Abstract
Understanding transcriptional changes engaged in stress resilience may reveal novel antidepressant targets. Here, we use gene co-expression analysis of RNA-sequencing data from brains of resilient mice to identify a gene network that is unique to resilience. Zfp189, which encodes a previously unstudied zinc finger protein, is the highest-ranked key driver gene in the network, and overexpression of Zfp189 in prefrontal cortical (PFC) neurons preferentially activates this network and promotes behavioral resilience. The transcription factor CREB is a predicted upstream regulator of this network and binds to the Zfp189 promoter. To probe CREB-Zfp189 interactions, we employ CRISPR-mediated locus-specific transcriptional reprogramming to direct CREB or G9a (a repressive histone methyltransferase) to the Zfp189 promoter in PFC neurons. Induction of Zfp189 with site-specific CREB is pro-resilient, whereas suppressing Zfp189 expression with G9a increases susceptibility. These findings reveal an essential role for Zfp189 and CREB-Zfp189 interactions in mediating a central transcriptional network of resilience.
Introduction
Stressful life events contribute to the risk for developing Major Depressive Disorder (MDD). However, most stress-exposed individuals do not develop MDD. Understanding the biological basis of such stress resilience may illuminate causal mechanisms underlying MDD and reveal novel therapeutic targets for this disorder.
The chronic social defeat stress (CSDS) paradigm in mice is a widely used and reliable model in which to explore the biological basis of stress resilience wherein a portion of stressed animals display behavior that mimics characteristics of MDD (termed ‘susceptible’), while the remainder do not (termed ‘resilient’)1,2. CSDS thus recapitulates the divergence in stress responses observed in humans. Importantly, resilience is not simply the absence of susceptibility, but rather an active homeostatic response to stress involving broad transcriptional changes across brain regions. In fact, far more early transcriptional changes are observed in stress-resilient than in stress-susceptible mice2,3. However, the relationship among different genes that have been found to be altered in the resilient brain, as well as their mechanistic regulation, have not been determined.
By clustering genes into units (modules) based on coordinated transcriptional regulation, network-based analytical methods such as weighted gene co-expression network analysis (WGCNA) provide an alternative approach to standard differential expression analysis of large transcriptional datasets. WGCNA has been used to define gene networks from RNA-sequencing (RNA-seq) of human post-mortem brain in complex disorders such as schizophrenia4, Parkinson’s disease5, Alzheimer’s disease6, autism7 and MDD8,9. However, because these studies involve in silico analysis of human brain RNA-seq data, the causality of the inferred relationships cannot be determined through in vivo experiments. Recent studies in mice have circumvented this problem by identifying stress-susceptibility networks dependent upon the expression of key module hub genes, which, when manipulated in vivo, affect both module gene expression and overall stress susceptibility3,8. However, the transcriptional organization of the quantitatively larger and translationally relevant resilient response remains entirely unexplored. Moreover, there is currently no understanding of how regulatory factors act upon central driver genes to activate key transcriptional networks. This knowledge may yield novel targets for more effective MDD therapeutics.
Here, we find that Zfp189, a gene that encodes a putative zinc-finger transcription factor not previously implicated in stress, controls a transcriptionally-active network unique to the resilient phenotype. We show that manipulating Zfp189 in prefrontal cortex (PFC) preferentially affects expression of this network as well as resilience behavior. We also employ CRISPR-dCas9 to illustrate the regulation of this network, wherein CREB, a well-studied transcription factor, binds to the Zfp189 locus to induce Zfp189, which triggers downstream gene network activity and increases resilience. Taken together, these data provide a functional characterization of the transcriptional changes involved in resilience, identify Zfp189 as an important molecular regulator of resilience, and demonstrate the higher-order mechanism by which upstream regulators interact with in-network key-driver genes to regulate large transcriptional networks that direct complex behavior.
Results
Zfp189 regulates a resilient-specific transcriptional network
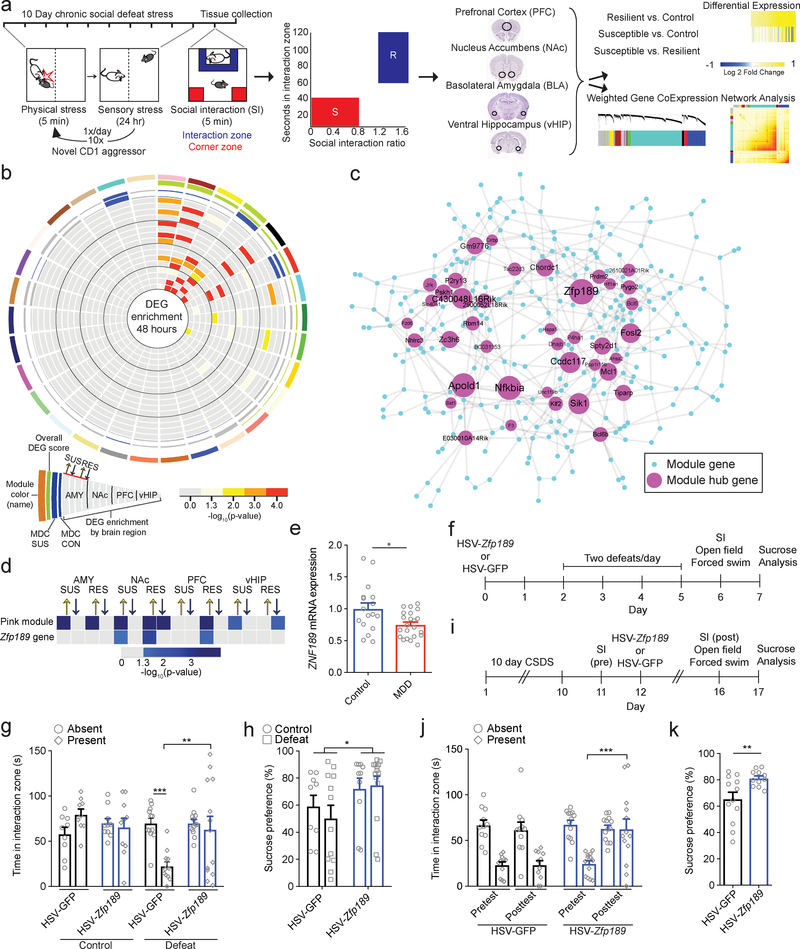
To identify transcriptional changes associated with stress resilience, we integrated WGCNA and differential expression data from our recently published3 RNA-seq dataset of mice following CSDS (Fig. 1a). WGCNA of the resilient phenotype, which had not been previously explored, revealed 30 modules present across four brain regions: PFC, ventral hippocampus (vHIP), nucleus accumbens (NAc), and basolateral amygdala (BLA) (Fig. 1b). To confirm the validity of this approach, we looked for the presence of known biological relationships within the modules via protein-protein interactions (PPI) and found 12 modules in which known PPIs were greater than expected by chance (Supplementary Fig. 1a). To determine the relevance of these networks to human MDD, we examined whether the CSDS-associated resilience modules are preserved in RNA-seq data from post-mortem MDD patients8. 56.6% of our modules are preserved in human brain, but—consistent with a role in resilience—more modules (11 vs. 2) show greater preservation in control conditions than in MDD (Supplementary Fig. 1b–e).
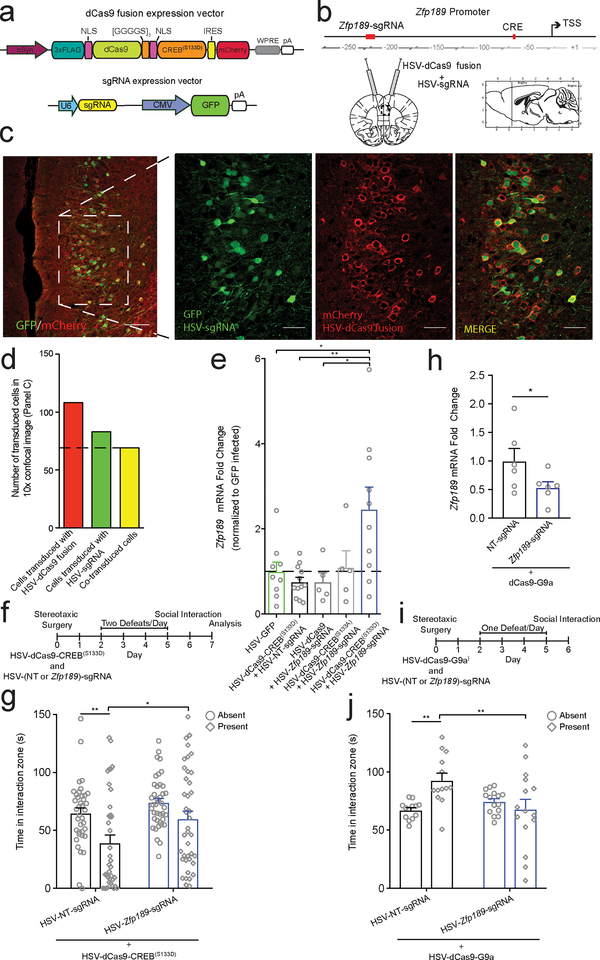
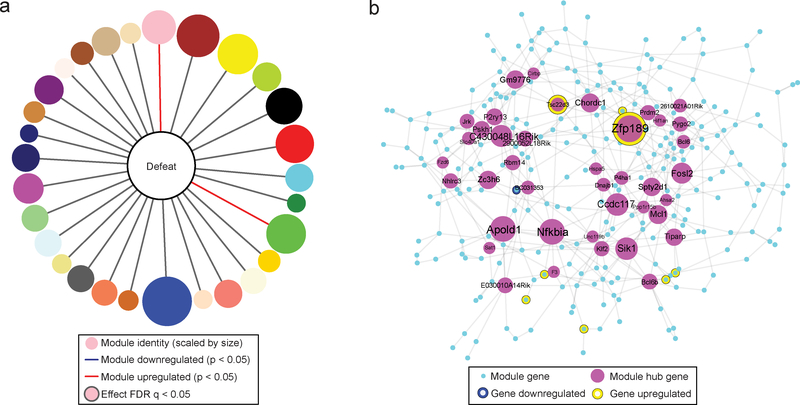
Fig 1. Identification of the resilient-specific pink module and its pro-resilient top key driver Zfp189.
