Abstract
Single cell RNA-sequencing (scRNA-seq) allows the measurement of transcriptomes from individual cells providing new insights into complex biological systems. scRNA-seq has enabled the identification of rare cell types, new cell states and intercellular communication networks that may be masked by traditional bulk transcriptional profiling. Researchers are increasingly using scRNA-seq to comprehensively characterize complex organs in health and disease. The diversity of immune cell types, some present at low frequency, in a transplanted organ undergoing rejection makes scRNA-seq ideally suited to characterize transplant pathologies because it can quantify subtle transcriptional differences between rare cell types. In this review we discuss single cell sequencing methods and their application in transplantation to date, current challenges and future directions. We believe that the remarkably rapid pace of technological development in this field makes it likely that single cell technologies such as scRNA-seq will have an impact in clinical transplantation within a decade.
Single cell RNA sequencing (scRNA-seq) technologies have developed rapidly since the the initial publication in 2009.1 Multicellular organisms are composed of individual cells capable of varied transcriptional output finely balanced to benefit the organism in health or to respond to injurious stimuli. scRNA-seq has provided a new means to comprehensively catalog the transcriptional landscape of each cell in a complex organism or tissue. The protocol itself follows several steps: Isolation of single cells, cell lysis, mRNA capture, reverse transcription, amplification, library generation and next-generation sequencing. Early ‘plate-based’ methods used manual handling to separate single cells into individual wells of a 96-well plate. A key innovation was the inclusion of a unique cellular barcode – a molecular tag composed of a random sequence of nucleotides - in the primers used to capture mRNA – used to assign the cell that the read (or mRNA) came from. Once a library unique to each cell was generated samples are multiplexed and next generation sequencing performed. More recent droplet based methods leverage microfluidic technologies to dramatically increase throughput. In this case, an individual cell is captured within a 2 nanoliter droplet that also contains lysis buffer and mRNA capture oligos with the unique barcode for that cell. These advances now allow researchers to generate 10 – 100,000 single cell transcriptomes in 1–3 days. Here we discuss the benefits and limitations of scRNA-seq to study complex diseases such as transplant rejection. We provide a review of published scRNA-seq studies from all commonly transplanted human solid organs. Much of the current published data comes from explanted or post mortem tissue samples with the largest datasets coming from kidney. Studies using biopsy tissue from functioning solid organs are limited.
The role of molecular genetics in transplantation
Bulk transcriptome analysis has historically been used by many groups in transplantation to investigate the patterns of gene expression occurring in dysfunctioning organs or in protocol biopsies.2–5 By comparing panels of genes differentially expressed in certain allograft pathologies, such as antibody mediated rejection (AMR), one can define classifier genes (gene sets agnostically defined by machine learning that can predict or ‘classify’ an outcome of interest) for each allograft pathology that form the basis for diagnostics. The Molecular Microscope uses this bulk transcriptome analysis approach and is being developed to aid in diagnosis of allograft biopsy pathology.6 Data from the hundreds of biopsies examined using this technology has been used to better understand complex allograft pathologies including AMR. This represents an important advance and a welcome addition to the clinicians tool box for the diagnosis of rejection in transplantation. AMR is the most common primary cause of late kidney allograft failure.7 This form of rejection is a relatively recent sub classification of rejection and intra-observer accuracy amongst nephropathologists is poor for key histologic lesions such as acute glomerulitis.8 Data suggests that the Molecular Microscope better predicts poor outcome known to be associated with the AMR diagnosis when compared to local histopathologic diagnosis and a positive AMR score on the Molecular Microscope has a PPV and NPV of 50% and 94%, respectively.9
Similar approaches using panels of genes sequenced from peripheral blood samples have been used to predict or diagnose rejection in kidney transplant recipients. Using microarrays the Salomon group identified 200 probe sets by multiple 3-way classifier tools that discriminated for acute rejection.10 Sarwal et al determined the expression of a predefined set of 43 genes by quantitative real-time PCR using the large cohort from the Assessment of Acute Rejection in Renal Transplantation (AART) study. A 17 gene panel set (now called kSORT) was able to predict acute rejection 3 months prior to detection by biopsy.11 Nanostring is another new technology that directly measures the number single RNA molecules in a sample using a light based capture and reporter probe system. All of these methods measure an averaged gene expression across the sample in question. Therefore cell to cell variation in gene expression is lost.
