Plant-specific TTL proteins function as positive regulators of BR responses likely by bringing together signaling components at the plasma membrane.
Abstract
Brassinosteroids (BRs) form a group of steroidal hormones essential for plant growth, development, and stress responses. BRs are perceived extracellularly by plasma membrane receptor-like kinases that activate an interconnected signal transduction cascade, leading to the transcriptional regulation of BR-responsive genes. TETRATRICOPEPTIDE THIOREDOXIN-LIKE (TTL) genes are specific for land plants, and their encoded proteins are defined by the presence of protein–protein interaction motives, that is, an intrinsic disordered region at the N terminus, six tetratricopeptide repeat domains, and a C terminus with homology to thioredoxins. TTL proteins thus likely mediate the assembly of multiprotein complexes. Phenotypic, molecular, and genetic analyses show that TTL proteins are positive regulators of BR signaling in Arabidopsis (Arabidopsis thaliana). TTL3 directly interacts with a constitutively active BRASSINOSTEROID INSENSITIVE1 (BRI1) receptor kinase, BRI1-SUPPRESSOR1 phosphatase, and the BRASSINAZOLE RESISTANT1 transcription factor and associates with BR-SIGNALING KINASE1, BRASSINOSTEROID INSENSITIVE2 kinases, but not with BRI1-ASSOCIATED KINASE1. A functional TTL3-green fluorescent protein (GFP) shows dual cytoplasmic plasma membrane localization. Depleting the endogenous BR content reduces plasma membrane localization of TTL3-GFP, while increasing BR content causes its plasma membrane relocalization, where it strengthens the association of BR signaling components. Our results reveal that TTL proteins promote BR responses and suggest that TTL proteins may function as scaffold proteins by bringing together cytoplasmic and plasma membrane BR signaling components.
INTRODUCTION
Brassinosteroids (BRs) are a family of growth-promoting hormones with essential roles in a wide range of developmental and physiological processes (Belkhadir and Jaillais, 2015; Chaiwanon et al., 2016; Jaillais and Vert, 2016). However, in addition to their well-established function in growth, essential roles in the trade-off between growth and tolerance to biotic and abiotic stress episodes are now being unveiled (Lozano-Durán and Zipfel, 2015; Zhang et al., 2016; Nolan et al., 2017; Tian et al., 2018). BRs are perceived at the plasma membrane by ligand-induced heterodimers of the receptor kinases BRASSINOSTEROID INSENSITIVE1 (BRI1) and SOMATIC EMBBRYOGENESIS RECEPTOR KINASE (SERK) protein family members, which activates an interconnected signal transduction cascade, leading to the transcriptional regulation of BR-responsive genes (Belkhadir and Jaillais, 2015). BRI1 KINASE INHIBITOR1 dissociates from activated BRI1, which phosphorylates the BR-SIGNALING KINASE (BSK) family proteins BSK1 and BSK3 and the CONSTITUTIVE DIFFERENTIAL GROWTH1, which in turn activate the BRI1-SUPPRESSOR1 (BSU1) phosphatase (Wang et al., 2014; Belkhadir and Jaillais, 2015; Ren et al., 2019). Next, the active (phosphorylated) BSU1 leads to dephosphorylation and inactivation of the glycogen synthase kinase3 (GSK3)–like BRASSINOSTEROID INSENSITIVE2 (BIN2). In the absence of BRs, BIN2 is active and phosphorylates the two homologous transcription factors BRASSINAZOLE RESISTANT1 (BZR1) and BRI1-ETHYL METHANESULFONATE SUPPRESSOR1 (BES1/BZR2), which results in their inactivation and degradation (Wang et al., 2014; Belkhadir and Jaillais, 2015). By contrast, when BR is present, BIN2 is inactivated and degraded by the proteasome mediated by the F-box E3 ubiquitin ligase KINK SUPPRESSED1 in bzr1-1D (Zhu et al., 2017), leading to both the stabilization and the activation of BZR1 and BES1 and therefore to transcriptional regulation of BR-responsive genes (Wang et al., 2014; Belkhadir and Jaillais, 2015).
In Arabidopsis (Arabidopsis thaliana), the TETRATRICOPEPTIDE THIOREDOXIN-LIKE (TTL) gene family is composed of four members (TTL1 to TTL4) and mutations in TTL1, TTL3, and TTL4 cause reduced growth under abiotic stresses such as salinity and drought (Rosado et al., 2006; Ceserani et al., 2009; Lakhssassi et al., 2012). This stress hypersensitivity is exacerbated in double and triple ttl mutants (Lakhssassi et al., 2012). TTL2 is specifically expressed in pollen grains and does not have a role in stress tolerance, but it is important for male sporogenesis (Lakhssassi et al., 2012). TTL genes encode proteins with a common modular architecture containing six tetratricopeptide repeat (TPR) domains distributed in specific positions throughout the sequence and a C-terminal sequence with homology to thioredoxins (Rosado et al., 2006; Lakhssassi et al., 2012). TPR domains are well-described protein–protein interaction modules; however, how TTL proteins function mechanistically in stress tolerance remains elusive.
Several lines of circumstantial evidence point to a role of TTL proteins in BR responses, which open the possibility of a direct link between stress tolerance and BR signaling by the TTL proteins. First, the TTL3 protein, whose gene is the most highly expressed among the TTL gene family, was identified as an interacting partner of the activated (phosphorylated) cytoplasmic domain of VASCULAR HIGHWAY1/BRI1-LIKE RECEPTOR KINASE2 (BRL2). BRL2 has a role in vascular development and belong to the BRI family (Ceserani et al., 2009), although BRL2 cannot bind BRs (Belkhadir and Jaillais, 2015). Second, a ttl3 mutant showed altered growth in the presence of exogenous BRs (Ceserani et al., 2009). Third, TTL proteins are predicted to interact and function as co-chaperones of Hsp90 (Prasad et al., 2010), which has been recently identified to have important roles in BR signaling by interacting with specific BR signaling components (Lachowiec et al., 2013; Samakovli et al., 2014; Shigeta et al., 2014, 2015). Fourth, a triple Arabidopsis line with T-DNA insertions in TTL1, TTL3, and TTL4 shows defects in vasculature development and male sporogenesis, hallmarks of BR-defective mutants (Yang et al., 2011; Lakhssassi et al., 2012). Finally, TTL1, TTL3, and TTL4 are specifically induced by BR application, but not by other hormones (Prasad et al., 2010).
Based on phenotypic, molecular, and genetic analyses, we show that TTL1, TTL3, and TTL4 genes, in addition to their reported role in abiotic stress tolerance, are positive regulators of BR signaling. The well-described TPR protein interaction modules of TTL proteins and their role in the assembly of multiprotein complexes (Blatch and Lässle, 1999; D’Andrea and Regan, 2003; Yang et al., 2005) led us to hypothesize that these proteins could function as a scaffold for BR signaling. Indeed, we show that TTL3 interacts with constitutively active BRI1, BSU1, and BZR1 and associates in vivo with most BR signaling components, with the exception of BRI1-ASSOCIATED KINASE1 (BAK1). We also show that a functional TTL3 tagged with a green fluorescent protein (GFP) shows a dual cytoplasmic and plasma membrane localization that is dependent on endogenous BR content. Furthermore, TTL3 greatly enhances the interaction between BSK1 and BZR1 at the plasma membrane and suppresses the BIN2-promoted cytoplasmic retention of BZR1-GFP. Together, we reveal that TTL proteins function as positive regulators of BR signaling, likely by bringing together signaling components at the plasma membrane.
RESULTS
Interaction Analysis of TTL3 with BRI1
The TTL3 protein, also known as VH1-interacting TPR-containing protein, has been identified as an interactor of the activated (phosphorylated) cytoplasmic domain of BRL2 (Ceserani et al., 2009), a receptor kinase of the BRI1 family with a role in vascular development (Caño-Delgado et al., 2004; Ceserani et al., 2009). TTL3 belongs to a family of four genes (TTL1 to TTL4) in Arabidopsis (Rosado et al., 2006; Lakhssassi et al., 2012). While TTL1, TTL3, and TTL4 show ubiquitous expression, TTL2 expression is restricted to pollen grains. We confirmed the reported defects in vein formation using a different ttl3 mutant allele (Supplemental Figure 1A) and showed that mutations in TTL1 and TTL4, but not TTL2, also caused venation defects that were markedly enhanced in a triple ttl1 ttl3 tt4 mutant (hereafter ttl134; Supplemental Figure 1A).
TTL3 has been proposed to be an adaptor protein of BRL2 that, through association with other proteins, modulates vein formation (Ceserani et al., 2009). TTL3, as for other TTL proteins from other plant species (Rosado et al., 2006; Lakhssassi et al., 2012), is characterized by the presence of six TPRs and a C-terminal domain with homology to thioredoxins. An in silico structural analysis of TTL3 predicts the presence of an intrinsically disordered region (IDR) at the N terminus (Supplemental Figure 2) with the rest of the protein forming a horseshoe-shaped structure composed of multiple helix-turn-helix motifs (Figure 1A). This structure is consistent with TTL3 being involved in protein–protein interactions and the assembly of multiprotein complexes (Blatch and Lässle, 1999; D’Andrea and Regan, 2003; Yang et al., 2005).
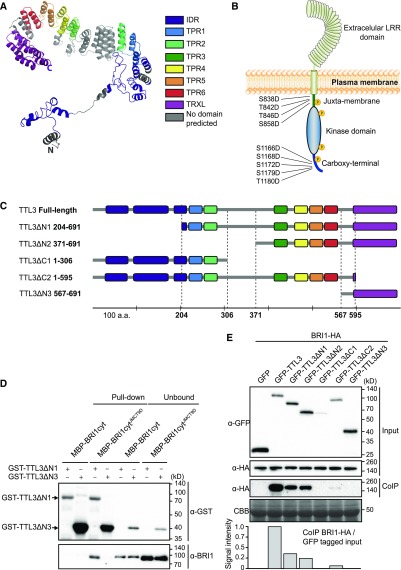
Figure 1.
TTL3 Interacts with BRI1 In Vivo and In Vitro.
(A) Structural model of TTL3 protein predicted in silico using I-TASSER server (Zhang, 2008) and processed by PyMOL (Schrödinger). C, C terminus; N, N terminus.
(B) Schematic representation of BRI1 protein and the nine Ser/Thr residues of the JM and CT domains that were substituted by Asp in the BAK1-independent, BRI1-constitutive (phosphomimetic) active form BRI1cytJMCT9D (Wang et al., 2008). LRR, leucine rich repeat. Yellow-circled P’s means that BRI1cytJMCT9D mimics the phosphorylated form of BRI1.
(C) Schematic representations of full-length and different truncated versions of TTL3 protein. Numbers indicate first and last amino acids (a.a.) of TTL3 truncated proteins. Domains and protein fragments interspacing the conserved domains are represented with the same color code as in (A).
(D) TTL3ΔN1 interacts with BRI1cytJMCT9D in vitro, as shown by a GST-pull-down assay. GST-TTL3ΔN1 and GST-TTL3ΔN3 were detected with anti-GST antibody. MBP-BRI1cyt and MBP-BRI1cytJMCT9D were detected using specific anti-BRI1 antibodies (Bojar et al., 2014). Pull-down reflects 20% of the total pulled down proteins. Unbound reflects 1% of the total unbound fraction.
(E) BRI1-HA co-IPs with GFP-TTL3 full-length and GFP-TTL3 truncated versions ΔN1, ΔN2, and ΔC1. Numbers indicate first and last amino acids of TTL3 truncated proteins. BRI1-HA was transiently coexpressed in N. benthamiana with GFP-TTL3 full-length and truncated versions, and GFP-tagged protein was immunoprecipitated using anti–GFP-Trap beads. Total (input), IP, and CoIP proteins were analyzed by immunoblotting. Equal loading was confirmed by Coomassie blue staining (CBB) of input samples. GFP- and HA-tagged proteins were detected with anti-GFP and anti-HA antibody, respectively. The amount of coIP BRI1-HA was normalized relative to the amount of GFP-tagged protein from the input, dividing the signal intensity of coIP BRI1-HA by the signal intensity of the each GFP-tagged protein from the input that coIP BRI1-HA.
The similarity between BRL2 and BRI1 kinase domains (Supplemental Figure 3) suggested that TTL3 could also interact with the BRI1 cytoplasmic domain. We therefore tested the direct interaction of TTL3 with the BRI1 cytoplasmic region that includes the juxta-membrane (JM), the kinase domain, and the carboxy-terminal (CT) domain (BRI1cyt; Figure 1B). While BRI1cyt was soluble when fused to an maltose binding protein (MBP) tag (Supplemental Figure 4), we were unable to produce full-length TTL3 protein fused to glutathione S-transferase (GST) despite many attempts. This low stability was probably caused by the presence of the IDR (Habchi et al., 2014). Accordingly, we produced in Escherichia coli two different fragments: TTL3 lacking the N terminus IDR (TTL3ΔN1) and TTL3 containing the TRLX domain (TTL3ΔN3; Figure 1C; Supplemental Figure 4). Using an in vitro GST pull-down assay, we did not detect an interaction between BRI1cyt and either TTL3ΔN1 or TTL3ΔN3 (Figures 1C and 1D). Because the activation of BRI1 is dependent on BAK1 transphosphorylation on specific residues at the JM and CT (Wang et al., 2008), we next used a BAK1-independent BRI1 constitutively active (phosphomimetic) form BRI1cytJMCT9D. In this mutant version, nine Ser and Thr have been substituted by Asp at the juxta-membrane (JM) andcarboxy-region CR domains (Figure 1B; Wang et al., 2008). In this case, BRI1cytJMCT9D was pulled down by TL3ΔN1, but not by TTL3ΔN3 (Figures 1C and 1D). This indicates that TTL3 predominantly interacts with active BRI1 and that this interaction occurs through the TPR domains, but not the thioredoxin-like (TRXL) domain of TTL3.
Next, we investigated this interaction in vivo by performing co-immunoprecipitation (Co-IP) assays after transient expression of tagged full-length TTL3 and BRI1 in Nicotiana benthamiana. After immunoprecipitation of GFP-TTL3 and free GFP using GFP-Trap beads, we detected a strong specific interaction between GFP-TTL3 and BRI1-HA (Figure 1E). Additional Co-IP experiments using a C-terminal GFP-tagged TTL3 protein (TTL3-GFP) coexpressed with C-terminal hemagglutinin (HA)-tagged BRI1 protein (BRI1-HA; Supplemental Figure 5A) and BRI1-GFP coexpressed with TTL3-HA (Supplemental Figure 5B) further confirmed the TTL3-BRI1 interaction and indicated that the position and tag used in the Co-IP experiments does not affect their in planta interaction.
