Abstract
The neuropathogenesis of HIV associated neurocognitive disorders (HAND) involves disruption of in mitochondrial homeostasis and increased neuroinflammation. However, it is unknown if alterations in mitochondrial biogenesis in the brain underlie the neuropathogenesis of HAND. In this study, neuropathological and molecular analyses of mitochondrial biogenesis and inflammatory pathways were performed in brain specimens from a well-characterized cohort of HIV+ cases that were on antiretroviral regimens. In vitro investigations using primary human astroglia and neurons were used to probe the underlying mechanisms of mitochondrial alterations. In frontal cortices from HAND brains compared to cognitive normal brains, total levels of transcription factors that regulate mitochondrial biogenesis, peroxisome proliferator-activated receptor γ coactivator 1-α (PGC-1α) and transcription factor A, mitochondrial (TFAM) were decreased. Immunohistochemical analyses revealed that TFAM was decreased in neurons and increased in astroglia. These changes were accompanied by decreased total mitochondrial DNA per cell and increased levels of messenger RNA for the proinflammatory cytokine interleukin (IL)-1β. To determine how IL-1β affects astroglial bioenergetic processes and mitochondrial activity, human astroglial cultures were exposed to recombinant IL-1β IL-1β induced mitochondrial activity within 30 minutes of treatment, altered mitochondrial related gene expression, altered mitochondrial morphology, enhanced ATP utilization and increased the expression of inflammatory cytokines. WIN55,212–2, an aminoalkylindole derivative and cannabinoid receptor agonist, blocked IL-1β-induced bioenergetic fluctuations and inflammatory gene expression in astroglia independent of CB1 receptors and PPARγ. A PPARα antagonist reversed the anti-inflammatory effects of WIN in human astroglia. These results show that mitochondrial biogenesis is differentially regulated in neurons and astroglia in HAND brains and that targeting astroglial bioenergetic processes may be a strategy to modulate neuroinflammation.
Introduction
HIV infection continues to be a major health concern in the Unites States and worldwide, with over 1.4 and 34 million HIV-infected individuals, respectively (Ledergerber and Battegay, 2014). HIV-associated neurocognitive disorders (HAND) persist in approximately 50% of these individuals despite effective antiretroviral therapy (ART) regimens (Heaton et al., 2011). Confounding this problem, HIV+ patients are living longer and are therefore facing age-related comorbidities along with the challenges of living with HIV infection and chronic exposure to ART (Boulias et al., 2016). Effective therapeutic strategies to prevent or reduce HAND in these patients could help millions of people, reduce dependence on healthcare services, and have a tremendous economic impact by extending lives and productivity.
The molecular mechanisms underlying the neuropathogenesis of HAND are likely numerous and varied depending upon genetic and environmental factors. Some possible mechanisms underlying HAND include premature activation of apoptosis (Kruman et al., 1998), dysregulated autophagy (Fields et al., 2015a), increased oxidative stress (Kruman et al., 1998), and altered calcium homeostasis (Bonavia et al., 2001). Importantly, mitochondrial function is related to all of these and many other key processes central to brain function (Nair et al., 2018). Indeed, disruptions in mitochondrial fission and fusion, mitophagy, mitochondrial transport, Ca2+ signaling and apoptosis have all been implicated as etiologies of HAND (Avdoshina et al., 2016; Fields et al., 2015b; Nooka and Ghorpade, 2017; Teodorof-Diedrich and Spector, 2018; Thangaraj et al., 2018). However, no studies have investigated the process of generating new mitochondria as an etiology underlying HAND.
To maintain healthy mitochondria, under a tightly regulated process, cells generate new mitochondria through a coordinated process of nuclear and mitochondrial gene expression, mtDNA replication, protein synthesis and lipid transfer (Ventura-Clapier et al., 2008). Mitochondrial biogenesis requires activation of transcription factors peroxisome proliferator-activated receptor γ coactivator 1-α (PGC-1α) and transcription factor A, mitochondrial (TFAM), which are activated when cells need energy (adenosine triphosphate [ATP]). Alterations in levels of PGC-1α and TFAM are implicated in several neurodegenerative diseases including Alzheimer’s Disease (AD), Huntington’s disease (HD), and Parkinson’s disease (PD) (Repunte-Canonigo et al., 2014; van der Walt et al., 2003; Wang et al., 2013; Ye et al., 2012). HIV infection (Casula et al., 2005), ART(Dalakas et al., 2001), and AD are also associated with reduced levels mitochondrial DNA (mtDNA) (Sheng et al., 2012). Enhancing mitochondrial biogenesis (Chaturvedi and Flint Beal, 2013; Repunte-Canonigo et al., 2014) is neuroprotective in rodent models of AD, HD, and PD. However, no studies have investigated PGC-1α in brains of HIV-infected individuals on cART.
While most studies have focused on how neuronal mitochondria are affected by HIV infection and ART, few studies have examined the role of mitochondrial activity in glia during HIV infection. Astroglia are immune-activated in HAND brains and in vitro stimulation of astroglia with HIV or relevant stimuli elicits inflammatory cytokine expression and astrogliosis (Avraham et al., 2014; Fields and Ghorpade, 2012; Kaul et al., 2001; Levine et al., 2016; Mamik et al., 2011; Tavazzi et al., 2014). Astroglia are intricately involved in providing neurons with energy substrates (Alberini et al., 2018). Hence, chronically stimulated astroglia may play a role in bioenergetic deficiencies and mitochondrial dysfunction in neurons during HAND. Recent studies show that increased mitochondrial activity in astroglia may negatively affect neuronal function in AD (Jiang and Cadenas, 2014; Yin et al., 2016) and HAND (Natarajaseenivasan et al., 2018). Shifting astroglial phenotype from inflammatory to anti-inflammatory may be a promising therapeutic strategy (Clarke et al., 2018; Liddelow and Barres, 2017; Liddelow et al., 2017) in neurodegenerative diseases. Therefore, our interest in astroglia is predicated upon inflammatory and metabolic roles in HAND and other neurodegenerative diseases.
Strategies to modulate astroglial activation may be protective against neurodegeneration. For example, agonists for cannabinoid receptors (CB) 1 and 2, peroxisome proliferator-activated receptor (PPAR) γ and PPARα have been shown to block inflammatory responses in astroglia (Drew et al., 2006; Janabi, 2002; Kozela et al., 2017; Pautz et al., 2010; Xu et al., 2006). The aminoalkylindole derivative, WIN 55,212–2 (WIN), which is in an agonist of CB1/2, PPARγ and PPARα (Payandemehr et al., 2015), blocks inflammatory responses in the brain of mice (Marchalant et al., 2007; Ramirez et al., 2005). WIN has been shown to be neuroprotective and improve memory in rodent models for AD and aging (Marchalant et al., 2008; Martin-Moreno et al., 2012; Ramirez et al., 2005). WIN also blocks inflammatory gene expression in in vitro models for astroglia (Aguirre-Rueda et al., 2015; Sheng et al., 2005). Anti-inflammatory molecules such as WIN provide tools to probe the role of metabolic processes in immune-stimulated astroglia in disease and develop therapeutic strategies.
In the present study, we investigated alterations in mitochondrial biogenesis in neurons and astroglia in postmortem brain specimens from a well-characterized cohort of HAND decedents that were on ART. Inflammatory stimuli were used to induce mitochondrial activity and inflammatory gene expression in human astroglia in vitro and WIN was used to test the effects of an anti-inflammatory agent on these processes. Selective antagonists for CB1, PPARγ and PPARα and siRNA targeting CB1 were used to identify the mechanisms by which WIN affects astroglia in vitro. Lastly, we tested the effect of immune activated astroglia on neuronal TFAM expression in vitro. Findings from these studies provide information on the status of mitochondrial biogenesis in HAND brains and explore the therapeutic potential of reducing immune activation of astroglia in neurodegenerative disease.
Methods
Study population
For the present study, we evaluated brain tissues from a total of 47 HIV+ donors (Table 1) from the National NeuroAIDS Tissue Consortium (NNTC) (Institutional Review Board [IRB] #080323). All studies adhered to the ethical guidelines of the National Institutes of Health and the University of California, San Diego. These cases had neuromedical and neuropsychological examinations within a median of 12 months before death. Subjects were excluded if they had a history of CNS opportunistic infections or non-HIV-related developmental, neurologic, psychiatric, or metabolic conditions that might affect CNS functioning (e.g., loss of consciousness exceeding 30 minutes, psychosis, etc). HAND diagnoses were determined from a comprehensive neuropsychological test battery administered according to standardized protocols (Woods et al., 2004).
Table 1.
Demographics and clinical data.
| Variables | Cognitively normal (n = 8) | HAND (n = 39) |
Cog. Normal vs HAND |
Cog. normal vs ANI vs MND vs HAD |
||||
|---|---|---|---|---|---|---|---|---|
| ANI (n=12) | MND (n=14) | HAD (n=13) | p-value (Cog. normal vs HAND; t-test) | Effect Size - Cohen’s d (Cog. norm vs HAND) | p-value (Cog. norm vs ANI, Cog. norm vs MND, Cog. norm vs HAD, ANI vs MND, ANI vs HAD, MND vs HAD; ANOVA) | Effect Size: overall (Cog. Norm vs ANI, Cog. norm vs MND, Cog. Norm vs HAD, ANI vs MND, ANI vs HAD, MND vs HAD) | ||
| Demographics | ||||||||
| Hispanic ethnicity (%) | 57.1 | 56.3 | 66.7 | 77.1 | N/A | N/A | N/A | N/A |
| Sex (f/m) | 0/8 | 3/9 | 2/12 | 1/12 | N/A | N/A | N/A | N/A |
| Years of education | 12.0 ± 2.09 | 13.41 ± 2.67 | 12.0 ± 3.37 | 9.0 ± 4.37 | 0.39 | 0.07 | 0.09 (> 0.99, > 0.99, 0.25, > 0.99, 0.12, 0.4) | 0.51 (0.14, 0.00, 0.3, 0.16, 0.49. 0.35) |
| Years of age | 37.3 ± 5.7 | 47.1 ± 9.0 | 44.5 ± 8.3 | 50.2 ± 10.0 | 0.0007*** | 1.29 | 0.01* (0.08, 0.25, 0.009, 0.87, 0.8, 0.33) | 0.51 (0.37, 0.28, 0.5, 0.11, 0.14, 0.26) |
| HIV disease characteristics | ||||||||
| Duration on ART regimen (months) | 68.83 ± 48.05 | 75.74 ± 83.68 | 68.7 ± 57.93 | 31.2 ± 32.9 | 0.12 | 0.41 | 0.102 (0.51, 0.35, > 0.99, > 0.99, 0.66, 0.45) | 0.37 (0.24, 0.17, 0.04, 0.10, 0.33, 0.24) |
| Plasma VL (log) | 4.3 ± 1.61 | 3.8 ± 1.55 | 4.2 ± 1.61 | 3.4 ± 1.59 | 0.46 | 0.32 | 0.65 (0.91, 0.99, 0.66, 0.93, 0.94, 0.64) | 0.21 (0.1, 0.03, 0.17, 0.09, 0.08, 0.18) |
| CD4 count | 147.7 ± 239.01 | 142.8 ± 198.21 | 30.1 ± 35.81 | 122.1 ± 122.8 | 0.41 | 0.27 | 0.204 (0.99, 0.36, 0.98, 0.25, 0.99, 0.43) | 0.29 (0.009, 0.23, 0.04, 0.25, 0.05, 0.21) |
Neuromedical and neuropsychological evaluation
Participants underwent a comprehensive neuromedical evaluation that included assessment of medical history, structured medical and neurological examinations, and the collection of blood, cerebrospinal fluid (CSF), and urine samples, as previously described (Heaton et al., 2010; Woods et al., 2004). Clinical data (plasma viral load [VL], postmortem interval, CD4 count, global, learning and motor deficit scores [GDS, LDS, and MDS]) were collected for the HAND donor cohorts.
