Abstract
Metastatic breast cancer (MBC) is a challenge for oncologists, and public efforts should focus on identifying additional molecular markers and therapeutic management to improve clinical outcomes. Among all diagnosed cases of breast cancer (BC; approximately 10%) involve metastatic disease; notably, approximately 40% of patients with early-stage BC develop metastasis within 5 years. The management of MBC consists of systemic therapy. Despite different treatment options, the 5-year survival rate is <20%, which may be due to a lack of response with de novo or acquired resistance. MicroRNAs (miRNAs or miRs) are promising biomarkers as they are readily detectable and have a broad spectrum and potential clinical applications. The aim of this study was to identify a miRNA profile for distinguishing patients with MBC who respond to systemic treatment. Patients with MBC were treated according to the National Comprehensive Cancer Network guidelines. We performed miRNA-Seq on 9 primary tumors using the Thermo Fisher Scientific Ion S5 system. To obtain global miRNA profiles, we carried out differentially expressed gene elimination strategy (DEGES) analysis between the responsive and non-responsive patients. The results identified a profile of 12 miRNAs associated with the response to systemic treatment. The data were validated in an independent cohort (TCGA database). Based on the results, the upregulation of miR-342-3p and miR-187-3p was associated with the response to systemic treatment, and with an increased progression-free survival (PFS) and overall survival (OS); by contrast, the downregulation of miR-301a-3p was associated with a higher PFS and OS. On the whole, the findings of this study indicate that these miRNAs may serve as biomarkers for the response to systemic treatment or the prognosis of patients with MBC. However, these data should be validated experimentally in other robust cohorts and using different specimens before implementing these miRNAs as biomarkers in clinical practice to benefit this group of patients.
Keywords: microRNAs, metastatic breast cancer, next-generation sequencing
Introduction
Breast cancer (BC) is the most prevalent type of cancer among women worldwide, with an estimated 1.7 million newly diagnosed cases in 2015. Although metastatic BC (MBC) is present in only 10% of all diagnosed patients, approximately 30-40% of early-stage BC cases will develop metastasis within 5 years (1,2). The management of MBC is not curative, and treatment consists of systemic therapy involving chemotherapy, hormonal agents and targeted therapy (3). Despite various treatment options, the 5-year survival ratio remains <20% [SEER Stat Fact Sheets: Breast Cancer. National Cancer Institute (4)]. This poor prognosis may be due to the fact that more than one-third of patients with MBC do not respond to chemotherapy (anthracyclines and taxanes), with a response rate to first-line treatment of only 20% [95% confidence index (CI) 11-28%]. Therefore, the progression of the disease occurs in <1 year. De novo or acquired resistance is the main reason for tumor relapse, contributing to a poor prognosis, a lack of therapeutic response and a fatal clinical outcome (5,6).
MicroRNAs (miRNAs or miRs) are small non-coding RNAs ~22 nt in length that negatively regulate gene expression through base pairing at 3′- or 5′-untranslated regions of messenger RNAs (mRNAs) (7). miRNAs are readily detected in formalin-fixed paraffin-embedded sections and in body fluids (e.g., blood, plasma, serum and saliva), and accordingly, these molecules have the potential to be used in clinical practice (8,9). A number of miRNAs play substantial roles in drug sensitivity/resistance in BC, and yet the majority of them have only been explored in early-stage disease (10).
Although MBC is not curable, the extent of survival with the quality of life is an important aspect for patients. In this regard, the aim of this study was to identify, through miRNA sequencing, a miRNA profile for patients with MBC who respond to systemic treatment. The results identified 12 miRNAs involved in response to systemic treatment (hormonotherapy and chemotherapy). In particular, the upregulation of miR-342-3p and miR-187-3p was associated with the response to systemic treatment, and with an improved progression-free survival (PFS) and overall survival (OS). Conversely, the downregulation of miR-301a-3p was associated with an increased PFS and OS. A further gene set enrichment analysis of putative targets of the identified miRNAs revealed their involvement in cancer, cytokine-cytokine receptor interaction, glioma, endocytosis and the mitogen-activated protein kinase (MAPK) signaling pathways. On the whole, we identified miRNAs associated with the response to systemic treatment, PFS and OS in patients with MBC. The results of this study confirm the importance of miRNAs as potential biomarkers of the response to treatment in patients with metastatic disease.
Patients and methods
Patient selection and treatment regimen
This prospective cohort study was approved by the Central Ethics and Scientific Committee at the National Cancer Institute in Mexico City (approval no. CEI/1001/16; 016/010/IBI). Informed consent was obtained for each patient enrolled. A total of 9 patients were enrolled diagnosed with MBC confirmed by positron emission tomography (PET) and computed tomography (CT) scans. All patients were treated according to the National Comprehensive Cancer Network (NCCN) guidelines (11). As first-line treatment, 44.4% of the patients received hormonal therapy and 55.5% platinum-based chemotherapy.
Outcome measurement
As mentioned above, all patients underwent PET or CT and were evaluated by The Response Evaluation Criteria in Solid Tumors (RECIST) at baseline and at 6 months (12). PFS was defined as the time from the commencement of treatment until disease progression or the last visit. OS was defined as the time from diagnosis until death or the last visit.
Preparation of tissue samples
A total of 9 MBC biopsies were collected from February, 2018 to November, 2018 at the time of diagnosis prior to any therapeutic procedures. Total RNA was extracted from the tumor samples using TRIzol reagent (cat. no. 15596-026, Invitrogen; Thermo Fisher Scientific;) and subsequently purified with the miRNeasy Mini kit (cat. no. 217004; Qiagen) according to the manufacturer's instructions. The RNA concentration was determined by Quibit 2.0 fluorometry using the Quibit RNA HS Assay kit (Thermo Fisher Scientific). After biopsies were obtained, pathological confirmation of at least 80% tumor cells was obtained. To construct a small RNA library, small RNAs ranging from 10 to 40 nt were assessed for quality and quantity using an Agilent 2100 bioanalyzer (Agilent Technologies).
Library preparation
Small RNA libraries were prepared using Ion Total RNA-Seq V2 (Life Technologies; Thermo Fisher Scientific; cat. nos. 4475936 and 44797789) according to the manufacturer's instructions. Following adapter ligation, first-strand cDNA synthesis and amplification were performed as follows: 94° for 2 min and 14 cycles at 94° 30 sec, 62° 30 sec and 68° 30 sec. Briefly, adapters were ligated to small RNAs (25 ng/sample), and libraries from 9 samples were pooled together in equimolar ratios (100 pM) for template preparation and chip loading using the Ion Chef System. The libraries were sequenced with Ion S5 using S530 chips, as recommended by the manufacturer (Life Technologies; Thermo Fisher Scientific; cat. nos. 4475936 and 44797789).
Data analysis
Raw reads were processed using the small RNA sequencing plugin provided in the Ion Torrent Suite (Thermo Fisher Scientific), and options were set to only retain reads between 17 and 35 nt. The trimmed reads were analyzed with Chimira using the default settings and aligned using the latest version of miRBase (13). The resulting miRNA counts were filtered, and only miRNAs with at least 1 count in every sample were retained. Normalization and differential expression analysis were carried out with the Bioconductor package DESeq2 (14), and miRNAs with a P-value <0.05 were selected for further analysis.
