Dysbiosis of the fecal microbiome is a common feature observed in ARV-treated people living with HIV. The degree to which HIV infection itself causes this dysbiosis remains unclear. Here, we demonstrate that medications used to treat HIV infection can influence the composition of the GI tract immune responses and its microbiome in the nonhuman primate SIV model.
KEYWORDS: HIV, SIV, antiretroviral agents, microbiome
ABSTRACT
Gastrointestinal (GI) immune system competency is dependent upon interactions with commensal microbiota, which can be influenced by wide-ranging pharmacologic interventions. In simian immunodeficiency virus (SIV)-infected Asian macaque models of human immunodeficiency virus (HIV) infection, we previously noted that initiation of antiretroviral therapy (ART) is associated with a specific imbalance (dysbiosis) of the composition of the intestinal bacteriome. To determine if ART itself might contribute to dysbiosis or immune dysfunction, we treated healthy rhesus macaques with protease, integrase, or reverse transcriptase inhibitors for 1 to 2 or for 5 to 6 weeks and evaluated intestinal immune function and the composition of the fecal bacterial microbiome. We observed that individual antiretrovirals (ARVs) modestly altered intestinal T-cell proinflammatory responses without disturbing total or activated T-cell frequencies. Moreover, we observed transient disruptions in bacterial diversity coupled with perturbations in the relative frequencies of bacterial communities. Shifts in specific bacterial frequencies were not persistent posttreatment, however, with individual taxa showing only isolated associations with T-cell proinflammatory responses. Our findings suggest that intestinal bacterial instability and modest immunological alterations can result from ART itself. These data could lead to therapeutic interventions which stabilize the microbiome in individuals prescribed ART.
IMPORTANCE Dysbiosis of the fecal microbiome is a common feature observed in ARV-treated people living with HIV. The degree to which HIV infection itself causes this dysbiosis remains unclear. Here, we demonstrate that medications used to treat HIV infection can influence the composition of the GI tract immune responses and its microbiome in the nonhuman primate SIV model.
INTRODUCTION
Combination antiretroviral therapy (ART) successfully inhibits lentiviral replication by preventing the de novo infection of activated CD4+ T cells. Although the widespread implementation of ART has been instrumental in reducing the morbidity and mortality of the human immunodeficiency virus (HIV) epidemic, the prolonged effects of antiretroviral (ARV) administration may not be inconsequential. In communities with widespread ART implementation, unresolved inflammation contributing to non-AIDS-related comorbidities is the leading cause of mortality in people living with HIV (1). While residual inflammation can itself be partially attributed to physical and immunological scarring following prolonged viral replication (2, 3), modest inflammation and immune dysfunction are similarly observed in patients who initiate ART in acute HIV infection (4–7).
Gastrointestinal (GI) complications are prevalent in ART-treated HIV-infected patients as well as in uninfected individuals taking ARVs as part of a preexposure prophylaxis (PreP) regimen (8), suggesting that ART might by itself contribute to inflammation. Although an estimated 70% of diarrhea-associated complications in HIV-infected, ARV-treated patients have been attributed to noninfectious causes (9–11), little is known about whether dysbiosis is a feature in these cases, as high-throughput microbial sequencing is not common clinical practice. In HIV infection, intestinal bacterial dysbiosis—an imbalance of the composition of the microbiome favoring enrichment of pathogen-associated taxa—has been shown to correlate with canonical markers of inflammation in both untreated and treated HIV infections (12–15). Although we along with others have demonstrated that these described dysbioses do not promote untreated lentiviral disease progression (16–19), a potential contribution of dysbiosis to comorbidities in ART-treated lentiviral infections has not been thoroughly assessed. Intriguingly, in Asian macaque models of HIV-1, we previously observed that the initiation of combination ART in simian immunodeficiency virus (SIV)-infected rhesus macaques potentiates intestinal bacterial instability (18).
The deleterious effects of prolonged antibiotic prescription on the commensal microbiome are well described and purported to underlie several chronic conditions (20). Similarly, though less well studied, the off-target effects of nonantibiotic pharmaceuticals have been shown to disrupt intestinal bacterial communities (21, 22). Disruptions to these communities by nonantibiotics can result not only from direct interference with commensal bacteria (23) but also from interference with conserved biochemical processes (24, 25), metabolic cascades (26), and coinhabitants of the commensal ecosystem (27, 28). Importantly, ARVs currently prescribed to inhibit HIV replication are not specific for HIV or SIV (29). Several reverse transcriptase (RT) inhibitors can be incorporated into the DNA of all replicating organisms (30), protease (PR) inhibitors target biochemically conserved aspartic proteases (31), and integrase (IN) inhibitors target catalytic domains conserved in some bacteriophages and prokaryotic transposases (32). PR inhibitors in particular have been shown to alter bacterial, fungal, and protozoal cultures in vitro (33–35).
