Summary
Argonaute-bound microRNAs silence mRNA expression in a dynamic and regulated manner to control organismal development, physiology and disease. We employed metabolic small RNA sequencing for a comprehensive view on intracellular microRNA kinetics in Drosophila. Based on absolute biogenesis- and decay-rates, microRNAs rank among the fastest produced and longest-lived cellular transcripts, disposing up to 105 copies per cell at steady-state. Mature microRNAs are produced within minutes, revealing tight intracellular coupling of biogenesis that is selectively disrupted by pre-miRNA-uridylation. Control over Argonaute protein homeostasis generates a kinetic bottleneck that cooperates with non-coding RNA surveillance to ensure faithful microRNA loading. Finally, regulated small RNA decay enables the selective rapid turnover of Ago1-bound microRNAs but not of Ago2-bound siRNAs, reflecting key differences in the robustness of small RNA silencing pathways. Time-resolved small RNA sequencing opens new experimental avenues to deconvolute the timescales, molecular features, and regulation of small RNA silencing pathways in living cells.
Introduction
MicroRNAs (miRNAs) control gene expression in plants and animals (Ghildiyal and Zamore, 2009). Loss of miRNA-mediated gene silencing is nearly always lethal in animals, certainly because miRNAs regulate diverse cellular pathways to sculpt organismal development and physiology (Bartel, 2018). Consistent with the view that many miRNAs act in confined physiological and developmental settings, their biogenesis and function is tightly controlled in a spatial and temporal manner (Michlewski and Caceres, 2019; Treiber et al., 2019).
MicroRNAs assemble into ribonucleoprotein complexes to silence complementary target RNAs via base-pairing. The central protein component is a member of the Argonaute (Ago) protein family (Hutvagner and Simard, 2008). In flies, small RNAs are sorted between Ago1 and Ago2: MicroRNAs usually bind to Ago1 and small interfering RNAs (siRNAs) to Ago2 (Kawamata and Tomari, 2010). The structure and thermodynamic properties of small RNA duplexes, together with the identity of the small RNA guide 5′ nucleotide, determine how a small RNA partitions between the two Ago proteins (Czech and Hannon, 2010). Most miRNAs have been evolutionarily selected to load only one of the two duplex-derived strands (miR), while the partner strand (miR*) is typically destroyed in the process of miR loading. But symmetric loading of either strand of a miRNA duplex may occur in a regulated manner (Ghildiyal et al., 2009); and miR* strands frequently assemble into Ago2 in flies, emphasizing the compound fate that small RNA duplexes may encounter in cells (Czech et al., 2009; Ghildiyal et al., 2009). In flies, a key step in the production of a functional Ago2-, but not Ago1-silencing complex, is the addition of a 2′ -O-methyl group to the 3′ end of the small RNA, a modification that stabilizes Ago2-bound small RNAs (Horwich et al., 2007; Ameres et al., 2010).
Small RNA abundance is a consequence of the relative contributions of small RNA production and decay, but the underlying kinetics remain obscure. MicroRNA production is a multi-step process that distributes across two cellular compartments (Kim et al., 2009): Biogenesis initiates co-transcriptionally at hairpin structures within primary transcripts (primary miRNAs; pri-miRNAs) (Y. Lee et al., 2004). Primary-miRNAs are sequentially processed by Drosha in the nucleus and Dicer-1 in the cytoplasm to produce mature miRNA duplexes (Kim et al., 2009). The production of small RNAs is tightly controlled at multiple levels, including miRNA transcription, processing and loading (Ha and Kim, 2014). Beyond these, the addition of nucleotides (predominantly uridines) to the 3´ end of pre-miRNAs recently emerged as a hallmark for the selective repression of dicing in metazoans (M. Lee et al., 2014). In flies, the cytoplasmic terminal uridylyltransferase (TUTase) Tailor specifically represses the maturation of mirtrons (Bortolamiol-Becet et al., 2015; Reimão-Pinto et al., 2015; 2016), a class of intron-derived miRNAs that are produced by splicing instead of Drosha-directed cropping (Okamura et al., 2007; Ruby et al., 2007). Notably, uridylation-triggered RNA decay by the 3´-to-5´ exoribonuclease Dis3l2 represents a cytoplasmic RNA surveillance pathway to prevent the accumulation of structured RNAs, including tRNA, rRNA, snoRNA and snRNA (Pirouz et al., 2016; Reimão-Pinto et al., 2016; Ustianenko et al., 2016; Łabno et al., 2016).
In contrast to miRNA biogenesis, the molecular principles and biological scope of small RNA turnover remain poorly defined. Individual miRNAs were reported to differ significantly in their stability: The cardiac-specific mammalian miR-208 exhibits a half-life of approximately 2 weeks (van Rooij et al., 2007), whereas individual miRNAs that exhibit cell-cycle-specific expression patterns are predicted to decay within hours or even minutes (Rissland et al., 2011), just like several miRNAs in the mouse retina in adaptation to light and darkness (Krol et al., 2010). These examples suggest that small RNA turnover may contribute significantly to the steady-state abundance of miRNAs.
A variety of biochemical processes have been proposed to diversify miRNA sequence and function, giving rise to miRNA variants that are commonly referred to as isomiRs (Ameres and Zamore, 2013; Bartel, 2018). The vast majority of heterogeneity concerns miRNA 3´ ends, because 5´ end precision defines the miRNA seed sequence and ensures unambiguous targeting (Seitz et al., 2008; Bartel, 2018). In flies, ~40% of Ago1-bound miRNAs are trimmed at their 3´ termini by the 3´-to-5´ exoribonuclease Nibbler (Nbr) (Han et al., 2011; Liu et al., 2011). Nbr acts predominantly on miRNAs that arise from pre-miRNAs that force Dicer or Drosha to generate an atypically long product. Those require further maturation after Ago1-binding, perhaps to ensure that their 3´ end settles in the Ago protein. Many more enzymatic activities may contribute to the formation of isomiRs (Ameres and Zamore, 2013), but their precise emergence remains vaguely defined, in part because currently available profiling approaches lack the temporal resolution to deconvolute their origin.
Understanding the molecular basis for the establishment of steady-state gene expression demands detailed insights into the relative kinetics of RNA biogenesis, processing and turnover in a transcript-specific and systematic manner (Rabani et al., 2014). Metabolic labeling strategies employing the incorporation of nucleotide analogs enable to track RNA species over time without interfering with cellular integrity. 4-thiouridine (4sU) provides unique physicochemical properties for thiol-specific reactivity and affinity, which enables biochemical isolation by reversible biotinylation (Cleary et al., 2005; Duffy et al., 2015; Miller et al., 2011). But the underlying protocols are laborious, require ample starting material, and typically exhibit low signal-to-noise performance because of limited biotinylation efficiency, particularly when studying RNAs of limited length (Duffy et al., 2015; Marzi et al., 2016). Recently described approaches for the chemical conversion of 4sU into cytidine analogs help to overcome these limitations by marking metabolically labeled RNA in the context of steady-state profiles generated by standard RNA sequencing protocols (Herzog et al., 2017; Riml et al., 2017; Schofield et al., 2018)
Here, we employed thiol-linked alkylation for the metabolic sequencing of small RNA (SLAMseq) to provide a comprehensive view on the timescales and molecular features of miRNA biogenesis, loading and turnover in living cells. Our results elucidate fundamental mechanistic principles that control the efficiency and specificity of miRNA-mediated gene regulation and open new experimental avenues for deconvoluting the intracellular dynamics of small RNA silencing pathways at unprecedented accessibility, efficiency and scalability.
Results
Intracellular kinetics of microRNA biogenesis
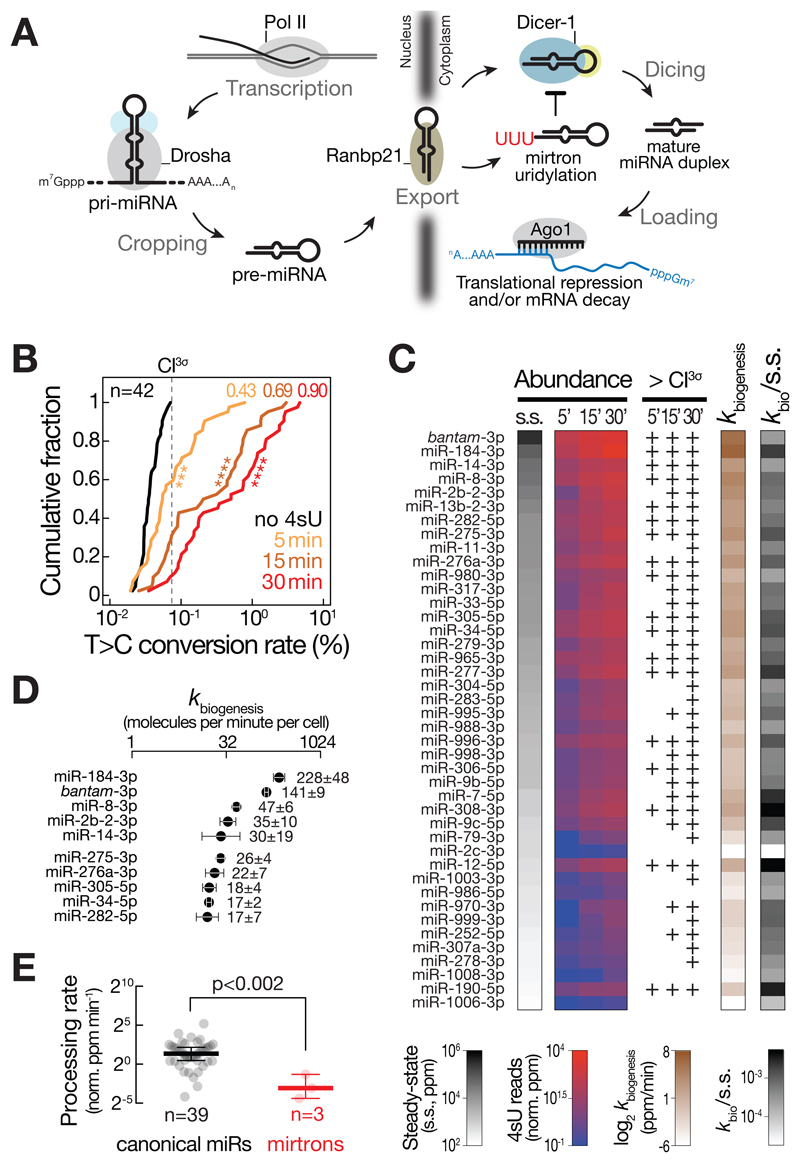
To investigate the intracellular kinetics of miRNA biogenesis (Figure 1A), we performed small RNA SLAMseq upon short pulse 4sU metabolic labeling in Drosophila S2 cells (for method validation see STAR Methods and Figure S1). To preempt confounding effects between the miRNA pathway and the RNAi pathway, we performed all following experiments in Drosophila S2 cells stably depleted of Ago2, a condition that left Ago1 levels unaltered (Figure S1P and Q). We determined the increase in T>C conversion rates of abundantly expressed miRNAs (>100 ppm at steady-state; n=42) in small RNA libraries prepared 5, 15, and 30 min after 4sU metabolic labeling and compared them to background signal detected in the absence of labeling (Figure 1B). Already 5 min of 4sU labeling resulted in a significant elevation in 4sU-labeled miRNAs (Kolmogorov-Smirnov test, p<10-3; Figure 1B). In fact, 43% of all miRNAs exhibited 4sU-labeling signal above the 3σ confidence interval (CI3σ) of background (no 4sU labeling; Figure 1B). At later time-points, the fraction of labeled miRNAs further increased to 69% (29 of 42) and 90% (38 of 42) after 15 and 30 min 4sU labeling, respectively (Figure 1B).
Figure 1. Intracellular kinetics and regulation of microRNA biogenesis.
(A) Overview of the miRNA pathway in flies. For details see text. (B) Cumulative distribution plot reports the mean T>C conversion rates for 42 abundantly expressed (> 100 ppm) miRNAs in small RNA libraries generated from total RNA of Drosophila S2 ago2ko cells treated with 4sU for the indicated time. Where applicable, T>C conversion rates were averaged between miR and miR*. P-values (Kolmogorov-Smirnov test; *** p<10-3; **** p<10-4) are shown. The fraction of miRs above 3σ confidence interval for T>C conversion in background (no 4sU) samples (dashed line; CI3σ) is indicated for each time-point. (C) Abundance of the indicated miRNAs at steady-state (s.s.; parts per million, ppm) and of newly produced miRNAs after the indicated time of 4sU metabolic RNA labeling (4sU reads; norm. ppm) is shown. T>C conversions relative to the CI3σ background-threshold are reported for each miRNA and 4sU labeling time-point. MicroRNA biogenesis rate as determined by linear regression are indicated in ppm per minute (kbiogenesis, ppm/min). kbiogenesis vs. steady-state ratios report the relative contribution of miRNA biogenesis rates to steady-state abundance. (D) Absolute biogenesis rates of abundantly produced miRNAs in molecules per minute per cell. Biogenesis rates were normalized to the absolute abundance of miRNAs in Drosophila S2 cells. Data represent mean ± stdev of two independent experiments. (E) Processing rates report transcriptional output-normalized mature miRNA biogenesis rates (in norm. ppm per minute) for 39 canonical miRNAs (canonical miRs; black) and three mirtrons (red) that are abundantly expressed (>100 ppm) in S2 ago2ko cells. P-value (Mann-Whitney test) is indicated. See also Figure S2.
