The AP2/ERF family transcription factor TINY is regulated by brassinosteroid signaling and coordinates brassinosteroid-regulated plant growth and drought responses.
Abstract
APETALA2/ETHYLENE RESPONSIVE FACTOR (AP2/ERF) family transcription factors have well-documented functions in stress responses, but their roles in brassinosteroid (BR)-regulated growth and stress responses have not been established. Here, we show that the Arabidopsis (Arabidopsis thaliana) stress-inducible AP2/ERF transcription factor TINY inhibits BR-regulated growth while promoting drought responses. TINY-overexpressing plants have stunted growth, increased sensitivity to BR biosynthesis inhibitors, and compromised BR-responsive gene expression. By contrast, tiny tiny2 tiny3 triple mutants have increased BR-regulated growth and BR-responsive gene expression. TINY positively regulates drought responses by activating drought-responsive genes and promoting abscisic acid–mediated stomatal closure. Global gene expression studies revealed that TINY and BRs have opposite effects on plant growth and stress response genes. TINY interacts with and antagonizes BRASSINOSTERIOID INSENSITIVE1-ETHYL METHANESULFONATE SUPRESSOR1 (BES1) in the regulation of these genes. Glycogen synthase kinase 3-like protein kinase BR-INSENSITIVE2 (BIN2), a negative regulator in the BR pathway, phosphorylates and stabilizes TINY, providing a mechanism for BR-mediated downregulation of TINY to prevent activation of stress responses under optimal growth conditions. Taken together, our results demonstrate that BR signaling negatively regulates TINY through BIN2 phosphorylation and TINY positively regulates drought responses, as well as inhibiting BR-mediated growth through TINY-BES1 antagonistic interactions. Our results thus provide insight into the coordination of BR-regulated growth and drought responses.
INTRODUCTION
Environmental challenges such as water deficit and extreme temperatures are associated with decreased plant growth and can cause severe crop losses (Fahad et al., 2017). Brassinosteroids (BRs) are a class of polyhydroxylated plant steroid hormones that play important roles in plant growth, development, and stress responses (Clouse et al., 1996; Nolan et al., 2017a). BRs are perceived through a receptor kinase, BRASSINOSTERIOID INSENSITIVE1 (BRI1), along with the coreceptor BRI1-ASSOCIATED RECEPTOR KINASE. BRs function through a cascade of signaling components including the negative regulator BRASSINOSTERIOID INSENSITIVE2 (BIN2), a glycogen synthase kinase 3-like kinase (He et al., 2002), to regulate transcription factors BRI1-ETHYL METHANESULFONATE SUPRESSOR1 (BES1) and BRASSINAZOLE-RESISITANT1 (BZR1; Clouse et al., 1996; Li and Chory, 1997; Li et al., 2002; Nam and Li, 2002; Wang et al., 2002; Yin et al., 2002; Gou et al., 2012).
BRs have been demonstrated to regulate drought, although there are mixed reports as to whether BRs promote or inhibit drought responses. Exogenous application of BRs can improve drought tolerance in Arabidopsis (Arabidopsis thaliana) and tomato (Solanum lycopersicum), supporting a positive role for BRs in drought responses (Kagale et al., 2007; Zhou et al., 2014). Consistent with this idea, overexpression of the vascular BR receptor BRI1-LIKE3 leads to increased drought tolerance (Fàbregas et al., 2018). However, BR biosynthesis and signaling loss-of-function mutants display increased survival under drought conditions (Northey et al., 2016; Nolan et al., 2017c; Ye et al., 2017). The BR signaling gain-of-function mutant bes1-D is hypersensitive to drought, indicating that BR signaling functions through BES1 to negatively regulate drought responses (Ye et al., 2017). Specifically, BES1 cooperates with WRKY46, WRKY54, and WRKY70 to promote plant growth–related gene expression but repress drought-responsive gene expression (Chen et al., 2017). Moreover, drought conditions promote the degradation of BES1 and WRKY54 to inhibit their effect on growth, leading to enhanced drought responses (Chen et al., 2017; Nolan et al., 2017c; Yang et al., 2017). One mechanism mediating the antagonism between BES1 and drought responses is mediated by the NO APICAL MERISTEM, ARABIDOPSIS THALIANA ACTIVATING FACTOR and CUP-SHAPED COTYLEDON (NAC) family transcription factor RESPONSIVE TO DESICCATION26 (RD26), which positively regulates drought survival and inhibits growth (Fujita et al., 2004). BES1 and RD26 bind to a common promoter element to inhibit each other’s transcriptional activity (Ye et al., 2017). In addition, BES1 and BZR1 regulate the expression of thousands of BR-responsive target genes including APETALA2/ETHYLENE RESPONSIVE FACTOR (AP2/ERF) transcription factors, which suggests that AP2/ERF transcription factors likely function along with BES1 to balance BR-regulated growth and stress responses (Sun et al., 2010; Yu et al., 2011; Guo et al., 2013).
The negative effect of BRs in drought responses is also linked to abscisic acid (ABA), a hormone that is induced during stress and promotes plant survival during drought (Kuromori et al., 2018). BR and ABA pathways antagonize one another through multiple signaling components. One notable point of crosstalk occurs at the GSK3-like protein kinase BIN2, which functions as a negative regulator in the BR pathway but is activated by ABA. The TYPE 2C PROTEIN PHOSPHATASES ABA INSENSITIVE1 (ABI1) and ABI2 dephosphorylate and inhibit BIN2 in the absence of ABA, but when ABA is present ABI1/ABI2 are inhibited to allow for BIN2 activation (Wang et al., 2018). BIN2, in turn, promotes ABA signaling through phosphorylation and activation of SNF1-RELATED PROTEIN KINASE2.2 and SNF1-RELATED PROTEIN KINASE2.3 kinases as well as downstream transcription factors such as ABI5 (Cai et al., 2014; Hu and Yu, 2014).
AP2/ERF transcription factors regulate plant drought responses as well as plant growth and development (Phukan et al., 2017; Xie et al., 2019). Many drought-tolerant plants generated by overexpressing stress-inducible AP2/ERF transcription factors displayed reduced plant growth (Sakuma et al., 2006; Karaba et al., 2007; Sharabi-Schwager et al., 2010); however, the mechanisms by which AP2/ERFs coordinate growth and stress responses have yet to be defined. TINY belongs to the DEHYDRATION-RESPONSIVE ELEMENT BINDING protein A4 subfamily of AP2/ERF family transcription factors that contains 17 members in Arabidopsis (Nakano et al., 2006). TINY was previously shown to inhibit plant growth (Wilson et al., 1996). Several lines of evidence point toward a role of TINY in stress responses in addition to regulating plant growth. TINY transcript levels are highly induced by various stresses such as dehydration, cold, and salt, and overexpression of TINY was associated with increased drought-responsive gene expression and hypersensitivity to ABA-mediated seed germination and root growth inhibition (Sun et al., 2008; Coego et al., 2014). Although TINY is known to be involved in controlling growth and stress programs, the specific pathways and mechanisms by which TINY mediates these responses remain to be established.
In this study, we found that TINY inhibits plant growth and promotes the drought response to alter the balance between BR-mediated plant growth and drought responses. TINY inhibits plant growth by negatively regulating BR signaling and BR-responsive gene expression. TINY interacts with and antagonizes BES1 on BR-induced genes involved in plant growth and BR-repressed genes implicated in drought responses. Furthermore, we demonstrate that TINY is phosphorylated and stabilized by BIN2, a negative regulator in the BR pathway but positive regulator in ABA and stress responses. TINY promotes ABA-induced stomatal closure, drought-responsive gene expression, and plant survival during drought conditions. Overall, these results provide a mechanism by which stress-inducible transcription factor TINY activates drought stress responses and shuts down BR-regulated growth through antagonistic interactions between TINY and BES1 on BR-responsive genes.
RESULTS
TINY Negatively Regulates Plant Growth by Inhibiting the BR Pathway
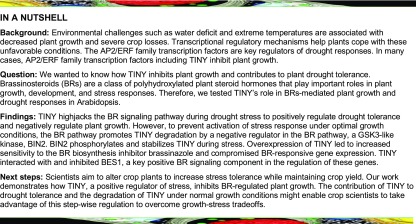
Previous studies indicated that TINY gain-of-function mutants display stunted growth, reduced hypocotyl elongation, and fertility defects (Wilson et al., 1996). Our BR-responsive transcriptome data sets revealed that a large number of transcription factors respond to BRs, including TINY (AT5G25810) and several of its homologs (TINYs; Supplemental Figure 1A; Yu et al., 2011). To confirm whether TINYs are regulated by BRs, TINY transcript levels were determined using the 4-week-old Arabidopsis wild-type and bes1-D plants treated with or without brassinolide (BL). The expression of TINY and TINY2 (At5G11590) was induced by BL in the wild type as well as in bes1-D (Supplemental Figure 1B); however, TINY3 (AT4G32800) was repressed by BL in the wild type and bes1-D. The regulation of TINYs by BRs as well as the growth phenotypes of TINY gain-of-function mutants suggests that TINY may be involved in BR-regulated plant growth.
To test this hypothesis, we first generated transgenic lines that overexpressed TINY (TINY-OE) in the wild-type Arabidopsis. Consistent with previous reports, TINY-OE transgenic lines displayed a stunted growth phenotype, the severities of which corresponded well with TINY transcript and TINY protein levels (Figure 1A; Supplemental Figure 1C). Additionally, we obtained T-DNA knockout mutants for TINY and its homologs (Supplemental Figure 1D). Single mutants of tiny, tiny2, or tiny3 did not show any obvious growth phenotypes under the tested conditions, which is likely due to genetic redundancy (Supplemental Figures 1E and 1F). We then generated tiny tiny2 tiny3 triple mutants and observed that tiny tiny2 tiny3 displayed enlarged growth with longer petioles (Figure 1B). Genetic complementation experiments with a construct under the control of the native TINY promoter (TINY:TINY-FLAG) were performed, and the results support that the phenotype of tiny tiny2 tiny3 is caused by loss of function of TINY and its close homologs (Supplemental Figure 1G).
Figure 1.
TINY Negatively Regulates Plant Growth by Inhibiting the BR Pathway.
(A) Growth phenotype of the 4-week-old wild-type (WT) and TINY-OE (lines 3 and 8) plants. The sixth leaves of plants were measured for petiole length. Data represent mean and sd, n = 30 to 36. *P < 0.05, **P < 0.01; Student’s t test.
(B) Phenotype of the 4-week-old wild-type (WT) and tiny tiny2 tiny3 plants. Data represent mean and SD, n = 15 to 22.
(C) and (D) BRZ sensitivity of TINY-OE and tiny tiny2 tiny3 plants. Seven-day-old seedlings were grown on 1/2 LS medium with 250 nM BRZ in dark (C). Hypocotyl was measured using ImageJ (D). Data represent mean and sd from three biological replicates (n = 3); each replicate contained 12 to 15 seedlings. *P < 0.05, **P < 0.01; two-way analysis of variance. WT, wild type.
(E) and (F) BR-regulated gene expression in TINY mutants. mRNA was extracted from 4-week-old plants for BR-induced genes (E) and BR-repressed genes (F) expression analysis. Data represent mean and sd from three biological replicates (n = 3). Each biological replicate was pooled tissue from three to four individual plants. *P < 0.05, **P < 0.01; Student’s t test.
