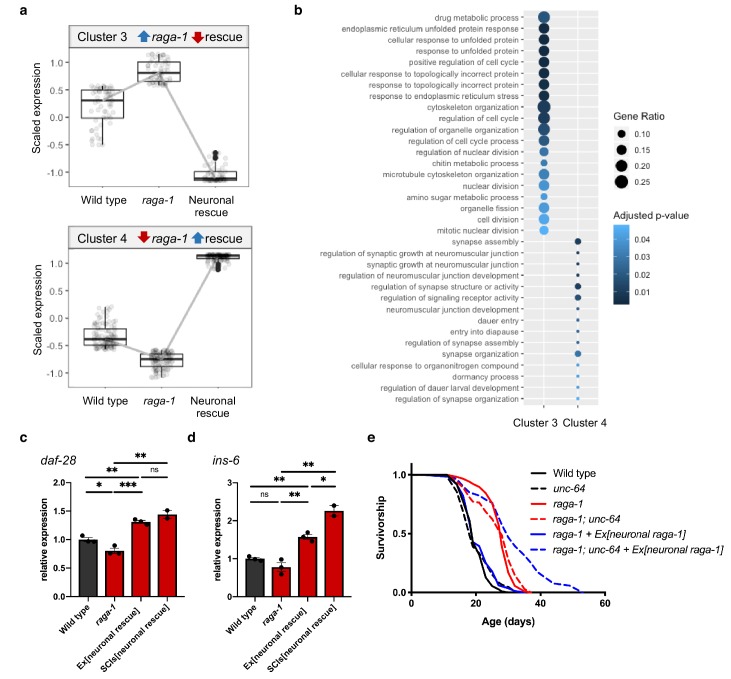
Figure 3. Neuronal TORC1 modulates aging via changes to organelle organization and neuropeptide signaling.
(a) Cluster analysis identified 59 genes that show increased expression in raga-1(ok386) that is reversed by neuronal rescue array (Cluster 3, top) and 107 genes that show decreased expression in raga-1 that is reversed by neuronal rescue (Cluster 4, bottom). (b) Over-representation analysis of the Gene Ontology biological process terms for genes comprising each of the clusters shown in panel (a). (c, d) qPCR to validate increased expression of daf-28 (c) and ins-6 (d) in independent samples. Samples were generated from L4 larvae to match the timepoint of RNA-Seq sample collection. Independent biological replicates were processed and amplified in parallel. ‘Ex[neuronal rescue]’ refers to raga-1(ok386) animals rescued by extrachromosomal array, ‘SCIs[neuronal rescue]’ refers to raga-1(ok386) animals rescued by single copy insertion. P values determined by 2-tailed t test. Source data are provided in Figure 3—source data 1. (e) The unc-64(e246) hypomorphic allele significantly extends lifespan in raga-1 neuronal rescue background (p<0.0001), but not in wild type (p=0.3449) or raga-1 mutant (p=0.2708). n = 3 independent biological replicates, sample size ranges between 64–121 deaths per treatment each replicate. P values are calculated with Log-rank (Mantel-Cox) test. Details on strains and lifespan replicates can be found in Supplementary file 6.
Figure 3—figure supplement 1. Gene clusters and differentially represented GO terms identified by analysis of RNA-seq.
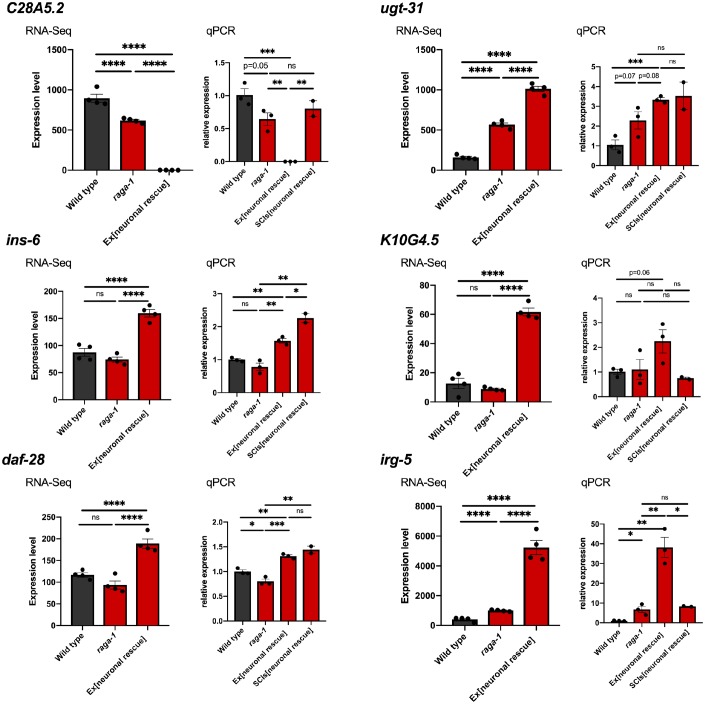
Figure 3—figure supplement 2. Validation of changes identified by RNA-seq in independent biological samples.
Figure 3—figure supplement 3. Neuronal raga-1 regulates ins-6 expression in adults.