Abstract
Sigma factors are key proteins that mediate the recruitment of RNA polymerase to the promoter regions of genes, for the initiation of bacterial transcription. Multiple sigma factors in a bacterium selectively recognize their cognate promoter sequences, thereby inducing the expression of their own regulons. In this paper, we report the crystal structure of the σ4 domain of Bacillus subtilis SigW bound to the -35 promoter element. Purine-specific hydrogen bonds of the -35 promoter element with the recognition helix α9 of the σ4 domain occurs at three nucleotides of the consensus sequence (G-35, A-34, and G’-31 in G-35A-34A-33A-32C-31C-30T-29). The hydrogen bonds of the backbone with the α7 and α8 of the σ4 domain occurs at G’-30. These results elucidate the structural basis of the selective recognition of the promoter by SigW. In addition, comparison of SigW structures complexed with the -35 promoter element or with anti-sigma RsiW reveals that DNA recognition and anti-sigma factor binding of SigW are mutually exclusive.
Introduction
Transcription in bacteria is initiated by sigma factors, which recruit the core RNA polymerase to a cognate promoter [1, 2]. Sigma factors selectively recognize promoter elements, -10 and -35 elements, and additional sequences, including extended -10 element and discriminator, which are present upstream of the transcription start site [3, 4]. During transcription initiation, the -10 element is strand-separated to form a transcription bubble [5, 6], whereas the -35 element is recognized by a helix-turn-helix (HTH) motif in the sigma factor, without strand separation [7, 8].
Sigma factors are categorized into two families, based on the sequence homology with Escherichia coli sigma factors: housekeeping σ70 required for bacterial homeostasis; and σ54 activated for nitrogen utilization [9]. The σ70 family is further sub-divided into five groups [4, 10]. Group I sigma factors are composed of σ1.1, σ2, σ3, and σ4 domains, which are responsible for the recognition of the discriminator, -10, extended -10, and -35 elements, respectively. The primary sigma factors, which regulate the transcription of housekeeping genes, belong to group I. Group II-V are classified depending on the presence or absence of the domains and motifs in group I. Alternative sigma factors, which are activated in response to a range of stress conditions, belong to groups II-V [10].
Group IV sigma factors are known as extracytoplasmic function (ECF) sigma factors, because they are activated in response to extracytoplasmic stresses [3, 10, 11]. They are the most divergent group of sigma factors, and almost all bacteria contain multiple ECF sigma factors. As an extreme case, Streptomyces coelicolor contains approximately 50 ECF sigma factors in its genome [10, 12]. The ECF sigma factors are composed of only σ2 and σ4 domains, which bind -10 and -35 elements, respectively [3, 13–15]. In many cases, promoter binding of ECF sigma factors is inhibited by binding to the cytoplasmic domain of transmembrane anti-sigma factors [10, 11, 16], and is activated to initiate the transcription of target genes in response to a specific stress signal by being released from the anti-sigma factor.
Bacillus subtilis contains at least seven ECF sigma factors: SigM, SigV, SigW, SigX, SigY, SigZ, and YlaC [17]. Of these sigma factors, SigW induces the transcription of its target regulon to counteract cell envelope stresses caused by antibiotics [18, 19], alkaline pH [20], and high salt concentration [21, 22]. In the absence of these stresses, SigW is downregulated by anti-sigma factor RsiW, which is localized in the plasma membrane [16, 23]. It is released from the anti-sigma factor under the stress conditions [24–27]. Under these conditions, transmembrane RsiW is sequentially cleaved by regulated intramembrane proteolysis by PrsW and RasP proteases; therefore, the cytoplasmic domain of RsiW is released together with SigW from the plasma membrane [25, 27]. Subsequently, the cytoplasmic domain of RsiW is completely degraded by ClpXP protease [28], allowing SigW to bind to the core RNA polymerase, and thereby to activate SigW-dependent transcription.
Even though crystal structures have been reported showing interactions between the σ4 domain and the -35 promoter element in group I and group IV sigma factors [3, 7, 8, 29, 30], structural information from diverse sigma factors is required to understand the selective recognition of the -35 element. In the work reported here, we determined the crystal structure of the σW4/-35 element complex, and analyzed the promoter binding mode of SigW. The structure reveals the unique regulation mode of SigW-dependent transcription, and the recognition specificity of the -35 element by the σ4 domain.
Materials and methods
Plasmid preparation and protein expression
DNA encoding the σ4 domain of B. subtilis SigW (σW4; residues 126–187) was amplified from the genome of B. subtilis 168 strain using polymerase chain reaction and inserted into pETDuet-1 vector (Merck Millipore, Billerica, MA, USA) expressing the N-terminal 6XHis tag and TEV protease cleavage site. The plasmid was transformed into E. coli strain BL21-star (DE3) (Thermo Fischer Scientific, Waltham, MA, USA) and cells were grown in Luria Broth media at 37°C. When the culture reached an OD600 of 0.6–0.7, the medium was cooled, and 0.4 mM isopropyl β-D-1-thiogalactopyranoside was added to the culture to induce σW4 expression. After overnight incubation at 15°C, the cells were harvested by centrifugation at 3,000 g for 10 min.
Purification of σW4/-35W
Cells expressing 6XHis-σW4 were resuspended in buffer A (20 mM HEPES pH 7.5, 1.0 M NaCl, and 10% (v/v) glycerol), lysed by sonication, and clarified by centrifugation at 20,000 g for 30 min, after the addition of DNase I and RNase A at a concentration of 10 μg/ml. σW4 was purified by immobilized metal affinity chromatography (IMAC) and size exclusion chromatography (SEC). Clarified cell lysate was loaded onto a 5 mL HisTrap nickel chelating column (GE Healthcare Bio-sciences, Uppsala, Sweden), and the resin was washed with buffer A, containing 80 mM imidazole. Proteins bound to the resin were eluted by an imidazole gradient (0.08–1.00 M imidazole). Fractions that contained 6XHis-σW4 were pooled and treated with TEV protease overnight at 25°C to cleave the His tag. After complete cleavage, the protein solution was dialyzed against buffer A for 3 h and passed through Ni-NTA resin to remove the 6XHis tag (Thermo Fisher Scientific, Rockford, IL, USA). σW4 was further purified by SEC using Superdex 75 preparatory grade column (GE Healthcare Biosciences) pre-equilibrated with buffer B (20 mM HEPES pH 7.5, 1.0 M NaCl, and 5% (v/v) glycerol).
