Abstract
Shigellosis is a public health threat in developed as well as developing countries like “India.” While antibiotic therapy is the mainstay of treatment for shigellosis, current emergence of multidrug-resistant strains of Shigella spp. has posed the problem more challenging. Lytic bacteriophages which destroy antibiotic resistant Shigella spp. have great potential in this context and hence their identification and detailed characterization is necessary. In this study we presented the isolation and a detailed characterization of a novel bacteriophage Sfin-1, which shows potent lytic activity against multidrug-resistant isolates of Shigella flexneri, Shigella dysenteriae, Shigella sonnei obtained from clinical specimens from shigellosis patients. It is also active against Escherichia coli C. The purified phage is lytic in nature, exhibited absorption within 5–10 min, a latent period of 5–20 min and burst size of ∼28 to ∼146 PFU/cell. The isolated phage shows stability in a broad pH range and survives an hour at 50°C. Genome sequencing and phylogenetic analyses showed that Sfin-1 is a novel bacteriophage, which is very closely related to T1-like phages (89.59% identity with Escherichia virus T1). In silico analysis indicates that Sfin-1 genome consists of double stranded linear DNA of 50,403 bp (GC content of 45.2%) encoding 82 potential coding sequences, several potential promoters and transcriptional terminators. Under electron microscopy, Sfin-1 shows morphology characteristics of the family Siphoviridae with an isometric head (61 nm) and a non-contractile tail (155 nm). This is most likely the first report of a lytic bacteriophage that is active against three of the most virulent multidrug-resistant Shigella species and therefore might have a potential role in phage therapy of patients infected with these organisms.
Keywords: bacteriophage, Shigella spp., phage therapy, genome sequencing, large terminase, LC-MS/MS
Introduction
Instances of Shigellosis infection were estimated to be about 170 million annually during the end of last century with about 1 million reported deaths in the developing countries. Although this number has decreased, shigellosis still presents itself as one of the most important pandemic diseases in the world (Sur et al., 2004).
Shigellosis is caused by the bacteria of the genus Shigella having four pathogenic serogroups (Shigella dysenteriae, Shigella flexneri, Shigella sonnei, and Shigella boydii). Infection is common by feco-oral route due to the intake of contaminated water and food (Baird-Parker, 1994). Although WHO recommends antibiotics like azithromycin, ciprofloxacin [a fluoroquinolone (FQ)] or one of the three second-line antibiotics, pivmecillinam, and ceftriaxone (a third-generation cephalosporin) for the shigellosis treatment, extensive use of these antibiotics has contributed to the emergence of FQ and multidrug-resistant Shigella species in several countries (Sivapalasingam et al., 2006; Von Seidlein et al., 2006; Yismaw et al., 2008; Nandy et al., 2010; Tariq et al., 2012; Muthuirulandi Sethuvel et al., 2017; Puzari et al., 2017). Thus, recurrent changes in antimicrobial resistance profile of Shigella isolates poses complications in recommending standard drugs for effective treatment of the disease. Currently, the most predominant species in endemic regions was S. flexneri followed by S. sonnei, S. boydii, and S. dysenteriae which may be responsible for causing diarrheal outbreak (Sengupta et al., 1990; Nandy et al., 2010).
Bacteriophages are specific bacterial viruses that at first attach to and then destroy their hosts through phage genome replication and bacterial lysis. These characteristics of phages hint at their applications as therapeutic agents against bacterial infections of humans and animals. The fact that, unlike antibiotics, phages can destroy target bacteria specifically without killing the normal microflora. It is crucial for the application of phages to inhibit specific bacterial growth, which can be a natural, non-toxic and active substitute for antibiotic therapy (O’flaherty et al., 2005; Gupta and Prasad, 2011). In view of the enormous rise in antibiotic resistance in several clinically significant bacterial species, use of lytic phages is gaining more and more attention as a therapeutic alternative in the place of antibiotics against infectious diseases.
However, numerous challenges have to face during application of phage therapy in human diseases due to the limited available information regarding the interaction between phage and the host (Housby and Mann, 2009). Again, for the application of phage therapy, it is necessary to isolate new phages that are yet to be cultivated, active against circulating strains and to determine their physiological and genomic characters in detail for assessing their suitability.
Several bacteriophages against Shigella spp. have been reported. Temperate bacteriophages such as SfI(38,389 bp), SfII(41,475 bp), Sf6(39,043 bp), SfIV(39,758 bp), SfV(37,074 bp), and SfX(37,355 bp) (Mavris et al., 1997; Guan et al., 1999; Allison et al., 2002; Casjens et al., 2004; Jakhetia et al., 2013; Sun et al., 2013) that are responsible in serotype conversion of S. flexneri have been reported. The lytic Shigella phages SF9, SP18 and ØSboM-AG3 are specific against S. dysenteriae, S. sonnei, and S. boydii, respectively. One broad spectrum Shigella phage pSf-1 was reported few years back that was specific against S. flexneri, S. sonnei, and S. boydii; another recently discovered phage vB_SsoS-ISF002 can infect both S. flexneri and S. sonnei but broad spectrum lytic phage that infects both S. flexneri and S. dysenteriae has not been reported yet (Allison and Verma, 2000; Faruque et al., 2003; Kim et al., 2010; Anany et al., 2011; Jun et al., 2013; Shahin et al., 2018).
Generally phages acquire and contribute genes, not only to other phage genomes but also to bacterial genomes and thus these are powerful factors in the evolution, physiology, and pathogenicity of the host bacteria. Here, we report the isolation and detailed characterization of a novel bacteriophage Sfin-1, which shows strong lytic activity against multidrug-resistant isolates of S. flexneri, S. dysenteriae, and S. sonnei. Discovery of new phage against Shigella spp. and their genomics would not only help to develop phage based therapy against the shigellosis, but also to understand the evolution strategy, the mosaic architecture of phages and the involvement of genes(s) if any, in host pathogenesis.
Materials and Methods
Bacterial Strains and Antimicrobial Resistance Tests
This study included a total number of 40 multidrug-resistant clinical isolates of S. flexneri, S. dysenteriae, S. boydii, S. sonnei, Salmonella enterica serovar Typhi, various Escherichia coli strains like K12, AG100, XL1 Blue, and E. coli C. All Shigella and Salmonella species were isolated from patient’s stool samples at Bacteriology Division of National Institute of Cholera and Enteric Diseases (NICED), “Kolkata” and reported thereafter (Nandy et al., 2010; Dutta et al., 2014). They were then grown in nutrient broth medium at 37°C for 24 h for further tests adhering to the biosecurity and institutional safety procedures under Biosafety Level II (BSL II).
Isolation and Purification of Bacteriophages
Water sample was collected from the Ganga River, near Serampore, Hooghly district, about 25 km from Kolkata, in the state of West Bengal, India. After the removal of particulate matters with filter paper (Whatman 1), the water sample was mixed with S. flexneri 2a strains, 10% (w/v) peptone and it was incubated at 37°C for 24 h to enrich the concentration of bacteriophages. The enriched culture was then mixed with 1% (w/v) chloroforms and was shaken well to remove the bacterial debris. The mixture was centrifuged and supernatant was filtered through 0.22 μm pore membrane (Millipore, United States). This supernatant was inoculated (10 μL) as spot on the soft agar plate mixed with S. flexneri 2a strains. Clear zone around the spot indicated the presence of specific bacteriophage in the water sample against S. flexneri 2a. Similarly, other isolated serotypes of Shigella spp. were also checked for determination of phage specificity.
Collected water sample was then subsequently used for plaque assay; 100 μL of filtrate and 200 μL S. flexneri 2a culture (OD600 = 0.3) was mixed with 3.5 mL soft agar (0.9%) and plated onto LB hard agar (1.8%) plate. This plate was incubated in 37°C for 24 h. Clear individual plaques formed on the plate were transferred to another S. flexneri 2a plate. A single plaque was transferred into 500 μL phage dilution medium (0.1% tryptone, 0.85% sodium chloride) and stored at 4°C for 24 h. The suspended phage solution was then allowed for another round of plaque assay. Thus a single plaque was transferred at least three times to purify the bacteriophage. After that, phage dilutions and plaque assay was performed to get confluent lysis plates. The soft agar layer was then scrapped out, suspended in cold phage dilution medium and kept on ice for 3 h. It was then centrifuged at 5,000 × g and the supernatant was collected. The phage lysate prepared by this procedure was then pelleted at 68,000 × g for 2 h at 4°C in an ultracentrifuge since phage precipitates produced thus has more titer values. For further purification, cesium chloride (CsCl) density gradient centrifugation was done (ρ = 1.3, 1.5, 1.7 g/mL) at 100,000 × g for 3 h at 4°C and the phage band obtained between 1.7 and 1.5 g/mL was collected. The phage particles were recovered from the band and were dialyzed against TM buffer (50 mM Tris–Cl, pH 8.0 containing 10 mM MgSO4) and stored at 4°C for further studies (Uchiyama et al., 2008).
Host Range
The strains to be tested were grown overnight in nutrient broth 3.5 mL of the molten soft agar (0.7% w/v) was mixed with 100 μL of the bacterial cell suspension and this mixture was overlaid onto the surface of solid basal LB Agar (1.5% w/v). 10 μL (about 1.0 × 1010 PFU/mL) of the phage suspension was spotted on the plate and it was then incubated at 37°C, overnight. Bacterial sensitivity to a bacteriophage was determined by bacterial lysis at the spot where the phage suspension was inoculated. Each test was repeated three times. According to the degrees of clarity, the spots were differentiated into two categories: clear (+), and no reaction (−) (Bennish et al., 1990; Chang et al., 2005).
Thermal and pH Stability
For thermal stability testing, 16 × 1012 Sfin-1 phage particles were incubated at 4, 37, 50, 60, 70, 80, 90°C in 1 mL and for each temperature aliquots (100 μL) were taken after 5, 15, 40, 60 min to be titered by the double layered agar plate method against S. flexineri 2a. For pH stability testing, 14 × 1010 Sfin-1 phage particles were kept in 1 mL of TM buffers (50 mM Tris, 10 mM MgSO4) at pH range of 2–12 (adjusted with HCl or NaOH for acidic or alkaline range, respectively) for 1 h at 37°C and then aliquots (100 μL) from each pH were titered by the double-layered agar plate method against S. flexineri 2a (Yoon et al., 2007; Ma and Lu, 2008).
Bacterial Challenge Test
Minimum inhibitory concentrations (MICs) of different antibiotics for three clinical Shigella isolates were determined. According to the CLSI guidelines (CLSI, 2016) the isolates were multi drug resistant. Bacteriolytic activity of phage Sfin-1 was determined as described by Wang et al. (2016) with some modifications. Cells of S. flexneri 2a (1A) (strain ID BCH5722, Table 1) and S. dysenteriae1 (1A) (strain ID BCH5762, Table 1) were allowed to grow in presence of ampicillin (32 μg/mL), chloramphenicol (32 μg/mL), tetracycline (16 μg/mL), cotrimoxazole (25 μg/mL), nalidixic acid (32 μg/mL), ciprofloxacin (4 μg/mL), norfloxacin (16 μg/mL), and ofloxacin (8 μg/mL), whereas S. sonnei (strain ID BCH7084, Table 1) was grown in presence of tetracycline (16 μg/mL), cotrimoxazole (25 μg/mL), and nalidixic acid (32 μg/mL). 20 mL of cultures (OD600 = 0.3) were harvested by centrifugation and resuspended in fresh 1 mL of LB. Thereafter phage Sfin-1 was added at an multiplicity of infection (MOI) of 0.1, 0.01, and 0.001, allowed for adsorption for 5 min (S. flexneri 2a and S. dysenteriae1) or 10 min (S. sonnei) at 37°C. Each mixture was then transferred to 20 mL of fresh LB. Aliquots were taken at specific time intervals for 5 h and the number of bacterial cells was monitored by spread plate method. As negative control bacterial cultures were inoculated with phage dilution medium and respective antibiotics.
TABLE 1.
