Abstract
Background
Haemoproteus (Parahaemoproteus) species (Haemoproteidae) are widespread blood parasites that can cause disease in birds, but information about their vector species, sporogonic development and transmission remain fragmentary. This study aimed to investigate the complete sporogonic development of four Haemoproteus species in Culicoides nubeculosus and to test if phylogenies based on the cytochrome b gene (cytb) reflect patterns of ookinete development in haemosporidian parasites. Additionally, one cytb lineage of Haemoproteus was identified to the species level and the in vitro gametogenesis and ookinete development of Haemoproteus hirundinis was characterised.
Methods
Laboratory-reared C. nubeculosus were exposed by allowing them to take blood meals on naturally infected birds harbouring single infections of Haemoproteus belopolskyi (cytb lineage hHIICT1), Haemoproteus hirundinis (hDELURB2), Haemoproteus nucleocondensus (hGRW01) and Haemoproteus lanii (hRB1). Infected insects were dissected at intervals in order to detect sporogonic stages. In vitro exflagellation, gametogenesis and ookinete development of H. hirundinis were also investigated. Microscopic examination and PCR-based methods were used to confirm species identity. Bayesian phylogenetic inference was applied to study the relationships among Haemoproteus lineages.
Results
All studied parasites completed sporogony in C. nubeculosus. Ookinetes and sporozoites were found and described. Development of H. hirundinis ookinetes was similar both in vivo and in vitro. Developing ookinetes of this parasite possess long outgrowths, which extend longitudinally and produce the apical end of the ookinetes. A large group of closely related Haemoproteus species with a similar mode of ookinete development was determined. Bayesian analysis indicates that this character has phylogenetic value. The species identity of cytb lineage hDELURB2 was determined: it belongs to H. hirundinis.
Conclusions
Culicoides nubeculosus is susceptible to and is a likely natural vector of numerous species of Haemoproteus parasites, thus worth attention in haemoproteosis epidemiology research. Data about in vitro development of haemoproteids provide valuable information about the rate of ookinete maturation and are recommended to use as helpful step during vector studies of haemosporidian parasites, particularly because they guide proper dissection interval of infected insects for ookinete detection during in vivo experiments. Additionally, in vitro studies readily identified patterns of morphological ookinete transformations, the characters of which are of phylogenetic value in haemosporidian parasites.
Keywords: Haemoproteus, Birds, Cytochrome b gene, Sporogony, Vectors, Culicoides nubeculosus
Background
Avian haemosporidian parasites (Plasmodium, Haemoproteus, Leucocytozoon and Fallisia) are widespread and remarkably diverse; this diversity frequently exceeds that of avian hosts [1–5]. During the past ten years, it has been shown that several Haemoproteus species can be harmful to their avian hosts, compromising their health and even causing mortality, especially in non-adapted birds [6–9]. This calls for additional studies aimed at a better understanding of pathogen transmission. Blood-sucking insects can also be negatively affected and even killed by Haemoproteus parasites after feeding on heavily infected blood (when > 1% of gametocytes are mature) [10–12].
Many studies have addressed parasite diversity and the phylogenetic relationships of haemosporidian parasites of various avian hosts [4, 5, 13–18], but information about vector competence and patterns of sporogonic development of these pathogens remains insufficient. Haemoproteus (Parahaemoproteus) species (Haemosporida: Haemoproteidae) are cosmopolitan in countries with warm and temperate climates [2, 3]. These blood parasites are transmitted by biting midges of the Ceratopogonidae [2]. Recently, several studies addressed Haemoproteus spp. sporogonic development in several species of Culicoides biting midges [19–23]. However, due to the big variety of Haemoproteus species and the scarcity of detailed information about patterns of sporogonic development in the majority of the described species, understanding haemoproteid parasite sporogony remains essential for better understanding the biology of haemosporidians. This calls for additional experimental and field vector research.
The presence of haemosporidian parasites in wild-caught insects has been frequently reported in many parts of the world using PCR-based diagnostics [24–30]. However, this methodology alone does not provide information about patterns of sporogonic development. Additionally, identification of PCR-positive insects does not necessarily show that sporogony is completed and sporozoites (infective stage for birds) reach salivary glands. On the other hand, the observation of infective sporozoite stages in salivary glands (accessed through insect dissection and microscopical analysis) provides stronger evidence about the possible vectorial capability of blood-sucking insects. Experimental infections not only allow one to follow parasite development and morphologically characterize each sporogonic stage, but also to confirm the presence of sporozoites in the salivary glands [22, 23, 31]. Laboratory-reared Culicoides nubeculosus can be used for conducting such experimental infections with Haemoproteus parasites. The methodology for rearing this biting midge in the laboratory was developed over 40 years ago [32], and so far, this insect has been shown to be susceptible to eight Haemoproteus species [22, 23].
Phylogenetic and genomics research are in progress in wildlife haemosporidians studies [33, 34]. However, it remains unclear if and how numerous available phylogenies, which are based on DNA sequences, reflect patterns in biology of haemosporidians. In other words, it remains insufficiently understood what the well-supported phylogenetic clades indicate in regard to the biology of the parasites. Phylogenies based on mitochondrial genes indicate haemosporidian parasite-vector relationships [22, 35] and it is probable that they also can be used for more delicate understanding of sporogony in haemosporidians. However, this issue has not been addressed in haemosporidian research.
Interestingly, the exflagellation, gametogenesis and development of ookinetes of Haemoproteus species can be readily induced in vitro when mature gametocytes are exposed to air. This process is easy to initiate with Haemoproteus parasites because they exflagellate in vitro without need of any vector-related gut factors, which is not the case in Plasmodium parasites of mammals [36] and birds [2]. This provides opportunities to access initial sporogonic stages (gametes, zygotes, ookinetes) in standard in vitro conditions for comparative studies. In vitro exflagellation experiments were successfully used to characterize gametogenesis and ookinete development of several Haemoproteus species [2, 37, 38]; they were used in haemosporidian parasite hybridization studies [39–41] and genomic research when large amount of pure parasite DNA is needed [42]. This methodology can also be applied in haemosporidian vector studies, since it provides information about ookinete morphological transformation and development rate. The latter character provides important guidance in determining the optimal dissection time of infected insects for the detection of mature ookinetes in vivo [2, 31].
Parasite distribution is influenced by numerous factors [2, 43]. In haemosporidians, not only parasite, vector and bird community factors are important in epidemiology [44], but the effect of environmental and other ecological variables are fundamental in parasite prevalence and distribution [45–47]. Identifying the factors, which limit parasite transmission, is important for better understanding disease epidemiology and development of preventive measures. This issue is particularly sensitive to address regarding several widespread Haemoproteus parasites, which are present and prevalent in European birds, but are not transmitted in Europe, for example Haemoproteus hirundinis, Haemoproteus payevskyi and Haemoproteus nucleocondensus [2, 48].
This study investigated the sporogonic development of four Haemoproteus (Parahaemoproteus) parasites: Haemoproteus belopolskyi (cytochrome b gene lineage hHIICT1), H. hirundinis (hDELURB2), H. nucleocondensus (hGRW01) and Haemoproteus lanii (hRB1). The main aim was to follow complete sporogony of these parasites in the biting midge Culicoides nubeculosus, which is widespread in Europe [49]. Additionally, we investigated the in vitro exflagellation, gametogenesis and ookinete development of H. hirundinis (hDELURB2) and compared this process with the same features reported in other avian haemoproteids. The phylogenetic relationships were inferred among Haemoproteus species, for which vectors and ookinete development in vitro have been identified. Available information about transmission of these parasites in Europe is discussed.
Methods
Blood sampling and microscopic analysis
Experiments were carried out at the Ventės Ragas Ornithological Station, Lithuania (55°20′28.1″N, 21°11′25.3″E) in May and June 2018. Birds were captured using mist nets, ‘Zigzag’ traps and a funnel trap. Blood samples were collected from the brachial vein (~30 µl) using heparinized microcapillaries. A small drop of blood was used to prepare blood smears which were immediately air-dried using a battery-powered fan, fixed in absolute methanol and stained with Giemsa [2]. Remaining blood was stored in SET buffer (0.05 M Tris, 0.15 M NaCl, 0.5 M EDTA, pH 8.0) for DNA extraction. Blood smears were examined using an Olympus BX-43 light microscope equipped with Olympus SZX2-FOF digital camera and QCapture Pro 6.0, Image Pro Plus (Olympus, Tokyo, Japan) imaging software. The analysis consisted of screening each slide for 15–20 min at low magnification (×400) and at least 100 fields at high magnification (×1000). Parasitaemia was determined by actual counting of the number of mature gametocytes per 1000 red blood cells or per 10,000 red blood cells when low parasitaemia was present (≤ 0.1%) [50]. Parasites were morphologically identified according to Valkiūnas [2] and available literature.
In vitro exflagellation and ookinete development rate
One individual northern house martin Delichon urbicum, with parasitaemia of mature gametocytes of approximately 0.2% was selected as a H. hirundinis (hDELURB2) gametocyte donor to observe in vitro exflagellation, gametogenesis and ookinete development. The experiment followed the methodology described by Valkiūnas [2] and Valkiūnas et al. [40]. Briefly, approximately 100 µl of blood was collected from the brachial vein and immediately mixed with a 3.7% solution of sodium citrate in a microtube Eppendorf type, in the proportion of 4 parts blood to 1 part sodium citrate solution. This mixture was maintained opened in a humid chamber at room temperature (~21 °C). Blood smears were prepared at intervals of 1, 3, 5, 10, 15, 30 and 45 min and at 1, 2, 3, 4, 6, 8, 10, 12 and 24 h after exposure of blood to air (EBA). The blood mixture was gently homogenised before blood smear preparation. Blood smears were air dried, fixed, stained and analysed as described for blood films above. The donor bird was released after blood collection. Representative preparations for examination of exflagellation, gametogenesis and ookinete development were deposited in the Nature Research Centre, Vilnius, Lithuania, under the accession numbers 49134–49147 NS.
Experimental design of sporogonic development in vivo
After microscopic examination of the blood smears, birds harbouring single Haemoproteus infections with low parasitaemia (≤ 0.1% mature gametocytes) were selected as donors (see Bukauskaitė et al. [20] for details of exposure). One individual of the following bird species was exposed to biting midges: the icterine warbler Hippolais icterina, the northern house martin Delichon urbicum, the great reed warbler Acrocephalus arundinaceus and the red-backed shrike Lanius collurio were infected with Haemoproteus belopolskyi (hHIICT1), Haemoproteus hirundinis (hDELURB2), Haemoproteus nucleocondensus (hGRW01) and Haemoproteus lanii (hRBS4), respectively. All birds were immediately released after the experiment.
Laboratory-reared Culicoides nubeculosus biting midges were kept in small cardboard boxes covered with fine silk mesh. Each box hosted approximately 80 biting midge individuals. Boxes with insects were gently pressed against the pectoral muscle of birds on a feather-free area. Insects were allowed to take a blood meal throughout the silk mesh for about 40 min. Biting midges where then transferred to a bigger cage (12 × 12 × 12 cm3) made of wire and fine silk mesh and males along with non-fed females were removed. Engorged females were kept at 24.8 ± 0.5 °C, 60 ± 4% humidity and controlled light-dark photoperiod (17:7 h). Insects were fed daily using cottons pads moistened with 10% sugar solution placed on the top of each cage.