Microarray datasets from biopsy tissue have been leveraged to infer disease mechanism.12–14 For example in AMR, top differentially expressed genes, or pathogenesis based transcripts, associated with the presence of donor specific antibodies (DSA) were assigned a cell of origin. The assigned origin of these transcripts were endothelial and NK cells, and a histologic diagnosis of AMR was associated with high expression of these transcripts.15 Another approach to understanding the cellular context of AMR is to examine the microarray expression of endothelial associated genes identified from the literature in study biopsies. High endothelial gene expression in biopsies associated with DSA was predictive of AMR.16
Some caution is necessary in imputing cell specific gene expression derived from a bulk transcriptional analysis. Our recent work showed that many transcripts that had been assigned to endothelial cells were in fact expressed by other cell types.17 This a priori approach to assigning cell origin for important disease related genes may be flawed as cell origin defined by in vitro methods may not be relevant in the diseased human allograft. The ideal approach to determining the source of important rejection associated genes would be to sequence the transcriptome of each individual cell in the sample. scRNA-seq allows for this – it identifies cell types in an unbiased manner without the need for predefined cell markers or assumptions. Computational approaches are used to visualize dynamic cellular processes such as disease progression, alterations in cell state and cellular differentiation.
Single cell methods and analysis
Massively parallel scRNA-seq is typically accomplished with droplet-based microfluidic methods such as DropSeq,18 InDrops19 or the commercial 10X Chromium system. The pros and cons of each system have been reviewed.20 A comparison of droplet based microfluidic scRNA-seq methods is summarized in Table 1. After cell encapsulation, mRNA is bound by oligo-dT, reverse transcribed and amplified. A unique oligonucleotide barcode is associated with each cell, and incorporated into each amplified transcript. Resulting cDNA libraries are sequenced using next generation sequencing. An alternative to scRNA-seq is single nucleus RNA-seq (snRNA-seq). This involves dissociating tissues such that intact nuclei are liberated into solution. Processing single nuclei using droplet-based methods results in the capture of unprocessed mRNA molecules that still contain introns. How this method compares to scRNA-seq is discussed below. The volume of raw sequence data resulting from a single experiment of modest size is considerable – approximately 20 gigabytes. As a consequence, some familiarity with coding and informatics is necessary for the proper interpretation of scRNA-seq data. Fortunately, an increasing variety of programs are simplifying this process which lowers the bar of entry for laboratories without coding experience.
Table 1.
A comparison of current high throughput droplet based single cell RNA-seq methods
| DropSeq | InDrops | 10X Chromium | |
|---|---|---|---|
| Year developed | 2015 | 2015 | 2016 |
| Commercial platform | No | 1CellBio | 10X Genomics |
| Full length sequence | No | No | No |
| 3’ or 5’ sequence | 3’ | 3’ | both |
| Capture efficiency | 12.8% | 90% | 65% |
|
Sensitivity (molecule detection limit) |
1e+1 | 1e+0.5 | 1e+2.5 |
| Accuracy (Pearson R) | >0.9 | >0.9 | >0.9 |
| Cell capacity per run | 10,000 | 10,000 | 80,000 |
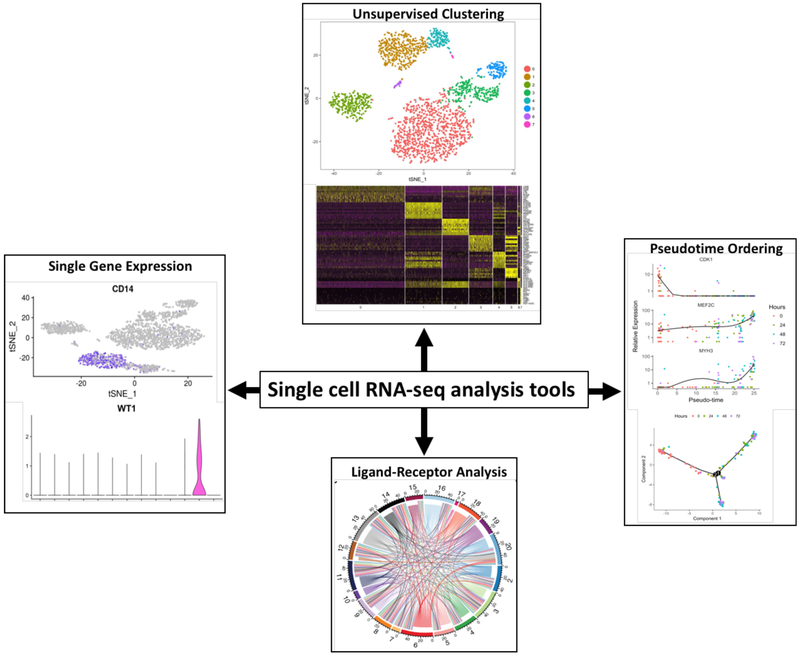
Although beyond the scope of this review, a variety of machine learning techniques allow for analysis and data visualization (Figure 1). Pseudotemporal ordering of cells is a method based on the hypothesis that a single cell experiment is a time-course experiment with each cell representing one point in time along a continuum.21 This allows for the examination of continuous biological processes such as disease progression. Branch point analysis stems from this hypothesis and can reveal important genes that determine cell fate or trajectory. Ligand-receptor analysis is another tool that is useful when applied to a rejecting allograft tissue sample given the complex immune interactions occurring between donor cells and immune cells during the alloimmune response. For example, the Jackson Laboratory demonstrated important ligand-receptor pairs in their dataset.22 Endothelial cell interactions with other cell types featured prominently in their analysis, and they demonstrate pericyte-endothelial cell interaction through PDGFB and PDGRFRB expression in endothelial cells and pericytes, respectively. Other intercellular communication networks included VEGFA and FLT1/KDR expression in podocytes and glomerular endothelial cells, and SLIT2 and ROBO4 expression in intercalated cells and endothelial cells, respectively. There are a myriad statistical and graphing packages available to present single cell data. Most groups in this field use the R programming language to run these packages but there are many more languages that can be used such as Python and Matlab.