The -35 element recognized by σW4 was prepared by mixing two complementary DNA fragments. Two single-stranded DNAs (5’-ATTGAAACCTTT-3’ and 5’-AAAAGGTTTCAA-3’) were synthesized (Biobasic, Seoul, South Korea), mixed at a 1:1 molar ratio in buffer B, and purified by SEC using a Superdex75 analytical column. The purified -35 element (-35W) was then mixed with σW4 at a 1.1:1 molar ratio. The mixture was dialyzed in buffer C (20 mM HEPES pH 7.5, 0.2 M NaCl, and 5% (v/v) glycerol) and concentrated to 15 mg/mL for crystal screening.
Crystallization, data collection, and structure determination
Crystallization of the σW4/-35W complex was performed using the micro-batch method at 20°C. The drop for crystal screening was prepared by mixing 1 μL of σW4/-35W (15 mg/mL) and 1 μL of crystallization solution under a layer of Al’s oil (Hampton Research, Aliso Viejo, Ca, USA). Crystals of σW4/-35W grew completely in a month under the conditions of Wizard Precipitant Synergy 127 (0.1M imidazole/hydrochloric acid pH 6.5, 30% (v/v) PEG1500, 10% (v/v) isopropanol, and 0.1 M CaCl2) (Rigaku, Tokyo, Japan). Crystals of σW4/-35W were picked using a cryo-loop (Hampton Research) and flash-frozen in a cold nitrogen stream. Diffraction data were collected at PLS-BL7A (Beam line 7A, Pohang Light Source, South Korea) [31] and were indexed, integrated, and scaled using MOSFLM [32].
The crystal structure of σW4/-35W was determined by the molecular replacement (MR) method using PHASER [33]. The structure of the E. coli σE4/-35 element (PDB ID: 2H27) was used as a template for MR. MR solution was found from the truncated σE4 (residues 127–186)/-35 element. Cycles of refinement and model building were performed at 3.1 Å resolution using PHENIX.refine [34] and COOT [35]. Final refinement resulted in R / Rfree values of 24.8 / 29.0% without residues in the disallowed region of the Ramachandran plot. The data collection and refinement statistics are summarized in Table 1. The final coordinates and structure factors were deposited in the Protein Data Bank (PDB ID: 6JHE). Structural alignment was performed using the DALI server [36]. Protein-ligand interactions were analyzed with LigPlot+ [37] and PDBePISA [38]. The free energy change (ΔG) of the protein caused by ligand binding was analyzed using PDBePISA [38]. The conversion of ΔG to a dissociation constant (Kd) was calculated using the equation Kd = e(ΔG/RT) (R = 1.987 cal/molK; T = 293K). DNA geometry was analyzed using w3DNA [39]. Surface charge distribution was calculated using APBS [40]. The figures were drawn using PyMOL [41] and ALSCRIPT [42].
Table 1. Data collection and refinement statistics.
| Data collection | ||
| Data set | σW4/-35W | |
| Space group | P6522 | |
| Unit cell | ||
| a, b, c (Å) | 61.61, 61.61, 119.96 | |
| α, β, γ (°) | 90.00, 90.00, 90.00 | |
| Resolution (Å) | 30.0–3.00 (3.18–3.00) | |
| Wavelength (Å) | 0.97933 | |
| Total/Unique reflections | 45479/3013 | |
| Completeness (%) | 99.4 (100.0) | |
| I/σ | 62.8 (16.4) | |
| Rmerge (%) | 10.4 (52.4) | |
| Refinement | ||
| Resolution | 30.0–3.10 | |
| No. reflections, working/free | 2715/140 | |
| Rwork/Rfree (%) | 24.8/29.0 | |
| No. atoms | 884 | |
| Protein | 436 | |
| DNA | 448 | |
| B factors | 69.0 | |
| RMSD | ||
| Bond length (Å) | 0.012 | |
| Bond angle (°) | 1.438 | |
| Ramachandran plot (%) | ||
| Favor | 94.1 | |
| Allowed | 5.9 | |
| Disallowed | 0.0 | |
Accession number
The final coordinates and structure factors were deposited in the Protein Data Bank (PDB ID: 6JHE for σW4/-35W).
Results and discussion
Overall structure
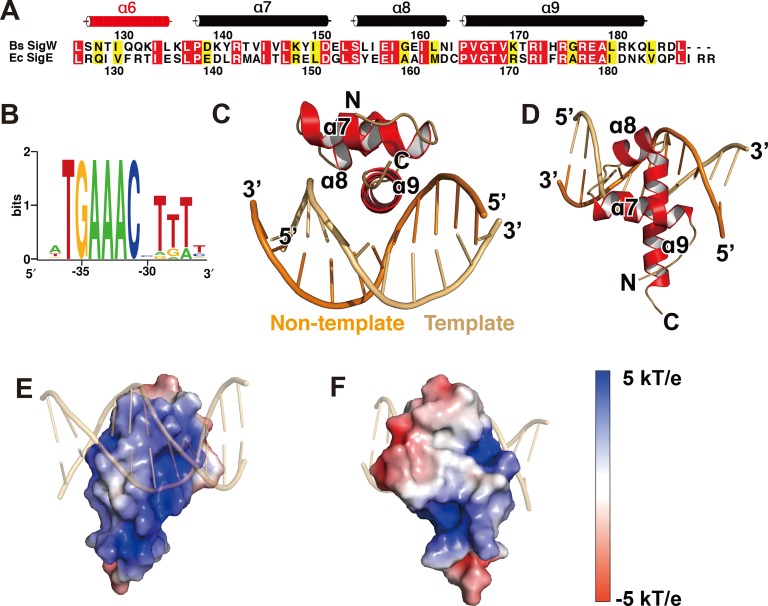
B. subtilis σW4 (residues 125–187) recognizes a cognate -35 promoter element (Fig 1A and 1B). Its consensus sequence is identified as T-36G-35A-34A-33A-32C-31X-30T-29T-28T-27, based on the promoter sequences of the SigW regulon [43]. σW4 was purified under high salt conditions (1 M NaCl) to minimize its instability, and dialyzed in low salt buffer together with the double-stranded DNA of A-38T-37T-36G-35A-34A-33A-32C-31C-30T-29T-28T-27 (-35W) to allow σW4 binding to -35W. Crystals of σW4/-35W belonging to a hexagonal space group grew under conditions containing PEG1500 and isopropanol as protein precipitants. Diffraction data were collected at a resolution of 3.1 Å (Table 1), and the structure was determined by molecular replacement using the structure of a truncated E. coli σE4/-35E (-35 promoter element for SigE binding) as a template [8].
