The role of antibody or virus-specific T-cell responses in control of acute Crimean-Congo hemorrhagic fever virus infection is largely unclear. This is a critical gap in our understanding of CCHF, and investigation of convalescence following severe acute CCHF has been limited by the lack of suitable small animal models. We report here a mouse model of CCHF in which infected mice develop severe disease but ultimately recover. Although mice developed an inflammatory immune response along with severe liver and spleen pathology, these mice also developed CCHFV-specific B- and T-cell responses and were protected from reinfection. This model provides a valuable tool to investigate how host immune responses control acute CCHFV infection and how these responses may contribute to the severe disease seen in CCHFV-infected humans in order to develop therapeutic interventions that promote protective immune responses.
KEYWORDS: Crimean-Congo hemorrhagic fever virus, adaptive immunity, animal models, inflammation, pathogenesis
ABSTRACT
Crimean-Congo hemorrhagic fever virus (CCHFV) is a cause of severe hemorrhagic fever. Its tick reservoir and vector are widely distributed throughout Africa, Southern and Eastern Europe, the Middle East, and Asia. Serological evidence suggests that CCHFV can productively infect a wide variety of species, but only humans develop severe, sometimes fatal disease. The role of the host adaptive immunity in control or contribution to the severe pathology seen in CCHF cases is largely unknown. Studies of adaptive immune responses to CCHFV have been limited due to lack of suitable small animal models. Wild-type mice are resistant to CCHFV infection, and type I interferon-deficient mice typically develop a rapid-onset fatal disease prior to development of adaptive immune responses. We report here a mouse model in which type I interferon-deficient mice infected with a clinical isolate of CCHFV develop a severe inflammatory disease but ultimately recover. Recovery was coincident with development of CCHFV-specific B- and T-cell responses that were sustained for weeks postinfection. We also found that recovery from a primary CCHFV infection could protect against disease following homologous or heterologous reinfection. Together this model enables study of multiple aspects of CCHFV pathogenesis, including convalescence, an important aspect of CCHF disease that existing mouse models have been unsuitable for studying.
IMPORTANCE The role of antibody or virus-specific T-cell responses in control of acute Crimean-Congo hemorrhagic fever virus infection is largely unclear. This is a critical gap in our understanding of CCHF, and investigation of convalescence following severe acute CCHF has been limited by the lack of suitable small animal models. We report here a mouse model of CCHF in which infected mice develop severe disease but ultimately recover. Although mice developed an inflammatory immune response along with severe liver and spleen pathology, these mice also developed CCHFV-specific B- and T-cell responses and were protected from reinfection. This model provides a valuable tool to investigate how host immune responses control acute CCHFV infection and how these responses may contribute to the severe disease seen in CCHFV-infected humans in order to develop therapeutic interventions that promote protective immune responses.
INTRODUCTION
Crimean-Congo hemorrhagic fever virus (CCHFV) is a member of the Nairoviridae family containing a negative-sense, tripartite genome that causes a severe febrile illness in humans, Crimean-Congo hemorrhagic fever (CCHF). The principal vector and reservoir for CCHFV are ticks of the Hyalomma genus. Humans can become infected with CCHFV via tick bites, during butchering of infected livestock, or during the care of infected patients in the health care setting (1). CCHF is characterized by a sudden onset of a nonspecific febrile illness that can rapidly progress to severe and often fatal hemorrhagic manifestations (1). Case fatality rates can be as high as 30% (1, 2). Currently, treatment is limited to supportive care, and although ribavirin is used clinically, evidence for clinical benefit is inconsistent (3, 4). Consequently, there is a critical need for vaccines and therapeutics that can effectively treat CCHFV infections.
Subclinical infections with CCHFV may be a majority of clinical outcomes following exposure to CCHFV (5), and most patients who develop overt clinical disease will ultimately recover. It is likely that host adaptive immune responses play a critical role in recovery from acute CCHF; however, our understanding of adaptive immune responses to CCHFV is limited. Although fatal human cases of CCHF often have absent antibody responses, survivors may not develop neutralizing antibody responses (6). In CCHFV-infected cynomolgus macaques, neither levels of CCHFV-specific antibody nor neutralizing capacity of serum correlated with disease outcome or severity (7). A subunit vaccine that induced high levels of neutralizing antibodies failed to protect against lethal CCHFV challenge in mice (8). Together, these data suggest that antibody responses against CCHFV may not correlate with disease outcome. Yet timely administration of neutralizing and nonneutralizing monoclonal antibodies could protect against lethal CCHFV challenge (9), demonstrating that antibodies can protect against fatal disease in mice. Lastly, humans infected with CCHFV have been shown to develop regulatory T-cell responses during acute disease (10), and fatal cases had elevated levels of CD8 T cells (11). Nevertheless, whether T cells contribute to control of acute CCHFV infection is also largely unknown.
Investigation of adaptive immune responses to primary CCHFV infection has been limited by the lack of suitable small animal models. Wild-type mice exhibit no overt clinical disease and restricted viral replication (12), limiting the usefulness of these mice for investigation of how host responses develop to severe CCHFV infection. Type I interferon (IFN)-deficient mice, in contrast, typically exhibit rapid-onset lethal disease prior to development of adaptive immune responses (12–15), limiting their usefulness for studying adaptive immunity to the virus.
In this study, we report a mouse model in which type I interferon-deficient mice (IFNAR−/−) inoculated subcutaneously (s.c.) with the clinical isolate CCHFV strain Hoti, develop severe clinical disease associated with high viral RNA loads, inflammatory immune responses, and liver and spleen pathology. However, in contrast to previously reported mouse models, most mice recovered from the infection. Recovery from acute CCHF correlated with development of CCHFV-specific B- and T-cell responses, and surviving mice were protected from rechallenge weeks postinfection. Cumulatively these data describe a mouse model in which we can study host responses that control severe acute infection with CCHFV.
RESULTS
Subcutaneous inoculation of IFNAR−/− mice results in severe clinical disease.
IFNAR−/− mice were inoculated s.c. with either 1 or 100 50% tissue culture infective doses (TCID50) of CCHFV strain Hoti. For comparison, we also inoculated mice with the same doses of CCHFV strain IbAr 10200. As previously reported (12), mice inoculated with either dose of CCHFV IbAr 10200 rapidly succumbed to the infection between days 5 and 7 postinfection (p.i.) (Fig. 1A and B). Mice inoculated with CCHFV strain Hoti developed progressively worsening weight loss that began on day 4 p.i. (Fig. 1A) and correlated with overt clinical disease, including lethargy, hunched posture, and piloerection. Despite exhibiting severe weight loss and overt clinical disease, nearly all mice survived the infection (1 TCID50, 5 of 6; 100 TCID50, 6 of 6). Recovery began about days 8 to 9 p.i. (Fig. 1A and B). By day 14 p.i., overt clinical signs had mostly resolved. We determined the 50% infective dose (ID50) of CCHFV Hoti in IFNAR−/− mice via the s.c. inoculation route to be 0.002 TCID50 (data not shown), indicating these mice are extremely susceptible to CCHFV Hoti. For the rest of our studies, we inoculated mice s.c. with 1 TCID50 of CCHFV Hoti, equivalent to 500 ID50.
FIG 1.
Adult IFNAR−/− mice inoculated subcutaneously with CCHFV strain Hoti develop severe disease and high viral RNA loads. Eight- to 12-week-old IFNAR−/− mice were inoculated with indicated dose of either CCHFV strain IbAr 10200 or strain Hoti via a single subcutaneous injection to the subscapular region. Mice were weighed daily (A) and monitored for clinical disease. Mice were humanely euthanized according to criteria described in Materials and Methods (B). n = 6 per group. (C and D) Eight- to 12-week-old IFNAR−/− mice were inoculated with 1 TCID50 of CCHFV strain Hoti. At the indicated time points, viral RNA loads in the serum (C) and tissues (D) were quantified by qRT-PCR. n = 5 to 8 per group. The dashed line indicates the limit of detection.