Figure 1. An example of single cell RNA-seq data analysis tools.
Unsupervised clustering of data and visualization using t-stochastic nearest neighbor embedding (tSNE) or heatmaps. Single gene expression using the Seurat FeaturePlot and VlnPlot functions.40 Pseudotime ordering using the Monocle.21 Ligand-receptor interactions visualized using Circos plot.41
Organ specific single cell studies
Single cell RNA-seq is now a foundational technique in a world-wide consortium that aims to generate a comprehensive human cell atlas (http://www.humancellatlas.org) describing every cell type in the human body in molecular terms. This map of all human cells will serve as a basis for understanding human health and disease. Groups have focused on defining the cellular and transcriptional heterogeneity of different organs in health, disease and during development. There are currently few scRNA-seq studies of normal or diseased human organs. Three groups have published single cell data from normal human kidney tissue. Young et al sequenced the transcriptomes from 42,747 cells from normal adult human nephrectomy samples.23 The 10X Chromium single cell 3’ kit (version 2 chemistry) was used in this study and aimed to capture 5000 cells/chip position. Proximal tubule cells made up 77% of the cells which is consistent with other studies.22,24 A subset of 4796 immune cells included 2 NK cell clusters, 2 NKT cell clusters, 3 mononuclear phagocyte clusters, B cells, CD8 T cells, helper T cells, mast cells, neutrophils, dendritic cells (DC) and 6 other unnamed clusters. This dataset demonstrates the immune cell heterogeneity in the normal human kidney and these immune cells represent 11% of all cells from the normal tissue samples. We have additionally reported 5524 adult human kidney nucleus transcriptomes, identifying 17 distinct cell types including podocytes, mesangium, proximal tubule cells (S1–S3), loop of Henle cells (descending and ascending), distal tubule cells, connecting segment cells, principal cells, and intercalated cells (type A and type B) and macrophages.25
Given the important role of immune cells in the alloimmune response, knowledge of the immune cell composition in normal kidney is a prerequisite to understanding the alloimmune response during rejection. Another important cell type central to antibody mediated rejection is the endothelial cell. The Jackson Laboratory group sequenced 22,469 individual transcriptomes from normal human kidney, revealing 27 different cell types.22 In their report 65% of the immune cells were lymphocytes (B cells, T cells and NKT cells) and they assumed these cells were from the peripheral circulation. They also found two CD68 positive clusters of myeloid cells differentiated by the expression of CD52. They assumed that CD52+ cells were from the peripheral circulation and that only CD52 negative cells can be tissue resident.26 CD68 positive CD52 negative cells in their dataset subclustered into classical and non classical monocytes in addition to interstitial macrophages and DCs. A cluster of 20 mast cells was also identified. The same study also identified 3 clusters of endothelial cells representing descending vasa recta capillary endothelium, ascending vasa recta capillary endothelium and afferent/efferent arteriolar endothelial cells. In contrast, the Young et al paper describes 4 endothelial cell clusters, glomerular, descending vasa recta and 2 ascending vasa recta clusters. The variation in the number of immune and endothelial cell subtypes found between these studies is likely due to biological and computational factors. For example, clustering analysis of the Young et al dataset was done after removal of all immune cells and proximal tubular cells which can affect cell clustering by increasing the number of cell subtypes defined using the same machine learning parameters. The Young et al dataset also included a greater number of total cells and source samples.