Fig 1. Structure of the σW4/-35W complex.
(A) Sequence alignment of σW4 and σE4. The secondary structure of the σW4 domain is displayed using a tube to indicate an α-helix. Identical residues are boxed in red, and similar residues are boxed in yellow. α6 disordered in the structure is displayed as a red tube, based on the crystal structure of the SigW/RsiW complex (PDB ID: 5WUQ). (B) Sequence logo for the -35 promoter element of the SigW regulon [44]. (C, D) Ribbon models of the σW4/-35W complex, drawn at two different orientations. α9 of σW4 is inserted into the major groove of -35W as a recognition helix of the HTH motif. (E, F) Surface model of σW4 with charge distribution. The electrostatic potential, from red (-5 kT/e) to blue (+5 kT/e), is plotted on the solvent-accessible surface calculated with a solvent probe radius of 1.4 Å.
The crystal structure contains a σW4 monomer and a double-stranded -35W in the asymmetric unit. Residues 134–186 of SigW and 11 nucleotide pairs of -35W were traced into the electron density (S1 Fig) and the final structure was refined at R / Rfree values of 24.8 / 29.0% (Table 1). σW4 is comprised of four α-helices (α6–α9) in the crystal structure of the SigW/RsiW complex [23]. However, residues 125–133, which correspond to α6, are disordered in the crystal structure of σW4/-35W (Fig 1A). The residues on α6 are likely to be flexible because they are not bound to DNA directly. α8–α9 of σW4 forms the HTH motif, and α9 is inserted into the major groove of -35W as a DNA recognition helix (Fig 1C and 1D) [7]. Positively-charged residues are distributed on the DNA binding surface of α8–α9, whereas hydrophobic patches are distributed on the opposite side that interacts with σW2 in the crystal structure of SigW/RsiW (Fig 1E and 1F) [23].
Interactions between σW4 and the -35 promoter element
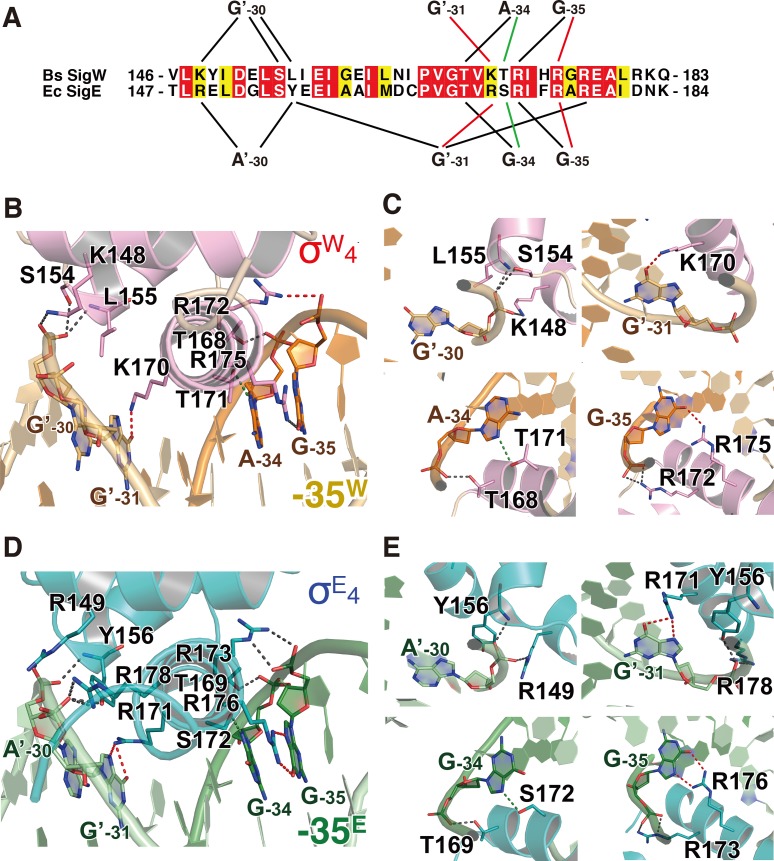
The recognition helix α9 mediates the major interactions between σW4 and -35W through hydrogen bonds and hydrophobic interactions. The bases and backbones of three purine nucleotides (G-35 and A-34 in the non-template strand and G’-31’ in the template strand; ‘ indicates the template strand of DNA) form hydrogen bonds with the residues on the N-terminal half of α9 in σW4 (Fig 2A–2C and S2 Fig). The guanine oxygen (O6) and backbone phosphate (OP2) of G-35 form hydrogen bonds with side chain amino groups (NH2) of R175 and R172, respectively. The purine nitrogen (N7) and backbone phosphate (OP2) of A-34 interact with the side chain oxygens (OG1) of T171 and T168. The guanine oxygen (O6) of G’-31 forms a hydrogen bond with the side chain amino group (NZ) of K170. The electron density map for the side chain of K170 is relatively weak; however, the most-preferred rotamer is at hydrogen bond distance to the O6 of G’-31 (S2B Fig). Hydrophobic interactions are observed between K170-G’-31T’-32, T171-A-34, H174-T’-32T’-33, and R175-T-36 (S3 Fig).
