Abstract
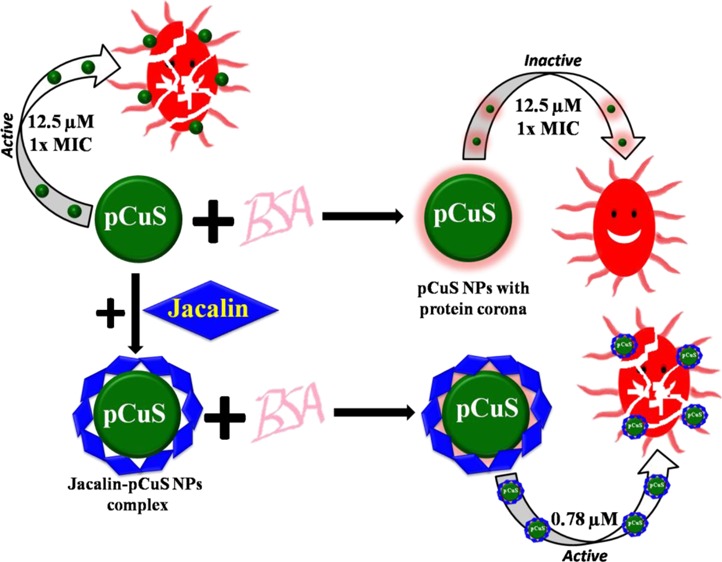
Herein, we report a new strategy based on jacalin functionalization to diminish the impact of biological fluids in the antibacterial applications of nanoparticles (NPs). Precoating pectin-capped copper sulfide NPs (pCuS) with bovine serum albumin produced a protein corona, which affects the antibacterial activity of pCuS. It was found that the minimum inhibitory concentration (MIC) increases fourfold because of the formation of the protein corona. Interestingly, the pCuS functionalized with jacalin enhance the targeting capabilities through bacterial cell surface glycan recognition with no interference from the protein corona. The MIC of pCuS decreases 16-fold on functionalization with jacalin. Mechanistic studies indicated that the pCuS functionalized with jacalin impede the protein corona interference and induce bacterial cell death by impairing the GSH/reactive oxygen species balance and disrupting the bacteria cell membrane. As a proof of concept, we used a bacteria-infected zebrafish animal model to demonstrate the interference of biological fluids in the antibacterial activity of NPs. Infected zebrafish treated with 1× MIC of pCuS failed to recover from the infection, but 4× MIC rescues the fish. The requirement of a high dose of NPs to treat the infection confirms the interference of biological fluids in nanotherapeutic applications. At the same time, the jacalin–pCuS complex rescues the infected fish at 16-fold lesser MIC. The results obtained from this study suggest that jacalin-mediated NP targeting may have broad implications in the development of future nanomedicine.
Introduction
Nanomaterials are believed to be next generation molecules with huge potential in diverse fields including medicine, catalysis, sensors, and so forth.1−3 The concerns over the nanotoxicity and poorly understood mechanism of action impede their development in health care.4 Moreover, the behavior of nanoparticles (NPs) in biological fluids remains enigmatic. Recent studies suggest that the surfaces of NPs are readily modified by proteins or other biomolecules present in biological fluids.5 The adsorption of the biomolecules leads to the formation of a shell on the NP surfaces, which is called as “corona”. The formation of biomolecular corona confers real physicochemical properties of the NPs in the biological milieu.6,7 Biomolecular corona has a strong influence on targeting efficacy, activity, biodistribution, clearance, and toxicity.8−10 The emerging evidence suggests that the protein coronas formed by the plasma protein affect the uptake and distribution of NPs. Monteiro-Riviere et al. showed that the uptake efficacy of silver NPs by HEK cells was reduced because of the association of human serum albumin, IgG, or transferrin.11 Lesniak et al. demonstrated that silica NPs have a stronger adhesion to the cell membrane and greater internalization efficiency in the absence of serum proteins.12 Hu et al. illustrated that the extremely high protein adsorption ability of graphene oxide nanosheets caused reduced cytotoxicity in the presence of 10% fetal bovine serum.13 Thus, the challenge imposed by the protein corona should be taken into account when evaluating the therapeutic potential of the NPs.
Antibacterial activity of NPs have received a great deal of attention because of their proposed ability to combat multi-drug resistance pathogens.14 To explore NPs as an antibacterial agent, it is important to consider the interference from protein corona and also find a method to mitigate the interference from biological fluids. Silver NPs has been recognized as an antibacterial agent, but the emerging evidence showed that silver has high toxicity as compared to silver sulfide NPs.15,16 Similarly, the cost-effective copper NPs have good antibacterial activity, but are highly toxic when tested in zebrafish.17 Recently, copper sulfide NPs (CuS NPs) have been intensively investigated in various fields including antibacterial, photothermal cancer therapy, biomolecule sensing, and molecular imaging.18 In this study, we have analyzed the ability of pectin-capped CuS NPs (pCuS) to form protein corona through studying the interaction between the NPs and bovine serum albumin (BSA). The choice of BSA is made because of its abundance in the serum and its ability to interact with NPs.19 Fluorescence spectroscopic analysis revealed that the pCuS binds to BSA with good affinities that are comparable to BSA–drug interactions reported in the literature. The NPs’ interaction with BSA results in the formation of protein corona on NPs’ surface. The differences imposed on the NPs’ surface leads to a substantial change in the antibacterial activity of pCuS.19
To mitigate the protein corona interference, we explored functionalization of the NPs with jacalin, a glycan-binding protein isolated from the edible seeds of jackfruit. Jacalin is a 66 kDa tetrameric protein with high specificity for galactose. The sugar specificity and exogenous ligand binding properties of jacalin have been studied extensively.20 Jacalin in complex with anticancer phytomoleules, shikonin, and silver NPs showed minimal requirement of the drug to induce cell death in human chronic myeloid leukemia.21 PEG phthalocyanine-gold NPs conjugated with jacalin have been employed to target specific glycans expressed on HT-29 colon adenocarcinoma cells.22 The glycan recognition properties of jacalin were reported for enhancing the antibacterial activity of N-lauryl tyramine-capped CuS NPs (NLTA-CuS NPs) against drug resistance bacteria.23 Herein, we demonstrated through a battery of biophysical and biochemical methods that the interference from the protein corona on the antibacterial activity of pCuS could be diminished by functionalizing the NPs with jacalin. To the best of our knowledge, this is the first study reporting a novel method based on lectins to curtail the protein corona interference in the antibacterial activity of NPs.