Characterizing the biochemical spectrum of off-target ARV effects is an essential step in ascribing a cause to an effect with regard to non-AIDS-related inflammation and comorbidities. Given the known potential for ARVs to influence the microbiome, we sought to understand the influence of representative classes of ARVs on the commensal bacteria and on intestinal immunity in vivo. Here, we treated healthy rhesus macaques with representative PR, IN, and RT inhibitors and characterized intestinal T-cell activation and fecal bacterial stability. We demonstrate that ART modestly alters intestinal T-cell proinflammatory responses and bacterial diversity. These findings form the framework for a holistic comprehension of the influence of ART on systemic immunity and, by extension, inflammatory complications and comorbidities associated with prolonged ART.
RESULTS
Study design.
Thirty-one SIV-uninfected, healthy, rhesus macaques were assigned to control (n = 6), PR inhibitor (n = 13), IN inhibitor (n = 6), or RT inhibitor (n = 6) groups and treated daily for 1 to 6 weeks. Animals treated with a PR inhibitor received darunavir (Prestiza) and ritonavir (Norvir), animals treated with IN inhibitor received L-870812 (L812), and animals treated with RT inhibitor received tenofovir (PMPA) or emtricitabine (FTC). An additional 2 animals were treated with an orally administered RT inhibitor, emtricitabine (TDF), in a pilot study. No differences were observed beyond those previously described for PMPA administration (data not shown), and, thus, this treatment was not further examined. Animals underwent peripheral mononuclear blood cell (PBMC) and jejunal (Jej) or rectal biopsy (RB) specimen sampling at 1 to 2 weeks posttreatment and, for a subset of PR inhibitor-treated and control animals, at 5 to 6 weeks posttreatment (Table 1). No changes in weight were observed posttreatment. Stool was collected longitudinally for 16S bacterial sequencing.
TABLE 1.
Characteristics of study animals
| Treatment and animal | Gender | Treatment initiation |
Wk 1–2 sampling |
Wk 5–6 sampling |
|||
|---|---|---|---|---|---|---|---|
| Datea | Wt (kg) | Date (no. of days posttreatment) | Wt (kg) | Date (no. of days posttreatment) | Wt (kg) | ||
| Control | |||||||
| Rh37034 | Male | 12.65 | 10-05-2016 | 11.97 | 11-02-2016 | 11.63 | |
| RhDFAi | Male | 9.38 | 10-05-2016 | 9.12 | 11-02-2016 | 8.45 | |
| RhDGGB | Male | 3.74 | 11-06-2017 | 3.76 | 12-04-2017 | 3.83 | |
| RhDGMh | Male | 3.39 | 11-06-2017 | 3.22 | 12-04-2017 | 3.42 | |
| RhDFK9 | Male | 7.32 | 1-30-2018 | 7.02 | 2-27-2018 | 7.15 | |
| RhDMEj | Female | 8.99 | 1-30-2018 | 8.65 | 2-27-2018 | 8.25 | |
| IN inhibitor | |||||||
| Rh37033 | Male | 9-27-2016 | 12.10 | 10-04-2016 (7) | 11.53 | ||
| RhDCKj | Male | 2-03-2015 | 9.61 | 2-10-2015 (7) | 9.63 | ||
| RhDCVf | Male | 2-03-2015 | 9.73 | 2-10-2015 (7) | 9.57 | ||
| RhDE1a | Male | 2-03-2015 | 11.85 | 2-10-2015 (7) | 11.67 | ||
| RhDE1aa | Male | 9-27-2016 | 6.21 | 10-04-2016 (7) | 6.05 | ||
| RhF64 | Female | 9-27-2016 | 7.73 | 10-04-2016 (7) | 7.71 | ||
| PR inhibitor | |||||||
| RhDCBC | Male | 3-09-2015 | 8.85 | 3-16-2015 (7) | 8.56 | ||
