Abstract
Objective
To explore the effect of cytosolic phospholipase A2α (cPLA2α) on hepatocellular carcinoma (HCC) cell adhesion and the underlying mechanisms.
Methods
Cell adhesion, detachment, and hanging-drop assays were utilized to examine the effect of cPLA2α on the cell-matrix and cell-cell adhesion. Downstream substrates and effectors of cPLA2α were screened via a phospho-antibody microarray. Associated signaling pathways were identified by the functional annotation tool DAVID. Candidate proteins were verified using Western blot and colocalization was investigated via immunofluorescence. Western blot and immunohistochemistry were used to detect protein expression in HCC tissues. Prognosis evaluation was conducted using Kaplan-Meier and Cox-proportional hazards regression analyses.
Results
Our findings showed that cPLA2α knockdown decreases cell-matrix adhesion but increases cell-cell adhesion in HepG2 cells. Microarray analysis revealed that phosphorylation of multiple proteins at specific sites were regulated by cPLA2α. These phosphorylated proteins were involved in various biological processes. In addition, our results indicated that the focal adhesion pathway was highly enriched in the cPLA2α-relevant signaling pathway. Furthermore, cPLA2α was found to elevate phosphorylation levels of FAK and paxillin, two crucial components of focal adhesion. Moreover, localization of p-FAK to focal adhesions in the plasma membrane was significantly reduced with the downregulation of cPLA2α. Clinically, cPLA2α expression was positively correlated with p-FAK levels. Additionally, high expression of both cPLA2α and p-FAK predicted the worst prognoses for HCC patients.
Conclusions
Our study indicated that cPLA2α may promote cell-matrix adhesion via the FAK/paxillin pathway, which partly explains the malignant cPLA2α phenotype seen in HCC.
Keywords: Hepatocellular carcinoma, cytosolic phospholipase A2α, cell-matrix adhesion, FAK, paxillin
Introduction
Hepatocellular carcinoma (HCC) is the most frequent primary liver malignancy. It is ranked the third-most common cause of cancer-related deaths worldwide1. In China, the incidence of liver cancer as well as deaths due to liver cancer are high, and they account for approximately 50% of the total cases and deaths across the globe2. Patients with HCC who are diagnosed at an early stage have the best chance of receiving effective treatments, such as resection, ablation, or transplantation, leading to long-term disease-free survival1. However, tumors are generally detected either at an intermediate or an advanced stage, which limits clinical therapeutic options3. The 5-year recurrence risk can be as high as 70%, even in patients who undergo surgical resection. Most cases of recurrence occur within the first 2 years following surgery, and are due to tumor invasion and metastasis4.
Cytosolic phospholipase A2α (cPLA2α, also known as group IVA PLA2) is a member of the intracellular phospholipase A2 family. It is known for its high selectivity in catalyzing membrane phospholipids containing arachidonic acid (AA) at the sn-2 position5. Free AA, which is released, is rapidly metabolized into various bioactive eicosanoids, including hydroxy eicosatetraenoic acids, prostaglandins, and leukotrienes6. These metabolites play vital roles in tumorigenesis, tumor metastasis, and progression7. Previous studies conducted by us demonstrated that cPLA2α could promote epithelial-mesenchymal transition (EMT), thereby contributing to tumor metastasis in HCC and breast cancer via AA production8, 9.
Cancer metastasis comprises a series of successive biological events. In the first step, cancer cells detach from the primary tumor and invade the surrounding extracellular matrix (ECM) and stromal cell layers10. Therefore, the migration capability of cancer cells assumes importance during metastasis. One prominent structure involved in cell migration is integrin-based focal adhesion (FA), which plays a crucial role in determining dynamic cell-matrix interactions11. FA kinase (FAK) is a nonreceptor tyrosine kinase that participates in FA complex formation. Its dysregulation is found in various types of cancer in relation to tumor metastasis12-15. Paxillin, which is a structural protein of the FA complex, also contributes to metastasis16. Although involvement of cPLA2 in cell-matrix adhesion in the immune system has been reported17, the role of cPLA2α in HCC cell adhesion as well as the involvement of FAK or paxillin in this biological process remains largely unknown.
In this study, we investigated the effect of cPLA2α on the cell-matrix and cell-cell adhesion of HCC cells. Using phospho-protein microarray technology, we analyzed the phosphoproteome profiles of cPLA2α-knockdown and cPLA2α-overexpressing HepG2 cells. We identified 2 proteins, FAK and paxillin, in the FA pathway as downstream molecules of cPLA2α. We also explored the prognostic role of cPLA2α and p-FAK in patients with HCC.
Materials and methods
Patients and follow-up
The tumor specimens used in the tissue microarray were obtained from 74 HCC patients who underwent surgical resection from January 2013 to January 2014 at the Tianjin Medical University Cancer Institute and Hospital. All tumor samples were histologically confirmed as HCC. All patients were staged in accordance with the 8th edition of TNM staging system based on AJCC. Informed consent was obtained from all patients involved. This study was conducted in accordance with the Declaration of Helsinki and approved by the Tianjin Medical University Cancer Institute and Hospital Ethics Committee. Post-surgical patient surveillance was conducted every 3 months via serum AFP and abdominal ultrasonography. Where recurrence was suspected, examination techniques were replaced with thoracoabdominal CT and abdominal magnetic resonance imaging (MRI) to confirm the diagnosis. Clinical data and follow-up results of these patients were recorded. No patient was lost during the follow-up period. The follow-up was updated to October 10, 2017. Eleven additional paired tumors and adjacent noncancerous tissues were collected from the HCC patients who had undergone surgical resection at our institute between 2014 and 2015, and used for western blot analysis.
Phospho-protein profiling by Phospho Explorer Antibody Array analysis
The Phospho Explorer Antibody Array (PEX100) was obtained from Full Moon Biosystems (Sunnyvale, CA, USA). Lysates of cPLA2α-knockdown as well as cPLA2α-overexpressing HepG2 cells were used as experimental samples. The detailed procedure was conducted as described previously18. The phosphorylation ratio of each phosphorylation site was calculated based on the following equation: phosphorylation ratio = phosphorylated molecules/unphosphorylated molecules. Phosphorylated proteins were considered as differentially expressed, once an increase (> 1.18) or a decrease (< 0.85) occurred in the expression level ratio in cPLA2α-knockdown HepG2 cells relative to cPLA2α-overexpressing HepG2 cells.
Bioinformatics analysis
The Database for Annotation, Visualization, and Integrated Discovery (DAVID) (https://david.ncifcrf.gov/) was used to identify significantly enriched Gene Ontology (GO) terms. We focused on the categories of biological process, molecular function, or cellular components, where those with a P < 0.01 and a false discovery rate (FDR) < 0.001 were classified as significantly enriched GO terms. The online functional annotation tool, DAVID, was also used for pathway enrichment analyses. Filtering criteria for significantly changed signaling pathways were a P < 0.01 and an FDR < 0.01. A map of the FA pathway showing annotated upregulation and downregulation of phosphorylated proteins was obtained using the Kyoto Encyclopedia of Genes and Genomes (KEGG) (https://www.kegg.jp/). Protein-protein interaction (PPI) network data were obtained using the Search Tool for Retrieval of Interacting Genes/Proteins (STRING) (http://www.string-db.org/) and then imported into Cytoscape v2.8.3 (http://www.cytoscape.org/) for network visualization.
