Abstract
The intention of this review was to condense ongoing findings on the use of circulating DNAs from bodily fluids (blood, serum and plasma) as cancer biomarkers in patients with oral squamous cell carcinoma. Studies were collected after searching databases: PubMed and Google library. Additional search was performed through cross-check on the bibliography of selected articles. After the selection process made by two of the authors, articles which met the inclusion criteria were included in the review. Results revealed that circulating DNAs from blood, serum or plasma appear as favorable candidates as cancer biomarkers in patients suffering from oral cancer. The possibility to forecast recurrences and metastases through follow-up by quantification of candidate DNAs serve as another possible characteristic to be directed in forthcoming studies. However, methodological standardization and even sampling are required to increase the power and accuracy of results.
Keywords: Blood, blood serum, cell-free DNA, oral cancer, oral squamous cell carcinoma biomarkers, plasma
INTRODUCTION
In modern oncology, genotyping of the tumor tissue is becoming a routine assay for decision-making clinically. Oral squamous cell carcinoma (OSCC) is one of the common malignancies of the oral cavity and tissue biopsies (histopathological analysis) remain gold standard for deciding the treatment protocol for OSCC. Conventional biopsies often give a concise picture mainly because of small region of tumor tissue which is studied under the microscope.[1] Many a times, OSCC is commonly diagnosed in the advanced stage. The absence of tumor is noticed commonly through traditional imaging after tumor resection. The use of biomarkers helps in early detection and better treatment of tumor. Tumor DNA is emerging biomarker in the diagnosis of OSCC. Biopsy is minimally invasive procedure, and hence, noninvasive procedures are preferred over biopsy. The detection of biomarkers using cell-free DNA (cfDNA) is one of the promising noninvasive methods.[2] Circulating tumor DNA (ct DNA) is present in the blood and other bodily fluid is derived from the DNA fragments shed by necrosis, apoptosis and phagocytosis of cells. These can serve as potential biomarkers for early detection of tumor, tumor profiling, to determine treatment, prognosis and disease recurrences. Thus, the aim of this review is to evaluate and assess the role of cfDNA as a biomarker in OSCC.[2,3]
TERMINOLOGIES
Liquid biopsy, circulating tumor DNA, cell-free DNA
Liquid biopsy is an upcoming diagnostic concept which is rapidly expanding the field in translational cancer research which comprises of circulating tumor cells (CTCs). CTCs are formed by detachment of cell from the primary tumor mass, permitting the migration of tumor cells to secondary sites through the lymphatic and blood system. Circulating cfDNA, extracellular fragments released from CTCs into the bloodstream as well as other fluids such as urine, serum, plasma, cerebrospinal fluid, seminal plasma and saliva to detect circulating molecules [Figure 1]. These fragments carry the same genetic alterations as of the original tumor cells.[4,5]
Figure 1.
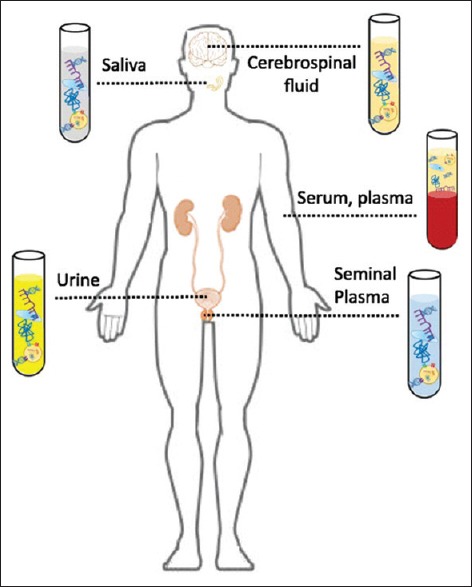
Circulating molecules in various biological fluids[4]
The sources of cfDNA in cancer patients are “healthy Cells” “malignant cells” and “tumor microenvironmental cells.” The origin of cfDNA is through apoptosis, necrosis, phagocytosis, oncosis, active secretions and netosis [Figure 2]. The size of the fragments of cfDNA is important, to know its presence or absence which differs at different blood levels of the body. The cfDNA from apoptotic cells are extremely fragmented, whereas DNA from necrotic cancer cells conclude to be longer DNA fragments and measures between 180 and 200 base pairs with half-life, ranging from 16 min to 13 h.[6]
Figure 2.