In order to determine whether the negative role of TINY on plant growth is related to the BR pathway, we performed BR response assays using the BR biosynthesis inhibitor brassinazole (BRZ) that reduces endogenous BRs and causes reduced hypocotyl elongation (Asami et al., 2000). TINY-OE plants had shorter hypocotyls and were more sensitive to BRZ compared with the wild type, while tiny tiny2 tiny3 plants were less sensitive to lower concentrations of BRZ (Figures 1C and 1D; Supplemental Figure 2A). TINY-OE plants were also more sensitive to the BR biosynthesis inhibitor propiconazole (Pcz; Hartwig et al., 2012) in soil-grown plants, whereas tiny tiny2 tiny3 mutants were less sensitive to Pcz treatment (Supplemental Figures 2B to 2D). Thus, the BR response phenotypes of TINY-OE and tiny tiny2 tiny3 are consistent with TINY negatively modulating BR-regulated growth. We then tested the expression of several BR-responsive marker genes (Zhang et al., 2009; Xie et al., 2011; Oh et al., 2012; Yang et al., 2017). BR-induced plant growth–related genes TOUCH4, SMALL AUXIN UPREGULATED RNA9, CELLULOSE SYNTHASE5 (CESA5), and INDOLE-3-ACETIC ACID INDUCIBLE19 (IAA19) were upregulated in tiny tiny2 tiny3 but downregulated in TINY-OE plants (Figure 1E), while BR-repressed genes involved in growth inhibition, Late Elongated Hypocotyl1 and ILI1 BINDING BHLH1, were reduced in tiny tiny2 tiny3 but increased in TINY-OE plants (Figure 1F). Taken together, these results indicate that TINY inhibits BR-regulated growth and BR-responsive gene expression.
BRs Promote TINY Degradation through the Inhibition of BIN2
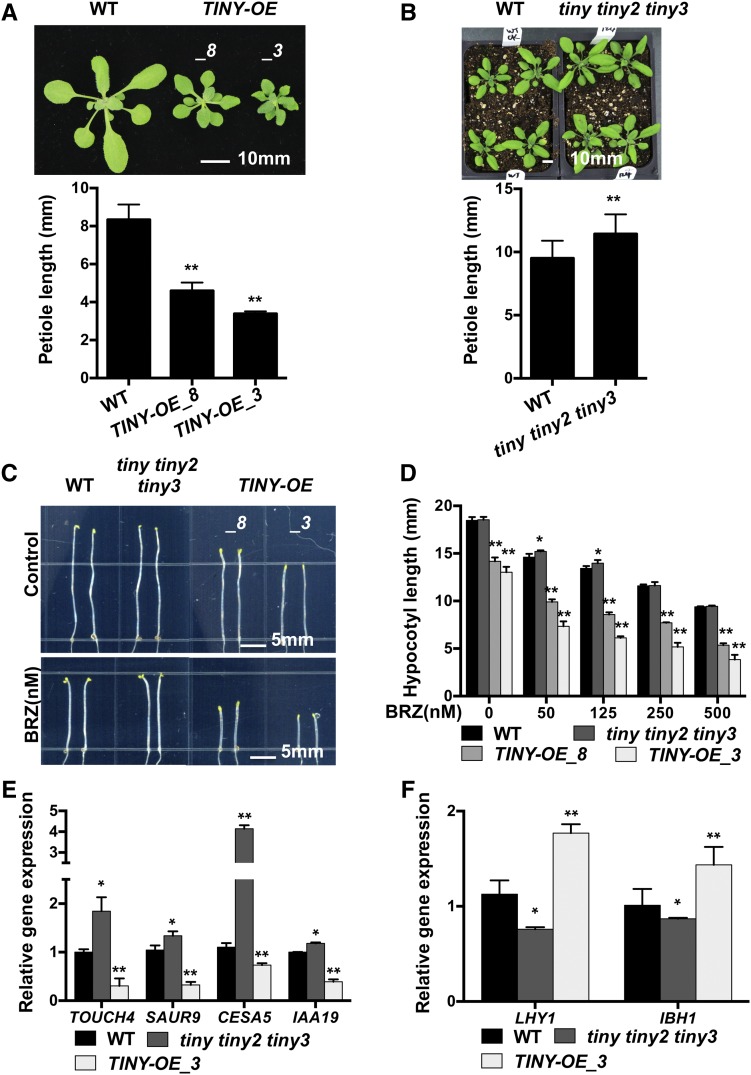
To characterize how BRs regulate TINY, we treated TINY-OE plants with BL and quantified the hypocotyl lengths. Surprisingly, we found that BL treatment completely rescued the hypocotyl elongation defects observed in TINY-OE plants (Figures 2A and 2B). This effect was also observed using transgenic lines in which TINY was under the control of an inducible promoter (XVE:TINY-FLAG; Supplemental Figures 3A and 3B), suggesting that the observed phenotype is not due to the effect of BL on TINY transcript levels. Since TINY negatively regulates plant growth, we then examined whether BR promotes TINY degradation to alleviate plant growth inhibition caused by TINY. In line with this idea, TINY protein was decreased with increasing BL concentrations (Figure 2C). Moreover, we used cycloheximide (CHX), which inhibits translation to block new protein synthesis, to test TINY protein half-life with or without BL. TINY protein had shorter half-life of ∼25 min with BL treatment compared with control, with a half-life of ∼50 min (Figures 2D and 2E). Furthermore, degradation of TINY could be inhibited by the proteasome inhibitor MG132 (Kisselev et al., 2012), suggesting that TINY stability is regulated by 26S proteasome–mediated degradation (Supplemental Figure 3C).
Figure 2.
TINY Is Phosphorylated and Stabilized by BIN2.
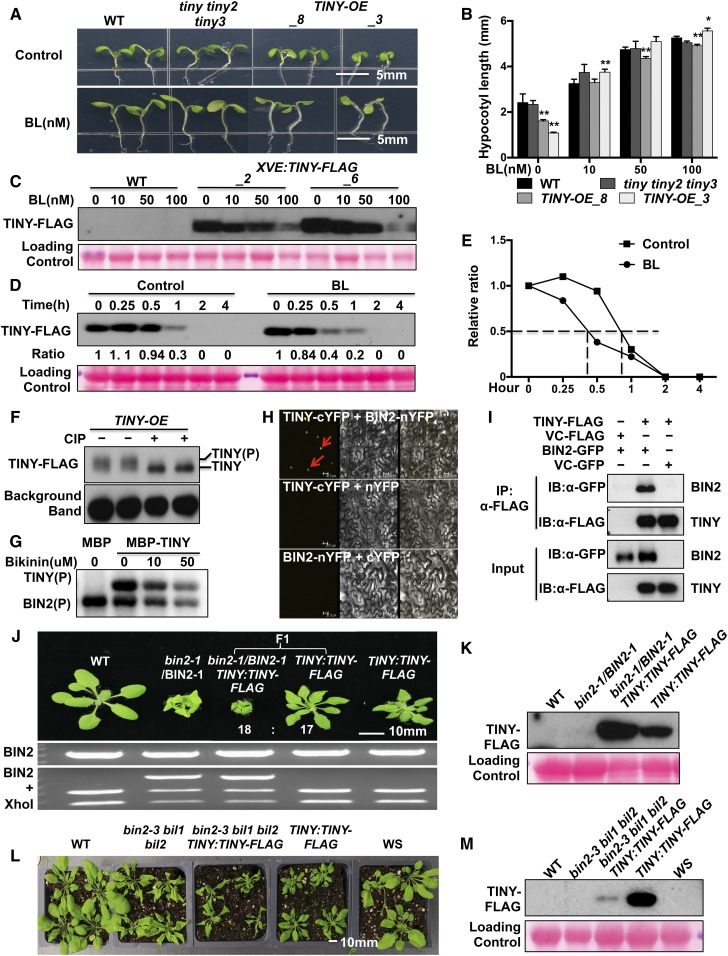
(A) and (B) BL responses of tiny mutants. Seven-day-old plants grown on 1/2 LS medium with or without 50 nM BL under light (A). Hypocotyl length was measured using ImageJ (B). Data represent mean and sd from three biological replicates (n = 3); each replicate contained 12 to 15 plants. *P < 0.05, **P < 0.01; two-way analysis of variance. WT, wild type.
(C) TINY-FLAG protein decreased after BL treatment. XVE:TINY-FLAG transgenic lines were treated with indicated concentration of BL and 10 μM β-estradiol for 7 d. Samples were collected to detect TINY protein with anti-FLAG antibody. WT, wild type.
(D) and (E) CHX treatment for TINY degradation. XVE:TINY-FLAG line 6 was grown on β-estradiol plates for 7 d. The seedlings were treated with 400 μM CHX in the absence or presence of 100 nM BL for the indicated time. Tissues were collected for immunoblotting analysis, and TINY-FLAG protein was detected with anti-FLAG antibody (D). Protein level was normalized with loading control and treatment at 0 h (E).
(F) TINY was phosphorylated in plants. TINY-FLAG protein was immunoprecipitated from 7-d-old TINY-OE seedlings (line 3 was used for most experiments unless otherwise specified) and treated with CIP. Phosphorylated TINY (P) was indicated.
(G) BIN2 phosphorylated TINY. MBP-TINY, but not MBP, was phosphorylated by BIN2 in the in vitro kinase assay. BIN2 phosphorylation of TINY was inhibited by indicated concentration of bikinin.
(H) TINY interacted with BIN2 in by BiFC. Cotransformation of TINY-cYFP and BIN2-nYFP led to the reconstitution of YFP activity in N. benthamiana nucleus under YFP filter, whereas coexpression of TINY-cYFP and nYFP or BIN2-nYFP and cYFP did not produce any positive YFP signal.
(I) Co-IP assay showed TINY and BIN2 interaction. TINY-FLAG and BIN2-GFP as well as control vectors were cotransformed into Arabidopsis protoplasts overnight. After 20 μM MG132 treatment for 1 h, protoplasts were collected and protein was immunoprecipitated with anti-FLAG and detected with anti-FLAG and anti-GFP antibodies.
(J) Phenotype of 3-week old bin2-1 TINY:TINY-FLAG double mutant (TINY:TINY-FLAG line 2 was used for crossing) in F1 generation (top). bin2-1 background was genotyped (Yan et al., 2009): BIN2 DNA was amplified by PCR (middle) and digested with XhoI (bottom).
(K) TINY protein level in bin2-1/BIN2-1 TINY:TINY-FLAG double mutant from (J).
(L) Phenotype of 4-week-old bin2-3 bil1 bil2 TINY:TINY-FLAG homozygous mutant (TINY:TINY-FLAG line 2 was used for crossing). Wassilewskija (WS) ecotype is used as control. WT, wild type.
(M) TINY protein level in bin2-3 bil1 bil2 TINY:TINY-FLAG from (L). WS, Wassilewskija; WT, wild type.
We also observed two bands corresponding to TINY protein in immunoblots (Figure 2C), indicating that TINY may be posttranslationally modified. To test whether the altered migration of TINY is due to phosphorylation, TINY-FLAG protein was immunoprecipitated from TINY-OE plants and treated with calf-intestinal alkaline phosphatase (CIP). CIP treatment caused TINY to shift from a higher molecular weight (phosphorylated) form to a lower molecular weight (unphosphorylated) form (Figure 2F), suggesting that TINY can be phosphorylated in plants.
BIN2 is a negative regulator in the BR pathway that phosphorylates and inhibits BES1 and BZR1 (He et al., 2002; Wang et al., 2002; Yin et al., 2002). BIN2 functions as a Ser/Thr kinase whose substrates typically contain repeats of a short consensus sequence (S/T-X-X-X-S/T, where X corresponds to any amino acid; Zhao et al., 2002). Phosphorylation by BIN2 often affects protein stability or activity (Youn and Kim, 2015). The TINY protein sequence contains 22 potential BIN2 phosphorylation sites (Supplemental Figure 3D). Therefore, we hypothesized that BIN2 phosphorylates TINY and affects its stability. An in vitro kinase assay was first conducted using glutathione S-transferase (GST)–tagged BIN2 (GST-BIN2) and maltose binding protein (MBP)-tagged TINY (MBP-TINY) proteins to test whether BIN2 phosphorylates TINY. We observed that TINY was phosphorylated by BIN2 and that this phosphorylation was inhibited by the BIN2-specific inhibitor bikinin (Figure 2G; De Rybel et al., 2009).
