Abstract
Background & Aims
CD44 variant 9 (CD44v9)-positive cancer stem-like cells strongly contribute to the development and recurrence of gastric cancer. However, the origin of CD44v9-positive cells is uncertain.
Methods
CD44v9, β-catenin, and epithelial splicing regulatory protein 1 signals were assessed by real-time reverse-transcription polymerase chain reaction, immunoblot analysis, or immunofluorescence microscopy. Capping actin protein of muscle Z-line α subunit 1 (CAPZA1) expression was assessed by immunoblot analysis or immunohistochemical analysis of Mongolian gerbils' gastric mucosa or human biopsy specimens. Levels of oxidative stress were assessed by measuring malondialdehyde and protein carbonylation. Histone H3 acetylation levels in the CAPZA1 proximal promoter region were measured by using chromatin immunoprecipitation analysis with an antibody against the acetylated histone H3 in human gastric carcinoma cell line (AGS) cells.
Results
CD44v9 is expressed in CAPZA1-overexpressing cells in human gastric cancer tissues. CAPZA1 overexpression enhanced expression of β-catenin, which is a transcription factor for CD44, and epithelial splicing regulatory protein 1, which increases alternative splicing of CD44 to generate CD44v9. CAPZA1-overexpressing cells after cytotoxin-associated gene A accumulation showed CD44v9 expression by inducing nuclear accumulation of β-catenin, concomitant with the enhancement of expression of Sal-like protein 4 and Krüppel-like factor 5, which encode reprogramming factors. Oxidative stress increased the CAPZA1 expression in AGS cells through the enhancement of histone H3 acetylation of CAPZA1 promoter. CAPZA1 expression was increased depending on oxidative stress in H pylori–infected gastric mucosa.
Conclusions
CD44v9 expression is evoked from CAPZA1-overexpressing cells after accumulation of cytotoxin-associated gene A. Our findings provide important insights into the mechanisms underlying the development of CD44v9-positive cells.
Keywords: Autophagy, CagA, Cancer Stem Cells, CD44v9, Eradication Therapy
Abbreviations used in this paper: AGS, human gastric carcinoma cell line; CAPZA1, capping actin protein of muscle Z-line α subunit 1; CagA, cytotoxin-associated gene A; cagPAI, cytotoxin-associated gene-pathogenicity island; CD44v9, CD44 variant 9; ChIP, chromatin immunoprecipitation; ESRP1, epithelial splicing regulatory protein 1; FBS, fetal bovine serum; KLF5, Krüppel-like factor 5; mRNA, messenger RNA; qPCR, quantitative polymerase chain reaction; SALL4, Sal-like protein 4; siRNA, small interfering RNA; WT-A10, pTet-off-cagA transfected MKN28 cells
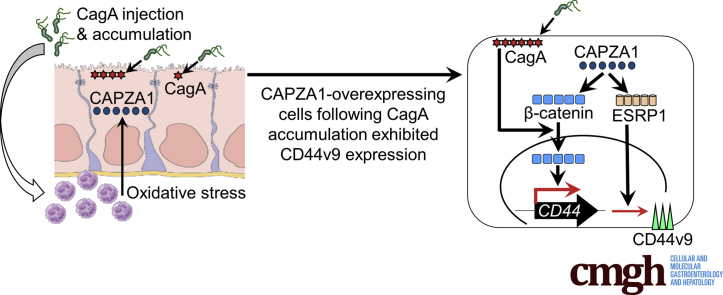
Graphical abstract
See editorial on page 528.
Summary.
Sal-like protein 4 and Krüppel-like factor 5 expression is strongly up-regulated in capping actin protein of muscle Z-line α subunit 1 (CAPZA1)-overexpressing cells after Helicobacter pylori infection. High epithelial splicing regulatory protein 1 expression in CAPZA1-overexpressing cells infected with H pylori induces CD44 variant 9 expression. CD44 variant 9–positive gastric cancer stem-like cells are developed from CAPZA1-overexpressing cells infected with H pylori.
Cancer stem cells are found in several cancer tissues and are considered to facilitate tumorigenesis because of their abilities to self-renew and transform into multiple cell types.1 CD44 is a cell-surface marker implicated in the development of cancer stem cells. Several isoforms of CD44 are generated by alternative splicing of its variant exons.2 CD44 variant 9 (CD44v9)-positive cancer stem-like cells show enhanced metastasis and treatment resistance.3, 4 These cells strongly contribute to the development and recurrence of gastric cancer.5, 6, 7 However, the origin of CD44v9-positive cells is uncertain.
Large-scale prospective cohort studies have shown a close relationship between chronic Helicobacter pylori infection and gastric carcinogenesis.8, 9 The H pylori–derived virulence factor cytotoxin-associated gene A (CagA) translocates into host epithelial cells via the bacterial type IV secretion system and functions as an oncogenic driver in gastric epithelial cells.10, 11, 12 CagA causes reprogramming and de-differentiation of fully differentiated cells into tissue stem-like precursor cells with cancer stem cell properties.13, 14, 15 However, translocated CagA usually is degraded by autophagy.16 Therefore, CagA must evade autophagic degradation and accumulate in host epithelial cells to promote their reprogramming and differentiation into tissue stem-like precursor cells with cancer stem cell properties. Indeed, our previous report showed that translocated CagA accumulates in CD44v9-positive cancer stem-like cells in human gastric cancer tissues by evading autophagic degradation.16
Capping actin protein of muscle Z-line α subunit 1 (CAPZA1) plays an important role in the regulation of actin polymerization and cell motility by binding to the barbed ends of actin filaments.17, 18, 19, 20 In addition to these functions, we previously showed that CAPZA1 negatively regulates autophagy by repressing expression of lysosomal-associated membrane protein 1.21 Consequently, the autophagic pathway is repressed because of inhibition of autolysosome formation in CAPZA1-overexpressing gastric epithelial cells infected with H pylori, resulting in accumulation of translocated CagA.21 Therefore, the presence of CAPZA1-overexpressing cells in H pylori–infected gastric mucosa is thought to increase the risk of gastric carcinogenesis. However, it is uncertain how accumulation of CagA in CAPZA1-overexpressing cells is associated with the development of gastric cancer. Here, we show that CAPZA1 enhances expression of β-catenin, which is a transcription factor for CD44, and epithelial splicing regulatory protein 1 (ESRP1), which increases alternative splicing of CD44 to generate CD44v9. Consequently, CAPZA1-overexpressing cells become CD44v9-positive cells after accumulation of CagA.
Results
CD44v9 Expression Is Induced Specifically in CAPZA1-Overexpressing Gastric Epithelial Cells on H pylori Infection
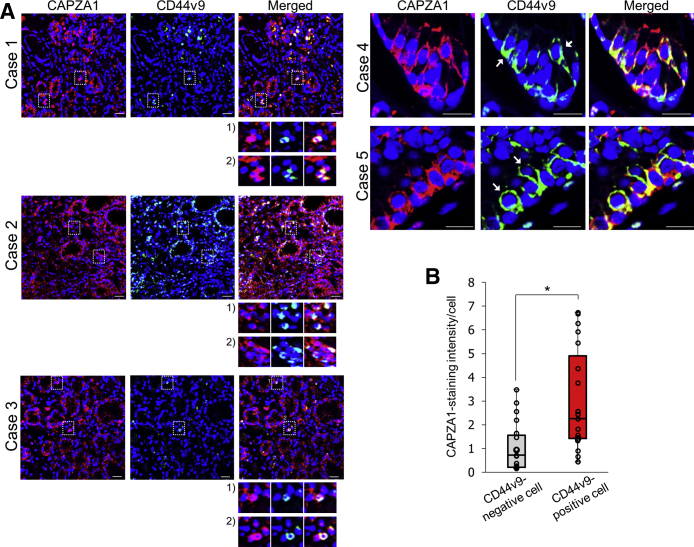
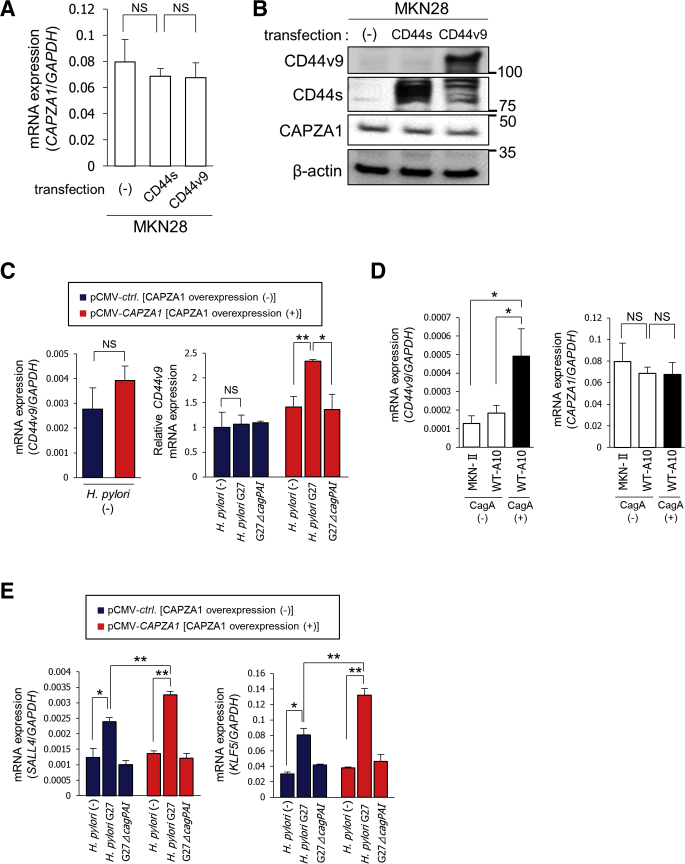