Neuropsychological evaluation was performed, and HAND diagnoses were determined via a comprehensive neuropsychological test battery, which was constructed to maximize sensitivity to neurocognitive deficits associated with HIV infection [see (Woods et al., 2004) for a list of tests]. Raw tests scores were transformed into demographically adjusted T-scores, including adjustments for age, education, gender and race. These demographically adjusted T-scores were converted to clinical ratings to determine presence and degree of neurocognitive impairment on seven neurocognitive domains, as previously described (Woods et al., 2004). As part of the neuropsychological battery, participants also completed self-report questionnaires of everyday functioning (i.e., Lawton and Brody Activities of Daily Living questionnaire; (Lawton and Brody, 1969), and/or Patient’s Assessment of Own Functioning; PAOFI; (Chelune and Baer, 1986; Chelune, 1986). Participant’s performance on the neuropsychological test battery and their responses to the everyday functioning questionnaires were utilized to assign HAND diagnoses following established criteria (Antinori et al., 2007), i.e., HIV-associated asymptomatic neurocognitive impairment (ANI), HIV-associated mild neurocognitive disorder (MND), and HIV-associated dementia (HAD).
ImmunoBlot
Frontal cortex tissues from human brains were homogenized and fractionated using a buffer that facilitates separation of the membrane and cytosolic fractions (1.0 mmol/L HEPES (Gibco, cat. no. 15630–080), 5.0 mmol/L benzamidine, 2.0 mmol/L 2-mercaptoethanol (Gibco, cat. no. 21985), 3.0 mmol/L EDTA (Omni pur, cat. no. 4005), 0.5 mmol/L magnesium sulfate, 0.05% sodium azide; final pH 8.8). In brief, as previously described (Fields et al., 2013), tissues from human brain samples (0.1 g) were homogenized in 0.7 ml of fractionation buffer containing phosphatase and protease inhibitor cocktails (Calbiochem, cat. no. 524624 and 539131). Samples were precleared by centrifugation at 5000 × g for 5 min at room temperature. Supernatants were retained and placed into appropriate ultracentrifuge tubes and were centrifuged at 436,000 × g for 1 h at 4°C in a TL-100 rotor (Beckman Coulter, Brea, CA). The supernatant was collected, as representing the cytosolic fraction, and the pellets were resuspended in 0.2 ml of buffer and re-homogenized for the membrane fraction.
After determination of the protein content of all samples by bicinchoninic acid assay (Thermo Fisher Scientific, cat. no. 23225), membrane fractions were loaded (20 μg total protein/lane) on 4–12% Bis-Tris gels (Invitrogen, cat. no. WG1402BX10) and electrophoresed in 5% HEPES running buffer and transferred onto PVDF membrane with iBlot transfer stacks (Invitrogen, cat. no. IB24001) using NuPage transfer buffer (ThermoFisher Scientific, cat. no NP0006). The membranes were blocked in 5% BSA in phosphate-buffered saline-tween 20 (PBST) for 1 h. Membranes were incubated overnight at 4 °C with primary antibodies. Following visualization, blots were stripped and probed with a mouse monoclonal antibody against β-actin (ACTB; Sigma Aldrich, cat. no. A5441) diluted 1:2000 in blocking buffer as a loading control. All blots were then washed in PBST, and then incubated with species-specific IgG conjugated to HRP (American Qualex, cat. no. A102P5) diluted 1:5000 in PBST and visualized with SuperSignal West Femto Maximum Sensitivity Substrate (ThermoFisher Scientific, cat. no. 34096). Images were obtained, and semi-quantitative analysis was performed with the VersaDoc gel imaging system and Quantity One software (Bio-Rad).
Immunohistochemistry and double immunofluorescence
Free-floating 40 μm thick vibratome sections of human brains were washed with phosphate buffered saline (PBS) 3 times, pre-treated for 20 min in 3% H2O2, and blocked with 2.5% horse serum (Vector Laboratories, cat. no. S-2012) for 1 h at room temperature. Sections were incubated at 4 °C overnight with the primary antibody, TFAM (Invitrogen, cat. no. PA5–23776) (Abcam, cat. no. ab23703) diluted in PBS. Sections were then incubated in secondary antibody, Immpress HRP Anti-rabbit IgG (Vector, cat. no. MP-7401) for 30 min, followed by NovaRED peroxidase (HRP) substrate made with NovaRED Peroxidase (HRP) Substrate Kit as per manufacturer’s instructions (Vector, cat. no. SK-4800). Control experiments consisted of incubation with secondary antibody only. Tissues were mounted on Superfrost plus slides (Fisherbrand, cat. no. 12–550-15) and coverslipped with cytoseal (Richard Allen Scientific, cat. no. 8310–16). Immunostained sections were imaged with a digital Olympus microscope and assessment of levels of TFAM immunoreactivity was performed utilizing the Image- Pro Plus program (Media Cybernetics, Silver Spring, MD). For each case a total of 3 sections (10 images per section) were analyzed in order to estimate the average number of immunolabelled cells per unit area (mm2) and the average intensity of the immunostaining (corrected optical density). Background levels were obtained in tissue sections immunostained in the absence of primary antibody. Therefore: corrected optical density = optical density − background.
Double immunolabelling studies were performed as previously described(Spencer et al., 2009) to determine the cellular localization of TFAM. For this purpose, vibratome sections of human brains were immunostained with antibodies against TFAM with GFAP (Sigma Aldrich, cat. no. G3893) and MAP2 (Santa Cruz Biotechnologies, cat# sc-32791). Sections were then reacted with fluorescent secondary antibodies, goat anti mouse IgG 488 (Invitrogen, cat. no. A11011) and goat anti rabbit IgG 568 (Invitrogen, cat. no. A11036). Sections were mounted on superfrost plus slides and cover-slipped with vectasheild (Vector, cat. no. 1000). Sections were imaged with a Zeiss 63X (N.A. 1.4) objective on an Axiovert 35 microscope (Zeiss) with an attached MRC1024 laser scanning confocal microscope system (BioRad, Hercules, CA). An examiner blinded to sample identification analyzed all immunostaining.
MtDNA isolation and quantification
To determine mitochondrial DNA (mtDNA) per cell, the copies of the mitochondrial gene Mt-ND2 were quantified and normalized to the numbers of RPP30 as previously described (Var et al., 2016; Vitomirov et al., 2017). Since RPP30 is a stable nuclear gene with an estimated two copies per cell, it is commonly used to control for cell numbers in tissues (Igarashi et al., 2017; Lin et al., 2016). MtDNA was isolated from the brain tissue of HAND donors using DNeasy Blood and Tissue kit (Qiagen, cat. nos. 69504 and 69506). Brain tissue was cut into small pieces and placed in a microcentrifuge tube. DNA was extracted from the tissue according to manufacturers’ instructions. DNA quantity and quality were determined using a spectrophotometer. RPP30 (Taqman, cat. no. Hs01124518) and Mt-ND2 (Taqman, cat. no. Hs02596874) gene expression assays were used. A master mix was prepared using 5 μl of 2x Fast Advanced Master Mix (Thermofisher Scientific, cat. no. 4444557), 0.5 μl each of 20x target primer and ACTB primer (Applied Biosystems, cat. no.1612290), and 2 μl of water per sample. To a microamp fast optical plate (Applied Biosystems, cat. no. 4346907), 8 μl of master mix and 2 μl of DNA were loaded into each well. The reactions were carried out at 48 °C for 30 min and 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Samples were analyzed in duplicate.
In vitro studies of human astrocytes
This study was approved by the University of California San Diego Human Research Protections Program. Astroglia and neurons were isolated from fetal human brain tissue from elective terminated pregnancy between 12 and 16 weeks of gestation, acquired from Advanced Bioscience Resources. Donors gave written informed consent for research-use of the cells and tissue. Tissue was fragmented and mechanically dissociated using a scalpel and washed 3 times with HBSS holding media (Gibco, cat. no.14175–095) with 1 mM Glutamax (Gibco, cat. no. 35050–061), 20 μg/mL Gentamicin (Gibco, cat. no. 15710–064) and 5 mM HEPES (Gibco, cat. no. 15630–080). The tissue was homogenized with the addition of 15 mL of 0.25% trypsin EDTA (Gibco, cat. no. 25200–056) for 5 min in a 37 °C incubator. After 5 min, 1 mL of a trypsin inhibitor (Roche, cat. no. 10109) and 24 mL of DMEM media (Gibco, cat. no. 11960–044) with human serum (Corning, cat. no. 35–060-cl) was added. The mixture was then centrifuged for 5 min at 4 °C to pellet the cells. Supernatant was removed and discarded, and the cells were resuspended in 5 ml of DMEM media and strained with a 70 μM strainer (Falcon, cat. no. 352350). The cell suspension was underlaid with 7 ml of a solution of filtered 8% BSA in PBS and cells were centrifuged at 1 × 104 rpm at 4 °C for 10 min. The supernatant was removed, and the cells were resuspended in DMEM media with human serum for astroglia or Neurobasal media (Gibco, cat. no. 21103–049) for neurons containing 2% B27 supplement (Gibco, cat. no. 17504044), 1 mM Glutamax, and 20 μg/ml Gentamicin. Astroglia were plated at a density of 1 × 107/T75 flask and cultured as adherent monolayers. Neurons were plated on coverslips coated with poly-ornithine (Sigma Aldrich, cat. no. P4957) at a density of 2 × 105 cells/well and cultured as adherent monolayers. After 1 week, the astroglia DMEM media with human serum was replaced with DMEM media with 10% fetal bovine serum (FBS) (Gibco, cat. no. 16000044) and 1% penicillin/ streptomycin (P/S) (Corning, cat. no. 30–001-CI-1). Every 3 days, a half media exchange was performed on each cell type.
Immunoblot of astroglial nuclear and cytoplasmic-enriched fractions
Cells were plated on 12 well plates at 800,000 cells per well and treated for 24 or 72 hours with Vehicle, WIN 20 μM, IL1β 10 ng/ml, or WIN and IL1β. On the day following treatment, cells were washed with sterile PBS and detached using 0.25% trypsin EDTA (Gibco, cat. no. 25200–056). Cells were collected using DMEM media with 10% fetal bovine serum (FBS) (Gibco, cat. no. 16000044) and 1% penicillin/ streptomycin (P/S) (Corning, cat. no. 30–001-CI-1). Cells were then centrifuged at 10,000 rpm for 30 seconds. The supernatant was discarded, and cells were washed with 700 μl of PBS followed by centrifugation at 10,000 rpm for 30 seconds. The PBS was removed and cells were lysed using a solution of 0.1% Triton-X in PBS with the addition of protease inhibitors. Cells were then centrifuged again at 10,000rpm for 30 seconds. The supernatant was retained as the cytosolic-enriched fraction and the pellet, the nuclear-enriched fraction, was resuspended in lysis buffer and sonicated. After protein concentration was determined using bicinchoninic acid assay (Thermo Fisher Scientific, cat. no. 23225) and samples were denatured in lamellae sample buffer (Bio Rad, cat. no. 1610747), cytosolic and nuclear fractions were loaded (10 μg total protein/lane) on 4–15% Criterion TGX stain free gels (Bio Rad, cat. no. 5678085) and electrophoresed in Tris/Glycine/SDS running buffer (Bio Rad, cat. no. 161–0772) and transferred onto LF PVDF membrane with Bio Rad transfer stacks and transfer buffer (Bio Rad, cat. no 1704275) using Bio Rad Trans Blot Turbo transfer system. After the transfer, total protein was imaged using Bio Rad ChemiDoc imager under the stain free blot setting for normalization purposes. The membranes were then blocked in 1% casein in tris - buffered saline (TBS) (Bio Rad, cat. no. 1610782) for 1 h. Membranes were incubated overnight at 4 °C with primary antibodies, TFAM 1:15000 (Invitrogen, cat. no. PA5–23776), PGC1-alpha 1:2000 (Abcam, cat. no. 54881), DRP1 1:1000 (Santa Cruz Biotechnologies, cat. no. sc32898), and GFAP 1:500 (Santa Cruz Biotechnologies, sc9065), diluted in blocking buffer. All blots were then washed in PBST, and then incubated with species-specific IgG conjugated to HRP (American Qualex, cat. no. A102P5) diluted 1:5000 in PBST and visualized with SuperSignal West Femto Maximum Sensitivity Substrate (ThermoFisher Scientific, cat. no. 34096). Images were obtained, and semi-quantitative analysis was performed with the ChemiDoc gel imaging system and Quantity One software (Bio-Rad).
