Abstract
Neuronal connectivity is dependent on size and shape of the dendritic arbor. However, mechanisms controlling dendritic arborization, especially in the peripheral nervous system, are not completely understood. Previous studies have shown that bone morphogenetic proteins (BMPs) are important initiators of dendritic growth in peripheral neurons. In this study, we examined the hypothesis that post-transcriptional regulation mediated by microRNAs (miRNAs) is necessary for BMP-7-induced dendritic growth in these neurons. To examine the role of miRNAs in BMP-7-induced dendritic growth, microarray analyses was used to profile miRNA expression in cultured sympathetic neurons from the superior cervical ganglia of embryonic day 21 rat pups at 6 and 24 h after treatment with BMP-7 (50 ng/mL). Our data showed that BMP-7 significantly regulated the expression of 43 of the 762 miRNAs. Of the 43 miRNAs, 22 showed robust gene expression; 14 were upregulated by BMP-7 and 8 were downregulated by BMP-7. The expression profile for miR-335, miR-664-1*, miR-21, and miR-23b was confirmed using qPCR analyses. Functional studies using morphometric analyses of dendritic growth in cultured sympathetic neurons transfected with miRNA mimics and inhibitors indicated that miR-664-1*, miR-23b, and miR-21 regulated early stages of BMP-7-induced dendritic growth. In summary, our data provide evidence for miRNA-mediated post-transcriptional regulation as important downstream component of BMP-7 signaling during early stages of dendritic growth in sympathetic neurons.
Keywords: MicroRNA, Dendrite, Bone morphogenetic proteins, Sympathetic neurons
Introduction
Dendrites are the primary sites of synapse formation in the nervous system, and the extent of the dendritic arbor correlates directly with synaptic density (Purves 1975; Purves and Hume 1981; Rubin 1985; Purves and Lichtman 1985) and tonic activity (De Castro et al. 1995). Changes in the number of dendrites and the size of the dendritic arbor are associated with neurodegenerative disease and neurodevelopmental disorders, including Alzheimer’s disease, Down’s syndrome, Rett’s syndrome, autism, and schizophrenia (Comery et al. 1997; Kaufmann and Moser 2000; Pardo and Eberhart 2007; Garey 2010; Penzes et al. 2011; Kulkarni and Firestein 2012; Supekar et al. 2013; Copf 2016). Therefore, understanding the mechanisms that influence the size and complexity of the dendritic arbor is crucial for understanding the development of neuronal connectivity and the functioning of the nervous system, and may provide insight regarding the pathogenesis and treatment of diverse neurological disorders.
Postganglionic sympathetic neurons provide a well-characterized model for studying dendritogenesis (Higgins et al. 1991, 1997; Lein et al. 2009). Sympathetic neurons initiate dendritic growth around E15 and continue to extend their dendritic arbor through early stages of postnatal life with continual growth and retraction of the dendritic arbor throughout adulthood (Purves et al. 1986). At maturity, mammalian sympathetic neurons have a relatively complex dendritic arbor with around 2–6 primary dendrites and elaborate branching patterns, and the complexity of the dendritic arbor is correlated to synaptic input and target size (Purves and Hume 1981; Purves and Lichtman 1985). Interestingly, these neurons do not extend a dendritic arbor when cultured in vitro in the absence of serum or ganglionic glia (Bruckenstein and Higgins 1988), but can be induced to extend dendrites in defined medium supplemented with bone morphogenetic proteins (BMPs) (Lein et al. 1995). The complexity of the dendritic arbor of sympathetic neurons exposed to BMP-7 for a month in culture is similar to that observed in vivo (Lein et al. 1995). Because it is possible to use BMPs to trigger dendritic growth in primary sympathetic neurons that lack dendrites, this culture system is an excellent model for studying the molecular mechanisms that regulate the initiation of dendritic arborization in these neurons.
BMPs mediate their effects on dendritic arborization by binding to their cognate serine-threonine kinase receptors, which causes the activation and nuclear translocation of SMAD transcription factors that regulate transcription of BMP-responsive genes (Garred et al. 2011; Guo et al. 2001; Wotton and Massague 2000). Recent studies have shown that in addition to controlling the expression of protein-coding genes, SMADs regulate expression of microRNA (miRNA)-coding genes, export of primary miRNA transcripts out of the nucleus, and processing to mature miRNAs (Reinke and Carthew 2008; Davis et al. 2010; Saj and Lai 2011). MicroRNAs (miRNAs) are small (20–22 nt) non-coding RNAs that play a crucial role in post-transcriptional gene regulation, either by tagging mRNA transcripts for degradation or by repressing mRNA translation (Carthew and Sontheimer 2009; Saj and Lai 2011; Tai and Schuman 2006). MiRNA genes are expressed as primary transcripts (pri-miRNA), which are processed by two RNAses—Drosha and Dicer—to generate mature miRNAs, which then regulate gene expression by formation of RNA-induced silencing complexes (RISC) consisting of multiple proteins, including Argonaute, that bind to the 3′UTR of target mRNA to regulate gene expression (Carthew and Sontheimer 2009). Interestingly, over 50% of known miRNAs are expressed in the nervous system, and in neurons, miRNAs are compartmentalized to dendrites, distal axons, and synapses (Konopka et al. 2011; Kosik 2006; Tai and Schuman 2006). Conditional knockout and genetic ablation of Dicer results in neuronal cell death or neurodegeneration-like phenotypes (Davis et al. 2008; Dorval et al. 2012), and several miRNAs are implicated in regulation of dendritic remodeling and synaptic plasticity in central neurons (Bicker et al. 2014; Irie et al. 2016; Konopka et al. 2011; Magill et al. 2010; Siegel et al. 2009; Tai and Schuman 2006). However, whether miRNAs are required for dendritic growth in peripheral neurons has yet to be determined.
Since BMPs are potent inducers of the developmental program for dendritic growth in sympathetic neurons (Lein et al. 1995; Higgins et al. 1997), and BMP signaling is known to modulate miRNA expression and function in other cell types (Reinke and Carthew 2008; Wang et al. 2010; Kang et al. 2012), the goal of this study was to test the hypothesis that miRNAs are necessary for BMP-induced dendritic growth in sympathetic neurons.
Materials and Methods
Materials
Recombinant human bone morphogenetic protein-7 (BMP-7) was a generous gift from Curis (Cambridge, MA). MirVana™ mimics and inhibitors for miR-21, miR-23b, miR-26a, miR-664-1, and miR-335-1, as well as negative controls for mimics and inhibitors and GAPDH siRNA were obtained from Thermo Fisher Scientific (Waltham, MA). All other tissue culture media components were obtained from Thermo Fisher Scientific and standard chemicals and supplies (ACS grade for general reagents and BioReagent for tissue culture use) were purchased from Millipore-Sigma (St. Louis, MO).
Animals
All protocols involving animals were approved by the University of California, Davis (Davis, CA) Institutional Animal Care and Use Committee. Timed-pregnant Sprague–Dawley rats were purchased from Charles River Laboratories (Hollister, CA) and housed in plastic cages in a temperature-controlled room (22 ± 2 °C) on a 12-h reverse light–dark cycle with food and water provided ad libitum. Pregnant dams were typically received from the vendor at embryonic day 18 (E18) and euthanized with CO2 on E21 to collect pups. Superior cervical ganglia (SCG) were harvest from euthanized pups for culture; no experimental manipulations were performed prior to euthanasia. CO2 has been shown to be effective method for euthanasia in rodents (Hackbarth et al. 2000; Ghogha et al. 2012; Boivin et al. 2017).
Culturing Sympathetic Neurons
Sympathetic neurons were dissociated from SCG of embryonic (E20/21) Sprague–Dawley rat pups according to previously described protocols (Ghogha et al. 2012). Typically, each rat litter had around 9–14 pups, resulting in 18–28 ganglia, which were dissociated using enzymatic digestion and mechanical trituration to yield a cell suspension that was plated onto different types of culture plates. The pups were not sexed and ganglia from both male and female pups were pooled to set up cultures. Each E20/21 SCG has 20,000 to 30,000 neurons (Davies 1978; Smolen et al. 1983); cultures were plated in 24-well plates with or without glass coverslips (Ted Pella, Redding, CA) at approximately 8000 cells/well for morphological analyses and in six well plates at 15,000–18,000 cells/well for RNA isolation. All wells were precoated with 100 μg/mL poly-d-lysine (BD Biosciences, San Jose, CA) prior to plating with cells. Each independent dissection of one litter of pups, consisting of cells isolated from pooled ganglia from all the pups in the litter, was considered a biological replicate.
Cultures were maintained in serum-free media (50:50 v/v DMEM:F12, Thermo Fisher Scientific) containing 100 ng/mL β-nerve growth factor (NGF (Harlan Laboratories, Indianapolis, IN), 500 μg/mL bovine serum albumin (BSA, Millipore-Sigma, Burlington, MA), 10 μg/mL bovine insulin (Thermo Fisher Scientific), and 10 μg/mL human transferrin (Thermo Fisher Scientific) at 35 °C in a humidified chamber containing 5% CO2. Cultures were treated with 2 μM cytosine-β-d-arabinofuranoside (Ara-C, Millipore-Sigma) for 48 h beginning 1 day after plating to eliminate non-neuronal cells prior to experimental treatments.
RNA Isolation and Analyses
Following the elimination of non-neuronal cells, cultured neurons were exposed to culture medium supplemented with BMP-7 (50 ng/mL) for 6 h or 24 h; negative controls were exposed to culture medium alone. This concentration of BMP-7 has been previously described to be maximally effective for triggering dendritic growth in SCG cultures (Lein et al. 1995). To ensure that the cells treated under the various conditions were lysed at the same time to collect RNA, the exposures were timed as follows: (i) at time = 0, BMP-7 was added to the wells for the 24 h exposure to BMP-7; (ii) 18 h later, BMP-7 was added to a separate set of wells for the 6 h exposure; (iii) 6 h later (24 h after addition of BMP-7 to the first set of wells), control cultures and BMP-7 exposed cultures were lysed and total RNA extracted using the miRNAeasy kit (Qiagen, Valencia, CA) per the manufacturer’s instructions. The exposure paradigm is similar to the one used for transcriptome analyses of BMP-induced dendritic growth (Garred et al. 2011). The isolated RNA was re-suspended in 30 μL of RNAse-free water, following which the quality and concentration of the isolated RNAs were assessed using a Nanodrop™ spectrophotometer. Samples with 260/230 ratio greater than 1.0 and a 260/280 ratio above 1.8 were considered to be suitable for microarray and quantitative PCR analyses (qPCR).
For microarray analysis, the total RNA from two independent dissections, each of which contained pooled RNA from multiple pups, was isolated and sent to LC Science (Houston, TX) for further processing and microarray analyses of the microRNA expression profiles using their proprietary µParaflo microfluidic chip technology, which has been utilized and validated in a number of publications (Zhou et al. 2012; Feng et al. 2013; Katchy and Williams 2014; Ge et al. 2015; Fay et al. 2018). The chip contained all the rat miRNAs that were listed in release 18.0 of the rat miRBase. Each chip included at least four technical replicates for each probe and 16 sets of control probes distributed throughout the array. RNA samples were labeled with Cy5 and the signals following hybridization with the microarray were detected using an Axon GenePix 4000B microarray scanner. The hybridization target signal was subtracted from the background signal and normalization was carried out using the local weighted scatterplot smoothing (LOWESS) method on the background subtracted data. To ensure that the differences in miRNA expression due to the various treatments were not canceled by the normalization, each RNA sample was spiked with a known ratio of spike-in positive control 20-mer RNA as an external normalization control. In order to detect differences between the control and BMP-7-treated samples, a one-way ANOVA was performed to detect differences in miRNA expression between negative control cultures and cultures exposed to BMP-7 for 6 or 24 h. The p value and power were calculated using a sample size of N = 2 for the two biological replicates from independent dissections.
