Abstract
Eukaryotic translation initiation factor 4 (eIF4E) has been demonstrated to promote tumorigenesis in different types of cancer; however, whether eIF4E is involved in the development of GBC is unclear. The present study aimed to explore the biological function of eIF4E in gallbladder cancer (GBC) and identified that the expression level of eIF4E was significantly increased in GBC tissues compared with that in normal gallbladder tissues. The overall survival (OS) was also shorter in the group of patients with GBC with increased eIF4E expression. Increased eIF4E was correlated with advanced stage and higher histologic grade. Knockdown of eIF4E significantly inhibited cell proliferation, colony formation and cell cycle-associated protein expression levels in 2 GBC cell lines. The weight of the tumors in the eIF4E knockdown group was remarkably decreased compared with the control group. It also was revealed that knockdown of eIF4E is associated with upregulating cyclin-dependent kinase inhibitor 1B and down-regulating the expression levels of cyclin E1 and cyclin D1 in vitro and in vivo. These data demonstrated that eIF4E is a novel prognostic marker in GBC and may serve a critical role in the regulation of cell proliferation.
Keywords: gallbladder cancer, eukaryotic translation initiation factor 4E, cell proliferation
Introduction
Gallbladder cancer (GBC) is the most common biliary tract malignancy and the fifth common gastrointestinal cancer, with an incidence of 2.5 per 100,000 population globally (1). The majority of GBC cases were patients who underwent exploration for cholelithiasis (2). Despite the collaboration within multidisciplinary teams of surgeons, oncologists, and endoscopy experts to provide the most appropriate treatment strategy for patients with GBC, the prognosis remains extremely poor (3,4). Therefore, it is essential to uncover the biological mechanism of GBC, in order to develop a novel therapeutic strategy for GBC.
The family of eukaryotic initiation factors (eIFs) includes at least 12 eukaryotic initiation factors, which are composed of several polypeptides (5). One of those polypeptides, namely eIF4E, was proposed by numerous previous studies to be a critical nexus for cancer development (6). Transcriptional profiling of metastatic human solid tumors revealed that eIF4E was one of the genes that was consistently up-regulated (7). However, whether eIF4E is associated with the human GBC remains unclear.
In present study, the protein level of eIF4E was notably increased in GBC tissues compared with normal gallbladder tissues by immunohistochemistry (IHC) analysis. Increased eIF4E levels were associated with a poorer prognosis. Subsequent results indicated that knockdown of eIF4E suppressed cell proliferation in vitro and in vivo.
Materials and methods
Patients and samples
The present study was approved by the Ethics Committee of the First Affiliated Hospital of Anhui Medical University (Hefei, China), and all patients provided written informed consent. A total of 76 GBC specimens were obtained from GBC patients who underwent radical cholecys-tectomy at the General Surgery Department between November of 2011 and November of 2016. All patients who were enrolled in the study did not receive any prior treatment. The control group included 58 normal gallbladder tissues, of which 21 specimens were obtained from patients who were diagnosed with hepatic hemangioma and underwent hepatectomy combined with cholecystectomy, and the other 37 gallbladder adenoma tissues obtained from patients with gallbladder adenoma who underwent laparoscopic cholecystectomy (Table I). All tissues were assessed by tissue microarrays (TMA). Anatomic and histologic grades were established according to the criteria by American Joint Committee on Cancer (AJCC) tumor node metastasis (TNM) Staging for Gallbladder Cancer (8).
Table I.
Expression level of eIF4E in normal gallbladder, gallbladder adenomas and GBC tissues.
| Variable | No. of cases <70/≥70 | No. of cases (male/female) | Optical density of eIF4E expression (x±s) | P-value |
|---|---|---|---|---|
| Normal gallbladder | 15/6 | 7/14 | 0.264 (0.145, 0.278) | 0.047a |
| Gallbladder adenomas | 21/16 | 13/24 | 0.341 (0.292, 0.374) | 0.010b |
| GBC | 49/27 | 24/52 | 0.401 (0.348, 0.453) | 3.610×10−7c |
The data of the average optical density of eIF4E expression in the three groups were analyzed by one-way analysis of variance and Bonferroni post-hoc test for multiple comparisons.
Normal gallbladder vs. gallbladder adenomas.
Gallbladder adenomas vs. GBC.
Normal gallbladder vs. GBC.