To derive the rate at which individual miRNAs were produced, we analyzed the number of library-depth-, sequence-content-, and 4sU-labeling-efficiency-normalized T>C conversion-containing reads at early time-points of the metabolic labeling experiment (i.e. 5, 15 and 30 min) by linear regression, providing highly-reproducible (Pearson’s correlation rP=0.98, p<10-4, Figure S2A) and quantitative information on miRNA biogenesis kinetics (in ppm/min; Figure 1C). Considering the concentration of individual miRNAs in total RNA (Figure S2B to F), the total RNA content per cell (Figure S2G), and the steady-state abundance of miRNAs in small RNA sequencing datasets (Figure 1C and S2F), our data estimated that each Drosophila S2 cell produces hundreds of miRNA molecules per minute (e.g. 228 ± 48 and 141 ± 9 molecules per minute for miR-184-3p and bantam-3p, respectively; and a total of 702 molecules per minute considering the 42 most abundant miRNAs; Figure 1D and S2H).
Global survey of miRNA biogenesis rates revealed that miRNA steady-state abundance generally correlated well with production rate, but individual small RNAs displayed comparably low steady-state abundance despite high biogenesis rates (e.g. miR-184-3p, miR-277-3p, or miR-12-5p), or vice versa (e.g. miR-14-3p, miR-304-5p, or miR-2c-3p; Figure 1C), indicating that the rate of production is not the sole determinant for intracellular miRNA abundance.
MicroRNA processing rates unveil the selective regulation of miRNA biogenesis
By inspecting miRNAs that share a common primary transcript, we observed very similar biogenesis rates among cistronic miRNAs (Figure S2I). The miR-11/998 cluster was a notable exception, because miR-998 accumulated at 7-fold lower rates compared to miR-11, perhaps as a consequence of the unusual miR-998 hairpin structure, which includes a large terminal loop that extends into the designated Dicer processing site (Figure S2I). Motivated by the observed example for regulated miRNA biogenesis, we normalized miRNA biogenesis rates to pri-miRNA transcription rates (determined by an optimized transient transcriptome sequencing protocol; TT-SLAMseq; see STAR Methods for details) in order to systematically quantify miRNA processing rates (Figure S2J) (Schwalb et al., 2016). Overall, we observed a significant, strong correlation between the rates of pri-miRNA transcription and mature miRNA biogenesis (Pearson’s correlation coefficient rP=0.75, p<10-3), consistent with the view that miRNA processing generally follows the law of mass action. However, individual miRNAs displayed comparably low biogenesis rates despite high transcriptional activity. A prominent example were mirtrons (i.e. miR-1003, miR-1006, or miR-1008): Compared to canonical miRNAs, mirtrons were processed at significantly lower rates (Mann-Whitney test, p<0.002; Figure 1E), while their primary transcriptional activity was indistinguishable (Mann-Whitney test, p>0.05; Figure S2K); and both mirtron and canonical miRNA hairpins accumulated to similar levels at steady-state (Mann-Whitney test, p>0.05; Figure S2L), indicating a selective inhibition of mirtron biogenesis after pre-miRNA formation. Consistent with a previously proposed model for the selective inhibition of mirtron biogenesis by 3´terminal RNA hairpin uridylation (Bortolamiol-Becet et al., 2015; Reimão-Pinto et al., 2015), we detected a moderate but statistically significant negative correlation between miRNA processing rates and pre-miRNA uridylation status (Pearson’s correlation coefficient rP=-0.45, p<0.03; Figure S2M). This indicates that pre-miRNA uridylation effectively disrupts the tight coupling of miRNA biogenesis.
Timescales, constraints, and regulation of miRISC formation
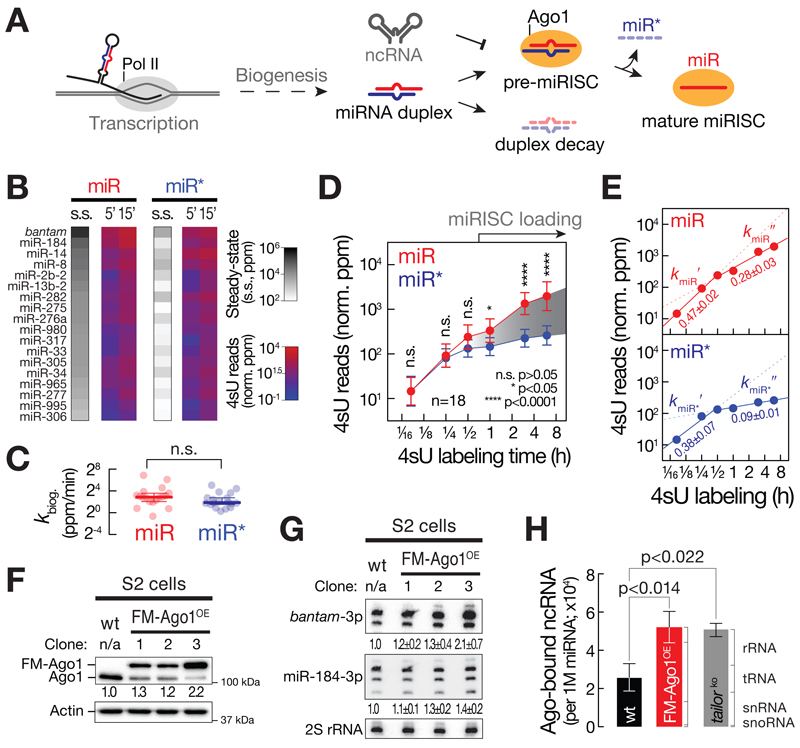
MicroRNA biogenesis initially yields a miRNA duplex before one of the two strands (the miR strand) is selectively stabilized by assembly into Ago1. In the process of miR-loading, the partner strand (the miR* strand) is expelled and degraded (Figure 2A). Alternatively, a miRNA duplex may be subjected to non-specific decay before encountering an Ago protein in a process that remains poorly characterized (Figure 2A). Small RNA SLAMseq experiments revealed that miRNA duplexes represent readily detectable intermediates of miRNA biogenesis in cells (Figure 2B to D): At early time-points after 4sU labeling (5 and 15 min), we observed no statistically significant difference in the relative abundance of 4sU reads (Figure 2B) or biogenesis rates (Figure 2C) of miR and miR* strands when analyzing 18 miRNAs for which both strands are readily detectable in small RNA libraries (Mann-Whitney test, p>0.05). Further progression through the labeling time course revealed a statistically significant, higher accumulation of 4sU-labeled miR compared to miR* strands (Mann-Whitney test, p<0.05; Figure 2D), indicating that – on average – the selective stabilization of miRs (presumably prompted by Ago-loading) occurs at much lower rates compared to miRNA duplex production. While individual miRNA duplexes exhibited a broad array of loading rates, the majority assembled only at 1h or later (Figure S3A). Further global kinetic modelling unveiled that the accumulation of miRs (and miR*s) was best described by a biphasic regression model (Figure 2E): At early time-points, 4sU-labeled miR and miR* strands accumulated at very similar rates (kmiR′=0.47±0.02 and kmiR*′=0.38±0.07; Figure 2E), reflecting the rapid biogenesis of miRNA duplexes (Figure 1 and 2B to D). Notably, the selective accumulation of miRs at later time-points (1 to 8 hours, Figure 2D) coincided with a pronounced decrease in the accumulation rate (kmiR′=0.28±0.03; Figure 2E), estimating that only ~60 % of produced miRs acquire a stable fate. Our data therefore predict that ~40% of miRNAs are turned over before Ago-loading, most certainly as a consequence of duplex decay. As expected, the second phase of miR* accumulation dropped much more severely (kmiR*′=0.09±0.01; Figure 2E), indicating that the vast majority of miR*s undergo rapid decay due to the combined action of miRISC loading and miRNA duplex turnover.
Figure 2. Timescales, constraints, and regulation of miRISC formation.
(A) Model for selective miRNA loading into Ago. See text for details. (B) Steady-state abundance (s.s.) and mean accumulation of T>C conversion containing reads normalized to 4sU-labeling efficiency (4sU reads in norm. ppm) of 18 abundantly expressed miR and miR* pairs at 5 and 15 min after 4sU metabolic labeling in ago2ko S2 cells. (C) Biogenesis rate (kbiog. in norm. ppm/min) of miR and miR* pairs (see Figure 1C and 2B). P-value (Mann-Whitney test; n.s., p>0.05) is shown. (D) Relative accumulation of metabolically labeled small RNAs upon 4sU labeling in ago2ko S2 cells. Mean ± 95% CI for 18 abundantly expressed miRNA duplexes partitioned in miR (red) and miR* (blue) strands is shown. P-values (Mann-Whitney test; n.s., p>0.05; *, p<0.05; ****, p<10-4) are indicated. Selective stabilization of miR versus miR* species is indicative of miRISC loading. (E) Linear regression of early time-points (5, 15 and 30 min) or late time-points (0.5, 1, 3, 6h) of experiment (D) are shown for miR (top, in red) and miR* strand (bottom, in blue). Slope ± stderr of the linear regressions is indicated. (F) Western blot analysis of endogenous (Ago1) and FLAG-MYC-tagged Ago1 (FM-Ago1) protein levels in wild-type S2 cells or three clones of expressing FLAG-MYC-tagged Ago1 (FM-Ago1OE). Actin indicates loading control. Relative Ago1 signal intensities are shown. (G) Northern hybridization assay probing for bantam-3p and miR-184-3p in wild-type S2 cells or three clones expressing FLAG-MYC-tagged Ago1 (FM-Ago1OE). Relative signal intensities of three independent experiments (mean ± stdev) normalized to 2S rRNA signal are indicated. (H) Ago-bound noncoding RNA levels (ncRNAs, normalized to 1 million miRNAs) in wild-type S2 cells (in black), S2 cells expressing FLAG-MYC-tagged Ago1 (clone 3) (in red, FM-Ago1OE) and S2 cells depleted of Tailor (tailorko) (Reimão-Pinto et al., 2016). Non-coding RNAs comprise ribosomal (rRNA), transfer (tRNA), small nuclear (snRNA) and small nucleolar (snoRNA) RNA species. Mean ± stdev of three (wt and FM-Ago1OE) or two (tailorko) replicate small RNA libraries are shown. P-values (two-tailed unpaired t-test) are indicated. See also Figure S3.
What causes such low yields in miRISC formation? Previous studies proposed that the availability of empty Ago-proteins may limit the accumulation of gene regulatory small RNAs in cancer cells (Diederichs et al., 2008; Martinez and Gregory, 2013). To address this in a non-transformed context, we overexpressed FLAG-tagged Ago1 in Drosophila S2 cells, selected single clones (Figure 2F and S3B), and determined the change in steady-state abundance of miRNAs by Northern hybridization (Figure 2G and S3C). Indeed, stepwise elevation of Ago1 levels correlated with a gradual increase in steady-state abundance of bantam-3p and miR-184-3p, resulting in up to 2-fold higher miRNA levels per cell compared to wild-type controls (Figure 2G and S3C). Because Ago1-availability limits the yields at which miRNA duplexes assemble into functional ribonucleoprotein complexes, miRISC formation represents a key kinetic bottleneck in cells. Notably, we detected a stepwise decrease in endogenous Ago1 protein levels upon overexpression of a FLAG-tagged transgene (Ago1 vs. FM-Ago1; Figure 2F). We therefore hypothesized that tight control over empty Ago protein levels may contribute to the selective loading of miRNAs and prevent abundant non-coding RNA decay intermediates from entering RISC. Indeed, high-throughput sequencing of Ago-bound small RNAs revealed a disproportional, statistically significant, >2-fold increase in ncRNAs (rRNA, tRNA, snRNA and snoRNA) relative to miRNAs upon overexpression of Ago1 (two-tailed unpaired t-test, p<0.014; Figure 2H and S3D), implying an important function of restricted Ago-availability in sustaining specificity of RISC formation. Finally, we tested if a similar imbalance in Ago-loading specificity can arise from elevated ncRNA decay intermediates. To this end, we focused on a dedicated cytoplasmic ncRNA quality control pathway, the TRUMP complex, which selectively removes aberrant, highly structured RNA polymerase III transcripts by uridylation-mediated RNA decay (Reimão-Pinto et al., 2016). Indeed, we detected a statistically significant, >2-fold increase in Ago-associated ncRNAs relative to miRNAs in S2 cells stably depleted of TRUMP complex, indicating that the selective removal of aberrant ncRNAs cooperates with Ago-protein homeostasis to ensure the specific loading of miRNAs (two-tailed unpaired t-test, p<0.022; Figure 2H).