Identification of putative miRNA targets
To identify possible targets of each miRNA, we first downloaded the TCGA Breast transcriptome dataset using Bioconductor package TCGA biolinks (15). Subsequently, we assessed differentially expressed (DE) genes between normal and stage IV primary tumor tissue with DESeq2, only selecting those mRNAs with P-adj <0.01 and a log2 Fold change ≤−1. As this would mean that the expression is at least 0.5-fold lower in the tumor samples, in this manner, we ensured that the putative mRNA targets of the miRNAs were downregulated in the tumor samples. Subsequently, we used these downregulated mRNAs with the Bioconductor package miRNAtap to predict targets in 5 databases (PicTar (pictar.mdc-berlin.de), TargetScan (targetscan.org), miRanda (microrna.org), DIANA (diana. imis.athena-innovation.gr) and miRDB (mirdb.org); we only considered predicted targets identified by at least 2 databases. Pearson's correlation coefficients were calculated between the predicted targets and miRNAs using the TCGA data.
Statistical analysis
For descriptive purposes, continuous variables are summarized as arithmetic means, medians and standard deviations. The Fisher-Pitman permutation test from the coin R package was employed using the normalized rlog counts (P<0.05). The Mann-Whitney U-test (non-parametric test) was used for inferential comparisons. Statistically significant and borderline significant variables (P<0.1) were included in multivariate logistic regression analysis. Survival results were analyzed using the Kaplan-Meier technique, and the log-rank test was employed for comparisons between subgroups. All variables were dichotomized for analyzing survival curves. Hazard ratios (HRs) were calculated along with their corresponding 95% CIs as a measure of association. Statistical significance was defined as P<0.05 using Student's t-test. Statistical analysis was conducted using R/Bioconductor.
Results
Clinical characteristics of patients with MBC and response rates to systemic treatment
Samples from 9 patients were sequenced. All patients were females and had stage IV disease. The mean of age of the patients was 53 years. All patients had an Eastern Cooperative Oncology Group (ECOG) performance status of 0 or 1. The most frequent metastasis sites were the cervical ganglia, mediastinum and lungs. The most frequent molecular subtype was luminal A. Four patients received hormone therapy (letrozol) and five patients received chemotherapy (docetaxel, paclitaxel, paclitaxel/trastuzumab or cyclophosphamide/adriamycin). According to RECIST, the overall response rate (ORR) was 44.44% (22.2% complete response and 22.2% partial response), stable disease was observed in 44.4%, and the disease progressed in 11.1% of patients (Table I).
Table I.
Clinicopathologic characteristics of the patients.
| Clinical parameters | Patients n=9 (100%) |
|---|---|
| Sex | |
| Female | 9 (100) |
| Age | |
| >50 | 6 (66.6) |
| ≤50 | 3 (33.3) |
| ECOG status | |
| 0-2 | 9 (100) |
| 3-4 | 0 (0) |
| Histopathology | |
| Infiltrating ductal carcinoma | 5 (55.5) |
| Classic lobular carcinoma | 1 (11.1) |
| Non-specific infiltrative | 2 (22.2) |
| Canalicular carcinoma | 1 (11.1) |
| Molecular subtype | |
| Luminal A | 5 (55.5) |
| Luminal B | 2 (22.2) |
| Triple negative | 2 (22.2) |
| Treatment | |
| Hormonotherapy | 4 (44.4) |
| Chemotherapy | 5 (55.5) |
| Metastatic | |
| Liver | 2 (22.2) |
| Lung | 3 (33.3) |
| Bones | 2 (22.2) |
| Cervical Ganglia | 5 (55.5) |
| Axillary Ganglia | 1 (11.1) |
| Mediastinum | 4 (44.4) |
| Response to systemic treatments | |
| Complete response | 2 (22.2) |
| Partial response | 2 (22.2) |
| Stable disease | 4 (44.4) |
| Progression | 1 (11.1) |
Identification of DE miRNAs in patients with MBC
To reveal putative miRNA biomarkers for the response to systemic treatment, we first assessed DE miRNAs between the responders and non-responder's groups. In total 12 miRNAs were found with P-values <0.05. It has been estimated that a sample size of >60 is needed to achieve an FDR of 10% (16); however, due to the nature of patients with MBC it is unlikely, that this number of samples will be obtained. Due to the small sample size in this study, the lowest FDR that was obtained was 0.25; thus, we opted for a P-value filtering and validation of our results in an independent and larger cohort to reduce the disadvantages of using a P-value.
This analysis revealed a panel of 12 miRNAs that separated the patients in these 2 groups. This panel included miRNAs with a P-value <0.05, among which 8 were upregulated (miR-7-5p, miR-141-3p, miR-187-3p, miR-200b-3p, miR-200c-3p, miR-301a-3p, miR-342-3p and miR-3182) and 4 were downregulated (miR-361-3p, miR-1273a, miR-4459 and miR-4485-3p) in the responders vs. the non-responders (Fig. 1). We then performed unsupervised hierarchical clustering of the 12 DE miRNAs using the Euclidian distance. As shown in Fig. 2, the tumors were clustered according to the respective response to treatment. Is important to mention that, in order to obtain the DE miRNAs, we employed DESeq2, an algorithm that assumes a negative binomial distribution (17,18), which is one of the best methods with which to analyze RNA-seq data due to its stringency, good control of false-positives, and improved sensitivity and specificity. Moreover, in addition to this, we employed the Fisher-Pitman permutation test on the 12 miRNAs. The results revealed that 6 miRNAs exhibited significant differences between the 2 groups (P<0.05) (miR-4485-3p, miR-1273a, miR-342-3p, miR-200c-3p, miR-200b-3p and miR-187-3p) and 1 miRNA was close to being significant (miR-301a-3p, P=0.055) (Table SI).
Figure 1.
(A-L) MicroRNAs associated with the response to systemic treatment in BC. Each bar plot shows the number of reads for each miRNA according to response (R) vs. no response (NR) to systemic treatment. *P<0.05. BC, breast cancer.
Figure 2.
Hierarchical clustering of miRNA expression. The BC samples were classified into 2 different groups (response vs. no response to systemic treatments) based on expression levels. Each column represents a patient with MBC, and each row represents a single miRNA. The expression of miRNAs is represented in blue (upregulated), red (downregulated) and white (no significant change or absence of data). BC, breast cancer; MBC, metastatic breast cancer.
miRNAs with clinical significance
To assess the clinical significance of the miRNA panel, we applied the non-parametric Mann-Whitney U test using clinical variables (Table II). The results revealed significant differences in the expression levels of some miRNAs according to the site of metastasis. For example, miR-4459 was overexpressed in patients with lung metastasis (P=0.02); this same miRNA was overexpressed in those with mediastinum metastasis (P=0.03). Patients without lung metastasis exhibited an upregulation of miR-342-3p and miR-200b-3p (P=0.02 and P=0.04, respectively), and miR-4485-3p, miR-1273a and miR-361-3p were overexpressed in those without cervical ganglia metastasis (P=0.03, P=0.01 and P=0.01, respectively). Notably, we found no significant differences in the expression levels of the 12 miRNAs in patients with liver or bone metastasis (Table II).
Table II.