| RhDE20 | Male | 3-09-2015 | 9.66 | 3-16-2015 (7) | 9.31 | ||
| RhDE2c | Male | 3-09-2015 | 8.70 | 3-16-2015 (7) | 8.48 | ||
| RhDE2w | Male | 3-09-2015 | 13.20 | 3-16-2015 (7) | 13.08 | ||
| Rh37073 | Female | 9-27-2016 | 11.48 | 10-05-2016 (8) | 11.33 | 11-02-2016 (36) | 11.56 |
| Rh37360 | Female | 9-27-2016 | 10.83 | 10-05-2016 (8) | 10.75 | 11-02-2016 (36) | 11.63 |
| RhZC08 | Female | 9-27-2016 | 7.02 | 10-05-2016 (8) | 6.95 | 11-02-2016 (36) | 6.62 |
| RhDF8d | Male | 10-23-2017 | 4.58 | 11-07-2017 (15) | 4.5 | 12-05-2017 (43) | 4.63 |
| RhDFPn | Male | 10-23-2017 | 6.17 | 11-07-2017 (15) | 6.06 | 12-05-2017 (43) | 6.19 |
| RhDGHi | Male | 10-23-2017 | 3.32 | 11-07-2017 (15) | 3.2 | 12-05-2017 (43) | 3.46 |
| RhDGMp | Male | 10-23-2017 | 3.43 | 11-07-2017 (15) | 3.45 | 12-05-2017 (43) | 3.67 |
| RhDFNj | Male | 1-17-2018 | 8.45 | 01-29-2018 (12) | 8.26 | 02-25-2018 (39) | 8.62 |
| RhDFZc | Male | 1-17-2018 | 7.13 | 01-29-2018 (12) | 7.37 | 02-25-2018 (39) | 7.73 |
| RT inhibitor (FTC) | |||||||
| RhCL7p | Male | 1-20-2015 | 13.43 | 1-27-2015 (7) | 13.18 | ||
| RhDCAV | Male | 1-20-2015 | 9.37 | 1-27-2015 (7) | 9.05 | ||
| RhDCJWA | Male | 1-20-2015 | 9.63 | 1-27-2015 (7) | 9.58 | ||
| RT inhibitor (PMPA) | |||||||
| RhCL4c | Male | 9-30-2014 | 11.31 | 10-07-2014 (7) | 11.2 | ||
| RhCL86 | Male | 9-30-2014 | 11.64 | 10-07-2014 (7) | 11.54 | ||
| RhF98 | Female | 9-30-2014 | 7.23 | 10-07-2014 (7) | 7.1 | ||
All dates are given in the form month-day-year.
ARV administration does not expand the activated T-cell pool in healthy macaques.
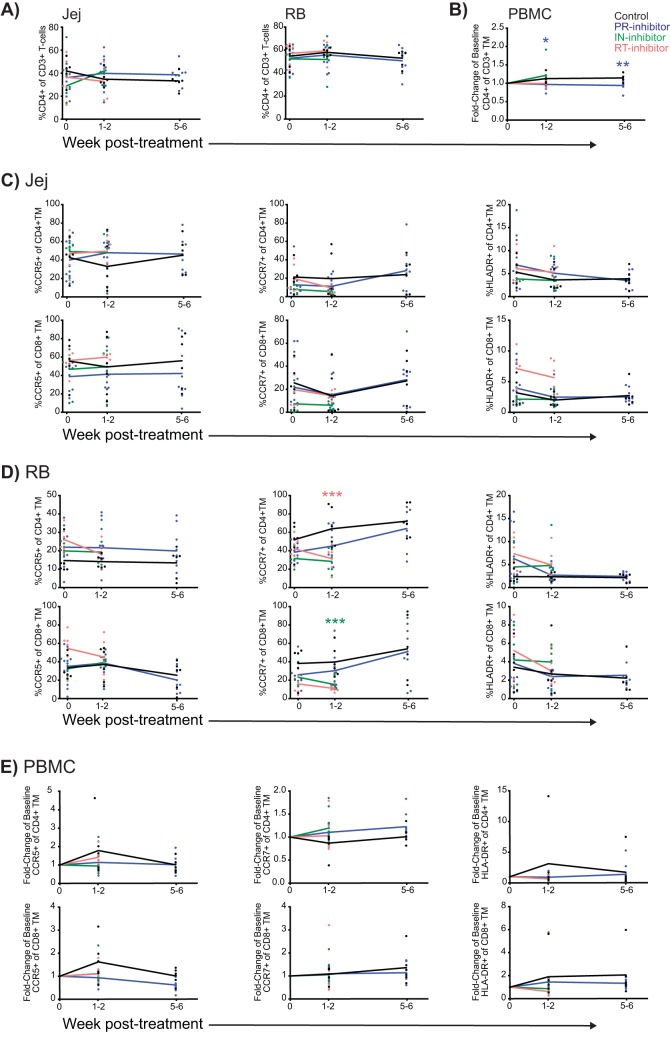
Lentiviral suppression following ART restores circulating CD4+ T cells; however, intestinal CD4+ T cells are not fully restored, and systemic lymphocytes exhibit signs of persistent activation. To determine if ART itself contributes to alterations in intestinal T-cell frequencies or activation, we quantified T-cell frequencies from intestinal biopsy specimen homogenates and PBMCs before and after ART by flow cytometry and further measured their expression of the following markers: CCR5, a lymphocyte mucosal homing marker and the HIV/SIV coreceptor; CCR7, which demarcates replenishable effector (CCR7−) and nonreplenishable central (CCR7+) memory lymphocytes; and HLA-DR, a major histocompatibility complex class II (MHC-II) receptor that is commonly upregulated on activated CD4+ as well as CD8+ T cells. No differences in CD4+ T-cell frequencies were observed in treated animals compared to levels in controls for either the small or large intestine (Fig. 1A); however, significantly lower CD4+ T-cell frequencies (fold change from baseline) were observed among PBMCs from the PR inhibitor-treated animals compared to levels in controls (Fig. 1B) (for weeks 1 to 2, P = 0.018; weeks 5 to 6, P = 0.005). No significant differences in CCR5 or HLA-DR expression by CD4+ or CD8+ memory T (TM) cells were observed between treated animals and controls at any of the measured time points though reduced expression of CCR7 in RB specimens was observed for CD4+ TM cells in RT inhibitor-treated animals (unpaired, two-way t test, P = 0.009) and for CD8+ TM cells in IN inhibitor-treated animals (P = 0.007) (Fig. 1C to E).