Dynamic emergence of 3´ isomiRs unravels molecular signatures of aging miRNAs
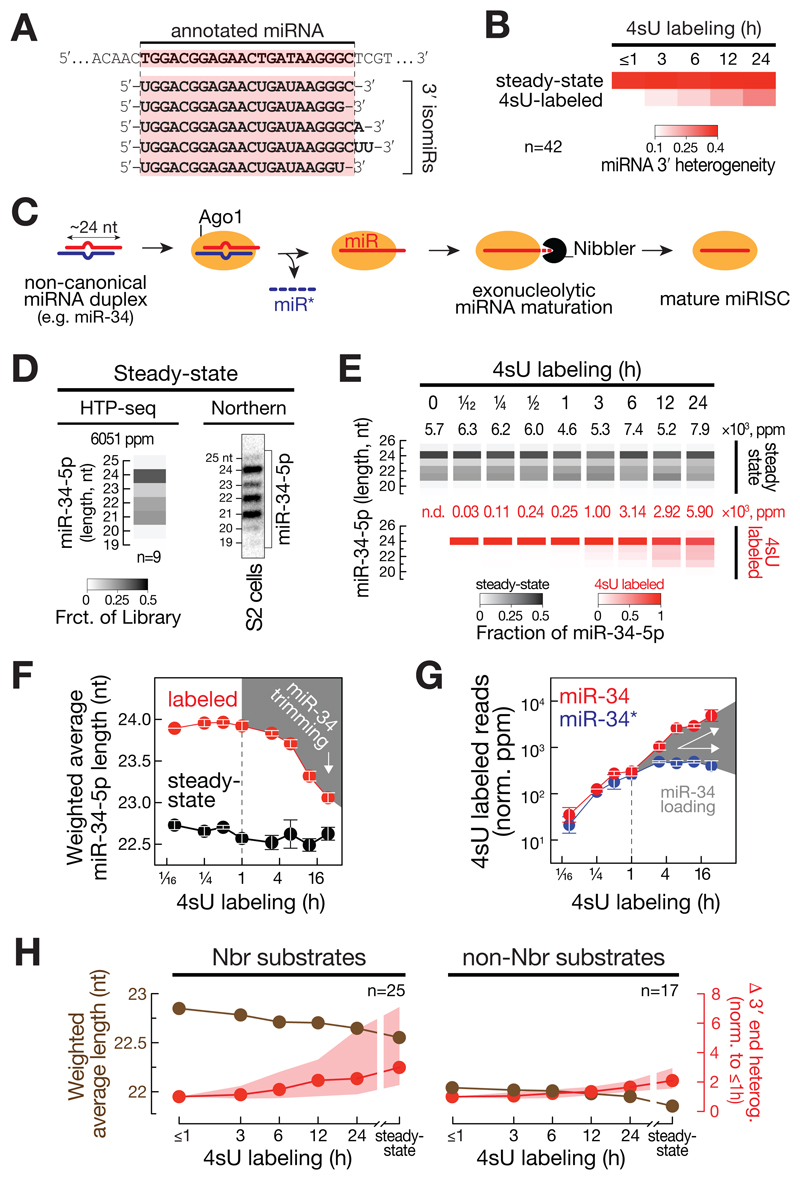
A variety of intracellular processes diversify small RNAs by modulating their length and terminal nucleotide content mostly at miRNA 3´ ends (3´ isomiRs; Figure 3A) (Ameres and Zamore, 2013). To address the dynamic emergence of isomiRs, we determined the median 3´ end heterogeneity of 4sU-labeled miRNAs (Figure 3B). While 3´ end heterogeneity of 42 abundant miRNAs (>100 ppm) remained constantly high at steady-state, 4sU-labeled miRNAs exhibited low heterogeneity at early time points and steadily diversified over time (Figure 3B).
Figure 3. Dynamic emergence of isomiRs unravels molecular signatures of aging miRNAs.
(A) Schematic representation of 3´ isomiRs. See text for details. (B) Median 3′ end heterogeneity of 42 abundantly expressed miRNAs in S2 ago2ko cells is shown for all (steady-state) or T>C conversion-containing (4sU-labeled) reads across a 4sU labeling time course. (C) Model for exonucleolytic miRNA maturation in Drosophila. See text for details. (D) Steady-state length distribution of miR-34-5p in Drosophila ago2ko S2 cells as determined by high-throughput sequencing of small RNAs (left, heat map represents mean of 9 measurements; average cloning count is indicated in parts per million, ppm) or Northern-hybridization experiments (right). (E) Length distribution of miR-34-5p in libraries prepared from Drosophila ago2ko S2 cells subjected to 4sU metabolic labeling for the indicated time. Length distribution of T>C conversion containing reads (4sU-labeled, red) and all reads (steady-state, black) are shown. Read count (in ppm) is indicated. (F) Weighted average length of miR-34-5p in libraries prepared from Drosophila ago2ko S2 cells subjected to 4sU metabolic labeling for the indicated time. Data represent mean ± stdev of T>C conversion-containing reads (labeled, red) and all reads (steady-state, black) from two independent biological replicates. Decrease in weighted average length of T>C conversion-containing reads indicates exonucleolytic trimming (highlighted by grey area). (G) Loading of miR-34-5p as determined by the relative abundance of T>C conversion-containing (4sU labeled) reads in miR-34-5p (miR strand, red) and miR-34-3p (miR* strand, blue) after 4sU metabolic labeling of Drosophila ago2ko S2 cells. Mean ± stdev of two independent experiments is shown. Loading is highlighted by grey area. (H) Mean weighted average nucleotide length of T>C conversion containing (4sU-labeled) reads (labeled in brown) or median and interquartile range of the change in 3´ end heterogeneity (relative to ≤1h, in red) is shown across a 4sU labeling time course or at steady-state in Drosophila ago2ko S2 cells for miRNAs classified as Nbr substrates (left, n=25) or non-Nbr substrates (right, n=17). See also Figure S4.
One well-established example for isomiR production in flies is the exonucleolytic maturation of miRNAs (e.g. miR-34) by Nbr (Figure 3C). In steady-state analyses, miR-34-5p exhibited a scattered length profile ranging from abundantly detected 24 to 21mer isoforms (Figure 3D). To disentangle the intracellular order of events that produce multiple miR-34-5p isoforms, we analyzed SLAMseq small RNA libraries (Figure 3E and F). In contrast to steady state small RNAs, which displayed compound length profile across the entire metabolic labeling time course (black heatmap, Figure 3E), 4sU-labeled miR-34-5p reads initially accumulated solely as 24mers (red heatmap, Figure 3E), consistent with the accurate processing of synthetic pre-miR-34 in vitro by recombinant Dcr-1 or fly lysate (Han et al., 2011). Only after 3h of 4sU labeling, we detected the emergence of shorter 4sU-labeled miR-34-5p 3´ isoforms (Figure 3E). From this time-point on, the weighted average length of miR-34-5p decreased continuously over time, slowly approaching the average length profile of miR-34-5p observed at steady-state (Figure 3F). Biochemical studies proposed that exonucleolytic maturation of miRNAs occurs only in the context of Ago1 after miR* strand removal, implying a single-strand-specific 3´-to-5´ exoribonucleolytic activity of Nbr (Han et al., 2011). We tested this hypothesis by comparing miR-34-5p trimming signal (Figure 3F) and miR-34 loading kinetics (Figure 3G), which revealed that trimming indeed co-occurred with the selective stabilization of miR-34-5p relative to miR-34-3p in SLAMseq libraries starting after 1 h of metabolic labeling (Figure 3G).
To globally inspect Nbr-directed 3´ isomiR formation, we focused on 25 miRNAs that significantly increased in weighted average length (two-tailed Student’s t-test, p<0.01) in small RNA libraries generated from S2 cells depleted of Nbr by CRISPR/Cas9 genome engineering, in the presence or absence of catalytic mutant Nbr transgene compared to wild-type S2 cells and Nbr-knockout S2 cells harboring a NbrWT rescue transgene (Figure S4A to C). These miRNAs comprised eight previously described and 17 novel Nbr substrates (Figure S4D). Consistent with the proposed role of Nbr in maturing unusually long miRNAs, 4sU-labeled Nbr-substrates were on average 0.8 nts longer (average weighted length: 22.8 nt) compared to non-Nbr-substrates (22.0 nts) at early time-points (≤1h) of a metabolic labeling experiment and decreased in size over the course of the experiment (Figure 3H). As a consequence, 3´ end heterogeneity of Nbr-substrates increased over time, revealing the dominant role of Nbr in the diversification of miRNA 3´ ends in flies, when compared to non-Nbr substrates (Figure 3H). Notably, even non-Nbr substrates were shortened over time, albeit to a lesser extent, establishing 3´ isomiR formation as a general hallmark of aging miRNAs (Figure 3H).
Contribution of stability to miRNA abundance
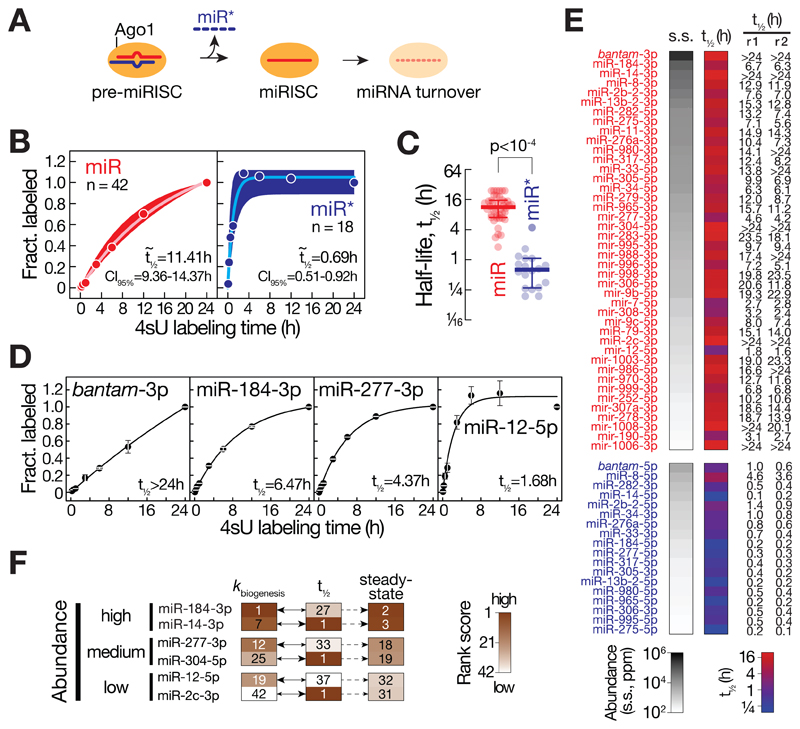
Understanding the contribution of half-life to steady-state miRNA profiles (Figure 4A) requires experimental approaches that rapidly and reproducibly report absolute and sequence content-normalized miRNA stabilities. To this end, we determined the 4sU-labeling rate averaged across individual positions for each miRNA in small RNA libraries prepared from total RNA of Drosophila S2 cells subjected to 4sU metabolic labeling for up to 24 h. The observed increase in median labeling rate over time was best described by single exponential kinetics, determining a highly reproducible median half-life of 11.41 h for the 42 abundantly expressed miR strands (Figure 4B and S5). Compared to miRs, miR*s exhibited a significantly shorter median half-life of 0.69 h (Mann-Whitney test, p<10-4; Figure 4C), consistent with the proposed rapid turnover of miR* during miRISC loading (Figure 2). Closer inspection of individual miRNAs revealed a broad distribution of stabilities that differed by more than one order of magnitude, ranging from half-lives of more than 24 h (e.g. bantam-3p) to less than two hours (e.g. miR-12-5p, Figure 4D and E). Notably, stability can profoundly impact steady-state small RNA abundance, because miRNAs accumulate to very similar steady-state levels as a consequence of high production rates) or high stability, respectively, across a broad array of steady-state abundances (Figure 4F).
Figure 4. Contribution of stability to miRNA abundance.