We previously reported that the H pylori–derived oncoprotein CagA, which usually is degraded by autophagy, is stabilized in CD44v9-positive cancer stem-like cells in human gastric cancer tissues.16 In addition, we recently reported that CagA evades autophagic degradation and accumulates in CAPZA1-overexpressing cells in human gastric cancer tissues, indicating that the presence of CAPZA1-overexpressing cells in H pylori–infected gastric mucosa increases the risk of gastric carcinogenesis.21 Based on our previous reports, we hypothesized that CD44v9 expression is enhanced in CAPZA1-overexpressing cells in H pylori–infected gastric mucosa. To test this hypothesis, we assessed whether CD44v9 is expressed in CAPZA1-overexpressing cells in gastric cancer tissues obtained from 5 cases of human gastric adenocarcinoma (cases 1, 2, and 3 showed poorly differentiated adenocarcinoma; cases 4 and 5 showed well-differentiated adenocarcinoma). CD44v9 expression was detected in cells strongly stained for CAPZA1 (CAPZA1-overexpressing cells) (Figure 1A). CAPZA1-overexpressing cells were detected consistently across the gastric cancer tumors, and CD44v9-positive cells selectively were detected among CAPZA1-overexpressing cells (Figure 1A). In addition, the staining intensity of CAPZA1 was significantly higher in CD44v9-positive cells than in adjacent CD44v9-negative cells based on analysis of 19 CD44v9-positive and CD44v9-negative cells in 10 regions of 5 human gastric cancer tissues (cases 1–5) (Figure 1B). These results suggest that CD44v9 and CAPZA1 are co-expressed in human gastric cancer tissues. We subsequently examined whether CD44 expression induces CAPZA1 expression by transfecting human gastric cancer cell line (MKN28) cells with an expression vector harboring the CD44 standard isoform (CD44s) or CD44v9.16 Overexpression of CD44s or CD44v9 did not increase messenger RNA (mRNA) or protein expression of CAPZA1 (Figure 2A and B). Subsequently, to examine the effect of overexpression of CAPZA1 on CD44v9 mRNA expression, we analyzed CD44v9 mRNA expression in pCMA-ctrl–transfected human gastric carcinoma cell line (AGS) cells (AGS cells) and mammalian expression vector (pCMV)-CAPZA1–transfected AGS cells (CAPZA1-overexpressing AGS cells). Overexpression of CAPZA1 did not increase CD44v9 mRNA expression significantly (Figure 2C). We subsequently infected CAPZA1-overexpressing AGS cells with the H pylori G27 strain or H pylori G27 cytotoxin-associated gene-pathogenicity island (cagPAI)-deleted isogenic mutant strain (H pylori G27 ΔcagPAI) for 5 hours and cultured them in antibiotic-containing medium for 24 hours, and then analyzed CD44v9 mRNA expression. H pylori G27 infection did not increase CD44v9 mRNA expression significantly in AGS cells (Figure 2C). By contrast, CD44v9 mRNA expression was significantly higher in CAPZA1-overexpressing AGS cells infected with H pylori G27 than in noninfected CAPZA1-overexpressing AGS cells (Figure 2C). H pylori G27 ΔcagPAI infection did not increase CD44v9 mRNA expression in CAPZA1-overexpressing cells (Figure 2C). Based on these findings, accumulation of CagA caused by H pylori infection of CAPZA1-overexpressing cells are hypothesized to increase CD44v9 mRNA expression. To determine whether CagA accumulation helps to induce CD44v9 mRNA expression, we examined CagA-expressing pTet-off-cagA transfected MKN28 cells (WT-A10) cells, in which forced CagA expression was induced by the pTet-off-cagA expression vector upon culture in the absence of doxycycline for 24 hours.16 Although the level of CD44v9 mRNA expression in WT-A10 cells without stable CagA expression was equivalent to that in MKN-II cells, it was increased significantly in CagA-expressing WT-A10 cells (Figure 2D). We subsequently examined whether CagA accumulation increases CAPZA1 expression. CAPZA1 mRNA expression was not increased in CagA-expressing WT-A10 cells (Figure 2D). These results indicate that accumulation of CagA resulting from H pylori infection in CAPZA1-overexpressing cells induces mRNA expression of CD44v9.
Figure 1.
CD44v9 expression is increased specifically in CAPZA1-overexpressing gastric epithelial cells upon H pylori infection. (A) Immunostaining of CAPZA1 and CD44v9 in human gastric adenocarcinoma. Case 1: 80-year-old man, poorly differentiated adenocarcinoma; case 2: 51-year-old man, poorly differentiated adenocarcinoma; and case 3: 35-year-old man, poorly differentiated adenocarcinoma. Cases 4 and 5 were a 75-year-old man and a 71-year-old man, respectively, who were H pylori–positive and had well-differentiated adenocarcinoma. Red indicates CAPZA1 and green indicates CD44v9. The white box contains CD44v9-positive cells and is enlarged at the bottom of each image. White arrowheads indicate CD44v9-positive cells. Nuclei (blue) were stained with 4′,6-diamidino-2-phenylindole (DAPI). Scale bar: 20 μm. (B) The intensity of CAPZA1 immunostaining per cell in CD44v9-positive cells and adjacent CD44v9-negative cells. By using 19 CD44v9-positive and CD44v9-negative cells detected in 10 regions of 5 human gastric tissues (cases 1–5), the intensity of CAPZA1 staining was quantified using ImageJ. The box-and-whisker plot presents the full range of variation, interquartile range, and median values. The data points indicate the CAPZA1 staining intensity of each cell. *P < .05 by the Student t test.
Figure 2.
Relationship between CD44v9 and CAPZA1 expression upon H pylori infection. (A) MKN28 cells were transfected with the pRC/CMV-CD44s or pRC/CMV-CD44v expression plasmid, and then CAPZA1 mRNA expression was determined by real-time quantitative PCR. Data are presented as the means ± SD of 3 independent assays. P values were calculated by a 1-way analysis of variance. (B) Expression of CAPZA1 in CD44s- and CD44v9-expressing MKN28 cells. (C) AGS cells were transfected with pCMV-ctrl (CAPZA1 overexpression [-]) or pCMV-CAPZA1 (CAPZA1 overexpression [+]), infected with H pylori G27 or H pylori G27 ΔcagPAI for 5 hours (multiplicity of infection, 50), and incubated in antibiotic-containing medium for 24 hours. mRNA expression of CD44v9 was measured by real-time qPCR. Data are presented as the means ± SD of 3 independent assays. ∗P < .05, **P < .01. P values were calculated by the Student t test (left panel) and a 1-way analysis of variance (right panel). (D) MKN28 cells were transfected with pTet-off-ctrl (MKN-II cells) or pTet-off-cagA (WT-A10 cells). CagA expression in WT-A10 cells was induced by culture in the absence of doxycycline for 24 hours. mRNA expression of CD44v9 and CAPZA1 was determined by real-time qPCR. Data are presented as the means ± SD of 3 independent assays. *P < .01. P values were calculated by a 1-way analysis of variance. (E) AGS cells were transfected with pCMV-ctrl or pCMV-CAPZA1, infected with H pylori G27 or H pylori G27 ΔcagPAI for 5 hours (multiplicity of infection, 50), and incubated in antibiotic-containing medium for 24 hours. mRNA expression of SALL4 and KLF5 was determined by real-time qPCR. Data are presented as the means ± SD of 3 independent assays. *P < .05, **P < .01. P values were calculated by a 1-way analysis of variance.
CagA reportedly mediates transactivation of Sal-like protein 4 (SALL4) and Krüppel-like factor 5 (KLF5), which are reprogramming factors that promote the acquisition of stem cell properties in gastric epithelial cells.22 We investigated whether expression of SALL4 and KLF5 was increased in CAPZA1-overexpressing cells infected with H pylori. Infection of H pylori G27 increased mRNA expression of SALL4 and KLF5 in CAPZA1-overexpressing AGS cells by 2.4- and 3.5-fold, respectively (Figure 2E). Furthermore, mRNA expression of SALL4 and KLF5 was significantly higher in CAPZA1-overexpressing AGS cells infected with H pylori G27 than in AGS cells infected with H pylori G27 (Figure 2E). On the other hand, mRNA expression of SALL4 and KLF5 was not increased in CAPZA1-overexpressing AGS cells infected with H pylori G27 ΔcagPAI (Figure 2E).