Insect dissection and parasite preparation
Experimentally-infected biting midges were dissected at set of intervals to prepare ookinete preparations (which were made between 4 and 12 h post-exposure), oocyst preparations (between 2 and 7 days post-exposure) and sporozoite preparations (between 6 and 9 days post-exposure). Dissection needles were disinfected after each dissection using fire to prevent contamination. Before dissection, insects were anesthetized using 96% ethanol vapour from a moistened cotton-wool ball.
For ookinete preparations, midguts were extracted and gently crushed on glass slides; thin smears were prepared, fixed and stained as described above for blood slides. To visualize oocysts, temporary preparations were made by isolation of midgut, which was placed on a glass slide and covered with a cover-slip. A drop of 2% mercurochrome solution was used to stain midguts and differentiate oocysts. To prepare sporozoite preparations, biting midge salivary glands were extracted and gently crushed to prepare small thin smears that were fixed in absolute methanol and stained with Giemsa using a 4% solution for 1 h. Residual parts of all dissected insects were fixed in 96% ethanol and used for PCR-based analysis to confirm the presence of corresponding parasite lineage.
All vector preparations were examined at high (×1000) magnification using the same equipment, as for examination of blood smears. Parasite images were collected to prepare measurement. All measurements are in micrometres. Statistical analyses were carried out using ‘R studio’ v.3.4.3. Representative preparations of vector stages were deposited in the Nature Research Centre, Vilnius, Lithuania, under the accession numbers 49126–49133 NS.
DNA extraction, PCR amplification, sequencing and sequence data analysis
Standard ammonium acetate protocol was applied for DNA extraction from blood and exposed biting midge samples [51]. A nested PCR protocol targeting a fragment of cytochrome b gene (cytb) was used [1, 52]. The first reaction was conducted with primers HaemNFI/HaemNR3 capable of amplifying Haemoproteus, Plasmodium and Leucocytozoon DNA. For the nested reaction, primers HaemF/HaemR2 capable of Haemoproteus and Plasmodium DNA amplification were used. Target DNA was amplified in 25 μl total volume including 50 ng of total genomic DNA template (2 μl), 12.5 μl of Dream Taq Master Mix (Thermo Fisher Scientific, Vilnius, Lithuania), 8.5 μl of nuclease-free water and 1 μl of each primer (10 μM concentration). One negative (nuclease-free water) and one positive control (a sample with Haemoproteus sp. infection positive in the slide) were used in every run. PCR products were evaluated using electrophoresis in a 2% agarose gel. Positive PCR products of ~480 bp were precipitated and sequenced from both strands using the Big Dye Terminator V3.1 Cycle Sequencing Kit (Thermo Fisher Scientific, Vilnius, Lithuania) and ABI PRISM™ 3100 capillary sequencing robot (Applied Biosystems, Foster City, CA, USA). Sequences were edited and aligned using BioEdit software [53] to create a consensus sequence. The presence of double peaks in electropherograms would have been considered as an indication of a mixed infection [54]. Consensus sequences obtained were aligned using BLAST (Basic Local Alignment Search Tool) with sequences from MalAvi (http://mbio-serv2.mbioekol.lu.se/Malavi/blast.html) and GenBank database (http//www.ncbi.nlm.nih.gov/BLAST). Sequences were deposited in GenBank (MN025422–MN025425).
Additionally, a longer cytb fragment (1001 bp) of each sample was amplified using a nested PCR protocol with primers AE298/AE299 for the first reaction, and primers AE064/AE065 for the second PCR reaction [55]. A fragment of caseinolytic protease (clpc) from the plastid genome (554 bp) was amplified using a nested protocol [56]. Sequences were deposited in the GenBank database under the accession numbers MK843310–MK843317. These data were not used in this study but are important for future taxonomic and phylogenetic work.
The possible presence of co-infections in parasite-donor birds was carefully checked, and the presence of co-infections was ruled out due to the absence of double peaks in sequence chromatograms and microscopic examination of blood films (see description of methods above). Importantly, presence of a single infection of corresponding parasite lineages was also supported by sequencing DNA isolated from insects when the sporogony was completed; double peaks in sequence chromatograms were absent.
Phylogenetic analysis
Phylogenetic relationships among Haemoproteus species in which vectors have been identified was inferred using the Bayesian algorithm implemented by MrBayes v.3.2.0 [57]. In total, 52 haemosporidian cytb sequences were used, 40 belonging to Haemoproteus species, 11 to Plasmodium spp. and one to Leucocytozoon sp. (lSISKIN2) was used as outgroup. Generalised time-reversible (GTR) evolutionary model was selected by MrModeltest2 [58]. Two simultaneous runs were conducted with a sample frequency of every 100th generation over 3 million generations. We discarded 25% of the trees as ‛burn-inʼ. The remaining trees were used to construct a majority rule consensus tree. The phylogeny was visualized using Fig Tree v.1.4 [59].
Preparations of in vitro zygotes and ookinetes of Haemoproteus species
From the 27 Haemoproteus lineages shown in sub-clades b1 and b2 (Fig. 1), in vitro zygote and ookinete preparations were available for 13 of them (Fig. 1, indicated with a star). This material was collected and deposited in the collection of the Nature Research Centre, Vilnius (Lithuania) during in vitro studies by Valkiūnas [2] and Valkiūnas et al. [39, 40]. These preparations (Additional file 1: Table S1) were used in this study to compare the development of corresponding Haemoproteus species in vitro. These materials were examined as described above for blood smears.
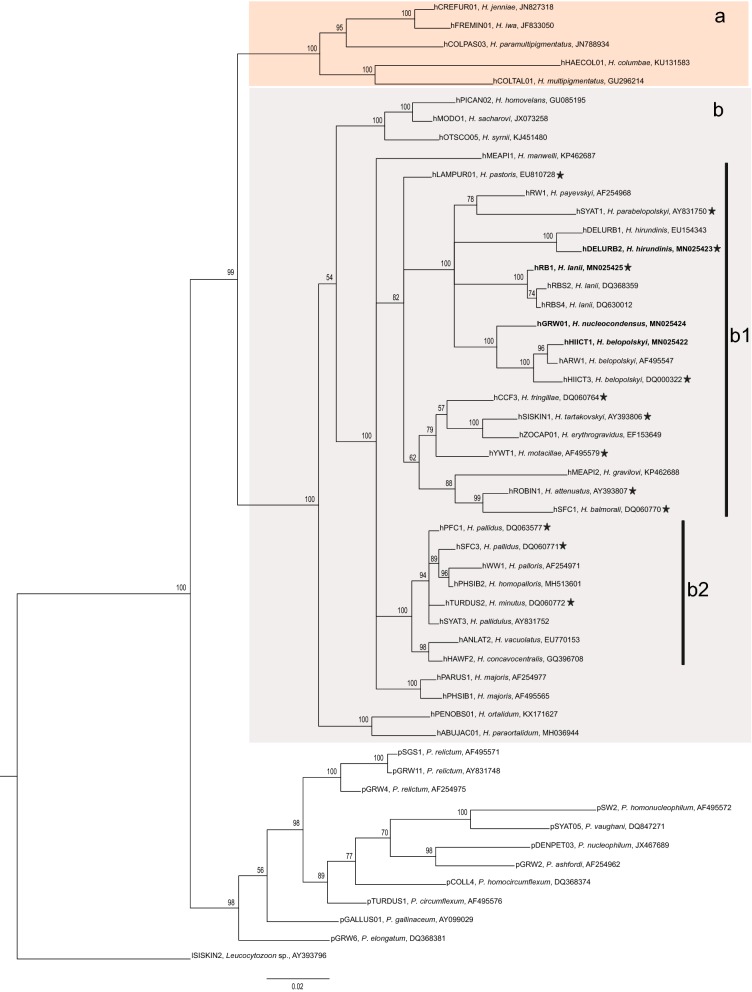
Fig. 1.
Bayesian phylogenetic inference of cytb lineages (478 bp) of 33 Haemoproteus species. The tree is rooted with Leucocytozoon sp. sequence (lSISKIN2). Clades A and B contain species of subgenera Haemoproteus and Parahaemoproteus, respectively. Sub-clade b1 contains species with slow development and relatively large-sized ookinetes, which produces a long outgrowth during development and possess prominent vacuoles in the cytoplasm. Sub-clade b2 contains species with fast development and relatively small ookinetes, which do not produce outgrowths and usually do not possess prominent vacuoles in the cytoplasm. MalAvi lineage codes are provided, followed by parasite species names and GenBank accession numbers. Nodal support values indicate Bayesian posterior probabilities. Names of the parasites, for which sporogony was investigated in this study are given in bold. Stars indicate parasite species, for which development in vitro has been investigated. Other explanations are given in the text
Results
The presence of single Haemoproteus infections was confirmed in all donor birds and these findings were supported both by microscopic examination of blood films and the absence of double-peaks in PCR-based testing. Exposed biting midges were also tested by the same PCR protocol used to test blood samples. It was possible to confirm the presence of the same parasite lineage in exposed insects and in blood samples, along with the absence of double-peaks in sequencing.
In vitro exflagellation and ookinete development of Haemoproteus hirundinis
Within 5 min after exposure of blood to air (EBA), mature macrogametocytes rounded up and exflagellation of microgametes was observed (Fig. 2a). First macrogametes (Fig. 2b) were observed between 10–15 min EBA, and at this time free microgametes were also observed (Fig. 2c). Zygotes were observed 60 min after EBA; it was possible to distinguish them due to the presence of a prominent vacuole located close to the nucleus (Fig. 2d). The initial stages of ookinete differentiation were detected 3 h after EBA, when a long finger-like outgrowth appeared; the latter locates tangentially to the main body of the differentiating ookinete (Fig. 2e). As the ookinete develops, this outgrowth extends longitudinally and forms the anterior or apical end of the ookinete. The first fully grown ookinetes were observed between 6–8 h after EBA (Fig. 2f). At this stage of development, parasite usually does not possess pigment granules anymore, and usually two prominent vacuoles (one at the anterior and another at apical pole) were visible in the cytoplasm (Fig. 2f). Mature ookinetes of H. hirundinis (n = 21) measured 12.8–18.9 µm (on average 15.3 ± 1.6 µm) in length, 1.9–3.2 (2.5 ± 0.3) µm in width and 21.0–43.4 (28.9 ± 5.2) µm2 in area.
Fig. 2.
