Abstract
Early life nutrition is a vital determinant of an individual’s life-long health and also directly influences the ecological and functional development of the gut microbiota. However, there are limited longitudinal studies examining the effect of diet on the gut microbiota development in early childhood. Here, up to seven stool samples were collected from each of 48 healthy children during their second year of life, and microbiota dynamics were assessed using 16S rRNA gene amplicon sequencing. Children’s dietary information was also collected during the same period using a validated food frequency questionnaire designed for this age group, over five time points. We observed significant changes in gut microbiota community, concordant with changes in the children’s dietary pattern over the 12-month period. In particular, we found differential effects on specific Firmicutes-affiliated lineages in response to frequent intake of either processed or unprocessed foods. Additionally, the consumption of fortified milk supplemented with a Bifidobacterium probiotic and prebiotics (synbiotics) further increased the presence of Bifidobacterium spp., highlighting the potential use of synbiotics to prolong and sustain changes in these lineages and shaping the gut microbiota community in young children.
Subject terms: Microbiology, Health sciences
Introduction
The gut microbiota co-evolved with the human host to develop a mutual symbiotic relationship1. The relationship between the host and resident microbes is vital for human development and health2. Gut microbiota colonisation and development takes place in early life and influences short and long-term health outcomes. Such outcomes include but are not limited to the development of overweight and obesity3–6, allergic diseases7–11 and neurological disorders12,13. Emerging evidence suggests that the microbial community is not yet mature in adolescents14, therefore, it is possible that gut microbiota development continues alongside human physiological development15. Thus, optimising early life conditions conducive to the development of symbiosis between the host and microbiota is important.
Diet is one of the most important factors that directly affects both the composition and metabolism of the gut microbiota16–18, principally through their colonisation and persistence19. For example, dietary diversity increases available substrates for the gut microbiota, thereby, increasing microbial diversity20, which has been linked to health status21. However, the increased reliance on processed foods in the last half-century, along with antibiotic use, and shifts in lifestyle and environment has challenged the symbiotic relationship established with the resident microbes22 and coincides with the apparent decrease in gut microbiota diversity in Western countries23.
The majority of research on the effect of early life nutrition on gut microbiota development has predominantly focused on breastfeeding and/or introduction of solid food during the first year24–34. However, important nutritional developments and dietary changes also occur in the second year35,36 when children transition from a predominantly milk-based diet to table foods. Given the paucity of longitudinal research on the effect of dietary intake on gut microbiota development in early life37, we aimed to longitudinally examine the effect of diet on the gut microbiota of children throughout their second year of life.
Results
Study subjects
The gut microbiota profiles were obtained from children in the Child Health and Resident Microbes (CHaRM) study which was run in adjunct to the Growing Up Milk ‘Lite’ (GUMLi) trial. The GUMLi trial was a double blind randomised controlled trial to investigate effects of toddler milk compared to unfortified cow’s milk in healthy (i.e. free of any known disease) children from the age of one to two years. GUMLi is a fortified milk supplemented with synbiotic; Bifidobacterium breve M-16V, long-chain galactooligosaccharides (GOS) and short-chain fructooligosaccharides (FOS). Of 52 children enrolled in the GUMLi trial in Brisbane, 51 consented to participate in the longitudinal CHaRM study, and 48 children (94%) completed the study. Table 1 outlines the characteristics of the CHaRM study subjects and samples collected. Among the CHaRM study subjects, there were no baseline differences between the trial milk groups (GUMLi vs control) for gender, birth order, gestation, mode of delivery, duration of any breastfeeding, current breastfeeding status, antibiotic exposure, daycare attendance, pet ownership or exposure to farm animals. The GUMLi group was, however, exclusively breastfed (i.e. received breastmilk only) longer (median 19.5 weeks, range 13.0–26.0) compared with the control group (median 15.2 weeks, range 1.5–20.6) (p = 0.051).
Table 1.
Summary of information for the CHaRM study subjects characteristics.
| Details | n (%) |
|---|---|
| Total number of children enrolled in the longitudinal CHaRM study | 51 |
| Female subjects enrolled in the study | 29 (56.8%) |
| Number of children withdrawn | 3 (5.8%) |
| Final number of children completing the CHaRM study | 48 (94.1%) |
| Female subjects completing the study | 27 (56.3%) |
| Median duration of exclusive breastfeeding | 17.3 weeks (6.5–26.0) |
| Median duration of any breastfeeding before the age of 2 years | 41.1 weeks (20.6–65.0) |
| Number of children exposed to antibiotics before the age of 2 years | 40 (78.4%) |
| Number of children completed the study who received GUMLi | 24 (50.0%) |
Gut microbiota characteristics
In total, 347 gut microbiota samples were collected, of which 345 were included for analyses. After quality scoring and filtering, the entire dataset was comprised of 126 different operational taxonomic units (OTUs) and 24 identified genera.
Shifts in gut microbiota from the age of one to two years
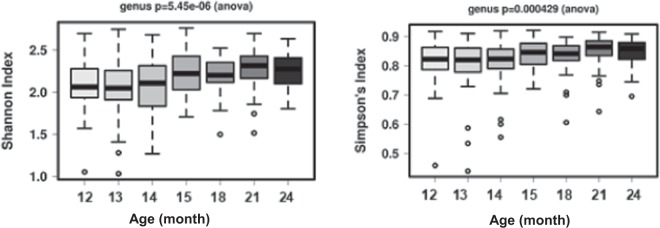
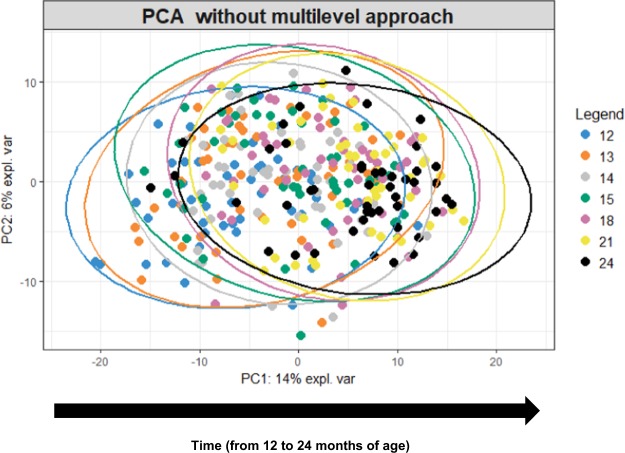
Alpha diversity scores significantly increased during the second year of life (p < 0.01). The overall richness and evenness of the microbial community expanded during this period irrespective of diet (Fig. 1). The Principal Components Analysis (PCA) highlighted a small but gradual shift of the gut microbial community with age (Fig. 2). We also observed significant changes (i.e. increase or decrease) in bacterial taxa from baseline (one year of age) to the end of study (two years of age) at phylum, family, genus and OTU levels as presented in Table S1. At the genus level, Eubacterium, Veillonella, Oscillospira, Streptococcus, Eggerthella and Akkermansia all significantly decreased during the second year of life (FDR < 0.05), while the relative abundance of Faecalibacterium increased during the same period. At the OTU level, the majority of those that significantly decreased in their relative abundance were assigned to the genera listed above, as well as Lachnospiraceae family. However, unspecified family members belonging to Lachnospiraceae and Erysipelotrichaceae and unspecified Blautia genus increased during this period.
