Abstract
The incidence of hepatocellular carcinoma (HCC) and the mortality resulting from HCC are both increasing. Most patients with HCC are diagnosed at advanced stages when curative treatments are impossible. Current drug therapy extends mean overall survival by only a short period of time. Genetic mutations associated with HCC vary widely. Therefore, transgenic and mutant animal models are needed to investigate the molecular effects of specific mutations, classify them as drivers or passengers, and develop targeted treatments. Cirrhosis, however, is the premalignant state common to 90% of HCC patients. Currently, no specific therapies are available to halt or reverse the progression of cirrhosis to HCC. Understanding the genetic drivers of HCC as well as the biochemical, mechanical, hormonal, and metabolic changes associated with cirrhosis could lead to novel treatments and cancer prevention strategies. Although additional therapies recently received Food and Drug Administration approval, significant clinical breakthroughs have not emerged since the introduction of the multikinase inhibitor sorafenib, necessitating alternate research strategies. Zebrafish (Danio rerio) are effective for disease modeling because of their high degree of gene and organ architecture conservation with human beings, ease of transgenesis and mutagenesis, high fecundity, and low housing cost. Here, we review zebrafish models of HCC and identify areas on which to focus future research efforts to maximize the advantages of the zebrafish model system.
Keywords: Cirrhosis, Tumor Microenvironment, Model Organisms, Inflammation, Drug-Induced Liver Injury, Autophagy
Abbreviations used in this paper: APAP, acetaminophen; DILI, drug-induced liver injury; DMBA, 7,12-dimethylbenz[a]anthracene; E2, 17β-estradiol; fabp10a, fatty acid binding protein 10a; GFP, green fluorescent protein; HBV, hepatitis B virus; HBx, hepatitis B x antigen; HCC, hepatocellular carcinoma; HCP, hepatitis C virus core protein; HCV, hepatitis C virus; HSC, hepatic stellate cell; ICC, intrahepatic cholangiocarcinoma; Lc3, Light Chain 3 Beta; lf:Yap, Tg[-2.8fabp10a:yap1-1βS87A; LSEC, liver sinusoidal endothelial cell; NAFLD, nonalcoholic fatty liver disease; ROS, reactive oxygen species; TAM, tumor-associated macrophage; TAN, tumor-associated neutrophil; TERT, telomerase reverse transcriptase; TGF, transforming growth factor; TME, tumor microenvironment; TP53, tumor protein 53; Wnt, Wingless; WT, wild-type; Yap, Yes-associated protein
Summary.
We review zebrafish models of hepatocellular carcinoma and highlight how the contributions using this model organism are unique. We focus on efforts to model the wide spectrum of genetic mutations found in hepatocellular carcinoma, the biochemical and hormonal changes associated with cirrhosis, the effects of the cancer microenvironment, and the role of metabolic processes such as glutamine and glucose metabolism, autophagy, and oxidative stress.
Hepatocellular carcinoma (HCC) is the fifth most commonly diagnosed cancer worldwide, however, HCC is the second leading cause of cancer death, causing almost 750,000 deaths in 2012.1 Mortality resulting from HCC approximates incidence, and incidence is increasing in most countries with an expected peak around 2021 in the United States.2 Because of advances in public health, chemotherapy treatment, and immunology, the mortality rates for many other types of cancer are decreasing in the United States. In stark contrast, however, liver cancer had the greatest increase in mortality between 1990 and 2009 among all cancers.3 When detected early, HCC can be treated curatively with surgical resection, transplantation, or occasionally with local ablative techniques.4 Transplantation is limited, however, because of the number of available donor organs, and the majority of HCC cases are detected after the point during which surgical or local ablative techniques are effective.5, 6
When surgery and local ablative therapies are not viable options, HCC patients are treated with sorafenib, a multikinase inhibitor with anti-angiogenesis properties. Treatment with sorafenib extends overall median survival by 3 months.7 Since its introduction more than a decade ago, researchers have sought to build on the success of the Sorafenib Hepatocellular Carcinoma Assessment and Randomization Protocol trial, but these efforts to iteratively improve upon this efficacy with new chemotherapeutics have largely been disappointing.8 Thus, some clinicians involved in the original Sorafenib Hepatocellular Carcinoma Assessment and Randomization Protocol trial suggested, “Continuing the drug development process in the same manner as we have in the past is unlikely to yield significant improvements.”9 Nevertheless, additional vascular endothelial growth factor receptor–targeting agents (lenvatinib, ramucirumab, cabozantinib, and regorafenib) recently were approved for treatment of HCC. As predicted, however, these agents only modestly improve mean overall survival.10, 11 Similarly, recently approved immuno-oncology therapies (nivolumab, pembrolizumab) enhance the treatment options for these patients, but have not yet led to a fundamental breakthrough.10, 11
Effective treatments for breast cancer and other solid tumors have been developed by targeting specific molecular abnormalities linked to diagnostic biomarkers. Recent technological innovations in DNA and RNA sequencing have spurred greater understanding of the genetic mutations and transcriptional abnormalities found in various cancers, including HCC. These efforts have shown many of the prevalent driver mutations present in HCC, including those in the telomerase reverse transcriptase (TERT) promoter, tumor protein 53 (TP53), and β-catenin (CTNNB1),12 however, the overall heterogeneity of HCC tumors is high compared with other solid tumors.13 Because of this variability, efforts to generate and study animal models of individual mutations and combinations of mutations are needed to develop targeted interventions.
The putative driver mutations for HCC are heterogeneous, and the initial liver injuries that precipitate mutagenesis vary from viral infection, to alcohol- or drug-induced liver injury, and to obesity and metabolic syndrome. Regardless of etiology, however, liver disease typically progresses through similar stages of inflammation and fibrosis to cirrhosis—the pathologic condition characterized by liver scarring formed by excessive collagen deposition and remodeling.14 More than 90% of HCC cases arise in patients with underlying cirrhosis,15 classifying cirrhosis as a premalignant condition and one of the greatest risk factors for developing cancer of any type. Currently, there are no available treatments to stop the progression of cirrhosis to HCC. Instead, clinicians rely on surveillance programs involving imaging and biomarkers (eg, α-fetoprotein) to screen patients’ cirrhotic livers periodically for newly developed tumors.16 Thus, a better understanding of the molecular and cellular links between fibrosis and HCC could lead to effective preventive treatments.
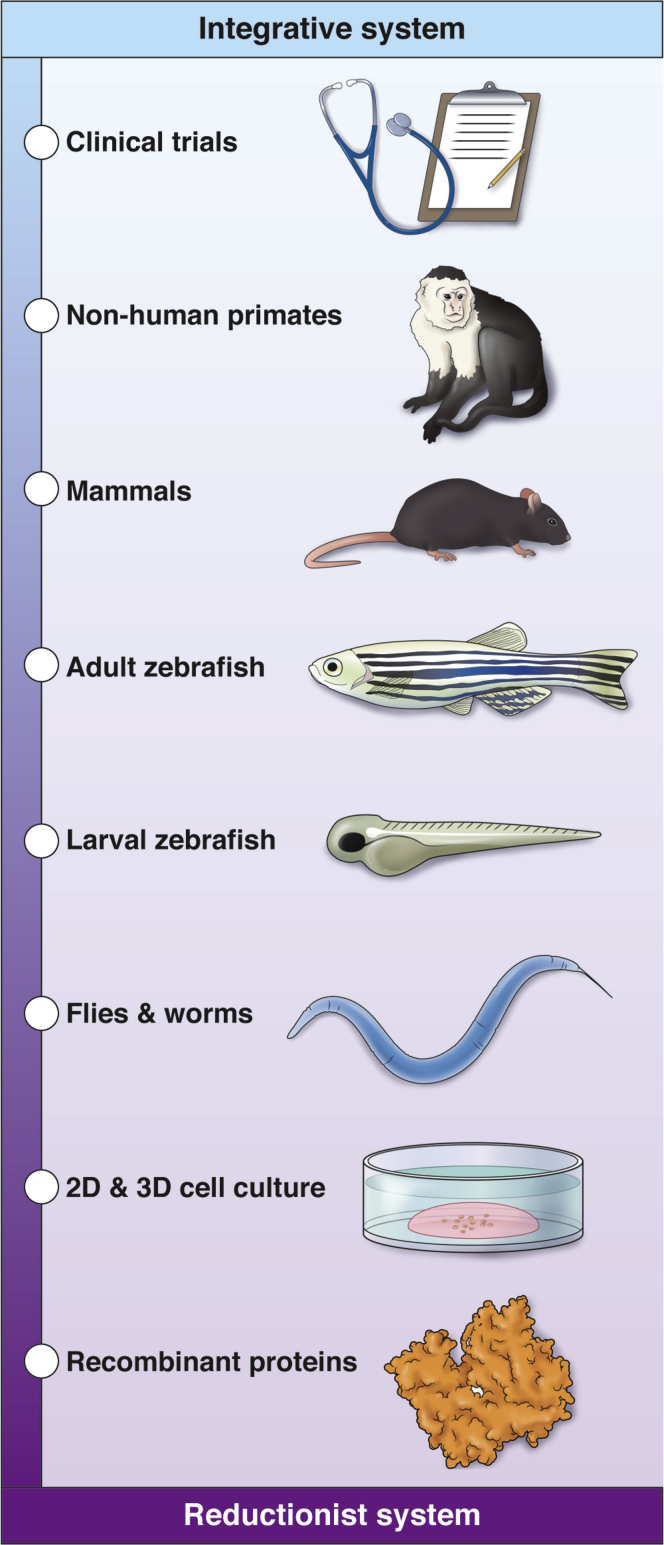
Biomedical research exists in a virtuous cycle of iterative gains in knowledge of disease from bedside to benchside back to bedside.3 These iterations, however, have been slow to yield transformative innovations for HCC treatment. This lack of progress suggests novelty and creativity must be applied at all points along the cycle. To wit, zebrafish cancer models are rapidly gaining in popularity and yielding clinically translatable insights.17, 18, 19 Zebrafish are vertebrates and have high conservation of both genes and organs with human beings,20 including the liver.21 Study of liver development in both fish and mammals can show new insight into HCC development. Many mechanisms of liver development and regeneration are shared in hepatic oncogenesis, and this important concept is reviewed elsewhere.21, 22, 23, 24 Zebrafish develop cancer when exposed to carcinogens or mutagens and in response to transgenic overexpression of oncogenes.18 Zebrafish model systems have several advantages compared with other animal models. Their high fecundity and low housing costs allow for forward genetic screening as well as large experimental sample sizes. They are transparent and develop externally, allowing for in vivo imaging and analysis of normal organogenesis mechanisms. New advances in sequencing and genome editing are further increasing the utility of zebrafish cancer models.25, 26, 27, 28 Small-molecule drugs can be added directly to the zebrafish water.29 If a larval-stage phenotype is found, then whole-organism, high-throughput chemical screening can be performed to identify the signaling pathways involved and assess potential molecular interventions30 (Figure 1).
Figure 1.
Zebrafish are a model system at the center of the reductionist to integrative experimental axis. Because they are amenable to live imaging and chemical and random mutagenesis screening studies, zebrafish larvae are an effective model system to study liver development and surrogate liver disease phenotypes. Because zebrafish develop liver cancer and are inexpensive to house, longitudinal studies using large cohorts of adult zebrafish can investigate the drivers of HCC. Thus, researchers using zebrafish have the advantages of both reductionist and integrative model systems. The protein crystal structure is TERT158 (PDB: 3DU5). 2D, 2-dimensional; 3D, 3 dimensional.
This review focuses on the current state of zebrafish HCC models, discussing their unique contributions and highlighting areas in which researchers should focus future efforts. We highlight efforts to use transgenic and mutant zebrafish to model the spectrum of genetic mutations and epigenetic changes found in human HCC. We describe the utility of investigating the molecular mechanisms of normal liver organogenesis to understand cancer mechanisms. We show how chemical screening experiments on zebrafish larvae yield insight into the biochemical, hormonal, and metabolic changes associated with cirrhosis. We review zebrafish studies that focus on the microenvironmental changes such as fibrosis, inflammation, and tumor-associated cell types. Finally, we assess zebrafish strategies to model cellular processes disrupted during liver disease progression including oxidative stress, drug-induced liver injury, and autophagy. Throughout, we highlight areas in which future zebrafish research would be impactful.
