Abstract
Copy number variation (CNV) is a main cause of male infertility, yet its influence still remains elusive in that of females. To investigate the correlation between CNV and female infertility, we applied whole-genome CNV analyses by next generation Sequencing (NGS), and analyzed 324 female infertility samples in Xinjiang Province, People's Republic of China. We identified 29 CNVs in total, of which 10 were novel CNVs. We found these CNVs mostly in chromosome X. The CNVs from one sample overlapped the POF1B gene that was related to premature ovarian failure (POF). The rest of these CNVs overlapped important functional genes related to neuropathy, brain, skin and retina, and the relationship between these CNVs and fertility needs to be studied further. We also found recurrent CNVs located on Xp22.31 and 22ql 1.21 in five and three cases, respectively. Our study first identified and characterized CNVs (CNVs preference, recurrent CNVs) in female infertility, also provided genetic evidence and references for future study and infertility etiology research.
Keywords: Copy number variations (CNVs), Female infertility, Next generation sequencing (NGS), Recurrent CNVs, Novel CNVs
Introduction
Copy number variations (CNVs) have been found to be responsible for a wide range of human diseases; CNVs on sex chromosomes are more likely to play key roles in germ cell development [1, 2, 3, 4,]. In the past two decades, CNVs causing male infertility have been widely reported, such as the sex chromosome CNVs (Y chromosomal microdeletions and X-link CNVs) [5, 6, 7, 8, 9, 10]. Furthermore, the role of CNVs in female infertility is still poorly understood. A few studies have reported that CNVs may disrupt key genes and pathways of ovaries, leading to premature ovarian failure (POF). Some CNVs are also related to genetic factors causing female infertility, such as the associated microdeletions/duplications in XY gonadal dysgenesis syndrome (XY-GD), MRKH (Mayer-Rokitansky-Küster-Hauser) syndrome [11, 12, 13, 14]. However, the correlation between female infertility and CNVs still remains elusive, especially for the female infertility in China.
Array comparative genomic hybridization (aCGH) is the well-accepted standard for identifying CNVs, but aCGH has disadvantages including restrictive requirement of DNA quality and relatively high cost. In recent years, next generation sequencing (NGS) technologies have been widely applied in chromosome aneuploidy testing and CNVs detection [7,15,16]. Next generation sequencing has several advantages: low cost, short period, high resolution, high accuracy and, small amount of input DNA. The microdeletion and microrepetition of chromosomal fragments can be found by analyzing the CNVs. Dong et al. [7] tested CNVs by high-throughput genome-wide sequencing on 33 male patients in China with spermatogenic failure. Liu et al. [15] found novel Y-chromosomal microdeletions were associated with non obstructive azoospermia using a high-throughput sequencing method. Wang et al. [16] showed that semiconductor sequencing is of equivalent efficacy as an aCGH in detecting CNVs of >1 Mb.
In this study, we investigated the correlation between the genome CNVs and female infertility by NGS. We further identified CNVs characteristics for Chinese female infertility samples and provide reference for female infertility etiology research.
Material and methods
Sample Collection and DNA Extraction. The study was approved by the Clinical Research Ethics Committee of the Xinjiang JiaYin Clinical Laboratory Center, Urumchi, Xinjiang Province, People's Republic of China (PRC). All patients signed written informed consent before participation. A total of 324 blood samples of female infertility patients (305 unexplained infertility, and 19 females with spontaneous abortions, including spontaneous abortion and recurrent spontaneous abortion) were collected from the Xinjiang JiaYin Clinical Laboratory Center in 2017. Two milliliters of peripheral blood samples were collected in vacutainers containing EDTA as anticoagulant, and DNA was extracted using a TIANamp Genomic DNA Kit (Item no: DP304; Tiangen Biochemical Reagent Co., Beijing, PRC). DNA quality was evaluated by Qubit®2.0 (ThermoFisher Scientific, Waltham, MA, USA) and Qubit™ ds DNA HS Assay kit (Catalog no. Q32854; ThermoFisher Scientific).