Figure 1.
Change in microbial community number (richness) and distribution (evenness) from baseline (12 months of age) to end of study (24 months of age).
Figure 2.
(a) Principal Component Analysis of all gut microbial OTUs collected over the 12 month period from baseline to end of study showing a gradual shift of the gut microbial community from the age of 12 to 24 months.
Shift in dietary pattern from the age of one to two years
The dietary patterns of children were initially explored with PCA38. First, analysis of all children enrolled in the GUMLi trial (n = 160) was performed to obtain robust results39 and clusters of food groups were visualised and identified with correlation circle plots (Fig. S1a). On the first component (explaining 16% of total variance), a shift in children’s dietary pattern was observed across time from one to two years of age (Fig. S1b). The most contributing factors to this shift is change from an “infant-like” diet represented by ‘baby’ foods at the first collection time point (yellow circle), to an “adult-like” diet by the last data collection time point. Furthermore, the analysis showed that the “adult-like” diet could be further subdivided in the second component to an ‘unhealthy’ diet represented by processed meat, savoury snacks, hot-chips/French fries and cakes (red circle); or a ‘healthy’ diet represented by meat/fish, fruit, vegetables, eggs/beans and bread/pasta (blue circle). We ran sub-group analysis of the CHaRM study participants only and found no difference in the dietary patterns.
Shift in gut microbiota with dietary pattern from the age of one to two years
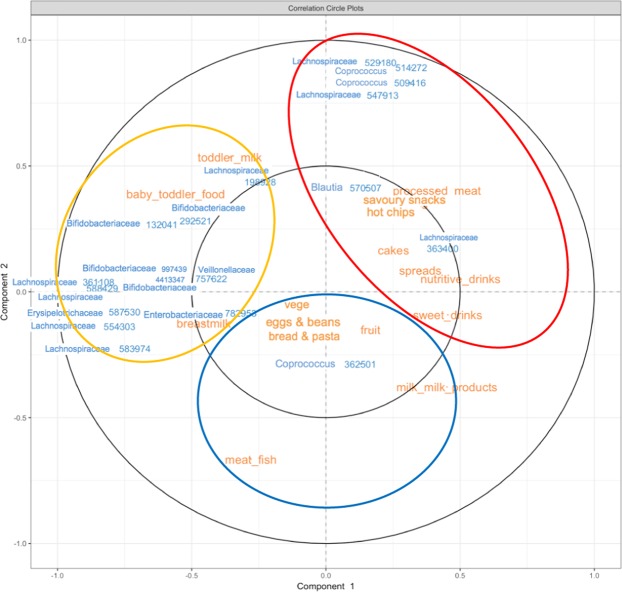
Data integration of food groups representing children’s dietary pattern with microbial OTUs from time points 0, 3, 6, 9, and 12 months was performed with sparse Partial Least Square (sPLS) analysis40, as shown in Fig. 3. The infant-like diet was correlated with unspecified genera belonging to Bifidobacterium and Ruminococcus as well as unknown members of Erysipelotrichaceae and Lachnospiraceae families (yellow circle). The ‘unhealthy’ adult-like diet correlated with unspecified members of Lachnospiraceae family and a Coprococcus genus (red circle). The ‘healthy’ adult-like diet correlated with a different unspecified Coprococcus genus (blue circle).
Figure 3.
Correlation circle plot showing global analysis of gut microbial OTUs and food groups. Ellipses showing clusters of food groups correlated with OTUs. Yellow circle = ‘baby’ foods, red circle = ‘unhealthy’ foods, and blue circle = ‘healthy’ foods.
Effect of children’s diet on their gut microbiota
In order to detect dynamics between specific microbial communities and food intake over the 12 months of the study period, we conducted sPLS analyses41 at each individual time point. The correlation between sPLS selected microbial OTUs and food groups are visualised as clustered image maps in Fig. S2. A summary of microbial taxa correlated with food groups over different time points is available in Table 2. Briefly, unspecified family members of Lachnospiraceae, a Ruminococcus genus, and a Bacteroides genus were positively associated with unprocessed foods (e.g. meat/fish, fruit) but negatively with processed food groups (e.g. processed meat, savoury snacks). Whereas, unspecified Lachnospiraceae family as well as Blautia and Clostridium genera were positively associated with processed food and negatively associated with unprocessed food groups. Unspecified Bifidobacterium genera were positively associated with GUMLi intake but negatively with other milk/milk products.
Table 2.
Summary of bacterial taxa associated with food groups over different time points.