The total RNA extracted from embryonic sympathetic neurons treated with control media, BMP-7, for 6 h or 24 h was also subjected to reverse transcription (RT) and qPCR analysis using the TaqMan® assay kits with specific microRNA reverse transcriptase (RT) primers and assays (Thermo Fisher Scientific) per the manufacturer’s instructions.
The RT primer and assay kits for the following miRNAs were used: rno-miR-21 (rno-miR-21-5p), rno-miR-23b (rno-miR-23b-3p), rno-miR-335, and rno-miR-664-1* (rno-miR-664-1-5p). Total RNA (6 ng/μL) was used as the starting material for the reactions; the manufacturer recommends from 1 to 10 ng, preferably closer to the lower end of the range. U6 mRNA, snoRNA, and 4.5S RNA(H) (Thermo Fisher Scientific) were used as controls. The qPCR reactions were run on total RNA samples obtained from at least two biological replicates, where each biological replicate was the pooled total RNA sample obtained from cultures of neurons dissociated from the SCG of all the pups from one dam. In each experiment, the RNA sample from each biological replicate was run as three technical replicates to determine reproducibility within each experiment. The qPCR run logs were then analyzed using the Bio-Rad CFX Manager software. Cq values for each miRNA sample were normalized against the controls, and a fold change was calculated (2−ΔΔCq).
Transfecting Embryonic Sympathetic Neurons
Primary cultures of sympathetic neurons are known to have a low efficiency (5–25%) for DNA transfection (Horbinski et al. 2001). Since the miRNA mimics and inhibitors used in this study were not fluorescently labeled, it was necessary to assess and optimize the transfection efficiency of small RNAs prior to mimic and inhibitor transfection. To optimize the transfection of small RNAs into these neurons, cultured embryonic sympathetic neurons were transfected with Silencer® FAM™ Labeled GAPDH siRNA using Lipofectamine® RNAiMAX siRNA Transfection Reagent (Thermo Fisher Scientific) per the manufacturer’s instructions with the following modifications—cells were transfected using a higher concentration of siRNA (30 pmol or 75 pmol instead of the recommended 10 pmol siRNA), and cells were incubated with the transfection mixture for only 4–5 h rather than the 1–3 days suggested by the manufacturer. These changes were made to increase efficiency of transfection while minimizing toxicity to the neurons.
The effectiveness of transfection was assessed by two different methods: (i) counting the number of FAM-labeled neurons 2 days following transfection; and (ii) examining the cultures for decreased GAPDH protein expression using immunofluorescence. To examine the expression of GAPDH, the transfected cultures were fixed with 4% paraformaldehyde (Spectrum Chemical Manufacturing Corporation, New Brunswick, NJ) and immunostained with the mouse monoclonal antibody 6C5 against GAPDH (AM 4300, 1 μg/mL Thermo Fisher Scientific) as previously described (Vigelsø et al. 2015; Reales et al. 2015). Antibody-antigen complexes were detected using Alexa-546-labeled anti-mouse secondary antibody (Thermo Fisher Scientific). The percentage of cells with green fluorescence was measured counted to determine the efficiency of transfection. Also, the percentage of cells showing a decreased expression of GAPDH protein as evidenced by decreased red fluorescence were quantitated to provide another measurement of transfection efficiency.
The optimized transfection protocol was used to transfect cells with unlabeled miRNA mimics and inhibitors. Validated mirVana® miRNA mimics for rno-miR-21 (rno-miR-21-5p), rno-miR-23b (rno-miR-23b-3p), and rno-miR-26a (rno-miR-26a-5p), and validated mirVana® miRNA inhibitors for rno-miR-335 and rno-miR-664-1* (rno-miR-664-1-5p) were obtained from Thermo Fisher Scientific; all of these have been used in previously published studies (Chen et al. 2017a, b; De Santi et al. 2017; Ishteiwy et al. 2012; Liu et al. 2015; Sandoval-Bórquez et al. 2017; Wu et al. 2014; Yuan et al. 2016).
To assess the role of individual miRNAs in BMP-7-induced dendritic growth, cultured sympathetic neurons were transfected with 75 pmol of one of the miRNA mimics or inhibitors, or with negative control mimics or inhibitors (Thermo Fisher Scientific). Cells were rinsed with control media 4–5 h following transfection and maintained in control media overnight. The transfected neurons were then maintained in the absence or presence of BMP-7 (50 ng/mL) for 5 days to induce dendritic growth, and then fixed and immunostained for morphological analyses as described below. The morphological analyses were performed on cultures obtained from at least two independent dissections (e.g., > 2 biological replicates) with each condition being run in triplicate for each biological replicate.
Quantification of Dendritic Morphology
Cultures were fixed with 4% paraformaldehyde in phosphate buffer and immunostained using a mouse monoclonal antibody against microtubule associated protein-2 (MAP2) IgG (1:2000-5000, BioLegend, San Diego, CA) and visualized using indirect immunofluorescence as previously described (Ghogha et al. 2012; Chandrasekaran et al. 2015). MAP2 is a cytoskeletal protein that is specifically localized to dendrites and is necessary for dendritic elongation (Caceres et al. 1984; Harada et al. 2002). Fluorescent images were acquired on the Nikon Optiphot fluorescent microscope using a SPOT camera. Dendritic morphology was assessed by counting the number of dendrites per neuron and measuring the total dendritic arbor of each neuron using ImageJ freeware (version 1.48, NIH), similar to the methods used in previous studies (Lein et al. 1995; Guo et al. 1999, 2001; Kim et al. 2002; Chandrasekaran et al. 2015). Dendritic number is a measure of the number of primary dendrites extended by a neuron—any primary dendrite that is longer than the longest diameter of the cell body is included in this measurement. Total dendritic arbor is measure of the length of the primary dendrites and all of its branches and provides a quantitative measure of dendritic growth initiation, extension, and branching. The experiment was repeated on cultures from at least two independent dissections and each experiment contained three technical replicates for each condition. Data are expressed as mean ± SEM and statistically significant differences between treatment groups were assessed using one-way ANOVA followed by Tukey’s test at p ≤ 0.05.
Statistical Analysis
Microarray Data
To identify statistically significant differences in miRNA expression between groups, the microarray data were analyzed using one-way and two-way ANOVA analyses. The two-way ANOVA was set up with Factor A being change in expression over time (0-h, 6-h, and 24-h BMP-7 exposure) and Factor B being different conditions (no BMP-7 exposure (Control) or BMP-7 exposure). Holm–Sidak test and Tukey’s test were used as post hoc tests for multiple comparison for the two-way ANOVA and one-way ANOVA, respectively. Any miRNA that had a p ≤ 0.05 for either the one-way ANOVA or for one of the factors for the two-way ANOVA was considered as statistically significant. The mean, standard deviation, and power of the statistical analyses were calculated using N = 2 (number of biological replicates used for the array).
qPCR Data
The qPCR experiments for rno-miR-23b, rno-miR-335, and rno-miR-664-1* were collected from two biological replicates and the qPCR analyses for rno-miR-21 were collected from three biological replicates. The fold changes in gene expression were calculated from the ΔΔCq values and expressed as mean ± SEM for each experiment, where the mean and standard deviation were calculated using the two biological replicates for each sample in the assay.
Morphological Analyses
The number of dendrites and the total dendritic arbor of the neuron are expressed as mean ± SEM, where the mean and SEM were calculated using the number of cells counted for each condition. Approximately 30–45 cells were counted per well/condition, resulting in over 100 cells/condition for each experiment. Specific N values for each condition are listed in the figure legends. Statistically significant differences between controls and experimental groups were assessed using one-way ANOVA with p ≤ 0.05, followed by post hoc tests. The mean, SEM, and the power of the statistical analyses were calculated using the specific N-value equal to the number of cells quantified for each condition.
Bioinformatics Analyses for Predicted miRNA Targets
For miRNAs potentially involved in dendritic growth regulation, validated targets were identified using miRTarBase, release 7 (http://mirtarbase.mbc.nctu.edu.tw/php/index.php), and the predicted targets were identified using miRDB (http://mirdb.org/). The predicted targets on miRDB with a target score of 85 or greater were further examined for relevance to dendritic growth or BMP signaling using UniProt and their GO annotation and the selective target lists were created.
Results
miRNome Profile of Sympathetic Neurons During Dendritogenesis
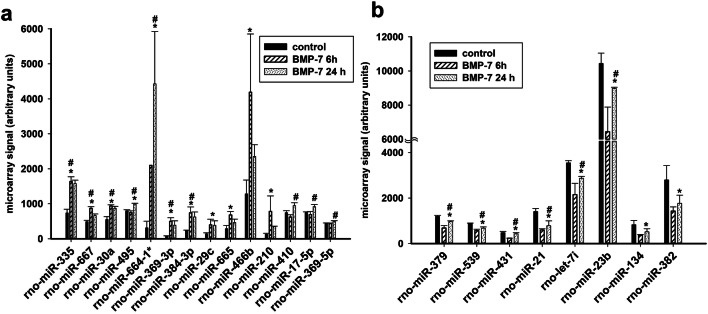
Previous studies have shown that BMP-7-triggered dendritic growth in cultured embryonic sympathetic neurons is dependent on transcriptional regulation during the first 24 h following exposure to BMP-7 with over 200 mRNAs being differentially regulated (Garred et al. 2011). To test the hypothesis that miRNAs are downstream effectors of BMP-7-induced dendritic growth and may be regulating the expression of some of these genes, the expression of miRNAs was compared between neurons maintained in culture medium versus neurons exposed to medium supplemented with BMP-7 at 50 ng/mL, which is a maximally effective concentration for triggering dendritic growth in these cultures (Lein et al. 1995). As determined by the miRNA microarray, BMP-7 (50 ng/mL) significantly changed the expression of 43 miRNAs out of 762 miRNAs in the rat miRNome (one-way ANOVA at p ≤ 0.05), with 11 miRNAs showing robust differences (p ≤ 0.01) between control and BMP-7-exposed neurons (Fig. 1). Although statistically significant, the overall levels for most miRNAs (as measured by the fluorescence intensity of the individual miRNAs on the microarray) was found to be under 5000 intensity units, with miR-23b showing one of the highest expression around 10000 intensity units, as determined using the Axon GenePix 4000B microarray scanner, which can detect maximum intensity value of 65535 units. These data suggest that most of these miRNAs were expressed at fairly low levels in these cells. Of the 43 differentially expressed miRNAs, 14 miRNAs were considered to have robust expression because their fluorescence intensity values for at least one of the conditions was > 500 units. Also, the fold changes for most miRNAs were maximum around two to threefold, which is much lower than those reported for the changes in mRNA expression of genes during BMP-induced dendritic growth in sympathetic neurons (Garred et al. 2011). Statistical analyses using two-way ANOVA (p ≤ 0.05) revealed eight additional miRNAs that showed significant differences in expression over time following BMP-7 exposure or when compared to control neurons (Table 1, Fig. 2). Of these 22 miRNAs, 14 miRNAs were increased following BMP-7 exposure (Fig. 2a), while eight miRNAs were decreased in expression following BMP-7 exposure (Fig. 2b), with 13 miRNAs showing significant differences between control and BMP-7-exposed neurons (p ≤ 0.05) over time and across conditions (Fig. 2, Table 1). Six miRNAs—miR-29c, miR-665, miR-466b, miR-210, miR-134, and miR-382—showed significant difference in expression between control and BMP-7-exposed neurons at the 6 h or 24 h time point but did not show clear temporal regulation of gene expression (Fig. 2, Table 1). On the other hand, miR-410, miR-17-5p, and miR-369-5p showed time-dependent regulation following BMP-7 exposure, but did not show a statistically significant difference between control and BMP-7 groups overall (Fig. 2, Table 1). Also, significant changes in the expression of 18 of the 22 miRNAs were observed within the first 6 h of BMP-7 exposure, suggesting that these miRNAs might be regulating expression of genes important for the initial stages of BMP-induced dendritic growth in sympathetic neurons.