Next, we tested the physical interaction between BIN2 and TINY. Yeast-two hybrid (Y2H) assays using TINY as bait and BIN2 as prey showed that TINY-BIN2 combinations specifically activate bacterial β-galactosidase (LacZ) reporter, while negative controls did not (Supplemental Figure 3E). The direct interaction between BIN2 and TINY was then confirmed using GST-pulldown assays. GST-tagged TINY (GST-TINY), but not GST alone, pulled down a significant amount of MBP-tagged BIN2 (MBP-BIN2) protein (Supplemental Figure 3F). We then confirmed the TINY-BIN2 interaction in planta using bimolecular fluorescence complementation (BiFC) and coimmunoprecipitation (Co-IP) assays. TINY-cYFP and BIN2-nYFP were cotransformed into Nicotiana benthamiana, and strong reconstituted yellow fluorescent protein (YFP) signal was observed in the nucleus (Figure 2H; Supplemental Figure 3G). However, no signal was observed in negative controls in which TINY-cYFP or BIN2-nYFP were cotransformed with N-terminal YFP (nYFP) or C-terminal YFP (cYFP), respectively. In Co-IP assays, FLAG antibody specifically pulled down BIN2-GFP coexpressed with TINY-FLAG in Arabidopsis protoplasts, further confirming the interaction between BIN2 and TINY (Figure 2I; Supplemental Figure 3H).
To investigate the effect of BIN2 phosphorylation of TINY, we generated TINY-FLAG lines under the control of the native TINY promoter (TINY:TINY-FLAG) and performed crosses with BIN2 gain-of-function bin2-1 mutants (Li et al., 2001) or loss-of-function bin2-3 bil1 bil2 triple mutants (Yan et al., 2009). All genotypes were confirmed before testing. TINY:TINY-FLAG plants displayed increased TINY transcript and protein levels as well as decreased plant growth (Figure 2J; Supplemental Figures 3I to 3K). We found that TINY protein accumulated in TINY:TINY-FLAG plants heterozygous for bin2-1 and hemizygous for the transgene (bin2-1/BIN2-1 TINY:TINY-FLAG; Figure 2K). Moreover, bin2-1/BIN2-1 TINY:TINY-FLAG plants had decreased growth compared with bin2-1/BIN2-1 or TINY:TINY-FLAG. These data suggest that TINY is stabilized by BIN2 and functions to inhibit plant growth. Next, we analyzed the growth phenotypes and TINY protein levels of TINY:TINY-FLAG in bin2-3 bil1 bil2 mutants. TINY:TINY-FLAG bin2-3 bil1 bil2 plants were only slightly smaller than bin2-3 bil1 bil2 (Figure 2L) and had reduced TINY-FLAG protein levels compared with the wild type (Figure 2M). Taken together, our molecular and genetic studies demonstrate that BIN2 phosphorylates and stabilizes TINY.
TINY Positively Regulates Drought Response
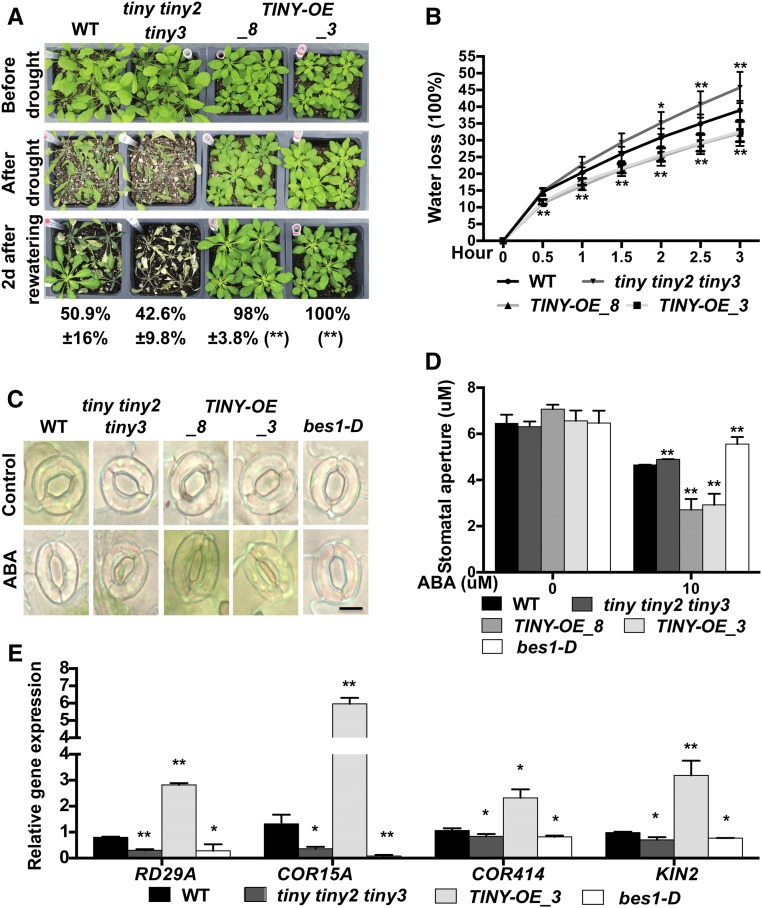
Given that TINY is induced by stresses such as dehydration, cold, and salt (Sun et al., 2008), we hypothesized that TINY promotes stress responses. We next examined the effect of TINY on drought response. TINY-OE plants had increased survival in water withholding assays compared with the wild type (Figure 3A). To further investigate how TINY promotes plant survival during water deficit stress, we conducted water loss assays. We found that detached leaves of TINY-OE lost water more slowly than the wild type but tiny tiny2 tiny3 lost water more quickly (Figure 3B). It was previously reported that overexpression of TINY led to hypersensitivity to ABA in seed germination and root elongation (Coego et al., 2014), suggesting TINY is a positive regulator of ABA responses.
Figure 3.
TINY Positively Regulates Drought Responses.
(A) Plant phenotypes for indicated genotypes after drought recovery assays. Phenotype of plants before drought (top), after drought (middle), and 2 d after rewatering (bottom). The survival rate after recovery was determined (bottom). Data represent mean and sd from four individual tray results (n = 4). Each genotype per tray contained 20 to 25 plants. **P < 0.01; two-way analysis of variance. WT, wild type.
(B) Detached leaf water loss assays. Leaves with similar developmental stages were detached and weighed at the indicated time. Water loss represents proportion of total weight lost compared with initial weight. Data represent mean and sd from four to five biological replicates (n = 4 to 5). Each replicate contained 10 to 15 leaves. *P < 0.05, **P < 0.01; two-way analysis of variance. WT, wild type.
(C) Representative stomata images of indicated genotypes. Epidermal peels of indicated genotypes were treated under light for 3h to make sure all stomatal were opened, then the peels were treated with or without 10 μM ABA for 3 h . Bars = 10 μm. WT, wild type.
(D) Stomatal apertures of indicated genotypes. Data represent mean and sd from three individual biological replicates (n = 3). Each replicate quantified 50 to 60 stomata from three individual leaves. *P < 0.05, **P < 0.01; two-way analysis of variance. WT, wild type.
(E) Drought-responsive gene expression levels in TINY mutants. mRNA was extracted from 4-week-old plants for gene expression analysis. Drought-responsive genes inculded RESPONSIVE TO DESICCATION 29A (RD29A), COLD REGULATED 15A (COR15A), COLD REGULATED 414 (COR414) and KINASE 2 (KIN2). Data represent mean and sd from three biological replicates (n = 3). *P < 0.05, **P < 0.01; Student’s t test.
Since ABA-regulated stomatal movement is highly related to drought tolerance (Qi et al., 2018), the effect of TINY on ABA-induced stomatal closure was measured. TINY-OE plants were more sensitive to ABA-induced stomatal closure than the wild type. By contrast, tiny tiny2 tiny3 mutants were slightly less sensitive to ABA-induced stomatal closure, similar to bes1-D, a BR gain-of-function mutant that exhibits ABA insensitivity and drought-susceptible phenotypes (Figures 3C and 3D; Supplemental Figure 4; Ryu et al., 2014; Nolan et al., 2017c; Ye et al., 2017). We also examined the expression of several drought-responsive marker genes (Verslues et al., 2006; Harb et al., 2010). These drought-responsive genes were increased in TINY-OE but decreased in tiny tiny2 tiny3 and bes1-D (Figure 3E). Taken together, these results demonstrate that TINY positively regulates ABA-mediated stomatal closure and drought-responsive genes to promote plant survival during drought.
TINY Negatively Regulates BR-Responsive Genes
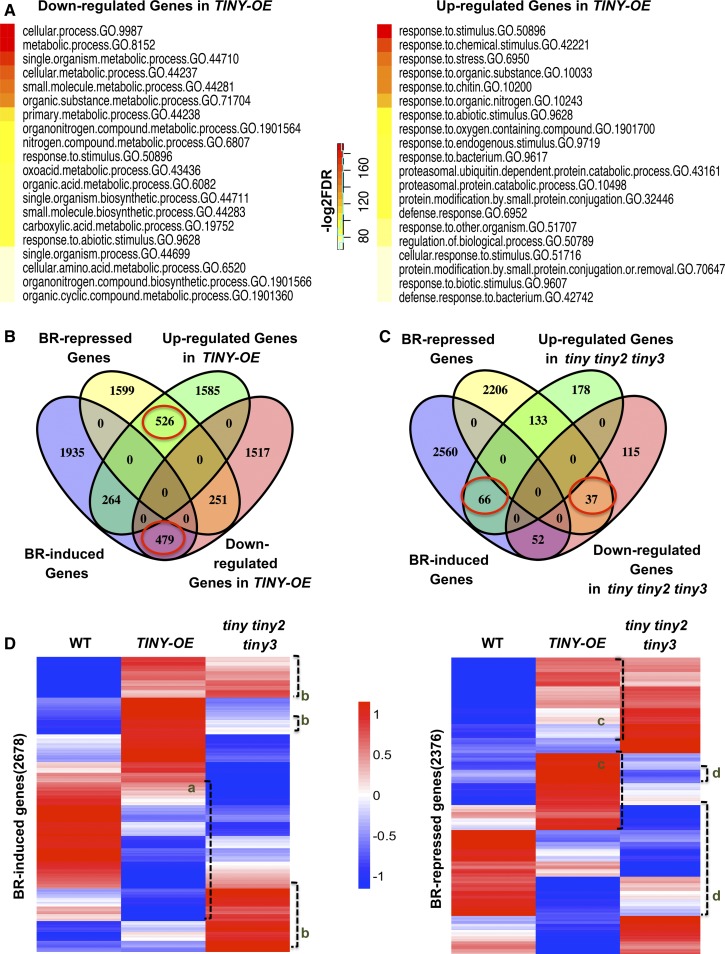
To study how TINY negatively regulates BR responses, we performed whole transcriptome RNA sequencing (RNA-seq) using the 4-week-old wild-type, TINY-OE, and tiny tiny2 tiny3 plants. Through this analysis, we identified 4622 genes that were differentially expressed (DEGs) in TINY-OE compared with the wild type, including 2375 upregulated genes and 2247 downregulated genes (Supplemental Data Set 1). TINY was previously reported to bind dehydration-responsive element (DRE)/C-repeat with a core sequence of A/GCCGAC as well as ethylene-responsive element with a core sequence of AGCCGCC (Sun et al., 2008). Promoter enrichment analysis identified promoter elements enriched in TINY-OE DEGs including DRE and several other elements (Supplemental Table 1).
Gene ontology (GO) enrichment analysis of DEGs in TINY-OE indicated that TINY-OE downregulated genes are implicated in growth-related processes such as cellular process and metabolic process, consistent with the inhibition of growth by TINY-OE (Figure 4A, left; Supplemental Data Set 2). On the other hand, TINY-OE–upregulated genes are enriched for GO terms associated with stress response including response to stimulus and response to stress (Figure 4A, right; Supplemental Data Set 2). We further compared TINY-regulated genes with published drought-responsive genes (Maruyama et al., 2009) and found that 324 (12.9%) of 2503 drought-induced and 430 (17%) of 2503 drought-repressed genes are regulated by TINY in the same direction (Supplemental Figure 5A), supporting a positive role for TINY in drought response.
Figure 4.
TINY Regulation on BR-Responsive Genes.
(A) List of the top 20 significant enriched GO terms about DEGs in TINY-OE as ranked by false discovery rate.
(B) and (C) Venn diagrams showing overlap among BR-regulated genes and DEGs in TINY-OE (B) and tiny tiny2 tiny3 (C) from QuantSeq. Red circle indicates genes differentially regulated by TINY and BR.