We further used Co-IP assays to map the TTL3 domains required for the interaction with BRI1. We performed this analysis in planta to determine the possible role of the IDR domain in the interaction, which was not possible using in vitro assays. We generated a series of truncated TTL3 fragments with deletions at the N terminus (TTL3ΔN1, TTL3ΔN2, TTL3ΔN3) and at the C terminus (TTL3ΔC1, TTL3ΔC2; Figure 1C), transcriptionally fused these fragments to GFP at the N terminus, and coexpressed these constructs with BRI1-HA in N. benthamiana leaves. All expressed proteins showed the expected molecular size (Figure 1E, input). TTL3ΔC1 and TTL3ΔC2 truncated proteins, both lacking the TRLX domain, showed lower accumulation than the others (Figure 1E, input), suggesting that TRLX is important for protein stabilization.
The full-length TTL3 and three of the five truncated TTL3 proteins, that is, GFP-TTL3ΔN1, GFP-TTL3ΔN2, and GFP-TTL3ΔC2, which all having in common TPR3 to TPR6 (Figure 1C), co-immunoprecipitated (CoIP) BRI1-HA with different efficiency, indicating that these domains are essential for the interaction, which is consistent with the in vitro data (Figure 1D). To quantify the interaction of the different TTL protein fragments and BRI1, the amount of CoIP BRI1-HA was normalized relative to the amount of protein input (Figure 1E). The strongest interaction occurs with the full-length TTL3 protein, indicating that all domains contribute to stabilize the interaction with BRI1. A lower but similar interaction was observed with GFP-TTL3ΔN1 and GFP-TTL3ΔN2, both containing the TRLX domain, indicating that this domain is important for stabilizing the interaction, but is not sufficient for the in vitro or in vivo interaction with BRI1 (Figures 1C and 1E). Consistent with this, removing the TRLX region in GFP-TTL3ΔC2 greatly reduced the interaction between TTL3 and BRI1 (Figures 1C to 1E).
Finally, the interaction between BRI1 and TTL3 was also investigated using bimolecular fluorescence complementation (BiFC) assays in N. benthamiana leaves, which provide additional information about the subcellular localization of the interaction. Coexpression of the TTL3-N-terminal half of YFP (nYFP) with the BRI1-C-terminal half of YFP (cYFP) or BRI1-nYFP with TTL3-cYFP (Supplemental Figure 6) reconstituted functional YFP proteins at the plasma membrane, confirming the interaction and consistent with the plasma membrane localization of BRI1.
BAK1, also known as SERK3, and other SERK proteins are transmembrane kinases that function as BR coreceptors (Ma et al., 2016). Similar Co-IP experiments using TTL3-GFP and BAK1 transiently coexpressed in N. benthamiana indicated that, contrary to BRI1, TTL3 does not associate in vivo with BAK1 (Supplemental Figure 5C). This result was verified by BiFC assays in N. benthamiana leaves since confocal microscopy analyses revealed that coexpression of TTL3-nYFP with BAK1-cYFP (Supplemental Figure 6A) and also BAK1-nYFP with TTL3-cYFP (Supplemental Figure 6B) did not reconstitute functional YFP proteins. To confirm that BiFC BAK1 constructs were functional, we performed a BiFC between BRI1 and BAK1 and observed weak but positive signals (Supplemental Figures 6A and 6B)
TTL Proteins Play a Positive Role in BR Signaling
The interaction of TTL3 with BRI1 supports a role for TTL3 in BR signaling. Furthermore, previous expression analyses indicate that TTL1, TTL3, and TTL4 transcripts are induced by BR (Prasad et al., 2010), which is also supported by public transcriptomic data (Supplemental Figure 7A). This upregulation of the TTL genes in response to BR was confirmed at the cellular level by analyzing transgenic plants transformed with the reporter β-glucuronidase gene driven by each of the TTL promoters (Supplemental Figure 7B).
We next analyzed the sensitivity to epibrassinolide (eBL) by measuring root growth in the presence or absence of exogenous 100 nM eBL in wild-type Columbia ecotype (Col-0), single ttl mutants, the triple ttl134 mutant, and bak1-4, a well-established mutant affected in BR responses (Chinchilla et al., 2007; He et al., 2007; Schwessinger et al., 2011; Gou et al., 2012). Single ttl mutants, the ttl134 mutant, and the bak1-4 mutant showed a similar root growth to Col-0 in control conditions (Figure 2A and Supplemental Figure 8A). However, bak1-4, ttl1, ttl3, and ttl4 show increased root length compared with the Col-0 control or ttl2 in the presence of eBL (Figure 2A and Supplemental Figure 8B). The lack of ttl2 sensitivity to BR is consistent with the pollen-specific expression of TTL2 (Lakhssassi et al., 2012). This decreased sensitivity to eBL of single ttl mutants was strongly enhanced in ttl134 (Figure 2A and Supplemental Figure 8B). Root growth sensitivity to eBL of the ttl134 mutant was then compared, in addition to bak1-4, to well-characterized genotypes affected in BR responses such as serk1-1 and the double serk1-1 bak1-4 mutant (Gou et al., 2012). In control conditions, all genotypes grew similar to Col-0, with the exception of serk1-1 bak1-4, which showed reduced root growth (Figure 2B and Supplemental Figure 8C), as reported previously (Du et al., 2012; van Esse et al., 2013). In the presence of 100 nM eBL, the root growth reduction of the Col-0 control was significantly higher than for the rest of the genotypes, including ttl134 (Figure 2B; Supplemental Figure 8D), while the serk1-1 bak1-4 double mutant was almost completely insensitive to eBL, as it showed a similar root growth in control and eBL-supplemented media.
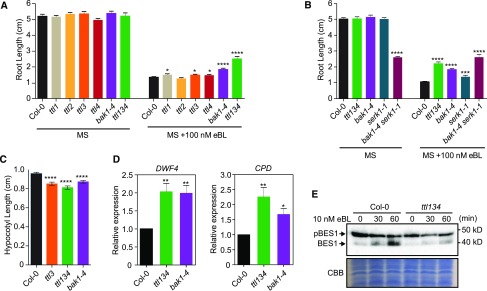
Figure 2.
TTL1, TTL3, and TTL4 Genes Play a Positive Role in BR Signaling.
(A) ttl1, ttl3, ttl4, and ttl134 show root growth hyposensitivity to BR. Statistical analysis of root length measurements of Col-0, ttl, and bak1-4 mutants in control conditions (MS) and in response to eBL. Seedlings were grown in long days for 4 d in half-strength MS agar solidified medium and then transferred to half-strength MS agar solidified medium (MS) or half-strength MS agar solidified medium supplemented with 100 nM eBL (MS + 100 nM eBL), and root length was measure 6 d later. Asterisks indicate statistical differences between mutant versus Col-0 determined by the unpaired t test (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 ****P ≤ 0.0001). Data represent mean values, error bars are sem, n ≥ 35 seedlings per experiment. The experiment was repeated three times with similar results.
(B) Root length responses to eBL of wild-type Col-0, ttl134, and BR perception mutants. Seedlings were grown and root length was analyzed as described in (A). Asterisks indicate statistical differences between mutant versus Col-0 as determined by the unpaired t test (***P ≤ 0.001, ****P ≤ 0.0001). Data represent mean values, error bars are sem, n = 30 seedlings per experiment. The experiment was repeated three times with similar results.
(C) Defective hypocotyl elongation in ttl mutants. Col-0, ttl3, ttl134, and bak1-4 seedlings were grown for 4 d in long-day photoperiod in half-strength MS agar solidified medium. Seedlings with the same size were then placed in the dark, and hypocotyl elongation was measured 3 d later. Asterisks indicate statistically significant differences between Col-0 and the indicated genotype as determined by the unpaired t test (****P ≤ 0.0001), values are mean, error bars are sem, n = 80 seedlings per experiment. The experiment was repeated twice with similar results.
(D) BR-responsive genes DWF4 and CPD show induced expression in ttl134 and bak1-4 relative to Col-0 seedlings. Seeds were germinated in half-strength MS agar solidified medium and grown vertically in long-day photoperiod conditions. Five-day-old seedlings were transferred to half-strength MS liquid medium and after 5 d of acclimation, the relative expression level of DWF4 and CPD was measured by RT-qPCR. The expression of DWF4 and CPD was first normalized to the expression of ACTIN2 and represented relative to the expression of Col-0. The data are shown as mean ± sem from at least three independent biological replicates (pool of 20 seedlings per biological replicate). Asterisks indicate statistically significant differences between the indicated genotype versus Col-0 as determined by the unpaired t test (*P ≤ 0.05, **P ≤ 0.01). The experiment was repeated three times with similar results.
(E) Phosphorylation status of BES1 in response to exogenously applied eBL in Arabidopsis Col-0 and ttl134. Ten-day-old seedlings pre-treated for 3 d with the BR biosynthetic inhibitor BRZ to deplete the endogenous pool of BRs were subjected to 10 nM eBL treatment for 0, 30, and 60 min. Total proteins from a pool of 20 seedlings were analyzed by an immunoblot assay with a specific anti-BES1 antibody (Yu et al., 2011). The top band corresponds to pBES1 and the bottom band to dephosphorylated BES1 (BES1). The experiment was repeated two times with similar results. CBB, Coomassie blue.
Hypocotyl elongation in the dark is dependent on active BR signaling (Bernardo-García et al., 2014); therefore, it was analyzed in ttl3, ttl134 and bak1-4 as a readout of defective BR signaling (Zhang et al., 2015). As reported previously, bak1-4 showed a reduction in hypocotyl elongation relative to Col-0 (Figure 2C; Supplemental Figure 8E; Li et al., 2002; Nam and Li, 2002). Similar to bak1-4, ttl3 and ttl134 mutants presented shorter hypocotyls than Col-0 (Figure 2C; Supplemental Figure 8E).
To investigate the contribution of TTL genes to BR responses at the molecular level, we first studied the expression of the BR-regulated genes CPD1 and DWF4 in Col-0, bak1-4, and the triple ttl134 mutants. As shown in Figure 2D, DWF4 and CDP1 expression was approximately twofold higher in ttl134 and bak1-4 compared with the Col-0 control. This increased CPD1 and DWF4 expression has been reported for BR signaling mutants such as bri1-5 (Wang et al., 2016), bri1-301 (Gou et al., 2012), and bik1 (Lin et al., 2013) and is caused by a lack of feedback regulation in the expression of these biosynthetic genes (Tanaka et al., 2005; Chung and Choe, 2013; Vriet et al., 2013). Second, we investigated the phosphorylation status of BES1 in Col-0 and the ttl134 mutant in response to eBL. Because the BR biosynthetic genes DWF4 and CDP1 are already induced in ttl134, to fully capture the BR signaling capacity of ttl134, we pretreated the seedlings with the BR biosynthesis inhibitor BRZ. Without BR treatment, a strong signal of phosphorylated BES1 (pBES1) and a weak signal of dephosphorylated (BES1) are present in Col-0 and ttl134 (Figure 2E). While 10 nM eBL caused an increase of dephosphorylated BES1 in Col-0 after 30 and 60 min, eBL caused little dephosphorylation of pBES1 in ttl134 seedlings (Figure 2E), confirming a defective BR signaling in ttl134.
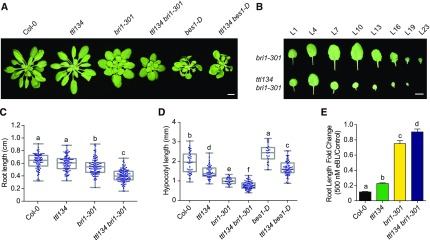
Despite the role exerted by TTL genes in BR responses, ttl134 adult plants do not exhibit obvious growth defects when under short-day or long-day photoperiod (Figure 3A and Supplemental Figure 9A). Phenotypic defects in BR sensitivity are often revealed in adult plants when mutations in potential BR signaling components are combined with weak bri1 alleles (Nam and Li, 2002; Kim and Wang, 2010; Schwessinger et al., 2011). Therefore, we generated and analyzed the phenotype of a quadruple homozygous ttl134 bri1-301 mutant. When grown in short days, the bri1-301 mutant displays a semi-dwarf cabbage-like rosette, a phenotype that was enhanced in the ttl134 bri1-301 quadruple mutant (Figure 3A). ttl134 bri1-301 plants have fewer leaves than bri1-301 plants (Supplemental Figure 9B), and the younger leaves were considerably smaller in ttl134 bri1-301 than in bri1-301 plants of the same age (Figure 3B). A stronger dwarf phenotype in ttl134 bri1-301 relative to the weak bri1-301 mutant was also observed in long-day-grown plants (Supplemental Figure 9A). ttl134 bri1-301 also shows enhanced defects in seedling root growth (Figure 3C; Supplemental Figure 9C) and hypocotyl elongation (Figure 3D) relative to bri1-301. Next, we determined the root growth BR sensitivity of ttl134 bri1-301 relative to Col-0, ttl134, and bri1-301 (Ren et al., 2019). The ttl134 bri1-301 seedlings exhibited less root growth inhibition than bri1-301 seedlings when grown on 500 nM eBL (Figure 3E).
Figure 3.
ttl134 Mutations Enhance the bri1-301–Defective Phenotype and Partially Reduce the BR Responses in bes1-D.
(A) Morphological phenotypes of 5-week-old plants grown in short days. Bar = 1 cm.
(B) Detached leaves of 5-week-old plants grown in short days (L1, oldest leaf; L23 youngest leaf). Bar = 1 cm.
(C) Root length of 3-d-old seedlings grown in long days in half-strength MS agar solidified medium. Dots represent individual measurements from three independent experiments. n denotes measurement of roots from independent seedlings: Col-0 (n = 92), ttl134 (n = 100), bri1-301 (n = 113), and ttl134 bri1-301 (n = 123). Box plots display the first and third quartiles, split by the median; whiskers extend to include the maximum and minimum values. Different lowercase letters indicate significant differences. Data were analyzed with one-way ANOVA and Tukey´s multiple comparison test; P < 0.05.
(D) Hypocotyl length of 7-d-old seedlings grown in long days in half-strength MS agar solidified medium. Dots represent individual measurements from three independent experiments. n denotes measurement of hypocotyls from independent seedlings: Col-0 (n = 44), ttl134 (n = 51), bri1-301 (n = 28), ttl134 bri1-301 (n = 73), bes1-D (n = 27), and ttl134 bes1-D (n = 71). Values are plotted and statistically analyzed as in (C).
(E) Root length fold changes of seedlings grown for 7 d in the absence (control) or presence of 500 nM eBL in long days. Values are mean, error bars are sem, and n = 22 seedlings per experiment. Different lowercase letters indicate significant differences. Data were analyzed with one-way ANOVA and Tukey´s multiple comparison test; P < 0.05. The experiment was repeated twice with similar results.