a) Overview of chronic social defeat stress (CSDS) protocol, social interaction test (SI) phenotyping, brain dissections, and RNA-sequencing analysis of four brain regions, prefrontal cortex (PFC), nucleus accumbens (Nac), basolateral amygdala (BLA) and ventral hippocampus (vHIP), used to identify resilient-specific transcriptional networks. b) Resilient modules (colored bars) identified by weighted gene co-expression network analysis (WGCNA). Modules, named with an arbitrary color (outer most ring) are ranked clockwise by overall differentially expressed gene (DEG) enrichment (p < 0.05, FC > 1.3). Phenotype specificity as determined by resilient module differential connectivity (MDC) in susceptible and control networks and DEG enrichment is displayed internally. Presence of MDC color denotes statistical significance (FDR q < 0.05). DEG enrichments are scaled by -log10(p-value) with only significant (p < 0.05) enrichments featured in color. The pink module (top) is the only module that shows DEGs across brain regions and MDC when compared to both susceptible and control mice. Modules were generated from n=44 RNA-seq libraries consisting of pooled brain samples with DEG enrichment assessed via a Fisher’s exact test with Benjamini-Hochberg FDR correction for multiple comparisons as indicated. c) Network structure of the pink module. Key drivers are featured and scaled in size according to number of connections in the network. Zfp189 is the top key driver gene. d) Correspondence between differential expression for the individual Zfp189 transcript and DEG enrichment for the pink module as a whole across phenotypes and brain areas. DEG enrichments are scaled by -log10(p-value) with only significant (p < 0.05) enrichments featured in color. In PFC, both Zfp189 and the pink module are only affected in animals resilient to CSDS (both upregulated). e) mRNA of ZNF189, the human ortholog of Zfp189, is reduced in post-mortem PFC from major depressive disorder (MDD) patients (two-tailed Mann Whitney, U = 110.0, p = 0.0291, n=17,22). f) Experimental timeline to characterize behavioral effects of virally overexpressing Zfp189 in PFC prior to CSDS. g) Pro-resilient behavioral effects of Zfp189 in SI. Mice injected intra-PFC with HSV-Zfp189 and exposed to CSDS spend more time in the interaction zone when a target mouse present than defeated HSV-GFP mice (mixed model ANOVA, interaction F1,40 = 8.501, p = 0.006, n=9,10,12,13 mice, Bonferroni post-test p < 0.01). h) Mice overexpressing Zfp189 in PFC have an elevated preference for sucrose relative to HSV-GFP mice (two-way ANOVA, F1,41 = 5.102, p = 0.029, n=9,12,10,14 mice). i) Experimental timeline to determine behavioral effects of overexpressing Zfp189 in PFC in CSDS-susceptible mice. j) Zfp189 reverses depression-like social withdrawal in susceptible mice. Susceptible mice injected intra-PFC with HSV-Zfp189 spend more time in the interaction zone when the target mouse is present in the post-injection post-test than the pre-injection pretest, but HSV-GFP injection does not change behavior (mixed model ANOVA, interaction F1,23 = 5.634, p = 0.026, n=11 HSV-GFP and 13 HSV-Zfp189 mice, Bonferroni post-test p < 0.001). k) Previously susceptible mice injected with HSV-Zfp189 have a higher sucrose preference than previously susceptible mice injected with HSV-GFP (two-tailed Mann Whitney, U = 29.0, p = 0.025, n=11,12 mice). *p < 0.05, **p < 0.01, ***p < 0.001. Bar graphs show mean ± SEM.
To identify modules that were specific to the resilient phenotype, we used module differential connectivity (MDC) analysis6 to analyze whether each resilient module was similarly structured when the module genes were analyzed using parallel RNA-seq data from susceptible mice and unstressed controls3. We also used enrichment analysis to determine the degree to which each resilient module contains differentially expressed genes (DEGs) in each of the four brain regions studied 48 hours after CSDS. To identify networks that were both resilient-specific and transcriptionally active, we combined these approaches (Fig. 1b). Only two modules, pink and brown, were enriched for DEGs across brain regions. While brown module connectivity is distinct from susceptible conditions (MDC = 0.9) but not from control conditions, pink module connectivity is distinct from both susceptible (MDC = 0.89) and control (MDC = 0.91). Therefore, of 30 modules generated from four brain regions of resilient mice, only the pink module is unique to the resilient phenotype and transcriptionally active across brain regions. Importantly, the pink module is both enriched for known molecular interactions and more strongly preserved in human controls (p = 1.07 × 10−17) than in human MDD (p = 1.14 × 10−15) (Supplementary Fig. 1).
To determine the structure of the pink module, we reconstructed the network based on individual gene-gene correlations and predicted gene regulators as previously described10,11 (Fig. 1c). 40 network genes met criteria for key drivers (Supplementary Table 1). Of these, the strongest key driver gene is Zfp189, which contains 52 defined connections and is tightly integrated in the core network (Supplementary Fig. 2a–b). To determine the relationship between key driver genes and the overall network response, we compared individual differential expression patterns of Zfp189 and the other top 10 key driver genes of the pink module to module DEGs across brain regions (Fig. 1d; Supplementary Fig. 2c). Only Zfp189 is differentially expressed (upregulated in PFC of resilient mice) and recapitulates the brain-region specific DEG enrichment profile of the pink module (also upregulated in PFC of resilient mice). As the PFC has been implicated in stress resilience12, Zfp189 may act in this brain region to drive stress resilience through the resilient-unique pink module. While Zfp189 has not been implicated previously in any neurobiological context, it is a Krüppel-associated box (KRAB) protein13 and its human ortholog ZNF189 has been shown to bind directly to DNA14, suggesting that Zfp189 may function as a transcription factor.
Zfp189 in PFC exerts pro-resilient and antidepressant-like actions
To explore Zfp189 in human resilience, we performed qPCR to evaluate expression of its ortholog ZNF189 in PFC of human post-mortem brains (Supplementary Table 2) and found that ZNF189 mRNA is reduced in human MDD compared to control subjects (Fig. 1e). By examining previous data showing RNA expression across mouse cortical cell types15, we found Zfp189 to be highly enriched in neurons. We corroborated these data by performing RNAscope on sections from mouse PFC, and observed that a large majority (80.3% ± 5.6; n=3) of cells expressing Zfp189 mRNA are neurons (Supplementary Fig. 3a). Therefore, to probe the causal role of Zfp189 in stress resilience, we used herpes simplex virus (HSV) vectors, which target neurons selectively (Supplementary Fig. 3b), to overexpress Zfp189 in PFC neurons and exposed mice to an accelerated social defeat paradigm (Fig. 1f; Supplementary Fig. 4a).
In the social interaction test (SI), defeated mice overexpressing Zfp189 in PFC spend more time interacting with a social target than defeated mice with control HSV-GFP (Fig. 1g). To determine whether this effect extends to other measures of behavior associated with depression- or anxiety-like phenotypes, we analyzed mice in the open field test (OFT), forced swim test (FST), and sucrose preference test. In OFT, unlike controls, HSV-Zfp189 mice do not show stress-induced anxiety-like effects (Supplementary Fig. 4b–c) and, while there are no FST differences (Supplementary Fig. 4d), HSV-Zfp189 mice consume significantly more sucrose than defeated controls (Fig. 1h). Together, these results are consistent with Zfp189 in PFC increasing resilience to CSDS.
Since antidepressant treatment is initiated after a diagnosis of MDD, molecular targets that reverse behavioral phenotypes seen in depression have higher therapeutic potential than targets which only prevent susceptibility. We therefore explored the antidepressant potential of Zfp189 by determining whether CSDS-induced alterations in behavior could be reversed (Fig. 1i). Delivering Zfp189 to the PFC after CSDS reversed stress-induced social deficits (Fig. 1j). While Zfp189 overexpression in susceptible mice did not affect OFT, locomotor testing, or FST (Supplementary Fig. 4e–g), HSV-Zfp189 mice displayed higher sucrose preference than HSV-GFP mice (Fig. 1k). Thus, Zfp189 in PFC exerts both pro-resilient and antidepressant-like actions.
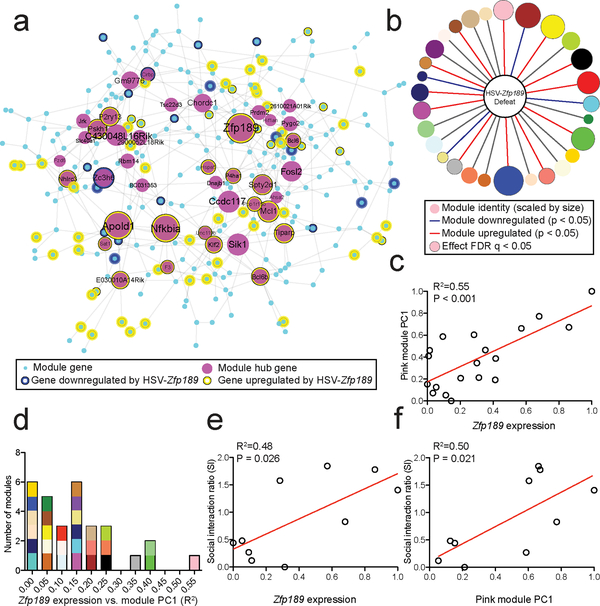
To test the effects of Zfp189 on gene expression profiles, we micro-dissected virally-infected tissue and performed RNA-seq. Overexpression of Zfp189 in PFC upregulated 33.1% (93) of pink module genes, including 47.5% (19) of key driver genes (Fig. 2a). Many of these genes are tightly integrated in the network structure, including Nfkbia and Apold1, the two top key driver genes after Zfp189. Downregulation was less prominent: 7.1% (20) of genes overall and only 5.0% (2) of hub genes. While Zfp189 overexpression in the PFC of previously susceptible mice significantly downregulates 4 and upregulates 12 resilient modules (Fig. 2b), in support of Zfp189 inducing behavioral resilience through the pink module specifically, Zfp189 overexpression in PFC significantly upregulates the pink module in both previously susceptible (FDR q = 1.67 × 10−12), and non-defeated control (FDR q = 2.63 × 10−4; Supplementary Fig. 5) mice.
Fig. 2. Antidepressant-like effects of Zfp189 associate with pink module expression changes.
a) Pink module genes are differentially expressed (p < 0.05, log2FC > |0.2|) in PFC following reversal of susceptibility with HSV-Zfp189. n=10 RNA-seq libraries consisting of unpooled PFC from 5 stressed HSV-GFP and 5 stressed susceptible to resilient HSV-Zfp189 mice. b) Module-wide enrichment for HSV-Zfp189 overexpression in PFC in previously susceptible mice. Enrichment is determined via multinomial logistic regression with Benjamini-Hochberg FDR correction for multiple comparisons as indicated. c) Variations in Zfp189 predict the first principal component of pink module expression. Linear regression, n = 19 RNA-seq libraries consisting of unpooled PFC from 5 stressed HSV-GFP, 4 unstressed HSV-GFP, 5 stressed susceptible to resilient HSV-Zfp189 and 5 unstressed HSV-Zfp189 mice. d) Histogram of the coefficient of determination (R2) from linear regression analysis examining the relationship between Zfp189 expression and the first principal component of each resilient module (modules are plotted accordingly and represented by color). Of all resilient modules, Zfp189 shows the strongest regulation of the pink module. e-f) Linear regression showing a positive relationship between resilient behavior and (e) Zfp189 levels in PFC of each mouse and (f) pink module expression (n=10 RNA-seq libraries consisting of 5 stressed HSV-GFP and 5 stressed susceptible to resilient HSV-Zfp189 mice).