Clinicopathologic characteristics related to miRNA expression in patients with metastatic breast cancer.
| Variables | Patients n=9 | miR-4485-3p Mean ± SD | miR-1273a Mean ± SD | miR-361-3p Mean ± SD | miR-342-3p Mean ± SD | miR-200c-3p Mean ± SD | miR-301a-3p Mean ± SD |
|---|---|---|---|---|---|---|---|
| Age | |||||||
| ≤53 | 6 | 7.94±6.51 | 22.28±12.20 | 10.67±11.77 | 35.55±35.39 | 34.025±47.18 | 17.93±25.33 |
| >53 | 3 | 3.33±3.85 | 13.13±6.63 | 7.59±7.68 | 31.14±13.43 | 23.85±7.68 | 6.29±8.76 |
| P-value | 0.38 | 0.39 | 0.9 | 0.71 | 0.71 | 0.54 | |
| Smoking | |||||||
| Positive | 3 | 10.32±6.39 | 28.57±9.99 | 21.86±8.35 | 18.56±8.50 | 18.67±5.81 | 6.12±3.82 |
| Negative | 6 | 4.44±5.16 | 14.46±8.92 | 3.53±1.91 | 41.84±32.97 | 36.61±46.36 | 17.28±25.78 |
| P-value | 0.16 | 0.09 | 0.02 | 0.38 | 0.9 | 1 | |
| Treatment | |||||||
| Hormone therapy | 4 | 7.56±5.077 | 20.23±8.33 | 7.57±6.105 | 39.70±41.39 | 41.19±58.93 | 18.26±32.83 |
| Chemotherapy | 5 | 5.47±7.02 | 18.31±13.95 | 6.105±11.30 | 29.58±18.01 | 22.18±7.65 | 9.79±7.18 |
| P-value | 0.55 | 0.55 | 1 | 0.9 | 0.55 | 0.73 | |
| HER2 | |||||||
| Positive | 2 | 9.79±11.03 | 23.89±22.54 | 16.24±21.56 | 29.99±29.31 | 24.01±1.20 | 13.54±4.74 |
| Negative | 7 | 5.43±4.70 | 17.815±8.47 | 7.75±6.49 | 35.25±30.99 | 32.52±43.48 | 13.56±24.42 |
| P-value | 0.5 | 1 | 1 | 1 | 0.5 | 0.33 | |
| Menarche | |||||||
| >12 years | 4 | 11.14±5.47 | 29.36±8.31 | 18.10±10.16 | 19.87±7.42 | 15.98±7.16 | 5.15±3.67 |
| ≤12 years | 5 | 2.61±2.86 | 11.00±3.12 | 2.87±1.16 | 45.45±35.51 | 42.35±49.39 | 20.28±27.62 |
| P-value | 0.03 | 0.01 | 0.01 | 0.41 | 0.41 | 0.7 | |
| Site of metastasis | |||||||
| Liver | |||||||
| Positive | 2 | 11.59±8.49 | 32.48±10.39 | 24.58±9.76 | 17.62±7.81 | 7.88±3.25 | 7.88±3.25 |
| Negative | 7 | 5.54±5.03 | 15.75±9.22 | 5.67±5.60 | 18.56±9.88 | 6.46±7.91 | 6.46±7.91 |
| P-value | 0.42 | 0.14 | 0.7 | 1 | 0.64 | 0.64 | |
| Lung | |||||||
| Positive | 2 | 10.29±6.41 | 26.78±12.30 | 17.55±14.00 | 12.70±6.74 | 14.08±8.26 | 5.49±4.74 |
| Negative | 7 | 4.46±5.18 | 15.35±9.29 | 5.68±5.59 | 44.77±30.05 | 38.91±44.91 | 17.59±25.54 |
| P-value | 0.26 | 0.16 | 0.26 | 0.02 | 0.26 | 0.71 | |
| Mediastinum | |||||||
| Positive | 2 | 2.97±3.23 | 12.59±5.88 | 6.20±6.93 | 49.60±36.43 | 48.23±53.98 | 21.75±31.39 |
| Negative | 7 | 9.14±6.49 | 24.42±12.04 | 12.39±12.25 | 21.67±15.39 | 16.55±11.03 | 7.00±6.34 |
| P-value | 0.19 | 0.11 | 0.55 | 0.11 | 0.28 | 0.73 | |
| Bone | |||||||
| Positive | 1 | 7.39±8.78 | 22.39±13.24 | 5.17±2.29 | 62.24±54.34 | 68.51±85.67 | 34.88±46.13 |
| Negative | 8 | 6.12±5.84 | 18.24±11.50 | 10.92±11.35 | 26.03±16.69 | 19.81±8.44 | 7.46±7.11 |
| P-value | 1 | 0.66 | 1 | 0.33 | 0.88 | 0.66 | |
| Cervical ganglia | |||||||
| Positive | 3 | 2.61±2.86 | 11.00±3.12 | 2.87±1.16 | 45.45±35.51 | 42.35±49.39 | 20.28±27.62 |
| Negative | 4 | 11.14±5.47 | 29.36±8.31 | 18.10±10.16 | 19.87±7.42 | 15.98±7.16 | 5.15±3.67 |
| P-value | 0.03 | 0.01 | 0.01 | 0.41 | 0.41 | 0.73 |
| Variables | miR−4459 Mean ± SD | miR-200b-3p Mean ± SD | miR-3182 Mean ± SD | miR −141-3p Mean ± SD | miR-7-5p Mean ± SD | miR-187-3p Mean ± SD |
|---|---|---|---|---|---|---|
| Age | ||||||
| ≤53 | 5.92±6.36 | 5.50±4.85 | 1,245.80±1,623.86 | 53.52±52.01 | 16.54±21.12 | 2.09±4.22 |
| >53 | 0.41±0.72 | 9.50±5.05 | 1,976.99±2,681.96 | 15.85±1.32 | 3.98±3.31 | 0.31±0.55 |
| P-value | 0.08 | 0.26 | 0.9 | 0.16 | 0.38 | 0.67 |
| Smoking | ||||||
| Positive | 8.65±8.80 | 3.85±2.99 | 903.56±338.08 | 24.04±7.53 | 4.86±1.54 | 0.30±0.53 |
| Negative | 1.80±1.73 | 8.32±5.34 | 1,782.52±2,318 | 49.09±54.96 | 16.10±21.50 | 2.10±4.21 |
| P-value | 0.35 | 0.38 | 0.54 | 0.9 | 0.54 | 0.47 |
| Treatment | ||||||
| Hormone therapy | 1.89±2.21 | 6.83±5.24 | 453.168±247.04 | 43.61±58.49 | 18.66±26.72 | 2.66±5.32 |
| Chemotherapy | 5.84±7.32 | 6.83±5.44 | 2,318.62±2,276.15 | 38.44±39.20 | 7.30±5.92 | 0.57±0.52 |
| P-value | 0.53 | 0.9 | 0.28 | 1 | 1 | 0.68 |
| HER2 | ||||||
| Positive | 9.79±11.03 | 5.83±3.00 | 2,565.25±2,691.15 | 69.44±53.67 | 10.76±8.67 | 0.96±0.04 |
| Negative | 2.46±3.12 | 7.12±5.61 | 1,182.18±1,752.23 | 32.54±43.72 | 12.80±20.36 | 1.65±3.98 |
| P-value | 0.29 | 0.88 | 0.5 | 0.22 | 0.66 | 0.26 |
| Menarche | ||||||
| >12 years | 7.34±7.65 | 3.73±2.45 | 760.75±397.20 | 24.26±6.16 | 6.77±4.00 | 0.23±0.46 |
| ≤12 years | 1.48±1.73 | 9.31±5.33 | 2,072.55±2,466.90 | 53.92±60.00 | 16.82±23.96 | 2.52±4.57 |