Fig. 1.
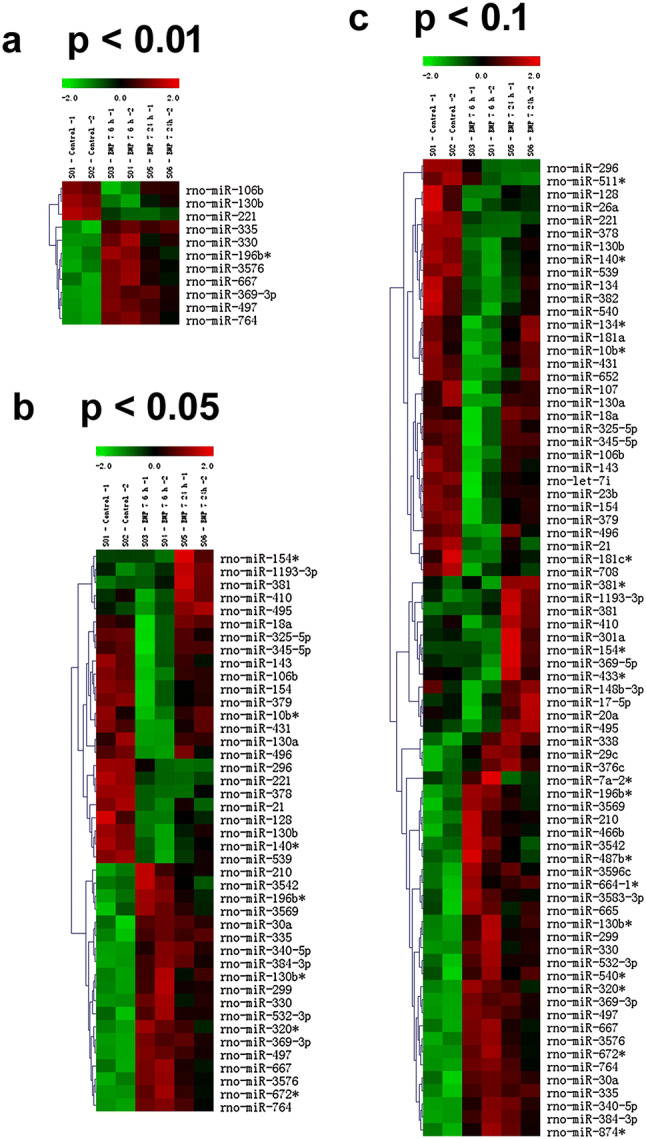
Cluster diagram showing miRNAs differentially regulated by BMP-7 in cultured sympathetic neurons. The columns are clustered based on experimental conditions, with rows showing the individual miRNA transcripts. The six columns in each panel represent two control samples, two samples from cultures exposed to BMP-7 for 6 h, and two samples from cultures exposed to BMP-7 at 24 h. The relative expression of the miRNAs is shown on a color scale where red represents the highest level of upregulation and green represents the lowest level of downregulation at different time points. Statistical significance was assessed by one-way ANOVA, followed by Tukey’s test. a–c show the miRNAs altered at p < 0.01, p < 0.05, and p < 0.1, respectively
Table 1.
Following miRNAs were found to have a statistically significant difference between control and BMP-7-treated neurons at different time points
| miRNA | Control | BMP-7 6 h | BMP-7 24 h | One-way ANOVA | Two-way ANOVA | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | p value | Power | Factor A: time BMP-7 0, 6 h, 24 h p value | Factor B: control versus BMP-7 p value | Power factor A/factor B | |
| A) MiRNAs that increase with BMP-7 treatment | |||||||||||
| rno-miR-335 | 735 | 108 | 1637 | 135 | 1579 | 97 | 0.007 | 1.0 | 0.002 | <0.001 | 0.99/1 |
| rno-miR-667 | 487 | 42 | 872 | 46 | 661 | 46 | 0.008 | 1.0 | 0.002 | <0.001 | 0.99/1 |
| rno-miR-30a | 548 | 85 | 939 | 31 | 873 | 53 | 0.014 | 0.9 | 0.020 | 0.001 | 0.74/1 |
| rno-miR-495 | 800 | 28 | 748 | 48 | 989 | 24 | 0.013 | 0.9 | 0.004 | 0.048 | 0.97/0.47 |
| rno-miR-29c | 139 | 31 | 396 | 162 | 384 | 130 | 0.020 | 0.17 | 0.147 | 0.02 | 0.23/0.74 |
| rno-miR-410 | 744 | 58 | 622 | 64 | 947 | 82 | 0.040 | 0.6 | 0.030 | 0.485 | 0.63/0.05 |
| rno-miR-664-1* | 310 | 191 | 2097 | 0 | 4425 | 1498 | 0.041 | 0.6 | 0.011 | 0.002 | 0.86/1 |
| rno-miR-369-3p | 78 | 5 | 497 | 102 | 386 | 126 | 0.043 | 0.6 | 0.010 | <0.001 | 0.86/1 |
| rno-miR-665 | 286 | 93 | 680 | 94 | 462 | 98 | 0.057 | 0.5 | 0.067 | 0.013 | 0.42/0.80 |
| rno-miR-384-3p | 240 | 8 | 740 | 170 | 623 | 142 | 0.060 | 0.5 | 0.019 | 0.001 | 0.75/1 |
| rno-miR-17-5p | 747 | 21 | 689 | 87 | 916 | 66 | 0.077 | 0.4 | 0.035 | 0.226 | 0.59/0.12 |
| rno-miR-369-5p | 446 | 5 | 434 | 14 | 492 | 25 | 0.081 | 0.4 | 0.037 | 0.162 | 0.58/0.18 |
| rno-miR-466b | 1282 | 396 | 4189 | 1664 | 2353 | 334 | 0.133 | 0.3 | 0.090 | 0.024 | 0.34/0.66 |
| rno-miR-210 | 130 | 25 | 781 | 442 | 346 | 15 | 0.170 | 0.2 | 0.106 | 0.033 | 0.3/0.58 |
| B) MiRNAs that decrease with BMP-7 treatment | |||||||||||
| rno-miR-379 | 1196 | 42 | 681 | 108 | 956 | 43 | 0.013 | 0.94 | 0.002 | <0.001 | 0.99/1 |
| rno-miR-539 | 875 | 26 | 572 | 57 | 678 | 56 | 0.018 | 0.87 | 0.004 | <0.001 | 0.97/1 |
| rno-miR-431 | 481 | 53 | 239 | 2 | 428 | 56 | 0.024 | 0.80 | 0.028 | 0.01 | 0.65/0.81 |
| rno-miR-21 | 1407 | 137 | 596 | 57 | 782 | 219 | 0.026 | 0.78 | 0.017 | 0.0001 | 0.777/1 |
| rno-let-7i | 3547 | 99 | 2152 | 494 | 2873 | 83 | 0.041 | 0.64 | 0.012 | 0.0001 | 0.84/1 |
| rno-miR-23b | 10439 | 609 | 6433 | 1451 | 8977 | 63 | 0.048 | 0.58 | 0.028 | 0.01 | 0.65/0.91 |
| rno-miR-134 | 829 | 189 | 360 | 38 | 517 | 138 | 0.088 | 0.38 | 0.205 | 0.03 | 0.16/0.57 |
| rno-miR-382 | 2798 | 629 | 1428 | 184 | 1768 | 356 | 0.099 | 0.35 | 0.251 | 0.04 | 0.13/0.51 |
Bold indicates p values that were less than 0.05
Fig. 2.
Microarray analyses of high abundance miRNAs differentially regulated by BMP-7. Relative microarray signal for 23 miRNAs in total RNA samples obtained from neurons exposed to BMP-7 at 50 ng/mL for 6 h or 24 h. a miRNAs upregulated by BMP-7. b miRNAs downregulated by BMP-7. Statistically significant differences were determined using two-way ANOVA (p < 0.05). * Significant difference between control and BMP-7-exposed group at p < 0.05; #Significant differences following BMP-7 treatment over time at p < 0.05
Of the 14 miRNAs whose expression was increased by BMP-7 (Fig. 2a), miR-335, miR-664-1*(miR-664-1-5p), and miR-466b had the highest expression in these neurons, and of these miR-335 and miR-664-1* showed significant changes in expression across time and between groups. miR-335 expression was significantly increased following 6 h of BMP-7 treatment and remained elevated at 24 h post-exposure (Figs. 1, 2a, Table 1), whereas expression of miR-664-1* increased at 6 h after addition of BMP-7 to the cultures and continued to increase at 24 h. Therefore, these two miRNAs were chosen for confirmation using qPCR and for functional studies.
Among the downregulated miRNAs, miR-23b and miR-21 were chosen for further characterization. miR-23b showed the highest expression and significant difference in expression over time and across groups (Fig. 2b, Table 1). Also, the expression of miR-23b and miR-21 has been previously reported to be regulated by TGF-β/BMP signaling in multiple cell types and shown to be important for neuronal function (Qin et al. 2009; Rogler et al. 2009; Ahmed et al. 2011; Strickland et al. 2011; Kang et al. 2012; Liu et al. 2016). Moreover, these miRNAs had shown to be expressed in the soma of sympathetic neurons and to be regulated by TGF-β family members in other cells (Li and Sun 2013; Natera-Naranjo et al. 2010; Rogler et al. 2009).
The expression profiles of miR-21, miR-23b, and miR-664-1* and miR-335 detected by qPCR analyses were similar to those observed using microarray analyses, although only the change in miR-21 expression was found to be statistically significant (Fig. 3). Due to the low expression of the miRNAs and the small fold changes in the expression between control and BMP-7-exposed cultures, the results from the qPCR analyses showed more variability between biological replicates and between different runs; however, profiles similar to the microarray analysis were observed for both replicates for miR-21, miR-23b, and miR-664-1* and for one of the two biological replicates for miR-335.
Fig. 3.
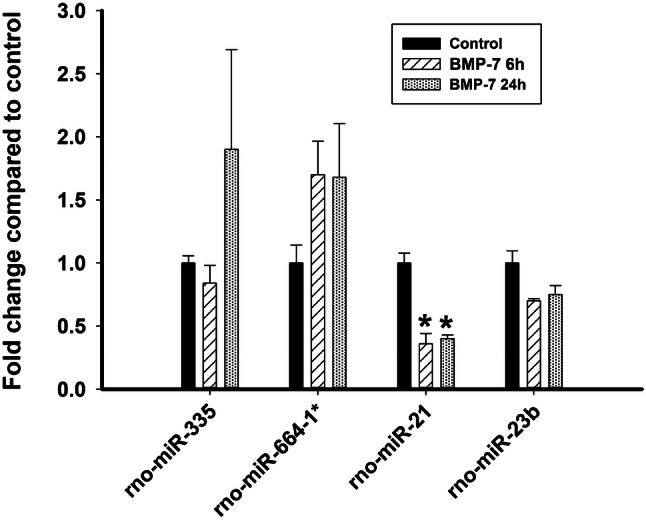
qPCR analysis of BMP-7 effects on miR-335, miR-664-1*, miR-21, miR-23b, and miR-26a expression. Total RNA extracted from cultured sympathetic neurons at 6 and 24 h after treatment with control medium or BMP-7 at 50 ng/mL was analyzed by qPCR to quantify expression of 5 miRNAs identified as differentially regulated by BMP-7 in the microarray screen. Expression of the target miRNA was normalized against snoRNA and U6 RNA for miR-335, miR-664-1*, snoRNA for miR-23b, and U6 RNA for miR-21. The figure shows data for the two biological replicates, each of which was run as 3 technical replicates. *Significantly different from control at p < 0.05 (N = 2)
Optimizing the Protocol for Transfecting Small RNAs into Cultured Sympathetic Neurons
To determine whether miRNAs play a functional role during BMP-7-induced dendritic growth, cultured sympathetic neurons were treated with either miRNA mimics or inhibitors to quantify the effects on BMP-induced dendritic growth of overexpression or inhibition of target miRNAs, respectively. Since cultured sympathetic neurons typically show low transfection efficiency (5–25%) for DNA transfections (Horbinski et al. 2001), and the miRNA mimics and inhibitors were unlabeled, the transfection protocol was optimized using Silencer® FAM™ Labeled GAPDH siRNA (Thermo Scientific) with the goal of identifying transfected neurons in culture.