Reagents and antibodies
The following reagents used in the present study were purchased from the following sources: Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), complete EDTA-free protease inhibitor cocktail (Roche Applied Science), sea plaque low melting temperature agarose (Lonza Group, Ltd.), GAPDH antibody (cat. no. sc-166545; 1:5,000; Santa Cruz Biotechnology, Inc.), eIF4E antibody (cat. no. sc-271480; 1:1,000; Santa Cruz Biotechnology, Inc.), cyclin-dependent kinase inhibitor 1B (p27) antibody (cat. no. P1484; 1:2,000; Sigma-Aldrich; Merck KGaA), cyclinE1 antibody (cat. no. AF0144; 1:1,000; Affinity Biosciences), cyclinD1 antibody (cat. no. AF0931; 1:1,000; Affinity Biosciences), Ki67 antibody (cat. no. AF0198, 1:200; Affinity Biosciences) and horseradish peroxidase-conjugated secondary antibodies (cat. nos. sc-2005 and sc-2004; 1:10,000; Santa Cruz Biotechnology, Inc.).
Cell culture
The 2 human GBC cell lines used in the present study were GBC-SD (Cell Bank of Type Culture Collection of the Chinese Academy of Sciences) and NOZ (Health Science Research Resources Bank). The GBC-SD cells were cultured in Dulbecco's modified Eagle medium (DMEM; Thermo Fisher Scientific, Inc.). The NOZ cells were cultured in William's medium E (Lonza Group, Ltd.) at 37°C in a humidified 5% CO2 incubator. Both media were supplemented with 10% fetal bovine serum (FBS, Gibco; Thermo Fisher Scientific, Inc.). The NOZ and GBC-SD cells were passaged for 9 times prior to use in the indicated functional experiments. All the cell lines were routinely examined for mycoplasma contamination prior to use.
RNA interference
To generate lentiviruses expressing the indicated shRNA, 293T cells (Americn Type Culture Collection) grown on a 6 cm dish were transfected with 2 µg shRNA (cloned in PLKO.1, Sigma-Aldrich; Merck KGaA), 2 µg pREV, 2 µg pGag/Pol/PRE, and 1 µg pVSVG. A total of 12 h after transfection using Lipofectamine® 2000, Invitrogen; Thermo Fisher Scientific, Inc.), the cells were cultured in DMEM containing 20% FBS for an additional 24 h. The culture medium containing lentivirus particles was filtered through a 0.45-lm polyvinylidene difluoride (PVDF) membrane filter (EMD Millipore). Then, 50% filtered DMEM containing lenti-virus particles with 50% fresh DMEM or William's medium E was incubated with GBC-SD or NOZ cells supplemented with 8 µg/ml polybrene (Sigma-Aldrich; Merck KGaA) for 24 h, followed by selection with 2 µg/ml puromycin for an additional 24 h. The knockdown efficiency was evaluated by western blot analysis. Cell flow was executed 96 h after transfection. The shRNA target sequences used were as follows: sh-control: CCT AAG GTT AAG TCG CCC TCG, sh-eIF4E-1 (human): CCA CTC TGT AAT AGT TCA GTA, sh-eIF4E-2 (human): CCA AAG ATA GTG ATT GGT TAT.
Cell proliferation assay
A total of 1×104 GBC-SD or NOZ cells expressing control shRNA or eIF4E shRNA suspended in 0.5 ml DMEM or William's medium E containing 10% FBS were plated in 24-well plates and incubated at 37°C for 7 days. The cells were counted using Countstar Automated Cell Counter (Simo Biotechnology Co., Ltd.) at 3rd, 5th and 7th days.
Colony formation assay
To measure colony formation on plates, 1×103 GBC-SD or NOZ cells expressing control shRNA or eIF4E shRNA suspended in 2 ml DMEM or William's medium E containing 10% FBS were plated in 6-well plates and incubated at 37°C for 3 weeks. The colonies were then fixed with 4% paraformaldehyde for 15 min at room temperature, stained with trypan blue (room temperature, 15 min), and scored using Image J (v1.8.0, National Institutes of Health). To measure the colony formation in soft agar, 1×104 GBC-SD or NOZ cells expressing control shRNA or eIF4E shRNA suspended in 1.5 ml DMEM or William's medium E containing 10% FBS and 0.3% SeaPlaque low melting temperature agarose were plated in 1 well of 6-well plates over a 1.5 ml layer of DMEM or William's medium E containing 10% FBS/0.6% agarose. Cells were incubated at 37°C for 3 weeks prior to fixing, staining and scoring, as aforementioned.
IHC
The expression level of eIF4E was detected in TMA slides of GBC, adenomas and normal gallbladder tissues as described previously (9). The average optical density of TMA staining was measured with Image Pro Plus 6.0 software (Media Cybernetics, Inc.).