(A) Model for the loading and turnover of small RNAs during assembly of the miRNA-induced silencing complex (miRISC). See main text for details. (B) The stability of miRNAs was determined by plotting the fraction of 4sU-labeling averaged across individual positions of small RNAs determined in a 4sU-metabolic labeling time course in Drosophila ago2ko S2 cells and normalized to the conversion rate measured at final time point (24h). The median 4sU-labeling fraction and interquartile range of 42 miR (red) or 18 miR*s (blue) is shown for individual time points. Data was fit to single exponential saturation kinetics to derive median half-lives (t½) and 95% confidence interval (CI95%). (C) Scatter dot plot reports the half-life of 42 miRs (red) and 18 miR*s (blue). Median and interquartile range are indicated. P-value (Mann-Whitney test) is shown. (D) Half-life of individual miRs (bantam-3p, miR-184-3p, miR-277-3p and miR-12-5p) was determined as described in (B). Values represent the mean ± stdev of two independent experiments. (E) Steady-state abundance and average half-life (t½) for the indicated miR (red) and miR* (blue). Individual half-life measurements for two independent experiments (r1 and r2) are reported. Half-life data that exceeded the maximum time of the measurement are indicated as > 24h. (F) MicroRNA biogenesis and turnover determine steady-state miRNA abundance in a miRNA-specific manner. MicroRNAs were ranked according to biogenesis rate (kbiogenesis), stability (t½) or steady-state abundance (steady-state) and the respective rank score (see heatmap reference) of three pairs of miRNAs that accumulate to similarly high, medium, or low levels at steady-state are shown. See also Figure S5.
MicroRNA decay kinetics resolve compound fates of small RNAs
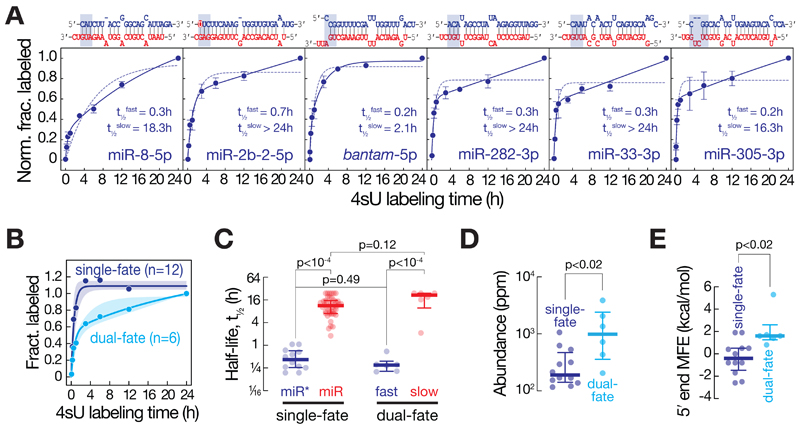
Most miRNA duplexes exhibit asymmetry in RISC loading, i.e. only one of the two strands assembles into Ago; but few duplexes have been proposed to load either strand (i.e. symmetric miRNA duplexes). In principle, small RNAs that originate from symmetric duplexes should display compound half-life kinetics to account for the dual fate of each strand (i.e. acting partially as stable guide or unstable miR*). Indeed, by carefully inspecting small RNA decay, we found that six out of 18 miR*s fit well to dual- but not single-phase kinetics (Figure 5A). The two half-lives observed for these small RNAs (median half-lives of t1/2fast = 0.30 h and t1/2slow= 21.4 h, Figure 5B) matched well those observed for miR*s (t1/2miR* = 0.42 h) and miRs (t1/2miR = 11.4 h), respectively (Figure 5C). In agreement with the partial loading of dual-fate miR*s, they also accumulated to significantly higher steady-state levels compared to miR*s that followed a simple single-exponential turnover model (Figure 5D). Consistent with the view that strand selection during RISC loading predominantly relies on the thermodynamic stability of small RNA duplex ends (Khvorova et al., 2003; Schwarz et al., 2003), dual-fate miR*s were significantly less paired at their 5´ end compared to single-fate miR*s (Mann-Whitney test, p<0.02; Figure 5E).
Figure 5. RNA decay kinetics resolve compound fates of small RNAs.
(A) The stability of miR* (blue) strands of the indicated duplexes was determined by plotting the fraction of 4sU-labeled small RNAs across a 4sU-metabolic labeling time course in Drosophila ago2ko S2 cells to single exponential (dotted line) or two-phase (solid line) saturation kinetics. Data represent mean ± stdev of two independent experiments. Half-lives determined from the dual-phase decay kinetics (t½fast and t½slow) are indicated. MicroRNA duplex structures are shown as previously predicted (Kozomara and Griffiths-Jones, 2010). The miR* strand (blue) is defined by lower steady-state abundance relative to the miR strand (red). Base-pairing of three 5´ terminal nucleotides of the miR* strand is highlighted in blue and was used to determine the minimal free energy (MFE) duplex stability (see panel E). The 5´U of miR-2b-2-5p is highlighted in red and may contribute to loading of the miR* strand. (B) Median and interquartile range of stability measurements of 18 miR*s that either follow single-exponential (dark blue, n=12) or dual-phase decay kinetics (light blue, n=6). (C) Half-lives of miRs (red) and miR*s (blue) that followed single-exponential decay kinetics (single-fate) are compared to the two half-lives (slow and fast) observed for miR*s that were best described by dual-phase kinetics (dual-fate). Median ± interquartile range is shown. P-values (Mann-Whitney test) are indicated. (D) Steady-state abundance of miR* that follow single-exponential decay kinetics (single-fate, n=12) or dual-phase kinetics (dual-fate, n=6) are compared. Median ± interquartile range is shown. P-values (Mann-Whitney test) are indicated. (E) Minimal free energy (MFE, kcal/mol) of the three 5´-terminal nucleotides of the miR* strand (highlighted by blue boxes in panel A for dual-fate duplexes) in a single-fate (dark blue, n=12) or dual-fate (light blue, n=6) duplex. Median ± interquartile range is shown. P-value (Mann-Whitney test) is shown.
Molecular determinants of miRNA turnover
To study the molecular principles of small RNA turnover, we sought an approach to selectively modulate the fate of miRNAs in a predictable manner. To this end, we exploited the fact that small RNAs partition into two Ago protein family members in flies, Ago1 and Ago2 (Förstemann et al., 2007; Tomari et al., 2007) (Figure 6A). We annotated miRNAs that accumulate exclusively in Ago1 or Ago2 in wild-type S2 cells by determining the relative abundance of small RNA in libraries before or after oxidation, a strategy to selectively clone 3´ methylated small RNAs (Figure S6A and B) (Ghildiyal et al., 2009). Next, we calculated the 4sU-labeling rate in small RNA SLAMseq libraries prepared from S2 cells depleted of Ago2 (Figure S1Q). Ago2-enriched small RNAs followed single exponential decay kinetics, exhibited a median half-life of 1h (Figure 6B), and failed to accumulate to high levels at steady state (Figure S6C), reminiscent of short-lived miR*s (t½=0.69 h; Figure 4B). In contrast, Ago1-enriched small RNAs strongly resembled decay kinetics of miRs (t½=11.41 h, Figure 4B) with a medium half-life of 18 h (Figure S6D). In SLAMseq experiments in wild-type S2 cells, Ago1-enriched small RNAs did not change in decay kinetics (t½=15.9 h; Figure S6D), but Ago2-bound small RNAs displayed two-phase kinetics (t½fast=0.2h, t½slow>24h; Figure 6B), consistent with their stabilization by partial Ago2-binding, resulting in a statistically significant higher steady-state accumulation when compared to S2 cells depleted of Ago2 (Mann-Whitney test, p<0.01; Figure S6C). Simultaneously, we prepared SLAMseq libraries from wild-type S2 cells but subjected total RNA to oxidation prior to small RNA cloning to enrich for Ago2-bound 3´ end methylated species. Here, Ago2-enriched small RNAs followed single-exponential decay kinetics with a medium half-life of more than 24h (Figure 6B). Together, these experiments reveal that compound small RNA half-lives observed in wild-type S2 cells are explained by the variable sorting of duplexes into specific Ago proteins. Finally, we compared the half-lives of 30 small RNAs that are most prominently associated with either Ago1 or Ago2 (Figure S6B) and found that small RNAs are significantly more stable when bound to Ago2 compared to Ago1 (Mann-Whitney test, p<10-4, Figure 6C).
Figure 6. Molecular determinants of miRNA turnover.
(A) In Drosophila, miRNAs preferentially load into Ago1 to form miRISC. At the same time, a subset of miR*s load onto Ago2 to form siRISC, prompting the methylation of Ago2-bound small RNAs at the 2´ position of the 3´-terminal ribose. If and how Argonaute protein identity influences small RNA stability remains unknown. (B) Median decay kinetics of Ago2-enriched miRNAs (n=8; classified in Figure S6B) in an 4sU metabolic labeling time course in wild-type (wt, black) or ago2ko S2 cells (red) or from wild-type S2 cells employing a cloning strategy that enriches for small RNAs with modified 3´ termini (wt oxidized, blue). Median and interquartile range of two-phase or one-phase saturation kinetics (as specified in main text) are shown. The half-lives (t½) as determined by curve-fitting are indicated. (C) Comparison of small RNA stabilities in the context of Ago1 and Ago2. Half-lives of the 30 most abundant Ago1-bound miRNAs (red, Ago1) compared to the most abundant miRs and miR*s in small RNA libraries employing a cloning strategy that enriches for small RNAs with modified 3´ termini (blue, Ago2). The median and interquartile range is indicated. P-value (Mann-Whitney test) is shown. (D) Half-lives of miRNAs subjected to exonucleolytic 3´ end trimming by Nbr (in red, n=25) and non-Nbr substrates (in black, n=17) as classified in Figure S4. Median and interquartile range are shown. P-value (Mann-Whitney test) is indicated. See also Figure S6.
The broad array of half-lives observed for Ago1-bound miRNAs (see Figure 4) suggested that factors other than Ago-protein identity specify the fate of miRNAs. Prompted by our observation that miRNAs continuously increased in 3´ end heterogeneity as they age (Figure 3H), we hypothesized that the enhanced exonucleolytic 3´ end trimming may contribute to miRNA turnover (Figure 4). Indeed, with a median half-life of 9.6 h, Nbr-substrates were significantly less stable compared to all other miRNAs (t1/2=14.8 h, Mann-Whitney test, p<0.03; Figure 6D), suggesting that miRNA 3´ end processing may act as a molecular driver for selective miRNA turnover.
Discussion
By establishing metabolic small RNA sequencing, we report absolute rates of synthesis and decay for 60 miRNAs (42 miR and 18 miR*) that are expressed at biologically relevant levels (>100 ppm at steady-state) in Drosophila S2 cells. Our data estimates that each cell produces up to 228±48 mature miRNA molecules per minute (for miR-184) and an average of 58±14 molecules per minute for the ten highest expressed miRNAs (Figure 1), a rate that is 29-times or 7-times higher, respectively, when compared to the highest previously reported production rate of any mRNA (up to 8 molecules per minute per cell) (Miller et al., 2011; Schwanhäusser et al., 2011). With a median half-life of 11.4 h (Figure 4), mature Ago-bound miRNAs are also on average 3.9-times more stable than mRNAs in the same cell type (median half-life of 2.9 h across 2132 transcripts; our unpublished data). Such unusual kinetic parameters explain how miRNAs can accumulate to high intracellular concentrations (up to 5.3 × 105 molecules per cell; Figure S2) in order to meet the biophysical requirements for effective target RNA binding and repression (Salomon et al., 2015; Wee et al., 2012).
Considering that at least 40 % of mature miRNAs are produced in less than 5 min, processing and transport of precursors must occur in a highly coordinated manner inside cells (Figure 1B). Consistent with the view that miRNA processing generally follows the law of mass action, mature miRNA biogenesis kinetics generally correlated well with the rates of pri-miRNA transcription (Figure S2J). But individual miRNAs exhibited processing rates that indicated a specific regulation during biogenesis (Figure S2J). Among these, the slow biogenesis of miR-998 (7-fold slower than its cistronic partner miR-11) represents an example for intrinsic regulation, since unpaired nucleotides at the designated processing site have previously been shown to interfere with efficient dicing in vitro (Figure S4I) (Tsutsumi et al., 2011). We also detected regulation of miRNA biogenesis in trans, as exemplified by mirtrons (i.e. miR-1003, miR-1006, or miR-1008; Figure S2J). Compared to canonical miRNAs, mirtrons were processed at significantly lower rates (Figure 1E), despite similar transcriptional activity (Figure S2K). Previous studies had already anticipated the selective dampening of mirtron biogenesis, because splicing-derived RNA hairpins (but not canonical pre-miRNAs) were selectively subjected to 3´ terminal uridylation by the TUTase Tailor and pre-miRNA 3´ end extensions effectively inhibited Dicer-mediated processing and triggered dmDis3l2-directed RNA decay (Bortolamiol-Becet et al., 2015; Reimão-Pinto et al., 2015; 2016). Consistent with this model, we detected a negative correlation between miRNA processing rates and pre-miRNA uridylation status (Figure S2M). Once assembled in Ago1, mirtrons do not differ in stability from canonical miRNAs (Figure 4E), highlighting the importance of regulation prior to assembly. Given the substantial contribution of miRNA biogenesis to the accumulation of mature miRNAs, 3´ uridylation of pre-miRNAs provides an effective strategy to tune miRNAs levels, perhaps explaining its prevalence across evolutionary distant species (Ha and Kim, 2014).