| Bacteria taxa (OTU ID) | Associated food group(s) | Association | Time point(s) |
|---|---|---|---|
| Lachnospiraceae (363400) | Processed meat | Positive | 0, 3, 6 |
| Hot chips (French fries) | Positive | 3, 6 | |
| Sweet drinks | Positive | 3, 6 | |
| Savoury snacks | Positive | 3, 6 | |
| Meat/fish | Negative | 0, 3 | |
| Lachnospiraceae (554303) | Eggs/beans | Positive | 9, 12 |
| Fruit | Positive | 9 | |
| Savoury snacks | Negative | 6 | |
| Hot chips (French fries) | Negative | 6 | |
| Nutritive drinks | Negative | 6 | |
| Sweet drinks | Negative | 6 | |
| Lachnospiraceae (588429) | Meat/fish | Positive | 3 |
| Eggs/beans | Positive | 9, 12 | |
| Fruit | Positive | 9, 12 | |
| Processed meat | Negative | 3 | |
| Blautia (546876) | Savoury snacks | Positive | 3 |
| Milk | Positive | 3 | |
| Processed meat | Positive | 3 | |
| Meat/fish | Negative | 3 | |
| Eggs/beans | Negative | 12 | |
| Ruminococcus (583398) | Meat/Fish | Positive | 3 |
| Processed meat | Negative | 3 | |
| Savoury snacks | Negative | 6 | |
| Nutritive drinks | Negative | 6 | |
| Coprococcus (362501) | Milk/milk products | Positive | 3, 6, 9 |
| Toddler milk | Negative | 6, 9 | |
| Bacteroides (305946) | Fruit | Positive | 9 |
| Eggs/beans | Positive | 9,12 | |
| Meat/fish | Positive | 9 | |
| Hot chips (French fries) | Negative | 6 | |
| Nutritive drinks | Negative | 6 | |
| Clostridium (317135) | Processed meat | Positive | 0 |
| Fruit | Positive | 0 | |
| Hot chips (French fries) | Positive | 6 | |
| Nutritive drinks | Positive | 6 | |
| Sweet drinks | Positive | 6 | |
| Savoury snacks | Positive | 6 | |
| Bifidobacterium (292521) | Breast milk | Positive | 0 |
| Nutritive drinks | Positive | 0 | |
| Toddler milk | Positive | 6, 9,12 | |
| Baby/toddler food | Positive | 9,12 | |
| Fruit | Negative | 0 | |
| Vegetables | Negative | 0 | |
| Milk/milk products | Negative | 6, 9, 12 | |
| Bifidobacterium (132041) | Toddler milk | Positive | 6, 9, 12 |
| Baby/toddler food | Positive | 9, 12 | |
| Milk/milk products | Negative | 6, 9, 12 |
Effect of synbiotic supplemented trial milk (GUMLi) on gut microbial community
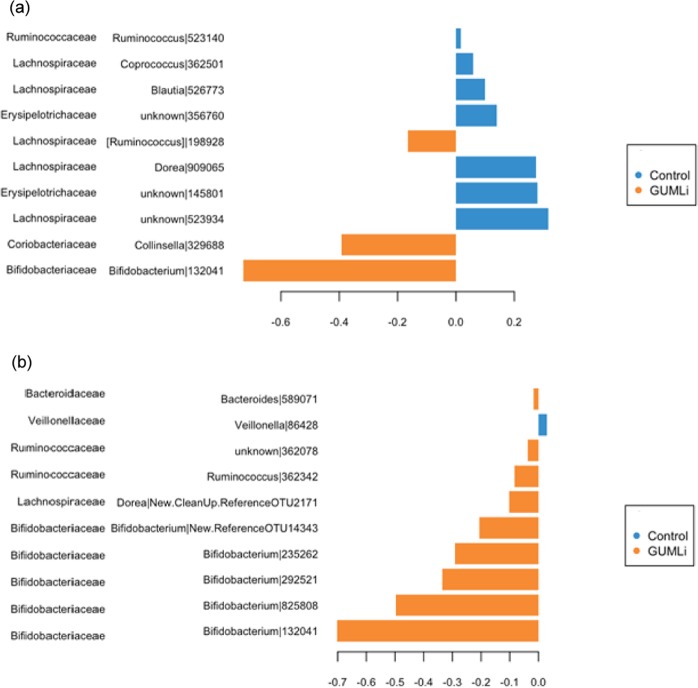
We performed sPLS-discriminant analysis (sPLS-DA)42 to identify the most discriminating microbial community between the trial milk groups. Three months after initiating the trial milk, the most discriminating OTUs between the milk groups were unspecified genus belonging to Bifidobacterium and Collinsella (Fig. 4a). At month 6 of the study, another Bifidobacterium genus discriminated the GUMLi group, which continued until month 9 of the study (data not shown). By the end of the study (month 12), five Bifidobacterium genera most discriminated the microbial community between the trial milk groups and these were associated with the GUMLi group (Fig. 4b).
Figure 4.
(a) sPLS-DA analysis of gut microbial community at month 3 of GUMLi trial and (b) at month 12 of GUMLi trial. The barplot highlight the most important OTUs (from bottom to top) selected by sPLS-DA, with colors indicating a maximum median abundance in a particular group.
Linear mixed models (LMM) were used to analyse the effect of different covariates on the Bifidobacterium community only, as the GUMLi was supplemented with a probiotic Bifidobacterium breve (B. breve) M-16V and FOS/GOS prebiotics (Table S2). Age most influenced the shift in the Bifidobacterium community. However, the strongest influence was the GUMLi intake, where OTU132041 increased the most but this OTU decreased with age. Breastfeeding duration had no impact on the Bifidobacterium community.
Phylogenetic analysis
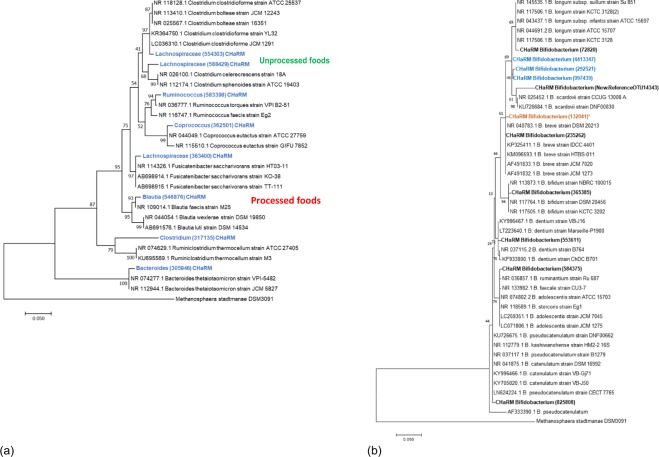
Next, we conducted phylogenetic analysis of bacterial taxa that were associated with processed or unprocessed food groups over different time points (Fig. 5a). Phylogenetic clusters were identified predominantly for Firmicutes but also a Bacteroidetes. For the Firmicutes phylum associated with unprocessed food groups, the first Lachnospiraceae was closely related to Clostridium clostridioforme or Clostridium bolteae, and another Lachnospiraceae with Clostridium celerecrescens and Clostridium sphenoides. The Ruminococcus was closely aligned with Ruminococcus torques and Ruminococcus faecis. The Bacteroides associated with unprocessed food was closely related to Bacteroides thetaiotaomicron. Among the OTUs associated with processed foods, the Lachnospiraceae aligned with Fusicatenibacter saccharivorans, the Blautia aligned with Blautia faecis, and the Clostridium aligned with Ruminoclostridium thermocellum. The Coprococcus closely aligned with Coprococcus eutactus, and this genus was positively associated with milk/milk product intake.
Figure 5.
(a) A maximum likelihood phylogenetic tree of the 16S rRNA gene sequence of the OTUs associated with food groups over different time points. OTUs associated with processed and unprocessed food groups are labelled for Firmictues phylum. The scale bar represents 5% sequence divergence with 1000 boot straps. Methanospheara stadtmanae DSM3091 was used as an outgroup. (b) A maximum likelihood phylogenetic tree of 11 Bifidobacterium OTUs found in the CHaRM study subjects. The scale bar represents 5% sequence divergence with 1000 bootstraps. Methanospheara stadtmanae DSM3091 was used as an outgroup. An OTU associated with GUMLi closely aligned with B. breve (orange, asterisc). A cluster is formed among OTUs associated with breastfeeding at baseline (blue).