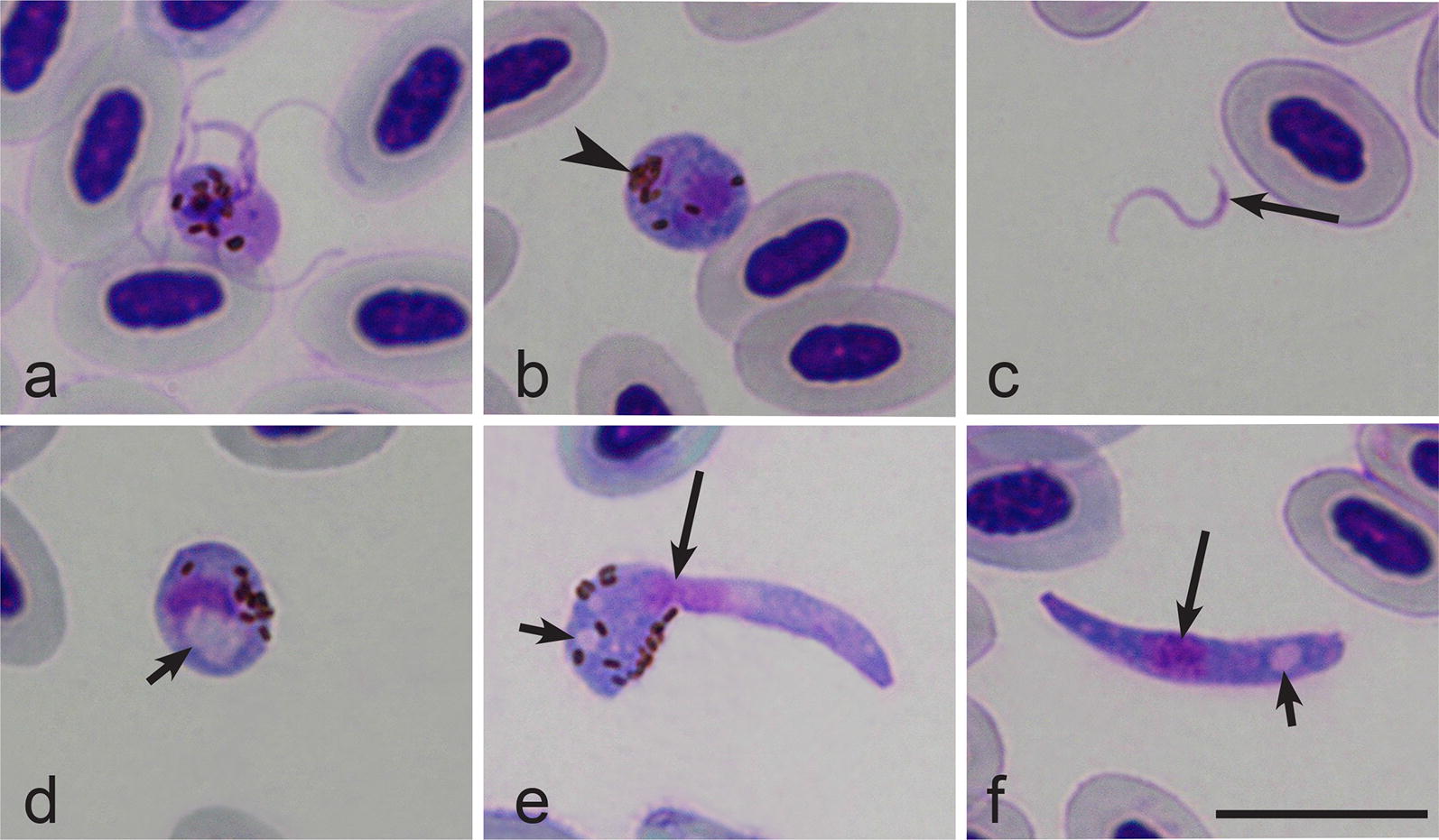
In vitro exflagellation (a), gametes (b, c), zygote (d) and ookinetes (e, f) of Haemoproteus hirundinis (hDELURB2). a Exflagellating microgametocyte. b Macrogamete. c Microgamete. d Zygote (note presence of a large vacuole in the cytoplasm). e Developing ookinete 3 h after exposure of mature gametocytes to air (note the presence of a long outgrowth and few vacuoles). f Mature ookinete with anterior end thinner than the posterior end (note the presence of a prominent vacuole, and absence of pigment granules). Long simple arrows, nuclei of parasites; short simple arrows, vacuoles; simple arrowhead, pigment granules. Methanol-fixed and Giemsa-stained thin films. Scale-bar: 10 µm
Sporogonic development in vivo
The capacity of C. nubeculosus to support the sporogonic development of H. belopolskyi (hHIICT1), H. hirundinis (hDELURB2), H. nucleocondensus (hGRW1) and H. lanii (hRB1) was confirmed by microscopic examination of corresponding bird blood stages and insect preparations (Fig. 3). Sporozoites of all these species were detected in salivary gland preparations (Fig. 3d, h, l, p).
Fig. 3.
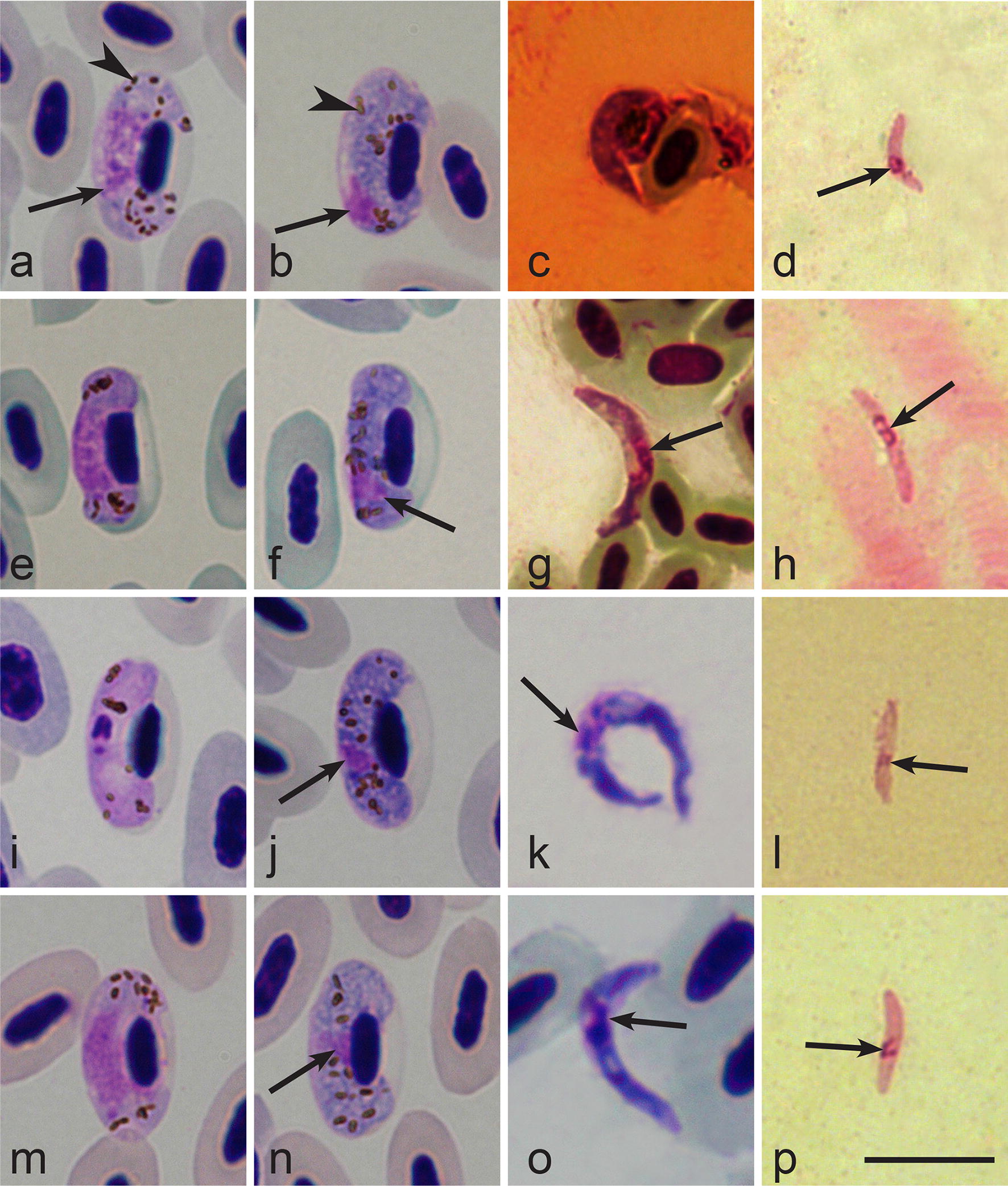
Mature gametocytes (a, b, e, f, i, j, m, n) from peripheral circulation of birds and ookinetes (c, g, k, o) and sporozoites (d, h, l, g) in Culicoides nubeculosus. a–d Haemoproteus belopolskyi (cytb lineage hHIICT1). E–h Haemoproteus hirundinis (hDELURB2). i–l Haemoproteus nucleocondensus (hGRW1). m–p Haemoproteus lanii (hRB1). Microgametocytes (a, e, i, m) and macrogametocytes (b, f, j, n) in the blood of Hippolais icterina (a, b), Delichon urbicum (e, f), Acrocephalus arundinaceus (i, j) and Lanius collurio (m, n). Long simple arrows, nuclei of parasites; simple arrowheads, pigment granules. Methanol-fixed and Giemsa-stained preparations. Scale-bar: 10 µm
Mature ookinetes of all examined species were observed in midgut preparations between 6 and 8 h post-exposure (hpe). They were observed 8 hpe in H. belopolskyi preparations (Fig. 3c), 6–8 hpe in H. hirundinis (Fig. 3g), 6 hpe in H. nucleocondensus (Fig. 3k) and 7 hpe in H. lanii (Fig. 3o). A few morphologically non-deformed ookinetes of studied parasite species were observed. Their length and width measured 15.5 × 2.0 µm in H. belopolskyi (n = 1); 9.4 × 2.5 µm in H. hirundinis (n = 1); 12.9–18.6 × 1.3–2.1 (mean 15.8 ± 1.5 by 1.6 ± 0.3) µm in H. nucleocondensus (n = 17) and 9.4–14.4 × 1.2–2.2 (mean 12.6 ± 1.5 by 1.6 ± 0.3) µm in H. lanii (n = 8). Ookinetes of all species were worm-like elongate bodies, with approximately centrally located nuclei and vacuolated cytoplasm; pigment granules were not observed. Haemoproteus nucleocondensus ookinetes were often observed in a circular position, with both anterior and posterior ends almost touching each other, resembling a circle (Fig. 3k).
Sporozoites were observed in salivary glands preparations between 7–9 days post-exposure (dpe). They were detected 7–9 dpe in H. belopolskyi and H. hirundinis (Fig. 3d and h, respectively), 7–8 dpe in H. nucleocondensus (Fig. 3l) and 6–9 dpe in H. lanii (Fig. 3p). Measurements of H. lanii (n = 15) sporozoites were 6.0–9.0 (mean 7.3 ± 1.0) µm in length, 0.9–1.4 (1.1 ± 0.1) µm in width and 2.4–4.2 (3.6 ± 0.5) µm2 in area. A small number of sporozoites were observed in other species. Their length and width measured 6.7–8.0 × 1.1–1.3 µm in H. belopolskyi (n = 4), 10.0 × 1.1 µm in H. hirundinis (n = 1) and 8.3 × 1.4 µm in H. nucleocondensus (n = 1). Sporozoites of all four studied species were fusiform bodies with pointed ends and slightly off-centre located nuclei.
Phylogenetic inference and its correspondence to patterns of ookinete development
All four Haemoproteus species investigated in this study clustered in a well-supported clade with other species transmitted by Culicoides insects (Fig. 1, Clade B). This clade contains parasites belonging to Parahaemoproteus subgenus. Haemoproteus (Haemoproteus) species, which are transmitted by louse flies (Hippoboscidae), clustered in a well-supported sister clade (Fig. 1, Clade A).