We next tested whether Zfp189 overexpression in PFC preferentially affects the pink network. To do this, we performed principal component analysis to determine pink module expression across HSV-Zfp189 and HSV-GFP samples in both unstressed and defeated groups. Because there is variability in the degree to which Zfp189 is overexpressed by our HSV (Supplementary Fig. 4a), we can leverage this variance to determine whether mice in which Zfp189 was expressed to a higher extent had stronger activation of the pink module and more resilient behavior. Notably, while this relationship between Zfp189 and the pink module is inherent in the RNA-seq data used to generate the pink module (Fig. 1b–c), there is no a priori relationship between the pink module in our HSV-Zfp189 sequencing data. As such, our finding that levels of Zfp189 expression in each PFC sample significantly explain variation in pink module expression as measured by the first principal component (PC1) (R2 = 0.55, p < 0.001; Fig. 2c), independently validates the inherent biological relationship between Zfp189 and pink module genes that we originally identified in our WGCNA analysis (Fig. 1c). Additionally, while WGCNA is undirected, in the current analysis overexpression of Zfp189 is a product of viral transduction, and establish a true causal link between Zfp189 expression and regulation of the pink module. To determine whether this causal relationship was unique to the pink module, we applied a similar approach to all other modules. Strikingly, Zfp189-induced expression changes drive expression of the pink module more than any other network (Fig. 2d). Further, both variation in Zfp189 expression and variation in pink module expression is significantly correlated with resilient behavior (R2 = 0.48, p = 0.026 and R2 = 0.50, p = 0.021 respectively; Fig. 2e–f). Together, these data validate our bioinformatic predictions and indicate that the pro-resilient effects of Zfp189 are preferentially associated with pink module gene expression in PFC.
CREB is an upstream regulator of the resilience module
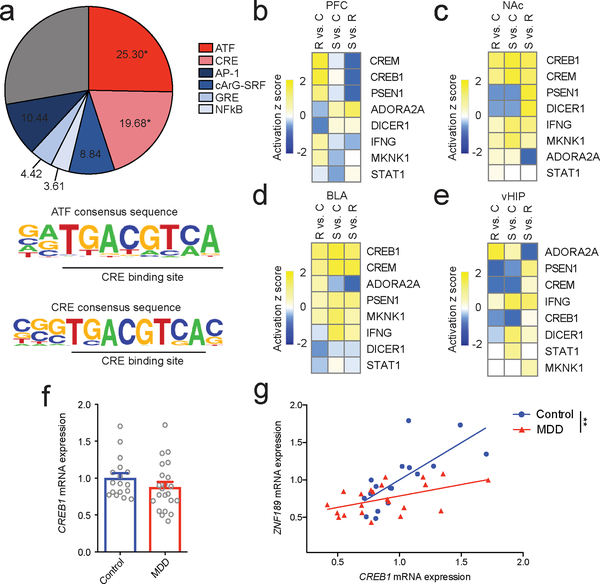
We next investigated how the pink module is regulated. We first used HOMER16 to evaluate pink module genes for overrepresentation of known binding motifs in silico. Both activating transcription factor 1 (ATF1) and cyclic AMP (cAMP) response element (CRE) binding protein (CREB) are predicted upstream regulators of pink module genes (FDR q = 0.027 and FDR q = 0.035; Fig. 3a and Supplementary Table 3). The closely related ATF1 and CREB predictions bind near-identical CRE-containing consensus sequences, indicating that it is the CRE site that is overrepresented in the pink module. CRE-containing consensus sequences are unique to the pink module, with statistical enrichment of any binding site apparent in only four of the 29 other networks (FDR q < 0.05; Supplementary Table 3), none of which contained this motif. Both ATF1 and CREB play important roles in cell survival17 and are ubiquitously activated by extracellular signals18, yet CREB has also been implicated extensively in both human MDD19,20 and animal models of depression21–24, while only non-ATF1 members of the ATF family have been implicated in stress responsivity25.
Fig 3. CREB is an upstream regulator of the pink module.
a) Binding motifs overrepresented (FDR q < 0.05) in the pink module (colored pink) with common non-overrepresented motifs (colored blue) included for comparison (top). Both significantly overrepresented binding motifs contain a CRE site (bottom). b-e) Upstream regulator analysis of transcriptional changes in pink module genes. CREB is an upstream regulator across brain regions and is predicted to be upregulated in resilience in PFC. f) mRNA levels of CREB1 in PFC from MDD patients and matched controls (two-tailed t test, t= 1.216, p = 0.232, n=17,22 mice). g) CREB1 and ZNF189 are correlated in the PFC of controls to a greater extent than in MDD subjects (ANCOVA, F1,35 = 7.702, p = 0.009, n = 17 control and 22 MDD). **p < 0.01. Bar graphs show mean ± SEM.
To complement this analysis, we performed an upstream regulator analysis using Ingenuity Pathways Analysis (IPA), which utilizes known interactions for a set of genes to determine upstream regulators based on downstream expression changes. Matching our results with HOMER, we identify CREB as a predicted upstream regulator of pink module transcription across brain regions (Fig. 3b–e; Supplementary Table 4). In contrast, our HOMER finding of ATF1 regulation is not reproduced by IPA. While CREB is predicted to be upregulated more in susceptibility than resilience in NAc, BLA, and vHIP in our dataset, the predicted regulation of CREB in PFC (up in resilient, no change in susceptible) mimics that of both pink module DEG enrichment and Zfp189 differential expression (Fig. 1d). Therefore, we hypothesized that CREB might regulate the pink module in PFC, in part, through direct interactions with Zfp189.
To investigate the role of CREB-Zfp189 interactions in pro-resilient regulation of the pink module, we analyzed a published ChIP-chip dataset of active, Ser133-phosphorylated CREB (pCREB) binding in NAc following CSDS, which reveals that pCREB binding to Zfp189 is reduced following CSDS, and reversed upon antidepressant treatment23. To determine human relevance, we performed qPCR analysis on MDD tissue from post-mortem PFC. While there is no effect of MDD on CREB1 expression (Fig. 3f), Creb1 mRNA levels are significantly correlated with Znf189 mRNA levels across samples in both control and MDD brains (R2 = 0.53, p < 0.001 and R2 = 0.25, p = 0.018 respectively; Fig. 3g). Moreover, there is a stronger positive relationship in control than MDD. As such, CREB-Zfp189 interactions may be reduced in both human MDD and mouse CSDS.
CREB-Zfp189 interactions regulate resilience
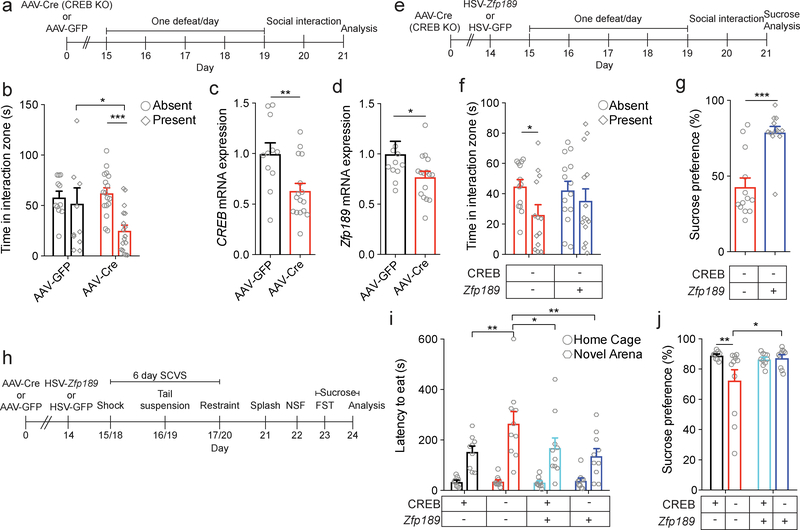
To evaluate our prediction that CREB drives resilience in the PFC, we injected CREBfl/fl mice with adeno-associated virus (AAV) expressing Cre recombinase plus GFP or GFP alone and exposed mice to a subthreshold social defeat (Fig. 4a). In accordance with our prediction, AAV-Cre mice (functionally a selective CREB KO in PFC neurons) developed behavioral abnormalities in response to this social stress but AAV-GFP control mice were unaffected (Fig. 4b). We next micro-dissected virally infected PFC and performed qPCR. Both Creb1 and Zfp189 levels are reduced by CREB KO (Fig. 4c–d), providing causal in vivo evidence of regulation of Zfp189 by CREB in PFC neurons.
Fig. 4. CREB knockout (KO) in PFC increases susceptibility but is rescued by Zfp189 overexpression.
a) Experimental timeline to evaluate behavioral effects of CREB KO in PFC. b) Local KO of CREB in PFC produces social avoidance in SI in a subthreshold social defeat procedure (mixed model ANOVA, interaction F1,26 = 4.656, p = 0.040, n=11,17 mice, Bonferroni post-test p < 0.001). c-d) CREB KO reduces mRNA levels of (c) Creb1 (two-tailed t test, t = 2.974, p = 0.006) and (d) Zfp189 (two-tailed Mann-Whitney, U = 45.0, p = 0.036), n=11,16 mice. e) Experimental timeline to determine whether Zfp189 overexpression in PFC reverses the deleterious effects of CREB KO. f) CREB KO in PFC produces social avoidance in the SI test but concurrent overexpression of Zfp189 reverses this deficit (mixed model ANOVA, target F1,25 = 5.690, p = 0.025, n=13,14 mice, Bonferroni post-test p < 0.05). g) Overexpression of Zfp189 in PFC increases sucrose preference in CREB KO mice (two-tailed t test, t = 5.176, p < 0.001, n=13 mice). h) Experimental schematic for female sub-chronic variable stress (SCVS) to investigate behavioral effects of CREB KO and Zfp189 overexpression in PFC. i) CREB KO in PFC increases latency to eat in the novel arena, but not when CREB KO is paired with Zfp189 overexpression (mixed model ANOVA, interaction F1,35 = 5.301, p = 0.027, n=9,11,10,10 mice, Bonferroni post-test compared to CREB−Zfp189- in novel arena, p < 0.01 for CREB+Zfp189-, p < 0.01 for CREB−Zfp189+, and p < 0.05 for CREB+Zfp18+). j) Mice with local KO of CREB in PFC have a lower sucrose preference than control mice, an effect blocked by concurrent Zfp189 overexpression (Kruskall-Wallis test, χ2(3) = 8.475, p = 0.037, n=9,11,10,10 mice, two-tailed Mann-Whitney post-test p = 0.009 for local KO of CREB and p = 0.029 for overexpression of Zfp189 with concurrent CREB KO). *p < 0.05, **p < 0.01, ***p < 0.001. Bar graphs show mean ± SEM.