Transfecting 30 pmol versus 75 pmol of GAPDH siRNA resulted in 43% versus 89% of the neurons expressing FAM, with no adverse effect on neuronal survival or health (data not shown). The transfection efficiency was also independently confirmed by examining the expression of GAPDH protein in the transfected neurons as detected using immunohistochemistry. Quantitative analysis of GADPH immunoreactivity in transfected cultures revealed that transfection with 30 pmol GAPDH siRNA decreased cytoplasmic GAPDH immunoreactivity in approximately 75% of the transfected neurons, whereas transfection with 75 pmol of GAPDH siRNA decreased cytoplasmic GAPDH immunoreactivity in approximately 99% of transfected neurons (Fig. 4a, b), suggesting that the optimized protocol using 75 pmol of small RNA resulted in a transfection efficiency of 75–100% with minimal toxicity. This optimized transfection protocol was then used to examine the effects on BMP-7-induced dendritic growth of transfection with specific miRNA modulators.
Fig. 4.
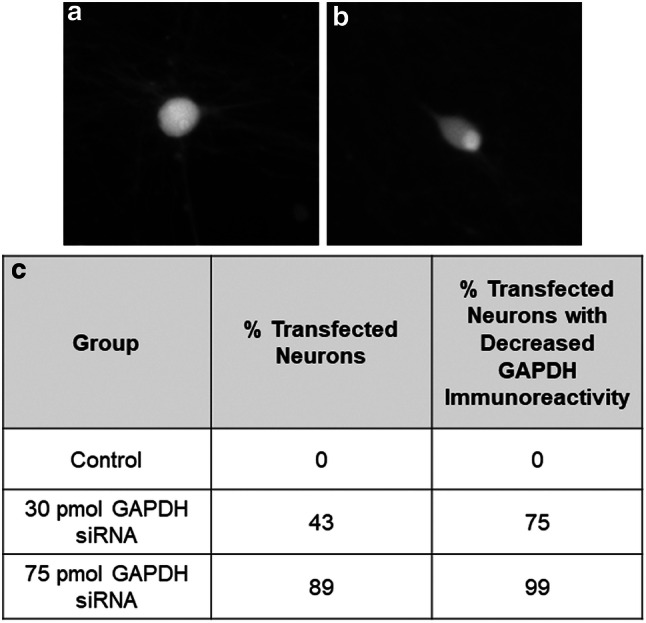
Optimizing small RNA transfection efficiency in cultured embryonic rat sympathetic neurons. a, b Representative photomicrographs of GAPDH protein in cultured sympathetic neurons transfected with either control siRNA (a) or 75 pmol of FAM-labeled siRNA for GAPDH (b). c Transfection efficiency in cultures transfected with 30 versus 75 pmol of FAM-labeled siRNA for GAPDH. N = 100 cells counted for each condition
Overexpression of miR-23b and miR-21 Inhibits BMP-7-induced Dendritic Growth
Previous studies have shown that miR-21 and miR-23b inhibit BMP signaling in many cell types including osteocytes, hepatocytes, keratinocytes (Du et al. 2009; Rogler et al. 2009; Ahmed et al. 2011; Liu et al. 2016). Since miR-21 and miR-23b were decreased in BMP-7-exposed neurons relative to neurons in control medium (Figs. 1, 2b, Table 1), we hypothesized that overexpressing these miRNAs would inhibit BMP-7-induced dendritic growth. To test this hypothesis, we quantified dendritic morphology in sympathetic neurons transfected with miRNA mimics of these specific miRNAs prior to treating cultures with BMP-7. Since miR-26a had been previously shown to be expressed in sympathetic neurons (Natera-Naranjo et al. 2010) but did not show significant differential expression in the presence of BMP-7, cultures were also treated with miR-26a mimic as another control.
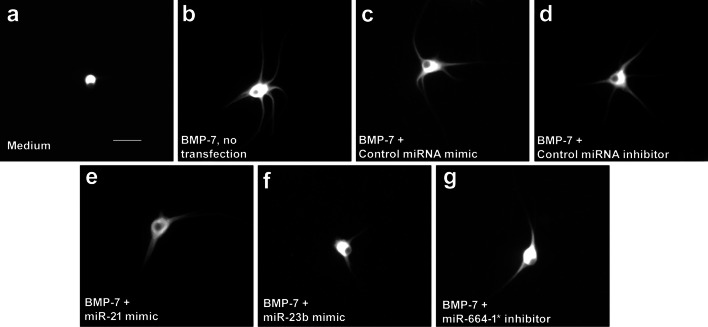
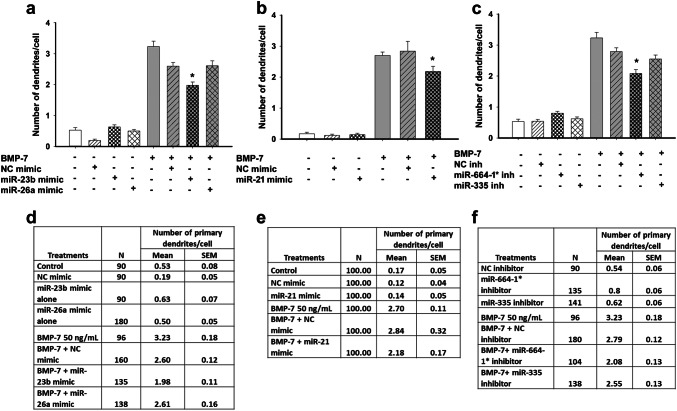
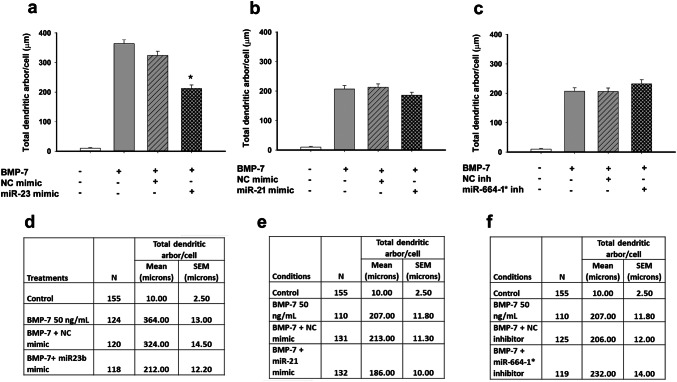
In agreement with previous studies (Lein et al. 1995; Chandrasekaran et al. 2015), rat sympathetic neurons cultured in defined medium in the absence of glial cells did not extend any dendrites (Figs. 5a, 6, 7). In contrast, neurons exposed to BMP-7 at a maximally effective concentration of 50 ng/mL extended multiple MAP-2 immunopositive processes whose shape was characteristic of dendrites (Fig. 5b). Transfection with Lipofectamine® RNAimax alone (data not shown) or with non-specific small RNA mimics or inhibitors (negative controls) had no significant effect on BMP-7-induced dendritic growth, as determined by the number of dendrites extended by neurons or the total length of the dendritic arbor compared to untransfected neurons treated with BMP-7 (Figs. 5c, d, 6, 7). Similarly, transfection with a rno-miR-26a mimic had no significant effect on dendritic number (Fig. 6a, d) or total dendritic arbor (data not shown) in the absence or presence of BMP-7. In the absence of BMP-7, transfection with rno-miR-21 mimic, or rno-miR-23b mimic, neither triggered nor inhibited dendritic growth (Fig. 6a, b, d, e). However, transfection with the miR-23b mimic significantly decreased, but did not completely block, BMP-7-induced dendritic growth, as evidenced by a significant reduction in both the number of dendrites (Figs. 5e, 6a, d, 7a, d) and the total dendritic arbor (Figs. 5e, 6d) compared to BMP-7-treated neurons transfected with the negative control mimic (Figs. 5c, 6a, d). Transfection with the miR-21 mimic significantly reduced the number of dendrites, but not the total dendritic arbor in BMP-7-treated cultures (Fig. 5d, 6b, e, 7b, e). Conversely, transfection with a miR-21 inhibitor had no effect on dendritic growth in the absence or presence of BMP-7 (data not shown).
Fig. 5.
Overexpression of miR-23b and miR-21 or inhibition of miR-664-1* altering BMP-7-induced dendritic growth in sympathetic neurons. Representative fluorescence micrographs of cultured sympathetic neurons immunostained for the dendrite-selective cytoskeletal protein, microtubule associated protein-2 (MAP-2). Neurons were grown in the absence (a) or presence (b–g) of BMP-7 at 50 ng/ml from DIV 5-10. BMP-7-induced dendritic growth was examined in untransfected neurons (b) or in neurons transfected with negative controls for miRNA mimics (c), negative controls for miRNA inhibitors (d), specific miRNA mimics (e, f), or specific mRNA inhibitors (g). Bar = 50 μM
Fig. 6.
Effect of miRNA modulation on number of primary dendrites in sympathetic neurons. Sympathetic neurons were transfected with mimics for miR-21, miR-23b, or miR-26a or inhibitors for miR-335 or miR-664-1* using Lipofectamine RNAimax. Controls were untransfected or transfected with negative controls (NC) for the miRNA mimics and inhibitors. Following transfection, cultures were treated with control medium or BMP-7 (50 ng/mL) for 5 days. The number of dendrites per neuron was quantified in cultures immunostained for MAP-2 to identify dendritic processes. Data are expressed as the mean ± SEM. a–c show the graphical representation of the data and d–f show the numerical data including the N for each condition, mean, and SEM. Statistical significance was assessed using one-way ANOVA, followed by Tukey’s post hoc test. *Significantly different from cultures transfected with respective negative controls in the presence of BMP-7 at p ≤ 0.05
Fig. 7.
Effect of miRNA modulation on total dendritic arbor of BMP-7-treated neurons. Sympathetic neurons were transfected with mimics for miR-21 or miR-23b or inhibitors for miR-664-1* using Lipofectamine RNAimax. Controls were untransfected or transfected with negative controls (NC) for the miRNA mimics and inhibitors. Following transfection, cultures were treated with control medium or BMP-7 (50 ng/mL) for 5 days. The total length of the dendritic arbor per neuron was quantified in cultures immunostained for MAP-2 to identify dendritic processes. Data are expressed as the mean ± SEM. a–c show the graphical representation of the data and d–f show the numerical values including N for each condition, mean, and SEM. Statistical significance was assessed using one-way ANOVA, followed by Tukey’s post hoc test. *Significantly different from cultures transfected with respective negative controls in the presence of BMP-7 at p ≤ 0.05
Inhibition of miR-664-1* Decreases the Number of Dendrites Induced by BMP-7
The expression levels of miR-335 and miR-664-1* were increased following BMP-7 treatment (Figs. 1, 2, 3). To determine whether the function of these miRNAs is required for BMP-7-induced dendritic growth, dendritic growth was quantified in sympathetic cultures transfected with miRNA inhibitors that specifically targeted these miRNAs. As observed with the miRNA mimics, transfection of sympathetic neurons with the miRNA inhibitors did not alter dendritic growth in the absence of BMP-7 (Fig. 6c, f). BMP-7-treated neurons transfected with the miR-664-1* inhibitor showed a significantly reduced dendritic number, but a total dendritic arbor that was comparable in length to that of BMP-7-treated neurons transfected with the negative control inhibitor (Fig. 5g, 6c, f, 7c, f). Transfection with the miR-335 inhibitor had no significant effect on BMP-7-induced dendritic growth with respect to either dendritic number (Fig. 6c, f) or total length of the dendritic arbor (data not shown).