Fourteen identified species of haemoproteids clustered together in our phylogenetic analysis (Fig. 1, sub-clade b1). Sporogonic development has been investigated in ten of these parasites, and data about their ookinete development are available; all these parasites are characterised by the presence of long outgrows during initial stages of ookinete development, the readily distinguishable character (Fig. 4b, e, h, k, n, q, t, w, z, cc). Additionally, mature ookinetes of these parasites develop relatively slowly in vitro; they appear approximately 6 h after EBA and even later at ~20 °C [2]. Interestingly, two Haemoproteus parasites (H. minutus and H. pallidus), which do not produce long outgrowth on initial stages of ookinete development (Fig. 5b, e, h), clustered together and appeared in a different well-supported sub-clade (Fig. 1, sub-clade b2) separately from parasites of sub-clade b1 (Fig. 1). Ookinetes of these two parasites develop relatively fast in vitro; they appear approximately between 2 and 3 h after EBA at same conditions [2]. Because (i) the sub-clades b1 and b2 are relatively well supported and (ii) the parasites appeared in these clades are readily distinguishable both by the rate of development and the mode of ookinete transformation at the initial stage of development, both these characters should be of important phylogenetic value. In other words, phylogenies based on partial cytb reflect patterns of ookinete development in haemosporidian parasites.
Fig. 4.
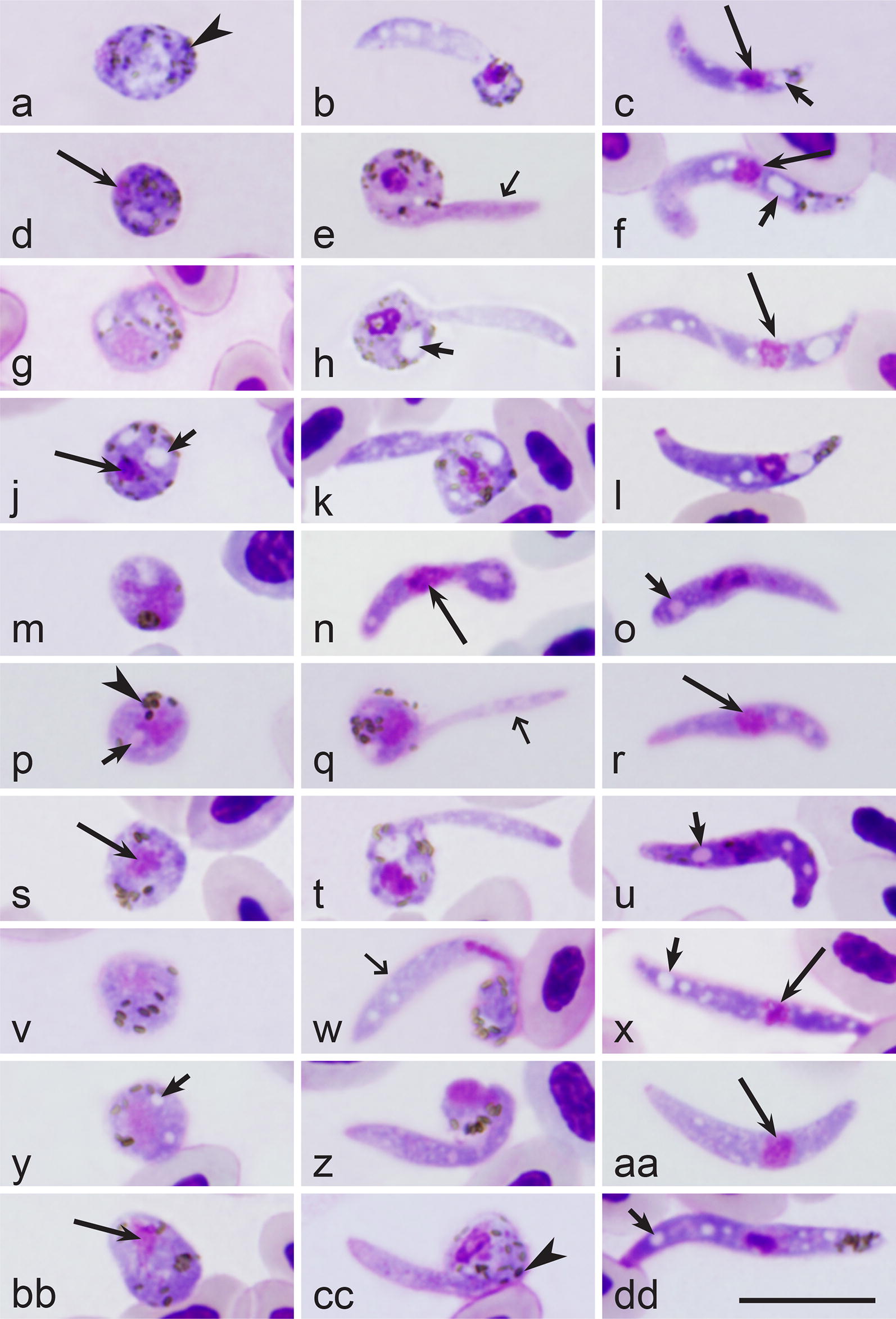
Zygotes (a, d, g, j, m, p, s, v, y, bb), growing ookinetes (b, e, h, k, n, q, t, w, z, cc) and mature ookinetes (c, f, i, l, o, r, u, x, aa, dd) of Haemoproteus attenuatus (cytb lineage hROBIN1, a–c), Haemoproteus balmorali (hSFC1, d–f), Haemoproteus belopolskyi (hHIICT3, g–i), Haemoproteus fringillae (hCCF3, j–l), Haemoproteus hirundinis (hDELURB2, m–o), Haemoproteus lanii (hRB1, p–r), Haemoproteus motacillae (hYWT1, s–u), Haemoproteus parabelopolskyi (hSYAT1, v–x), Haemoproteus pastoris (hLAMPUR1, y–aa) and Haemoproteus tartakovskyi (hSISKIN1, bb–dd) during development in vitro. Note that all these parasites produce long outgrowths during initial stages off ookinete development, and their ookinetes are markedly vacuolated; these parasites appeared in one relatively well-supported sub-clade b1 in the phylogenetic tree (Fig. 1). Long simple arrows, nuclei of parasites; short simple arrows, vacuoles; simple arrowheads, pigment granules; short simple wide arrows, outgrowths of developing ookinetes. Methanol-fixed and Giemsa-stained thin blood films. Scale-bar: 10 µm
Fig. 5.
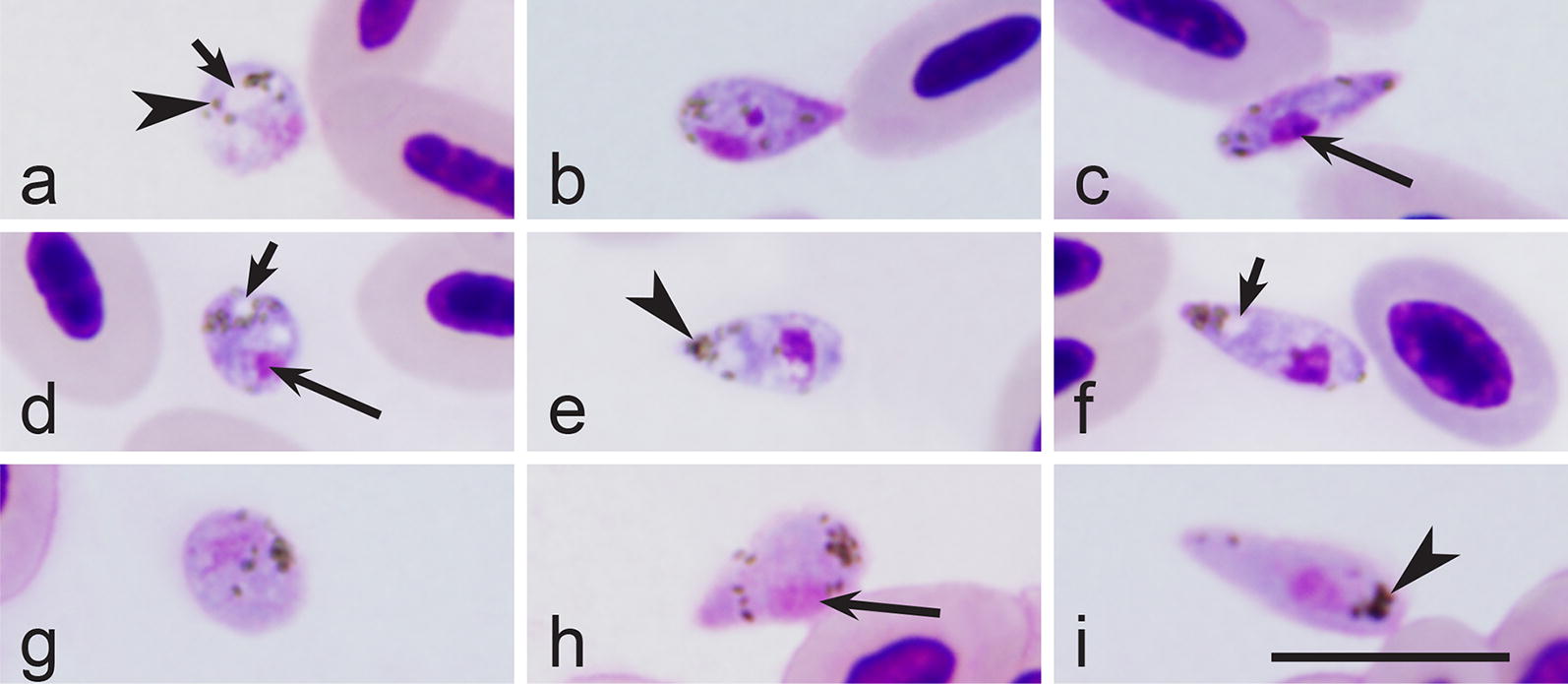
Zygotes (a, d, g), growing ookinetes (b, e, h) and mature ookinetes (c, f, i) of Haemoproteus minutus (hTURDUS2, a–c), Haemoproteus pallidus (hSFC3, d–f) and Haemoproteus pallidus (hPFC1, g–i) during development in vitro. Note that all these parasites do not produce any outgrowths during ookinete development, and their ookinetes do not contain vacuoles (c, d) or are only slightly vacuolated (f); these parasites appeared in one well-supported sub-clade b2 in the phylogenetic tree (Fig. 1). Long simple arrows, nuclei of parasites; short simple arrows, vacuoles; simple arrowheads, pigment granules. Methanol-fixed and Giemsa-stained thin blood films. Scale-bar: 10 µm
Discussion
This study adds four species of Haemoproteus to the list of avian haemoproteids that are transmitted by biting midges of the Ceratopogonidae and complete sporogonic development in laboratory-reared C. nubeculosus. The presence of sporozoites of H. belopolskyi (hHIICT1), H. hirundinis (hDELURB2), H. nucleocondensus (hGRW1) and H. lanii (hRB1) in the salivary glands of C. nubeculosus biting midges is evidence that this insect likely can act as a natural vector of these parasites.
Eight species of haemoproteids were recorded completing sporogonic development and producing sporozoites in C. nubeculosus [22, 23]. This insect readily supports sporogony of haemoproteids and is easy to maintain in the laboratory; we recommend using C. nubeculosus in experimental avian Haemoproteus research. This insect has a wide distribution across Eurasia [49] and can feed on the blood of many mammalian and avian species [20, 60–63].