SC inoculation of IFNAR−/− mice results in high viral RNA loads and persistence in multiple tissues.
To evaluate viral dissemination and replication in this model, at serial time points p.i. with CCHFV Hoti, mice were euthanized, and tissues were collected for quantitative reverse transcription-PCR (qRT-PCR) analysis of viral burdens. Serum viremia rapidly peaked at day 4 p.i. (mean, 105.8 RNA copies/μl) and thereafter declined below the limit of detection of our assay by day 14 p.i. (Fig. 1C). We found that CCHFV Hoti replicated in every tissue analyzed, with viral titers typically peaking at days 4 to 8 p.i. (Fig. 1D). The tissues with the highest viral titers during acute disease were the liver (peak mean, 106.5 RNA copies/μg) and lymphoid tissue (peak mean, 105.7 to 106.2 RNA copies/μg). However, high viral RNA loads were also observed in the lungs, brain, and kidney. Interestingly, we were able to detect viral RNA in all tissues at 4 weeks p.i., suggestive of viral persistence. To further evaluate the kinetics of clearance of CCHFV in these mice, we evaluated viral RNA burdens at day +67 p.i. (9.5 weeks p.i. [wpi]). We found that levels of viral RNA in the spleen, cervical lymph nodes, and kidney were nearly identical to viral RNA loads observed at 4 weeks p.i. (Fig. 1D). These data suggest CCHFV establishes viral persistence in these tissues. In contrast, viral RNA loads in the liver, brain, and lungs were declining (Fig. 1D), suggesting the virus was gradually being cleared from these tissues. In addition to qRT-PCR analysis, we analyzed infectious viral titers in the liver and serum of mice through day 14 p.i. by focus-forming assay. We found that infectious virus was readily detected in both the serum and liver at day 4 p.i., but infectious virus titers declined rapidly, with most mice negative for infectious virus at day 8 p.i. and all mice negative at day 14 p.i. (data not shown).
Mice inoculated s.c. with CCHFV Hoti develop severe liver pathology.
The liver is a key target of CCHFV in infected humans, and a common biomarker for CCHF in human patients is elevated levels of liver enzymes, such as aspartate aminotransferase (AST) and alanine aminotransferase (ALT). Levels of these enzymes often negatively correlate with disease outcome in CCHF patients (1, 16). We therefore analyzed these parameters in CCHFV Hoti-infected mice. We found that both AST and ALT levels increased following CCHFV infection, peaking on day 8 p.i. and declining thereafter (Fig. 2A and B). ALT levels were significantly increased as early as day 4 p.i. and remained elevated until at least day 14 p.i. Levels of both enzymes had returned to baseline by day 21 p.i. (Fig. 2A and B).
FIG 2.
IFNAR−/− mice inoculated with CCHFV Hoti exhibit liver pathology. IFNAR−/− mice were inoculated with 1 TCID50 of CCHFV strain Hoti. At the indicated time points, levels of aspartate aminotransferase (AST) (A) and alanine aminotransferase (ALT) (B) in the blood were quantified using Preventive Care disks in a Vetscan2 analyzer. Statistical comparison to mock-infected mice was performed using an ordinary one-way analysis of variance (ANOVA) with the Holm-Sidak multiple-comparison test. *, P < 0.05; **, P < 0.01; ****, P < 0.0001. n = 5 to 9 per group. Formalin-fixed sections of liver tissue were stained with hematoxylin and eosin (C) and evaluated for pathology. To identify viral antigen, formalin-fixed sections were stained with rabbit anti-CCHFV NP (D). Representative images are shown at ×10 magnification (C and D) and ×40 magnification (insets in panel D). Black arrows indicate lesions at 14 and 28 dpi.
To further investigate the liver pathology caused by CCHFV infection in these mice, we evaluated formalin-fixed sections of liver tissue at multiple time points p.i. CCHFV Hoti-infected mice developed severe pathology in the liver, with acute hepatitis and necrosis beginning by day 4, correlating with early clinical signs of disease, and peaking on day 8 p.i. (Fig. 2C), consistent with our AST and ALT data. Interestingly, necrotic lesions in the liver were still observed at day 14 p.i. (Fig. 2C), a time point at which mice had largely recovered from overt clinical disease, but consistent with the elevated ALT levels seen at day 14 p.i. (Fig. 2B). Mild necrosis was still observed at day 28 p.i. (Fig. 2C), suggesting resolution of pathology in the liver following CCHFV infection is prolonged. We also stained sections for viral antigen and at day 4 p.i. found viral antigen predominantly in Kuppfer cells, endothelial cells, and hepatocytes (Fig. 2D). Interestingly, despite severe pathology at day 8 p.i., viral antigen was only rarely found in stained sections at this time point (Fig. 2D), and after day 14 p.i., no viral antigen could be detected in this tissue (Fig. 2D). These data demonstrate that CCHFV Hoti-infected mice develop severe liver pathology, consistent with human CCHF cases, and that resolution of pathology in the liver is prolonged.
CCHFV infection results in severe spleen pathology.
The spleen had high viral RNA loads during acute disease, and we therefore evaluated the consequence of CCHFV infection in this tissue. Grossly, CCHFV infection resulted in splenomegaly, with spleen weights significantly increasing by day 8 p.i., remaining enlarged at day 14 p.i., and similar to mock infected by day 28 p.i. (data not shown). Histological analysis showed that CCHFV Hoti-infected mice developed inflammatory white pulp necrosis on day 4 p.i. (Fig. 3A). This was followed by severe follicular loss with histiocytic and reticuloendothelial hyperplasia at day 8 p.i. (Fig. 3A). By day 14 p.i., most mice had resolved this pathology; however, at day 28 p.i., the spleens exhibited follicular hyperplasia, indicating ongoing pathology in the spleen. Immunohistochemistry showed that at day 4 p.i., CCHFV antigen was found in mononuclear cells of the red and white pulp. By day 8 p.i., viral antigen was only rarely found in stained sections, and thereafter viral antigen was unable to be detected (Fig. 3B) Together these data indicate that CCHFV infection results in substantial and extended pathology in the spleen.
FIG 3.
IFNAR−/− mice inoculated with CCHFV Hoti exhibit severe spleen pathology. IFNAR−/− mice were inoculated with 1 TCID50 of CCHFV strain Hoti. At the indicated time points, the spleen was collected and fixed with formalin. Formalin-fixed sections of spleen tissue were stained with hematoxylin and eosin (A) and evaluated for pathology. To identify viral antigen, formalin-fixed sections were stained with rabbit anti-CCHFV NP (B). Representative images are shown at ×10 magnification (A and B) and ×40 magnification (inset B). Black arrows at 4 dpi indicate white pulp necrosis, and those at 28 dpi indicate a hyperplastic follicle.
CCHFV infection results in an inflammatory immune response.
