Abstract
Purpose:
To differentiate metastatic lesions in the spine originated from primary lung cancer and other cancers using radiomics and deep learning, compared to traditional hot-spot ROI analysis.
Methods:
In a retrospective review of clinical spinal MRI database with a dynamic contrast enhanced (DCE) sequence, a total of 61 patients without prior cancer diagnosis and later confirmed to have metastases (30 lung; 31 non-lung cancers) were identified. For hot-spot analysis, a manual ROI was placed to calculate three heuristic parameters from the wash-in, maximum, and wash-out phases in the DCE kinetics. For each case, the 3D tumor mask was generated by using the normalized-cut algorithm. Radiomics analysis was performed to extract histogram and texture features from three DCE parametric maps. Deep learning was performed using these maps as inputs into a conventional convolutional neural network (CNN), as well as using all 12 sets of DCE images into a convolutional long short term memory (CLSTM) network.
Results:
For hot-spot ROI analysis, mean wash-out slope was 0.25 ± 10% for lung metastases and −9.8 ± 12.9% for other tumors. CHAID classification using a wash-out slope of −6.6% followed by wash-in enhancement ratio of 98% achieved a diagnostic accuracy of 0.79. Radiomics analysis using features representing tumor heterogeneity only reached the highest accuracy of 0.71. Classification using CNN achieved a mean accuracy of 0.71 ± 0.043, whereas a CLSTM improved accuracy to 0.81 ± 0.034.
Conclusions:
DCE-MRI machine-learning analysis methods have potential to predict lung cancer metastases in the spine, which may be used to guide subsequent workup for confirmed diagnosis.
Keywords: DCE-MRI, Radiomics, Deep learning, Spinal metastases
1. Introduction
Patients presenting with pain in the spine are often suspected to have lesions compressing the spinal cord, and MRI is usually performed for diagnosis. The most common malignancy in the spine is metastatic cancer, and approximately 30% of patients present with an unknown primary [1–3]. In these patients, a final diagnosis is needed to proceed with treatment. If the origin of the cancer in the spine can be accurately predicted, this can narrow the search field and help determine the most appropriate imaging method to locate the primary tumor without the need of performing invasive spinal biopsy.
In Western world with established health care systems, PET/CT is the most commonly used imaging for diagnosis of primary cancer and whole-body staging when the metastatic cancer in the spine is suspected. However, the patient may have to wait for insurance approval and delay the diagnosis. In the developing countries, PET/CT and the 18F-FDG tracer are limited and very expensive, and thus this exam may not be available to many patients. If other cheaper imaging examinations can be used to locate the primary tumor, it will provide a cost-effective management approach to help patients. Among all patients presenting with spinal pain with an unknown primary cancer site, lung metastasis is the most prevalent [3]. If this primary can be accurately predicted by MRI, subsequent workup can be focused to pulmonary imaging, e.g. using CT, which is easily doable and much cheaper.
While conventional MRI can easily detect metastasis in the spine, cancers from different primary appear similar and often indistinguishable. Many studies have shown that dynamic contrast-enhanced MRI (DCE-MRI) can provide additional information for further characterization of the detected spinal lesions [4–11], but only a few tried to differentiate metastasis from different primary cancers [5,6].
For diagnosis using DCE-MRI, the most common method is to measure the signal intensity time course from a manually-placed region of interest (ROI) to evaluate DCE kinetic parameters. A radiologist can also evaluate the morphological presentation of the tumor, which can be combined with the DCE parameters to make a diagnosis. Additionally, computer-aided or radiomics-based analysis are commonly utilized to extract quantitative parameters from the entire segmented tumor, and that can be used for a thorough evaluation of morphological and DCE kinetic features to aid in diagnosis [12–15]. Very recently, deep learning has been demonstrated as a feasible, albeit powerful method to automatically evaluate the entire lesion for diagnosis [16–18], or lesion detection [19,20], without use of pre-defined metrics. All available images can be used as inputs for the algorithm to achieve the best diagnostic accuracy. Each of these three methods has their own pros and cons, which is an active research area for diagnosis.
The purpose of this study is to differentiate metastatic cancer in the spine originated from lung cancer and other non-lung tumors, by using the conventional ROI-based method and the more sophisticated machine-learning based methods, including radiomics and deep learning. The diagnostic results and limitations of these methods were compared.
2. Materials and methods
2.1. Patients
This study was approved by the Ethics Committee of our hospital, and the informed consent was waived. In a retrospective review of spinal clinical MRI database in our hospital that included a DCE sequence from 2011 to 2015, a total of 61 patients with confirmed osseous spinal metastases originating from a known primary tumor were identified. The cases were selected by identifying patients who had pain in the supine and came to our hospital for diagnosis using MRI. All of them did not have prior history of any cancer diagnosis. Information regarding primary cancer source was obtained from review of medical records. Distribution of primary cancer sites included: 30 patients confirmed with lung cancer (mean age 56); 9 with breast cancer (mean age 54); 7 with thyroid cancer (mean age 50); 6 with prostate cancer (mean age 72); 6 with liver cancer (mean age 52); 3 with renal cancer (mean age 65). The age and sex distribution between the lung cancer (16 males, 14 females, mean age 56) and other cancer (16 males, 15 females, mean age 57) groups were about the same.
2.2. MR imaging protocol
MR scans were performed on a 3 T Siemens or 3 T GE scanner with a consistent protocol. The conventional imaging sequences included transverse T2WI, sagittal T2WI without and with fat suppression, and sagittal T1WI acquired by using the fast spin echo pulse sequence. After the abnormal region was identified on sagittal view, DCE-MRI was performed using the three-dimensional (3D) volume interpolated breath-hold examination (3D VIBE) sequence in the transversal plane to further examine that region. The imaging parameters were: repetition time TR = 4.1 ms, echo time TE = 1.5 ms, flip angle = 10°, acquisition matrix = 256 × 192 and field of view FOV = 250 × 250 mm. Approximately 30 slices with 3-mm thickness were prescribed to cover the abnormal vertebrae. The temporal resolution varied from 10 to 14 s. The contrast agent, 0.1 [mmol/kg] Gd-DTPA, was injected after one set of pre-contrast images were acquired, by using an Ulrich power injector at a rate of 2 ml/s followed by 20 cc saline flush at the same rate. A total of 12 frames were acquired, so the total DCE-MRI acquisition time period ranged from 120 to 168 s. When the DCE study was done using the GE scanner, the LAVA (Liver Acceleration Volume Acquisition) pulse sequence with similar spatial and temporal resolution was used. Fig. 1 shows two case examples, one from lung and the other from thyroid cancer. The corresponding DCE-MRI, including the pre- and post-contrast images and the subtraction enhancement maps are shown in Fig. 2.