Possible Targets for miR-21, miR-23b, and miR-664-1*
MiRNAs are known to not only modulate the expression of individual target genes within the cell, but also to coordinately regulate the expression of a number of protein-coding genes (Carthew and Sontheimer 2009; Tai and Schuman 2006). Therefore, we used bioinformatics to generate a list of potential target mRNAs that could be regulated by these three miRNAs during BMP-7-induced dendritic growth. Since there are few validated targets of these miRNAs, we used not only miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/php/index.php) to identify validated targets with target scores of 85 or higher, but also miRDB (http://mirdb.org/) to predict targets for these miRNAs. We identified 17 validated targets and 47 predicted targets for miR-21, two validated targets and 141 predicted targets for miR-23b, and 78 predicted targets for miR-664-1*. The genes coded for proteins involved in a wide variety of cellular processes including membrane transport, cytoskeleton organization, tyrosine kinase signaling, cytokine signaling, RNA processing, transcriptional regulation, and protein degradation. A subset of these genes (Table 2) are known to regulate dendritic growth, dendritic spine formation, axonal growth, and TGF-β/BMP signaling in other cell types (Garred et al. 2011; Guo et al. 2001; Pavlopoulos et al. 2011; Wotton and Massague 2000). Therefore, these genes are potential candidates for downstream targets of miR-21, miR-23b, and miR-664-1* during dendritic growth regulation.
Table 2.
Selected candidate targets for miR-21, miR-23b, and miR-664-1* that may regulate BMP-7-induced dendritic growth in sympathetic neurons
| Target mRNA | Accession ID | Go annotation |
|---|---|---|
| Rno-miR-21-5p validated targets | ||
| Programmed cell death 4 (Pdc4) | NM_022265 | Apoptotic signaling, BMP signaling |
| Tropomyosin 1 (Tpm1) | NM_001034069 | Actin binding, cytoskeletal protein binding |
| Potassium voltage-gated channel subfamily J member 16 (Kcnj16) | NM_053314 | Regulation of transmembrane ion transport |
| Phosphatase and tensin homolog (Pten) | NM_031606 | Brain morphogenesis, dendritic spine morphogenesis, neuronal differentiation |
| Pellino E3 ubiquitin protein ligase 1 (Peli1) | NM_001100565 | Protein ubiquitination, Toll signaling |
| Integrin subunit beta 1 (ITGB1) | NM_001039254 | Axon extension, dendrite morphogenesis, integrin binding, laminin binding |
| ELAV-like RNA binding protein 4 (Elavl4) | NM_001077651 | Dendrite morphogenesis, learning |
| SKI proto-oncogene (Ski) | XM_017603153 | SMAD binding, negative regulator of BMP signaling, myelination in peripheral nervous system |
| TIMP metallopeptidase inhibitor 3 (Timp3) | NM_012886 | Negative regulation of ERK signaling, positive regulation of apoptotic signaling pathway |
| SMAD family member 7 (Smad7) | NM_030858 | Negative regulator of BMP signaling |
| T-cell lymphoma invasion and metastasis 1 (Tiam1) | NM_001100558.1 | BDNF signaling pathway, dendritic spine morphogenesis, axogenesis |
| Rno-miR-21-5p predicted targets | ||
| Transforming growth factor, beta-induced (Tgfbi) | NM_053802 | Chondrocyte differentiation, cell adhesion, angiogenesis |
| Cytoplasmic polyadenylation element binding protein 3 (Cpeb3) | XM_017590345.1. | Regulation of dendritic spine development, synaptic plasticity |
| Kruppel-like factor 3 (basic) (Klf3) | NM_001105742 | Negative transcriptional regulator |
| Neurotrophin 3 (Ntf3) | NM_001270869 | Axon guidance, nervous system development |
| G protein-coupled receptor 64 (Gpr64) | NM_001270872 | Orphan receptor, cell signaling |
| Rho GTPase activating protein 24 (Arhgap24) | NM_001012032 | Cell differentiation, GTPase activity |
| Eph receptor A4 (Epha4) | NM_001162411 | Axon guidance, dendritic spine morphogenesis, neuron projection fasciculation |
| A kinase (PRKA) anchor protein 6 (Akap6) | NM_022618 | cAMP-dependent PKA activity, regulation of epinephrine response |
| Jagged 1 (Jag1) | NM_019147 | Notch signaling, negative regulator of neuronal differentiation |
| Rno-miR-23b-3p predicted targets | ||
| G protein-coupled receptor 64 (Gpr64) | NM_001270872 | G protein receptor signaling |
| FERM domain-containing 5 (Frmd5) | XM_230513 | Cytoskeletal structure, protein kinase binding, integrin binding, regulation of cellular migration, cell adhesion, cell motility |
| Protein kinase (cAMP-dependent, catalytic) inhibitor alpha (Pkia) | NM_053772 | Negative regulator of cAMP-dependent protein kinase activity, negative regulator of protein import into the nucleus |
| Inturned planar cell polarity protein (Intu) | XM_006232293 | Smoothened signaling, cilium assembly, limb development, keratinocyte development |
| Casein kinase 1, gamma 3 (Csnk1g3) | NM_022855 | Wnt signaling, cell shape, endocytosis, protein phosphorylation |
| Latrophilin 2 (Lphn2) | NM_001190475 | Synapse assembly, PDZ domain binding |
| Mastermind-like 2 (Maml2) | XM_001072241 | Notch signaling pathway |
| Mediator complex subunit 12 (Med12) | NM_001193292 | Wnt signaling, protein ubiquitination, spinal cord development |
| Transforming growth factor, beta receptor III (Tgfbr3) | NM_017256 | TGF-beta signaling, BMP signaling, response to hypoxia, |
| E74-like factor 2 (Elf2) | NM_001033909 | Transcriptional regulation |
| Ubiquitin-like modifier-activating enzyme 6 (Uba6) | NM_001107213 | Protein ubiquitination, hippocampal development, dendritic spine morphogenesis |
| Lysophosphatidic acid receptor 1 (Lpar1) | NM_053936 | Inhibiting G protein-coupled signaling pathway, positive regulator of dendritic spine development, apoptosis, NF-kappaB pathway |
| SIX homeobox 4 (Six4) | NM_001106739 | Negative regulator of neuronal apoptosis, regulator of synaptic growth |
| Zinc finger protein 423 (Zfp423) | NM_053583 | BMP signaling, olfactory neuron development |
| Rho-associated coiled-coil containing protein kinase 2 (Rock2) | NM_013022 | Actin cytoskeleton reorganization, dendrite morphogenesis, Rho signal transduction |
| Cysteine-rich PDZ-binding protein (Cript) | NM_019907 | Microtubule organization, regulation of post synaptic density protein 95 clustering |
| SMAD-specific E3 ubiquitin protein ligase 2 (Smurf2) | NM_001107061 | SMAD binding, negative regulator of TGF-beta signaling pathway |
| Nedd4 binding protein 1 (N4bp1) | XM_001068071 | Negative regulation of protein ubiquitination |
| BMP-2-inducible kinase (Bmp2 k) | XM_006250707 | Regulation of bone mineralization, positive regulation of Notch signaling pathway, regulation of clathrin-dependent endocytosis |
| Nedd4 family-interacting protein 2 (Ndfip2) | NM_001108390 | Positive regulation of protein ubiquitination, positive regulation of I-kappaB kinase/NF-kappaB signaling |
| Rno-miR-664-1* (rno-miR-664-1-5p) predicted targets | ||
| Rho guanine nucleotide exchange factor (GEF) 15 (Arhgef15) | NM_001105789 | Negative regulation of synapse maturation |
| Protein tyrosine kinase 2 beta (Ptk2b) | NM_017318 | Negative regulator of apoptosis, regulator of neuronal projection, positive regulator of ROS |
| Synaptotagmin I (Syt1) | NM_001033680 | Positive regulation synaptic transmission, dendrite regulation |
| Reticulon 4 receptor-like 1 (Rtn4rl1) | NM_181377 | Cytokine signaling pathway, negative regulation of axon regulation |
| Serum response factor (Srf) | NM_001109302 | Cell–cell adhesion, neuron migration, hippocampus development |
| Arginine-glutamic acid dipeptide (RE) repeats (Rere) | NM_053885 | Dendrite morphogenesis, nerve branching morphogenesis |
| Protein tyrosine phosphatase, receptor type F (Ptprf) | NM_019249 | Neurite outgrowth, axon regeneration, dendrite morphogenesis |
| Sterile alpha and TIR motif-containing 1 (Sarm1) | NM_001105817 | Dendrite morphogenesis, neuron death |
| GLI family zinc finger 2 (Gli2) | NM_001107169 | Anterior–posterior patterning, axon guidance, neuronal development, hindbrain development |
| Calcium/calmodulin-dependent protein kinase ID (Camk1d) | NM_001107365 | Dendritic development, neuronal differentiation |
| SIX homeobox 3 (Six3) | NM_023990 | Brain development, negative regulation of neuronal differentiation |
| 3-phosphoinositide-dependent protein kinase-1 (Pdpk1) | NM_031081 | Negative regulator of TGF-beta signaling, neuron apoptosis, focal adhesion assembly |
| Josephin domain-containing 1 (Josd1) | NM_001025009 | Protein ubiquitination (deubiquitination) |
| Ankyrin repeat domain 1 (Ankrd1) | NM_013220 | TGF-beta signaling pathway, cellular response to hypoxia, cardiac tissue morphogenesis |
| Ubiquitin-specific peptidase 49 (Usp49) | NM_001136470 | Protein deubiquitination |
| Glutathione peroxidase 7 (Gpx7) | NM_001106673 | Response to oxidative stress, peroxidase activity |
| Nedd4 binding protein 1 (N4bp1) | XM_001068071 | Negative regulation of protein ubiquitination |
Discussion
Our data support the hypothesis that miRNA are important downstream mediators of BMP-7-induced dendritic growth in sympathetic neurons. Specifically, we demonstrated that (1) BMP-7 differentially regulates expression of numerous miRNAs; and (2) BMP-7-induced dendritic growth is significantly reduced by either overexpressing specific miRNAs downregulated by BMP-7 or inhibiting specific miRNAs upregulated by BMP. These findings extend previous studies showing that miRNAs play a critical role during neuronal differentiation, dendritic growth, dendritic spine development, and synaptic plasticity in the central nervous system (Impey et al. 2010; Konopka et al. 2011; Hong et al. 2013; Maciotta et al. 2013). Specifically miR-134 has been shown to reduce dendritic complexity in hippocampal neurons, whereas miR-132, miR-195, and miR-9 are necessary to maintain dendritic complexity in these central neurons (Bicker et al. 2014; Chen et al. 2017a, b; Magill et al. 2010; Meza-Sosa et al. 2014; Tai and Schuman 2006). While miRNAs have been localized to the axonal compartment and neuronal cell bodies of rat sympathetic neurons (Natera-Naranjo et al. 2010), their function in peripheral neurons is not fully understood. Our study provides the first insight into the importance of miRNAs in regulating dendritic growth in sympathetic neurons.