The gain-of-function bes1-D mutant shows constitutive BR responses, including higher elongation of hypocotyl and petioles due to increased levels of BES1 protein (Figures 3A and 3D; Yin et al., 2002). A ttl134 bes-1D quadruple mutant displays shorter hypocotyls and petioles than bes1-D (Figures 3A and 3D; Supplemental Figure 9D). This indicates that mutations in TTL genes reduce BR responses of bes1-D. Together, these results indicate that the TTLs play a positive role in the regulation of BR responses.
BRs Regulate the Cytoplasmic/Plasma Membrane Localization of TTL3
To further explore how TTL3 functions in BR signaling, we analyzed its subcellular localization. Although the BiFC interaction of TTL3 with BRI1 suggests a plasma membrane localization of TTL3, expression of a C-terminal GFP-tagged TTL3 in N. benthamiana indicated a predominant cytoplasmic localization in basal conditions (Supplemental Figure 10A). However, plasmolyzed cells show the presence of GFP-TTL3 in Hechtian strands, indicating that TTL3 also associated with the plasma membrane (Supplemental Figure 10B). To gain further insight into TTL3 localization, a genomic fragment including a 1.7-kb TTL3 promoter region upstream of the start codon was transcriptionally fused to GFP to generate the TTL3p:TTL3g-GFP construct and transformed into both ttl3 and ttl134 mutants using Agrobacterium tumefaciens. After confocal analysis of a large number of independent stable transgenic lines, we selected two homozygous lines, one in the ttl3 background (hereafter TTL3-GFP 1.2) and the other in the ttl134 background (TTL3-GFP 2.4), which presented noticeable fluorescence signals. Venation defects of ttl3 and ttl134 were restored to levels similar to Col-0 in TTL3-GFP 1.2 and TTL3-GFP 2.4 (Supplemental Figure 1B). Furthermore, root growth of TTL3-GFP 1.2 (Supplemental Figures 11A and 11B) and TTL3-GFP 2.4 (Figures 4A to 4C) were restored to wild-type levels in the presence of eBL, indicative of a functional TTL3-GFP protein.
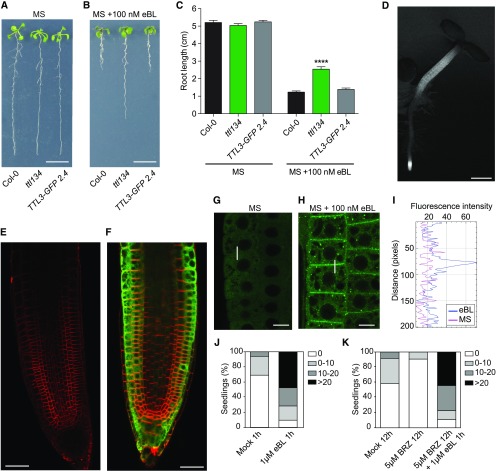
Figure 4.
BRs Regulate the Cytoplasmic/Plasma Membrane Localization of TTL3.
(A) to (C) The root growth responses to eBL of the ttl134 triple mutant are complemented in TTL3-GFP 2.4. Seedlings were grown for 4 d in half-strength MS agar solidified medium and then transferred to half-strength MS agar solidified medium (A) or half-strength MS agar solidified medium supplemented with 100 nM brassinolide (B) and root length was measured (C). (A) and (B) Representative photographs of seedlings, 6 d after being transferred to control or eBL treatment. Bar in (A) and (B) = 1 cm. (C) Statistical analysis of root length of Col-0, ttl134, and the complementation line TTL3-GFP 2.4. Asterisks indicate statistically significant differences between the indicated genotype versus Col-0 as determined by the unpaired t test (****P ≤ 0.0001). Data represent mean values, error bars are sem, and n = 30 seedlings per experiment. The experiment was repeated three times with similar results.
(D) Expression pattern of TTL3-GFP in 3-d-old TTL3-GFP 2.4 Arabidopsis seedlings. Images were captured using conventional wide field fluorescence microscopy with a GFP filter. Bar = 500 µm.
(E) and (F) Longitudinal median section of root tips of a 3-d-old Col-0 (E) and TTL3-GFP 2.4 seedling as observed by laser scanning confocal microscopy (F). Images are a merge of green channel showing TTL3-GFP expression and red channel showing plasma membrane stained with FM4-64. Bar = 20 µm.
(G) and (H) Confocal images showing localization of TTL3-GFP in epidermal cells from the root meristematic zone in 4-d-old Arabidopsis TTL3-GFP 2.4 in half-strength MS agar solidified medium, in control conditions (1-h treatment with eBL solvent) (G), or after 1 h of 1 µM eBL treatment (H) in half-strength MS agar liquid medium. Bar = 10 µm (horizontal bar). (I) Quantification of fluorescent protein signal in plasma membrane versus cytoplasm. Line scan measurements spanning membrane and cytoplasm were performed (represented in [G] and [H] as a vertical white line), and representative plot profiles of sample measurements are presented.
(J) and (K) Quantification of the cytoplasmic and plasma membrane localization of TTL3-GFP in 4-d-old Arabidopsis TTL3-GFP 2.4 seedlings treated for 1 h with 1 µM eBL (J) or pre-treated for 12 h with 5 µM BRZ prior to 1 µM eBL application for 1 h (K). The number of cells with dual cytoplasmic/plasma membrane localization in meristematic and transition zone was counted for each analyzed root using confocal microscopy. Seedlings were grouped in categories according to the number of cells that presented this dual localization, and the percentage of seedlings displaying each category depicted in the key was calculated. Represented categories in the key indicate the number of cells per seedling with dual cytoplasmic/plasma membrane localization. At least 16 seedlings per treatment, and ∼200 cells (cell from epidermis, cortex, and endodermis all combined) per seedling of the meristematic region of the root tip were analyzed.
We then used TTL3-GFP 2.4 (which showed a stronger fluorescence signal than TTL3-GFP 1.2) to analyze the cellular and subcellular localization of TTL3. Examination under a stereomicroscope indicated that TTL3-GFP accumulated mainly at the root tip and the hypocotyl of Arabidopsis seedlings (Figure 4D). This accumulation coincides with cells that undergo strong BR signaling, leading to active growth, and highly resembles the accumulation pattern of BRI1-GFP (Geldner et al., 2007; Wilma van Esse et al., 2011; Fàbregas et al., 2013). Cellular analysis using confocal microscopy was performed in 3-day-old roots, simultaneously localizing TTL3-GFP with the FM4-64, a lipophilic red dye that labels the plasma membrane and tracks plasma membrane-derived endosomes (Vida and Emr, 1995). In Col-0 control roots, no GFP signal was detected (Figure 4E), while analysis of TTL3-GFP 2.4 revealed the presence of TTL3-GFP in all cell files of the root apical meristem (Figure 4F). Further up, in the meristematic region, TTL3-GFP showed a predominant localization in the outer cell layers (epidermis and cortex; Figure 4F).
At the subcellular level, TTL3-GFP mostly showed a cytoplasmic localization in the root meristematic cells (Figure 4G). However, we sometimes observed seedlings that, in addition to the cytoplasmic GFP localization, showed GFP signal at the plasma membrane. Quantification of the plasma membrane localization of TTL3-GFP (see Figure 4G legend and “Methods” for details) in control growth conditions indicated that in ∼30% of the seedlings some cells showed TTL3-GFP localization at the plasma membrane (Figure 4J). Treatment with 1 μM eBL, a concentration previously used to analyze short-term BRI1 KINASE INHIBITOR1 dynamics (Wang and Chory, 2006), increased the amount of TTL3-GFP protein (Supplemental Figures 12A and 12B) consistent with the increased expression of TTL3 by BRs. Interestingly, exogenous eBL treatment enhanced the relocalization of TTL3-GFP from the cytoplasm to the plasma membrane (Figures 4H to 4J; Supplemental Figures 13A and 13B). eBL treatment also caused the appearance of GFP-labeled intracellular structures (Figure 4H). These intracellular TTL3-GFP structures do not colocalize with FM4-64 (Supplemental Figure 13A), eliminating the possibility that they correspond to plasma membrane–derived endosomes; thus, their identity remains elusive.
Consistent with the possibility that the plasma membrane localization of TTL3-GFP in seedlings grown in control medium was caused by endogenous BRs, the percentage of seedlings with plasma membrane signal decreased from ∼30 to ∼5% after treatment with BRZ (Figure 4K). Further treatment of these seedlings with eBL reverted this effect and increased the plasma membrane localization of TTL3-GFP (Figure 4K).
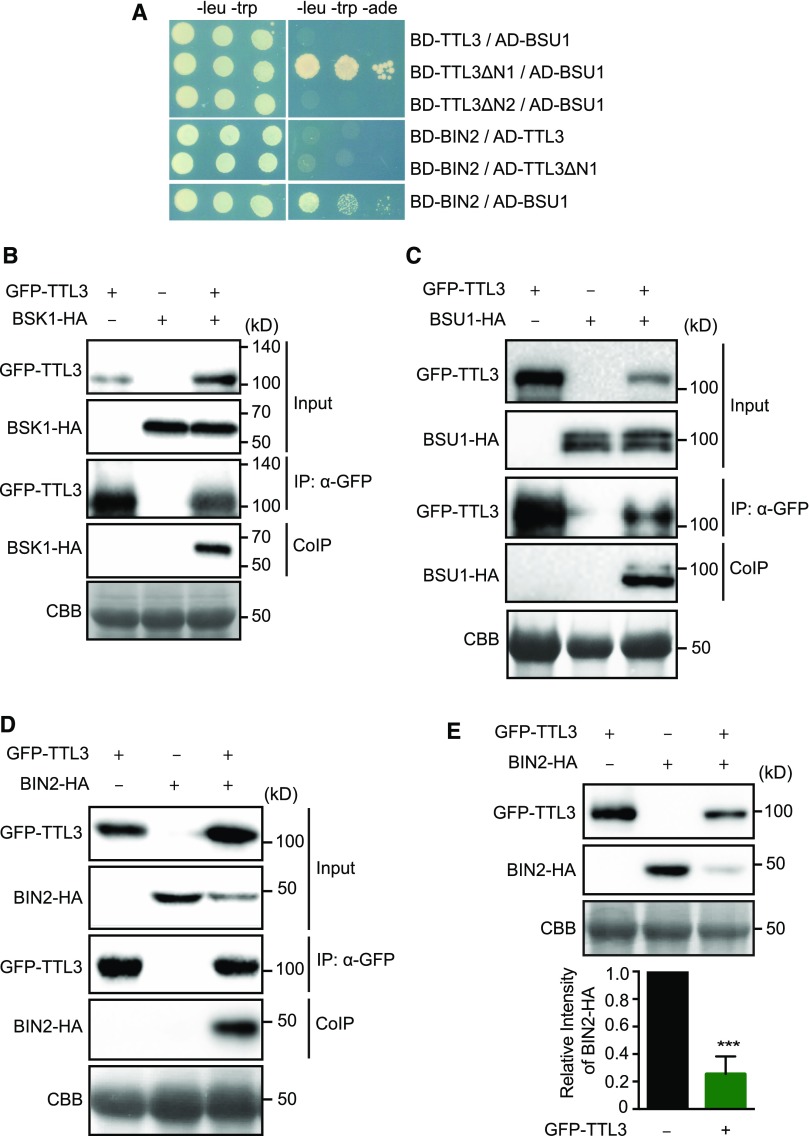
TTL3 Associates with the BR Signaling Components BSK1, BSU1, and BIN2 and Directly Interacts with BSU1
Our previous analyses indicate that TTL proteins are required for BR signaling. Considering the structure of these proteins and the interaction of TTL3 with BRI1, one possibility is that these TTL proteins function as a scaffold of additional BR signaling components. We first investigated possible direct interactions between TTL3 and the cytoplasmic BR signaling components BSK1, BSU1, and BIN2, using yeast two-hybrid assays. Using a full-length TTL3 protein, we did not identify interactions with any of the investigated BR components (Figure 5A). However, immunoblot analysis indicated that BD-TTL3 fusion protein was not produced (Supplemental Figure 14), similar to what previously occurred in E. coli. Therefore, we generated additional yeast two-hybrid constructs using the TTL3ΔN1 and TTL3ΔN2 fragments (Figure 1C). As shown in Figure 5A, TTL3ΔN1, but not TTL3ΔN2, interacted with BSU1, indicating that the six TPR domains are required for the interaction. In contrast to BSU1, BIN2 and BSK1 did not interact with TTL3ΔN1 (Figure 5A). As a control, we used the positive interaction of BIN2 with BSU1 as described previously (Kim et al., 2009).
Figure 5.
TTL3 Associates with BSK1 and BIN2 and Directly Interacts with BSU1.
(A) Yeast-two-hybrid assays to determine the interaction of full-length TTL3, the TTL3 fragment TTL3ΔN1 (amino acids 204 to 691), and the TTL3 fragment TTL3ΔN2 (amino acids 371 to 691) with BIN2 and BSU1. Growth on plasmid-selective media (left column) and interaction-selective media (lacking adenine [-ade], right column) are shown.
(B) BSK1 co-immunoprecipitates with TTL3. BSK1-HA, and GFP-TTL3 were transiently expressed in N. benthamiana. GFP-TTL3 was IP with anti-GFP Trap beads. Total (input), IP, and CoIP proteins were analyzed by immunoblotting. Equal loading was confirmed by Coomassie blue staining (CBB) of input samples. GFP-TTL3 and BSK1-HA were detected with anti-GFP and anti-HA antibody, respectively.
(C) BSU1 co-immunoprecipitates with TTL3. GFP-TTL3 and BSU-HA proteins were transiently expressed in N. benthamiana, IP, and analyzed as described in (B). GFP-TTL3 and BSU1-HA were detected with anti-GFP and anti-HA antibodies, respectively.
(D) BIN2 co-immunoprecipitates with TTL3. BIN2-HA and GFP-TTL3 proteins were expressed in N. benthamiana, IP, and analyzed as described in (B). GFP-TTL3 and BSU1-HA were detected with anti-GFP and anti-HA, respectively.
(E) TTL3 promotes BIN2 depletion. BIN2-HA with and without GFP-TTL3 was expressed in N. benthamiana. Protein extracts were analyzed by immunoblotting. Equal loading was confirmed by Coomassie blue staining (CBB) of input samples. GFP-TTL3 and BIN2-HA were detected with anti-GFP and anti-HA antibody, respectively. Bottom graph represents the signal density of BIN2-HA coexpressed with or without GFP-TTL3 in N. benthamiana was quantified based on the six biological repeats. The immunoblot signal intensity of BIN2-HA coexpressed with GFP-TTL3 was normalized to the immunoblot signal intensity of BIN2-HA coexpressed with an empty vector. Asterisks indicate statistical differences as determined by the unpaired t test (***P ≤ 0.001).