Library Construction and Sequencing. We enzymatically fragmented 300ng genomic DNA with an average size of 150 bp, followed by library construction according to the Life Technology Ion Xpress Plus Fragment Library preparation guide (Life Technologies, Carlsbad, CA, USA). The constructed library was quantified using Qubit® 2.0 (ThermoFisher Scientific). The libraries were mixed in proportion, and pooled P1 chip to ensure an average sequencing depth of ~0.1 x for each sample.
Data Analysis. All sequencing reads were aligned to the reference human genome National Center for Biotechnology Information (NCBI) Build37/hgl9 by TMAP software (www.tmap.net) Meanwhile, duplicate sequences were removed by using Picard software (https://broadinstitute.github.io/picard/) Each chromosome was divided into 40 kb non overlapped bins, and the number of reads mapped to each bin was calculated. We normalized the GC percentages in each bin by LOWESS regression. Circular binary segmentation (CBS), a reliable algorithm that is widely used in the analysis of comparative genomic hybridization arrays, allowed us to precisely define changed points by partitioning chromosomes into regions of equal copy numbers. The CNVs obtained by analysis were compared to the Database of Genomic Variants (DGV), International Standards for Cytogenomic Arrays (ISCA), DECIPHER databases (DEC), and the normal polymorphism CNVs were filtered out.
Results
Classification Statistics and Clinical Phenotypes of Infertility. One-hundred and thirty of the 324 infertile females had primary infertility, and the average age of infertility was 32.2±4.6, and infertile mean years were 4.3 ± 3.2. There were 194 patients with secondary infertility, with the average age at 34.8 ± 5.7, and the infertile mean years were 3.5 ± 3.2. Primary infertility and secondary infertility showed significant differences in average age and infertile mean years, but there were no significant difference in CNV variation rates between the primary infertility group and secondary infertility group [χ2 test with 95% confidence interval (95% CI)]. (No significant difference, data not shown.)
The results of Copy Number Variations. After filtration using DGV, we detected 29 CNVs in 29 samples (Table 1). All the chromosome abnormalities were CNVs, no abnormal chromosome numbers were found. As shown in Figure 1, CNVs were distributed in different regions of the chromosomes. Copy number variations occurred 10 times on chromosome X with the highest frequency. One sample has duplication CNV on Xq21.1 -q21.2 involved in POF1B, this patient had two miscarriages and one spontaneous abortion. The duplication CNV (9q22.32-q22.33) containing the HSD17B3 gene was found in another sample, this patient had normal sex hormones and a normal uterus. According to DGV, 10 novel CNVs were screened out. In this study, we found five recurring CNVs located on Xp22.31, and three times on 22q11.21.
Table 1.
Copy number variations detected and their descriptions. (The bolded CNVs indicate high frequencies.)
| CNVs Type | Code | Chromosome Location | Start-End Position | Size (Mb) | Number of RefSeq Genes | Included in DGV, ISCA, DEC | Disease-Related Gene in OMIM | Phenotype |
|---|---|---|---|---|---|---|---|---|
| Deletion | R1701 | 7q31.31 | 120420001-1205-80000 | 0.16 | 1 | no | TSPAN12 | primary 4 years; infertility no symptoms |
| R1702 | 8p23.3-p23.2 | 160001-3880000 | 3.72 | 10 | yes | CLN8; ARHGEF10 | primary 4 years; infertility no symptoms | |