Western blot analysis
The cells were lysed in radioimmunopre-cipitation assay buffer (Beyotime of Institute of Biotechnology) in an ice bath for 30 min, and centrifuged at 12,000 x g for 20 min at 4°C. The supernatant was stored at −80°C until analysis. The protein concentration was measured using the bicinchoninic acid (BCA) assay kit (Beyotime of Institute of Biotechnology). The equal amount (30 µg) of proteins was loaded onto 10% SDS-PAGE and transferred by electroblotting to a PVDF membrane (EMD Millipore). The membrane was blocked with 5% bovine serum albumin (Sangon Biotech Co., Ltd.) for 1 h at room temperature. Primary antibodies were incubated at 4°C overnight. The secondary antibody was added to the membrane at room temperature for 1 h. Image J (v1.8.0) was used for density analysis.
Xenograft model
A total of 5×106 NOZ cells expressing control shRNA or eIF4E shRNA were individually injected into the dorsal flanks of 4-week-old male athymic nude mice (Nanjing SLAC Laboratory Animal Co. Ltd.) (n=5 per group). The mice were randomly assigned to groups in the experiment (sh-control: 5 nude mice, sh-eIF4E-1: 5 nude mice). A total of 1 week after injection, tumors were measured per 4 days with Vernier calipers and calculated using the following equation: volume=length x width2 ×0.52. Then, 4 weeks after injection, the mice were sacrificed, and the tumors were excised and weighed. During measurement of the weight of the tumors, the experimentalists were blinded to the information of tumor tissues. The excised tumors were homogenized, and proteins were extracted for western blot analysis, as aforementioned. Studies on animals were conducted on the basis of approval from the Animal Research Ethics Committee of the University of Science and Technology of China (approval no. PXTG-MYD2017102611).
Statistical analysis
The statistical analysis was conducted by using SPSS 17.0 software (IBM Corp.). Data were presented as the mean ± standard deviation. The skewness data are expressed as the mean with 1st and 3rd interquartile ranges, and were determined using a Mann-Whitney U test. The mortality risks of GBC were analyzed using Kaplan-Meier estimator for univariate survival analysis followed by log-rank test. The patient's age, sex, anatomic stage, histological grade and eIF4E status indicators were analyzed by the Cox proportional hazards model. The data from the in vitro experiments were analyzed using one-way analysis of variance followed by Bonferroni's correction post-hoc test.
Results
Overexpressing eIF4E indicates a poor prognosis in patients with GBC
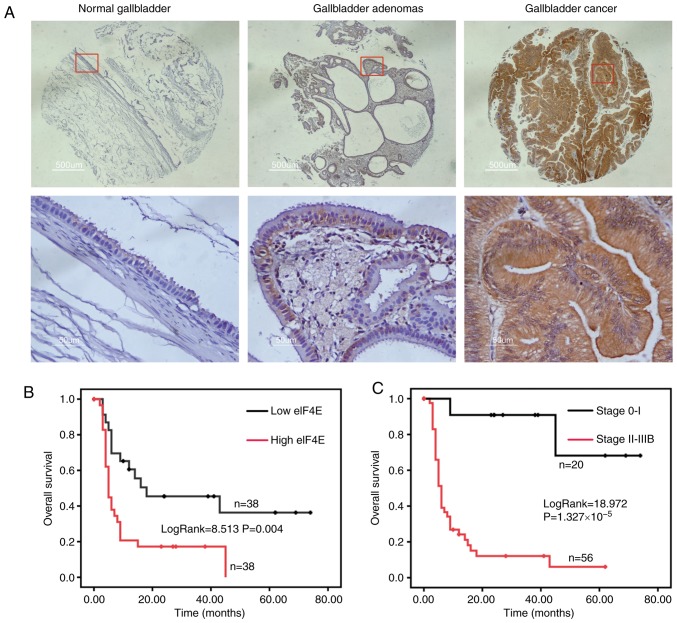
eIF4E expression was assessed in normal gallbladder, gallbladder adenomas and GBC by IHC. The expression of eIF4E was significantly increased in GBC compared with gallbladder adenomas, and normal gallbladder tissue exhibited significantly decreased eIF4E expression compared with gallbladder adenomas (Fig. 1A; Table I). The expression of eIF4E was notably increased in advanced stage GBC (anatomic stage III and IV) compared with early stage GBC (anatomic stage I and II) (P=0.022; Table II). Furthermore, the eIF4E expression was markedly associated with histological grade (P=0.004; Table II).
Figure 1.
eIF4E overexpression is associated with a poor prognosis in patients with GBC. (A) Representative photos of immunohistochemical staining of eIF4E in normal gallbladder, gallbladder adenomas and GBC. Scale bar, 500 µm. (B) The Kaplan-Meier curves based on eIF4E expression via immunohistochemical analysis in patients with GBC. The overall survival of the high eIF4E group (n=38; optical density of tissue microarray staining was ≥0.3955) was significantly decreased compared with that in the low eIF4E group (n=38; log-rank=8.513; P=0.004). (C) The Kaplan-Meier curves based on Tumor Node Metastasis stage in patients with GBC. The overall survival of the early stage group (n=20) was significantly increased compared with that in the advanced stage group (n=56; log-rank=18.972; P=1.327×10−5). eIF4E, eukaryotic translation initiation factor 4; GBC, gallbladder cancer.