Modeling Genetic Mutations Found in HCC
HCC arises from the accumulation of multiple genetic mutations resulting in changes to the genomic landscape13 (Figure 2). Recent studies using whole-genome and exome sequencing enabled identification and analysis of the genetic alterations contained in HCC patient tumors.31 Further efforts to analyze these data sets will uncover recurrent and novel HCC driver mutations. Personalized medicine is a term used to describe clinical approaches that match a molecular treatment to individual differences in genetic make-up. The canonical example of a successful personalized medicine strategy is treating human epidermal growth factor receptor 2 (HER2)-positive breast cancer patients with trastuzumab, which specifically targets HER2.32 In HCC, the mean number of mutations per tumor is between 35 and 80, and it is estimated that 4–8 of these mutations are truly oncogenic drivers.33 Identifying potential driver mutations is an important first step toward personalized medicine for HCC, but to truly be effective, a treatment must address a bona fide dominant oncogenic addiction loop, be tied to an extant and reliable biomarker to identify suitable patients, and use an efficacious and safe therapeutic agent.34 Animal models are needed for preclinical testing of each of these conditions.
Figure 2.
Genetic mutational landscape of HCC. Font size and gene name frequency correlate to the frequency of mutations found in tumors from patients with HCC based on data from Lee31 and Zucman-Rossi et al.34 Green-colored genes have well-established zebrafish HCC models. Orange-colored genes have zebrafish HCC models of closely related genes or pathways, and red-colored genes have no published zebrafish HCC model.
Between 47% and 60% of HCC cases present with activating mutations in the TERT promoter, which lead to hyperactivation of TERT and genomic instability.31, 35 Typically, TERT expression is repressed in somatic cells including hepatocytes, which restricts the total number of cell divisions. In contrast, TERT expression often is high in self-renewing cells such as stem cells, and a recent study identified a population of hepatocytes that express high levels of TERT and can repopulate the liver during both homeostasis and after injury.36 Cancer cells hijack this stem cell mechanism to avoid the Hayflick limit and proliferate unrestrained. Inactivating mutations in TP53 are the second most common mutations found in HCC patients, occurring in more than 30% of cases.34 Known as the guardian of the genome, TP53 is a potent tumor suppressor that acts by initiating cell-cycle arrest or apoptosis in response to various cellular stressors including DNA damage and is thought to be undruggable.8 Frequent mutations leading to activation of the Wingless (Wnt) pathway, especially β-catenin (CTNNB1) and axin,37 and Ras pathways38 also are observed in HCC. Epigenetic changes occur as well, including global DNA hypomethylation and mutations in epigenetic regulators, such as the chromatin remodelers ARID1A, ARID2A, and KMT family genes.9 Aggressive cases of HCC often contain mutations in UHRF1, an essential regulator of DNA methylation.39
Zebrafish develop cancer, including HCC, spontaneously, after exposure to carcinogens, or as a result of genetic mutagenesis.18, 40 Zebrafish livers are similar in structure to mammalian livers, consisting of polarized hepatocytes supported by biliary epithelial cells, liver sinusoidal endothelial cells (LSECs), hepatic stellate cells (HSCs), and various immune cells, and they develop HCC, which is histologically similar to mammalian systems.19 Transgenic and mutant zebrafish have been developed to model aspects of Wnt and Ras signaling and the epigenetic underpinnings of HCC. Zebrafish models of HCC begin with mutation or transgenic overexpression of a known or suspected oncogene. Transgenesis often is accomplished by using the Tol2 transposon system derived from Medaka fish.41, 42 Several kits have been developed to enable rapid Tol2-plasmid construction using Gateway cloning methods to combine tissue-specific promoters, various transgenes, and fluorescent reporters in a modular way.43, 44 Many of the zebrafish models of HCC use the fatty acid binding protein 10a (fabp10a) promoter to drive hepatocyte-specific transgene expression.45 Furthermore, because of their large clutch size, zebrafish have long been used for forward genetic screening using random mutagenesis to induce and identify mutations and are responsible for important developmental phenotypes.46, 47, 48, 49 These approaches and, more recently, the Sanger Center’s Zebrafish Mutation Project generated mutant alleles, which cover much of the zebrafish genome.50 Furthermore, approaches utilizing clustered regularly interspaced short palindromic repeats - CRISPR-associated protein 9 (CRISPR-Cas9) are becoming the dominant method to create additional mutations.51 High-efficiency CRISPR-mediated mutagenesis protocols enable phenotypic analysis in the F0 generation,26 and tissue-specific knockouts are possible using loxP insertions and Cre recombinase-mediated excision.52, 53, 54 This plethora of genetic tools allows flexibility and creativity in generating zebrafish cancer models.
In some cases, disrupting a single signal is sufficient to induce and model HCC, however, strategies to potentiate HCC formation often are used in parallel. The 2 most fruitfully used zebrafish cancer potentiation assays are to cross the novel transgenic or mutant line into the tp53 null background55 or to compare tumorigenesis rates in the fish of interest with wild-type siblings after treatment with a chemical carcinogen, such as 7,12-dimethylbenz[a]anthracene (DMBA). DMBA is metabolized primarily in the liver and induces a baseline level of liver tumorigenesis.56 In both cases, comparing cohorts of transgenic or mutant fish with cohorts of their wild-type siblings can yield insight into tumor initiation rate as well as HCC progression. These tumor-induction strategies have distinct benefits: the tp53-/- strategy allows for consistent tumor generation with a single tumor-suppressor mutation present in both control and experimental cohorts, whereas the DMBA carcinogen model induces a variety of mutations, which might better model the broad spectrum of mutations found in human disease.
Cell-Cycle Pathway and Epigenome Modifiers
Zebrafish mutants for tp53 develop malignant peripheral nerve sheath tumors at a moderate rate of approximately 28% by 16.5 months.55 Although these mutants do not develop HCC spontaneously, tp53 mutant zebrafish develop liver tumors if a secondary insult is present. Thus, one effective method to determine if a low-penetrance allele is truly a driver mutation is to compare liver tumor rates between fish bearing the novel allele and the tp53 inactivating mutation with fish bearing the tp53 mutation alone. For example, ubiquitin-like with plant homeodomain and Really Interesting New Gene finger domains 1 (UHRF1) is expressed in many cancers and functions to regulate DNA methylation. Although mutations in UHRF1 sometimes are found in human HCC, it was not known whether these were driver or passenger mutations. To address this question, Mudbhary et al39 overexpressed human UHRF1 in zebrafish and observed DNA hypomethylation and decreased liver size during development. When combined with tp53 haploinsufficiency, UHRF1 overexpression induced liver cancer at an accelerated rate, and confirmed UHRF1 as a bona fide oncogene.39
Wnt Signaling
A large subgroup of HCC cases is classified by activation of the Wnt signaling pathway: approximately 19% of human HCC cases carry activating mutations in CTNNB1, and 10% of cases carry inactivating mutations in AXIN1.37 Developmental studies in zebrafish have shown critical roles for Wnt signaling during normal liver organogenesis.57 Inactivating mutation of zebrafish adenomatous polyposis coli (apc) activated Wnt/β-catenin signaling and induced spontaneous formation of intestinal, pancreatic, and liver tumors.58 The extent of tumorigenesis was increased when combined with the chemical carcinogen DMBA. A complementary transgenic model of Wnt-activated HCC uses a hepatocyte-specific promoter to drive an activated form of β-catenin.59 These fish showed an enlarged liver phenotype during larval stages and developed spontaneous liver tumors by 3 months post fertilization (mpf). The Stainier group used this as a surrogate phenotype to screen for chemicals that might prevent Wnt-driven HCC, discovering c-Jun N-terminal kinase (JNK) inhibitors and serotonin reuptake inhibitors rescued the large liver phenotype.59 Thus, the molecular and cellular mechanisms by which the Wnt/β-catenin signaling pathway operates during normal development and HCC progression were illuminated and potential treatment avenues identified.
Ras Signaling and Cross-Talk
Up to 50% of HCC tumors present with some form of activation of the Ras signaling pathway,37 and approximately 7% of HCC cases have mutations in KRAS.60 Nguyen et al61 developed several transgenic zebrafish lines expressing activated forms of Ras in hepatocytes. Transgenic zebrafish expressing zebrafish krasV12 developed liver tumors by 65–90 dpf.61 The rate of tumor formation was accelerated when bred into the tp53 mutant background but with no increase in overall tumor formation. To tune the levels of Ras activation, an inducible, hepatocyte-specific krasV12 transgenic fish line also was generated.62 A chemical screen in this line showed that simultaneous treatment with inhibitors targeting the Raf-MEK-ERK and the PI3K-AKT-mTOR pathways reduced liver size at 7 dpf.62
Subsequent iterations of the transgene overexpression strategy generated models of HCC by expressing murine Myc,63 or a fish-specific oncogene xmrk, a hyperactive mutant epidermal growth factor receptor homolog.64, 65, 66, 67 Crossing these lines facilitated analysis of cooperation between the signaling pathways. Zebrafish studies have shown that hepatocyte-specific expression of a dominant-negative form of RhoA caused increased mortality in the krasV12 HCC model.68 Similarly, cross-talk between the epidermal growth factor and kras pathways was explored using xmrk- and krasV12-expressing zebrafish.67 Expanding these models by taking advantage of larval-stage phenotypes correlating to HCC development (ie, enlarged livers by 5–7 dpf), chemical inhibition studies were performed to examine how differentially driven tumors respond to treatments. Small-molecule inhibition of vascular endothelial growth factor/fibroblast growth factor pathway reversed the krasV12- and Myc-induced liver enlargement phenotype, whereas Wnt pathway suppression rescued the krasV12 phenotype but not the Myc phenotype.69 Expansion of this strategy could enable development of molecular targeted therapies based on which signaling pathways are activated in patient tumors.
Hepatitis B and C
Hepatitis B virus (HBV) and hepatitis C virus (HCV) can contribute to HCC by directly integrating into and disrupting cancer-related genes such as TERT. Chronic HBV or HCV infection also causes inflammation and oxidative stress, which can lead to fibrosis and cirrhosis (discussed later). Furthermore, expression of viral core proteins contributes to HCC. Cell culture and mouse model experiments have shown that expression of hepatitis B x antigen (HBx) and viral envelope proteins dysregulated cellular transcription and proliferation (reviewed by Levrero and Zucman-Rossi70). Likewise, cell culture and mouse models indicated that expression of HCV core protein and the nonstructural proteins NS3 and NS5A contributed similarly to oncogenic transformation (reviewed by Arzumanyan et al71). Transgenic expression of HBx in the zebrafish liver induced steatosis, increased expression of lipogenic genes, and induced liver hyperplasia.72 A second study crossed hepatocyte-specific HBx transgenic fish into the tp53 mutant background and found HCC present in 44% of fish by 11 mph.73 In previous work, the Yuh group found HBx transgene up-regulated the src tyrosine kinase signaling pathway in mice,74 and they verified transgenic fish overexpressing src also developed HCC at an increased rate in the tp53 mutant background.73 Future zebrafish studies should focus on overexpression of HBV preS/S envelop proteins or model integration of HBV into specific genomic loci such as the tert promoter.
Transgenic zebrafish expressing HCV core protein (HCP) developed HCC at twice the rate of wild-type controls when exposed to the carcinogen thioacetamide.75 A second set of studies developed zebrafish models to investigate how HCP transcription and translation are regulated in vivo.76, 77 Such models could be used for chemical screening to identify drugs capable of suppressing HCV replication. Interestingly, transgenic expression of both HBx and HCP in hepatocytes—a model of HBV and HCV co-infection—caused fish to develop severe liver fibrosis and intrahepatic cholangiocarcinoma (ICC).78 HBx and HCP co-expression activated the transforming growth factor (TGF)-β–dependent JNK/pSmad3L pathway, and tgfb1 knockdown mitigated fibrosis and ICC development.78 In sum, zebrafish models are useful for investigating specific mechanisms by which HBV and HCV induce liver cancer.
In these ways, zebrafish models offer advantages to study and validate oncogenic driver mutations found in HCC. The tp53 mutant background allows for discovering the effects of other mutations for driver potential.55 The apc mutants57, 58 and β-catenin transgenic line59 enable modeling Wnt-driven HCC. Epigenetic regulators frequently mutated in HCC are delineated by dmnt mutant fish79 and the transgenic line overexpressing UHRF1.39 Ras, Myc, and epidermal growth factor signaling, pathways that frequently are up-regulated in diverse mutational landscapes, are modeled by transgenic lines expressing activated forms.61, 80 Transgenic overexpression of HBV and HCV proteins facilitate modeling virus-induced HCC and ICC.73, 78 Cumulatively, these tools allow for reductionist approaches to probe molecular mechanisms driving different types of HCC tumors and offer opportunities to discover druggable weaknesses and develop targeted therapies.