Results and Discussion
Preparation of Pectin-Capped Copper Sulfide NPs
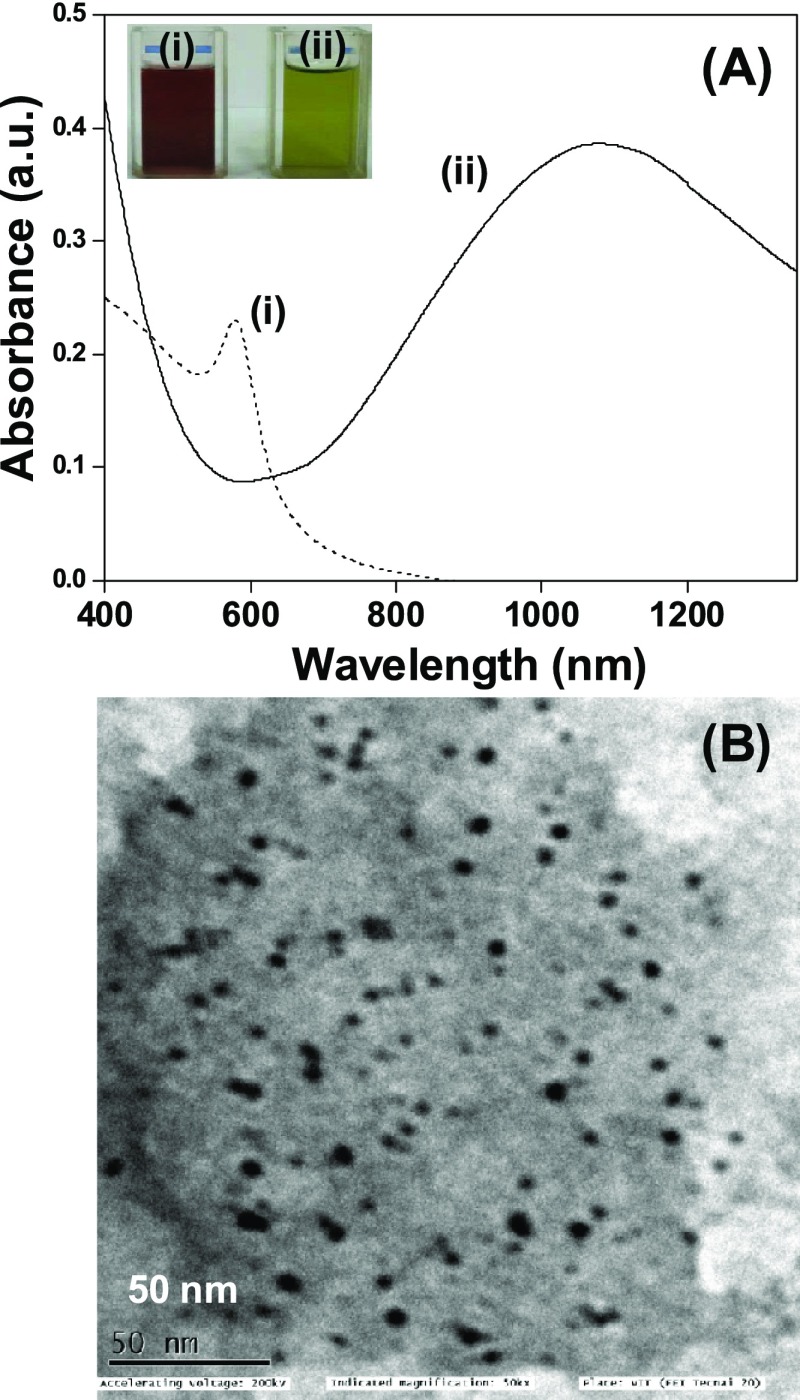
pCuS were synthesized by reacting copper NPs with sodium sulfide. It is seen that the brown color copper NPs turn olive green and show the characteristic broad band of CuS in the near infrared region, peaking at 1080 nm (Figure 1A). The absorption peak was assigned to an electron-acceptor state lying within the band gap.24,25 The loss of surface plasmon resonance maximum at 580 nm of pCuNPs indicates the complete conversion of CuNPs to pCuS. The absence of absorption peaks correspond to the Cu2S phase at 475 nm, suggests that the preparation has high-quality pCuS.26 The powder X-ray diffraction of pCuS is shown in Figure S1A. The diffractions peaks at 2θ values of 21.63°, 27.48°, 29.35°, 31.50°, 32.45°, 47.58°, and 59.23 were assigned to the (004), (101), (102), (103), (006), (107), and (116) reflections. The diffraction pattern is matched with the diffraction library of CuS (JCPDS 74-1234) and indexed for the single covellite phase with a hexagonal crystalline structure. The broadening of the diffraction peaks suggests the formation of nanodimensional particles. Transmission electron microscope images revealed that the particles are spherical with a size range from 2 to 10 nm (Figure 1B). The selected area diffraction patterns account for the (1 0 2) and (1 0 3) lattice planes of a hexagonal structure of pCuS (Figure S1B). Energy-dispersive X-ray analysis revealed the presence of Cu and S in the preparation (Figure S1C).
Figure 1.
(A) UV–visible spectra of (i) Cu NPs and (ii) CuS NPs. The inset corresponds to photographs of (i) Cu NPs and (ii) pCuS. (B) Transmission electron microscopy image of pCuS.
BSA–pCuS Interaction
The capability of BSA to form a protein corona on pCuS surfaces was first evaluated through the interaction studies. It is noted that the intrinsic fluorescence of BSA was quenched by the addition of pCuS, suggesting the interaction between BSA and pCuS (Figure 2).
Figure 2.
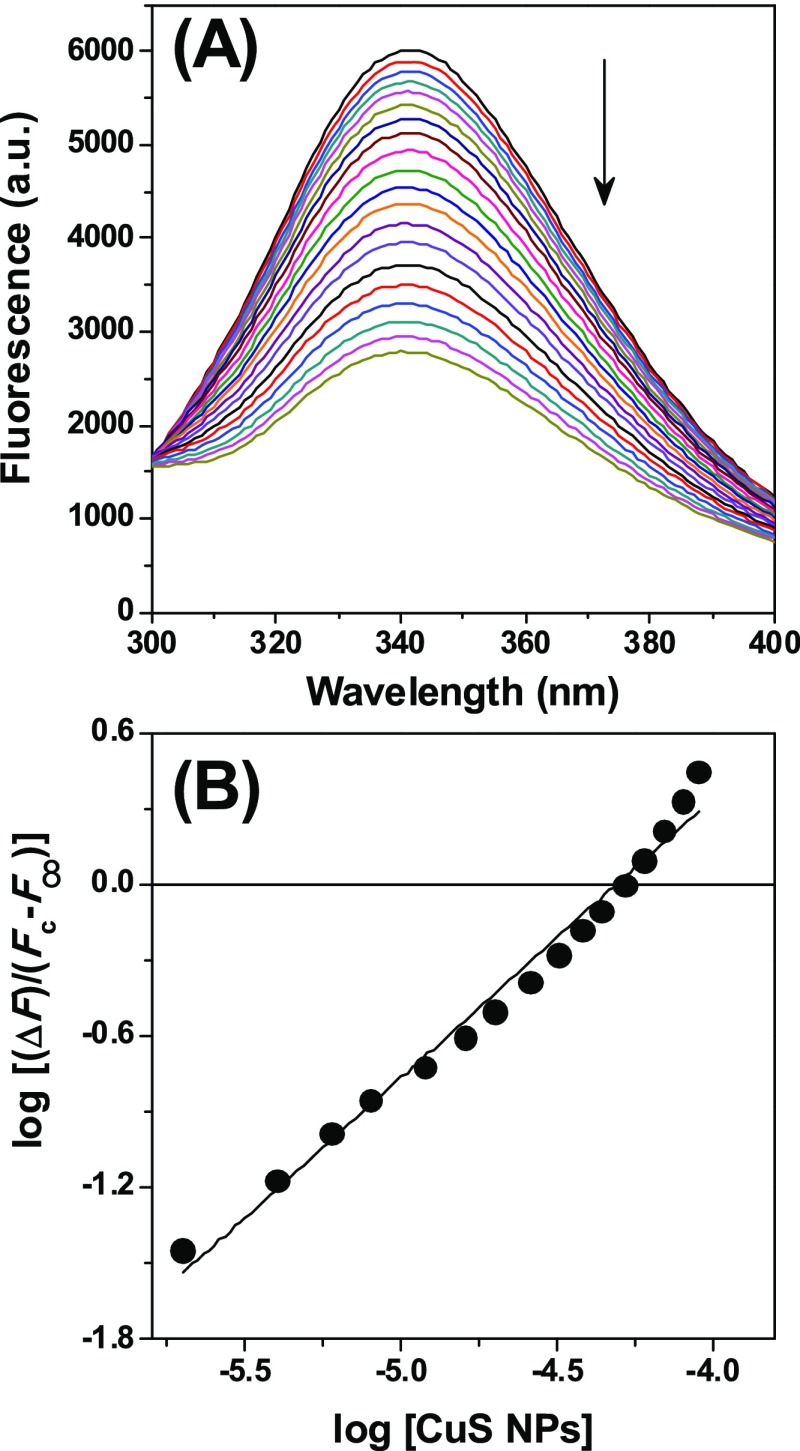
Fluorescence titration. (A) Addition of pCuS to BSA quenches the intrinsic fluorescence. Excitation wavelength = 280 nm. (B) Double logarithmic plot for the interaction of pCuS with BSA. The abscissa intercept of this plot yielded the pKa value of the BSA–pCuS interaction.