Increased Expression of ESRP1 and β-Catenin in CAPZA1-Overexpressing Cells, in Combination With Accumulation of CagA, Induces CD44v9 Expression
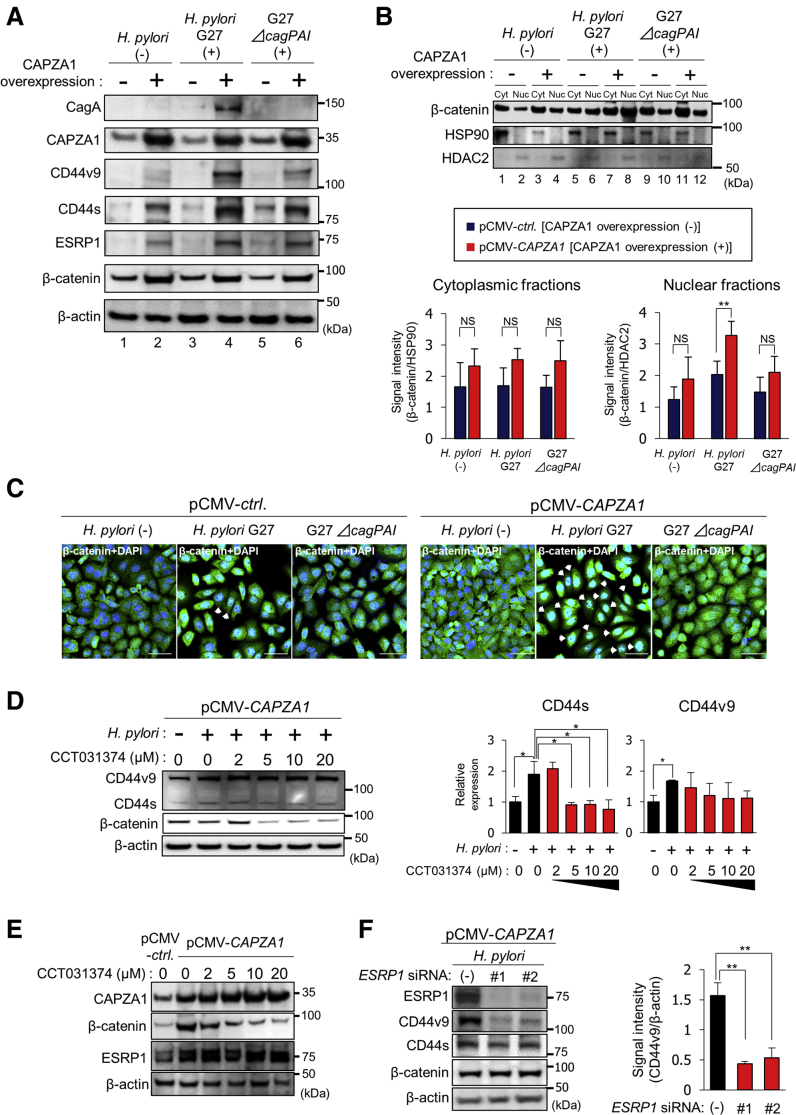
Translocated CagA was strongly detected in CAPZA1-overexpressing cells infected with H pylori G27 and was not detected in H pylori G27-infected AGS cells or H pylori G27 ΔcagPAI-infected cells, confirming that translocated CagA accumulates in CAPZA1-overexpressing AGS cells (Figure 3A). Overexpression of CAPZA1 did not induce CD44v9 expression (Figure 3A, lanes 1 and 2). In addition, H pylori G27 infection did not induce CD44v9 expression in AGS cells (Figure 3A, lanes 1, 3, and 5). By contrast, H pylori G27 infection significantly enhanced CD44v9 expression in CAPZA1-overexpressing AGS cells (Figure 3A, lanes 2 and 4). Expression of CD44s in CAPZA1-overexpressing AGS cells was higher than that in AGS cells and was increased further by H pylori G27 infection (Figure 3A). CD44v9 is generated by alternative splicing of CD44 mediated by ESRP.23 Intriguingly, basal ESRP1 expression was higher in CAPZA1-overexpressing AGS cells than in AGS cells (Figure 3A, lanes 1 and 2). CD44 is a target gene of nuclear β-catenin.24 Basal expression of β-catenin also was higher in CAPZA1-overexpressing AGS cells than in AGS cells (Figure 3A, lanes 1 and 2). CagA is reported to enhance nuclear accumulation of β-catenin.25, 26 H pylori G27 infection tended to increase nuclear accumulation of β-catenin in AGS cells (Figure 3B, lanes 2 and 6, and C). Nuclear accumulation of β-catenin in CAPZA1-overexpressing AGS cells was enhanced further by H pylori G27 infection, but not by H pylori G27 ΔcagPAI infection (Figure 3B and C). This is thought to be because CagA is degraded by autophagy in AGS cells, but escapes from autophagic degradation in CAPZA1-overexpressing AGS cells and thereby strongly induces nuclear translocation of β-catenin. To investigate whether this increase in nuclear accumulation of β-catenin was associated with the induction of CD44 expression, we reduced the nuclear and cytosolic levels of β-catenin in AGS cells via CCT031374 hydrobromide treatment.27 Treatment with CCT031374 at concentrations higher than 5 μmol/L reduced the total β-catenin level in CAPZA1-overexpressing AGS cells infected with H pylori and, consequently, H pylori–mediated induction of CD44 expression was suppressed significantly (Figure 3D). By contrast, CCT031374 treatment did not attenuate the increase in CD44v9 expression significantly (Figure 3D). We subsequently examined ESRP1 expression in CAPZA1-overexpressing AGS cells treated with CCT031374. Treatment with CCT031374 scarcely affected the high expression of ESRP1 in CAPZA1-overexpressing AGS cells (Figure 3E). Based on these results, we hypothesized that up-regulation of CD44v9 expression in CAPZA1-overexpressing cells infected with H pylori is mediated mainly by ESRP1. To test this hypothesis, we assessed the effect of ESRP1 knockdown on up-regulation of CD44v9 expression. To rule out any off-target effects of the small interfering RNA (siRNA), we used 2 different ESRP1-targeting siRNAs (ESRP1 siRNA 1 and ESRP1 siRNA 2). ESRP1 knockdown attenuated the up-regulation of CD44v9 expression in CAPZA1-overexpressing AGS cells infected with H pylori in comparison with control siRNA (Figure 3F). These results indicate that up-regulation of CD44v9 expression in CAPZA1-overexpressing cells is owing mainly to increased expression of ESRP1, but not of β-catenin. Collectively, our findings suggest that CAPZA1 overexpression predisposes cells to develop into CD44v9-positive cells via accumulation of CagA upon H pylori infection.
Figure 3.
Enhanced expression of ESRP1 and β-catenin in CAPZA1-overexpressing cells contributes to the development of CD44v9-positive cells. (A) AGS cells were transfected with pCMV-ctrl or pCMV-CAPZA1, infected with H pylori G27 or H pylori G27 ΔcagPAI for 5 hours (multiplicity of infection, 50), and incubated in antibiotic-containing medium for 24 hours. The intracellular CagA level and expression of CD44v9, CD44s, ESRP1, and β-catenin were examined by Western blot. (B) AGS cells were transfected with pCMV-ctrl or pCMV-CAPZA1, infected with H pylori G27 or H pylori G27 ΔcagPAI for 5 hours (multiplicity of infection, 50), and incubated in antibiotic-containing medium for 24 hours. β-catenin localization in the cytoplasmic (Cyt) and nuclear (Nuc) fractions was examined. Data are presented as the means ± SD of 3 independent assays. **P < .01. P values were calculated by 1-way analysis of variance. (C) AGS cells were transfected with pCMV-ctrl or pCMV-CAPZA1, infected with H pylori G27 or H pylori G27 ΔcagPAI for 5 hours (multiplicity of infection, 50), incubated in antibiotic-containing medium for 24 hours, and stained for β-catenin. Scale bar: 20 μm. (D) AGS cells were transfected with pCMV-CAPZA1, infected with H pylori ATCC700392 for 5 hours (multiplicity of infection, 50), and incubated in antibiotic- and CCT031374-containing medium for 24 hours. Expression of CD44v9 and CD44s was examined. Data are presented as the means ± SD of 3 independent assays. *P < .05. P values were calculated by 1-way analysis of variance. (E) AGS cells were transfected with pCMV-ctrl or pCMV-CAPZA1, and incubated in CCT031374-containing medium for 24 hours. Expression of CAPZA1, β-catenin, and ESRP1 was examined. (F) AGS cells were transfected with pCMV-CAPZA1, subsequently transfected with control siRNA (-) or 2 different ESRP1 siRNAs 1 and 2, infected with H pylori ATCC700392 for 5 hours, and incubated in antibiotic-containing medium for 24 hours. Expression of ESRP1, CD44v9, CD44s, and β-catenin was examined. Data represent the means ± SD of 3 independent assays. **P < .01. P values were calculated by 1-way analysis of variance.
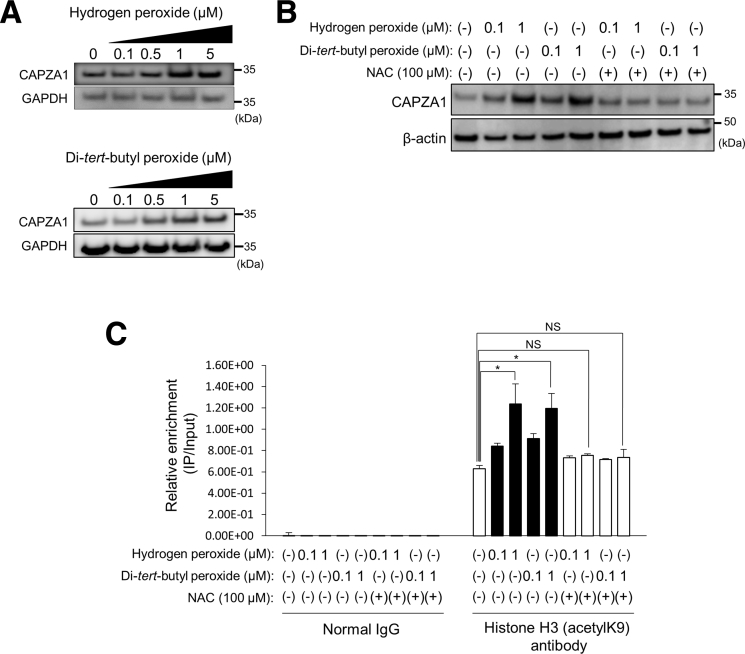
H pylori Infection–Induced Oxidative Stress Increases CAPZA1 Expression in the Gastric Mucosa
Our previous report showed that chronic H pylori infection significantly increases CAPZA1 expression in the gastric mucosa of Mongolian gerbils and that CAPZA1-overexpressing cells are detected throughout the gastric mucosal epithelium from gastric glands to gastric pits.21 By contrast, H pylori infection does not up-regulate CAPZA1 expression in AGS cells in vitro, indicating that it does not directly induce CAPZA1 expression in gastric epithelial cells.21 In the present study, we investigated how CAPZA1 expression is induced in H pylori–infected gastric mucosal epithelium. The inflammatory response in H pylori–infected gastric mucosa, which is induced via reactive oxygen species and reactive nitrogen species generated by neutrophils and monocytes, contributes to the development of H pylori–related disease.28 On the other hand, CD44v9-positive cells show enhanced resistance to reactive oxygen species owing to stabilization of the glutamate-cystine transporter the glutamate-cystine transporter (xCT) at the cytoplasmic membrane.3 Ishimoto et al29 proposed that chronic inflammation induced by H pylori infection leads to the generation of CD44v9-positive cells. We thus hypothesized that this inflammatory response contributes to the induction of CAPZA1 expression, leading to development of CD44v9-positive cells. To investigate this, we examined changes in CAPZA1 expression in AGS cells upon treatment with hydrogen peroxide or di-tert-butyl peroxide. Treatment with hydrogen peroxide or di-tert-butyl peroxide increased CAPZA1 expression in a dose-dependent manner (Figure 4A). In addition, these increases were abrogated by N-acetylcysteine (NAC) treatment (Figure 4B). We previously reported that alteration in the chromatin structure by acetylated histone H3 is involved in induction of CAPZA1 expression.21 We subsequently analyzed alterations in the chromatin structure of the CAPZA1 proximal promoter region in AGS cells treated with hydrogen peroxide or di-tert-butyl peroxide by performing chromatin immunoprecipitation (ChIP) with an antibody against acetylated histone H3. We used quantitative polymerase chain reaction (qPCR) primers that specifically targeted 1 kb downstream of the transcription start site of CAPZA1. The level of acetylated histone H3 was increased significantly upon hydrogen peroxide or di-tert-butyl hydroperoxide treatment, but was unaltered upon NAC treatment (Figure 4C). These observations suggest that oxidative stress induces CAPZA1 expression via enhanced histone H3 acetylation of the CAPZA1 promoter.