Fig 2. Interactions between σ4 domain and -35 promoter element.
(A) Schematic diagram representing the hydrogen bonds between σ4 and the -35 element. The sequences of the -35 element-binding interface in B. subtilis SigW and E. coli SigE are aligned. Black lines indicate the backbone interactions, green lines show the purine-specific interactions, and red the guanine-specific interactions. (B, C) Hydrogen bonds between σW4 and -35W. Residues and nucleotides, which form hydrogen bonds, are drawn as pink and orange stick models, respectively. Dotted lines indicate hydrogen bonds between σW4 and -35W. (D, E) Interactions between σE4 and -35E (PDB ID: 2H27) [8]. Residues and nucleotides, which form hydrogen bonds, are drawn as teal and dark green stick models. The ribbon models are drawn at the same orientations as those in (B) and (C). Dotted lines indicate hydrogen bonds between σE4 and -35E. The dotted lines in (B-E) are colored by the same scheme as the lines in (A).
In addition to α9, α7-α8 contributes to -35W binding without base specificity. The backbone atoms (OP1, OP2, and OP2) of G’-30 form hydrogen bonds with the side chain amino group (NZ) of K148, the side chain oxygen (OG) of S154, and the backbone nitrogen of L155, respectively (Fig 2B and 2C). Altogether, α9 in σW4 specifically recognizes -35W, and α7-α8 provides additional contacts, leading to a tighter interaction.
Structural comparison of σW4/-35W and σE4/-35E
SigE is an E. coli ECF sigma factor activated in response to envelope stress and induces transcription of heat shock proteins [45]. Its σ4 domain (σE4) recognizes the -35 element, of which the consensus sequence is G-35G-34A-33A-32C-31T-30T-29 (-35E) [46]. The σE4 structure is highly similar to σW4 [8]. σW4 is superimposed on σE4 with a root mean square deviation (RMSD) value of 1.8 Å for 53 Cα atoms (S4A Fig). The overall fold is conserved between σW4 and σE4 and the main differences are observed at the N- and C-termini (S4A and S4B Fig).
Overall, the interactions of σE4 and σW4 with the corresponding -35 elements are conserved. Three nucleotides, G-35, G-34, and G’-31, in -35E, which correspond to G-35, A-34, and G’-31 in -35W, mediate purine nucleotide-specific interactions with the recognition α-helix (Fig 2A). The backbone and base of G-35 (G-35 in σW4) form hydrogen bonds with R173 (R172 in σW4) and R176 (R175 in σW4). The backbone and base of G-34 (A-34 in -35W) form hydrogen bonds with T169 (T168 in σW4) and S172 (T171 in σW4). Although the -34 position is not identical between -35E and -35W (G-34 in -35E and A-34 in -35W), hydrogen bonds mediated by the backbone phosphate and purine N7 are conserved. The base of G’-31 forms a hydrogen bond with R171 (R170 in σW4). The backbone phosphate of G’-31 also forms a hydrogen bond with R178, which is not observed in the σW4/-35W structure. A’-30 in-35E mediates backbone interactions similarly to G’-30 in -35W (Fig 2). The backbone phosphate of A’-30 forms hydrogen bonds with the NH1 of R149 (K148 in σW4) and the backbone nitrogen of Y156 (L155 in σW4) (Fig 2D and 2E). Interaction between S154 and A’-30 in σW4/-35W is missing in the σE4/-35E structure. In summary, purine base-specific hydrogen bonds in the structures of σW4/-35W and σE4/-35E are conserved, whereas the hydrogen bonds with the nucleotide backbone are slightly different.
Hydrophobic interactions between σE4 and -35E are mostly conserved in the σW4/-35W structure (S3 Fig). Residues R171, F175, and R176 in σE4 (K170, H174, and R175 in σW4) interact with T’-32, T’-33, and C-36 in -35E (T’-32, T’-33, and T-36 in -35W), respectively. Hydrophobic interactions between P166/G168/T169 and G-34 and between Y156 and A’-30 are observed only in σE4/-35E, whereas the interaction between K170 and G’-31 in σW4/-35W is observed only in σE4/-35E. A cation-π interaction is observed between R176 and the pyrimidine ring of C’-34 in the structure of σE4/-35E. However, the distance between the base (T-36) and the corresponding residue (R175) is too far to form a cation-π interaction in the structure of σW4/-35W (S5 Fig).
DNA geometry of -35W
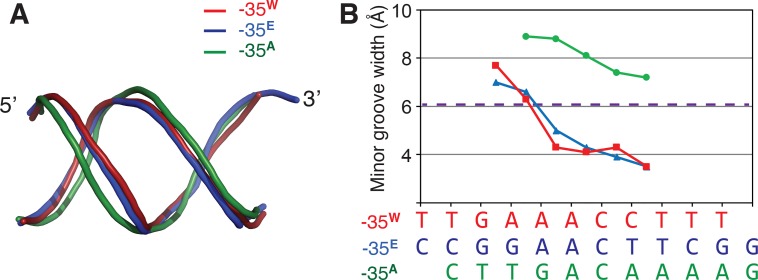
Nucleotides A-33A-32 in -35E (G-35G-34A-33A-32C-31T-30T-29) do not form hydrogen bonds with σE4, although these nucleotides are conserved among the -35 elements of the SigE regulon, and mutating these nucleotides in the Salmonella enterica serovar Typhimurium SigE has been shown to lead to defective transcription [47]. These nucleotides are involved in characteristic oligo(dA)/oligo(dT)-like DNA geometry that is rigid and straight with a narrow minor groove [8, 48]. A previous structural study of the σE4/-35E complex suggested that the geometry of the narrowed minor groove is critical for σE4 recognition [8], and we show that -35W also displays a narrowed minor groove (Fig 3). Like -35E, the narrowing of the minor groove of -35W begins at A-33A-32 and is stabilized downstream of the -35 element, even though the downstream sequence of -35W has an insertion of two cytosines (A-33A-32C-31C-30T-29) (Fig 3). In contrast, -G-33A-32 in -35A (the -35 element of Thermus aquaticus SigA) has a wider minor groove than normal B-DNA (Fig 3). The crystal structure of σW4/-35W supports the suggestion that the A-33A-32 conservation in the -35 element for group IV sigma factors is critical for the formation of the narrow minor groove [8].