The mechanism of quenching may be static or dynamic; thus, we analyzed the quenching data with the Stern–Volmer equation.27
| 1 |
| 2 |
where F0 and Fc are the relative fluorescence intensities of BSA at 340 nm in the absence and presence of pCuS, respectively, Ksv is the Stern–Volmer fluorescence quenching constant, Kq is the biomolecular quenching constant, and τ0 is the average fluorescence lifetime of BSA. From the linear Stern–Volmer plot, Ksv (7.577 × 103 M–1) was determined (Figure S2A). The Kq (1.51 × 1012 L/mol s) calculated for the BSA–pCuS interaction was higher than the Kq (1010 L/mol s) obtained for the diffusion-controlled quenching process. These results suggest that the quenching process is static and a nonfluorescent complex is likely formed between BSA and pCuS. From the quenching study, the binding constant (Ka) was calculated by the following expression.28,29
| 3 |
where F∞ is the change in fluorescence intensity at infinite pCuS concentration, n is the number of binding sites, and Ka is the association constant. F∞ was obtained from the ordinate intercept of the plot of F0/ΔF∞ versus 1/[pCuS]. A double logarithmic plot for the interaction of pCuS with BSA is shown in Figure 2B. The Ka calculated for the BSA–pCuS interaction is 2.04 (±0.43) × 104 M–1, which is comparable to those observed generally for BSA–drug interactions.30,31
The binding of BSA with pCuS was further characterized by measuring the hydrodynamic diameter and zeta potential of the BSA–pCuS complex and pCuS. Compared to pCuS, the hydrodynamic diameter of the BSA–pCuS complex increases from 122.6 to 162.4 nm (Figure S3). The increase in the size is attributed to the presence of BSA coating (protein corona) on the NPs. The negative zeta potential of pCuS (−21.3 mV) and the BSA–pCuS complex (−17.4 mV) suggest that the surface were negatively charged (Figure S4), though the BSA–pCuS complex was lesser negative than pCuS. However, it is clear from this study that the formation of BSA corona on the NPs’ surface had no significant differences on the surface charge of the NPs.
BSA Interference on the Antibacterial Activity of pCuS
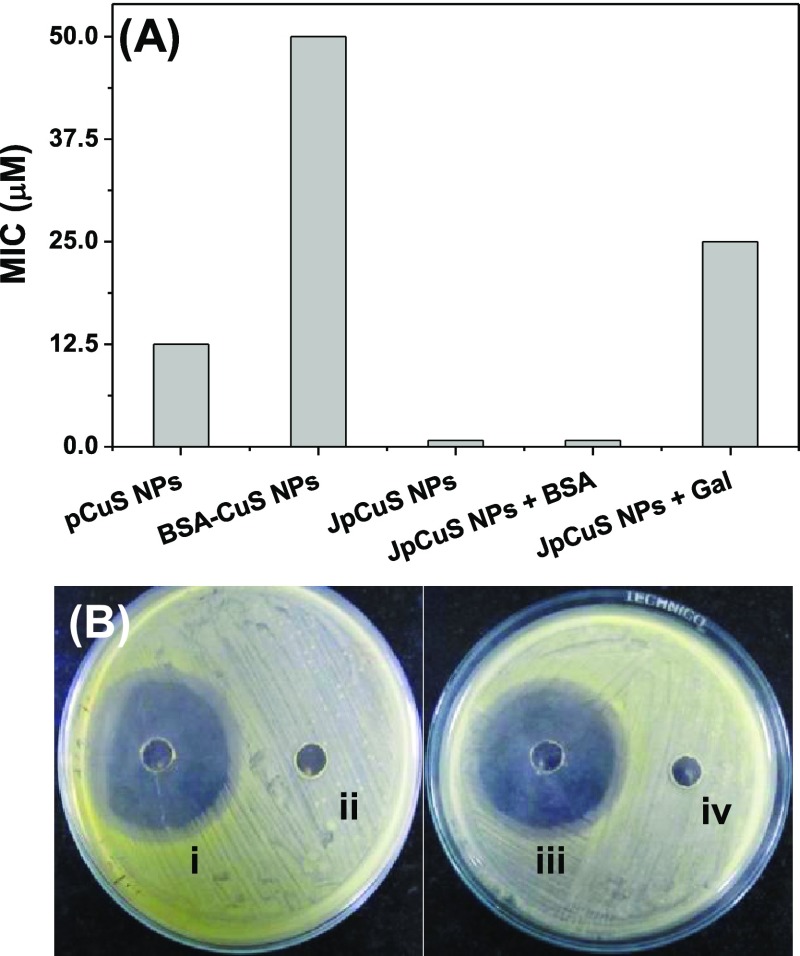
Having studied the protein corona formation with pCuS, we set out to determine the minimum inhibitory concentration (MIC) of pCuS to kill Gram-negative (Escherichia coli, Pseudomonas aeruginosa) and Gram-positive (Bacillus subtilis, Staphylococcus aureus) bacteria by resazurin microtiter assay (REMA). In this method, live cells appear pink in color because of the reduction of resazurin to resarufin, whereas dead cells appear blue in color because of lack of respiration.32 From Figure 3A, the MIC of pCuS required to kill the bacteria was determined as 12.5 μM. Strikingly, the addition of BSA to pCuS increases the MIC four-fold from 12.5 to 50 μM, which indicates that the protein corona formed by BSA interfered with the antibacterial activity. A further evidence for the protein corona interference was evaluated by the conventional zone of inhibition (ZOI) assay. Figure 3B shows the ZOI around the well loaded with 12.5 μM pCuS, but there is no zone formed around the well loaded with the BSA–pCuS complex. The diameter of the zone formed around the well treated alone with pCuS reflects the inhibition of bacteria growth. These results are consistent with REMA and indicate that the antibacterial activity of pCuS was hindered by the formation of BSA corona.
Figure 3.