A previous study showed that inhibition of transcription within the first 24 h of BMP-7 treatment inhibited dendritic growth but there was no effect of transcriptional inhibition on dendritic growth if the inhibitor was added later than 48 h after BMP-7 treatment (Garred et al. 2011). Also, many mRNAs were modulated by BMP-7 within 6 h (Garred et al. 2011). Since the focus of the study was to elucidate the importance of miRNAs for the initiation of dendritic growth and to identify miRNAs that were among the early response genes, a similar time course of 6 h and 24 h was used for this study. Further studies with later time points might reveal additional function for miRNAs in synaptogenesis and dendritic remodeling at later stages.
Our microarray analyses identified 22 of 762 mature rat miRNAs that showed detectable expression and were differentially regulated by BMP-7 at a concentration known to trigger dendritic growth in sympathetic neurons. The overall microarray signal intensity of many of these differentially regulated miRNAs was low, suggesting that these are generally present in low abundance in sympathetic neurons. Of these 22 miRNAs, 9 (miR-335, miR-30a, miR-210, miR-17, miR-29c, miR-21, miR-134, miR-let-7i, and miR-23b) have been reported to be expressed in the somatodendritic compartment of hippocampal neurons (van Spronsen et al. 2013), and miR-23b has been differentially localized between the soma or axons in superior cervical ganglia neurons derived from 3-day-old rat pups (Natera-Naranjo et al. 2010). miR-134 and miR-21 have been implicated in the regulation of dendritic growth or neurite outgrowth in various neuronal cell types (Strickland et al. 2011; van Spronsen et al. 2013; Bicker et al. 2014). Interestingly, many of these miRNAs showed significantly increased or decreased gene expression within the first 6 h following BMP-7 exposure. The observation that these genes are regulated during the early stages of BMP-induced dendritic growth suggests that these miRNAs could be important signaling molecules linking activation of BMP receptors to increased dendritogenesis in sympathetic neurons.
Four of the miRNAs identified by microarray analyses as being differentially regulated by BMP-7, i.e., miR-664-1*, miR-335, miR-23b, and miR-21, exhibited similar patterns of gene expression when analyzed by qPCR. However, there were differences in the fold changes observed across the biological replicates; thus, with the exception of miR-21, the changes in miRNA expression over time or across treatments were not found to be statistically significant. Also, although the overall profile of the changes in miRNA across treatments was similar between qPCR and the microarray, there were differences between the microarray and qPCR analyses in observed fold changes in gene expression across conditions. It has been previously reported that there is low correlation between microarray and qPCR even when using the same RNA sample with high variability among biological replicates for low abundance miRNAs (Chen et al. 2009). Also, another study showed that using different platforms for validating miRNA expression can lead to different results with respect to profiles of differentially regulated miRNA and fold changes in expression, especially when using qPCR to analyze samples with low miRNA expression (Mestdagh et al. 2014). Therefore, it is possible that the lack of concordance between the microarray and qPCR data in our experiment is due to one or more of the following reasons: (1) low total RNA yield; (2) low abundance of individual miRNAs in these neurons; or (3) the small fold changes in gene expression observed following BMP-7 treatment.
Our data demonstrated that BMP-7 decreased expression of miR-21 and miR-23b in sympathetic neurons, and overexpressing miR-23b or miR-21 in BMP-7-treated neurons inhibited BMP-7-induced dendritic growth. A previous study on the transcriptional changes elicited by BMP-7 showed that the majority of the changes in gene regulation occurred within the first 24 h after addition of BMP-7 to cultured sympathetic neurons (Garred et al. 2011); however, BMP-7 effects on dendritic growth persist beyond 24 h. Similarly, although the miRNAs investigated in this study are altered in the first 6–24 h after addition of BMP-7 to the cultures, it is likely that their target genes maintain altered expression levels over a longer duration of time, which could explain the observed effects over a 5-day period.
Our data suggest that miR-23b and miR-21 prevent sympathetic neurons from extending dendrites in the absence of BMP signaling. These data are in agreement with previous studies showing that miR-21 and miR-23b are downregulated by BMP-7 and inhibit BMP signaling in multiple cell types (Luzi et al. 2008; Rogler et al. 2009; Ahmed et al. 2011; Diotel et al. 2015; Su et al. 2015). Thus, our study provides further evidence supporting the role of miR-23b and miR-21 as downstream targets of BMP signaling. In addition, overexpression of miR-23b decreased both dendritic number and total dendritic arbor, whereas inhibition of miR-21 only decreased dendrite number. These studies suggest that extension of primary dendrites and growth and elongation or branching of existing dendrites are differentially regulated in sympathetic neurons.
A unique finding of our study was the observation that miR-644-1* was significantly upregulated by BMP-7 in sympathetic neurons. Inhibition of miR-664-1* inhibited BMP-7-induced dendritic growth. These data are the first reported evidence of a role for miR-664-1* in the BMP signaling pathway and in the developing nervous system. Previous studies have demonstrated the ability to observe dendritic retraction in cultured neurons over time (Guo et al. 1997, 1999). Further studies to examine the changes in dendritic number and arbor over time following modulation of the miRNAs and to identify targets of these miRNAs during dendritogenesis will help us better understand the mechanisms underlying miRNA-mediated regulation of dendrite number and dendrite length in these neurons.
Despite observing that BMP-7 strongly upregulated miR-335 expression in the microarray analyses in sympathetic neurons, inhibiting miR-355 had no effect on BMP-7-induced dendritic growth. Previous studies have shown that expression of miR-335 is altered in the central nervous system following NMDA stimulation and in dorsal root ganglia neurons following peripheral nerve injury (Kye et al. 2011; Norcini et al. 2014). miR-335 has also been shown to function as an anti-mitotic and pro-apoptotic in mouse cortical neurons (Sathyan et al. 2007). It is, therefore, possible that miR-335 is involved in aspects of other than dendritic growth that are regulated by BMP-7 in these neurons. This is a question that warrants further investigation.
Although overexpressing miRNAs downregulated by BMP-7 or inhibiting miRNA upregulated by BMP-7 had statistically significant effects on BMP-7-induced dendritic growth, experimental modulation of any individual miRNA did not completely block BMP-induced dendritic growth or robustly reduce the size of the dendritic arbor. Also, we were unable to induce or enhance dendritic growth by modulating the levels of an individual miRNA in control neurons, in the absence of BMP-7. One possible explanation is that modulating just one miRNA is not sufficient to affect all the targets needed for dendritic growth. In support of this hypothesis, target mRNAs have been shown to contain binding sites for multiple miRNAs (Saj and Lai 2011). Moreover, the study by Garred et al. (2011) identified many genes upregulated by BMP-7 during the induction of dendritic growth in sympathetic neurons, at least some of which are likely direct targets of BMP-7 signaling and do not involve miRNA as an intermediate. Previous studies have also shown that altering just one pathway or one particular target gene is not sufficient to induce dendritic growth in the absence of BMP-7 (Drahushuk et al. 2002; Kim et al. 2004; Lein et al. 2007), suggesting that altering the expression of a single gene or pathway downstream of BMP-7 might not be sufficient to trigger the entire program of dendritic growth in control neurons. Further studies are necessary to better understand the interplay between the BMP-7-regulated miRNAs during dendritic growth regulation.
MicroRNAs mediate their effects through binding to the 3′UTR of target mRNAs. The bioinformatics analyses revealed potential targets for miR-21, miR-23b, and miR-664-1* that could be downstream of BMP-7 signaling and involved in dendritic growth regulation. These targets were chosen because of their connection to BMP signaling or their role in processes associated with BMP signaling or dendritic growth such as cytoskeletal reorganization, neuronal differentiation, brain morphogenesis, and protein degradation (Lein et al. 1995; Guo et al. 2001).
These bioinformatics studies provide a starting point for further biochemical and genetic studies for identifying genes that are regulated by these miRNAs and are necessary for BMP-7-induced dendritic growth in sympathetic neurons. Previous studies have shown that the expression of two of the predicted target genes for miR-21—Jagged 1 and Elavl4—is increased in cultured sympathetic neurons following BMP-7 exposure with both mRNAs showing strong upregulation in gene expression at 24 h of BMP-7 treatment (Garred et al. 2011). Since the timing of decrease of the expression of miR-21 observed in our study precedes the increase in the expression of these genes in the Garred et al. study, it is possible that downregulation of miR-21 expression might be causing the increase in the expression of these genes. Further biochemical studies are required to determine if Elaval4 and Jagged 1 are targets of miR-21 in sympathetic neurons and whether the presence of these mRNAs in BMP-7-treated neurons is necessary for BMP-7-induced dendritic growth.
Similarly, previous studies have shown that proteasome activity is necessary for BMP-7-induced dendritic growth in sympathetic neurons (Guo et al. 2001). Our bioinformatics analysis showed predicted target genes for miR-21 and miR-23b included a number of genes associated with protein ubiquitination—Pellino E3 ubiquitin protein ligase 1 (Peli1), Ubiquitin-like modifier-activating enzyme 6 (Uba6), Nedd4 family-interacting protein 2 (Ndfip2), and SMAD-specific E3 ubiquitin protein ligase 2 (Smurf2), while putative miR-664-1* targets included proteins involved in protein deubiquitination such as Ubiquitin-specific peptidase 49 (Usp49) and Nedd4 binding protein 1 (N4bp1). Further biochemical studies are necessary to determine whether the expression levels of these genes are altered by the knockout or overexpression of these miRNAs in sympathetic neurons and if these genes are mediators of BMP-7 signaling during dendritic growth.
Dendritic hypertrophy of sympathetic neurons in the SCG has been observed in spontaneously hypertensive rat and is thought to contribute to pathogenesis of hypertension in animal models (Kondo et al. 1990; Peruzzi et al. 1991; Jackson et al. 2013). Similarly, statins, which decrease blood pressure, have been shown to reduce sympathetic activity and decrease dendritic growth in sympathetic neurons (Kishi et al. 2003; Kim et al. 2009). Some of the miRNAs identified in our study such as miR-21, miR-382, miR-210, and miR-23b are modulated in hypertensive subjects and are being explored as potential markers for hypertension (Wei et al. 2013; Shi et al. 2015; Li et al. 2018). Also, autonomic dysfunction has been observed in neurodegenerative diseases such as Parkinson’s disease and is correlated with disease progression and mortality (De Pablo-Fernandez et al. 2017; Merola et al. 2018). MicroRNAs are being explored as biomarkers and therapeutic targets for Parkinson’s disease and some of miRNAs identified in our study including miR-335, miR-21, let-7i, miR-134, and miR-410 have been shown to be differentially regulated in the brain or blood cells in Parkinson’s disease patients (Maciotta et al. 2013; Shinde et al. 2015; Leggio et al. 2017). Further studies are needed to fully understand if and how the changes in dendritic growth induced by these miRNAs contribute to blood pressure regulation and neurodegenerative diseases.
Conclusions
In summary, we identified a subset of microRNAs in the rat genome that are regulated by BMP-7 during the initiation of dendritic growth in sympathetic neurons. Of these differentially regulated miRNAs, we demonstrated that miR-23b and miR-21 inhibit BMP-7-induced dendritic growth, whereas miR-664-1 is important for regulating dendritic number in these neurons. Collectively, these data provide the first evidence that miRNAs are important downstream mediators of BMP-induced dendritic growth in sympathetic neurons.
Acknowledgements
We thank Donald Bruun and Hao Chen at UC Davis for assistance in setting up and maintaining sympathetic cultures.