Despite the simple morphology of sporozoites and few morphological characters visible under the light microscope in Haemoproteus species, it is possible to notice morphological differences among them on parasite species level [20–22]. Sporozoites of some species can be readily distinguished from each other by length, as is the case in Haemoproteus noctuae and Haemoproteus syrnii during development in Culicoides impunctatus; mainly, sporozoites of the former parasite are significantly longer [20]. In the present study, sporozoites of H. hirundinis seem to be longer than in other species as well, while the width was similar in all studied species. However, this information should be carefully interpreted because sporozoites of Haemoproteus parasites were few in preparations during this study. Only preparations of H. lanii contained sufficient numbers of sporozoites to perform statistical analysis, providing opportunity to measure and analyse their morphological features in more detail.
It is interesting to note that only a few sporogonic stages were found in the majority of insect preparations of almost all investigated parasite species in this study. This prevented a detailed statistical comparison of reported ookinetes, oocysts and sporozoites. The low intensity of sporogonic stages is likely due to the selection of donor birds with light parasitaemia of mature gametocytes (approximately 0.1%) for exposure of insects in this study. Haemoproteus parasites are highly virulent and even lethal to C. nubeculosus and other biting midges and even mosquitoes when they feed on a blood meal with gametocytaemia over 2–3% [11, 12]. Numerous ookinetes of Haemoproteus parasites develop rapidly, then migrate to midguts and kill insects due to midgut damage after feeding on heavily infected blood. It is thus essential to avoid such conditions during experiments aimed at studying complete sporogonic development of Haemoproteus parasites in vectors. Several studies used donor birds with gametocytaemia ranging between 0.2% and approximately 2% [10, 63] to expose biting midges. Mortality of exposed insects occurred, but some insects survived and more ookinetes, oocysts and sporozoites were observed in insect preparations for analysis. Thus, it is important to determine optimal parasitaemia, which can be used in experimental exposure of insects: numerous insects would survive, but parasite sporogonic stages might be difficult to find and visualize if the parasitaemia is too light (0.1% in this study). Conversely, the majority of insects would die soon after exposure if the parasitaemia is relatively high (> 1%), but more sporogonic stages would be detected in the few surviving insects. In each vector exposure experiment, the optimal parasitaemia in donor birds should be determined. Based on available information, it is likely between 0.5 and 1% in many tested Haemoproteus and Culicoides species. Actually, this is close to Haemoproteus parasitaemia, which predominates in wildlife and might be optimal for natural transmission [2]. Because the rate of ookinete development and ookinete size are markedly different in different Haemoproteus species [23], virulence of haemosporidian parasites to their insect vectors probably depends on certain parasite and insect species. However, this issue remains insufficiently understood. Further studies using birds with different levels of parasitaemia should be encouraged to determine the best parasitaemia range for experimental infections, so that insects would survive during the experiment and sufficient number of ookinetes, oocysts and sporozoites can be obtained for research in insect preparations depending on each study aim.
Only a small portion of Plasmodium spp. sporozoites reach salivary glands after maturation of oocysts. In Plasmodium gallinaceum, an avian malarial parasite, about 10–20% of sporozoites released from oocysts were located in Aedes aegypti salivary glands. This migration happened during eight hours after sporozoites were released into the haemocoel [64]. After this period, the sporozoites persisted in the haemocoel for a short period until degradation [64, 65]. It is possible that the same pattern of sporozoite survival and salivary gland invasion occurs in Haemoproteus parasites, but such observations remain insufficient in haemoproteids. It is known that almost all insects fed on birds infected with Haemoproteus species can support ookinetes development, but only about 20% of them support oocysts’ development and less than half of infected insects that survived until termination of sporogony possessed sporozoites in salivary gland preparations of C. impunctatus [10]. Because a significantly lower number of sporozoites (usually < 100) develop in oocysts of Haemoproteus (Parahaemoproteus) parasites than in avian Plasmodium species (usually > 1000) [2], few of the former probably reach salivary glands, resulting in difficulties to visualize them in preparations. This also can explain the low number of sporozoites observed in exposed C. nubeculosus in this study.
In vitro exflagellation and development of ookinetes have been examined in several Haemoproteus species [2, 37–39, 41]. The present study adds information about in vitro development of H. hirundinis, a common parasite of swallows. Regarding experimental vector studies, these data are important because they show the rate of mature ookinete development, indicating the most optimal time interval when insects should be dissected for ookinete detection in vivo during experimental research. After maturation, ookinetes of some Haemoproteus species rapidly escape from midgut contents and penetrate epithelial cells of the gut, so they can be overlooked in the midgut in in vivo preparations if ookinete preparations were made too late and the ookinetes had already moved from the midguts [31]. The rate of ookinete development is markedly different in different Haemoproteus species: it ranges from ~2 hpe in H. minutus to 6–12 hpe and even more in H. tartakovskyi (between 6 and 24 hpe) at ~20 °C [2, 63]. As a result, ookinetes of H. minutus are absent from midgut preparations prepared 4 hpe and later after exposure and cannot be detected, but ookinetes of H. tartakovskyi are still developing and would be readily visible. Our study shows that ookinete preparations of H. hirundinis should be prepared approximately 6 hpe at 20 °C. For many examined species, the most optimal periods for visualization of Haemoproteus parasites in vectors are 6–8 hpe for ookinetes, 3–5 dpe for oocysts and 6–8 dpe for sporozoites [12, 22, 23, 63].
Sporogonic development of H. belopolskyi (hHIICT1) has been investigated in wild-caught biting midges C. impuctatus at 15–18 °C; ookinetes were detected 1 dpe and sporozoites 7 dpe [31]. In the present study, the sporogonic development of the same parasite lineage was more rapid: mature ookinetes were recorded 8 hpe and sporozoites were observed 6 dpe. This difference might be due to the different temperature conditions for sporogony; the insects were maintained at 24–25 °C during this study. Numerous studies addressed the relationship of sporogony in Plasmodium parasites and temperature conditions [66], but information about Haemoproteus species remain insufficient.
In a previous study with H. lanii, ookinetes and sporozoites were observed in experimentally exposed C. impunctatus 1–2 and 5–8 dpe, respectively; the exposed insects were kept at natural wildlife temperature conditions ranging between 14 °C at night and 18 °C during the daytime [10]. In the present study, ookinetes of H. lanii hRB1 were found 6 hpe and sporozoites detected 6–9 dpe at 24–25 °C. In both studies the development rate of sporozoites was similar, despite the difference in temperature conditions. The reported difference between the sporogony rate in these two studies of H. lanii is likely due to different intervals of exposed insect dissections: sporozoites were detected 5 dpe in the former study, but dissections of insects for sporozoites were performed one day later in this study. However, it is important to note that three lineages of H. lanii have been identified, all of which are present in the same species of avian host (Lanius collurio) [67]. The study by Valkiūnas & Iezhova [10] was carried out using an unidentified lineage of H. lanii, so it is difficult to rule out that observed differences are due to different parasite lineages used in the experiments. Futher detailed experimental research is needed to understand Haemoproteus parasite sporogony rate in different parasite lineages belonging to the same species.
As expected, all four parasite lineages used in this study clustered together (Fig. 1, sub-clade b1) in our phylogenetic analysis and appeared with other Haemoproteus (Parahaemoproteus) species that are transmitted by biting midges (Fig. 1, Clade B) [20]. All species belonging to the subgenus Haemoproteus and transmitted by louse flies of the Hippoboscidae [2] were placed in a well-supported sister clade (Fig. 1, Clade A). This phylogenetic analysis is in accordance with studies, which showed that phylogenies using cytb indicate parasite-vector relationships. This gene is important for the metabolism of haemosporidians in vectors and is of phylogenetic value in evolutionary studies aiming determination of major Dipteran insect groups, which are involved in haemosporidian parasite transmission [22, 68]. Phylogenies based on this gene readily distinguish parasites belonging to subgenera Haemoproteus and Parahaemoproteus [20–22, 56, 69]. It is interesting to note that the differences between species of Haemoproteus and Parahaemoproteus are not only due to inhabiting different groups of dipteran insects (louse flies and biting midges, respectively), but also are manifested in the size of oocysts, the number of developing germinate centres in oocysts and the number of sporozoites produced in each of these groups [2]. In addition, our study shows that phylogenies based on partial cytb also indicate patterns of ookinete development in haemosporidian parasites (Fig. 1).
Maturing H. hirundinis ookinetes possess long finger-like outgrowths (Fig. 2e). This readily distinguishable character has been observed in developing ookinetes of several haemoproteid parasites [2, 37, 40] such as Haemoproteus attenuatus (hROBIN1) (Fig. 4a–c), Haemoproteus balmorali (hSFC1) (Fig. 4d–f), H. belopolskyi hHIICT3 (Fig. 4g–i), Haemoproteus fringillae hCCF3 (Fig. 4j–l), H. hirundinis (hDELURB2) (Fig. 4m–o), H. lanii (hRB1) (Fig. 4p–r), Haemoproteus motacillae (hYWT1) (Fig. 4s–u), Haemoproteus parabelopolskyi (hSYAT1) (Fig. 4v–x), Haemoproteus pastoris (hLAMPUR1) (Fig. 4y–aa) and Haemoproteus tartakovskyi (hSISKIN1) (Fig. 4bb–dd). Marked vacuolization of the cytoplasm also seems to be a character of all species listed above (Fig. 4c, f, i, l, o, r, u, x, aa, dd); however, this feature does not seem to be a unique for parasites of this clade (Fig. 1, sub-clade b1), since vacuoles were also present in ookinetes of H. pallidus (lineage hSFC3) (Fig. 1, sub-clade b2 and Fig. 5e–f).
This study not only strengthens the conclusion of former studies that phylogenies based on partial cytb sequences readily distinguish lineages belonging to parasites of the subgenera Haemoproteus and Parahaemoproteus [20–22, 56, 69], but also shows that well-supported clades in such phylogenetic trees have important biological meaning. In particular, our study suggests that patterns of ookinete transformation from zygote stage can be forecasted based exclusively on such phylogenies, which are easy to develop. Additionally, the relatively small density of cellular organelles in macrogametocytes is indicated by pale staining of the cytoplasm in these sexually-important cells, and the pale staining is readily visible under a light microscope. This feature is likely of important phylogenetic value because all investigated haemoproteids possessing the same character cluster together in available phylogenetic trees, as has been reported in several previous studies [23, 70, 71]. Interestingly, ookinetes of parasites of this clade (H. minutus, H. pallidus) develop fast (≤ 4 hpe) at a temperature of approximately 18–20 °C. Our study extends this conclusion and shows that 14 parasite species, which appeared in a relatively well-supported sub-clade b1 (Fig. 1) also are similar due to patterns of their ookinete development. In general, in all these species (i) long finger-like outgrowths appear at early stage of ookinete development (Figs. 2e; 4b, e, h, k, n, q, t, w, z, cc); and (ii) ookinetes mature relatively slowly (≥ 6 hpe) at a temperature of approximately 18–20 °C. In summary, our study shows that phylogenies based on cytb and mitochondrial genomes not only indicate large groups of vectors, which transmit parasites of certain groups of haemosporidians [22, 69], but also group haemosporidian species based on delicate patterns of their sporogony [23].