Single cell studies from other commonly transplanted solid organs is more limited. Reyfman et al sequenced 48,937 transcriptomes using droplet based microfluidics from 12 human lung specimens 6 of which were donor lung.27 They corrected for batch effects using canonical correlation analysis to compare the single cell landscape in normal human lung with diseased lung (interstitial lung disease). Thirteen cell clusters were identified including all the major immune cell types, T cells, NK cells, monocytes/macrophages, B cells, plasma cells, dendritic cells and mast cells. See et al studied cardiac myocytes from healthy adult human heart using the Fluidigm C1 single cell Auto Prep System.28 Single nuclei were studied in this case as source tissue was snap frozen from samples taken historically. The authors do not report the number of nuclei studied in this report. However, one can assume the number was low as the Fluidigm system can only capture a maximum of 800 cells or nuclei per chip. Finally, scRNA-seq has been studied in liver donor samples. MacParland et al studied 8444 cells from the caudate lobes of 5 livers procured and subsequently transplanted.29 They identified 20 discrete cell populations of hepatocytes, endothelial cells, cholangiocytes, hepatic stellate cells, B cells, conventional and non conventional T cells, NK-like cells, and distinct intrahepatic monocyte/macrophage populations. Their analysis included cells with up to 50% mitochondrial gene expression per cell which is higher than the standard 20% cut off used by other groups. This suggests significant technical artifact and limits the number of other genes sequenced and available to define cell clusters, reducing sensitivity to identify new cell types or subtypes.
Tissue dissociation: A major challenge in scRNA-seq
A major challenge to the study of solid organs using scRNA-seq is tissue dissociation. The most common methods for dissociating cells include a combination of mechanical and warm enzymatic digestion to create a single cell suspension. There are three major limitations with this approach. Proteolytic digestion at 37 C induces transcriptional stress responses that confound analysis.30 It also causes dissociation bias – because some cells are easily released from the tissue and resistant to cell death (for example, leukocytes), whereas others are fragile or difficult to release, and may be entirely absent from the analysis (for example, podocytes). Finally, single cell dissociation is incompatible with archival, frozen tissue.
We have determined that snRNA-seq solves many of these limitations. In head to head comparison of adult mouse kidney, we compared scRNA-seq data generated using DropSeq with snRNA-seq data generated from nuclei using sNuc-DropSeq, DroNc-seq and 10X Chromium.31 The scRNA-seq dataset identified 12 cell types but glomerular cell types were absent and one cluster expressed artifactual dissociation-induced stress response genes. By contrast, snRNA-seq from all three platforms captured a diversity of kidney cell types including 20-fold more podocytes as well as mesangial cells and endothelial cells that were not represented in the single cell dataset. No stress response genes were detected since the entire nucleus dissociation protocol is carried out on ice. Even though the nucleus contains much less mRNA than the whole cell, gene detection sensitivity was equivalent between cell and nucleus platforms. We could also generate snRNA-seq from snap frozen kidney which allows investigators to perform snRNA-seq of biobanked tissue. Our analysis revealed that single nucleus RNA-seq of adult kidney offers reduced dissociation bias, can be successfully performed on frozen samples including inflamed and fibrotic tissue, eliminates dissociation-induced transcriptional stress responses and has comparable gene detection compared to scRNA-seq. These results will allow for the banking of allograft specimens for future snRNA-seq analysis.
scRNA-seq in transplantation.