Figure 4.
Biological oxidants induce CAPZA1 expression by enhancing histone H3 acetylation of the CAPZA1 promoter. (A) AGS cells were incubated with hydrogen peroxide or di-tert-butyl peroxide for 2 hours, and then the CAPZA1 expression level was analyzed. (B) AGS cells were incubated with NAC for 24 hours before treatment with hydrogen peroxide or di-tert-butyl peroxide, and then CAPZA1 expression was analyzed. (C) AGS cells were incubated with NAC for 24 hours before treatment with hydrogen peroxide or di-tert-butyl peroxide. Thereafter, these cells were analyzed by a ChIP assay with an anti–histone H3 (acetyl K9) antibody or IgG obtained from rabbit serum. Real-time PCR showed the relative enrichment of the CAPZA1 promoter in the DNA fragments pulled down by an anti–histone H3 (acetyl K9) antibody. Data are presented as the means ± SD of 3 independent assays. *P < .05. P values were calculated by a 1-way analysis of variance. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
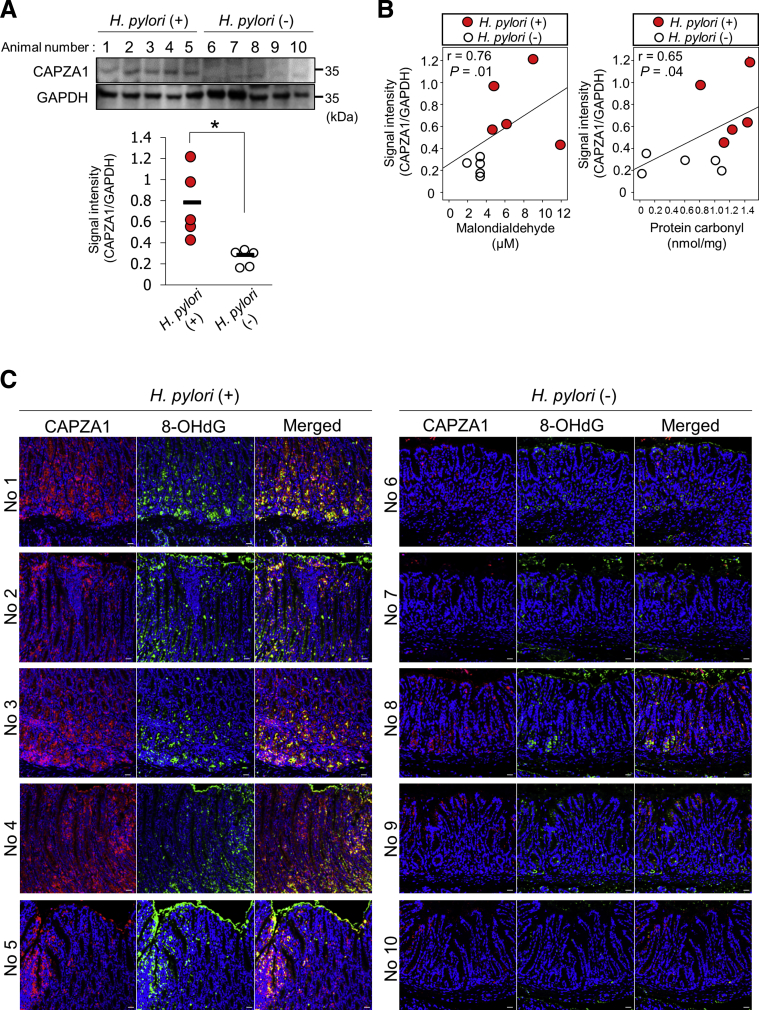
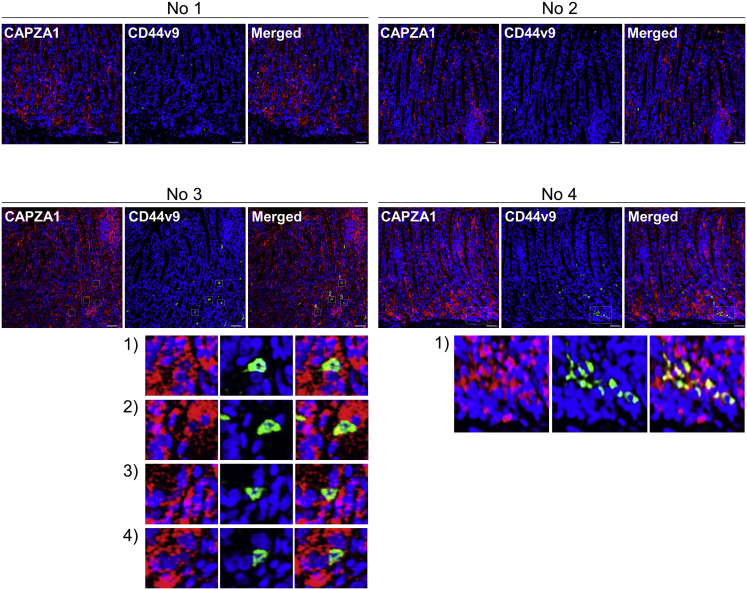
H pylori infection significantly increased CAPZA1 expression in the gastric mucosa of Mongolian gerbils (Figure 5A). Malondialdehyde and protein carbonyl levels are used as biomarkers of oxidative stress. There was a significant linear correlation between CAPZA1 expression and lipid peroxidation (r = 0.76; P = .01), and between CAPZA1 expression and protein carbonylation (r = 0.65; P = .04) (Figure 5B). To visually assess the relationship between oxidative stress and CAPZA1 overexpression in H pylori–infected gastric epithelium, we performed immunostaining of 8-hydroxy-2'-deoxyguanosine, which is produced upon oxidative damage of the nucleoside deoxyguanosine and then excreted extracellularly. CAPZA1-overexpressing cells in H pylori–infected gastric mucosa mainly co-localized with 8-hydroxy-2'-deoxyguanosine–positive cells (Figure 5C, no. 1–4). These results suggest that oxidative stress induced by H pylori infection increases CAPZA1 expression. We subsequently examined whether CD44v9 expression was induced in CAPZA1-overexpressing cells in H pylori–infected gastric mucosa. In gastric mucosa of 2 of 5 Mongolian gerbils, CD44v9-positive cells were detected among CAPZA1-overexpressing cells, indicating that CD44v9 expression is induced not only by CAPZA1 overexpression (Figure 6). When considered together with the in vitro H pylori infection data, adherence of H pylori to CAPZA1-overexpressing cells leading to CagA accumulation in H pylori–infected gastric mucosa is considered to be required for the development of CD44v9-positive cells.
Figure 5.
The CAPZA1 expression level correlates with the level of oxidative stress injury induced by H pylori infection in the gastric mucosa. (A) Mongolian gerbils were inoculated with H pylori KS strains (8 × 108 colony-forming units/mL) (n = 5) or vehicle (n = 5). Seven months later, the animals were killed and their stomachs were excised. Western blot of protein extracts for CAPZA1 was performed. The CAPZA1 signal intensity in each mouse was analyzed using ImageJ. The data points indicate the CAPZA1 signal intensity in each animal. *P < .05. P values were calculated by the Student t test. (B) Levels of malondialdehyde and protein carbonylation in gastric tissues of Mongolian gerbils infected with H pylori KS strains or vehicle were measured. Correlation coefficients between CAPZA1 expression and levels of malondialdehyde and protein carbonylation were determined by correlation analysis. (C) Immunostaining analysis of CAPZA1 and 8-hydroxy-2'-deoxyguanosine (8-OHDG) in gastric mucosa of 10 Mongolian gerbils infected with or without H pylori (5 per group). Red indicates CAPZA1 and green indicates 8-OHDG. Scale bar: 20 μm. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Figure 6.
Representative immunostaining results for co-localization of CAPZA1 and CD44v9 in gastric mucosa of Mongolian gerbils infected with H pylori. CD44v9-positive cells were detected in gastric mucosa of 2 of 5 Mongolian gerbils infected with H pylori (no. 3 and 4). Red indicates CAPZA1 and green indicates CD44v9. The white box contains CD44v9-positive cells and is enlarged at the bottom of each panel. Scale bar: 20 μm.