The simple characters of haemosporidian ookinete development, which are readily recognisable under a light microscope (rate and mode of transformation) should be genetically determined, particularly due to the following reasons. First, complex morphological and functional changes precede each ookinete development, including building of organelles of the apical complex, endoplasmic reticulum, crystalloid particles and others [2, 72]. Transformation of ookinetes via development of long finger-like outgrowths is related to ultrastructural changes in zygotes; this process is genetically determined [73]. Secondly, the rate of ookinete development depends on many internal and external (temperature, insect gut-factors) structural circumstances mentioned above, and is also likely determined genetically. The present study shows that phylogenies based on mitochondrial genomes are worth more attention in regard to better understanding patterns of sporogonic development in haemosporidian parasites. Additionally, it is possible that cytb phylogenetic inferences are more likely to reflect haemosporidian parasite development in vectors rather than in blood and tissue stages. This can be explained by the critical role of mitochondrial complex during the development of haemosporidians in insect vectors [68].
The presence of more or less prominent vacuoles in the cytoplasm of ookinetes (Figs. 4 and 5) is a common feature of parasites in sub-clade b1 (Fig. 1). Such vacuoles also were observed in ookinetes of parasites belonging to subgenus Haemoproteus (Fig. 1, clade A) [38]. These cytoplasmic vacuole-like inclusions are in fact the gatherings of an amorphous dense material called crystalloid material [2, 74]. The crystalloid material is likely washed out during fixation with alcohols and looks like an empty space, which is usually described as a ‘vacuole’ in stained preparations under a light microscope. The crystalloid likely performs energy functions and takes part in the metabolism of lipids, which is genetically determined [2, 73–75]. However, there is a variation in the presence of these vacuoles even among different lineages of the same parasite species. This can be observed in H. pallidus (hSFC3) that possess vacuoles (Fig. 5d–f) and H. pallidus (hPFC1) that does not possess visible vacuoles in the ookinetes (Fig. 5g–i). The lack of vacuoles was registered in H. minutus (Fig. 5a–c). Available data indicated that ookinetes of parasites of sub-clade b1 (Figs. 1 and 4) are more heavily vacuolated than ookinetes of sub-clade b2 (Figs. 1 and 5). However, only two Haemoproteus species (H. pallidus and H. minutus) have been investigated in regard to this character so far, and additional studies are needed to prove or reject this hypothesis.
The transmission of H. hirundinis and H. nucleocondensus have not been reported in Europe [2, 48, 76]. These are common parasites of the northern house martin D. urbicum and the great reed-warbler A. arundinaceus, respectively. These birds become infected at African wintering grounds, but juveniles are free of these infections in Europe before seasonal migration. It has been speculated that absence of susceptible vectors might interrupt transmission. The present study rejects this hypothesis because sporogony of both parasites was completed in C. nubeculosus, which is widespread in Europe and readily bites birds ([49]; this study). It is interesting that the same epidemiology has been reported for Plasmodium relictum (pGRW4) infection in temperate regions of Europe. In general, vector and sufficient temperature conditions for sporogony of this parasite are present, but transmission is absent in Europe [41]. It is probable that other ecological factors, which are related to vector ecology, might restrict transmission of these haemosporidian infections at bird breeding grounds. The first factor might be associated with the nesting biology of these bird species. The northern house martin builds closed nests with a small entrance; such nests might be non-attractive for biting midges. However, information about this aspect of the biology of Culicoides biting midges remains insufficient. It is known that biting midges can visit artificially-made nest-boxes of forest birds [77], but it is unclear if the same happens in northern house martin nests. These birds often build the nests below human-made structures such as bridges and houses in densely populated human settlements at different ecological conditions. This is not the case for the great reed-warbler: this bird builds open nests. However, both the northern house martin and the great reed-warbler prefer to nest in relatively open areas either close to human settlements or in reeds, respectively [78, 79]. The latter bird species also nests in environments with windy conditions, which do not favour the long flight of fragile biting midges [80]. These ecological factors might minimize the contact of Culicoides biting midges with these two-bird species resulting in a low probability for infection transmission from parent birds to offspring. Additionally, the prepatent development of H. hirundinis and H. nucleocondensus remain non-investigated; there is no information about longevity of prepatent period during these infections. In some Haemoproteus species the prepatent period varies between two and three weeks [2]. The majority of northern house martin and great reed warbler populations leave for wintering grounds early (in August) in Europe when the majority of H. hirundinis and H. nucleocondensus infections might be still non-patent, and that might prevent determining infected juveniles before seasonal migrations, at least in some of them. The present study indicates that factors other than availability of vectors might restrict transmission of H. hirundinis and H. nucleocondensus in Europe. Ecological factors preventing transmission of avian haemosporidians need more research. Further detailed studies on parasite life-cycles and their vector ecology are needed for a better understanding the transmission of these infections and the epidemiology of haemoproteosis.
Partial sequences of cytb gene have often been used to barcode haemosporidians [67] and are also often used to develop phylogenies, which distinguish haemosporidians belonging to different families and genera [3, 15, 17, 37, 70. 71]. However, information about other gene sequences of avian haemosporidians remains insufficiently developed and difficult to use in phylogenetic analysis but is important in biodiversity research. This is particularly true for apicoplast gene sequences, which provide valuable information in distinguishing morphologically similar haemosporidian species, which differ just in a few nucleotides in partial cytb sequences [81, 82]. We added information about partial sequences of the clpc genes along with cytb gene sequences, which extend molecular characterization of studies haemosporidian species and might be helpful for future research.
Conclusions
This study adds four species of avian haemoproteids (H. belopolskyi hHIICT1, H. hirundinis hDELURB2, H. nucleocondensus hGRW01 and H. lanii hRB1) to the list of parasites, which complete sporogony in C. nubeculosus. Due to the high virulence of avian haemoproteids in blood-sucking arthropods, parasite donor birds with light parasitaemia (of approximately 0.5–1%) are recommended to be used during experimental exposure of insects. Phylogenies based on partial sequences of cytb indicate patterns of sporogony of certain lineages. This study identified two clades of Haemoproteus lineages, which differ markedly in the rate of ookinete development and morphological patterns of ookinete transformation at initial stages of development. Ecological factors other than vector availability and temperature conditions should be identified to understand mechanisms preventing distribution of H. hirundinis and H. nucleocondensus infections and other haemosporidian parasites of tropical origin in temperate Europe.
Supplementary information
Additional file 1: Table S1. Parasites, host species and accession numbers of preparations, which were used for comparisons of in vitro ookinete development in this study.
Acknowledgments
We thank V. Jusys and V. Eigirdas (Ventės Ragas Ornithological Station, Lithuania) for help with bird catching and identification during field work and Dr Rasa Binkienė, for participation in fieldwork. Culicoides nubeculosus were provided by the Pirbright Institute as part of a grant funded by the Biotechnology and Biological Sciences Research Council (BBS/E/I/00001701), UK.
Abbreviations
- cytb
mitochondrial cytochrome b
- DNA
deoxynucleic acid
- dpe
days post-exposure
- EBA
exposure of blood to air
- hpe
hours post-exposure
- PCR
polymerase chain reaction
Authors’ contributions
Experimental conception and design: CRFC, DB and GV. Biting midge maintenance, experiments, dissection and laboratory maintenance: CRFC, DB and RB. Donor bird collection and testing: MI, TI and GV. Morphological analysis CRFC, DB, TI and GV. Phylogenetic analysis and its discussion: CRFC and GV. Paper writing: CRFC and GV. All authors read and approved the final manuscript.
Funding
This study was funded by the Research Council of Lithuania (No. DOTSUT-137-09.3.3-LMT-K-712-02-0004) and also supported by the Open Access to Research Infrastructure of the Nature Research Centre under the Lithuanian Open Access Network Initiative.
Availability of data and materials
The data supporting the findings of this study are included within the article and its additional file. Representative preparations of blood (Accession Numbers 49134NS-49147NS) and vector stages (49126NS-49133NS) were deposited in the Nature Research Centre, Vilnius, Lithuania. The sequences were deposited in the GenBank database under the Accession Numbers MN025422–MN025425 and MK843310–MK843317.
Ethics approval and consent to participate
The experiments described herein comply with the current laws of Lithuania, have been performed by licenced researchers and were approved by the Lithuania and Environmental Protection Agency, Vilnius (2018-04-13, No. 24). None of the experimental birds suffered apparent injury during sampling and all birds were released.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Carolina Romeiro Fernandes Chagas, Email: crfchagas@gmail.com.
Dovilė Bukauskaitė, Email: dovilebu7@gmail.com.
Mikas Ilgūnas, Email: ilgunasmikas@gmail.com.
Rasa Bernotienė, Email: rasa.bernotiene@gamtc.lt.
Tatjana Iezhova, Email: tatjana.jezova@gamtc.lt.
Gediminas Valkiūnas, Email: gediminas.valkiunas@gamtc.lt.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13071-019-3679-1.