Statistical analysis
All statistical analyses were conducted using SPSS 19.0 (SPSS Inc., Chicago, IL). Continuous variables were compared between two groups via unpaired t-tests, while categorical variables were compared via the Mann-Whitney U test. Overall survival (OS) and disease-free survival (DFS) were calculated using the Kaplan-Meier method. The log-rank test was used for univariate analysis. Multivariable analysis for OS and DFS were performed by including all significant variables of the univariate analysis in a Cox proportional hazards regression analysis. All statistical tests were two tailed and statistical significance was set at P < 0.05.
Details related to the other assays are described in the supplementary materials and methods.
Results
cPLA2α modulates cell-matrix and cell-cell adhesion in HepG2 cells
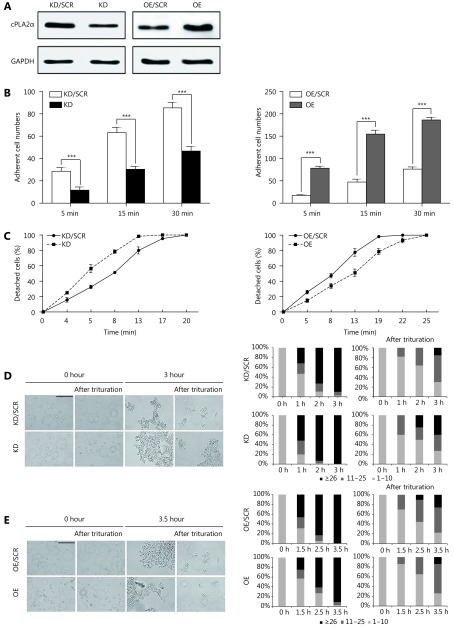
First, we successfully established stable suppression and overexpression of cPLA2α in HepG2 cell lines via the lentiviral infection system (Figure 1A). It was observed that adherence to the plastic cell culture dish surface was significantly impaired by cPLA2α knockdown. Therefore, we inferred that cPLA2α may regulate the interaction between cells and the ECM. For verification purposes, we performed a cell adhesion assay, which revealed that the attachment rate of cPLA2α-downregulated HepG2 cells to fibronectin, at 5, 15, and 30 min was remarkably reduced compared to that of the control cells (Figure 1B, left panel). In contrast, the number of adherent cells significantly increased in the cPLA2α-overexpressing HepG2 cells compared with the number in the control group (Figure 1B, right panel).
1.
cPLA2α modulates cell-matrix and cell-cell adhesion in HepG2 cells. (A) The knockdown and overexpression efficiency of cPLA2α in HepG2 cells was examined by western blot. The abbreviation of cPLA2α-knockdown and cPLA2α-overexpressing HepG2 cells were KD and OE, respectively. The abbreviation of negative control of cPLA2α-knockdown and cPLA2α-overexpressing HepG2 cells were KD/SCR and OE/SCR, respectively. (B) KD/SCR, KD, OE/SCR and OE cells were seeded on twelve-well plates precoated with fibronectin. The number of adherent cells was counted in five random fields on every coverslip under microscopy (200 ×) at 5, 15, 30 min (***P < 0.001). (C) The strength of cell attachment to the substratum was measured by the percentage of detached cells at different time points with trypsin treatment. (D) The hanging-drop assay was performed on KD/SCR and KD cells with or without trituration. Cell aggregates of different sizes (represented by the number of cells per aggregate) were counted before and after trituration at the indicated incubation time points, and their proportions in each sample were shown in the bar graphs. Scale bar: 200 μm. (E) The hanging-drop assay was performed on OE/SCR and OE cells with or without trituration. Cell aggregates of different sizes (represented by the number of cells per aggregate) were counted before and after trituration at the indicated incubation time points, and their proportions in each sample were shown in the bar graphs. Scale bar, 200 μm. Data are represented as the mean ± SD. All experiments were repeated at least three times.
Furthermore, we determined the effect of cPLA2α on the strength of HepG2 cell anchorage to a Matrigel-coated substratum via a cell detachment assay. The results indicated that cPLA2α-knockdown HepG2 cells detached after 13 min of exposure to trypsin whereas control cells detached only after 20 min of exposure to trypsin (Figure 1C, left panel). By contrast, cPLA2α-overexpressing HepG2 cells detached following 25 min of exposure to trypsin, compared to the control cells that detached following 19 min of exposure (Figure 1C, right panel).
Changes in cell-matrix adhesion are frequently accompanied by the opposite alteration in cell-cell adhesion during cancer progression19. Therefore, a hanging-drop assay was conducted to examine the cell-cell adhesion rate and strength of cPLA2α-knockdown and cPLA2α-overexpressing HepG2 cells. Larger aggregates were more quickly formed by HepG2 cells than by control cells, due to decreased cPLA2α expression in the former (Figure 1D, left bar charts). Following 3 h of incubation, approximately 40% of cPLA2α-knockdown cells remained as large aggregates after trituration (≥ 26 cells), which was substantially higher than that of the control cells (~15%; Figure 1D, right bar charts). Conversely, cPLA2α overexpression tended to suppress cell clustering because the percentage of large aggregates (≥ 26 cells) was lower than that in control cells at different incubation time points before and after trituration (Figure 1E).
Taken together, these data suggest that cPLA2α plays an important role in cell-matrix and cell-cell adhesion in HepG2 cells.
Characterization of differentially expressed phospho-proteins in cPLA2α-knockdown and cPLA2α-overexpressing HepG2 cells
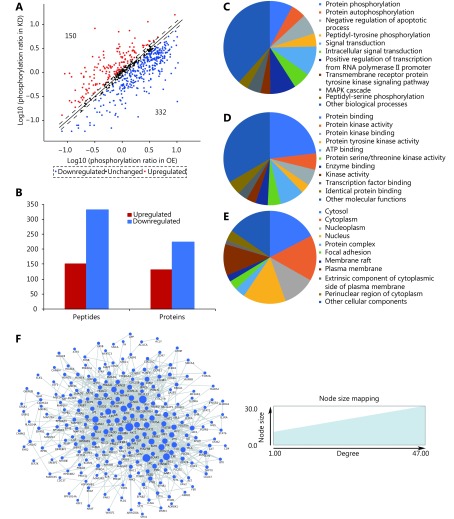
We attempted to identify the molecular mechanism(s) underlying cPLA2α-mediated cell adhesion via a high-throughput phospho-protein array technique. This commercial array, which contains 1,318 site-specific and phospho-specific antibodies, enables efficient profiling of protein phosphorylation20. This method facilitates the detection of differentially expressed phosphorylated proteins and potential cPLA2α downstream substrates and effectors that participate in cell adhesion. Our study applied cell lysates from cPLA2α-knockdown and cPLA2α-overexpressing HepG2 cells. Log phosphorylation ratio was calculated for each screened protein (phosphorylation ratio = phosphorylated molecules/unphosphorylated molecules). For each protein, the ratio (logs) for cPLA2α-knockdown cells was plotted against the ratio (logs) for cPLA2α-overexpressing HepG2 cells (Figure 2A). Fold ratios of > 1.18 and < 0.85 were defined as cutoff criteria. The array revealed 150 upregulated phosphorylation sites in 130 proteins, and 332 downregulated phosphorylation sites in 224 proteins in cPLA2α-knockdown cells relative to cPLA2α-overexpressing cells ( Figure 2A and 2B). Next, a GO analysis was conducted to obtain a system-level view of these differentially expressed, phosphorylated proteins. The result showed that the targets were enriched in many biological processes, molecular functions, and cellular components (Figure 2C, 2D, and 2E), indicating that cPLA2α may play various roles in different processes. These differentially expressed phosphorylated proteins were searched against the STRING database for interaction information and imported to Cytoscape for PPI network construction. A widely connected network composed of 268 proteins and 983 connections was mapped (Figure 2F).