Next, we used Co-IP and BiFC in N. benthamiana to investigate possible associations of TTL3 with BSK1, BSU1, and BIN2. TTL3 strongly associates with BSK1 in both Co-IP (Figure 5B) and BiFC assays (Supplemental Figure 6A). BiFC between TTL3 and BSK1 was also obtained when we exchanged nYFP and cYFP tags (Supplemental Figure 6B) and, consistent with the plasma membrane localization of BSK1, the BiFC signal for BSK1-TTL3 was observed at the plasma membrane. TTL3 also associated with BSU1 and BIN2 in both Co-IP and BiFC assays (Figures 5C and 5D; Supplemental Figures 6A and 6B). Although BSU1 and BIN2 present a dual nuclear and cytoplasmic localization (Vert and Chory, 2006; Maselli et al., 2014), BiFC signals were only observed in the cytoplasm for both TTL3-BSU1 and TTL3-BIN2, which is consistent with the lack of TTL3 protein in the nucleus. A cytoplasmic BiFC signal was also obtained when YFP halves were interchanged among TTL3-BSU1 and TTL3-BIN2 (Supplemental Figure 2B). Interestingly, we consistently observed that when TTL3 was coexpressed with BIN2, there was a depletion of BIN2 protein amount in the total protein extract (Figure 5D, input). This was further quantified using six biological replicates, confirming that upon TTL3 expression the levels of BIN2 were reduced by ∼80% (Figure 5E). This was unique for BIN2 since we did not find the same effect with any other protein of the BR pathway (Figures 1E, 5B, and 5C) and suggests that TTL3 might function, at least in part, in BR signaling, by negatively regulating the amount of BIN2.
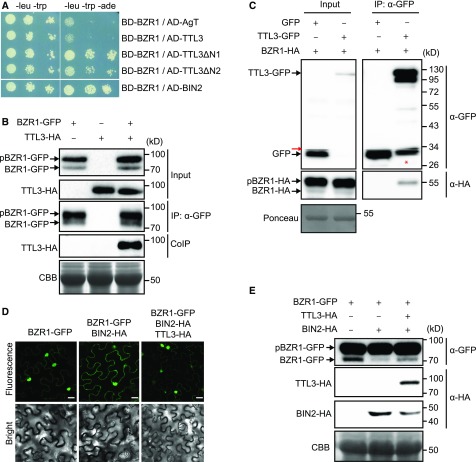
TTL3 Interacts with the Transcription Factors BZR1 and Affects Its Cytoplasmic/Nuclear Localization
In the absence of BRs, BIN2 phosphorylates and inactivates BZR1 and BES1, the two major transcription factors mediating BR-induced transcriptional changes (Belkhadir and Jaillais, 2015). We performed a yeast two-hybrid assay between TTL3 and the transcription factor BZR1 (Figure 6A). As a positive control (Figure 6A), we used the reported interaction between BZR1 with BIN2 (He et al., 2002). As expected, based on the lack of accumulation of the full-length TTL3 protein in yeast (Supplemental Figure 14), no interaction with BZR1 was obtained (Figure 6A). However, we did detect an interaction between TTL3ΔN1 and TTL3ΔN2 (Figure 1C) and BZR1 (Figure 6A), indicating that the TPR3-to-TPR6 region of TTL3 is sufficient for its interaction with BZR1.
Figure 6.
TTL3 Interacts with BZR1 and Regulates Its Cytoplasmic/Nuclear Localization.
(A) Yeast-two-hybrid assays to determine the interaction of BZR1 with TTL3, the TTL3 fragment TTL3ΔN1 (amino acids 204 to 691), the TTL3 fragment TTL3ΔN2 (amino acids 371 to 691), and BIN2. Interaction of BZR1 with a fragment of Simian virus 40 large T-antigen (AD-AgT) was also included to show BD-BZR1 self-activation capacity. Growth on plasmid-selective media (left column) and interaction-selective media (lacking adenine, -ade], right column) are shown.
(B) TTL3 co-immunoprecipitates with BZR1. TTL3-HA and BZR1-GFP were transiently expressed in N. benthamiana. BZR1-GFP was IP with anti-GFP Trap beads. Total (input), IP, and CoIP proteins were analyzed by immunoblotting. Equal loading was confirmed by Coomassie blue staining (CBB) of input samples. BZR1-GFP and TTL3-HA were detected with anti-GFP and anti-HA, respectively. The top band corresponds to phosphorylated BZR1 (pBZR1-GFP) and the bottom band to dephosphorylated BZR1 (BZR1-GFP).
(C) Co-IP of BZR1-HA with TTL3-GFP expressed in transfected Arabidopsis Col-0 protoplasts. Samples were analyzed as in (B). Protoplasts cotransfected with free GFP and BRI1-HA, were used as a negative control for Co-IP. Equal loading was confirmed by Ponceau staining of input samples. TTL3-GFP and free GFP were detected with anti-GFP antibody and BRI1-HA was detected with anti-HA antibody. Asterisk indicates GFP that results from proteolytic cleavage of TTL3-GFP. Red arrow indicates an artifact from imaging the blot with high sensitivity using an Azure c300 Chemiluminescent Western Blot Imaging System.
(D) TTL3 abolishes the cytoplasmic retention of BZR1 by BIN2. Subcellular localization of BZR1-GFP alone, coexpressed with BIN2-HA, and with BIN2-HA and TTL3-HA in N. benthamiana leaves. Images of the GFP signal were obtained using laser scanning confocal microscopy. Images show a single equatorial plane in N. benthamiana leaves. Bar = 20 μm. The experiment was repeated three times with similar results.
(E) Immunoblot analysis of the BZR1-GFP proteins transiently expressed alone, coexpressed with BIN2-HA, and coexpressed with BIN2-HA and TTL3-HA in N. benthamiana leaves observed by confocal microscopy in (D). Proteins were analyzed by immunoblotting. Equal loading was confirmed by Coomassie blue staining (CBB) of input samples. BZR1-GFP was detected with anti-GFP antibody, while TTL3-HA and BIN2-HA were detected with anti-HA antibody. In the anti-GFP blot, the top band corresponds to phosphorylated BZR1 (pBZR1-GFP) and the bottom band to dephosphorylated BZR1 (BZR1-GFP).
TTL3 also associates with BZR1 in Co-IP experiments in N. benthamiana (Figure 6B) and in Arabidopsis mesophyll protoplasts (Figure 6C). Phosphorylated and dephosphorylated BZR1 proteins show a marked difference in mobility in SDS-PAGE upon expression in N. benthamiana (Figure 6B; Supplemental Figure 15A) and in Arabidopsis protoplasts (Figure 6C; Gampala et al., 2007; Ryu et al., 2007). Interestingly, only the pBZR1 was CoIP with TTL3 (Figure 6C; Supplemental Figure 15A), suggesting a preferential association of TTL3 with pBZR1. BiFC assays further confirmed the in vivo association of BZR1 with TTL3 (Supplemental Figures 6A and 6B). We also observed that while the BiFC signal of TTL3 with plasma membrane BR components results in a smooth YFP fluorescence signal, the BiFC signal of TTL3 with the cytoplasmic components appears punctate (Supplemental Figures 6A and 6B). A similar punctate BiFC signal has been reported previously for BZR1 with BRZ-SENSITIVE-SHORT HYPOCOTYL1 (Shimada et al., 2015) or BES1 with DOMINANT SUPPRESSOR OF KAR2 (Nolan et al., 2017), although its significance remains unknown.
We then analyzed the effect of TTL3 on the nuclear and cytoplasmic localization of BZR1-GFP. As reported previously, BZR1-GFP in N. benthamiana is mainly localized in the nucleus (Figure 6D), while coexpression of BIN2 together with BZR1-GFP promotes its phosphorylation and its cytoplasmic retention (Figure 6D; Kim et al., 2009). Coexpressing TTL3-HA with BZR1-GFP and BIN2-HA suppressed the cytoplasmic retention of BZR1-GFP promoted by BIN2 (Figure 6D). We also used Arabidopsis plants expressing the salicylate hydroxylase (NahG) gene, as these plants are efficiently transiently transformed using A. tumefaciens (Rosas-Díaz et al., 2016). Similar to N. benthamiana, coexpressing BIN2-HA together with BZR1-GFP increased its cytoplasmic accumulation, which was further abolished by TTL3-HA (Supplemental Figure 15B). This BZR1 nuclear/cytoplasmic localization is associated with the dephosphorylation status of BZR1 (Figure 6E), indicating that TTL3 negatively regulates BIN2 phosphorylation of BZR1 and regulates its activity, likely by promoting BIN2 depletion (Figure 5E).
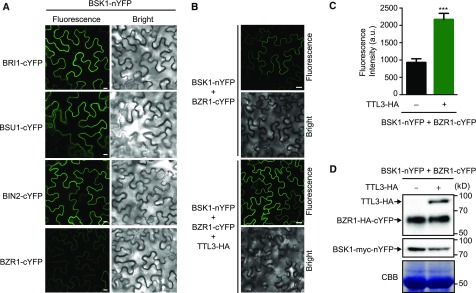
TTL3 Reinforces BSK1-BZR1 Association at the Plasma Membrane
The search for BZR1 interacting proteins in Arabidopsis through tandem affinity purification allowed the identification of BSK1 as a BZR1 interactor (Wang et al., 2013), a result that we confirmed using Co-IP (Supplemental Figure 16). This led the authors to suggest that BR signaling components exist in a multiprotein complex at the plasma membrane. The formation of a multiprotein complex for BR components at the plasma membrane has been recently proposed, with BSK3 (a protein that contain TPR domains) acting as a possible scaffold (Ren et al., 2019). Therefore, we aimed to investigate whether TTL3 could function as an additional scaffold in BR signaling. For this, we determined whether TTL3 expression affects the association at the plasma membrane of BSK1 with cytoplasmic BR components using BiFC. As shown in Figure 7A, a strong BiFC signal was obtained for BSK1 with BRI1, BSU1, and BIN2, while a weak signal was obtained with BZR1. The strong BiFC signal detected for BSK1 with BRI1 and with BSU1 is expected, since these BR signaling components directly interact with BSK1 (Kim et al., 2009). BIN2, although mainly localizing at the nucleus and cytosol, also localizes at the plasma membrane (Vert and Chory, 2006) and directly interacts with several plasma membrane–localized BSKs (Sreeramulu et al., 2013; Ren et al., 2019). Importantly, the BSK1-BZR1 BiFC signal was strongly enhanced when we coexpressed with TTL3-HA (Figures 7B and 7C), indicating that TTL3 increases the association between BSK1 and BZR1 at the plasma membrane. Further immunoblot analysis confirmed that the increase in BSK1-BZR1 BiFC fluorescence was not due to a differential expression of the BiFC proteins caused by TTL3 expression (Figure 7D).
Figure 7.
Coexpression of TTL3 Enhances pBZR1-BSK1 Interaction.
(A) BiFC shows strong association of BSK1 with BRI1, BSU1, and BIN2 and weak association with BZR1. N. benthamiana leaves were co-agroinfiltrated with the Agrobacterium strains harboring a construct to express the BSK1 protein fused to the N-terminal half of YFP and the BRI1, BSU1, BIN2, or BZR1 proteins fused to the C-terminal half of YFP and observed under a laser scanning confocal microscope. Strong fluorescence signals are observed when BSK1-nYFP is coexpressed with BRI1-cYFP, BSU1-cYFP, or BIN2-cYFP. A faint YFP signal is observed when BSK1-nYFP is coexpressed with BZR1-cYFP. From left to right columns, images show BiFC YFP fluorescence in green and bright-field. Bar = 20 µm. The experiment was repeated two times with similar results.
(B) Expression of TTL3 increases the weak BiFC association of BSK1 and BZR1. N. benthamiana leaves were co-agroinfiltrated with the Agrobacterium strains harboring the corresponding constructs to express the BSK1 protein fused to the N-terminal half of YFP and the BZR1 protein fused to the C-terminal half of YFP. N. benthamiana leaves were pre-treated with 5 µM eBL for 3 h before confocal imaging analysis. Coexpression of TTL3-HA together with BSK1-nYFP and BZR1-cYFP highly enhances the GFP signal. From left to right columns, BiFC YFP fluorescence in green and bright field. Bar = 20 µm. The experiment was repeated three times with similar results.
(C) Quantification of the BiFC fluorescence intensity of BSK1 and BZR1 in the presence or absence of TTL3-HA described in (B). Asterisks indicate statistical differences between BiFC fluorescence intensity of BSK1 and BZR1 in the absence or presence of TTL3-HA determined by the unpaired t test (***P ≤ 0.001). Data represent mean values, error bars are sem, and n = 5 randomly chosen regions of infiltrated leaves. The experiment was repeated three times with similar results. a.u., arbitrary units.
(D) Immunoblot analysis reveals similar amounts of BSK1-nYFP and BZR1-cYFP when coexpressed with or without TTL3-HA. Proteins were transiently expressed as described in (B). Equal loading was confirmed by Coomassie blue staining (CBB) of total proteins. BSK1-nYFP contains a myc tag (BSK1-myc-nYFP) and was detected using anti-myc antibody, while BZR1-cYFP contains a HA tag (BZR1-HA-cYFP) and was detected using an anti-HA antibody. TTL3-HA was also detected with an anti-HA antibody. The experiment was repeated three times with similar results.
DISCUSSION
The expression of TTL genes is induced by BRs and TTL3 shows its highest expression at the root elongation zone and at the hypocotyl, areas of high BR activity (González-García et al., 2011; Bernardo-García et al., 2014). Individual ttl1, ttl3, and ttl4, and particularly the triple ttl134 mutants, are hyposensitive to BR in root growth assays and show reduced hypocotyl elongation. Further lines of genetic evidence supporting the function of TTL genes in BR signaling come from phenotypic analyses of the quadruple mutants of ttl genes with either bri1-301 or bes1-D. At the molecular level, ttl134 shows increased expression of BR-repressed genes, whereas BR-induced dephosphorylation of the transcription factor BES1 is strongly reduced. At the cellular level, a functional TTL3-GFP shows a dual localization in the cytoplasm and plasma membrane in untreated seedlings. Treatment with eBL caused TTL3-GFP relocalization from the cytoplasm to the plasma membrane, while treatment with a BR biosynthesis inhibitor had the opposite effect. Furthermore, coexpression of TTL3 together with BZR1 and BIN2 abolishes the BIN2-dependent BZR1 cytoplasmic retention in Arabidopsis and N. benthamiana (Gampala et al., 2007; Ryu et al., 2007; Kim et al., 2009; Tang et al., 2011). Overall, this study reveals that plant-specific TTL proteins function as positive regulators of BR signaling.