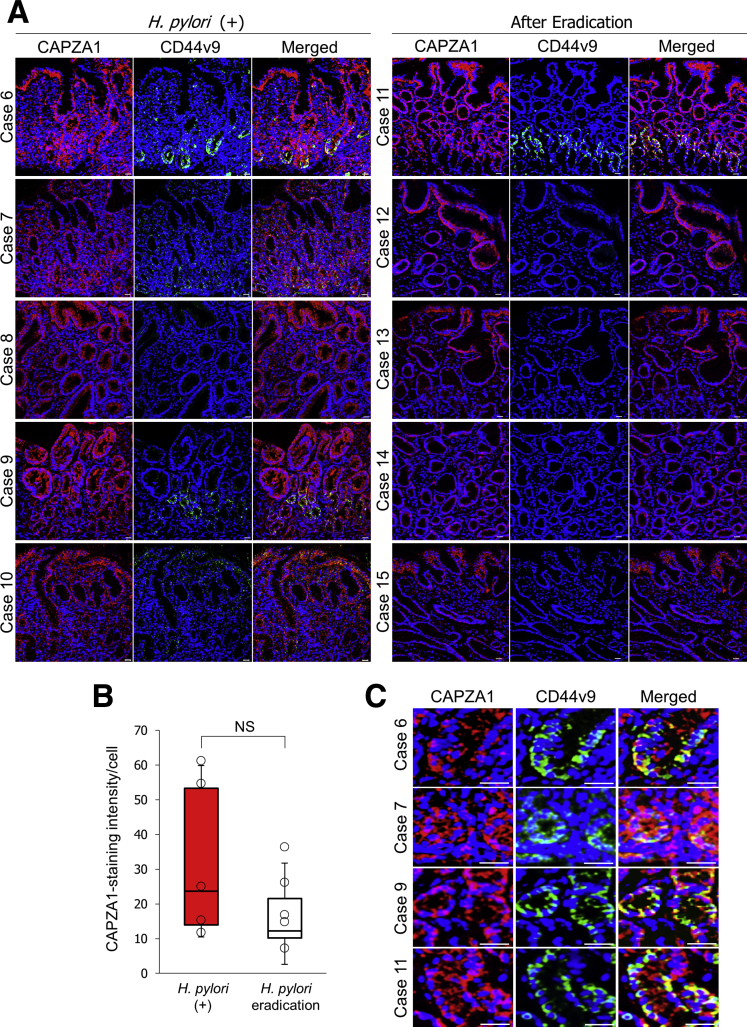
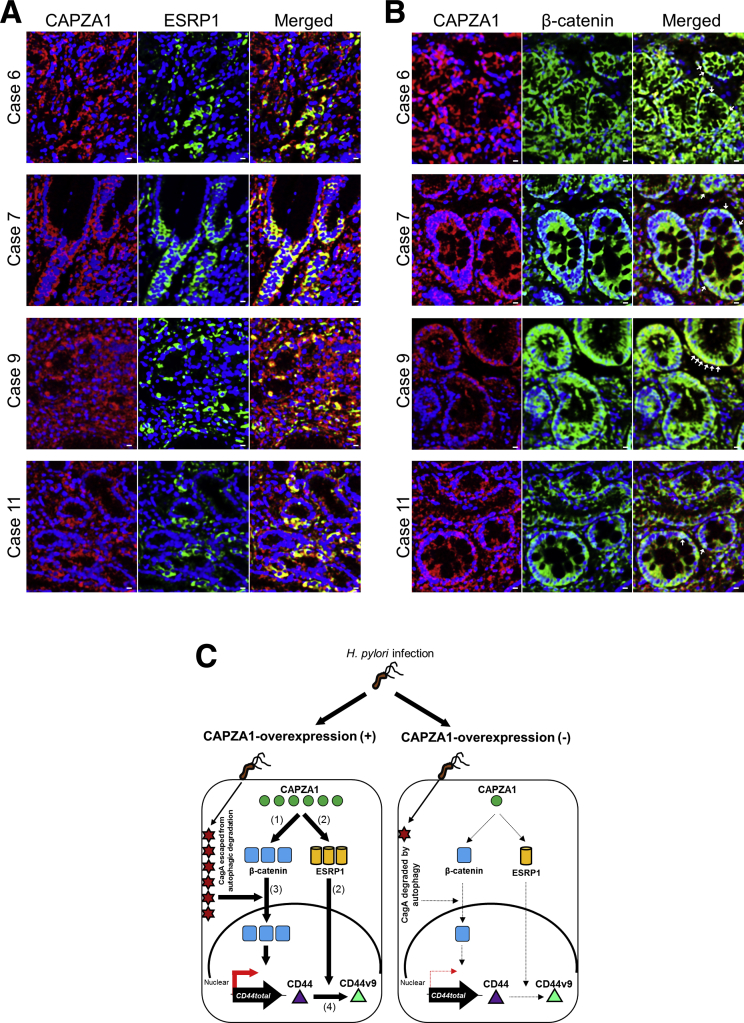
We subsequently investigated the effect of eradication therapy on the level of CAPZA1 expression by examining 5 patients with H pylori infection and 5 patients who had received eradication therapy. CAPZA1-overexpressing cells were detected in tissues not only of H pylori–infected patients, but also of H pylori–eradicated patients (Figure 7A). By using individual images acquired from 10 patients (5 per group), we assessed the staining intensity of CAPZA1 per cell. The data points in Figure 7B represent the average CAPZA1 staining intensity per cell in each individual image. The level of CAPZA1 expression per cell tended to be reduced in gastric biopsy specimens from patients who had received H pylori eradication therapy; however, this difference was not statistically significant (Figure 7B). These biopsy specimens were taken from patients with a mild-to-moderate level of neutrophil infiltration. Therefore, continuation of the inflammatory response after eradication therapy is thought to contribute to induction of CAPZA1 overexpression. CAPZA1 overexpression was detected mainly in gastric pits and gastric gland ductal epithelium, but also was detected in other cell types (Figure 7A). Although these cell types were not identified, we showed that CD44v9 expression was detected in CAPZA1-overexpressing cells in gastric epithelium with or without H pylori eradication therapy and that the expression patterns of CD44v9 and CAPZA1 co-localized (Figure 7A and C, cases 6, 7, 9, and 11). These results suggest that CAPZA1 overexpression in gastric epithelium contributes to the development of CD44v9-positive cells in H pylori–infected human gastric mucosa. Moreover, expression of ESRP1 was detected in gastric tissues containing CD44v9-positive cells and the expression patterns of CAPZA1 and ESRP1 co-localized (Figure 8A). In addition, nuclear localization of β-catenin was detected in CAPZA1-overexpressing cells (Figure 8B). These biopsy specimens were taken from inflamed tissue that did not show dysplasia, and these patients had not progressed to gastric cancer. Nevertheless, CD44v9 expression was detected in CAPZA1-overexpressing cells of tissue that was inflamed owing to H pylori infection (Figure 7). In addition, CD44v9 expression was detected in CAPZA1-overexpressing cells of H pylori–eradicated tissue (Figure 7A and C, case 11), suggesting that accumulation of CagA caused by H pylori infection contributes to induction of CD44v9 expression after eradiation therapy. These findings show that analysis of CAPZA1 expression levels after eradication therapy can provide important insights into the risk of gastric cancer after H pylori eradication.
Figure 7.
Effect of eradication therapy on CAPZA1 expression. (A) Immunostaining analysis of CAPZA1 and CD44v9 in human gastric tissues. Cases 6, 7, 8, 9, 10, 11, 12, 13, 14, and 15 were a 51-year-old woman, a 62-year-old man, a 60-year-old woman, a 60-year-old man, a 54-year-old man, a 68-year-old woman, a 66-year-old man, an 80-year-old woman, a 39-year-old man, and a 74-year-old woman, respectively. Cases 6–10 were H pylori–positive. Cases 11–15 had received eradication therapy. Red indicates CAPZA1 and green indicates CD44v9. Scale bar: 20 μm. (B) The CAPZA1 staining intensity per cell in human gastric mucosa was quantified using image analysis software (Tissue Quest version 4.0; TissueGnostics). The data points indicate the average CAPZA1 staining intensity per cell in each individual image. The box-and-whisker plot presents the full range of variation, interquartile range, and median values. P values were calculated by the Student t test. (C) Representative enlarged fields containing CD44v9-positive cells in human gastric mucosa of cases 6, 7, 9, and 11. Scale bar: 20 μm.
Figure 8.
Relationships between CAPZA1 and ESRP1 expression and nuclear translocation of β-catenin in human gastric mucosa. (A) Immunostaining of CAPZA1 and ESRP1 in human gastric tissues (cases 6, 7, 9, and 11; CD44-positive gastric mucosa). Red indicates CAPZA1 and green indicates ESRP1. Scale bar: 20 μm. (B) Immunostaining of CAPZA1 and β-catenin in human gastric tissues. Red indicates CAPZA1 and green indicates β-catenin. White arrowheads indicate nuclear β-catenin. Scale bar: 20 μm. (C) Schematic representation of the proposed mechanism by which CD44v9 expression is induced in CAPZA1-overexpressing cells infected with H pylori. (1) Overexpression of CAPZA1 enhances expression of β-catenin. (2) Overexpression of CAPZA1 induces high expression of ESRP1. (3) Infection of CAPZA1-overexpressing cells with H pylori aberrantly enhances nuclear translocation of β-catenin owing to accumulation of CagA, leading to induction of CD44total expression. (4) High expression of ESRP1 owing to overexpression of CAPZA1 promotes alternative splicing of CD44total to generate CD44v9.
These findings show that H pylori infection induces CD44v9 expression in CAPZA1-overexpressing cells as follows: (1) overexpression of CAPZA1 enhances expression of β-catenin, (2) overexpression of CAPZA1 induces high ESRP1 expression, (3) H pylori infection of CAPZA1-overexpressing cells enhances nuclear translocation of β-catenin owing to accumulation of CagA, and (4) ESRP1 overexpression promotes alternative splicing of CD44total to generate CD44v9 (Figure 8C). By contrast, in H pylori–infected cells not overexpressing CAPZA1, CD44v9 expression is not induced because CagA is degraded by autophagy, and β-catenin and ESRP1 are lowly expressed (Figure 8C).
Discussion
The present study makes a significant contribution to the field of carcinogenesis in H pylori–infected gastric mucosa. Specifically, our results show a novel and pivotal role of CAPZA1 in CD44v9 expression and show that the presence of CAPZA1-overexpressing cells induces expansion of CD44v9-positive cells. The induction of CD44v9 expression in CAPZA1-overexpressing cells infected with H pylori is caused by aberrant β-catenin signaling via intracellularly accumulated CagA and by alternative splicing of CD44total via overexpression of ESRP1. Accumulation of CagA in CAPZA1-overexpressing cells also aberrantly induced SALL4 and KLF5 expression. These findings show that overexpression of CAPZA1, in combination with accumulation of CagA, predisposes cells to develop into CD44v9-positive cells in H pylori–infected gastric mucosa.
Increased expression of SALL4 and KLF5 induced by CagA promotes an intestinal stem/progenitor-like state and thereby is implicated in intestinal metaplasia of the gastric mucosa.22 This suggests that acquisition of stem cell properties in gastric epithelial cells via CagA-mediated up-regulation of SALL4 and KLF5 is associated with the development of gastric cancer. Here, H pylori infection increased mRNA expression of SALL4 and KLF5 in CAPZA1-overexpressing cells more than in pCMV6-ctrl–transfected cells (Figure 2E). Therefore, H pylori was considered to robustly induce stem cell properties in CAPZA1-overexpressing gastric epithelial cells.