2.
Characterization of differentially expressed phospho-proteins in cPLA2α-knockdown and cPLA2α-overexpressing HepG2 cells. (A) For each phosphorylation site analyzed, the ratio (in logs) of phosphorylated to unphosphorylated protein in KD cells was plotted against the same ratio (in logs) in OE cells. Each phosphorylation site of which ratio was > 1.18 or < 0.85 was marked as a red or blue plot, respectively. Otherwise, the sites were marked as black plots. (B) The bar chart demonstrated that 150 phosphorylation sites in 130 proteins were upregulated and 332 phosphorylation sites in 224 proteins were downregulated in KD cells when compared to OE cells. (C-E) Gene oncology analysis of differentially expressed phosphoproteome in terms of biological process (C), molecular function (D) and cellular component (E). (F) The PPI network of differentially expressed phosphoproteome and their interactions were represented as nodes and edges. The node size reflected the indicated interaction degree.
In summary, the phospho-antibody microarray technique indicated that changes in cPLA2α may lead to multiple phosphorylation events at specific sites.
KEGG pathway analysis reveals the enrichment of FA pathway in differentially expressed phosphorylated proteins
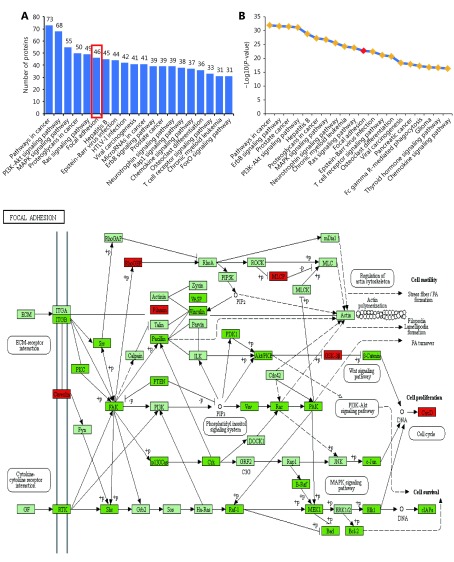
In order to further explore key pathways associated with these differentially expressed phosphorylated proteins, we applied the online functional annotation tool, DAVID, to identify significant pathway categories linked to cPLA2α. Our analysis indicated that these target proteins were heavily involved in various biological pathways. Based on screening criteria (P < 0.01 and FDR < 0.01), we enriched 82 KEGG pathway categories with these phospho-proteins ( Supplementary Table S1). The top 20 signaling pathways based on the number of relevant phosphorylated proteins are shown (Figure 3A). We observed that the FA pathway, which is key to controlling cell-matrix adhesion, ranked sixth among the related pathways. This result was highly consistent with the finding that cPLA2α may enhance cell-matrix adhesion in HepG2 cells. The FA pathway was also within the top 20 pathway terms based on P-values (Figure 3B and Supplementary Table S1). The online functional annotation tool, KEGG, was used to conduct the KEGG pathway mapping analysis to intuitively visualize each differentially expressed phosphorylated protein in the FA pathway (Figure 3C). The map displayed concordant changes in cPLA2α and critical cell adhesion molecules, including FAK and paxillin21, as they were downregulated (highlighted in dark green) in cPLA2α-knockdown cells compared with those in cPLA2α-overexpressing cells.
S1.
KEGG pathway enrichment of differentially expressed phosphorylated proteins by DAVID
3.
KEGG pathway analysis reveals the enrichment of FA pathway in differentially expressed phosphorylated proteins. (A, B) The top 20 enriched signaling pathways of differentially expressed phosphorylated proteins. X axis, KEGG term; (A) Y axis, number of phosphorylated proteins; (B) Y axis, negative logarithm (-log10) of P-value. The red frame (A) and rhombus (B) represented the focal adhesion pathway. (C) The KEGG pathway map of focal adhesion. Red rectangles indicated the upregulated phosphorylated proteins; dark green rectangles indicated the downregulated phosphorylated proteins; and aqua rectangles indicated the unchanged phosphorylated proteins.
Considered as a whole the data indicate that components in the FA pathway play a role in cPLA2α-mediated cell-matrix adhesion.
cPLA2α regulates the phosphorylation and colocalization of FAK and paxillin
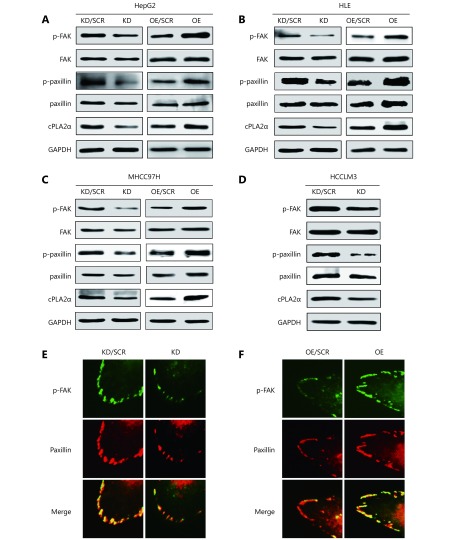
The above pathway analyses denoted that a wide spectrum of phosphorylated proteins in the FA pathway were involved in cPLA2α-regulated cell-matrix adhesion. Among the proteins in the FA complex, FAK, a signaling protein which plays a pivotal role in integrin-mediated signals, initiates the first step in FAK activation, which is autophosphorylation at Tyr-39722. Paxillin is a FA-associated structural protein that can be phosphorylated at Tyr-118 by FAK23. Therefore, these two specific phosphorylation sites were selected for further analysis. Immunoblot analysis confirmed that phosphorylation of FAK (Tyr-397) and paxillin (Tyr-118) in cPLA2α-knockdown HepG2 cells was reduced (Figure 4A). We further investigated the phosphorylation status of these 2 proteins in 3 other HCC cell lines, namely HLE, MHCC97H, and HCCLM3, and obtained similar results. These results demonstrated that targeted downregulation of cPLA2α significantly inhibited the expression of p-FAK and p-paxillin (Figure 4B-4D).