(D) Clustering analysis of genes defferential expressed in TINY-OE and tiny tiny2 tiny3. The color legend indicates normalized gene expression value among genotypes. Cluster a indicates BR-induced genes downregulated in TINY-OE, cluster b indicates BR-induced genes upregulated in tiny tiny2 tiny3 mutant. Cluster c indicates BR-repressed genes upregulated in TINY-OE, cluster d indicates BR-repressed genes downregulated in tiny tiny2 tiny3 mutant. WT, wild type.
Since TINY functions to inhibit BR-induced plant growth, we hypothesized that TINY-regulated genes are regulated by BRs in an opposite manner. To test this idea, we compared DEGs in TINY-OE to BR-induced and BR-repressed genes previously identified by RNA-seq (Supplemental Data Set 1; Yu et al., 2011; Wang et al., 2014; Ye et al., 2017). The results of these comparisons showed that BR-regulated genes were largely regulated by TINY in the opposite way (Figure 4B). Specifically, 479 (17.8%) of 2678 BR-induced genes were downregulated in TINY-OE and 526 (22.1%) of 2376 BR-repressed genes were upregulated in TINY-OE. By contrast, a smaller proportion of BR-induced genes were induced in TINY-OE (264 [9.8%] of 2678 genes) or repressed by BRs in TINY-OE (251 [10.5%] of 2376 genes). Additionally, we identified 581 DEGs in tiny tiny2 tiny3 mutants including 377 upregulated genes and 204 downregulated genes (Supplemental Data Set 1). This smaller number of DEGs is consistent with the relatively subtle phenotype of tiny tiny2 tiny3 and supports the idea that TINY functions redundantly with other AP2/ERFs. A large portion of tiny tiny2 tiny3 DEGs was affected by BRs, although the pattern of regulation between BR and tiny tiny2 tiny3 DEGs appears to be complex (Figure 4C).
To further investigate how TINY-OE and tiny tiny2 tiny3 affect BR-related gene expression, we performed clustering analysis using BR-regulated genes (Figure 4D). It confirmed that many BR-induced genes showed lower expression patterns in TINY-OE (cluster a) and higher expression in tiny tiny2 tiny3 (cluster b), while BR-repressed genes had higher expression in TINY-OE (cluster c) and lower expression in tiny tiny2 tiny3 (cluster d). Overall, our transcriptome analyses support a role of TINY in modulating BR-responsive gene expression largely in an antagonistic manner.
Since many BR-responsive genes are directly regulated by BES1/BZR1 (Sun et al., 2010; Yu et al., 2011), we compared TINY-OE DEGs with BES1/BZR1 target genes (Supplemental Data Set 1): 2110 (45.6%) of 4622 TINY-OE DEGs (P-value, 1.73 × 10−6, Fisher’s exact test) are BES1/BZR1 targets (Supplemental Figure 5B). TINY DNA affinity purification sequencing targets (O’Malley et al., 2016) also showed a high degree of overlap with BES1/BZR1 target genes, with 793 (49.5%) of 1602 TINY DNA affinity purification targets bound by BES1/BZR1 (P-value, 4.9 × 10−9, Fisher’s exact test; Supplemental Figure 5C). Together, these results suggest that TINY and BES1 likely share a common set of target genes.
TINY and BES1 Physically Interact
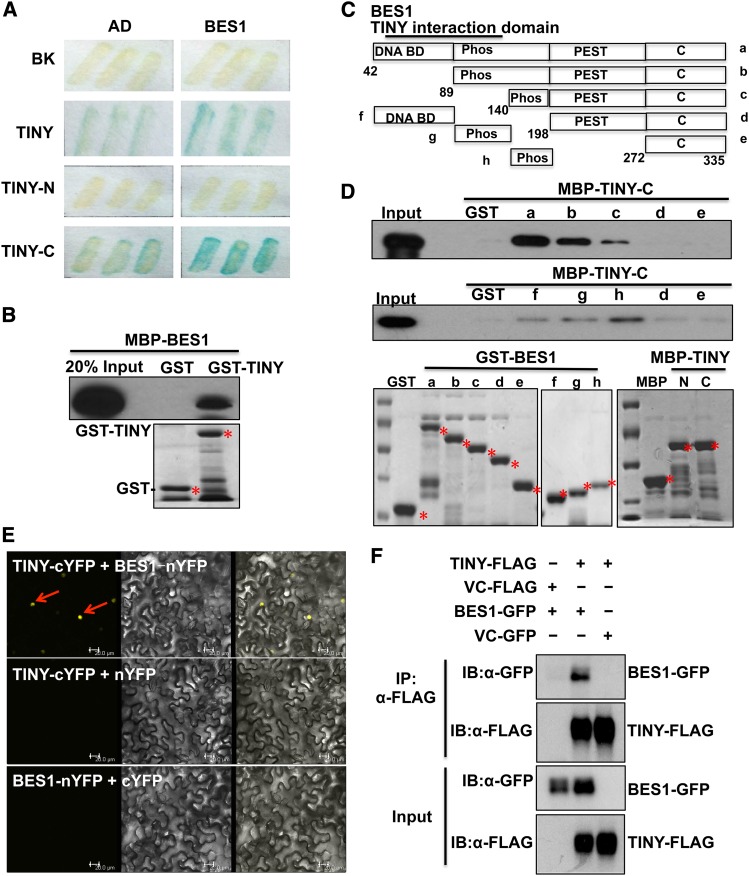
We then tested whether TINY and BES1 interact to modulate BR-responsive gene expression. TINY and BES1 interacted directly in Y2H assays (Figure 5A). TINY has an N-terminal DNA binding domain and a C-terminal activation domain (Nakano et al., 2006). Y2H indicated that TINY, especially its C-terminal activation domain, interacted with BES1 in yeast (Figure 5A). GST pull-down assay also confirmed TINY and BES1 interaction (Figure 5B). We next mapped the domain(s) responsible for BES1-TINY interactions. Several truncated GST-BES1 fragments were used to map which domain of BES1 was required for the interaction with TINY C-terminal (Figure 5C). A truncated BES1 fragment consisting of amino acids 198 to 335 did not interact with TINY, while the domain from 42 to 198 of BES1 was important for the interaction (Figure 5D). Moreover, BiFC confirmed that TINY and BES1 interact in the nucleus (Figure 5E; Supplemental Figure 6A). Finally, TINY-FLAG pulled down BES1-GFP coexpressed in Arabidopsis protoplasts in Co-IP assays, but BES1-GFP was not enriched in control samples (Figure 5F; Supplemental Figure 6B). These data strongly suggest that BES1 and TINY physically interact.
Figure 5.
TINY Interacts with BES1.
(A) TINY interacted with BES1 in yeast as detected by β-galactosidase (LacZ) activity. TINY N-terminal DNA binding domain (TINY-N) and its C-terminal activation domain (TINY-C) indicate TINY N-terminal DNA binding domain and C-terminal activation domain (AD), respectively.
(B) TINY interacted with BES1 in GST pull-down assay. Approximately equal amounts of GST, GST-TINY, and MBP-BES1 were used in the assays, as shown by a Coomassie-stained gel (bottom). Asterisks indicated the desired protein. BES1 was detected by immunoblotting using anti-MBP antibody.
(C) Schematic diagram of BES1 used for GST pull-down assay. DNA DB indicated BES1 DNA binding domain; Phos represented the phosphorylation domain of BES1; PEST represented protein sequence enriched in proline, glutamic acid, Ser, and Thr. C indicated BES1 C-terminus. (D) TINY interacted with BES1 in GST pull-down assay. Approximately equal amounts of proteins were used in the assays, as shown by a Coomassie-stained gel (bottom). TINY was detected by immunoblotting with anti-MBP antibody. Asterisks showed the desired protein.
(E) TINY interacted with BES1 by BiFC. Cotransfection of TINY-cYFP and BES1-nYFP led to the reconstitution of YFP activity in N. benthamina, whereas coexpression of TINY-cYFP and nYFP or BES1-nYFP and cYFP did not produce any positive YFP signal.
(F) Co-IP assay showed TINY and BES1 interaction. TINY-FLAG and BES1-GFP as well as control vectors were cotransformed into Arabidopsis protoplasts overnight. After 20 μM MG132 treatment, protoplasts were collected and protein was immunoprecipitated with anti-FLAG and detected with anti-FLAG and anti-GFP antibodies.
TINY and BES1 Antagonistically Control BR-Repressed Drought-Responsive Genes
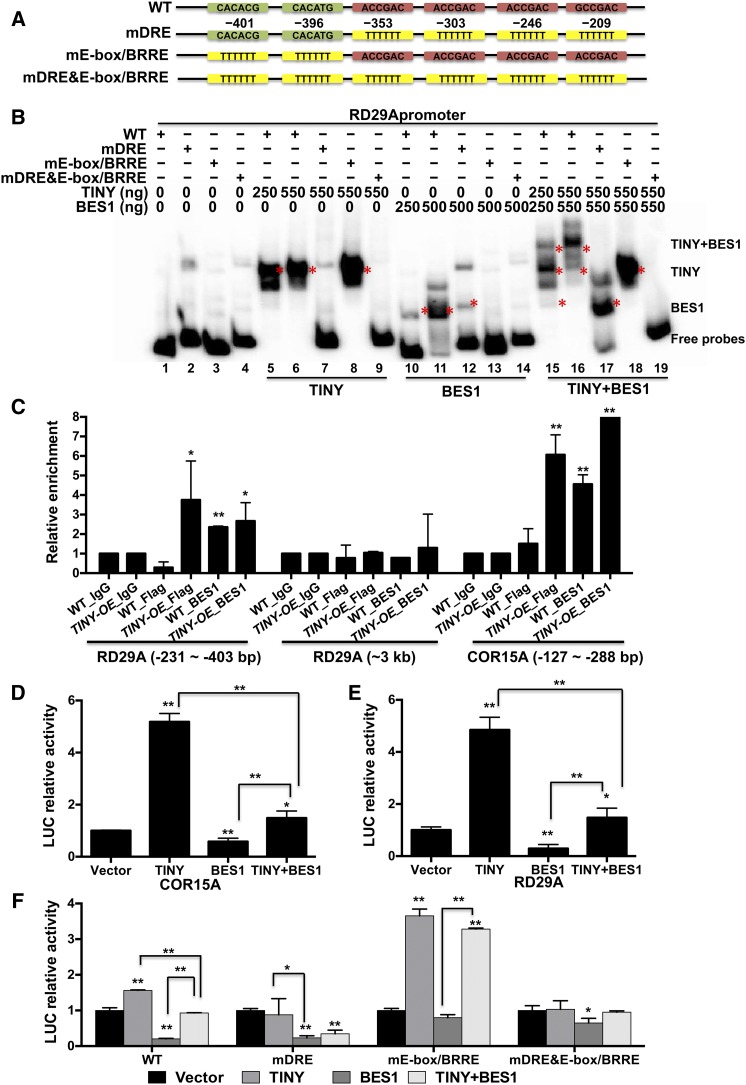
To further explore the mechanisms by which TINY and BES1 regulate BR-responsive genes, we selected the drought-responsive marker gene RD29A, which is induced by TINY but repressed by BES1 (Figure 3E). BES1 was previously reported to bind conserved DNA binding elements: E-box with sequence CANNTG (with N corresponding to any nucleotide) and BRRE with CGTG(T/C)G and C(A/G)CACG sequence, whereas TINY binds to sites such as DRE elements (Sun et al., 2008, 2010; Yu et al., 2011). Consistent with the idea that BES1 and TINY regulate RD29A, sequence analysis showed that the RD29A promoter contained E-box, BRRE, and DRE elements (Figure 6A). Next, we conducted electrophoretic mobility shift assay (EMSA) experiments using recombinant TINY and BES1 proteins along with a 272-bp RD29A promoter fragment containing E-box, BRRE, and DRE elements as DNA probes (Figure 6A). These experiments showed that TINY and BES1 bind to the wild-type RD29A probes, while TINY and BES1 binding was abolished upon mutation of DRE (mDRE) or E-box and BRRE (mE-box/BRRE) promoter elements, respectively (Figure 6B, lanes 5 to 14).
Figure 6.
TINY and BES1 Had Reciprocal Inhibition on Drought-Responsive Genes.