In cells, miRNAs initially accumulate as duplexes, because we detected no significant difference in the initial production rates of miR and miR* (Figure 2C). In fact, Ago-loading (as inferred from the selective accumulation of miR over miR*) commences with an average delay of one hour, implying that a process that guides miRNAs to Ago1 is rate limiting or absent (Figure 2D). The latter is indirectly supported by an in vivo small RNA tagging approach using the small RNA duplex-specific methyltransferase HEN1 from Arabidopsis thaliana, which demands a significant fraction of miRNA duplexes to be accessible (Alberti et al., 2018). Consistent with a physical uncoupling of miRNA biogenesis and Ago loading, miRNA duplexes were previously shown to assemble into Ago1 or Ago2 irrespective of the pathway that produces them (Förstemann et al., 2007).
In quantitative terms, RISC assembly represents a critical kinetic bottleneck for the accumulation of mature miRNAs in cells (Figure 2). Based on dual phase kinetics of miRs at early metabolic labeling time-points, we estimate that ~40 % of miRNAs decay as duplexes before they encounter Ago1 (Figure 2E). While such low yields appear wasteful at first glance, the kinetic constriction of miRISC formation ensures the specific loading of miRNAs and the omission of RNAs originating from tRNAs, rRNAs, snoRNAs or snRNAs. Overt elevation of Ago1 protein levels overcomes this constriction, but disproportionally enhances the appearance of ncRNA-derived small RNAs in Ago1 (Figure 2G and H). Limited Ago1 levels that enforce the selective loading of miRNAs in wild-type cells are presumably achieved by the selective ubiquitination and proteolytic decay of empty Ago1 (Figure 2F) (Derrien et al., 2012; Kobayashi et al., 2019; Martinez and Gregory, 2013; Smibert et al., 2013). Notably, we observed a similar disproportional accumulation of ncRNA-derived small RNAs in Ago1 upon elevating aberrant non-coding RNAs by interfering with ncRNA surveillance (Figure 2H) (Reimão-Pinto et al., 2016). In this respect, our data is reminiscent of the spurious entry of RNAs into the nuclear siRNA pathway upon depletion of TRAMP-mediated ncRNA surveillance pathway in fission yeast (Bühler et al., 2008). Therefore, Ago1 protein homeostasis and ncRNA surveillance cooperate to ensure the fidelity of RISC-mediated gene silencing. More generally, these results imply an important and long-standing selection against the accumulation of random RNA fragments that may evoke the inappropriate formation of gene regulatory complexes (Houseley and Tollervey, 2009).
In the course of their life-time, miRNAs are subjected to significant changes in nucleotide composition at their 3´ end (Figure 3). Consistent with a precise ruler function of Drosha and Dicer (Kim et al., 2009), mature miRNAs varied little in sequence composition shortly after they were born. But 3´ heterogeneity increases significantly as miRNAs age, and most isomiRs emerged at time-points that followed Ago1-loading (Figure 3B). Our data therefore warrants caution when extracting precise cropping and dicing positions from currently available miRNA annotations that derive mostly from bulk small RNA sequencing data (Kozomara and Griffiths-Jones, 2014). Mechanistically, most 3´ heterogeneity is introduced by the 3´-to-5´ exoribonuclease Nbr. In fact, our refined annotation revealed that 60% of miRNAs are subjected to Nbr-mediated trimming, albeit to different extent (Figure S4). Time-resolved profiling of Nbr-substrates confirmed many of the mechanistic aspects of miRNA 3´ end trimming in vitro (Figure 3) (Han et al., 2011; Liu et al., 2011): Trimming is most obvious among miRNAs with unusual length, occurs after Ago1 loading, and requires miR* strand removal. But miRNAs trimmed by Nbr were also significantly less stable compared those that were not (Figure 6D), implying an additional role of Nbr in defining small RNA fate. Clarifying the relationship between 3´ resections and small RNA turnover demands further investigations, particularly given its recently expanded role in the maturation of other small RNA classes, such as Piwi-interacting RNAs (piRNAs) (Hayashi et al., 2016).
Many miRNAs rank among the most stable cellular transcripts and collectively they approximate half-lives that compare to stable non-coding RNA classes, such as tRNAs (Hopper, 2013). But individual miRNA species exhibit a broad distribution of stabilities ranging from 5 min, as observed for many miR*s, to >24 h for miRs (Figure 4E). A broad stability range also applied to Ago1-bound miRNAs (t½=1.8 h to >24 h), with profound consequences for steady-state miRNA abundance across a broad range of intracellular concentrations (Figure 4F). Notably, many miRNAs face complex fates, as most evident for select miR*s that undergo biphasic exponential decay (Figure 5). In such cases, small RNA decay kinetics most certainly reflect the compound contribution of Ago-loading and rapid turnover of symmetric miRNA duplexes as determined by thermodynamic stability of small RNA duplex ends (Khvorova et al., 2003; Schwarz et al., 2003). Overall, the major determinant for small RNA stability is the assembly of RISC itself, which prompts the selective stabilization of miRs upon specific binding of 5´ and 3´ ends by Ago’s Mid and PAZ domain, respectively (Sasaki and Tomari, 2012), and the acceleration of miR* turnover upon duplex unwinding (Kawamata and Tomari, 2010) (Figure 4). In flies, miRNAs are sorted into functionally distinct Ago complexes (Förstemann et al., 2007), as shown by the selective stabilization of Ago2-bound small RNAs in wild-type cells compared to cells depleted of Ago2 (Figure 6). Notably, Ago-protein identity profoundly effects small RNA half-lives, since Ago2-bound small RNAs were almost twice as stable compared those in Ago1 (Figure 6C). Such differences in stability may reflect the specific needs of the miRNA and the siRNA pathway, respectively: While Ago1 must enable the selective and rapid turnover of miRNAs to account for the changing regulatory requirements during organismal development, cellular differentiation and changing environmental conditions (Bartel, 2018), Ago2-directed antiviral defense most certainly benefits from a long-term memory arising from stable, virus-derived siRNA in RISC (Ghildiyal and Zamore, 2009). At the molecular level, 3´ end trimming (see above) and/or target-directed decay represent possible routes to decay for select miRNAs (Ameres et al., 2010; Bitetti et al., 2018; Cazalla et al., 2010; Kleaveland et al., 2018; la Mata et al., 2015; Marcinowski et al., 2012; Xie et al., 2012), and protection against those by 2´O-methylation at the 3´ terminal ribose stabilizes Ago2-bound siRNAs in vitro and in vivo (Ameres et al., 2010).
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Stefan Ameres (stefan.ameres@imba.oeaw.ac.at).
Experimental Model and Subject Details
Cell culture
Drosophila Schneider 2 (S2) cells were obtained from the Zamore lab and cultured in Schneider’s Drosophila Medium containing 10% FBS. Cells were maintained at 27°C and passaged every 3 days. Cells have been validated (by genome, mRNA, and small RNA sequencing) and are of male origin.
Method Details
HPLC
HPLC analysis was essentially performed as described by (Spitzer et al., 2014; Andrus and Kuimelis, 2001): Briefly, 8 µg of total RNA was incubated overnight (> 16h) with 1.6 U Bacterial Alkaline Phosphatase (Invitrogen) and 0.2 U Phosphodiesterase I (Worthington Biochemical) in a final volume of 130 µl containing 0.1 mM DTT, 15 mM MgCl2 and 35 mM Tris-HCl (pH 7.5) to digest into single nucleosides. Single nucleosides were precipitated by adding 6 µl 3M NaOAc (pH 5.2), 150 µl ice-cold 100% EtOH and 30 µl 10 mM DTT and incubating 10 min at -80°C. Samples were spun down at 12,500 x g for 5 min at room temperature. Supernatant was transferred to a new tube and precipitation was repeated by adding 30 µl 10 mM DTT and 270 µl ice-cold 100% EtOH, incubating for 10 min at -80°C and spinning down at 12,500 x g for 5 min at room temperature. Supernatant was transferred to a new tube and evaporated in a speed-vac machine to complete dryness. Samples were dissolved in 200 µl H2O and half of the sample was loaded on HPLC on a Supelco Discovery C18 (bonded phase silica 5 µM particle, 250×4.6 mm) reverse phase column. HPLC buffer B (90% acetonitrile) was prepared and ribonucleosides were separated by using an isocratic gradient of 0% buffer B for 10 min, 0-10% of buffer B for 25 min, 10-100% buffer B for 10 min, 100 % buffer B for 5 min (wash), 100-0% buffer B for 2 min, and 0% buffer B for 18 min (equilibration). Uridine and 4sU standards were prepared ranging from 250-8000 ng for uridine and 3.125-100 ng for 4sU and analyzed in the same HPLC run as the samples of interest.
CRISPR/Cas9 genome editing
Ago2 and Nbr genes were disrupted using CRISPR/Cas9 as previously described (Reimão-Pinto et al., 2016) (see Figure S1P, Q and S4). In brief, Drosophila S2 cells were transfected with a mix of pAc-sgRNA-Cas9 plasmids each encoding for Cas9 and a single sgRNA targeting the second exon Ago2 or first exon of Nbr using Cellfectin II (Gibco) according to the manufacturer’s instructions. For guide RNA sequences see Supplementary Table 1. Cells were selected for transgene expression using 5 µg/ml puromycin for 9 days, followed by serial dilution under non-selecting conditions and expanded to single-cell line clones. Individual clones were tested for successful targeting by Surveyor assay, and positive selection was verified by Western blot analysis (Figure S1Q and S4B). Knockout of nbr was additionally confirmed by Northern Blot analysis (Figure S4C). The genetic deletion alleles determined by Sanger sequencing are described in Figure S1P and S4A.
Generation of overexpression clonal cell lines
To generate Ago1OE cell lines, wild-type and ago2ko S2 cells were co-transfected with 400 ng of pAFMW-Ago1 and 100 ng pHygro plasmids. To generate Nbr-wtOE or Nbr-CMOE cells in nbrko background, nbrko S2 cells were co-transfected with 400 ng of either pAFMW-Nbr-WT or pAFMW-Nbr-CM (D435A, E437A) (Han et al., 2011) and 100 ng pHygro plasmids. The generation of clonal cell lines was performed as described previously (Reimão-Pinto et al., 2016; 2015). Briefly, the respective expression plasmids were added to 50 µl of FBS-free Schneider’s Drosophila media supplemented with 3.5µl FuGENE HD Transfection Reagent (Promega). After incubating this for 5 minutes, the mix was added to 2 ml of a 106 cells/ml cell suspension and plated. The following day, 300 µg of hygromycin B was added per ml of Schneider’s Drosophila media containing 10% FBS. After two weeks of hygromycin selection, a dilution series of 20000, 2000 and 200 cells were plated and colonies emerging from single-clones were picked using a sterile tip under the microscope after another two weeks when cells were forming individual visible clonal populations.
Western blotting
S2 cells were pelleted and lysed on ice with lysis buffer (30mM HEPES KOH, pH 7.4; 100mM KOAc; 2mM MgOAc; 5mM DTT; 0.5% NP-40; 5% Glycerol). Protein lysates were loaded on 8% SDS PAGE, transferred to Immobilon-P PVDF membrane (Millipore), and probed over night at 4°C. Antibodies were used at a dilution of 1:5 for anti-Ago2 (mouse; kind gift from M. Siomi), 1:10’000 for anti-Ago1 (mouse; kind gift from M. Siomi) (Miyoshi, 2005), 1:1’000 for anti-Nibbler (mouse; produced in-house as described in (Hayashi et al., 2016)), and 1:10’000 for anti-Actin (A2066, rabbit, Sigma-Aldrich) and detected by secondary HRP-antibody-conjugates G21040 (Invitrogen; dilution 1:10,000). Images were acquired on a ChemiDoc MP Imaging System (BioRad) using ImageLab v5.1.1 (BioRad).