A separate phylogenetic analysis of Bifidobacterium was conducted (Fig. 5b). Our phylogenetic analysis is comparable to the comparative genomics study of Bifidobacterium43. Bifidobacterium genera that were associated with the GUMLi group were related to all B. breve strains used in the phylogenetic analysis. However, because B. breve M-16V 16S rRNA sequence has not been released to the public, we were not able to determine if these B. breve detected in the CHaRM cohort were a B. breve M-16V strain. Other Bifidobacterium genera grouped with B. longum and B. scardovii. Another Bifidobacterium genus grouped with several species, including B. catenulatum, B. kashiwanohense and B. pseudocatenulatum. Bifidobacterium genera that positively correlated with frequent breast milk intake at baseline formed a cluster in a phylogenetic analysis.
Discussion
This study is the first to examine how diet during a child’s second year of life affects the gut microbiota. We observed correlations between dietary pattern and the bacterial community. Children’s dietary patterns shifted across time from infant-like to adult-like diet, regardless of children being involved in a trial to investigate the effect of fortified milk. The adult-like diet further deviated to either ‘healthy’ diet characterised by meat/fish, fruit, vegetables, eggs/beans and bread/pasta, or ‘unhealthy’ diet characterised by processed meat, savoury snacks, hot-chips/French fries and cakes. Such dietary patterns in young children are in line with findings from previous studies on diet of similar aged children44,45. We did not observe strong correlation between bacterial OTUs and breastfeeding except at the baseline. This is likely due to a considerable drop in breastfeeding rates from the age of one to two years, which is generally observed during the second year of life. These findings also suggest that the effects of diet in the second year of life have a stronger selective pressure on the gut microbiota than breastfeeding.
The gut microbiota shifted over time, demonstrated by changes in gut microbial community members and increased α-diversity indices. We observed increases in relative abundances of Lachnospiraceae and Ruminococcaceae taxa. Previous studies identified that Lachnospiraceae was a marker of microbiota maturation from infant-like to an adult-like community occurring in the second year of life34,46,47. Laursen and colleagues34 reported that introduction of family foods shifted the gut microbiota composition and α-diversity in nine-month-old children, suggesting increased intake of nutritionally diverse foods rich in fibre and proteins may be the main driver of gut microbial α-diversity development. We also observed that Faecalibacterium was the only genus, which significantly increased from the age of one to two years. F. prausnitzii, currently the sole species identified in the Faecalibacterium genus48 was previously reported as one of the indicators of gut microbiota maturity in young children46,47. They are important butyrate producers and have anti-inflammatory potential49 and F. prausnitzii is often the most abundant gut bacterium found in healthy adults50. An increase in Faecalibacterium abundance with age in this otherwise healthy CHaRM study cohort further suggests F. prausnitzii as one of the markers for gut microbiota maturity during the developmental period.
We observed trends in association between gut microbiota with food groups, particularly with members of the Firmicutes phylum. An unspecified member of Lachnospiracea family has shown persistent positive correlations with processed foods and negative correlations with unprocessed foods over several study time points (0, 3, 6 month of study). A phylogenetic analysis showed that this OTU closely aligned with Fusicatenibacter saccharivorans, a novel species of the Lachnospiraceae family isolated from human faeces51. Processed food is an epitome of a modern ‘Western’ diet. The increased consumption of processed foods equates to increased exposure to food additives52, and a number of studies have identified the negative effect of food additives on gut microbiota53–55. In contrast, some OTUs positively correlated with unprocessed foods, but negatively with processed food, suggesting that these bacteria may have a capacity for better growth in the presence of protein rich foods, rather than processed carbohydrate/sugar rich foods. Another unspecified Lachnospiraceae closely aligned with C. bolteae and C. clostridioforme in the phylogenetic analysis. Higher abundance of C. bolteae has been remarked upon as part of cross-sectional studies of autism-spectrum disorder56. Whilst we did not specifically quantify this particular Lachnospiraceae, its relative abundance decreased over time (Table S1). Another Lachnospiraceae most closely aligned with C. celecrescens and C. sphenoides, were also found to decrease over time. Changes in the relative abundances of C. sphenoides have been observed with a decrease in cholesterol intake in obese adults57, suggesting their potential role as microbial biomarkers for dietary responsiveness in otherwise healthy children.
Bifidobacteria are widely used as probiotics at all ages. The CHaRM study subjects were enrolled in a randomised controlled trial investigating the effect of GUMLi, a fortified milk supplemented with a B. breve M-16V probiotic and GOS/FOS prebiotics. Therefore, additional analyses were carried out to specifically investigate how the Bifidobacterium community may be affected by diet and/or type of milk products consumed. Generally, the absolute and relative abundance of Bifidobacterium is greater for breastfed infants58, and the Bifidobacterium community also “matures” with age59. We observed decreased abundance of a certain Bifidobacterium genus as children aged, whereas, other Bifidobacterium genera increased their abundance. There were positive correlations between a few Bifidobacterium genera with breast milk intake (i.e. breastfeeding) and these genera clustered closely in a phylogenetic analysis (Fig. 5b) but there were no close relatives to assign these strains in our analysis. Although children in the GUMLi group had a longer exclusive-breastfeeding duration, overall, breastfeeding had no impact on Bifidobacterium community during the second year of life, when adjusted for other covariates. A Bifidobacterium genus which significantly increased with GUMLi intake was closely related to B. breve strains in the phylogenetic analysis, and further studies using quantitative PCR is required to determine the presence of the probiotic B. breve M-16V. In addition, whether the effect on Bifidobacterium community in the GUMLi group was specifically due to the probiotic B. breve M-16V, or the potential role of the GOS and FOS prebiotic mixture that may have contributed to this, requires further investigation.
The CHaRM study has made novel contributions to research in early life gut microbiota development, however, there are a number of study limitations. Further studies would benefit from the following improvements. Due to the small sample size of this study, there was insufficient statistical power to define significance at the OTU level of classification in the longitudinal modelling. However, our observation of increased abundance of bacterial families, including Lachnospiraceae and Ruminococcaceae, as well as the genus Faecalibacterium are in line with previously reported gut microbiota profiles of similar aged children. For the dietary assessment, this study used a FFQ, designed to reflect long-term dietary patterns. Ideally, detailed dietary-records over several days are better to assess the effect of diet on gut microbiota37, but the respondent burden is larger and time consuming60. Researchers must choose a balance between participant (parents) burden and the data collected61 and while parents and caregivers in this 12-month study did comply with the FFQ over five time points, it was burdensome and could not be expanded.