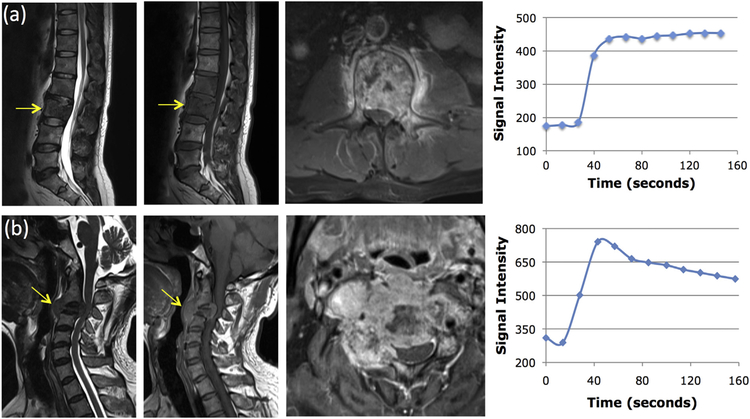
Fig. 1.
Two case examples. From left to right: the sagittal T2-weighted, T1-weighted, axial contrast-enhanced images, and the signal intensity time course measured from a region of interest (ROI) manually placed on the strongest enhancement area. Top panel: (a) A 45-year-old man with metastatic lung cancer, showing the plateau DCE kinetic pattern. Bottom panel: (b) A 55-year-old man with metastatic thyroid cancer, showing the wash-out DCE kinetic pattern.
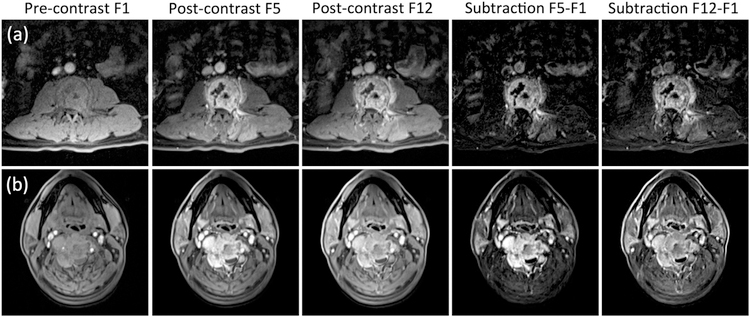
Fig. 2.
The DCE-MRI of two cases shown in Fig. 1. From left to right: the pre-contrast image acquired in Frame-1, the post-contrast images acquired in Frame-5 (50–60 s after injection) and the last Frame-12 (140–150 s after injection), and the subtraction images (F5-F1) and (F12-F1). Top panel: (a) The metastatic lung cancer showing similar enhancements in F5 and F12 and similar subtraction images without a clear wash-out. Bottom panel: (b) The metastatic thyroid cancer showing a stronger enhancement in F5 than in F12, and the subtraction images also clearly demonstrate contrast wash-out.
2.3. Hot-spot ROI-based DCE kinetic analysis
For each case, an ROI was manually placed on an area that demonstrated avid enhancement and excluded regions with cystic lesions, calcification, necrosis, and hemorrhage, as illustrated in a previous publication [6]. The signal intensity time course was measured and evaluated to find the pre-contrast signal intensity (S0), two adjacent time points that showed the largest difference in their signal intensities (S2 and S1) during the wash-in phase, and the maximum intensity (Smax). In the lung mets case shown in Fig. 1, the S1 and S2 were DCE frames #3 and #4; in the thyroid mets case, the difference between #3 and #4 was slightly greater than between #2 and #3, and thus used in the calculation. The Smax was DCE frame #12 in the lung mets case, and #4 in the thyroid mets case. Two heuristic parameters were calculated as:
For cases with a clearly visible peak enhancement occurring approximately 60 s after injection, the wash-out slope was calculated using the peak (Speak) and the signal intensity at the last time point (Slast). For cases that did not show a peak before 85 s, in order to catch the increasing intensities in the DCE time course, the slope between the signal intensities at the 67 s (S67seconds) and the last time point was calculated. This method was developed based on the analysis of various lesions in the spine using DCE-MRI, as described in two previous studies [10,11]. Therefore, the wash-out slope was calculated as:
These three measured parameters were used to differentiate between lung cancer and other cancers by utilizing the logistic regression and Chi-square Automatic Interaction Detector (CHAID) decision tree classification method. In addition to the heuristic analysis, a two-compartmental pharmacokinetic analysis was applied to obtain the influx transport constant Ktrans and the out-flux rate constant kep ([1/min]), by using the methods reported previously [11]. The pharmacokinetic parameters were highly correlated with heuristic parameters, thus they were not independent parameters, and not further analyzed.