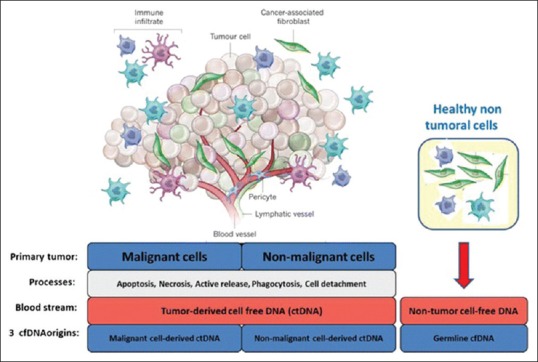
Distinct cellular origins of circulating DNA found in the blood of cancer patients[6]
HISTORICAL BACKGROUND
In the year 1940s Mandel and Metais first described the presence of cfDNA in human blood samples.[7] After a decade of this discovery, characteristics and the origins of the cfDNA were studies. Two decades later, Tan et al. in the year 1966 observed the increased presence of cfDNA in the blood of systemic lupus erythematosus versus healthy individuals leading to the formation of anti-dsDNA antibodies. Similar observations were made by Koffler et al. in the year 1973. They observed increased levels of cfDNA in other conditions also such as leukemia, rheumatic arthritis and malignant tumors. It was in the year 1977 Leon et al. revealed cfDNA in the field of oncology. They found that cfDNA in the blood samples of cancer patients were notably higher than healthy individuals. This attracted the importance of cfDNA opening the way to potential biomedical applications in oncology. Later, it was found by Stroun et al., that cancer patients harbor tumor-specific molecular alterations cfDNA.[8] They found cfDNA which appeared in circulation were part of tumor origin, and it was double stranded which were specific to the tumor DNA.[6,9] Since then the concept of liquid biopsy was began. Recently, emerging concept is that these cfDNA and ctDNA can also be detected in urine of cancer patients with bladder cancer, Non-small cell lung cancer (NSCLC), bladder cancer, urothelial cancer and hepatocellular cancer. Zhang et al. reported cfDNA fragments in female recipients of renal transplants from male donor.[10]
BIOLOGICAL PROPERTIES OF CELL-FREE DNA
Size of cell-free DNA
The size of the cfDNA is of great interest to know the structures and origins which leads to higher diagnostic value. There are many controversies in the literature regarding clear conclusion in this area. Different methods have been used to find out the fragment size of cfDNA. With regard to cancer Stroun et al. isolated and characterized cfDNA size in plasma from patients with advanced cancer. They found purified DNA as double-stranded composed of fragment length ranging about <0.5–21 kbp, and it was confirmed by gel electrophoresis method.[8] Giacona observed the size of cfDNA in blood plasma from pancreatic cancer patients showed autoradiographic bands at sizes 185–200 bp.[11] The quantitative polymerase chain reaction (qPCR) methods have been applied to measure cell death phenomena in prostrate and testicular cancer. The amplicons indicated cfDNA in plasma varied from 200 bp to 250 bp. The principle of such analysis is concluded that the fragments with greater length are associated with necrosis and the smaller length with apoptosis phenomena.[12] The atomic force microscopic study on colorectal cancer and healthy individuals showed the length of cfDNA varied from 135 bp to 180 bp, respectively.[13] Investigators have explored that changes in the size of cfDNA molecules are associated with types of cancer and found that there is a higher proportion of longer DNA in cancer.[14] Mouliere et al. in their qPCR study, examined size distribution of DNA in plasma from metastatic colorectal cancer patients and healthy individuals and suggested higher fragments characterizes the tumor-derived cfDNA, and these are shorter than nontumor derived tissues. The overall size profile was found to be correlated to the presence of DNA in plasma. The larger amount of DNA in plasma showed the presence of short cfDNA in plasma of cancer patients and the lower amount of DNA in plasma showed longer cfDNA in plasma.[15]
Content
cfDNA contains epigenomic and genomic, as well as mitochondrial and viral DNA. Authors have conducted epigenomic studies using methylation markers SHOX2 and SPEPT9 in OSCC.[3] Genomic studies using markers such as epidermal growth factor receptor (EGFR), T53, p16, PIK3CA, CDKN2A, HRAS and NRAS have been of great value in screening and early detection of cancers.[16,17,18,19] Mitochondrial markers have expressed a potential link between circulating cell-free mtDNA (CfmtDNA) content and cancers. CfmtDNA serves as a major approach for initial cancer diagnosis with some exclusive advantages over nuclear cfDNA.[20] Viral cfDNA has been studied in cancer for the presence of human papillomavirus (HPV) and Epstein-Barr virus and cfDNA was useful in detection and treatment of cancer.[21]
Concentration
There is much difference in the concentration of cfDNA between plasma or serum samples. In literatures, it is found that the range vary from 0 and >1000 ng/mL of blood, with an average of 180 ng/mL cfDNA. Healthy individuals the concentrations varied between 0 and 100 ng/mL cfDNA of blood, with an average of 30 ng/mL cfDNA. In cancer patients, it varies because of different methods applied for the detection of cfDNA.[22] However, overlapping DNA concentrations was noticed in healthy individuals compared to the patients with benign and malignant disease. The variability of cfDNA levels in cancer patients is likely to be coupled with tumor burden, stage, vascularity, cellular turnover and response to therapy. The quantification of cfDNA concentrations alone does not appear to be useful in a diagnostic setting. The consideration of cfDNA concentration might justify in combination with other blood tumor biomarkers.[23]