Susceptibility test for various clinically isolated antibiotic resistant strains to the phage Sfin-1, isolated from river Ganges water samples Kolkata, India.
| Sl. no | Strain ID | Bacterial isolates with different serotypes | Antimicrobial resistance profile by disc diffusion method | Lysis by Sfin-1 |
| 1 | BCH5722 | Shigella flexneri 2a (1A) | ACTQNaCipNorOfx | + |
| 2 | BCH4025 | Shigella flexneri 2a (2A) | ACQ | + |
| 3 | BCH3651 | Shigella flexneri 2a (3A) | ACTQ | + |
| 4 | BCH3557 | Shigella flexneri 2a (4A) | CTQNa | + |
| 5 | BCH7151 | Shigella flexneri 2a (5A) | ACTQNaCipNorOfx | + |
| 6 | BCH5762 | Shigella dysenteriae 1(1A) | ACTQNaCipNorOfx | + |
| 7 | BCH5848 | Shigella dysenteriae 1(2A) | ACTQNaCipNorOfx | + |
| 8 | BCH5859 | Shigella dysenteriae 1(3A) | ACTQNaCipNorOfx | + |
| 9 | BCH5912 | Shigella dysenteriae 1(4A) | ACTQNaCipNorOfx | + |
| 10 | BCH5946 | Shigella dysenteriae 1(5A) | ACTQNaCipNorOfx | + |
| 11 | BCH7084 | Shigella sonnei (1) | TQNa | + |
| 12 | BCH7264 | Shigella sonnei (2) | TQNa | + |
| 13 | BCH3143 | Shigella boydii (1) | TQNa | − |
| 14 | BCH4324 | Shigella boydii (2) | TQNa | − |
| 15 | BCR62 | Salmonella enterica serovar Typhi (1) | NaCipNorAzm | − |
| 16 | BCR43 | Salmonella enterica serovar Typhi (2) | NaAzm | − |
| 17 | Escherichia coli K12 | − | ||
| 18 | Escherichia coli C | + | ||
| 19 | Escherichia coli AG100 | − | ||
| 20 | XL1 Blue | − |
A, ampicillin; C, chloramphenicol; T, tetracycline; Q, cotrimoxazole; Na, nalidixic acid; Cip, ciprofloxacin; Nor, norfloxacin; Ofx, ofloxacin; Azm, azithromycin.
One Step Growth Curve
One step growth experiments were carried out by a method described elsewhere (Chang et al., 2005; Malek et al., 2009) with modification. Briefly, Shigella spp. (S. flexneri 2a (1A), S. dysenteriae 1 (1A), S. sonnei) were grown in LB medium at 37°C. Then 20 mL of Shigella culture (OD600 = 0.3) was harvested by centrifugation (5,000 × g, 4°C, 10 min). The pellet was resuspended in 1 mL of fresh LB and phage Sfin-1 was added to Shigella culture at a MOI of 0.01. The mixture was incubated for maximum adsorption (5 min for S. flexneri 2a and S. dysenteriae 1, 7 min for S. sonnei) at 37°C followed by 104-fold of dilution at final volume of 10 mL. During incubation at 37°C, 100 μL of aliquots were taken at different time intervals up to 100 min, mixed with 200 μL of S. flexineri 2a culture, plated on double-layered agar for phage titration. Three independent experiments were run for each Shigella spp. Burst size was determined as a ratio of the average bacteriophage particles produced after the burst and the average number of phage particles adsorbed.
Genome Analysis of Phage Sfin-1
Bacteriophage DNA was isolated from purified phages (procedure described above), using the phage DNA isolation kit (Norgen, Canada) according to the manufacturer’s instruction with modifications (Berg et al., 2016). Whole genome sequencing of Sfin-1 was performed by ION Xpress (S5-00205) version 5.0.4. The count of read was 67,228 with the average length of 322 bp per read. The sequence data could be assembled into a single contig of 50,403 bp using SPAdes 3.8.0 (Bankevich et al., 2012). The nucleotide sequence of the genome of Sfin-1 was submitted at GenBank under accession number MF468274. The nucleotide sequences of the phage Sfin-1 were auto annotated by GeneMarkS1 (Ver 3.26) (Besemer et al., 2001) and the function of the proteins encoded by the coding sequences (CDSs) were predicted based on BLASTp program and conserved domain search2. The probable replication origin of Sfin-1 was predicted by GenSkew program3. Putative promoter regions were predicted using Neural Network Promoter Prediction tool of the Berkeley Drosophila Genome Project4 (minimum promoter score: 0.9). Rho-independent transcription terminators were identified using ARNOLD terminator finding program (Lesnik et al., 2001). tRNA scan–SE search program5 was used to detect Putative tRNAs, if any (Lowe and Chan, 2016). Whole genome comparisons were made with Mauve6. Sequences of tail fiber proteins were compared by using the online Protein Predict Tool (Yachdav et al., 2014).
Electron Microscopy
The phage suspension (about 1 × 1012 PFU/mL) was negatively stained with 2% (w/v) uranyl-acetate and then examined under a FEI Tecnai 12 Bio Twin Transmission Electron Microscope at an operating voltage of 200 kV.
Identification of Proteins Associated With Sfin-1 Virions
A total of 100 μL (1 × 1018 PFU/mL) purified phage was diluted with 50 mM NH4HCO3. It was then treated with 100 mM DTT at 95°C for 1 h followed by 250 mM iodoacetamide at room temperature in dark for 45 min. The sample was then digested with Trypsin and incubated overnight at 37°C. The peptides were extracted in 0.1% formic acid and incubated at 37°C for 45 min. The solution thus prepared was centrifuged at 10,000 × g and the supernatant was vacuum dried and dissolved in 20 μL of 0.1% formic acid in water. 10 μL of it was subjected to ACQUITY UPLC BEH C18 column (Waters, United Kingdom) for separation of peptides; the peptides separated on the column were directed to Waters Synapt G2 Q-TOF instrument (Waters, United Kingdom) for MS and MS/MS analysis. The raw data was processed using MassLynx 4.1 WATERS. The individual peptides MS/MS spectra were matched to the database sequence for protein identification on PLGS software, Waters. The obtained spectrometry information was analyzed with PLGS software 3.0.2 (Waters, United Kingdom) using the National Centre for Biotechnology Information (NCBI) non-redundant database and the specific database created in this study based on the predicted CDSs of phage Sfin-1. The important parameter settings for the PLGS analysis were as follows: peptide mass tolerance (ppm), 50; fragment mass tolerance (ppm), 100; maximal missed cleavages, 2.
Determination of the Bacteriophage Genome Ends
The identification of phage packaging strategies and genome ends of a bacteriophage can be obtained by comparative analysis of phylogenetic relationships of amino acid sequences of terminase large subunit of phage with the other phages of known packaging strategies (Wittmann et al., 2014; Amarillas et al., 2017). The phylogenetic tree was thus reconstructed using the large terminase amino acid sequence of the phages and the relationships among the phage Sfin-1 and with the other phages were analyzed.
The predicted amino acid sequences of the large terminase subunits genes of the phages were collected from NCBI and were used for phylogenetic analysis. The bacteriophages involved into this study have been molecularly analyzed and contains well characterized ds DNA bacteriophages with different types of packaging strategies dependent on terminase actions (headful, 5′-extended cos ends, 3′-extended cos ends and direct terminal repeats). All the sequences were aligned using ClustalW in MEGA7 with default parameters. Phylogenetic tree was built using the neighbor-joining method and phylogenies were determined by bootstrap analysis of 1,000 replicates in MEGA 7.0 version (Filipski et al., 2014).
Additionally the genome ends were determined as described by Amarillas and Leon-Felix (Amarillas et al., 2017). To detect the presence of circularly permutated terminally redundant genome ends, approximately 1 μg phage DNA was digested with specific restriction enzymes (BglII, MluI) according to the manufacturer’s guidelines (NEB, United States). After 2 h of incubation at 37°C the digests were heated to 80°C for 15 min and then cooled fast in ice or slow at room temperature and allowed to run on agarose gel (0.8% w/v) in TAE electrophoresis buffer. The gel was stained with ethidium bromide and visualized with UV illumination. GeneRuler 1 kb Plus DNA Ladder (Thermo Fisher Scientific, United States) was used as DNA molecular weight marker.
Phage Receptor Identification
The receptor properties of Sfin-1 were determined as describe previously with some modifications (Kiljunen et al., 2011). To check the effect of proteinase K on adsorption of phage Sfin-1, S. flexneri 2a, S. dysenteriae 1, and S. sonnei cultures (OD600 = 0.3) were treated with proteinase K (250 mg/mL, SRL) at 55°C for 2 h, washed and allowed for adsorption assay at an MOI of 0.0001 as described above. In order to study whether periodate can inhibit Sfin-1 and hosts interaction, the following experiment was performed. S. flexneri 2a, S. dysenteriae1, and S. sonnei cultures were centrifuged at 5,000 × g for 5 min, bacterial pellets were resuspended into sodium acetate (50 mM, pH 5.2) in presence or absence of 200 mM NaIO4 and incubated for 2 h (protected from light). After incubation the cells were washed and performed adsorption assay.
To ensure that the possible effect was not due to the incubation of host cells at 55°C, a control experiment without proteinase K was also performed. LB medium was used as a non-absorbing control for both assays. The phage titer in the control supernatant was set to 100%.
Statistical Analysis
For the thermal stability assays, the difference in the titer values taken between 0 and 60 min were calculated for each temperature. Then the titer value difference for each temperature was compared to that of 4°C using Student’s t-test. Two way ANOVA test was performed to analyze the data of Bacterial challenge test. Student’s t-tests were performed to analyze the data of phage receptor identification. All statistical analysis were performed by software GraphPad Prism 7.0.
Results and Discussion
Isolation of Bacteriophage
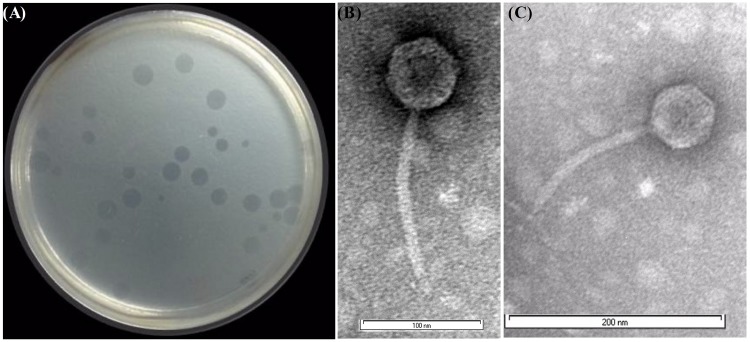
Several environmental water samples from ponds, creeks, streams, and canal ways were collected and the presence of phage that infects Shigella spp. was tested according to the method described in “Materials and Methods” section. The water sample from the River Ganges, Kolkata (located near Serampore, Hooghly of West Bengal, India) was found to contain phage, designated as Sfin-1 that could grow in antibiotic resistant various strains of Shigella spp. and resulted in clear lytic plaques with size ranging from 1.5 to 2.0 mm in diameter and well defined boundaries in the lawn of the bacteria (Figure 1A). Purification of the virion was done by CsCl density gradient ultracentrifugation after plaque purification and phage propagation as described in “Materials and Methods” section. The transmission electron microscopy (TEM) photos and the genome organization (described below) suggest that only one type of phage was present into the purified sample. If more than one bacteriophage were present there, then more genes of specific phage proteins such as tape measure protein and large terminase subunit would have been expected.
FIGURE 1.
Shgella spp. specific phage Sfin-1 and its morphology. (A) Plaques of Sfin-1 in the lawn of Shigella flexneri 2a. The phage particles were prepared, negatively stained and examined by electron microscope as described in “Materials and Methods” section. (B,C) The electron micrograph presented as broad view of the phage in 100 and 200 nm scale, respectively.
Broad Host Range
The ability to lyse various pathogenic Shigella spp. of newly isolated phage was analyzed by the spot test. The strains belonging to the Shiga toxin producing clinical isolates of S. flexneri, S. dysenteriae, S. boydii, S. Sonnei were used. Other enteropathogens like Salmonella typhi and various E. coli strains like K12, AG100, XL1 Blue and E. coli C. were also checked. The Shigella serovars are resistant to multiple antibiotics (amoxicillin, chloramphenicol, tetracycline, ciprofloxacin, norfloxacin, nalidixic acid, cotrimoxazole, ofloxacin, and azithromycin) (Amezquita-Lopez et al., 2014) (Table 1). Phage suspensions produced clear zones of lysis in case of various serotypes of S. flexneri, S. dysenteriae, S. sonnei, and E. coli C., but no reaction was observed for other species.
Our host range studies suggest that Sfin-1 is a newly isolated phage with a broad lytic spectrum. The wide host range infectivity against various Shigella isolates shows that Sfin-1 is a polyvalent phage on Shigella, known human and animal pathogen and E. coli C. Although phages are commonly very specific infecting only a single species of bacteria, some polyvalent phages have also been described (Hamdi et al., 2017).
From the Ganges River that flows through the city of Kolkata, we isolated the phage Sfin-1 near Serampore, Hooghly. Since Shigella spp. is communicated to human by fecal-oral route, the isolation of phage Sfin-1 indicates fecal contamination of the river. This lytic phage is active against a broad host range of antibiotic resistant strains of S. flexneri, S. dysenteriae, and S. sonnei. This is the first report of bacteriophage that is lytic against three pathogenic Shigella spp. and E. coli C. Therefore, it can be an excellent choice for phage therapy against antibiotic resistant Shigellosis.