Since our data indicate that CREB is upstream of Zfp189, that Zfp189 promotes resilience, and that Zfp189 acts through pink module expression changes, we reasoned that CREB KO in PFC neurons is pro-susceptible at least in part through the consequent reduction in Zfp189. If this is the case, concurrent overexpression of Zfp189 should rescue the pro-susceptible effects of CREB KO. To evaluate this, we injected AAV-Cre in PFC of CREBfl/fl mice while overexpressing either HSV-Zfp189 or HSV-GFP and exposed mice to a subthreshold defeat (Fig. 4e). Importantly, all cells infected by HSVs were previously infected by AAVs, indicating that Zfp189 is overexpressed in neurons that lack CREB (Supplementary Fig. 6). Consistent with our previous findings, CREB KO mice injected with HSV-GFP show social avoidance, whereas CREB KO mice supplemented with HSV-Zfp189 do not (Fig. 4f). Zfp189 overexpressing CREB KO mice also have higher sucrose preference than GFP overexpressing CREB KO mice (Fig. 4g), further supporting the capacity of Zfp189 overexpression to mitigate the pro-susceptible effects of CREB KO in PFC.
While the pink module was identified from a dataset of male mice3 and transcriptional effects of stress exhibit sex differences8,26–28, we recently found that gene manipulations can induce similar behavioral effects even in the context of sex-specific transcriptional changes29. Therefore, we manipulated PFC of female CREBfl/fl mice with AAV-Cre or -GFP plus either HSV-Zfp189 or HSV-GFP and exposed mice to six days of sub-chronic variable stress (SCVS), a protocol that reliably induces depression-like behavior in female mice8,26,27 (Fig. 4h). In the novelty suppressed feeding (NSF) test, CREB KO mice show the longest latency to feed in the novel arena (Fig. 4i). While there is no difference in grooming time in the splash test or latency to immobility in the FST (Supplementary Fig. 7), CREB KO mice also display significantly diminished sucrose preference (Fig. 4j). Zfp189 overexpression blocks these effects of CREB KO, thus demonstrating that CREB KO increases stress susceptibility in females that, similar to males, can be rescued by concurrent Zfp189 overexpression.
CRISPR-mediated CREB-Zfp189 interactions promote resilience
Although our data suggest a functional relationship between CREB and Zfp189 expression (Fig. 4c–d), indirect interactions between the two factors could be responsible. To more definitively evaluate the role of CREB-Zfp189 interactions in resilience, and to probe the causal consequence of direct CREB action at the Zfp189 gene in PFC, we used CRISPR technology as a tool for locus-specific editing: to target recruitment of CREB selectively to the Zfp189 gene promoter in PFC neurons. We fused the nuclease-dead, RNA-guided, DNA-binding protein Cas9 to a constitutively active, phosphomimetic mutant form of CREBS133D (dCas9-CREBS133D) and designed sgRNAs to target near the consensus CRE motif in the Zfp189 promoter. The construct design is shown in Fig. 5a. In vitro validation of sgRNAs shows that the top Zfp189-targeting sgRNA (Zfp189-sgRNA) induces Zfp189 expression only when the active CREBS133D, and not the phospho-null CREBS133A or dCas9 with no functional moiety, is recruited to the Zfp189 locus (U = 1.0, p = 0.009, n = 5–6; Supplementary Fig. 8). Zfp189-sgRNA targets dCas9-CREBS133D ~150 bp upstream of the CRE motif in the Zfp189 promoter (Fig. 5b). With the exception of the Zfp189 locus, there is no complementary site in the mouse genome with fewer than three base mismatches, suggesting a low in silico probability of off-target effects (Supplementary Table 5). Our control non-targeting (NT-) sgRNA is also predicted to target no specific sequence in the mouse genome (Supplementary Table 6).
Fig 5. CRISPR-mediated, locus-specific modulation of Zfp189 with CREB or G9a bidirectionally controls resilient behavior.
a) Schematic of the CRISPR vectors. Variable dCas9 functional moiety in orange. Variable gene-targeting single guide RNA (sgRNA) in yellow. b) Location of Zfp189-targeting sgRNA binding site relative to other features in the Zfp189 promoter (in red). CRISPR vectors were packaged in HSV and delivered as a viral cocktail bilaterally to PFC. Hashed box denotes field of confocal imaging for Panel C, left. c) Immunohistological staining shows high degree of colocalization of HSV-sgRNA expression vector (GFP) and HSV-dCas9 fusion expression vector (mCherry) in PFC neurons. Left: 10x objective, scale bar = 100 μm. Right: 20x objective, scale bar = 50 μm. Repeated with similar results in three animals. d) Quantification of virus colocalization. e) CRISPR-mediated targeting of active, pseudo-phosphorylated CREB(S133D) to Zfp189 is sufficient to increase mRNA expression in PFC relative to HSV-GFP, un-targeted dCas9-CREBS133D, and dCas9 with no functional domain targeted to Zfp189 (Kruskall-Wallis test, χ2(5) = 10.27, p = 0.036, n=9,12,5,5,19 mice, two-tailed Mann-Whitney post-test p = 0.035, p = 0.004, and p = 0.040 respectively). Targeting dominant negative CREB(S133A) to Zfp189 has no effect. f) Experimental timeline to determine effect of CRISPR-mediated placement of CREB at the Zfp189 promoter in PFC neurons. g) Pro-resilient effects of CRISPR-dependent CREB-Zfp189 interactions. dCas9-CREBS133D delivered with Zfp189-targeting sgRNA increases time in the interaction zone when a target mouse is present relative to dCas9-CREBS133D with non-targeted (NT) sgRNA (Mixed model ANOVA, virus F1,76 = 6.235, p = 0.015, n=38,40 mice, Bonferroni post-test p < 0.05). h) Targeting dCas9 with G9a to the Zfp189 promoter reduces Zfp189 expression (two-tailed t test, t = 2.835, p = 0.037, n=6 mice). i) Experimental timeline to determine effect of CRISPR-mediated localization of G9a to the Zfp189 promoter in PFC neurons. j) Pro-susceptible effects Zfp189-targeted repression with G9a. dCas9-G9a delivered with Zfp189-targeting sgRNA decreases time in the interaction zone when a target mouse is present relative to dCas9-G9a with non-targeted (NT) sgRNA (Mixed model ANOVA, interaction F1,26 = 9.844, p = 0.0042, n=13,15 mice, Bonferroni post-test p < 0.01). *p < 0.05, **p < 0.01. Bar graphs show mean ± SEM.
We independently packaged our sgRNA expression vectors and our dCas9 fusion protein expression vectors in HSVs, and co-delivered these vectors to the mouse PFC. We observe that the two HSVs infect predominantly the same neurons (Fig. 5c–d), and targeting dCas9-CREBS133D to the Zfp189 promoter increases Zfp189 expression in mouse PFC (Fig. 5e). These data demonstrate the ability to harness the physiologically-relevant mechanism of CREB-mediated induction of Zfp189 expression with CRISPR-mediated transcriptional reprogramming in mouse brain.
We next determined whether direct CREB-mediated activation of Zfp189 is sufficient to promote resilience. We injected HSV-dCas9-CREBS133D paired with either HSV-NT-sgRNA or HSV-Zfp189-sgRNA into PFC and exposed mice to an accelerated social defeat stress procedure (Fig. 5f). In SI, mice with dCas9-CREBS133D targeted to Zfp189 show significantly increased resistance to CSDS-induced social avoidance relative to mice with untargeted dCas9-CREBS133D (Fig. 5g). Thus, inducing this single interaction between pCREB and Zfp189 in PFC neurons is sufficient to increase resilience to social defeat.
We next determined whether an epigenetic modification that suppresses Zfp189 expression prevents resilient-like behavior. To do this, we fused dCas9 to the repressive histone methyltransferase G9a (dCas9-G9a) and injected HSV-dCas9-G9a paired with either HSV-NT-sgRNA or HSV-Zfp189-sgRNA into PFC. Directing G9a to the Zfp189 promoter reduces Zfp189 expression in the PFC (Fig. 5h). To determine the behavioral effects of this manipulation, we exposed mice to a sub-threshold defeat of one 10-minute defeat each day over 4 days and examined behavior in SI (Fig. 5i). As predicted, mice with dCas9-G9a targeted to Zfp189 show significantly decreased social interaction relative to mice with untargeted dCas9-G9a (Fig. 5j).
To test our hypothesis that these effects are associated with changes in pink module gene expression, we micro-dissected virally-infected tissue from the dCas9-CREBS133D experiment and performed RNA-seq (Supplementary Fig. 9a). Both the pink and green resilient modules are activated in response to CRISPR-mediated, CREB-Zfp189 interactions in the context of social defeat (p = 0.010 and p = 0.048 respectively; Fig. 6a). Relative to traditional overexpression (Fig. 2a), we observe fewer regulated pink module genes overall (Fig. 6b) which likely is due to the more physiologically-relevant levels of Zfp189 induction (~2.5-fold Zfp189 induction with CRISPR-mediated recruitment of CREB vs. ~20-fold Zfp189 induction with HSV overexpression).
Fig 6. CRISPR-mediated induction of CREB-Zfp189 interactions activates the pink module.
a) Module overlap for PFC DEGs (p < 0.05, log2FC > |0.2|) resulting from Zfp189-targeted dCas9-CREB(S133D) compared to NT-dCas9-CREB(S133D) in mice exposed to social defeat. Enrichment is determined via multinomial logistic regression with Benjamini-Hochberg FDR correction for multiple comparisons as indicated. b) Pink module genes differentially expressed in defeated mice (p < 0.05, log2FC > |0.2|). n=20 RNA-seq libraries consisting of unpooled samples from 8 Zfp189-targeted dCas9-CREB(S133D) and 12 NT-dCas9-CREB(S133D) mice.
To determine whether inducing CREB-Zfp189 interactions in PFC neurons of unstressed controls is sufficient to produce similar changes in pink module genes, we repeated our dCas9-CREBS133D injections in mice not exposed to CSDS (Supplementary Fig. 9). Again, the pink module is significantly affected when CREB is directed to the Zfp189 promoter (FDR q = 0.047; Supplementary Fig. 9c), with a predominant upregulation of pink module genes (Supplementary Fig. 9d). Importantly, we observe no regulation of genes nearest to the 49 most homologous off-target sites of the Zfp189-sgRNA across the mouse genome in unstressed control animals, and regulation of only one off target gene (Tsc22d3) in the defeat cohort, which is likely a result of the defeat experience itself (Supplementary Fig. 9e). These data substantiate the specificity of our CRISPR approach to direct CREB action at Zfp189 alone.