FIG 1.
ART does not induce significant T-cell activation healthy macaques. (A) Individual and mean (arithmetic) longitudinal frequencies in Jej and RB specimens of CD4+ T cells in control animals and animals treated with PR inhibitor, IN inhibitor, and RT inhibitor. (B) Individual and mean CD4+ T-cell fold change from baseline frequencies in PBMCs. Asterisks denote significance of values for the ART groups compared to those of the controls, color coded by ART treatment. (C and D) Frequency in Jej and RB specimens of CD4+ and CD8+ TM cells expressing CCR5, CCR7, and HLA-DR, as indicated. (E) Fold change from baseline frequencies of CD4+ and CD8+ TM cells expressing CCR5, CCR7, and HLA-DR, as indicated, in PBMCs. Statistical significance between groups was assessed by an unpaired, two-way t test as follows: in PBMCs from PR inhibitor-treated animals, fold change in frequency of CD4+ TM cells at weeks 1 to 2, P = 0.018, and at week 5 to 6, P = 0.005; in RB specimens from RT inhibitor-treated animals frequency of CCR7+ CD4+ TM cells at weeks 1 to 2, P = 0.009, and from IN inhibitor-treated animals, frequency of CCR7+ CD8+ TM cells at weeks 1 to 2, P = 0.007.
A modest proinflammatory response follows ARV administration in healthy macaques.
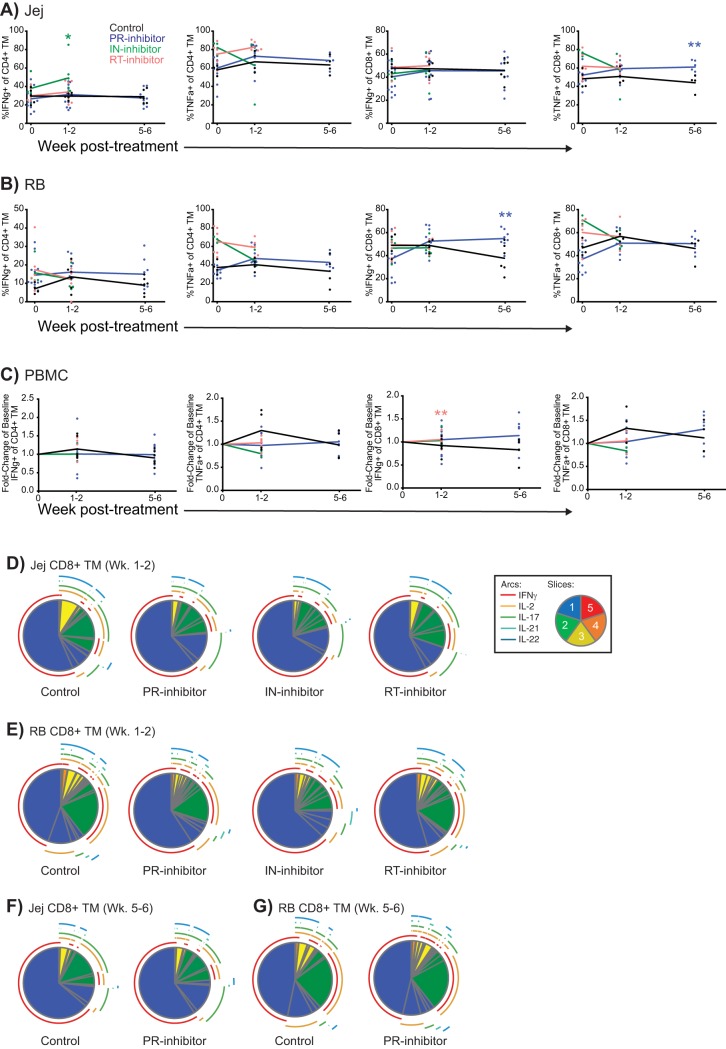
Residual immune dysfunction in ARV-treated individuals includes a proinflammatory milieu which is known to contribute to non-AIDS comorbidities. Although we did not observe an expansion of activated T cells following ART initiation, an increase in proinflammatory T cells from within the activated T-cell pool could contribute to inflammation. To assess proinflammatory cytokine production by intestinal T cells, we mitogenically stimulated intestinal biopsy specimen single-cell suspensions and PBMCs and measured the frequency of T cells expressing either gamma interferon (IFN-γ) or tumor necrosis factor alpha (TNF-α). Among small intestinal lymphocytes isolated from IN inhibitor-treated animals at 1 to 2 weeks post-ART, we observed increased IFN-γ-expressing CD4+ TM cells compared to levels in controls (unpaired, two-way t test, P = 0.03), and in PR inhibitor-treated animals at 5 to 6 weeks post-ART, we noted increased TNF-α-expressing CD8+ TM cells (P = 0.009) (Fig. 2A). In biopsy specimens of the large intestine, the frequency of CD8+ TM cells expressing IFN-γ at 5 to 6 weeks of treatment initiation was significantly increased compared to that of controls (P = 0.008) (Fig. 2B). Among PBMCs, the fold change from baseline expression of IFN-γ among CD8+ TM cells was significantly higher at 1 to 2 weeks posttreatment in RT inhibitor-treated animals than in controls (P = 0.005) (Fig. 1C).