Fig 3. DNA conformation of the -35 promoter element.
(A) DNA backbone geometry. -35 elements from the structures of σW4/-35W, E.coli σE4/-35E (PDB ID: 2H27), and Thermus aquaticus σA4/-35A (PDB ID: 1KU7) are superposed. The 5′ and 3′-ends of the non-template strands are labeled. -35W, 35E, and -35A are colored red, blue, and green, respectively. (B) Plot showing the minor groove width of the -35 elements. The sequences of the -35 elements are aligned with the plot. -35W and -35E have narrower minor grooves than a normal B-DNA, while -35A contains a wider minor groove. The dashed purple line indicates the minor groove width of standard B-form DNA.
Structural comparison of σW4/-35W and SigW/RsiW
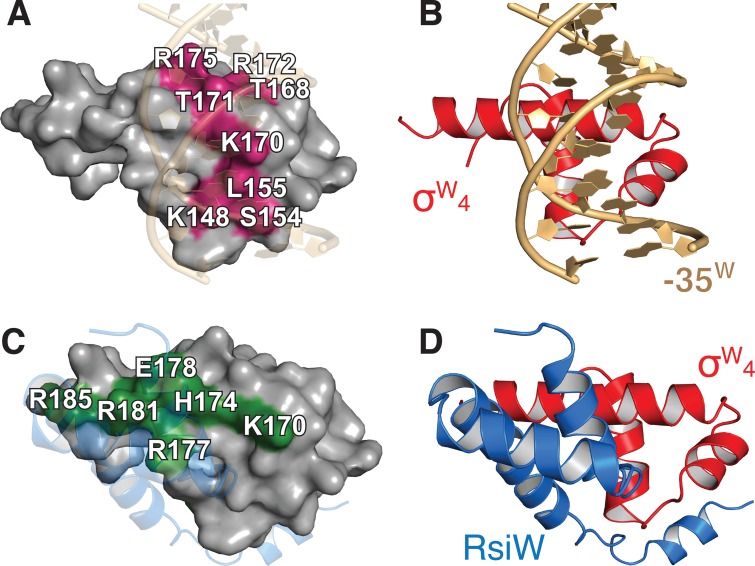
The crystal structure of SigW complexed with anti-sigma RsiW was previously reported [23]. The structures of σW4 under the binding of -35W and RsiW superimpose with an RMSD value of 1.6 Å for 53 Cα atoms (S4B Fig). Although conformational differences of σW4 in the structures bound to -35W and RsiW are minor, σW4 bound to -35W exists in a slightly compact conformation. σW4 interacts with -35W through the residues K148, S154, L155, T168, K170, T171, R172, and R175, with a surface area of 621.2 Å2 buried at the binding interface (Fig 4A and 4B). σW4 interacts with RsiW through the residues I150, K170, H174, R177, E178, R181, R185, and L187, these interactions result in an approximately 50% larger burial of surface area (915.2 Å2) (Fig 4C and 4D). The surface area of σW4 that binds -35W and RsiW partially overlaps with residue K170 on σW4 (Fig 4A and 4C), indicating that the interactions of -35W and RsiW with σW4 are mutually exclusive.
Fig 4. Structural comparison of σW4/-35W and SigW/RsiW.
(A) Surface model of σW4 bound to -35W. The magenta surface indicates the -35W binding interface. Residues on the binding surface are labeled. (B) Ribbon model of σW4/-35W structure. (C) Surface model of σW4 bound to RsiW. The RsiW binding surface is colored green, and the residues are labeled. (D) Ribbon model of σW4 bound to RsiW. σW4 structures in (A)-(D) are drawn at the same orientation.
In the crystal structure of the SigW/RsiW complex, the -10 element-binding surface of σW2 is buried in the surface of σW4 [23], whereas the -35 element-binding surface of σW4 is directly blocked by RsiW (Fig 4), suggesting that SigW inhibition by RsiW competes with -35 element binding in the promoter. The binding of the -35 element to σW4 results in a ΔG of -10.5 kcal/mol, which corresponds to a Kd value of 1.47 x 10−8 M. RsiW binding to SigW results in a surface burial of 1619.6 Å2, and a ΔG of -17.85 kcal/mol. The free energy change corresponds to a Kd value of 4.84 x 10−14 M. These observations indicate that DNA binding of SigW is structurally repressed in the presence of RsiW.
Role of conserved residues in σW4
B. subtilis contains multiple ECF sigma factors that respond to diverse environmental stresses. The positions L137, L147, and E157 of σW4 are highly conserved in B. subtilis ECF sigma factors (Fig 5A and 5B). The highly conserved residues are likely to be involved in the intrinsic folding of SigW, but not in DNA binding. L137 and L147 stabilize σW4 as part of the central innermost hydrophobic cluster (S6A and S6B Fig). E157 is associated with σW2 binding in the crystal structure of the SigW/RsiW complex (S6C and S6D Fig).
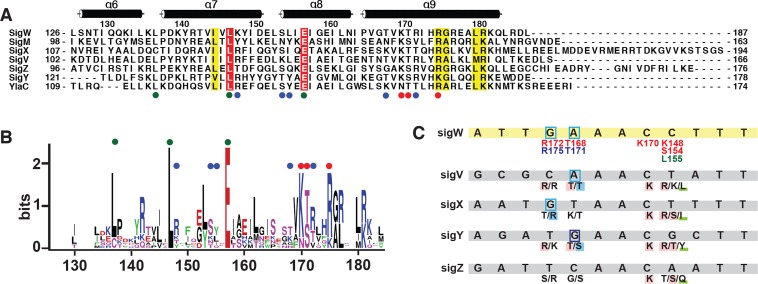
Fig 5. Conserved residues in σW4.
