949–956. [DOI] [PubMed] [Google Scholar]
- Vojta L., Soll J., Bölter B. (2007). Protein transport in chloroplasts—targeting to the intermembrane space. FEBS J. 274: 5043–5054. [DOI] [PubMed] [Google Scholar]
- Voulhoux R., Bos M.P., Geurtsen J., Mols M., Tommassen J. (2003). Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299: 262–265. [DOI] [PubMed] [Google Scholar]
- Weidberg H., Amon A. (2018). MitoCPR—A surveillance pathway that protects mitochondria in response to protein import stress. Science 360: 360. [DOI] [PMC free article] [PubMed] [Google Scholar]