Author Contributions
KP did most of the qpCR analysis and the functional studies on individual miRNAs, and helped draft the manuscript. KW completed the functional studies, ST did the microarray studies, and TB and KT helped with the optimization of the qPCR analysis and the functional studies using GAPDH siRNA. PL provided the funding and resources for animal work, and VC designed the study, provided some of the funding for the study, isolated neurons for the study, supervised the students during experimental setup and data analysis, and wrote the manuscript.
Funding
This work was supported by the Saint Mary’s College Summer Research Program (KP, ST, TB) Faculty Development Fund (VC), Faculty Research Grant (VC), and by the National Institute of Environmental Health Sciences (grants ES014901 and ES017592 to PJL). The funding agencies were not involved in the study design, in the collection, analysis, or interpretation of data, in the writing of the report, or in the decision to submit the paper for publication.
Compliance with Ethical Standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical Approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. All procedures performed in studies involving animals were in accordance with the ethical standards of University of California at Davis and Saint Mary’s College of California, where these studies were conducted. This article does not contain any studies with human participants performed by any of the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Ahmed MI, Mardaryev AN, Lewis CJ et al (2011) MicroRNA-21 is an important downstream component of BMP signalling in epidermal keratinocytes. J Cell Sci 124:3399–3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicker S, Lackinger M, Weiß K, Schratt G (2014) MicroRNA-132, -134, and -138: a microRNA troika rules in neuronal dendrites. Cell Mol Life Sci. 10.1007/s00018-014-1671-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boivin GP, Hickman DL, Creamer-Hente MA et al (2017) Review of CO2 as a Euthanasia agent for laboratory rats and mice. J Am Assoc Lab Anim Sci 56:491–499 [PMC free article] [PubMed] [Google Scholar]
- Bruckenstein DA, Higgins D (1988) Morphological differentiation of embryonic rat sympathetic neurons in tissue culture. Dev Biol 128:337–348. 10.1016/0012-1606(88)90296-5 [DOI] [PubMed] [Google Scholar]
- Caceres A, Banker G, Steward O et al (1984) MAP2 is localized to the dendrites of hippocampal neurons which develop in culture. Brain Res 315:314–318 [DOI] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ (2009) Origins and mechanisms of miRNAs and siRNAs. Cell 136:642–655. 10.1016/j.cell.2009.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran V, Lea C, Sosa JC et al (2015) Reactive oxygen species are involved in BMP-induced dendritic growth in cultured rat sympathetic neurons. Mol Cell Neurosci 67:116–125. 10.1016/j.mcn.2015.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Gelfond JAL, McManus LM, Shireman PK (2009) Reproducibility of quantitative RT-PCR array in miRNA expression profiling and comparison with microarray analysis. BMC Genom 10:407. 10.1186/1471-2164-10-407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Qiu F, Zhou K et al (2017a) Pathogenic role of microRNA-21 in diabetic retinopathy through downregulation of PPARα. Diabetes 66:1671–1682. 10.2337/db16-1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Jiang X-M, Zhao L-J et al (2017b) MicroRNA-195 prevents dendritic degeneration and neuron death in rats following chronic brain hypoperfusion. Cell Death Dis 8:e2850. 10.1038/cddis.2017.243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comery TA, Harris JB, Willems PJ et al (1997) Abnormal dendritic spines in fragile X knockout mice: maturation and pruning deficits. Proc Natl Acad Sci USA 94:5401–5404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copf T (2016) Impairments in dendrite morphogenesis as etiology for neurodevelopmental disorders and implications for therapeutic treatments. Neurosci Biobehav Rev 68:946–978. 10.1016/J.NEUBIOREV.2016.04.008 [DOI] [PubMed] [Google Scholar]
- Davies DC (1978) Neuronal numbers in the superior cervical ganglion of the neonatal rat. J Anat 127:43–51 [PMC free article] [PubMed] [Google Scholar]
- Davis TH, Cuellar TL, Koch SM et al (2008) Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J Neurosci 28:4322–4330. 10.1523/JNEUROSCI.4815-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BN, Hilyard AC, Nguyen PH et al (2010) Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha. Mol Cell 39:373–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Castro F, Sánchez-Vives MV, Muñoz-Martínez EJ, Gallego R (1995) Effects of postganglionic nerve section on synaptic transmission in the superior cervical ganglion of the guinea-pig. Neuroscience 67:689–695. 10.1016/0306-4522(95)00079-X [DOI] [PubMed] [Google Scholar]
- De Pablo-Fernandez E, Tur C, Revesz T et al (2017) Association of autonomic dysfunction with disease progression and survival in Parkinson disease. JAMA Neurol 74:970. 10.1001/jamaneurol.2017.1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santi C, Vencken S, Blake J et al (2017) Identification of MiR-21-5p as a functional regulator of mesothelin expression using MicroRNA capture affinity coupled with next generation sequencing. PLoS ONE 12:e0170999. 10.1371/journal.pone.0170999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diotel N, Beil T, Strähle U, Rastegar S (2015) Differential expression of id genes and their potential regulator znf238 in zebrafish adult neural progenitor cells and neurons suggests distinct functions in adult neurogenesis. Gene Expr Patterns 19:1–13. 10.1016/j.gep.2015.05.004 [DOI] [PubMed] [Google Scholar]
- Dorval V, Smith PY, Delay C et al (2012) Gene network and pathway analysis of mice with conditional ablation of Dicer in post-mitotic neurons. PLoS ONE 7:e44060. 10.1371/journal.pone.0044060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drahushuk K, Connell TD, Higgins D (2002) Pituitary adenylate cyclase-activating polypeptide and vasoactive intestinal peptide inhibit dendritic growth in cultured sympathetic neurons. J Neurosci 22:6560–6569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Yang S, An D et al (2009) BMP-6 inhibits microRNA-21 expression in breast cancer through repressing delta EF1 and AP-1. Cell Res 19:487–496 [DOI] [PubMed] [Google Scholar]
- Fay MJ, Alt LAC, Ryba D et al (2018) Cadmium nephrotoxicity is associated with altered MicroRNA expression in the rat renal cortex. Toxics 6:16. 10.3390/toxics6010016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Liang Y, Wang F, Chen J (2013) Detection of genetically modified tomato using PCR coupled with muParaflo microfluidics microarrays. J Nanosci Nanotechnol 13:8266–8274 [DOI] [PubMed] [Google Scholar]
- Garey L (2010) When cortical development goes wrong: schizophrenia as a neurodevelopmental disease of microcircuits. J Anat 217:324–333. 10.1111/j.1469-7580.2010.01231.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garred MM, Wang MM, Guo X et al (2011) Transcriptional responses of cultured rat sympathetic neurons during BMP-7-induced dendritic growth. PLoS ONE 6:e21754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X, Zheng L, Huang M et al (2015) MicroRNA expression profiles associated with acquired gefitinib-resistance in human lung adenocarcinoma cells. Mol Med Rep 11:333–340. 10.3892/mmr.2014.2757 [DOI] [PubMed] [Google Scholar]
- Ghogha A, Bruun DA, Lein PJ (2012) Inducing dendritic growth in cultured sympathetic neurons. J Vis Exp 61:e3546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Metzler-Northrup J, Lein P et al (1997) Leukemia inhibitory factor and ciliary neurotrophic factor regulate dendritic growth in cultures of rat sympathetic neurons. Dev Brain Res 104:101–110. 10.1016/S0165-3806(97)00142-9 [DOI] [PubMed] [Google Scholar]
- Guo X, Chandrasekaran V, Lein P et al (1999) Leukemia inhibitory factor and ciliary neurotrophic factor cause dendritic retraction in cultured rat sympathetic neurons. J Neurosci 19:2113–2121. 10.1523/JNEUROSCI.19-06-02113.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Lin Y, Horbinski C et al (2001) Dendritic growth induced by BMP-7 requires Smad1 and proteasome activity. J Neurobiol 48:120–130 [PubMed] [Google Scholar]
- Hackbarth H, Küppers N, Bohnet W (2000) Euthanasia of rats with carbon dioxide-animal welfare aspects. Lab Anim 34:91–96. 10.1258/002367700780578055 [DOI] [PubMed] [Google Scholar]
- Harada A, Teng J, Takei Y et al (2002) MAP2 is required for dendrite elongation, PKA anchoring in dendrites, and proper PKA signal transduction. J Cell Biol 158:541–549. 10.1083/jcb.200110134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins D, Lein P, Osterhout DJ, Johnson M (1991) Tissue culture of autonomic neurons. In: Banker G, Goslin K (eds) Culturing nerve cells. MIT Press, Cambridge, pp 177–205 [Google Scholar]
- Higgins D, Burack M, Lein P, Banker G (1997) Mechanisms of neuronal polarity. Curr Opin Neurobiol 7:599–604. 10.1016/S0959-4388(97)80078-5 [DOI] [PubMed] [Google Scholar]
- Hong J, Zhang H, Kawase-Koga Y, Sun T (2013) MicroRNA function is required for neurite outgrowth of mature neurons in the mouse postnatal cerebral cortex. Front Cell Neurosci 7:151. 10.3389/fncel.2013.00151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horbinski C, Stachowiak MK, Higgins D, Finnegan SG (2001) Polyethyleneimine-mediated transfection of cultured postmitotic neurons from rat sympathetic ganglia and adult human retina. BMC Neurosci 2:2. 10.1186/1471-2202-2-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impey S, Davare M, Lasiek A et al (2010) An activity-induced microRNA controls dendritic spine formation by regulating Rac1-PAK signaling. Mol Cell Neurosci 43:146–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie K, Tsujimura K, Nakashima H, Nakashima K (2016) MicroRNA-214 promotes dendritic development by targeting the Schizophrenia-associated gene Quaking (Qki). J Biol Chem 291:13891–13904. 10.1074/jbc.M115.705749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishteiwy RA, Ward TM, Dykxhoorn DM, Burnstein KL (2012) The microRNA -23b/-27b cluster suppresses the metastatic phenotype of castration-resistant prostate cancer cells. PLoS ONE 7:e52106. 10.1371/journal.pone.0052106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson KL, Marques FZ, Watson AMD et al (2013) A novel interaction between sympathetic overactivity and aberrant regulation of renin by miR-181a in BPH/2 J genetically hypertensive mice. Hypertension 62:775–781. 10.1161/HYPERTENSIONAHA.113.01701 [DOI] [PubMed] [Google Scholar]
- Kang H, Davis-Dusenbery BN, Nguyen PH et al (2012) Bone morphogenetic protein 4 promotes vascular smooth muscle contractility by activating microRNA-21 (miR-21), which down-regulates expression of family of dedicator of cytokinesis (DOCK) proteins. J Biol Chem 287:3976–3986. 10.1074/jbc.M111.303156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katchy A, Williams C (2014) Profiling of estrogen-regulated microRNAs in breast cancer cells. J Vis Exp. 10.3791/51285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann WE, Moser HW (2000) Dendritic anomalies in disorders associated with mental retardation. Cereb Cortex 10:981–991 [DOI] [PubMed] [Google Scholar]