2.4. Lesion segmentation using normalized cut and region growing
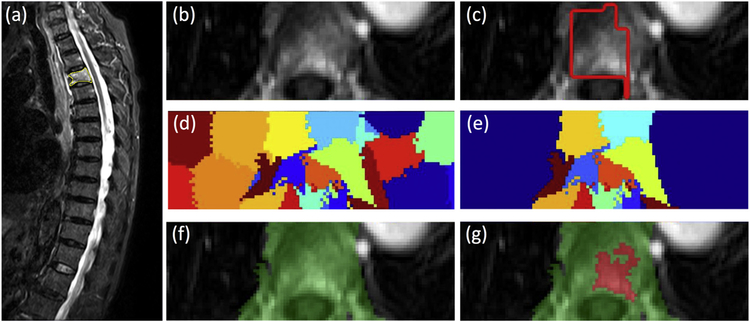
Three-dimensional lesion segmentation was performed for all patients in this study. Since DCE-MRI was acquired in the axial plane based on the abnormal region identified on the sagittal acquisition, an automatic segmentation method was developed following this same approach. The detailed methods are illustrated in Fig. 3. The abnormal area on sagittal T2W images was first manually outlined by a radiologist and then transformed to the axial view DCE-MRI for tumor segmentation using a normalized cut algorithm with region growing [21]. The global coordinates of all voxels outlined on sagittal slices (Fig. 3a) were transformed to axial DCE (Fig. 3b) and used as the initial search area (Fig. 3c). In order to cover the entire lesion, the left and right boundary of the initial search box was expanded by a factor of 5 (Fig. 3d). The normalized cut algorithm was utilized to divide the expanded search area on each slice into partitions, and those overlapping with the initial transformed area were kept (Fig. 3e). Then, the remaining partitions on all DCE slices were combined into a 3D mask (Fig. 3f), and the most strongly enhancing voxel was identified as the seed point for region growing to find the lesion boundary inside this 3D mask (Fig. 3g).
Fig. 3.
Identification and segmentation of the enhanced tumor on Axial DCE images based on the diseased segment drawn on Sagittal T2W images using the normalized cut algorithm. (a) Sagittal T2W image with marked diseased segment. (b) Axial view contrast-enhanced image. (c) The diseased segment in yellow in (a) is transferred to the axial image, shown as red box. (d) The clusters generated using the normalized cut. (d) The clusters containing any part of the red box ROI. (f) The vertebral region combining all remaining clusters. (g) The enhanced tumor lesion (in red) is generated by region growing within the green field. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.5. Differential diagnosis using radiomics analysis
Radiomics analysis was performed to extract DCE kinetic parameters and texture features within the segmented lesion. The analysis was done on three computed DCE parametric maps corresponding to the ROI-based analysis, including the steepest wash-in SE map, maximum SE map, and wash-out slope map. These maps were generated on a pixel-by-pixel basis, using the formula described above for hot-spot analysis. In each case, the DCE frames for S1, S2, Smax and Speak were the same as those used in the hot-spot analysis. No apparent patient motion was noted in the short DCE acquisition period of 120–168 s, and as such between-frame registration was not needed. The color-coded maps from the lung cancer and thyroid cancer cases are shown in Fig. 4. On each map, 20 gray-level co-occurrence matrix (GLCM) texture features were calculated according to Haralick et al. [22], including autocorrelation, cluster prominence, cluster shade, contrast, correlation, dissimilarity, energy, entropy, homogeneity 1, homogeneity 2, maximum probability, sum average, information measure of entropy, sum variance, sum entropy, difference variance, correlation, difference entropy, information measure of correlation 1, and information measure of correlation 2.
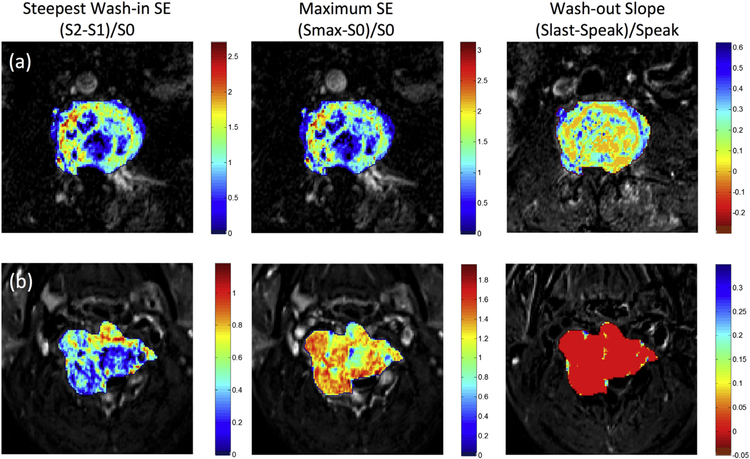
Fig. 4.
The generated DCE maps from the two case examples shown in Fig. 1 for (a) metastatic lung cancer and (b) metastatic thyroid cancer. The map is generated using the equation for all voxels, but only the voxels within the tumor ROI are shown by color according to the color maps. The entire thyroid cancer shows a clear wash-out pattern in red color; in contrast, the lung cancer mainly shows the plateau pattern in orange to green color. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Furthermore, the histogram or the population distribution curve of all voxels within the tumor ROI on each map was generated, and a total of 13 parameters were obtained, including the 10%, 20%… 80% to 90% percentile values, mean, standard deviation, kurtosis and skewness. On each map, 20 texture and 13 histogram features were calculated, and a total of 99 quantitative features from three maps were obtained for each patient.
The best radiomics model was generated in three steps: 1) ranking features; 2) selecting combination of features; 3) building a final model based on selected features. The features were first properly normalized. Due to the limited number of cases (total N = 61), a random forest algorithm was used to select 3–5 features to form the diagnostic classifier [23]. We first selected parameters with highest significance scores from DCE histograms only, texture features only, and then selected parameters from combined histogram and texture features. The random forest with 500 trees was applied for classification of bootstrap samples randomly selected from 61 patients, and based on these results the discriminating capability of features could be assessed and ranked. Approximately 60% of cases were selected randomly in each run, and the significance of a feature could be assessed as the loss of accuracy after this feature was removed. Then, according to the ranking, the top 3, 4, 5, … 10 features were selected to build the diagnostic model by using logistic regression. The discrimination accuracy was evaluated by the receiver operating characteristic (ROC) analysis using 10-fold stratified cross-validation. In each fold, 3 cases from lung metastasis (lung mets) group and 3 cases from the non-lung metastasis (non-lung mets) group were used as the testing set, and the remaining cases used for training. This process was repeated many times using different combination of selected features (3, 4, 5 … 10) and the results were used to find the best model according to the highest AUC. After the features included in the best model were decided, they were used to build a final diagnostic classifier with logistic regression, and the accuracy was evaluated in the entire dataset of 61 cases.