| R1703 | 16pl3.2-pl3.11 | 14720001-15300000 | 1.04 | 20 | yes | ND1; MYH11; ABCC6 | primary 5 years; infertility no symptoms | |
| R1704 | Xq21.33 | 93900001-95140000 | 1.24 | 0 | yes | primary infertility 5 years; no symptoms | ||
| R1705 | Xp22.31 | 6420001-8160000 | 1.68 | 6 | yes | STS | primary 6 years; infertility no symptoms | |
| Duplication | R1707 | 3q27.3-q28 | 187560001-188880000 | 1.32 | 2 | no | LPP | primary 5 years; infertility no symptoms |
| R1708 | 4q34.3 | 178120001-179840000 | 1.72 | 4 | no | AGA | secondary 1 year; no infertility symptoms | |
| R1709 | 5q21.3-q22.1 | 108900001-110020000 | 1.12 | 3 | no | primary infertility 4 years; no symptoms | ||
| R1710 | 6ql6.3 | 102480001-103860000 | 1.36 | 1 | yes | (most of) GRIK2 | secondary infertility 5 years; no symptoms | |
| R1711 | 7q21.13 | 88920001-90440000 | 1.20 | 7 | yes | CLDN12; GTPBP10 | primary 1 year; no infertility symptoms | |
| R1712 | 9q22.32-q22.33 | 98480001-99560000 | 1.08 | 8 | no | HSD17B3 | secondary 1 year; no infertility symptoms | |
| R1713 | 11q24.2 | 124580001-126980000 | 2.40 | 36 | no | ROBO3; FOXRED1; HEPACAM; HYLS1; TIRAP; KIRREL3 | primary infertility 3 years; no symptoms | |
| R1714 | 12pl3.33 | 640001-1720000 | 1.08 | 7 | no | WNK1 | primary 3 years; infertility no symptoms | |
| R1715 | 15qll.2 | 20840001-22780000 | 1.74 | 5 | yes | NBEAP1 | primary 1 year; no infertility symptoms | |
| R1716 | 15qll.2 | 20840001-22780000 | 1.74 | 5 | yes | NBEAP1 | primary infertility 5 years; no symptoms | |
| R1717 | 16pl3.11-pl2.3 | 15580001-18620000 | 2.12 | 15 | yes | NDE1; MYH11; ABCC6; XYLT1 | secondary infertility 1 year; no symptoms | |
| R1718 | 17pl2 | 14610001-15440000 | 1.28 | 6 | yes | PMP22 | secondary 9 years; no infertility symptoms | |
| R1719 | 18pll.21 | 13880001-14040000 | 0.16 | 1 | yes | MC2R | secondary 1 year; no infertility symptoms | |
| R1720 | 18pll.21 | 18620001-21820000 | 2.20 | 55 | yes | RTN4R; SERPIND1; PIBB; SCARF2; COMPT; PRODH | secondary infertility 5 years; no symptoms | |
| R1721 | 22qll.21 | 18960001-211000000 | 1.68 | 37 | yes | RTN4R; TBX1; GP1BB; SCARF2; COMT | primary infertility 1 year; no symptoms | |
| R1722 | 22qll.21 | 18920001-21420000 | 2.04 | 47 | yes | RTN4R; TBX1; SERPIND1; GP1BB; SCARF2;COMT; PRODH; SNAP29 | primary infertility 1 year; no symptoms | |
| R1723 | Xql2-ql3.1 | 67060001-68060000 | 1.00 | 4 | no | OPHN1 | primary infertility 5 years; no symptoms | |
| R1724 | Xql3.3 | 74180001-75380000 | 1.20 | 4 | no | ABCB7; ZDHHC15 | secondary infertility 1 year; one spontaneous abortion | |
| R1725 | Xp21.1 | 33040001-34720000 | 1.68 | 3 | yes | DMD | primary infertility 1 year; no symptoms | |
| R1726 | Xq21.1-q21.2 | 83580001-84780000 | 1.20 | 5 | no | POF1B | secondary infertility 1 one year; spontaneous two miscarriages; abortion | |
| R1727 | Xp22.31 | 6420001-8160000 | 1.68 | 6 | yes | STS | secondary infertility 8 years; no symptoms | |
| R1728 | Xp22.31 | 6420001-8160000 | 1.76 | 6 | yes | STS | secondary infertility 4 years; no symptoms | |
| R1729 | Xp22.21 | 6420001-8160000 | 1.72 | 5 | yes | STS | secondary infertility 3 years; no symptoms | |
| R1730 | Xp22.31 | 6420001-8160000 | 1.68 | 6 | yes | STS | secondary infertility 1 one year; spontaneous one miscarriage; abortion |
RefSeq genes: Reference Sequence genes; DGV: Database of Genomic Variants; ISCA: International Standards of Cytogenomic Arrays; DEC: DECIPHER; OMIM: Online Mendelian Inheritance in Man.
Figure 1.