Despite these constraints, our study still revealed trends in the dietary change and associated microbial OTUs, providing new insights into the influence of diet on the development of the gut microbiota during the second year of life. We believe this is the first study to assess the gut microbiota profile together with dietary information collected throughout the second year of life. It, thereby, provides a valuable time-series dataset allowing for monitoring the trends in microbial community shifts in early life along with dietary intake. A strength of this research is that we applied a range of multivariate projection-based methods (i.e. PCA, sPLS and sPLS-DA) to investigate the effect of diet on the gut microbial community, and at different time points. The analysis enabled us to capture diversity within Firmicutes phylum members showing different capacities to grow with either processed or unprocessed foods. The synbiotic-supplemented milk appears to have facilitated recruitment and expansion of Bifidobacterium community members among children who consumed this milk over the 12-month period.
Materials and Methods
Subject and sample collection and analysis
The stool samples analysed and reported here were collected from children participating in the CHaRM study conducted in Brisbane, Australia. These children were recruited in a multi-centre trial (the GUMLi trial) investigating the effect of growing up milk (i.e. fortified milk for young children) compared to unfortified cow’s milk on various outcomes in childhood.
Details of the study methodology are available in Supplementary Information. Briefly, the intervention milk (GUMLi) was a micronutrient fortified milk with reduced energy and protein content compared to other GUM available in the market, and supplemented with probiotic B. breve M-16V and prebiotics, long-chain GOS and short-chain FOS. The control milk was an unfortified cow’s milk and both milks were in powder form and unidentifiable. Stool samples were collected from the Brisbane GUMLi trial participants who agreed to partake the CHaRM study by their mother or caregiver and collected at 0 (baseline), 1, 2, 3, 6, 9 and 12 months into the trial.
We used the Eating Assessment in Toddlers Food Frequency Questionnaire (EAT FFQ)38 to assess the dietary intake of the GUMLi trial participants at 0, 3, 6, 9, and 12 months of the study. The common food groups used to explore dietary patterns are detailed in Table S3. Other information such as breastfeeding, mode of delivery and antibiotic usage, as well as other demographic and relevant data were obtained from mothers/caregivers during the GUMLi trial data collection.
Ethics, consent and permission
Ethical approvals were obtained from the University of Queensland Human Research Ethics Committee (reference 2014001318) and the Northern B Health and Disability Ethics Committee of the Ministry of Health, New Zealand (HDEC reference number 14/NTB/152). Written informed consent was obtained from parents/guardians on the participating child’s behalf, prior to enrolment in the trial. Additional consent was obtained for the participation in the CHaRM study in Brisbane. All experiments were performed in accordance with relevant guidelines and regulations.
Gut microbiota DNA extraction from stool samples
Gut microbiota DNA was extracted using the repeated beating and a column technique62 adapted for use with the automated Maxwell 16 MDx system (Promega).
Sub-samples of stool (0.15 g) were transferred into a 2 mL screw-capped tube containing 0.4 g of sterile zirconia beads (0.1 mm and 1 mm diameter). Into this tube, 600 μL of lysis buffer (500 mM NaCl, 50 mM TRIS-HCl (pH 8.0), 50 mM EDTA and 4% [w/v] sodium dodecyl sulfate) was added and homogenised in the Precellys 24 homogeniser (Bertin Corp) at 5000 rpm, for 3 × 60 second intervals. The homogenised mixture was then incubated at 70 °C for 15 minutes, with gentle shaking by hand every 5 minutes. After incubation, the mixtures were then centrifuged at 4 °C/RT for 5 minutes at 16,363 rcf. The supernatant was transferred to a fresh 1.5 ml micro centrifuge tube and 30 μL of Proteinase K was added to the supernatant and then vortexed for 30 seconds, then incubated at 56 °C for 20 minutes. The mixtures were then transferred to the well of Maxwell 16 MDx cartridges, and 65 μL of elution buffer (Promega, catalogue no. AS1290) was added to elution tubes. The non-template control (NTC) was placed as a quality control measure, for each new batch of lysis buffer and elution buffer. After the automated DNA purification was completed, purified DNA in elution buffer were placed on a magnetic stand to remove magnetic particles, and the supernatant carefully transferred to a new micro-centrifuge tube. To each sample, 2 μL of RNase (10 mg/ml) was added then incubated at 37 °C for 15 minutes. The DNA concentrations were measured using a Nano-Drop Lite Spectrophotometer (Thermo Fisher Scientific). DNA samples were then normalised to a concentration of 5 ng/μL and checked for their quality by PCR.
The PCR was carried out with a total volume of 25 uL comprised of 12.5 μL 2XMango Mix (Bioline), 9 μL H2O, 1.5 μL MgCl2 50 mM, 0.5 μL 10 μM primers 341F-CCTACGGGNGGCWGCAG and 805R-GACTACHVGGGTATCTAATCC and 1 μL of 5 ng/μL template. This primer pair was chosen for its coverage and reduced bias, based on an experimental evaluation of 512 primer pairs63. The thermo-cycling condition was 1 cycle of 3 minutes at 95 °C, followed by 25 cycles of 30 seconds each at 95 °C, 55 °C, 72 °C, and then 1 cycle of 5 minutes at 72 °C and hold at 4 °C. The PCR products were then analysed with 1% agarose gel electrophoresis (1% agarose + 1 × TAE buffer). Any samples with unsuccessful PCR were repeated at different concentrations (1, 5, 0.5 and 10 ng/μL) until amplification was achieved. Four DNA samples that did not amplify were cleaned using the phenyl chloroform extraction method and purified using the ethanol precipitation method. DNA samples were stored at −30 °C prior to 16S rRNA sequencing. The amplicon libraries were created from the V3/V4 hypervariable regions of the bacterial 16S rRNA gene using 341F and 805R primers before being subjected to 16S rRNA sequencing with the MiSeq platform (Illumina).
Bioinformatics
Raw 16S rRNA sequences were joined, demultiplexed and quality controlled using the Quantitative Insights Into Microbial Ecology (QIIME) version 1.9.1 pipeline64. Details of scripts are available in Supplementary Information. The chimera check and removal was conducted using USEARCH version 6.1.54465. The OTU was aligned using PyNAST66 with a 97% sequence similarity threshold against the Greengenes database version 13.867 using open reference picking method in QIIME. The OTUs with less than 0.1% of total sequence were filtered and samples with less than 1000 read counts were discarded. Two samples produced less than 1000 reads and one sample produced 1448 reads, with the remaining samples producing more than 2000 reads (median 12318 reads per sample). Based on these findings we chose to work with the data produced from 345 samples. The data were normalised by Total Sum Scaling (TSS) and then transformed using the Centred Log Ratio (TSS + CLR) for downstream analysis of composition data68 in both mixOmics69 and Calypso70. The read counts produced from the four no template controls were very low (7–41) and thereby not considered further.