2.6. Differential diagnosis using deep learning
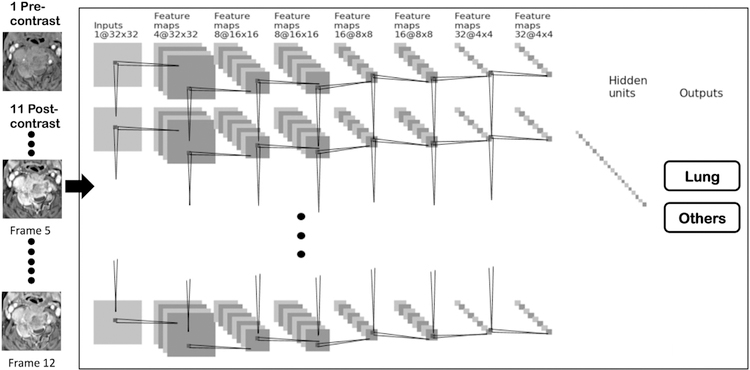
Deep learning was used to investigate the diagnostic accuracy that can be achieved using a fully automated approach without manual hand-crafted features. Due to the small case number, the dataset was first augmented by 20 times using a random affine transformation with combination of rotation, translation, scaling and shearing [24]. Detailed augmentation methods have been reported before in Chang et al. [18]. Two separate convolutional neural network (CNN) architectures were applied. First, the three DCE parametric maps were used as independent inputs in a conventional feed-forward CNN. Second, to incorporate time-dependent information from the entire 12 sets of DCE images, we applied used a convolutional long short term memory (CLSTM) network [25,26], by inputting the 12 DCE datasets into the network in a time order as shown in Fig. 5. Each 2D imaging slice was used as an independent input. For each case, the smallest bounding box containing the segmented tumor was used as input. The segmented tumor ROI’s from all slices were projected together, and the smallest bounding box to cover the outer boundary of projected ROI’s was used for this case. Since only the tumor ROI was considered, the pixels outside the tumor in the box were set to zero. The 12 sets of DCE images were normalized together to a mean = 0 and standard deviation = 1.
Fig. 5.
Diagram of the recurrent CNN. The architecture uses 7 serial convolutional LSTM layers via 3 × 3 filters followed by the ReLU nonlinear activation function. All 12 sets of DCE images are used as inputs, and the signal intensity is normalized using the same scale. Dropout at 50% is applied to all convolutional and fully-connected layers after the second layer. Feature maps are down sampled to 25% of the previous layer by convolutions with a stride length of two. The last dense layer is obtained by flattening the convolutional output feature maps from all states. Softmax is used as the activation function of the last fully connected layer.
Detailed procedures were described in Chang et al. [18]. For the conventional CNN architecture, the underlying network was composed by a strided convolution in every other layer (i.e. 2nd, 4th, and 6th) to reduce the spatial resolution to 25% of the previous resolution. Each convolutional operation was followed by a nonlinear rectified linear (ReLU) activation function [27,28]. This function was chosen because of its well-documented advantages including stable gradients at the extreme values of optimization. Dropout at 50% was applied to all convolutional and fully-connected layers to limit over-fitting and add stochasticity to the training process [29,30]. The 7th layer output feature maps from all cells were flattened to a one-dimensional vector. The softmax activation function was used for final classification, with a threshold of 0.5. For the CLSTM network, 7 stacked convolutional long short term memory layers were fed into a final fully connected layer before output, as in the architecture shown in Fig. 5.
The algorithm was implemented with a standard cross entropy loss function and the Adam optimizer with an initial learning rate of 0.001, which was kept as a constant throughout the training [31]. The software code was written in Python 3.5 using the open-source TensorFlow 1.0 library (Apache 2.0 license). Experiments were performed on a GPU-optimized workstation with a single NVIDIA GeForce GTX Titan X (12GB, Maxwell architecture). A forward pass for the classification test of a new patient could be achieved in < 0.01 s. The results were evaluated using 10-fold cross validation. The range and the mean value with standard deviation were calculated to show the prediction accuracy.
3. Results
3.1. Hot-spot ROI-Based DCE parameters
Table 1 summarizes the mean ± standard deviation of 5 characteristic DCE parameters measured from the manually placed ROI on the hot spot. The wash-out slope and kep showed a significant difference between the lung cancer and other primary tumors. The mean wash-out slope was 0.25% in lung cancer, indicating most lung cancers showed the plateau DCE kinetic pattern. The mean wash-out slope was −9.8% for other tumors, indicating that many of them showed the wash-out DCE kinetic pattern. In the two examples shown in Fig. 1, the DCE time course shows a plateau pattern for the lung cancer, and a clear wash-out pattern for the thyroid cancer. In the DCE images shown in Fig. 2, the signal intensity is similar between Frame-5 and Frame-12 for the lung cancer. For the thyroid cancer, the intensity in Frame-12 is clearly lower compared to Frame-5, and the degree of enhancement is lower in the subtraction image of (Frame 12 – Frame 1) compared to that of (Frame 5 – Frame 1). Among all non-lung mets, the breast (−12.9%) and thyroid (−15.6%) cancers had the most prominent wash-out.
Table 1.
The DCE parameters analyzed from the ROI manually placed on the strongly enhanced tissue, data shown is [mean ± standard deviation].
| Tumor origin | Maximum SE (%) | Wash-in SE (%) | Wash-out Slope (%) | Ktrans (1/min) | kep (1/min) | |
|---|---|---|---|---|---|---|
| Lung cancer | Lung (N = 30) | 243 ± 89 | 146 ± 60 | 0.25 ± 10 | 0.10 ± 0.04 | 0.39 ± 0.16 |
| Other tumors | Total (N = 31) | 220 ± 109 | 142 ± 76 | −9.8 ± 12.9 | 0.10 ± 0.06 | 0.58 ± 0.24 |
| Breast (N = 9) | 299 ± 134 | 197 ± 95 | −12.9 ± 8.24 | 0.14 ± 0.07 | 0.60 ± 0.16 | |
| Thyroid (N = 7) | 210 ± 59 | 130 ± 47 | −15.6 ± 14.4 | 0.10 ± 0.03 | 0.68 ± 0.28 | |
| Prostate (N = 6) | 165 ± 77 | 126 ± 74 | −7.7 ± 12.9 | 0.08 ± 0.05 | 0.56 ± 0.29 | |
| Liver (N = 6) | 196 ± 111 | 118 ± 55 | −5.9 ± 16.2 | 0.08 ± 0.05 | 0.52 ± 0.27 | |
| Kidney (N = 3) | 164 ± 80 | 87 ± 37 | 1.3 ± 11.3 | 0.07 ± 0.04 | 0.42 ± 0.26 | |
| P value* | 0.199 | 0.614 | 0.001 | 0.634 | 0.001 |
Significant with P < 0.05 in the comparison between lung cancer and other tumors.