Table II.
Expression of eIF4E and clinicopathological characteristics of patients with gallbladder cancer.
| Variable | N | Optical density of eIF4E expression
|
||
|---|---|---|---|---|
| Value, mean (1st and 3rd interquartile ranges) | Z-value | P-value | ||
| Age | −0.613 | 0.540 | ||
| <70 | 49 | 0.401 (0.334, 0.455) | ||
| ≥70 | 27 | 0.398 (0.357, 0.452) | ||
| Sex | −0.995 | 0.320 | ||
| Male | 24 | 0.383 (0.348, 0.429) | ||
| Female | 52 | 0.408 (0.347, 0.467) | ||
| Primary tumor (T) | −2.194 | 0.028 | ||
| Tis-T1 | 22 | 0.350 (0.278, 0.440) | ||
| T2-T4 | 54 | 0.394 (0.357, 0.460) | ||
| Regional lymph nodes (N) | −0.235 | 0.814 | ||
| N0 | 52 | 0.391 (0.342, 0.467) | ||
| N1 | 24 | 0.408 (0.353, 0.447) | ||
| Anatomic stage | −2.283 | 0.022 | ||
| 0-I | 20 | 0.349 (0.288, 0.435) | ||
| II-IIIB | 56 | 0.413 (0.357, 0.457) | ||
| Histologic grade | −2.908 | 0.004 | ||
| G1-G2 (well-moderately) | 51 | 0.387 (0.326, 0.431) | ||
| G3-G4 (poorly-undifferentiated) | 25 | 0.441 (0.377, 0.521) | ||
Statistical analyses were performed using Mann-Whitney U test. eIF4E, eukaryotic translation initiation factor 4.
Then, the Kaplan-Meier analysis indicated that the group with increased eIF4E expression (the optical density of TMA staining was ≥0.3955) was significantly associated with poor overall survival (OS) in patients with GBC (log-rank=8.513; P=0.004; Fig. 1B). However, the OS of the early stage (0-I) group was notably improved compared with that in the advanced stage (II-IIIB) group (log-rank=18.972; P=1.327×10−5; Fig. 1C). The median OS for the high eIF4E expression group was 5 months, whilst it was 18 months for the low eIF4E expression group (Table III).
Table III.
Multivariate Cox regression analysis of the anatomic stage, histologic grade and optical density of eukaryotic translation initiation factor 4 expression in gallbladder cancer cells.
| Variable | B | SE B | Wald | Relative risk (95% confidence interval) | P-value |
|---|---|---|---|---|---|
| Anatomic stage | 2.532 | 0.762 | 11.041 | 12.575 (2.825-55.986) | 0.001 |
| Histologic grade | 0.794 | 0.353 | 5.067 | 2.212 (1.108-4.416) | 0.024 |
| Optical density of eIF4E expression | 1.054 | 0.374 | 7.937 | 2.868 (1.378-5.969) | 0.005 |
Furthermore, multivariate Cox regression analysis confirmed that overexpression of eIF4E was negatively correlated with post-operative OS, whereas it was positively associated with anatomic stage and historical grade, indicating that eIF4E expression is an independent prognostic factor in patients with GBC.
Knockdown of eIF4E inhibits the proliferation and colony formation of GBC and NOZ cells in vitro
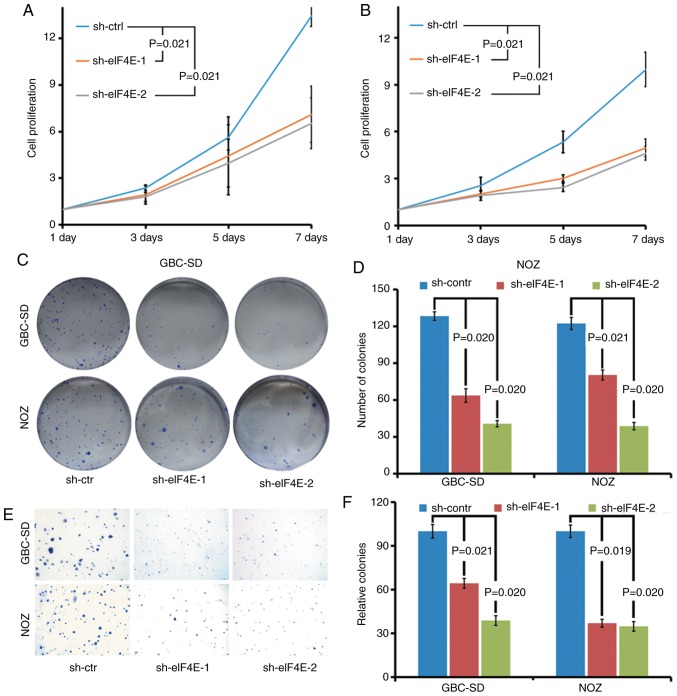
The biological function of eIF4E in GBC was investigated using eIF4E-specific shRNA knockdown cells. The effects of eIF4E on proliferation of GBC cells were determined by cell growth curve and colony formation assays. As demonstrated in Fig. 2A and B, the cell growth curve indicated that the knockdown of eIF4E significantly inhibited the proliferation of GBC-SD and NOZ cells. Consistent with the observations of the cell growth curve, the colony forming ability of GBC-SD and NOZ cells was markedly decreased in eIF4E shRNA transfected cells (sh-eIF4E group; Fig. 2C and D). Furthermore, the number of the colonies in the sh-eIF4E group was significantly decreased compared with that of the sh-control group in soft agar (Fig. 2E and F).