Northern hybridization
Northern hybridization experiments were performed as described previously (Han et al., 2011). For Northern probes see Supplementary Table 1. For quantitative determination of miR-184 and bantam, synthetic RNA-oligonucleotides were loaded at defined absolute quantities (1000, 500, 100, 50, 10, 5, 1 fmole) (Supplementary Table 1). The levels of synthetic or endogenous miRNA levels were detected using a probe against 22-mer miR-184-3p and the abundant 23-mer for bantam-3p (Supplementary Table 1). Northern blots were subsequently imaged using a Typhoon PhosphorImager (Amersham), quantified using ImageQuant TL and Excel.
Small RNA SLAMseq in Drosophila S2 cells
S2 cells were seeded at a density of 3×106 cells/ml the day before the experiment in 35 mm dishes. 4sU-metabolic labelling was performed by incubating S2 cells in Schneider’s Drosophila Medium containing 10% FBS supplemented with 500 µM 4sU (Sigma-Aldrich). During the metabolic labeling of wild-type S2 cells, 4sU containing medium was exchanged every 3h hours for the duration of the pulse. At respective time-points, cells were harvested using a cell scraper, pelleted for 5 min at 200 x g (4°C) and resuspended in TRIzol® (Ambion). RNA extraction and carboxyamidomethylation of total RNA was performed as described previously (Herzog et al., 2017). Briefly, RNA extraction was performed following manufacturers instruction, with the modification of performing precipitation and washes with 0.1 mM DTT supplemented isopropanol and ethanol. Finally, RNA was resuspended in 1 mM DTT. 20 µg of total RNA harvested from ago2ko cells, and 40 µg of total RNA from wt S2 cells was treated with 10 mM iodoacetamide under optimal reaction conditions (50 mM NaPi, pH 8; 50% DMSO; 15 min at 50°C), subsequently precipitated and used for small RNA library preparation.
Small RNA libraries
Small RNA libraries were prepared as previously described (Jayaprakash et al., 2011). In brief, total RNA was resolved on a 15% denaturing polyacrylamide gel, 18 – 30nt long RNA was excised, gel-eluted and subjected to 2S rRNA depletion as previously described (Hayashi et al., 2016). Size selected and 2S rRNA depleted RNA was subjected to 3ʹ and 5ʹ adapter ligation. Both adaptors contain 4 random nucleotides at the ligation interfaces, to reduce ligation biases. Note, that ligation steps need to proceed overnight (on ice) to prevent the underrepresentation of s4U-labeling at miRNA termini due to ligation biases. Ligation products were subjected to reverse transcription using SuperScript III (Invitrogen), according to the manufacturer’s instructions, PCR amplified and sequenced on an Illumina HiSeq 2500 on SR50 mode. Total RNA from wild-type cells was split in half after the initial size selection, one of which underwent oxidation by incubating in borate buffer (pH 8.6) containing 25mM NaIO4 at room temperature for 30 min as described previously (Seitz et al., 2008). Ago-bound small RNA libraries were generated by biochemical purification of Argonaute ribonucleoprotein complexes prior to 3ʹ and 5ʹ adapter ligation.
TT-SLAMseq
To determine the immediate transcriptional output of miRNA precursor transcripts, a modified protocol of transient transcriptome sequencing (TTseq) (Schwalb et al., 2016) was combined with SLAMseq (Herzog et al., 2017). Note, that by combining SLAMseq with TTseq, unlabeled transcripts that frequently contaminate RNA libraries generated by 4sU-capturing approaches, such as TT-seq, particularly when investigating transient transcription products in short pulse labeling setups, can be bioinformatically discarded by inspecting 4sU nucleotide conversion introduced by SLAMseq.
For short-pulse metabolic labeling, 3.6 × 107 ago2ko S2 cells were subjected to 4sU-labeling for 5 min using 1 mM 4sU. The labeling was performed in Falcon tubes to allow immediate cell pelleting and lysis after the short labeling period. Cell pelleting, lysis and RNA isolation was performed as described above (Small RNA SLAMseq in Drosophila S2 cells). 50 µg total RNA was fragmented in 20 µl 1x RNA Fragmentation buffer for 1 min at 94°C (NEBNext® Magnesium RNA Fragmentation Module), resulting in ~1 kb fragments. The reaction was immediately placed on ice and stopped by adding 2 µl Fragmentation stop solution. The Fragmentation buffer was removed by ethanol precipitation using solutions supplemented with 0.1 mM DTT. Immediately after resuspending the RNA pellet in 350 µl RNase free water without DTT, the biotinylation reaction was performed essentially as described in (Duffy et al., 2015): MTSEA biotin-XX (Biotium) was freshly prepared in DMF to achieve a stock solution with a concentration of 50 µg/ml. 5 µg of MTSEA biotin-XX, 20 mM HEPES (pH 7.5) and 1 mM EDTA (pH 8) was added to the fragmented RNA to reach a final volume of 500 µl. The reaction was incubated for 30 min at room temperature on a rotating wheel and protected from light. The unbound biotin fraction was removed by Acid-Phenol:Chloroform extraction (125:24:1, Ambion), followed by isopropanol precipitation without the addition of DTT. After resuspending the pellet in 100 µl RNase free water without DTT, the RNA sample was denatured at 65°C for 10 min and rapidly cooled on ice for 5 min. For enrichment of 4sU labeled transcript fragments, RNA was mixed with 100 µl µMACS streptavidin beads (Miltenyi) and incubated for 15 min at room temperature on a rotating wheel and protected from light. In the meantime, the µMACS column (Milenyi) was fixed to the magnetic stand and equilibrated once with 100 µl nucleic acid equilibration buffer (Miltenyi) and twice with 100 µl 1x biotinylation buffer (20 mM HEPES and 1 mM EDTA). The RNA bound to the beads was transferred to the column and thereby magnetically fixed. After three washing steps with 1 ml washing buffer (100 mM Tris-HCl pH 7.5, 10 mM EDTA pH 8, 1 M NaCl, 0.1% Tween20), the captured RNA was eluted using 100 µl 100 mM DTT. The elution step was repeated after 5 min and the eluates were pooled. The collected RNA was purified and concentrated via RNeasy MinElute Spin Column (Qiagen) according to manufacturer’s instruction and eluted with 14 µl 1 mM DTT.
The whole pulldown sample was subjected to iodoacetamide treatment as described previously (Herzog et al., 2017), followed by TURBO DNase treatment (2 units per sample, Invitrogen) to remove potential DNA contaminants. The RNA-sequencing library was prepared using NEBNext® Ultra™ II Directional RNA Library Prep Kit for Illumina® (NEB) according to manufacturer’s protocol but the fragmentation time prior to first strand synthesis was shortened to 8 min due to the pre-fragmentation of RNA before biotinylation. The libraries were sequenced on an Illumina HiSeq 2500 on SR100 mode.
General bioinformatic procedure
Barcoded libraries were demultiplexed using Picard Tools BamIndexDecoder v1.13 and converted to fastq using Picard Tools SamToFastq v1.82. Sequencing adapters were trimmed with cutadapt v1.16. For small RNA sequencing data, seqtk (v1.2-r101) was used to trim the 4 random nucleotides from both ends. The libraries were then mapped using bowtie v1.2.2 (Langmead et al., 2009), allowing for 3 mismatches to dm6 (r6.12) and, separately, all Drosophila pre-miRNA hairpins described in mirbase 21 extended downstream by 20 basepairs (Kozomara and Griffiths-Jones, 2014).
All reads mapping to 5p and 3p arms of a miRNA hairpin locus were reduced to only retain those starting at the most frequent starting positions for both arms. For the example locus, identically mapping reads were then collapsed while retaining read counts and only reads that are represented with a frequency of at least 1% of the most abundant 5p/3p read are displayed.
A nextflow analysis pipeline (Di Tommaso et al., 2017) was established to map and evaluate miRNAs and their mutations (available at https://github.com/AmeresLab/smRNAseq): It first identifies miRNAs mapping to the most common 5ʹ isoforms for both 5p and 3p arms of the miRNA hairpin. Then mutation rates for each possible mutation are extracted using Rsamtools (1.28.0), restricting the analysis to the first 18 nucleotides of each locus. Mutations are then normalized to the frequency of each nucleobase within this region. Finally, any further analysis is restricted to reads mapping with a frequency above 100 ppm normalized to all small RNA mapping reads. T>C containing reads are only counted from alignments to miRNA hairpins, if the mutation is within the first 18nt and passes a base calling score threshold of 27. Resulting read counts are then normalized to the numbers of T’s within the first 18nt.
For TT-SLAMseq data, the adapter-clipped reads were trimmed by 10nt from the 5´ end and reverse complemented using seqtk. Pre-processed reads were further processed using Digital Unmasking of Nucleotide conversion-containing k-mers (DUNK), SLAM-DUNK v0.3.4, a T>C aware alignment software package based on NextGenMap (Sedlazeck et al., 2013) developed to recover T>C conversions from SLAMseq data sets (Neumann et al., 2019). Briefly, reads were aligned to the full reference genome (dm6) and only uniquely mapping reads were retained. SNPs exceeding a coverage cutoff of 10x and a variant fraction cutoff of 0.2 were called using VarScan2.4.1 (Koboldt et al., 2012) using default parameters. Non-SNP overlapping T>C conversions with a base quality of Phred score >26 were identified. T>C containing reads and total reads aligning within the custom defined pri-miRNA producing loci were reported. The pri-miRNA producing loci (bed files provided in GEO datasets) were defined by overlapping the miRNA coordinates with annotated coding and non-coding transcripts. These transcription units were manually curated by inspecting published GRO-seq and Pol II-ChIP-seq data sets (Core et al., 2012).
Statistical analyses were performed in R v3.4.1, Prism v7.0 (GraphPad) or Excel v15.22 (Microsoft).
Small RNAseq data analysis
Demultiplexed libraries are aligned to the Drosophila genome in a step wise manner, to Drosophila viral genomes, the rRNA precursor, the mitochondrial genome, and then the full genome, all using bowtie v1.2.2. Unique mapping is used in the first two steps and up to 3 mismatches are allowed for the latter two steps. Alignment to the Drosophila genome is done in two passes, once only allowing unique mappers, and a second time using the remaining unmapped reads, allowing for multiple mapping locations. The final combined alignment was evaluated in a hierarchical manner, recovering the reads of non-coding RNAs (tRNA, mitochondrial, snRNAs and snoRNAs) and coding RNAs (structured RNAs, cisNATs, miRNA, genic reads, TEs and regions without annotation). Structured RNAs were previously defined (Okamura et al., 2008), whereas cisNAT regions were determined manually as any coordinates with overlapping 3´ UTRs greater than 21 nucleotides in length. Reads mapping to coding RNAs were then summarized as small RNA mapping reads and used as normalization factor for microRNA mapping reads in subsequent analyses. For global analyses of all miRNAs, the reads of two replicates in ago2ko cells were merged for each time-point prior to alignment. Individual examples miRNAs were analyzed and displayed independently for each replicate.
Transcriptional Output of miRNA precursor transcripts
To assess the transcriptional output of miRNA precursor transcripts, T>C containing reads of each pri-miRNA producing locus from the TT-SLAMseq samples were normalized to sequencing depth, length of the genomic interval and T-content (by dividing by the relative coverage across T positions within the genomic interval).
MiRNA Biogenesis rate and half-life calculation
Conversion rates were determined by investigating the occurrence of any given mutation within nucleotide 1-18 of each miRNA 5´ isoform, normalizing to genome matching instances at this position, and averaging to the total number of reference nucleotides from position 1-18 for each 5´ isoform. To analyze production of miR and miR* sequences, only reads containing high base calling quality (Phred score >27) T>C conversions were considered. The total counts were corrected by subtracting the baseline, normalized to labelling efficiency (see below) and divided by U-content in positions 1-18. Biogenesis rates (kbiogenesis) were determined as the slope of a linear fit of T>C containing reads at 5, 15, and 30 minutes. The transcriptional output-normalized miRNA biogenesis rates (i.e. miRNA processing rates) was determined by dividing the biogenesis rate of each miRNA by the transcriptional output of its precursor transcript (see above). Half-lives were calculated by applying exponential fits (one and two-phase associations in GraphPad Prism v7.0) to baseline subtracted T>C conversion rates for each miR or miR*. One-phase associations follow an exponential equation similar to first-order kinetics (1), and two-phase associations are described by a composite of two exponential equations (2).
| (1) |
| (2) |
Here, y is given by the background subtracted T>C conversion rate, and t is the time of each time-point; P is an asymptotic plateau for the equation; k, kFast and kSlow are the rates, describing either total stability, the Fast or Slow component of a given miR/miR*; fFast and fSlow describe the proportion of species following the Fast/Slow kinetics. The half-life is subsequently calculated as defined for first-order kinetic reactions (3):
| (3) |
Curve fitting was performed according to the integrated rate law for a first-order reaction or second-order-reaction where applicable in R v3.4.1.