| P-value | 0.26 | 0.19 | 0.9 | 0.73 | 0.9 | 0.22 |
| Site of metastasis | ||||||
| Liver | ||||||
| Positive | 12.98±6.52 | 2.31±1.96 | 976.16±443.83 | 27.84±5.15 | 5.57±1.33 | 0.46±0.65 |
| Negative | 1.80±1.73 | 7.11±4.50 | 1,820.28±2,295.30 | 30.11±38.51 | 7.00±6.59 | 0.32±0.50 |
| P-value | 0.06 | 0.28 | 0.85 | 0.42 | 1 | 1 |
| Lung | ||||||
| Positive | 10.05±6.85 | 2.47±1.41 | 714.16±551.74 | 19.49±14.91 | 3.942.96 | 0.300.53 |
| Negative | 1.10±1.39 | 9.01±4.72 | 1,877.22±2,250.58 | 51.36±52.90 | 16.56±21.13 | 2.104.21 |
| P-value | 0.02 | 0.04 | 0.9 | 0.71 | 0.26 | 0.47 |
| Mediastinum | ||||||
| Positive | 0.49±0.99 | 11.10±4.28 | 1,469.77±2,016.29 | 67.36±60.17 | 19.83±26.40 | 3.15±5.02 |
| Negative | 6.96±6.48 | 3.42±1.92 | 1,505±2,027.359 | 19.44±11.26 | 6.36±4.29 | 0.18±0.41 |
| P-value | 0.03 | 0.01 | 0.9 | 0.41 | 0.73 | 0.08 |
| Bone | ||||||
| Positive | 1.70±2.40 | 8.80±7.64 | 432.07±141.03 | 77.61±74.47 | 35.25±32.21 | 5.32±7.53 |
| Negative | 4.77±6.37 | 6.27±4.69 | 1,791.66±2,071.41 | 30.20±35.18 | 5.81±5.52 | 0.411±0.51 |
| P-value | 0.65 | 0.88 | 0.5 | 0.22 | 0.11 | 0.63 |
| Cervical ganglia | ||||||
| Positive | 1.48±1.73 | 9.31±5.33 | 2,072.55±2,466.90 | 53.92±60.00 | 16.82±23.96 | 2.52±4.57 |
| Negative | 7.34±7.65 | 3.73±2.45 | 760.75±397.20 | 24.26±6.16 | 6.77±4.00 | 0.23±0.46 |
| P-value | 0.26 | 0.19 | 0.9 | 0.73 | 0.9 | 0.22 |
Values in bold font indicate statistically significant differences (P<0.05).
Validation of the panel of miRNAs in TCGA in patients with stage IV disease
We further evaluated this miRNA panel in an independent cohort using TCGA. All the clinical data of the patients with MBC included in the database were obtained using the clinical variable 'Person neoplasm cancer status', and only those patients with complete information of their clinical outcome were selected (follow-up median, 2.6 years). Through this analysis, we identified 17 patients with MBC from the 1,060 patients with BC included in this cohort. A patient was excluded due to the unavailability of clinical response data. We then classified these patients into 2 groups according to their response to systemic treatment: Response vs. no response (in terms of TCGA: Tumor-free and with tumor, respectively). Only 2 patients of 14 presented a response to systemic treatment (Table SII).
The validation of the miRNA panel obtained from our cohort in these 2 TCGA groups (responders vs. no responders), revealed that TCGA patients expressed 8 of the 12 miRNAs (miR-200c-3p, miR-301a-3p, miR-342-3p, miR-361-3p, miR-7-5p, miR-200b-3p, miR-187-3p and miR-141-3p) (Fig. 3). A trend of the upregulation of miR-200c-3p, miR-342-3p, miR-200b-3p, miR-187-3p and miR-141-3p was evident in the response group (Fig. 3A, C, F and G), which was in accordance with the results from our cohort (Fig. 1A, C, F and G).
Figure 3.
(A-H) MicroRNAs associated with the response to systemic treatment in BC in the TCGA cohort. The bar plot shows the numbers of reads for each miRNA according to response (R) vs. no response (NR) to systemic treatment.
miRNAs associated with the survival of patients in TCGA with stage IV disease
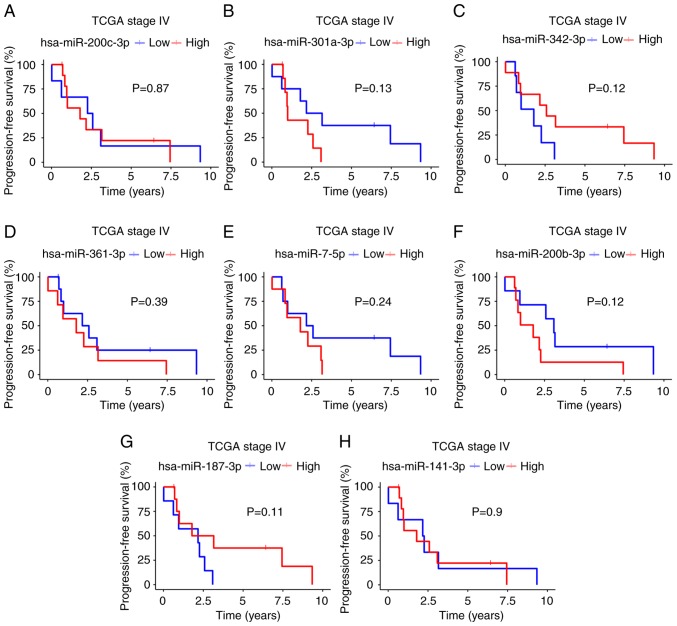
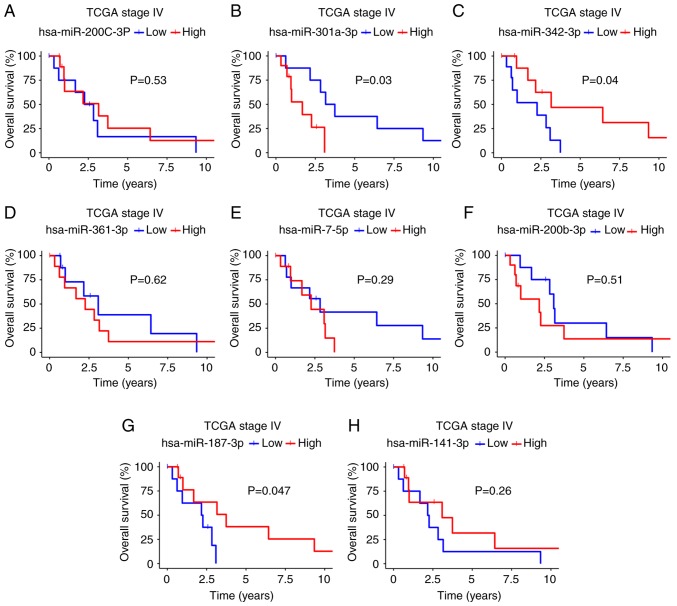
Using the available TCGA clinical data, we determined the impact of the 8 validated miRNAs on survival by associating miRNA expression (high or low) with PFS and OS (Figs. 4 and 5). Kaplan-Meier analysis revealed that patients with a better PFS had a high expression of miR-342-3p and miR-187-3p (median survival: miR-342-3p, 1.79 vs. 2.58 years, P=0.12; miR-187-3p, 2.17 vs. 2.47; P=0.11; Fig. 4C and G), but a low expression of miR-301a-3p (2.63 vs. 0.9; P=0.13; Fig. 4B). Moreover, this trend was also observed for OS: A high expression of miR-342-3p and miR-187-3p was found in the patients with a better OS (miR-342-3p, 2.26 vs. 3.16 years, P=0.04; miR-187-3p, 3.74 vs. 2.22, P=0.047; Fig. 5C and G), with a low expression of miR-301a-3p (3.45 vs. 1.68, P= 0.03; Fig. 5B). According to univariate analysis of these 3 miRNAs, only miR-342-3p was associated with OS (HR, 0.25; 95% CI, 0.006-1.2; P>0.056), although borderline in multivariate regression Cox analyses (HR, 0.08; 95% CI, 0.006-1.2; P=0.068). According to univariate analysis, miR-187-3p was associated with PFS (HR, 0.25; 95% CI, 0.062-0.98; P>0.046) and none miRNA was associated with PFS in multivariate regression Cox analyses (Tables III and SIII).