(A) Schematic diagram of the promoter region of RD29A. Wild type (WT) with DRE (in red) and E-box/BRRE (in green) were indicated. mDRE&mE-box/BRRE, the mutation of DRE, E-box, and BRRE elements.
(B) TINY and BES1 bind to RD29A promoter as revealed by EMSA. The DNA sequences that contained the wild-type (WT) or mutated forms from (A) were used as probes for DNA binding assay. WT or mutated probes were labeled with [γ-32P]ATP, and the indicated amount (ng) of proteins were used for reaction. Asterisks show the corresponding bands indicated on the right.
(C) TINY and BES1 enriched on RD29A and COR15A promoter in ChIP assay. The wild-type (WT) and TINY-OE plants were used to prepare chromatin and ChIP with antibodies against FLAG, BES1, or IgG as control. The ChIP products were used to detect promoters containing DRE, E-box, and BRRE and about upstream 3 kb of the transcriptional start site of RD29A as control. Data represent mean and sd from three biological replicates (n = 3). Each biological replicate was pooled tissue from three to four individual plants. *P < 0.05, **P < 0.01; Student’s t test.
(D) to (F) Transient LUC reporter assays with indicated promoter:LUC reporters were performed in N. benthamiana leaves with TINY and/or BES1 effector. LUC relative activities were normalized with total protein and control. Data represent mean and sd from three to five biological replicates (n = 3 to 5). Each biological replicate was comprised of five 7-mm leaf discs from one leaf. *P < 0.05, **P < 0.01; Student’s t test. WT, wild type.
Interestingly, when TINY and BES1 were added together, a higher molecular weight band was observed (Figure 6B, lanes 15 and 16), suggesting that BES1 and TINY bind to the RD29A promoter simultaneously. Conversely, TINY and BES1 could not form a complex when probes containing mDRE or mE-box/BRRE or when both E-box/BRRE and DRE element mutations were combined (Figure 6B, lanes 17 to 19). To test whether the higher molecular weight band corresponded to the TINY and BES1 complex, we fixed either TINY or BES1 protein concentrations and gradually increased the concentration of the other protein. The formation of the higher molecular weight band corresponding to TINY and BES1 was stronger with increasing amount of the other protein added (Supplemental Figure 7A, lanes 2 to 10). Moreover, the addition of either TINY or BES1 antibody caused the TINY-BES1 complex to super shift (Supplemental Figure 6A, lanes 11 and 13), suggesting that the higher molecular weight band corresponds to the TINY-BES1 complex.
We also performed chromatin immunoprecipitation (ChIP) assays in the wild-type and TINY-OE plants with FLAG and BES1 antibodies. TINY-FLAG was associated with the RD29A promoter region (−231 to ∼ −403 bp) containing E-box, BRRE, and DRE elements in TINY-OE plants, but not in the wild-type plants (Figure 6C, columns 3 and 4). The RD29A promoter is also enriched in BES1 ChIP from both the wild-type and TINY-OE plants (Figure 6C, columns 5 and 6). By contrast, such enrichment was not detected in the more upstream 3-kb (3-kb) promoter region, which lacks TINY and BES1 binding sites (Figure 6C, columns 7 to 12). Similar enrichment was also observed on the drought-responsive COLD-REGULATED15A (COR15A) promoter, which contains TINY and BES1 binding sites (Figure 6C, columns 13 to 18; Supplemental Figure 7B). Taken together, our results indicate that TINY interacts with BES1 and binds to drought-responsive genes in plants.
To test how TINY and BES1 affect RD29A and COR15A gene expression, we generated promoter:LUC reporters for these genes by fusing 1-kb promoter sequence with a luciferase (LUC) reporter. Transient expression of TINY or BES1 with the reporter in Nicotiana benthamiana showed that TINY alone activated RD29A and COR15A promoters, while BES1 alone had an inhibitory effect (Figures 6D and 6E; Supplemental Figure 7C). When TINY and BES1 were coexpressed with reporters, TINY and BES1 mutually inhibited each other’s activity on the promoters of these target genes. We further examined the regulation of TINY and BES1 by generating mutated RD29A promoter variants: mDREmE-box/BRRE (Figure 6F; Supplemental Figure 7D). Mutation of TINY binding sites (mDRE) on RD29A compromised TINY-mediated activation. Similarly, mutation of BES1 binding sites (mE-box/BRRE) compromised BES1-mediated repression. However, mutation of both TINY and BES1 binding sites compromised TINY and most of BES1 effect on these promoters. Taken together, these results indicate that TINY and BES1 interact and antagonize each other’s transcriptional activities on drought-responsive genes by binding to their corresponding target sites.
TINY and BES1 Antagonistically Control BR-Induced Growth-Related Genes
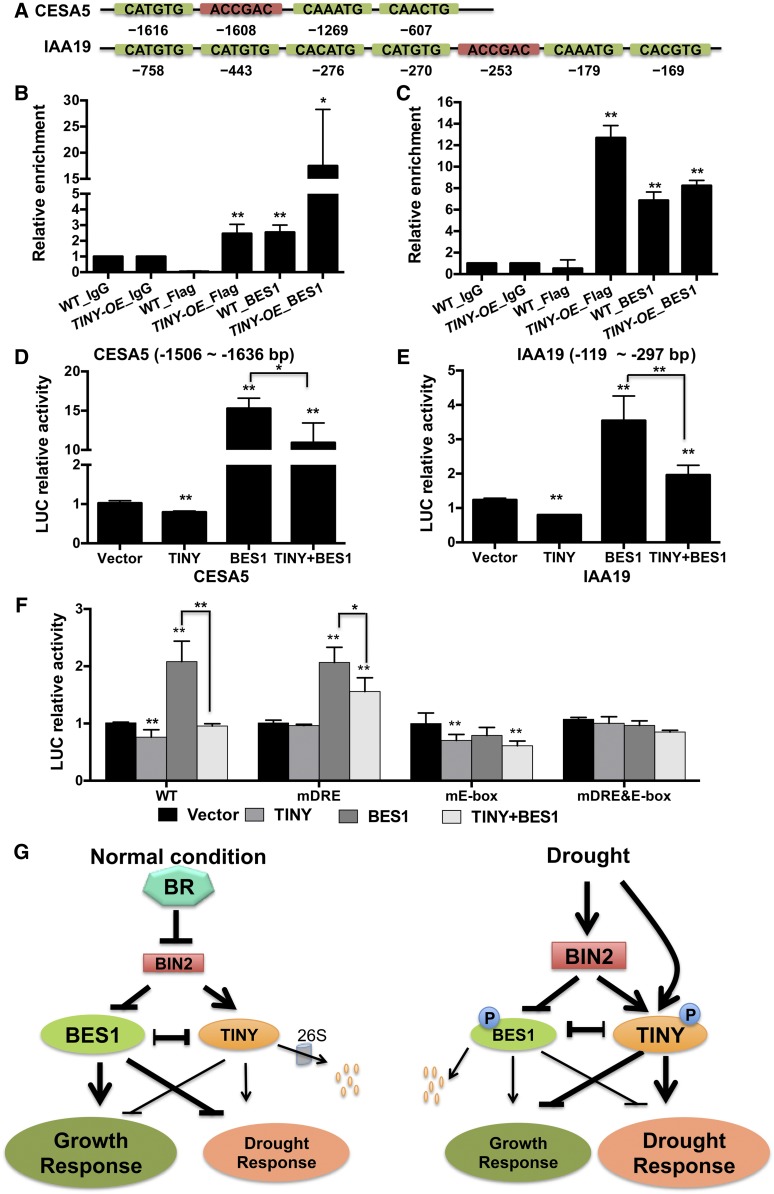
To test how TINY affects BR-induced growth-related genes, we selected two BR-induced genes that are downregulated by TINY: CESA5 and IAA19 (Figure 1E). The promoters of these genes contain several DRE and E-box elements (Figure7A). ChIP assays indicated that CESA5 and IAA19 promoters were enriched after BES1 or TINY were pulled down (Figures 7B and 7C). Additionally, promoter:LUC reporter analysis indicated that TINY alone repressed both promoters, while BES1 alone activated both promoters (Figures 7D and 7E; Supplemental Figure 8A). Similarly, when TINY and BES1 were coexpressed with reporters, TINY and BES1 showed an antagonistic effect on CESA5 and IAA19 promoter activity, respectively (Figures 7D and 7E; Supplemental Figure 8A).
Figure 7.
TINY and BES1 Antagonistically Regulate BR-Induced Growth-Related Genes.
(A) Schematic diagram of the promoter region of CESA5 and IAA19.
(B) and (C) TINY and BES1 enriched on CESA5 (B) and IAA19 (C) promoter in ChIP assay. Data represent mean and sd from three biological replicates (n = 3), with each replicate containing pooled tissue from three to four individual plants. *P < 0.05, **P < 0.01; Student’s t test. WT, wild type.
(D) to (F) Transient LUC reporter assays with indicated promoter:LUC reporters were performed in N. benthamiana leaves with TINY and/or BES1 effector. LUC relative activities were normalized with total protein and control. Data represent mean and sd from three to five biological replicates (n = 3 to 5). Each biological replicate was comprised of five 7-mm leaf discs from one leaf. *P < 0.05, **P < 0.01; Student’s t test.
(G) A working model for TINY in BR-regulated plant growth and drought response. TINY inhibits plant growth and promotes drought response. Under normal condition (left), BR promotes TINY degradation although BIN2 phosphorylation and 26S proteasome pathway to alleviate TINY’s inhibitory effect on BR-induced plant growth. BR-induced accumulation of BES1 also inhibits TINY activation of drought-responsive genes to further facilitate plant growth. Under drought condition (right), TINY is induced at transcriptional level to promote drought response and inhibit plant growth by inhibiting BES1 functions. Stresses and ABA-activated BIN2, in turn, stabilize and enforce TINY function.
TINY and BES1 regulation of IAA19 was further confirmed using a mutated IAA19 promoter with DRE motifs (mDRE) and E-box (mEbox). Similar to RD29A, mutation of TINY binding sites (mDRE) compromised TINY-mediated repression and mutation of BES1 binding sites (mE-box) compromised BES1-mediated activation (Figure 7F; Supplemental Figure 8B). These results indicate that TINY and BES1 antagonize each other’s transcriptional activities on BR-induced growth-related genes by binding to different promoter elements.
DISCUSSION
Plants need to adjust growth and stress response under different environmental conditions. In this study, we demonstrated that AP2/ERF family transcription factor TINY promotes drought tolerance and inhibits plant growth through interactions with BES1 in the BR pathway (Figure 7G). Under drought conditions, stress-induced TINY promotes drought responses by activating drought-responsive gene expression and alleviating BES1 repression of these genes. BIN2, which is activated by stresses and ABA (Youn and Kim, 2015; Wang et al., 2018), stabilizes TINY to reinforce the function of TINY in stress responses. At the same time, TINY inhibits BR-regulated plant growth by repressing the expression of BR-induced genes related to growth. Under normal growth conditions, BR signaling inhibits TINY function through several mechanisms. First, BR signaling reduces TINY protein level by inhibiting the activity of BIN2 required for TINY accumulation. Second, TINY activation on drought-responsive genes and TINY repression on BR-induced/growth-related genes are inhibited by BES1 that accumulates in response to BRs. Our results thus establish that TINY is regulated by BR signaling, modulates BR-regulated gene expression, and promotes drought response and inhibits plant growth.
Our results revealed a mechanism for TINY in conferring drought tolerance. Consistent with previous reports that TINY is induced during dehydration and overexpression of TINY leads to upregulated drought-responsive gene expression (Sun et al., 2008), our results demonstrate that TINY contributes to plant drought response by promoting ABA-induced stomatal closure, directly binding to and activating drought-responsive gene expression (Figures 3 and 6).