TTL proteins contain several defined domains involved in protein–protein interactions and assembly of multiprotein complexes. Consistent with this structure, TTL3 protein associates in vivo with all core BR signaling components, with the exception of BAK1, and interacts directly with BRI1, BSU1, and BZR1. Mapping the interaction domains of TTL3 with BRI1 indicates that the last four TPRs are essential for this interaction and that both TRLX and the IDR contribute to strengthen the interaction. The presence of an IDR in TTL proteins can provide additional advantages in their scaffolding and regulatory function since IDRs allow their interaction with a large number of partners due to their ability to adopt different conformations, thus allowing the assembly of multiple proteins (Soutourina, 2018). We also found that interaction of TTL3 with BSU1 requires all six TPRs, while only the last four TPR domains are required for the interaction with BZR1.
How does TTL function mechanistically in BR signaling? The BR-dependent plasma membrane relocalization of TTL3, probably through interaction with phosphorylated BRI1 and the association with other BR components, suggests that these proteins function as a scaffold by bringing them together at the plasma membrane. This is supported by the finding that a weak BZR1 and BSK1 association at the plasma membrane is greatly enhanced by TTL3 overexpression. Our data indicate that TTL3 promotes dephosphorylation and nuclear localization of BZR1, while TTL3 increased BiFC interaction of BSK1-BZR1 at the plasma membrane. These seemingly contradictory results can be explained by the irreversible assembly of the two YFP halves, an intrinsic feature of BiFC. It is possible that the stability of the BiFC complex formed between BSK1 and BZR1 facilitates the visualization of this likely transient interaction but will also hinder the dynamics of this protein complex formation (Kudla and Bock, 2016).
Although in current models of BR signaling phosphorylation/dephosphorylation of transcription factors take place exclusively in the cytoplasm and the nucleus (Wang et al., 2012; Belkhadir and Jaillais, 2015), a survey of the literature provides evidence that the plasma membrane could be an active site of BR signaling, from perception of the hormone to dephosphorylation of the transcription factors: (1) A significant amount of phosphorylated BZR1 located at the plasma membrane is greatly reduced upon BR treatment (Gampala et al., 2007). (2) BSK1 has been identified as an interactor of BZR1 using nontargeted proteomics, which led the authors to propose that BR signaling components exist in the plasma membrane as a multiprotein complex (Wang et al., 2013). (3) Several BSKs that are plasma membrane bound interact with BIN2, suggesting that dephosphorylation of BZR1 and BIN2 is also taking place at the plasma membrane (Sreeramulu et al., 2013; Maselli et al., 2014). In a recent report, BSK3 has been shown to directly interact with BRI1, BSU1, and BIN2 (Ren et al., 2019) and is proposed to function as a scaffold that positively regulates BR signaling by locating BSU1 and BIN2 at the plasma membrane and enhancing their interaction.
The basic function of scaffolding proteins is the assembly of signaling components that enhance the efficiency of the signaling cascade by increasing their local concentrations as well as the localization of the signaling reaction to a specific area of the cell. This could be particularly important in BR signal components because some of these proteins are expressed at vanishingly low levels, like BSU1 and BIN2 (Mora-García et al., 2004; Peng et al., 2008). This scaffolding function of TTL proteins might also have a role in enhancing signaling specificity by preventing spurious interactions of BR signaling components. This is important because some BR signaling components participate in pathways other than BR. For example, BAK1 and related SERK coreceptors are involved in numerous responses (Ma et al., 2016) in addition to their role in BR signaling, and BIN2 shows multiple targets that result in different signaling outcomes (Kim et al., 2012; Cai et al., 2014). The importance of scaffold proteins to generate signaling specificity of BIN2 (and related GSK3-line kinases) was recently highlighted by the identification of the plant-specific protein POLAR (Houbaert et al., 2018). POLAR acts in concert with BREAKING OF ASYMMETRY IN THE STOMATAL LINEAGE as a stomatal lineage scaffold for GSK3-like kinases that confines them to specific subcellular localizations during stomatal differentiation (Houbaert et al., 2018). Another example is BSK1, originally identified in BR signaling by proteomic studies (Tang et al., 2008) but later found also to regulate immunity (Shi et al., 2013). Because TTL1, TTL3, and TTL4 were previously reported to play a role in abiotic stress tolerance and there is increasing evidence for the coordination of BR-promoted growth and abiotic stress responses (Zhang et al., 2016; Nolan et al., 2017; Tian et al., 2018), we cannot exclude that the function of TTL3 (and probably other TTLs) as a scaffold of BR signaling components contribute to this crosstalk.
Our work uncovers TTL proteins as components of BR signaling and places the BR pathway in a spatial context. BSK3 kinase has been show to function in early BR signaling by acting as a scaffold at the plasma membrane that mediates the assembly of BR components (Ren et al., 2019). Further characterization of these scaffold proteins will reveal how components of the BR cascade are spatially assembled so that their localization and local concentration are controlled for optimal signaling.
METHODS
Plant Material
All Arabidopsis (Arabidopsis thaliana) plants generated in this study are of the Col-0 ecotype. Arabidopsis mutants lines used in this study have been described previously: ttl1 (AT1G53300) Salk_063943; ttl2 (AT3G14950) Salk_106516; ttl3 (AT2G42580) Sail_193_B05; ttl4 (AT3G58620) Salk_026396; ttl134: ttl1 ttl3 ttl4 triple mutant (Lakhssassi et al., 2012); bak1-4 (SALK_116202; Chinchilla et al., 2007); serk1-1 (SALK_044330; Albrecht et al., 2005); serk1-1 bak1-4 double mutant (obtained by crossing serk1-1 with bak1-4); bri1-301 (Xu et al., 2008); ttl134 bri1-301 quadruple mutant (obtained by crossing ttl134 with bri1-301); bes1-D (Yin et al., 2002); and ttl134 bes1-D quadruple mutant (obtained by crossing ttl134 with bes1-D). Transgenic lines TTL1p:GUS; TTL2p:GUS; TTL3p:GUS, and TTL4p:GUS (Lakhssassi et al., 2012) were also described previously. Generation of transgenic lines TTL3-GFP 1.2 (TTL3p:TTL3g-GFP line 1.2 in ttl3 background) and TTL3-GFP 2.4 (TTL3p:TTL3g-GFP line 2.4 in ttl134 background) is described in the section “Generation of Transgenic Plants.”
Plant Manipulation and Growth Conditions
Arabidopsis standard handling procedures and conditions were used to promote seed germination and growth. Seeds were surface sterilized and cold treated for 3 d at 4°C. Next, seeds were sowed onto half-strength Murashige and Skoog (MS) agar solidified medium (0.6% [w/v] agar for horizontal growth and 1% [w/v] for vertical growth) containing 1.5% [w/v] Suc, unless otherwise stated. Plates were placed either vertically or horizontally in a culture chamber at 22 ± 1°C, under cool-white light (at 120 µmol photon m−2 s−1) with a long-day photoperiod (at 16-h light/8-h dark cycle) unless otherwise stated. When required, seedlings were transferred to soil after 7 d of in vitro growth and watered every 2 d. In soil, plants were grown in a mixture of organic substrate and vermiculite (4:1 [v/v]) under controlled conditions at 23 ± 1°C, 16-h light/8-h dark cycle (at ∼120 µmol photon m−2 s−1). Freshly harvested seeds were used for all phenotypic analyses.
Plasmid Constructs
A genomic fragment spanning the 1.7-kb TTL3 promoter (TTL3p) region upstream of the start codon and the TTL3 genomic region (TTL3g) without stop codon was PCR amplified using the primers detailed in Supplemental Table 1 and cloned into the pCR8 ENTRY vector (Invitrogen).
The coding DNA sequence (CDS) without the stop codon of TTL3 and BSK1, as well as the CDS with the stop codon of the wild-type BRI1 cytoplasmic domain (residues 814 to 1196), BRI1 cytoplasmic domain JMCT9D (BRI1cytJMCT9D residues 250 to 662), and TTL3 truncated version TTL3ΔN1 (residues 204 to 691), TTL3ΔN2 (residues 371 to 691), TTL3ΔN3 (residues 567 to 691), TTL3ΔC1 (residues 1 to 306), and TTL3ΔC2 (residues 1 to 595) was PCR amplified using the primers detailed in Supplemental Table 1 and cloned into the pENTR/D-TOPO vector using the pENTR Directional TOPO cloning kit (Invitrogen). The pUNI51 (Salk Institute) cDNA clone was used as template to PCR amplify the TTL3 CDS without the stop codon. Total RNA from Arabidopsis Col-0 was used to generate cDNA that was then used as a template to PCR amplify BSK1 CDS without the stop codon. The expression clone pMAC-flag-BRI1-CD-JMCT9D (Wang et al., 2008) was used as template for PCR amplification of BRI1cytJMCT9D, and it was a gift from Xiaofeng Wang (College of Horticulture Northwest, AandF University, Yangling Shaanxi).
pENTR vectors including CDS without the stop codon of BRI1, BAK1, BIN2, BSU1, and BZR1 were obtained by Gateway BP-reaction (Invitrogen) using an expression clone for each gene of interest (containing attB sites) and the pDONR/Zeo vector. Expression clones, used as templates for cloning BRI1, BAK1, and BZR1 in the pENTR/D-Topo by Gateway BP-reaction, were published previously (Schwessinger et al., 2011; Lozano-Durán et al., 2014). The expression clones used to clone BSU1 (Mora-García et al., 2004) and BIN2 (Bernardo-García et al., 2014) in pENTR/D-Topo by the Gateway BP-reaction were a gift from Santiago Mora Garcia (Fundación Instituto Leloir and Instituto de Investigaciones Bioquímicas de Buenos Aires) and Salomé Prat (Centro Nacional de Biotecnología-Consejo Superior de Investigaciones Cientificas), respectively.
All the resulting pENTR clones were verified by diagnostic PCR, restriction analysis, and sequencing. These pENTR clones in combination with the appropriate destination vectors (pDEST) were used to create the final Gateway-expression constructs by LR-reaction (Invitrogen). The pETG-30A and pETG-30A vectors were provided by the European Molecular Biology Laboratory (EMBL) and were used as pDEST to generate GST and MBP N-terminal fusion proteins for GST-pull-down assays. The pGWB4, pGWB5, pGWB6, and pGWB14, from the pGWB vector series, were provided by Tsuyoshi Nakagawa (Department of Molecular and Functional Genomics, Shimane University; Nakagawa et al., 2007) and were used as pDEST for the transient expression in Nicotiana Benthamiana in the Co-IP and coexpression assays (pGWB5, pGWB6, and pGWB14) or for generating stable transgenic Arabidopsis lines (pGWB4). pDEST-GW-VYNE and pDEST-GW-VYCE (Gehl et al., 2009) were used for BiFC assays. The Gateway destination vector pUC19(35S:GW-GFP) and pBSSK(35S:GW-HA) were used to transfect protoplasts for transient expression and Co-IP assays. The pUC19(35S:GW-GFP) vector was provided by José Alonso (Department of Plant and Microbial Biology, North Carolina State University) and contains the pGWB5 cassette between HindIII-SacI restriction sites in the pUC19 vector backbone. The pBSSK(35S:GW-HA) vector was generated in this work by cloning the pGWB14 cassette between HindIII-SacI in the pBSSK vector backbone. The pGADT7(GW) and pGBKT7(GW) destination vectors were provided by Salomé Prat (Nacional de Biotecnología-Consejo Superior de Investigaciones Cientificas) and used for yeast two-hybrid assay. All the expression clones were verified by diagnostic PCR and restriction analysis.
Generation of Transgenic Plants
Expression constructs were transformed into Agrobacterium tumefaciens strain GVG3101::pMP90 through electroporation and confirmed by diagnostic PCR. The pGWB4 harboring the TTL3p:TTL3g-GFP construct was transformed into Arabidopsis plants by floral dip (Clough and Bent, 1998) to generate stable transgenic plants. TTL3p:TTL3g-GFP was transformed into both the ttl3 single mutant and the ttl134 triple mutant. T3 or T4 homozygous transgenic plants were used in this study.
Venation Pattern Phenotype
Cotyledons (embryonic leaves) from 2-week-old seedlings were cleared and observed under a Nikon AZ100 Multizoom light microscope to analyze vascular patterning, and the percentage of cotyledons displaying each venation pattern categories is depicted in Supplemental Figure 1. Approximately 200 cotyledons per genotype were analyzed. Representative images of each observed venation pattern categories were acquired.
For clearing cotyledons, the 2-week-old seedlings were immersed sequentially in 50% ethanol for 1 h, 99% ethanol overnight, and 50% ethanol for 1 h, and finally transferred to double distilled water. Seedlings were mounted on slides in 50% glycerol and visualized under a light microscope or using the Nikon AZ100 Multizoom microscope system.
Morphological Characterization of Seedlings
Seedlings were grown vertically in long-day photoperiod for 3 d (root analysis) or 7 d (hypocotyl analysis) in half-strength MS agar solidified medium supplemented with 1.5% (w/v) Suc. The root and hypocotyl length of seedlings grown vertically was measured, and the data were analyzed as described in under “Quantification and Statistical Analysis.”
eBL Sensitivity Determined by Root Growth Inhibition
Two different BR-inhibition root growth assays were performed to measure BR sensitivity. In the first assay, seedlings were grown vertically in a long-day photoperiod for 4 or 5 d in half-strength MS agar solidified medium supplemented with 1.5% (w/v) Suc and then transferred to half-strength MS agar solidified medium supplemented with 1.5% (w/v) Suc containing either mock (eBL solvent as control) or 100 nM eBL (PhytoTechnology Laboratories) and photographed 6 or 8 d later. In the second assay, seedling were grown vertically in a long-day photoperiod for 7 d in half-strength MS agar solidified medium supplemented with 1.5% (w/v) Suc, containing either mock (eBL solvent as control) or 500 nM eBL (PhytoTechnology Laboratories) and photographed. Root length of seedlings grown for 7 d in the presence of 500 nM eBL was divided by the mean of root length of seedlings grown for 7 d in the absence of BL to calculate the root length fold change.
The eBL (PhytoTechnology Laboratories) was added from a 5 mM stock solution freshly prepared in 80% (v/v) ethanol.