- Kim I-J, Beck HN, Lein PJ, Higgins D (2002) Interferon gamma induces retrograde dendritic retraction and inhibits synapse formation. J Neurosci 22:4530–4539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I-J, Drahushuk KM, Kim W-Y et al (2004) Extracellular signal-regulated kinases regulate dendritic growth in rat sympathetic neurons. J Neurosci 24:3304–3312. 10.1523/JNEUROSCI.3286-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W-Y, Gonsiorek EA, Barnhart C et al (2009) Statins decrease dendritic arborization in rat sympathetic neurons by blocking RhoA activation. J Neurochem 108:1057–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi T, Hirooka Y, Mukai Y et al (2003) Atorvastatin causes depressor and sympatho-inhibitory effects with upregulation of nitric oxide synthases in stroke-prone spontaneously hypertensive rats. J Hypertens 21:379–386. 10.1097/01.hjh.0000052443.12292.22 [DOI] [PubMed] [Google Scholar]
- Kondo M, Terada M, Shimizu D et al (1990) Morphometric study of the superior cervical and stellate ganglia of spontaneously hypertensive rats during the prehypertensive stage. Virchows Arch B Cell Pathol Incl Mol Pathol 58:371–376 [DOI] [PubMed] [Google Scholar]
- Konopka W, Schütz G, Kaczmarek L (2011) The microRNA contribution to learning and memory. Neuroscientist 17:468–474 [DOI] [PubMed] [Google Scholar]
- Kosik KS (2006) The neuronal microRNA system. Nat Rev Neurosci 7:911–920 [DOI] [PubMed] [Google Scholar]
- Kulkarni VA, Firestein BL (2012) The dendritic tree and brain disorders. Mol Cell Neurosci 50:10–20. 10.1016/J.MCN.2012.03.005 [DOI] [PubMed] [Google Scholar]
- Kye MJ, Neveu P, Lee Y-S et al (2011) NMDA mediated contextual conditioning changes miRNA expression. PLoS ONE 6:e24682. 10.1371/journal.pone.0024682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leggio L, Vivarelli S, L’Episcopo F et al (2017) microRNAs in Parkinson’s disease: from pathogenesis to novel diagnostic and therapeutic approaches. Int J Mol Sci 1:2. 10.3390/ijms18122698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein P, Johnson M, Guo X et al (1995) Osteogenic protein-1 induces dendritic growth in rat sympathetic neurons. Neuron 15:597–605 [DOI] [PubMed] [Google Scholar]
- Lein PJ, Guo X, Shi GX et al (2007) The novel GTPase Rit differentially regulates axonal and dendritic growth. J Neurosci 27:4725–4736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein PJ, Fryer A, Higgins D (2010) Cell culture: autonomic and enteric neurons. In: Squire LR (ed) Encyclopedia of Neuroscience. Elsevier Ltd, California, pp 625–632 [Google Scholar]
- Li B, Sun H (2013) miR-26a promotes neurite outgrowth by repressing PTEN expression. Mol Med Rep 8:676–680. 10.3892/mmr.2013.1534 [DOI] [PubMed] [Google Scholar]
- Li X, Wei Y, Wang Z (2018) microRNA-21 and hypertension. Hypertens Res 41:649–661. 10.1038/s41440-018-0071-z [DOI] [PubMed] [Google Scholar]
- Liu FJ, Kaur P, Karolina DS et al (2015) MiR-335 regulates Hif-1α to reduce cell death in both mouse cell line and rat ischemic models. PLoS ONE 10:e0128432. 10.1371/journal.pone.0128432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Hao W, Wang X, Su H (2016) miR-23b targets Smad 3 and ameliorates the LPS-inhibited osteogenic differentiation in preosteoblast MC3T3-E1 cells. J Toxicol Sci 41:185–193. 10.2131/jts.41.185 [DOI] [PubMed] [Google Scholar]
- Luzi E, Marini F, Sala SC et al (2008) Osteogenic differentiation of human adipose tissue-derived stem cells is modulated by the miR-26a targeting of the SMAD1 transcription factor. J Bone Miner Res 23:287–295. 10.1359/jbmr.071011 [DOI] [PubMed] [Google Scholar]
- Maciotta S, Meregalli M, Torrente Y (2013) The involvement of microRNAs in neurodegenerative diseases. Front Cell Neurosci 7:265. 10.3389/fncel.2013.00265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magill ST, Cambronne XA, Luikart BW et al (2010) microRNA-132 regulates dendritic growth and arborization of newborn neurons in the adult hippocampus. Proc Natl Acad Sci USA 107:20382–20387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merola A, Romagnolo A, Rosso M et al (2018) Autonomic dysfunction in Parkinson’s disease: a prospective cohort study. Mov Disord 33:391–397. 10.1002/mds.27268 [DOI] [PubMed] [Google Scholar]
- Mestdagh P, Hartmann N, Baeriswyl L et al (2014) Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat Methods 11:809–815. 10.1038/nmeth.3014 [DOI] [PubMed] [Google Scholar]
- Meza-Sosa KF, Pedraza-Alva G, Perez-Martinez L (2014) microRNAs: key triggers of neuronal cell fate. Front Cell Neurosci 8:175. 10.3389/fncel.2014.00175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natera-Naranjo O, Aschrafi A, Gioio AE, Kaplan BB (2010) Identification and quantitative analyses of microRNAs located in the distal axons of sympathetic neurons. RNA 16:1516–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norcini M, Sideris A, Martin Hernandez LA et al (2014) An approach to identify microRNAs involved in neuropathic pain following a peripheral nerve injury. Front Neurosci 8:266. 10.3389/fnins.2014.00266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo CA, Eberhart CG (2007) The neurobiology of Autism. Brain Pathol 17:434–447. 10.1111/j.1750-3639.2007.00102.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlopoulos E, Trifilieff P, Chevaleyre V et al (2011) Neuralized1 activates CPEB3: a function for nonproteolytic ubiquitin in synaptic plasticity and memory storage. Cell 147:1369–1383. 10.1016/j.cell.2011.09.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzes P, Cahill ME, Jones KA et al (2011) Dendritic spine pathology in neuropsychiatric disorders. Nat Neurosci 14:285–293. 10.1038/nn.2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peruzzi D, Hendley ED, Forehand CJ (1991) Hypertrophy of stellate ganglion cells in hypertensive, but not hyperactive, rats. Am J Physiol 261:R979–R984. 10.1152/ajpregu.1991.261.4.R979 [DOI] [PubMed] [Google Scholar]
- Purves D (1975) Functional and structural changes in mammalian sympathetic neurones following interruption of their axons. J Physiol 252:429–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purves D, Hume RI (1981) The relation of postsynaptic geometry to the number of presynaptic axons that innervate autonomic ganglION. J Neurosci 1:441–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purves D, Lichtman JW (1985) Geometrical differences among homologous neurons in mammals. Science 228:298–302 [DOI] [PubMed] [Google Scholar]
- Purves D, Hadley RD, Voyvodic JT (1986) Dynamic changes in the dendritic geometry of individual neurons visualized over periods of up to three months in the superior cervical ganglion of living mice. J Neurosci 6:1051–1060. 10.1523/JNEUROSCI.06-04-01051.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin W, Zhao B, Shi Y et al (2009) BMPRII is a direct target of miR-21. Acta Biochim Biophys Sin (Shanghai) 41:618–623 [DOI] [PubMed] [Google Scholar]
- Reales E, Bernabé-Rubio M, Casares-Arias J et al (2015) The MAL protein is crucial for proper membrane condensation at the ciliary base, which is required for primary cilium elongation. J Cell Sci 128:2261–2270. 10.1242/jcs.164970 [DOI] [PubMed] [Google Scholar]
- Reinke C, Carthew R (2008) BMP signaling goes posttranscriptional in a microRNA sort of way. Dev Cell 15:174–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogler CE, LeVoci L, Ader T et al (2009) MicroRNA-23b cluster microRNAs regulate transforming growth factor-beta/bone morphogenetic protein signaling and liver stem cell differentiation by targeting Smads. Hepatology 50:575–584. 10.1002/hep.22982 [DOI] [PubMed] [Google Scholar]
- Rubin E (1985) Development of the rat superior cervical ganglion: initial stages of synapse formation. J Neurosci 5:697–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saj A, Lai EC (2011) Control of microRNA biogenesis and transcription by cell signaling pathways. Curr Opin Genet Dev 21:504–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval-Bórquez A, Polakovicova I, Carrasco-Véliz N et al (2017) MicroRNA-335-5p is a potential suppressor of metastasis and invasion in gastric cancer. Clin Epigenetics 9:114. 10.1186/s13148-017-0413-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathyan P, Golden HB, Miranda RC (2007) Competing interactions between micro-RNAs determine neural progenitor survival and proliferation after ethanol exposure: evidence from an ex vivo model of the fetal cerebral cortical neuroepithelium. J Neurosci 27:8546–8557. 10.1523/JNEUROSCI.1269-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Liao J, Liu B et al (2015) Mechanisms and therapeutic potential of microRNAs in hypertension. Drug Discov Today 20:1188–1204. 10.1016/j.drudis.2015.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinde S, Mukhopadhyay S, Mohsen G, Khoo SK (2015) Biofluid-based microRNA biomarkers for Parkinson’s disease: an overview and update. Peer Rev Artic 2:15–25 [Google Scholar]
- Siegel G, Obernosterer G, Fiore R et al (2009) A functional screen implicates microRNA-138- dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat Cell Biol 11:705. 10.1038/ncb1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolen AJ, Wright LL, Cunningham TJ (1983) Neuron numbers in the superior cervical sympathetic ganglion of the rat: a critical comparison of methods for cell counting. J Neurocytol 12:739–750. 10.1007/BF01258148 [DOI] [PubMed] [Google Scholar]
- Strickland IT, Richards L, Holmes FE et al (2011) Axotomy-induced miR-21 promotes axon growth in adult dorsal root ganglion neurons. PLoS ONE 6:e23423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Liao L, Shuai Y et al (2015) MiR-26a functions oppositely in osteogenic differentiation of BMSCs and ADSCs depending on distinct activation and roles of Wnt and BMP signaling pathway. Cell Death Dis 6:e1851–e1851. 10.1038/cddis.2015.221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supekar K, Uddin LQ, Khouzam A et al (2013) Brain hyperconnectivity in children with autism and its links to social deficits. Cell Rep 5:738–747. 10.1016/j.celrep.2013.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai H-C, Schuman EM (2006) MicroRNA: microRNAs Reach out into Dendrites. Curr Biol 16:R121–R123. 10.1016/J.CUB.2006.02.006 [DOI] [PubMed] [Google Scholar]
- van Spronsen M, van Battum EY, Kuijpers M et al (2013) Developmental and activity-dependent miRNA expression profiling in primary hippocampal neuron cultures. PLoS ONE 8:e74907. 10.1371/journal.pone.0074907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigelsø A, Dybboe R, Hansen CN et al (2015) GAPDH and β-actin protein decreases with aging, making Stain-Free technology a superior loading control in Western blotting of human skeletal muscle. J Appl Physiol 118:386–394. 10.1152/japplphysiol.00840.2014 [DOI] [PubMed] [Google Scholar]
- Wang J, Greene SB, Bonilla-Claudio M et al (2010) Bmp signaling regulates myocardial differentiation from cardiac progenitors through a MicroRNA-mediated mechanism. Dev Cell 19:903–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C, Henderson H, Spradley C et al (2013) Circulating miRNAs as potential marker for pulmonary hypertension. PLoS ONE 8:e64396. 10.1371/journal.pone.0064396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wotton D, Massague J (2000) Transcriptional control by the TGF- b/Smad signaling system. EMBO J 19:1745–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Chen W, Liu S et al (2014) Huaier suppresses proliferation and induces apoptosis in human pulmonary cancer cells via upregulation of miR-26b-5p. FEBS Lett 588:2107–2114. 10.1016/j.febslet.2014.04.044 [DOI] [PubMed] [Google Scholar]
- Yuan M, Tang Y, Zhou C et al (2016) Elevated plasma CaM expression in patients with acute cerebral infarction predicts poor outcomes and is inversely associated with miR-26b expression. Int J Neurosci 126:408–414. 10.3109/00207454.2015.1020537 [DOI] [PubMed] [Google Scholar]
- Zhou X et al (2012) MicroRNA Profiling Using µParaflo Microfluidic Array Technology. In: Fan JB (ed) Next-generation MicroRNA expression profiling technology. Humana Press, New York, pp 153–182 [Google Scholar]