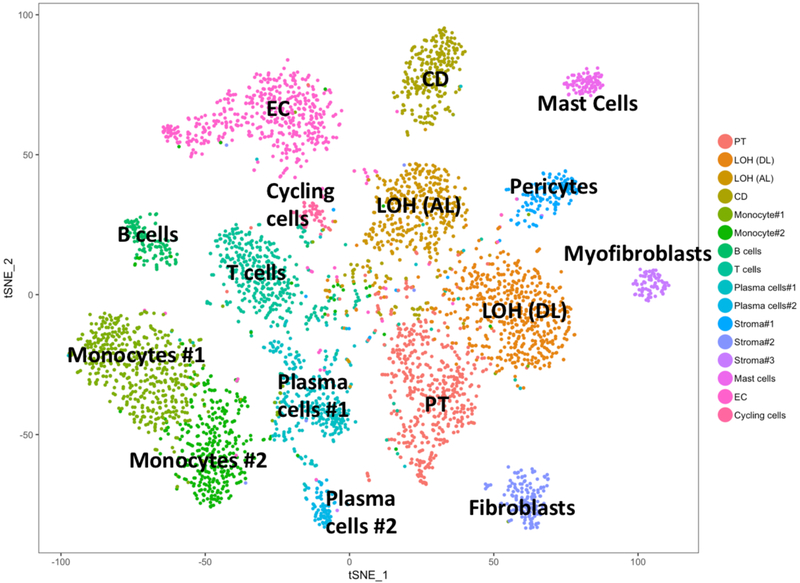
Currently there is limited published scRNA-seq data in the solid organ transplant field. We published the first report of scRNA-seq of a kidney transplant undergoing mixed rejection.17 This study sequenced cells from a single 16G core taken at the time of diagnostic biopsy. The InDrops method was used and the libraries generated were sequenced to a depth of ~50,000 mapped reads per cell. A total of 4487 cells passed quality filters and on average 1481 transcripts from 827 different genes were detected per cell. Unsupervised clustering analysis using t-stochastic nearest neighbor embedding (tSNE) identified 16 cell clusters. These included 3 tubular cell types, 3 leukocyte populations, 4 lymphocyte cells types, 3 stromal cell types, endothelial cell types and cycling cells (Figure 2). Readers can query this dataset and others directly at http://humphreyslab.com/SingleCell/. All the major immune cell groups were represented in this dataset as well as most donor kidney cell types. Glomerular cell types were not identified.
Figure 2. tSNE of 4487 cells from a human kidney transplant biopsy with mixed rejection.
Analysis of 4487 cells from a human kidney transplant with mixed rejection using tSNE revealed 16 cell clusters. These included 3 tubular cell types, 3 leukocyte populations, 4 lymphocyte cells types, 3 stromal cell types, endothelial cell types and cycling cells.
Our analysis allowed several interesting conclusions to be made. Firstly, two different monocyte clusters were identified by differential gene expression between clustered cell types (FCN1-positive cells versus CD16-positive) and were confirmed by immunohistochemistry staining of mixed rejection tissue samples. Staining was sparse or absent in normal kidney and intermediate in AMR biopsy tissue. Secondly, a subclustering analysis defined 3 separate endothelial cell types: resting, angiogenic and activated. Gene expression from the resting subcluster correlates with external scRNA-seq datasets of normal human endothelium in brain and pancreas. This paper also confirms the importance of the endothelium in AMR. Most of the top genes found in association with AMR from microarray studies are expressed uniquely in the endothelium. However, genes described as endothelial associated in previous microarray studies were mostly expressed in non endothelial cells in this single cell dataset. The genes expressed in microarray studies were assumed to come from endothelial cells based on gene expression studies of human endothelial cells in vitro.32,33 This highlights a major limitation of the microarray based approach to the investigating of allograft pathologies. It must be noted that the studies by Wu et al are based on a single human kidney transplant biopsy. These findings are still to be confirmed in other biopsies and by other groups.
There are limited studies of disease in non renal solid organ transplants. In lung transplantation Ferguson et al used the 10X Genomics single cell method to study an explanted lung allograft with chronic lung allograft dysfuntion.34 This report was in abstract form and no details regarding the scRNA-seq data were presented. Habal and colleagues performed a limited single cell study of cardiac chronic allograft vasculopathy.35 They digested coronary artery from an explanted cardiac allograft and isolated 81 T cells using color flow sorting for T cell markers. As tissue preservation and dissociation methods improve we are likely to see more scRNA-seq studies across all solid organ transplantation.