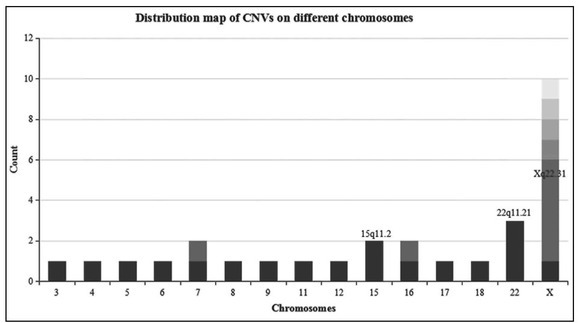
The figure shows the distribution map of CNVs on different chromosomes. The x-axis represents the number of chromosome and the y-axis represents the count of different CNVs in the 324 samples. Different color depth means different CNVs on the same chromosome. Copy number variations of Xq22.31, 22qll.21 and 15qll.2 show high frequencies in this study.
Discussion
This research showed the CNV preference for chromosome X in the 324 female infertility patients. The X chromosome is the key chromosome for female sexual organ development, and the long arm of the X chromosome was a key area of gonad development. The variation in chromosome X was prone to cause abnormal developments of gonad and infertility [17]. The presence of CNVs in the female genome may result in abnormalities in gamete
formation and meiosis of oocyte, thereby influence fertility. One sample exhibited CNV on Xq21.1-q21.2, which involved a gene, POF1B. Loss of function in POF1B could lead to exaggerated germ cell apoptosis and POF [this gene included by the Online Mendelian Inheritance in Man (OMIM)], but this CNV was duplication instead of deletion [18, 19]. Combined with clinical phenotype and female sex hormone levels detection, we believed that this CNV duplication may influence fertility. For CNV (9q22.32-q22.33) duplication, it contained HSD17B3 gene. The HSD17B3 gene mutations could impair testosterone bio-synthesis and causing male under masculinization [20], but its function has not been reported in females. For the rest of these CNVs, some contain protein coding genes related to diseases in the OMIM database, but they are mostly associated with other genetic disorders such as neuropathy, brain, skin and retina. Thus, the association between female infertility and CNVs are still unclear. Our data showed that CNVs may be not the main cause of unexplained infertility. In these 29 CNVs, there were five deletions and 24 duplications (Table 1). Our method cannot detect the inversions and translocations, which may lead to the low detection rate of the actual CNVs.
We also found five recurring CNVs on Xp22.31, and three times on 22q11.21. The average size of Xp22.31 CNVs was about 1.6 M, including one deletion and four duplications. For clinical phenotypes, four patients with normal phenotype (sex hormone level and normal uterus), and one patient had one miscarriages and one spontaneous abortion. Xp22.31 contained the encoding genes of STS, VCX2, VCX, VCX3A,PNPLA4,HDHD1, etc. It was reported that the deletion, reduplication, and rearrangement of Xp22.31 can lead to X-linked ichthyosis. These five patients all showed no X-linked ichthyosis clinical phenotype. In 2010, Krausz et al. [21] found the CNVs on Xp22.31 for two male infertility patients. So far, there has been no report about Xp22.31 on female infertility.
The Xp22.31 exhibits a frequency of 0.15% in a healthy population [22]. Thus, the correlation between Xp22.31 and female infertility needs to be further investigated.
Three of the female infertility patients had CNVs on 22q11.21 duplications. The average size of Xp22.31 CNVs was about 2 M. The clinical phenotypes of these three patients were unexplained primary infertility, and their husbands exhibited normal sperm. The deletion CNV (22ql1.21) was also found in two types IMRKH (Mayer-Rokitansky-Küster-Hauser) syndrome [14, 23]. The association between 22ql 1.21 deletions and Müllerian aplasia has been reported [24, 25]. Moreover, our patients have normal uterus and menstruation.
There are many factors that influence female infertility including POF, leiomyomas, endometriosis and polycystic ovarian syndrome (PCOS), etc. For the first time, our study explored the correlation between CNVs and female infertility using NGS technology, and also provided genetic evidence and references for future study and infertility etiology research.
Footnotes
Declaration of Interest. The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Funding. This study was supported by Natural Science funds of Xinjiang Uygur Autonomous Region [2017D01B10].