Figure 2.
Knockdown of eIF4E inhibits the proliferation and colony formation of GBC and NOZ cells in vitro. (A) GBC-SD and (B) NOZ cells were transfected with lentiviruses expressing control shRNA or eIF4E shRNA. A total of 48 h after infection, cell numbers were counted at the indicated time points. (C) Colony formation assays of GBC and NOZ cells after shRNAs transfection. A total of 1×103 cells/well were seeded and incubated at 37°C for 3 weeks. (D) Quantitative analysis of colony forming assay. (E) The colony formation assay of GBC and NOZ cells in soft agar following shRNAs transfection. A total of 1×104 cells/agar were seeded and incubated at 37°C for 3 weeks. (F) Quantitative analysis of colony forming assay. All the experiments were repeated 3 times. eIF4E, eukary-otic translation initiation factor 4; shRNA, short hairpin RNA; ctr/ctrl/contr, control. Statistical analyses were performed using one-way analysis of variance followed by Bonferroni's correction post-hoc test.
Knockdown of eIF4E inhibits the cell cycle of GBC and NOZ cells in vitro
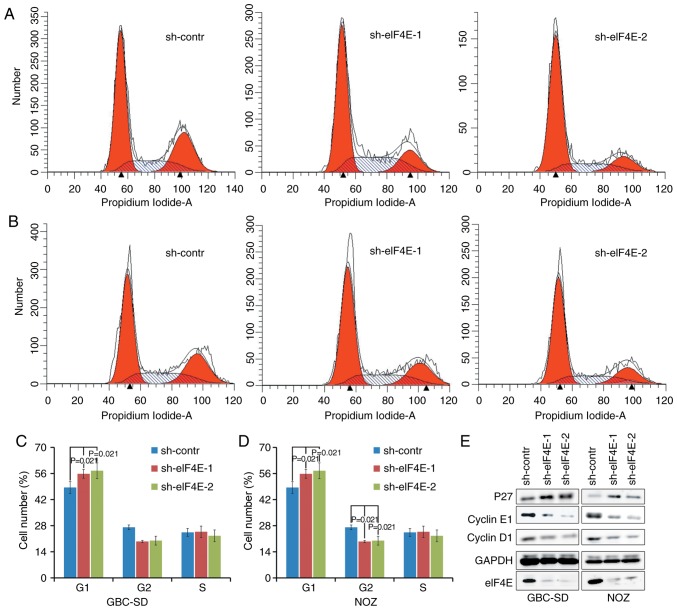
Subsequent analyses were conducted to evaluate the effects of eIF4E on cell cycle using flow cytometry. A total of 96 h after lentivirus infection, an increase in the fraction of cells in the G0/G1 phase was observed in sh-eIF4E group compared with that in the sh-control group in GBC-SD and NOZ cells. The fraction of cells in the G2/M phase decreased in knocking down eIF4E shRNA-transfected NOZ cells (Fig. 3A-D). In addition, whether eIF4E was able to regulate the expression levels of cell cycle-associated proteins was investigated using western blot analysis. As indicated in Fig. 3E, knockdown of eIF4E resulted in a decrease in the expression levels of cyclin D and cyclin E, in addition to an increase in the expression level of p27 in GBC-SD and NOZ cells compared with those in the control group. Collectively, these data demonstrated that eIF4E serves a pivotal role in regulating proliferation of GBC cells by modulating the expression of cell cycle-associated proteins.
Figure 3.