Figure 4.
(A-H) Progression-free survival of patients with stage IV disease in TCGA based on 8 validated miRNAs. The high expression of miRNAs is shown by a red line. A blue line denotes miRNA downregulation.
Figure 5.
(A-H) Overall survival of patients with stage IV disease in TCGA based on 8 validated miRNAs. A high expression of miRNAs is shown by a red line. A blue line denotes miRNA downregulation.
Table III.
Overall survival in patients expressing the miRNA panel.
| Overall survival
|
||||
|---|---|---|---|---|
| Univariate analysis
|
Multivariate analysis
|
|||
| HR (95% CI) | P-value | HR (95% CI) | P-value | |
| Age | ||||
| <60 vs. >60 years | 0.36 (0.084-1.5) | 0.17 | 0.10 (0.004-2.46) | 0.162 |
| Progesterone status | ||||
| Positive vs. negative | 0.51 (0.13-1.9) | 0.32 | 3.73 (0.277-50.34) | 0.32 |
| hsa-miR-187-3p | ||||
| High vs. low | 0.22 (0.042-1.2) | 0.082 | 1.19 (0.08-17.89) | 0.89 |
| hsa-miR-301a-3p | ||||
| High vs. low | 4.1 (0.75-23) | 0.1 | 0.72 (0.07-6.82) | 0.77 |
| hsa-miR-342-3p | ||||
| High vs. low | 0.25 (0.062-1) | 0.056 | 0.08 (0.006-1.200) | 0.068 |
Values in bold font indicate statistically significant differences (P<0.05).
Identification of mRNA targets of miRNAs and their participation in pathways of cancer
We focused in the identification of the mRNA targets for miR-342-3p, miR-187-3p and miR-301a-3p as we observed that their expression was associated with the response to systemic treatment and with PFS, both in our data and in the TCGA data. Patients with a higher expression of miR-342-3p and miR-187-3p exhibited a trend for a longer PFS, although none of the survival curves were statistically significant due to the small size of the cohort (Fig. 4C and G). To achieve this goal, we analyzed the TCGA breast transcriptome dataset, as described in the Patients and methods section. We reasoned that, since these 3 miRNAs were overexpressed in patients with response to systemic treatment, they may thus act as tumor-suppressor miRNAs, probably inhibiting key oncogenes. Thus, to obtain their putative targets, we only focused on the downregulated mRNAs.
We found 8,116 downregulated mRNAs in the tumor samples. Using 5 databases (PicTar, TargetScan, miRanda, DIANA and miRDB), we identified 416 mRNAs predicted to be potential targets of the 3 miRNAs (Table SIV). To achieve further insight into the biological functions of these miRNAs, we performed KEGG pathway analysis of the 416 predicted targets in WEB-based Gene SET Analysis Toolkit. We only selected the top altered pathways per miRNA, considering those that had a P-value <0.05 and ranked them by their P-value, and identified 16 dysregulated signaling pathways related to miR-342-3p, miR-187-3p and miR-301a-3p (Fig. 6). Moreover, the results revealed miR-301a-3p and miR-342-3p affected the same pathways in cancer, cytokine-cytokine receptor interaction, glioma and endocytosis. We also observed this effect between miR-342-3p and miR-187-3p, which affect the MAPK signaling pathway (Fig. 6).
Figure 6.
KEGG pathway heatmap. Functional annotation of 3 miRNAs using the KEGG pathway database with adjusted P-values showed the top altered pathways dysregulated by miR-342-3p, miR-187-3p and miR-301a-3p.
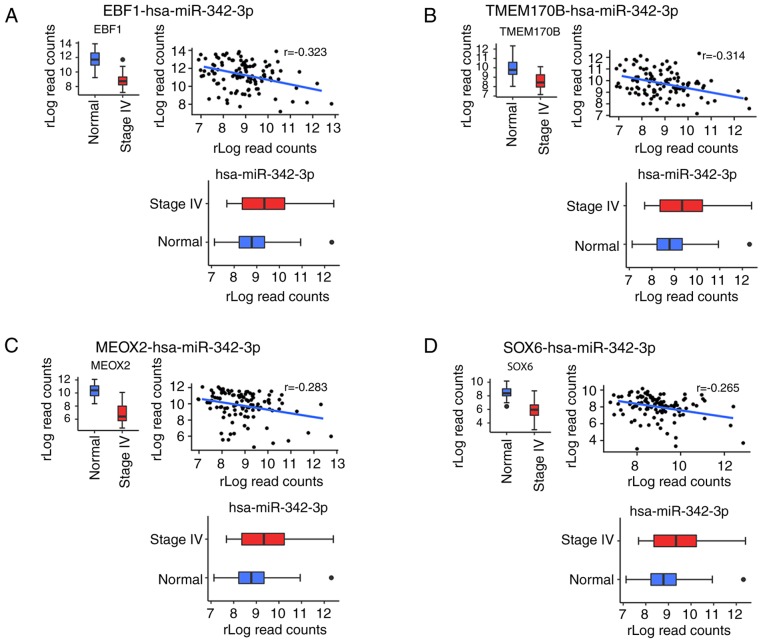
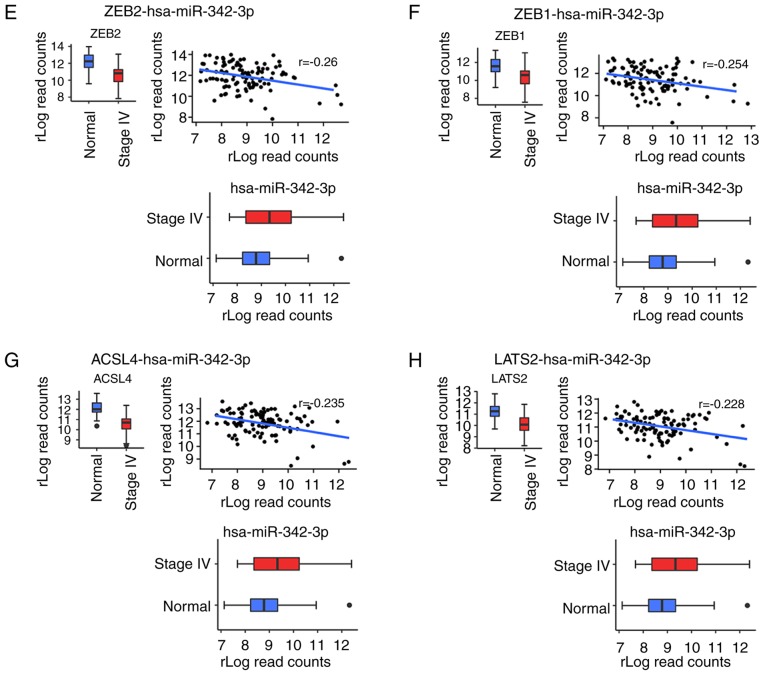
Identification of main miR-342-3p targets
To investigate the main predicted targets, we focused on miR-342-3p as it was associated with a lack of lung metastasis and response to treatment and was a prognostic factor for OS in both the Kaplan-Meier and Cox multivariate analyses. Pearson's correlation analysis was performed between miR-342-3p and the predicted targets, only considering interactions with a negative coefficient and with a P-value <0.05. Based on these criteria, the number of mRNAs targets was reduced from 56 to 15 (Table SV), and the predicted targets were ranked according their correlation coefficient. In total 8 mRNAs were found to be the main targets of miR-342-3p (Fig. 7). The most negatively correlated target was the nucleic transcription factor early B cell factor1 (EBF1) (Fig. 7A). Other transcription factors were also determined as targets, such as mesenchyme homeobox 2 (MEOX2), zinc finger E-box binding homeobox 1 and 2 (ZEB1/ZEB2) (Fig. 7C, E and F). In addition, tumor suppressors were related to this miRNA, including transmembrane protein 170B (TMEM170B), SRY-box 6 (SOX6) and large tumor suppressor kinase 2 (LATS2). Acetyl-CoA synthetase long chain family member 4 (ACSL4) is another identified target involved in arachidonic acid metabolism (Fig. 7G).