References
- 1.Stankiewicz P, Lupski JR. Structural variation in the human genome and its role in disease. Annu Rev Med. 2010;61(1):437–455. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- 2.Mukherjee A, Dass G, Mohanarao GJ, Katneni VK, Banerjee D, Das TK. Copy number differences of Υ chromosomal genes between superior and inferior quality semen producing crossbred (Bos taurus x Bos indicus) bulls. Anim Biotechnol. 2015;26(1):65–72. doi: 10.1080/10495398.2014.887020. et al. [DOI] [PubMed] [Google Scholar]
- 3.Knauff EAH, Blauw HM, Pearson PL, Kok K, Wijmenga C, Veldink JH. Copy number variants on the X chromosome in women with primary ovarian insufficiency. Fertil Steril. 2011;95(5):1584–1588. doi: 10.1016/j.fertnstert.2011.01.018. et al. [DOI] [PubMed] [Google Scholar]
- 4.Yan J, Fan L, Zhao Y, You L, Wang L, Zhao H. DYZ1 copy number variation, Υ chromosome polymorphism and early recurrent spontaneous abortion/ early embryo growth arrest. Eur J Obstet Gynecol Reprod Biol. 2011;159(2):371–374. doi: 10.1016/j.ejogrb.2011.07.033. et al. [DOI] [PubMed] [Google Scholar]
- 5.Tüttelmann F, Simoni M, Kliesch S, Ledig S, Dworniczak B, Wieacker P. Copy number variants in patients with severe oligozoospermia and Sertolicell-only syndrome. PloS One. 2011;6(4):el9426. doi: 10.1371/journal.pone.0019426. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eggers S, Deboer KD, Bergen JVD, Gordon L, White SJ, Jamsai D. Copy number variation associated with meiotic arrest in idiopathic male infertility. Fertil Steril. 2015;103(1):214–219. doi: 10.1016/j.fertnstert.2014.09.030. et al. [DOI] [PubMed] [Google Scholar]
- 7.Dong Y, Pan Y, Wang R, Zhang Z, Xi Q, Liu RZ. Copy number variations in spermatogenic failure patients with chromosomal abnormalities and unexplained azoospermia. Genet Mol Res. 2015;14(4):16041–16049. doi: 10.4238/2015.December.7.17. [DOI] [PubMed] [Google Scholar]
- 8.Simoni M, Bakker E, Krausz C. EAA/EMQN best practice guidelines for molecular diagnosis of y-chromosomal microdeletions. State of the art 2004. Int J Androl. 2004; 27(4):240–249. doi: 10.1111/j.1365-2605.2004.00495.x. [DOI] [PubMed] [Google Scholar]
- 9.Yang Y, Zhang SZ, Peng LM, Ding XP, Lin L, Wang J. Studies on molecular epidemiology of Υ chromosome azoospermia factor microdeletions in Chinese patients with idiopathic azoospermia or severe oligozoospermia. Chinese J Med Genet. 2003;20(5):385–389. et al. [PubMed] [Google Scholar]
- 10.Lo GD, Chianese C, Ars Ε, Ruiz-Castañé Ε, Forti G, Krausz C. Recurrent X chromosome-linked deletions: Discovery of new genetic factors in male infertility. J Med Genet. 2014;51(5):340–344. doi: 10.1136/jmedgenet-2013-101988. [DOI] [PubMed] [Google Scholar]
- 11.Ledig S, Röpke A, Wieacker P. Copy number variants in premature ovarian failure and ovarian dysgenesis. Sex Dev. 2010;4(4-5):225–232. doi: 10.1159/000314958. [DOI] [PubMed] [Google Scholar]
- 12.Tšuiko O, Nõukas M, Žilina O, Hensen K, Tapanainen JS, Mägi R. Copy number variation analysis detects novel candidate genes involved in follicular growth and oocyte maturation in a cohort of premature ovarian failure cases. Hum Reprod. 2016;31(8):1913–1925. doi: 10.1093/humrep/dew142. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ledig S, Hiort O, Scherer G, Wolff G, Morlot S, Kuechler A. Array-CGH analysis in patients with syndromic and non-syndromic XY gonadal dysgenesis: Evaluation of array CGH as diagnostic tool and search for new candidate loci. Hum Reprod. 2010;25(10):2637–2646. doi: 10.1093/humrep/deq167. et al. [DOI] [PubMed] [Google Scholar]