Statistical analysis
To verify differences in factors that may potentially influence gut microbiota profiles among the CHaRM study subjects, t-test was used for normally distributed or Mann-Whitney test for non-normally distributed continuous variables. Comparison between categorical variables were performed using a Chi-square test or Fisher’s exact test when appropriate. These tests were carried out in Stata (version 13.1, StataCorp).
Gut microbiota analysis
To determine trends in α-diversity indices, we used Calypso’s Diversity Page for analysis of microbial diversity. Changes (i.e. increase or decrease) in median bacterial taxa abundance from baseline to the end of study at phylum, family, genus and OTU levels investigated using Wilcoxon sign rank test in Calypso’s ‘Stats Page’ for statistical comparison of sample groups. P-values were adjusted for multiple testing using false discovery rate (FDR) and FDR < 0.05 was considered as statistically significant.
We used the mixOmics R package69 to explore the effect of diet on gut microbial community. The gut microbial community, as well as the children’s dietary patterns over the 12-month period were initially visualised with PCA. We applied the sPLS method to explore relationships between microbial OTUs and food groups at corresponding dietary data collection (i.e. 0, 3, 6, 9, and 12 month of study). The sPLS ‘canonical mode’ was used to identify the most correlated bacterial OTU and diet and visualised with clustered image maps40. The sPLS-DA, an extension of sPLS, enables the selection of most discriminative variables (i.e. trial milk) to classify the samples42 was used to investigate the effect of trial milk on gut microbiota. Cross-validation (5 fold cross-validation repeated 50 times) was used to select the optimum number of parameters (i.e. the number of components and the number of variables to select on each component) based on classification performance.
As the trial synbiotic milk (GUMLi) contained a Bifidobacterium probiotic, we fitted Linear Mixed Model (LMM)71 using the R package ‘nlme’72 to analyse the effect of GUMLi and other covariates on Bifidobacterium OTUs. The following covariates were chosen based on the relevance with the gut microbial community: age, trial milk group, duration of any breastfeeding, antibiotic exposure since birth, and dietary pattern. Previously, we identified that breastfeeding was the most significant factor that altered the gut microbial community at one year of age (p < 0.05)73. Breastfeeding status indicated by duration (in weeks) fitted this modelling better, rather than the breastfeeding status (i.e. yes or no) at each time point. We used dietary pattern scores to analyse the overall effect of diet on the microbial community. Dietary pattern 1 represents a shift from baby-like to adult-like diet, whereas, dietary pattern 2 represents a shift from unhealthy to healthy diet. P-values were adjusted for multiple testing using Benjamini Hochberg FDR74, and FDR < 0.05 was considered statistically significant.
Phylogenetic analysis
Sequences related to OTUs associated with food groups over different time points were run through blastn standard nucleotide BLAST75 to identify closely related organisms. The 16S rRNA sequences of these organisms were retrieved from NCBI and used for phylogenetic reconstruction. These were aligned with Arb SILVA (https://www.arb-silva.de/) and imported into MEGA776 for phylogenetic analysis. The maximum likelihood method based on the Kimura 2-parameter model77 was used to infer the evolutionary tree, evaluated with 1000 bootstrap replications.
Supplementary information
Dietary intake influences gut microbiota development of healthy Australian children from the age of one to two years - supplementary information
Acknowledgements
M. Matsuyama was supported by the Australian government Research and Training Program scholarship. KALC is funded by National Health and Medical Research Council (NHMRC) Career Development fellowship (GNT1087415). The authors thank members of the Morrison Laboratory at the University of Queensland, Dr. Emily Hoedt and Ms Nida Murtaza for their assistance in phylogenetic analysis and Mr. Jing Jie Teh for the detection and removal of chimeric sequences. The authors thank Dr. Anne Bernard from QFAB Bioinformatics at the University of Queensland for biostatistics support provided through the Child Health Research Centre.
Author Contributions
Misa M. designed and conducted the CHaRM study and wrote the manuscript. S.P., K.A.L.C. and Misa M. analysed the data. Mark M., P.S.W.D. and R.J.H. provided supervision for Misa M., C.W. and A.L. provided the full dietary data. All authors contributed, read, and approved the final manuscript.
Data Availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Competing Interests
The CHaRM study was partially funded by Nutricia Australia Pty Ltd. The funder had no involvement in the collection, analysis and interpretation of data or the writing of the manuscript. P.S.W.D. has received honoraria for presentations and consultations from Danone, Nurticia, Nestle, Mead Johnson and Aspen Nutritionals. R.J.H. has received honoraria for presentations and consultation from Danone Nutricia, Nestle and Aspen Nutritionals. C.W. has received honoraria for presentations and consultations from Danone, Nutricia, Pfizer and Fonterra. Misa M., Mark M., K.A.L.C., S.P., A.L. declare no potential conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-48658-4.