In agreement with the general notion that stress tolerance in plants is often associated with reduced growth rates and productivity (Bechtold and Field, 2018), TINY functions to inhibit plant growth. Although TINY was found to inhibit growth more than 20 years ago (Wilson et al., 1996), the mechanisms by which TINY functions in plant growth have not been established. Our genetic and physiological results demonstrated that TINY inhibits plant growth by interrupting BR-induced plant growth. The upregulation of TINY transcripts by BRs might represent a feedback mechanism for BR signaling (Figure 1; Supplemental Figures 1B and 2). TINY overexpression led to a dwarf phenotype resembling BR loss-of-function mutants with increased sensitivity to the BR biosynthesis inhibitor BRZ and Pcz in hypocotyl elongation and petiole length assays, respectively. This was further supported by the observation that tiny tiny2 tiny3 triple mutants displayed increased growth and reduced sensitivity to BRZ and Pcz treatments. Additionally, TINY negatively regulates BR-responsive genes involved in cell elongation (Figures 1E and 1F). The negative relationship between TINY and the BR pathway is also supported by our global gene expression studies (Figure 4). A large proportion of BR-induced genes were downregulated in TINY-OE, and many BR-repressed genes were upregulated in TINY-OE. The negative effects of TINY on BR-regulated growth support the notion that inhibition of plant growth is dependent on the repression of genes that are required for cell elongation and division (Claeys and Inzé, 2013; Kudo et al., 2018).
We established the mechanism by which BES1 and TINY inhibit each other’s activity on BR-induced plant growth genes and BR-repressed drought-responsive genes. Global gene expression studies indicated that TINY and BES1 regulate a significant set of genes in opposite ways (Figure 4). Approximately 50% of TINY-OE DEGs were direct BES1/BZR1 targets (Supplemental Figures 5B and 5C). Further molecular studies indicated that BES1 and TINY could bind to different DNA binding sites and inhibit each other’s activities (Figure 5 to 7). Consistent with BES1-TINY interactions (Figure 5), these two factors functionally interact with each other on drought-induced or growth-related gene promoters (Figures 6 and 7). While TINY activates and BES1 represses drought-induced genes, they act in opposite ways for growth related genes (i.e., BES1 activates and TINY represses). In both cases, TINY and BES1 have opposite effects in the regulation of drought-induced or growth-related genes (Figures 6 and 7). TINY and BES1 binding to DNA and their interaction appear to be important as mutations of one of the binding sites usually reduce but do not abolish their subtractive functional interactions (Figures 6 and 7). Further studies are needed to define the detailed mechanisms by which BES1 and TINY antagonize each other’s activities.
This finding expands our understanding the role of BRs in transcriptional regulation of drought response. We have previously found that drought and starvation promote BES1 degradation to reduce BR-regulated plant growth (Chen et al., 2017; Nolan et al., 2017c; Yang et al., 2017). We also revealed that BR negatively regulates drought-responsive genes by antagonizing transcriptional activity of drought-induced transcription factor RD26 (Ye et al., 2017). Unlike BES1 and RD26 that bind to the same promoter element to antagonize each other’s function (Ye et al., 2017), TINY and BES1 bind to different binding sites to inhibit each other (Figures 6 and 7). This reciprocal transcriptional inhibition provides additional insight into the mechanisms that plants use to balance BR-induced growth and drought stress responses.
It is worth noting that BR and drought do not antagonize each other all the times. A recent study showed that overexpression of the vascular-specific BR BRI1-LIKE RECEPTOR3 conferred drought tolerance without penalizing plant growth (Fàbregas et al., 2018). It is possible that BRs and drought have different interactions depending on tissue specificities.
In this study, we showed that the function of TINY is negatively regulated by BR signaling through BIN2 (Figure 2). BIN2 is a negative regulator in the BR pathway and phosphorylates BES1/BZR1 as well as many substrates involved in plant development and stress responses (Youn and Kim, 2015). BIN2 phosphorylates and stabilizes TINY as TINY protein was more abundant in gain-of-function bin2-1 mutant but less abundant in BIN2 loss-of-function bin2-3 bil1 bil2 mutant. The results reinforce the idea that BIN2 phosphorylation have different effects on its substrates (Ye et al., 2012; Bernardo-García et al., 2014; Zhang et al., 2014; Chen et al., 2017). These results indicate that BR signaling inhibits TINY accumulation, thereby preventing TINY from unwanted activation of drought responses under normal growth conditions. Recent studies showed that BIN2 activity is activated by ABA and stresses (Youn and Kim, 2015; Wang et al., 2018). BIN2 phosphorylation and stabilization of TINY provide a mechanism for activation of drought responses.
In summary, this study revealed the function of TINY in BR-regulated growth and drought response. TINY functions to promote drought response and inhibit plant growth under drought conditions, whereas BR signaling inhibits TINY to prevent unnecessary stress response under normal conditions. This coordination of BR-regulated growth and stress responses is achieved through BIN2 phosphorylation of TINY and antagonism between TINY and BES1 on growth- and drought-regulated genes. Future identification of TINY interacting partners and target genes could further our understanding of the mechanisms by which TINY controls BR-regulated growth and drought responses.
METHODS
Plant Materials, Growth Conditions, and Hormone Responses
Arabidopsis (Arabidopsis thaliana) accession Columbia was used as the wild type along with the following previously described mutants: bes1-D (Yin et al., 2002), bri1-301 (Xu et al., 2008), bin2-3 bil1 bil2 (Yan et al., 2009), and bin2-1 (Li et al., 2001). T-DNA insertion mutants tiny (AT5G25810, SALK_206788), tiny2 (AT5G11590, SALK_202794), and tiny3 (AT4G32800, SALK_149004) were obtained from Arabidopsis Biological Resource Center and confirmed before use. Seeds were sterilized using 70% (v/v) ethanol containing 0.1% (v/v) Triton X-100 and grown on one half Linsmaier and Skoog (LS) plates (LSP03-1LT, Caisson Laboratories). Seedlings were transferred into soil (SS#1-F1P, SunGro) in a growth chamber setting with long day (16 h light/8 h dark) or short day (8 h light/16 h dark). The light intensity and temperature in the growth chamber was ∼120 to 150 µmol m−2 s−1 (bulbs from Philips, F32T8/TL741) and 22°C, respectively.
BRZ and BL response was performed with sterilized seeds on various concentrations of BRZ (Asami et al., 2000) and BL (Li et al., 2010) plates. Approximately 12 to 15 seeds of each genotype were spread on three individual BRZ and BL plates per concentration. BRZ and BL plates with seeds were placed at 4°C for 3 d for stratification. BRZ plates then were exposed to light for 8 h and put in the dark for 7 d at room temperature. BL plates were put in light for 7 d at room temperature. Plates were scanned using a flatbed scanner, and the hypocotyl was measured using ImageJ. Statistical analysis was performed, and variances of populations (sd) were calculated from three individual plates’ results.
For Pcz (Hartwig et al., 2012) treatment in soil, Pcz (Syngenta) was directly dissolved in water. All tested genotypes were grown in the same tray and randomly assigned with or without 250 μM Pcz from the beginning of transferring into soil to the end of leaf measurement. Four trays were included for each trial experiment. The sixth leaves of plants were measured to assess petiole length.
Plasmid Constructs and Generation of Transgenic Plants
TINY coding region cloned from the wild type, including BES1 promoter and 2034 bp of its native promoter, was fused with FLAG tag into pZP211 vector (Yin et al., 2002). TINY coding region was also cloned into pMDC7 to generate estradiol (Jung et al., 2015)-inducible overexpression lines. All constructs were generated using standard restriction enzyme digestion and confirmed by DNA sequencing.
For transgenic plants, plasmid constructs were transferred into Agrobacterium tumefaciens strain GV3101 and transformed into plants by the floral dip method. Transgenic lines were screened on 1/2 LS plates plus 75 mg/mL gentamycin (Omega) or 25 mg/mL hygromycin B (Thermo Fisher Scientific). The transgene expression was confirmed by immunoblotting using anti-FLAG antibody (F7425, Sigma-Aldrich) at 1:1000 dilution. BES1:TINY-FLAG line 3 (represented as TINY-OE) was used for most experiments unless otherwise specified. T3 homozygous plants were used for all experiments. Other antibodies were as follows: anti-FLAG M2 (M8823, Sigma-Aldrich), anti-MYC (AV38156, Sigma-Aldrich) using 1:2000 dilution, anti-BES1 (Yin et al., 2002), anti-GFP (Nolan et al., 2017c), anti-IgG (12-370, Millipore Sigma), anti-MBP (New England Biolabs [NEB]) using 1:1000 dilution, and anti-TINY (generated ourselves). HERK1 antibody (Guo et al., 2009) served as internal control. All primers used in this study are provided in Supplemental Table 2.
Water Deficit Assays
Water deficit stress experiments were performed as described previously (Chen et al., 2017), with minor modifications. Seven-day-old seedlings were transferred into weighted soil. All genotypes we tested were grown in the same tray and randomly assigned and then grown for three more weeks in the short day growth chamber withholding water. Four trays were included for each trail experiment. After rewatering the plants for 2 d, the numbers of survived plants were counted and the tests were repeated three times.
For detached leaf water loss assays, rosette leaves at the same developmental stages from 4-week-old plants were excised from roots and placed in open Petri dishes without lids for the indicated time. The fresh weights were monitored at each time point. Water loss represents proportion of total weight lost compared with initial fresh weight.
For ABA-mediated stomatal closure, 4-week-old short day–grown, fully expanded leaves were floated with abaxial side up in MES/KOH buffer (50 mM KCl and 10 mM MES-KOH, pH 6.15) at room temperature under 120 μmol·m−2·s−1 of light for 3 h. Once the stomata were fully open, 10 μM ABA and control solution were added to buffer for another 3 h under light conditions. The epidermal strips were immediately peeled from the abaxial surface of leaves, and stomatal aperture images were taken using a BX40 microscope (Olympus) and analyzed by ImageJ. To avoid any rhythmic effects on stomatal closure, experiments were started at the same time during the day. Five images per leaf and three leaves per genotype containing ∼50 to 60 stomata in total were used for data analysis. The experiments were repeated three times for statistical analysis.
Gene Expression and QuantSeq Analysis
For gene expression studies, total RNA was extracted from different genotypes of 4-week-old, long day–grown plants using the RNeasy Mini kit (Qiagen). cDNA generated using iScript Reverse Transcription Supermix (Bio-Rad) was used for gene expression studies. SYBR GREEN PCR Master Mix (Applied Biosystems) and Mx4000 multiplex Quantitative CR System (Stratagene) were used for quantitative real-time PCR (qPCR) analysis with two technical replicates and three biological replicates. Statistical analysis was performed, and variances of population (sd) were calculated from three biological replicates. UBQ5 was used as the internal control.
For RNA sequencing analysis, total RNA was extracted from 4-week-old, long day–grown plants using Zymo DirecZol kit (Zymo Research). RNA concentrations and quality were analyzed using an AATI Fragment Analyzer with Standard Sensitivity RNA Analysis kit (DNF-489-0500). Approximately 500 ng of RNA was used for library construction via the QuantSeq 3′ mRNA-Seq Library Prep FWD kit (Illumina) and sequenced on an Illumina HiSeq 2500 system (50-bp single end reads). FASTQ files for each sample were subject to quality control and trimming and mapped to The Arabidopsis Information Resource 10 (TAIR10) genome using the BlueBee Arabidopsis (TAIR10) Lexogen QuantSeq 2.2.2 FWD pipeline. Principal component analysis was used to examine the data, and the wild-type control sample 2 was identified as an outlier and excluded from further analysis. All raw and processed RNA-seq data described in this study have been deposited in the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/). For differential expression analysis, the R package DEseq2 (https://bioconductor.org/packages/release/bioc/html/DESeq2.html) was used to test the null hypothesis that expression of a given gene is not different between two genotypes. This null hypothesis was tested using a model with a negative binomial distribution. P-values of all statistical tests were converted to adjusted P-values (q-values; Benjamini et al., 2001). A false discovery rate of 10% (q-value) was used to account for multiple testing.
Venny (http://bioinfogp.cnb.csic.es/tools/venny/index.html) was used to perform the comparisons of DEGs. Clustering analysis used the “aheatmap” function of the NMF package in R (https://cran.r-project.org/web/packages/NMF/index.html). BINGO (Maere et al., 2005) was used for GO term enrichment analysis. Promoter analysis was conducted using DREME (Bailey, 2011) with 1-kb upstream sequences downloaded from https://www.arabidopsis.org/tools/bulk/sequences/index.jsp. All TAIR10 promoters were used as background when determining enrichment in promoter analysis.