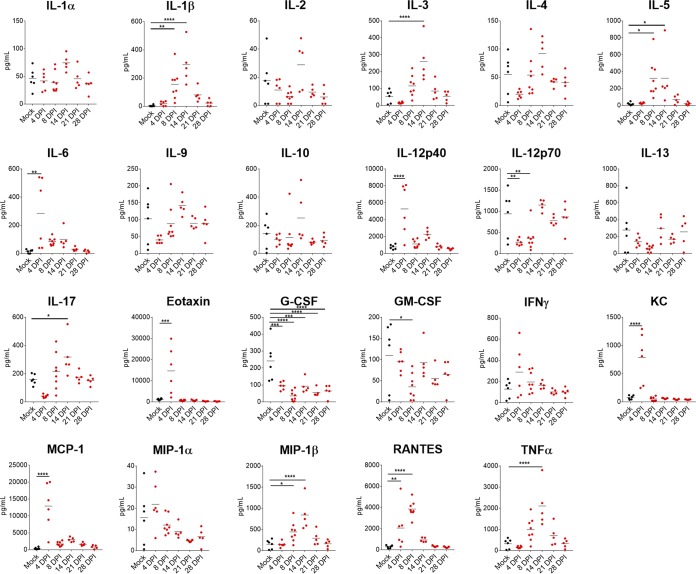
CCHFV infection of humans often results in elevated inflammatory cytokines, such as gamma interferon (IFN-γ), interleukin 1 (IL-1), IL-6, and tumor necrosis factor α (TNF-α) (1, 17). We therefore evaluated the cytokine response in the serum of CCHFV Hoti-infected mice (Fig. 4). We found that consistent with human cases of CCHF, inflammatory cytokine IL-6 was upregulated at day 4 p.i. Interestingly, levels of proinflammatory TNF-α, IL-1β, and IL-17 were elevated as late as day 14 p.i. suggesting prolonged inflammatory responses to the infection in these mice. Although levels of IFN-γ trended higher at day 4 p.i., this was not statistically significant compared to mock-infected mice. Cytokines IL-12p40, monocyte chemoattractant protein 1 (MCP-1), macrophage inflammatory protein 1β (MIP-1β), and RANTES (“regulated on activation, normal T-cell expressed and secreted”) were also elevated following CCHFV infection. These cytokines are associated with inflammatory immune conditions and promote recruitment of immune cells to the site of inflammation (18, 19), and their elevation further suggests a largely inflammatory immune response to CCHFV infection. Cumulatively, these data suggest that CCHFV infection in these mice results in a strong inflammatory immune response driven by cytokines associated with proliferation, activation, and recruitment of macrophages and granulocytes.
FIG 4.
IFNAR−/− mice inoculated with CCHFV Hoti develop an inflammatory immune response to the infection. IFNAR−/− mice were inoculated with 1 TCID50 of CCHFV strain Hoti. At the indicated time points, serum was collected and irradiated, and cytokine levels were quantified with a 23-plex mouse cytokine kit. Statistical comparison to mock-infected mice was performed using an ordinary one-way ANOVA with the Holm-Sidak multiple-comparison test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. n = 5 to 8 per group. Horizontal bars indicate the mean.
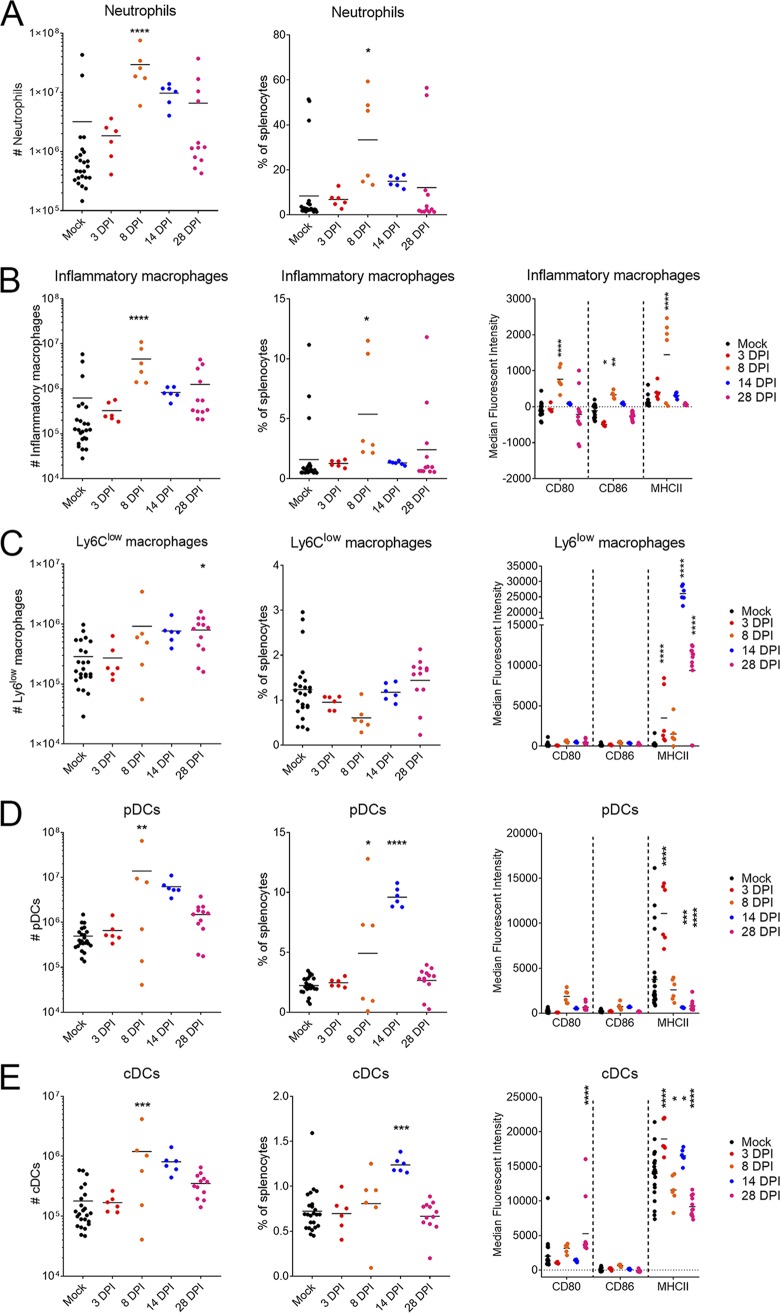
Our cytokine analysis indicated upregulation of multiple cytokines associated with macrophage and granulocytic responses. To further characterize this response, we evaluated splenocytes just prior to onset of clinical disease (day 3 p.i.), at peak clinical disease (day 8 p.i.), at resolution of acute disease (day 14 p.i.), and with persistence (day 28 p.i.) by flow cytometry. We identified an expansion in both absolute number and percentage of neutrophils (CD11b+ CD11clo/− Ly6Cint Ly6Ghi) and inflammatory macrophages (CD11b+ CD11clo/− Ly6Chi) at day 8 p.i. (Fig. 5A and B). Inflammatory macrophages from CCHFV-infected mice upregulated surface expression of activation markers major histocompatibility complex II (MHC-II), CD86, and CD80 at day 8 p.i. (Fig. 5B). The numbers and percentages of neutrophils remained slightly elevated at day 14 p.i. but this was not significant compared to mock-infected mice (Fig. 5A). By day 14 p.i., inflammatory macrophages in CCHFV-infected mice were similar to those of mock-infected mice in numbers, percentages, and expression of surface activation markers (Fig. 5B). Interestingly, we saw significant increases in surface expression of MHC-II on Ly6Clow macrophages (CD11b+ CD11clo/− Ly6Clow) at days 14 and 28 p.i. (Fig. 5C). However, these cells did not upregulate the costimulatory molecules CD80 and CD86, suggesting these cells were not fully activated in response to the infection (Fig. 5C). Plasmacytoid dendritic cells (pDCs) (CD11b− CD11cint) increased in number at day 8 p.i. and in percentages at days 8 and 14 p.i. (Fig. 5D). We also saw increases in numbers of classical dendritic cells (cDCs) (CD11chi) in the spleen at day 8 p.i. and percentage at day 14 p.i. (Fig. 5E). pDCs upregulated MHC-II at day 3 p.i., but expression of MHC-II was significantly reduced at days 14 and 28 p.i. compared to mock-infected mice. These cells also failed to significantly upregulate CD80 and CD86 compared to mock-infected mice (Fig. 5D). cDCs upregulated MHC-II at 3 dpi, and thereafter levels of MHC-II slightly but significantly fluctuated compared to mock-infected mice, although expression remained high at all time points (Fig. 5E). Although cDCs upregulated CD80 at day 28 p.i., they failed to upregulate CD80 at other time points, and levels of CD86 remained similar to those of mock-infected mice at all time points (Fig. 5E). In 3 of the 24 mock-infected mice, we observed markedly higher numbers of neutrophils and inflammatory macrophages in the spleen (Fig. 5A and B). It was unclear the reason for this response in these mice as these mice had no clinical signs of disease at the time of scheduled euthanasia. Nevertheless, these data indicate that CCHFV infection results in significant immune activation within the spleen characterized by expansion of neutrophils and inflammatory macrophages, which is then followed by an expansion of pDCs and cDCs.