Baseline subtraction of T>C conversion containing reads
The background T>C conversion rate was estimated from non 4sU-labeled samples and subtracted from the T>C conversion rates in each 4sU labeling time-point in an isoform-specific manner. This subtraction yields the background subtracted T>C conversion containing reads for each 18, 19, 20, etc.-mers of each miRNA.
Labelling efficiency normalization
Labelling efficiency is described as the average T>C conversion rate at saturation of metabolic 4sU labeling (24h). This is calculated from the sequencing data by fitting one-phase associations (described above) and averaging the plateau of the 10 fastest miR species. Dividing T>C conversion containing reads by this labelling efficiency yields the labelling efficiency normalization.
Determination of length characteristics
The sum of baseline subtracted, labelling efficiency normalized reads for each length isoform was used to calculate the weighted arithmetic mean length of both steady state levels and T>C conversion containing reads. From the length distributions, 3´ heterogeneity is calculated as previously described (Seitz et al., 2008). In brief, the abundance of each length isoform was subtracted from the abundance of the maximum length isoform and used to calculate the weighted arithmetic mean. To determine the 3´ heterogeneity of early time-points (≤ 1h), the reads from the 5 min, 10 min, 15 min, 30min and 60 min were merged prior to the analysis of 3′ end heterogeneity.
Precursor-miRNA analysis
Precursor-miRNA abundance (sum of prefix- and genome-matching reads) and uridylation status was determined for pre-miRNA-mapping reads of 40-100 nt size selected total RNA libraries generated from wild-type S2 cells as previously described (Reimão-Pinto et al., 2015), but considering only mono or homopolymeric uridine-additions.
Minimal Free Energy calculations
The minimal free energy between each miR:miR* pair was derived for the structure of nt 1, 2, and 3 of a miR with the corresponding target region on the miR* strand, and vice versa. The binding status of each nucleotide was assessed based on their duplex structures and using RNAeval (v2.4.11) allowed calculation of the minimal free energy in these states.
Definition of Nbr substrates
Nbr substrates were defined by a significant increase in weighted average length (two-tailed Student’s t-test, p<0.01) in small RNA libraries generated from S2 cells, in which Nbr was functionally depleted (nbrko or nbrko;FM-Nbr-CMOE), compared to cells containing functional Nbr protein (wt or nbrko;FM-Nbr-WTOE).
Small RNA Slamseq Method Validation
To validate small RNA SLAMseq, we subjected Drosophila S2 cells to 4sU treatment for 24 h, followed by total RNA extraction and thiol-linked alkylation for the metabolic sequencing of small RNAs (SLAMseq; Figure S1A) (Herzog et al., 2017). In resulting cDNA libraries, we observed the accumulation of T>C conversions, a feature that was not frequently observed in libraries prepared in the same way from unlabeled cells (exemplified for the miR-184 locus; Figure S1B). We systematically inspected the frequency and specificity of this feature by aligning reads to annotated microRNA loci in the Drosophila genome and compared T>C conversions to any other given nucleotide conversion (Figure S1C): In the absence of 4sU metabolic labeling, we observed an overall median nucleotide conversion rate of less than 0.1%, a rate that is consistent with Illumina-reported sequencing error rates. After 4sU labeling, we observed a statistically significant (p<10-4, Mann-Whitney test), 75-fold increase in T>C conversions, while all other conversions remained below the expected rate of sequencing errors (Figure S1C). More specifically, we measured a median 4sU-incorporation of 2.24% (corresponding to one 4sU incorporation in every 45 uridines), consistent with incorporation rates determined by HPLC measurements of total RNA-derived nucleosides (Figure S1D). Metabolic labeling under unperturbed cellular conditions (final concentration of 500 µM 4sU) therefore caused the vast majority (>95%) of 4sU-labeled small RNAs to exhibit at most one 4sU incorporation event (Figure S1E). Note, that single-site incorporations are readily detected by SLAMseq but insufficient to support quantitative biochemical recovery by reversible affinity purification approaches even when applying advanced reversible coupling chemistry (Duffy et al., 2015). As expected, treatment of unlabeled total RNA with iodoacetamide did not impact abundance or nucleotide conversion rates in small RNA libraries (Figure S1F to H).
The quantitative recovery of 4sU incorporations at single nucleotide resolution by SLAMseq enabled us to systematically evaluate any adverse impact of metabolic labeling on microRNA processing and loading: To this end, we first determined the relative abundance of small RNAs in SLAMseq libraries generated from S2 cells before and 24 h after 4sU metabolic RNA labeling, which revealed a statistically significant, strong correlation (Pearson’s correlation coefficient rP=0.99, p<10-4, Figure S1I), regardless of miRNA U-content (Figure S1J). In agreement with the assumption that 4sU labeling reached steady-state after 24h, we observed no global or specific change in miRNA abundance for both total and 4sU-labeled small RNAs (Spearman correlation rS=0.98, p<10-4, Figure S1K and L). Note, that normalization to U-content is required to prevent systematic errors in the quantification of labeled small RNAs after 4sU-metabolic labeling (Figure S1M and N). In a more rigorous test for position-dependent effects on miRNA processing or loading, we determined the over- or underrepresentation of 4sU-labeling-derived T>C conversions at individual positions of any given small RNA that is either derived from the 5p- or 3p arm of a miRNA precursor, or constitutes a miR or miR* strand, as defined by selective Ago-loading. We examined the 71 abundantly expressed (>100 ppm) miRNAs (corresponding to 34 5p- and 37 3p-miRNAs, or 45 miR and 26 miR*) but observed no significant alteration in relative T>C conversion rates at any given position (Figure S1O). We concluded that 4sU-metabolic labeling does not affect miRNA biogenesis, loading, or steady-state abundance, thus enabling the examination of intracellular miRNA kinetics in unperturbed conditions.
Quantification and Statistical Analysis
All software used in this study is listed in the Key Resources Table and referred to in the Method Details section. The statistical details of experiments including statistical tests used, number of replicates and the value of n are denoted in the relevant figure legends and in the main text. All statistical tests, Spearman rank and Pearson’s r correlations were analyzed in Prism v7.0 (GraphPad) or Excel (Microsoft). A p Value of < 0.05 was considered to be statistically significant. Asterisks define the degree of significance and is presented as * p < 0.05, ** p < 0.01, *** p < 0.001, or **** p < 0.0001. Linear regression and fitting of exponential curves was done in R v3.4.1 and Prism v7.0, using the according functions as detailed in Methods section and in relevant figure legends.
Data and Software Availability
The Nextflow analysis pipeline that was established to map small RNA reads and evaluate their nucleotide conversion rates is available at https://github.com/AmeresLab/smRNAseq. The pipeline also includes the R script used to calculate fits (for half-lives and biogenesis rates), background subtraction, labelling efficiency normalization and 3´ heterogeneity. Raw sequence reads, processed data and bed files of the annotated pri-miRNA loci have been deposited at GEO under the accession number GSE129600.
Supplementary Material
Acknowledgements
We thank M. Siomi for kindly sharing Ago1 and Ago2 antibodies; Gabriela Krssakova and Karl Mechtler for HPLC analysis; and all members of the Ameres laboratory for technical support and helpful discussions. Small RNA sequencing was performed at the VBCF NGS Unit (https://www.viennabiocenter.org/facilities). Research in the Ameres lab is supported by the Austrian Academy of Sciences, the Austrian Science Fund FWF (Y-733-B22 START, W127-B09, and F4322) and the European Research Council (ERC-StG-338252 miRLIFE and ERC-PoC-825710 SLAMseq).
Footnotes
Author contributions
B.R., V.A.H., and S.L.A. conceived the research, designed and performed the experiments, and analyzed the data. R.A.M. and I.S. generated and validated genome-engineered S2 cell lines. N.F. performed TT-SLAMseq experiments. I.S. assisted with small RNA cloning. V.A.H. and S.L.A. wrote the paper with input from all authors.
Declaration of Interests
VAH, BR and SLA declare competing financial interest. A patent application related to this work has been filed.
References
- Alberti C, Manzenreither RA, Sowemimo I, Burkard TR, Wang J, Mahofsky K, Ameres SL, Cochella L. Cell-type specific sequencing of microRNAs from complex animal tissues. Nat Methods. 2018;15:283–289. doi: 10.1038/nmeth.4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, Zamore PD. Target RNA-Directed Trimming and Tailing of Small Silencing RNAs. Science. 2010;328:1534–1539. doi: 10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol. 2013;14:475–488. doi: 10.1038/nrm3611. [DOI] [PubMed] [Google Scholar]
- Andrus A, Kuimelis RG. Analysis and purification of synthetic nucleic acids using HPLC. Curr Protoc Nucleic Acid Chem. 2001;Chapter 10:Unit 10.5–10.5.13. doi: 10.1002/0471142700.nc1005s01. [DOI] [PubMed] [Google Scholar]
- Bartel DP. Metazoan MicroRNAs. Cell. 2018;173:20–51. doi: 10.1016/j.cell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett AR, Tibbit C, Ponting CP, Liu JL. Mutagenesis and homologous recombination in Drosophila cell lines using CRISPR/Cas9. Biology Open. 2014;3:42–49. doi: 10.1242/bio.20137120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitetti A, Mallory AC, Golini E, Carrieri C, Carreño Gutiérrez H, Perlas E, Pérez-Rico YA, Tocchini-Valentini GP, Enright AJ, Norton WHJ, Mandillo S, et al. MicroRNA degradation by a conserved target RNA regulates animal behavior. Nature Structural & Molecular Biology. 2018;25:244–251. doi: 10.1038/s41594-018-0032-x. [DOI] [PubMed] [Google Scholar]
- Bortolamiol-Becet D, Hu F, Jee D, Wen J, Okamura K, Lin C-J, Ameres SL, Lai EC. Selective Suppression of the Splicing-Mediated MicroRNA Pathway by the Terminal Uridyltransferase Tailor. Mol Cell. 2015;59:217–228. doi: 10.1016/j.molcel.2015.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bühler M, Spies N, Bartel DP, Moazed D. TRAMP-mediated RNA surveillance prevents spurious entry of RNAs into the Schizosaccharomyces pombe siRNA pathway. Nature Structural & Molecular Biology. 2008;15:1015–1023. doi: 10.1038/nsmb.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalla D, Yario T, Steitz JA. Down-Regulation of a Host MicroRNA by a Herpesvirus saimiri Noncoding RNA. Science. 2010;328:1563–1566. doi: 10.1126/science.1187197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary MD, Meiering CD, Jan E, Guymon R, Boothroyd JC. Biosynthetic labeling of RNA with uracil phosphoribosyltransferase allows cell-specific microarray analysis of mRNA synthesis and decay. Nat Biotechnol. 2005;23:232–237. doi: 10.1038/nbt1061. [DOI] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Gilchrist DA, Fargo DC, Kwak H, Adelman K, Lis JT. Defining the Status of RNA Polymerase at Promoters. Cell Rep. 2012;2:1025–1035. doi: 10.1016/j.celrep.2012.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Hannon GJ. Small RNA sorting: matchmaking for Argonautes. Nat Rev Genet. 2010;12:19–31. doi: 10.1038/nrg2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Zhou R, Erlich Y, Brennecke J, Binari R, Villalta C, Gordon A, Perrimon N, Hannon GJ. Hierarchical Rules for Argonaute Loading in Drosophila. Mol Cell. 2009;36:445–456. doi: 10.1016/j.molcel.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien B, Baumberger N, Schepetilnikov M, Viotti C, De Cillia J, Ziegler-Graff V, Isono E, Schumacher K, Genschik P. Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Proc Natl Acad Sci USA. 2012;109:15942–15946. doi: 10.1073/pnas.1209487109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Tommaso P, Chatzou M, Floden EW, Barja PP, Palumbo E, Notredame C. Nextflow enables reproducible computational workflows. Nat Biotechnol. 2017;35:316–319. doi: 10.1038/nbt.3820. [DOI] [PubMed] [Google Scholar]