H pylori mainly adheres to mucus pit cells, which are terminally differentiated cells with a short lifespan, and injects CagA via the bacterial type IV secretion system.30 Injected CagA usually is degraded by autophagy.16 Therefore, CagA must evade autophagic degradation to promote reprogramming of mucus pit cells and their de-differentiation into tissue stem-like precursor cells. We previously reported that CAPZA1 also is overexpressed in mucus pit cells and that CagA evades autophagic degradation in CAPZA1-overexpressing cells.21 Therefore, intestinal transdifferentiation of gastric epithelial cells and expansion of CD44v9-positive cells are thought to occur when CAPZA1-overexpressing pit cells are abundant in H pylori–infected gastric mucosa.
H pylori infection induces a chronic inflammatory response by the host.28 Chronic oxidative stress is thought to increase the risk of gastric carcinogenesis by increasing the level of DNA damage and by preventing DNA repair.31 Here, we showed that the levels of CAPZA1 expression and oxidative damage in H pylori–infected gastric mucosa were correlated significantly (Figures 4 and 5). Therefore, eradication therapy is thought to decrease the level of CAPZA1-overexpressing cells in gastric mucosa because it effectively reduces chronic inflammation caused by H pylori infection. In the present study, CAPZA1 expression was decreased after eradication therapy, but this effect was not significant (Figure 7B). The reason for this may be that these biopsy specimens were taken from patients with a mild-to-moderate level of neutrophil infiltration even after eradication therapy. These findings suggest that continuation of the inflammatory response after eradication therapy contributes to formation of CD44v9-positive cells owing to survival of CAPZA1-overexpressing cells with accumulated CagA. In fact, we observed CD44v9-positive cells in gastric tissues from a patient who had received eradiation therapy (Figure 7A and C). Fukase et al32 reported that H pylori infection and gastric cancer are closely related, and that eradication therapy helps to prevent metachronous gastric carcinogenesis. However, the annual incidence of metachronous gastric cancer is up to 2.5%, indicating that gastric cancer develops even after bacterial eradication.33 Our findings suggest that CAPZA1-overexpressing cells remaining in the gastric mucosa after eradication therapy increase the risk of metachronous gastric cancer and that reduction of CAPZA1 expression by amelioration of chronic inflammation after eradication therapy is important to prevent the development of gastric cancer.
The present study focused on whether CAPZA1-overexpressing cells are progenitors of CD44v9-positive cells in H pylori–infected gastric mucosa. CAPZA1-overexpressing cells were detected in both poorly differentiated (cases 1, 2, and 3) and well-differentiated (cases 4 and 5) adenocarcinoma, and these cells expressed CD44v9 (Figure 1). Overall, our data show that CD44v9-positive cells arise from CAPZA1-overexpressing cells in H pylori–infected gastric mucosa. Lee et al34 reported that the appearance of CAPZA1-overexpressing cells is related to well-differentiated tumors, but there is no difference in terms of intestinal- and diffuse-type adenocarcinomas. In addition, Go et al35 showed that the positive expression rate of CD44v9 is increased significantly in well-differentiated adenocarcinoma. Based on these reports, the positive expression rate of CD44v9 may correlate with the appearance of CAPZA1-overexpressing cells, consistent with our results.
Cancer stem cells, which have similar characteristics to tissue stem/progenitor cells,36 are believed to be key tumor-initiating cells and underlie cancer recurrence.37 We previously reported that the rate of gastric cancer recurrence among patients who have undergone endoscopic treatment for primary early gastric cancer is significantly higher in the CD44v9-positive cohorts than in the CD44v9-negative cohorts.5 Here, we show that CAPZA1-overexpressing cells are progenitors of CD44v9-positive cells. Thus, the present study provides important insights into the mechanisms underlying gastric carcinogenesis in H pylori–infected gastric mucosa, which can be used to prevent de novo development and metachronous recurrence of gastric cancer.
Materials and Methods
Cell Culture
The human gastric cancer cell line AGS was purchased from American Type Culture Collection (Rockville, MA) and cultured in RPMI 1640 medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS). WT-A10 and MKN-II cells were maintained in RPMI 1640 medium supplemented with 10% FBS, 10 mmol/L HEPES, 1 mmol/L sodium pyruvate, 0.1 mmol/L nonessential amino acids, 2 mmol/L L-glutamine, 500 μg/mL G418, 100 μg/mL hygromycin B, and 0.2–1 μg/mL doxycycline.16 CagA expression in WT-A10 cells was induced by culture in the absence of doxycycline for 24 hours.16 MKN28 cells cultured in RPMI 1640 medium (Invitrogen) were supplemented with 10% FBS.16
Bacterial Strains and Culture Conditions
H pylori strains ATCC700392, G27, G27 cagPAI-deleted isogenic mutant, KS0651, KS0706, KS0635, KS0691, and KS0791 were cultured on Brucella Agar (Becton-Dickinson, Franklin Lakes, NJ) containing 7% sheep blood and 7% FBS for 2 days at 37°C under microaerobic conditions, which were maintained using AnaeroPack MicroAero (Mitsubishi Gas, Tokyo, Japan). KS numbered strains were isolated from gastric biopsy specimens and stored at -80°C in Brucella Broth (Becton-Dickinson) containing 25% glycerol.
Drugs and Antibodies
CCT031374 hydrobromide (4675; R&D Systems, Minneapolis, MN) was used to inhibit beta-catenin/T-cell factor (TCF)-dependent transcription. Hydrogen peroxide (081-04215; Wako, Tokyo, Japan) and di-tert-butyl peroxide (D3411; Tokyo Kasei Kogyo Co, Tokyo, Japan) were used as reactive oxygen species. N-acetylcysteine (NAC) (A7250; Sigma-Aldrich, St. Louis, MO) was used as a membrane-permeant aminothiol capable of scavenging reactive oxygen species. The following antibodies were used for Western blot: anti–F-actin–capping protein subunit α-1 (AB6016, 1:2000; Merck Millipore, Billerica, MA), anti-CD44v9 (CAC-LKG-M001, 1:1000; Cosmo Bio, Tokyo, Japan), anti-CD44 (HPA005785, 1:1000; Sigma-Aldrich), anti-ESRP1 (HPA023720, 1:1000; Sigma-Aldrich), anti–β-catenin (610154, 1:1000; BD Biosciences, San Diego, CA), anti-heat shock protein 90 (HSP90) (ADI-SPS-771-D, 1:1000; Enzo Life Science, Farmingdale, NY), and anti-HDAC2 (515100, 1:1000; Thermo Fisher Scientific, Waltham, MA). Anti-CD44v9 (CAC-LKG-M001, 1:500; Cosmo Bio), anti–8 hydroxyguanosine (ab10802, 1:200; Abcam, Cambridge, MA), anti-ESRP1 (HPA023720, 1:200; Sigma-Aldrich), anti–β-catenin (610154, 1:200; BD Biosciences), anti-CAPZA1 (OTI2G4) (MA5-25093, 1:500; Thermo Fisher Scientific), and anti–F-actin–capping protein subunit α-1 (AB6016, 1:500; Merck Millipore) antibodies were used for immunohistochemistry, and an anti–β-catenin antibody (610154, 1:50; BD Biosciences) was used for fluorescence immunocytochemistry. Cells were grown to subconfluence and transfected with ESRP1 siRNA 1 (SI04337956, Hs_RBM35A_3; Qiagen, Valencia, CA) and ESRP1 siRNA 2 (SI04178944, Hs_RBM35A_1; Qiagen) using Lipofectamine 2000 (11668027; Thermo Fisher Scientific) according to the manufacturer’s instructions. As a nonsilencing control cells were transfected with AllStars Negative Control siRNA (SI03650318; Qiagen).
In Vitro Infection of H pylori
Cells were incubated with H pylori ATCC700392, G27 (H pylori G27) or G27 cagPAI-deleted isogenic mutant strain (H pylori G27 ΔcagPAI) for 5 hours (multiplicity of infection, 50), and bacteria were killed by incubation in RPMI 1640 medium containing 400 mg/mL kanamycin for 24 hours.
In Vivo Infection of H pylori
Six-week-old Mongolian gerbils (MON/Jms/Gbs) were purchased from Japan SLC Inc (Shizuoka, Japan) and then were inoculated with H pylori KS0651, KS0706, KS0635, KS0691, or KS0791 (8 × 108 colony-forming units/mL each). Seven months later, the animals were killed and their stomachs were excised. To confirm H pylori infection, the number of viable colony-forming units was determined by plating samples on Nissui Helicobacter agar (Nissui Pharmaceutical, Tokyo, Japan). Levels of malondialdehyde and protein carbonylation were measured using a TBARS Assay Kit (Cayman, Ann Arbor, MI) and an OxiSelect Protein Carbonyl Enzyme-Linked Immunosorbent Assay Kit (Cell Biolabs, San Diego, CA), respectively.
ChIP Assay
ChIP was performed using a Simple ChIP Plus Sonication Chromatin IP Kit (56383S; Cell Signaling Technology, Beverly, MA) following the manufacturer’s instructions. Cross-linking was performed using a 1% formaldehyde solution in phosphate-buffered saline. Before immunoprecipitation, each input fraction was saved and used as a positive control. The supernatants were immunoprecipitated with anti–histone H3 (acetyl K9) (ab4441; Abcam) antibodies or IgG from rabbit serum (18140; Sigma-Aldrich) at 4°C overnight. Then, the resulting enriched genomic DNA samples were measured by qPCR using the EpiTect ChIP-qPCR Primer Assay kit for CAPZA1 (GPH100073(-)01A; Qiagen; -1 kb from the transcription start site). These ChIP qPCR primers were predesigned, and qPCR assays were optimized to measure genomic DNA in the region -1 kb from the transcription start site of CAPZA1.
Human Tissue Specimens
Human gastric adenocarcinoma tissue specimens (case 1: 80-year-old man, poorly differentiated adenocarcinoma; case 2: 51-year-old man, poorly differentiated adenocarcinoma; and case 3: 35-year-old man, poorly differentiated adenocarcinoma) were obtained from ReproCELL Incorporated (Yokohama, Japan). Human early gastric adenocarcinoma tissue specimens were obtained from a 75-year-old man (case 4) and a 71-year-old man (case 5), both of whom underwent endoscopic submucosal dissection at Keio University Hospital. The pathologic diagnosis of both specimens was well-differentiated adenocarcinoma according to the Japanese Gastric Cancer Association classification of gastric carcinoma (14th edition). Human gastric biopsy specimens (antrum and body) were obtained via endoscopy. H pylori infection was evaluated by a [13C] urea breath test and histopathologic diagnosis.38 The cut-off value for a negative urea breath test was <2.5%39 (cases 6–10: H pylori–positive; cases 11–15: H pylori–negative). Biopsy specimens were examined histologically using H&E. The severity of neutrophil infiltration was classified into 4 grades (absent, mild, moderate, and severe) according to the updated Sydney system.40 CAPZA1 expression was evaluated immunohistologically. The staining intensity of CAPZA1 per cell was analyzed using image analysis software (Tissue Quest version 4.0; TissueGnostics, Vienna, Austria).