4.
cPLA2α regulates the phosphorylation and colocalization of FAK and paxillin. (A) Western blot analysis of cPLA2α, p-FAK (Tyr-397), FAK, p-paxillin (Tyr-118) and paxillin in cPLA2α KD/SCR, KD, OE/SCR and OE HepG2 cells. (B) Western blot analysis of cPLA2α, p-FAK (Tyr-397), FAK, p-paxillin (Tyr-118) and paxillin in cPLA2α KD/SCR, KD, OE/SCR and OE HLE cells. (C) Western blot analysis of cPLA2α, p-FAK (Tyr-397), FAK, p-paxillin (Tyr-118) and paxillin in cPLA2α KD/SCR, KD, OE/SCR and OE MHCC97H cells. (D) Western blot analysis of cPLA2α, p-FAK (Tyr-397), FAK, p-paxillin (Tyr-118) and paxillin in cPLA2α KD/SCR and KD HCCLM3 cells. (E) Immunofluorescence staining of p-FAK (Tyr-397) and paxillin as well as their colocalization in cPLA2α KD/SCR and KD HepG2 cells. (F) Immunofluorescence staining of p-FAK (Tyr-397) and paxillin as well as their colocalization in cPLA2α OE/SCR and OE HepG2 cells. Original magnifications, 400 ×.
Additionally, ectopic expression of cPLA2α may lead to increased phosphorylation of FAK (Tyr-397) and paxillin (Tyr-118) in HepG2 cells (Figure 4A). Similarly, phosphorylation levels of p-FAK and p-paxillin were markedly increased in HLE and MHCC97H cells with stably overexpressed cPLA2α (Figure 4B and 4C). Considering that FAK is recruited and activated at FA sites where paxillin localizes24, we next examined colocalization of activated FAK (Tyr-397) and paxillin via immunofluorescence. The results indicated that decreased cPLA2α expression levels may hinder FAK phosphorylation at FA sites. Therefore, colocalization of p-FAK and paxillin was significantly reduced (Figure 4E). Reciprocally, upregulation of cPLA2α promoted p-FAK localization at FA sites (Figure 4F).
Collectively, these results suggest that cPLA2α may mediate cell-matrix adhesion through the FAK/paxillin pathway in HCC cells.
Expression of cPLA2α and p-FAK in HCC samples and their correlation with clinical prognosis
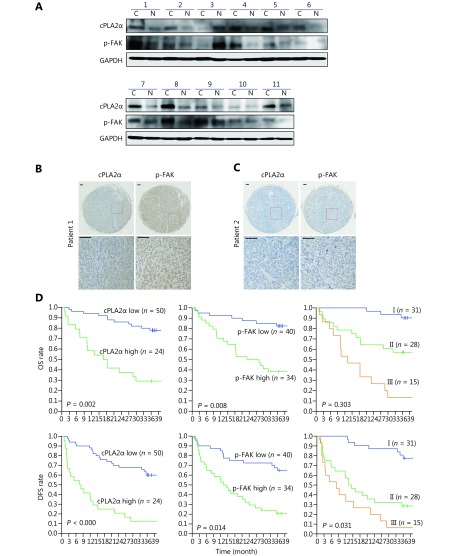
Our study demonstrated the association between cPLA2α and p-FAK via in vitro experiments. We investigated expression patterns of cPLA2α and p-FAK in human HCCs. Eleven pairs of HCC tumors and adjacent noncancerous tissues were examined via Western blot analysis. The expression levels of cPLA2α and p-FAK were significantly higher in HCC tissues than in peritumoral tissues (9/11 and 10/11, respectively; Figure 5A). Interestingly, overexpression of cPLA2α was accompanied by p-FAK upregulation, and even exceptional one-paired tissues (sample 3) shared the same pattern. We subsequently performed an immunohistochemical analysis of cPLA2α and p-FAK expression via a tissue microarray composed of primary tumor tissues from 74 HCC patients. Immunostaining indicated cytoplasmic staining for cPLA2α and nuclear and cytoplasmic staining for p-FAK (Figure 5B and 5C). Correlation analysis confirmed correlation between cPLA2α and p-FAK (P < 0.05; Table 1). Clinicopathological analysis showed that the cPLA2α expression was significantly associated with macrovascular invasion (P < 0.0001), and that p-FAK expression was significantly linked to tumor size ( P = 0.005), number of tumors (P = 0.040), and satellite nodes (P = 0.049); (Table 1). In regard to prognostic value, HCC patients with high expression levels of cPLA2α or p-FAK had lower OS and DFS rates than the group with low expression levels (Figure 5D, left and middle line charts). We further evaluated the combined effect of cPLA2α and p-FAK on clinical prognosis. The result showed that the cPLA2αHigh/p-FAKHigh group had the lowest DFS rate among the 3 groups of patients (P = 0.031), indicating that this was the group that was most susceptible to metastasis and recurrence of HCC. However, the expression levels of cPLA2α and p-FAK had no impact on the OS rate (P = 0.303); (Figure 5D, right line charts). Multivariate analysis revealed that cPLA2α or p-FAK alone, as well as a combination of these two, were independent prognostic factors for HCC patients (Table 2).
5.
Expression of cPLA2α and p-FAK in HCC samples and their correlation with clinical prognosis. (A) Western blot analysis of cPLA2α and p-FAK expression in 11 paired cancer tissues (C) and adjacent noncancerous tissues (N). (B, C) Representative images (upper, 50 ×; lower, 200 ×) of cPLA2α and p-FAK expression by immunohistochemistry. (B) High expression of both cPLA2α and p-FAK; (C) low expression of both cPLA2α and p-FAK. (D) Kaplan-Meier analyses of overall survival (OS) and disease-free survival (DFS) rate according to cPLA2α and p-FAK expression. I, cPLA2αLow/p-FAKLow; II, cPLA2αLow/p-FAKHigh and cPLA2αHigh/p-FAKLow; III, cPLA2αHigh/p-FAKHigh.
1.
Correlation between factors and clinicopathological characteristics in the 74 patients with HCC.