FIG 2.
ART modestly alters proinflammatory cytokine expression in healthy macaques. (A and B) Individual and mean frequencies of IFN-γ- and TNF-α-expressing CD4+ TM cells and CD8+ TM cells, as indicated, in Jej and RB specimens. Asterisks denote significance of values for ART groups compared to those of the controls, color coded by ART treatment. (C) Fold change from baseline frequencies of IFN-γ- and TNFα-expressing CD4+ TM cells and CD8+ TM cells, as indicated, in PBMCs. (D to G) Pie charts depicting the relative expression levels of IFN-γ, IL-2, IL-17, IL-21, and/or IL-22 in CD8+ TM cells of Jej (D and F) or RB (E and G) specimens at 1 to 2 or at 5 to 6 weeks (Wk) posttreatment in the indicated groups. Pie arcs represent individual cytokines expressed, and pie slices represent the number of coexpressed cytokines, as designated in the legend. Statistical significance between groups in panels A to C was assessed by unpaired, two-way t test: in Jej specimens of IN inhibitor-treated animals, frequency of IFN-γ+ CD4+ TM cells at weeks 1 to 2, P = 0.03; in Jej specimens of PR inhibitor-treated animals, frequency of TNFα+ CD8+ TM cells at weeks 5 to 6, P = 0.009; in RB specimens of PR inhibitor-treated animals, frequency of IFN-γ+ CD8+ TM cells at weeks 5 to 6, P = 0.008; PBMCs of RT inhibitor-treated animals, fold change in frequency of IFN-γ+ CD8+ TM cells at weeks 1 to 2, P = 0.005. Significance for polyfunctional data in panels D to G was assessed by a Spice permutation test.
In addition to skewed frequencies of proinflammatory T cells, infected ARV-treated patients and macaques commonly display an accumulation of monofunctional cells, i.e., those capable of expressing only a single cytokine (36). To determine whether ART alone may skew functionality, we assessed the ability of intestinal T cells to simultaneously express IFN-γ, interleukin-2 (IL-2), IL-17, IL-21, and IL-22 in response to mitogenic stimulation ex vivo. In all cases, a dominant mono-IFN-γ response was evident, with no significant differences observed in functional profiles observed between groups or time points for either CD4+ (data not shown) or CD8+ TM cells (Fig. 2C to G).
ARV administration is associated with perturbations of the bacteriome in healthy macaques.