Knockdown of eIF4E inhibits the cell cycle of GBC and NOZ cells in vitro. (A) GBC-SD and (B) NOZ cells were transfected with shRNA and incubated for 96 h, and the cell cycle distribution was evaluated by flow cytometry. (C and D) Representative histograms and the percentage of (C) GBC-SD and (D) NOD cells in each phase are shown (mean ± standard deviation; n=3). (E) Western blot analysis was performed to examine the cell-cycle proteins, including p27, cyclin D1 and cyclin E1. eIF4E, eukaryotic translation initiation factor 4; shRNA, short hairpin RNA; contr, control; p27, cyclin-dependent kinase inhibitor 1B. Statistical analyses were performed using one-way analysis of variance followed by Bonferroni's correction post-hoc test.
Knockdown of eIF4E inhibits the GBC tumor growth in vivo
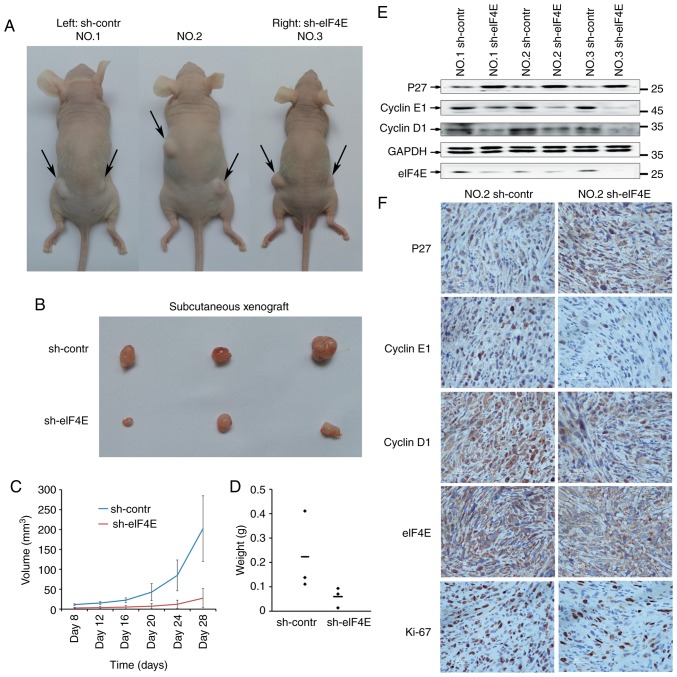
To additionally confirm the function of eIF4E in GBC development, subcutaneous xenograft models were established in BALB/C nude mice using NOZ cells. As demonstrated in Fig. 4A-D, the tumor growth was significantly suppressed in sh-eIF4E group compared with that in the sh-control group, as evidenced by decrease of volume and weight of xenografts.
Figure 4.
Knockdown of eIF4E inhibits the GBC tumor growth in vivo. (A) A total of 5×106 NOZ cells expressing control shRNA and eIF4E shRNA were individually injected to the left flank and right flank of nude mice. Then, 4 weeks after injection, the mice were sacrificed (n=3). (B) Representative tumor tissues excised from mice. (C and D) The tumor volume and weight of the tumor in mice were measured and calculated. (E) The expression levels of cell cycle-associated proteins and eIF4E were evaluated by western blot analysis among the indicated groups. (F) The immunohistochemical staining of cell cycle-associated proteins, Ki 67 and eIF4E, in tumors. Scale bar, 50 µm. eIF4E, eukaryotic translation initiation factor 4; shRNA, short hairpin RNA; contr, control; p27, cyclin-dependent kinase inhibitor 1B.
Additionally, the expression levels of p27, cyclin D1 and cyclin E1 in harvested subcutaneous xenografts was detected using western blot analysis, and the expression levels of P27, cyclin D1, cyclin E1 and Ki-67 by IHC assays. As demonstrated in Fig. 4E and F, knockdown of eIF4E resulted in the decreased expression levels of cyclin D1, cyclin E1 and Ki-67, while increased expression of p27. Taken together, these results indicated that knockdown of eIF4E inhibits the GBC tumor growth in vivo.
Discussion
At present, the biological mechanism of eIF4E in GBC is not well understood. In the present study, it was revealed that the expression of eIF4E was associated with the prognosis of GBC and its role in the proliferation of GBC cells.
Several previous studies have verified that eIF4E is overexpressed or deregulated in different types of cancer, including breast cancer (10), melanoma (11), and prostate (12), lung (13) and colorectal (14) cancer. It also has been demonstrated that a subset of mRNAs encoding cancer-associated proteins, such as c-MYC and cyclin D1, were identified to be sensitive to the activity of the eIF4F complex (15) and may serve as a convergence point for hyperactive signaling pathways to promote tumorigenesis (16). The present study confirmed that knockdown of eIF4E in GBC cells inhibited cell proliferation, oncogenic potential and cell cycle arrest, which is consistent with the previously described data (17,18).