In vitro treatments
Primary astroglia were treated with WIN 55–212-2 (WIN; R&D Systems, cat. no. 1038) and IL-1β (InvivoGen, cat. no. rcyec-hil1b). Treatments were solubilized in DMSO at concentrations of 10 mM and 20 ng/μl, respectively to create stock solutions. Treatments were diluted in DMEM media with P/S 1% (100 IU) and FBS 10% to desired concentrations. Astroglia treated with vehicle were treated with an equivalent amount of DMSO as was present in the sample treated with the highest amount of solubilized WIN/ IL-1β. Primary neurons were treated with conditioned media obtained from astrocytes treated with Vehicle, WIN, IL-1β, and WIN+ IL-1β. Conditioned media was used to treat the primary neurons at a 10% concentration for 72 h before fixation.
Treatment of astroglia with CB1, PPARγ and PPARα selective inhibitors
Astrocytes were split into 12 well plates at 500,000 cells/well on the day prior to treatment. To inhibit CB1, Rimonabant, (SR 141716A), (Sigma Aldrich, cat. no. SML0800) was used at 10 μM. To inhibit PPAR gamma, GW9662 (Sigma Aldrich, cat. no. M6191) was used at a concentration of 10 μM. To inhibit PPAR alpha, GW6471 was used at a concentration of 10 μM. Inhibitors were added to the cells to pretreat for 30 minutes followed by WIN 55,212–2 20uM for one hour and then IL1B at 10 ng/ml. Astrocytes were treated for 6 hours prior to RNA isolation.
Transfection of astroglia with siRNA for CB1
Astrocytes were split into 12 well plates at 500,000 cells/well on the day prior to transfection. Astrocytes were transfected using Lipofectamine 3000 (Thermo Fischer Scientific, cat. no. L3000075) without the use of the P3000 reagent. Lipofectamine 3000 and siRNA were diluted separately in Opti-Mem Media and then mixed together at a 1:1 ratio and left to incubate for 15 minutes at room temperature. After 15 minutes, the si-RNA-lipid complexes were added to the cells. CB1 siRNA (Santa Cruz Biotechnologies, cat. no. sc 39910) was used to knock down CB1 in the astrocytes. For controls, we used a control siRNA, (Santa Cruz Biotechnologies, cat. no. sc-37007) as well as siRNA diluent only. Cells were transfected on day one and day three before being harvested on day 5 for RNA or protein.
Seahorse Assay
Astroglia were split into a seahorse plate at 3 × 104 cells/well and treated with Vehicle, WIN, IL-1β, and WIN+ IL-1β for 24 h. On the following day, cultures were incubated in a non-CO2 incubator at 37 °C to equilibrate for approximately 30 minutes prior to assay. Baseline measurements of ECAR were taken prior to addition of oligomycin (2 μM), followed by a titrated concentration of FCCP, and then rotenone (500 nM) together with antimycin (1 μM) (Sigm a-Aldrich, cat no. A8674). After each addition of mitochondrial inhibitor, three readings were taken before injection of the subsequent inhibitor. ECAR was automatically calculated and recorded by the Seahorse XFe96 software. Rates were calculated by the Seahorse analyzer, reported as log of H+ production rate and then normalized to control for presentation. Samples were run in biological replicates of five in four independent experiments.
ATP Assay
Astroglia were split into a 96 well plate at 3 × 104 cells/well. After treatment with Vehicle, WIN, IL-1β, and WIN + IL-1β for 24 h, half of the media (50 μl) from each well was removed. To the remaining half media and cells, 50 μl of Cell Titer Glo 2.0 (Promega, cat. no. G9242) reagent was added. The plate was agitated for 2 min using a shaker and left to equilibrate for 10 min. Luminescence was recorded with Tecan i-control plate reader with an integration and settle time of 1 s each.
Electron Microscopy
Astroglia were plated on coverslips (Fischer, cat. no. 12–545-81) at a density of 2 × 104 cells/coverslip. After 24 h, cells were treated with Vehicle, WIN, IL-1β or IL-1β + WIN. After 24h of treatment, cells were fixed in glutaraldehyde, then fixed in osmium tetroxide and embedded in Epon Araldite. Blocks with the cells were detached from the coverslips and mounted for sectioning with an ultramicrotome (Leica). Cells were randomly acquired from three grids. Grids were analyzed with a Zeiss OM 10 electron microscope (Rockenstein et al., 2001). Electron micrographs were obtained at a magnification of 25,000X.
RNA isolation and TaqMan® human inflammation array and real-time reverse transcription polymerase chain reaction (RT2PCR)
Astroglia were split into 12 well plates at 5 × 105 cells/well for RNA isolation. After treatment for 6 h with vehicle, WIN, IL-1β, or WIN+ IL-1β, media were removed and the cells were washed once with (PBS). RNA was extracted with RNeasy plus mini kit (Qiagen, cat. no. 74136) according to manufacturer’s instructions and analyzed for purity and concentration with a spectrophotometer. RNA was reverse transcribed into cDNA with a high capacity cDNA Reverse Transcription Kit (Life technologies, cat. no. 4358813) as per manufacturer’s instructions. Gene expression was determined using RT2PCR and TaqMan gene expression assays. Taqman gene expression assays were performed using the StepOnePlus sequence-detection system (life technologies), using primers specific to IL-6 (Taqman, cat. no. hs00174131), CB1 (Taqman, cat. no.hs1038522), and ActB (Applied Biosystems, cat. no.1612290). A master mix was made using 5 μl of 2x Fast advanced master mix (Thermofisher Scientific, cat. no. 4444557), 0.5 μl of 20X primers, and 2 μl of water per reaction well. To each well of a microamp fast optical plate (Applied Biosystems, cat. no. 4346907), 8 μl of master mix and 2 μl of cDNA was added (The reactions were carried out at 48 °C for 30 min and 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min). Samples were analyzed in duplicate. Fold changes were calculated using the comparative CT method.
For the RT2PCR, RNA was reverse transcribed into cDNA using the RT2 first strand kit (Qiagen, cat. no. 330401). The cDNA was used on RT2 Profiler PCR array, Human Mitochondria (Qiagen, cat. no. PAHS-087Z) plate in combination with RT2 SYBR Green qPCR Mastermix (Qiagen, cat. no. 330529). CT values were exported to an Excel file and then uploaded to http://www.qiagen.com/geneglobe. Samples were assigned to either control or test groups and normalized to a reference gene. The data analysis center then calculated fold change using the comparative CT method. The data analysis center then generated scatter plots and heat maps, which were exported for visualization of gene expression alterations.
Immunocytochemistry
Neurons were plated on coverslips in a 24 well plate at a density of 2 × 105 cells/well and exposed to 20% conditioned media from astroglia. After 72 h, media was removed, and cells were washed 3 times with PBS. Cells were fixed with 4% paraformaldehyde for 20 min at 4 °C and then washed 3 times with PBS. Cells were blocked with blocking buffer (5% NGS, 0.2% Triton-X, 1% BSA, diluted in PBS) for 1 h and incubated overnight with primary antibodies: TFAM (Invitrogen, cat. no. PA5–23776) and MAP2 (Santa Cruz Biotechnologies, cat# sc-32791) diluted in blocking buffer. Control wells were incubated in blocking buffer overnight. The following day, primary antibodies were removed, and cells were washed with PBST 3 times. Secondary antibodies, goat anti mouse IgG 488 (Invitrogen, cat. no. A11011) and goat anti rabbit IgG 568 (Invitrogen, cat. no. A11036), were diluted 1:500 in blocking buffer and added to the cells for 30 min. Secondary antibodies were removed and Hoechst 3342 (Invitrogen, cat. no. H3570) was added, 1:5,000 for 5 min. Coverslips were washed with PBS 3 times and mounted on slides using Vectashield (Vector, H-1000). Fluorescence was visualized using confocal microscopy.
Cytokine Quantification
Astroglia cultures were treated with vehicle, WIN, IL-1β, or WIN+IL-1β in 6 well plates for 24 h. Supernatant was collected and analyzed for inflammatory cytokines, TNF-α, IL-6, IL-12p70, interferon (IFN) γ, IL-4, and IL-10 using V-PLEX Proinflammatory Panel 1 Human Kit (Mesoscale, cat. no. K15049D-4) as per manufacturer’s instructions. Samples were tested in duplicate.
Results
Mitochondrial biogenesis proteins are reduced in HAND brains on ART
Abnormal mitochondrial biogenesis is associated with multiple neurodegenerative diseases, as shown by altered levels of PGC-1α and TFAM (Calkins et al., 2011; Tain et al., 2009; Zuchner et al., 2004). Additionally, previous studies have shown that HIV alters mitochondrial metabolic processes leading to oxidative stress and apoptosis (Parikh et al., 2015; Thomas et al., 2009). PGC-1α acts as a master regulator of transcription of genes necessary for mitochondrial biogenesis (Roy Chowdhury et al., 2012) including TFAM. Once TFAM is translated to protein, it is translocated to mitochondria where it participates in mtDNA replication and mtDNA gene transcription (Escriva et al., 1999). To determine the expression levels of mitochondrial biogenesis proteins in brains of HIV+ donors, we analyzed frontal lobe lysates from HAND cases as well as Non-HAND cases (Table 1). Brain lysate membrane fractions were analyzed for PGC-1α, TFAM and ACTB levels by immunoblot. In brain lysates from HAND cases, PGC-1α and TFAM protein band intensity decreased with increasing neurocognitive impairment (NCI), with the least being detected in HIV-associated dementia (HAD) samples (Fig. 1 A). Densitometry analyses of PGC-1α and TFAM bands show protein levels were decreased by 40% (Fig. 1 B) and 50% (Fig. 1 C), respectively, in brains from HAD cases compared to Non-HAND cases. PGC-1α and TFAM band signals were reduced in HAND cases compared to Cognitive Normal (CN) cases. Densitometry analysis showed a 30% and 50% decrease in PGC-1α and TFAM levels (p<0.05) (Fig. 1, D and E) in postmortem brains of HAND compared to CN HIV+ patients. These results suggest that transcriptional regulators of mitochondrial biogenesis are deficient in brains of decedents that were diagnosed with HAND.
Figure 1. Mitochondrial biogenesis proteins are reduced in HAND brains on ART.
(A) Immunoblot of HAND donor frontal lobe lysates with antibodies specific for PGC-1α and TFAM, normalized to ACTB. (B and C) Quantification of PGC-1α and TFAM band intensity stratified by HAND diagnosis. (D and E) Quantification of PGC-1α and TFAM band intensity stratified by CN vs. HAND. Statistical significance was determined by an unpaired t test. *, p<0.05, **, p<0.01.