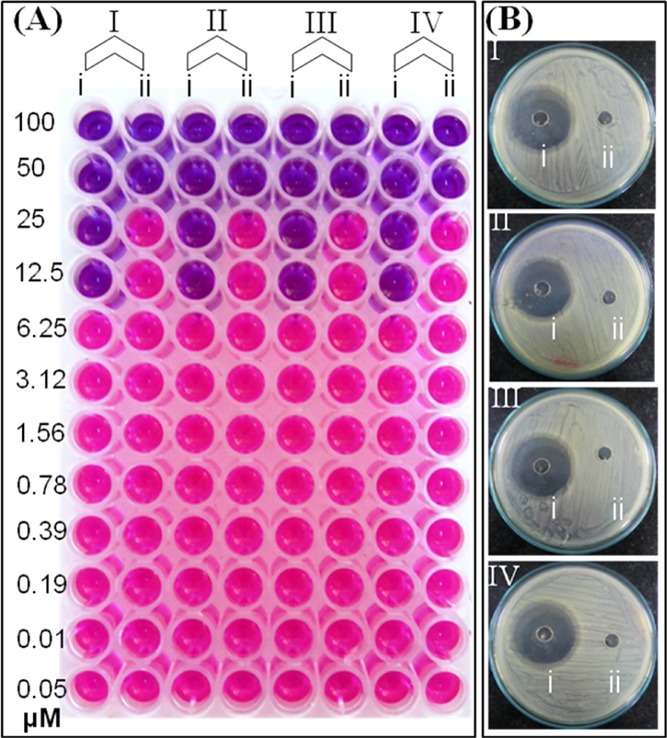
Antibacterial activity of pCuS. (A) MIC was discerned from REMA. Blue color indicate dead cells, pink color denotes live cells, where (i)—pCuS and (ii)—BSA + pCuS. (B) Zone of inhibition assay was performed at 12.5 μM pCuS, where (i)—pCuS and (ii)—BSA + pCuS. I—E. coli, II—P. aeruginosa, III—B. subtilis, and IV—S. aureus. BSA—50 μM; jacalin—50 μM.
Jacalin to Mitigate Protein Corona Interference
Previous studies showed that jacalin improves the antibacterial activity of NPs through recognizing bacterial cell surface glycan.23 Thus, we hypothesized that jacalin could also diminish the interference from serum proteins. To test this hypothesis, we evaluated the interaction between jacalin and pCuS using the intrinsic fluorescence of jacalin. The titration of jacalin with pCuS is shown in Figure S5A. The observed fluorescence quenching caused by pCuS was attributed to the formation of the non-fluorescent jacalin–pCuS complex. Analyzing the quenching data by the Stern–Volmer plot yielded the Kq for the jacalin–pCuS interaction as 1.38 × 1013 L/mol s, which suggests that the quenching mechanism is static [Figure S5B]. Using eq 3, Ka calculated for the binding of jacalin and pCuS is 1.91 (±0.62) × 104 M–1 [Figure S5C], which is comparable to those observed for jacalin–sugar interaction and jacalin interaction with other ligands.20 The interference of pCuS with the glycan recognition site of jacalin was evaluated by estimating the binding constant in the presence of specific sugar, galactose [Figure S6]. The Ka value derived for the binding of jacalin and pCuS in the presence of galactose is 1.41 (±0.36) × 104 M–1, which is in closer agreement with the Ka value estimated without galactose. These results suggest that jacalin forms a stable complex with pCuS without affecting the glycan recognition site. Having demonstrated the complex formation between pCuS and jacalin without affecting the glycan binding site, we set out to determine the influence of BSA on the MIC of jacalin–pCuS (JpCuS) against Gram-positive and Gram-negative bacteria by REMA. The MIC of pCuS was determined in the presence of varying concentrations of jacalin. It was noted that, about 50 μM jacalin is sufficient to get the lowest MIC (Figure S7). The MIC of JpCuS against E. coli is 0.78 μM, which is much lesser than the uncomplex pCuS, indicating that the glycan recognition nature of jacalin improves the antibacterial activity of pCuS at very low concentrations (Figures 4A and S8). It is noteworthy that the MIC of JpCuS was not affected by the presence of BSA, suggesting that jacalin diminished the serum protein interference in the antibacterial activity of NPs. However, blocking the sugar binding site of jacalin with galactose reverses the MIC from 0.78 to 25 μM, indicating the importance of a galactose-binding site of jacalin for bacterial recognition.23
Figure 4.
Representative antibacterial activity of JpCuS against E. coli. (A) MIC discerned from REMA, and (B) ZOI performed at (i) 12.5 μM JpCuS, (ii) BSA + 12.5 μM pCuS, (iii) BSA + 12.5 μM JpCuS, and (iv) 12.5 μM JpCuS + galactose. As noted, BSA has no interference with JpCuS, but galactose interferes with the antibacterial activity of JpCuS. BSA—50 μM; jacalin—50 μM.
Additionally, the ZOI experiment showed a nearly equal zone around the well loaded with either JpCuS or JpCuS containing BSA (Figure 4B). Unlike JpCuS, the galactose-containing JpCuS were unable to inhibit the proliferation of E. coli at the MIC of pCuS (Figure 4B), complementing the results of REMA. A similar trend was observed for P. aeruginosa, B. subtilis, and S. aureus with JpCuS. In all cases, the MIC of pCuS was lowered by 16-fold on complexing with jacalin (Table S1). However, the addition of galactose to JpCuS hinders their antibacterial efficacy, as evidenced by the MIC reversal (Table S1). It is noteworthy that the diameter of the zone observed with JpCuS is bigger than pCuS, indicating superior activity [Table 1]. The most reasonable explanation for the higher antibacterial of JpCuS is attributed to the increase in recognizing bacteria cell surface through the jacalin sugar-binding site. Our findings were consistent with the previous study where jacalin-functionalized NLTA-CuS NPs were more toxic to the tested bacterial strains than NLTA-CuS NPs.23 Taken together, our study signifies the importance of jacalin to mitigate the protein corona interference and also derive higher efficacy from pCuS against the tested bacterial strains.
Table 1. ZOI in Diameter (mm).
| bacteria | pCuS | JpCuS | JpCuS + BSA |
|---|---|---|---|
| E. coli | 22 | 36 | 37 |
| P. aeruginosa | 23 | 39 | 39 |
| B. subtilis | 20 | 37 | 38 |
| S. aureus | 19 | 40 | 38 |
Antibacterial Mechanism
To understand the antibacterial mechanism of the pCuS, we first examine the membrane integrity of the bacteria using acridine orange (AO)/propidium iodide (PI) dual staining method. AO is a cell membrane-permeable dye that binds to all nucleic acid and fluoresces green, whereas PI is a membrane impermeable dye that only enters into dead cells with compromised membranes and fluoresces red.33
To analyze the membrane integrity, we treated bacteria with different compositions of NPs at the MIC of pCuS for 3 h, followed by AO/PI staining to examine the integrity of the bacteria membrane. As shown in Figure 5, bacteria without NPs treatment fluoresces green, indicating that the membrane was seen as intact and cells are live. The cells treated with 12.5 μM pCuS fluoresces red, suggesting the integrity of the membrane was indeed disturbed and cells are killed by pCuS. However, the cells treated with 12.5 μM pCuS–BSA complexes shows green fluorescence, indicating that the BSA coating on the NPs’ surface passivates the interaction of pCuS and bacterial membranes, leading to decreased membrane disruption. Interestingly, strong red fluorescence was observed from the cells treated with pCuS–jacalin complexes even in the presence of BSA, suggesting that jacalin diminishes the protein corona interference and facilitates pCuS to disrupt the membranes. Additional evidence for the membrane damages was confirmed by a scanning electron microscopy (SEM) study [Figure S9].