Whether the knockdown of eIF4F is an appropriate strategy for cancer treatment is an important avenue of study. By generating an eIF4F haploinsufficient mouse model, Truitt et al (19) identified that decreasing the level of eIF4E inhibited the oncogenic potential of human KRAS-driven lung cancer cells, and that this was not detrimental to normal mammalian physiological processes. In addition, the eIF4E must be phosphorylated to promote tumor development (20). The androgen receptor is a negative regulator of phosphorylation of serine 209 in eIF4F, indicating a potential therapeutic target in prostate cancer (21). Based on preclinical data demonstrating that eIF4F regulates the translation of mRNAs involved in cell survival and chemotherapy resistance, a clinical trial (clinical trial no. 01675128) that combined ISIS 183750, a second-generation antisense oligonucleotide designed to inhibit the production of the eIF4E protein, and irinotecan in patients with irinotecan-refractory metastatic colorectal cancer and did not result in objective clinical responses (22). However, Robichaud et al (23) attempted to provide a rationale for targeting eIF4E phosphorylation in cancer cells and cells that comprise the tumor microenvironment to halt metastasis, demonstrating the efficacy of this strategy using merestinib, representing a therapeutic strategy for cancer.
The present study identified that the expression of eIF4E was markedly upregulated in GBC tissues compared with gallbladder adenomas samples or normal gallbladder tissues, and that high eIF4E expression levels in GBC tissues were associated with advanced stage, higher histologic grade and poorer prognosis.
In summary, the results from the present study demonstrated that eIF4E served a critical role in the regulation of cell proliferation, and may function as an independent prognostic maker for GBC.
Acknowledgments
The authors would like to thank Dr Chengfeng Wang from the Chinese Academy of Science Key Laboratory of Innate Immunity and Chronic Disease, School of Life Sciences and Medical Center, University of Science and Technology of China for his scrupulous and skillful assistance in generating the subcutaneous xenograft models in BALB/C nude mice.
Funding
The present study was financially supported by Natural Science Research Project of Anhui Medical University (grant no. 2018xkj057).
Availability of data and materials
All data generated during this study are included in this published article.
Authors' contributions
XG and DF made substantial contributions to the design of the present study; DF, JP, GW, and DZ performed the experiments; DF and JP analyzed the data; DF wrote the manuscript. All authors approved the final version of the manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of the First Affiliated Hospital of Anhui Medical University (Hefei, China), and all patients provided written informed consent. Studies on animals were conducted on the basis of approval from the Animal Research Ethics Committee of the University of Science and Technology of China (approval no. PXTG-MYD2017102611).
Patient consent for publication
All patients provided written informed consent.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Randi G, Franceschi S, La Vecchia C. Gallbladder cancer worldwide: Geographical distribution and risk factors. Int J Cancer. 2006;118:1591–1602. doi: 10.1002/ijc.21683. [DOI] [PubMed] [Google Scholar]
- 2.Pitt SC, Jin LX, Hall BL, Strasberg SM, Pitt HA. Incidental gallbladder cancer at cholecystectomy: When should the surgeon be suspicious? Ann Surg. 2014;260:128–133. doi: 10.1097/SLA.0000000000000485. [DOI] [PubMed] [Google Scholar]
- 3.Elmasry M, Lindop D, Dunne DF, Malik H, Poston GJ, Fenwick SW. The risk of malignancy in ultrasound detected gallbladder polyps: A systematic review. Int J Surg. 2016;33(Pt A):28–35. doi: 10.1016/j.ijsu.2016.07.061. [DOI] [PubMed] [Google Scholar]
- 4.Dasari BVM, Ionescu MI, Pawlik TM, Hodson J, Sutcliffe RP, Roberts KJ, Muiesan P, Isaac J, Marudanayagam R, Mirza DF. Outcomes of surgical resection of gallbladder cancer in patients presenting with jaundice: A systematic review and meta-analysis. J Surg Oncol. 2018;118:477–485. doi: 10.1002/jso.25186. [DOI] [PubMed] [Google Scholar]
- 5.Aitken CE, Lorsch JR. A mechanistic overview of translation initiation in eukaryotes. Nat Struct Mol Biol. 2012;19:568–576. doi: 10.1038/nsmb.2303. [DOI] [PubMed] [Google Scholar]