Clearance
Plasma nucleases were found to be only partially contributed to the degradation of plasma cfDNA in humans. Liver and the reticuloendothelial system are likely to have the involvement in clearing of cfDNA. Studies have shown that kidney might not be important for cfDNA clearance because of negative charge of cfDNA. However, urine contains plasma-derived cfDNA.[24,25] Celec et al. suggested minimal involvement of clearance of cfDNA from kidney with unknown external clearance mechanism.[26]
DIFFERENT DETECTION METHOD OF CELL-FREE DNA
Different technologies for cfDNA detection have been developed during the last years, allowing it to be analyzed from the level of a point mutation to that of the entire genome. Classical methods of analyzing cfDNA are Sanger sequencing, q-PCR-based methods, fluorescent assays, chromatographic-based methods (e.g., spectrophotometric strategies).[5] Digital PCR (dPCR)-based technologies are extremely sensitive techniques designed for the detection of specific point mutations, copy-number variations, short indels and gene fusions. These technologies include allele-specific q-PCR, digital q-PCR, droplet-PCR, microfluidic systems for parallel PCR, and BEAMing (beads, emulsions, amplification and magnetic).[27] Deep sequencing, next-generation sequencing (NGS) technologies are the other alternative for cfDNA characterization. These technologies allow high-throughput and relatively low-cost analyses to identify cfDNA alterations across wide genomic regions and have the advantage of not requiring prior knowledge of the genetic alterations of the tumor.[8]
Sanger sequencing
Sanger sequencing is gold standard for mutation detection for the tissues. In liquid biopsies, this technique has two disadvantages – one is low amount of cf DNA in sample will prevent sequential analysis of targeted genes and the another one is specific alleles cannot be identified if mutated allele frequencies are below 20%. This will lead to false-positive results and will result in inaccurate treatment decision.[28,29]
Quantitative polymerase chain reaction
There are many methods under qPCR, Peptide nucleic acid-clamp PCR, locked nucleic acid/DNA-PCR, amplification refractory mutation system and COLD-PCR. Recently, some of the qPCR commercial kits have been applied for the detection of cfDNA which are useful in clinical studies such as Cobas (Roche) and Therascreen (Qiagen). These kits have been approved by the Food and Drug Administration (FDA) for EGFR mutations on exon 19 deletions, exon 21 L858R and T790M in NSCLC patients. The limitations of these commercially available kits are sensitivity of these kits is not sufficient to the reliability of patients for precision medicine treatment because the detection of mutations in liquid biopsy occurs at lower allele frequencies. Furthermore, the selected mutation panel is not covered so there will be 40%–50% loss of positivity in patients’ sample.[30,31]
Digital polymerase chain reaction and digital droplet polymerase chain reaction
To improve the analytical sensitivity of conventional PCR dPCR was introduced in the field of circulating DNA. These PCR utilizes oil-based droplet systems such as droplet dPCR (Bio-Rad), picoliter droplet-based dPCR (RainDance), BEAMING and microfluidic systems, i.e., parallel PCR reactions (Fluidigm).[32,33,34] The sensitivity of dPCR is found to be 64%–82% and this method detects allele prevalence as low as 0.1%. In case of low frequencies due to the absence of mutated cfDNA replication in each replicate sequential analysis and triplicate analysis is required which is a disadvantage of this technique.[35,36]
Deep sequencing
NGS or massive parallel sequencing (NGS)is currently applied to all types of tumor tissue profiling as a routine method. Different assays are used for clinical applications such as AmpliSeq Cancer Panel, Thermo Fisher; RAS Panel, Illumina. In NSCLC, patients’ oncomine-targeted sequencing kit which is approved by FDA, confirmed 77% of detection of cfDNA for EGFR mutations. Similar results were procured with AmpliSeq used on cancer panel. Other NGS methods are Tam-Seq, SAFE-sequencing system, Guardent 360 digital sequencing test, CAncer Personalized Profiling-sequencing, iDES and PARE. Whole exome sequencing or whole genome sequencing is some of the deep sequencing methods. The advantage of such techniques is that there is no need of prior knowledge of molecular alteration, but it requires longer time and needs bioinformatics expertise [Figure 3].[5,37,38]
Figure 3.