Discussion
Here, we elucidate the higher-order organization of the transcriptional response to stress across limbic brain regions and demonstrate that a single, causal regulatory interaction can control activity of a phenotype-specific transcriptional network with definitive effects on complex behavior. Consequently, these data provide a mechanistic bridge between individual neuronal genes and the large-scale transcriptional response observed in resilience2,3. Taken together, our data show that the resilient-specific pink module is preferentially activated by Zfp189—the module’s strongest driver gene, and that Zfp189 (and thereby the pink module) is regulated by CREB in PFC neurons. As CREB-Zfp189 interactions are impeded in both chronically stressed mice23 and depressed humans (Fig. 3g), and promote resilience to CSDS (Fig. 5g,j), pharmacological manipulations of Zfp189, either directly or through CREB-dependent mechanisms, have the potential to regulate an entire network of pro-resilient genes and may be an attractive target for MDD therapeutics.
Because we show that manipulations of either Zfp189 (Fig. 2b) or CREB-Zfp189 interactions (Fig. 6a) significantly affect the resilient-specific pink module, our findings are distinct from investigations of causal regulators of resilience to date that have focused on individual genes or molecular pathways2,22,30,31. Notably, our data support the existence of a transcriptional hierarchy as we repeatedly show that manipulation of one putative transcription factor (Zfp189), either directly or through epigenetic manipulations related to its upstream regulator CREB, is sufficient to increase resilience in multiple contexts (Fig. 2g–k; Fig. 4f–g,i–j; Fig. 5g) by activating a network of uniquely resilient genes. Even so, it is unlikely that the 281 genes in the pink module are the sole contributors to CSDS resilience, especially given the thousands of genes involved in the resilient response2,3. Consequently, there is likely some redundancy whereby multiple genes produce similar effects, and that the ability of the CREB-Zfp189-pink module axis to promote resilience occurs in the context of other molecular changes yet to be identified. Future studies are needed to address these possibilities.
Although our manipulations were initiated in neurons (Supplementary Fig. 3), some of the genes in the pink module are enriched in other cell types. For example, Apold1, which is the third-highest ranking key driver gene and is upregulated following HSV-Zfp189 injection into PFC neurons (Fig. 2a), is endothelial-cell specific32. These observations show that manipulations of Zfp189 in neurons only cause downstream transcriptional effects in non-neuronal cell types through cell-cell signaling. While changes in the endothelium have been shown to play a role in CSDS susceptibility33, the role of endothelial cells in resilience has not yet been defined. These findings illustrate the advantages of whole-tissue RNA-seq, as transcriptional relationships between cell types can be observed.
Our proposed mechanism for regulation of the pink module involves CREB activating Zfp189 by binding to the Zfp189 promoter. While we show that CREB KO in PFC neurons increases susceptibility, and that this can be mitigated with concurrent overexpression of Zfp189 in the same cells (Fig. 4), this approach affects hundreds or thousands of genes, and it is impossible to discern whether our observed effects are due to altered CREB-Zfp189 interactions or simply the pro-resilient effects of Zfp189 obscuring the pro-susceptible effects of CREB KO. Most studies to date have relied on overexpression/knockdown strategies, which are limited in defining the exact causal mechanisms driving the progression of neuropsychiatric disease. To circumvent this problem and more definitively characterize the role of regulatory interactions in controlling transcriptional networks, we employed CRISPR. While neuroepigenetic editing has been employed to answer questions related to neuropsychiatric disease34–36, CRISPR technology provides a cheaper, more modular, more easily employable approach that can answer a wider array of relevant research questions and has been used previously to induce locus-specific epigenetic modifications in mammalian brain37,38. Our data demonstrate that CRISPR-dependent locus-specific epigenetic modifications can be used to mimic and block endogenously identified interactions and affect both downstream gene expression (Fig. 6; Supplementary Fig. 9c) and behavior (Fig. 5g,j).
Together, we describe a vital single molecular interaction of the known transcriptional regulator CREB and the novel downstream transcription factor Zfp189 that is capable of activating a network of genes in PFC neurons to mediate stress resilience. These findings elucidate the molecular mechanisms involved in stress resilience, as well as provide a broad molecular framework for the hierarchical organization and regulation of gene co-expression networks and their relationship to complex behavior.
Contact For Reagent and Resource Sharing
Further information and requests for resources and reagents, including custom R scripts used in this study, should be directed to and will be fulfilled by Eric Nestler (eric.nestler@mssm.edu).
Methods
Animals
Experimental mice were either C57BL/6J mice or C57BL/6J mice genetically modified for a conditional brain region-specific Cre-dependent CREB knockout by insertion of loxP sites flanking Creb1 exon 222. In addition, 6-month-old CD1 aggressor mice were used as aggressors to induce social stress in the chronic social defeat stress (CSDS) procedure, but were not included in any analysis. Wildtype C57BL/6J mice were 8 weeks old at the time of experimentation. CREBfl/fl mice were used between 8 weeks and 4 months of age due to breeding considerations, and age was counterbalanced across experimental conditions. C57BL/6J mice were housed 5 per cage; CD1 mice were single-housed. C57BL/6J mice undergoing CSDS were single-housed following the final defeat and C57BL/6J mice undergoing SCVS were single-housed following the final stressor. In order to maintain consistent study design, unstressed controls were single-housed at the same time as stressed mice. Once mice were single-housed, they remained as such until the end of experimentation. For all experiments, mice were randomized to experimental groups and only healthy, well-appearing mice were selected for experimentation. All mice were maintained on a 12-hour light-dark cycle with lights on at 7:00 AM and a controlled temperature range of 22–25°C. Food and water were provided ad libitum except for the 24 hours preceding novelty suppressed feeding (NSF) testing, when food was removed. All experiments conformed to the Institutional Animal Care and Use Committee (IACUC) guidelines at Mount Sinai, which approved the animal experiments in the study. Behavioral testing took place during the animals’ light cycle. In cases where non-automated analysis was used, experimenter was blinded to experimental group. Order of testing in behavioral experiments was counterbalanced and assignment to experimental groups was random.
Methods
Stress protocols and behavioral testing
CSDS and social interaction tests were performed according to established protocols1,2. CD1 retired breeder mice were screened for aggression in three-minute intervals over the course of three days. CD1 mice consistently attacking 8-week-old male C57BL/6J screener mice were included as aggressors for CSDS. On the first day of stress, CD1 mice were placed on one side of a large hamster cage separated by a perforated Plexiglas divider and an 8-week-old male C57BL/6J was placed on the other side. Importantly, this Plexiglas divider allows for sensory, but not physical, contact between CD1 and C57BL/6J mice. During each defeat, C57BL/6J mice were placed in the same side of the cage as the CD1 aggressor for a period of 7.5–10 minutes. This duration was kept constant throughout each experiment and was predetermined to titrate the defeat based on the overall aggression in the CD1 cohort during screening. During this time, the CD1 aggressor physically attacked the C57BL/6J mouse. Following the physical bout, C57BL/6J mice were returned to the other (empty) side of the divider where they remained in sensory contact with the CD1 aggressor that had just attacked them, but could not be further harmed physically. For each consecutive defeat session (24 hours later for CSDS and subthreshold defeat and 12 hours later for accelerated defeat), the C57BL/6J mouse was exposed to a new CD1 aggressor in a different hamster cage and the procedure was repeated. Control C57BL/6J mouse were double-housed in a mouse cage separated by a perforated divider for the length of stress. To control for handling effects, control mice were moved to the adjacent half cage whenever a stress occurred for experimental mice. CSDS took place for a duration of 10 days, accelerated defeat took place over the course of 4 days with 2 defeats per day (to coincide with the time course of HSV-mediated transgene expression), and subthreshold defeat took place over the course of 5 days.
SCVS was performed as previously described26 with three different hour-long stresses performed twice over a total of six days. Briefly, 8-week-old female C57BL/6J mice were exposed to foot shock (0.45 mA) on days one and four, tail suspension on days two and five, and restraint stress (in a 50 ml Falcon tube in the home cage) on days three and six. Mice were group housed (five mice/cage) when they were not being stressed and control mice remained in their home cages throughout.
Behavioral tests occurred in a behavior suite different from where stress exposure was performed. In cases where a test was repeated following a manipulation, a different behavior room was used for the second test. Mice were given one hour to habituate to the behavioral room prior to behavioral testing. Due to the timeline of HSV expression39, multiple behaviors occurred on the same day when HSV vectors were used. In order to minimize spillover effects from one test to another, tests were separated by a minimum interval of 2 hours. Behavioral analysis for social interaction, locomotion and open field test (OFT) was performed automatically by video tracking software (Ethovision 10.0, Noldus), FST was analyzed manually on pre-recorded video by investigators blind to study design and sucrose preference test and NSF were analyzed manually in real time. To ensure adequate power, sample sizes were chosen in accordance with number of mice needed to show statistical significance in CSDS and SCVS as defined by previous studies3,26.
Social interaction testing was performed under red light 24 hours after the last social defeat stress. C57BL/6J mice were placed into an open arena with an empty wire cage at one side (interaction zone). Mice were given 2.5 minutes to explore the arena and then removed. A novel CD1 aggressor to which the C57BL/6J mouse had never been exposed to was placed in the cage (interaction zone) and the procedure was repeated. Time in the interaction zone was recorded automatically with video tracking software. Data were analyzed as time spent in the interaction zone when the aggressor was absent compared to time spent in the interaction zone when the aggressor was present. In cases where mice were subset into resilient and susceptible phenotypes, defeated mice with social interaction ratio scores > 1.2 that spent > 60 seconds in the interaction zone when the target was present were determined to be resilient. Defeated mice with SI ratio scores < 0.8 that spent < 40 seconds in the interaction zone when the target was present were determined to be susceptible.
OFT was performed by allowing mice 10 minutes to explore an open arena under red light. Although nothing was physically placed in the arena, center and periphery were defined in video tracking software. Total time in center was recorded and utilized for analysis. In addition, total distance moved during this time was analyzed to determine locomotor effects.
Forced swim test (FST) was always performed last in the sequence of behavior. FST was performed in 4L Pyrex beakers filled with two liters of 25°C (± 1°) water. Mice were placed into the water and recorded by a front-facing camera for a period of six minutes. Investigators blind to study design scored FST videos by recording latency to the first immobility state.
NSF was performed following 24 hours of food deprivation. Female mice were placed in a novel arena with corncob bedding and a single piece of food in the center of the arena. Time to feed was recorded manually under white light. Mice were given a maximum of 10 minutes to eat, after which the trial was ended and latency of 600 seconds was recorded. After the mouse ate in the novel arena, the mouse was returned to the home cage where a single piece of food was located in the center, and time to eat in the home cage was recorded. Data were analyzed as latency to eat in the novel arena and latency to eat in the home cage.