Phage Morphology
The phage Sfin-1 which is specific against various antibiotic resistant Shigella spp. was purified and subjected to TEM. The TEM study suggested that this phage has an isometric head (61 nm in diameter) and a non-contractile tail (approximately 155 nm) with which a basal tuft is attached (Figures 1B,C). The mature phage does not have neck, base plate, spikes, or fiber. According to the guidelines of the International Committee on Taxonomy of Viruses (Fauquet and Fargette, 2005), which is based on virion morphological features, the phage Sfin-1 was assigned to the family Siphoviridae in the order Caudovirales. More than 95% of the phages reported so far grouped into the order Caudovirales (tailed phages). According to the classification system of Ackermann (1998), most (60%) of the bacteriophages belong to the family Siphoviridae with flexible and long tails. So the phage Sfin-1 belongs to this taxonomic classification.
Bacterial Challenge Test
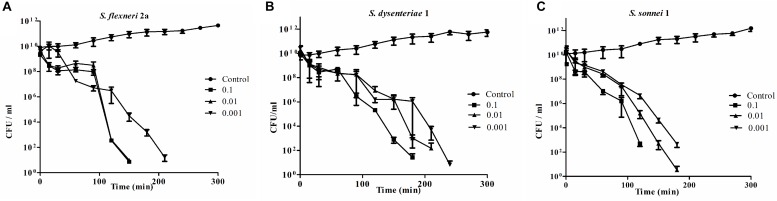
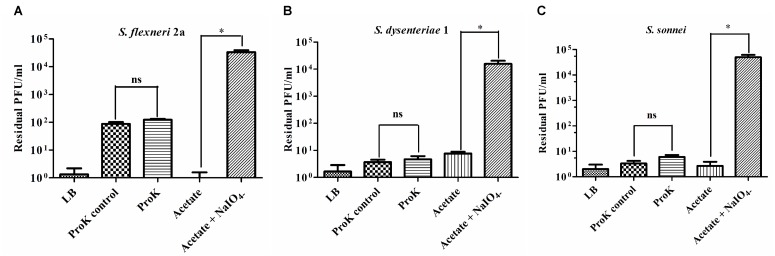
With the addition of phage Sfin-1 at an MOI of 0.001, 0.01, and 0.1 to mid-exponential phase cells (OD600 = 0.3) in in vitro culture conditions, challenge tests were performed to investigate the ability of phage Sfin-1 to lyse multidrug-resistant S. flexneri 2a (1A) (Strain Id BCH5722, Table 1) in presence of antibiotics ampicillin, chloramphenicol, tetracycline, cotrimoxazole, ciprofloxacin, norfloxacinand ofloxacin (Figure 2A). In each case, control experiment was performed where bacterial cells were grown only in presence of respective antibiotics and phage suspension buffer without phage particles. The viability of bacterial cells was significantly decreased when infected with MOI of 0.1, 0.01, and 0.001. The two way ANOVA tests for multiple comparisons showed that the mean differences between the cell lysis data for all three MOIs and control are highly significant (P < 0.0001). Within one and half hours after phage addition, cells were started to decrease rapidly and complete lysis occurred within two and half hours in case of MOI of 0.1 and 0.01. But in case of 0.001, complete lysis occurs after 3 h. Similar results were observed for S. dysenteriae1 (1A) (Strain ID BCH5762, Table 1) in presence of the same antibiotics as mentioned above (Figure 2B) and S. sonnei (Strain ID BCH7084) cells in presence of tetracycline, cotrimoxazole, and nalidixic acid (Figure 2C). The in vitro challenge tests established that the phage Sfin-1 could be used to inactivate the multidrug-resistant pathogenic strains of Shigella and therefore, it could be useful as biocontrol agent. The efficacy of this phage in controlling Shigella infection however has to be determined by in vivo studies. It is important to mention that a host population known as bacterial insensitive mutants (BIMs) that can resist lysis may emerge and grow again long after phage treatment (Yordpratum et al., 2011; Yamaki et al., 2014; Amarillas et al., 2017). Use of phage cocktail with more than one phage that follows different infection mechanisms may solve this problem (Yamaki et al., 2014).
FIGURE 2.
Bacterial challenge test of phage Sfin-1 on different clinical isolates of Shigella spp. Clinically isolated species of (A) Shigella flexneri 2a, (B) Shigella dysenteriae 1, and (C) Shigella sonnei were grown (OD600 = 0.3) in 20 mL LB in presence of several antibiotics as described in “Materials and Methods” section, hervested by centrifugation, resuspended in 1 mL LB medium and infected with Sfin-1 at an MOI of 0.1, 0.01, and 0.001. After adsorption, the cultures were diluted 21 fold in LB and incubated for 5 h with shaking at 37°C. At different time intervals, viability of Shigella spp. was determined by spread plate method. As negative control Shigell spp. were grown in absence of Sfin-1 in presence of antibiotics. Two way ANOVA indicated significant difference between control and Sfin-1 infected sets (P < 0.0001, n = 3).
So the result of host cell lysis caused by phage Sfin-1 showed that MOI is directly related to cell death. If higher number of phages are applied on cells, destabilization of the outer membrane occurs, which in turn causes cell lysis. Since it is not the result of phage replication and release, it is called “lysis from without” (Brown and Bidle, 2014; Amarillas et al., 2017). In Supplementary Table S1 phage Sfin-1 is compared with other 44 Shigella phages so far reported in the NCBI database. Only 26% of the isolated Shigella phages have been characterized till date.
Infectivity of Sfin-1
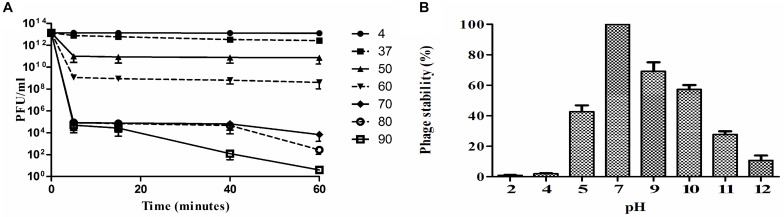
Thermal stability test was carried out with Sfin-1 at pH 7.0 in order to investigate the heat resistant capability of the phage. The activity of phage Sfin-1 remained moderately same when warmed at 37°C or 50°C for 5 min. Activity decreased significantly into 0.1–0.01% when incubated at 60°C or 70°C for 5 min (P < 0.005). When heated at 80°C or 90°C for 5 min only 0.001% activity was retained (P < 0.005). Most of the phages remained active even after 60 min incubation at 37°C or 50°C, whereas only 0.1 and 0.01% phages were active after 60 min incubation at 60 and 70°C, respectively (P < 0.005), the phage activity remarkably decreased at 80°C or 90°C after 60 min incubation (P < 0.0005). This result suggests that the phage Sfin-1 is moderately stable toward heat stress at both 37 and 50°C (Figure 3A).
FIGURE 3.
Stability of phage Sfin-1 in wide temperature and pH range. (A) Thermal stability of phage Sfin-1 at various temperatures as indicated. Sfin-1 phage particles (16 × 1012) were incubated at different temperatures in 1 mL and for each temperature the number of infectious phage particles was determined from 100 μL aliquots from various time points by plaque assay against S. flexineri 2a. Result was plotted as mean ± SD (n = 3). (B) pH stability of phage Sfin-1. In 1 mL of TM buffer having different pH Sfin-1 phage particles (14 × 1010) were incubated at 37°C for 1 h and the number of infectious phage particles from each sample was determined with 100 μL aliquots by plaque assay against S. flexineri 2a. Result was plotted as mean ± SD (n = 3).
The Shigella infection commonly happens in intestine where the pH is somewhat acidic. In order to control Shigella with Sfin-1 in human intestine, it is therefore, essential to know its pH stability. Highest activity was observed after 1 h incubation at pH 7.0 at 37°C, while reduction of activity was observed at different pH. Around 42.7% or 10.8% recovery of infectious phage Sfin-1 was found at pH 5.0 and 12.0, respectively. This result suggested that extreme pH as well as lower pH though affect the phage stability but a remarkable fraction of Sfin-1 remained active (Figure 3B). Rapid absorption, moderate thermal and pH stability therefore indicate that this phage may be applied for therapeutic purpose. However, in order to achieve therapeutic efficacy the phages must be delivered to the small intestine through encapsulation otherwise it may not survive at low gastric pH (Vinner et al., 2017).
One-Step Growth Curve
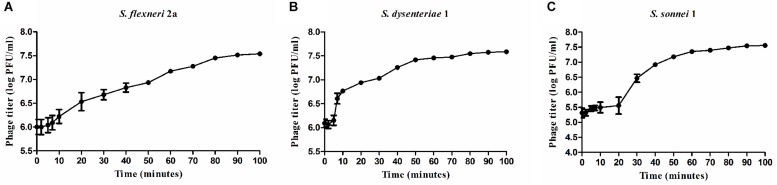
The one step growth curve performed at 37°C for Sfin-1 propagated on S. flexneri 2a and S. dysenteriae1 showed a latent period of about 5 min and the average burst size was 27–28 PFU/cell (Figure 4). While in case of S. sonnei, the latent period was 10 min and the average burst size was estimated to be 146 PFU/cell. A phage with a large burst size can have practical advantage in therapy, because within a short period of time phage population can increase its initial dose by several 100 folds (Gallet et al., 2011).
FIGURE 4.
One step growth curve of bacteriophage Sfin-1. Shigella flexneri 2a, Shigella dysenteriae 1, Shigella sonnei were infected with Sfin-1 at 37°C at an MOI of 0.01. After phage absorption, the cultures were diluted 104-fold, incubated at 37°C and the titers in PFU per mL of Sfin-1 from the infected cultures at different time points were determined. Result was plotted as mean ± SD (n = 3). (A–C) Present one step growth curves of Sfin-1 in Shigella flexneri 2a, Shigella dysenteriae 1, and Shigella sonnei, respectively.
Whole Genome Phylogenetic and Synteny Study
For better understanding of phage Sfin-1 biology, its genome was sequenced. The whole genome sequencing study by Ion torrent reveals that the Sfin-1 genome size is 50,403 bp with 45.20% GC content, close to 50.9% of the host’s chromosome (Wei et al., 2003). The Genome of Sfin-1 shows total 82 protein CDSs after auto-annotation with GeneMarkS; 19 of which are rightward in orientation while others are leftward (Figure 5A). Among them 23 had annotated function (Table 2). No tRNA was found in the Sfin-1 genome; this suggests that upon entry into the host, phage is completely dependent on the host tRNA for its protein synthesis.
FIGURE 5.
Genome organization of Sfin-1. (A) The Sfin-1 genome map was schematically presented. The predicted CDSs are indicated as arrows, the orientation of which shows the transcription. With different colors predicted molecular function for CDS of virion morphogenesis (green arrows), DNA metabolism and replication (red arrows), DNA packaging (yellow arrows), cell lysis (violet arrows), hypothetical proteins (blue arrows), putative promoters (pink) are denoted. (B) Comparative genomic maps of phage Sfin-1, pSf-2, Shfl1 was constructed using the Mauve progressive alignments to determine conserved sequence regions. This alignment resulted into two large synteny locally collinear blocks (LCBs) with 28,894 bp (red) and 16,173 bp (green), one small LCB with 5,334 bp (sky), indicating DNA regions which are homologous among the genomes. Graphs inside the blocks show high similarity between the genomes. There are some non-identical genome regions which are denoted with white color inside the blocks. Although there seems to be genomic rearrangement, the block sequence remains the same across the genomes of all phages.
TABLE 2.