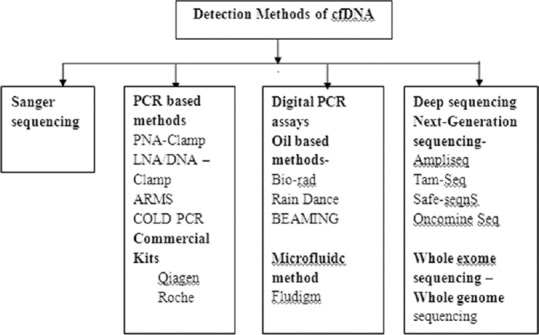
Different detection method of cell-free DNA
Cell-free DNA in oral squamous cell carcinoma patients
In this review, the literature search on cfDNA in OSCC showed the following English articles from 2005 to 2017 [Table 1].
Table 1.
Cell-free DNA in oral squamous cell carcinoma patients
| Name of the author, years, references | CfDNA | Sample type | Size of sample | Detection method | Observations |
|---|---|---|---|---|---|
| cfDNA as a detection and diagnostic markers | |||||
| Shukla et al., 2013[2] | CfDNA | Saliva and plasma | 390 patients (90 potentially malignant lesions, 150 OSCCs and 150 posttreatment OSCCs) | Spectrophotometry | No significant difference between groups |
| Mazurek et al., 2016[39] | HPV 16/18 KRAS, EGFR | Plasma | 200 HNSCC patients | qPCR | 96.4% positivity for HPV. No somatic mutation detection |
| Wang. Y et al., 2015[19] | TP53, PIK3CA, CDKN2A, HRAS, NRAS, HPV 16-18 | Saliva and plasma | 93 HNSCC | Digital PCR | HPV 16 positivity 76% in saliva 86% in plasma |
| Schröck et al., 2017[3] | Methylation markers SOX2 and SPET9 | Saliva and plasma | 649 HNSCC | qPCR | 59% positivity |
| cfDNA as a prognostic and metastatic markers | |||||
| Lin et al., 2018[40] | cfDNA | Plasma | 121 OSCC and 50 matched control | Spectometry | cfDNA associated with tumoral size and poor prognosis of OSCC |
| Hamana et al., 2005[41] | Nine microsatellite markers | Tissue and serum | 64 SCC patients | PCR | Allelic serum imbalance was 44% and 20% in pre-and post-operation |
| Kakimoto et al.,2008[42] | Microsatellite markers | Tissue and serum | 20 OSCC patients | PCR | 90% alleilic imbalance in serum and associated with poor prognosis of patients |
OSCCs: Oral squamous cell carcinomas, cfDNA: Cell-free DNA, HNSACC: Head and neck squamous cell carcinomas, qPCR: Quantitative polymerase chain reaction, HPV: Human papillomavirus, KRAS: Kirsten rat sarcoma 2 viral oncogene homolog, EGFR: Epidermal growth factor receptor, HRAS: Harvey rat sarcoma viral oncogene homolog, NRAS: Neuroblastoma RAS viral oncogene homolog
Cell-free DNA as diagnostic marker
To date, many studies have tested cfDNA analyses in OSCCs. Shukla et al. cfDNA analyzed by spectrophotometry the amount of deoxyribonucleic acid within the plasma of 390 patients (90 potentially malignant lesions, 150 OSCCs and 150 posttreatment OSCCs) and 150 healthy controls, but no significant differences were noticed between the groups. The possible reason could be cfDNA is prevented from entering the bloodstream due to the rich lymphatic drainage of the oral mucosa.[2]
Mazurek et al. observed HPV detection using cfDNA found to be of great value for OSCC. They analyzed the cfDNA levels in 200 head and neck squamous cell carcinomas (HNSCC) by examining HPV16/18, KRAS and EGFR mutations through q-PCR. A higher level of the total cfDNA was observed in patients with oropharyngeal squamous cell carcinoma as compared to different HNSCC. The level of cfDNA increased in patients with both greater lymph node affectation and tumoral stage. From all patients, 14% showed positivity for HPV, most of whom were HPV16 positive (96.4%), while somatic EGFR and KRAS mutations were not detected. These results showed that HPV cfDNA tests can be used for detection and observation of HPV-positive HNSCC.[39]
A cohort study conducted by Wang et al., on 93 HNSCC to identify somatic mutations (TP53, PIK3CA, CDKN2A, HRAS and NRAS) or HPV genes. Authors searched for the presence of either HPV type 16 (HPV16) or HPV18 sequences in tumor DNA in both plasma and saliva samples. dPCR was used to quantify HPV. Of 93, 30 patients were positive for HPV16 in primary head and neck tumors, but none showed HPV18. Tumor DNA was found in both saliva and plasma of oral cavity cancer patients. Tumor DNA in the saliva 76% (n = 93) and plasma 86% (n = 47) appeared to be a potentially valuable biomarker for detection of HNSCC.[19]