One aspect of HCC that is not well understood in any model organism is TERT activity. This is despite the fact that TERT-activating mutations are the most commonly observed. Zebrafish with liver-specific TERT overexpression would be a clinically relevant background upon which to study how other driver mutations function—similar to the tp53 mutation synergy strategy. To complicate matters, TERT has context-dependent roles in cirrhosis and HCC; although TERT activation is a hallmark of HCC, TERT inactivation is linked to cirrhosis, and reactivation of TERT can reduce cirrhosis in mouse models.81 Furthermore, the identification of a population of TERThigh hepatocytes, which repopulate the liver both during homeostasis and after injury, raises questions as to whether this population of cells is present in humans and disrupted in liver disease.36 Clearly, the continued generation of novel HCC models is imperative to develop treatment and prevention strategies for HCC. Many of the druggable target gene mutations (ie, protein kinases and other enzymes) found in other cancer types, however, are mutated infrequently in HCC.31 This argues both for increased investment in developing new classes of drugs to target difficult mutant proteins and for investment into investigating changes associated with cirrhosis to develop novel HCC prevention strategies.
Modeling Microenvironmental, Biochemical, and Inflammatory Changes Associated With Cirrhosis
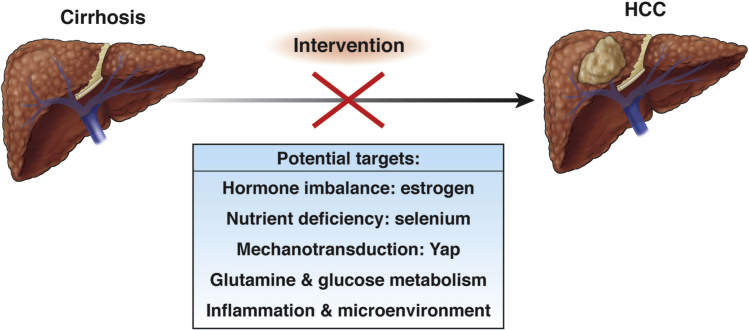
More than 90% of HCC tumors arise in the background of a cirrhotic liver. Thus, cirrhosis is a highly relevant indicator of cancer risk. Despite this clear risk, the molecular links between cirrhosis and tumor formation are largely unknown. In contrast, much is known about the biochemical and physical changes in the cirrhotic liver, including increased estrogen levels,82, 83 decreased selenium levels,84 and increased physical stiffness.85, 86, 87 To understand how individual changes such as these may be causative to liver cancer development, reductionist in vivo models are needed.
Hormone Imbalance
In most countries, both liver disease and HCC are sexually dimorphic: more frequent in males than females with ratios ranging from 2:1 to >5:1.88 Sex hormone imbalance occurs in males with alcoholic cirrhosis as a decrease in testosterone and increase in free estrogen.89 Furthermore, estrogen levels increase in patients during liver regeneration after resection, indicating a potential role of estrogen in liver cell growth.90 To model the effects of estrogen on liver health in zebrafish, our group treated zebrafish with 17β-estradiol (E2) and found increased liver size in both adults and larvae.91 Transcriptomic analysis showed increased gene expression of cell-cycle regulators, particularly in male livers. Furthermore, metabolomic analysis showed estrogen exposure induced changes in pyrimidine and purine metabolism, again predominantly in male livers. To identify the receptor by which estrogen mediates its effect, we leveraged the larvae liver size phenotype: chemical inhibition of canonical estrogen receptors 1 and 2 had no effect on liver size. In contrast, when an inhibitor of the G-protein–coupled estrogen receptor (gper) was present, E2 treatment failed to increase liver size. Thus, E2 exerts its effect via noncanonical estrogen signaling through gper.
We tested the molecular mechanisms downstream of gper and showed that phosphoinositide 3-kinase (PI3K) and mechanistic target of rapamycin (mTOR) signaling were necessary because inhibitors of several enzymes in this cascade reduced or ablated the ability of E2 to increase larval liver size. gper-/- zebrafish showed significantly reduced DMBA-induced tumor formation. In an additional tumorigenesis study, wild-type (WT) fish treated with E2 developed more tumors than controls and co-treatment with a chemical GPER inhibitor decreased mortality and tumor growth. Importantly, this effect was evident almost entirely in the male cohort.91 This study highlights 2 of the most important advantages of zebrafish cancer models: the ability to sex-stratify longitudinal cancer studies while maintaining cohorts of an appropriate number for meaningful statistical analyses, and the ability to use surrogate, larval-stage phenotypes to uncover necessary molecular receptors and efficacious chemical inhibitors.
Nutrient Deficiencies
Selenium is an essential nutrient found to be deficient in some cirrhotic patients.84, 92 Selenium is incorporated into and essential for the function of selenoproteins, a class of proteins shown to have context-dependent tumor-preventing or tumor-promoting activity.93 To investigate the role of selenium in HCC formation, our group began with the observation that zebrafish with a mutation in selenoprotein H (seph; ortholog of human SELH) had severe defects in endodermal organogenesis. Transcriptomic profiling of seph mutants showed up-regulation of an inflammatory gene signature and strong induction of Tp53 target genes.94 To show genetic linkage between tp53 and seph, we compared DMBA-induced gastrointestinal tumor formation in the progeny of tp53-/-;seph+/- fish and found that seph deficiency greatly increased tumor formation over the course of 1 year.94 This study provides molecular and genetic evidence for decreased selenoprotein H function as a mechanism by which selenium deficiency in cirrhotic patients leads to HCC. It will be important to apply zebrafish disease modeling strategies to other micronutrient deficiencies associated with nonalcoholic fatty liver disease (NAFLD) and liver disease.95
Mechanotransduction and Glutamine and Glucose Metabolism
Cirrhosis is an advanced stage of liver fibrosis. Fibrosis is the result of excess extracellular matrix production and cross-linking mediated primarily by HSCs, the liver’s resident fibroblasts, in response to chronic injury.96 The regenerative capacity of the liver allows fibrosis to accumulate over decades and create a unique cellular microenvironment that might promote the accumulation of oncogenic mutations and survival of transformed cells.97 An additional consequence of massive fibrosis is increased physical stiffness. Within cirrhotic patients, liver stiffness, which can be measured via ultrasound or magnetic resonance elastography in the clinic, correlates with increased HCC risk, but whether stiffness is causative for HCC development is unknown.85 Thus, animal models of liver stiffness and molecular signaling pathways associated with mechanotransduction are needed.
Altered activity of Yes-associated protein (Yap), the transcriptional regulator central to the Hippo pathway, recently has emerged as one mechanism by which cells respond to mechanical cues such as substrate stiffness.98 Although mutations in Yap or Hippo signaling pathway genes are not frequent in HCC, immunohistochemical analysis of HCC samples showed nuclear Yap staining, indicative of active Yap signaling, in approximately 60% of cases, implicating transcriptional or post-transcriptional misregulation of Yap.99
To investigate how Yap influences HCC, we examined the Tg(-2.8fabp10a:yap1-1βS87A) transgenic line (referred to as lf:Yap) that expresses an activated form of Yap specifically in hepatocytes.100 Hepatocyte-specific overexpression of activated Yap induced hepatomegaly in both adult and larval zebrafish. Importantly, liver tumor formation after DMBA exposure was accelerated in lf:Yap fish compared with WT sibling controls. Transcriptomic analysis indicated increased expression of glutamine synthetase genes (glula and glulb) in lf:Yap livers. Metabolomic analyses and in vivo isotope-labeled nitrogen incorporation assays confirmed that Yap acts to reprogram nitrogen metabolism via glutamine synthetase and promote the formation of glutamine as a precursor for nucleotide biosynthesis. Disruption of glula and glulb by antisense morpholino injection returned lf:Yap livers to WT size, and chemical inhibition of glutamine synthetase rescued the hepatomegaly phenotype in both larval- and adult-stage zebrafish.100 An additional study showed that Yap similarly regulates expression of glucose transporter glut1, modulating glucose uptake.101 Treating lf:Yap larvae with either of 2 separate small-molecule Glut1 inhibitors normalized the large liver phenotype.101 Thus, using a transgenic zebrafish model in tumorigenesis studies followed by mechanistic analysis using both adult- and larval-stage surrogate phenotypes identified Yap signaling as a driver of HCC, illuminated novel biochemical and molecular mechanisms, and validated novel chemical inhibition strategies.
The earlier-described examples illuminate strategies used and contributions made to understanding genetic and environmental drivers of HCC using zebrafish models. In the remaining sections, we discuss how zebrafish are being used to understand emerging mechanisms that sustain and influence HCC (Figure 3).
Figure 3.
Biochemical and physiological changes associated with cirrhosis are potential targets for HCC prevention.
Modeling Microenvironmental Changes Associated With Inflammation
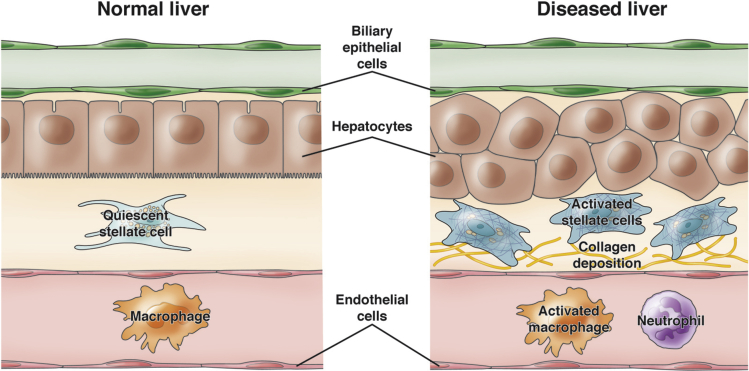
The cellular composition of the zebrafish liver is similar to that of the mammalian liver (Figure 4). The zebrafish liver has 3 lobes: 2 lateral and 1 ventral. The hexagonal lobule architecture of mammalian livers, however, is not present in zebrafish; instead, hepatocytes are arranged in tubules consisting of 2 rows of hepatocytes.19 The apical side of each hepatocyte faces bile ductules, and the basolateral side of each hepatocyte faces LSECs.102, 103 Zebrafish bile canaliculi exist within each hepatocyte rather than between hepatocytes, as found in the mammalian liver.104 Thus, the functional polarity of the hepatocyte is conserved. The zebrafish liver also contains HSCs, which are activated in response to injury.105, 106 The presence of liver-resident macrophages remains controversial, with some pathology analyses indicating that the normal liver might not contain resident macrophages.107 Nonetheless, populations of macrophages and neutrophils can be found in the liver, especially after injury,108 and unpublished observations from our group suggest an important role for macrophages during liver development.
Figure 4.
Zebrafish liver cellular architecture and tumor microenvironment. The zebrafish liver contains the same types of cells as the mammalian liver: hepatocytes, biliary epithelial cells (cholangiocytes), liver sinusoidal endothelial cells, stellate cells, and macrophages. Similar to the mammalian liver, hepatocytes are polarized between blood vessels and bile ductules. The bile canaliculi in zebrafish exist within each hepatocyte rather than between hepatocytes as observed in mammals. Upon injury or onset of liver disease, stellate cells and macrophages take on activated phenotypes, and other immune cells such as neutrophils invade the liver. Transgenic zebrafish lines labeling each of these cell types (see text) enable studies focusing on interactions between normal, activated, and transformed cells within the tumor niche.
Much effort in the zebrafish community has focused on generating transgenic reporter lines using the promoter or enhancer regions of tissue-specific genes to drive expression of fluorescent proteins: fabp10a for hepatocytes,45 kdrl or flk1 for LSECs,109, 110 krt18111 or a Notch-responsive Epstein–Barr virus tp1 enhancer103 for cholangiocytes/biliary epithelial cells, hand2 for HSCs,105 mpeg1.1112 and mfap4113 for macrophages, lyz114 and mpx (also called mpo)114, 115 for neutrophils. Most studies have used these reporters to probe the mechanisms of normal liver development during larval stages, however, the following examples applied these reporters to adult zebrafish HCC models and probe the influence of tumor microenvironment (TME) cell types on tumorigenesis and HCC progression.
HCC develops most frequently in the context of chronic fibrosis and inflammation. These unique aspects of the TME are maintained by supportive cell types such as fibroblasts and immune cells. Investigating the mechanisms by which these supportive cells interact with hepatocytes and maintain the TME is vital.
Fibrosis is produced initially by liver resident fibroblasts—HSCs.116 Liver inflammation is complex and involves many types of immune cells. The liver houses the largest percentage of macrophages in the body, and liver resident macrophages (ie, Kupffer cells) respond to many injury cues.117 After a tumor is formed, immune and stromal cells become important for maintaining the TME. These include tumor-associated fibroblasts (reviewed by Kubo et al,118 Affo et al,119 and Huang et al120), neutrophils (TANs) (reviewed by Ringelhan et al121), macrophages (TAMs) (reviewed by Krenkel and Tacke,117 Ringelhan et al,121 and Shirabe et al122), and other immune cells. Understanding the interactions and co-dependencies between tumor cells and cell types in the TME as well as the signaling pathways involved could lead to new therapeutic targets or avenues.