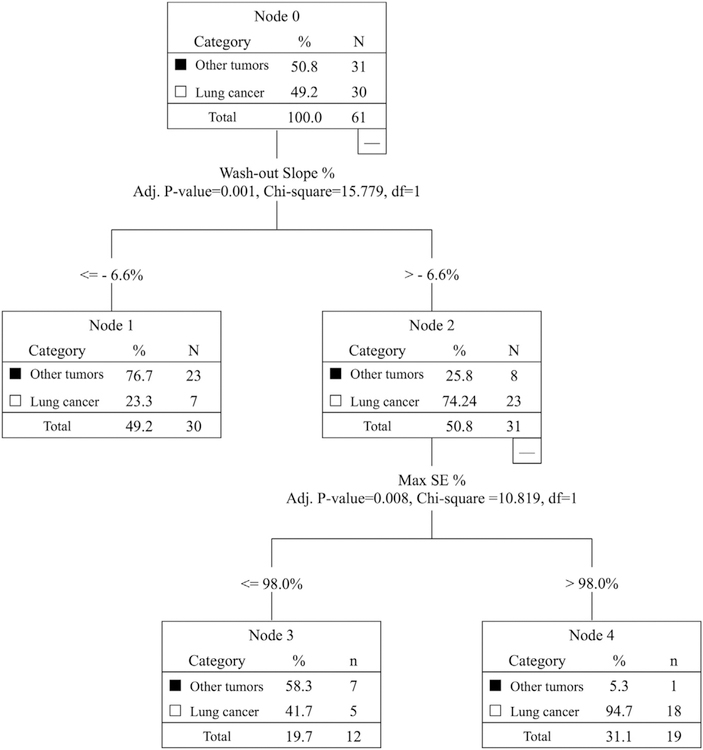
Classification was done by using the logistic regression and CHAID decision tree based on the three heuristic parameters (Steepest wash-in SE, Max SE, Wash-out slope). The accuracy obtained using logistic regression was 0.74, and that by using CHAID with the wash-out slope of −6.6% followed by maximum SE of 98% was 0.79, as shown in Fig. 6. True Positive (TP) for diagnosis of lung cancer = 18/30 cases; False Negative (FN) = 12/30 cases; True Negative (TN) for diagnosis of other tumors = 30/31 cases; and False Positive (FP) = 1/31 case. The Sensitivity = 60%; Specificity = 96.8%; Positive Predicting Value = 94.7%; and Negative Predicting Value = 71.4%.
Fig. 6.
The diagnostic results analyzed using the Chi-square Automatic Interaction Detector (CHAID) decision tree classification method. The overall diagnostic accuracy of 0.79 is achieved by using the first threshold of wash-out slope - 6.6%, followed by the second threshold of wash-in SE 98%. True Positive (TP) for diagnosis of lung cancer = 18 cases; False Negative (FN) = 12 cases; True Negative (TN) for diagnosis of other tumors = 30 cases; and False Positive (FP) = 1 case. The Sensitivity = 60%; Specificity = 96.8%; Positive Predicting Value = 94.7%; and Negative Predicting Value = 71.4%.
3.2. Radiomics using DCE histogram parameters and texture
In the color maps shown in Fig. 4, almost all voxels in the entire thyroid cancer show the wash-out pattern (in red color), but the voxels in the lung cancer are more heterogeneous with most of them showing plateau (in orange to green color). Based on the 10-fold cross-validation results, we found that by increasing the number of features from 3 to 4 to 5 the accuracy improved slightly, but then the accuracy did not increase further with more features; therefore, we only reported the results using 3 and 5 features. The diagnostic accuracy and the selected histogram and texture features are listed in Table 2. It was noted that the accuracy obtained using the texture features only (0.59–0.62) was lower compared to that analyzed using histogram only (0.67–0.68), or histogram+texture (0.68–0.71). The accuracy of radiomics analysis was worse compared to the hot-spot ROI-based analysis of 0.79.
Table 2.
Accuracy in differentiating lung metastases from other cancers based on selected features in the radiomics analysis.
| Accuracy | Histogram + texture | Histogram features only | Texture features only |
|---|---|---|---|
| 3 features | 0.68 (90% value and kurtosis from wash-out map, information measure of entropy from max SE map) |
0.67 (90% value and kurtosis from wash-out map, mean from max SE map) |
0.59 (information measure of entropy from max SE map, entropy and dissimilarity from steepest wash-in map) |
| 5 features | 0.71 (90% value, kurtosis and autocorrelation from wash-out map, information measure of entropy from max SE map, entropy from steepest wash-in map) |
0.68 (90% value and kurtosis from wash-out map, mean and kurtosis from max SE map, 50% value from steepest wash-in map) |
0.62 (information measure of entropy from max SE map, entropy and dissimilarity from steepest wash-in map, dissimilarity and contrast from wash-out map) |
3.3. Deep learning using convolutional neural network
Classification was performed using two different deep learning approaches, evaluated by 10-fold cross-validation. The accuracy achieved using three generated DCE parametric maps as inputs in a conventional CNN was 0.61–0.74, mean 0.71 with standard deviation of 0.043. The accuracy achieved using all 12 sets of DCE images as inputs in a CLSTM network was 0.75–0.84, mean 0.81 with standard deviation of 0.034. The accuracy achieved by CLSTM was significantly higher than that achieved using CNN. The sensitivity for detecting lung mets was 0.60 ± 0.07 for CNN, 0.75 ± 0.07 for CLSTM; and the specificity was 0.76 ± 0.06 for CNN, 0.83 ± 0.06 for CLSTM. To provide a clear comparison of these three different analysis approaches, the essential methods, the number of analyzed parameters, the diagnostic evaluation methods, and the obtained results are summarized in Table 3.