| Clinicopathological variables | Total | p-FAK† | P | cPLA2α† | P | |||
| Low (%) | High (%) | Low (%) | High (%) | |||||
| †Final staining score ≤ 3 was defined as low expression; final staining score > 3 was defined as high expression. *P < 0.05 was considered statistically significant. Abbreviations: AFP, alpha fetoprotein; ALB, albumin; HBV, hepatitis B virus; HCV, hepatitis C virus; MVI, microvascular invasion; MAVI, macrovascular invasion. | ||||||||
| Age (years) | ≤ 55 | 43 | 25 (58.1%) | 18 (41.9%) | 0.409 | 31 (72.1%) | 12 (27.9%) | 0.331 |
| > 55 | 31 | 15(48.4%) | 16 (51.6%) | 19 (61.3%) | 12 (38.7%) | |||
| Gender | Male | 60 | 31(51.7%) | 29 (48.3%) | 0.397 | 43 (71.7%) | 17 (28.3%) | 0.121 |
| Female | 14 | 9(64.3%) | 5 (35.7%) | 7 (50.0%) | 7 (50.0%) | |||
| AFP (μg/L) | ≤ 400 | 47 | 27(57.4%) | 20 (42.6%) | 0.443 | 33 (70.2%) | 14 (29.8%) | 0.524 |
| > 400 | 27 | 13(48.1%) | 14 (51.9%) | 17 (63.0%) | 10 (37.0%) | |||
| ALB (g/L) | ≤ 40 | 25 | 14(56.0%) | 11 (44.0%) | 0.812 | 17 (68.0%) | 8 (32.0%) | 0.955 |
| > 40 | 49 | 26 (53.1%) | 23 (46.9%) | 33 (67.3%) | 16 (32.7%) | |||
| Tumor size (cm) | ≤ 5 | 35 | 25 (71.4%) | 10 (28.6%) | 0.005* | 27 (77.1%) | 8 (22.9%) | 0.098 |
| > 5 | 39 | 15 (38.5%) | 24 (61.5%) | 23 (59.0%) | 16 (41.0%) | |||
| Tumor number | ≤ 1 | 58 | 35 (60.3%) | 23(39.7%) | 0.040* | 41 (70.7%) | 17 (29.3%) | 0.278 |
| > 1 | 16 | 5 (31.3%) | 11(68.7%) | 9 (56.3%) | 7 (43.7%) | |||
| HBV | Absent | 21 | 9 (42.9%) | 12 (57.1%) | 0.227 | 17 (81.0%) | 4 (19.0%) | 0.124 |
| Present | 53 | 31 (58.5%) | 22 (41.5%) | 33 (62.3%) | 20 (37.7%) | |||
| HCV | Absent | 68 | 39 (57.4%) | 29 (42.6%) | 0.057 | 47 (69.1%) | 21 (30.9%) | 0.341 |
| Present | 6 | 1 (16.7%) | 5 (83.3%) | 3(50.0%) | 3 (50.0%) | |||
| Liver cirrhosis | Absent | 38 | 18 (47.4%) | 20 (52.6%) | 0.239 | 23 (60.5%) | 15 (39.5%) | 0.187 |
| Present | 36 | 22 (61.1%) | 14 (38.9%) | 27 (75.0%) | 9 (25.0%) | |||
| Tumor encapsulation | Absent | 70 | 37 (52.9%) | 33 (47.1%) | 0.391 | 46 (65.7%) | 24 (34.3%) | 0.157 |
| Present | 4 | 3 (75.0%) | 1 (25.0%) | 4 (100%) | 0 (0%) | |||
| Tumor thrombus | Absent | 26 | 14 (53.8%) | 12 (46.2%) | 0.979 | 18 (69.2%) | 8 (30.8%) | 0.823 |
| Present | 48 | 26 (54.2%) | 22 (45.8%) | 32 (66.7%) | 16 (33.3%) | |||
| Satellite node | Absent | 48 | 30 (62.5%) | 18 (37.5%) | 0.049* | 33 (68.8%) | 15 (31.2%) | 0.769 |
| Present | 26 | 10 (38.5%) | 16 (61.5%) | 17 (65.4%) | 9 (34.6%) | |||
| MVI | Absent | 70 | 37 (52.9%) | 33 (47.1%) | 0.391 | 47 (67.1%) | 23 (32.9%) | 0.746 |
| Present | 4 | 3 (75.0%) | 1 (25.0%) | 3 (75.0%) | 1 (25.0%) | |||
| MAVI | Absent | 68 | 37 (54.4%) | 31 (45.6%) | 0.836 | 50 (73.5%) | 18 (26.5%) | 0.000* |
| Present | 6 | 3 (50.0%) | 3 (50.0%) | 0 (0%) | 6 (100%) | |||
| p-FAK | Low | 40 | 31 (77.5%) | 9 (22.5%) | 0.049* | |||
| High | 34 | 19 (55.9%) | 15 (44.1%) | |||||
| cPLA2α | Low | 50 | 31 (62.0%) | 19 (38.0%) | 0.049* | |||
| High | 24 | 9 (37.5%) | 15 (62.5%) | |||||
2.
Univariate and multivariate analysis of prognostic factors associated with OS and DFS in 74 patients with HCC.
| Variables | Number | Univariate analysis | Multivariate analysis | Univariate analysis | Multivariate analysis | |||||||
| 3-year
OS (%) |
P | HR
(95% CI) |
P | 3-year
DFS (%) |
P | HR
(95% CI) |
P | |||||
| Data are presented as HR (95% CI) and P-value. †I, cPLA2αLow/p-FAKLow; II, cPLA2αLow/p-FAKHigh and cPLA2αHigh/p-FAKLow; III, cPLA2αHigh/p-FAKHigh. *P < 0.05 was considered statistically significant. Abbreviations: OS, overall survival; DFS, disease free survival; AFP, alpha fetoprotein; ALB, albumin; HBV, hepatitis B virus; HCV, hepatitis C virus; MVI, microvascular invasion; MAVI, macrovascular invasion; N/Y, no/yes; L/H, low/high. | ||||||||||||
| Age (years)
(≤ 55/> 55) |
43/31 | 65.1/58.1 | 0.665 | 51.2/35.5 | 0.290 | |||||||
| Gender (male/female) | 60/14 | 60.0/71.4 | 0.503 | 46.7/35.7 | 0.369 | |||||||
| AFP (μg/L)
(≤ 400/> 400) |
47/27 | 66.0/55.6 | 0.343 | 48.9/37.0 | 0.249 | |||||||
| ALB (g/L)
(≤ 40/> 40) |
25/49 | 64.0/61.2 | 0.926 | 44.0/44.9 | 0.808 | |||||||
| Tumor size
(cm) (≤ 5/> 5) |
35/39 | 71.4/53.8 | 0.116 | 57.1/33.3 | 0.055 | |||||||
| Tumor number
(≤ 1/> 1) |
58/16 | 67.2/43.8 | 0.088 | 46.6/37.5 | 0.428 | |||||||
| HBV (N/Y) | 21/53 | 57.1/64.2 | 0.643 | 42.9/45.3 | 0.869 | |||||||
| HCV (N/Y) | 68/6 | 66.2/16.7 | 0.009* | 1.81
(0.65, 5.02) |
0.256 | 47.1/16.7 | 0.123 | |||||
| Liver cirrhosis (N/Y) | 38/36 | 55.3/69.4 | 0.310 | 34.2/55.6 | 0.112 | |||||||
| Tumor
encapsulation (N/Y) |
70/4 | 60.0/0 | 0.155 | 41.4/0 | 0.062 | |||||||
| Tumor
thrombus (N/Y) |
26/48 | 61.5/62.5 | 0.793 | 50.0/41.7 | 0.626 | |||||||
| Satellite
nodule (N/Y) |
48/26 | 66.7/53.8 | 0.179 | 45.8/42.3 | 0.670 | |||||||
| MVI (N/Y) | 70/4 | 62.9/50.0 | 0.491 | 44.3/50.0 | 0.820 | |||||||
| MAVI (N/Y) | 68/6 | 66.2/16.7 | 0.000* | 1.93
(0.67, 5.61) |
0.226 | 48.5/0 | 0.000* | 1.79
(0.68, 4.71) |
0.239 | |||
| p-FAK (L/H) | 40/34 | 82.5/38.2 | 0.000* | 3.34
(1.37, 8.18) |
0.008* | 65.0/20.6 | 0.000* | 2.28
(1.18, 4.38) |
0.014* | |||
| cPLA2α (L/H) | 50/24 | 78.0/29.2 | 0.000* | 4.20
(1.68, 10.46) |
0.002* | 60.0/12.5 | 0.000* | 3.97
(1.97, 8.01) |
0.000* | |||
| p-FAK +
cPLA2α† |
0.000* | 1.59
(0.66, 3.83) |
0.303 | 0.000* | 2.05
(1.07, 3.93) |
0.031* | ||||||
| I | 31 | 90.3 | 77.4 | |||||||||
| II | 28 | 57.1 | 28.6 | |||||||||
| III | 15 | 13.3 | 6.7 | |||||||||
Thus, we concluded that, apart from cPLA2α expression alone, p-FAK and cPLA2α expressions combined with p-FAK were also associated with poor prognosis in HCC patients.