Figure 7.
(A-H) Correlation analysis between miR-342-3p and the main predicted targets. Eight mRNAs were found to be the main targets of miR-342-3p.
Discussion
In the era of personalized therapy, it has been reported that more than one-third of patients with MBC do not respond to systemic treatment (1,6). However, there is a permanent effort on the part of the scientific community to identify biomarkers that can help patients with MCB who will benefit from systemic therapy (5,6,10). This study is a pioneer in this regard, and our findings highlight the important role of miRNAs as predictors of response to systemic treatment in MBC.
Is well known that chemotherapy and hormonotherapy act through different mechanisms; this is the reason why systemic treatment for patients with BC is decided according to the molecular subtype (3). In spite of this, it has been reported that the detection of a single miRNA or group of these can predict resistance to multiple therapeutic strategies; however, the majority of studies are still preclinical (10). In this regard, we focused on identifying miRNAs associated with response to systemic treatments in patients with MBC that could be considered biomarkers of response to systemic treatments in the near future, irrespective of the molecular subtype.
Through a comprehensive sequencing approach, we obtained a panel of 12 miRNAs that separated patients with MBC into 2 groups: Responders vs. non-responders (Figs. 1 and 2). In an effort to provide information about biomarkers for the response to treatment in patients with MBC, we validated our results in a large clinical cohort using TCGA. Previous studies have confirmed that TCGA cohorts represent a robust and external independent means of validating genomic data (19,20). Indeed, this genomic database symbolizes a revolution in the acquisition of information about tumors. The TCGA dataset has established validated quality standards ensuring homogeneity across data generated by multiple research groups; a precise strict set of criteria to be used for advanced genomic analysis and sequencing technologies of tissue samples exist. Moreover, TCGA includes data from a treatment-naïve cohort of 1,060 patients with BC (19). For our analysis, we retrieved genomic data for 16 patients with stage IV disease, 2 of whom presented a response to systemic treatment.
We validated our miRNA dataset in these patients with stage IV disease from TCGA and found that the upregulation of miR-342-3p and miR-187-3p was associated with a response to systemic treatment. Moreover, miR-342-3p, miR-187-3p and miR-301a-3p were associated with PFS and OS. Although we found clinical relevance for miR-187-3p and miR-301a-3p, there is little information about the roles and clinical relevance of these molecules in BC. A study published in 2012 reported that miR-187 expression was associated with an aggressive, invasive phenotype and a poor outcome in BC, which is contrary to the results of the present study; however, the cohort consisted of patients with early and locally advanced stages, and the authors did not validate their data using TCGA (21). Nonetheless, miR-187 expression has been shown to inhibit the proliferation, migration and invasion of osteosarcoma by targeting ZEB2 (22). Similarly, two other studies reported that miR-187-3p expression induces apoptosis and inhibits cellular migration, invasion and metastasis in non-small cell lung cancer and hepatocellular carcinoma (23,24).
In this study, we found miR-301a-3p downregulation to be associated with an improved PFS and OS; similarly, a recent study demonstrated that this miRNA plays an oncogenic role and that its expression is associated with metastasis and a poor prognosis of patients with estrogen receptor-positive BC. Furthermore, the expression of miR-301a-3p contributes to the development of estrogen independence and a highly invasive phenotype of BC (25). In hepatocellular carcinoma, higher levels of miR-301a-3p expression have been associated with a poor prognosis and chemoresistance, as demonstrated by an in vitro analysis (26). Overall, these findings provide us with an opportunity to enhance our knowledge about the roles of miR-187-3p and miR-301a-3p as treatment response or prognosis biomarkers for MBC and other neoplasia.
In this study, the upregulation of miR-342-3p was associated with the absence of lung metastasis and the response to systemic therapy, and was found to be a prognostic factor for OS. It has also been reported that high levels of miR-342-expression may act as a biomarker for tamoxifen sensitivity in estrogen receptor-alpha-positive BC (27). Similarly, miR-342 downregulation has been reported to be related to recurrence as it contributes to tamoxifen resistance by regulating genes involved in apoptosis and cell cycle progression, both processes that influence breast tumori-genesis and tumor response to therapeutic intervention (28). These findings are supported by the results in the study by Young et al, who reported that a high expression of miR-342 was related to tamoxifen sensitivity and a better survival in a TCGA cohort (29).
In this study, we identified the main targets of miR-342-3p, and by integrating this information, we found that some of these targets have been implicated in treatment resistance mechanisms. For instance, EBF1 is involved in drug resistance and may be a potential prognostic marker in leukemia (30). The overexpression of different transcription factors, such as MEOX2 and ZEB1 contributes to chemoresistance in lung and breast cancers, respectively (31,32) and the overexpression of ZEB2 appears to be involved in resistance to cisplatin and epidermal growth factor inhibitors in gastric and bladder cancers, respectively (33,34). Another target identified was ACSL4, which was recently associated with drug resistance in BC through the involvement of ABC transporters (35). Further characterization of miR-342-3p targets revealed a negative correlation with the tumor suppressor TMEM170B; although it has been reported that its overexpression in BC promotes inhibition of the WNT/ß-catenin pathway, its association with resistance to treatment has not been evaluated (36). Taken together, the results highlight the importance and participation of miR-342-3p in regulating several traits involved in tumor progression and resistance to treatment.
The results of this study indicate that miR-342-3p in coordination with miR-301a-3p participates in cytokine-cytokine receptor interaction, glioma and endocytosis pathways, which are involved in drug resistance and associated with a poor prognosis (Fig. 6). Recent RNA-Seq data demonstrate that the cytokine-cytokine receptor interaction pathway is a key factor in treatment resistance of triple-negative BC (37), concordant with studies describing that cytokines secreted via cancer stroma cells are crucial for conferring resistance to chemo therapy (38,39). Endocytosis is another process that when altered, is involved in resistance to trastuzumab in BC through Src-mediated phosphorylation of CAV1 (40). Finally, we found that miR-342-3p and miR-187-3p regulate the MAPK cascade, which is involved in several important physiological functions such as proliferation, survival and chemoresistance, as well as a poor outcome of patients with BC (41,42).