Features of the protein coding sequences of bacteriophage Sfin-1 and homology to protein database.
| CDSs | start | Stop | Length (bp) | Predicted Functions | Best BLASTp match and Identity (%) | Conserved protein domain family | |
| CDS1 | 703 | 2 | 702 | Hypothetical protein | Hypothetical protein ISF001_0035 [Shigella phage vB_SsoS-ISF002] | 100 | |
| CDS2 | 1240 | 764 | 477 | Phage protein | Hypothetical protein Shfl1p34[Shigella virus Shfl1] | 92 | |
| CDS3 | 2364 | 1252 | 1113 | Phage capsid and scaffold | Major capsid protein [Shigella phage SH6] | 99 | pfam09979 COG3566 |
| CDS4 | 3026 | 2367 | 639 | Phage minor capsid protein | Minor capsid protein [Shigella phage SH6] | 99 | COG2369 pfam04233TIGR01641 |
| CDS5 | 3127 | 3011 | 117 | Phage minor capsid protein | Minor capsid protein [Shigella phage SH6] | 97 | |
| CDS6 | 4400 | 3117 | 1284 | Phage portal protein | Putative portal protein [Shigella virus Shfl1] | 98 | pfam06381 TIGR01555COG3567 |
| CDS7 | 6025 | 4457 | 1569 | Phage terminase, large subunit | Putative terminase large subunit [Shigella virus Shfl1] | 99 | COG5410 TIGR0160COG5362 pfam03237 |
| CDS8 | 6589 | 6065 | 525 | Phage terminase, small subunit | Putative terminase small subunit [Shigella virus Shfl1] | 99 | pfam16677 |
| CDS9 | 6886 | 6674 | 213 | Phage protein | Hypothetical protein JMPW1_023 [Escherichia phage JMPW1] | 97 | |
| CDS10 | 7087 | 6902 | 186 | Phage protein | Hypothetical protein pSf2_045 [Shigella phage pSf-2] | 98 | |
| CDS11 | 7229 | 7068 | 162 | Phage protein | Hypothetical protein pSf2_046 [Shigella phage pSf-2] | 98 | |
| CDS12 | 7597 | 7394 | 204 | Phage protein | Hypothetical protein pSf2_047 [Shigella phage pSf-2] | 97 | |
| CDS13 | 7827 | 7597 | 231 | Phage protein | Hypothetical protein JMPW1_019 [Escherichia phage JMPW1] | 100 | |
| CDS14 | 8171 | 7827 | 345 | Phage protein | Hypothetical protein B508_00140 [Escherichia phage ADB-2] | 95 | |
| CDS15 | 8377 | 8168 | 210 | Phage protein | Hypothetical protein JMPW2_017 [Escherichia phage JMPW2] | 99 | |
| CDS16 | 9022 | 8450 | 573 | JK_59P | Hypothetical protein T1p62 [Escherichia virus T1] | 97 | |
| CDS17 | 9134 | 9024 | 111 | Phage protein | Hypothetical protein [Escherichia phagevB_EcoS_SH2] | 92 | |
| CDS18 | 9607 | 9131 | 477 | 3′-phosphatase, 5′-polynucleotide kinase, phage-associated | 3′-phosphatase, 5′-polynucleotide kinase [Escherichia phagevB_EcoS_SH2] | 95 | cd07502 PHA02530 pfam03767TIGR01675 |
| CDS19 | 10193 | 9747 | 447 | Phage protein | hypothetical protein SH6_0017 [Shigella phage SH6] | 85 | |
| CDS20 | 11283 | 10627 | 657 | Phage protein | Hypothetical protein [Escherichia phagevB_EcoS_SH2] | 78 | |
| CDS21 | 11567 | 11280 | 288 | Hypothetical protein | Hypothetical protein pSf2_056 [Shigella phage pSf-2] | 97 | |
| CDS22 | 11811 | 11551 | 261 | hypothetical protein | Hypothetical protein B508_00105 [Escherichia phage ADB-2] | 96 | |
| CDS23 | 12075 | 11926 | 150 | Phage protein | Hypothetical protein [Escherichia phagevB_EcoS_SH2] | 96 | |
| CDS24 | 12560 | 12075 | 486 | Phage protein | Hypothetical protein T1p70[Escherichia virus T1] | 88 | |
| CDS25 | 13135 | 12632 | 504 | Phage protein | Hypothetical protein B508_00095 [Escherichia phage ADB-2] | 88 | |
| CDS26 | 13338 | 13132 | 207 | Hypothetical protein | Hypothetical protein Shfl1p10 [Shigella virus Shfl1] | 99 | |
| CDS27 | 13600 | 13409 | 192 | Hypothetical protein | Hypothetical protein JMPW2_006 [Escherichia phage JMPW2] | 96 | |
| CDS28 | 13783 | 13610 | 174 | Phage protein | Hypothetical protein pSf2_062 [Shigella phage pSf-2] | 98 | |
| CDS29 | 13890 | 13780 | 111 | Phage protein | Hypothetical protein SH6_0007 [Shigella phage SH6] | 100 | |
| CDS30 | 14114 | 13887 | 228 | Phage protein | Hypothetical protein B508_00085 [Escherichia phage ADB-2] | 99 | |
| CDS31 | 14351 | 14121 | 231 | Phage protein | Hypothetical protein B508_00080 [Escherichia phage ADB-2] | 91 | |
| CDS32 | 14900 | 14430 | Phage protein | Hypothetical protein [Escherichia phagevB_EcoS_SH2] | 90 | ||
| CDS33 | 15097 | 14903 | 195 | Phage protein | Hypothetical protein Shfl1p02 [Shigella virus Shfl1] | 100 | |
| CDS34 | 15703 | 15188 | 516 | Phage protein | Hypothetical protein Shfl1p01 [Shigella virusShfl1] | 90 | |
| CDS35 | 16391 | 16603 | 213 | Hypothetical protein | Hypothetical protein [Escherichia phagevB_EcoS_SH2] | 100 | |
| CDS36 | 16600 | 17304 | 705 | Phage protein | DNA methylase [Shigella phage SH6] | 97 | COG0270 |
| CDS37 | 17370 | 17705 | 336 | Hypothetical protein | Hypothetical protein SH6_0079 [Shigella phage SH6] | 93 | |
| CDS38 | 17779 | 17940 | 162 | Hypothetical protein | Hypothetical protein Shfl1p82 [Shigella virus Shfl1] | 92 | |
| CDS39 | 17970 | 18386 | 417 | Phage protein | Hypothetical protein JMPW1_074 [Escherichia phage JMPW1] | 93 | |
| CDS40 | 18379 | 18579 | 201 | Phage protein | Hypothetical protein T1p02 [Escherichia virus T1] | 98 | |
| CDS41 | 18596 | 18949 | 354 | Phage protein | Hypothetical protein pSf2_078 [Shigella phage pSf-2] | 99 | |
| CDS42 | 19133 | 19357 | 225 | Phage protein | Hypothetical protein [Escherichia phagevB_EcoS_SH2] | 97 | |
| CDS43 | 19361 | 19570 | 210 | Phage protein | Hypothetical protein B508_00010 [Escherichia phage ADB-2] | 100 | |
| CDS44 | 19651 | 20067 | 417 | Phage protein | Hypothetical protein B508_00005 [Escherichia phage ADB-2] | 98 | |
| CDS45 | 20144 | 21712 | 1569 | Phage protein | Hypothetical protein [Escherichia phagevB_EcoS_SH2] | 100 | |
| CDS46 | 21717 | 22124 | 408 | Phage protein | Hypothetical protein [Escherichia phagevB_EcoS_SH2] | 99 | |
| CDS47 | 22314 | 22529 | 216 | hypothetical protein | Hypothetical protein SH6_0068 [Shigella phage SH6] | 94 | |
| CDS48 | 22946 | 22542 | 405 | Phage protein | Hypothetical protein T1p11 [Escherichia virus T1 | 89 | |
| CDS49 | 23431 | 22943 | 489 | Phage lysin (EC 3.2.1.17) | Endolysin [Shigella phage SH6] | 99 | cd00737 COG3772 pfam00959 |
| CDS50 | 23709 | 23431 | 279 | Phage holin | Hypothetical protein T1p14 [Escherichia virus T1] | 100 | |
| CDS51 | 23941 | 23765 | 177 | Phage protein | Hypothetical protein B508_00370 [Escherichia phage ADB2] | 99 | |
| CDS52 | 25156 | 24005 | 1152 | Phage protein | Hypothetical protein pSf2_07 [Shigella phage pSf-2] | 97 | |
| CDS53 | 25540 | 25235 | 306 | JK_65P | Hypothetical protein pSf2_07 [Shigella phagepSf-2] | 94 | |
| CDS54 | 25926 | 25717 | 210 | Phage protein | Hypothetical protein B508_00360 [Escherichia phage ADB-2] | 94 | |
| CDS55 | 26244 | 25993 | 252 | Phage protein | Hypothetical protein B508_00355 [Escherichia phage ADB-2] | 99 | |
| CDS56 | 26954 | 26241 | 714 | DNA adenine methyltransferase, phage-associated | DNA N-6-adenine-methyltransferase [Escherichia phage ADB-2] | 98 | TIGR01712pfam05869 |
| CDS57 | 27438 | 27022 | 417 | Phage protein | VRR-NUC domain-containing protein [Escherichia phage ADB-2] | 100 | pfam08774 smart00990 |
| CDS58 | 29453 | 27435 | 2019 | DNA helicase, phage-associated | Putative ATP-dependent helicase [Shigella virus Shfl1] | 99 | COG 1062 cd18799 pfam 0851 |
| CDS59 | 29548 | 30000 | 453 | Phage protein | Hypothetical protein Shfl1p58 [Shigella virus Shfl1] | 99 | pfam1549 |
| CDS60 | 30061 | 30996 | 936 | DNA primase | DNA primase/helicase [Escherichia phage JMPW1] | 99 | smart00778 pfam08273 COG4643 pfam02655 |
| CDS61 | 31097 | 32881 | 1785 | Phage tail fiber protein | Putative tail fiber [Shigella virus Shfl1] | 94 | pfam 13884 PHA 00430 |
| CDS62 | 33323 | 32910 | 414 | Phage protein | Hypothetical protein Shfl1p55 [Shigella virus Shfl1] | 98 | |
| CDS63 | 34017 | 33370 | 648 | Phage-associated recombinase | Putative recombination protein [Shigella phage vB_SsoS-ISF002 | 98 | |
| CDS64 | 35156 | 34092 | 1065 | Phage exonuclease | Hypothetical protein B508_00305 [Escherichia phage ADB-2] | 99 | pfam12684 |
| CDS65 | 35683 | 35913 | 231 | Hypothetical protein | Hypothetical protein pSf2_020 [Shigella phage pSf-2] | 97 | |
| CDS66 | 35916 | 36872 | 957 | Phage protein | Hypothetical protein pSf2_021 [Shigella phage pSf-2] | 99 | |
| CDS67 | 40349 | 36900 | 3450 | Tail fiber protein | Putative tail fiber protein [Shigella virus Shfl1] | 98 | COG 4733 pfam13550 |
| CDS68 | 41026 | 40427 | 600 | Phage tail assembly protein | Tail assembly protein [Escherichia phageJMPW1] | 100 | COG 4723 pfam 06805 |
| CDS69 | 41757 | 41023 | 735 | Phage tail assembly protein | Tail assembly protein [Escherichia phageJMPW1] | 99 | cd 8073 pfam 00877 |
| CDS70 | 42566 | 41754 | 813 | Minor tail protein | Putative minor tail protein [Escherichia phage JMPW2] | 99 | pfam05100 COG 4672 TIGR01600 |
| CDS71 | 42968 | 42615 | 354 | Tail fiber protein | Tail fiber protein [Escherichia phage JMPW1] | 99 | pfam 05939 COG4718 |
| CDS72 | 43038 | 42955 | 84 | Phage tail length tape-measure protein 1 | Putative tail tape measure protein [Shigella virus Shfl1] | 96 | pfam06791COG5281 TIGR01541pfam09718 PRK03918 |
| CDS73 | 45842 | 43035 | 2808 | Phage tail length tape-measure protein 1 | Putative tail tape measure protein [Shigella virus Shfl1] | 98 | pfam06791COG5281 TIGR01541pfam09718 PRK03918 |
| CDS74 | 46152 | 45883 | 270 | Phage protein | Hypothetical protein pSf2_028 [Shigella phage pSf-2] | 99 | |
| CDS75 | 46517 | 46200 | 318 | Phage protein | Hypothetical protein ISF001_0027 [Shigella phage vB_SsoS-ISF002] | 99 | |
| CDS76 | 47299 | 46631 | 669 | Phage tail fibers | Tail fiber protein [Escherichia phage JMPW1] | 97 | pfam08813 |
| CDS77 | 47699 | 47301 | 399 | JK_18P | Hypothetical protein pSf2_031 [Shigella phage pSf-2] | 99 | |
| CDS78 | 48132 | 47689 | 444 | Phage protein | Hypothetical protein pSf2_032 [Shigella phage pSf-2] | 97 | |
| CDS79 | 48496 | 48125 | 372 | JK_22P | Hypothetical protein pSf2_033 [Shigella phage pSf-2] | 98 | |
| CDS80 | 48906 | 48496 | 411 | JK_23P | Hypothetical protein B508_00220 [Escherichia phage ADB-2] | 98 | |
| CDS81 | 49238 | 48951 | 288 | JK_24P | Hypothetical protein ISF001_0033 [Shigella phage vB_SsoS-ISF002] | 97 | |
| CDS82 | 50247 | 49288 | 960 | Phage protein | Hypothetical protein B508_00210 [Escherichia phage ADB-2] | 98 | |
The whole genome Basic Local Alignment Search Tool (BLAST) analysis of Sfin-1 against the NCBI data base showed that Sfin-1 is related to two phages i.e., Shigella phage Shfl1 (GenBank accession number: NC_015456) and Shigella phage pSf-2 (GenBank accession number: KP085586). The phage Sfin-1 genome sequence shares 91 and 92% nucleotide identity with Shigella phages Shfl1 and pSf-2, respectively. Genomic features of three phages ware compared in Supplementary Table S2. Though the genome sizes, GC contents, number of transcription terminator sequences and CDSs are quite similar and genes of predicted structural and functional proteins share high degree of homology, they are differently arranged and their orientations are sometimes opposite (Supplementary Figure S1). Maximum differences present into their hypothetical proteins which are uncharacterized according to database. Around 75% genes of Sfin-1 are of unknown functions while most of them have >80% homology with their counterparts in Shfl1 and Psf-2 genome. Since these phages were isolated from different geographical locations, the high degree of similarity probably appears from their complex evolutionary relationships with their common host S. flexineri (Shen et al., 2016). Since the genome of Sfin-1 diverges from the other phage genomes, with similarity searches only a fraction (28.04%) of protein functions could be predicted emphasizing the novelty of this phage. Therefore, a thorough investigation is needed to fully understand its biology. The mauve alignment of Sfin-1, Shfl1 and pSf-2 resulted into two large synteny locally collinear blocks (LCBs) with 28,894 bp (red) and 16,173 bp (green), one small LCB with 5,334 bp (sky), indicating DNA regions which are homologous among the genomes. Graphs inside the blocks show high similarity between the genomes. There are some non-identical genome regions which are denoted with white color inside the blocks. Although there seems to be genomic rearrangement, the block sequence remains the same across the genomes of all phages.