TFAM is reduced in neuronal processes and increased in astroglia in HAND frontal cortices compared to tissue from cognitively normal individuals
TFAM is a nuclear encoded factor that stimulates mtDNA transcription and is required for mitochondrial biogenesis to proceed. To determine if the biochemical alterations in TFAM levels are associated with neurocognitive impairment, we performed immunostaining for TFAM in the frontal cortex of CN HIV+ and HAND patients. TFAM signal was reduced and showed less signal throughout the apical dendrites of neurons in HAND cases compared to CN HIV+ cases (Fig. 2 A). Optical density analysis revealed a 35% decrease (p<0.05) of TFAM protein in neurons of HAND cases (n=6) compared to Non-HAND cases (n=6) (Fig. 2 B). To better understand the alteration in TFAM levels the brains of HAND decedents, we performed double immunolabeling from TFAM and two cell-specific markers: GFAP for astroglia and MAP2 for neurons. Double-immunolabeling with TFAM (red) and MAP2 (green) was performed in frontal cortex sections from CN HIV+ cases (n=4) and HAND cases (n=4). The signals for TFAM and MAP2 were robust throughout the neuron soma and processes in CN HIV+ brains; however, TFAM and MAP2 signals were less intense in brains from HAND decedents (Fig. 2 C). Quantification of TFAM and MAP2 signal revealed a significant decrease in HAND brains compared to CN subjects (Fig. 2 D and E). Merged images revealed a higher number of co-immunostained (yellow) regions for the CN HIV+ patient. Quantification analysis revealed 70% colocalization of TFAM and MAP2 for HIV+ and only 25% for HAND (Fig. 2 F), suggesting reduced TFAM in neuronal processes in HAND tissues. In vibratome sections of frontal cortex, the signal for GFAP antibody binding was increased in HAND samples compared to CN, as expected (Fig. 2 G). The levels of the signal for GFAP were increased ~80% in brains from HAND decedents compared to those from CN decedents (Fig. 2 H). Interestingly, the signal for TFAM co-localization with GFAP was increased by 50% in brains from HAND decedents compared to those from CN decedents (Fig. 2 I). These data suggest reduced mitochondrial biogenesis signaling in neuronal processes is concurrent with increased mitochondrial biogenesis in astroglia.
Figure 2. TFAM is reduced in neuronal processes and increased in astroglia in HAND frontal cortices compared to tissue from cognitively normal individuals.
(A and B) Immunostaining of TFAM in the frontal cortices of HAND (n=6) and CN (n=6) brain tissue and its quantification. (C) Double-immunostaining of TFAM and MAP2 in frontal cortices of HAND (n=4) and CN (n=4) brain tissue. (D-F) Quantification of TFAM signal, MAP2 signal, and % colocalization of TFAM/MAP2 signal. (G) Double-immunostaining of GFAP and TFAM in the frontal cortices of HAND and CN brain tissue. (H and I) Quantification of GFAP and % TFAM/GFAP colocalization. Statistical significance was determined by an unpaired t test. *, p<0.05, **, p<0.01.
mtDNA is decreased and IL-1β is increased in HAND brains exposed to ART
Decreased TFAM expression in MAP2-positive processes along with increased TFAM in GFAP-expressing cells suggests reduced neuronal mitochondrial biogenesis concomitant with metabolically activated, pro-inflammatory astroglia. As TFAM is directly involved in mtDNA replication, the reductions in TFAM observed in neurons may indicate reduced levels of mtDNA in these brains. Therefore, we assessed mitochondrial-to-nuclear DNA ratios to quantify mtDNA per cell. We measured the levels of NADH dehydrogenase subunit MT-ND2 to quantify mtDNA and compared this to the nuclear gene RPP30, the quantities of which correlate with cell count. We discovered by RT-PCR that levels of mtDNA were significantly reduced in minor neurocognitive disorder (MND) and HAD brains when compared to the levels of mtDNA/cell in CN brains as shown by normalized MT-ND2 (Fig. 3 A). The combined levels of mtDNA/cell in all HAND brains was reduced by ~40% compared to levels of mtDNA/cell in CN brains, but these differences did not reach statistical significance (Fig. 3 B).
Figure 3. mtDNA is decreased and IL-1β is increased in HAND brains exposed to ART.
DNA was isolated from HAND donors’ brains and mtDNA and IL1β was measured. (A and B) Quantification of mtDNA copies/cell stratified by HAND diagnosis and CN vs. HAND. (C and D) RNA was isolated from HAND donors’ brains and reverse transcribed to cDNA. Fold change of IL-1β was measured using RT2PCR with ACTB as a normalization control. The fold change in IL-1β expression stratified by HAND diagnosis and CN v. HAND was quantified. Statistical significance was determined by an unpaired t test. *, p<0.05.
To monitor the level of inflammatory astroglia, we quantified the mRNA for IL-1B using ACTB as a normalization control using RT-PCR. The levels of mRNA for IL-1β were significantly increased in all HAND subjects compared to CN brains (Fig. 3 C). The levels of mRNA from all HAND groups combined was 3-fold higher than in the CN group, however, the differences did not reach significance, due to high inter-patient variability (Fig. 3 D). Nevertheless, these data suggest that the IL-1β pathway was active in the HAND brains despite the ART regimens.
Inflammatory cytokine (IL-1β) stimulates mitochondrial energy metabolism in astroglial cell cultures, and this is muted by treatment with WIN, independent of CB1.
Synchronous and biologically linked shifts in metabolism and immune processes are normally thought to occur in cells of the immune system, such as macrophages, T-cells, and natural killer cells (Lee et al., 2018; Van den Bossche et al., 2017). To determine if alterations in metabolic function occur in immune-stimulated astroglia, we exposed primary human astroglia to increasing doses of IL-1β (100 pg, 1 ng, 10 ng, and 20 ng/ml) for 1 h. To determine the effects of pre-exposure of these astroglia to an anti-inflammatory molecule, we treated cells with WIN (10 μM) for 1 h prior to IL-1β treatment. After 1h exposure to IL-1β, astroglia were analyzed for oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) to determine shifts in oxidative phosphorylation and glycolysis, respectively. IL-1β increased OCR and ECAR of astroglia in a dose dependent manner (Fig. 4 A and B). Astroglia treated with WIN (10 μM) for 1 h prior to treatment with IL-1β (10 ng/ml) for 1h showed no increase in OCR or ECAR compared to vehicle-treated cells (Fig. 4 C and D). These acute changes in IL-1β-treated astroglia suggest a metabolic shift is involved in the inflammatory response and that an anti-inflammatory molecule can block this metabolic shift.
Figure 4. Inflammatory cytokine (IL-1β) stimulates mitochondrial energy metabolism in astroglial cell cultures, and this is muted by treatment with WIN, independent of CB1.
(A and B) OCR and ECAR maximum levels in astroglia after exposure for 30 minutes to an increasing dose response of IL-1β. (C and D) OCR and ECAR maximum levels in astroglia after they were treated with treated with IL-1β or WIN alone or treated with WIN before IL-1β treatment. (E and F) OCR and ECAR maximum levels after pretreatment with CB1 antagonist (SR 141716A) and then treatment with WIN and IL-1β. (G and H) Quantification of lumincescence using ATP assay on astroglial lysates treated with increasing doses of IL-1b. (H) Quantification of lumincescence using ATP assay on astroglial lysates treated with vehicle, IL-1β or WIN alone or with WIN and then IL-1β. (I) Electron micrograph images of astroglia treated with vehicle, IL-1β or WIN alone or with WIN and then IL-1β. (J) Quantification of average mitochondrion length. All graphs plot mean ± SEM. All experiments were performed in three independent experiments using at least two astroglial lines with different genetic background. Data were analyzed by one-way ANOVA.
WIN is a CB1/2 agonist and CB1 has been reported to be localized to and active on mitochondria (Gutierrez-Rodriguez et al., 2018). To determine if WIN activities to block IL-1b-induced mitochondrial activity are dependent on CB1, astroglia were treated with the CB1 antagonist SR 141716A (SR) (10 μM) for one hour before treatment with WIN. After treatment with IL-1β, astroglia were analyzed for OCR and ECAR. Treatment with SR had no effect on the ability of WIN to block IL-1β induced increase in OCR or ECAR (Fig. 4 E and F). These data suggest that CB1 is not involved in WIN function to block IL-1β induced increase in OCR or ECAR.
To determine if changes in metabolic activity induced by inflammatory stimuli yielded changes in ATP, we treated astroglia with increasing doses of IL-1β and measured cellular ATP levels in cell lysates. IL-1β induced a significant reduction in ATP levels at concentrations of 10 and 20ng/ml (Fig. 4 G). Pretreatment of the astroglia with WIN had no effect on basal ATP levels, but prevented the reduction in ATP in cells exposed to IL-1β (Fig. 4 H). Taken together, these data show that immune stimulated astroglia have an altered metabolic phenotype associated with increased mitochondrial metabolism, and that pretreatment with a cannabinoid agonist can prevent these alterations.
IL-1β and WIN induced alterations in mitochondrial activity. To see if these treatments caused changes in mitochondrial morphology, we performed ultrastructural analyses of the astroglia using transmission electron microscopy (TEM). Astroglia were plated on coverslips and treated with vehicle, WIN, IL-1β, or WIN+ IL-1β. Treatment with IL-1β caused mitochondrial elongation compared to vehicle treated cells, but pretreatment with WIN prevented this morphological change (Fig. 4 I). When mitochondrial length was quantified, treatment with IL-1β showed a significant increase in mitochondrial length compared to the vehicle. Pretreatment with WIN prevented this increase in mitochondrial length (Fig. 4 J).
IL-1β induces PGC1α expression in astroglia and it is blocked by WIN
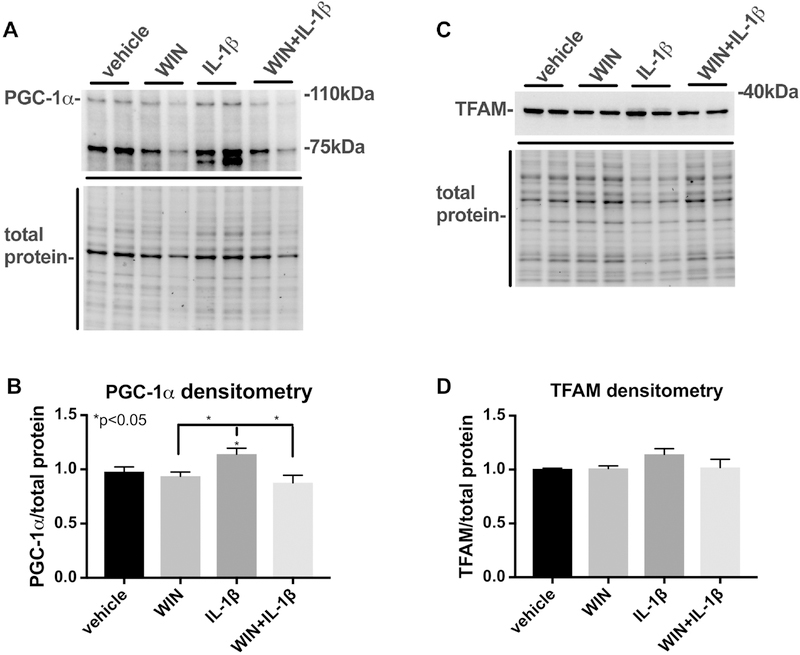
Little is known about the role of mitochondrial biogenesis in immune activation of astroglia. To determine the effects of IL-1β and WIN on PGC1α and TFAM expression, astroglia were exposed to IL-1β after pretreatment with WIN for 1 h. One day later nuclear and cytoplasmic lysates were isolated from the astroglial culutres. Nuclear-enriched lysates were analyzed for PGC1α protein expression and cytoplasmic-enriched lysates were analyzed for TFAM protein levels by immunoblot. The band corresponding to PGC1α migrated to approximately 100 kDa and the intensity of this band was quantified for densitometry analysis and normalized to the total protein transferred to the membrane (Fig. 5 A). A stronger band appeared at 75kDa, though the function of a 75kDa isoform of PGC1α, to our knowledge, has not been reported. Densitometry analyses of blots from three independent astroglia lines showed that IL-1β induced a significant increase levels of PGC1α compared to levels in vehicle, WIN or WIN+ IL-1β treated astroglia (Fig. 5 B). The immunoblot for TFAM revealed a strong band at approximately 30 kDa that was consistently more intense than this band in vehicle, WIN or WIN + IL-1β treated astroglia (Fig. 5 C). However, densitometry analyses showed no significant difference in TFAM compared to vehicle, WIN or WIN_IL-1β treated astroglia (Fig. 5 D).