The limitations of this study were the following: i) The small number of patients with MBC (only 2 of whom in the TCGA cohort presented a response to treatment) and; ii) from the patients in TCGA, only one was Hispanic and the remainder Caucasian; thus, considering ethnicity, it could be considered that BC is a disease with similar transcriptomic and genomic alterations. Nonetheless, on the whole, our results suggest that miRNA-342-3p and miRNA-187-3p may serve as good biomarkers for a response to systemic treatment in patients with MBC and that miR-301-3p may constitute a prognosis biomarker. However, these data should be validated experimentally in other robust cohorts and using different specimens before implementing these miRNAs as biomarkers in clinical practice to benefit this group of patients.
Supplementary Data
Acknowledgments
The authors would like to thank Tumor Bank of the Instituto Nacional de Cancerologia (INCan) of México City for providing some biological samples. The authors would also like to thank Dr Alfonso Méndez Tenorio from Escuela Nacional de Ciencias Biológicas, Instituto Politécnico Nacional for the support with ThermoFisher Scientific Ion S5 equipment. In addition, the authors would like to thank Histotechnology Eduardo Bautista Nava for its support in the performance of immunohistochemistry and the interns Gerardo Cuamani Mitznahuatl and Jaime Jesús Ordónez Adán, for their support in the collection of clinical data. Antonio Daniel Martinez Gutierrez is a doctoral student from Programa de Doctorado en Ciencias Biomedicas, Universidad Nacional Autónoma de México (UNAM) and was supported by CONACYT (628988).
Funding
This study was supported by the Council for Science and Technology (CONACyT) (SALUD-2015-1-262044, PN-249020).
Availability of data and materials
All data generated or analyzed during this study are included in this published article or are available from the corresponding author on reasonable request.
Authors' contributions
ADCP and CPP. were the principal investigators and contributed to the conception and design of the study. ADMG contributed to all bioinformatics analyses. OMC CLC, LTOM and ADCP were involved in the sequencing. FIPR contributed to the pathological analysis of the samples. RVR, AAM, FLM, JEBR, LAH and DCDL recollected samples and were responsible for all clinical data of the patients. ADCP, ADMG and CPP wrote, drafted, revised the manuscript. All authors contributed to the conception of the article; all authors revised the final manuscript and agreed to its submission to this journal.
Ethics approval and consent to participate
This study complies with ethical standards. The study was approved by Instituto Nacional de Cancerología of México City ethics committees. The participants provided consent, including patient information, after they received verbal and written explanation. Individual consent was signed by each individual.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Gonzalez-Angulo AM, Morales-Vasquez F, Hortobagyi GN. Overview of resistance to systemic therapy in patients with breast cancer. Adv Exp Med Biol. 2007;608:1–22. doi: 10.1007/978-0-387-74039-3_1. [DOI] [PubMed] [Google Scholar]
- 2.Visovsky C. Treatment considerations for the management of patients with hormone receptor-positive metastatic breast cancer. J Adv Pract Oncol. 2014;5:321–330. [PMC free article] [PubMed] [Google Scholar]
- 3.Berthold D. Third consensus on medical treatment of metastatic breast cancer. Ann Oncol. 2010;21:665. doi: 10.1093/annonc/mdp556. author reply 655-656. [DOI] [PubMed] [Google Scholar]
- 4.Cancer Stat Facts: Female Breast Cancer. https://seer.cancer.gov/statfacts/html/breast.html. Accessed January 31, 2019.
- 5.Haynes B, Sarma A, Nangia-Makker P, Shekhar MP. Breast cancer complexity: Implications of intratumoral heterogeneity in clinical management. Cancer Metastasis Rev. 2017;36:547–555. doi: 10.1007/s10555-017-9684-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bakker JL, Wever K, van Waesberghe JH, Beeker A, Meijers-Heijboer H, Konings IR, Verheul HMW. What is the benefit of treatment with multiple lines of chemotherapy for patients with metastatic breast cancer? A retrospective cohort study. Cancer Epidemiol. 2015;39:848–853. doi: 10.1016/j.canep.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 7.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bovell L, Shanmugam C, Katkoori VR, Zhang B, Vogtmann E, Grizzle WE, Manne U. miRNAs are stable in colorectal cancer archival tissue blocks. Front Biosci (Elite Ed) 2012;4:1937–1940. doi: 10.2741/e514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Occhipinti G, Giulietti M, Principato G, Piva F. The choice of endogenous controls in exosomal microRNA assessments from biofluids. Tumour Biol. 2016;37:11657–11665. doi: 10.1007/s13277-016-5164-1. [DOI] [PubMed] [Google Scholar]
- 10.Campos-Parra AD, Mitznahuatl GC, Pedroza-Torres A, Romo RV, Reyes FIP, López-Urrutia E, Pérez-Plasencia C. Micro-RNAs as potential predictors of response to breast cancer systemic therapy: Future clinical implications. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18061182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gradishar WJ, Anderson BO, Balassanian R, Blair SL, Burstein HJ, Cyr A, Elias AD, Farrar WB, Forero A, Giordano SH, et al. NCCN guidelines insights: Breast cancer, version 1. 2017. J Natl Compr Cancer Netw. 2017;15:433–451. doi: 10.6004/jnccn.2017.0044. [DOI] [PubMed] [Google Scholar]
- 12.Watanabe H, Okada M, Kaji Y, Satouchi M, Sato Y, Yamabe Y, Onaya H, Endo M, Sone M, Arai Y. New response evaluation criteria in solid tumours-revised RECIST guideline (version 1.1) Gan To Kagaku Ryoho. 2009;36:2495–2501. In Japanese. [PubMed] [Google Scholar]
- 13.Vitsios DM, Enright AJ. Chimira: Analysis of small RNA sequencing data and microRNA modifications. Bioinformatics. 2015;31:3365–3367. doi: 10.1093/bioinformatics/btv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, Sabedot TS, Malta TM, Pagnotta SM, Castiglioni I, et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44:e71. doi: 10.1093/nar/gkv1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tibshirani R. A simple method for assessing sample sizes in microarray experiments. BMC Bioinformatics. 2006;7:106. doi: 10.1186/1471-2105-7-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rapaport F, Khanin R, Liang Y, Pirun M, Krek A, Zumbo P, Mason CE, Socci ND, Betel D. Erratum to: Comprehensive evaluation of differential gene expression analysis methods for RNA-seq data. Genome Biol. 2015;16:261. doi: 10.1186/s13059-015-0813-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Costa-Silva J, Domingues D, Lopes FM. RNA-Seq differential expression analysis: An extended review and a software tool. PLoS One. 2017;12:e0190152. doi: 10.1371/journal.pone.0190152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chin L, Andersen JN, Futreal PA. Cancer genomics: From discovery science to personalized medicine. Nat Med. 2011;17:297–303. doi: 10.1038/nm.2323. [DOI] [PubMed] [Google Scholar]