(A) Sequence alignment of σ4 domains of B. subtilis ECF sigma factors. The highly conserved K137, L148, and E157 are indicated by green circles. Residues that form purine- and backbone-specific hydrogen bonds are indicated by red and blue circles, respectively. (B) Sequence logo for the σ4 domains of B. subtilis ECF sigma factors. (C) The sequences of the -35 promoter elements for B. subtilis ECF sigma factors. Residues that form hydrogen bonds with the -35 promoter element are predicted based on the interactions between σW4 and -35W and the sequence alignment in (A).
The residues involved in DNA binding are less conserved than those involved in intrinsic folding. Residues K170, T171, and R175, which mediate purine-specific hydrogen bonds in σW4, are aligned to K/R, S/T, and K/R in B. subtilis ECF sigma factors, as well as E. coli SigE (Fig 5A and 5B). The conservation of these residues correlates with the three nucleotides that mediate purine-specific interactions (G-35, A-34, and G’-31 in -35W). For example, A-34 in the SigV promoter interacts with T144 and T147, as does A-34 in -35W, and T168/T171 in σW4, whereas C-34 in the SigZ promoter is aligned with G138 and S141. These observations suggest that sequence variation in the DNA-binding interface confers the specificity required to discriminate between -35 elements (Fig 5C). In summary, conserved residues in B. subtilis ECF σ4 are mainly involved in intramolecular stability and interactions with the -35 element. Slight sequence variations in the residues involved in the binding of the -35 element may contribute to the fine-tuning of the promoter selectivity and binding affinity for individual ECF sigma factors.
Conclusions
Group IV ECF sigma factor is activated in response to environmental stress and initiates transcription of its own regulon to counter that stress. The crystal structure of σW4/-35W shows that SigW selectively recognizes cognate -35 promoter elements which have a narrowed minor groove, in a similar manner to the E. coli SigE. Comparison with the SigW/RsiW structure shows that SigW binding to the -35 promoter element and anti-sigma RsiW is mutually exclusive. These results provide the structural basis for the mechanism of SigW activation, and improve our understanding of the selective interactions between σ4 domains and their cognate -35 promoter elements.
Supporting information
Red and orange stick models indicate σW4 and -35W, respectively. 2Fo-Fc maps are drawn at two different orientations. The contour level is set to 1.0 σ.
(TIF)
2Fo-Fc maps are drawn at same scale and orientation as those for models in Fig 2C and arranged in the same order as the panels in Fig 2C. The contour level is set to 1.0 σ.
(TIF)
(A, B) Hydrophobic interactions between σW4 and -35W shown at two different orientations. Residues and nucleotides, which are associated with hydrophobic bonds, are drawn as red and orange stick models. Dotted lines indicate hydrophobic interactions. (C, D) 2Fo-Fc electron density maps in (C) and (D) are shown at same scale and orientation as those for models in S2A and S2B Fig, respectively. The contour level is set to 1.0 σ. (E, F) Hydrophobic interactions between σE4 and -35E. Residues and nucleotides which are associated with hydrophobic bonds are drawn as purple and green stick models.
(TIF)
(A) Superposition of σE4/-35E and σW4/-35W structures. N- and C-termini of σW4 are labeled. (B) Distribution of root-mean-square values between Cα positions of superimposed σE4 and σW4 structures. (C) Superposition of σW4 domains from the structures of σW4/-35W and SigW/RsiW.
(TIF)
(A) The green dotted line indicates the cation-π interaction between R176 of σE4 (cyan model) and C-36 of 35E (green). The corresponding residue in σW4 (R175) and base in -35W (T-36) are drawn as a stick model. The distance between the Arg and pyrimidine is labeled.
(TIF)
The residues that participate in intramolecular interactions with L137 (A), L147 (B), or E157 (C) are drawn as stick models and the residue number is labeled. The helix and loop in σW4 are colored pink and light brown. (D) σW4 in SigW/RsiW structure (PDB ID: 5WUQ) is superposed onto that of the σW4/-35W structure. SigW in SigW/RsiW structure is colored green. E157 in σW4 interacts with T71 in σW2 and does not participate in DNA binding.
(TIF)
Data Availability
The final coordinates and structure factors were deposited in Protein Data Bank (PDB ID: 6JHE).