FIG 5.
CCHFV Hoti infection results in expansion of neutrophils and inflammatory macrophages. IFNAR−/− mice were inoculated with 1 TCID50 of CCHFV strain Hoti. At the indicated time points, the spleen was collected and stained with fluorescently conjugated antibodies for flow cytometry. Cells were gated to exclude debris, doublets, and nonviable cells. Neutrophils (A), inflammatory macrophages (B), Ly6Clow macrophages (C), peripheral dendritic cells (pDCs) (D), and classical dendritic cells (cDCs) (E) were identified according to the gating strategy outlined in the text (data not shown). Horizontal bars indicate the mean. Samples from mock-infected mice were collected at each time point and pooled for analysis. n = 24 for mock, 6 each for days 3, 8, and 14 p.i., and 12 for day 28 p.i. Statistical comparison to mock-infected mice was performed using an ordinary one-way ANOVA with the Holm-Sidak multiple-comparison test for cell numbers and percentages and two-way ANOVA with Dunnett’s multiple-comparison test for median fluorescent intensity of activation markers. *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
Mice infected with CCHFV develop CCHFV-specific B- and T-cell responses.
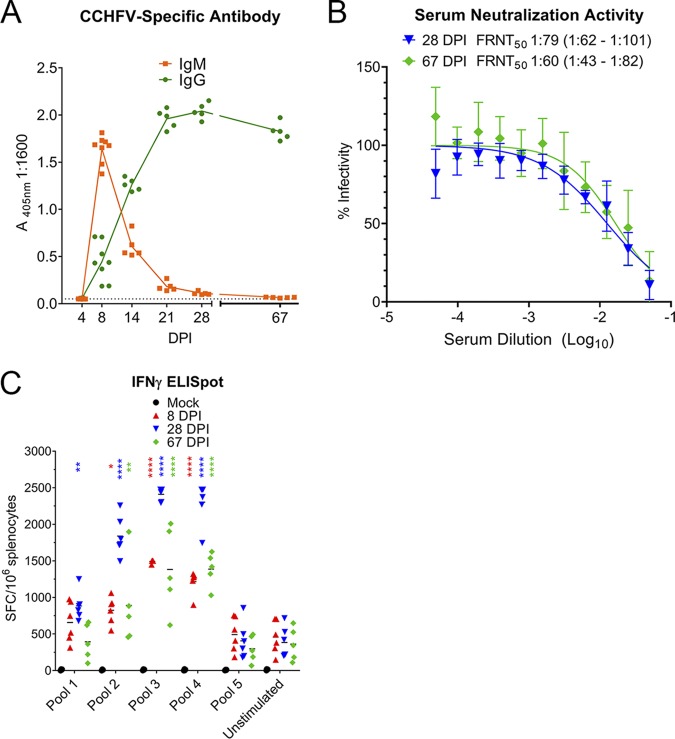
In this model, the majority of Hoti-infected mice survived the acute infection, and this provided the ability to study the host responses during recovery from acute CCHF. We found that CCHFV-specific antibody responses developed rapidly in infected mice, with CCHFV-specific IgM developing as early as day 8 p.i. (Fig. 6A). Levels of CCHFV-specific IgM declined thereafter, whereas levels of CCHFV-specific IgG increased and were sustained to at least 9 weeks p.i. (Fig. 6A). Mice developed marginally neutralizing antibody responses by day 28 p.i. (50% focus reduction neutralization test [FRNT50], 1:79) (Fig. 6B). The neutralization capacity of the serum did not substantially improve by 9.5 weeks p.i. (FRNT50, 1:60). As determined by IFN-γ enzyme-linked immunosorbent spot (ELISpot) assay, CCHFV-infected mice developed T-cell responses against the CCHFV nucleoprotein by day 8 p.i. The T-cell response peaked at day 28 p.i. but was sustained for at least 9 weeks p.i. (Fig. 6C). Compared to unstimulated splenocytes, the T-cell response was largely directed against peptides in pools 3 and 4, corresponding to amino acids 201 through 411 of the CCHFV nucleoprotein (NP) (Fig. 6C). Together these data demonstrate that CCHFV Hoti-infected mice develop CCHFV-specific adaptive immune responses by 8 days postinfection (dpi), and these responses are sustained for weeks postinfection.
FIG 6.
IFNAR−/− mice inoculated with CCHFV Hoti develop CCHFV-specific B- and T-cell responses. IFNAR−/− mice were inoculated with 1 TCID50 of CCHFV strain Hoti. At the indicated time points, serum was collected, and the amount of CCHFV-specific antibody was quantified by ELISA (A). The 405-nm absorbance of a 1:1,600 dilution of serum is shown. The dashed line indicates background absorbance determined from wells receiving no serum. n = 5 to 8 per group. Neutralizing capacity of serum was quantified by focus reduction neutralization test (B). FRNT50 was calculated by nonlinear regression, and the 95% confidence interval is shown. n = 5 mice per time point. Each serum sample was measured in triplicate, and error bars indicate standard deviation. CCHFV-specific T-cell responses in the spleen were quantified using an IFN-γ ELIspot (C). Cells were stimulated with overlapping peptides derived from the CCHFV nucleoprotein pooled at 18 to 25 peptides per pool. Horizontal bars indicate the mean. Statistical comparison to unstimulated splenocytes was performed using a two-way ANOVA with Dunnett’s multiple-comparison test at each respective time point. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Hoti-infected mice develop durable immunity to CCHFV.
Our data indicate that mice that survive acute CCHFV infection sustain adaptive immune responses against the virus for weeks p.i. We therefore wanted to evaluate whether mice that survived a primary infection with CCHFV Hoti were protected against reinfection with CCHFV. The significant genetic diversity of CCHFV, particularly in the viral glycoprotein precursor (2), could result in incomplete protection against reinfection with a heterologous strain of CCHFV. To test the hypothesis that recovery from CCHFV infection would provide durable protection against reinfection with CCHFV, we either mock infected or infected mice with CCHFV strain Hoti and allowed mice to recover from the primary infection. To test homologous reinfection, 9 weeks later, mice were rechallenged intraperitoneally (i.p.) with CCHFV Hoti. When inoculated i.p., CCHFV Hoti results in lethal disease in naive mice (13). To test a heterologous rechallenge, we similarly inoculated a group of recovered mice i.p. with CCHFV strain UG3010. Strain UG3010 differs from CCHFV strain Hoti at the amino acid level by 5% in the S and L segments and 25% in the M segment and is highly virulent in naive mice (data not shown). Mice that were mock infected on day 0 and infected with CCHFV Hoti or UG3010 i.p. 9 weeks later developed severe disease with high viral RNA loads, and all but one Hoti-infected mouse ultimately succumbed (Fig. 7A to C). In contrast, mice that had survived primary CCHFV Hoti infection were protected from disease after reinfection with CCHFV Hoti or CCHFV UG3010, exhibiting no weight loss or clinical signs of disease (Fig. 7A and B). Viral RNA loads in mice that received a primary CCHFV Hoti infection and then were reinfected with CCHFV Hoti or UG3010 were similar to those in mice that received a mock secondary challenge (Fig. 7C), suggesting these mice effectively controlled the secondary infection. Two mice that received a Hoti primary infection and UG3010 secondary infection had markedly higher viral RNA loads in the liver and kidney than mice that received a mock or Hoti secondary infection (Fig. 7C). These data could suggest that protection against heterologous CCHFV strains may be incomplete. However, the differences in viral RNA loads were not significant between these groups, nor did we observe differences in clinical disease or survival between groups reinfected with either virus. Cumulatively, these data demonstrate that recovery from primary CCHFV infection can provide significant protection against disease upon reinfection by both homologous and heterologous strains of CCHFV.
FIG 7.