To determine the eBL sensitivity of Col-0 and mutants, the root length of seedlings grown vertically was measured, and the data were analyzed as described under “Quantification and Statistical Analysis.”
eBL Sensitivity Determined by Phosphorylation Status of BES1
Seedlings were grown vertically in a long-day photoperiod for 7 d in half- strength MS agar solidified medium supplemented with 1.5% (w/v) Suc and then transferred to half-strength MS liquid medium supplemented with 1.5% (w/v) Suc containing 2.5 µM BRZ (TCI Europe) and grown for three more days. To determine the eBL sensitivity of Col-0 and ttl134, the seedlings were treated with either mock (eBL solvent as control) or 10 nM eBL (PhytoTechnology Laboratories) and frozen in liquid nitrogen after 0, 30, and 60 min of treatment. Total protein was extracted as described under “Extraction of Total Protein from Arabidopsis” and analyzed by immunoblotting using an anti-BES1 antibody (dilution 1:500; Yu et al., 2011) as described under “Immunoblot.”
Phenotypic Analysis of Hypocotyl Elongation in Dark
Freshly harvested seeds were surface sterilized and cold treated for 3 d at 4°C. Next, seeds were sowed individually onto half-strength MS 1% (p/v) agar solidified medium containing 1.5% Suc for vertical growth. Seedlings were grown for 4 d in long-day photoperiod and then placed in dark conditions (vertical growth in a culture chamber at 22 ± 1°C). Seedlings were photographed, and hypocotyl length was measured 3 d after placing plates in dark conditions.
Total RNA Extraction and Quantitative PCR Analysis
Ten-day-old seedlings (10 seedlings per biological replicate) grown for 5 d on half-strength MS agar solidified medium were transferred to half-strength MS liquid medium supplemented with 1% (w/v) Suc (grown for five extra days) and were used for total RNA extraction. Plant tissue was ground to a fine powder in liquid nitrogen. Approximately 100 mg of ground tissue per sample was homogenized in 1 mL of the commercial reagent TRIsure (Bioline), and total RNA was extracted following the manufacturer’s instructions. The RNA concentration and purity were determined spectrophotometrically (Nanodrop ND-1000 spectrophotometer). RNA samples (10 µg per sample) were DNase treated with Turbo DNA-free DNase (Ambion), and 1 µg of RNA per sample was run on a 1% agarose gel to confirm RNA integrity. First-strand cDNA was synthesized from 1 μg of RNA using the iScript cDNA synthesis kit (Bio-Rad), according to the manufacturer’s instructions. cDNAs were amplified in triplicate by quantitative PCR (qPCR) using SsoFast EvaGreen supermix (Bio-Rad) and the MyiQ Thermal cycler (Bio-Rad). The relative expression values were determined using ACTINE2 as a reference gene and plotted relative to Col-0 expression level. Primers used for qPCR are listed in Supplemental Table 2.
Transient Expression in N. benthamiana
For transient expression in N. benthamiana, A. tumefaciens (GV3101::pMP90) carrying the different constructs was used together with the p19 strain for infiltration into 4- to 5-week-old N. benthamiana leaves at the abaxial side of the leaf lamina. After infiltration, all plants were kept in the greenhouse and analyzed 2 d later. Agrobacteria cultures were grown overnight in Luria-Bertani medium containing rifampicin (50 µg/mL), gentamycin (25 µg/mL), and the construct-specific antibiotic. Cells were then harvested by centrifugation (for 15 min at 3000g in 50-mL falcon tubes) at room temperature, pellets were resuspended in agroinfiltration solution (10 mM MES, pH 5.6, 10 mM MgCl2, and 1 mM acetosyringone), and incubated for 2 h in dark conditions at room temperature. For double infiltration experiments, Agrobacterium strains were infiltrated at OD600 of 0.4 for the constructs and 0.2 for the p19 strain. For triple infiltration experiments, Agrobacterium strains were infiltrated at OD600 of 0.26 for the constructs and at OD600 of 0.2 for the p19 strain. An Agrobacterium strain harboring an empty vector (or β-glucuronidase [GUS]-HA expressing vector) was used to obtain a total OD600 of approximately 1 in all the infiltration experiments.
For eBL treatment analysis, leaves were pre-treated with 5 µM BL for 3 h prior to sample collection. N. benthamiana leaves were infiltrated with water or 5 µM eBL (PhytoTechnology Laboratories) infiltration solution (10 μL eBL 5mM stock solution in 10 mL of water), made from a 5 mM stock solution freshly prepared in 80% (v/v) ethanol.
Transient Expression in Arabidopsis NahG Plants
A. tumefaciens–mediated expression in Arabidopsis NahG plants (Rosas-Díaz et al., 2016) was performed as described for transient expression in N. benthamiana, with some modifications. Agrobacterium strains were resuspended with an equal OD600 in infiltration solution to obtain a total OD600 of 0.05 for injection into the abaxial leaf sides of 4- to 5-week-old Arabidopsis plants. At least six plants per co-infiltration mixture and four leaves per plant were used per experiment.
Recombinant Protein Purification and In Vitro Pull-Down Assay
The coding sequences of wild-type BRI1 cytoplasmatic domain (residues 814 to 1196), BRI1 cytoplasmatic domain JMCT9D (residues 250 to 662), TTL3ΔN1 (residues 204 to 691), and TTL3ΔN3 (residues 567 to 691) were cloned as described under “Plasmid Constructs” to generate MBP-BRI1cyt, MBP-BRI1cytJMCT9D, GST-TTL3ΔN1, and GST-TTL3ΔN3 constructs. Recombinant proteins were expressed in Escherichia coli strain BL21 (DE3) and extracted using buffer A (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, and 1% Triton X-100, pH 8, supplemented with 1 mM phenylmethylsulfonyl fluoride [PMSF], 0.2 µL/10 mL Benzonase Nuclease [Sigma-Aldrich], and 1 mg/mL Lisozyme). MBP and GST fusion proteins were purified with glutathione Sepharose 4B GST-tagged protein purification resin (GE Healthcare) or MBP binding protein coupled to agarose beads (MBP-Trap_A, Chromotek), respectively, according to the manufacturers’ instructions.
To investigate protein–protein interactions, the GST-tagged proteins were first captured by the glutathione agarose–coated beads and then incubated with the MBP-tagged proteins in dilution/wash buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10% glycerol, 10 mM EDTA, pH 8, 10 mM DTT, 0.5 mM PMSF, and 1% [v/v] P9599 protease inhibitor cocktail [Sigma-Aldrich]) at 4°C for 1 h in an end-over-end rocker. Protein–protein interaction complexes bound to the glutathione agarose–coated beads were pulled down, washed three times with the dilution/wash buffer and analyzed by immunoblot as described under “Immunoblot.”
Immunoblotted GST- and MBP-tagged protein was detected using an anti-GST antibody (catalog no. G7781, Sigma-Aldrich; dilution 1:10,000) and a specific anti-BRI1 antibody (dilution 1:2000; Bojar et al., 2014) as described in the section “Immunoblot.”
Protein Extraction and Co-IP in N. benthamiana
Protein extraction and Co-IP in N. benthamiana were performed as described previously (Kadota et al., 2016), with some modifications. Briefly, 4-week-old N. benthamiana plants were used for transient expression assays as described under “Transient Expression in N. benthamiana.” Leaves were ground to fine powder in liquid nitrogen. Approximated 0.5 g of ground leaves per sample was used, and total proteins were then extracted with extraction buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10% glycerol, 10 mM EDTA, pH 8, 1 mM NaF, 1 mM Na2MoO4·2H2O, 10 mM DTT, 0.5 mM PMSF, 1% [v/v] P9599 protease inhibitor cocktail [Sigma-Aldrich]); Nonidet P-40, CAS: 9036-19-5 [USB Amersham Life Science] 0.5% (v/v) for Co-IP involving transmembrane proteins BRI1 and BAK1, and 0.2% [v/v] for the rest of Co-IP) added at 2 mL/g powder using an end-over-end rocker for 30 min at 4°C. Samples were centrifuged 20 min at 4°C and 9000 rpm (9056g). Supernatants (∼4 mg/mL protein) were filtered by gravity through Poly-Prep chromatography columns (731-1550, Bio-Rad), and 100 μL was reserved for immunoblot analysis as input. The remaining supernatants were incubated for 2 h at 4°C with 15 μL of GFP-Trap coupled to agarose beads (Chromotek) in an end-over-end rocker. During incubation of protein samples with GFP-Trap beads, the final concentration of detergent (Nonidet P-40) was adjusted to 0.2% (v/v) in all cases to avoid unspecific binding to the matrix as recommended by the manufacturer. Following incubation, the beads were collected and washed four times with the wash buffer (similar to extraction buffer but without detergent). Finally, beads were resuspended in 75 μL of 2× concentrated Laemmli sample buffer and heated at 60°C for 30 min (for Co-IP involving transmembrane proteins BRI1 and BAK1) or at 70°C for 20 min (for the remaining Co-IPs) to dissociate immunocomplexes from the beads. Total (input), immunoprecipitated (IP), and CoIP proteins were separated in a 10% SDS-PAGE gel and analyzed as described in the section “Immunolot.”
BiFC Assays
Leaves were co-agroinfiltrated as described in the section “Agrobacterium-Mediated Transient Expression in N. benthamiana” with the Agrobacterium strain harboring a construct to express a given protein (Protein A) fused to the N-terminal half of YFP (Protein A-nYFP) and the BiFC partner protein (Protein B) fused to the C-terminal half of YFP (Protein B-cYFP), and vice versa (Protein A-cYFP and Protein B-nYFP) to test both BiFC directions. Leaves were observed using a confocal microscope 2 d after infiltration, as described in the section “Confocal Imaging of Arabidopsis and N. benthamiana.”
Confocal Imaging of Arabidopsis and N. benthamiana
Arabidopsis seedlings were germinated in half-strength MS agar solidified medium (1% [w/v] agar for vertical growth) supplemented with 1.5% Suc. For eBL treatment analysis, 4-d-old seedlings were incubated in 2 mL of half-strength MS medium supplemented with 1.5% (w/v) Suc containing either mock (eBL solvent as control) or 1 µM eBL (PhytoTechnology Laboratories). For BRZ/eBL treatment analysis, 3-d-old seedlings were incubated in 2 mL of half-strength MS medium supplemented with 1% (w/v) Suc, containing either mock (BRZ solvent as control) or 5 µM BRZ (TCI Europe) for 12 h (overnight). The next morning, samples were further treated with mock or 1 µM eBL (PhytoTechnology Laboratories) for another 1 h before being analyzed by confocal microcopy. The eBL (PhytoTechnology Laboratories) and BRZ (TCI Europe) were added from a 5 mM stock solution freshly prepared in 80% (v/v) ethanol. For visualization of plasma membrane, seedlings were incubated in 1 mL of double distilled water containing 1 µg/mL FM4-64 (Invitrogen Molecular Probes) prepared from a 1 mg/mL stock solution for 3 to 4 min, rinsed in double distilled water to remove the excess stain, and visualized under confocal microscopy.
For confocal imaging of N. benthamiana leaves in coexpression and BiFC experiments, GFP or YFP fluorescence of the lower epidermis of the leaf was visualized with the confocal 2 d after infiltration.
Confocal imaging of Arabidopsis NahG plants was performed as described for N. benthamiana, but in this case, images are a maximum Z-projection of seven 1-μm spaced confocal planes from the cell equatorial plane to the cell surface.
All confocal images were obtained using a Leica TCS SP5 II confocal microscope equipped with a 488-nm argon laser for GFP and YFP and a 561-nm He-Ne laser for FM4-64. Leica LAS AF Lite platform and the Java-based image-processing program FIJI (Schindelin et al., 2012; Schneider et al., 2012) were used in the processing of all microscopy images.
Stereo Microscopy of Arabidopsis Seedlings
Representative images of Arabidopsis seedlings were acquired using the Nikon Eclipse Ti basic fluorescence microscope system with a filter for GFP or ZEISS SteREO Discovery V12 with digital cam Axiocam 503 color (excitation wavelengths 488 and emission wavelengths 498 to 550). Wild-type Col-0 Arabidopsis seedlings were used as a negative control for chlorophyll fluorescence.
GUS Staining Assay
Four-day-old seedlings were transferred to a medium containing 0.2 µM eBL (PhytoTechnology Laboratories) for 24 h and then stained for GUS activity. Plant tissues were immersed in histochemical GUS staining buffer (100 mM NaPO4, pH 7, 0.5 mM K3[Fe(CN)6], 0.5 mM K4[Fe(CN)6], 20% methanol, 0.3% Triton X-100, and 2 mM 5-bromo-4-chloro-3-indoxyl-β-d-glucuronide cyclohexylammonium [Gold Biotechnology]) in multi-well plates, vacuum infiltrated (60 cm Hg) for 10 min three times, and then wrapped in aluminum foil and incubated at 37°C for 12 h. Samples were then washed several times with 95% ethanol until complete tissue clarification, stored in 50% glycerol, and photographed using the Nikon AZ100 Multizoom microscope system.
Protoplast Transient Expression Assays
Protoplast extraction and transfection were performed as described previously (Yoo et al., 2007). Briefly, leaves from 5-week-old Arabidopsis Col-0 plants grown under a 10-h daylight photoperiod were cut into very small strips and digested for 3 h in the darkness at room temperature. Protoplasts were then washed and resuspended to a concentration of 5 × 105 protoplasts/mL before polyethylene glycol–mediated transfection for 10 min. Twenty microliters of plasmid expressing GFP or 100 μL of plasmid expressing TTL3-GFP/BZR1-HA was used to transfect 2 mL of protoplasts for each transfection. All the plasmids were used at a concentration of 1 µg/µL. The transfected protoplasts were incubated for 6 h at room temperature and collected for protein extraction and immunoprecipitation, as described for N. benthamiana samples.
Yeast Two-Hybrid Assay
The Gal4-based yeast two-hybrid system (Clontech Laboratories) was used for testing the interaction between TTL3 and different components of the BR signaling pathway. The bait and prey constructs are explained under “Plasmid Constructs.” The bait and prey plasmids were transformed into Saccharomyces cerevisiae strain AH109 as described previously (Gietz and Schiestl, 1995), and transformants were grown on plasmid-selective media (synthetic defined (SD)/−Trp−Leu). Plates were incubated at 28°C for 4 d and independent colonies for each bait–prey combination were resuspended in 200 μL of sterile water. Tenfold serial dilutions were made and 5 μL of each dilution were spotted onto three alternative interaction-selective medium (SD/−Trp−Leu−His+3-AT (for 3-amino-1,2,4-triazole, 2mM), SD/−Trp−Leu−Ade, and SD/−Trp−Leu−Ade+3-AT). Plates were incubated at 28°C and photographed 3 or 7 d later.