Funding Statement
DYK was supported by the Yeungnam University Research Grant. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Burgess RR, Travers AA, Dunn JJ, Bautz EK. Factor stimulating transcription by RNA polymerase. Nature. 1969;221(5175):43–6. Epub 1969/01/04. 10.1038/221043a0 . [DOI] [PubMed] [Google Scholar]
- 2.Saecker RM, Record MT Jr., Dehaseth PL. Mechanism of bacterial transcription initiation: RNA polymerase—promoter binding, isomerization to initiation-competent open complexes, and initiation of RNA synthesis. J Mol Biol. 2011;412(5):754–71. Epub 2011/03/05. 10.1016/j.jmb.2011.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feklistov A, Sharon BD, Darst SA, Gross CA. Bacterial sigma factors: a historical, structural, and genomic perspective. Annu Rev Microbiol. 2014;68:357–76. 10.1146/annurev-micro-092412-155737 . [DOI] [PubMed] [Google Scholar]
- 4.Paget MS. Bacterial Sigma Factors and Anti-Sigma Factors: Structure, Function and Distribution. Biomolecules. 2015;5(3):1245–65. Epub 2015/07/02. 10.3390/biom5031245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feklistov A, Darst SA. Structural basis for promoter-10 element recognition by the bacterial RNA polymerase sigma subunit. Cell. 2011;147(6):1257–69. 10.1016/j.cell.2011.10.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campagne S, Marsh ME, Capitani G, Vorholt JA, Allain FH. Structural basis for -10 promoter element melting by environmentally induced sigma factors. Nat Struct Mol Biol. 2014;21(3):269–76. 10.1038/nsmb.2777 . [DOI] [PubMed] [Google Scholar]
- 7.Campbell EA, Muzzin O, Chlenov M, Sun JL, Olson CA, Weinman O, et al. Structure of the bacterial RNA polymerase promoter specificity sigma subunit. Mol Cell. 2002;9(3):527–39. . [DOI] [PubMed] [Google Scholar]
- 8.Lane WJ, Darst SA. The structural basis for promoter -35 element recognition by the group IV sigma factors. PLoS Biol. 2006;4(9):e269 10.1371/journal.pbio.0040269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wigneshweraraj S, Bose D, Burrows PC, Joly N, Schumacher J, Rappas M, et al. Modus operandi of the bacterial RNA polymerase containing the sigma54 promoter-specificity factor. Mol Microbiol. 2008;68(3):538–46. Epub 2008/03/12. 10.1111/j.1365-2958.2008.06181.x . [DOI] [PubMed] [Google Scholar]
- 10.Helmann JD. The extracytoplasmic function (ECF) sigma factors. Adv Microb Physiol. 2002;46:47–110. Epub 2002/06/21. . [DOI] [PubMed] [Google Scholar]
- 11.Osterberg S, del Peso-Santos T, Shingler V. Regulation of alternative sigma factor use. Annu Rev Microbiol. 2011;65:37–55. Epub 2011/06/07. 10.1146/annurev.micro.112408.134219 . [DOI] [PubMed] [Google Scholar]
- 12.Staron A, Sofia HJ, Dietrich S, Ulrich LE, Liesegang H, Mascher T. The third pillar of bacterial signal transduction: classification of the extracytoplasmic function (ECF) sigma factor protein family. Mol Microbiol. 2009;74(3):557–81. Epub 2009/09/10. 10.1111/j.1365-2958.2009.06870.x . [DOI] [PubMed] [Google Scholar]
- 13.Lonetto M, Gribskov M, Gross CA. The sigma 70 family: sequence conservation and evolutionary relationships. J Bacteriol. 1992;174(12):3843–9. Epub 1992/06/01. 10.1128/jb.174.12.3843-3849.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haugen SP, Berkmen MB, Ross W, Gaal T, Ward C, Gourse RL. rRNA promoter regulation by nonoptimal binding of sigma region 1.2: an additional recognition element for RNA polymerase. Cell. 2006;125(6):1069–82. Epub 2006/06/17. 10.1016/j.cell.2006.04.034 . [DOI] [PubMed] [Google Scholar]
- 15.Haugen SP, Ross W, Manrique M, Gourse RL. Fine structure of the promoter-sigma region 1.2 interaction. Proc Natl Acad Sci U S A. 2008;105(9):3292–7. Epub 2008/02/22. 10.1073/pnas.0709513105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshimura M, Asai K, Sadaie Y, Yoshikawa H. Interaction of Bacillus subtilis extracytoplasmic function (ECF) sigma factors with the N-terminal regions of their potential anti-sigma factors. Microbiology. 2004;150(Pt 3):591–9. 10.1099/mic.0.26712-0 . [DOI] [PubMed] [Google Scholar]
- 17.Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390(6657):249–56. 10.1038/36786 . [DOI] [PubMed] [Google Scholar]
- 18.Cao M, Wang T, Ye R, Helmann JD. Antibiotics that inhibit cell wall biosynthesis induce expression of the Bacillus subtilis sigma(W) and sigma(M) regulons. Mol Microbiol. 2002;45(5):1267–76. 10.1046/j.1365-2958.2002.03050.x . [DOI] [PubMed] [Google Scholar]
- 19.Pietiainen M, Gardemeister M, Mecklin M, Leskela S, Sarvas M, Kontinen VP. Cationic antimicrobial peptides elicit a complex stress response in Bacillus subtilis that involves ECF-type sigma factors and two-component signal transduction systems. Microbiology. 2005;151(Pt 5):1577–92. 10.1099/mic.0.27761-0 . [DOI] [PubMed] [Google Scholar]
- 20.Wiegert T, Homuth G, Versteeg S, Schumann W. Alkaline shock induces the Bacillus subtilis sigma(W) regulon. Mol Microbiol. 2001;41(1):59–71. 10.1046/j.1365-2958.2001.02489.x . [DOI] [PubMed] [Google Scholar]
- 21.Hahne H, Mader U, Otto A, Bonn F, Steil L, Bremer E, et al. A comprehensive proteomics and transcriptomics analysis of Bacillus subtilis salt stress adaptation. J Bacteriol. 2010;192(3):870–82. 10.1128/JB.01106-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steil L, Hoffmann T, Budde I, Volker U, Bremer E. Genome-wide transcriptional profiling analysis of adaptation of Bacillus subtilis to high salinity. J Bacteriol. 2003;185(21):6358–70. Epub 2003/10/18. 10.1128/JB.185.21.6358-6370.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Devkota SR, Kwon E, Ha SC, Chang HW, Kim DY. Structural insights into the regulation of Bacillus subtilis SigW activity by anti-sigma RsiW. PLoS One. 2017;12(3):e0174284 Epub 2017/03/21. 10.1371/journal.pone.0174284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Asai K. Anti-sigma factor-mediated cell surface stress responses in Bacillus subtilis. Genes Genet Syst. 2018;92(5):223–34. Epub 2018/01/19. 10.1266/ggs.17-00046 . [DOI] [PubMed] [Google Scholar]
- 25.Schobel S, Zellmeier S, Schumann W, Wiegert T. The Bacillus subtilis sigmaW anti-sigma factor RsiW is degraded by intramembrane proteolysis through YluC. Mol Microbiol. 2004;52(4):1091–105. 10.1111/j.1365-2958.2004.04031.x . [DOI] [PubMed] [Google Scholar]