Recovery from primary CCHFV Hoti infection protects against heterologous and homologous reinfection. Eight- to 12-week-old IFNAR−/− mice were mock infected or inoculated with 1 TCID50 of CCHFV strain Hoti, and mice were allowed to recover from the infection. At 9.5 weeks later, mice were either mock infected or infected with 10 TCID50 of CCHFV strain Hoti or strain UG3010 via i.p. injections. Mice were weighed daily, monitored for disease, and humanely euthanized according to criteria listed in Materials and Methods (A and B). n = 8 to 12 per group. On day +4, a subset of mice were euthanized, and the liver, spleen, and kidney were collected for evaluation of viral RNA loads by qRT-PCR (C). Horizontal bars indicate the mean, and dashed lines indicate the limit of detection. Statistical comparison to mock-infected mice was performed using an ordinary one-way ANOVA with the Holm-Sidak multiple-comparison test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
DISCUSSION
We report here a mouse model of CCHF in which mice develop severe disease characterized by overt clinical signs, high viral RNA loads, inflammatory cytokine responses, and organ pathology. However, in contrast to previously reported models of CCHF, the majority of infected mice recover from the infection. Recovery from infection correlated with long-lasting antibody and T-cell responses to CCHFV. We have previously reported that IFNAR−/− mice inoculated i.p. with CCHFV Hoti develop a severe disease with near-uniform lethality (13). In contrast, data reported here demonstrate that s.c. inoculation of the same CCHFV strain results in most mice surviving the infection. These distinct outcomes suggest the route of inoculation of CCHFV can impact disease outcome. However, only slight differences in time to death were seen when comparing routes of inoculation with CCHFV IbAr 10200 (12), suggesting that route of inoculation may have strain-specific consequences (clinical isolate CCHFV strain Hoti versus mouse-passaged strain IbAr 10200). However, when mice were inoculated i.p. with CCHFV strain Oman or Matin, mice became ill, but all survived (data not shown), suggesting that i.p. inoculation of CCHFV can also result in survivable disease. The slower clinical progression of CCHFV Hoti when inoculated s.c. may also provide sufficient time for host adaptive immune responses to be engaged and control the infection. It is also possible that s.c. inoculation of CCHFV Hoti results in more efficient priming of protective innate and adaptive immune responses that are otherwise avoided in i.p. injections (20).
Cumulatively we observed a largely inflammatory immune activation in response to CCHFV infection, consistent with human cases of CCHF (21). Notably, levels of IL-6 and TNF-α were elevated following CCHFV infection in our model, and these cytokines have been found to negatively correlate with disease outcome in CCHFV-infected humans (17, 21, 22). Although we did not identify the in vivo source of these cytokines, it has been shown that macrophages were productively infected with CCHFV in vitro and secreted IL-6, MCP-1, RANTES, MIP-1α, and TNF-α in response to CCHFV infection (23), cytokines that were systemically upregulated in our model. Consistent with the observed cytokine profile in response to CCHFV infection, we also observed an expansion of neutrophils and inflammatory macrophages within the spleen during peak acute disease. These data suggest that accumulation of these cell types in the spleen of CCHFV-infected mice may support viral replication and inflammatory immune responses. In a previous report evaluating CCHFV-infected STAT1−/− mice, dendritic cells and CD11b+ and F480+ macrophages were found to upregulate CD80 and CD86, but only CD11b+ macrophages upregulated MHC-II (14), suggesting some antigen-presenting cells failed to upregulate necessary molecules for T-cell activation. Although we utilized a different identification scheme within our studies, we found that inflammatory macrophages, which significantly expanded at day 8 p.i., upregulated MHC-II, CD80, and CD86, suggesting that these cell types were upregulating necessary proteins for antigen presentation to T cells. In contrast, although we saw an expansion in cDC and pDC populations, these cells largely failed to upregulate costimulatory CD80 and CD86, suggesting these cells may be inefficiently stimulating T cells. These data together suggest antigen presentation to T cells following CCHFV infection may occur through inflammatory macrophages, although further studies are needed to confirm this hypothesis.
Although humans develop CCHFV-specific antibody and memory T-cell responses, it is unknown if these responses can protect against reinfection by CCHFV. The significant genetic heterogeneity among CCHFV strains (2) invites the possibility that immune responses against a specific strain of CCHFV, induced by either natural infection or vaccination, may be incomplete against heterologous CCHFV strains. Furthermore, our data indicated that even weeks p.i., antibody responses in mice remained poorly neutralizing against a homologous strain of CCHFV, suggesting that neutralizing activity against a heterologous strain could be further reduced. However, our data indicate that primary infection with CCHFV Hoti provided protection against disease upon reinfection with a highly divergent strain of CCHFV. Although the glycoproteins of CCHFV exhibit the greatest genetic diversity, we detected sustained CCHFV-specific T-cell responses against the more-conserved CCHFV nucleoprotein. Vaccine-induced responses solely against the nucleoprotein can be protective (24, 25), and it is therefore likely that immune responses against the more-conserved CCHFV nucleoprotein could provide cross protection. Studies with monoclonal antibodies against the glycoproteins have shown that although there exist antigenic differences among diverse strains of CCHFV, there are neutralizing epitopes conserved among divergent strains (26, 27). Encouragingly, our data suggest that immunity to CCHFV infection may be broadly protective, an important consideration for ongoing vaccine development.
Despite detecting sustained adaptive immune responses to CCHFV, we observed persistence of viral RNA in multiple tissues for weeks p.i. This was coincident with persistent pathology in the liver and spleen. We have previously reported persistence of viral RNA in CCHFV-infected favipiravir-treated mice and late-onset development of lethal CCHFV disease following cessation of antiviral treatment (13). The data reported here further suggest that viral persistence and pathology may be a common outcome in CCHFV-infected mice that survive acute infection. However, the relevance of viral persistence in mice to human infections requires further studies as the long-term sequelae following recovery from acute CCHF are poorly studied. Persistence of viral RNA has been detected in lymphoid tissue of CCHFV-infected cynomolgus macaques (own unpublished data), suggesting viral persistence may not be an artifact of the mouse model. We also cannot exclude the possibility that the persistent viral RNA we detected for weeks p.i. represents noninfectious and/or nonreplicating viral RNA. We were unable to detect infectious virus in the liver after day 8 p.i. by focus-forming assay despite high levels of viral RNA in this tissue. Similarly, immunohistochemistry for viral antigen failed to detect viral antigen after day 8 p.i. in either the liver or spleen. Further studies into this phase of CCHF may be warranted as viral persistence is increasingly appreciated for other viral hemorrhagic fevers, such as those caused by Lassa virus and Ebola virus (28–30).
To date, mouse models have been unsuitable for studying recovery from severe CCHF. Mice either exhibited severely restricted viral replication and no disease (wild-type mice) or rapidly succumbed to lethal disease within days p.i. (type I interferon-deficient mice). Although the cynomolgus macaque model for CCHFV represents an immunocompetent model of CCHF in which animals develop a spectrum of disease outcomes (7), ethical and practical considerations limit the usefulness of this model for initial investigations into the host and viral determinants of CCHFV pathogenesis. An important gap in our understanding of CCHF is what host responses are necessary to control acute CCHFV. These data are important as most CCHF patients will recover from the infection, and understanding how these patients control the infection may help design therapeutic strategies to promote a protective immune response to the virus. Our data demonstrated that by day 8 p.i., mice develop robust CCHFV-specific B- and T-cell responses. These responses were coincident with a rapid resolution of clinical signs of disease after day 8 p.i. The antibody response in these mice, characterized by a rapid onset but poorly neutralizing response even weeks p.i., is consistent with human CCHFV infections (6). Our data also demonstrated that mice rapidly developed CCHFV nucleoprotein-specific T cells suggesting these responses may also contribute to control of acute CCHFV infection. Interestingly, at day 8 p.i., severe pathology was observed in the liver and spleen, despite nearly undetectable viral antigen. These data suggest that host responses, although effective in controlling viral replication, may contribute to the severe tissue pathology seen in these mice. However, studies in mice deficient in adaptive immune responses demonstrated that mice could develop severe tissue pathology in the absence of cytolytic T-cell activity, suggesting this host response is dispensable for tissue damage following CCHFV infection (31). Further studies are needed to determine whether host responses to CCHFV infection contribute to the pathogenesis of CCHFV.