Protein–Protein Interaction Experiments
For the Y2H assays, TINY and its fragments were cloned into bait pGBKT7 vectors (Clontech), while BES1 and BIN2 were cloned into prey vector pGADT7 (Clontech). The constructs were transformed into yeast strain Y187 and screened using medium lacking Trp and Leu. The LacZ reporter activity was measured using 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside according to the manufacturer’s protocol (Clontech).
GST pull-down assays were conducted using TINY, BES1, and BIN2 that were cloned into pET42a (Novagen) or pET-MBP-H (Nolan et al., 2017c) vectors to generate GST- or MBP-tagged protein, respectively. The recombinant proteins were purified using either glutathione beads (Sigma-Aldrich) or amylose resin (NEB). Approximately 2 μg of proteins was mixed into 1 mL of pull-down buffer (50 mM Tris-HCl, pH 7.5, 200 mM NaCl, 0.5% Triton X-100, 0.5 mM β-mercaptoethanol, and proteinase inhibitor cocktail) and incubated at room temperature for 2 h. The incubation reaction mixture was then pulled down by glutathione-Sepharose beads for another 1 h and followed with eight times wash using the same pull-down buffer. The pull-down protein was separated on SDS-PAGE gel and detected by anti-MBP antibody at 1:1000 dilution. The experiments repeated three times.
BiFC experiments were conducted as previously described (Ye et al., 2012). Briefly, TINY and BES1 cDNAs were cloned into the N or C terminus of enhanced YFP vectors. Agrobacteria were grown in Luria-Bertani medium containing 0.2 M acetosyringone overnight. Collected cells were washed and resuspended to OD600 of 0.5 with one half LS infiltration medium. Combinations of Agrobacterium were transient expressed in Nicotiana benthamiana leaves. YFP signals were detected with an SP5 X MP confocal microscope (Leica Microsystems) after 1.5 d of infiltration. The protein accumulation of BiFC combinations was detected using anti-GFP antibody (A-11122, Invitrogen) at 1:1000 dilution.
For the Co-IP experiments, Arabidopsis protoplasts transformed with tested vectors (35S:TINY-FLAG, 35S:BIN2-GFP, and 35S:BES1-GFP), as well as control vectors overnight, were treated with 20 μM MG132 1 h before harvest. The harvested cells were homogenized in Co-IP buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 10% (v/v) glycerol, 0.1% (v/v) Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride, 20 μM MG132, and cocktail protease inhibitors) for 1 h at 4°C. FLAG M2 antibody (5 µg) (M8823, Sigma-Aldrich) was pre-bound to protein G Dynabeads (10003D, Fisher) for 30 min in PBS buffer with 0.02% Tween 20, and the beads were washed once with the same PBS buffer for immunoprecipitation (IP). After protein extraction, 10 μL of anti-FLAG pre-bound Dynabeads was added to total proteins for another 1.5 h at 4°C. Dynabeads was precipitated using DynaMagnetic rack (12321D, Thermo Fisher Scientific) and washed twice with Co-IP buffer with Nonidet P-40 and twice with Co-IP buffer without Nonidet P-40. After adding 2× SDS protein-loading buffer and boiling for 5 min, the IP products were used for immunoblotting with anti-GFP (Nolan et al., 2017c) and FLAG (F7425, Sigma-Aldrich) antibody at 1:1000 dilution.
Kinase and CIP Assay
Approximately 1 μg of MBP, MBP-TINY, and GST-BIN2 were incubated into 20 μL of kinase buffer (20 mM Tris, pH 7.5, 100 mM NaCl, and 12 mM MgCl2) containing 10 μCi of [γ-32P]ATP. Reaction was incubated at 37°C for 1 h and then stopped by adding 20 μL of 2× SDS buffer. After boiling at 94°C for 5 min, proteins were resolved on SDS-PAGE gel and dried. Phosphorylation images were detected using a Typhoon FLA 9500 system.
For CIP treatment, TINY-FLAG was first immunoprecipitated from TINY-OE transgenic plants. Approximately 1 g of homogenized plants tissue was incubated with 5 mL of IP buffer (100 mM Tris-HCl, pH 7.4, 75 mM NaCl, 1 mM EDTA, 0.05% SDS, 10% glycerol, 0.1% Triton X-100, and proteinase inhibitor cocktail) for 30 to 40 min, followed by adding 20 μL of anti-FLAG magnetic beads (M8823, Sigma-Aldrich) and incubating for 1 h. The collected beads were washed with IP buffer three times and saved for CIP treatment. Approximately 1 μg of immunoprecipitated TINY-FLAG proteins was treated with 10 units of CIP (NEB) at 37°C for 0.5 h. The reaction was stopped by adding 20 μL of 2× SDS buffer, 5 min of boiling, and resolved on SDS-PAGE gel.
EMSA Experiments
EMSA experiments were performed as described previously (Yin et al., 2005). Approximately 200 ng of PCR-generated RD29A promoter fragments was labeled with [γ-32P]ATP using T4 polynucleotide kinase (NEB). Specifically, 20 units of polynucleotide kinase and 50 μCi of [γ-32P]ATP were mixed at 37°C for 1 h. The labeled probes were purified using a gel extraction kit (Qiagen) following the manufacturer’s instructions, and labeling specificity was determined by using a scintillation counter. For binding reaction, in 20 μL of reaction binding buffer (25 mM HEPES-KOH, pH 8.0, 1 mM DTT, 50 mM KCl, and 10% glycerol), ∼0.2 ng of probes and the indicated amount of purified proteins were mixed to incubate on ice for 30 to 40 min. The reaction mixtures were resolved on 5% native polyacrylamide gels with 1× TGE buffer (3.3 g/L Tris, 14.3 g/L Gly, and 0.39 g/L EDTA, pH 8.7).
Chromatin Immunoprecipitation
ChIP was performed as described previously (Nolan et al., 2017b), with modifications. Briefly, 3 g of 2-week-old seedlings were fixed in 1% formaldehyde for nuclei and chromatin isolation. The chromatin was sheared with 14 cycles of 10 s on and 1 min off in an ice water bath using a Diagenode Bioruptor Sonication System. Next, 5 μg of anti-FLAG (F7425, Sigma-Aldrich), anti-BES1, or anti-IgG was used to immunoprecipitate chromatin, which was collected with 30 μL of protein A Dynabeads (10003D, Fisher). The enrichment of specific transcription factors was examined by qPCR with primers from indicated regions. qPCR analysis with two technical repeats were used to calculate enrichment folds compared with anti-IgG control. The averages and sd were derived from three biological replicates.
Promoter Activity Analysis
The promoter transient expression analysis was performed as described previously (Ye et al., 2017). Promoters of tested genes were cloned into LUC reporter construct to generate reporter. The reporters were coexpressed with effectors such as TINY (35S:TINY-MYC) and/or BES1 (35S:BES1) into N. benthamiana leaves as well as empty control vectors. Agrobacteria were grown in Luria-Bertani medium containing 0.2 M acetosyringone overnight. Collected cells were washed and resuspended to OD600 of 0.5 with 1/2 LS infiltration medium. Equal amount of individual Agrobacteria culture or the combination of that was infiltrated into five N. benthamiana leaves at the same developmental stage. All the combination tests were included on the same leaves to avoid differential expression. Five punched bands (diameter was 7 mm) per leaf were collected after 1.5 d of incubation. The LUC reporter activities were measured following the manufacturer’s instructions (Promega) and using a Berthold Centro LB960 luminometer. The LUC activity was normalized to the total protein content measured by Bradford assay. The relative LUC activity was calculated compared with control expression. Immunoblotting was used to ensure proper expression of effectors such as BES1 or TINY.
Accession Numbers
QuantSeq data from this article can be found in the Gene Expression Omnibus (GSE128946). The accession numbers for the studied genes are as follows: TINY, AT5G25810; TINY2, AT5G11590; TINY3, AT4G32800; RD29A, AT5G52310; COR15A, AT2G42540; CESA5, AT5g09870; IAA19, AT3G15540.
Supplemental Data
Supplemental Figure 1. TINY inhibits plant growth.
Supplemental Figure 2. TINY negatively regulates BR signaling.
Supplemental Figure 3. BR promotes TINY protein degradation by BIN2.
Supplemental Figure 4. BR signaling negatively regulates drought response.
Supplemental Figure 5. TINY positively regulate drought revealed by global gene expression.
Supplemental Figure 6. TINY and BES1 interaction revealed by BiFC and Co-IP assay.
Supplemental Figure 7. TINY and BES1 bind and regulate to RD29A expression.
Supplemental Figure 8. TINY and BES1 protein transient expression level in N. benthamiana.
Supplemental Table 1. Putative TINY binding elements.
Supplemental Table 2. Primers, promoter and probe sequences used in this study.
Supplemental Data Set 1. Genes regulated by BR, TINY, and drought.
Supplemental Data Set 2. GO term enrichments by TINY.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Mark Estelle for providing the pMDC7 vector and Nam-Hai Chua for the inducer system. We thank Hongqing Guo for helpful discussions and editing the article and Yong Gao for assistance with genotyping. The work is supported by the National Science Foundation (MCB-1818160), the National Institutes of Health (1R01GM120316-01A1), and the Plant Sciences Institute at Iowa State University. Z.X. was partially supported by a China Scholarship Council fellowship.
AUTHOR CONTRIBUTIONS
Z.X. and Y.Y. conceived the project. Z.X. designed and performed most of the experiments, with the following exceptions. H.J. performed CHX treatment, EMSA experiments, purified the recombinant proteins, prepared protoplasts for Co-IP, immunoblotting for BiFC and performed drought survival assays. T.N., Z.X., M.Z., and Z.L. conducted QuantSeq and T.N. analyzed the QuantSeq data. T.N. assisted with confocal microscopy for BiFC assays. B.T. genotyped tiny mutants. Z.X., T.N., and Y.Y. wrote the article.
Footnotes
Articles can be viewed without a subscription.