- 26.Heinrich J, Hein K, Wiegert T. Two proteolytic modules are involved in regulated intramembrane proteolysis of Bacillus subtilis RsiW. Mol Microbiol. 2009;74(6):1412–26. 10.1111/j.1365-2958.2009.06940.x . [DOI] [PubMed] [Google Scholar]
- 27.Ellermeier CD, Losick R. Evidence for a novel protease governing regulated intramembrane proteolysis and resistance to antimicrobial peptides in Bacillus subtilis. Genes Dev. 2006;20(14):1911–22. 10.1101/gad.1440606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zellmeier S, Schumann W, Wiegert T. Involvement of Clp protease activity in modulating the Bacillus subtilissigmaw stress response. Mol Microbiol. 2006;61(6):1569–82. 10.1111/j.1365-2958.2006.05323.x . [DOI] [PubMed] [Google Scholar]
- 29.Li L, Fang C, Zhuang N, Wang T, Zhang Y. Structural basis for transcription initiation by bacterial ECF sigma factors. Nat Commun. 2019;10(1):1153 Epub 2019/03/13. 10.1038/s41467-019-09096-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hubin EA, Fay A, Xu C, Bean JM, Saecker RM, Glickman MS, et al. Structure and function of the mycobacterial transcription initiation complex with the essential regulator RbpA. Elife. 2017;6 Epub 2017/01/10. 10.7554/eLife.22520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park SY, Ha SC, Kim YG. The protein crystallography beamlines at the pohang light source II. Biodesign. 2017;5:30–4. [Google Scholar]
- 32.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):271–81. Epub 2011/04/05. 10.1107/S0907444910048675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40(Pt 4):658–74. Epub 2007/08/01. 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68(Pt 4):352–67. 10.1107/S0907444912001308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holm L, Laakso LM. Dali server update. Nucleic Acids Res. 2016;44(W1):W351–5. Epub 2016/05/01. 10.1093/nar/gkw357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laskowski RA, Swindells MB. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model. 2011;51(10):2778–86. 10.1021/ci200227u . [DOI] [PubMed] [Google Scholar]
- 38.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–97. 10.1016/j.jmb.2007.05.022 . [DOI] [PubMed] [Google Scholar]
- 39.Lu XJ, Olson WK. 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat Protoc. 2008;3(7):1213–27. Epub 2008/07/05. 10.1038/nprot.2008.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jurrus E, Engel D, Star K, Monson K, Brandi J, Felberg LE, et al. Improvements to the APBS biomolecular solvation software suite. Protein Sci. 2018;27(1):112–28. Epub 2017/08/25. 10.1002/pro.3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schrodinger, LLC. The PyMOL Molecular Graphics System, Version 1.8. 2015.
- 42.Barton GJ. ALSCRIPT: a tool to format multiple sequence alignments. "Protein Engineering, Design and Selection". 1993;6(1):37–40. 10.1093/protein/6.1.37 [DOI] [PubMed] [Google Scholar]
- 43.Huang X, Fredrick KL, Helmann JD. Promoter recognition by Bacillus subtilis sigmaW: autoregulation and partial overlap with the sigmaX regulon. J Bacteriol. 1998;180(15):3765–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14(6):1188–90. Epub 2004/06/03. 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Erickson JW, Gross CA. Identification of the sigma E subunit of Escherichia coli RNA polymerase: a second alternate sigma factor involved in high-temperature gene expression. Genes Dev. 1989;3(9):1462–71. Epub 1989/09/01. 10.1101/gad.3.9.1462 . [DOI] [PubMed] [Google Scholar]
- 46.Rhodius VA, Suh WC, Nonaka G, West J, Gross CA. Conserved and variable functions of the sigmaE stress response in related genomes. PLoS Biol. 2006;4(1):e2 10.1371/journal.pbio.0040002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miticka H, Rezuchova B, Homerova D, Roberts M, Kormanec J. Identification of nucleotides critical for activity of the sigmaE-dependent rpoEp3 promoter in Salmonella enterica serovar Typhimurium. FEMS Microbiol Lett. 2004;238(1):227–33. Epub 2004/09/01. 10.1016/j.femsle.2004.07.039 . [DOI] [PubMed] [Google Scholar]
- 48.Haran TE, Mohanty U. The unique structure of A-tracts and intrinsic DNA bending. Q Rev Biophys. 2009;42(1):41–81. Epub 2009/06/11. 10.1017/S0033583509004752 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Red and orange stick models indicate σW4 and -35W, respectively. 2Fo-Fc maps are drawn at two different orientations. The contour level is set to 1.0 σ.
(TIF)
2Fo-Fc maps are drawn at same scale and orientation as those for models in Fig 2C and arranged in the same order as the panels in Fig 2C. The contour level is set to 1.0 σ.
(TIF)
(A, B) Hydrophobic interactions between σW4 and -35W shown at two different orientations. Residues and nucleotides, which are associated with hydrophobic bonds, are drawn as red and orange stick models. Dotted lines indicate hydrophobic interactions. (C, D) 2Fo-Fc electron density maps in (C) and (D) are shown at same scale and orientation as those for models in S2A and S2B Fig, respectively. The contour level is set to 1.0 σ. (E, F) Hydrophobic interactions between σE4 and -35E. Residues and nucleotides which are associated with hydrophobic bonds are drawn as purple and green stick models.
(TIF)
(A) Superposition of σE4/-35E and σW4/-35W structures. N- and C-termini of σW4 are labeled. (B) Distribution of root-mean-square values between Cα positions of superimposed σE4 and σW4 structures. (C) Superposition of σW4 domains from the structures of σW4/-35W and SigW/RsiW.
(TIF)
(A) The green dotted line indicates the cation-π interaction between R176 of σE4 (cyan model) and C-36 of 35E (green). The corresponding residue in σW4 (R175) and base in -35W (T-36) are drawn as a stick model. The distance between the Arg and pyrimidine is labeled.
(TIF)
The residues that participate in intramolecular interactions with L137 (A), L147 (B), or E157 (C) are drawn as stick models and the residue number is labeled. The helix and loop in σW4 are colored pink and light brown. (D) σW4 in SigW/RsiW structure (PDB ID: 5WUQ) is superposed onto that of the σW4/-35W structure. SigW in SigW/RsiW structure is colored green. E157 in σW4 interacts with T71 in σW2 and does not participate in DNA binding.
(TIF)
Data Availability Statement
The final coordinates and structure factors were deposited in Protein Data Bank (PDB ID: 6JHE).