Figure 5. IL-1β induces PGC-1α expression in astroglia and this is blocked by WIN.
Astroglia were treated with vehicle, IL-1β or WIN alone or with WIN and then IL-1β for one day and then nuclear and cytoplasmic-enriched lysates were analyzed by immunoblot for PGC1α and TFAM expression, respectively. (A) Image of immunoblot of PGC1a in nuclear-enriched lysates and image of the total protein transferred to the membrane. (B). Quantification of PGC-1α band densitometry normalized to total protein densitometry. (C) Image of immunoblot of TFAM in cytoplasmic-enriched lysates and image of the total protein transferred to the membrane. (D) Quantification of PGC-1α band densitometry normalized to total protein densitometry. All graphs plot mean ± SEM. All experiments were performed in three independent experiments using at least two astroglial lines with different genetic background. Data were analyzed by one-way ANOVA.
IL-1β alters mitochondrial gene expression that is reversed by WIN
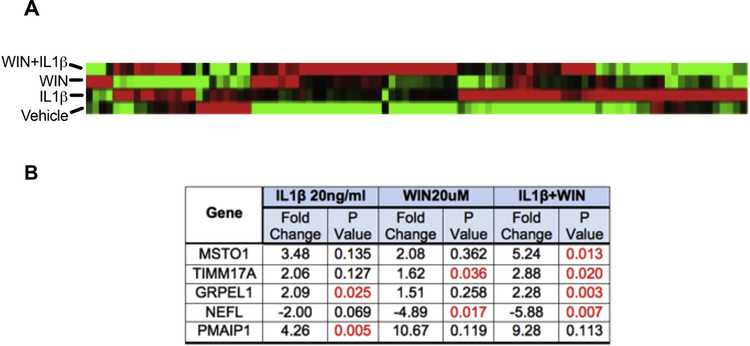
Mitochondrial biogenesis requires the regulated expression of many mtDNA- and nuclear-encoded genes. To determine if and to what extent IL-1β, WIN, or the combination of the two alter mitochondrial gene expression along with TFAM, we assayed for expression of 84 mitochondrial-associated genes in RNA isolated from astroglia treated with vehicle, WIN, IL1β or WIN + IL1β The gene list included metabolite transporters, mitochondrial fission/fusion proteins, proteins of the electron transport chain, and many others (supplementary data). The cluster gram reveals the genes that were altered by over 2-fold versus vehicle-treated cells (Fig. 6 A). In astroglia treated with IL-1β, there were two significantly upregulated genes, GRPEL1, involved in mitochondrial protein import from the inner membrane to the mitochondrial matrix, and PMAIP1, which promotes efflux of apoptogenic proteins from the mitochondria (Fig. 6 B). In astroglia treated with WIN, TIMM17A, which is also involved in mitochondrial protein import across the mitochondrial inner membrane was significantly increased, and NEFL, which is a neurofilament light chain protein was significantly reduced in expression (Fig 6 B). In astroglia treated with WIN and IL-1β, MSTO1, which is involved in the regulation of mitochondrial distribution and morphology, TIMM17A, and GRPEL1 were all significantly upregulated (Fig. 6 B). These data indicate that gene expression of mitochondrial as well as other proteins is altered in astroglia treated with WIN, IL-1β, and a combination of the two.
Figure 6. IL-1β alters mitochondrial gene expression, and this is reversed by WIN.
Astroglia were treated with Vehicle, WIN, IL-1β, and WIN+IL-1β for or 24 h. RNA was isolated from the astroglia and analyzed for 84 mitochondrial genes using Human Mitochondria RT2 Profiler PCR array. (A) A heat map of gene expression alterations when astroglia are treated with vehicle, WIN, IL-1β, and WIN+IL-1β. (B) A table of genes with the most significant expression changes in all three of the treatment groups (significant p values indicated in red) indicates common points of gene regulation. Data were analyzed by one-way ANOVA.
WIN-mediated inhibition of IL-1β-induced inflammatory gene expression is reversed by a PPARα antagonist
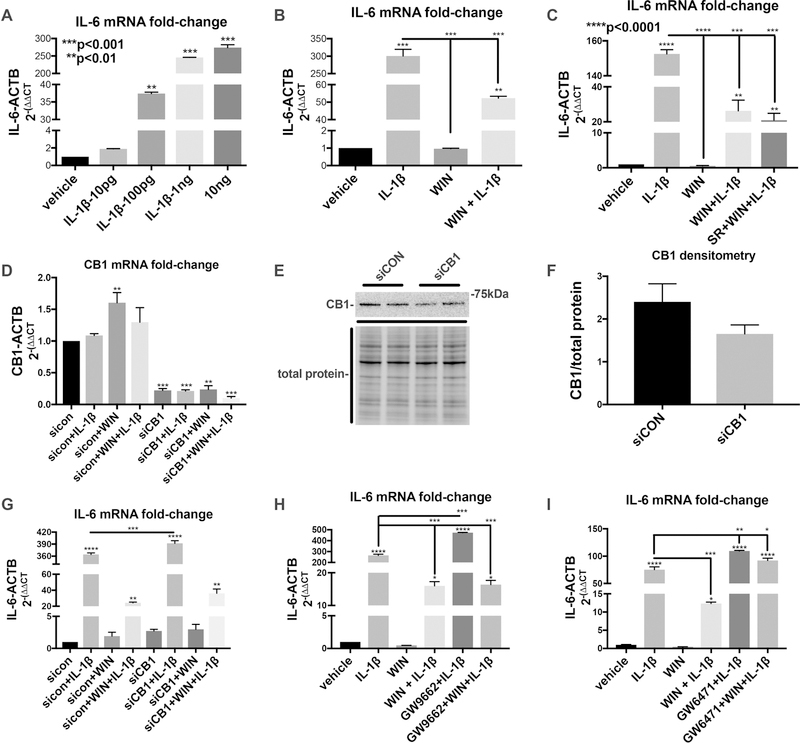
To investigate how IL-1β and WIN affect expression of inflammatory genes known to be involved in neurodegenerative diseases, using the experiments described above, we assayed for mRNA expression of IL-6 a prototypical inflammatory cytokine. As expected, IL-1β induced expression of mRNA for IL-6 in a dose dependent manner (Fig. 7 A) and WIN pretreatment reduced IL-1β-induced expression of mRNA for IL-6 (Fig. 7 B). To determine the pathway through which WIN is acting, we first treated astroglia with the CB1 antagonist, SR 141716A (SR) (10 μM) for one hour and then with WIN for one hour before treating with IL-1β for one day. SR had no effect on the levels of IL-6 mRNA in astroglia treated with WIN and IL-1β (Fig. 1 C). Next, astroglia were transfected with siRNA to knock down CB1 expression. CB1 mRNA levels were knocked down by at least 80% compared to cells transfected with control siRNA (siCON) (Fig. 7 D). CB1 protein levels were reduced, though not proportional with the level of mRNA reduction (Fig. 7 E). Densitometry analysis revealed a decrease of about 30% compared to astroglia transfected with siCON, though this was not a significant difference (Fig. 7 F). To determine if this moderate reduction in CB1 protein levels affect the activity of WIN to inhibit IL-1β induction of inflammatory genes, IL-6 mRNA was measured. There was no difference in levels of IL-6 mRNA between WIN-treated astroglia that were transfected with siCON versus those transfected with siCB1 (Fig. 7 G).
Figure 7. WIN-mediated inhibition of IL-1β-induced inflammatory gene expression is reversed by PPARα antagonist.
(A) IL-6 mRNA fold-change after 6 hours in astroglia treated with an increasing dose response of IL-1β. (B) IL-6 mRNA fold-change after 6 hours in astroglia treated with vehicle, WIN, IL-1β, and WIN+IL-1β. (C) IL-6 mRNA fold-change after 6 hours in astroglia treated with with vehicle, WIN, IL-1β, WIN+IL-1β and SR+WIN+ IL-1β. (D) IL-6 mRNA fold-change after 6 hours in astroglia first transfected with control siRNA or CB1-specific siRNA and then vehicle, WIN, IL-1β, and WIN+IL-1β. (E) Immunoblot for CB1 in astroglia first transfected with control siRNA or CB1-specific siRNA. (F) Quantification of CB1 band densitometry normalized to total protein densitometry. (G) IL-6 mRNA fold-change after 6 hours in astroglia first transfected with control siRNA or CB1-specific siRNA and then vehicle, WIN, IL-1β, and WIN+IL-1β. (H) IL-6 mRNA fold-change after 6 hours in astroglia treated with with vehicle, WIN, IL-1β, WIN+IL-1β and GW9662+WIN+ IL-1β. (I) IL-6 mRNA fold-change after 6 hours in astroglia treated with vehicle, WIN, IL-1β, WIN+IL-1β and GW6471+WIN+ IL-1β. All graphs plot mean ± SEM. All experiments were performed in three independent experiments using at least two astroglial lines with different genetic background. Data were analyzed by one-way ANOVA.
To test for the involvement of PPARγ and PPARα in the anti-inflammatory effects of WIN, astroglia were pretreated with GW9662 and GW6471, respectively. Pretreatment of astroglia with GW9662 followed by treatment with IL-1β caused a significant increase in IL-6 mRNA compared to treatment with IL-1β alone, consistent with an anti-inflammatory role of PPARγ in astroglia (Fig. 7 H). However, GW9662 had no effect on the anti-inflammatory effect of WIN (Fig. 7 H). Pretreatment of astroglia with GW6471 followed by treatment with IL-1β caused a significant increase in IL-6 mRNA compared to treatment with IL-1β alone, consistent with an anti-inflammatory role of PPARα in astroglia (Fig. 7 I). Pretreatment with GW6471 completely restored IL-1b-induced IL6 mRNA levels in WIN treated astroglia compared to those treated with WIN and no GW6471 (Fig. 7 I). These data suggest that WIN is signaling through PPARα to block IL-6 mRNA expression.
WIN blocks IL-1β-induced expression of multiple inflammatory cytokines in astroglia.
To determine if these changes in mRNA are reflected in the proteins being secreted by the astroglia, we collected supernatants and analyzed them for expression of 6 cytokines by MesoScale Electro-chemiluminescent Assay. IL-1β induced expression of all cytokines tested. IL-1β induced a ~1000-fold increase in IL-6 protein levels, but WIN pretreatment reduced these levels by ~95% (Fig. 8 A). TNF-α protein levels climbed 30-fold with IL-1β, and WIN pretreatment reduced these levels by ~60% (Fig. 8 B). IL-1β induced a 14-fold increase in IL-12p70 protein levels, while WIN inhibited this by ~70% (Fig. 8 C). Although IL-1β induced a 800-fold increase in IFNy protein levels, WIN pretreatment further increased IFNy by ~30% (Fig. 8 D). IL-1β induced a six-fold increase in IL-4 protein levels, that WIN reduced by ~65% (Fig. 8 E). IL-10 protein levels increased by 3-fold with IL-1β, and WIN pretreatment reduced this by ~40% (Fig. 8 F). These data suggest that along with reducing mitochondrial activity, WIN reduces inflammatory and anti-inflammatory gene expression with the exception of INFy, further supporting the role of immune-stimulated astroglia in inflammation.
Figure 8. IL-1β alters inflammatory gene expression, and this is reversed by WIN.
Astroglia were treated with vehicle, WIN, IL-1β, and WIN+IL-1β for one day and then supernatants were analyzed by Mesoscale Discovery electrochemiluminescent assay. (A-F) Levels of cytokines in the supernatant: IL-6 (A), TNF-α (B), IL-12p70 (C), IFNγ (D), IL-4 (E) and IL-10 (F). All graphs plot mean ± SEM. All experiments were performed in three independent experiments using at least two astroglial lines with different genetic background. Data were analyzed by one-way ANOVA.