Moreover the alignment of the three phages also demonstrated that some regions are highly homologous with significant rearrangements (Figure 5B). This indicated that these phages share a common genome organization although positions of the genes are different.
Module Analysis
The annotated proteins of Sfin-1 can be categorized into the following functional groups: DNA metabolism and replication proteins; this module presents in the middle part of the Sfin-1 genome. They are 3′-phosphatase, 5′-polynucleotide kinase/CDS18, Phage associated N-6-DNA adenine-methyl transferase/CDS56, DNA helicase/CDS58, DNA primase/helicase/CDS60, and phage associated recombinase/CDS63, phage exonuclease/CDS64). The 3′-phosphatase, 5′-polynucleotide kinase belongs to the family that includes the C-terminal domain of the bifunctional enzyme T4 polynecleotide kinase/phosphatase PNKP. The PNKP phosphatase domain can catalyze the hydrolytic elimination of the 3′-phosphoryl group of DNA, RNA and deoxynucleoside 3′-monophosphates. The enzyme N-6-DNA adenine-methyl transferase (DAM) is involved in methylation of GATC sequence of its own DNA to protect it from exonuclease. The counterpart of this enzyme of Sfin-1 is present in Escherichia phage ADB-2 (99% identity). There are two helicase coding genes in the Sfin-1 phage genome; one is ATP dependent and has 99% identity with Shigella phage Shfl1 helicase and another is primase associated having 99% identity with Escherichia phage JMPW1. The primase/helicase protein has a zinc finger motif at its N terminal region and ATP binding region at its C-terminal part with origin recognition property. CDS63 contains a protein which encodes phage associated recombinase domain that is commonly found associated with Pfam04404 of ERF superfamily. The family includes single strand annealing proteins (SSAPs), such as Rad52, ERF, Red-beta, and RecT that function in RecA independent and RecA dependent DNA recombination pathways. This type of phage encoded recombinase are mainly involved in horizontal gene transfer by homologous recombination, thus promotes gene shuffling among phages which accelerates evolution. The phage exonuclease acts together with recombinase and involves into replication process from fork structures as well as in nucleotide metabolism. CDS64 encodes an exonuclease VIII that is related to PDDEXK superfamily. Thus 3′-phosphatase, 5′-polynucleotide kinase, phage recombinase, exonuclease are involved in DNA metabolism and recombination process of the phage genome after entering into host cells.
Sequenced based prediction of the phage Sfin-1 genome identified that upstream and downstream cluster genes are involve in viral head morphogenesis and tail component formation, respectively. CDS3, CDS4, and CDS5 which are likely to produce phage capsid and scaffold protein belong to Phage Mu F like protein family. Members of this family are required for viral head morphogenesis. CDS6 encodes head and tail junction portal protein that is believed to form the pore through which genome is packaged into the prohead and is also a part of the packaging motor (Lokareddy et al., 2017). CDS7, CDS8 encode phage large and small subunit, respectively, which are involved in packaging of the concatameric DNA in phage capsids (Mobberley et al., 2008). Apart from upstream genes, the downstream genes CDS61, CDS67, CDS70, CDS71, CDS72, CDS73, and CDS 76 probably encode the tail component whereas CDS68 and CDS69 direct the synthesis of protein responsible for tail assembly. CDS72 and CDS73 that encodes tail tape measure protein is the second largest gene of the phage genome. Although genomes of phages Sfin-1, Shfl1 and pSf-2 were found to be highly similar as discussed above, they infect different species (Supplementary Table S2). As tail fiber proteins are involved in host range determination (Yang et al., 2018), the sequence and predicted features of three phages’ tail fiber proteins were compared. Four tail fiber proteins of Sfin-1 are (A) CDS61, (B) CDS67, (C) CDS71, and (D) CDS76. Their counterparts in Shfl1 are CDS53 (93.60% identity), CDS47 (98% identity), CDS43 (95.73% identity), CDS39 (96.85% identity), respectively and in pSf-2 are CDS16 (97.31% identity), CDS22 (98% identity), CDS26 (94.02% identity), CDS30 (100% identity), respectively. The tail fiber proteins of these phages not only share high degree of nucleotide sequence homology, the positions of helix and strands in them are also very similar.
However, 69, 116, 239, 277, 313, 345, 380, 415, 483, and 545 of tail fiber protein A, 43, 48, 72, 101, 152, 158, 190, 198, 199, and 1,089 for tail fiber protein B, 8, 61, and 67 for tail fiber protein C amino acid residues of Sfin-1 sequence differ to those in phages Shfl1 and Psf-2, which might result in differences in their protein binding regions (Supplementary Figures S2–S5) and ultimately determine the changes in host specificity. No difference of amino acid residue of tail fiber protein D sequence has been noticed.
Tail length of the lambdoid phages corresponds to the length of the tail tape measure protein where single amino acid is equivalent to ∼0.15 nm (Katsura, 1990). According to this hypothesis the tail length of the phage Sfin-1 is approximately about 144 nm long which is approximately close to our measured length 155 nm. Most of the virion morphogenesis genes encode proteins which show similarity to either Shigella phage psf-2 or Shigella virus Shfl1 encoded proteins. However, gene order of Shfl1 is totally reverse to the Sfin-1 while psf-2 possesses the same gene orientation but at different positions. The terminase subunits and CDS7-CDS8, the DNA packaging genes of Sfin-1 have counterparts in Shigella phage Shfl1 with 99% identity. These proteins are mainly involved in ATP dependent DNA packaging system.
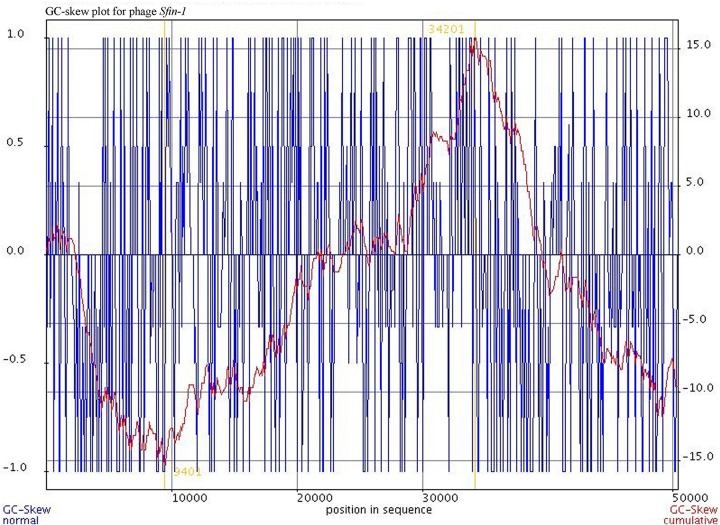
Cell lysis proteins, phage lysine/CDS49 and holin/CDS50 are present in Sfin-1 phage genome. These genes are crucial for host cell destruction during burst step of phage life cycle. Once the new phage progeny has been assembled, most of the phages lyse their host by using a dual lysis system, which contains a pore forming protein holin and cell wall degrading enzyme phage lysozyme or endolysin. CDS49 and CDS50 located contagiously at the middle part of the Sfin-1 genome that are involved in cell lysis. CDS49 encodes 162 amino acids long phage lysozyme/endolysin belonging to the pfam00959 family found in dsDNA phages. Members of this family in conjunction with holin (CDS50) cleave the ß1,4-glycosidiclinkage of polysaccharide present in the bacterial membrane (Ziedaite et al., 2005). BLASTp analysis also indicates the presence of one DNA binding transcriptional regulatory cro protein encoded by CDS59 belonging to the HTH_XRE superfamily. Sfin-1 may use this protein to regulate transcriptional timing in the gene expression. So presence of lysis genes but no lysogeny related genes into the Sfin-1 genome clearly indicates that this phage is a potent lytic phage. The GeneSkew program, an application for computing and plotting nucleotide skew data predicted the probable replication origin of Sfin-1. The GC-skew plot (Figure 6) indicates that the replication origin could be the region around the nucleotide 34201 (close to CDS64).
FIGURE 6.
Cumulative GC skew analysis of Sfin-1 genome sequence. The cumulative graph displays the global minimum and maximum. The window size of 1,000 bp and a step size of 100 bp were used to calculate the global minimum and maximum. The blue and red lines represent the GC-skew and the cumulative GC-skew, respectively. The putative origin of replication (9,401 nt) and the putative terminus location (34,201 nt) can be predicted from the minimum and maximum of a GC-skew.
Proteomic Analysis
The purified phage of Sfin-1 was analyzed by liquid chromatography/tandem mass spectrometry (LC-MS/MS). The LC-MS/MS analysis detected 22 phage proteins including 16 structural with a coverage ranging from 10 to 25% and 6 functional with a coverage ranging from 16 to 23% (Table 3). Among 16 structural proteins, the phage capsid and scaffold protein, minor capsid protein, tail fiber protein, phage lysin, phage holin, minor tail protein, tail assembly protein, terminase large and small subunit are associated with virion morphogenesis and phage packaging functions while the other 6 functional proteins are phage helicase, exonuclease, recombinase, 5′ polynucleotide kinase, DNA adenine methyltransferase, primase. These proteins are involved in DNA metabolism and replication functions of the phage genome after entering into host cells. The phage encoded recombinase is mainly involved in horizontal gene transfer by homologous recombination process which helps in gene rearrangement among phages resulting in acceleration of evolution.
TABLE 3.