Fluorescence Immunocytochemistry
AGS cells were infected with H pylori for 5 hours, incubated in RPMI 1640 culture medium containing 400 mg/mL kanamycin for 24 hours, washed in phosphate-buffered saline, fixed with 4% paraformaldehyde, and incubated with an anti–β-catenin antibody (Cell Signaling Technology). Alexa Fluor 488–conjugated anti-rabbit IgG (Invitrogen) was used as the secondary antibody. Nuclei (blue) were stained with 4′,6-diamidino-2-phenylindole. Samples were examined using an LSM710 Zeiss (Carl Zeiss Meditec, Jena, Germany) confocal microscope.
Immunohistochemistry
Tissue sections were fixed in 4% paraformaldehyde, depleted of paraffin, and rehydrated using a graded ethanol series. Thereafter, the sections were subjected to antigen retrieval by heating for 10 minutes at 105°C in Target Retrieval Solution (pH 9.0; Dako, Tokyo, Japan) and incubated overnight at 4°C with a primary antibody. Immunoreactivity was detected using Alexa Fluor 568–conjugated anti-rat IgG and Alexa Fluor 488–conjugated anti-rabbit IgG (Invitrogen). Samples were examined using an LSM710 Zeiss confocal microscope.
RNA Isolation and Quantitative Reverse-Transcription PCR
Total RNA was isolated using an SV Total RNA Isolation System (Promega, Madison, WI). Reverse transcription was performed using a PrimeScript Reverse Transcription reagent Kit (Takara, Ohtsu, Japan) in accordance with the manufacturer's guidelines. PCR was performed using a SYBR Premix Ex Taq Perfect Real Time kit (Takara), a Thermal Cycler Dice Real Time System (Takara), and the following primers: CD44total: forward: 5'-CCGCTATGTCCAGAAAGGA-3’, and reverse: 5'-CTGTCTGTGCTGTCGGTGAT-3’; CD44v9: forward: 5'-GGCTTGGAAGAAGATAAAGACC-3’, and reverse: 5'-TGCTTGATGTCAGAGTAGAAGTTG-3’; SALL4: forward: 5'-TCGTCTGCTAGCGCTCTTCAGATC-3’, and reverse: 5'-CGGCGGGCTGAGTTATTGTTCG-3’; KLF5: forward: 5'-TAACCCCGATTTGGAGAAAC-3’, and reverse: 5'-TGGCTTTTCACCAGTGTGAG-3’; CAPZA1: forward: 5'-ACAATCTCCTCAGGGAAGGGG-3’, and reverse: 5'-TGCTTCTTTCCGTAAGTGGTCAAA-3’; and GAPDH: forward: 5'-GACATCAAGAAGGTGGTGAAGCAG-3’, and reverse: 5'-ATACCAGGAAATGAGCTTGACAAA-3’. GAPDH was used as the internal control.
Western Blot
Total protein (10 μg/lane) was separated on a 4%–12% NuPAGE gradient gel (Invitrogen). An anti–β-actin antibody (Sigma-Aldrich) was used as the loading control. Immunoreactive bands were detected by chemiluminescence using ECL Prime (GE Healthcare, Piscataway, NJ). Signals were quantified using ImageJ software (National Institutes of Health, Bethesda, MD).
Study Approval
This study was performed in accordance with the principles of the Declaration of Helsinki. Isolation of H pylori strains from participants and biopsy of gastric mucosal tissues for research use were approved by the Ethics Committee of the Keio University School of Medicine (20130040, 20070068, and 20140102). All participants provided written informed consent. Animal experiments and procedures were approved by the Keio University Animal Research Committee (08080-12).
Statistics
All values are expressed as the means ± SD. Differences between 2 groups were statistically evaluated by the Student t test with JSTAT (Tokyo, Japan) statistical software (version 8.2). For multiple comparisons, a 1-way analysis of variance was used. P < .05 was considered significant. Correlation analysis was performed using SPSS version 22 for Windows (SPSS Inc, Chicago, IL).
Acknowledgments
The authors are grateful to the Collaborative Research Resources, School of Medicine, Keio University, for technical assistance.
Footnotes
Author contributions Hitoshi Tsugawa and Hidekazu Suzuki conceived the study and planned the experiments; Hitoshi Tsugawa, Chihiro Kato, Hideki Mori, and Kaori Kameyama performed the experiments; Hitoshi Tsugawa, Hideki Mori, Juntaro Matsuzaki, Hideyuki Saya, Masanori Hatakeyama, Makoto Suematsu, and Hidekazu Suzuki analyzed the data; and Hitoshi Tsugawa, Hideki Mori, Juntaro Matsuzaki, and Hidekazu Suzuki wrote the manuscript.
Conflicts of interest This author discloses the following: Hidekazu Suzuki has received scholarship funds from Daiichi-Sankyo Co, EA Pharma Co, Otsuka Pharmaceutical Co Ltd, and Tsumura Co to conduct research, and has received service honoraria from Astellas Pharm, Inc, Astra-Zeneca KK, Daiichi-Sankyo Co, EA Pharma Co, Otsuka Pharmaceutical Co Ltd, Takeda Pharmaceutical Co Ltd, Tsumura Co, Mylan EPD Co, and Zeria Pharmaceutical Co Ltd. The remaining authors disclose no conflicts.
Funding This work was supported by Grants-in-Aid sfor Scientific Research (C) 16K08349 and 19K07068 (H.T.), the Yakult Bio-Science Foundation (H.T.), a Grant-in-Aid for Scientific Research (B) 16H05291 (H.Su.), the Ministry of Education, Culture, Sports, Science and Technology (MEXT)-supported program for the Strategic Research Foundation at Private Universities S1411003 (H.Su.), and Keio Gijuku Academic Development Funds (H.Su.).
References
- 1.Reya T., Morrison S.J., Clarke M.F., Weissman I.L. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 2.Dalerba P., Dylla S.J., Park I.K., Liu R., Wang X., Cho R.W., Hoey T., Gurney A., Huang E.H., Simeone D.M., Shelton A.A., Parmiani G., Castelli C., Clarke M.F. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci U S A. 2007;104:10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ishimoto T., Nagano O., Yae T., Tamada M., Motohara T., Oshima H., Oshima M., Ikeda T., Asaba R., Yagi H., Masuko T., Shimizu T., Ishikawa T., Kai K., Takahashi E., Imamura Y., Baba Y., Ohmura M., Suematsu M., Baba H., Saya H. CD44 variant regulates redox status in cancer cells by stabilizing the xCT subunit of system xc(-) and thereby promotes tumor growth. Cancer Cell. 2011;19:387–400. doi: 10.1016/j.ccr.2011.01.038. [DOI] [PubMed] [Google Scholar]
- 4.Miyoshi S., Tsugawa H., Matsuzaki J., Hirata K., Mori H., Saya H., Kanai T., Suzuki H. Inhibiting xCT improves 5-fluorouracil resistance of gastric cancer induced by CD44 variant 9 expression. Anticancer Res. 2018;38:6163–6170. doi: 10.21873/anticanres.12969. [DOI] [PubMed] [Google Scholar]
- 5.Hirata K., Suzuki H., Imaeda H., Matsuzaki J., Tsugawa H., Nagano O., Asakura K., Saya H., Hibi T. CD44 variant 9 expression in primary early gastric cancer as a predictive marker for recurrence. Br J Cancer. 2013;109:379–386. doi: 10.1038/bjc.2013.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kodama H., Murata S., Ishida M., Yamamoto H., Yamaguchi T., Kaida S., Miyake T., Takebayashi K., Kushima R., Tani M. Prognostic impact of CD44-positive cancer stem-like cells at the invasive front of gastric cancer. Br J Cancer. 2017;116:186–194. doi: 10.1038/bjc.2016.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamakawa Y., Kusuhara M., Terashima M., Kinugasa Y., Sugino T., Abe M., Mochizuki T., Hatakeyama K., Kami K., Yamaguchi K. CD44 variant 9 expression as a predictor for gastric cancer recurrence: immunohistochemical and metabolomic analysis of surgically resected tissues. Biomed Res. 2017;38:41–52. doi: 10.2220/biomedres.38.41. [DOI] [PubMed] [Google Scholar]
- 8.Uemura N., Okamoto S., Yamamoto S., Matsumura N., Yamaguchi S., Yamakido M., Taniyama K., Sasaki N., Schlemper R.J. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med. 2001;345:784–789. doi: 10.1056/NEJMoa001999. [DOI] [PubMed] [Google Scholar]
- 9.Hsu P.I., Lai K.H., Hsu P.N., Lo G.H., Yu H.C., Chen W.C., Tsay F.W., Lin H.C., Tseng H.H., Ger L.P., Chen H.C. Helicobacter pylori infection and the risk of gastric malignancy. Am J Gastroenterol. 2007;102:725–730. doi: 10.1111/j.1572-0241.2006.01109.x. [DOI] [PubMed] [Google Scholar]
- 10.Ohnishi N., Yuasa H., Tanaka S., Sawa H., Miura M., Matsui A., Higashi H., Musashi M., Iwabuchi K., Suzuki M., Yamada G., Azuma T., Hatakeyama M. Transgenic expression of Helicobacter pylori CagA induces gastrointestinal and hematopoietic neoplasms in mouse. Proc Natl Acad Sci U S A. 2008;105:1003–1008. doi: 10.1073/pnas.0711183105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hatakeyama M. Oncogenic mechanisms of the Helicobacter pylori CagA protein. Nat Rev Cancer. 2004;4:688–694. doi: 10.1038/nrc1433. [DOI] [PubMed] [Google Scholar]
- 12.Hatakeyama M. Helicobacter pylori CagA and gastric cancer: a paradigm for hit-and-run carcinogenesis. Cell Host Microbe. 2014;15:306–316. doi: 10.1016/j.chom.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 13.Bagnoli F., Buti L., Tompkins L., Covacci A., Amieva M.R. Helicobacter pylori CagA induces a transition from polarized to invasive phenotypes in MDCK cells. Proc Natl Acad Sci U S A. 2005;102:16339–16344. doi: 10.1073/pnas.0502598102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bessede E., Staedel C., Acuna Amador L.A., Nguyen P.H., Chambonnier L., Hatakeyama M., Belleannee G., Megraud F., Varon C. Helicobacter pylori generates cells with cancer stem cell properties via epithelial-mesenchymal transition-like changes. Oncogene. 2014;33:4123–4131. doi: 10.1038/onc.2013.380. [DOI] [PubMed] [Google Scholar]