Table 3.
The comparison of hot-spot, radiomics, and deep learning classification methods and the obtained results
| Method | Number of parameters | Evaluation method | Final cases used in accuracy test | Accuracy |
|---|---|---|---|---|
| Hot-spot using manually drawn ROI | 3 | CHIAD decision tree | Entire dataset (30 lung mets and 31 others) | 0.79 |
| Radiomics using segmented 3D tumor | 99 | Random forest + logistic regression | Entire dataset (30 lung mets and 31 others) | 0.71 (best model from Table 2) |
| Deep learning (CNN) using 3 parametric maps | Not-defined | Not-defined | 10-Fold cross-validation (6−7 test cases in each run) | 0.71 ± 0.043 (range 0.61−0.74) |
| Deep learning (CLSTM) using 12 DCE images | Not-defined | Not-defined | 10-Fold cross-validation (6−7 test cases in each run) | 0.81 ± 0.034 (range 0.75−0.84) |
4. Discussion
In the present study, three different analysis methods based on hotspot, radiomics and deep learning were applied to differentiate the spinal mets coming from lung cancer and other tumors. The pros and cons of each method were described, and the achieved accuracy was compared. The results showed that the DCE kinetic measure of washout slope from a hot-spot was the best parameter to differentiate primary lung cancer from other tumors. A CHAID decision tree using the wash-out slope followed by maximum SE could achieve an accuracy of 0.79. In comparison, the radiomics analysis performed from the segmented whole tumor in 3D could only achieve the highest accuracy of 0.71, while the deep learning CLSTM network using the entire sets of DCE images reached an accuracy of 0.81.
The cause of death in most cancer patients occurs due to metastasis and complications. Thus, early detection of metastasis is critical, as it can be better controlled. The most common metastatic cancer site is the liver, followed by the lung, and then bones, where the spine is the most vulnerable site to be invaded by metastases in the skeletal system. Patients without a known history of cancer often seek medical attention due to nerve compression and back pain. When metastatic cancer was suspected, finding and confirming the primary lesion became the most important task for treatment planning. While in the Western world PET/CT is the standard of care for such patients, it is expensive; in developing countries the availability of PET/CT system and the [18F]-FDG isotope tracer maybe limited. Since lung mets are the most common primary, if it is suspected, a CT scan can be performed quickly at a low cost. Even in the Western world, this study also has a good clinical value, to help patients with lung mets to be diagnosed early without relying on the PET/CT, which needs insurance approval and causes delay. Therefore, in this study we tried to predict origin of spinal mets that come from lung cancer and other cancers.
The appearance of many spinal lesions was similar on conventional MRI [32–36]. Osteolytic lesion was the most common abnormality seen in the spine, and metastatic lesion was often accompanied with soft tissue mass. The imaging presentation may vary substantially due to many factors, including local myelofibrosis, infarction, edema, pathological compression fracture and infection, adding to the many challenges of the differential diagnosis. DCE-MRI has been proven as a valuable technique for assessing tumor angiogenesis, and it has been widely applied for diagnosis and pre-operative staging for many cancers. For the spine, DCE-MRI has been applied to differentiate various diseases, including primary bone tumors such as myeloma, lymphoma, chordoma [10,11,37,38]; benign lesions such as tuberculosis, giant cell tumor of the bone [9,10]; as well as metastatic cancers of different origins, e.g. hypervascular renal vs. hypovascular prostate [8], and cancers of different origins [5,6]. For surgical planning, the information of blood supply may predict intraoperative blood loss, which can be used to plan for embolization before surgery [39–41].
Many different analysis methods can be applied to extract DCE parameters, either from a hot spot or from the whole tumor. In this study we first measured the signal intensity time course from an ROI manually placed on a strongly enhanced area. The wash-out slope was the best predictor to differentiate the two groups. The CHAID classification accuracy using the wash-out slope of −6.6% followed by maximum SE of 98% was 0.79.
Morphological presentation of spinal lesions could be evaluated using a scoring system based on pre-defined features, as used in a previous study to differentiate chordoma from giant cell tumor of the bone [10]. In recent several years, radiomics analysis has been widely applied to extract thorough information from medical images, for performing many clinical tasks such as differential diagnosis of benign and malignant lesions or subtype cancers [13–15], and prediction of therapeutic response or prognosis [42–44]. The tumor was first segmented, and then the histogram-based parameters and high-level texture features extracted using quantitative algorithms were measured. All these parameters were then combined for feature selection to build an optimal diagnostic/predictive classifier. Radiomics was commonly applied to analyze images acquired in different sequences (e.g. T2, T1 pre- and post-contrast, diffusion weighted imaging, FLAIR, etc.), not multiple sets of post-contrast images acquired using DCE-MRI. Therefore, in this study, we analyzed features on three DCE parametric maps generated corresponding to the hot spot analysis: the wash-in SE map, maximum SE map, and wash-out slope map. To avoid using a “black-box” classification method, we used random forest algorithm for feature selection, not for the final classification. A similar approach was used in Gallego-Ortiz et al. [45]. The accuracy in the combined histogram+texture analysis was 0.68 by using 3 features and 0.71 by using 5 features. The results showed that the accuracy was inferior to that of hot-spot analysis, and that the texture (i.e. heterogeneity within the tumor) did not add much value for improving differential diagnosis. Although metastatic cancers are highly prevalent in liver, lung, brain and spine, there are few studies trying to predict the origins based on imaging analysis. In a very recent study by Ortiz-Ramón et al., they tried to differentiate brain metastasis coming from 27 lung cancer, 23 melanoma and 17 breast cancer patients [46].