An important limitation of this model is the lack of a type I interferon response in the mice. Although this results in impaired innate immunity, type I interferon responses are also involved in host adaptive immune responses (32, 33). For example, type I interferon can both suppress virus-specific antibody responses (34) and enhance early T-independent antibody responses (35). Similarly, type I interferon signaling can both promote and suppress antiviral T-cell responses—either indirectly through modulation of antigen-presenting cells or directly by acting on the T cells themselves (36). In particular, blockade or absence of type I interferon signaling can inhibit DC maturation and antigen presentation to T cells, as evidenced by reduced expression of MHC-II and costimulatory molecules (37, 38). Nevertheless, our ELISA and ELIspot data indicate adaptive immune responses to CCHFV do not strictly require type I interferon.
In conclusion, this report describes a mouse model for the study of CCHFV pathogenesis that can be used to study control and recovery from acute CCHFV infection. In contrast to previously described acutely lethal mouse models, in our model, mice exhibit severe disease, but ultimately most recover from the infection. This model recapitulates many aspects of the acute and convalescence phases of CCHF in humans. Similar to human infections, we found that CCHFV-infected mice developed an inflammatory immune response against the virus that was associated with severe pathology in the liver and spleen. However, these mice developed early adaptive immune responses to CCHFV that were sustained for weeks p.i. Consistent with these findings, mice that survived a primary CCHFV infection were protected against disease upon reinfection with homologous or heterologous strains of CCHFV. Together these findings describe a mouse model that can be used to study all aspects of CCHF, including early acute disease, severe acute disease, and convalescence. Ongoing studies are using this model to evaluate how host innate and adaptive responses develop to CCHFV, which responses are necessary to control acute CCHFV infection, and whether these responses are protective or contribute to the severe pathology seen in CCHF.
MATERIALS AND METHODS
Biosafety and ethics.
All procedures with infectious CCHFV were conducted under biosafety level 4 (BSL4) conditions in accordance with operating procedures approved by the Rocky Mountain Laboratories institutional biosafety committee. Animal experiments were approved by the institutional animal care and use committee and performed by experienced personnel under veterinary oversight. Mice were group housed in HEPA-filtered cage systems and acclimatized to BSL4 conditions prior to the start of the experiment. They were provided with nesting material and food and water ad libitum.
Mice.
IFNAR−/− mice on the C57BL/6 background were from an in-house breeding colony. Mixed-sex 8- to 12-week-old mice were used for all studies. Mice were inoculated with indicated doses of CCHFV via a 100-μl subcutaneous injection to the subscapular region. Virus was diluted in sterile Dulbecco’s modified Eagle’s medium (DMEM) without additives. Mice were humanely euthanized according to the following criteria: weight loss of >25%; ataxia; extreme lethargy (animal unresponsive to touch); bloody discharge from nose, mouth, rectum, or urogenital area; tachypnea; dyspnea; or paralysis of the limbs. Although we comprehensively evaluated the mice for any of these clinical signs, mice were typically euthanized for weight loss or extreme lethargy. The ID50 for Hoti was calculated with the Reed and Muench method (39) by evaluating mice inoculated subcutaneously with 10-fold dilutions of CCHFV Hoti from 100 to 0.001 TCID50. Any mice that succumbed following inoculation were considered infected. Mice that survived to the planned study endpoint on day 21 or 28 postinfection were confirmed infected by ELISA for CCHFV-specific antibody. Mice without detectable antibody were considered uninfected for ID50 calculations.
Virus.
The CCHFV strains Hoti and IbAr 10200 used in this study were cultured, and the titer was determined by SW13 cell 50% tissue culture infectious dose as previously described (13). CCHFV strain UG3010 was generously provided by Eric Bergeron, Centers for Disease Control and Prevention. UG3010 was passaged once on SW13 cells in our laboratory to generate viral stocks, and the titer was determined as previously described (13).
Flow cytometry.
Single-cell suspensions of mouse spleens were prepared by passaging of tissue through a 70-μm-pore strainer (Corning), and red blood cells were lysed with ACK lysis buffer (Gibco). Fc receptors were blocked with mouse TruStain FcX (Biolegend), and cells were stained with fluorophore-conjugated anti-CD11b (clone M1/70), anti-CD11c (clone N418), anti-Ly6C (clone HK1.4), anti-Ly6G (clone 1A8), anti-IA/IE (MHC-II clone M5/114.15.2), anti-CD80 (clone 16-10A1), and anti-CD86 (clone GL-1). All were purchased from Biolegend. Zombie Aqua viability dye (Biolegend) was also utilized. Data were acquired using a FACSymphony instrument (BD), and analysis was performed using FlowJo v10.5.3 software. Cells were gated by forward and side scatter to exclude debris and then gated to remove doublets and nonviable cells. Cell populations were further identified as described in the text.
Quantitative PCR.
For studies utilizing CCHF Hoti, viral RNA was quantified as previously described (13). For the study in which mice were rechallenged with CCHFV Hoti or UG3010, viral quantification was performed as previously described with modified primers and cycling conditions. Briefly, a one-step reverse transcription-PCR was set up with forward primer 5′-AAAATGAAGAAGGCACTCCTGAG-3′, reverse primer 5′-GCAGACACCCATTTCACTGATTCT-3′, and two probes mixed 1:1: 5′-CCAATGAAGTGGGGGAAGAA-3′ and 5′-CCAATGAAGTGGGGAAAGAA-3′. The probes were labeled with 5′ 6-carboxyfluorescein (FAM) and 3′ black hole quencher 1 (BHQ). Primers and probes were sourced from Integrated DNA Technologies. qRT-PCR was performed with Quantifast one-step enzyme mix (Qiagen), and cycling was performed on a QuantStudio5 instrument (Thermo Fisher) as follows: 50°C for 10 min and 95°C for 5 min, followed by 40 cycles of 95°C for 10 s, 50°C for 20 s, and 72°C for 1 s with single acquisition. Absolute quantification with an internal standard curve was performed as previously described (13). No-template controls were included in parallel to ensure specificity.
Blood chemistry and cytokine analysis.
For blood chemistry analysis, blood was collected from deeply anesthetized mice via cardiac puncture into lithium heparin tubes (BD) and analyzed on a Vetscan VS2 analyzer with Preventive care profile disks (Abaxis). For cytokine analysis, serum was collected and subjected to 8 megarads of gamma irradiation (40). Serum was then diluted 1:4, and cytokines were quantified using the Bio-Plex Pro mouse cytokine 23-plex assay (Bio-Rad) according to the kit instructions.
CCHFV-specific ELISA.
CCHFV-specific antibody was quantified by ELISA as previously described (7) with horseradish peroxidase-conjugated anti-mouse IgM or anti-mouse IgG (Southern Biotech) used as indicated to detect bound antibody.
CCHFV neutralization assay.
The neutralization capacity of mouse serum was quantified by an SW13 cell focus reduction neutralization test. Serum was heat inactivated at 56°C for 30 min. Serum was then diluted in L-15–2% fetal bovine serum (FBS) culture medium. Challenge virus was diluted in L-15 culture medium and mixed 1:1 with diluted serum. The mixture was incubated at 37°C for 1 h. The remaining infectious virus was quantified by SW13 cell focus-forming assay as previously described (13). Foci were visualized with goat anti-mouse IgG conjugated to Alexa Fluor 488 (Thermo Fisher) and counted on an S6 Universal analyzer (Cellular Technology, Ltd.).
ELIspot.