Yeast Two-Hybrid Protein Extraction
For immunoblot analysis, one or two independent yeast cotransformants (a and b) for each bait–prey plasmid combination were grown in 50 mL of SD/−Leu−Trp to an OD600 of 0.7 to 1. Cultures were centrifuged at 4000 rpm for 3 min. The resulting pellet was washed once with cold water and resuspended in 200 μL of RIPA buffer (2 mM sodium phosphate buffer, pH 7, 0.2% Triton X-100, 0.02% [w/v] SDS, 0.2 mM EDTA, pH 8, and 10 mM NaCl) containing protease inhibitor (1 tablet/10 mL, cOmplete, Mini, EDTA-free Protease Inhibitor Cocktail, Roche). Glass beads (500 μL, 425 to 600 μm, Sigma-Aldrich) were added, and the sample was vortexed in FastPrepTM FP120 (BIO 101) at a power setting of 5.5 for two 15-s intervals separated by 1-min interval on ice. Next, 400 μL of RIPA buffer with protease inhibitors was added, and the sample was vigorously vortexed. The supernatant was recovered, and the protein concentration was determined using Bradford assays. Total protein (50 μg) was resolved on 10% polyacrylamide/SDS gels and analyzed by immunoblotting using an anti-Myc Tag (1:2000, Abgent), which is transcriptionally fused to Gal4BD, as described in the section “Immunoblot.”
Extraction of Total Protein from Arabidopsis
Arabidopsis tissue was ground to a fine powder in liquid nitrogen. Approximately 100 mg of ground tissue per sample was used for total protein extraction. Denatured protein extracts were obtained by homogenizing and incubating plant material in 200 μL of 2× Laemmli buffer (125 mM Tris-HCl, pH 6.8, 4% [w/v] SDS, 20% [v/v] glycerol, 2% [v/v] β-mercaptoethanol, and 0.01% [w/v] bromophenol blue) for 5 min at 95°C, centrifuged (for 5 min at 20,000g) at room temperature, and the total proteins from supernatant were separated in a 10% SDS-PAGE gel and analyzed as described in the section “Immunoblot.”
Immunoblot
Proteins separated by SDS-PAGE polyacrylamide gel electrophoresis were electroblotted using Trans-blot Turbo Transfer System (Bio-Rad) onto polyvinylidene difluoride (PVDF) membranes (Immobilon-P, Millipore) following instructions by the manufacturer (preprogramed protocols optimized for the molecular weight of the proteins of interest). PVDF membranes, containing electroblotted proteins, were then incubated with the appropriate primary antibody followed by the appropriate secondary peroxidase-conjugated antibody. In addition to the primary antibodies described in the previous methods section, the following primary antibodies were used for detection of epitope-tagged proteins: mouse monoclonal anti-GFP clone B-2 (1:600, catalog no. sc-9996, lot no. C0619, Santa Cruz Biotechnology), mouse monoclonal anti-HA clone HA-7 (1:3000, catalog no. H3663, Sigma-Aldrich), and mouse monoclonal anti-myc clone 9E10 (1:2000, catalog no. AM1007a, Abgent). The secondary antibodies used in this study were as follows: anti-mouse IgG whole molecule−Peroxidase (1:80,000; catalog no. A9044, lot no. 031M4752, Sigma-Aldrich) and anti-rabbit IgG whole molecule−Peroxidase (1:80,000; catalog no. A0545, lot no. 026M4782V, Sigma-Aldrich).
Proteins and epitope-tagged proteins on immunoblots were detected using the Clarity ECL Western Blotting Substrate or SuperSignal West Femto Maximum Sensitivity Substrate according to the manufacturer’s instructions, and images of different time exposures were acquired using the Chemidoc XRS+System (Bio-Rad). SDS-PAGE polyacrylamide gels and immunoblotted PVDF membranes were stained with Coomassie Brilliant Blue R 250 to confirm equal loading of the different samples in a given experiment.
QUANTIFICATION AND STATISTICAL ANALYSIS
Arabidopsis eFP Browser Data Analysis
Gene expression level data from hormone responses were retrieved from the Arabidopsis eFP Browser (Hormone Series) website (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi; Winter et al., 2007). Data used for the analysis were obtained from 7-d-old wild-type seedlings. Differential expression was calculated by dividing the expression value of each gene in a given hormone treatment by the corresponding mock control (fold change of hormone treatment relative to the mock). The hormone gene expression response was calculated and the heatmap was created using Excel (Microsoft). In the heatmap, red represents induction and blue represents repression as response to the indicated hormone.
Quantification of Fluorescent Protein Signal
For quantification of fluorescent protein signal in plasma membrane versus cytoplasm, all images were analyzed using FIJI software (Schindelin et al., 2012; Schneider et al., 2012). To measure the ratio between nuclear and cytoplasmic signals, a small area of fixed size (8 pixels) was drawn, and measurements of integrated densities were taken from representative areas within the plasma membrane and cytoplasm of each cell. To delineate the plasma membrane area, FM4-64 was used to stain the cells. Average ratios between plasma membrane and cytoplasmic signal intensities were calculated based on measurements from three cells per plant; n = 10 plants analyzed (three cells per plant). This experiment was repeated twice with similar results.
Additionally, for quantification of fluorescent protein signal, line scan measurements spanning membrane and cytoplasm were performed from images using FIJI software (Schindelin et al., 2012; Schneider et al., 2012), and representative plot profiles of sample measurements are presented in Figure 4I.
The quantification the BiFC fluorescence intensity of BSK1 and BZR1 in the presence or absence of TTL3-HA (Figure 7B) was performed using FIJI software (Schindelin et al., 2012; Schneider et al., 2012). The area in pixels and raw integrated density of the fluorescence signal were measured in at least five randomly chosen regions of infiltrated leaves per experiment. Fluorescence intensity (arbitrary units) was calculated by dividing the raw integrated density by the area in pixels of the fluorescence signal.
Statistics
Band intensity quantification of protein signal detected by immunoblot, integrated densities from representative areas within the plasma membrane and cytoplasm of each cell analyzed by confocal imaging, and Arabidopsis root and hypocotyl lengths were measured from images using FIJI software (Schindelin et al., 2012; Schneider et al., 2012). The data for qRT-PCR were gathered with MyiQ optical system software (Bio-Rad). For statistical analysis, unpaired t test or one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test (P < 0.05) was performed using Prism 6.00 for Mac (GraphPad Software, www.graphpad.com). ANOVA and/or t test results for the data presented in each figure are detailed in the Supplemental Data Set. Asterisks indicate statistical differences between mutant versus Col-0, unless otherwise specified, as determined by the unpaired t test (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001). Different lowercase letters in the graphs indicate significant differences. Data represent mean values and error bars are sem. In figure legends, n means number of plants for phenotypic analysis, numbers of biological replicates (three technical replicates per biological replicate) for qPCR analysis, or number of cells (three independent measurements performed per cell) analyzed for quantification of fluorescent protein signal in plasma membrane versus cytoplasm. The experiments were repeated at least three times with similar results.
In Silico Three-Dimensional Structural Model of TTL3
The in silico protein structure prediction for TTL3 protein was built by submitting primary sequences to the I-TASSER server (Zhang, 2008) and processed by PyMOL (Schrödinger). IDRs were predicted using GlobPlot 2 (http://globplot.embl.de/). TPR and thioredoxin-like (TPRX) domains were predicted using the SMART/Pfam server and were described previously (Lakhssassi et al., 2012).
Accession Numbers
Sequence data from this article can be found in the EMBL/GenBank data libraries under the following accession numbers: AT1G53300 for TTL1, AT3G14950 for TTL2, AT2G42580 for TTL3, AT3G58620 for TTL4, AT4G33430 for BAK1, AT1G71830 for SERK1, AT4G39400 for BRI1, AT2G01950 for BRL2, AT4G35230 for BSK1, AT1G03445 for BSU1, AT4G18710 for BIN2, AT1G75080 for BZR1, AT1G19350 for BES1.
Supplemental Data
Supplemental Figure 1. TTL genes are required for cotyledon vein pattern formation. Supports Figure 1.
Supplemental Figure 2. TTL3 presents an intrinsically disorder region (IDR) at the N terminus. Supports Figure 1.
Supplemental Figure 3. Protein sequence alignment of BRI1 and BRL2 cytoplasmic domain. Supports Figure 1.
Supplemental Figure 4. Purified GST- and MBP-fused proteins used for the GST Pull-down assays. Supports Figure 1.
Supplemental Figure 5. TTL3 specifically associates with BRI1, but not with BAK1 or free GFP. Supports Figure 1.
Supplemental Figure 6. TTL3 associates with BSK1, BSU1, BIN2, and BZR1 in BiFC experiments. Supports Figures 1, 5, and 6.
Supplemental Figure 7. The expression of TTL1, TTL3, and TTL4 are specifically induced by BRs. Supports Figures 2 and 3.
Supplemental Figure 8. TTL1, TTL3, and TTL4 genes play a positive role in BR signaling. Supports Figure 2.
Supplemental Figure 9. Mutations in TTL1, TTL3, and TTL4 aggravate the weak bri1-301 phenotype and alleviate part of the bes1-D phenotypes. Supports Figure 3.
Supplemental Figure 10. TTL3 presents a cytoplasmic/plasma membrane sub-cellular localization. Supports Figure 4.
Supplemental Figure 11. TTL3-GFP 1.2 complements the root length phenotype of ttl3 in response to eBL treatment. Supports Figure 4.
Supplemental Figure 12. Immunoblot analyses revel that eBL treatment induces TTL3-GFP protein stabilization and that there are no degradation-products of TTL3-GFP in the Arabidopsis TTL3-GFP 2.4 line. Supports Figure 4.
Supplemental Figure 13. BRs regulate the cytoplasmic/plasma membrane localization of TTL3. Supports Figure 4.
Supplemental Figure 14. TTL3 N terminus negatively affects the stabilization of TTL3 in yeast heterologous system. Supports Figures 5 and 6.
Supplemental Figure 15. TTL3 preferentially associates with the phosphorylated form of BZR1 by Co-IP and regulates its cytoplasmic/nuclear localization. Supports Figure 6.
Supplemental Figure 16. BSK1 co-immunoprecipitates with BZR1. Supports Figure 7.
Supplemental Table 1. List and description of primers used for cloning into pENTR.
Supplemental Table 2. List of primers used for quantitative RT-PCR.
Supplemental Data Set. Statistical report of t tests and ANOVAs results for the data presented in each figure.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Yanhai Yin and Michael Hothorn for generously providing the anti-BES1 and the anti-BRI1 antibodies, respectively. We thank Salomé Prat (BIN2 expression clone), Santiago Mora Garcia (BSU1 expression clone), José Alonso (GFP-tagged GW vector for expression in protoplasts), and Xiaofeng Wang (pMAC-flag-BRI1-CD-JMCT9D, BRI1 phosphomimetic mutant) for providing expression clones and vectors.
This work was supported by the Ministerio de Economía y Competitividad (cofinanced by the European Regional Development Fund) (grant BIO2017-82609-R to M.A.B.) and by a Formación del Personal Investigador Fellowship from the Ministerio de Economía y Competitividad (FPI-BES 2015-071256 to A.G.-M.), by the Shanghai Center for Plant Stress Biology (Chinese Academy of Sciences), Chinese 1000 Talents Program (to A.P.M.), and by the Gatsby Charitable Foundation and the European Research Council (grant 309858 “PHOSPHinnATE” to C.Z.). Support was also provided by the Ramón y Cajal Program (RYC-2013-12699 MINECO-Universidad de Málaga, Spain to D.P.); by a fellowship from Agencia Española de Cooperación Internacional para el Desarrollo (Ministerio de Asuntos Exteriores y Cooperacion); and by the Ministerio de Educacion y Ciencia (Universidad de Málaga, Spain) to N.L.
AUTHOR CONTRIBUTIONS
All authors (V.A.-S., A.G.-M., A.G.C., N.L., J.P.-S., Y.L., A.E.d.V., D.P., J.P.-R., A.P.M., V.V., O.B., and C.Z.) designed the experiments. V.A.-S., A.G.-M., A.G.C., N.L., J.P.-S., Y.L., A.E.d.V., D.P., J.P.-R., and A.P.M. performed the experiments and analyzed the data. V.A.-S., A.P.M., and M.A.B. wrote the article. All authors commented on the article.