Conditioned media from IL-1β-treated astroglia reduces neuronal TFAM and MAP2 expression, and this is reversed by WIN
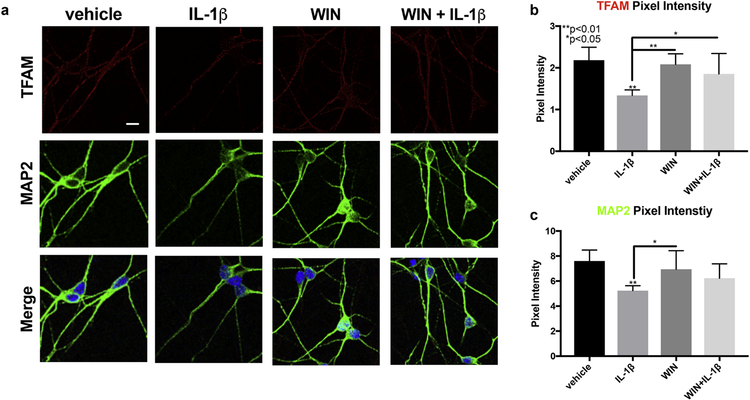
Several reports have shown that inflammatory cytokine-activated astroglia are toxic to neurons (Sama et al., 2008; Ye et al., 2013). However, the effect that IL-1β-activated astroglia have on mitochondrial biogenesis in neurons has not been explored. To test if IL-1β-activated astroglia may affect neuronal mitochondrial biogenesis, and further, if WIN may block this effect, we treated primary human neurons with the conditioned media collected from astroglia treated with WIN, IL1β or WIN + IL-1β for 72 hours. Neurons were exposed to 20% CM for 72 h, fixed, and then immunostained for TFAM (red) and MAP2 (green) and then analyzed by confocal microscopy. The signal for TFAM was punctate throughout the neuronal processes in vehicle-treated cells, however, the signal for TFAM was much weaker in neurons exposed to CM from IL-1β-treated astrocytes (Fig 9 A). No changes in TFAM intensity compared to vehicle-treated cells were apparent in cells treated with WIN or WIN+IL-1β (Fig. 9 A). The signal for MAP2 was also reduced in IL-1β-treated cells (Fig. 9 A). Quantification of the pixel intensity for TFAM showed that CM from IL-1β-treated astroglia reduced TFAM signal by ~40%, while WIN blocked this effect (Fig. 9 B). Similarly, quantification of the pixel intensity for MAP2 showed that CM from IL-1β-treated astroglia reduced TFAM signal by ~30%, and WIN also blocked this effect (Fig. 9 C). These data suggest that IL-1β-activated astroglia in culture can reduce neuronal TFAM and possibly mitochondrial biogenesis, and exposure to WIN blocks this effect.
Figure 9. Conditioned media from IL-1β-treated astroglia reduces neuronal TFAM and MAP2 expression, and this is reversed by WIN.
(A) Astroglia were treated with vehicle, WIN, IL-1β, and WIN+IL-1β for one day. Astroglial conditioned media was collected and used to treat primary neurons that were grown on glass coverslips, which were then fixed and immunostained for TFAM (red) and MAP2 (green). (B) Signal quantification of TFAM in primary neurons. (C) Signal quantification of MAP2 in primary neurons. All experiments were performed in three independent experiments using at least two astroglial lines and neuron lines with different genetic background. Data were analyzed by one-way ANOVA.
Discussion
This study provides evidence of a mechanistic link between reduced mitochondrial biogenesis in neurons and increased metabolic activity and inflammation in astroglia in brains from HIV+ individuals that were on ART regimens and diagnosed with HAND (Schematic). This mechanism was further supported by an overall decrease in the quantity of mtDNA per cell and increases in inflammatory gene expression in HAND brains when compared to CN. In vitro experiments showed that human astroglia undergo rapid and robust shifts in metabolic activity in response to inflammatory stimuli that are commonly expressed in HAND. IL-1β induced increased energy demand in astroglia as indicated by alterations in OCR, ECAR, ATP production, PGC1α expression. These changes in metabolic function are accompanied by increased expression of genes for mitochondrial, metabolic, and inflammatory pathways, and morphological changes. Importantly, exposing the astroglia to an anti-inflammatory molecule, WIN, blocks the increases in metabolic activity and alterations in mitochondrial morphology as well as inflammatory gene expression (Schematic). The anti-inflammatory activity of WIN was inhibited by a PPARα antagonist. Moreover, WIN blocks neurotoxicity and reverses reductions in the expression TFAM in neurons exposed to CM from stimulated astroglia, suggesting that molecules that block increases in astroglial metabolic function and inflammatory gene expression may have a therapeutic role against neurotoxicity and alterations in neuronal bioenergetic capacity. These findings identify alterations that are consistent with multiple studies that suggest a mechanism in which mitochondrial biogenesis and bioenergetic capacity of neurons is compromised in neurodegenerative diseases with strong neuroinflammatory signatures (Jiang and Cadenas, 2014; Yin et al., 2016).
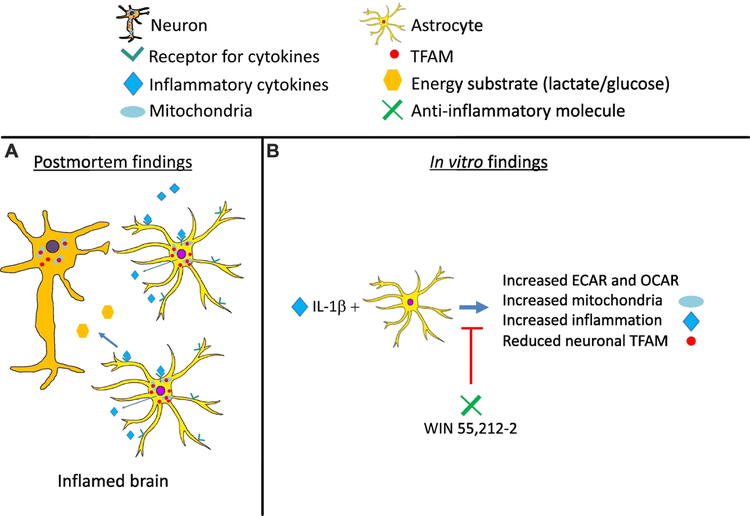
Schematic:
(A) Findings from postmortem brain specimens from HAND cases reveal overall reduced levels of molecules associated with mitochondrial biogenesis. However, mitochondrial biogenesis may be increased in activated astroglia along with inflammatory cytokines in the brain. Studies by other groups (Jiang and Cadenas, 2014; Natarajaseenivasan et al., 2018; Yin et al., 2016) have shown that activated astroglia, compared in non-activated astroglia, may produce less energy substrate to neurons and this may be neurotoxic. (B) Analyses of metabolic and mitochondrial changes in cytokine-activated astroglia, revealed human rapid changes in ECAR and OCAR and increased mitochondrial biogenesis concurrent with increased inflammatory gene expression. Similar to the findings in brain tissues, TFAM was reduced in neurons treated with CM from these IL-1β-activated astroglia. Importantly, a known anti-inflammatory molecule, WIN inhibited the metabolic and mitochondrial changes in the astroglia and the reduction is neuronal TFAM. WIN also inhibited inflammatory gene expression in the IL-1β-activated astroglia via PPARα. These studies provide support for targeting astroglia to restore neuronal bioenergetics.
Previous studies have shown that dysfunctional mitochondrial dynamics (fission and fusion), mitochondrial transport, mitophagy and the production of reactive oxygen species are implicated in HAND during the ART era (Avdoshina et al., 2016; Bonavia et al., 2001; Fields et al., 2015b; Huang et al., 2012; Kitayama et al., 2008; McArthur et al., 2010; Teodorof-Diedrich and Spector, 2018; Thangaraj et al., 2018). However, no studies have investigated the role mitochondrial biogenesis in HAND brains. The findings presented here are consistent with studies in other neurodegenerative diseases such as AD and HD, in which mitochondrial biogenesis proteins TFAM and PGC1α are reduced in the brains (Repunte-Canonigo et al., 2014; van der Walt et al., 2003; Wang et al., 2013; Ye et al., 2012),. Due to the alterations in mitochondrial biogenesis, mitochondrial dynamics and mitophagy, mitochondria provide promising therapeutic targets for preventing neurocognitive decline in HAND in the ART era.
Astroglia, the most numerous cell type in the brain (Tower and Young, 1973), regulate metabolism (Alberini et al., 2018), inflammation (Sofroniew, 2015), and neuronal function (Alberini et al., 2018; Khakh and Sofroniew, 2015). Astrogliosis remains a hallmark of the HIV-infected brain in the cART era (Levine et al., 2016). However, little is known about how metabolic changes regulating astroglial function may contribute to the neuropathogenesis of HIV. The findings presented here are consistent with recent studies (Jiang and Cadenas, 2014; Natarajaseenivasan et al., 2018) that aged, activated, and immune-stimulated astroglia consume more energy than non-activated astroglia (Jiang and Cadenas, 2014; Yin et al., 2016), and switch from primarily depending on glycolysis to oxidative phosphorylation in the context of AD (Jiang and Cadenas, 2014; Natarajaseenivasan et al., 2018). Our findings show that these changes in astroglia may depend upon fluctuations in the expression of metabolic and mitochondrial genes. Moreover, similar to our findings, the metabolic switch to oxidative phosphorylation also proved to be toxic to neurons that were exposed to CM from activated astroglia (Natarajaseenivasan et al., 2018). The current findings that TFAM was reduced in neurons and increased in astroglia in HAND brains are consistent with a reduction in metabolic activity in neurons and an increase in metabolic activity in astroglia. IL-1β-induced expression of PGC1α in astroglia is consistent with a the findings of a study that showed lipopolysaccharide and interferon-γ induce mitochondrial biogenesis (Wang et al., 2014). These observations of enhanced mitochondrial biogenesis support increased bioenergetic needs in astoglia, which is consistent with our findings that intracellular ATP levels are reduced as ECAR and OCR are increased concomitant with increased cytokine production. Astroglia may be using the increased ATP more rapidly that it is produced through oxidative phosphorylation. This metabolic switch by astroglia may be comparable, though not perfectly analogous to changes in macrophages, which undergo metabolic shifts that precede differentiation into an inflammatory or anti-inflammatory phenotype (Van den Bossche et al., 2017). Indeed, the anti-inflammatory molecule WIN blocked IL-1β-induced metabolic changes. While these studies do not link the increases in astroglial metabolism to increased inflammatory signaling, they do provide evidence that they two are activated simultaneously an inhibited simultaneously by WIN. Hence, metabolic regulation of astroglia may serve as a novel therapeutic target to protect bioenergetic capacity of neurons in inflammatory environments. However, the mechanisms through which IL-1β induces and WIN blocks increased metabolism in astroglia deserves further investigation.