We foresee numerous applications of scRNA-seq to transplantation research. The ability to demonstrate receptor-ligand expression between different cell types within a sample has been demonstrated as discussed above. Such an analysis in rejecting allograft tissue could lead to increases in our understanding of the complex interactions between immune cells and donor cells in transplant rejection. Wu et al demonstrate stromal cell (pericytes, fibroblasts and myofibroblasts) expression of the chemokine CXCL12 which promotes lymphocyte and monocyte chemotaxis through its cognate receptor CXCR4, expressed in T cells, monocytes and mast cells in their rejecting biopsy sample.17 Collecting duct epithelial cells expressed KITLG (stem cell factor) the product of which binds to the receptor encoded by KIT, which was expressed in mast cells. This is further evidence for the complex interactions occurring in the rejecting allograft.
Biomarker discovery is another area facilitated by scRNA-seq studies in transplantation. Recent advances have seen the introduction of tests designed to diagnose allograft rejection with better sensitivity and specificity than the current gold standards, histology, DSA screening, proteinuria and creatinine. A number of non invasive molecular biomarkers have been proposed with varying negative (NPV) and positive predictive values (PPV), important parameters to consider in the assessment of biomarker suitability. Panels of various urine and blood mRNAs, miRNAs and proteins have been considered as biomarkers in transplantation. A number of such tests are used in clinical practice. AlloMap is a peripheral blood test that uses a panel of 11 genes to predict T cell mediated rejection in heart transplantation.36 The kSORT test panel of 17 genes from peripheral blood is used to predict acute rejection in kidney transplant patients.11
Allosure is a peripheral blood test for predicting rejection in kidney transplant patients.37 The Allosure test measures the percentage of circulating donor derived cell free DNA to predict allograft pathology. More in depth analyses of this subject can be found in a review by Dharnidharka.38 Limitations of current biomarker tests, such as the Allosure test, are their non specificity for rejection or subtype of rejection. The Allosure test simply reflects dead donor cells. However, Allosure is a noninvasive diagnostic test used by many as an alternative or adjunct to biopsy for the diagnosis of rejection. Single cell RNA-seq has the potential to stimulate the development of many new biomarkers. Single cell RNA-seq can identify markers of cell states specific to allograft pathologies such as AMR or T cell mediated rejection. Single cell RNA-seq approaches also have the potential to uncover novel targets for therapy something that is needed for AMR, a major cause of allograft failure that has no good efficacious treatments. However, scRNA-seq methods are in their infancy and the costs are still very high. How this technology might be harnessed to use as a practical tool in clinical transplantation is not yet known.
We predict that the roles played by donor and recipient immune cells in the alloimmune response will be a productive avenue of investigation using scRNA-seq in the years to come. Prior to scRNA-seq, to identify cell origin from a mixture of cells originating from more than one individual a priori knowledge of each cell source was required. For example, fluorescence in situ hybridization analysis of Y chromosome positive cells could be used to identify cells from a male recipient who received a female donor organ. But this does not measure mRNA levels at the same time in those cells. By contrast, scRNA-seq can measure the natural genetic variation present in mRNA transcripts – called expressed single nucleotide variants – that exist between two non identical individuals. With knowledge of these differences by exome sequencing of host and donor, these variations in mRNA sequence can be used to assign a host vs. donor origin for each cell type in a scRNA-seq library.39 This powerful approach will allow investigators to ask new questions concerning rare but devastating conditions such as graft-vs-host disease in solid organ transplantation, where persistence of donor-derived leukocytes is already known to play a role in pathology. But whether persisting donor-derived cells play roles in other diseases like rejection is unknown. Whether donor-derived dendritic cells persist long term in a kidney allograft, and if they do, whether they modulate the immune response in rejection are the kinds of questions that scRNA-seq may be able to address.
Conclusions
Single cell RNA sequencing is a powerful technology that will revolutionize our understanding of cell biology. It is an unbiased approach that allows for the discovery of novel cell types, cell states and dynamics. The ability to examine individual cells from clinically relevant human tissue samples will hopefully lead to a breakthrough in the study of difficult to treat transplant pathologies. To date progress has been limited to a priori approaches to the study of human disease and molecular genetics approaches were previously limited by averaging effects. Now rare but potentially significant cell types can be studied and important genes networks uncovered allowing for the identification of new therapeutic targets. The field of transplantation is poised to benefit substantially from the application of this transformative technology to transplantation research.