References
- 1.Ley RE, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moeller AH, et al. Cospeciation of gut microbiota with hominids. Science. 2016;353:380–382. doi: 10.1126/science.aaf3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collado MC, Isolauri E, Laitinen K, Salminen S. Distinct composition of gut microbiota during pregnancy in overweight and normal-weight women. Am. J. Clin. Nutr. 2008;88:894–899. doi: 10.1093/ajcn/88.4.894. [DOI] [PubMed] [Google Scholar]
- 4.Vael C, Verhulst SL, Nelen V, Goossens H, Desager KN. Intestinal microflora and body mass index during the first three years of life: an observational study. Gut Pathog. 2011;3:8. doi: 10.1186/1757-4749-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalliomaki M, Collado MC, Salminen S, Isolauri E. Early differences in fecal microbiota composition in children may predict overweight. Am. J. Clin. Nutr. 2008;87:534–538. doi: 10.1093/ajcn/87.3.534. [DOI] [PubMed] [Google Scholar]
- 6.Luoto R, et al. Initial Dietary and Microbiological Environments Deviate in Normal-weight Compared to Overweight Children at 10 Years of Age. J Pediatr Gastroenterol Nutr. 2011;52:90–95. doi: 10.1097/MPG.1090b1013e3181f3457f. [DOI] [PubMed] [Google Scholar]
- 7.Abrahamsson TR, et al. Low gut microbiota diversity in early infancy precedes asthma at school age. Clin. Exp. Allergy. 2014;44:842–850. doi: 10.1111/cea.12253. [DOI] [PubMed] [Google Scholar]
- 8.Dzidic M, et al. Aberrant IgA responses to the gut microbiota during infancy precede asthma and allergy development. J. Allergy Clin. Immunol. 2017;139:1017–1025 e1014. doi: 10.1016/j.jaci.2016.06.047. [DOI] [PubMed] [Google Scholar]
- 9.Arslanoglu S, et al. Early dietary intervention with a mixture of prebiotic oligosaccharides reduces the incidence of allergic manifestations and infections during the first two years of life. J. Nutr. 2008;138:1091–1095. doi: 10.1093/jn/138.6.1091. [DOI] [PubMed] [Google Scholar]
- 10.Arslanoglu S, et al. Early neutral prebiotic oligosaccharide supplementation reduces the incidence of some allergic manifestations in the first 5 years of life. J Biol Regul Homeost Agents. 2012;26:49–59. [PubMed] [Google Scholar]
- 11.Penders J, Stobberingh EE, Brandt PAVD, Thijs C. The role of the intestinal microbiota in the development of atopic disorders. Allergy. 2007;62:1223–1236. doi: 10.1111/j.1398-9995.2007.01462.x. [DOI] [PubMed] [Google Scholar]
- 12.Adams JB, Johansen LJ, Powell LD, Quig D, Rubin RA. Gastrointestinal flora and gastrointestinal status in children with autism–comparisons to typical children and correlation with autism severity. BMC Gastroenterol. 2011;11:22. doi: 10.1186/1471-230X-11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finegold SM, et al. Pyrosequencing study of fecal microflora of autistic and control children. Anaerobe. 2010;16:444–453. doi: 10.1016/j.anaerobe.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 14.Agans R, et al. Distal gut microbiota of adolescent children is different from that of adults. FEMS Microbiol. Ecol. 2011;77:404–412. doi: 10.1111/j.1574-6941.2011.01120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nylund L, et al. Microarray analysis reveals marked intestinal microbiota aberrancy in infants having eczema compared to healthy children in at-risk for atopic disease. BMC Microbiol. 2013;13:12. doi: 10.1186/1471-2180-13-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flint HJ, Scott KP, Louis P, Duncan SH. The role of the gut microbiota in nutrition and health. Nat Rev Gastroenterol Hepatol. 2012;9:577–589. doi: 10.1038/nrgastro.2012.156. [DOI] [PubMed] [Google Scholar]
- 17.David LA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scott KP, Gratz SW, Sheridan PO, Flint HJ, Duncan SH. The influence of diet on the gut microbiota. Pharmacol. Res. 2013;69:52–60. doi: 10.1016/j.phrs.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 19.Shanahan Fergus, van Sinderen Douwe, O’Toole Paul W, Stanton Catherine. Feeding the microbiota: transducer of nutrient signals for the host. Gut. 2017;66(9):1709–1717. doi: 10.1136/gutjnl-2017-313872. [DOI] [PubMed] [Google Scholar]
- 20.Heiman ML, Greenway FL. A healthy gastrointestinal microbiome is dependent on dietary diversity. Mol Metab. 2016;5:317–320. doi: 10.1016/j.molmet.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Renz H, et al. An exposome perspective: Early-life events and immune development in a changing world. J. Allergy Clin. Immunol. 2017;140:24–40. doi: 10.1016/j.jaci.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 23.Segata N. Gut Microbiome: Westernization and the Disappearance of Intestinal Diversity. Curr. Biol. 2015;25:R611–613. doi: 10.1016/j.cub.2015.05.040. [DOI] [PubMed] [Google Scholar]
- 24.Azad MB, et al. Gut microbiota of healthy Canadian infants: profiles by mode of delivery and infant diet at 4 months. Can. Med. Assoc. J. 2013;185:385–394. doi: 10.1503/cmaj.121189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bäckhed F, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015;17:690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 26.Bokulich NA, et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med. 2016;8:343ra382. doi: 10.1126/scitranslmed.aad7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan W, et al. Diversity of the intestinal microbiota in different patterns of feeding infants by Illumina high-throughput sequencing. World J. Microbiol. Biotechnol. 2013;29:2365–2372. doi: 10.1007/s11274-013-1404-3. [DOI] [PubMed] [Google Scholar]
- 28.Madan JC, et al. Association of Cesarean Delivery and Formula Supplementation With the Intestinal Microbiome of 6-Week-Old Infants. Jama Pediatr. 2016;170:212–219. doi: 10.1001/jamapediatrics.2015.3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roger LC, McCartney AL. Longitudinal investigation of the faecal microbiota of healthy full-term infants using fluorescence in situ hybridization and denaturing gradient gel electrophoresis. Microbiology. 2010;156:3317–3328. doi: 10.1099/mic.0.041913-0. [DOI] [PubMed] [Google Scholar]
- 30.Fallani M, et al. Determinants of the human infant intestinal microbiota after the introduction of first complementary foods in infant samples from five European centres. Microbiology. 2011;157:1385–1392. doi: 10.1099/mic.0.042143-0. [DOI] [PubMed] [Google Scholar]
- 31.Koenig JE, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. USA. 2011;108(Suppl 1):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stewart CJ, et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature. 2018;562:583–588. doi: 10.1038/s41586-018-0617-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yassour M, et al. Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci Transl Med. 2016;8:343ra381. doi: 10.1126/scitranslmed.aad0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laursen MF, et al. Infant gut microbiota development is driven by transition to family foods independent of maternal obesity. mSphere. 2016;1:e00069–00015. doi: 10.1128/mSphere.00069-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baird J, et al. Being big or growing fast: systematic review of size and growth in infancy and later obesity. Brit Med J. 2005;331:929–931. doi: 10.1136/bmj.38586.411273.E0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siega-Riz AM, et al. Food consumption patterns of infants and toddlers: where are we now? J. Am. Diet. Assoc. 2010;110:S38–S51. doi: 10.1016/j.jada.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 37.Laursen MF, Bahl MI, Michaelsen KF, Licht TR. First Foods and Gut Microbes. Front Microbiol. 2017;8:356. doi: 10.3389/fmicb.2017.00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mills VC, et al. Relative validity and reproducibility of a food frequency questionnaire for identifying the dietary patterns of toddlers in New Zealand. J Acad Nutr Diet. 2015;115:551–558. doi: 10.1016/j.jand.2014.09.016. [DOI] [PubMed] [Google Scholar]