Sucrose preference test was performed as a two-bottle choice test. One bottle was filled with water and the other bottle was filled with 2% sucrose. Initial weights of each bottle were recorded and bottle weights were recorded each morning and evening over the sucrose preference period. Sucrose preference was calculated as change in weight of the sucrose bottle/change in weight of both bottles X 100. Total sucrose preference was used for analysis.
In order to allow for peak viral expression at the time of behavior, adeno-associated virus (AAV) infusions and behavior were separated by four weeks and herpes simplex virus (HSV) viral infusions and behavior were separated by 4–5 days as described in reported experimental timelines.
Tissue collection
Mice were euthanized 24 hours after final behavior via cervical dislocation. In order to acquire only infected tissue within prefrontal cortex (PFC), microdissection was performed under a fluorescent microscope. Brain slices containing PFC were either visualized on blade and punched directly or suspended in cold phosphate buffered saline (PBS) prior to tissue dissection. PFC was collected as either a single midline 12-gauge punch or bilateral 14-gauge punches, which varied according to virus spread. However, only PFC was included in dissections. Mice were excluded from analysis when the PFC was not correctly targeted. Dissected tissue was immediately frozen on dry ice. Since our HSV and AAV vectors express green fluorescent protein (GFP), there was no way to distinguish expression of the two viruses. As such, for experiments in which HSV and AAV vectors were both injected, PFC was collected with a 12-gauge punch and downstream quantitative reverse-transcription polymerase chain reaction (qPCR) was used to validate virus effects. In cases where dual 14-gauge punches were used, two punches (bilateral) from each mouse were combined, but samples were never pooled between mice.
Viral reagents
We overexpressed Zfp189 using bicistronic p1005 HSV expressing GFP alone or GFP plus Zfp189. This involves a dual promoter approach whereby GFP expression is driven by a cytomegalovirus (CMV) promoter but Zfp189 expression is driven by IE4/5. Zfp189 was inserted into the p1005 plasmid from a plasmid containing the mouse Zfp189 gene (Origene MR209370), which was packaged into HSV.
We overexpressed Cre recombinase in CREBfl/fl mice using AAV serotype 2 AAV-CMV-Cre-GFP virus from the University of North Carolina Vector core. Similar to expression of Zfp189, this virus induces Cre expression via the CMV promoter. In CREBfl/fl mice where we intended to preserve CREB expression, we injected AAV-CMV-GFP (serotype 2).
We repurposed the CRISPR system in order to target CREB binding to the Zfp189 promoter. We cloned and synthesized fusion constructs of a phosphomimetic mutant form of CREB or G9a fused to nuclease-dead Staphylococcus pyogenes Cas9 protein (dCas9-CREBS133D and dCas9-G9a, respectively), which can localize to specific sites along the genome based on the complementarity of a specific single guide RNA (sgRNA) sequence. sgRNA sequences for Zfp189 were determined first in silica based on the sequence of the Zfp189 promotor and a suite of 10 sgRNAs were designed to bind to distinct DNA sequences proximal to the CRE motif within the promoter. To promote specificity, sgRNA off-target effects were predicted at crispr.mit.edu according to their published algorithm40, and candidate sgRNAs with complementary sequences in the mouse genome with fewer than three base mismatches were excluded. The sequence of our non-targeting sgRNA (protospacer sequence: GCGAGGTATTCGGCTCCGCG) was identified from the GeCKO v2 libraries and validated to ensure that no genomic site was targeted. All 10 candidate sgRNAs were de novo synthesized as gBlocks from Integrated DNA Technologies containing a U6 promoter, variable target sequence, guide RNA scaffold and termination signal. These were sub-cloned into a p1005 variant plasmid for HSV packaging. All Zfp189-sgRNAs were first validated in N2A cells to identify the most effective Zfp189-targeting sgRNA. Cells were transfected and lysed after 48 hours and mRNA expression was analyzed with qPCR. The top sgRNA (protospacer sequence: GTGTCTCGGTTAGCAAGAAG) was packaged into HSV and tested in vivo.
In vivo confirmation of Zfp189 induction was validated via qPCR on dissected PFC tissue. In order to minimize between-mouse variability, a hemispheric approach was utilized wherein the test constructs were injected into one hemisphere while control constructs was injected into the other hemisphere of the same mouse.
Viral-mediated gene transfer
Stereotaxic surgeries targeting PFC were performed as previously described3,41. Mice were anesthetized with an intraperitoneal injection of ketamine (100 mg/kg) and xylazine (10 mg/kg) dissolved in sterile water. Subsequently, mice were placed in a small-animal stereotaxic device (Kopf Instruments) and the skull surface was exposed. 33-gauge needles (Hamilton) were utilized to infuse 0.5 μL of virus at a rate of 0.1 μL/minute followed by a five-minute rest period to prevent backflow. For CREBfl/fl and CRISPR experiments, 1 μL of virus at a rate of 0.2 μL/minute was utilized to maximize viral spread within the PFC. The following coordinates were utilized for PFC (from Bregma: anterior/posterior +1.8 mm, medial/lateral +0.75 mm, dorsal/ventral −2.7 mm; 15° angle). While PFC injections targeted the infralimbic cortex, virus spread sometimes extended beyond these anatomical boundaries to other PFC regions.
Immunohistochemistry
Mice were anesthetized with an intraperitoneal injection of ketamine (100 mg/kg) and xylazine (10 mg/kg) and transcardially perfused with a fixative solution containing 4% paraformaldehyde (PFA) (w/v) in 0.1 M Na2HPO4/ Na2HPO4, pH 7.5 at 4°C delivered at 20 ml/minute for 5 minutes with a peristaltic pump. Brains were post-fixed for 24 hours in 4% PFA at 4°C. Sections of 30 μm thickness were cut in the coronal plane with a vibratome (Leica, Nussloch, Germany) and stored at −20°C in a solution containing 30% ethylene glycol (v/v), 30% glycerol (v/v) and 0.1 M phosphate buffer. Free-floating sections were processed for immunohistochemistry as follows. On day 1, sections were rinsed three times for 10 minutes in PBS before permeabilization for 15 minutes in PBS containing 0.2% Triton X-100 (Fisher). Sections were then rinsed three times in PBS followed by a blocking step of 1 hour incubation in PBS containing 3% bovine serum albumin. Primary antibodies against GFP (Aves Lab, GFP-1020, Polyclonal: IgY Lot: GFP879484, 1:500 dilution), mCherry (Abcam, ab125096, Clone: 1C51, Lot: GR3201780–3, 1:500 dilution),CD34 (Abcam, ab81289, Clone: EP373Y, Lot: GR201207–37, dilution 1:250) and NeuN (Abcam, ab104224, Clone: 1B7, Lot: GR3215839–1, dilution 1:500) were diluted in blocking solution and sections incubated overnight at 4°C with gentle shaking. Sections were then washed three times in PBS and incubated with secondary antibodies (donkey anti-chicken Alexa Fluor 488, donkey anti-rabbit Cy3, donkey anti-mouse Alexa Fluor 594; Jackson ImmunoResearch, 1:500 dilution) for 2 hours at room temperature. After three rinses in PBS, sections were incubated for 5 minutes with DAPI (Sigma, D9542), washed again three times in PBS and then mounted in Vectashield (Vector Labs).
GFP and NeuN expression was assessed in the PFC using a LSM 710 laser-scanning confocal microscope (Carl Zeiss) imaged using a 10X, 20X or 40X oil immersion objective with a 1.0 digital zoom.
RNAscope
Fluorescent in situ hybridization (FISH) for Rbfox3 (NeuN) mRNA (#313311) and Zfp189 mRNA (#569561) was performed using the RNAscope® Fluorescent Multiplex 2.0 assay as per the manufacturer’s instructions (Advanced Cell Diagnostics, Hayward, CA, USA)42. Briefly, fresh whole mouse brains were embedded in OCT medium and quickly frozen in 2-methylbutane chilled to −80°C. 20 μm cryosections of PFC were than prepared and mounted on SuperFrost Plus slides. Sections were fixed and pre-treated according to the RNAscope® guide for fresh frozen tissue. After pre-treatment, sections were hybridized with FISH probes using the HybEZ Hybridization System. After several amplification sets, the sections were counterstained with DAPI and mounted using Prolong Gold. Reactive cells were analyzed bilaterally in the NAc. Confocal images were acquired on a LSM710 confocal microscope (Carl Zeiss) using a x40 or x63 oil immersion objective.
RNA isolation, qPCR, library preparation, and sequencing
Total RNA was isolated from frozen dissected PFC tissue using QIAzol lysis reagent and purified using the miRNAeasy mini kit (Qiagen). Following isolation, RNA to be utilized in qPCR was quantified by Nano Drop (Thermo Fisher) and converted to cDNA with iScript (Bio-Rad). qPCR samples were analyzed in triplicate using the standard ΔΔCT method. For non-CRISPR experiments, hypoxanthine phosphoribosyltransferase 1 (Hprt1) was utilized for normalization for both mice and humans, and for CRISPR experiments, gene expression was normalized to the geometric mean of Hprt1, dCas9, and GFP transcripts to account for both cell health and appropriate delivery of tool components. For RNA being utilized for RNA-seq, RNA integrity (RIN) was assayed using an Aligent 2100 Bioanalyzer (Aligent, Santa Clara CA). Average RIN values were above nine and samples with RIN values less than eight were excluded from analysis. Each sample consisted of PFC punches from the same animal with no pooling between animals. Not all mice within each experiment were processed for RNA-seq. Samples utilized for sequencing were determined in all cases by quality of the viral targeting and quality of the resulting RNA (as determined by RIN value). Additionally, in cases where the transcriptional effects of a particular behavior were being analyzed, samples that represented the group phenotype (overall effect) were selected for sequencing. For CRISPR experiments, only samples in which the effect of the CRISPR manipulation could be demonstrated were included in the analysis. RNA sequencing of the effects of Zfp189 expression included five samples per group (five HSV-Zfp189 and five HSV-GFP) for previously susceptible mice and 4–5 samples per group (five HSV-Zfp189 and four HSV-GFP) for unstressed controls. RNA sequencing of CRISPR-infected tissue included three samples per group (three dCas9-CREB(S133D) + NT-sgRNA and three dCas9-CREB(S133D) + Zfp189-sgRNA) for undefeated controls and 8–12 samples per group (12 dCas9-CREB(S133D) + NT-sgRNA and eight dCas9-CREB(S133D) + Zfp189-sgRNA) for defeated mice. Libraries were prepared using the TruSeq RNA Sample Prep Kit v2 (Illumina, San Diego CA). Briefly, mRNA was polyA selected from the total RNA pool. mRNA was then fragmented and converted to cDNA with reverse transcriptase followed by cDNA size selection and purification with AMPure XP beads (Beckman Coulter, Brea CA). To identify each sample, strand-specific adapters were ligated to adenylated 3’ ends and an additional size selection step was performed. The cDNA library was then amplified using polymerase chain reaction. During this step, barcodes of six base pairs were added to the adaptors. Library quality and concentration was measured by the Bioanalyzer before sequencing. Libraries were sequenced by either the Genomics Core Facility of the Icahn School of Medicine at Mount Sinai (Zfp189 overexpression) using an Illumina HiSeq 2500 System with v3 chemistry and 100 base pair single-end reads or GENEWIZ (CRISPR sequencing) using an Illumina HiSeq System with 150 base pair paired-end reads. Multiplexing was performed to ensure minimum reads of 20 million for each sample.