To explore the involvement of neutrophils in HCC progression, Yan et al used the krasV12-induced HCC model in conjunction with lyz:dsRed zebrafish, which labels neutrophils.108 Upon krasV12 induction, they found rapid recruitment of neutrophils into the liver, and pharmacologic activation of neutrophils accelerated HCC progression. Transcriptional analysis of isolated hepatocytes implicated tgfβ signaling activation, and analysis of isolated neutrophils showed a decrease in antitumor cytokine transcripts, which could be reversed by pharmacologic inhibition of Tgfβ.108 Thus, neutrophil–hepatocyte interactions should be explored further in cases of Ras-driven HCC as well as other HCC classes.
A study by de Oliveira et al123 focused on how diet and macrophages influence HCC development. They used the activated β-catenin zebrafish HCC model59 in combination with a high-cholesterol diet, which increased activated tumor necrosis factor-α+ macrophages in the livers of juvenile zebrafish. Treatment with metformin, a drug used to treat diabetes and NAFLD, or ablation of macrophages, reduced the effect of the high-cholesterol diet.123 This study links dietary effects and macrophage function to inflammation and Wnt-driven HCC.
As discussed previously, HCC is a sex-biased disease, occurring far more frequently in males.88 A study found that the kras-induced HCC model induced tumors more frequently in male fish compared with female fish.124 Furthermore, tumors in male fish contained more invading TANs and TAMs than tumors in female fish.125 Ablation of either neutrophils or macrophages during development led to decreased tumor size. Furthermore, cortisol was more highly expressed in male fish than in female fish, corresponding to a cohort of male HCC patients compared with females. Inhibition of glucocorticoid signaling resulted in decreased TAN and TAM recruitment and decreased tumor severity in male livers, whereas hydrocortisone treatment—a cortisol analog—increased TAN and TAM recruitment and induced more aggressive tumors in female livers.125
An additional study used the TgBAC(hand2:GFP) (Green fluorescent protein) line and found that male tumors in the krasV12-induced HCC model also had increased HSC infiltration and activation compared with female tumors.126 Serotonin, which has been linked to mouse and human HSC activation and liver disease progression in mice,127 also was increased in male zebrafish livers. Activation of serotonin biosynthesis increased the severity of tumors in female fish; inhibition of serotonin biosynthesis decreased the severity of tumors in male fish. Transcriptional analysis of isolated HSCs implicated tgfβ expression and secretion.126 Male-biased tumorigenesis as well as cortisol activation of TANs and TAMs and serotonin-induced activation of HSCs also were observed in the xmrk and myc overexpression zebrafish liver tumorigenesis models.67 This suggests that such mechanisms could be relevant in HCC cases carrying different driver mutations. These studies add to growing literature on the effects of sex hormones on HCC and show the practicality of zebrafish HCC studies, which can use cohorts of sufficient number to stratify studies based on sex and maintain statistical power.
This series of studies explored the HCC tumor microenvironment and used transcriptional analyses and pharmacologic interventions to investigate the molecular mechanisms responsible. Such zebrafish TME studies will be enhanced as the CRISPR technology improvements in the zebrafish community enable more tractable tissue- and cell type-specific knockout strategies. Future zebrafish liver cancer studies should focus on tumor vascularization, biliary tumors, and the influence of the adaptive immune system.
Current and Future Efforts
This final section focuses specifically on scientific areas we have identified that have been linked to HCC in other models, have tools or assays developed in zebrafish, and these tools have not been applied extensively to zebrafish HCC studies.
Aging
Because cancer frequency increases with age, animal models of aging are vital. Zebrafish have been used in aging research as a vertebrate model of very gradual senescence.128 Zebrafish show some indicators of aging, but they live approximately 50% longer than laboratory mice,129 which makes aging studies difficult. The African turquoise killifish Northobranchius furzeri, a similarly sized teleost, has a life span of approximately 8 weeks (depending on the strain), and it develops spontaneous liver and kidney neoplasias as a function of age.130 Developing killifish HCC models similar to the zebrafish models described here should be a priority.
Modeling Autophagy
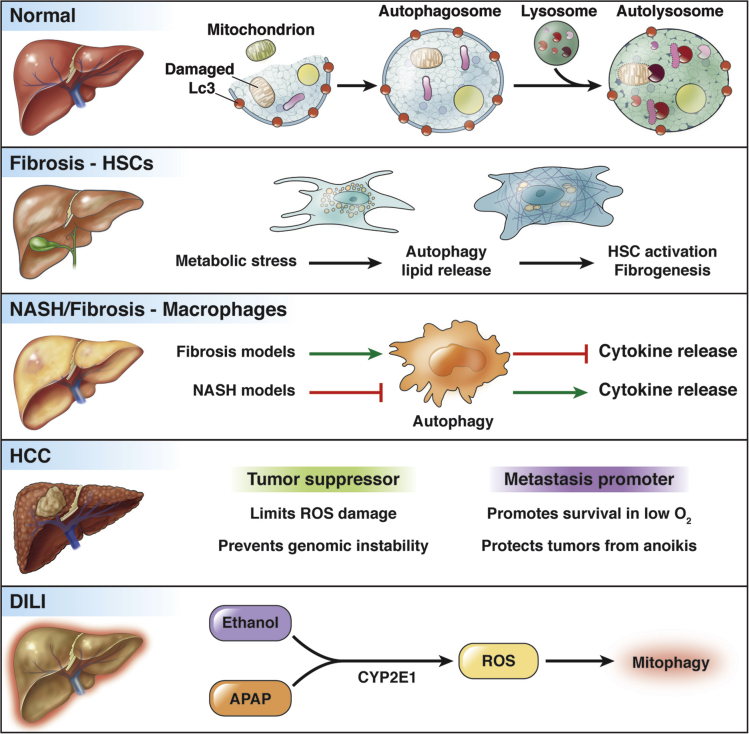
Autophagy is a lysosomal degradation pathway that maintains cellular homeostasis by degrading dysfunctional or unneeded cellular components and recycling them. As a highly metabolic organ, the liver uses autophagy for many normal processes, and dysregulation of autophagy is thought to play a role in many forms of liver disease including α1-antitrypsin deficiency, NAFLD/alcoholic-liver disease, HBV and HCV infection, fibrosis, and HCC131 (Figure 5).
Figure 5.
Autophagy in normal, diseased, and injured liver. The autophagy pathway sequesters damaged organelles and cytosolic components in the Lc3-coated autophagosome, which then fuses with a lysosome to form an autolysosome where the cargo is degraded and recycled. During liver disease, autophagy plays varied roles in different liver cell types. Metabolic stress can activate autophagy in HSCs, causing their activation and contributing to fibrosis. Macrophages use autophagy to regulate cytokine release depending on the injury model (fibrosis or nonalcoholic steatohepatitis [NASH]). In HCC, autophagy limits tumor formation and growth but also can promote survival of metastases. In DILI, mitophagy functions to clear ROS-damaged mitochondria to limit liver damage. CYP2E1, Cytochrome P450 2E1.
Core autophagy proteins function to prevent liver cancer development: mosaic deletion of Atg5 or liver-specific deletion of Atg7 caused mice to develop liver tumors,132, 133 as did Beclin1 haploinsufficiency.134 However, the tumors formed in Atg7 or Atg5 null livers do not metastasize, which indicates that autophagy might be necessary for metastasis.132, 133 Autophagy plays a role in fibrosis by governing the activation of HSCs. Oxidative stress induces endoplasmic reticulum stress and triggers the unfolded protein response135 through the X-box binding protein 1 (XBP1) pathway,136 inducing autophagy. Autophagy in HSCs functions to enable lipid-droplet degradation as part of the HSC activation process, which leads to liver fibrosis.137 Liver-resident macrophages use autophagy differently, depending on the liver injury mechanism. In the carbon tetrachloride–induced liver fibrosis mouse model, liver-resident macrophages use autophagy to prevent release of inflammatory cytokines, which can activate HSCs and exacerbate liver fibrosis.138 In a diet-induced mouse model of nonalcoholic steatohepatitis, however, saturated fatty acid (palmitic acid) treatment induced a maladaptive impairment of macrophage autophagy via hypoxia-inducible factor signaling, which led to release of proinflammatory cytokines.139 Autophagy is a homeostasis-maintaining mechanism central to normal liver function and liver disease progression with overlapping mechanisms and cell type- and context-dependent functions.
Because of the conflicting roles of autophagy found in different liver cell types, model systems able to re-create and probe these interactions in vivo are vital to advancing our understanding. A clear advantage of zebrafish when compared with other model organisms is their suitability for live imaging, and several zebrafish autophagy reporter lines have been generated. The process of autophagy begins with recruitment of a double-membrane structure—the autophagosome—to engulf the targeted protein or organelle. The autophagosome then fuses with a lysosome, forming an autolysosome, where the contents are degraded and recycled. A commonly used autophagy reporter uses transgenic overexpression of GFP-Lc3, which tracks the localization of Microtubule Associated Protein 1 Light Chain 3 Beta (Map1lc3b or Lc3), a protein necessary for autophagosome formation. When autophagy levels in a cell are low, GFP-Lc3 remains diffuse in the cytoplasm, but when autophagy is activated, puncta of GFP can be observed and quantified because Lc3 clusters on nascent autophagosomes.140 He et al141 generated transgenic zebrafish ubiquitously expressing GFP-Lc3 and observed high levels of autophagy in various tissues during embryonic and larval developmental stages. A hepatocyte-specific GFP-Lc3 reporter zebrafish line also was generated,142 and both the ubiquitous and hepatocyte-specific lines showed increased autophagy in response to inhibition of Tor signaling.
To enable more quantitative measurement of autophagy in vivo, the Mizushima group developed a ratiometric autophagy biosensor GFP-LC3-RFP-LC3ΔG. This probe is cleaved by endogenous autophagy-related peptidases and releases both GFP-LC3 and red fluorescent protein (RFP) fused to a mutant LC3 lacking C-terminal glycine (LC3ΔG), which is not targeted to autophagosomes and thus remains in the cytoplasm functioning as an internal control.143 The ratio of GFP/RFP is therefore a carefully calibrated measure of autophagic flux. Injection of messenger RNA encoding this construct into zebrafish embryos allowed for characterization of autophagy in early embryonic stage zebrafish and several larval tissues including skeletal muscle, retina, and lens. They found that zebrafish null for FIP200—a gene known to be essential for autophagosome formation144—have drastically lower basal levels of autophagic flux.143 This probe has not yet been applied to studying the role of autophagy in liver development or disease.
Drug-Induced Liver Injury, Oxidative Stress, and Mitochondria
Mitophagy is the selective degradation of mitochondria via autophagy in response to mitochondrial damage or stress.145 Inappropriate mitophagy is linked to aging, neurodegenerative diseases, drug-induced liver injury (DILI), and cancer—including hepatocellular carcinoma.131 Specifically, mitophagy is thought to be protective against both alcohol-induced146 and acetaminophen (APAP)-induced147, 148 liver injury. Because both ethanol and APAP metabolism lead to an increase in reactive oxygen species (ROS), and ROS damage mitochondria, selective removal of mitochondria after severe oxidative stress is hepatoprotective. The study of molecular mechanisms governing mitophagy in the context of liver disease is an emerging area of research; early indications from mouse models implicate Parkin147 and Nrf2149 signaling. Undeniably, more work must be done to illuminate the role of mitophagy in liver disease to develop new treatment options.
Zebrafish have been used in various ways to investigate liver injury mechanisms induced by oxidative stress. Zebrafish are a well-established model system for studying both alcohol-150 and APAP-induced151 liver injury. The Sadler group performed pioneering studies establishing zebrafish larvae as a model for alcohol toxicity.152, 153 Specifically, using larval ethanol exposures, they linked oxidative stress to the unfolded protein response and the development of hepatic steatosis.152, 153 Adult zebrafish models of alcohol toxicity also show promise for future work on alcohol’s influence on fibrosis and HCC.154, 155 In collaboration with North et al,151 our laboratory developed both larval and adult zebrafish models of APAP toxicity. A targeted chemical screen identified prostagalndin E2 treatment reduced APAP-induced liver toxicity to a similar extent as N-acetylcysteine, the only treatment currently available. Furthermore, co-treatment with prostaglandin E2 and N-acetylcysteine increased the time window for successful intervention.151
Recently, Jain et al156 performed a CRISPR screen using cells treated with respiratory chain inhibitors, which leads to massive ROS production, as a model for mitochondrial disease. Guides targeted to von Hippel-Lindau factor effectively suppressed the mitochondrial disease phenotype. We verified this finding in vivo, showing vhl null fish are resistant to respiratory chain inhibition–induced death. Chemical activation of hypoxia-inducible factor signaling in WT fish also alleviated death caused by respiratory chain inhibition. Future studies should explore the links between hypoxia, ROS, and liver damage, as well as the links between DILI, oxidative stress, and mitophagy.