The methylation markers SHOX2 and SPEPT9 were analyzed by Schröck et al., in a cohort of 649 head and neck cancer patients showing 59% of patients with methylation positive. Methylation levels correlated with survival rate which showed importance in monitoring treatment efficiency and also emerged as promising biomarkers for tumor diagnosis. cfDNA also assessed aberrant DNA methylation patterns precisely in these patients.[3]
Cell-free DNA as a prognosis and metastatic markers
Lin et al. measured plasma cfDNA in 121 patients with OSCC and 50 matched controls by spectrometry. They found plasma cfDNA was significantly raised in patients with OSCC compared to controls. Plasma cfDNA levels related to larger tumor size, cervical lymph node metastasis and late stage. Higher plasma cfDNA levels were identified with a poor prognosis of OSCC. The association between high cfDNA concentration and poor prognosis could be linked to tumor burden and/or comorbidities.[40]
Hamana et al. studied nine microsatellite markers (D5s178, D9S104, IFNA, D11S910, D11S1356, D13S273, TP53, D18S46 and D22S274) OSCC patients. Microsatellite instability analyses were conducted using cfDNA on tissue and serum samples at three different time points: preoperatively, postoperatively and 4 weeks after surgery. The occurrence of allelic imbalance patterns in serum were related with the allelic imbalance in paired tumor tissue which mirrored the characteristics of the tumor. The serum cfDNA in the form of allelic imbalances were detected both in pre- and post-operative samples showing 44% and 20%, respectively. Four weeks’ postsurgery, patients with an allelic imbalance showed metastasis, but no recurrence was found. IFNA at the 9p21 locus was found to be the most common altered allelic imbalance in plasma samples. This suggested that the microsatellite markers are useful as a predictive and prognostic marker.[41]
A study by Kakimoto et al. analyzed panel microsatellite markers in blood and tumor tissue samples before and 1 month after surgery. An allelic imbalance was observed to be 90% in tumor DNA in the serum of patients. The existence of allelic imbalance in postoperative patient serum was coupled with a poor prognosis. These results give hope that microsatellite analysis using cfDNA could help to assess risk of recurrence, metastasis and death in OSCC.[42]
SOURCE OR FACTORS AFFECTING CELL-FREE DNA
Alterations in cell-free DNA
Standardization is one of the major problems in detecting cfDNA. The preanalytic problems could be low sensitivity due to low concentration of cfDNA, there will be a loss of sampled material which may reduce the sensitivity of molecular profiling. The factors which can be considered for preanalytical problems range from sample collection, storage and the fixatives used. The following are few of the common problems: (1) Blood collection: samples should be collected in ethylenediaminetetraacetic acid, some of the studies have shown PCR inhibition in heparinized blood, (2) blood centrifugation done within few hours of blood withdrawn to avoid cell lyses and release of germline DNA which may dilute the cfDNA, (3) proper storage should be done with stabilized fixatives, (4) diurnal variations and accurate clinical conditions need to be better defined before comparisons and clinical utility can be validated and (5) efficacy of the extraction methods such as phenol–chloroform-based methods, affinity colum-based, magnetic bead-based and polymer-based methods have implications in detection and another key issue is quantification before assessment on specific assay platforms [Figure 4].[30]
Figure 4.