RNA-seq
Raw reads obtained from PFC were mapped to mm10 using HISAT243. SAM files were converted to BAM files and were sorted according to chromosome number using SAMTools44. Counts of reads mapped to genes were obtained using HTSeq-count45 against Ensembl v90 annotation. Differential expression analysis was carried out using the R package DESeq246. For virally infected tissue, sequenced samples in which the overexpressed transgene could not be adequately detected (Zfp189 for HSV-Zfp189 or dCas9, sgRNA, or Zfp189 for CRISPR studies) were considered to be a result of experimenter error (viral targeting or tissue selection) and were removed from analysis.
Identification of resilient-specific co-expression networks
In order to identify gene networks implicated in resilience, we utilized a previously published dataset from our group that reported weighted gene co-expression network analysis (WGCNA) modules and differentially expressed genes following CSDS3. Briefly, WGCNA modules and differential expression profiles were generated from RNA-seq data with tissue taken at 3 time-points after 10 days of CSDS. Mice were exposed to stress, phenotyped in the social interaction test, and the PFC, nucleus accumbens (Nac), ventral hippocampus (vHIP), and basolateral amygdala (BLA) were dissected to for RNA-seq. Differential expression comparisons were region-specific but brain regions were pooled prior to WGCNA. As such, reported WGCNA networks represent relationships of genes across brain regions.
Resilient modules presented herein are identical to the resilient modules presented in the supplementary material of Bagot et al.3 Also, DEGs from the supplementary material were utilized for enrichment analysis, and unprocessed data were utilized to generate module structures with additional analyses as necessary.
Our approach to identifying resilient-specific co-expression networks was predicated on our hypothesis that resilient-specific modules would be both unique to the resilient phenotype and transcriptionally active immediately following CSDS. As such, we first examined the 30 resilient modules for module differential connectivity (MDC)6. MDC is a measure of how connectedness amongst a set of genes is altered in the same genes in a different condition. This was performed as previously described3 and modules with FDR q < 0.05 are reported as significantly differentially connected. However, while previous studies from our group3,8 identified networks for functional validation based on MDC in a single condition, gene expression3,23 and circuit47 alterations in the resilient phenotype are distinct from that of susceptibility or control and therefore, only networks that showed MDC when compared to both phenotypes were considered resilient-specific.
Module enrichment analysis
In order to identify statistical overlap between previously identified differentially expressed genes (Gene Expression Omnibus database: GSE72343)3 and modules, we utilized the Super Exact Test (SET) R package48. The SET evaluates multi-set interactions to determine the difference between observed and expected overlap, which is then quantified statistically as an enrichment p-value and fold change. Expected overlap for two gene sets is dependent on the size of the gene sets and the number of total variables in the dataset (background number of genes). Higher than expected overlaps are indicated by larger fold change values, which represents the ratio of observed overlap to expected overlap. SET is advantageous in this setting as enrichment for upregulated and downregulated genes can be determined separately. Protein-protein interactions were determined using STRING version 10.549.
Human brain analysis
To identify whether our resilient mouse modules were preserved in human brain, we used the modulePreservation function of R package WGCNA to compare module identity in mouse to RNA sequencing data from human control and human MDD (GEO dataset: GSE102556)8. All brain regions included in the human analysis were included, and male and female subjects were combined. To evaluate the difference between preservation in human controls and human MDD, we compared module preservation p-values in each condition. A 10-fold change in p-value was considered to be a detectable deviation.
To evaluate CREB1 and ZNF189 in human brain, we used qPCR on reverse-transcribed mRNA from BA25, the PFC region most homologus to the ventral medial PFC targeted in our mouse studies. Samples from individuals with alcohol in their blood at the time of death were excluded due to possible effects on the transcriptome in PFC. Demographics of final cohort are shown in Supplementary Table 2. There were no significant differences in any demographic between MDD and control. Experiments were conducted in accordance with the guidelines of the Douglas Institute Research Ethics Board. However, no specific ethical approval or guidance was provided for analysis of existing post-mortem tissue.
Identification of the pink module network structure
We generated a network structure for the pink module using the Algorithm for the Reconstruction of Accurate Cellular Networks (ARACNE)10. Critically, however, ARACNE is not a directed analysis, so connections between genes are still based on correlations. In order to attempt to resolve regulatory genes within the pink module, we next performed Key Driver Analysis (KDA)11 on the ARACNE reconstructed network. KDA is predicated on the understanding that more important regulatory genes will have a larger effect on other genes in the module. Therefore, more important genes should forge more direct connections in the final module structure than less important genes. We analyzed the pink module structure at a threshold of 2 layers to identify the most important regulatory genes. Key driver genes had a number of connections significantly above the average value for the network and were considered for further in vivo analysis.
Identification of the relationship between Zfp189 and the pink module
To establish the relationship between Zfp189 and the WGCNA modules, we first obtained gene expression from DESeq2’s regularized log-transformation. Each gene’s expression was then standardized to have zero mean and unit standard deviation across all samples. PCA was performed on each module (excluding Zfp189) to obtain the first principal component after weighing as the “eigen-gene” for each module. The Pearson’s correlation analysis was then performed between Zfp189 and each module’s eigen-gene to get the R2 score.
Determination of pink module upstream regulators
To probe the pink module for specific regulatory binding sites, we utilized HOMER motif analysis16. HOMER examines binding sites within a gene set to see if a specific binding motif is significantly enriched compared to what would be expected in a background gene set (in our case, the entire mouse genome). HOMER motif analysis was performed on all resilient modules. As such, we utilized a FDR cutoff of p < 0.05 for significant upstream regulators. To reduce the detection of false positives, we limited our candidates to known binding motifs.
To complement motif analysis, we performed upstream regulator analysis using the upstream regulator tool in QIAGEN’s Ingenuity® Pathway Analysis (QIAGEN Redwood City, www.qiagen.com/ingenuity) (IPA). This function predicts the identity and direction of change of known upstream regulators for a given differential expression signature from the magnitude and scale of gene expression changes in a dataset. Predictions used in this study were based on experimentally observed interactions within all datasets in IPA with the stringent filter setting applied. Reported p-value calculations were determined from the Ingenuity Knowledge Base reference set considering both direct and indirect relationships. Any regulator for which there was sufficient evidence to generate an activation/inhibition prediction is reported. Input to IPA was prepared from transcriptional changes determined by RNA sequencing in 48 hours after CSDS (Gene Expression Omnibus database: GSE72343)3. Data were filtered for protein-coding genes. Three separate comparisons (susceptible vs. resilient, resilient vs. control, and susceptible vs. control) were utilized and fold change values for all pink module genes were included as input.
Evaluation of overlap between RNA-seq and resilient modules
To test the overlap between RNA-seq differential list and the resilient modules, we used multinomial logistic regression to predict the module membership using gene expression changes as regressors. The gene expression changes are log fold changes (LFCs) from the differential analysis. To control for noise, we combined p-values with LFCs by forcing them to zero when the p-value is less than 0.05 to derive a so-called p-value adjusted LFC (PLFC). To control for covariates, such as gene length, GC content, etc., that may affect the differential analysis, we included the log-transformed basal gene expression (LBGE) as a covariate in the logistic regression. The LBGE is standardized to have zero mean and the same standard deviation as PLFC to facilitate optimization. In multinomial logistic regression, we used the turquoise module as the reference since it is the largest module and does not show overlap with the differential list. The coefficient of the PLFC from the regression analysis can be interpreted as the significance of the overlap while the coefficient of the LBGE indicates the bias of the covariates.
Gene ontology
Gene Ontology for Biological Pathways (GOBP) was determined in EnrichR with gene identities of differentially expressed genes50.
Statistics
Statistics were performed in Prism versions 5.0 and 8.0 for Mac (GraphPad Software, La Jolla CA) and SPSS Statistics version 22 (IBM Corp, Armonk NY). No statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those reported in previous publications2,3. Unless otherwise stated, measurements were taken from distinct samples. For all behavioral analyses, outlier detection was performed using a Grubbs test with an alpha value of 0.05 and statistical outliers were excluded from analysis. Because there is no non-parametric equivalent for repeated measures tests, in accordance with the literature all SI and NSF data were analyzed using a mixed model analysis of variance (ANOVA) where one of the factors (i.e., target present, target absent) was treated as a repeated measure. A three-factor analysis model was employed when there were either two behavioral conditions (control vs. stress or pre-test vs. post-test) or two viruses (HSV and AAV) in addition to the repeated measure. In other cases, a two-factor analysis model was employed. For all tests other than SI and NSF, Levene’s test of variance was first utilized to ensure that the data met the assumptions necessary for parametric statistics. In cases where data met the assumptions necessary for parametric statistics, data from two groups were analyzed using a two-tailed students t-test and data from three or more groups were analyzed with a one-way or two-way ANOVA. When data were from the same animal, paired or repeated measures were utilized. In cases where the data did not meet assumptions for parametric statistics, data from two groups were analyzed using an independent samples Mann-Whitney and data from three or more groups were analyzed with a Kruskal-Wallis test. In these cases different groups were compared by independent samples Mann-Whitney tests as post-tests. For parametric statistics, Bonferroni tests were used as post-tests. For comparisons of two continuous variables, we used linear regression analysis, in which case the coefficient of determination (R2) is reported. To compare linear regressions, we used analysis of covariance (ANCOVA). Data that survives multiple comparisons correction have been indicated, and analyses presented with correction are specified in the text with “FDR”. Additional information can be found in the Life Sciences Reporting Summary.
Data availability
RNA-seq data reported in the paper is deposited in GEO with the accession number: GSE118317. Other data that support the findings of this study are available from the corresponding author upon request.
Code availability
Scripts and code utilized in the analysis of study data are available from the corresponding author upon request.
Supplementary Material
Supplementary Table 1. Pink module genes and key drivers.
List of pink module genes and status as a key driver from KDA (key driver analysis). Key driver genes are ranked by number of connections two layers deep in network structure, which is included.