- 20.Mulrane L, Madden SF, Brennan DJ, Gremel G, McGee SF, McNally S, Martin F, Crown JP, Jirström K, Higgins DG, et al. miR-187 is an independent prognostic factor in breast cancer and confers increased invasive potential in vitro. Clin Cancer Res. 2012;18:6702–6713. doi: 10.1158/1078-0432.CCR-12-1420. [DOI] [PubMed] [Google Scholar]
- 21.Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee AV, et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173:400.e11–416.e11. doi: 10.1016/j.cell.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fei D, Zhao K, Yuan H, Xing J, Zhao D. MicroRNA-187 exerts tumor-suppressing functions in osteosarcoma by targeting ZEB2. Am J Cancer Res. 2016;6:2859–2868. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Sun C, Li S, Yang C, Xi Y, Wang L, Zhang F, Li D. MicroRNA-187-3p mitigates non-small cell lung cancer (NSCLC) development through down-regulation of BCL6. Biochem Biophys Res Commun. 2016;471:82–88. doi: 10.1016/j.bbrc.2016.01.175. [DOI] [PubMed] [Google Scholar]
- 24.Dou C, Liu Z, Xu M, Jia Y, Wang Y, Li Q, Yang W, Zheng X, Tu K, Liu Q. MiR-187-3p inhibits the metastasis and epithelial-mesenchymal transition of hepatocellular carcinoma by targeting S100A4. Cancer Lett. 2016;381:380–390. doi: 10.1016/j.canlet.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 25.Lettlova S, Brynychova V, Blecha J, Vrana D, Vondrusova M, Soucek P, Truksa J. MiR-301a-3p suppresses estrogen signaling by directly inhibiting ESR1 in ERα positive breast cancer. Cell Physiol Biochem. 2018;46:2601–2615. doi: 10.1159/000489687. [DOI] [PubMed] [Google Scholar]
- 26.Hu J, Ruan J, Liu X, Xiao C, Xiong J. MicroRNA-301a-3p suppressed the progression of hepatocellular carcinoma via targeting VGLL4. Pathol Res Pract. 2018;214:2039–2045. doi: 10.1016/j.prp.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 27.He YJ, Wu JZ, Ji MH, Ma T, Qiao EQ, Ma R, Tang JH. MiR-342 is associated with estrogen receptor-α expression and response to tamoxifen in breast cancer. Exp Ther Med. 2013;5:813–818. doi: 10.3892/etm.2013.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cittelly DM, Das PM, Spoelstra NS, Edgerton SM, Richer JK, Thor AD, Jones FE. Downregulation of miR-342 is associated with tamoxifen resistant breast tumors. Mol Cancer. 2010;9:317. doi: 10.1186/1476-4598-9-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Young J, Kawaguchi T, Yan L, Qi Q, Liu S, Takabe K. Tamoxifen sensitivity-related microRNA-342 is a useful biomarker for breast cancer survival. Oncotarget. 2017;8:99978–99989. doi: 10.18632/oncotarget.21577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liao D. Emerging roles of the EBF family of transcription factors in tumor suppression. Mol Cancer Res. 2009;7:1893–1901. doi: 10.1158/1541-7786.MCR-09-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ávila-Moreno F, Armas-López L, Álvarez-Moran AM, López-Bujanda Z, Ortiz-Quintero B, Hidalgo-Miranda A, Urrea-Ramírez F, Rivera-Rosales RM, Vázquez-Manríquez E, Peña-Mirabal E, et al. Overexpression of MEOX2 and TWIST1 is associated with H3K27me3 levels and determines lung cancer chemoresistance and prognosis. PLoS One. 2014;9:e114104. doi: 10.1371/journal.pone.0114104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X, Zhang Z, Zhang Q, Zhang Q, Sun P, Xiang R, Ren G, Yang S. ZEB1 confers chemotherapeutic resistance to breast cancer by activating ATM. Cell Death Dis. 2018;9:57. doi: 10.1038/s41419-017-0087-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang T, Dong P, Li L, Ma X, Xu P, Zhu H, Wang Y, Yang B, Liu K, Liu J, et al. MicroRNA-200c regulates cisplatin resistance by targeting ZEB2 in human gastric cancer cells. Oncol Rep. 2017;38:151–158. doi: 10.3892/or.2017.5659. [DOI] [PubMed] [Google Scholar]
- 34.Adam L, Zhong M, Choi W, Qi W, Nicoloso M, Arora A, Calin G, Wang H, Siefker-Radtke A, McConkey D, et al. MiR-200 expression regulates epithelial-to-mesenchymal transition in bladder cancer cells and reverses resistance to epidermal growth factor receptor therapy. Clin Cancer Res. 2009;15:5060–5072. doi: 10.1158/1078-0432.CCR-08-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orlando UD, Castillo AF, Medrano MAR, Solano AR, Maloberti PM, Podesta EJ. Acyl-CoA synthetase-4 is implicated in drug resistance in breast cancer cell lines involving the regulation of energy-dependent transporter expression. Biochem Pharmacol. 2019;159:52–63. doi: 10.1016/j.bcp.2018.11.005. [DOI] [PubMed] [Google Scholar]
- 36.Li M, Han Y, Zhou H, Li X, Lin C, Zhang E, Chi X, Hu J, Xu H. Transmembrane protein 170B is a novel breast tumori-genesis suppressor gene that inhibits the Wnt/β-catenin pathway. Cell Death Dis. 2018;9:91. doi: 10.1038/s41419-017-0128-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shaheen S, Fawaz F, Shah S, Büsselberg D. Differential expression and pathway analysis in drug-resistant triple-negative breast cancer cell lines using RNASeq analysis. Int J Mol Sci. 2018;19 doi: 10.3390/ijms19061810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones VS, Huang RY, Chen LP, Chen ZS, Fu L, Huang RP. Cytokines in cancer drug resistance: Cues to new therapeutic strategies. Biochim Biophys Acta. 2016;1865:255–265. doi: 10.1016/j.bbcan.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 39.Senthebane DA, Rowe A, Thomford NE, Shipanga H, Munro D, Mazeedi MA, Almazyadi HA, Kallmeyer K, Dandara C, Pepper MS, et al. The role of tumor microenvironment in chemo-resistance: To survive, keep your enemies closer. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18071586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sung M, Tan X, Lu B, Golas J, Hosselet C, Wang F, Tylaska L, King L, Zhou D, Dushin R, et al. Caveolae-mediated endocytosis as a novel mechanism of resistance to trastuzumab emtansine (T-DM1) Mol Cancer Ther. 2018;17:243–253. doi: 10.1158/1535-7163.MCT-17-0403. [DOI] [PubMed] [Google Scholar]
- 41.De Luca A, Maiello MR, D'Alessio A, Pergameno M, Normanno N. The RAS/RAF/MEK/ERK and the PI3K/AKT signalling pathways: Role in cancer pathogenesis and implications for therapeutic approaches. Expert Opin Ther Targets. 2012;16(Suppl 2):S17–S27. doi: 10.1517/14728222.2011.639361. [DOI] [PubMed] [Google Scholar]
- 42.Generali D, Buffa FM, Berruti A, Brizzi MP, Campo L, Bonardi S, Bersiga A, Allevi G, Milani M, Aguggini S, et al. Phosphorylated ERalpha, HIF-1alpha, and MAPK signaling as predictors of primary endocrine treatment response and resistance in patients with breast cancer. J Clin Oncol. 2009;27:227–234. doi: 10.1200/JCO.2007.13.7083. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article or are available from the corresponding author on reasonable request.