References
- Asami T., Min Y.K., Nagata N., Yamagishi K., Takatsuto S., Fujioka S., Murofushi N., Yamaguchi I., Yoshida S. (2000). Characterization of brassinazole, a triazole-type brassinosteroid biosynthesis inhibitor. Plant Physiol. 123: 93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T.L. (2011). DREME: Motif discovery in transcription factor ChIP-seq data. Bioinformatics 27: 1653–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold U., Field B. (2018). Molecular mechanisms controlling plant growth during abiotic stress. J. Exp. Bot. 69: 2753–2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Drai D., Elmer G., Kafkafi N., Golani I. (2001). Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 125: 279–284. [DOI] [PubMed] [Google Scholar]
- Bernardo-García S., de Lucas M., Martínez C., Espinosa-Ruiz A., Davière J.M., Prat S. (2014). BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth. Genes Dev. 28: 1681–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z., Liu J., Wang H., Yang C., Chen Y., Li Y., Pan S., Dong R., Tang G., Barajas-Lopez Jde.D., Fujii H., Wang X. (2014). GSK3-like kinases positively modulate abscisic acid signaling through phosphorylating subgroup III SnRK2s in Arabidopsis. Proc. Natl. Acad. Sci. USA 111: 9651–9656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Nolan T.M., Ye H., Zhang M., Tong H., Xin P., Chu J., Chu C., Li Z., Yin Y. (2017). Arabidopsis WRKY46, WRKY54, and WRKY70 transcription factors are involved in brassinosteroid-regulated plant growth and drought responses. Plant Cell 29: 1425–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claeys H., Inzé D. (2013). The agony of choice: How plants balance growth and survival under water-limiting conditions. Plant Physiol. 162: 1768–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse S.D., Langford M., McMorris T.C. (1996). A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 111: 671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coego A., Brizuela E., Castillejo P., Ruíz S., Koncz C., del Pozo J.C., Piñeiro M., Jarillo J.A., Paz-Ares J., León J., Consortium T.; TRANSPLANTA Consortium (2014). The TRANSPLANTA collection of Arabidopsis lines: A resource for functional analysis of transcription factors based on their conditional overexpression. Plant J. 77: 944–953. [DOI] [PubMed] [Google Scholar]
- De Rybel B., et al. (2009). Chemical inhibition of a subset of Arabidopsis thaliana GSK3-like kinases activates brassinosteroid signaling. Chem. Biol. 16: 594–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fàbregas N., et al. (2018). Overexpression of the vascular brassinosteroid receptor BRL3 confers drought resistance without penalizing plant growth. Nat. Commun. 9: 4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahad S., et al. (2017). Crop production under drought and heat stress: Plant responses and management options. Front. Plant Sci. 8: 1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M., Fujita Y., Maruyama K., Seki M., Hiratsu K., Ohme-Takagi M., Tran L.S., Yamaguchi-Shinozaki K., Shinozaki K. (2004). A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 39: 863–876. [DOI] [PubMed] [Google Scholar]
- Gou X., Yin H., He K., Du J., Yi J., Xu S., Lin H., Clouse S.D., Li J. (2012). Genetic evidence for an indispensable role of somatic embryogenesis receptor kinases in brassinosteroid signaling. PLoS Genet. 8: e1002452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Li L., Ye H., Yu X., Algreen A., Yin Y. (2009). Three related receptor-like kinases are required for optimal cell elongation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 106: 7648–7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Li L., Aluru M., Aluru S., Yin Y. (2013). Mechanisms and networks for brassinosteroid regulated gene expression. Curr. Opin. Plant Biol. 16: 545–553. [DOI] [PubMed] [Google Scholar]
- Harb A., Krishnan A., Ambavaram M.M., Pereira A. (2010). Molecular and physiological analysis of drought stress in Arabidopsis reveals early responses leading to acclimation in plant growth. Plant Physiol. 154: 1254–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig T., Corvalan C., Best N.B., Budka J.S., Zhu J.Y., Choe S., Schulz B. (2012). Propiconazole is a specific and accessible brassinosteroid (BR) biosynthesis inhibitor for Arabidopsis and maize. PLoS One 7: e36625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J.X., Gendron J.M., Yang Y., Li J., Wang Z.Y. (2002). The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 99: 10185–10190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Yu D. (2014). BRASSINOSTEROID INSENSITIVE2 interacts with ABSCISIC ACID INSENSITIVE5 to mediate the antagonism of brassinosteroids to abscisic acid during seed germination in Arabidopsis. Plant Cell 26: 4394–4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C., Zhao P., Seo J.S., Mitsuda N., Deng S., Chua N.H. (2015). PLANT U-BOX PROTEIN10 regulates MYC2 stability in Arabidopsis. Plant Cell 27: 2016–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagale S., Divi U.K., Krochko J.E., Keller W.A., Krishna P. (2007). Brassinosteroid confers tolerance in Arabidopsis thaliana and Brassica napus to a range of abiotic stresses. Planta 225: 353–364. [DOI] [PubMed] [Google Scholar]
- Karaba A., Dixit S., Greco R., Aharoni A., Trijatmiko K.R., Marsch-Martinez N., Krishnan A., Nataraja K.N., Udayakumar M., Pereira A. (2007). Improvement of water use efficiency in rice by expression of HARDY, an Arabidopsis drought and salt tolerance gene. Proc. Natl. Acad. Sci. USA 104: 15270–15275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisselev A.F., van der Linden W.A., Overkleeft H.S. (2012). Proteasome inhibitors: An expanding army attacking a unique target. Chem. Biol. 19: 99–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo M., Kidokoro S., Yoshida T., Mizoi J., Kojima M., Takebayashi Y., Sakakibara H., Fernie A.R., Shinozaki K., Yamaguchi-Shinozaki K. (2018). A gene-stacking approach to overcome the trade-off between drought stress tolerance and growth in Arabidopsis. Plant J. 97: 240–256 [DOI] [PubMed] [Google Scholar]
- Kuromori T., Seo M., Shinozaki K. (2018). ABA transport and plant water stress responses. Trends Plant Sci. 23: 513–522. [DOI] [PubMed] [Google Scholar]
- Li J., Chory J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90: 929–938. [DOI] [PubMed] [Google Scholar]
- Li J., Nam K.H., Vafeados D., Chory J. (2001). BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 127: 14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wen J., Lease K.A., Doke J.T., Tax F.E., Walker J.C. (2002). BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110: 213–222. [DOI] [PubMed] [Google Scholar]
- Li L., Ye H., Guo H., Yin Y. (2010). Arabidopsis IWS1 interacts with transcription factor BES1 and is involved in plant steroid hormone brassinosteroid regulated gene expression. Proc. Natl. Acad. Sci. USA 107: 3918–3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S., Heymans K., Kuiper M. (2005). BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21: 3448–3449. [DOI] [PubMed] [Google Scholar]
- Maruyama K., et al. (2009). Metabolic pathways involved in cold acclimation identified by integrated analysis of metabolites and transcripts regulated by DREB1A and DREB2A. Plant Physiol. 150: 1972–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T., Suzuki K., Fujimura T., Shinshi H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140: 411–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam K.H., Li J. (2002). BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110: 203–212. [DOI] [PubMed] [Google Scholar]
- Nolan T.M., Brennan B., Yang M., Chen J., Zhang M., Li Z., Wang X., Bassham D.C., Walley J., Yin Y. (2017c). Selective autophagy of BES1 mediated by DSK2 balances plant growth and survival. Dev. Cell 41: 33–46.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan T., Chen J., Yin Y. (2017a). Cross-talk of brassinosteroid signaling in controlling growth and stress responses. Biochem. J. 474: 2641–2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan T., Liu S., Guo H., Li L., Schnable P., Yin Y. (2017b). Identification of brassinosteroid target genes by chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) and RNA-sequencing. Methods Mol. Biol. 1564: 63–79. [DOI] [PubMed] [Google Scholar]
- Northey J.G., Liang S., Jamshed M., Deb S., Foo E., Reid J.B., McCourt P., Samuel M.A. (2016). Farnesylation mediates brassinosteroid biosynthesis to regulate abscisic acid responses. Nat. Plants 2: 16114. [DOI] [PubMed] [Google Scholar]
- O’Malley R.C., Huang S.C., Song L., Lewsey M.G., Bartlett A., Nery J.R., Galli M., Gallavotti A., Ecker J.R. (2016). Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 166: 1598. [DOI] [PubMed] [Google Scholar]
- Oh E., Zhu J.Y., Wang Z.Y. (2012). Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 14: 802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phukan U.J., Jeena G.S., Tripathi V., Shukla R.K. (2017). Regulation of Apetala2/ethylene response factors in plants. Front. Plant Sci. 8: 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J., Song C.P., Wang B., Zhou J., Kangasjärvi J., Zhu J.K., Gong Z. (2018). Reactive oxygen species signaling and stomatal movement in plant responses to drought stress and pathogen attack. J. Integr. Plant Biol. 60: 805–826. [DOI] [PubMed] [Google Scholar]
- Ryu H., Cho H., Bae W., Hwang I. (2014). Control of early seedling development by BES1/TPL/HDA19-mediated epigenetic regulation of ABI3. Nat. Commun. 5: 4138. [DOI] [PubMed] [Google Scholar]
- Sakuma Y., Maruyama K., Osakabe Y., Qin F., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2006). Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18: 1292–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharabi-Schwager M., Samach A., Porat R. (2010). Overexpression of the CBF2 transcriptional activator in Arabidopsis counteracts hormone activation of leaf senescence. Plant Signal. Behav. 5: 296–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., et al. (2010). Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell 19: 765–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Yu J.P., Chen F., Zhao T.J., Fang X.H., Li Y.Q., Sui S.F. (2008). TINY, a dehydration-responsive element (DRE)-binding protein-like transcription factor connecting the DRE- and ethylene-responsive element-mediated signaling pathways in Arabidopsis. J. Biol. Chem. 283: 6261–6271. [DOI] [PubMed] [Google Scholar]
- Verslues P.E., Agarwal M., Katiyar-Agarwal S., Zhu J., Zhu J.K. (2006). Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J. 45: 523–539. [DOI] [PubMed] [Google Scholar]
- Wang H., Tang J., Liu J., Hu J., Liu J., Chen Y., Cai Z., Wang X. (2018). Abscisic acid signaling inhibits brassinosteroid signaling through dampening the dephosphorylation of BIN2 by ABI1 and ABI2. Mol. Plant 11: 315–325. [DOI] [PubMed] [Google Scholar]
- Wang X., Chen J., Xie Z., Liu S., Nolan T., Ye H., Zhang M., Guo H., Schnable P.S., Li Z., Yin Y. (2014). Histone lysine methyltransferase SDG8 is involved in brassinosteroid-regulated gene expression in Arabidopsis thaliana. Mol. Plant 7: 1303–1315. [DOI] [PubMed] [Google Scholar]
- Wang Z.Y., Nakano T., Gendron J., He J., Chen M., Vafeados D., Yang Y., Fujioka S., Yoshida S., Asami T., Chory J. (2002). Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2: 505–513. [DOI] [PubMed] [Google Scholar]
- Wilson K., Long D., Swinburne J., Coupland G. (1996). A dissociation insertion causes a semidominant mutation that increases expression of TINY, an Arabidopsis gene related to APETALA2. Plant Cell 8: 659–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L., Yang C., Wang X. (2011). Brassinosteroids can regulate cellulose biosynthesis by controlling the expression of CESA genes in Arabidopsis. J. Exp. Bot. 62: 4495–4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Nolan T.M., Jiang H., Yin Y. (2019). AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Front. Plant Sci. 10: 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Huang J., Li B., Li J., Wang Y. (2008). Is kinase activity essential for biological functions of BRI1? Cell Res. 18: 472–478. [DOI] [PubMed] [Google Scholar]
- Yan Z., Zhao J., Peng P., Chihara R.K., Li J. (2009). BIN2 functions redundantly with other Arabidopsis GSK3-like kinases to regulate brassinosteroid signaling. Plant Physiol. 150: 710–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Li C., Cai Z., Hu Y., Nolan T., Yu F., Yin Y., Xie Q., Tang G., Wang X. (2017). SINAT E3 ligases control the light-mediated stability of the brassinosteroid-activated transcription factor BES1 in Arabidopsis. Dev. Cell 41: 47–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H., et al. (2017). RD26 mediates crosstalk between drought and brassinosteroid signalling pathways. Nat. Commun. 8: 14573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H., Li L., Guo H., Yin Y. (2012). MYBL2 is a substrate of GSK3-like kinase BIN2 and acts as a corepressor of BES1 in brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 20142–20147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Wang Z.Y., Mora-Garcia S., Li J., Yoshida S., Asami T., Chory J. (2002). BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191. [DOI] [PubMed] [Google Scholar]
- Yin Y., Vafeados D., Tao Y., Yoshida S., Asami T., Chory J. (2005). A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120: 249–259. [DOI] [PubMed] [Google Scholar]
- Youn J.H., Kim T.W. (2015). Functional insights of plant GSK3-like kinases: Multi-taskers in diverse cellular signal transduction pathways. Mol. Plant 8: 552–565. [DOI] [PubMed] [Google Scholar]
- Yu X., Li L., Zola J., Aluru M., Ye H., Foudree A., Guo H., Anderson S., Aluru S., Liu P., Rodermel S., Yin Y. (2011). A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 65: 634–646. [DOI] [PubMed] [Google Scholar]
- Zhang L.Y., et al. (2009). Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 21: 3767–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Ye H., Guo H., Johnson A., Zhang M., Lin H., Yin Y. (2014). Transcription factor HAT1 is phosphorylated by BIN2 kinase and mediates brassinosteroid repressed gene expression in Arabidopsis. Plant J. 77: 59–70. [DOI] [PubMed] [Google Scholar]
- Zhao J., Peng P., Schmitz R.J., Decker A.D., Tax F.E., Li J. (2002). Two putative BIN2 substrates are nuclear components of brassinosteroid signaling. Plant Physiol. 130: 1221–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Wang J., Li X., Xia X.J., Zhou Y.H., Shi K., Chen Z., Yu J.Q. (2014). H2O2 mediates the crosstalk of brassinosteroid and abscisic acid in tomato responses to heat and oxidative stresses. J. Exp. Bot. 65: 4371–4383. [DOI] [PMC free article] [PubMed] [Google Scholar]