- Diederichs S, Jung S, Rothenberg SM, Smolen GA, Mlody BG, Haber DA. Coexpression of Argonaute-2 enhances RNA interference toward perfect match binding sites. Proc Natl Acad Sci USA. 2008;105:9284–9289. doi: 10.1073/pnas.0800803105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy EE, Rutenberg-Schoenberg M, Stark CD, Kitchen RR, Gerstein MB, Simon MD. Tracking Distinct RNA Populations Using Efficient and Reversible Covalent Chemistry. Mol Cell. 2015;59:858–866. doi: 10.1016/j.molcel.2015.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förstemann K, Horwich MD, Wee LM, Tomari Y, Zamore PD. Drosophila microRNAs Are Sorted into Functionally Distinct Argonaute Complexes after Production by Dicer-1. Cell. 2007;130:287–297. doi: 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Xu J, Seitz H, Weng Z, Zamore PD. Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. RNA. 2009;16:43–56. doi: 10.1261/rna.1972910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- Han BW, Hung J-H, Weng Z, Zamore PD, Ameres SL. The 3′-to-5′ Exoribonuclease Nibbler Shapes the 3′ Ends of MicroRNAs Bound to Drosophila Argonaute1. Current Biology. 2011;21:1878–1887. doi: 10.1016/j.cub.2011.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi R, Schnabl J, Handler D, Mohn F, Ameres SL, Brennecke J. Genetic and mechanistic diversity of piRNA 3'-end formation. Nature. 2016;539:588–592. doi: 10.1038/nature20162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog VA, Reichholf B, Neumann T, Rescheneder P, Bhat P, Burkard TR, Wlotzka W, von Haeseler A, Zuber J, Ameres SL. Thiol-linked alkylation of RNA to assess expression dynamics. Nat Methods. 2017;14:1198–1204. doi: 10.1038/nmeth.4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper AK. Transfer RNA post-transcriptional processing, turnover, and subcellular dynamics in the yeast Saccharomyces cerevisiae. Genetics. 2013;194:43–67. doi: 10.1534/genetics.112.147470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, Zamore PD. The Drosophila RNA Methyltransferase, DmHen1, Modifies Germline piRNAs and Single-Stranded siRNAs in RISC. Current Biology. 2007;17:1265–1272. doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- Houseley J, Tollervey D. The Many Pathways of RNA Degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- Jayaprakash AD, Jabado O, Brown BD, Sachidanandam R. Identification and remediation of biases in the activity of RNA ligases in small-RNA deep sequencing. Nucleic Acids Research. 2011;39:e141–e141. doi: 10.1093/nar/gkr693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata T, Tomari Y. Making RISC. Trends Biochem Sci. 2010;35:368–376. doi: 10.1016/j.tibs.2010.03.009. [DOI] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Kleaveland B, Shi CY, Stefano J, Bartel DP. A Network of Noncoding Regulatory RNAs Acts in the Mammalian Brain. Cell. 2018;174:350–362.e17. doi: 10.1016/j.cell.2018.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Shoji K, Kiyokawa K, Negishi L, Tomari Y. Iruka Eliminates Dysfunctional Argonaute by Selective Ubiquitination of Its Empty State. Mol Cell. 2019;73:119–129.e5. doi: 10.1016/j.molcel.2018.10.033. [DOI] [PubMed] [Google Scholar]
- Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Research. 2014;42:D68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Research. 2010;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, Busskamp V, Markiewicz I, Stadler MB, Ribi S, Richter J, Duebel J, Bicker S, Fehling HJ, Schübeler D, Oertner TG, et al. Characterizing Light-Regulated Retinal MicroRNAs Reveals Rapid Turnover as a Common Property of Neuronal MicroRNAs. Cell. 2010;141:618–631. doi: 10.1016/j.cell.2010.03.039. [DOI] [PubMed] [Google Scholar]
- la Mata de M, Gaidatzis D, Vitanescu M, Stadler MB, Wentzel C, Scheiffele P, Filipowicz W, Grosshans H. Potent degradation of neuronal miRNAs induced by highly complementary targets. EMBO Rep. 2015;16:500–511. doi: 10.15252/embr.201540078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Kim B, Kim VN. Emerging Roles of RNA Modification: m 6 A and U-Tail. Cell. 2014;158:980–987. doi: 10.1016/j.cell.2014.08.005. [DOI] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. The EMBO Journal. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Abe M, Sabin LR, Hendriks GJ, Naqvi AS, Yu Z, Cherry S, Bonini NM. The Exoribonuclease Nibbler Controls 3′ End Processing of MicroRNAs in Drosophila. Current Biology. 2011;21:1888–1893. doi: 10.1016/j.cub.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R, Bernhart SH, Höner Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL. ViennaRNA Package 2.0. Algorithms Mol Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcinowski L, Tanguy M, Krmpotic A, Rädle B, Lisnić VJ, Tuddenham L, Chane-Woon-Ming B, Ruzsics Z, Erhard F, Benkartek C, Babic M, et al. Degradation of Cellular miR-27 by a Novel, Highly Abundant Viral Transcript Is Important for Efficient Virus Replication In Vivo. PLoS Pathog. 2012;8:e1002510. doi: 10.1371/journal.ppat.1002510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet j. 2011;17:10. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- Martinez NJ, Gregory RI. Argonaute2 expression is post-transcriptionally coupled to microRNA abundance. RNA. 2013;19:605–612. doi: 10.1261/rna.036434.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzi MJ, Ghini F, Cerruti B, de Pretis S, Bonetti P, Giacomelli C, Gorski MM, Kress T, Pelizzola M, Muller H, Amati B, et al. Degradation dynamics of microRNAs revealed by a novel pulse-chase approach. Genome Res. 2016;26:554–565. doi: 10.1101/gr.198788.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michlewski G, Caceres JF. Post-transcriptional control of miRNA biogenesis. RNA. 2019;25:1–16. doi: 10.1261/rna.068692.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C, Schwalb B, Maier K, Schulz D, Dumcke S, Zacher B, Mayer A, Sydow J, Marcinowski L, Dolken L, Martin DE, et al. Dynamic transcriptome analysis measures rates of mRNA synthesis and decay in yeast. Mol Syst Biol. 2011;7:458–458. doi: 10.1038/msb.2010.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev. 2005;19:2837–2848. doi: 10.1101/gad.1370605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann T, Herzog VA, Muhar M, von Haeseler A, Zuber J, Ameres SL, Rescheneder P. Quantification of experimentally induced nucleotide conversions in high-throughput sequencing datasets. BMC Bioinformatics. 2019;20:258. doi: 10.1186/s12859-019-2849-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Chung W-J, Ruby JG, Guo H, Bartel DP, Lai EC. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008;453:803–806. doi: 10.1038/nature07015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The Mirtron Pathway Generates microRNA-Class Regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirouz M, Du P, Munafò M, Gregory RI. Dis3l2-Mediated Decay Is a Quality Control Pathway for Noncoding RNAs. Cell Rep. 2016;16:1861–1873. doi: 10.1016/j.celrep.2016.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabani M, Raychowdhury R, Jovanovic M, Rooney M, Stumpo DJ, Pauli A, Hacohen N, Schier AF, Blackshear PJ, Friedman N, Amit I, et al. High-Resolution Sequencing and Modeling Identifies Distinct Dynamic RNA Regulatory Strategies. Cell. 2014;159:1698–1710. doi: 10.1016/j.cell.2014.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimão-Pinto MM, Ignatova V, Burkard TR, Hung J-H, Manzenreither RA, Sowemimo I, Herzog VA, Reichholf B, Fariña-Lopez S, Ameres SL. Uridylation of RNA Hairpins by Tailor Confines the Emergence of MicroRNAs in Drosophila. Mol Cell. 2015;59:203–216. doi: 10.1016/j.molcel.2015.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimão-Pinto MM, Manzenreither RA, Burkard TR, Sledz P, Jinek M, Mechtler K, Ameres SL. Molecular basis for cytoplasmic RNA surveillance by uridylation‐triggered decay in Drosophila. The EMBO Journal. 2016;35:2417–2434. doi: 10.15252/embj.201695164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riml C, Amort T, Rieder D, Gasser C, Lusser A, Micura R. Osmium-Mediated Transformation of 4-Thiouridine to Cytidine as Key To Study RNA Dynamics by Sequencing. Angew Chem Int Ed. 2017;56:13479–13483. doi: 10.1002/anie.201707465. [DOI] [PubMed] [Google Scholar]
- Rissland OS, Hong SJ, Bartel DP. MicroRNA Destabilization Enables Dynamic Regulation of the miR-16 Family in Response to Cell-Cycle Changes. Mol Cell. 2011;43:993–1004. doi: 10.1016/j.molcel.2011.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon WE, Jolly SM, Moore MJ, Zamore PD, Serebrov V. Single-Molecule Imaging Reveals that Argonaute Reshapes the Binding Properties of Its Nucleic Acid Guides. Cell. 2015;162:84–95. doi: 10.1016/j.cell.2015.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki HM, Tomari Y. The true core of RNA silencing revealed. Nature Structural & Molecular Biology. 2012;19:657–660. doi: 10.1038/nsmb.2302. [DOI] [PubMed] [Google Scholar]
- Schofield JA, Duffy EE, Kiefer L, Sullivan MC, Simon MD. TimeLapse-seq: adding a temporal dimension to RNA sequencing through nucleoside recoding. Nat Methods. 2018;15:221–225. doi: 10.1038/nmeth.4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwalb B, Michel M, Zacher B, Frühauf K, Demel C, Tresch A, Gagneur J, Cramer P. TT-seq maps the human transient transcriptome. Science. 2016;352:1225–1228. doi: 10.1126/science.aad9841. [DOI] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the Assembly of the RNAi Enzyme Complex. Cell. 2003;115:199–208. doi: 10.1016/S0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Sedlazeck FJ, Rescheneder P, Haeseler von A. NextGenMap: fast and accurate read mapping in highly polymorphic genomes. Bioinformatics. 2013;29:2790–2791. doi: 10.1093/bioinformatics/btt468. [DOI] [PubMed] [Google Scholar]
- Seitz H, Ghildiyal M, Zamore PD. Argonaute Loading Improves the 5′ Precision of Both MicroRNAs and Their miRNA* Strands in Flies. Current Biology. 2008;18:147–151. doi: 10.1016/j.cub.2007.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert P, Yang JS, Azzam G, Liu JL, Lai EC. Homeostatic control of Argonaute stability by microRNA availability. Nature Structural & Molecular Biology. 2013;20:789–795. doi: 10.1038/nsmb.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer J, Hafner M, Landthaler M, Ascano M, Farazi T, Wardle G, Nusbaum J, Khorshid M, Burger L, Zavolan M, Tuschl T. Laboratory Methods in Enzymology: Protein Part B, Methods in Enzymology. Elsevier; 2014. PAR-CLIP (Photoactivatable Ribonucleoside-Enhanced Crosslinking and Immunoprecipitation) pp. 113–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari Y, Du T, Zamore PD. Sorting of Drosophila Small Silencing RNAs. Cell. 2007;130:299–308. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treiber T, Treiber N, Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat Rev Mol Cell Biol. 2019;20:5–20. doi: 10.1038/s41580-018-0059-1. [DOI] [PubMed] [Google Scholar]
- Tsutsumi A, Kawamata T, Izumi N, Seitz H, Tomari Y. Recognition of the pre-miRNA structure by Drosophila Dicer-1. Nature Structural & Molecular Biology. 2011;18:1153–1158. doi: 10.1038/nsmb.2125. [DOI] [PubMed] [Google Scholar]
- Ustianenko D, Pasulka J, Feketova Z, Bednarik L, Zigackova D, Fortova A, Zavolan M, Vanacova S. TUT‐DIS3L2 is a mammalian surveillance pathway for aberrant structured non‐coding RNAs. The EMBO Journal. 2016;35:2179–2191. doi: 10.15252/embj.201694857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of Stress-Dependent Cardiac Growth and Gene Expression by a MicroRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- Wee LM, Flores-Jasso CF, Salomon WE, Zamore PD. Argonaute Divides Its RNA Guide into Domains with Distinct Functions and RNA-Binding Properties. Cell. 2012;151:1055–1067. doi: 10.1016/j.cell.2012.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Ameres SL, Friedline R, Hung JH, Zhang Y, Xie Q, Zhong L, Su Q, He R, Li M, Li H, et al. Long-term, efficient inhibition of microRNA function in mice using rAAV vectors. Nat Methods. 2012;9:403–409. doi: 10.1038/nmeth.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Łabno A, Warkocki Z, Kuliński T, Krawczyk PS, Bijata K, Tomecki R, Dziembowski A. Perlman syndrome nuclease DIS3L2 controls cytoplasmic non-coding RNAs and provides surveillance pathway for maturing snRNAs. Nucleic Acids Research. 2016 doi: 10.1093/nar/gkw649. 2010, gkw649. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Nextflow analysis pipeline that was established to map small RNA reads and evaluate their nucleotide conversion rates is available at https://github.com/AmeresLab/smRNAseq. The pipeline also includes the R script used to calculate fits (for half-lives and biogenesis rates), background subtraction, labelling efficiency normalization and 3´ heterogeneity. Raw sequence reads, processed data and bed files of the annotated pri-miRNA loci have been deposited at GEO under the accession number GSE129600.