Virion proteins detected by LC-MS/MS.
| CDS | Start | End | Mass (Da) | Predicted function | BLASTp (best match)1 | Identity (%)2 | LC-MS/MS3 |
| 03 | 2364 | 1252 | 40117 | Phage capsid and scaffold | WP_022638357/Lactobacillus plantarum | 10/24 (42) | 25 |
| 04 | 3026 | 2367 | 24234 | Minor capsid | WP_002314920/Enterococcus faecium | 23/89 (26) | 15 |
| 05 | 3127 | 3011 | 432991 | Phage minor capsid | WP_024636649/Enterococcus faecium | 10/31 (32) | 13 |
| 06 | 4400 | 3117 | 48011 | Portal protein | WP_009841304/Vibrio spp. AND4 | 15/38 (39) | 25 |
| 07 | 6025 | 4457 | 60287 | Terminase large subunit | YP_006119910/Escherichia coli O83 | 28/115 (24) | 14 |
| 08 | 6589 | 6065 | 19320 | Terminase small subunit | YP_006780198/Escherichia coli O104 | 11/24 (46) | 13 |
| 18 | 9607 | 9131 | 17985 | 3′-phosphatase, 5′-polynucleotide kinase | NP_268935/Streptococcus phage 370.1 | 9/12 (75) | 16 |
| 49 | 23431 | 22943 | 18276 | Phage lysin | YP_535193/Lactobacillus salivarius UCC118 | 5/15 (33) | 19 |
| 50 | 23709 | 23431 | 7577 | Phage holin | WP_061631936/Streptococcus pneumonia | 6/15 (40) | 12 |
| 56 | 26954 | 26241 | 27014 | DNA adenine methyltransferase | NP_268935/Streptococcus phage 370.1 | 7/22 (32) | 16 |
| 58 | 29453 | 27435 | 76000 | DNA helicase | WP_058540300/Pseudomonas putida | 29/98 (30) | 14 |
| 60 | 30061 | 30996 | 34612 | DNA primase | WP_023241409/Salmonella enterica | 27/76 (36) | 23 |
| 61 | 31097 | 32881 | 64478 | Tail fiber protein | NP_857641/Yersinia pestis | 13/40 (33) | 10 |
| 63 | 34017 | 33370 | 23935 | Phage associated recombinase | WP_060441855/Serratia marcescens | 32/126 (25) | 16 |
| 64 | 35156 | 34092 | 40165 | Phage exonuclease | YP_004243626/Bacillus subtilis | 16/50 (32) | 18 |
| 67 | 40349 | 36900 | 127462 | Tail fiber protein | WP_010939430/Desulfovibrio vulgaris | 109/484 (30) | 21 |
| 68 | 41026 | 40427 | 20880 | Tail assembly protein | WP_023658790/Pseudomonas fluorescens | 23/73 (32) | 20 |
| 69 | 41757 | 41023 | 28345 | Tail assembly protein | WP_005987870/Desulfovibrio africanus | 17/70 (24) | 17 |
| 70 | 42566 | 41754 | 29021 | Minor Tail protein | WP_007364011/Opitutaceae bacterium TAV1 | 7/23 (30) | 18 |
| 71 | 42968 | 42615 | 13041 | Tail fiber protein | WP_010939430/Desulfovibrio vulgaris | 5/11 (45) | 21 |
| 73 | 45842 | 43035 | 101408 | Tail-length tape measure protein | WP_063259367/Enterobacter cloacae | 63/284 (25) | 22 |
| 76 | 47299 | 46631 | 24077 | Tail fiber protein | NP_857641/Yersinia pestis | 9/27 (33) | 10 |
1The matched homologs of LC-MS/MS data. 2The percentage identity is calculated based on the number of identical amino acid residues (numerator) over the number of compared residues (denominator) and shown in parentheses. 3The sequence coverage (%) determined by mass spectrometry of the protein is indicated.
Determination of the Bacteriophage Genome Ends
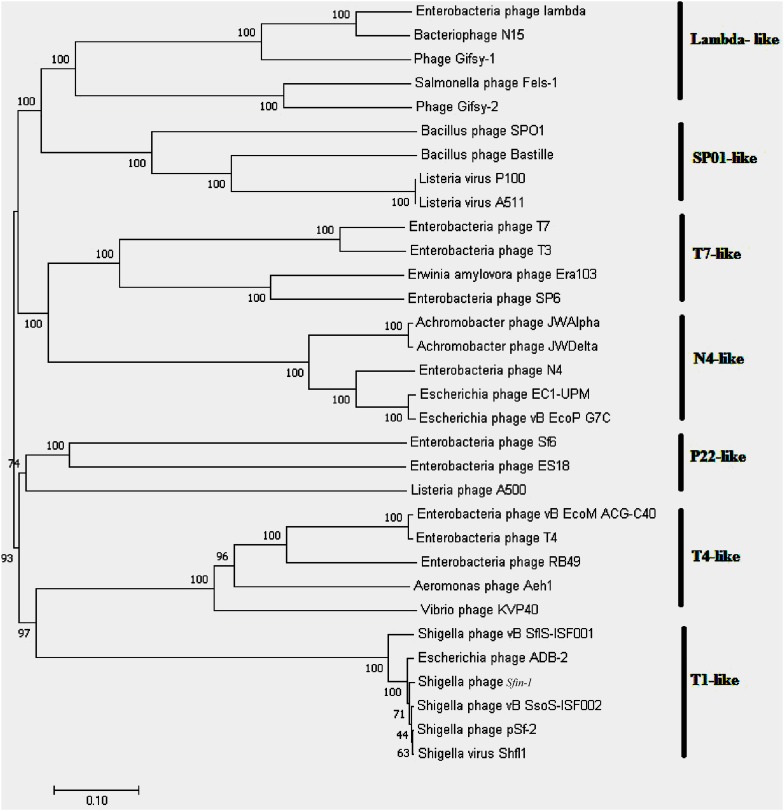
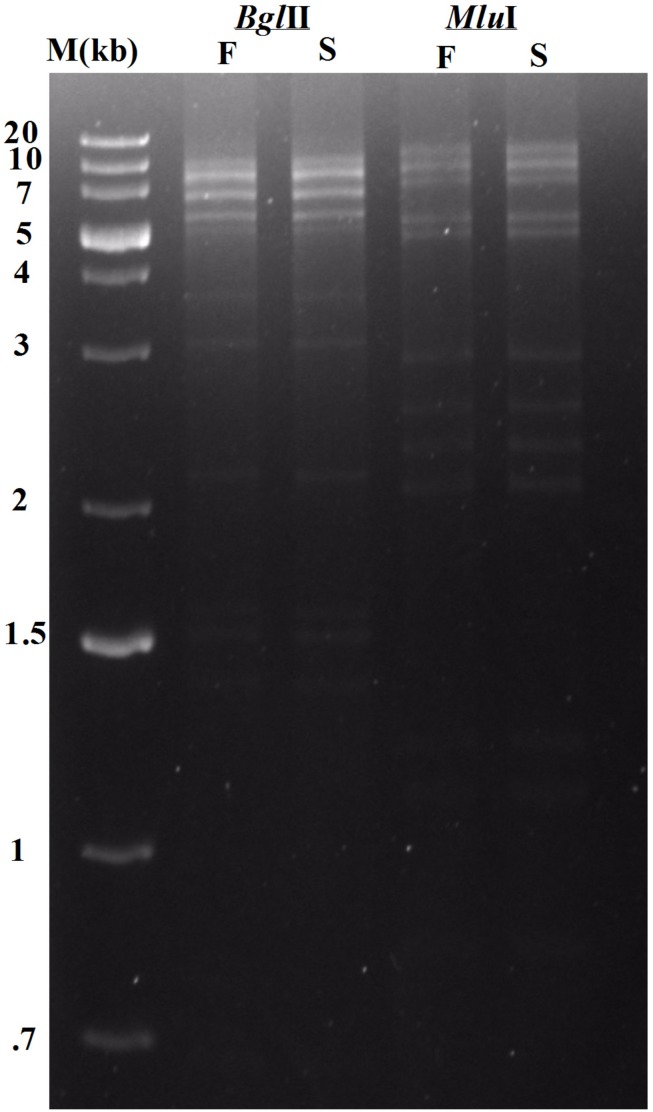
Whole genome sequencing followed by assembly of Sfin-1 genome initially generated a linear 50,530 bp fragment with a 127 bp terminal repeat at both of its ends. The linear genome is expected as in tailed bacteriophages within the channel of portal protein only one dsDNA can pass and therefore, the head contains linear genome. However, linear phage genome may have different types of ends. It is known that phage terminase enzyme creates the virion DNA ends and this enzyme is one of the most conserved phage proteins within the group. Therefore, comparative analysis of terminase amino acid sequence of a phage clusters it with others that generate similar ends. According to the phylogenetic analysis of the large terminase subunit, Sfin-1 was clustered with the terminase of E. coli phage ADB-2, Shigella phage Shfl1 and psf-2 which belong to T1 family of phage (Figure 7). This family has double stranded terminal repeats in their chromosome ends. Based on its close relationship with the T1 like phages, it is predicted that Sfin-1 genome is possibly circularly permutated with direct terminal repeats. For a circularly permuted headful packaging phage chromosome, the site of initiation cleavage is not precise. So, alternative initiation cuts are spread over regions on the concatemers. As a result, chromosome lengths of individual virions are imprecise. The undigested phage DNA as well as restriction pattern of these type of phages are expected to contain all the fragments from a circular genome along with a submolar pac fragment as happened with P22 genome (Casjens and Gilcrease, 2009) and in case of imprecise series initiation cleavage like phage sf6 and ES18, the pac fragment may not be detected. Instead a blur background will be observed because of the variable lengths of terminal fragments (Casjens and Gilcrease, 2009). When Sfin-1 genomic DNA was digested with restriction endonucleases BglII and MluI, the result was in agreement with the predicted result based on a circular Sfin-1 genome (Figure 8). Restriction digests were warmed at 80°C and then cooled down slowly or rapidly. After slow cooling single stranded cohesive ends of the phage genome are expected to anneal and appear as a longer fragment in gel electrophoresis. But no difference was noticed between slow and fast cooled sets for both the enzymes indicating absence of cohesive ends in the Sfin-1 genome. Additionally blur background was observed in electrophoresis gel. This result indicates that Sfin-1 is a T1 like headful packaging phage.
FIGURE 7.
Phylogenetic tree of terminase large subunit. Phages with known packaging mechanisms were only included. Bootstrap analysis was performed with 1,000 repetitions. The terminase large subunits were compared in the MEGA 7.0 version using neighbor-joining method.
FIGURE 8.
Enzymatic analysis of Sfin-1 genomic DNA. Phage DNA was completely digested with BglII and MluI and the products were analyzed by 0.8% agarose gel electrophoresis, Lane M indicates the 1 kb Plus DNA Ladder. F and S indicate that the digests were heated to 80°C for 15 min and then cooled fast on ice or slow at room temperature, respectively.
Identification of Host Receptor
The crucial step in the phage infection is its adsorption to the surface of the host through receptor. As the Shigella spp. belongs to the class of gram negative bacteria with complex lipopolysaccharide (LPS) and protein at its cell surface, the outer membrane carbohydrate (LPS) or protein may act as the specific receptor for phage infection. Therefore, it is very important to find out whether LPS or protein is the recognition site of phage Sfin-1 during infection. To determine the actual receptor of the phage Sfin-1, outer membrane LPS and proteins of S. flexneri 2a, S. dysenteriae 1, and S. sonnei were degraded by periodate and proteinase K respectively prior to the infection. Sfin-1 showed no change in infection efficiency with or without proteinase K treatment to the above mentioned hosts. In contrast, high number of phage particles remained unabsorbed when hosts were pre-treated with periodates (Figure 9). So, this experiment suggests that the adsorption of phage Sfin-1 to the host is mediated by the outer membrane complex LPS structure. When LPS is used as phage receptor, phage shows strain specificity. Here, we see Sfin-1, which uses LPS as receptor to infect three Shigella spp. This apparent anomaly may be attributed to the close relationship between Shigella spp. Although there is heterogeneity among LPS’s O-antigens of Shigella spp. they share extensive similarity (Liu et al., 2008).
FIGURE 9.
Sfin-1 infections on proteinase K and periodate treated host. The effect of proteinase K and sodium periodate on adsorption of phage Sfin-1. Shigella flexneri 2a, Shigella dysenteriae 1, and Shigella sonnei cultures (OD600 = 0.3) were treated with proteinase K (250 mg/mL) or sodium periodate (200 mM NaIO4) followed by Sfin-1 (MOI 0.0001) infection. Upon centrifugation, the phage titer in supernatant was determined as described in “Materials and Methods” section. Cells suspended in LB, cells incubated at 55°C in LB and cells in acetate buffer were used as control. The results are shown as residual PFU percentages. The phage titer in the control supernatant was set to 100%. Mean ± SD of three independent experiments are indicated. To determine the significance of the differences between group means, unpaired t-tests were performed between the controls and the tests. ∗Significance level, i.e., P < 0.05, “ns” indicates non-significant. (A–C) Results of S. flexneri 2a, S. dysentariae1, and S. sonnei, respectively.
Concluding Remarks
Multidrug-resistant Shigella infection has already become a critical problem in many countries. Against the backdrop of widespread antibiotic resistance, phages are re-emerging as promising therapeutic agent to control bacterial infections. In the current study we have characterized a novel thermostable and wide range pH tolerant Siphoviridae phage Sfin-1 that infects and lyses the important antibiotic resistant enteropathogens Shigella spp. This is the first reported phage which infects both S. flexneri and S. dysenteriae. The article presents complete physical characterizations, sequence analysis and detailed genome annotation of phage Sfin-1. Phage structural proteins have also been identified through LC-MS/MS study. The Genomic data are important resources to study and use phages to control specific bacteria species. Phylogenetic analysis concludes that Sfin-1 belongs to the T1-like bacteriophage and thus it may be packaged by headful packaging method. Further analysis of phage Sfin-1 cell wall receptor revealed that, bacteriophage Sfin-1 recognizes LPS O-antigen as its primary receptor for adsorption. Further studies on Sfin-1 phage will be useful to apply it for therapeutic purpose against multidrug-resistant shigellosis.