Footnotes
The authors have no financial conflicts related to this work.
References
- 1. Tang F, Barbacioru C, Wang Y, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6(5):377–382. [DOI] [PubMed] [Google Scholar]
- 2. Famulski KS, Broderick G, Einecke G, et al. Transcriptome analysis reveals heterogeneity in the injury response of kidney transplants. Am J Transplant. 2007;7(11):2483–2495. [DOI] [PubMed] [Google Scholar]
- 3. O’Connell PJ, Zhang W, Menon MC, et al. Biopsy transcriptome expression profiling to identify kidney transplants at risk of chronic injury: a multicentre, prospective study. Lancet. 2016;388(10048):983–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Flechner SM, Kurian SM, Head SR, et al. Kidney transplant rejection and tissue injury by gene profiling of biopsies and peripheral blood lymphocytes. Am J Transplant. 2004;4(9):1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Menon MC, Keung KL, Murphy B, et al. The Use of Genomics and Pathway Analysis in Our Understanding and Prediction of Clinical Renal Transplant Injury. Transplantation. 2016;100(7):1405–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Halloran PF, Reeve J, Akalin E, et al. Real time central assessment of kidney transplant indication biopsies by microarrays: The INTERCOMEX Study. Am J Transplant. 2017;17(11)2851–2862. [DOI] [PubMed] [Google Scholar]
- 7. Sellarés J, de Freitas DG, Mengel M, et al. Understanding the causes of kidney transplant failure: the dominant role of antibody-mediated rejection and nonadherence. Am J Transplant. 2012;12(2):388–399. [DOI] [PubMed] [Google Scholar]
- 8. Furness PN, Taub N, Assmann KJ, et al. International variation in histologic grading is large, and persistent feedback does not improve reproducibility. Am J Surg Pathol. 2003;27(6):805–810. [DOI] [PubMed] [Google Scholar]
- 9. Halloran PF, Pereira AB, Chang J, et al. Microarray diagnosis of antibody-mediated rejection in kidney transplant biopsies: an international prospective study (INTERCOM). Am J Transplant. 2013;13(11):2865–2874. [DOI] [PubMed] [Google Scholar]
- 10. Kurian SM, Williams AN, Gelbart T, et al. Molecular classifiers for acute kidney transplant rejection in peripheral blood by whole genome gene expression profiling. Am J Transplant. 2014;14(5):1164–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Roedder S, Sigdel T, Salomonis N, et al. The kSORT assay to detect renal transplant patients at high risk for acute rejection: results of the multicenter AART study. PLoS Med. 2014;11(11):e1001759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Halloran PF, Reeve J, Aliabadi AZ, et al. Exploring the cardiac response to injury in heart transplant biopsies. JCI Insight. 2018;3(20). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lande JD, Patil J, Li N, et al. Novel insights into lung transplant rejection by microarray analysis. Proc Am Thorac Soc. 2007;4(1):44–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Parkes MD, Halloran PF, Hidalgo LG. Evidence for CD16a-Mediated NK Cell Stimulation in Antibody-Mediated Kidney Transplant Rejection. Transplantation. 2017;101(4):e102–e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hidalgo LG, Sis B, Sellares J, et al. NK cell transcripts and NK cells in kidney biopsies from patients with donor-specific antibodies: evidence for NK cell involvement in antibody-mediated rejection. Am J Transplant. 2010;10(8):1812–1822. [DOI] [PubMed] [Google Scholar]
- 16. Sis B, Jhangri GS, Bunnag S, et al. Endothelial gene expression in kidney transplants with alloantibody indicates antibody-mediated damage despite lack of C4d staining. Am J Transplant. 2009;9(10):2312–2323. [DOI] [PubMed] [Google Scholar]
- 17. Wu H, Malone AF, Donnelly EL, et al. Single-Cell Transcriptomics of a Human Kidney Allograft Biopsy Specimen Defines a Diverse Inflammatory Response. J Am Soc Nephrol. 2018;29(8):2069–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Macosko EZ, Basu A, Satija R, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161(5):1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Klein AM, Mazutis L, Akartuna I, et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161(5):1187–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Malone AF, Wu H, Humphreys BD. Bringing Renal Biopsy Interpretation Into the Molecular Age With Single-Cell RNA Sequencing. Semin Nephrol. 2018;38(1):31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Trapnell C, Cacchiarelli D, Grimsby J, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32(4):381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sivakamasundari V, Bolisetty M, Sivajothi S, et al. Comprehensive Cell Type Specific Transcriptomics of the Human Kidney. BioRxiv. 2017. 10.1101/238063. [DOI] [Google Scholar]