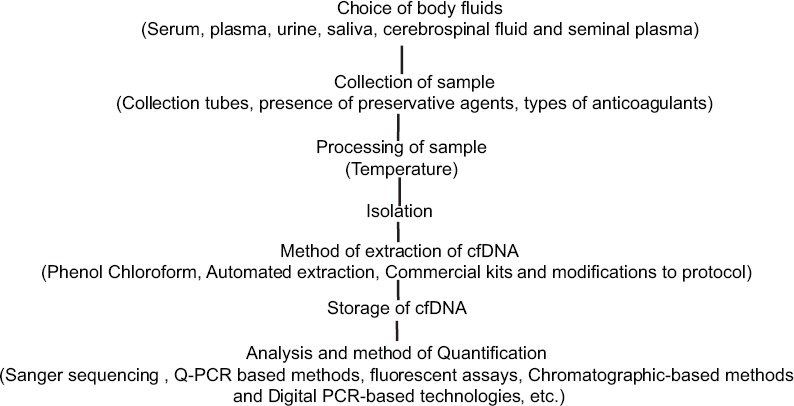
Factors affecting cell-free DNA
FUTURE PERSPECTIVES OF CELL-FREE DNA
The biomarkers cfDNA has emerged as a potential noninvasive approach in precision, personalized medicine. The detection and analysis of cfDNA represent a promising biomarker for early cancer detection, detection of minimal residual disease, recurrence and metastasis. In oral cancer, the role of cfDNA in the clinical usage is still inadequate as compared to other cancer anatomic locations thus require additional research for effective implementation. The discovery of a sturdy panel of sensitive and specific current biomarkers for oral cancer at different stages would facilitate clinicians to improve deciding the protocol and prognosis and mark the beginning of personalized medicine. Better information of the biology and origin of current biomarkers would be the key for the event for effective therapies and also for the management of oral cancer. Research efforts should be addressed to perform large, prospective multicenter studies with standard protocol followed by all to investigate the role of cfDNA in oral cancer.
CONCLUSION
cfDNA holds as one of the promising biomarkers for new generation as a molecular diagnostics for early cancer detection, molecular profiling analysis, monitoring of the treatment response and demonstration of minimal residual disease and relapse in OSCC. It is a potential noninvasive approach in precision, personalized medicine. The size of the cfDNA more likely has an impact in diagnostic application. Compared to other cancers, in oral cancer, the impact of cfDNA in the clinical setting is still limited. Further research is required for effective implementation.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.Mascitti M, Orsini G, Tosco V, Monterubbianesi R, Balercia A, Putignano A, et al. An overview on current non-invasive diagnostic devices in oral oncology. Front Physiol. 2018;9:1510. doi: 10.3389/fphys.2018.01510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shukla D, Kale AD, Hallikerimath S, Yerramalla V, Subbiah V. Can quantifying free-circulating DNA be a diagnostic and prognostic marker in oral epithelial dysplasia and oral squamous cell carcinoma? J Oral Maxillofac Surg. 2013;71:414–8. doi: 10.1016/j.joms.2012.04.039. [DOI] [PubMed] [Google Scholar]
- 3.Schröck A, Leisse A, de Vos L, Gevensleben H, Dröge F, Franzen A, et al. Free-circulating methylated DNA in blood for diagnosis, staging, prognosis, and monitoring of head and neck squamous cell carcinoma patients: An observational prospective cohort study. Clin Chem. 2017;63:1288–96. doi: 10.1373/clinchem.2016.270207. [DOI] [PubMed] [Google Scholar]
- 4.Di Meo A, Bartlett J, Cheng Y, Pasic MD, Yousef GM. Liquid biopsy: A step forward towards precision medicine in urologic malignancies. Mol Cancer. 2017;16:80. doi: 10.1186/s12943-017-0644-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siravegna G, Marsoni S, Siena S, Bardelli A. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol. 2017;14:531–48. doi: 10.1038/nrclinonc.2017.14. [DOI] [PubMed] [Google Scholar]
- 6.Thierry AR, El Messaoudi S, Gahan PB, Anker P, Stroun M. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. 2016;35:347–76. doi: 10.1007/s10555-016-9629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gold B, Cankovic M, Furtado LV, Meier F, Gocke CD. Do circulating tumor cells, exosomes, and circulating tumor nucleic acids have clinical utility? A report of the association for molecular pathology. J Mol Diagn. 2015;17:209–24. doi: 10.1016/j.jmoldx.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stroun M, Anker P, Lyautey J, Lederrey C, Maurice PA. Isolation and characterization of DNA from the plasma of cancer patients. Eur J Cancer Clin Oncol. 1987;23:707–12. doi: 10.1016/0277-5379(87)90266-5. [DOI] [PubMed] [Google Scholar]
- 9.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426–37. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 10.Zhang J, Tong KL, Li PK, Chan AY, Yeung CK, Pang CC, et al. Presence of donor-and recipient-derived DNA in cell-free urine samples of renal transplantation recipients: Urinary DNA chimerism. Clin Chem. 1999;45:1741–6. [PubMed] [Google Scholar]