References
- Albrecht C., Russinova E., Hecht V., Baaijens E., de Vries S. (2005). The Arabidopsis thaliana SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASES1 and 2 control male sporogenesis. Plant Cell 17: 3337–3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhadir Y., Jaillais Y. (2015). The molecular circuitry of brassinosteroid signaling. New Phytol. 206: 522–540. [DOI] [PubMed] [Google Scholar]
- Bernardo-García S., de Lucas M., Martínez C., Espinosa-Ruiz A., Davière J.-M., Prat S. (2014). BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth. Genes Dev. 28: 1681–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatch G.L., Lässle M. (1999). The tetratricopeptide repeat: A structural motif mediating protein-protein interactions. Bioessays 21: 932–939. [DOI] [PubMed] [Google Scholar]
- Bojar D., Martinez J., Santiago J., Rybin V., Bayliss R., Hothorn M. (2014). Crystal structures of the phosphorylated BRI1 kinase domain and implications for brassinosteroid signal initiation. Plant J. 78: 31–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z., Liu J., Wang H., Yang C., Chen Y., Li Y., Pan S., Dong R., Tang G., Barajas-Lopez J. de D., Fujii H., Wang X. (2014). GSK3-like kinases positively modulate abscisic acid signaling through phosphorylating subgroup III SnRK2s in Arabidopsis. Proc. Natl. Acad. Sci. USA 111: 9651–9656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caño-Delgado A., Yin Y., Yu C., Vafeados D., Mora-García S., Cheng J.-C., Nam K.H., Li J., Chory J. (2004). BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development 131: 5341–5351. [DOI] [PubMed] [Google Scholar]
- Ceserani T., Trofka A., Gandotra N., Nelson T. (2009). VH1/BRL2 receptor-like kinase interacts with vascular-specific adaptor proteins VIT and VIK to influence leaf venation. Plant J. 57: 1000–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaiwanon J., Wang W., Zhu J.-Y., Oh E., Wang Z.-Y. (2016). Information integration and communication in plant growth regulation. Cell 164: 1257–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D., Zipfel C., Robatzek S., Kemmerling B., Nürnberger T., Jones J.D.G., Felix G., Boller T. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500. [DOI] [PubMed] [Google Scholar]
- Chung Y., Choe S. (2013). The regulation of brassinosteroid biosynthesis in Arabidopsis. Crit. Rev. Plant Sci. 32: 396–410. [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- D’Andrea L.D., Regan L. (2003). TPR proteins: The versatile helix. Trends Biochem. Sci. 28: 655–662. [DOI] [PubMed] [Google Scholar]
- Du J., Yin H., Zhang S., Wei Z., Zhao B., Zhang J., Gou X., Lin H., Li J. (2012). Somatic embryogenesis receptor kinases control root development mainly via brassinosteroid-independent actions in Arabidopsis thaliana. J. Integr. Plant Biol. 54: 388–399. [DOI] [PubMed] [Google Scholar]
- Fàbregas N., Li N., Boeren S., Nash T.E., Goshe M.B., Clouse S.D., de Vries S., Caño-Delgado A.I. (2013). The brassinosteroid insensitive1-like3 signalosome complex regulates Arabidopsis root development. Plant Cell 25: 3377–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gampala S.S., et al. (2007). An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev. Cell 13: 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehl C., Waadt R., Kudla J., Mendel R.R., Hänsch R. (2009). New GATEWAY vectors for high throughput analyses of protein-protein interactions by bimolecular fluorescence complementation. Mol. Plant 2: 1051–1058. [DOI] [PubMed] [Google Scholar]
- Geldner N., Hyman D.L., Wang X., Schumacher K., Chory J. (2007). Endosomal signaling of plant steroid receptor kinase BRI1. Genes Dev. 21: 1598–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R.D., Schiestl R.H. (1995). Transforming yeast with DNA (invited chapter). Methods Mol. Cell. Biol. 5: 255–269. [Google Scholar]
- González-García M.-P., Vilarrasa-Blasi J., Zhiponova M., Divol F., Mora-García S., Russinova E., Caño-Delgado A.I. (2011). Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development 138: 849–859. [DOI] [PubMed] [Google Scholar]
- Gou X., Yin H., He K., Du J., Yi J., Xu S., Lin H., Clouse S.D., Li J. (2012). Genetic evidence for an indispensable role of somatic embryogenesis receptor kinases in brassinosteroid signaling. PLoS Genet. 8: e1002452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habchi J., Tompa P., Longhi S., Uversky V.N. (2014). Introducing protein intrinsic disorder. Chem. Rev. 114: 6561–6588. [DOI] [PubMed] [Google Scholar]
- He J.X., Gendron J.M., Yang Y., Li J., Wang Z.Y. (2002). The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 99: 10185–10190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He K., Gou X., Yuan T., Lin H., Asami T., Yoshida S., Russell S.D., Li J. (2007). BAK1 and BKK1 regulate brassinosteroid-dependent growth and brassinosteroid-independent cell-death pathways. Curr. Biol. 17: 1109–1115. [DOI] [PubMed] [Google Scholar]
- Houbaert A., et al. (2018). POLAR-guided signalling complex assembly and localization drive asymmetric cell division. Nature 563: 574–578. [DOI] [PubMed] [Google Scholar]
- Jaillais Y., Vert G. (2016). Brassinosteroid signaling and BRI1 dynamics went underground. Curr. Opin. Plant Biol. 33: 92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota Y., Macho A.P., Zipfel C. (2016). Immunoprecipitation of plasma membrane receptor-like kinases for identification of phosphorylation sites and associated proteins. In Methods in Molecular Biology (New York: Springer; ), pp. 133–144. [DOI] [PubMed] [Google Scholar]
- Kim T.-W., Wang Z.-Y. (2010). Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu. Rev. Plant Biol. 61: 681–704. [DOI] [PubMed] [Google Scholar]
- Kim T.-W., Guan S., Sun Y., Deng Z., Tang W., Shang J.-X., Sun Y., Burlingame A.L., Wang Z.-Y. (2009). Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol. 11: 1254–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.-W., Michniewicz M., Bergmann D.C., Wang Z.-Y. (2012). Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 482: 419–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla J., Bock R. (2016). Lighting the way to protein-protein interactions: Recommendations on best practices for bimolecular fluorescence complementation analyses. Plant Cell 28: 1002–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachowiec J., Lemus T., Thomas J.H., Murphy P.J.M., Nemhauser J.L., Queitsch C. (2013). The protein chaperone HSP90 can facilitate the divergence of gene duplicates. Genetics 193: 1269–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakhssassi N., Doblas V.G., Rosado A., del Valle A.E., Posé D., Jimenez A.J., Castillo A.G., Valpuesta V., Borsani O., Botella M.A. (2012). The Arabidopsis tetratricopeptide thioredoxin-like gene family is required for osmotic stress tolerance and male sporogenesis. Plant Physiol. 158: 1252–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wen J., Lease K.A., Doke J.T., Tax F.E., Walker J.C. (2002). BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110: 213–222. [DOI] [PubMed] [Google Scholar]
- Lin W., Lu D., Gao X., Jiang S., Ma X., Wang Z., Mengiste T., He P., Shan L. (2013). Inverse modulation of plant immune and brassinosteroid signaling pathways by the receptor-like cytoplasmic kinase BIK1. Proc. Natl. Acad. Sci. USA 110: 12114–12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Durán R., Zipfel C. (2015). Trade-off between growth and immunity: Role of brassinosteroids. Trends Plant Sci. 20: 12–19. [DOI] [PubMed] [Google Scholar]
- Lozano-Durán R., Bourdais G., He S.Y., Robatzek S. (2014). The bacterial effector HopM1 suppresses PAMP-triggered oxidative burst and stomatal immunity. New Phytol. 202: 259–269. [DOI] [PubMed] [Google Scholar]
- Ma X., Xu G., He P., Shan L. (2016). SERKing coreceptors for receptors. Trends Plant Sci. 21: 1017–1033. [DOI] [PubMed] [Google Scholar]
- Maselli G.A., Slamovits C.H., Bianchi J.I., Vilarrasa-Blasi J., Caño-Delgado A.I., Mora-García S. (2014). Revisiting the evolutionary history and roles of protein phosphatases with Kelch-like domains in plants. Plant Physiol. 164: 1527–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-García S., Vert G., Yin Y., Caño-Delgado A., Cheong H., Chory J. (2004). Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 18: 448–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41. [DOI] [PubMed] [Google Scholar]
- Nam K.H., Li J. (2002). BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110: 203–212. [DOI] [PubMed] [Google Scholar]
- Nolan T.M., Brennan B., Yang M., Chen J., Zhang M., Li Z., Wang X., Bassham D.C., Walley J., Yin Y. (2017). Selective autophagy of BES1 mediated by DSK2 balances plant growth and survival. Dev. Cell 41: 33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng P., Yan Z., Zhu Y., Li J. (2008). Regulation of the Arabidopsis GSK3-like kinase BRASSINOSTEROID-INSENSITIVE 2 through proteasome-mediated protein degradation. Mol. Plant 1: 338–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad B.D., Goel S., Krishna P. (2010). In silico identification of carboxylate clamp type tetratricopeptide repeat proteins in Arabidopsis and rice as putative co-chaperones of Hsp90/Hsp70. PLoS One 5: e12761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H., Willige B.C., Jaillais Y., Geng S., Park M.Y., Gray W.M., Chory J. (2019). BRASSINOSTEROID-SIGNALING KINASE 3, a plasma membrane-associated scaffold protein involved in early brassinosteroid signaling. PLoS Genet. 15: e1007904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado A., Schapire A.L., Bressan R.A., Harfouche A.L., Hasegawa P.M., Valpuesta V., Botella M.A. (2006). The Arabidopsis tetratricopeptide repeat-containing protein TTL1 is required for osmotic stress responses and abscisic acid sensitivity. Plant Physiol. 142: 1113–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas-Díaz T., Cana-Quijada P., Amorim-Silva V., Botella M.A., Lozano-Durán R., Bejarano E.R. (2016). Arabidopsis NahG plants as a suitable and efficient system for transient expression using Agrobacterium tumefaciens. Mol. Plant. Mol. Plant 10: 353–356. [DOI] [PubMed] [Google Scholar]
- Ryu H., Kim K., Cho H., Park J., Choe S., Hwang I. (2007). Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell 19: 2749–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samakovli D., Margaritopoulou T., Prassinos C., Milioni D., Hatzopoulos P. (2014). Brassinosteroid nuclear signaling recruits HSP90 activity. New Phytol. 203: 743–757. [DOI] [PubMed] [Google Scholar]
- Schindelin J., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwessinger B., Roux M., Kadota Y., Ntoukakis V., Sklenar J., Jones A., Zipfel C. (2011). Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet. 7: e1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Shen Q., Qi Y., Yan H., Nie H., Chen Y., Zhao T., Katagiri F., Tang D. (2013). BR-SIGNALING KINASE1 physically associates with FLAGELLIN SENSING2 and regulates plant innate immunity in Arabidopsis. Plant Cell 25: 1143–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeta T., Zaizen Y., Asami T., Yoshida S., Nakamura Y., Okamoto S., Matsuo T., Sugimoto Y. (2014). Molecular evidence of the involvement of heat shock protein 90 in brassinosteroid signaling in Arabidopsis T87 cultured cells. Plant Cell Rep. 33: 499–510. [DOI] [PubMed] [Google Scholar]
- Shigeta T., Zaizen Y., Sugimoto Y., Nakamura Y., Matsuo T., Okamoto S. (2015). Heat shock protein 90 acts in brassinosteroid signaling through interaction with BES1/BZR1 transcription factor. J. Plant Physiol. 178: 69–73. [DOI] [PubMed] [Google Scholar]
- Shimada S., Komatsu T., Yamagami A., Nakazawa M., Matsui M., Kawaide H., Natsume M., Osada H., Asami T., Nakano T. (2015). Formation and dissociation of the BSS1 protein complex regulates plant development via brassinosteroid signaling. Plant Cell 27: 375–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutourina J. (2018). Transcription regulation by the Mediator complex. Nat. Rev. Mol. Cell Biol. 19: 262–274. [DOI] [PubMed] [Google Scholar]
- Sreeramulu S., Mostizky Y., Sunitha S., Shani E., Nahum H., Salomon D., Hayun L.B., Gruetter C., Rauh D., Ori N., Sessa G. (2013). BSKs are partially redundant positive regulators of brassinosteroid signaling in Arabidopsis. Plant J. 74: 905–919. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Asami T., Yoshida S., Nakamura Y., Matsuo T., Okamoto S. (2005). Brassinosteroid homeostasis in Arabidopsis is ensured by feedback expressions of multiple genes involved in its metabolism. Plant Physiol. 138: 1117–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., et al. (2011). PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat. Cell Biol. 13: 124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Kim T.-W., Oses-Prieto J.A., Sun Y., Deng Z., Zhu S., Wang R., Burlingame A.L., Wang Z.-Y. (2008). BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321: 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y., et al. (2018). Hydrogen peroxide positively regulates brassinosteroid signaling through oxidation of the BRASSINAZOLE-RESISTANT1 transcription factor. Nat. Commun. 9: 1063–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Esse W., van Mourik S., Albrecht C., van Leeuwen J., de Vries S. (2013). A mathematical model for the coreceptors SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 and SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE3 in BRASSINOSTEROID INSENSITIVE1-mediated signaling. Plant Physiol. 163: 1472–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G., Chory J. (2006). Downstream nuclear events in brassinosteroid signalling. Nature 441: 96–100. [DOI] [PubMed] [Google Scholar]
- Vida T.A., Emr S.D. (1995). A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128: 779–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriet C., Russinova E., Reuzeau C. (2013). From squalene to brassinolide: The steroid metabolic and signaling pathways across the plant kingdom. Mol. Plant 6: 1738–1757. [DOI] [PubMed] [Google Scholar]
- Wang C., et al. (2013). Identification of BZR1-interacting proteins as potential components of the brassinosteroid signaling pathway in Arabidopsis through tandem affinity purification. Mol. Cell. Proteomics 12: 3653–3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Chory J. (2006). Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science 313: 1118–1122. [DOI] [PubMed] [Google Scholar]
- Wang R., Liu M., Yuan M., Oses-Prieto J.A., Cai X., Sun Y., Burlingame A.L., Wang Z.-Y., Tang W. (2016). The brassinosteroid-activated BRI1 receptor kinase is switched off by dephosphorylation mediated by cytoplasm-localized PP2A B’ subunits. Mol. Plant 9: 148–157. [DOI] [PubMed] [Google Scholar]
- Wang W., Bai M.-Y., Wang Z.-Y. (2014). The brassinosteroid signaling network-a paradigm of signal integration. Curr. Opin. Plant Biol. 21: 147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Kota U., He K., Blackburn K., Li J., Goshe M.B. (2008). Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev. Cell 15: 220–235. [DOI] [PubMed] [Google Scholar]
- Wang Z.-Y., Bai M.-Y., Oh E., Zhu J.-Y. (2012). Brassinosteroid signaling network and regulation of photomorphogenesis. Annu. Rev. Genet. 46: 701–724. [DOI] [PubMed] [Google Scholar]
- Wilma van Esse G., Westphal A.H., Surendran R.P., Albrecht C., van Veen B., Borst J.W., de Vries S.C. (2011). Quantification of the brassinosteroid insensitive1 receptor in planta. Plant Physiol. 156: 1691–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J. (2007). An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Huang J., Li B., Li J., Wang Y. (2008). Is kinase activity essential for biological functions of BRI1? Cell Res. 18: 472–478. [DOI] [PubMed] [Google Scholar]
- Yang C.-J., Zhang C., Lu Y.-N., Jin J.-Q., Wang X.-L. (2011). The mechanisms of brassinosteroids’ action: From signal transduction to plant development. Mol. Plant 4: 588–600. [DOI] [PubMed] [Google Scholar]
- Yang J., Roe S.M., Cliff M.J., Williams M.A., Ladbury J.E., Cohen P.T.W., Barford D. (2005). Molecular basis for TPR domain-mediated regulation of protein phosphatase 5. EMBO J. 24: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Wang Z.-Y., Mora-García S., Li J., Yoshida S., Asami T., Chory J. (2002). BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191. [DOI] [PubMed] [Google Scholar]
- Yoo S.-D., Cho Y.-H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Yu X., Li L., Zola J., Aluru M., Ye H., Foudree A., Guo H., Anderson S., Aluru S., Liu P., Rodermel S., Yin Y. (2011). A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 65: 634–646. [DOI] [PubMed] [Google Scholar]
- Zhang Y. (2008). I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu Z., Wang J., Chen Y., Bi Y., He J. (2015). Brassinosteroid is required for sugar promotion of hypocotyl elongation in Arabidopsis in darkness. Planta 242: 881–893. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Zhu J.-Y., Roh J., Marchive C., Kim S.-K., Meyer C., Sun Y., Wang W., Wang Z.-Y. (2016). TOR signaling promotes accumulation of BZR1 to balance growth with carbon availability in Arabidopsis. Curr. Biol. 26: 1854–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.-Y., Li Y., Cao D.-M., Yang H., Oh E., Bi Y., Zhu S., Wang Z.-Y. (2017). The F-box protein KIB1 mediates brassinosteroid-induced inactivation and degradation of GSK3-like kinases in Arabidopsis. Mol. Cell 66: 648–657. [DOI] [PMC free article] [PubMed] [Google Scholar]