The IFN-γ ELIspot was performed using a colorimetric mouse IFN-γ ELIspot kit from Cellular Technology, Ltd., according to the manufacturer’s instructions. Splenocytes were plated at 300,000 to 500,000 cells per well of the ELIspot plate. Fifteen-mer peptides overlapping by 11 amino acids derived from the Hoti nucleoprotein sequence were synthesized (Genscript). Peptides were dissolved in dimethyl sulfoxide (DMSO; Hybrimax grade [Sigma]) and pooled with 19 to 25 peptides per pool. Peptides were used at a final concentration of 1 μg/ml each in CTL-Test medium. For the positive control, cells were stimulated with concanavalin A (Life Technologies), and for negative controls, cells were stimulated with CTL-Test medium containing DMSO vehicle alone. Cells were incubated for 20 h at 37°C with 5% CO2. All measurements were performed in duplicate for each mouse. After incubation, plates were developed according to the kit instructions and fixed with formalin according to approved procedures for removal of samples under BSL4 conditions, and then spots were counted with an S6 Universal analyzer (Cellular Technology, Ltd.). The upper limit of detection was set at 750 spots per well.
Histology and immunohistochemistry.
Tissues were fixed in 10% neutral buffered formalin with two changes for a minimum of 7 days. Tissues were placed in cassettes and processed with a Sakura VIP-6 Tissue Tek on a 12-h automated schedule, using a graded series of ethanol, xylene, and ParaPlast Extra. Embedded tissues were sectioned at 5 μm and dried overnight at 42°C prior to staining. Specific anti-CCHF immunoreactivity was detected using a rabbit anti-CCHF NP (Open Biosystems) specific for NP peptide (KDEMNRWFEEFKKGNGLVD) at a 1:1,500 dilution as the primary antibody and Biogenex SS rabbit link-biotinylated anti-rabbit immunoglobulin (catalog no. HK336-9R) neat as the secondary antibody. The tissues were then processed for immunohistochemistry using the Ultra automated processor and Ventana/Roche Tissue Diagnostics Discovery DAB Map detection kit (catalog no. 760-124ms) with a DABMap kit (Ventana Medical Systems).
Statistics.
The indicated statistical analyses were performed in GraphPad Prism v7.04.
ACKNOWLEDGMENTS
This research was supported by the Intramural Research Program of the NIH, NIAID. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We wish to thank Shelly Robertson and Aaron Carmody, NIH/NIAID, for helpful discussions on flow cytometry.
REFERENCES
- 1.Ergonul O. 2006. Crimean-Congo haemorrhagic fever. Lancet Infect Dis 6:203–214. doi: 10.1016/S1473-3099(06)70435-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bente DA, Forrester NL, Watts DM, McAuley AJ, Whitehouse CA, Bray M. 2013. Crimean-Congo hemorrhagic fever: history, epidemiology, pathogenesis, clinical syndrome and genetic diversity. Antiviral Res 100:159–189. doi: 10.1016/j.antiviral.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 3.Hawman DW, Feldmann H. 2018. Recent advances in understanding Crimean-Congo hemorrhagic fever virus. F1000Res 7:1715. doi: 10.12688/f1000research.16189.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson S, Henschke N, Maayan N, Mills I, Buckley BS, Kakourou A, Marshall R. 2018. Ribavirin for treating Crimean Congo haemorrhagic fever. Cochrane Database Syst Rev 6:CD012713. doi: 10.1002/14651858.CD012713.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bodur H, Akinci E, Ascioglu S, Öngürü P, Uyar Y. 2012. Subclinical infections with Crimean-Congo hemorrhagic fever virus, Turkey. Emerg Infect Dis 18:640–642. doi: 10.3201/eid1804.111374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shepherd AJ, Swanepoel R, Leman PA. 1989. Antibody response in Crimean-Congo hemorrhagic fever. Rev Infect Dis 11(Suppl 4):S801–S806. doi: 10.1093/clinids/11.Supplement_4.S801. [DOI] [PubMed] [Google Scholar]
- 7.Haddock E, Feldmann F, Hawman DW, Zivcec M, Hanley PW, Saturday G, Scott DP, Thomas T, Korva M, Avsic-Zupanc T, Safronetz D, Feldmann H. 2018. A cynomolgus macaque model for Crimean-Congo haemorrhagic fever. Nat Microbiol 3:556–562. doi: 10.1038/s41564-018-0141-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kortekaas J, Vloet RP, McAuley AJ, Shen X, Bosch BJ, de Vries L, Moormann RJ, Bente DA. 2015. Crimean-Congo hemorrhagic fever virus subunit vaccines induce high levels of neutralizing antibodies but no protection in STAT1 knockout mice. Vector Borne Zoonotic Dis 15:759–764. doi: 10.1089/vbz.2015.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bertolotti-Ciarlet A, Smith J, Strecker K, Paragas J, Altamura LA, McFalls JM, Frias-Stäheli N, García-Sastre A, Schmaljohn CS, Doms RW. 2005. Cellular localization and antigenic characterization of Crimean-Congo hemorrhagic fever virus glycoproteins. J Virol 79:6152–6161. doi: 10.1128/JVI.79.10.6152-6161.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gazi U, Yapar D, Karasartova D, Gureser AS, Akdogan O, Unal O, Baykam N, Taylan Ozkan A. 2018. The role of Treg population in pathogenesis of Crimean Congo hemorrhagic fever. Virus Res 250:1–6. doi: 10.1016/j.virusres.2018.04.003. [DOI] [PubMed] [Google Scholar]
- 11.Akıncı E, Yılmaz M, Bodur H, Öngürü P, Bayazıt FN, Erbay A, Özet G. 2009. Analysis of lymphocyte subgroups in Crimean-Congo hemorrhagic fever. Int J Infect Dis 13:560–563. doi: 10.1016/j.ijid.2008.08.027. [DOI] [PubMed] [Google Scholar]
- 12.Zivcec M, Safronetz D, Scott D, Robertson S, Ebihara H, Feldmann H. 2013. Lethal Crimean-Congo hemorrhagic fever virus infection in interferon α/β receptor knockout mice is associated with high viral loads, proinflammatory responses, and coagulopathy. J Infect Dis 207:1909–1921. doi: 10.1093/infdis/jit061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hawman DW, Haddock E, Meade-White K, Williamson B, Hanley PW, Rosenke K, Komeno T, Furuta Y, Gowen BB, Feldmann H. 2018. Favipiravir (T-705) but not ribavirin is effective against two distinct strains of Crimean-Congo hemorrhagic fever virus in mice. Antiviral Res 157:18–26. doi: 10.1016/j.antiviral.2018.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bente DA, Alimonti JB, Shieh WJ, Camus G, Stroher U, Zaki S, Jones SM. 2010. Pathogenesis and immune response of Crimean-Congo hemorrhagic fever virus in a STAT-1 knockout mouse model. J Virol 84:11089–11100. doi: 10.1128/JVI.01383-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oestereich L, Rieger T, Neumann M, Bernreuther C, Lehmann M, Krasemann S, Wurr S, Emmerich P, de Lamballerie X, Ölschläger S, Günther S. 2014. Evaluation of antiviral efficacy of ribavirin, arbidol, and T-705 (favipiravir) in a mouse model for Crimean-Congo hemorrhagic fever. PLoS Negl Trop Dis 8:e2804. doi: 10.1371/journal.pntd.0002804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swanepoel R, Gill DE, Shepherd AJ, Leman PA, Mynhardt JH, Harvey S. 1989. The clinical pathology of Crimean-Congo hemorrhagic fever. Rev Infect Dis 11(Suppl 4):S794–S800. doi: 10.1093/clinids/11.Supplement_4.S794. [DOI] [PubMed] [Google Scholar]
- 17.Ergonul O, Tuncbilek S, Baykam N, Celikbas A, Dokuzoguz B. 2006. Evaluation of serum levels of interleukin (IL)-6, IL-10, and tumor necrosis factor-alpha in patients with Crimean-Congo hemorrhagic fever. J Infect Dis 193:941–944. doi: 10.1086/500836. [DOI] [PubMed] [Google Scholar]