Figure 5.
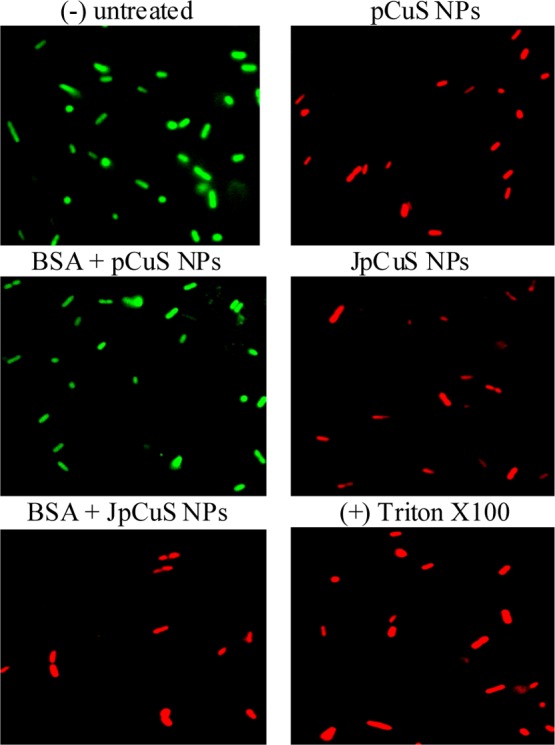
Representative membrane integrity assay. E. coli was treated with NPs, followed by staining with AO/PI and imaged in fluorescence microscopy. Untreated cells serve as negative (−) control and cells treated with Triton X100 served as positive (+) control. Red fluorescence indicates that cells have compromised membranes. BSA—50 μM; jacalin—50 μM. Magnification—100×.
The loss of membrane integrity may not be the only antibacterial mechanism; thus, we examined the oxidative stress caused by pCuS. Oxidative stress refers to excessive generation of reactive oxygen species (ROS) that cause damage to membranes, proteins, and DNA.33 Having observed the membrane damage, we analyzed the ROS production by dichlorofluorescein diacetate (H2DCFDA) method. The NP-induced ROS production by bacterial cells can oxidize the H2DCFDA to green fluorescent dichlorofluorescein (DCF).33 As shown in Figure 6, we only observed strong green fluorescence from bacterial cells treated with pCuS and not from pCuS–BSA, indicating the pCuS-induced ROS generation was prevented by the BSA corona. Remarkably, the addition of jacalin to pCuS overcomes the protein corona interference and generates excess ROS to oxidize H2DCFDA to green fluorescent DCF. In a biological system, ROS generation and clearance are well orchestrated by redox enzymes. Reduced glutathione (GSH) is an important antioxidant tripeptide that protects the cells from oxidative stress by scavenging ROS.34,35 Hence, the total GSH levels in the cells treated with pCuS were quantified by the (dithio-bis(2-nitrobenzoic acid)) method.35 As shown in Figure S10, the amount of GSH depleted largely in the cells treated with pCuS and JpCuS as compared to the cells treated with BSA–pCuS, indicating that the JpCuS trigger extensive ROS generation and deplete the antioxidant GSH level. These observations correlate well with membrane integrity studies where JpCuS shows excellent membrane damaging activities even in the presence of BSA (Figure 6). Collectively, our study demonstrated that JpCuS induces bacterial cell death independent of the interference from protein corona by impairing GSH/ROS balance and damaging the bacteria cell membrane.
Figure 6.
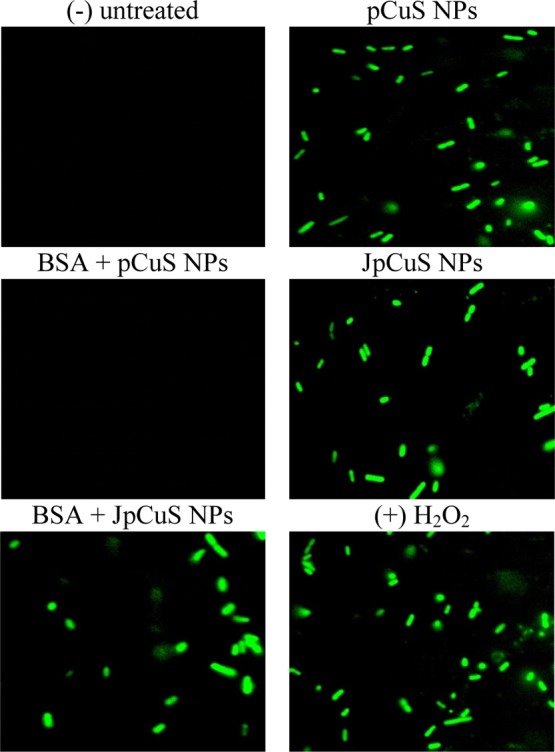
Representative ROS assay. E. coli was treated with NPs, followed by staining with H2DCFDA and imaged in fluorescence microscopy. Untreated cells serve as negative (−) control and cells treated with 12.5 μM H2O2 served as positive (+) control. Green fluorescence indicates the generation of excess ROS by treated cells. BSA—50 μM; jacalin—50 μM. Magnification—100×.
In Vivo Study
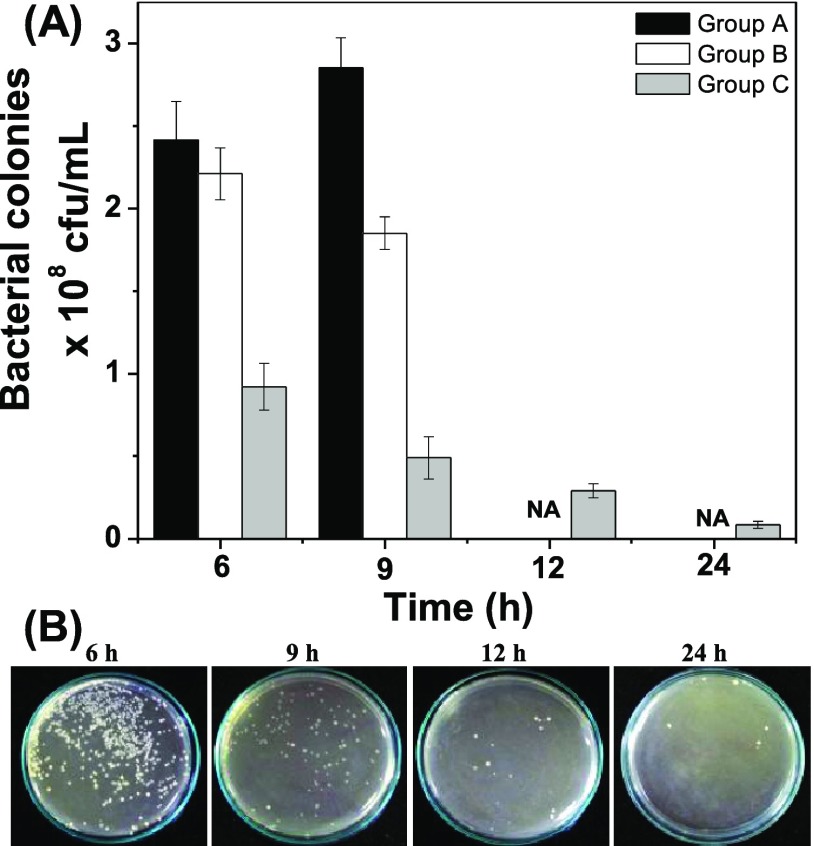
Zebrafish is a good animal model for preliminary drug testing because 80% of zebrafish genome resembles that of humans.36,37 To prove that biological fluids interfere with therapeutic efficacy of NPs, we studied the antibacterial activity of pCuS against E. coli-infected zebrafish. About 30 animals were intramuscularly infected with E. coli and divided into three groups. Group A serves as infected control. After 3 h of infection, Group B and Group C were treated with pCuS. According to REMA data, the MIC of pCuS increase four-fold in the presence of serum albumin. Thus, the Group B infected fish were treated with 1× MIC (12.5 μM) and Group C infected fish were treated with 4× MIC (50 μM). It is noted that Group C fish survived because of treatment, whereas Group A and Group B succumbed to the infection in 8–10 h. Bacterial load in each group at different time points after infection and treatment was determined by the LB-agar plate method. As shown in Figure 7A, a higher number of bacterial colonies were observed in Group A at 6 and 9 h of infection. At the same time, Group B fish treated with 1× MIC showed small decrement in the bacterial colonies at 6 and 9 h, but not as less as Group C. Hence, the severity of the infections in Group A and Group B kills the animal. It is noteworthy that 1× MIC pCuS is insufficient to decrease the bacterial load drastically, suggesting that the biological fluids prevent the action of NPs. However, treatment with high dose (4× MIC) can overcome the interference from biological fluids and rescue the fish by eliminating the bacterial load over time (Figure 7A). These results are consistent with the above in vitro analysis.