- 39.Floyd FJ, Widaman KF. Factor analysis in the development and refinement of clinical assessment instruments. Psychol Assessment. 1995;7:286–299. doi: 10.1037/1040-3590.7.3.286. [DOI] [Google Scholar]
- 40.González I, Lê Cao K-A, Davis M, Déjean S. Insightful graphical outputs to explore relationships between two ‘omics’ data sets. BioData Mining. 2013;5:19. doi: 10.1186/1756-0381-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lê Cao, K.-A., Rossouw, D., Robert-Granié, C. & Besse, P. A sparse PLS for variable selection when integrating omics data. Stat Appl Genet Mol Biol7 (2008). [DOI] [PubMed]
- 42.Lê Cao KA, Boitard S, Besse P. Sparse PLS discriminant analysis: biologically relevant feature selection and graphical displays for multiclass problems. BMC Bioinformatics. 2011;12:253. doi: 10.1186/1471-2105-12-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lugli GA, et al. Investigation of the evolutionary development of the genus Bifidobacterium by comparative genomics. Appl. Environ. Microbiol. 2014;80:6383–6394. doi: 10.1128/AEM.02004-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bell LK, Golley RK, Daniels L, Magarey AM. Dietary patterns of Australian children aged 14 and 24 months, and associations with socio-demographic factors and adiposity. Eur J Clin Nutr. 2013;67:638–645. doi: 10.1038/ejcn.2013.23. [DOI] [PubMed] [Google Scholar]
- 45.Pryer JA, Rogers S. Dietary patterns among a national sample of British children aged 1 1/2-4 1/2 years. Public Health Nutr. 2009;12:957–966. doi: 10.1017/S1368980008003364. [DOI] [PubMed] [Google Scholar]
- 46.Subramanian S, et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature. 2014;510:417–421. doi: 10.1038/nature13421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blanton LV, et al. Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science. 2016;351:aad3311. doi: 10.1126/science.aad3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Benevides L, et al. New Insights into the Diversity of the Genus Faecalibacterium. Front Microbiol. 2017;8:1790. doi: 10.3389/fmicb.2017.01790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sommer F, Anderson JM, Bharti R, Raes J, Rosenstiel P. The resilience of the intestinal microbiota influences health and disease. Nat. Rev. Microbiol. 2017;15:630–638. doi: 10.1038/nrmicro.2017.58. [DOI] [PubMed] [Google Scholar]
- 50.Miquel S, et al. Faecalibacterium prausnitzii and human intestinal health. Curr. Opin. Microbiol. 2013;16:255–261. doi: 10.1016/j.mib.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 51.Takada T, Kurakawa T, Tsuji H, Nomoto K. Fusicatenibacter saccharivorans gen. nov., sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 2013;63:3691–3696. doi: 10.1099/ijs.0.045823-0. [DOI] [PubMed] [Google Scholar]
- 52.Carrigan A, et al. Contribution of food additives to sodium and phosphorus content of diets rich in processed foods. J Ren Nutr. 2014;24(13–19):19e11. doi: 10.1053/j.jrn.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suez J, et al. Artificial sweeteners induce glucose intolerance by altering the gut microbiota. Nature. 2014;514:181–186. doi: 10.1038/nature13793. [DOI] [PubMed] [Google Scholar]
- 54.Roberts CL, et al. Translocation of Crohn’s disease Escherichia coli across M-cells: contrasting effects of soluble plant fibres and emulsifiers. Gut. 2010;59:1331–1339. doi: 10.1136/gut.2009.195370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chassaing B, et al. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature. 2015;519:92–96. doi: 10.1038/nature14232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Song Y, Liu C, Finegold SM. Real-time PCR quantitation of clostridia in feces of autistic children. Appl. Environ. Microbiol. 2004;70:6459–6465. doi: 10.1128/AEM.70.11.6459-6465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Korpela K, et al. Gut microbiota signatures predict host and microbiota responses to dietary interventions in obese individuals. PLoS One. 2014;9:e90702. doi: 10.1371/journal.pone.0090702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lau, A. S., Xiao, J. Z. & Liong, M. T. In Beneficial Microorganisms in Medical and Health Applications 39–72 (Springer, 2015).
- 59.Matsuki T, Watanabe K, Tanaka R, Fukuda M, Oyaizu H. Distribution of bifidobacterial species in human intestinal microflora examined with 16S rRNA-gene-targeted species-specific primers. Appl. Environ. Microbiol. 1999;65:4506–4512. doi: 10.1128/aem.65.10.4506-4512.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shim JS, Oh K, Kim HC. Dietary assessment methods in epidemiologic studies. Epidemiol Health. 2014;36:e2014009. doi: 10.4178/epih/e2014009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Patel MX, Doku V, Tennakoon L. Challenges in recruitment of research participants. Adv Psychiatr Treat. 2003;9:229–238. doi: 10.1192/apt.9.3.229. [DOI] [Google Scholar]
- 62.Yu Z, Morrison M. Improved extraction of PCR-quality community DNA from digesta and fecal samples. BioTechniques. 2004;36:808–812. doi: 10.2144/04365ST04. [DOI] [PubMed] [Google Scholar]
- 63.Klindworth A, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Edgar RC, Flyvbjerg H. Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics. 2015;31:3476–3482. doi: 10.1093/bioinformatics/btv401. [DOI] [PubMed] [Google Scholar]
- 66.Caporaso JG, et al. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.DeSantis TZ, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lê Cao K-A, et al. MixMC: a multivariate statistical framework to gain insight into microbial communities. PloS one. 2016;11:e0160169. doi: 10.1371/journal.pone.0160169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rohart F, Gautier B, Singh A, Le Cao K. A. mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput Biol. 2017;13:e1005752. doi: 10.1371/journal.pcbi.1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zakrzewski M, et al. Calypso: a user-friendly web-server for mining and visualizing microbiome-environment interactions. Bioinformatics. 2017;33:782–783. doi: 10.1093/bioinformatics/btw725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Laird NM, Ware JH. Random-effects models for longitudinal data. Biometrics. 1982;38:963–974. doi: 10.2307/2529876. [DOI] [PubMed] [Google Scholar]
- 72.Pinheiro, J. et al. nlme: Linear and Nonlinear Mixed Effects Models. R package version v. 3.1–131, https://CRAN.R-project.org/package=nlme (2017).
- 73.Matsuyama, M. et al. Breastfeeding: a key modulator of gut microbiota characteristics in late infancy. Journal of developmental origins of health and disease, 1–8 (2018). [DOI] [PubMed]
- 74.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- 75.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 76.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kimura M. A Simple Method for Estimating Evolutionary Rates of Base Substitutions through Comparative Studies of Nucleotide-Sequences. J. Mol. Evol. 1980;16:111–120. doi: 10.1007/Bf01731581. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dietary intake influences gut microbiota development of healthy Australian children from the age of one to two years - supplementary information
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.