Data Availability
The datasets generated for this study can be found in the GenBank under accession number MF468274.
Author Contributions
SA and NG conceived and designed the whole study. SD supplied the clinical samples. SA and BR performed the experiments. AG analyzed the phage structure. SA, UB, and NG analyzed the results. BB performed the statistical analysis. SA, UB, SD, BB, and NG prepared the manuscript. All authors wrote, read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Sujoy K. Dasgupta and Dr. W. Ghosh (Department of Microbiology, Bose Institute) for infrastructural support and important suggestions on the work. Laboratory work of Mr. Arindam Ganai, Technical Officer of the NICED is gratefully acknowledged. We also thank Drs. Santanu Roy, Prajna Mandal, and Saktibrata Bhowmik (Acharya Prafulla Chandra College) for their support to this work.
Funding. This work was supported by the Department of Science and Technology (SERB), India.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01876/full#supplementary-material
Comparative genomic analysis of sfin-1 with Shfl1 and pSf-2. Comparison of Sfin-1 genome with that of (A) Shfl1 and (B) pSf-2 are schematically presented. Green (virion morphogenesis), yellow (DNA packaging), red (DNA metabolism and replication), and gray (cell lysis) colors represent different groups of homologous proteins of known functions. Numbers within parentheses indicate degree of homology as calculated via tblastx. The violet arrow heads indicate Sfin-1-specific genes which are absent in shfl1 and pSf-2. Black arrowheads denote genes of Shfl1 and pSf-2 absent in Sfin-1. Hypothetical proteins having more than 90% homology are indicated by saffron color and less than 90% by sky blue color.
The comparison of tail fiber protein sequence A (CDS61) of Sfin-1, CDS53 of Shfl1 and CDS16 of pSf-2. (A) The predicted features comparison of three proteins. The red diamond represents protein binding region, the red and blue rectangles represent the helix and strand of proteins, respectively. (B) The comparison of amino acid sequences of three proteins by ClustalW. ∗Represents same amino acid and blank represents different amino acids.
The comparison of tail fiber protein sequence B (CDS67) of Sfin-1, CDS47 of Shfl1 and CDS22 of pSf-2. (A) The predicted features comparison of three proteins. The red diamond represents protein binding region, the red and blue rectangles represent the helix and strand of proteins, respectively. (B) The comparison of amino acid sequences of three proteins by ClustalW. ∗Represents same amino acid and blank represents different amino acids.
The comparison of tail fiber protein sequence C (CDS71) of Sfin-1, CDS43 of Shfl1 and CDS26 of pSf-2. (A) The predicted features comparison of three proteins. The red diamond represents protein binding region, the red and blue rectangles represent the helix and strand of proteins, respectively. (B) The comparison of amino acid sequences of three proteins by ClustalW. ∗Represents same amino acid and blank represents different amino acids.
The comparison of tail fiber protein sequence D (CDS76) of Sfin-1, CDS39 of Shfl1 and CDS30 of pSf-2. (A) The predicted features comparison of three proteins. The red diamond represents protein binding region, the red and blue rectangles represent the helix and strand of protein, respectively. (B) The comparison of amino acid sequences of three proteins by ClustalW. ∗Represents same amino acid and blank represents different amino acids.
Comparative analysis of Sfin-1 with different other Shigella phages available in NCBI database. NA∗ = no data available.
Comparative analysis of Sfin-1 with pSf-2 and Shfl1 genomes.
References
- Ackermann H. W. (1998). Tailed bacteriophages: the order caudovirales. Adv. Virus Res. 51 135–201. 10.1016/s0065-3527(08)60785-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison G. E., Angeles D., Tran-Dinh N., Verma N. K. (2002). Complete genomic sequence of SfV, a serotype-converting temperate bacteriophage of Shigella flexneri. J. Bacteriol. 184 1974–1987. 10.1128/jb.184.7.1974-1987.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison G. E., Verma N. K. (2000). Serotype-converting bacteriophages and O-antigen modification in Shigella flexneri. Trends Microbiol. 8 17–23. 10.1016/s0966-842x(99)01646-7 [DOI] [PubMed] [Google Scholar]
- Amarillas L., Rubi-Rangel L., Chaidez C., Gonzalez-Robles A., Lightbourn-Rojas L., Leon-Felix J. (2017). Isolation and characterization of phiLLS, a novel phage with potential biocontrol agent against multidrug-resistant Escherichia coli. Front. Microbiol. 8:1355. 10.3389/fmicb.2017.01355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amezquita-Lopez B. A., Quinones B., Lee B. G., Chaidez C. (2014). Virulence profiling of shiga toxin-producing Escherichia coli recovered from domestic farm animals in Northwestern Mexico. Front. Cell Infect. Microbiol. 4:7. 10.3389/fcimb.2014.00007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anany H., Lingohr E. J., Villegas A., Ackermann H. W., She Y. M., Griffiths M. W., et al. (2011). A Shigella boydii bacteriophage which resembles Salmonella phage ViI. Virol. J. 8:242. 10.1186/1743-422X-8-242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird-Parker A. C. (1994). 1993 fred griffith review lecture. Foods and microbiological risks. Microbiology 140(Pt 4), 687–695. 10.1099/00221287-140-4-687 [DOI] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennish M. L., Salam M. A., Haider R., Barza M. (1990). Therapy for shigellosis. II. Randomized, double-blind comparison of ciprofloxacin and ampicillin. J. Infect. Dis. 162 711–716. 10.1093/infdis/162.3.711 [DOI] [PubMed] [Google Scholar]
- Berg J. A., Merrill B. D., Crockett J. T., Esplin K. P., Evans M. R., Heaton K. E., et al. (2016). Characterization of five novel brevibacillus bacteriophages and genomic comparison of brevibacillus phages. PLoS One 11:e0156838. 10.1371/journal.pone.0156838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besemer J., Lomsadze A., Borodovsky M. (2001). GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 29 2607–2618. 10.1093/nar/29.12.2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C. M., Bidle K. D. (2014). Attenuation of virus production at high multiplicities of infection in Aureococcus anophagefferens. Virology 466-467 71–81. 10.1016/j.virol.2014.07.023 [DOI] [PubMed] [Google Scholar]
- Casjens S., Winn-Stapley D. A., Gilcrease E. B., Morona R., Kuhlewein C., Chua J. E., et al. (2004). The chromosome of Shigella flexneri bacteriophage Sf6: complete nucleotide sequence, genetic mosaicism, and DNA packaging. J. Mol. Biol. 339 379–394. 10.1016/s0022-2836(04)00377-8 [DOI] [PubMed] [Google Scholar]
- Casjens S. R., Gilcrease E. B. (2009). Determining DNA packaging strategy by analysis of the termini of the chromosomes in tailed-bacteriophage virions. Methods Mol. Biol. 502 91–111. 10.1007/978-1-60327-565-1_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H. C., Chen C. R., Lin J. W., Shen G. H., Chang K. M., Tseng Y. H., et al. (2005). Isolation and characterization of novel giant Stenotrophomonas maltophilia phage phiSMA5. Appl. Environ. Microbiol. 71 1387–1393. 10.1128/aem.71.3.1387-1393.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H. W., Kim K. H. (2011). Comparative genomic analysis of bacteriophage EP23 infecting Shigella sonnei and Escherichia coli. J. Microbiol. 49 927–934. 10.1007/s12275-011-1577-0 [DOI] [PubMed] [Google Scholar]
- CLSI (2016). Performance Standards for Antimicrobial Susceptibility Testing. CLSI document M100. Wayne, PA: CLSI. [Google Scholar]
- Dutta S., Das S., Mitra U., Jain P., Roy I., Ganguly S. S., et al. (2014). Antimicrobial resistance, virulence profiles and molecular subtypes of Salmonella enterica serovars typhi and paratyphi a blood isolates from Kolkata, India during 2009-2013. PLoS One 9:e101347. 10.1371/journal.pone.0101347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faruque S. M., Chowdhury N., Khan R., Hasan M. R., Nahar J., Islam M. J., et al. (2003). Shigella dysenteriae type 1-specific bacteriophage from environmental waters in Bangladesh. Appl. Environ. Microbiol. 69 7028–7031. 10.1128/aem.69.12.7028-7031.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauquet C. M., Fargette D. (2005). International committee on taxonomy of viruses and the 3,142 unassigned species. Virol. J. 2:64. 10.1186/1743-422X-2-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipski A., Murillo O., Freydenzon A., Tamura K., Kumar S. (2014). Prospects for building large timetrees using molecular data with incomplete gene coverage among species. Mol. Biol. Evol. 31 2542–2550. 10.1093/molbev/msu200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallet R., Kannoly S., Wang I. N. (2011). Effects of bacteriophage traits on plaque formation. BMC Microbiol. 11:181. 10.1186/1471-2180-11-181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George D. T., Stephenson D. P., Tran E., Morona R., Verma N. K. (2013). Complete genome sequence of SfII, a serotype-converting bacteriophage of the highly prevalent Shigella flexneri serotype 2a. Genome Announc. 1:e626-13. 10.1128/genomeA.00626-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M. D., Lampel K. A., Strockbine N. A., Fernandez R. E., Melton-Celsa A. R., Maurelli A. T. (2014). Clinical isolates of Shiga toxin 1a-producing Shigella flexneri with an epidemiological link to recent travel to Hispaniola. Emerg. Infect. Dis. 20 1669–1677. 10.3201/eid2010.140292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan S., Bastin D. A., Verma N. K. (1999). Functional analysis of the O antigen glucosylation gene cluster of Shigella flexneri bacteriophage SfX. Microbiology 145(Pt 5), 1263–1273. 10.1099/13500872-145-5-1263 [DOI] [PubMed] [Google Scholar]
- Gupta R., Prasad Y. (2011). Efficacy of polyvalent bacteriophage P-27/HP to control multidrug resistant Staphylococcus aureus associated with human infections. Curr. Microbiol. 62 255–260. 10.1007/s00284-010-9699-x [DOI] [PubMed] [Google Scholar]
- Hamdi S., Rousseau G. M., Labrie S. J., Tremblay D. M., Kourda R. S., Ben Slama K., et al. (2017). Characterization of two polyvalent phages infecting Enterobacteriaceae. Sci. Rep. 7:40349. 10.1038/srep40349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Housby J. N., Mann N. H. (2009). Phage therapy. Drug Discov. Today 14 536–540. 10.1016/j.drudis.2009.03.006 [DOI] [PubMed] [Google Scholar]
- Jakhetia R., Talukder K. A., Verma N. K. (2013). Isolation, characterization and comparative genomics of bacteriophage SfIV: a novel serotype converting phage from Shigella flexneri. BMC Genomics 14:677. 10.1186/1471-2164-14-677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun J. W., Giri S. S., Kim H. J., Yun S. K., Chi C., Chai J. Y., et al. (2016a). Bacteriophage application to control the contaminated water with Shigella. Sci. Rep. 6:22636. 10.1038/srep22636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun J. W., Kim H. J., Yun S. K., Chai J. Y., Lee B. C., Park S. C. (2016b). Isolation and comparative genomic analysis of T1-Like Shigella bacteriophage pSf-2. Curr. Microbiol. 72 235–241. 10.1007/s00284-015-0935-2 [DOI] [PubMed] [Google Scholar]
- Jun J. W., Kim J. H., Shin S. P., Han J. E., Chai J. Y., Park S. C. (2013). Characterization and complete genome sequence of the Shigella bacteriophage pSf-1. Res. Microbiol. 164 979–986. 10.1016/j.resmic.2013.08.007 [DOI] [PubMed] [Google Scholar]
- Jun J. W., Yun S. K., Kim H. J., Chai J. Y., Park S. C. (2014). Characterization and complete genome sequence of a novel N4-like bacteriophage, pSb-1 infecting Shigella boydii. Res. Microbiol. 165 671–678. 10.1016/j.resmic.2014.09.006 [DOI] [PubMed] [Google Scholar]
- Katsura I. (1990). Mechanism of length determination in bacteriophage lambda tails. Adv. Biophys. 26 1–18. 10.1016/0065-227x(90)90004-d [DOI] [PubMed] [Google Scholar]