Summary and Conclusions
Zebrafish have livers that function and develop HCC similarly to human livers and offer unique advantages for researchers aiming to understand and develop novel treatments for liver disease. The ease of transgenesis and loss-of-function mutagenesis studies combined with the inexpensive housing costs facilitate the analysis of putative driver mutations found in HCC. Chemical screening combined with larval liver-size phenotypes enables identification and analysis of biochemical modifiers of liver growth and HCC development. Their small size and transparency promote live imaging studies to understand interactions between the liver, the microenvironment, and the immune system during development, liver disease, DILI, and oxidative stress. In these ways, zebrafish models complement more traditional rodent models in their ability to both dissect how specific genetic or biochemical insults influence liver health and also allow for unbiased screening for unknown genetic or biochemical modulators of liver biology.
There are some limitations of zebrafish HCC models. Mammalian hepatocytes have functional differences based on zonation between the central vein and the portal triads. No studies have identified comparable physical zonation in the zebrafish liver. Future studies should analyze the microarchitecture of the zebrafish liver for any indication of functional zonation or heterogeneity. Zebrafish and their livers are very small. In some cases, this is an advantage (eg, for live imaging and low housing costs), but in other situations this is a disadvantage (eg, less tissue for analysis by sectioning, transcriptomics, or Western blot). Antibody-based assays are somewhat fickle, likely owing to the limited identification and validation of antibodies able to recognize zebrafish antigens. Finally, tissue-specific knockouts using Cre-Lox in zebrafish lag behind the tools available for mice. The lack of a tractable zebrafish embryonic stem cell culture system has limited progress toward tissue-specific knockout generation, but improved genome editing tools and practices promise to overcome this hurdle. As more research groups adopt the zebrafish model organism, many of these problems will be overcome.
The premise of this review is that the current pipeline of HCC therapeutics has not produced major breakthroughs since the introduction of sorafenib. This raises the question, at which point in the pipeline can zebrafish models of HCC contribute? The clearest advantage of zebrafish models is in vivo chemical screening studies, which accelerate the identification of potentially efficacious compounds. How to proceed once these compounds are identified is a vital question with which the HCC community must engage. In cases in which the compounds identified are already Food and Drug Administration–approved for some other use, perhaps it is appropriate to move directly to Phase I clinical trials. This is occurring for some compounds identified using screens of other zebrafish disease models.157 In cases in which the compounds identified have not yet been tested in patients, the pharmacokinetics and toxicology must be worked out in the appropriate and accepted mammalian models before Phase I trials. However, it is our opinion that verifying the efficacy of identified compounds in mammalian HCC models is not necessary and could be counterproductive given the poor track record of translation from murine models to clinical success.
Acknowledgments
The authors thank Chad Walesky, Arkadi Shwartz, Olivia Weeks, and Eleanor Quenzer for critical reading of the manuscript.
Footnotes
Author contributions Paul J. Wrighton and Wolfram Goessling reviewed the literature, drafted and critically revised the manuscript, and designed Figure 1, Figure 2, Figure 3, and 5; and Isaac M. Oderberg designed Figure 4 and contributed to the associated text.
Conflicts of interest This authors discloses the following: Wolfram Goessling is a consultant and scientific advisory board member of Camp4 Therapeutics. The remaining authors disclose no conflicts.
Funding Supported by the American Liver Foundation and by National Institutes of Health grant F32AA025271 (P.W.); by National Institutes of Health grant F32AA027135 (I.O.); and by National Institutes of Health grants R01DK090311, R01DK105198, and R24OD017870, and the Claudia Adams Barr Program in Innovative Basic Cancer Research (W.G.). Wolfram Goessling is also a Pew Scholar in the Biomedical Sciences.
References
- 1.Ferlay J., Soerjomataram I.I., Dikshit R., Eser S., Mathers C., Rebelo M., Parkin D.M., Forman D.D., Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2014;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Beste L.A., Leipertz S.L., Green P.K., Dominitz J.A., Ross D., Ioannou G.N. Trends in burden of cirrhosis and hepatocellular carcinoma by underlying liver disease in US veterans, 2001-2013. Gastroenterology. 2015;149:1471–1482.e5. doi: 10.1053/j.gastro.2015.07.056. [DOI] [PubMed] [Google Scholar]
- 3.Baselga J., Bhardwaj N., Cantley L.C., DeMatteo R., DuBois R.N., Foti M., Gapstur S.M., Hahn W.C., Helman L.J., Jensen R.A., Paskett E.D., Lawrence T.S., Lutzker S.G., Szabo E. AACR cancer progress report 2013. Clin Cancer Res. 2013;21:S1–S128. doi: 10.1158/1078-0432.CCR-15-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruix J., Reig M., Sherman M. Evidence-based diagnosis, staging, and treatment of patients with hepatocellular carcinoma. Gastroenterology. 2016;150:836–853. doi: 10.1053/j.gastro.2015.12.041. [DOI] [PubMed] [Google Scholar]
- 5.Goh B.K.P., Teo J.Y., Chan C.Y., Lee S.Y., Jeyaraj P., Cheow P.C., Chow P.K.H., Ooi L.L.P.J., Chung A.Y.F. Importance of tumor size as a prognostic factor after partial liver resection for solitary hepatocellular carcinoma: implications on the current AJCC staging system. J Surg Oncol. 2016;113:89–93. doi: 10.1002/jso.24099. [DOI] [PubMed] [Google Scholar]
- 6.Roayaie S., Jibara G., Tabrizian P., Park J.W., Yang J., Yan L., Schwartz M., Han G., Izzo F., Chen M., Blanc J.F., Johnson P., Kudo M., Roberts L.R., Sherman M. The role of hepatic resection in the treatment of hepatocellular cancer. Hepatology. 2015;62:440–451. doi: 10.1002/hep.27745. [DOI] [PubMed] [Google Scholar]
- 7.Llovet J.M., Ricci S., Mazzaferro V., Hilgard P., Gane E., Blanc J.F., Cosme de Oliveira A., Santoro A., Raoul J.L., Forner A., Schwartz M., Galle P.R., Seitz J.F., Borbath I., Häussinger D., Giannaris T., Shan M., Moscovici M., Voliotis D., Bruix J., SHARP Investigators Study Group Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 8.Llovet J.M., Hernandez-Gea V. Hepatocellular carcinoma: reasons for phase III failure and novel perspectives on trial design. Clin Cancer Res. 2014;20:2072–2079. doi: 10.1158/1078-0432.CCR-13-0547. [DOI] [PubMed] [Google Scholar]
- 9.Llovet J.M., Villanueva A., Lachenmayer A., Finn R.S. Advances in targeted therapies for hepatocellular carcinoma in the genomic era. Nat Rev Clin Oncol. 2015;12:408–424. doi: 10.1038/nrclinonc.2015.103. [DOI] [PubMed] [Google Scholar]
- 10.Greten T.F., Lai C.W., Li G., Staveley-O’Carroll K.F. Targeted and immune-based therapies for hepatocellular carcinoma. Gastroenterology. 2019;156:510–524. doi: 10.1053/j.gastro.2018.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dawkins J., Webster R.M. The hepatocellular carcinoma market. Nat Rev Drug Discov. 2018;18:13–14. doi: 10.1038/nrd.2018.146. [DOI] [PubMed] [Google Scholar]
- 12.Farazi P.A., DePinho R.A. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer. 2006;6:674–687. doi: 10.1038/nrc1934. [DOI] [PubMed] [Google Scholar]
- 13.Vogelstein B., Papadopoulos N., Velculescu V.E., Zhou S., Diaz L.A., Jr., Kinzler K.W. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohen J.C., Horton J.D., Hobbs H.H. Human fatty liver disease: old questions and new insights. Science. 2011;332:1519–1523. doi: 10.1126/science.1204265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forner A., Llovet J., Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 16.Kansagara D., Papak J., Pasha A.S., O’Neil M., Freeman M., Relevo R., Quiñones A., Motu’apuaka M., Jou J.H. Screening for hepatocellular carcinoma in chronic liver disease: a systematic review. Ann Intern Med. 2014;161:261–269. doi: 10.7326/M14-0558. [DOI] [PubMed] [Google Scholar]
- 17.Hwang K.L., Goessling W. Baiting for cancer: using the zebrafish as a model in liver and pancreatic cancer. Adv Exp Med Biol. 2016;916:391–410. doi: 10.1007/978-3-319-30654-4_17. [DOI] [PubMed] [Google Scholar]
- 18.White R., Rose K., Zon L. Zebrafish cancer: the state of the art and the path forward. Nat Rev Cancer. 2013;13:624–636. doi: 10.1038/nrc3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goessling W., Sadler K.C. Zebrafish: an important tool for liver disease research. Gastroenterology. 2015;149:1361–1377. doi: 10.1053/j.gastro.2015.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Santoriello C., Zon L.I. Hooked! Modeling human disease in zebrafish. J Clin Invest. 2012;122:2337–2343. doi: 10.1172/JCI60434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.North T.E., Goessling W. Endoderm specification, liver development, and regeneration. Methods Cell Biol. 2011;101:205–223. doi: 10.1016/B978-0-12-387036-0.00010-4. [DOI] [PubMed] [Google Scholar]
- 22.Goessling W., Stainier D.Y. Endoderm specification and liver development. Biophys Methods Cell Biol. 2016;134:463–483. doi: 10.1016/bs.mcb.2016.03.042. [DOI] [PubMed] [Google Scholar]
- 23.Cox A.G., Goessling W. The lure of zebrafish in liver research: regulation of hepatic growth in development and regeneration. Curr Opin Genet Dev. 2015;32:153–161. doi: 10.1016/j.gde.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shin D., Monga S.P.S. Cellular and molecular basis of liver development. Compr Physiol. 2013;3:799–815. doi: 10.1002/cphy.c120022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu L.Y., Fox C.S., North T.E., Goessling W. Functional validation of GWAS gene candidates for abnormal liver function during zebrafish liver development. Dis Model Mech. 2013;6:1271–1278. doi: 10.1242/dmm.011726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burger A., Lindsay H., Felker A., Hess C., Anders C., Chiavacci E., Zaugg J., Weber L.M., Catena R., Jinek M., Robinson M.D., Mosimann C. Maximizing mutagenesis with solubilized CRISPR-Cas9 ribonucleoprotein complexes. Development. 2016;143:2025–2037. doi: 10.1242/dev.134809. [DOI] [PubMed] [Google Scholar]
- 27.Ablain J., Durand E.M., Yang S., Zhou Y., Zon L.I. A CRISPR/Cas9 vector system for tissue-specific gene disruption in zebrafish. Dev Cell. 2015;32:756–764. doi: 10.1016/j.devcel.2015.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yin L., Maddison L.A., Li M., Kara N., Lafave M.C., Varshney G.K., Burgess S.M., Patton J.G., Chen W. Multiplex conditional mutagenesis using transgenic expression of Cas9 and sgRNAs. Genetics. 2015;200:431–441. doi: 10.1534/genetics.115.176917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peterson R.T., Link B.A., Dowling J.E., Schreiber S.L. Small molecule developmental screens reveal the logic and timing of vertebrate development. Proc Natl Acad Sci U S A. 2000;97:12965–12969. doi: 10.1073/pnas.97.24.12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rennekamp A.J., Peterson R.T. 15 years of zebrafish chemical screening. Curr Opin Chem Biol. 2014;24:58–70. doi: 10.1016/j.cbpa.2014.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee J.-S. The mutational landscape of hepatocellular carcinoma. Clin Mol Hepatol. 2015;21:220–229. doi: 10.3350/cmh.2015.21.3.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harbeck N., Gnant M. Breast cancer. Lancet. 2017;389:1134–1150. doi: 10.1016/S0140-6736(16)31891-8. [DOI] [PubMed] [Google Scholar]