References
- 1.Bensch S, Stjernman M, Hasselquist D, Ostman O, Hansson B, Westerdahl H, et al. Host specificity in avian blood parasites: a study of Plasmodium and Haemoproteus mitochondrial DNA amplified from birds. Proc R Soc. 2000;267:1583–1589. doi: 10.1098/rspb.2000.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Valkiūnas G. Avian malaria parasites and other haemosporidia. Boca Raton: CRC Press; 2005. [Google Scholar]
- 3.Clark NJ, Clegg SM, Lima MR. A review of global diversity in avian haemosporidians (Plasmodium and Haemoproteus: Haemosporida): new insights from molecular data. Int J Parasitol. 2014;44:329–338. doi: 10.1016/j.ijpara.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 4.Chagas CR, Valkiūnas G, de Oliveira Guimarães L, Monteiro EF, Guida FJ, Simões RF, et al. Diversity and distribution of avian malaria and related haemosporidian parasites in captive birds from a Brazilian megalopolis. Malar J. 2017;16:83. doi: 10.1186/s12936-017-1729-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schumm YR, Wecker C, Marek C, Wassmuth M, Bentele A, Willems H, et al. Blood parasites in Passeriformes in central Germany: prevalence and lineage diversity of Haemosporida (Haemoproteus, Plasmodium and Leucocytozoon) in six common songbirds. PeerJ. 2019;6:e6259. doi: 10.7717/peerj.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olias P, Wegelin M, Zenker W, Freter S, Gruber AD, Klopfleisch R. Avian malaria deaths in parrots, Europe. Emerg Infect Dis. 2011;17:950–952. doi: 10.3201/eid1705.101618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunn JC, Goodman SJ, Benton TG, Hamer KC. Avian blood parasite infection during the non-breeding season: an overlooked issue in declining populations? BMC Ecol. 2013;13:30. doi: 10.1186/1472-6785-13-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valkiūnas G, Pendl H, Olias P. New Haemoproteus parasite of parrots, with remarks on the virulence of haemoproteids in naive avian hosts. Acta Trop. 2017;176:256–262. doi: 10.1016/j.actatropica.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 9.Ortiz-Catedral L, Brunton D, Stidworthy MF, Elsheikha HM, Pennycott T, Schulze C, et al. Haemoproteus minutus is highly virulent for Australasian and South American parrots. Parasit Vectors. 2019;12:40. doi: 10.1186/s13071-018-3255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valkiūnas G, Iezhova TA. The transmission of Haemoproteus belopolskyi (Haemosporida: Haemoproteidae) of blackcap by Culicoides impunctatus (Diptera: Ceratopogonidae) J Parasitol. 2004;90:196–198. doi: 10.1645/GE-3223RN. [DOI] [PubMed] [Google Scholar]
- 11.Valkiūnas G, Kazlauskienė R, Bernotienė R, Bukauskaitė D, Palinauskas V, Iezhova TA. Haemoproteus infections (Haemosporida, Haemoproteidae) kill bird-biting mosquitoes. Parasitol Res. 2014;113:1011–1018. doi: 10.1007/s00436-013-3733-4. [DOI] [PubMed] [Google Scholar]
- 12.Bukauskaitė D, Bernotienė R, Iezhova TA, Valkiūnas G. Mechanisms of mortality in Culicoides biting midges due to Haemoproteus infection. Parasitology. 2016;143:1748–1754. doi: 10.1017/S0031182016001426. [DOI] [PubMed] [Google Scholar]
- 13.Santiago-Alarcon D, Palinauskas V, Schaefer HM. Diptera vectors of avian haemosporidian parasites: untangling parasite life cycles and their taxonomy. Biol Rev Camb Philos Soc. 2012;87:928–964. doi: 10.1111/j.1469-185X.2012.00234.x. [DOI] [PubMed] [Google Scholar]
- 14.Lacorte GA, Félix GM, Pinheiro RR, Chaves AV, Almeida-Neto G, Neves FS, et al. Exploring the diversity and distribution of neotropical avian malaria parasites—a molecular survey from southeast Brazil. PLoS ONE. 2013;8:e57770. doi: 10.1371/journal.pone.0057770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neto JM, Pérez-Rodríguez A, Haase M, Flade M, Bensch S. Prevalence and diversity of Plasmodium and Haemoproteus parasites in the globally-threatened aquatic warbler Acrocephalus paludicola. Parasitology. 2015;142:1183–1189. doi: 10.1017/S0031182015000414. [DOI] [PubMed] [Google Scholar]
- 16.Chagas CR, Guimarães L, Monteiro EF, Valkiūnas G, Katayama MV, Santos SV, et al. Hemosporidian parasites of free-living birds in the São Paulo Zoo, Brazil. Parasitol Res. 2016;115:1443–1452. doi: 10.1007/s00436-015-4878-0. [DOI] [PubMed] [Google Scholar]
- 17.Ciloglu A, Yildirim A, Duzlu O, Onder Z, Dogan Z, Inci A. Investigation of avian haemosporidian parasites from raptor birds in Turkey, with molecular characterisation and microscopic confirmation. Folia Parasitol (Praha). 2016;17:63. doi: 10.14411/fp.2016.023. [DOI] [PubMed] [Google Scholar]
- 18.Ivanova K, Zehtindjiev P, Mariaux J, Dimitrov D, Georgiev BB. Avian haemosporidians from rain forests in Madagascar: molecular and morphological data of the genera Plasmodium, Haemoproteus and Leucocytozoon. Infect Genet Evol. 2018;58:115–124. doi: 10.1016/j.meegid.2017.12.017. [DOI] [PubMed] [Google Scholar]
- 19.Garvin MC, Greiner EC. Ecology of Culicoides (Diptera: Ceratopogonidae) in southcentral Florida and experimental Culicoides vectors of the avian hematozoan Haemoproteus danilewskyi Kruse. J Wildl Dis. 2003;39:170–178. doi: 10.7589/0090-3558-39.1.170. [DOI] [PubMed] [Google Scholar]
- 20.Bukauskaitė D, Žiegytė R, Palinauskas V, Iezhova TA, Dimitrov D, Ilgūnas M, et al. Biting midges (Culicoides, Diptera) transmit Haemoproteus parasites of owls: evidence from sporogony and molecular phylogeny. Parasit Vectors. 2015;8:303. doi: 10.1186/s13071-015-0910-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Žiegytė R, Markovets MY, Bernotienė R, Mukhin A, Iezhova TA, Valkiūnas G, Palinauskas V. The widespread biting midge Culicoides impunctatus (Ceratopogonidae) is susceptible to infection with numerous Haemoproteus (Haemoproteidae) species. Parasit Vectors. 2017;10:397. doi: 10.1186/s13071-017-2317-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bukauskaitė D, Iezhova TA, Ilgūnas M, Valkiūnas G. High susceptibility of the laboratory-reared biting midges Culicoides nubeculosus to Haemoproteus infections, with review on Culicoides species that transmit avian haemoproteids. Parasitology. 2018;146:333–341. doi: 10.1017/S0031182018001373. [DOI] [PubMed] [Google Scholar]
- 23.Chagas CRF, Bukauskaitė D, Ilgūnas M, Iezhova T, Valkiūnas G. A new blood parasite of leaf warblers: molecular characterization, phylogenetic relationships, description and identification of vectors. Parasit Vectors. 2018;11:538. doi: 10.1186/s13071-018-3109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishtiaq F, Guillaumot L, Clegg SM, Phillimore AB, Black RA, Owens IPF, et al. Avian haematozoan parasites and their associations with mosquitoes across Southwest Pacific Islands. Mol Ecol. 2008;17:4545–4555. doi: 10.1111/j.1365-294X.2008.03935.x. [DOI] [PubMed] [Google Scholar]
- 25.Kim KS, Tsuda Y, Sasaki T, Kobayashi M, Hirota Y. Mosquito blood-meal analysis for avian malaria study in wild bird communities: laboratory verification and application to Culex sasai (Diptera: Culicidae) collected in Tokyo. Jpn Parasitol Res. 2009;105:1351–1357. doi: 10.1007/s00436-009-1568-9. [DOI] [PubMed] [Google Scholar]
- 26.Njabo KY, Cornel AJ, Bonneaud C, Toffelmier E, Sehgal RN, Valkiūnas G, et al. Nonspecific patterns of vector, host and avian malaria parasite associations in a central African rainforest. Mol Ecol. 2011;20:1049–1061. doi: 10.1111/j.1365-294X.2010.04904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Puente J, Martinez J, Aguilar J, Herrero J, Merino S. On the specificity of avian blood parasites: revealing specific and generalist relationships between haemosporidians and biting midges. Mol Ecol. 2011;20:3275–3287. doi: 10.1111/j.1365-294X.2011.05136.x. [DOI] [PubMed] [Google Scholar]
- 28.Ferraguti M, Puente J, Ruiz S, Soriguer R, Figuerola J. On the study of the transmission network of blood parasites from SW Spain: diversity of avian haemosporidians in the biting midge Culicoides circumscriptus and wild birds. Parasit Vectors. 2013;6:208. doi: 10.1186/1756-3305-6-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bobeva A, Zehtindjiev P, Ilieva M, Dimitrov D, Mathis A, Bensch S. Host preferences of ornithophilic biting midges of the genus Culicoides in the eastern Balkans. Med Vet Entomol. 2015;29:290–296. doi: 10.1111/mve.12108. [DOI] [PubMed] [Google Scholar]
- 30.Martin E, Chu E, Shults P, Golnar A, Swanson DA, Benn J, et al. Culicoides species community composition and infection status with parasites in an urban environment of east central Texas, USA. Parasit Vectors. 2019;12:39. doi: 10.1186/s13071-018-3283-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Žiegytė R, Palinauskas V, Bernotienė R, Iezhova TA, Valkiūnas G. Haemoproteus minutus and Haemoproteus belopolskyi (Haemoproteidae): complete sporogony in the biting midge Culicoides impunctatus (Ceratopogonidae), with implications on epidemiology of haemoproteosis. Exp Parasitol. 2014;145:74–79. doi: 10.1016/j.exppara.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 32.Boorman J. The maintenance of laboratory colonies of Culicoides variipenniss (Coq.), C. nubeculosus (Mg.) and C. riethi Kieff (Diptera, Ceratopogonidae) Bull Entomol Res. 1974;64:371–377. doi: 10.1017/S0007485300031254. [DOI] [Google Scholar]
- 33.Galen SC, Borner J, Martinsen ES, Schaer J, Austin CC, West CJ, et al. The polyphyly of Plasmodium: comprehensive phylogenetic analyses of the malaria parasites (order Haemosporida) reveal widespread taxonomic conflict. R Soc Open Sci. 2018;5:171780. doi: 10.1098/rsos.171780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Videvall E. Genomic advances in avian malaria research. Trends Parasitol. 2019;35:254–266. doi: 10.1016/j.pt.2018.12.005. [DOI] [PubMed] [Google Scholar]
- 35.Pacheco MA, Matta NE, Valkiūnas G, Parker PG, Mello B, Stanley CE, Jr, et al. Mode and rate of evolution of haemosporidian mitochondrial genomes: timing the radiation of avian parasites. Mol Biol Evol. 2018;35:383–403. doi: 10.1093/molbev/msx285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sherman IW. Malaria: Parasite biology, pathogenesis, and protection. Washington DC: American Society for Microbiology Press; 1998. [Google Scholar]
- 37.Dimitrov D, Valkiūnas G, Zehtindjiev P, Ilieva M, Bensch S. Molecular characterization of haemosporidian parasites (Haemosporida) in yellow wagtail (Motacilla flava), with description of in vitro ookinetes of Haemoproteus motacillae. Zootaxa. 2013;3666:369–381. doi: 10.11646/zootaxa.3666.3.7. [DOI] [PubMed] [Google Scholar]
- 38.Coral AA, Valkiūnas G, González AD, Matta NE. In vitro development of Haemoproteus columbae (Haemosporida: Haemoproteidae), with perspectives for genomic studies of avian haemosporidian parasites. Exp Parasitol. 2015;157:163–169. doi: 10.1016/j.exppara.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 39.Valkiūnas G, Iezhova TA, Križanauskienė A, Palinauskas V, Bensch S. In vitro hybridization of Haemoproteus spp.: an experimental approach for direct investigation of reproductive isolation of parasites. J Parasitol. 2008;94:1385–1394. doi: 10.1645/GE-1569.1. [DOI] [PubMed] [Google Scholar]