Figure 7.
In vivo study. (A) Treatment of E. coli-infected zebrafish with pCuS. Group A—no treatment, Group B—treated with 1× MIC pCuS, and Group C—treated with 4× MIC pCuS. Because of infection, fish died after 9 h in Group A and Group B and not available (NA) for the assay. (B) Treatment of E. coli-infected zebrafish with 0.78 μM JpCuS. Muscle tissues were collected at different time points and cultured on an LB-agar plate. The number of colonies formed in the plate were calculated and reported in cfu.
In order to demonstrate the jacalin ability to diminish the biological fluids’ interference, we treated the infected zebrafish with 0.78 μM of JpCuS. Strikingly, the treated fish survive the infection and restore the animal to normal life. Bacterial colony count assay revealed that the bacterial load was drastically eliminated over time as a result of treatment with JpCuS [Figure 7B]. It is noteworthy that the concentration of JpCuS used in the treatment was 16-fold lesser than the MIC of pCuS, suggesting that the pCuS exhibits superior antibacterial activity as a result of jacalin functionalization.
Conclusions
In conclusion, we found that pCuS have good antibacterial activity against Gram-negative and Gram-positive bacteria. However, their efficiency was impaired in the presence of serum protein. To mitigate the interference of biomolecules like serum protein, we developed a strategy based on the lectin, jacalin. The results suggest that jacalin empowers the pCuS to attach to the bacterial membranes irrespective of the interference from BSA and exert superior antibacterial activity. A mechanistic study revealed that the bacterial cell death was a result of loss of membrane integrity and weakened GSH/ROS balance. Additionally, to demonstrate the antibacterial activity of JCuS NPs in complex biological medium, a bacteria-infected zebrafish model has been proposed, where the biomolecules interference was predicted. It was proven that pCuS is insufficient to rescue the fish at 1× MIC, because of the biomolecular interference. However, JCuS NPs could reduce the bacterial load and rescues the fish at 16-fold lower MIC. The results from this study may open the way toward the design of NPs with lectins to overcome the interference of biomolecules in nanomedicine.
Experimental Section
Synthesis of Copper Sulfide NPs
Briefly, for the synthesis of copper sulfide NPs, 50 mg of pectin and 1 mM CuCl2 was dissolved in 50 mL of double distilled water. Then, 200 μL of ammonium hydroxide solution was added to form blue color copper–ammonia complex. After 5 min, 400 μL of hydrazine hydride was added and the reaction mixture was allowed to stand at room temperature for another 3 h. The appearance of reddish-brown color indicates the formation of copper NPs. After that, 1 mM Na2S was added to react with copper NPs. The solution changed from reddish-brown to olive green color in 3 h, indicating the formation of pCuS.
Characterization
UV–vis spectroscopy of the NPs was monitored by a Jasco-UV–visible–NIR spectrophotometer from 400 to 1300 nm. The crystalline nature of pCuS was characterized by an XRD-Bruker D8 ADVANCE X-ray diffractometer using monochromatic Cu Kα radiation. High-resolution transmission electron microscope (Tecnai, G2 20 S-Twin) image was recorded with NPs deposited onto the copper grid at room temperature. Hydrodynamic size and zeta potential of the samples were measured by a Malvern zetasizer version 6.20.
Interaction Studies
Jacalin for interaction studies was purified from the seeds of jackfruit using the reported procedure and their purity confirmed by SDS-PAGE and their activity by hemeagglutination assay. Fatty acid free BSA was purchased from Sigma-Aldrich. Protein solution was prepared in PBS buffer (10 mM sodium phosphate, 150 mM NaCl, pH 7.4). All the interaction studies were performed with the protein samples of OD280 ≈ 0.1. The intrinsic fluorescence of the proteins was recorded on a JASCO-FP8200 spectrofluorimeter. The excitation was set at 280 nm and the emission was recorded from 300 to 400 nm. Interaction studied was carried out by titrating 3 mL of protein solution with small aliquots of pCuS (1 mM). The change in the fluorescence because of binding was measured after an equilibration period of 2 min. All titrations were performed in triplicate to arrive at average values.
Antibacterial Activity
The sample of pCuS with BSA or jacalin was prepared by incubating the NPs with 50 μM of proteins for 2 h at 4 °C. The strains used for antibacterial studies are Gram negative (E. coli—MTCC723; P. aeruginosa—MTCC1688) and Gram positive (S. aureus—MTCC3160; B. subtilis—MTCC441). Typically, bacteria were grown in LB media and tested as reported in our previous paper.23 The MIC of NPs was assessed from REMA. Briefly, 100 μL of NPs (100 μM) was added into a 96-well plate and serially diluted. About 100 μL of bacteria (1 × 105 cfu/mL) was added to each well and cultured for 24 h at 37 °C. Then, 30 μL of resazurin solution (0.01 % wt/vol) was added to each well and cultured further for 2 h at 37 °C. The fluorescence intensity in each well was measured by fluorescence microplate reader (Biotech, synergy H1, Japan), where the excitation and emission were set at 530 and 580 nm, respectively. The antibacterial activity of the NPs was further evaluated by the ZOI method. Briefly, log phase bacterial culture was swabbed uniformly on LB agar plates using sterile cotton swabs. About 10 mm diameter wells were created using gel puncture and the wells were loaded with defined concentrations of the NPs. The plates were incubated at 37 °C for 12 h. The formation of zones around the well was measured with a ruler.