- 23. Young MD, Mitchell TJ, Vieira Braga FA, et al. Single-cell transcriptomes from human kidneys reveal the cellular identity of renal tumors. Science. 2018;361(6402):594–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Park J, Shrestha R, Qiu C, et al. Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science. 2018;360(6390):758–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wu H, Uchimura K, Donnelly EL, et al. Comparative Analysis and Refinement of Human PSC-Derived Kidney Organoid Differentiation with Single-Cell Transcriptomics. Cell Stem Cell. 2018;23(6):869–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Buggins AG, Mufti GJ, Salisbury J, et al. Peripheral blood but not tissue dendritic cells express CD52 and are depleted by treatment with alemtuzumab. Blood. 2002;100(5):1715–1720. [PubMed] [Google Scholar]
- 27. Reyfman PA, Walter JM, Joshi N. Single-Cell Transcriptomic Analysis of Human Lung Reveals Complex Multicellular Changes During Pulmonary Fibrosis. BioRxiv. 2018. 10.1101/296608. [DOI] [Google Scholar]
- 28. See K, Tan WLW, Lim EH, et al. Single cardiomyocyte nuclear transcriptomes reveal a lincRNA-regulated de-differentiation and cell cycle stress-response in vivo. Nat Commun. 2017;8(1):225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. MacParland SA, Liu JC, Ma XZ, et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat Commun. 2018;9(1):4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Adam M, Potter AS, Potter SS. Psychrophilic proteases dramatically reduce single cell RNA-seq artifacts: A molecular atlas of kidney development. Development. 2017;144(19):3625–3632. doi: 10.1242/dev.151142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wu H, Kirita Y, Donnelly EL, et al. Advantages of single-nucleus over single-cell RNA-sequencing of adult kidney: rare cell types and novel cell states revealed in fibrosis. J Am Soc Nephrol In Press. 2019;30(1):23–32. doi: 10.1681/ASN.2018090912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ho M, Yang E, Matcuk G, et al. Identification of endothelial cell genes by combined database mining and microarray analysis. Physiol Genomics. 2003;13(3):249–262. [DOI] [PubMed] [Google Scholar]
- 33. Sengoelge G, Luo W, Fine D, et al. A SAGE-based comparison between glomerular and aortic endothelial cells. Am J Physiol Renal Physiol. 2005;288(6):F1290–1300. [DOI] [PubMed] [Google Scholar]
- 34. Ferguson A, Iasella C, Chen W, et al. Gene Expression Profiling of Lung Transplant Patients Using Next-Generation Sequencing to Identify Biomarkers for Chronic Lung Allograft Dysfunction [Abstract A4732]. Am J Respir Crit Care. 2018;197:A4732. [Google Scholar]
- 35. Habal M, Myung A, Yan H, et al. Single-Cell Analysis of Graft Infiltrating T-Cells in Cardiac Allograft Vasculopathy [Abstract]. Am J Transplant. 2017;17(suppl 3). [Google Scholar]
- 36. Deng MC, Eisen HJ, Mehra MR, et al. Noninvasive discrimination of rejection in cardiac allograft recipients using gene expression profiling. Am J Transplant. 2006;6(1):150–160. [DOI] [PubMed] [Google Scholar]
- 37. Bloom RD, Bromberg JS, Poggio ED, et al. Cell-Free DNA and Active Rejection in Kidney Allografts. J Am Soc Nephrol. 2017;28(7):2221–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dharnidharka VR, Malone A. Biomarkers to detect rejection after kidney transplantation. Pediatr Nephrol. 2018;33(7):1113–1122. [DOI] [PubMed] [Google Scholar]
- 39. Kang HM, Subramaniam M, Targ S, et al. Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat Biotechnol. 2018;36(1):89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Butler A, Hoffman P, Smibert P, et al. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Krzywinski MI, Schein JE, Birol I, et al. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]