Discussion
In mammalian cells, cPLA2α is a widely expressed phospholipase. Activation of cPLA2α is calcium dependent. In normal physiological processes and pathophysiological reactions, its regulatory functions are mainly dependent on its glycerophospholipid hydrolysates, AA, and lysophospholipids6, 25. Reportedly, metabolites of both these hydrolysates are associated with inflammation, carcinogenesis, and cancer progression7, 26. Previously, our studies confirmed the promalignant role of cPLA2α in breast cancer and HCC8, 9. In addition to the alteration of migratory, invasive, and metastatic abilities, attachment to the plastic surface of cell culture dishes changed with the downregulation or upregulation of cPLA2α expression. We performed a cell adhesion and detachment assay to ascertain whether knocking down cPLA2α would significantly weaken cell-matrix adhesion in HepG2 cells. By contrast, heterotypic adhesion was enhanced when cPLA2α expression was increased. We also observed tighter connections between cancer cells in cPLA2α-knockdown HepG2 cells than in control cells and reduced cell-cell adhesion in cPLA2α-overexpressing HepG2 cells. These results were consistent with the role played by cPLA2α in promoting the EMT process, which allows epithelial cancer cells to acquire a mesenchymal phenotype; these cells are then able to migrate and metastasize easily8, 27. To our knowledge, our research is among the first studies to investigate the effect of cPLA2α on cell adhesion in HCC.
Protein arrays based on antigen-antibody reaction (antibody arrays) is a form a high-throughput technology used for proteome profiling. This technology effectively interprets biological functions of candidate proteins based on known identities of previously profiled proteins18, 28. We used the Phospho Explorer Antibody Array, an ELISA-based antibody array that detects protein phosphorylation on a large scale29. This phospho-antibody array has been widely applied to improve our understanding of cancer pathogenesis. For example, Jiang et al.30, conducted a quantitative phosphoproteomic analysis using this array, to comprehensively explore the role of SSBP1 in breast cancer metastasis. We identified multiple proteins, the phosphorylation status of which were modulated by cPLA2α in HCC cells. KEGG pathway analysis of these differentiated phospho-proteins revealed that the PI3K/Akt, MAPK, and Ras signaling pathways were within the top 5 related pathways (Supplementary Table S1). The dysregulation of these pathways has been widely characterized and well recognized in cancer development and progression31, 32. These results are consistent with revealed functions of cPLA2α in HCC8, 33.
The FA pathway was the sixth top relevant pathway with 46 associated proteins. Such significant enrichment precisely confirmed our present findings that cPLA2α may promote cell-matrix adhesion in HCC cells. This pathway regulates dynamic interaction between cells and the ECM through FA formation and disassembly. Such interaction is crucial for cancer cell migration and metastasis34. FAK is a key regulator of integrin-mediated adhesion. FAK can be auto-phosphorylated at Tyr-397 upon recruitment of this kinase to FA sites following the binding of the transmembrane integrin receptor to ECM. An activated FAK provides both signal transduction and scaffolding functions19, 35. Paxillin is an important FA-associated cytoskeletal adaptor protein that provides a docking site for FAK. In turn, FAK can phosphorylate paxillin at Tyr-118 to regulate its function24. Phosphorylation of both proteins is required for FA formation, cell motility, and metastasis36. The current study indicated that cPLA2α may stimulate the phosphorylation of FAK and paxillin at Tyr-397 and Tyr-118, respectively. Reduced p-FAK colocalization with the FA marker paxillin in cPLA2α-knockdown HepG2 cells strongly decreased the activity of the integrin-mediated FA pathway37, whereas increased colocalization was observed in cPLA2α-overexpressing HepG2 cells. These observations suggested that cPLA2α may promote cell-matrix adhesion through activation of the FAK/paxillin pathway in HCC. To the best of our knowledge, ours is the first study to investigate the mechanism underlying cell-matrix adhesion regulated by cPLA2α in HCC. The positive association between cPLA2α and the FAK/paxillin pathway may provide a possible explanation for the cancer-promoting role of cPLA2α in HCC. However, further research is needed to confirm this finding.
Here, Western blot and immunohistochemistry related techniques were utilized to investigate the association between cPLA2α expression and p-FAK expression in 11 paired clinical tissue samples and another cohort of 74 HCC patients, respectively. Similar to our previous results, our present findings showed that cPLA2α expression was higher in primary HCC tissues than in the adjacent nontumor tissues. This study was the first to reveal that cPLA2α overexpression was accompanied by an increase in p-FAK levels. This result was consistent with in vitro experiments showing that p-FAK (Tyr-397) expression could be accelerated by cPLA2α upregulation. Furthermore, we estimated the prognostic role of cPLA2α and p-FAK in HCC patients. We divided the 74 patients into 3 subclasses, cPLA2αHigh/p-FAKHigh, cPLA2αHigh/p-FAKLow or cPLA2αLow/p-FAKHigh, and cPLA2αLow/p-FAKLow, based on expression levels. Among these 3 groups, the group with higher expression levels of cPLA2α and p-FAK exhibited the poorest disease-free survival outcomes. Thus, cPLA2αHigh/p-FAKHigh was more sensitive than cPLA2αHigh or p-FAKHigh alone in predicting the prognosis of HCC patients.
Conclusions
Overall, the current study showed that cPLA2α may enhance cell-matrix adhesion via the FAK/paxillin pathway in HCC. This function of cPLA2α may be essential for its prometastatic role in HCC. However, further in-depth research may be required to verify this notion. In addition, the expression of cPLA2α or p-FAK alone, or the co-index (cPLA2α/p-FAK), may serve as prognostic factors for HCC patients. These findings could contribute to the development of new treatment strategies for HCC.
Acknowledgments
This work was supported by grants from Key Project of Tianjin Natural Science Foundation (Grant No. 18JCZDJC35200), NSFC-FRQS program (Grant No. 81661128009), The Science & Technology Development Fund of Tianjin Education Commission for Higher Education (Grant No. 2017KJ202), and Scientific Research Foundation for Returned Scholars and Doctoral Program of Tianjin Medical University Cancer Institute and Hospital (Grant No. B1703).
Conflict of interest statement
No potential conflicts of interest are disclosed.
Supplementary materials and methods
Cell lines and cell culture
MHCC97H and HCCLM3 cells were bought from the Liver Cancer Institute, Zhongshan Hospital, Fudan University, Shanghai, China. HepG2 cells were obtained from American Type Culture Collection (ATCC; Manassas, VA, USA). HLE cells from the Health Science Research Resources Bank (Osaka, Japan) were also utilized. MHCC97H, HCCLM3 and HLE cells were cultured in Dulbecco’s modified Eagle’s medium (Gibco, Carlsbad, CA, USA), and HepG2 cells were incubated in Eagle’s minimum essential medium (Gibco), containing 10% fetal bovine serum (FBS; HyClone Laboratories Inc., Novato, CA, USA) and 1% penicillin-streptomycin solution (PS; Hyclone Laboratories Inc.) at 37 °C under a 5% CO2 atmosphere.