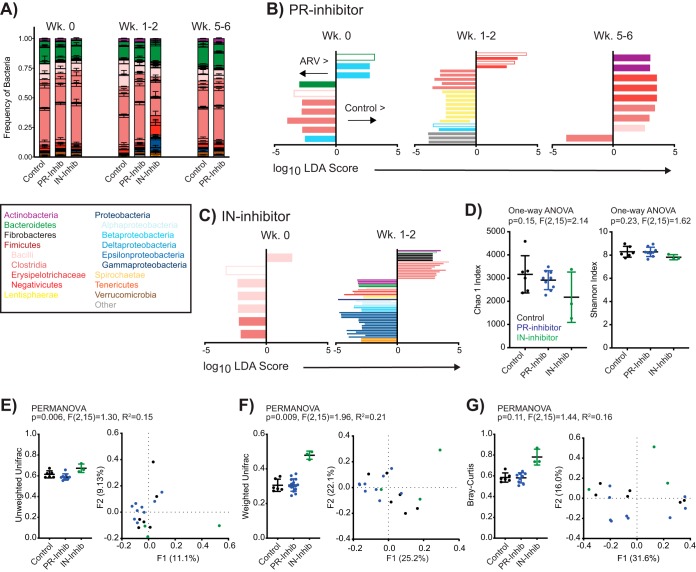
In people living with HIV, GI tract bacterial dysbiosis has been broadly characterized by an overall decline in bacterial richness, including a coincident enrichment for intestinal Proteobacteria at the expense of Firmicutes (12–14). We have similarly observed that the induction of ART in SIV-infected macaques results in intestinal bacterial dysbiosis (18). To determine if the proinflammatory alterations observed in IN and PR inhibitor-treated animals (Fig. 2A to C) could be attributed to bacterial dysbiosis, we isolated fecal bacterial DNA from a subset of these macaques and measured relative bacterial frequencies by next-generation sequencing of 16S rRNA genes. Characterization of taxa of interest revealed that gross community structures were preserved following the administration of ART and compared to those of control animals (Fig. 3A). To determine if alterations in specific taxa were significantly different between control and ARV-treated animals, we next compared frequencies of all identified bacterial taxa between animals utilizing the linear discriminant analysis (LDA) effect size (LEfSe) algorithm (37). Of 402 uniquely identified taxa, 100 features (taxa identified at any phylogenetic level) were significantly different in ARV-treated animals than in controls at either 1 to 2 weeks or 5 to 6 weeks posttreatment but not at baseline or at baseline but not at either posttreatment time point (Fig. 3B and C; see also Table S1 in the supplemental material). Five differentially abundant taxa were identified in both PR and IN inhibitor comparisons: Prevotellaceae NK3B31 species, the genera Blautia of the family Lachnospiraceae and Solobacterium of the family Erysipelotrichaceae, and orders Bacillales (of which Bacillaceae was the only identified family) and Burkholderiales. Importantly, although these taxa were identified in both therapeutic comparisons at a single time point, no features were longitudinally differentially abundant within a therapeutic group.
FIG 3.
ART significantly alters intestinal bacterial diversity absent persistent changes in the stool microbiome. (A) Relative distribution of mean (± standard error of the mean) bacterial frequencies of interest within the stool of control, PR inhibitor-treated, and IN inhibitor-treated animals at designated time points. Bacteria are color coded according to the legend. (B and C) LDA scores for features identified as differentially abundant and biologically relevant by LEfSe in PR inhibitor- and IN inhibitor-treated animals compared to values for the controls at week 0 but not posttreatment (Wk 0) or at weeks 1 to 2 or 5 to 6 posttreatment but not at baseline. Identified taxa are distinguished and color coded at the level of class as in panel A and differentially scored such that taxa enriched in controls are identified on the positive axis and those enriched in ARV-treated rhesus macaques are on the negative axis. Open bars denote identified taxa that were identified in both PR and IN inhibitor comparisons at any time point. (D) Individual and mean (± standard error of the mean) alpha-diversity measures Chao1 and Shannon at 1 to 2 weeks posttreatment, color coded by treatment. (E to G) Beta-diversity measures unweighted UniFrac, weighted UniFrac, and Bray-Curtis analyses, as indicated, of fecal bacteria in rhesus macaques at 1 to 2 weeks posttreatment, color coded by treatment. Data are shown as individual and mean (± standard error of the mean) within-group measures (left) where individual data points represent the mean distance of individual animals to all other animals within the same group, and by PCoA (right). Significance was determined by LEfSe in panels B and C, by one-way ANOVA in panel D, and by PERMANOVA in panels E to G.