- Kiljunen S., Datta N., Dentovskaya S. V., Anisimov A. P., Knirel Y. A., Bengoechea J. A., et al. (2011). Identification of the lipopolysaccharide core of Yersinia pestis and Yersinia pseudotuberculosis as the receptor for bacteriophage phiA1122. J. Bacteriol. 193 4963–4972. 10.1128/JB.00339-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. H., Chang H. W., Nam Y. D., Roh S. W., Bae J. W. (2010). Phenotypic characterization and genomic analysis of the Shigella sonnei bacteriophage SP18. J. Microbiol. 48 213–222. 10.1007/s12275-010-0055-4 [DOI] [PubMed] [Google Scholar]
- Lesnik E. A., Sampath R., Levene H. B., Henderson T. J., Mcneil J. A., Ecker D. J. (2001). Prediction of rho-independent transcriptional terminators in Escherichia coli. Nucleic Acids Res. 29 3583–3594. 10.1093/nar/29.17.3583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Knirel Y. A., Feng L., Perepelov A. V., Senchenkova S. N., Wang Q., et al. (2008). Structure and genetics of Shigella O antigens. FEMS Microbiol. Rev. 32 627–653. 10.1111/j.1574-6976.2008.00114.x [DOI] [PubMed] [Google Scholar]
- Lokareddy R. K., Sankhala R. S., Roy A., Afonine P. V., Motwani T., Teschke C. M., et al. (2017). Portal protein functions akin to a DNA-sensor that couples genome-packaging to icosahedral capsid maturation. Nat. Commun. 8:14310. 10.1038/ncomms14310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T. M., Chan P. P. (2016). tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44 W54–W57. 10.1093/nar/gkw413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y. L., Lu C. P. (2008). Isolation and identification of a bacteriophage capable of infecting Streptococcus suis type 2 strains. Vet. Microbiol. 132 340–347. 10.1016/j.vetmic.2008.05.013 [DOI] [PubMed] [Google Scholar]
- Malek W., Wdowiak-Wrobel S., Bartosik M., Konopa G., Narajczyk M. (2009). Characterization of phages virulent for Robinia pseudoacacia Rhizobia. Curr. Microbiol. 59 187–192. 10.1007/s00284-009-9421-z [DOI] [PubMed] [Google Scholar]
- Mavris M., Manning P. A., Morona R. (1997). Mechanism of bacteriophage SfII-mediated serotype conversion in Shigella flexneri. Mol. Microbiol. 26 939–950. 10.1046/j.1365-2958.1997.6301997.x [DOI] [PubMed] [Google Scholar]
- Mobberley J. M., Authement R. N., Segall A. M., Paul J. H. (2008). The temperate marine phage PhiHAP-1 of Halomonas aquamarina possesses a linear plasmid-like prophage genome. J. Virol. 82 6618–6630. 10.1128/JVI.00140-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthuirulandi Sethuvel D. P., Devanga Ragupathi N. K., Anandan S., Veeraraghavan B. (2017). Update on: Shigella new serogroups/serotypes and their antimicrobial resistance. Lett. Appl. Microbiol. 64 8–18. 10.1111/lam.12690 [DOI] [PubMed] [Google Scholar]
- Nandy S., Mitra U., Rajendran K., Dutta P., Dutta S. (2010). Subtype prevalence, plasmid profiles and growing fluoroquinolone resistance in Shigella from Kolkata, India (2001-2007): a hospital-based study. Trop. Med. Int. Health 15 1499–1507. 10.1111/j.1365-3156.2010.02656.x [DOI] [PubMed] [Google Scholar]
- O’flaherty S., Ross R. P., Flynn J., Meaney W. J., Fitzgerald G. F., Coffey A. (2005). Isolation and characterization of two anti-staphylococcal bacteriophages specific for pathogenic Staphylococcus aureus associated with bovine infections. Lett. Appl. Microbiol. 41 482–486. 10.1111/j.1472-765x.2005.01781.x [DOI] [PubMed] [Google Scholar]
- Puzari M., Sharma M., Chetia P. (2017). Emergence of antibiotic resistant Shigella species: a matter of concern. J. Infect. Public Health 11 451–454. 10.1016/j.jiph.2017.09.025 [DOI] [PubMed] [Google Scholar]
- Sengupta P. G., Mandal S., Sen D., Das P., Deb B. C., Pal S. C. (1990). Multidrug resistant epidemic shigellosis in a village in west Bengal, 1984. Indian J. Public Health 34 15–19. [PubMed] [Google Scholar]
- Shahin K., Bao H., Komijani M., Barazandeh M., Bouzari M., Hedayatkhah A., et al. (2019). Isolation, characterization, and PCR-based molecular identification of a siphoviridae phage infecting Shigella dysenteriae. Microb. Pathog. 131 175–180. 10.1016/j.micpath.2019.03.037 [DOI] [PubMed] [Google Scholar]
- Shahin K., Bouzari M. (2018). Bacteriophage application for biocontrolling Shigella flexneri in contaminated foods. J. Food Sci. Technol. 55 550–559. 10.1007/s13197-017-2964-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahin K., Bouzari M., Wang R. (2018). Isolation, characterization and genomic analysis of a novel lytic bacteriophage vB_SsoS-ISF002 infecting Shigella sonnei and Shigella flexneri. J. Med. Microbiol. 67 376–386. 10.1099/jmm.0.000683 [DOI] [PubMed] [Google Scholar]
- Shen M., Le S., Jin X., Li G., Tan Y., Li M., et al. (2016). Characterization and comparative genomic analyses of Pseudomonas aeruginosa Phage PaoP5: new members assigned to PAK_P1-like viruses. Sci. Rep. 6:34067. 10.1038/srep34067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivapalasingam S., Nelson J. M., Joyce K., Hoekstra M., Angulo F. J., Mintz E. D. (2006). High prevalence of antimicrobial resistance among Shigella isolates in the United States tested by the national antimicrobial resistance monitoring system from 1999 to 2002. Antimicrob. Agents Chemother. 50 49–54. 10.1128/aac.50.1.49-54.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Lan R., Wang Y., Wang J., Wang Y., Li P., et al. (2013). Isolation and genomic characterization of SfI, a serotype-converting bacteriophage of Shigella flexneri. BMC Microbiol. 13:39. 10.1186/1471-2180-13-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sur D., Ramamurthy T., Deen J., Bhattacharya S. K. (2004). Shigellosis: challenges & management issues. Indian J. Med. Res. 120 454–462. [PubMed] [Google Scholar]
- Tariq A., Haque A., Ali A., Bashir S., Habeeb M. A., Salman M., et al. (2012). Molecular profiling of antimicrobial resistance and integron association of multidrug-resistant clinical isolates of Shigella species from Faisalabad, Pakistan. Can. J. Microbiol. 58 1047–1054. 10.1139/w2012-085 [DOI] [PubMed] [Google Scholar]
- Uchiyama J., Rashel M., Maeda Y., Takemura I., Sugihara S., Akechi K., et al. (2008). Isolation and characterization of a novel Enterococcus faecalis bacteriophage phiEF24C as a therapeutic candidate. FEMS Microbiol. Lett. 278 200–206. [DOI] [PubMed] [Google Scholar]
- Vinner G. K., Vladisavljevic G. T., Clokie M. R. J., Malik D. J. (2017). Microencapsulation of Clostridium difficile specific bacteriophages using microfluidic glass capillary devices for colon delivery using pH triggered release. PLoS One 12:e0186239. 10.1371/journal.pone.0186239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Seidlein L., Kim D. R., Ali M., Lee H., Wang X., Thiem V. D., et al. (2006). A multicentre study of Shigella diarrhoea in six Asian countries: disease burden, clinical manifestations, and microbiology. PLoS Med. 3:e353. 10.1371/journal.pmed.0030353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Zheng P., Ji W., Fu Q., Wang H., Yan Y., et al. (2016). SLPW: a virulent bacteriophage targeting methicillin-resistant Staphylococcus aureus In vitro and In vivo. Front. Microbiol. 7:934. 10.3389/fmicb.2016.00934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J., Goldberg M. B., Burland V., Venkatesan M. M., Deng W., Fournier G., et al. (2003). Complete genome sequence and comparative genomics of Shigella flexneri serotype 2a strain 2457T. Infect. Immun. 71 2775–2786. 10.1128/iai.71.5.2775-2786.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann J., Dreiseikelmann B., Rohde M., Meier-Kolthoff J. P., Bunk B., Rohde C. (2014). First genome sequences of Achromobacter phages reveal new members of the N4 family. Virol. J. 11:14. 10.1186/1743-422X-11-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yachdav G., Kloppmann E., Kajan L., Hecht M., Goldberg T., Hamp T., et al. (2014). PredictProtein – an open resource for online prediction of protein structural and functional features. Nucleic Acids Res. 42 W337–W343. 10.1093/nar/gku366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaki S., Omachi T., Kawai Y., Yamazaki K. (2014). Characterization of a novel Morganella morganii bacteriophage FSP1 isolated from river water. FEMS Microbiol. Lett. 359 166–172. 10.1111/1574-6968.12560 [DOI] [PubMed] [Google Scholar]
- Yang C., Wang H., Ma H., Bao R., Liu H., Yang L., et al. (2018). Characterization and genomic analysis of SFPH2, a novel T7virus infecting Shigella. Front. Microbiol. 9:3027. 10.3389/fmicb.2018.03027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yismaw G., Negeri C., Kassu A. (2008). A five-year antimicrobial resistance pattern of Shigella isolated from stools in the Gondar university hospital, northwest Ethiopia. Trop. Doct. 38 43–45. 10.1258/td.2007.060215 [DOI] [PubMed] [Google Scholar]
- Yoon S. S., Barrangou-Poueys R., Breidt F., Jr., Fleming H. P. (2007). Detection and characterization of a lytic Pediococcus bacteriophage from the fermenting cucumber brine. J. Microbiol. Biotechnol. 17 262–270. [PubMed] [Google Scholar]
- Yordpratum U., Tattawasart U., Wongratanacheewin S., Sermswan R. W. (2011). Novel lytic bacteriophages from soil that lyse Burkholderia pseudomallei. FEMS Microbiol. Lett. 314 81–88. 10.1111/j.1574-6968.2010.02150.x [DOI] [PubMed] [Google Scholar]
- Ziedaite G., Daugelavicius R., Bamford J. K., Bamford D. H. (2005). The Holin protein of bacteriophage PRD1 forms a pore for small-molecule and endolysin translocation. J. Bacteriol. 187 5397–5405. 10.1128/jb.187.15.5397-5405.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparative genomic analysis of sfin-1 with Shfl1 and pSf-2. Comparison of Sfin-1 genome with that of (A) Shfl1 and (B) pSf-2 are schematically presented. Green (virion morphogenesis), yellow (DNA packaging), red (DNA metabolism and replication), and gray (cell lysis) colors represent different groups of homologous proteins of known functions. Numbers within parentheses indicate degree of homology as calculated via tblastx. The violet arrow heads indicate Sfin-1-specific genes which are absent in shfl1 and pSf-2. Black arrowheads denote genes of Shfl1 and pSf-2 absent in Sfin-1. Hypothetical proteins having more than 90% homology are indicated by saffron color and less than 90% by sky blue color.
The comparison of tail fiber protein sequence A (CDS61) of Sfin-1, CDS53 of Shfl1 and CDS16 of pSf-2. (A) The predicted features comparison of three proteins. The red diamond represents protein binding region, the red and blue rectangles represent the helix and strand of proteins, respectively. (B) The comparison of amino acid sequences of three proteins by ClustalW. ∗Represents same amino acid and blank represents different amino acids.
The comparison of tail fiber protein sequence B (CDS67) of Sfin-1, CDS47 of Shfl1 and CDS22 of pSf-2. (A) The predicted features comparison of three proteins. The red diamond represents protein binding region, the red and blue rectangles represent the helix and strand of proteins, respectively. (B) The comparison of amino acid sequences of three proteins by ClustalW. ∗Represents same amino acid and blank represents different amino acids.
The comparison of tail fiber protein sequence C (CDS71) of Sfin-1, CDS43 of Shfl1 and CDS26 of pSf-2. (A) The predicted features comparison of three proteins. The red diamond represents protein binding region, the red and blue rectangles represent the helix and strand of proteins, respectively. (B) The comparison of amino acid sequences of three proteins by ClustalW. ∗Represents same amino acid and blank represents different amino acids.
The comparison of tail fiber protein sequence D (CDS76) of Sfin-1, CDS39 of Shfl1 and CDS30 of pSf-2. (A) The predicted features comparison of three proteins. The red diamond represents protein binding region, the red and blue rectangles represent the helix and strand of protein, respectively. (B) The comparison of amino acid sequences of three proteins by ClustalW. ∗Represents same amino acid and blank represents different amino acids.
Comparative analysis of Sfin-1 with different other Shigella phages available in NCBI database. NA∗ = no data available.
Comparative analysis of Sfin-1 with pSf-2 and Shfl1 genomes.
Data Availability Statement
The datasets generated for this study can be found in the GenBank under accession number MF468274.