Cell transfection
The lentiviral vectors carrying cPLA2α shRNA, cPLA2α, and their respective scrambled controls (Genechem Co., Shanghai, China) were separately transfected into cells by using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) in accordance with the manufacturer’s instructions. Stably transfected clones were selected using a puromycin (Gibco)-containing medium. The transfection efficiency was validated through western blot assay.
Western blot assay
The cell lysates were separated through sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The separated products were transferred to polyvinylidene difluoride membranes (Immobilon-P, Millipore, Billerica, MA, USA). The following primary antibodies were used: cPLA2α (1:500, GTX110218) (GeneTex, Irvine, TX, USA); FAK (1:100, H-1: sc-1688), paxillin (1:200, D-9: sc-365174), and GAPDH (1:1000, 0411: sc-47724) (Santa Cruz Biotechnology, CA, USA); p-FAK (1:1000, ET1610-34) (Hangzhou HuaAn Biotechnology Co., Zhejiang, China); and p-paxillin (1:500, 2541) (Cell Signaling Technologies, Danvers, MA, USA).
Cell adhesion assay
In brief, 2 × 105 cells/well were added to a 12-well plate containing glass coverslips pretreated with 10 μg/mL fibronectin (R&D Systems Inc., Minneapolis, MN, USA). After 5, 15, and 30 min of incubation, the cells were gently washed twice with cold PBS and then fixed with 4% paraformaldehyde for 5-10 min. Five random fields (200 ×) were chosen to count the number of attached cells. All the samples were tested in triplicate and the data are expressed as the mean ± S.D.
Cell detachment assay
The cells were seeded at 1 × 105 cells/well in a 12-well plate, which was pretreated with 1.2 mg/mL Matrigel (BD Biosciences, Franklin Lakes, NJ, USA). After 48 h, the cells were washed with PBS and then trypsinized with 0.25% trypsin (Hyclone Laboratories Inc.) at 20 °C with gentle agitation. The number of detached cells was counted at different time points, and the total number of cells/well was determined after trypsinization was completed. One well was used for each time point, and each experiment was independently performed at least three times.
Hanging-drop adhesion assay
Briefly, a 30 μL drop of the cells (1 × 105/mL) in culture medium was placed on the inner surface of a 35 mm dish lid. The lid was placed on a dish to ensure that the drops with suspended cells hung from the lid. Approximately 2 mL of the medium was added to the dish to prevent evaporation. Then, the dishes were incubated at 37 °C in a 5% CO2 atmosphere. The lid was then inverted, and cell trituration was performed with a 20 μL pipette tip for 20 times. Three random fields of each drop were photographed, and > 1000 cells were counted per experiment.
Immunofluorescence
Approximately 4 × 104/well cells were seeded in a 12-well plate containing sterile coverslips. After the cells attached, they were fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100 in PBS, and incubated with the primary antibodies p-FAK (1:50) (Hangzhou HuaAn Biotechnology Co.) and paxillin (1:10) (Santa Cruz Biotechnology) overnight at 4 °C. The secondary antibodies used in this study included Alexa Fluor 488-conjugated and 594-conjugated antibodies (Invitrogen, Carlsbad, CA, USA). Cell nuclei were counterstained with diamidino phenylindole (Solarbio, Beijing, China). The fluorescence intensity of the cells was visualized under a fluorescence microscope (Zeiss, Oberkochen, BW, Germany).
Immunohistochemistry
Paraffin-embedded tissue slides were deparaffinized and rehydrated. Microwave antigen retrieval was conducted using 10 mM citrate (pH = 6.0) (Zhongshan Golden Bridge Biotechnology, Beijing, China), and the primary antibodies against cPLA2α (1:80, N-216: sc-438) (Santa Cruz Biotechnology) and p-FAK (1:25) (Hangzhou HuaAn Biotechnology Co.) were applied to the slides and incubated at 4 °C overnight. Next, the tissues were incubated with the secondary anti-rabbit antibody (PV-6002) (Zhongshan Golden Bridge Biotechnology) for 1 h at 37 °C, stained with DAB (Zhongshan Golden Bridge Biotechnology), and counter-stained with 10% Mayer's hematoxylin (Solarbio).
The staining intensity of the cPLA2α expression was evaluated as follows: 0, no staining; 1, weak staining; and 2, strong staining. The percentage of positive cells was scored as follows: 0, < 10% positive cells; 1, 10%–40% positive cells; 2, 40%–70% positive cells; and 3, 70%–100% positive cells. The sum of the two scores was graded as low (score = 0–3) or high (score = 4–5). For p-FAK, the criteria of intensity evaluation were as follows: 0, negative staining; 1, weak staining; 2, moderate staining; and 3, strong staining. The extent of positive cells was scored as follows: 0, < 5%; 1, 5%–25%; 2, 26%–50%; 3, 51%–75%; and 4, > 75%. The sum of the two scores was classified as low (score = 0–3) or high (score = 4–7).
S1.