- 11.Giacona MB, Ruben GC, Iczkowski KA, Roos TB, Porter DM, Sorenson GD, et al. Cell-free DNA in human blood plasma: Length measurements in patients with pancreatic cancer and healthy controls. Pancreas. 1998;17:89–97. doi: 10.1097/00006676-199807000-00012. [DOI] [PubMed] [Google Scholar]
- 12.Ellinger J, Wittkamp V, Albers P, Perabo FG, Mueller SC, von Ruecker A, et al. Cell-free circulating DNA: Diagnostic value in patients with testicular germ cell cancer. J Urol. 2009;181:363–71. doi: 10.1016/j.juro.2008.08.118. [DOI] [PubMed] [Google Scholar]
- 13.Binnig G, Quate CF, Gerber C. Atomic force microscope. Phys Rev Lett. 1986;56:930–3. doi: 10.1103/PhysRevLett.56.930. [DOI] [PubMed] [Google Scholar]
- 14.Ellinger J, Bastian PJ, Haan KI, Heukamp LC, Buettner R, Fimmers R, et al. Noncancerous PTGS2 DNA fragments of apoptotic origin in sera of prostate cancer patients qualify as diagnostic and prognostic indicators. Int J Cancer. 2008;122:138–43. doi: 10.1002/ijc.23057. [DOI] [PubMed] [Google Scholar]
- 15.Mouliere F, El Messaoudi S, Gongora C, Guedj AS, Robert B, Del Rio M, et al. Circulating cell-free DNA from colorectal cancer patients may reveal high KRAS or BRAF mutation load. Transl Oncol. 2013;6:319–28. doi: 10.1593/tlo.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gedvilaitė V, Schveigert D, Cicėnas S. Cell-free DNA in non-small cell lung cancer. Acta Med Litu. 2017;24:138–44. doi: 10.6001/actamedica.v24i2.3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pantel K. TP53 mutations on circulating cell-free DNA. EBioMedicine. 2016;10:15–6. doi: 10.1016/j.ebiom.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dace P, Olita H, Ludmila E, Ingrida D. Tumour suppressor gene CDKN2A/p16 germline mutations in melanoma patients with additional cancer and cancer in their family history. Acta Univ Latviensis. 2003;662:25–32. [Google Scholar]
- 19.Wang Y, Springer S, Mulvey CL, Silliman N, Schaefer J, Sausen M, et al. Detection of somatic mutations and HPV in the saliva and plasma of patients with head and neck squamous cell carcinomas. Sci Transl Med. 2015;7:293ra104. doi: 10.1126/scitranslmed.aaa8507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kohler C, Radpour R, Barekati Z, Asadollahi R, Bitzer J, Wight E, et al. Levels of plasma circulating cell free nuclear and mitochondrial DNA as potential biomarkers for breast tumors. Mol Cancer. 2009;8:105. doi: 10.1186/1476-4598-8-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vo JH, Nei WL, Hu M, Phyo WM, Wang F, Fong KW, et al. Comparison of circulating tumour cells and circulating cell-free Epstein-Barr virus DNA in patients with nasopharyngeal carcinoma undergoing radiotherapy. Sci Rep. 2016;6:13. doi: 10.1038/s41598-016-0006-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fleischhacker M, Schmidt B. Circulating nucleic acids (CNAs) and cancer – A survey. Biochim Biophys Acta. 2007;1775:181–232. doi: 10.1016/j.bbcan.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 23.Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, et al. DNA fragments in the blood plasma of cancer patients: Quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61:1659–65. [PubMed] [Google Scholar]
- 24.de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008;14:6302–9. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 25.Yu SC, Lee SW, Jiang P, Leung TY, Chan KC, Chiu RW, et al. High-resolution profiling of fetal DNA clearance from maternal plasma by massively parallel sequencing. Clin Chem. 2013;59:1228–37. doi: 10.1373/clinchem.2013.203679. [DOI] [PubMed] [Google Scholar]
- 26.Celec P, Vlková B, Lauková L, Bábíčková J, Boor P. Cell-free DNA: The role in pathophysiology and as a biomarker in kidney diseases. Expert Rev Mol Med. 2018;20:e1. doi: 10.1017/erm.2017.12. [DOI] [PubMed] [Google Scholar]
- 27.Volckmar AL, Sültmann H, Riediger A, Fioretos T, Schirmacher P, Endris V, et al. A field guide for cancer diagnostics using cell-free DNA: From principles to practice and clinical applications. Genes Chromosomes Cancer. 2018;57:123–39. doi: 10.1002/gcc.22517. [DOI] [PubMed] [Google Scholar]