- 6.Pelletier J, Graff J, Ruggero D, Sonenberg N. Targeting the eIF4F translation initiation complex: A critical nexus for cancer development. Cancer Res. 2015;75:250–263. doi: 10.1158/0008-5472.CAN-14-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- 8.Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A. AJCC Cancer Staging Manual. 7th ed. Springer; New York, NY: 2010. [Google Scholar]
- 9.Lu Li M, Zhang J, Li F, Zhang H, Wu B, Tan X, Zhang Z, Gao L, Mu GJ, et al. Yes-associated protein 1 (YAP1) promotes human gallbladder tumor growth via activation of the AXL/MAPK pathway. Cancer Lett. 2014;355:201–209. doi: 10.1016/j.canlet.2014.08.036. [DOI] [PubMed] [Google Scholar]
- 10.Yin X, Kim RH, Sun G, Miller JK, Li BD. Overexpression of eukaryotic initiation factor 4E is correlated with increased risk for systemic dissemination in node-positive breast cancer patients. J Am Coll Surg. 2014;218:663–671. doi: 10.1016/j.jamcollsurg.2013.12.020. [DOI] [PubMed] [Google Scholar]
- 11.Khosravi S, Tam KJ, Ardekani GS, Martinka M, McElwee KJ, Ong CJ. eIF4E is an adverse prognostic marker of melanoma patient survival by increasing melanoma cell invasion. J Invest Dermatol. 2015;135:1358–1367. doi: 10.1038/jid.2014.552. [DOI] [PubMed] [Google Scholar]
- 12.Kwegyir-Afful AK, Bruno RD, Purushottamachar P, Murigi FN, Njar VC. Galeterone and VNPT55 disrupt Mnk-eIF4E to inhibit prostate cancer cell migration and invasion. FEBS J. 2016;283:3898–3918. doi: 10.1111/febs.13895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu F, Wang X, Li J, Gu K, Lv L, Zhang S, Che D, Cao J, Jin S, Yu Y. miR-34c-3p functions as a tumour suppressor by inhibiting eIF4E expression in non-small cell lung cancer. Cell Prolif. 2015;48:582–592. doi: 10.1111/cpr.12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiegering A, Uthe FW, Jamieson T, Ruoss Y, Hüttenrauch M, Küspert M, Pfann C, Nixon C, Herold S, Walz S, et al. Targeting translation initiation bypasses signaling crosstalk mechanisms that maintain high MYC levels in colorectal cancer. Cancer Discov. 2015;5:768–781. doi: 10.1158/2159-8290.CD-14-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siddiqui N, Sonenberg N. Signalling to eIF4E in cancer. Biochem Soc Trans. 2015;43:763–772. doi: 10.1042/BST20150126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wan J, Shi F, Xu Z, Zhao M. Knockdown of eIF4E suppresses cell proliferation, invasion and enhances cisplatin cytotoxicity in human ovarian cancer cells. Int J Oncol. 2015;47:2217–2225. doi: 10.3892/ijo.2015.3201. [DOI] [PubMed] [Google Scholar]
- 18.Chen B, Zhang B, Xia L, Zhang J, Chen Y, Hu Q, Zhu C. Knockdown of eukaryotic translation initiation factor 4E suppresses cell growth and invasion, and induces apoptosis and cell cycle arrest in a human lung adenocarcinoma cell line. Mol Med Rep. 2016;13:3384. doi: 10.3892/mmr.2016.4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Truitt ML, Conn CS, Shi Z, Pang X, Tokuyasu T, Coady AM, Seo Y, Barna M, Ruggero D. Differential requirements for eIF4E dose in normal development and cancer. Cell. 2015;162:59–71. doi: 10.1016/j.cell.2015.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robichaud N, del Rincon SV, Huor B, Alain T, Petruccelli LA, Hearnden J, Goncalves C, Grotegut S, Spruck CH, Furic L, et al. Phosphorylation of eIF4E promotes EMT and metastasis via translational control of SNAIL and MMP-3. Oncogene. 2015;34:2032–2042. doi: 10.1038/onc.2014.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D'Abronzo LS, Bose S, Crapuchettes ME, Beggs RE, Vinall RL, Tepper CG, Siddiqui S, Mudryj M, Melgoza FU, Durbin-Johnson BP, et al. The androgen receptor is a negative regulator of eIF4E phosphorylation at S209: Implications for the use of mTOR inhibitors in advanced prostate cancer. Oncogene. 2017;36:6359–6373. doi: 10.1038/onc.2017.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duffy AG, Makarova-Rusher OV, Ulahannan SV, Rahma OE, Fioravanti S, Walker M, Abdullah S, Raffeld M, Anderson V, Abi-Jaoudeh N, et al. Modulation of tumor eIF4E by antisense inhibition: A phase I/II translational clinical trial of ISIS 183750-an antisense oligonucleotide against eIF4E-in combination with irinotecan in solid tumors and irinotecan-refractory colorectal cancer. Int J Cancer. 2016;139:1648–1657. doi: 10.1002/ijc.30199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robichaud N, Hsu BE, Istomine R, Alvarez F, Blagih J, Ma EH, Morales SV, Dai DL, Li G, Souleimanova M, et al. Translational control in the tumor microenvironment promotes lung metastasis: Phosphorylation of eIF4E in neutrophils. Proc Natl Acad Sci USA. 2018;115:E2202–E2209. doi: 10.1073/pnas.1717439115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated during this study are included in this published article.