To quantify the influence of ART on fecal bacterial communities, we next compared indices of alpha diversity (Chao1 richness and Shannon evenness) and beta diversity (UniFrac and Bray-Curtis dissimilarity) across our treatment groups. At 1 to 2 weeks posttreatment, these analyses revealed no differences across groups for alpha diversity (Fig. 3D). Significant differences in phylogenetic beta diversity were seen across treatment groups, as measured by unweighted UniFrac (permutational multivariate analysis of variance [PERMANOVA], P = 0.006) (Fig. 3E) and weighted UniFrac (P = 0.009) (Fig. 3F). No differences were apparent by nonphylogenetic Bray-Curtis (Fig. 3G) analysis, and posttest analyses (individual PERMANOVA) did not identify a specific post-ART group as significantly different from controls.
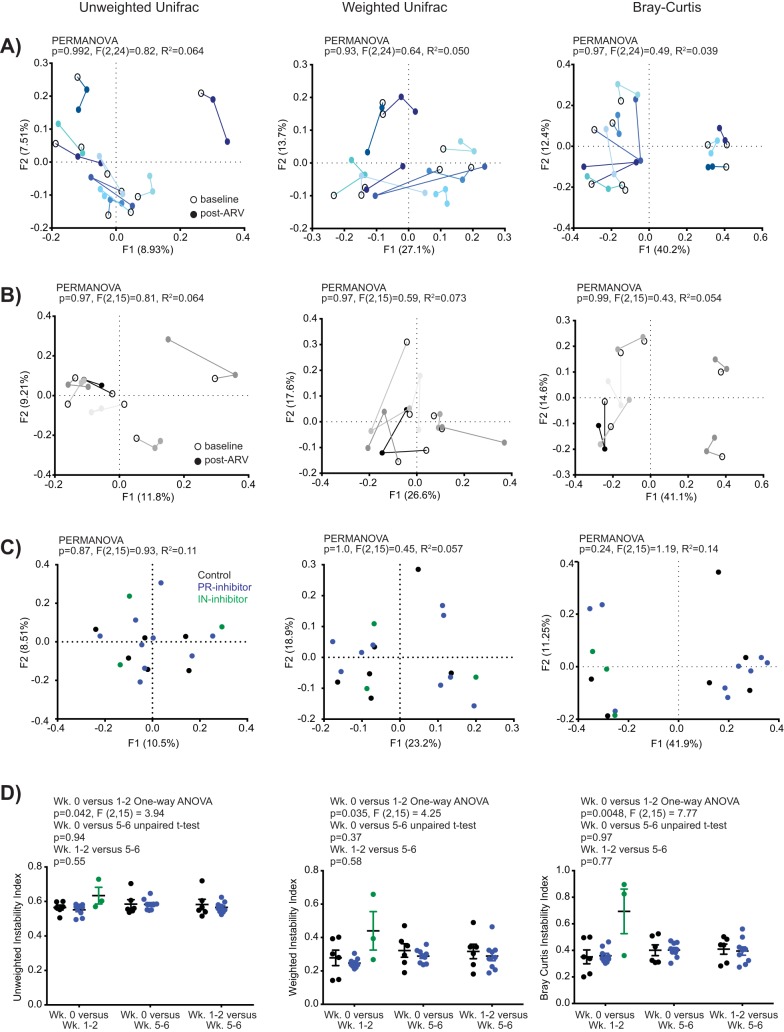
The observed differences in phylogenetic beta diversity absent changes in alpha diversity or persistent shifts in taxon abundance over time may indicate that ART induces bacterial community instability. Indeed, although temporal shifts in community beta diversity (all measures) were evident for each PR-treated animal by principal-coordinate analysis (PCoA) (Fig. 4A), these communities did not segregate by time post-ART. Temporal shifts in control animals were more subdued (Fig. 4B), with variability most evident by weighted UniFrac, and no segregation by assigned treatment group was evident at baseline (Fig. 4C). To assess bacterial community instability, we utilized an instability index whereby each index (value) is derived from the comparison of a single animal’s beta diversity measurement across two time points. For all beta-diversity measurements, the instability index comparing baseline to values at weeks 1 to 2 posttreatment was considered significantly different across treatment groups (Fig. 4D) (unweighted UniFrac one-way analysis of variance [ANOVA], P = 0.042; weighted UniFrac, P = 0.035; Bray-Curtis, P = 0.0048). These differences did not persist at weeks 5 to 6 post-ART.
FIG 4.
ART promotes transient intestinal bacterial instability in the stool microbiome of healthy rhesus macaques. (A to C) Unweighted UniFrac, weighted UniFrac, and Bray-Curtis measures, as indicated, of beta diversity by PCoA for individual PR inhibitor-treated animals across time (A), individual control animals across time (B), and individual animals at baseline (C, color-coded by treatment group). (D) Instability indices comparing beta-diversity measures (as in panel C) across individual time points. Significance was assessed by two-way unpaired t test, one-way ANOVA, or PERMANOVA, as indicated.