Since DCE-MRI is not a standard procedure for evaluation of spinal lesions, the case number reported in all published studies is small. Furthermore, the heterogeneity from complicated anatomic structures and the vascular bone marrow might limit the prediction accuracy of radiomics analysis. In this study, we also implemented a deep learning network to evaluate the accuracy that could be achieved. Most deep learning methods use images acquired in different sequences as inputs, and currently, there is no established network specifically designed for the full set of pre- and post-contrast images acquired in DCE-MRI [47]. In our protocol we had a total of 12 sets of images. In order to consider the change of signal intensity over time, all DCE images were normalized together. The Long Short Term Memory (LSTM) network, one of the Recurrent Neural Network (RNN), can connect previous information to the present task. The LSTM is explicitly designed to avoid long-term dependency and focus primarily on short-term memory. We used images from different DCE time points as independent inputs to LSTM, but also considered the changes of signal intensity in these different sets of images. Meanwhile, the same hierarchical features were calculated from each time frame, and thus more information from all DCE frames could be used for prediction of diagnosis. The range of accuracy in 10-fold cross validation was 0.75–0.84, with the mean of 0.81 ± 0.034, slightly better than the 0.79 achieved in hot spot ROI analysis. To compare the results using LSTM with the conventional CNN, we also used the three generated parametric maps as inputs for the CNN, and found that the mean accuracy was only 0.71 ± 0.043, similar to the accuracy of radiomics analysis. The results suggest that LSTM is an appropriate network to consider the entire sets of DCE images and track the change of signal intensity in a time sequence.
The major limitation of this study was the small case number. However, although not optimal for radiomics or deep learning, the results obtained using three different methods could still give insights to their value in solving this very challenging problem. The spine lesion segmentation method and the three different DCE analysis methods presented in this study may be applied to other studies to further investigate their clinical value in predicting diagnosis or further in prognosis.
In conclusion, we have shown that a simple hot-spot ROI analysis could be applied to characterize DCE kinetics of the metastatic cancer in the spine and differentiate the primary from lung cancer and other tumors. We have implemented deep learning and shown the potential in this clinical application. The recurrent neural network using CLSTM could track the change of signal intensity in pre- and post-contrast images in the DCE-MRI, with accuracy comparable to the hot-spot analysis, and better compared to conventional CNN and radiomics. For patients suspected to have metastatic cancer in the spine, DCE-MRI may help to predict the primary cancer source coming from lung, and that may help to reach an early confirmed diagnosis using CT alone without having to wait for the expensive PET/CT.
Acknowledgments
Funding support
This study was supported in part by NIH R01 CA127927, the National Natural Science Foundation of China (81701648, 81871326), and the Key Clinical Projects of the Peking University Third Hospital (BYSY2018007).
References
- [1].Sciubba DM, Petteys RJ, Dekutoski MB, et al. Diagnosis and management of metastatic spine disease. J Neurosurg Spine 2010;13:94–108. [DOI] [PubMed] [Google Scholar]
- [2].Robson P Metastatic spinal cord compression: a rare but important complication of cancer. Clin Med (Lond) 2014;14:542–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Piccioli A, Maccauro G, Spinelli MS, Biagini R, Rossi B. Bone metastases of unknown origin: epidemiology and principles of management. J Orthop Traumatol 2015;16:81–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Zha Y, Li M, Yang J. Dynamic contrast enhanced magnetic resonance imaging of diffuse spinal bone marrow infiltration in patients with hematological malignancies. Korean J Radiol 2010;11:187–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Khadem NR, Karimi S, Peck KK, Yamada Y, Lis E, Lyo J, et al. Characterizing hypervascular and hypovascular metastases and normal bone marrow of the spine using dynamic contrast-enhanced MR imaging. AJNR Am J Neuroradiol 2012;33:2178–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lang N, Su MY, Yu HJ, Lin M, Hamamura MJ, Yuan H. Differentiation of myeloma and metastatic cancer in the spine using dynamic contrast-enhanced MRI. Magn Reson Imaging 2013;31:1285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Dutoit JC, Vanderkerken MA, Verstraete KL. Value of whole body MRI and dynamic contrast enhanced MRI in the diagnosis, follow-up and evaluation of disease activity and extent in multiple myeloma. Eur J Radiol 2013;82:1444–52. [DOI] [PubMed] [Google Scholar]
- [8].Saha A, Peck KK, Lis E, Holodny AI, Yamada Y, Karimi S. Magnetic resonance perfusion characteristics of hypervascular renal and hypovascular prostate spinal metastases: clinical utilities and implications. Spine 2014;39:E1433–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lang N, Su MY, Yu HJ, Yuan H. Differentiation of tuberculosis and metastatic cancer in the spine using dynamic contrast-enhanced MRI. Eur Spine J 2015;24:1729–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lang N, Su MY, Xing X, Yu HJ, Yuan H. Morphological and dynamic contrast enhanced MR imaging features for the differentiation of chordoma and giant cell tumors in the axial skeleton. J Magn Reson Imaging 2017;45:1068–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lang N, Yuan H, Yu HJ, Su MY. Diagnosis of spinal lesions using heuristic and pharmacokinetic parameters measured by dynamic contrast-enhanced MRI. Acad Radiol 2017;24:867–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kumar V, Gu Y, Basu S, Berglund A, Eschrich SA, Schabath MB, et al. Radiomics: the process and the challenges. Magn Reson Imaging 2012;30:1234–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014;5:4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology 2016;278:563–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 2017;14:749–62. [DOI] [PubMed] [Google Scholar]
- [16].Antropova N, Huynh BQ, Giger ML. A deep feature fusion methodology for breast cancer diagnosis demonstrated on three imaging modality datasets. Med Phys 2017;44:5162–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Le MH, Chen J, Wang L, Wang Z, Liu W, Cheng KT, et al. Automated diagnosis of prostate cancer in multi-parametric MRI based on multimodal convolutional neural networks. Phys Med Biol 2017;62:6497–514. [DOI] [PubMed] [Google Scholar]
- [18].Chang P, Grinband J, Weinberg BD, Bardis M, Khy M, Cadena G, et al. Deep learning convolutional neural networks accurately classify genetic mutations in gliomas. AJNR Am J Neuroradiol 2018;39:1201–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wang J, Fang Z, Lang N, Yuan H, Su MY, Baldi P. A multi-resolution approach for spinal metastasis detection using deep Siamese neural networks. Comput Biol Med 2017;84:137–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhu Y, Wang L, Liu M, Qian C, Yousuf A, Oto A, et al. MRI-based prostate cancer detection with high-level representation and hierarchical classification. Med Phys 2017;44:1028–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Huang SH, Chu YH, Lai SH, Novak CL. Learning-based vertebra detection and iterative normalized-cut segmentation for spinal MRI. IEEE Trans Med Imaging 2009;28:1595–605. [DOI] [PubMed] [Google Scholar]
- [22].Haralick RM, Shanmugam K. Textural features for image classification. IEEE Trans Syst Man Cybern 1973:610–21.