Membrane Integrity
The integrity of the bacterial membrane was judged by a dual-staining method using the fluorescence probe, AO and PI. Briefly, 1 × 105 cfu/mL was cultured with a defined concentration of NPs for 12 h at 37 °C. Then, the cells were collected by centrifugation at 6000 rpm for 3 min at 4 °C and stained with AO and PI for 2 h in the dark. About 10 μL of the stained cells was placed on a glass slide with a coverslip and imaged under a fluorescence microscope (Nikon Eclipse). Green and red filters were used for AO and PI, respectively. Cells treated with TritonX-100 were used as the positive control.
Generation of Reactive Oxygen Species
ROS generation was assessed by the fluorescence probe, dichlorofluorescein diacetate (DCFH2-DA). Briefly, 1 × 105 cfu/mL was cultured with a defined concentration of NPs for 12 h at 37 °C. Then, the cells were collected by centrifugation at 6000 rpm for 3 min at 4 °C and treated with DCFH2-DA for 30 min in the dark. About 10 μL of the treated cells was placed on a glass slide with a coverslip and imaged under a fluorescence microscope (Nikon Eclipse). A green filter were used to image cells with excess ROS. Cells treated with H2O2 were used as the positive control.
Animal Study
The interference of the biological fluids in the therapeutic activity of the NPs was tested using a zebrafish model. Healthy adult zebrafish (Danio rerio) weighing ∼300 mg were procured from the local aquarium and allowed to acclimatize in the lab environment for 1 week. Bacterial infections to zebrafish were done as reported in our previous paper.37 Briefly, 10 μL of 0.1 OD660nm bacteria cultures was injected intramuscularly and the infection allowed to spread for the next 3 h. Then, the fish was divided into three groups. Group A serves as an infected control, Group B was treated through injecting intramuscularly 10 μL of 12.5 μM pCuS. Group C was treated through injecting intramuscularly 10 μL of 50 μM pCuS. At a defined time point, fish from each group were sacrificed and the muscle tissue was dissected and homogenized in 1 mL of PBS buffer. The homogenate was diluted 104 times and plated in triplicate on sterile LB-agar plates. The number of bacterial colonies formed was counted manually after 24 h of incubation at 37 °C. In parallel to this experiment, about 10 infected fish were treated with 0.78 μM JpCuS. The bacterial load in the muscle tissue was determined as described above. All animal experiments were performed in compliance with the CPCSEA guidelines for laboratory animal facilities (Central Act 26 of 1982) and approved by the Institutional Animal Ethics Committee (CPCSEA-493/SASTRA/IAEC/RPP) of SASTRA Deemed University, India.
Acknowledgments
S.B.S. gratefully acknowledges Teaching Assistantship from SASTRA University.
Glossary
Abbreviations
- pCuS
pectin-capped copper sulfide nanoparticles
- BSA
bovine serum albumin
- JpCuS
complex of jacalin and pCuS
- LB
Luria–Bertani
- MIC
minimum inhibitory concentration
- REMA
resazurin microtiter assay
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.9b01886.
pXRD, SAED pattern, and EDAX of pCuS; Stern–Volmer plot; particle size; zeta potential; binding study; SEM analysis; and GSH assay (PDF)
Author Contributions
S.B.S. and S.V. equally contributed. The paper was written through contributions of all the authors. All the authors have given approval to the final version of the paper.
We thank DST FIST (SB/FST/ETI-331/2013) and the central research facility (R&M/0021/SCBT-007/2012-13), SASTRA University for the infrastructure.
The authors declare no competing financial interest.
Supplementary Material
References
- Elahi N.; Kamali M.; Baghersad M. H. Recent biomedical applications of gold nanoparticles: a review. Talanta 2018, 184, 537–556. 10.1016/j.talanta.2018.02.088. [DOI] [PubMed] [Google Scholar]
- Liu L.; Corma A. Metal Catalysts for Heterogeneous Catalysis: From Single Atoms to Nanoclusters and Nanoparticles. Chem. Rev. 2018, 118, 4981–5079. 10.1021/acs.chemrev.7b00776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G.; Roy I.; Yang C.; Prasad P. N. Nanochemistry and Nanomedicine for Nanoparticle-based Diagnostics and Therapy. Chem. Rev. 2016, 116, 2826–2885. 10.1021/acs.chemrev.5b00148. [DOI] [PubMed] [Google Scholar]
- Sharifi S.; Behzadi S.; Laurent S.; Laird Forrest M.; Stroeve P.; Mahmoudi M. Toxicity of Nanomaterials. Chem. Soc. Rev. 2012, 41, 2323–2343. 10.1039/c1cs15188f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoudi M.; Lynch I.; Ejtehadi M. R.; Monopoli M. P.; Bombelli F. B.; Laurent S. Protein-nanoparticle interactions: opportunities and challenges. Chem. Rev. 2011, 111, 5610–5637. 10.1021/cr100440g. [DOI] [PubMed] [Google Scholar]
- Shannahan J. H.; Lai X.; Ke P. C.; Podila R.; Brown J. M.; Witzmann F. A. Silver nanoparticle protein corona composition in cell culture media. PLoS One 2013, 8, e74001 10.1371/journal.pone.0074001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes-Rius A.; de Puig H.; Kah J. C. Y.; Borros S.; Hamad-Schifferli K. Optimizing the properties of the protein corona surrounding nanoparticles for tuning payload release. ACS Nano 2013, 7, 10066–10074. 10.1021/nn404166q. [DOI] [PubMed] [Google Scholar]
- Salvati A.; Pitek A. S.; Monopoli M. P.; Prapainop K.; Bombelli F. B.; Hristov D. R.; Kelly P. M.; Åberg C.; Mahon E.; Dawson K. A. Transferrin-functionalized nanoparticles lose their targeting capabilities when a biomolecule corona adsorbs on the surface. Nat. Nanotechnol. 2013, 8, 137–143. 10.1038/nnano.2012.237. [DOI] [PubMed] [Google Scholar]
- Mirshafiee V.; Mahmoudi M.; Lou K.; Cheng J.; Kraft M. L. Protein corona significantly reduces active targeting yield. Chem. Commun. 2013, 49, 2557–2559. 10.1039/c3cc37307j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z.-J.; Posati T.; Moyano D. F.; Tang R.; Yan B.; Vachet R. W.; Rotello V. M. The interplay of monolayer structure and serum protein interactions on the cellular uptake of gold nanoparticles. Small 2012, 8, 2659–2663. 10.1002/smll.201200794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro-Riviere N. A.; Samberg M. E.; Oldenburg S. J.; Riviere J. E. Protein binding modulates the cellular uptake of silver nanoparticles into human cells: implications for in vitro to in vivo extrapolations?. Toxicol. Lett. 2013, 220, 286–293. 10.1016/j.toxlet.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesniak A.; Fenaroli F.; Monopoli M. P.; Åberg C.; Dawson K. A.; Salvati A. Effects of the presence or absence of a protein corona on silica nanoparticle uptake and impact on cells. ACS Nano 2012, 6, 5845–5857. 10.1021/nn300223w. [DOI] [PubMed] [Google Scholar]