The anti-inflammatory activity of WIN has been reported in models for astroglia and microglia in in vitro and in vivo settings. However, there is some inconsistency in the studies regarding the signaling pathways through which WIN acts. Our report confirms findings from other groups that show WIN blocks the expression of inflammatory genes in astroglia exposed to recombinant IL-1β (Curran et al., 2005; Sheng et al., 2005) and a similar effect is also observed in astroglia treated with recombinant amyloid (A)β1–42 (Aguirre-Rueda et al., 2015). Curran et al found, similar to our results, that WIN inhibits IL-1β-induced mRNA expression of IL-8, intercellular adhesion molecule (ICAM) 1, and vascular cell adhesion molecule 1 independent of CB1 or CB2 receptors (Curran et al., 2005). These findings are inconsistent with another study that showed CB1 and CB2 antagonists partially suppressed the effect of WIN to block IL-1β induction of inducible NO synthase (iNOS) (Sheng et al., 2005). It may be that the IL-1β signaling pathway leading to iNOS expression is independent of the pathway leading to IL-1β induced IL-8, ICAM and VCAM expression. The findings by Sheng et al also differed from Curran et al and our study by showing strong CB2 expression in human fetal astroglia (Curran et al., 2005; Sheng et al., 2005), as CB2 was undetectable in all of the astrocyte cultures used in our study as measured by RT2PCR and immunostaining. Another study showed that WIN prevents the effects of Aβ1–42 on primary astrocytes and increases expression of PPARγ, although the astrocytes were derived from rats (Aguirre-Rueda et al., 2015). Consistent with the anti-inflammatory effect of PPARγ, we found that a PPARγ antagonist increased IL-1β-induced IL-6 expression but had no effect on the function of WIN. Our studies provide the first evidence that PPARα is involved in the anti-inflammatory effects of WIN on IL-1β activated human astroglia. These findings suggest that PPARα in astroglia maybe a promising therapeutic target in HAND and other neurodegenerative diseases involving IL-1β. More detailed mechanistic studies that knockdown or overexpress PPARα in astroglia will help to better understand the role of PPARα in neuroinflammation. Neuropathological studies of PPARα expression in astroglia and microglia in control and diseased brains could be useful to develop therapeutic strategies that target the nuclear receptor.
Activated astroglia have been shown to be neurotoxic in number of different model systems and especially in HIV-associated neurological disorders (Natarajaseenivasan et al., 2018; Sama et al., 2008; Ton and Xiong, 2013). The neurotoxicity of IL-1β treated astroglia was reported by Sama et al to act through the calcineurin/NFAT pathway (Sama et al., 2008). Importantly, PPARα has been shown to interact with calcineurin and NFAT, though in cardiac cells (Li et al., 2007; Schaeffer et al., 2004). No studies, to our knowledge, have investigated the effect that activated astroglia have on neuronal mitochondrial biogenesis and specifically TFAM. Our findings are consistent with a study that found that IL-1β and TNFα treated astrocytes are neurotoxic through glutamate production (Ye et al., 2013). Our findings also indicate for the first time that pre-treating astroglial cultures with WIN in vitro can reduce the neurotoxicity and prevent loss of TFAM signal after exposure to CM from IL-1β-treated astroglia. This finding could have implications for HAND given the clear reduction of neuronal TFAM observed in postmortem brain tissues from HIV+ people with neurocognitive impairment. Future studies are needed to identify the factors secreted from (neurotoxic factors) or sequestered by (bioenergetic substrate) astroglia that result in TFAM down regulation in neurons and whether that precedes loss of MAP2 staining and dendritic simplification.
The cannabinoids ∆9− tetrahydrocannabinol (THC) and cannabidiol (CBD) have shown neuroprotective and anti-inflammatory properties and also interact with PPAR receptors (reviewed in (Maroon and Bost, 2018)). THC has been shown to increase transcriptional activity of PPARα (Takeda et al., 2014) and selective antagonists of PPARα blocked THC-induced effects (Fishbein-Kaminietsky et al., 2014). THC and CBD bind directly to PPARγ (Granja et al., 2012) and induce PPARγ transcriptional activity (O’Sullivan and Kendall, 2010; O’Sullivan et al., 2009; O’Sullivan et al., 2005). These studies suggest that THC and CBD may also serve as probes to better understand neurodegenerative mechanisms involving inflammation and therapeutic strategies targeting PPARs.
This study is limited in that the findings in the postmortem human brain tissues are largely associative and the mechanisms underlying reduced neuronal TFAM have not been identified. It is also noteworthy that while the postmortem tissues analyzed here were from cases that were exposed to ART, regimens change rapidly as newer drugs have come to market. Also, many of these cases may have lived with HIV before the wide-spread implementation of combination ART, which is not the situation for many people currently living with HIV and the findings must be viewed in this context. However, in vitro models showed that astroglia activated by IL-1β can induce changes similar to those observed in the human tissues (reduced neuronal TFAM). Importantly, other brain cells, mainly microglia, are involved in neuroimmune signaling in the brain and this study only focuses on the role of astroglia. While these studies do implicate PPARα as a mediator of the anti-inflammatory activity of WIN, how WIN modulates metabolic and mitochondrial activity in astroglia is still unknown. This mechanism will be important when developing therapeutics targeting metabolic function in immune activated astroglia.
Our study identifies alterations in mitochondrial biogenesis as a potential mechanism contributing to the neuropathogenesis of HAND in people exposed to ART. Importantly, these shifts in metabolic activity may be cell dependent and indicate that therapies designed to target PPARα in astroglia may be neuroprotective HAND patients. These findings support the recent interest in astroglia as mediators of neurodegenerative diseases (Clarke et al., 2018; Hamby and Sofroniew, 2010). Future studies are needed to develop therapies aimed at astroglia to reduce neuroinflammation and restore immunometabolic balance in the brain.
Supplementary Material
Highlights.
Mitochondrial biogenesis is reduced in brains of HIV+ cases with neurocognitive impairment compared to cognitive normal controls from cART era
Transcription factor A, mitochondrial colocalization with neurons is reduced but colocalization with astroglia is increased in HIV+ brains with neurocognitive impairment
IL-1β increases PGC-1α expression, oxygen consumption rate and extracellular acidification rate in human astroglia
An aminoalkylindole derivative, WIN 55–212-2, blocks IL-1β-induced oxygen consumption and glycolysis in astroglia in the presence of a CB1 antagonist
WIN 55–212-2 blockage of IL-1β-induced inflammatory gene expression in human astroglia is dependent upon PPARα
Acknowledgements
This work was supported by National Institutes of Health (NIH) grants MH115819 (JAF), NS105177 (JAF), MH105319 (CA) and MH062512 (IG) from the National Institute on Mental Health (NIMH) and DA026306 (IG) from National Institute on Drug Abuse. This publication was also made possible from NIH funding through the NIMH and National Institute on Neurological Diseases and Stroke by the following grants:
Manhattan HIV Brain Bank (MHBB): U24MH100931
Texas NeuroAIDS Research Center (TNRC): U24MH100930
National Neurological AIDS Bank (NNAB): U24MH100929
California NeuroAIDS Tissue Network (CNTN): U24MH100928
Data Coordinating Center (DCC): U24MH100925
Its contents are solely the responsibility of the authors and do not necessarily represent the official view of the NNTC or NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguirre-Rueda D, et al. , 2015. WIN 55,212–2, agonist of cannabinoid receptors, prevents amyloid beta1–42 effects on astrocytes in primary culture. PLoS One 10, e0122843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberini CM, et al. , 2018. Astrocyte glycogen and lactate: New insights into learning and memory mechanisms. Glia 66, 1244–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antinori A, et al. , 2007. Updated research nosology for HIV-associated neurocognitive disorders. Neurology 69, 1789–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avdoshina V, et al. , 2016. The HIV Protein gp120 Alters Mitochondrial Dynamics in Neurons. Neurotox Res 29, 583–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avraham HK, et al. , 2014. The cannabinoid CB(2) receptor agonist AM1241 enhances neurogenesis in GFAP/Gp120 transgenic mice displaying deficits in neurogenesis. Br J Pharmacol 171, 468–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonavia R, et al. , 2001. HIV-1 Tat causes apoptotic death and calcium homeostasis alterations in rat neurons. Biochem Biophys Res Commun 288, 301–8. [DOI] [PubMed] [Google Scholar]
- Boulias K, et al. , 2016. An Epigenetic Clock Measures Accelerated Aging in Treated HIV Infection. Mol Cell 62, 153–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calkins MJ, et al. , 2011. Impaired mitochondrial biogenesis, defective axonal transport of mitochondria, abnormal mitochondrial dynamics and synaptic degeneration in a mouse model of Alzheimer’s disease. Hum Mol Genet 20, 4515–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casula M, et al. , 2005. Infection with HIV-1 induces a decrease in mtDNA. J Infect Dis 191, 1468–71. [DOI] [PubMed] [Google Scholar]
- Chaturvedi RK, Flint Beal M, 2013. Mitochondrial diseases of the brain. Free Radic Biol Med 63, 1–29. [DOI] [PubMed] [Google Scholar]
- Chelune GJ, Baer RA, 1986. Developmental norms for the Wisconsin Card Sorting test. J Clin Exp Neuropsychol 8, 219–28. [DOI] [PubMed] [Google Scholar]
- Chelune GJ, Heaton RK, & Lehman RA, 1986. Neuropsychological and personality correlates of patients’ complaints of disability
- Clarke LE, et al. , 2018. Normal aging induces A1-like astrocyte reactivity. Proc Natl Acad Sci U S A 115, E1896–E1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran NM, et al. , 2005. The synthetic cannabinoid R(+)WIN 55,212–2 inhibits the interleukin-1 signaling pathway in human astrocytes in a cannabinoid receptor-independent manner. J Biol Chem 280, 35797–806. [DOI] [PubMed] [Google Scholar]
- Dalakas MC, et al. , 2001. Mitochondrial alterations with mitochondrial DNA depletion in the nerves of AIDS patients with peripheral neuropathy induced by 2’3’-dideoxycytidine (ddC). Lab Invest 81, 1537–44. [DOI] [PubMed] [Google Scholar]
- Drew PD, et al. , 2006. Peroxisome proliferator-activated receptor agonist regulation of glial activation: relevance to CNS inflammatory disorders. Neurochem Int 49, 183–9. [DOI] [PubMed] [Google Scholar]
- Escriva H, et al. , 1999. Expression of mitochondrial genes and of the transcription factors involved in the biogenesis of mitochondria Tfam, NRF-1 and NRF-2, in rat liver, testis and brain. Biochimie 81, 965–71. [DOI] [PubMed] [Google Scholar]
- Fields J, et al. , 2015a. HIV-1 Tat alters neuronal autophagy by modulating autophagosome fusion to the lysosome: implications for HIV-associated neurocognitive disorders. J Neurosci 35, 1921–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields J, et al. , 2013. Age-dependent molecular alterations in the autophagy pathway in HIVE patients and in a gp120 tg mouse model: reversal with beclin-1 gene transfer. J Neurovirol 19, 89–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields J, Ghorpade A, 2012. C/EBPbeta regulates multiple IL-1beta-induced human astrocyte inflammatory genes. J Neuroinflammation 9, 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields JA, et al. , 2015b. HIV alters neuronal mitochondrial fission/fusion in the brain during HIV-associated neurocognitive disorders. Neurobiol Dis [DOI] [PMC free article] [PubMed]
- Fishbein-Kaminietsky M, et al. , 2014. Ultralow doses of cannabinoid drugs protect the mouse brain from inflammation-induced cognitive damage. J Neurosci Res 92, 1669–77. [DOI] [PubMed] [Google Scholar]
- Granja AG, et al. , 2012. A cannabigerol quinone alleviates neuroinflammation in a chronic model of multiple sclerosis. J Neuroimmune Pharmacol 7, 1002–16. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Rodriguez A, et al. , 2018. Localization of the cannabinoid type-1 receptor in subcellular astrocyte compartments of mutant mouse hippocampus. Glia 66, 1417–1431. [DOI] [PubMed] [Google Scholar]