- 28.Endris V, Penzel R, Warth A, Muckenhuber A, Schirmacher P, Stenzinger A, et al. Molecular diagnostic profiling of lung cancer specimens with a semiconductor-based massive parallel sequencing approach: Feasibility, costs, and performance compared with conventional sequencing. J Mol Diagn. 2013;15:765–75. doi: 10.1016/j.jmoldx.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 29.Oh JE, Lim HS, An CH, Jeong EG, Han JY, Lee SH, et al. Detection of low-level KRAS mutations using PNA-mediated asymmetric PCR clamping and melting curve analysis with unlabeled probes. J Mol Diagn. 2010;12:418–24. doi: 10.2353/jmoldx.2010.090146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wan JC, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C, et al. Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017;17:223–38. doi: 10.1038/nrc.2017.7. [DOI] [PubMed] [Google Scholar]
- 31.Xu T, Kang X, You X, Dai L, Tian D, Yan W, et al. Cross-platform comparison of four leading technologies for detecting EGFR mutations in circulating tumor DNA from non-small cell lung carcinoma patient plasma. Theranostics. 2017;7:1437–46. doi: 10.7150/thno.16558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83:8604–10. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Domínguez-Vigil IG, Moreno-Martínez AK, Wang JY, Roehrl MH, Barrera-Saldaña HA. The dawn of the liquid biopsy in the fight against cancer. Oncotarget. 2018;9:2912–22. doi: 10.18632/oncotarget.23131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yung TK, Chan KC, Mok TS, Tong J, To KF, Lo YM, et al. Single-molecule detection of epidermal growth factor receptor mutations in plasma by microfluidics digital PCR in non-small cell lung cancer patients. Clin Cancer Res. 2009;15:2076–84. doi: 10.1158/1078-0432.CCR-08-2622. [DOI] [PubMed] [Google Scholar]
- 35.Sacher AG, Paweletz C, Dahlberg SE, Alden RS, O’Connell A, Feeney N, et al. Prospective validation of rapid plasma genotyping for the detection of EGFR and KRAS mutations in advanced lung cancer. JAMA Oncol. 2016;2:1014–22. doi: 10.1001/jamaoncol.2016.0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Busser B, Lupo J, Sancey L, Mouret S, Faure P, Plumas J, et al. Plasma circulating tumor DNA levels for the monitoring of melanoma patients: Landscape of available technologies and clinical applications. Biomed Res Int. 2017;2017:5986129. doi: 10.1155/2017/5986129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rachiglio AM, Esposito Abate R, Sacco A, Pasquale R, Fenizia F, Lambiase M, et al. Limits and potential of targeted sequencing analysis of liquid biopsy in patients with lung and colon carcinoma. Oncotarget. 2016;7:66595–605. doi: 10.18632/oncotarget.10704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu S, Lou F, Wu Y, Sun DQ, Zhang JB, Chen W, et al. Circulating tumor DNA identified by targeted sequencing in advanced-stage non-small cell lung cancer patients. Cancer Lett. 2016;370:324–31. doi: 10.1016/j.canlet.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mazurek AM, Rutkowski T, Fiszer-Kierzkowska A, Małusecka E, Składowski K. Assessment of the total cfDNA and HPV16/18 detection in plasma samples of head and neck squamous cell carcinoma patients. Oral Oncol. 2016;54:36–41. doi: 10.1016/j.oraloncology.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 40.Lin LH, Chang KW, Kao SY, Cheng HW, Liu CJ. Increased plasma circulating cell-free DNA could be a potential marker for oral cancer. Int J Mol Sci. 2018;19 doi: 10.3390/ijms19113303. pii: E3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hamana K, Uzawa K, Ogawara K, Shiiba M, Bukawa H, Yokoe H, et al. Monitoring of circulating tumour-associated DNA as a prognostic tool for oral squamous cell carcinoma. Br J Cancer. 2005;92:2181–4. doi: 10.1038/sj.bjc.6602635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kakimoto Y, Yamamoto N, Shibahara T. Microsatellite analysis of serum DNA in patients with oral squamous cell carcinoma. Oncol Rep. 2008;20:1195–200. [PubMed] [Google Scholar]