- 18.Arango Duque G, Descoteaux A. 2014. Macrophage cytokines: involvement in immunity and infectious diseases. Front Immunol 5:491–491. doi: 10.3389/fimmu.2014.00491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cooper AM, Khader SA. 2007. IL-12p40: an inherently agonistic cytokine. Trends Immunol 28:33–38. doi: 10.1016/j.it.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 20.Heath WR, Carbone FR. 2013. The skin-resident and migratory immune system in steady state and memory: innate lymphocytes, dendritic cells and T cells. Nat Immunol 14:978. doi: 10.1038/ni.2680. [DOI] [PubMed] [Google Scholar]
- 21.Papa A, Tsergouli K, Çağlayık DY, Bino S, Como N, Uyar Y, Korukluoglu G. 2016. Cytokines as biomarkers of Crimean-Congo hemorrhagic fever. J Med Virol 88:21–27. doi: 10.1002/jmv.24312. [DOI] [PubMed] [Google Scholar]
- 22.Saksida A, Duh D, Wraber B, Dedushaj I, Ahmeti S, Avsic-Zupanc T. 2010. Interacting roles of immune mechanisms and viral load in the pathogenesis of Crimean-Congo hemorrhagic fever. Clin Vaccine Immunol 17:1086–1093. doi: 10.1128/CVI.00530-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peyrefitte CN, Perret M, Garcia S, Rodrigues R, Bagnaud A, Lacote S, Crance J-M, Vernet G, Garin D, Bouloy M, Paranhos-Baccalà G. 2010. Differential activation profiles of Crimean-Congo hemorrhagic fever virus- and Dugbe virus-infected antigen-presenting cells. J Gen Virol 91:189–198. doi: 10.1099/vir.0.015701-0. [DOI] [PubMed] [Google Scholar]
- 24.Zivcec M, Safronetz D, Scott DP, Robertson S, Feldmann H. 2018. Nucleocapsid protein-based vaccine provides protection in mice against lethal Crimean-Congo hemorrhagic fever virus challenge. PLoS Negl Trop Dis 12:e0006628. doi: 10.1371/journal.pntd.0006628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aligholipour Farzani T, Hanifehnezhad A, Foldes K, Ergunay K, Yilmaz E, Hashim Mohamed Ali H, Ozkul A. 2019. Co-delivery effect of CD24 on the immunogenicity and lethal challenge protection of a DNA vector expressing nucleocapsid protein of Crimean Congo hemorrhagic fever virus. Viruses 11:E75. doi: 10.3390/v11010075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zivcec M, Guerrero LIW, Albarino CG, Bergeron E, Nichol ST, Spiropoulou CF. 2017. Identification of broadly neutralizing monoclonal antibodies against Crimean-Congo hemorrhagic fever virus. Antiviral Res 146:112–120. doi: 10.1016/j.antiviral.2017.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahmed AA, McFalls JM, Hoffmann C, Filone CM, Stewart SM, Paragas J, Khodjaev S, Shermukhamedova D, Schmaljohn CS, Doms RW, Bertolotti-Ciarlet A. 2005. Presence of broadly reactive and group-specific neutralizing epitopes on newly described isolates of Crimean-Congo hemorrhagic fever virus. J Gen Virol 86:3327–3336. doi: 10.1099/vir.0.81175-0. [DOI] [PubMed] [Google Scholar]
- 28.Deen GF, Broutet N, Xu W, Knust B, Sesay FR, McDonald SLR, Ervin E, Marrinan JE, Gaillard P, Habib N, Liu H, Liu W, Thorson AE, Yamba F, Massaquoi TA, James F, Ariyarajah A, Ross C, Bernstein K, Coursier A, Klena J, Carino M, Wurie AH, Zhang Y, Dumbuya MS, Abad N, Idriss B, Wi T, Bennett SD, Davies T, Ebrahim FK, Meites E, Naidoo D, Smith SJ, Ongpin P, Malik T, Banerjee A, Erickson BR, Liu Y, Liu Y, Xu K, Brault A, Durski KN, Winter J, Sealy T, Nichol ST, Lamunu M, Bangura J, Landoulsi S, Jambai A, Morgan O, Wu G, Liang M, Su Q, Lan Y, Hao Y, Formenty P, Ströher U, Sahr F. 2017. Ebola RNA persistence in semen of Ebola virus disease survivors—final report. N Engl J Med 377:1428–1437. doi: 10.1056/NEJMoa1511410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McElroy AK, Akondy RS, Harmon JR, Ellebedy AH, Cannon D, Klena JD, Sidney J, Sette A, Mehta AK, Kraft CS, Lyon MG, Varkey JB, Ribner BS, Nichol ST, Spiropoulou CF. 2017. A case of human Lassa virus infection with robust acute T-cell activation and long-term virus-specific T-cell responses. J Infect Dis 215:1862–1872. doi: 10.1093/infdis/jix201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacobs M, Rodger A, Bell DJ, Bhagani S, Cropley I, Filipe A, Gifford RJ, Hopkins S, Hughes J, Jabeen F, Johannessen I, Karageorgopoulos D, Lackenby A, Lester R, Liu RSN, MacConnachie A, Mahungu T, Martin D, Marshall N, Mepham S, Orton R, Palmarini M, Patel M, Perry C, Peters SE, Porter D, Ritchie D, Ritchie ND, Seaton RA, Sreenu VB, Templeton K, Warren S, Wilkie GS, Zambon M, Gopal R, Thomson EC. 2016. Late Ebola virus relapse causing meningoencephalitis: a case report. Lancet 388:498–503. doi: 10.1016/S0140-6736(16)30386-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindquist ME, Zeng X, Altamura LA, Daye SP, Delp KL, Blancett C, Coffin KM, Koehler JW, Coyne S, Shoemaker CJ, Garrison AR, Golden JW. 2018. Exploring Crimean-Congo hemorrhagic fever virus-induced hepatic injury using antibody-mediated type I interferon blockade in mice. J Virol 92. doi: 10.1128/JVI.01083-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kadowaki N, Antonenko S, Lau J-N, Liu Y-J. 2000. Natural interferon alpha/beta-producing cells link innate and adaptive immunity. J Exp Med 192:219–226. doi: 10.1084/jem.192.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le Bon A, Tough DF. 2002. Links between innate and adaptive immunity via type I interferon. Curr Opin Immunol 14:432–436. doi: 10.1016/S0952-7915(02)00354-0. [DOI] [PubMed] [Google Scholar]
- 34.Moseman EA, Wu T, de la Torre JC, Schwartzberg PL, McGavern DB. 2016. Type I interferon suppresses virus-specific B cell responses by modulating CD8+ T cell differentiation. Sci Immunol 1:eaah3565. doi: 10.1126/sciimmunol.aah3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swanson CL, Wilson TJ, Strauch P, Colonna M, Pelanda R, Torres RM. 2010. Type I IFN enhances follicular B cell contribution to the T cell-independent antibody response. J Exp Med 207:1485–1500. doi: 10.1084/jem.20092695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crouse J, Kalinke U, Oxenius A. 2015. Regulation of antiviral T cell responses by type I interferons. Nat Rev Immunol 15:231. doi: 10.1038/nri3806. [DOI] [PubMed] [Google Scholar]
- 37.Simmons DP, Wearsch PA, Canaday DH, Meyerson HJ, Liu YC, Wang Y, Boom WH, Harding CV. 2012. Type I IFN drives a distinctive dendritic cell maturation phenotype that allows continued class II MHC synthesis and antigen processing. J Immunol 188:3116–3126. doi: 10.4049/jimmunol.1101313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Longhi MP, Trumpfheller C, Idoyaga J, Caskey M, Matos I, Kluger C, Salazar AM, Colonna M, Steinman RM. 2009. Dendritic cells require a systemic type I interferon response to mature and induce CD4+ Th1 immunity with poly IC as adjuvant. J Exp Med 206:1589–1602. doi: 10.1084/jem.20090247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Epidemiol 27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- 40.Feldmann F, Shupert WL, Haddock E, Twardoski B, Feldmann H. 2019. Gamma irradiation as an effective method for inactivation of emerging viral pathogens. Am J Trop Med Hyg 100:1275–1277. doi: 10.4269/ajtmh.18-0937. [DOI] [PMC free article] [PubMed] [Google Scholar]