- 14.Ledig S, Schippert C, Strick R, Hoffmann M, Wolff G, Morlot S. Recurrent aberrations identified by array-CGH in patients with Mayer-Rokitansky-Küster-Hauser syndrome. Fertil Steril. 2011;95(5):1589–1594. doi: 10.1016/j.fertnstert.2010.07.1062. et al. [DOI] [PubMed] [Google Scholar]
- 15.Liu X, Li ZS, Su Z, Zhang JJ, Li HG, Xie J. Novel Y-chromosomal microdeletions associated with non-obstructive azoospermia uncovered by high throughput sequencing of sequence-tagged sites (STSs) Sci Rep. 2016;6:21831. doi: 10.1038/srep21831. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang MZ, Lin FQ, Li M, He D, Yu QH, Yang XX. Semiconductor sequencing analysis of chromosomal copy number variations in spontaneous miscarriage. Med Sci Monit. 2017;23:5550–5557. doi: 10.12659/MSM.905094. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gardner RJM, Sutherland GR, Shaffer LG. Chromosome abnormalities and genetic counseling. Am J Hum Genet. 1997;60(6):1567. [Google Scholar]
- 18.Bione S, Rizzolio F, Sala C, Ricotti R, Goegan M, Manzini MC. Mutation analysis of two candidate genes for premature ovarian failure, DACH2 and POF1B. Hum Reprod. 2004;19(12):2759–2766. doi: 10.1093/humrep/deh502. et al. [DOI] [PubMed] [Google Scholar]
- 19.Lacombe A, Lee H, Zahed L, Choucair M, Muller JM, Nelson SF. Disruption of POF1B binding to nonmuscle actin filaments is associated with premature ovarian failure. Am J Hum Genet. 2006;79(1):113–119. doi: 10.1086/505406. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindqvist A, Hughes IA, Andersson S. Substitution mutation C268Y causes 17 β-hydroxysteroid dehydrogenase 3 deficiency. J Clin Endocr Metab. 2001;86(2):921–923. doi: 10.1210/jcem.86.2.7172. [DOI] [PubMed] [Google Scholar]
- 21.Krausz C, Giachini C, Lo GD, Daguin F, Chianese C, Ars Ε. High resolution X chromosome-specific array-CGH detects new CNVs in infertile males. PloS One. 2012;7(10):e44887. doi: 10.1371/journal.pone.0044887. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li F, Shen Y, Köhler U, Sharkey FH, Menon D, Coulleaux L. Interstitial microduplication of Xp22.31: Causative of intellectual disability or benign copy number variant? Eur J Med Genet. 2010;53(2):93–99. doi: 10.1016/j.ejmg.2010.01.004. et al. [DOI] [PubMed] [Google Scholar]
- 23.Cheroki C, Krepischi-Santos AC, Rosenberg C, Jehee FS, Mingroni-Netto RC, Pavanello FI. Report of a del22q11 in a patient with Mayer-Rokitansky-Küster-Hauser (MRKH) anomaly and exclusion of WNT-4, RAR-γ, and RXR-a, as major genes determining MRKH anomaly in a study of 25 affected women. Am J Med Genet A. 2006;140A(12):1339–1342. doi: 10.1002/ajmg.a.31254. et al. [DOI] [PubMed] [Google Scholar]
- 24.Cheroki C, Rrepischi-Santos AC, Szuhai K, Brenner V, Kim CA, Otto PA. Genomic imbalances associated with mullerian aplasia. J Med Genet. 2008;45(4):228–232. doi: 10.1136/jmg.2007.051839. et al. [DOI] [PubMed] [Google Scholar]
- 25.Sundaram UT, McDonald-McGinn DM, Huff D, Emanuel BS, Zackai EH, Driscoll DA. Primary amenorhhea and absent uterus in the 22q11.2 deletion syndrome. Am J Med Genet A. 2007;143A(17):216–218. doi: 10.1002/ajmg.a.31736. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]