- [23].Breiman L Random forests. Mach Learn 2001;45:5–32. [Google Scholar]
- [24].Hazewinkel M Affine transformation. Encyclopedia of mathematics Springer; 2001. [Google Scholar]
- [25].Hochreiter S, Schmidhuber J. Long short-term memory. Neural Comput 1997;9:1735–80. [DOI] [PubMed] [Google Scholar]
- [26].Xingjian SH, Chen Z, Wang H, Yeung DY, Wong WK, Woo WC. Convolutional LSTM network: a machine learning approach for precipitation nowcasting. Advances in neural information processing systems 2015. p. 802–10.
- [27].Nair V, Hinton GE. Rectified linear units improve restricted boltzmann machines. Proceedings of the 27th International Conference on International Conference on Machine Learning. Haifa, Israel: Omnipress; 2010:807–814. [Google Scholar]
- [28].Krizhevsky Alex, Sutskever Ilya, Hinton Geoffrey E.. ImageNet classification with deep convolutional neural networks. in Advances in neural information processing systems, proceedings of the neural information processing systems conference, 2012. p 1097–1105. [Google Scholar]
- [29].Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res 2014;15:1929–58. [Google Scholar]
- [30].Baldi P, Sadowski P. The dropout learning algorithm. Artif Intell 2014;210:78–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kingma D, Ba J. Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980 2014.
- [32].Erlemann R, Reiser MF, Peters PE, Vasallo P, Nommensen B, Kusnierz-Glaz CR, et al. Musculoskeletal neoplasms: static and dynamic Gd-DTPA enhanced MR imaging. Radiology 1989;171:767–73. [DOI] [PubMed] [Google Scholar]
- [33].Hermann G, Abdelwahab LF, Miller TT, Klein MJ, Lewis MM. Tumour and tumour-like conditions of the soft tissue: magnetic resonance imaging features differentiating benign from malignant masses. Br J Radiol 1992;65:14–20. [DOI] [PubMed] [Google Scholar]
- [34].Moulton JS, Blebea JS, Dunco DM, Braley SE, Bisset GS 3rd, Emery KH. MR imaging of soft-tissue masses: diagnostic efficacy and value of distinguishing between benign and malignant lesions. Am J Roentgenol 1995;164:1191–9. [DOI] [PubMed] [Google Scholar]
- [35].May DA, Good RB, Smith DK, Parsons TW. MR imaging of musculoskeletal tumors and tumor mimickers with intravenous gadolinium: experiences with 242 patients. Skeletal Radiol 1997;26:2–15. [DOI] [PubMed] [Google Scholar]
- [36].Kim HJ, Ryu KN, Choi WS, Choi BK, Choi JM, Yoon Y. Spinal involvement of hematopoietic malignancies and metastasis. differentiation using MR imaging Clin Imaging 1999;23:125–33. [DOI] [PubMed] [Google Scholar]
- [37].Lang P, Honda G, Roberts T, Vahlensieck M, Johnston JO, Rosenau W, et al. Musculoskeletal neoplasm: perineoplastic edema versus tumor on dynamic post-contrast MR imaging with spatial mapping of instantaneous enhancement rates. Radiology 1995;197:831–9. [DOI] [PubMed] [Google Scholar]
- [38].Moulopoulos LA, Maris TG, Papanikolaou N, Panagi G, Vlahos L, Dimopoulos MA. Detection of malignant bone marrow involvement with dynamic contrast-enhanced magnetic resonance imaging. Ann Oncol 2003;14:152–8. [DOI] [PubMed] [Google Scholar]
- [39].Kato S, Hozumi T, Takaki Y, Yamakawa K, Goto T, Kondo T. Optimal schedule of preoperative embolization for spinal metastasis surgery. Spine (Phila Pa 1976) 2013;38:1964–9. [DOI] [PubMed] [Google Scholar]
- [40].Qiao Z, Jia N, He Q. Does preoperative transarterial embolization decrease blood loss during spine tumor surgery? Interv Neuroradiol 2015;21:129–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Jiang L, Liu XG, Wang C, et al. Surgical treatment options for aggressive osteoblastoma in the mobile spine. Eur Spine J 2015;24:1778–85. [DOI] [PubMed] [Google Scholar]
- [42].Nie K, Shi L, Chen Q, Hu X, Jabbour SK, Yue N, et al. Rectal cancer: assessment of neoadjuvant chemoradiation outcome based on radiomics of multiparametric MRI. Clin Cancer Res 2016;22:5256–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Braman NM, Etesami M, Prasanna P, Dubchuk C, Gilmore H, Tiwari P, et al. Intratumoral and peritumoral radiomics for the pretreatment prediction of pathological complete response to neoadjuvant chemotherapy based on breast DCE-MRI. Breast Cancer Res 2017;19:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wang G, He L, Yuan C, Huang Y, Liu Z, Liang C. Pretreatment MR imaging radiomics signatures for response prediction to induction chemotherapy in patients with nasopharyngeal carcinoma. Eur J Radiol 2018;98:100–6. [DOI] [PubMed] [Google Scholar]
- [45].Gallego-Ortiz C, Martel AL. Improving the accuracy of computer-aided diagnosis for breast MR imaging by differentiating between mass and nonmass lesions. Radiology 2016;278:679–88. [DOI] [PubMed] [Google Scholar]
- [46].Ortiz-Ramón R, Larroza A, Ruiz-España S, Arana E, Moratal D. Classifying brain metastases by their primary site of origin using a radiomics approach based on texture analysis: a feasibility study. Eur Radiol 2018. May 14 10.1007/s00330-018-5463-6. [Epub ahead of print]. [DOI] [PubMed]
- [47].Antropova N, Abe H, Giger ML. Use of clinical MRI maximum intensity projections for improved breast lesion classification with deep convolutional neural networks. J Med Imaging (Bellingham) 2018;5:014503. [DOI] [PMC free article] [PubMed] [Google Scholar]