Supplementary Table 2. Human cohort demographics.
Gender, ethnicity, history of abuse, cause of death, age, post-mortem interval (PMI), brain pH, and presence of antidepressants, alcohol, and drugs of abuse in blood of human samples used in analysis.
Supplementary Table 3. Upstream motifs of module genes.
HOMER-defined upstream regulators for genes in the pink, blue, gold, red, and turquoise modules, which are the only modules with significant (FDR q < 0.05) upstream regulators. Included are the probed DNA sequence, number and percent of in-module genes containing sequence, number and percent of background genes containing sequence, and significance of overlap.
Supplementary Table 4. IPA associations of CREB and pink module genes.
IPA-annotated associations between CREB and genes within the pink module used to designate CREB as an upstream regulator.
Supplementary Table 5. Zfp189-targeting sgRNA predicted binding sites throughout Mus musculus genome.
Except for at the targeted Zfp189 locus, there are no high-affinity binding sites for our Zfp189-targeting sgRNA across the entire mouse genome. Of the 141 predicted off-targets, the top sites with “scores”>0.3 are represented here. “Score” ranges from 0–100 and denotes the predicted on-target faithfulness of our synthesized sgRNA acting at the mismatched locus. The closest “matches” exhibit >3 mismatched nucleotides.
Supplementary Table 6. Non-targeting sgRNA protospacer predicted binding sites throughout Mus musculus genome.
There are no high-affinity binding sites for our NT-sgRNA across the entire mouse genome. All of the 15 total low-probability sites and “scores” are presented above. “Score” ranges from 0–100 and denotes the predicted on-target faithfulness of our synthesized sgRNA acting at the mismatched locus.
Acknowledgments
This work was supported by National Institutes of Health grants F30MH110073 (Z.S.L.), T32GM007280 (Z.S.L.), T32MH096678 (Z.S.L.), K99DA045795 (P.J.H), U01AG046170 (B.Z.), R01AG057907 (B.Z.), RF1AG057440 (B.Z.), P50MH096890, and R01MH051399 (E.J.N.) and the Hope for Depression Research Foundation.
Footnotes
Competing Interests Statement
All authors declare no competing interests.
Accession codes
RNA-seq data reported in the paper is deposited in GEO with the accession number: GSE118317.
References
- 1.Berton O et al. Essential role of BDNF in the mesolimbic dopamine pathway in social defeat stress. Science 311, 864–868 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Krishnan V et al. Molecular Adaptations Underlying Susceptibility and Resistance to Social Defeat in Brain Reward Regions. Cell 131, 391–404 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Bagot RC et al. Circuit-wide Transcriptional Profiling Reveals Brain Region-Specific Gene Networks Regulating Depression Susceptibility. Neuron 90, 969–983 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maschietto M et al. Co-expression network of neural-differentiation genes shows specific pattern in schizophrenia. BMC Med. Genomics 8, 23 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yue Z et al. Repositioning drugs by targeting network modules: a Parkinson’s disease case study. BMC Bioinformatics 18, 532 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang B et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell 153, 707–720 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parikshak NN et al. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell 155, 1008–1021 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Labonte B et al. Sex-specific transcriptional signatures in human depression. Nat. Med 23, 1102–1111 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malki K et al. Identification of genes and gene pathways associated with major depressive disorder by integrative brain analysis of rat and human prefrontal cortex transcriptomes. Transl. Psychiatry 5, e519 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Margolin AA et al. ARACNE: An Algorithm for the Reconstruction of Gene Regulatory Networks in a Mammalian Cellular Context. BMC Bioinformatics 7, S7 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang B & Zhu J Identification of Key Causal Regulators in Gene Networks. Proc. World Congr. Eng II, 5–8 (2013). [Google Scholar]
- 12.Covington HE 3rd et al. Antidepressant effect of optogenetic stimulation of the medial prefrontal cortex. J. Neurosci 30, 16082–16090 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Odeberg J et al. Cloning and characterization of ZNF189, a novel human Kruppel-like zinc finger gene localized to chromosome 9q22-q31. Genomics 50, 213–221 (1998). [DOI] [PubMed] [Google Scholar]
- 14.Najafabadi HS et al. C2H2 zinc finger proteins greatly expand the human regulatory lexicon. Nat. Biotechnol 33, 555–562 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Zeisel A et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347, 1138–1142 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Heinz S et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bleckmann SC et al. Activating transcription factor 1 and CREB are important for cell survival during early mouse development. Mol. Cell. Biol 22, 1919–1925 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shaywitz AJ & Greenberg ME CREB: A Stimulus-Induced Transcription Factor Activated by A Diverse Array of Extracellular Signals. Annu. Rev. Biochem 68, 821–861 (1999). [DOI] [PubMed] [Google Scholar]
- 19.Xiao X et al. The cAMP responsive element-binding (CREB)-1 gene increases risk of major psychiatric disorders. Mol. Psychiatry (2017). doi: 10.1038/mp.2017.243 [DOI] [PubMed] [Google Scholar]
- 20.Juhasz G et al. The CREB1-BDNF-NTRK2 pathway in depression: multiple gene-cognition-environment interactions. Biol. Psychiatry 69, 762–771 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Carlezon WAJ, Duman RS & Nestler EJ The many faces of CREB. Trends Neurosci 28, 436–445 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Covington HE 3rd et al. A role for repressive histone methylation in cocaine-induced vulnerability to stress. Neuron 71, 656–670 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilkinson MB et al. Imipramine treatment and resiliency exhibit similar chromatin regulation in the mouse nucleus accumbens in depression models. J. Neurosci 29, 7820–7832 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen AC, Shirayama Y, Shin KH, Neve RL & Duman RS Expression of the cAMP response element binding protein (CREB) in hippocampus produces an antidepressant effect. Biol. Psychiatry 49, 753–762 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Green TA et al. Induction of activating transcription factors (ATFs) ATF2, ATF3, and ATF4 in the nucleus accumbens and their regulation of emotional behavior. J. Neurosci 28, 2025–2032 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hodes GE et al. Sex Differences in Nucleus Accumbens Transcriptome Profiles Associated with Susceptibility versus Resilience to Subchronic Variable Stress. J. Neurosci 35, 16362–76 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LaPlant Q et al. Role of nuclear factor kappaB in ovarian hormone-mediated stress hypersensitivity in female mice. Biol. Psychiatry 65, 874–80 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gray AL, Hyde TM, Deep-Soboslay A, Kleinman JE & Sodhi MS Sex differences in glutamate receptor gene expression in major depression and suicide. Mol Psychiatry (2015). [DOI] [PubMed] [Google Scholar]
- 29.Lorsch ZS et al. Estrogen receptor α drives pro-resilient transcription in mouse models of depression. Nat. Commun 9, 1116 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vialou V et al. DeltaFosB in brain reward circuits mediates resilience to stress and antidepressant responses. Nat. Neurosci 13, 745–752 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dias C et al. β-catenin mediates stress resilience through Dicer1/microRNA regulation. Nature 516, 51–55 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y et al. Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron 89, 37–53 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Menard C et al. Social stress induces neurovascular pathology promoting depression. Nat. Neurosci 20, 1752–1760 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heller E a et al. Locus-specific epigenetic remodeling controls addiction- and depression-related behaviors. Nat. Neurosci 17, 1720–1727 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamilton PJ et al. Cell-Type-Specific Epigenetic Editing at the Fosb Gene Controls Susceptibility to Social Defeat Stress. Neuropsychopharmacology 43, 272–284 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heller EA et al. Targeted Epigenetic Remodeling of the Cdk5 Gene in Nucleus Accumbens Regulates Cocaine- and Stress-Evoked Behavior. J. Neurosci 36, 4690–4697 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu XS et al. Rescue of Fragile X Syndrome Neurons by DNA Methylation Editing of the FMR1 Gene. Cell 172, 979–992.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu XS et al. Editing DNA Methylation in the Mammalian Genome. Cell 167, 233–247.e17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only references
- 39.Neve RL, Neve KA, Nestler EJ & Carlezon WAJ Use of herpes virus amplicon vectors to study brain disorders. Biotechniques 39, 381–391 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Hsu PD et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol 31, 827–832 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hamilton PJ, Lim CJ, Nestler EJ & Heller EA Viral Expression of Epigenome Editing Tools in Rodent Brain Using Stereotaxic Surgery Techniques. Methods Mol. Biol 1767, 205–214 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang F et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn 14, 22–29 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim D, Langmead B & Salzberg SL HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anders S, Pyl PT & Huber W HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Friedman AK et al. Enhancing depression mechanisms in midbrain dopamine neurons achieves homeostatic resilience. Science 344, 313–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang M, Zhao Y & Zhang B Efficient Test and Visualization of Multi-Set Intersections. Sci. Rep 5, 16923 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Szklarczyk D et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 45, D362–D368 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen EY et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Pink module genes and key drivers.
List of pink module genes and status as a key driver from KDA (key driver analysis). Key driver genes are ranked by number of connections two layers deep in network structure, which is included.
Supplementary Table 2. Human cohort demographics.
Gender, ethnicity, history of abuse, cause of death, age, post-mortem interval (PMI), brain pH, and presence of antidepressants, alcohol, and drugs of abuse in blood of human samples used in analysis.
Supplementary Table 3. Upstream motifs of module genes.
HOMER-defined upstream regulators for genes in the pink, blue, gold, red, and turquoise modules, which are the only modules with significant (FDR q < 0.05) upstream regulators. Included are the probed DNA sequence, number and percent of in-module genes containing sequence, number and percent of background genes containing sequence, and significance of overlap.
Supplementary Table 4. IPA associations of CREB and pink module genes.
IPA-annotated associations between CREB and genes within the pink module used to designate CREB as an upstream regulator.
Supplementary Table 5. Zfp189-targeting sgRNA predicted binding sites throughout Mus musculus genome.
Except for at the targeted Zfp189 locus, there are no high-affinity binding sites for our Zfp189-targeting sgRNA across the entire mouse genome. Of the 141 predicted off-targets, the top sites with “scores”>0.3 are represented here. “Score” ranges from 0–100 and denotes the predicted on-target faithfulness of our synthesized sgRNA acting at the mismatched locus. The closest “matches” exhibit >3 mismatched nucleotides.
Supplementary Table 6. Non-targeting sgRNA protospacer predicted binding sites throughout Mus musculus genome.
There are no high-affinity binding sites for our NT-sgRNA across the entire mouse genome. All of the 15 total low-probability sites and “scores” are presented above. “Score” ranges from 0–100 and denotes the predicted on-target faithfulness of our synthesized sgRNA acting at the mismatched locus.
Data Availability Statement
RNA-seq data reported in the paper is deposited in GEO with the accession number: GSE118317. Other data that support the findings of this study are available from the corresponding author upon request.