- Hamby ME, Sofroniew MV, 2010. Reactive astrocytes as therapeutic targets for CNS disorders. Neurotherapeutics 7, 494–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton RK, et al. , 2010. HIV-associated neurocognitive disorders persist in the era of potent antiretroviral therapy: CHARTER Study. Neurology 75, 2087–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton RK, et al. , 2011. HIV-associated neurocognitive disorders before and during the era of combination antiretroviral therapy: differences in rates, nature, and predictors. J Neurovirol 17, 3–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CY, et al. , 2012. HIV-1 Vpr triggers mitochondrial destruction by impairing Mfn2-mediated ER-mitochondria interaction. PLoS One 7, e33657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi Y, et al. , 2017. Single Cell-Based Vector Tracing in Patients with ADA-SCID Treated with Stem Cell Gene Therapy. Mol Ther Methods Clin Dev 6, 8–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janabi N, 2002. Selective inhibition of cyclooxygenase-2 expression by 15-deoxy-Delta(12,14)(12,14)-prostaglandin J(2) in activated human astrocytes, but not in human brain macrophages. J Immunol 168, 4747–55. [DOI] [PubMed] [Google Scholar]
- Jiang T, Cadenas E, 2014. Astrocytic metabolic and inflammatory changes as a function of age. Aging Cell 13, 1059–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kau M., et al. , 2001. Pathways to neuronal injury and apoptosis in HIV-associated dementia. Nature 410, 988–94. [DOI] [PubMed] [Google Scholar]
- Khakh BS, Sofroniew MV, 2015. Diversity of astrocyte functions and phenotypes in neural circuits. Nat Neurosci 18, 942–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitayama H, et al. , 2008. Human immunodeficiency virus type 1 Vpr inhibits axonal outgrowth through induction of mitochondrial dysfunction. J Virol 82, 2528–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozela E, et al. , 2017. Modulation of Astrocyte Activity by Cannabidiol, a Nonpsychoactive Cannabinoid. Int J Mol Sci 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruman II, et al. , 1998. HIV-1 protein Tat induces apoptosis of hippocampal neurons by a mechanism involving caspase activation, calcium overload, and oxidative stress. Exp Neurol 154, 276–88. [DOI] [PubMed] [Google Scholar]
- Lawton MP, Brody EM, 1969. Assessment of older people: self-maintaining and instrumental activities of daily living. Gerontologist 9, 179–86. [PubMed] [Google Scholar]
- Ledergerber B, Battegay M, 2014. [Epidemiology of HIV]. Ther Umsch 71, 437–41. [DOI] [PubMed] [Google Scholar]
- Lee YS, et al. , 2018. An Integrated View of Immunometabolism. Cell 172, 22–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AJ, et al. , 2016. Multilevel analysis of neuropathogenesis of neurocognitive impairment in HIV. J Neurovirol 22, 431–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, et al. , 2007. Activation of peroxisome proliferator-activated receptor-alpha prevents glycogen synthase 3beta phosphorylation and inhibits cardiac hypertrophy. FEBS Lett 581, 3311–6. [DOI] [PubMed] [Google Scholar]
- Liddelow SA, Barres BA, 2017. Reactive Astrocytes: Production, Function, and Therapeutic Potential. Immunity 46, 957–967. [DOI] [PubMed] [Google Scholar]
- Liddelow SA, et al. , 2017. Neurotoxic reactive astrocytes are induced by activated microglia. Nature 541, 481–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HT, et al. , 2016. Application of Droplet Digital PCR for Estimating Vector Copy Number States in Stem Cell Gene Therapy. Hum Gene Ther Methods 27, 197–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamik MK, et al. , 2011. HIV-1 and IL-1beta regulate astrocytic CD38 through mitogen-activated protein kinases and nuclear factor-kappaB signaling mechanisms. J Neuroinflammation 8, 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchalant Y, et al. , 2008. Cannabinoid receptor stimulation is anti-inflammatory and improves memory in old rats. Neurobiol Aging 29, 1894–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchalant Y, et al. , 2007. Anti-inflammatory property of the cannabinoid agonist WIN-55212–2 in a rodent model of chronic brain inflammation. Neuroscience 144, 1516–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroon J, Bost J, 2018. Review of the neurological benefits of phytocannabinoids. Surg Neurol Int 9, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Moreno AM, et al. , 2012. Prolonged oral cannabinoid administration prevents neuroinflammation, lowers beta-amyloid levels and improves cognitive performance in Tg APP 2576 mice. J Neuroinflammation 9, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur JC, et al. , 2010. Human immunodeficiency virus-associated neurocognitive disorders: Mind the gap. Ann Neurol 67, 699–714. [DOI] [PubMed] [Google Scholar]
- Nair RR, et al. , 2018. Impaired mitochondrial fatty acid synthesis leads to neurodegeneration in mice. J Neurosci [DOI] [PMC free article] [PubMed]
- Natarajaseenivasan K, et al. , 2018. Astrocytic metabolic switch is a novel etiology for Cocaine and HIV-1 Tat-mediated neurotoxicity. Cell Death Dis 9, 415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nooka S, Ghorpade A, 2017. HIV-1-associated inflammation and antiretroviral therapy regulate astrocyte endoplasmic reticulum stress responses. Cell Death Discov 3, 17061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan SE, Kendall DA, 2010. Cannabinoid activation of peroxisome proliferator-activated receptors: potential for modulation of inflammatory disease. Immunobiology 215, 611–6. [DOI] [PubMed] [Google Scholar]
- O’Sullivan SE, et al. , 2009. Time-dependent vascular effects of Endocannabinoids mediated by peroxisome proliferator-activated receptor gamma (PPARgamma). PPAR Res 2009, 425289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan SE, et al. , 2005. Novel time-dependent vascular actions of Delta9-tetrahydrocannabinol mediated by peroxisome proliferator-activated receptor gamma. Biochem Biophys Res Commun 337, 824–31. [DOI] [PubMed] [Google Scholar]
- Parikh NI, et al. , 2015. Lipoprotein concentration, particle number, size and cholesterol efflux capacity are associated with mitochondrial oxidative stress and function in an HIV positive cohort. Atherosclerosis 239, 50–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pautz A, et al. , 2010. Regulation of the expression of inducible nitric oxide synthase. Nitric Oxide 23, 75–93. [DOI] [PubMed] [Google Scholar]
- Payandemehr B, et al. , 2015. Involvement of PPAR receptors in the anticonvulsant effects of a cannabinoid agonist, WIN 55,212–2. Prog Neuropsychopharmacol Biol Psychiatry 57, 140–5. [DOI] [PubMed] [Google Scholar]
- Ramirez BG, et al. , 2005. Prevention of Alzheimer’s disease pathology by cannabinoids: neuroprotection mediated by blockade of microglial activation. J Neurosci 25, 1904–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repunte-Canonigo V, et al. , 2014. Gene expression changes consistent with neuroAIDS and impaired working memory in HIV-1 transgenic rats. Mol Neurodegener 9, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockenstein E, et al. , 2001. Early formation of mature amyloid-beta protein deposits in a mutant APP transgenic model depends on levels of Abeta(1–42). J Neurosci Res 66, 573–82. [DOI] [PubMed] [Google Scholar]
- Roy Chowdhury SK, et al. , 2012. Impaired adenosine monophosphate-activated protein kinase signalling in dorsal root ganglia neurons is linked to mitochondrial dysfunction and peripheral neuropathy in diabetes. Brain 135, 1751–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sama MA, et al. , 2008. Interleukin-1beta-dependent signaling between astrocytes and neurons depends critically on astrocytic calcineurin/NFAT activity. J Biol Chem 283, 21953–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer PJ, et al. , 2004. Calcineurin and calcium/calmodulin-dependent protein kinase activate distinct metabolic gene regulatory programs in cardiac muscle. J Biol Chem 279, 39593–603. [DOI] [PubMed] [Google Scholar]
- Sheng B, et al. , 2012. Impaired mitochondrial biogenesis contributes to mitochondrial dysfunction in Alzheimer’s disease. J Neurochem 120, 419–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng WS, et al. , 2005. Synthetic cannabinoid WIN55,212–2 inhibits generation of inflammatory mediators by IL-1beta-stimulated human astrocytes. Glia 49, 211–9. [DOI] [PubMed] [Google Scholar]
- Sofroniew MV, 2015. Astrocyte barriers to neurotoxic inflammation. Nat Rev Neurosci 16, 249–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer B, et al. , 2009. Beclin 1 gene transfer activates autophagy and ameliorates the neurodegenerative pathology in alpha-synuclein models of Parkinson’s and Lewy body diseases. The Journal of neuroscience : the official journal of the Society for Neuroscience 29, 13578–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tain LS, et al. , 2009. Rapamycin activation of 4E-BP prevents parkinsonian dopaminergic neuron loss. Nat Neurosci 12, 1129–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S, et al. , 2014. Delta(9)-THC modulation of fatty acid 2-hydroxylase (FA2H) gene expression: possible involvement of induced levels of PPARalpha in MDA-MB-231 breast cancer cells. Toxicology 326, 18–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazzi E, et al. , 2014. Brain inflammation is a common feature of HIV-infected patients without HIV encephalitis or productive brain infection. Curr HIV Res 12, 97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teodorof-Diedrich C, Spector SA, 2018. Human Immunodeficiency Virus Type 1 gp120 and Tat Induce Mitochondrial Fragmentation and Incomplete Mitophagy in Human Neurons. J Virol 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangaraj A, et al. , 2018. HIV-1 TAT-mediated microglial activation: role of mitochondrial dysfunction and defective mitophagy. Autophagy 14, 1596–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S, et al. , 2009. Mitochondria influence Fas expression in gp120-induced apoptosis of neuronal cells. Int J Neurosci 119, 157–65. [DOI] [PubMed] [Google Scholar]
- Ton H, Xiong H, 2013. Astrocyte Dysfunctions and HIV-1 Neurotoxicity. J AIDS Clin Res 4, 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tower DB, Young OM, 1973. The activities of butyrylcholinesterase and carbonic anhydrase, the rate of anaerobic glycolysis, and the question of a constant density of glial cells in cerebral cortices of various mammalian species from mouse to whale. J Neurochem 20, 269–78. [DOI] [PubMed] [Google Scholar]
- Van den Bossche J, et al. , 2017. Macrophage Immunometabolism: Where Are We (Going)? Trends Immunol 38, 395–406. [DOI] [PubMed] [Google Scholar]
- van der Walt JM, et al. , 2003. Mitochondrial polymorphisms significantly reduce the risk of Parkinson disease. Am J Hum Genet 72, 804–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Var SR, et al. , 2016. Mitochondrial injury and cognitive function in HIV infection and methamphetamine use. AIDS 30, 839–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura-Clapier R, et al. , 2008. Transcriptional control of mitochondrial biogenesis: the central role of PGC-1alpha. Cardiovasc Res 79, 208–17. [DOI] [PubMed] [Google Scholar]
- Vitomirov A, et al. , 2017. Random shearing as an alternative to digestion for mitochondrial DNA processing in droplet digital PCR. Mitochondrion 32, 16–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DB, et al. , 2013. Declines in Drp1 and parkin expression underlie DNA damage-induced changes in mitochondrial length and neuronal death. J Neurosci 33, 1357–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, et al. , 2014. Mitochondrial biogenesis of astrocytes is increased under experimental septic conditions. Chin Med J (Engl) 127, 1837–42. [PubMed] [Google Scholar]
- Woods SP, et al. , 2004. Interrater reliability of clinical ratings and neurocognitive diagnoses in HIV. J Clin Exp Neuropsychol 26, 759–78. [DOI] [PubMed] [Google Scholar]
- Xu J, et al. , 2006. Peroxisome proliferator-activated receptor-alpha and retinoid X receptor agonists inhibit inflammatory responses of astrocytes. J Neuroimmunol 176, 95–105. [DOI] [PubMed] [Google Scholar]
- Ye L, et al. , 2013. IL-1beta and TNF-alpha induce neurotoxicity through glutamate production: a potential role for neuronal glutaminase. J Neurochem 125, 897–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, et al. , 2012. The early events of Alzheimer’s disease pathology: from mitochondrial dysfunction to BDNF axonal transport deficits. Neurobiol Aging 33, 1122 e1–10. [DOI] [PubMed] [Google Scholar]
- Yin F, et al. , 2016. Energy metabolism and inflammation in brain aging and Alzheimer’s disease. Free Radic Biol Med 100, 108–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuchner S, et al. , 2004. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat Genet 36, 449–51. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.