- 33.Schulze K., Imbeaud S., Letouzé E., Alexandrov L.B., Calderaro J., Rebouissou S., Couchy G., Meiller C., Shinde J., Soysouvanh F., Calatayud A.L., Pinyol R., Pelletier L., Balabaud C., Laurent A., Blanc J.F., Mazzaferro V., Calvo F., Villanueva A., Nault J.C., Bioulac-Sage P., Stratton M.R., Llovet J.M., Zucman-Rossi J. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015;47:505–511. doi: 10.1038/ng.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zucman-Rossi J., Villanueva A., Nault J.C., Llovet J.M. Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology. 2015;149:1226–1239. doi: 10.1053/j.gastro.2015.05.061. [DOI] [PubMed] [Google Scholar]
- 35.Nault J.C., Zucman-Rossi J. TERT promoter mutations in primary liver tumors. Clin Res Hepatol Gastroenterol. 2016;40:9–14. doi: 10.1016/j.clinre.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 36.Lin S., Nascimento E.M., Gajera C.R., Chen L., Neuhöfer P., Garbuzov A., Wang S., Artandi S.E. Distributed hepatocytes expressing telomerase repopulate the liver in homeostasis and injury. Nature. 2018;556:244–248. doi: 10.1038/s41586-018-0004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Villanueva A., Llovet J.M. Targeted therapies for hepatocellular carcinoma. Gastroenterology. 2011;140:1410–1426. doi: 10.1053/j.gastro.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Newell P., Toffanin S., Villanueva A., Chiang D.Y., Minguez B., Cabellos L., Savic R., Hoshida Y., Lim K.H., Melgar-Lesmes P., Yea S., Peix J., Deniz K., Fiel M.I., Thung S., Alsinet C., Tovar V., Mazzaferro V., Bruix J., Roayaie S., Schwartz M., Friedman S.L., Llovet J.M. Ras pathway activation in hepatocellular carcinoma and anti-tumoral effect of combined sorafenib and rapamycin in vivo. J Hepatol. 2009;51:725–733. doi: 10.1016/j.jhep.2009.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mudbhary R., Hoshida Y., Chernyavskaya Y., Jacob V., Villanueva A., Fiel M.I., Chen X., Kojima K., Thung S., Bronson R.T., Lachenmayer A., Revill K., Alsinet C., Sachidanandam R., Desai A., SenBanerjee S., Ukomadu C., Llovet J.M., Sadler K.C. UHRF1 overexpression drives DNA hypomethylation and hepatocellular carcinoma. Cancer Cell. 2014;25:196–209. doi: 10.1016/j.ccr.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lam S.H., Wu Y.L., Vega V.B., Miller L.D., Spitsbergen J., Tong Y., Zhan H., Govindarajan K.R., Lee S., Mathavan S., Murthy K.R.K., Buhler D.R., Liu E.T., Gong Z. Conservation of gene expression signatures between zebrafish and human liver tumors and tumor progression. Nat Biotechnol. 2006;24:73–75. doi: 10.1038/nbt1169. [DOI] [PubMed] [Google Scholar]
- 41.Suster M.L., Kikuta H., Urasaki A., Asakawa K., Kawakami K. Transgenesis in zebrafish with the Tol2 transposon system. Methods Mol Biol. 2009;561:41–63. doi: 10.1007/978-1-60327-019-9_3. [DOI] [PubMed] [Google Scholar]
- 42.Felker A., Mosimann C. Contemporary zebrafish transgenesis with Tol2 and application for Cre/lox recombination experiments. Methods Cell Biol. 2016;135:219–244. doi: 10.1016/bs.mcb.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 43.Kwan K.M., Fujimoto E., Grabher C., Mangum B.D., Hardy M.E., Campbell D.S., Parant J.M., Yost H.J., Kanki J.P., Chien C Bin. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- 44.Don E.K., Formella I., Badrock A.P., Hall T.E., Morsch M., Hortle E., Hogan A., Chow S., Gwee S.S.L., Stoddart J.J., Nicholson G., Chung R., Cole N.J. A Tol2 gateway-compatible toolbox for the study of the nervous system and neurodegenerative disease. Zebrafish. 2017;14:69–72. doi: 10.1089/zeb.2016.1321. [DOI] [PubMed] [Google Scholar]
- 45.Her G.M., Chiang C.-C., Chen W.-Y., Wu J.-L. In vivo studies of liver-type fatty acid binding protein (L-FABP) gene expression in liver of transgenic zebrafish (Danio rerio) FEBS Lett. 2003;538:125–133. doi: 10.1016/s0014-5793(03)00157-1. [DOI] [PubMed] [Google Scholar]
- 46.van Eeden F.J.M., Granato M., Odenthal J., Haffter P. Developmental mutant screens in the zebrafish. Methods Cell Biol. 1998;60:21–41. doi: 10.1016/s0091-679x(08)61892-0. [DOI] [PubMed] [Google Scholar]
- 47.Haffter P., Nüsslein-Volhard C. Large scale genetics in a small vertebrate, the zebrafish. Int J Dev Biol. 1996;40:221–227. [PubMed] [Google Scholar]
- 48.Amsterdam A., Burgess S., Golling G., Chen W., Sun Z., Townsend K., Farrington S., Haldi M., Hopkins N. A large-scale insertional mutagenesis screen in zebrafish. Genes Dev. 1999;13:2713–2724. doi: 10.1101/gad.13.20.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leshchiner I., Alexa K., Kelsey P., Adzhubei I., Austin-Tse C.A., Cooney J.D., Anderson H., King M.J., Stottmann R.W., Garnaas M.K., Ha S., Drummond I.A., Paw B.H., North T.E., Beier D.R., Goessling W., Sunyaev S.R. Mutation mapping and identification by whole-genome sequencing. Genome Res. 2012;22:1541–1548. doi: 10.1101/gr.135541.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kettleborough R.N.W., Busch-Nentwich E.M., Harvey S.A., Dooley C.M., De Bruijn E., Van Eeden F., Sealy I., White R.J., Herd C., Nijman I.J., Fényes F., Mehroke S., Scahill C., Gibbons R., Wali N., Carruthers S., Hall A., Yen J., Cuppen E., Stemple D.L. A systematic genome-wide analysis of zebrafish protein-coding gene function. Nature. 2013;496:494–497. doi: 10.1038/nature11992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hwang W.Y., Fu Y., Reyon D., Maeder M.L., Tsai S.Q., Sander J.D., Peterson R.T., Yeh J.-R.J., Joung J.K. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kimura Y., Hisano Y., Kawahara A., Higashijima S.I. Efficient generation of knock-in transgenic zebrafish carrying reporter/driver genes by CRISPR/Cas9-mediated genome engineering. Sci Rep. 2014;4:6545. doi: 10.1038/srep06545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Irion U., Krauss J., Nusslein-Volhard C. Precise and efficient genome editing in zebrafish using the CRISPR/Cas9 system. Development. 2014;141:4827–4830. doi: 10.1242/dev.115584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li M., Zhao L., Page-McCaw P.S., Chen W. Zebrafish genome engineering using the CRISPR–Cas9 system. Trends Genet. 2016;32:815–827. doi: 10.1016/j.tig.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Berghmans S., Murphey R.D., Wienholds E., Neuberg D., Kutok J.L., Fletcher C.D.M., Morris J.P., Liu T.X., Schulte-Merker S., Kanki J.P., Plasterk R., Zon L.I., Look A.T. Tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc Natl Acad Sci U S A. 2005;102:407–412. doi: 10.1073/pnas.0406252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spitsbergen J.M., Tsai H.-W., Reddy A., Miller T., Arbogast D., Hendricks J.D., Bailey G.S. Neoplasia in zebrafish (Danio rerio) treated with 7,12-dimethylbenz[a]anthracene by two exposure routes at different developmental stages. Toxicol Pathol. 2000;28:705–715. doi: 10.1177/019262330002800511. [DOI] [PubMed] [Google Scholar]
- 57.Goessling W., North T.E., Lord A.M., Ceol C., Lee S., Weidinger G., Bourque C., Strijbosch R., Haramis A.P., Puder M., Clevers H., Moon R.T., Zon L.I. APC mutant zebrafish uncover a changing temporal requirement for wnt signaling in liver development. Dev Biol. 2008;320:161–174. doi: 10.1016/j.ydbio.2008.05.526. [DOI] [PubMed] [Google Scholar]
- 58.Haramis A.P.G., Hurlstone A., van der Velden Y., Begthel H., van den Born M., Offerhaus G.J.A., Clevers H.C. Adenomatous polyposis coli-deficient zebrafish are susceptible to digestive tract neoplasia. EMBO Rep. 2006;7:444–449. doi: 10.1038/sj.embor.7400638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Evason K.J., Francisco M.T., Juric V., Balakrishnan S., Lopez Pazmino M. del P., Gordan J.D., Kakar S., Spitsbergen J., Goga A., Stainier D.Y.R. Identification of chemical inhibitors of beta-catenin-driven liver tumorigenesis in zebrafish. PLoS Genet. 2015;11:e1005305. doi: 10.1371/journal.pgen.1005305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Karnoub A.E., Weinberg R.A. Ras oncogenes: split personalities. Nat Rev Mol Cell Biol. 2008;9:517–531. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nguyen A.T., Emelyanov A., Koh C.H.V., Spitsbergen J.M., Lam S.H., Mathavan S., Parinov S., Gong Z. A high level of liver-specific expression of oncogenic KrasV12 drives robust liver tumorigenesis in transgenic zebrafish. Dis Model Mech. 2011;4:801–813. doi: 10.1242/dmm.007831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nguyen A.T., Emelyanov A., Koh C.H.V., Spitsbergen J.M., Parinov S., Gong Z. An inducible krasV12 transgenic zebrafish model for liver tumorigenesis and chemical drug screening. Dis Model Mech. 2012;5:63–72. doi: 10.1242/dmm.008367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li Z., Zheng W., Wang Z., Zeng Z., Zhan H., Li C., Zhou L., Yan C., Spitsbergen J.M., Gong Z. A transgenic zebrafish liver tumor model with inducible Myc expression reveals conserved Myc signatures with mammalian liver tumors. Dis Model Mech. 2013;6:414–423. doi: 10.1242/dmm.010462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li Z., Luo H., Li C., Huo X., Yan C., Huang X., Al-Haddawi M., Mathavan S., Gong Z. Transcriptomic analysis of a transgenic zebrafish hepatocellular carcinoma model reveals a prominent role of immune responses in tumour progression and regression. Int J Cancer. 2014;135:1564–1573. doi: 10.1002/ijc.28794. [DOI] [PubMed] [Google Scholar]
- 65.Li Z., Zheng W., Li H., Li C., Gong Z. Synergistic induction of potential Warburg effect in zebrafish hepatocellular carcinoma by co-transgenic expression of Myc and xmrk oncogenes. PLoS One. 2015;10:1–19. doi: 10.1371/journal.pone.0132319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li Z., Huang X., Zhan H., Zeng Z., Li C., Spitsbergen J.M., Meierjohann S., Schartl M., Gong Z. Inducible and repressable oncogene-addicted hepatocellular carcinoma in Tet-on xmrk transgenic zebrafish. J Hepatol. 2012;56:419–425. doi: 10.1016/j.jhep.2011.07.025. [DOI] [PubMed] [Google Scholar]
- 67.Yang Q., Yan C., Gong Z. Activation of liver stromal cells is associated with male-biased liver tumor initiation in xmrk and Myc transgenic zebrafish. Sci Rep. 2017;7:10315. doi: 10.1038/s41598-017-10529-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chew T.W., Liu X.J., Liu L., Spitsbergen J.M., Gong Z., Low B.C. Crosstalk of Ras and Rho: activation of RhoA abates Kras-induced liver tumorigenesis in transgenic zebrafish models. Oncogene. 2013;33:1–11. doi: 10.1038/onc.2013.240. [DOI] [PubMed] [Google Scholar]
- 69.Yan C., Yang Q., Huo X., Li H., Zhou L., Gong Z. Chemical inhibition reveals differential requirements of signaling pathways in krasV12- and Myc- induced liver tumors in transgenic zebrafish. Sci Rep. 2017;7:45796. doi: 10.1038/srep45796. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 70.Levrero M., Zucman-Rossi J. Mechanisms of HBV-induced hepatocellular carcinoma. J Hepatol. 2016;64:S84–S101. doi: 10.1016/j.jhep.2016.02.021. [DOI] [PubMed] [Google Scholar]
- 71.Arzumanyan A., Reis H.M.G.P.V., Feitelson M.A. Pathogenic mechanisms in HBV- and HCV-associated hepatocellular carcinoma. Nat Rev Cancer. 2013;13:123–135. doi: 10.1038/nrc3449. [DOI] [PubMed] [Google Scholar]
- 72.Shieh Y.-S., Chang Y.-S., Hong J.-R., Chen L.-J., Jou L.-K., Hsu C.-C., Her G.M. Increase of hepatic fat accumulation by liver specific expression of Hepatitis B virus X protein in zebrafish. Biochim Biophys Acta. 2010;1801:721–730. doi: 10.1016/j.bbalip.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 73.Lu J.W., Yang W.Y., Tsai S.M., Lin Y.M., Chang P.H., Chen J.R., Wang H.D., Wu J.L., Jin S.L.C., Yuh C.H. Liver-specific expressions of HBx and src in the p53 mutant trigger hepatocarcinogenesis in zebrafish. PLoS One. 2013;8:e76951. doi: 10.1371/journal.pone.0076951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lu J.W., Hsia Y., Yang W.Y., Lin Y.I., Li C.C., Tsai T.F., Chang K.W., Shieh G.S., Tsai S.F., Wang H.D., Yuh C.H. Identification of the common regulators for hepatocellular carcinoma induced by hepatitis B virus X antigen in a mouse model. Carcinogenesis. 2012;33:209–219. doi: 10.1093/carcin/bgr224. [DOI] [PubMed] [Google Scholar]