- 40.Valkiūnas G, Palinauskas V, Križanauskienė A, Bernotienė R, Kazlauskienė R, Iezhova TA. Further observations on in vitro hybridization of hemosporidian parasites: patterns of ookinete development in Haemoproteus spp. J Parasitol. 2013;99:124–136. doi: 10.1645/GE-3226.1. [DOI] [PubMed] [Google Scholar]
- 41.Valkiūnas G, Iezhova TA, Palinauskas V, Ilgūnas M, Bernotienė R. The evidence for rapid gametocyte viability changes in the course of parasitemia in Haemoproteus parasites. Parasitol Res. 2015;144:2903–2909. doi: 10.1007/s00436-015-4491-2. [DOI] [PubMed] [Google Scholar]
- 42.Palinauskas V, Križanauskienė A, Iezhova TA, Bolshakov CV, Jönsson J, Bensch S, et al. A new method for isolation of purified genomic DNA from haemosporidian parasites inhabiting nucleated red blood cells. Exp Parasitol. 2013;133:275–280. doi: 10.1016/j.exppara.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 43.Atkinson CT, Thomas NJ, Hunter BC. Parasitic diseases of wild birds. Ames: Wiley-Blackwell; 2008. [Google Scholar]
- 44.Garcia-Longoria L, Marzal A, de Lope F, Garamszegi L. Host-parasite interaction explains variation in the prevalence of avian haemosporidians at the community level. PLoS ONE. 2019;14:e0205624. doi: 10.1371/journal.pone.0205624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fallon SM, Bermingham E, Ricklefs RE. Island and taxon effects in parasitism revisited: avian malaria in the Lesser Antilles. Evolution. 2003;57:606–615. doi: 10.1111/j.0014-3820.2003.tb01552.x. [DOI] [PubMed] [Google Scholar]
- 46.Loiseau C, Iezhova T, Valkiūnas G, Chasar A, Hutchinson A, Buermann W, et al. Spatial variation of haemosporidian parasite infection in African rainforest bird species. J Parasitol. 2010;96:21–29. doi: 10.1645/GE-2123.1. [DOI] [PubMed] [Google Scholar]
- 47.Pulgarín-R PC, Gómez JP, Robinson S, Ricklefs RE, Cadena CD. Host species, and not environment, predicts variation in blood parasite prevalence, distribution, and diversity along a humidity gradient in northern South America. Ecol Evol. 2018;8:3800–3814. doi: 10.1002/ece3.3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Križanauskienė A, Iezhova T, Palinauskas V, Chernetsov N, Valkiūnas G. Haemoproteus nucleocondensus n. sp. (Haemosporida, Haemoproteidae) from a Eurasian songbird, the great reed warbler Acrocephalus arundinaceus. Zootaxa. 2012;3441:36–46. doi: 10.11646/zootaxa.3441.1.3. [DOI] [Google Scholar]
- 49.Mathieu B, Cêtre-Sossah C, Garros C, Chavernac D, Balenghien T, Carpenter S, et al. Development and validation of IIKC: an interactive identification key for Culicoides (Diptera: Ceratopogonidae) females from the Western Palaearctic region. Parasit Vectors. 2012;5:137. doi: 10.1186/1756-3305-5-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Godfrey RD, Fedynich AM, Pence DB. Quantification of hematozoa in blood smears. J Wildl Dis. 1987;23:558–565. doi: 10.7589/0090-3558-23.4.558. [DOI] [PubMed] [Google Scholar]
- 51.Richardson DS, Jury FL, Blaakmeer K, Komdeur J, Burke T. Parentage assignment and extra-group paternity in a cooperative breeder: the Seychelles warbler (Acrocephalus sechellensis) Mol Ecol. 2001;10:2263–2273. doi: 10.1046/j.0962-1083.2001.01355.x. [DOI] [PubMed] [Google Scholar]
- 52.Hellgren O, Waldenström J, Bensch S. A new PCR assay for simultaneous studies of Leucocytozoon, Plasmodium and Haemoproteus from avian blood. J Parasitol. 2004;90:797–802. doi: 10.1645/GE-184R1. [DOI] [PubMed] [Google Scholar]
- 53.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 54.Pérez-Tris J, Bensch S. Diagnosing genetically diverse avian malarial infections using mixed-sequence analysis and TA-cloning. Parasitology. 2005;131:15–23. doi: 10.1017/S003118200500733X. [DOI] [PubMed] [Google Scholar]
- 55.Pacheco MA, García-Amado MA, Manzano J, Matta NE, Escalante AA. Blood parasites infecting the hoatzin (Opisthocomus hoazin), a unique Neotropical folivorous bird. PeerJ. 2019;7:e6361. doi: 10.7717/peerj.6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martinsen ES, Perkins SL, Schall JJ. A three-genome phylogeny of malaria parasites (Plasmodium and closely related genera): evolution of life-history traits and host switches. Mol Phylogenet Evol. 2008;47:261–273. doi: 10.1016/j.ympev.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 57.Ronquist F, Heulsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 58.Nylander JAA. MrModeltest v2. Program distributed by the author: Evolutionary Biology Centre, Uppsala University; 2004. [Google Scholar]
- 59.Rambaut A. FigTree: Tree figure drawing tool version 1.4.0. 2006–2012. Institute of Evolutionary Biology, University of Edinburgh. http://tree.bio.ed.ac.uk/software/figtree/.
- 60.Jennings DM, Mellor PS. The vector potential of British Culicoides species for bluetongue virus. Vet Microbiol. 1988;17:1–10. doi: 10.1016/0378-1135(88)90074-0. [DOI] [PubMed] [Google Scholar]
- 61.Pages N, Bréard E, Urien C, Talavera S, Viarouge C, Lorca-Oro C, et al. Culicoides midge bites modulate the host response and impact on bluetongue virus infection in sheep. PLoS ONE. 2014;9:e83683. doi: 10.1371/journal.pone.0083683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Svobodová M, Dolnik OV, Čepička I, Rádrová J. Biting midges (Ceratopogonidae) as vectors of avian trypanosomes. Parasit Vectors. 2017;10:224. doi: 10.1186/s13071-017-2158-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Žiegytė R, Bernotienė R, Palinauskas V, Valkiūnas G. Haemoproteus tartakovskyi (Haemoproteidae): Complete sporogony in Culicoides nubeculosus (Ceratopogonidae), with implications for avian haemoproteid experimental research. Exp Parasitol. 2016;160:17–22. doi: 10.1016/j.exppara.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 64.Hillyer JF, Barreau C, Vernick KD. Efficiency of salivary gland invasion by malaria sporozoites is controlled by rapid sporozoite destruction in the mosquito haemocoel. Int J Parasitol. 2006;37:673–681. doi: 10.1016/j.ijpara.2006.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mueller AK, Kohlhepp F, Hammerschmidt C, Michel K. Invasion of mosquito salivary glands by malaria parasites: prerequisites and defense strategies. Int J Parasitol. 2010;40:1229–1235. doi: 10.1016/j.ijpara.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reiner RC, Jr, Geary M, Atkinson PM, Smith DL, Gething PW. Seasonality of Plasmodium falciparum transmission: a systematic review. Malar J. 2015;14:343. doi: 10.1186/s12936-015-0849-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bensch S, Hellgren O, Pérez-Tris J. MalAvi: a public database of malaria parasites and related haemosporidians in avian hosts based on mitochondrial cytochrome b lineages. Mol Ecol Resour. 2009;9:1353–1358. doi: 10.1111/j.1755-0998.2009.02692.x. [DOI] [PubMed] [Google Scholar]
- 68.Hino A, Hirai M, Tanaka TQ, Watanabe Y, Matsuoka H, Kita K. Critical roles of the mitochondrial complex II in oocyst formation of rodent malaria parasite Plasmodium berghei. J Biochem. 2012;152:259–268. doi: 10.1093/jb/mvs058. [DOI] [PubMed] [Google Scholar]
- 69.Pacheco MA, Cepeda AS, Bernotienė R, Lotta IA, Matta NE, Valkiūnas G, et al. Primers targeting mitochondrial genes of avian haemosporidians: PCR detection and differential DNA amplification of parasites belonging to different genera. Int J Parasitol. 2018;48:657–670. doi: 10.1016/j.ijpara.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Križanauskienė A, Pérez-Tris J, Palinauskas V, Hellgren O, Bensch S, Valkiūnas G. Molecular phylogenetic and morphological analysis of haemosporidian parasites (Haemosporida) in a naturally infected European songbird, the blackcap Sylvia atricapilla, with description of Haemoproteus pallidulus sp. nov. Parasitology. 2010;137:217–227. doi: 10.1017/S0031182009991235. [DOI] [PubMed] [Google Scholar]
- 71.Dimitrov D, Iezhova TA, Zehtindjiev P, Bobeva A, Ilieva M, Kirilova M, et al. Molecular characterisation of three avian haemoproteids (Haemosporida, Haemoproteidae), with the description of Haemoproteus (Parahaemoproteus) palloris n. sp. Syst Parasitol. 2016;93:431–449. doi: 10.1007/s11230-016-9638-8. [DOI] [PubMed] [Google Scholar]
- 72.Aikawa M, Carter R, Ito Y, Nijhout MM. New observations on gametogenesis, fertilization, and zygote transformation in Plasmodium gallinaceum. J Protozool. 1984;31:403–413. doi: 10.1111/j.1550-7408.1984.tb02987.x. [DOI] [PubMed] [Google Scholar]
- 73.Sinden RE. Gametogenesis and sexual development. In: Sherman IW, editor. Malaria: Parasite biology, pathogenesis, and protection. Washington DC: American Society for Microbiology Press; 1998. pp. 25–48. [Google Scholar]
- 74.Atkinson CT. Haemoproteus. In: Atkinson CT, Thomas NJ, Hunter BC, editors. Parasitic diseases of wild birds. Ames: Wiley-Blackwell; 2008. pp. 13–35. [Google Scholar]
- 75.Desser SS, Bennett GF. The genera Leucocytozoon, Haemoproteus and Hepatocystis. In: Kreir JP, Baker JR, editors. Parasitic protozoa. 2. New York: Academic Press; 1993. pp. 273–307. [Google Scholar]
- 76.Bensch S, Waldenström J, Jonzén N, Westerdahl H, Hansson B, Sejberg D, et al. Temporal dynamics and diversity of avian malaria parasites in a single host species. J Anim Ecol. 2007;76:112–122. doi: 10.1111/j.1365-2656.2006.01176.x. [DOI] [PubMed] [Google Scholar]
- 77.Votýpka J, Synek P, Svobodova M. Endophagy of biting midges attacking cavity-nesting birds. Medic Vet Entomol. 2009;23:277–280. doi: 10.1111/j.1365-2915.2009.00800.x. [DOI] [PubMed] [Google Scholar]
- 78.BirdLife International. Delichon urbicum (amended version of 2016 assessment). The IUCN Red List of Threatened Species. 2017. 10.2305/IUCN.UK.2017-3.RLTS.T103811886A118748864.en. Accessed 20 Mar 2019. [DOI]
- 79.BirdLife International. Acrocephalus arundinaceus (amended version of 2016 assessment). The IUCN Red List of Threatened Species. 2017. 10.2305/IUCN.UK.2017-1.RLTS.T104317670A111179363.en. Accessed 20 Mar 2019. [DOI]
- 80.Elbers ARW, Koenraadt CJM, Meiswinkel R. Mosquitoes and Culicoides biting midges: vector range and the influence of climate change. Rev Sci Tech Off Int Epiz. 2015;34:123–137. doi: 10.20506/rst.34.1.2349. [DOI] [PubMed] [Google Scholar]
- 81.Valkiūnas G, Ilgūnas M, Bukauskaitė D, Chagas CRF, Bernotienė R, Himmel T, et al. Molecular characterization of six widespread avian haemoproteids, with description of three new Haemoproteus species. Acta Trop. 2019;197:105051. doi: 10.1016/j.actatropica.2019.105051. [DOI] [PubMed] [Google Scholar]
- 82.Levin II, Valkiūnas G, Iezhova TA, O’Brien SL, Parker PG. Novel Haemoproteus species (Haemosporida: Haemoproteidae) from the swallow–tailed gull (Lariidae), with remarks on the host range of hippoboscid-transmitted avian hemoproteids. J Parasitol. 2012;98:847–854. doi: 10.1645/GE-3007.1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Parasites, host species and accession numbers of preparations, which were used for comparisons of in vitro ookinete development in this study.
Data Availability Statement
The data supporting the findings of this study are included within the article and its additional file. Representative preparations of blood (Accession Numbers 49134NS-49147NS) and vector stages (49126NS-49133NS) were deposited in the Nature Research Centre, Vilnius, Lithuania. The sequences were deposited in the GenBank database under the Accession Numbers MN025422–MN025425 and MK843310–MK843317.