- Hu W.; Peng C.; Lv M.; Li X.; Zhang Y.; Chen N.; Fan C.; Huang Q. Protein corona-mediated mitigation of cytotoxicity of graphene oxide. ACS Nano 2011, 5, 3693–3700. 10.1021/nn200021j. [DOI] [PubMed] [Google Scholar]
- Wang L.; Hu C.; Shao L. The antimicrobial activity of nanoparticles: present situation and prospects for the future. Int. J. Nanomed. 2017, 12, 1227–1249. 10.2147/ijn.s121956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza L. R. R.; da Silva V. S.; Franchi L. P.; de Souza T. A. J. Toxic and Beneficial Potential of Silver Nanoparticles: The Two Sides of the Same Coin. Adv. Exp. Med. Biol. 2018, 1048, 251–262. 10.1007/978-3-319-72041-8_15. [DOI] [PubMed] [Google Scholar]
- Devi G. P.; Ahmed K. B. A.; Varsha M. K. N. S.; Shrijha B. S.; Lal K. K. S.; Anbazhagan V.; Thiagarajan R. Sulfidation of silver nanoparticle reduces its toxicity in zebrafish. Aquat. Toxicol. 2015, 158, 149–156. 10.1016/j.aquatox.2014.11.007. [DOI] [PubMed] [Google Scholar]
- Dharsana U. S.; Varsha M. K. N. S.; Behlol A. A. K.; Veerappan A.; Thiagarajan R. Sulfidation modulates the toxicity of biogenic copper nanoparticles. RSC Adv. 2015, 5, 30248–30259. 10.1039/c4ra17322h. [DOI] [Google Scholar]
- Goel S.; Chen F.; Cai W. Synthesis and biomedical applications of copper sulfide nanoparticles: from sensors to theranostics. Small 2014, 10, 631–645. 10.1002/smll.201301174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnanadhas D. P.; Ben Thomas M.; Thomas R.; Raichur A. M.; Chakravortty D. Interaction of silver nanoparticles with serum proteins affects their antimicrobial activity in vivo. Antimicrob. Agents Chemother. 2013, 57, 4945–4955. 10.1128/aac.00152-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komath S. S.; Kavitha M.; Swamy M. J. Beyond carbohydrate binding: new directions in plant lectin research. Org. Biomol. Chem. 2006, 4, 973–988. 10.1039/b515446d. [DOI] [PubMed] [Google Scholar]
- Ahmed K. B. A.; Mahapatra S. K.; Raja M. R. C.; Subramaniam S.; Sengan M.; Rajendran N.; Das S. K.; Haldar K.; Roy S.; Sivasubramanian A.; Anbazhagan V. Jacalin-capped silver nanoparticles minimize the dosage use of the anticancer drug, shikonin derivatives, against human chronic myeloid leukemia. RSC Adv. 2016, 6, 18980–18989. 10.1039/c5ra27952f. [DOI] [Google Scholar]
- Obaid G.; Chambrier I.; Cook M. J.; Russell D. A. Targeting the oncofetal Thomsen-Friedenreich disaccharide using jacalin-PEG phthalocyanine gold nanoparticles for photodynamic cancer therapy. Angew. Chem., Int. Ed. Engl. 2012, 51, 6158–6162. 10.1002/anie.201201468. [DOI] [PubMed] [Google Scholar]
- Ahmed K. B. A.; Subramaniyan S. B.; Banu S. F.; Nithyanand P.; Veerappan A. Jacalin-copper sulfide nanoparticles complex enhance the antibacterial activity against drug resistant bacteria via cell surface glycan recognition. Colloids Surf., B 2018, 163, 209–217. 10.1016/j.colsurfb.2017.12.053. [DOI] [PubMed] [Google Scholar]
- Kalanur S. S.; Seo H. Tuning plasmonic properties of CuS thin films via valence band filling. RSC Adv. 2017, 7, 11118–11122. 10.1039/c6ra27076j. [DOI] [Google Scholar]
- Xie Y.; Riedinger A.; Prato M.; Casu A.; Genovese A.; Guardia P.; Sottini S.; Sangregorio C.; Miszta K.; Ghosh S.; Pellegrino T.; Manna L. Copper sulfide nanocrystals with tunable composition by reduction of covellite nanocrystals with Cu+ ions. J. Am. Chem. Soc. 2013, 135, 17630–17637. 10.1021/ja409754v. [DOI] [PubMed] [Google Scholar]
- Haram S. K.; Mahadeshwar A. R.; Dixit S. G. Synthesis and Characterization of Copper Sulfide Nanoparticles in Triton-X 100 Water-in-Oil Microemulsions. J. Phys. Chem. 1996, 100, 5868–5873. 10.1021/jp952391n. [DOI] [Google Scholar]
- Lakowicz J. R.Principles of Fluorescence Spectroscopy, 3rd ed.; Springer: New York, 2006. [Google Scholar]
- Huang D.; Geng F.; Liu Y.; Wang X.; Jiao J.; Yu L. Biomimetic interactions of proteins with functionalized cadmium sulfide quantum dots. Colloids Surf., A 2011, 392, 191–197. 10.1016/j.colsurfa.2011.09.054. [DOI] [Google Scholar]
- Ayaz Ahmed K. B.; Reshma E.; Mariappan M.; Anbazhagan V. Spectroscopic investigation on the interaction of ruthenium complexes with tumor specific lectin, jacalin. Spectrochim. Acta, Part A 2015, 137, 1292–1297. 10.1016/j.saa.2014.09.047. [DOI] [PubMed] [Google Scholar]
- Liu J.; He Y.; Liu D.; He Y.; Tang Z.; Lou H.; Huo Y.; Cao X. Characterizing the binding interaction of astilbin with bovine serum albumin: a spectroscopic study in combination with molecular docking technology. RSC Adv. 2018, 8, 7280–7286. 10.1039/c7ra13272g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wani T. A.; Bakheit A. H.; Zargar S.; Hamidaddin M. A.; Darwish I. A. Spectrophotometric and molecular modelling studies on in vitro interaction of tyrosine kinase inhibitor linifanib with bovine serum albumin. PLoS One 2017, 12, e0176015 10.1371/journal.pone.0176015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengan M.; Subramaniyan S. B.; Arul Prakash S.; Kamlekar R.; Veerappan A. Effective elimination of biofilm formed with waterborne pathogens using copper nanoparticles. Microb. Pathog. 2019, 127, 341–346. 10.1016/j.micpath.2018.12.025. [DOI] [PubMed] [Google Scholar]
- Subramaniyan S. B.; Ramani A.; Ganapathy V.; Anbazhagan V. Preparation of self-assembled platinum nanoclusters to combat Salmonella typhi infection and inhibit biofilm formation. Colloids Surf., B 2018, 171, 75–84. 10.1016/j.colsurfb.2018.07.023. [DOI] [PubMed] [Google Scholar]
- Aquilano K.; Baldelli S.; Ciriolo M. R. Glutathione: new roles in redox signaling for an old antioxidant. Front. Pharmacol. 2014, 5, 196. 10.3389/fphar.2014.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman I.; Kode A.; Biswas S. K. Assay for quantitative determination of glutathione and glutathione disulfide levels using enzymatic recycling method. Nat. Protoc. 2006, 1, 3159–3165. 10.1038/nprot.2006.378. [DOI] [PubMed] [Google Scholar]
- Sullivan C.; Kim C. H. Zebrafish as a model for infectious disease and immune function. Fish Shellfish Immunol. 2008, 25, 341–350. 10.1016/j.fsi.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Ahmed K. B. A.; Raman T.; Anbazhagan V. Platinum nanoparticles inhibit bacteria proliferation and rescue zebrafish from bacterial infection. RSC Adv. 2016, 6, 44415. 10.1039/c6ra03732a. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.