- 75.Rekha R.D., Amali A.A., Her G.M., Yeh Y.H., Gong H.Y., Hu S.Y., Lin G.H., Wu J.L. Thioacetamide accelerates steatohepatitis, cirrhosis and HCC by expressing HCV core protein in transgenic zebrafish Danio rerio. Toxicology. 2008;243:11–22. doi: 10.1016/j.tox.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 76.Zhao Y., Qin W., Zhang J.P., Hu Z.Y., Tong J.W., Ding C.B., Peng Z.G., Zhao L.X., Song D.Q., Jiang J.D. HCV IRES-mediated core expression in zebrafish. PLoS One. 2013;8:e56985. doi: 10.1371/journal.pone.0056985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ding C.-B., Zhao Y., Zhang J.-P., Peng Z.-G., Song D.-Q., Jiang J.-D. A zebrafish model for subgenomic hepatitis C virus replication. Int J Mol Med. 2015;35:791–797. doi: 10.3892/ijmm.2015.2063. [DOI] [PubMed] [Google Scholar]
- 78.Liu W., Chen J.R., Hsu C.H., Li Y.H., Chen Y.M., Lin C.Y., Huang S.J., Chang Z.K., Chen Y.C., Lin C.H., Gong H.Y., Lin C.C., Kawakami K., Wu J.L. A zebrafish model of intrahepatic cholangiocarcinoma by dual expression of hepatitis B virus X and hepatitis C virus core protein in liver. Hepatology. 2012;56:2268–2276. doi: 10.1002/hep.25914. [DOI] [PubMed] [Google Scholar]
- 79.Mudbhary R., Sadler K.C. Epigenetics, development, and cancer: zebrafish make their ARK. Birth Defects Res Part C. 2011;93:194–203. doi: 10.1002/bdrc.20207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zheng W., Li Z., Nguyen A.T., Li C., Emelyanov A., Gong Z. Xmrk, Kras and Myc transgenic zebrafish liver cancer models share molecular signatures with subsets of human hepatocellular carcinoma. PLoS One. 2014;9:e91179. doi: 10.1371/journal.pone.0091179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rudolph K.L., Change S., Millard M., Schreiber-Agus N., DePinho R.A. Inhibition of experimental liver cirrhosis in mice by telomerase gene delivery. Science. 2000;287:1253–1258. doi: 10.1126/science.287.5456.1253. [DOI] [PubMed] [Google Scholar]
- 82.Hudson B., Connor S.O., Paulsen A., Purcell N., Rennie G.C., Seah S., Taft H.P., Wang C. A study of the endocrine manifestations of hepatic cirrhosis. Q J Med. 1976;177:145–178. [PubMed] [Google Scholar]
- 83.Kent J.R., Scaramuzzi R.J., Lauwers W., Parlow A.F., Hill M., Penardi R., Hilliard J. Plasma testosterone, estradiol, and gonadotrophins in hepatic insufficiency. Gastroenterology. 1973;64:111–116. [PubMed] [Google Scholar]
- 84.Burk R.F., Hill K.E., Motley A.K., Byrne D.W., Norsworthy B.K. Selenium deficiency occurs in some patients with moderate-to-severe cirrhosis and can be corrected by administration of selenate but not selenomethionine: a randomized controlled trial. Am J Clin Nutr. 2015;102:1126–1133. doi: 10.3945/ajcn.115.110932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mueller S., Sandrin L. Liver stiffness: a novel parameter for the diagnosis of liver disease. Hepat Med. 2010;25:49–67. doi: 10.2147/hmer.s7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Georges P.C., Hui J.-J., Gombos Z., McCormick M.E., Wang A.Y., Uemura M., Mick R., Janmey P.A., Furth E.E., Wells R.G. Increased stiffness of the rat liver precedes matrix deposition: implications for fibrosis. Am J Physiol Gastrointest Liver Physiol. 2007;293:G1147–G1154. doi: 10.1152/ajpgi.00032.2007. [DOI] [PubMed] [Google Scholar]
- 87.Olsen A.L., Bloomer S.A., Chan E.P., Gaça M.D., Georges P.C., Sackey B., Uemura M., Janmey P.A., Wells R.G. Hepatic stellate cells require a stiff environment for myofibroblastic differentiation. Am J Physiol Gastrointest Liver Physiol. 2011;301:G110–G118. doi: 10.1152/ajpgi.00412.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.El-Serag H.B., Rudolph K.L. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 89.Guéchot J., Peigney N., Ballet F., Vaubourdolle M., Giboudeau J., Poupon R. Sex hormone imbalance in male alcoholic cirrhotic patients with and without hepatocellular carcinoma. Cancer. 1988;62:760–762. doi: 10.1002/1097-0142(19880815)62:4<760::aid-cncr2820620420>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 90.Francavilla A., Panella C., Polimeno L., Giangaspero A., Mazzaferro V., Pan C.-E., Van Thiel D.H., Starzl T.E. Hormonal and enzymatic parameters of hepatic regeneration in patients undergoing major liver resections. Hepatology. 1990;12:1134–1138. doi: 10.1002/hep.1840120510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chaturantabut S., Shwartz A., Evason K.J., Cox A.G., Labella K., Schepers A.G., Yang S., Aravena M., Houvras Y., Mancio-Silva L., Romano S., Gorelick D.A., Cohen D.E., Zon L.I., Bhatia S.N., North T.E., Goessling W. Estrogen activation of G protein-coupled estrogen receptor 1 regulates phosphoinositide 3-kinase and mTOR signaling to promote liver growth in zebrafish and proliferation of human hepatocytes. Gastroenterology. 2019;156:1788–1804. doi: 10.1053/j.gastro.2019.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Burk R.F., Early D.S., Hill K.E., Palmer I.S., Boeglin M.E. Plasma selenium in patients with cirrhosis. Hepatology. 1998;27:794–798. doi: 10.1002/hep.510270322. [DOI] [PubMed] [Google Scholar]
- 93.Hatfield D.L., Yoo M.H., Carlson B.A., Gladyshev V.N. Selenoproteins that function in cancer prevention and promotion. Biochim Biophys Acta. 2009;1790:1541–1545. doi: 10.1016/j.bbagen.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cox A.G., Tsomides A., Kim A.J., Saunders D., Hwang K.L., Evason K.J., Heidel J., Brown K.K., Yuan M., Lien E.C., Lee B.C., Nissim S., Dickinson B., Chhangawala S., Chang C.J., Asara J.M., Houvras Y., Gladyshev V.N., Goessling W. Selenoprotein H is an essential regulator of redox homeostasis that cooperates with p53 in development and tumorigenesis. Proc Natl Acad Sci U S A. 2016;113:E5562–E5571. doi: 10.1073/pnas.1600204113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pickett-Blakely O., Young K., Carr R.M. Micronutrients in nonalcoholic fatty liver disease pathogenesis. Clin Mol Gastroenterol Hepatol. 2018;6:451–462. doi: 10.1016/j.jcmgh.2018.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wallace M.C., Friedman S.L. Hepatic fibrosis and the microenvironment: fertile soil for hepatocellular carcinoma development. Gene Expr. 2014;16:77–84. doi: 10.3727/105221614X13919976902057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hernandez-Gea V., Toffanin S., Friedman S.L., Llovet J.M. Role of the microenvironment in the pathogenesis and treatment of hepatocellular carcinoma. Gastroenterology. 2013;144:512–527. doi: 10.1053/j.gastro.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dupont S., Morsut L., Aragona M., Enzo E., Giulitti S., Cordenonsi M., Zanconato F., Le Digabel J., Forcato M., Bicciato S., Elvassore N., Piccolo S. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 99.Xu M.Z., Yao T.J., Lee N.P.Y., Ng I.O.L., Chan Y.T., Zender L., Lowe S.W., Poon R.T.P., Luk J.M. Yes-associated protein is an independent prognostic marker in hepatocellular carcinoma. Cancer. 2009;115:4576–4585. doi: 10.1002/cncr.24495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cox A.G., Hwang K.L., Brown K.K., Evason K.J., Beltz S., Tsomides A., O’Connor K., Galli G.G., Yimlamai D., Chhangawala S., Yuan M., Lien E.C., Wucherpfennig J., Nissim S., Minami A., Cohen D.E., Camargo F.D., Asara J.M., Houvras Y., Stainier D.Y.R., Goessling W. Yap reprograms glutamine metabolism to increase nucleotide biosynthesis and enable liver growth. Nat Cell Biol. 2016;18:886–896. doi: 10.1038/ncb3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cox A.G., Tsomides A., Yimlamai D., Hwang K.L., Miesfeld J., Galli G.G., Fowl B.H., Fort M., Ma K.Y., Sullivan M.R., Hosios A.M., Snay E., Yuan M., Brown K.K., Lien E.C., Chhangawala S., Steinhauser M.L., Asara J.M., Houvras Y., Link B., Vander Heiden M.G., Camargo F.D., Goessling W. Yap regulates glucose utilization and sustains nucleotide synthesis to enable organ growth. EMBO J. 2018;37:e100294. doi: 10.15252/embj.2018100294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Matthews R.P., Lorent K., Russo P., Pack M. The zebrafish onecut gene hnf-6 functions in an evolutionarily conserved genetic pathway that regulates vertebrate biliary development. Dev Biol. 2004;274:245–259. doi: 10.1016/j.ydbio.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 103.Lorent K., Moore J.C., Siekmann A.F., Lawson N., Pack M. Reiterative use of the Notch signal during zebrafish intrahepatic biliary development. Dev Dyn. 2010;239:855–864. doi: 10.1002/dvdy.22220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ellis J.L., Bove K.E., Schuetz E.G., Leino D., Valencia C.A., Schuetz J.D., Miethke A., Yin C. Zebrafish abcb11b mutant reveals strategies to restore bile excretion impaired by bile salt export pump deficiency. Hepatology. 2018;67:1531–1545. doi: 10.1002/hep.29632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yin C., Evason K.J., Maher J.J., Stainier D.Y.R. The basic helix-loop-helix transcription factor, heart and neural crest derivatives expressed transcript 2, marks hepatic stellate cells in zebrafish: analysis of stellate cell entry into the developing liver. Hepatology. 2012;56:1958–1970. doi: 10.1002/hep.25757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Huang M., Chang A., Choi M., Zhou D., Anania F.A., Shin C.H. Antagonistic interaction between Wnt and Notch activity modulates the regenerative capacity of a zebrafish fibrotic liver model. Hepatology. 2014;60:1753–1766. doi: 10.1002/hep.27285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Menke A.L., Spitsbergen J.M., Wolterbeek A.P.M., Woutersen R.A. Normal anatomy and histology of the adult zebrafish. Toxicol Pathol. 2011;39:759–775. doi: 10.1177/0192623311409597. [DOI] [PubMed] [Google Scholar]
- 108.Yan C., Huo X., Wang S., Feng Y., Gong Z. Stimulation of hepatocarcinogenesis by neutrophils upon induction of oncogenic kras expression in transgenic zebrafish. J Hepatol. 2015;63:420–428. doi: 10.1016/j.jhep.2015.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sakaguchi T.F., Sadler K.C., Crosnier C., Stainier D.Y.R. Endothelial signals modulate hepatocyte apicobasal polarization in zebrafish. Curr Biol. 2008;18:1565–1571. doi: 10.1016/j.cub.2008.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jin S.W., Beis D., Mitchell T., Chen J.N., Stainier D.Y.R. Cellular and molecular analyses of vascular tube and lumen formation in zebrafish. Development. 2005;132:5199–5209. doi: 10.1242/dev.02087. [DOI] [PubMed] [Google Scholar]
- 111.Wilkins B.J., Gong W., Pack M. A novel keratin18 promoter that drives reporter gene expression in the intrahepatic and extrahepatic biliary system allows isolation of cell-type specific transcripts from zebrafish liver. Gene Expr Patterns. 2014;14:62–68. doi: 10.1016/j.gep.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ellett F., Pase L., Hayman J.W., Andrianopoulos A., Lieschke G.J. Mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood. 2011;117:e49–e56. doi: 10.1182/blood-2010-10-314120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Walton E.M., Cronan M.R., Beerman R.W., Tobin D.M. The macrophage-specific promoter mfap4 allows live, long-term analysis of macrophage behavior during mycobacterial infection in zebrafish. PLoS One. 2015;10:e0138949. doi: 10.1371/journal.pone.0138949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hall C., Flores M., Storm T., Crosier K., Crosier P. The zebrafish lysozyme C promoter drives myeloid-specific expression in transgenic fish. BMC Dev Biol. 2007;7:1–17. doi: 10.1186/1471-213X-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mathias J.R., Perrin B.J., Liu T.-X., Kanki J., Look A.T., Huttenlocher A. Resolution of inflammation by retrograde chemotaxis of neutrophils in transgenic zebrafish. J Leukoc Biol. 2006;80:1281–1288. doi: 10.1189/jlb.0506346. [DOI] [PubMed] [Google Scholar]