KEGG pathway enrichment of differentially expressed phosphorylated proteins by DAVID
References
- 1.Forner A, Reig M, Bruix J Hepatocellular carcinoma. Lancet. 2018;391:1301–14. doi: 10.1016/S0140-6736(18)30010-2. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Chen MN, Wei L, Law CT, Tsang FH, Shen JL, Cheng CL, et al RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67:2254–70. doi: 10.1002/hep.29683. [DOI] [PubMed] [Google Scholar]
- 4.Forner A, Llovet JM, Bruix J Hepatocellular carcinoma. Lancet. 2012;379:1245–55. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 5.Murakami M, Taketomi Y, Miki Y, Sato H, Hirabayashi T, Yamamoto K Recent progress in phospholipase A2 research: from cells to animals to humans . Prog Lipid Res. 2011;50:152–92. doi: 10.1016/j.plipres.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 6.Leslie CC Cytosolic phospholipase A2: physiological function and role in disease . J Lipid Res. 2015;56:1386–402. doi: 10.1194/jlr.R057588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang DZ, Dubois RN Eicosanoids and cancer. Nat Rev Cancer. 2010;10:181–93. doi: 10.1038/nrc2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu H, He YC, Qi LS, Chen L, Luo Y, Chen LW, et al cPLA2α activates PI3K/AKT and inhibits Smad2/3 during epithelial-mesenchymal transition of hepatocellular carcinoma cells. Cancer Lett. 2017;403:260–70. doi: 10.1016/j.canlet.2017.06.022. [DOI] [PubMed] [Google Scholar]
- 9.Chen L, Fu H, Luo Y, Chen LW, Cheng RF, Zhang N, et al cPLA2α mediates TGF-β-induced epithelial-mesenchymal transition in breast cancer through PI3k/Akt signaling . Cell Death Dis. 2017;8:e2728. doi: 10.1038/cddis.2017.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valastyan S, Weinberg RA Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147:275–92. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zaidel-Bar R, Itzkovitz S, Ma'ayan A, Iyengar R, Geiger B Functional atlas of the integrin adhesome. Nat Cell Biol. 2007;9:858–67. doi: 10.1038/ncb0807-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miyazaki T, Kato H, Nakajima M, Sohda M, Fukai Y, Masuda N, et al FAK overexpression is correlated with tumour invasiveness and lymph node metastasis in oesophageal squamous cell carcinoma. Br J Cancer. 2003;89:140–5. doi: 10.1038/sj.bjc.6601050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lahlou H, Sanguin-Gendreau V, Zuo DM, Cardiff RD, McLean GW, Frame MC, et al Mammary epithelial-specific disruption of the focal adhesion kinase blocks mammary tumor progression. Proc Natl Acad Sci USA. 2007;104:20302–7. doi: 10.1073/pnas.0710091104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu HG, Nam JO, Miller NL, Tanjoni I, Walsh C, Shi L, et al p190RhoGEF (Rgnef) promotes colon carcinoma tumor progression via interaction with focal adhesion kinase . Cancer Res. 2011;71:360–70. doi: 10.1158/0008-5472.CAN-10-2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao JH, Guan JL Signal transduction by focal adhesion kinase in cancer. Cancer Metastasis Rev. 2009;28:35–49. doi: 10.1007/s10555-008-9165-4. [DOI] [PubMed] [Google Scholar]
- 16.Deakin NO, Turner CE Distinct roles for paxillin and Hic-5 in regulating breast cancer cell morphology, invasion, and metastasis. Mol Biol Cell. 2011;22:327–41. doi: 10.1091/mbc.e10-09-0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sano H, Zhu XD, Sano A, Boetticher EE, Shioya T, Jacobs B, et al Extracellular signal-regulated kinase 1/2-mediated phosphorylation of cytosolic phospholipase A2 is essential for human eosinophil adhesion to fibronectin . J Immunol. 2001;166:3515–21. doi: 10.4049/jimmunol.166.5.3515. [DOI] [PubMed] [Google Scholar]
- 18.Kang SM, Elf S, Lythgoe K, Hitosugi T, Taunton J, Zhou W, et al p90 ribosomal S6 kinase 2 promotes invasion and metastasis of human head and neck squamous cell carcinoma cells. J Clin Invest. 2010;120:1165–77. doi: 10.1172/JCI40582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang WG, Sanders AJ, Katoh M, Ungefroren H, Gieseler F, Prince M, et al Tissue invasion and metastasis: Molecular, biological and clinical perspectives. Semin Cancer Biol. 2015;35 Suppl:S244–75. doi: 10.1016/j.semcancer.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 20.Eke I, Schneider L, Förster C, Zips D, Kunz-Schughart LA, Cordes N EGFR/JIP-4/JNK2 signaling attenuates cetuximab-mediated radiosensitization of squamous cell carcinoma cells. Cancer Res. 2013;73:297–306. doi: 10.1158/0008-5472.CAN-12-2021. [DOI] [PubMed] [Google Scholar]
- 21.Wu J, Li Y, Dang YZ, Gao HX, Jiang JL, Chen ZN HAb18G/CD147 promotes radioresistance in hepatocellular carcinoma cells: a potential role for integrin β1 signaling. Mol Cancer Ther. 2015;14:553–63. doi: 10.1158/1535-7163.MCT-14-0618. [DOI] [PubMed] [Google Scholar]
- 22.Frame MC, Patel H, Serrels B, Lietha D, Eck MJ The FERM domain: organizing the structure and function of FAK. Nat Rev Mol Cell Biol. 2010;11:802–14. doi: 10.1038/nrm2996. [DOI] [PubMed] [Google Scholar]
- 23.López-Colomé AM, Lee-Rivera I, Benavides-Hidalgo R, López E Paxillin: a crossroad in pathological cell migration. J Hematol Oncol. 2017;10:50. doi: 10.1186/s13045-017-0418-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schaller MD Paxillin: a focal adhesion-associated adaptor protein. Oncogene. 2001;20:6459–72. doi: 10.1038/sj.onc.1204786. [DOI] [PubMed] [Google Scholar]
- 25.Niknami M, Patel M, Witting PK, Dong QH Molecules in focus: cytosolic phospholipase A2-α . Int J Biochem Cell Biol. 2009;41:994–7. doi: 10.1016/j.biocel.2008.07.017. [DOI] [PubMed] [Google Scholar]
- 26.Gardell SE, Dubin AE, Chun J Emerging medicinal roles for lysophospholipid signaling. Trends Mol Med. 2006;12:65–75. doi: 10.1016/j.molmed.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 27.Kalluri R, Weinberg RA The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–8. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanchez-Carbayo M Antibody arrays: technical considerations and clinical applications in cancer. Clin Chem. 2006;52:1651–9. doi: 10.1373/clinchem.2005.059592. [DOI] [PubMed] [Google Scholar]
- 29.Havel LS, Kline ER, Salgueiro AM, Marcus AI Vimentin regulates lung cancer cell adhesion through a VAV2-Rac1 pathway to control focal adhesion kinase activity. Oncogene. 2015;34:1979–90. doi: 10.1038/onc.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang HL, Sun HF, Gao SP, Li LD, Huang S, Hu X, et al SSBP1 suppresses TGFβ-driven epithelial-to-mesenchymal transition and metastasis in triple-negative breast cancer by regulating mitochondrial retrograde signaling. Cancer Res. 2016;76:952–64. doi: 10.1158/0008-5472.CAN-15-1630. [DOI] [PubMed] [Google Scholar]
- 31.Fruman DA, Rommel C PI3K and cancer: lessons, challenges and opportunities. Nat Rev Drug Discov. 2014;13:140–56. doi: 10.1038/nrd4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rovida E, Stecca B Mitogen-activated protein kinases and Hedgehog-GLI signaling in cancer: A crosstalk providing therapeutic opportunities? Semin Cancer Biol. 2015;35:154–67. doi: 10.1016/j.semcancer.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 33.Xu L, Han C, Lim K, Wu T Cross-talk between peroxisome proliferator-activated receptor δ and cytosolic phospholipase A2α/cyclooxygenase-2/prostaglandin E2 signaling pathways in human hepatocellular carcinoma cells . Cancer Res. 2006;66:11859–68. doi: 10.1158/0008-5472.CAN-06-1445. [DOI] [PubMed] [Google Scholar]
- 34.Eke I, Cordes N Focal adhesion signaling and therapy resistance in cancer. Semin Cancer Biol. 2015;31:65–75. doi: 10.1016/j.semcancer.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 35.Lechertier T, Hodivala-Dilke K Focal adhesion kinase and tumour angiogenesis. J Pathol. 2012;226:404–12. doi: 10.1002/path.v226.2. [DOI] [PubMed] [Google Scholar]
- 36.Lee BY, Timpson P, Horvath LG, Daly RJ FAK signaling in human cancer as a target for therapeutics. Pharmacol Ther. 2015;146:132–49. doi: 10.1016/j.pharmthera.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 37.Sulzmaier FJ, Jean C, Schlaepfer DD FAK in cancer: mechanistic findings and clinical applications. Nat Rev Cancer. 2014;14:598–610. doi: 10.1038/nrc3792. [DOI] [PMC free article] [PubMed] [Google Scholar]