- 15.Saito Y., Murata-Kamiya N., Hirayama T., Ohba Y., Hatakeyama M. Conversion of Helicobacter pylori CagA from senescence inducer to oncogenic driver through polarity-dependent regulation of p21. J Exp Med. 2010;207:2157–2174. doi: 10.1084/jem.20100602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsugawa H., Suzuki H., Saya H., Hatakeyama M., Hirayama T., Hirata K., Nagano O., Matsuzaki J., Hibi T. Reactive oxygen species-induced autophagic degradation of Helicobacter pylori CagA is specifically suppressed in cancer stem-like cells. Cell Host Microbe. 2012;12:764–777. doi: 10.1016/j.chom.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 17.Matsuo K., Xiang Y., Nakamura H., Masuko K., Yudoh K., Noyori K., Nishioka K., Saito T., Kato T. Identification of novel citrullinated autoantigens of synovium in rheumatoid arthritis using a proteomic approach. Arthritis Res Ther. 2006;8:R175. doi: 10.1186/ar2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hopmann R., Cooper J.A., Miller K.G. Actin organization, bristle morphology, and viability are affected by actin capping protein mutations in Drosophila. J Cell Biol. 1996;133:1293–1305. doi: 10.1083/jcb.133.6.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takeda S., Minakata S., Koike R., Kawahata I., Narita A., Kitazawa M., Ota M., Yamakuni T., Maeda Y., Nitanai Y. Two distinct mechanisms for actin capping protein regulation--steric and allosteric inhibition. PLoS Biol. 2010;8:e1000416. doi: 10.1371/journal.pbio.1000416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bombardier J.P., Eskin J.A., Jaiswal R., Correa I.R., Jr., Xu M.Q., Goode B.L., Gelles J. Single-molecule visualization of a formin-capping protein ‘decision complex' at the actin filament barbed end. Nat Commun. 2015;6:8707. doi: 10.1038/ncomms9707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsugawa H., Mori H., Matsuzaki J., Sato A., Saito Y., Imoto M., Suematsu M., Suzuki H. CAPZA1 determines the risk of gastric carcinogenesis by inhibiting Helicobacter pylori CagA-degraded autophagy. Autophagy. 2019;15:242–258. doi: 10.1080/15548627.2018.1515530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujii Y., Yoshihashi K., Suzuki H., Tsutsumi S., Mutoh H., Maeda S., Yamagata Y., Seto Y., Aburatani H., Hatakeyama M. CDX1 confers intestinal phenotype on gastric epithelial cells via induction of stemness-associated reprogramming factors SALL4 and KLF5. Proc Natl Acad Sci U S A. 2012;109:20584–20589. doi: 10.1073/pnas.1208651109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yae T., Tsuchihashi K., Ishimoto T., Motohara T., Yoshikawa M., Yoshida G.J., Wada T., Masuko T., Mogushi K., Tanaka H., Osawa T., Kanki Y., Minami T., Aburatani H., Ohmura M., Kubo A., Suematsu M., Takahashi K., Saya H., Nagano O. Alternative splicing of CD44 mRNA by ESRP1 enhances lung colonization of metastatic cancer cell. Nat Commun. 2012;3:883. doi: 10.1038/ncomms1892. [DOI] [PubMed] [Google Scholar]
- 24.Wielenga V.J., Smits R., Korinek V., Smit L., Kielman M., Fodde R., Clevers H., Pals S.T. Expression of CD44 in Apc and Tcf mutant mice implies regulation by the WNT pathway. Am J Pathol. 1999;154:515–523. doi: 10.1016/S0002-9440(10)65297-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murata-Kamiya N., Kurashima Y., Teishikata Y., Yamahashi Y., Saito Y., Higashi H., Aburatani H., Akiyama T., Peek R.M., Jr., Azuma T., Hatakeyama M. Helicobacter pylori CagA interacts with E-cadherin and deregulates the beta-catenin signal that promotes intestinal transdifferentiation in gastric epithelial cells. Oncogene. 2007;26:4617–4626. doi: 10.1038/sj.onc.1210251. [DOI] [PubMed] [Google Scholar]
- 26.Franco A.T., Israel D.A., Washington M.K., Krishna U., Fox J.G., Rogers A.B., Neish A.S., Collier-Hyams L., Perez-Perez G.I., Hatakeyama M., Whitehead R., Gaus K., O'Brien D.P., Romero-Gallo J., Peek R.M., Jr. Activation of beta-catenin by carcinogenic Helicobacter pylori. Proc Natl Acad Sci U S A. 2005;102 doi: 10.1073/pnas.0504927102. 10646–10451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ewan K., Pajak B., Stubbs M., Todd H., Barbeau O., Quevedo C., Botfield H., Young R., Ruddle R., Samuel L., Battersby A., Raynaud F., Allen N., Wilson S., Latinkic B., Workman P., McDonald E., Blagg J., Aherne W., Dale T. A useful approach to identify novel small-molecule inhibitors of Wnt-dependent transcription. Cancer Res. 2010;70:5963–5973. doi: 10.1158/0008-5472.CAN-10-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzuki H., Nishizawa T., Tsugawa H., Mogami S., Hibi T. Roles of oxidative stress in stomach disorders. J Clin Biochem Nutr. 2012;50:35–39. doi: 10.3164/jcbn.11-115SR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishimoto T., Izumi D., Watanabe M., Yoshida N., Hidaka K., Miyake K., Sugihara H., Sawayama H., Imamura Y., Iwatsuki M., Iwagami S., Baba Y., Horlad H., Komohara Y., Takeya M., Baba H. Chronic inflammation with Helicobacter pylori infection is implicated in CD44 overexpression through miR-328 suppression in the gastric mucosa. J Gastroenterol. 2015;50:751–757. doi: 10.1007/s00535-014-1019-y. [DOI] [PubMed] [Google Scholar]
- 30.Hatakeyama M. Structure and function of Helicobacter pylori CagA, the first-identified bacterial protein involved in human cancer. Proc Jpn Acad Ser B Phys Biol Sci. 2017;93:196–219. doi: 10.2183/pjab.93.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Butcher L.D., den Hartog G., Ernst P.B., Crowe S.E. Oxidative stress resulting from Helicobacter pylori infection contributes to gastric carcinogenesis. Cell Mol Gastroenterol Hepatol. 2017;3:316–322. doi: 10.1016/j.jcmgh.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fukase K., Kato M., Kikuchi S., Inoue K., Uemura N., Okamoto S., Terao S., Amagai K., Hayashi S., Asaka M., Japan Gast Study Group Effect of eradication of Helicobacter pylori on incidence of metachronous gastric carcinoma after endoscopic resection of early gastric cancer: an open-label, randomised controlled trial. Lancet. 2008;372:392–397. doi: 10.1016/S0140-6736(08)61159-9. [DOI] [PubMed] [Google Scholar]
- 33.Nakajima T., Oda I., Gotoda T., Hamanaka H., Eguchi T., Yokoi C., Saito D. Metachronous gastric cancers after endoscopic resection: how effective is annual endoscopic surveillance? Gastric Cancer. 2006;9:93–98. doi: 10.1007/s10120-006-0372-9. [DOI] [PubMed] [Google Scholar]
- 34.Lee Y.J., Jeong S.H., Hong S.C., Cho B.I., Ha W.S., Park S.T., Choi S.K., Jung E.J., Ju Y.T., Jeong C.Y., Kim J.W., Lee C.W., Yoo J., Ko G.H. Prognostic value of CAPZA1 overexpression in gastric cancer. Int J Oncol. 2013;42:1569–1577. doi: 10.3892/ijo.2013.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Go S.I., Ko G.H., Lee W.S., Kim R.B., Lee J.H., Jeong S.H., Lee Y.J., Hong S.C., Ha W.S. CD44 variant 9 serves as a poor prognostic marker in early gastric cancer, but not in advanced gastric cancer. Cancer Res Treat. 2016;48:142–152. doi: 10.4143/crt.2014.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mani S.A., Guo W., Liao M.J., Eaton E.N., Ayyanan A., Zhou A.Y., Brooks M., Reinhard F., Zhang C.C., Shipitsin M., Campbell L.L., Polyak K., Brisken C., Yang J., Weinberg R.A. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zavros Y. Initiation and maintenance of gastric cancer: a focus on CD44 variant isoforms and cancer stem cells. Cell Mol Gastroenterol Hepatol. 2017;4:55–63. doi: 10.1016/j.jcmgh.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vaira D., Malfertheiner P., Megraud F., Axon A.T., Deltenre M., Hirschl A.M., Gasbarrini G., O'Morain C., Garcia J.M., Quina M., Tytgat G.N. Diagnosis of Helicobacter pylori infection with a new non-invasive antigen-based assay. HpSA European study group. Lancet. 1999;354:30–33. doi: 10.1016/s0140-6736(98)08103-3. [DOI] [PubMed] [Google Scholar]
- 39.Kato M., Asaka M., Ohara S., Toyota T. Clinical studies of 13C-urea breath test in Japan. J Gastroenterol. 1998;33(Suppl 10):36–39. [PubMed] [Google Scholar]
- 40.Dixon M.F., Genta R.M., Yardley J.H., Correa P. Classification and grading of gastritis. The updated Sydney System. International Workshop on the Histopathology of Gastritis, Houston 1994. Am J Surg Pathol. 1996;20:1161–1181. doi: 10.1097/00000478-199610000-00001. [DOI] [PubMed] [Google Scholar]