- 116.Wells R.G., Schwabe R.F. Origin and function of myofibroblasts in the liver. Semin Liver Dis. 2015;35:97–106. doi: 10.1055/s-0035-1550061. [DOI] [PubMed] [Google Scholar]
- 117.Krenkel O., Tacke F. Liver macrophages in tissue homeostasis and disease. Nat Rev Immunol. 2017;17:306–321. doi: 10.1038/nri.2017.11. [DOI] [PubMed] [Google Scholar]
- 118.Kubo N., Araki K., Kuwano H., Shirabe K. Cancer-associated fibroblasts in hepatocellular carcinoma. World J Gastroenterol. 2016;22:6841–6850. doi: 10.3748/wjg.v22.i30.6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Affo S., Yu L.-X., Schwabe R.F. The role of cancer-associated fibroblasts and fibrosis in liver cancer. Annu Rev Pathol Mech Dis. 2017;12:153–186. doi: 10.1146/annurev-pathol-052016-100322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Huang L., Xu A.M., Liu S., Liu W., Li T.J. Cancer-associated fibroblasts in digestive tumors. World J Gastroenterol. 2014;20:17804–17818. doi: 10.3748/wjg.v20.i47.17804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ringelhan M., Pfister D., O’Connor T., Pikarsky E., Heikenwalder M. The immunology of hepatocellular carcinoma. Nat Immunol. 2018;19:222–232. doi: 10.1038/s41590-018-0044-z. [DOI] [PubMed] [Google Scholar]
- 122.Shirabe K., Mano Y., Muto J., Matono R., Motomura T., Toshima T., Takeishi K., Uchiyama H., Yoshizumi T., Taketomi A., Morita M., Tsujitani S., Sakaguchi Y., Maehara Y. Role of tumor-associated macrophages in the progression of hepatocellular carcinoma. Surg Today. 2012;42:1–7. doi: 10.1007/s00595-011-0058-8. [DOI] [PubMed] [Google Scholar]
- 123.de Oliveira S., Houseright R.A., Graves A.L., Golenberg N., Korte B.G., Miskolci V., Huttenlocher A. Metformin modulates innate immune-mediated inflammation and early progression of NAFLD-associated hepatocellular carcinoma in zebrafish. J Hepatol. 2019;70:710–721. doi: 10.1016/j.jhep.2018.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Li Y., Li H., Spitsbergen J.M., Gong Z. Males develop faster and more severe hepatocellular carcinoma than females in krasV12 transgenic zebrafish. Sci Rep. 2017;7:1–10. doi: 10.1038/srep41280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yan C., Yang Q., Gong Z. Tumor-associated neutrophils and macrophages promote gender disparity in hepatocellular carcinoma in zebrafish. Cancer Res. 2017;77:1395–1407. doi: 10.1158/0008-5472.CAN-16-2200. [DOI] [PubMed] [Google Scholar]
- 126.Yang Q., Yan C., Yin C., Gong Z. Serotonin activated hepatic stellate cells contribute to sex disparity in hepatocellular carcinoma. Clin Mol Gastroenterol Hepatol. 2017;3:484–499. doi: 10.1016/j.jcmgh.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ebrahimkhani M.R., Oakley F., Murphy L.B., Mann J., Moles A., Perugorria M.J., Ellis E., Lakey A.F., Burt A.D., Douglass A., Wright M.C., White S.A., Jaffré F., Maroteaux L., Mann D.A. Stimulating healthy tissue regeneration by targeting the 5-HT2B receptor in chronic liver disease. Nat Med. 2011;17:1668–1673. doi: 10.1038/nm.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tsai S.B., Lin M.C., Baughman A.M., Kishi S., Goto T., Uchiyama J. The zebrafish as a vertebrate model of functional aging and very gradual senescence. Exp Gerontol. 2003;38:777–786. doi: 10.1016/s0531-5565(03)00108-6. [DOI] [PubMed] [Google Scholar]
- 129.Gerhard G.S., Cheng K.C. A call to fins! Zebrafish as a gerontological model. Aging Cell. 2002;1:104–111. doi: 10.1046/j.1474-9728.2002.00012.x. [DOI] [PubMed] [Google Scholar]
- 130.Di Cicco E., Tozzini E.T., Rossi G., Cellerino A. The short-lived annual fish Nothobranchius furzeri shows a typical teleost aging process reinforced by high incidence of age-dependent neoplasias. Exp Gerontol. 2011;46:249–256. doi: 10.1016/j.exger.2010.10.011. [DOI] [PubMed] [Google Scholar]
- 131.Czaja M.J., Ding W.X., Donohue T.M., Friedman S.L., Kim J.S., Komatsu M., Lemasters J.J., Lemoine A., Lin J.D., Ou J.H.J., Perlmutter D.H., Randall G., Ray R.B., Tsung A., Yin X.M. Functions of autophagy in normal and diseased liver. Autophagy. 2013;9:1131–1158. doi: 10.4161/auto.25063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Inami Y., Waguri S., Sakamoto A., Kouno T., Nakada K., Hino O., Watanabe S., Ando J., Iwadate M., Yamamoto M., Lee M.S., Tanaka K., Komatsu M. Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J Cell Biol. 2011;193:275–284. doi: 10.1083/jcb.201102031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Takamura A., Komatsu M., Hara T., Sakamoto A., Kishi C., Waguri S., Eishi Y., Hino O., Tanaka K., Mizushima N. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Qu X., Yu J., Bhagat G., Furuya N., Hibshoosh H., Troxel A., Rosen J., Eskelinen E., Mizushima N., Ohsumi Y., Cattoretti G., Levine B. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hernández-Gea V., Hilscher M., Rozenfeld R., Lim M.P., Nieto N., Werner S., Devi L.A., Friedman S.L. Endoplasmic reticulum stress induces fibrogenic activity in hepatic stellate cells through autophagy. J Hepatol. 2013;59:98–104. doi: 10.1016/j.jhep.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kim R.S., Hasegawa D., Goossens N., Tsuchida T., Athwal V., Sun X., Robinson C.L., Bhattacharya D., Chou H., Zhang D.Y., Fuchs B.C., Lee Y., Hoshida Y., Friedman S.L. The XBP1 arm of the unfolded protein response induces fibrogenic activity in hepatic stellate cells through autophagy. Sci Rep. 2016;6:39342. doi: 10.1038/srep39342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hernndezgea V., Ghiassinejad Z., Rozenfeld R., Gordon R., Fiel M.I., Yue Z., Czaja M.J., Friedman S.L. Autophagy releases lipid that promotes fibrogenesis by activated hepatic stellate cells in mice and in human tissues. Gastroenterology. 2012;142:938–946. doi: 10.1053/j.gastro.2011.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lodder J., Denaës T., Chobert M.N., Wan J.H., El-Benna J., Pawlotsky J.M., Lotersztajn S., Teixeira-Clerc F. Macrophage autophagy protects against liver fibrosis in mice. Autophagy. 2015;11:1280–1292. doi: 10.1080/15548627.2015.1058473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang X., Ribeiro M., Iracheta-Vellve A., Lowe P., Ambade A., Satishchandran A., Bukong T., Catalano D., Kodys K., Szabo G. Macrophage-specific HIF-1α contributes to impaired autophagic flux in non-alcoholic steatohepatitis. Hepatology. 2018;69:545–563. doi: 10.1002/hep.30215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mizushima N., Yamamoto A., Matsui M., Yoshimori T., Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.He C., Bartholomew C.R., Zhou W., Klionsky D.J. Assaying autophagic activity in transgenic GFP-Lc3 and GFP-Gabarap zebrafish embryos. Autophagy. 2009;5:520–526. doi: 10.4161/auto.5.4.7768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Cui J., Sim T.H.F., Gong Z., Shen H.M. Generation of transgenic zebrafish with liver-specific expression of EGFP-Lc3: a new in vivo model for investigation of liver autophagy. Biochem Biophys Res Commun. 2012;422:268–273. doi: 10.1016/j.bbrc.2012.04.145. [DOI] [PubMed] [Google Scholar]
- 143.Kaizuka T., Morishita H., Hama Y., Tsukamoto S., Matsui T., Toyota Y., Kodama A., Ishihara T., Mizushima T., Mizushima N. An autophagic flux probe that releases an internal control. Mol Cell. 2016;64:835–849. doi: 10.1016/j.molcel.2016.09.037. [DOI] [PubMed] [Google Scholar]
- 144.Hara T., Takamura A., Kishi C., Iemura S.I., Natsume T., Guan J.L., Mizushima N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Anding A.L., Baehrecke E.H. Cleaning house: selective autophagy of organelles. Dev Cell. 2017;41:10–22. doi: 10.1016/j.devcel.2017.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ding W., Li M., Chen X., Ni H., Lin C., Gao W., Lu B., Stolz D.B., Clemens D.L., Yin X. Autophagy reduces acute ethanol-induced hepatotoxicity and steatosis in mice. Gastroenterology. 2010;139:1740–1752. doi: 10.1053/j.gastro.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Williams J.A., Ni H.M., Haynes A., Manley S., Li Y., Jaeschke H., Ding W.X. Chronic deletion and acute knockdown of Parkin have differential responses to acetaminophen-induced mitophagy and liver injury in mice. J Biol Chem. 2015;290:10934–10946. doi: 10.1074/jbc.M114.602284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ni H.M., Williams J.A., Jaeschke H., Ding W.X. Zonated induction of autophagy and mitochondrial spheroids limits acetaminophen-induced necrosis in the liver. Redox Biol. 2013;1:427–432. doi: 10.1016/j.redox.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gao Y., Chu S., Zhang Z., Zuo W., Xia C., Ai Q., Luo P., Cao P., Chen N. Early stage functions of mitochondrial autophagy and oxidative stress in acetaminophen-induced liver injury. J Cell Biochem. 2016;118:3130–3141. doi: 10.1002/jcb.25788. [DOI] [PubMed] [Google Scholar]
- 150.Howarth D.L., Passeri M., Sadler K.C. Drinks like a fish: using zebrafish to understand alcoholic liver disease. Alcohol Clin Exp Res. 2011;35:826–829. doi: 10.1111/j.1530-0277.2010.01407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.North T.E., Babu I.R., Vedder L.M., Lord A.M., Wishnok J.S., Tannenbaum S.R., Zon L.I., Goessling W. PGE2-regulated wnt signaling and N-acetylcysteine are synergistically hepatoprotective in zebrafish acetaminophen injury. Proc Natl Acad Sci U S A. 2010;107:17315–17320. doi: 10.1073/pnas.1008209107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tsedensodnom O., Vacaru A.M., Howarth D.L., Yin C., Sadler K.C. Ethanol metabolism and oxidative stress are required for unfolded protein response activation and steatosis in zebrafish with alcoholic liver disease. Dis Model Mech. 2013;6:1213–1226. doi: 10.1242/dmm.012195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Passeri M.J., Cinaroglu A., Gao C., Sadler K.C. Hepatic steatosis in response to acute alcohol exposure in zebrafish requires sterol regulatory element binding protein activation. Hepatology. 2009;49:443–452. doi: 10.1002/hep.22667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lin J.-N., Chang L.-L., Lai C.-H., Lin K.-J., Lin M.-F., Yang C.-H., Lin H.-H., Chen Y.-H. Development of an animal model for alcoholic liver disease in zebrafish. Zebrafish. 2015;12:271–280. doi: 10.1089/zeb.2014.1054. [DOI] [PubMed] [Google Scholar]
- 155.Dlugos C.A., Brown S.J., Rabin R.A. Gender differences in ethanol-induced behavioral sensitivity in zebrafish. Alcohol. 2011;45:11–18. doi: 10.1016/j.alcohol.2010.08.018. [DOI] [PubMed] [Google Scholar]
- 156.Jain I.H., Zazzeron L., Goli R., Alexa K., Schatzman-Bone S., Dhillon H., Goldberger O., Peng J., Shalem O., Sanjana N.E., Zhang F., Goessling W., Zapol W.M., Mootha V.K. Hypoxia as a therapy for mitochondrial disease. Science. 2016;352:54–61. doi: 10.1126/science.aad9642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lam P.-Y., Peterson R.T. Developing zebrafish disease models for in vivo small molecule screens. Curr Opin Chem Biol. 2019;50:37–44. doi: 10.1016/j.cbpa.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Gillis A.J., Schuller A.P., Skordalakes E. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature. 2008;455:633–637. doi: 10.1038/nature07283. [DOI] [PubMed] [Google Scholar]