Immunological alterations in ARV-treated animals are associated with specific bacterial perturbations.
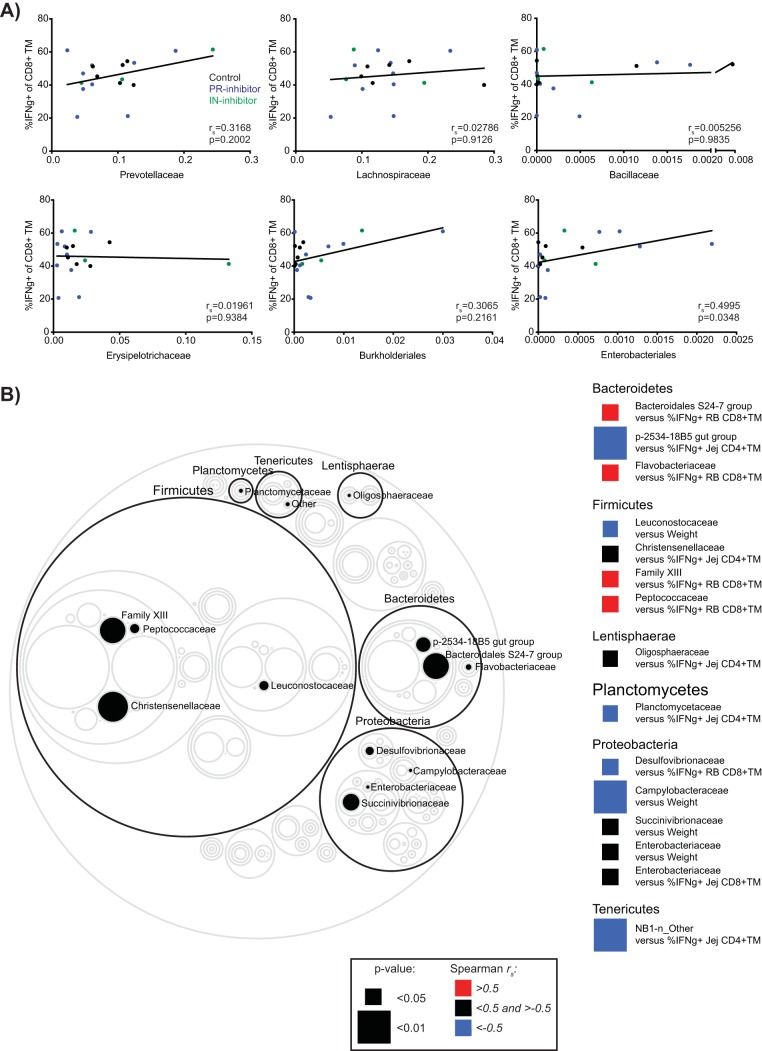
Although the differences in taxon abundances between control and ARV-treated animals were not maintained longitudinally, these differences coincided with differences in proinflammatory cytokine secretion and, thus, may be causally related. In order to investigate whether such a relationship might exist, we assessed whether Prevotellaceae, Bacillaceae, Lachnospiraceae, Erysipelotrichaceae or Burkholderiales, orders pertaining to the differentially abundant operational taxonomic units (OTUs) shared in both the PR and IN inhibitor LEfSe analyses (Fig. 3B and C), correlated with small or large intestine CD8+ TM cell IFN-γ expression at 1 to 2 weeks posttreatment (Fig. 5A). We similarly assessed whether the order Enterobacteriales, commonly identified as a strong correlate of immune dysfunction in HIV-infected individuals, might associate with IFN-γ expression in this time frame. Of the examined relationships, only Enterobacteriales (of which Enterobacteriaceae was the only identified family) showed a significant correlation with Jej CD8+ TM cell IFN-γ expression (P = 0.0348). No correlations were observed in relation to CD8+ IFN-γ expression in RB specimens (data not shown). We further extended our analysis by comparing frequencies of all identified bacterial families in all animals at weeks 1 to 2 post-ART to IFN-γ expression by intestinal T cells as well as to animal weight (Fig. 5B). Isolated associations were identified as significantly different, with the most correlations emerging from among the Clostridiales and Enterobacteriales orders; however, no clear pattern emerged between familial abundance and IFN-γ expression or weight.
FIG 5.
Associations between fecal bacterial taxa perturbed by ART and the expression of T-cell proinflammatory cytokines or animal weight. (A) Association between the frequency of IFN-γ-expressing Jej CD8+ TM cells and relative fecal frequencies of Prevotellaceae, Lachnospiraceae, Bacillaceae, Erysipelotrichaceae, Burkholderiales, and Enterobacteriales at weeks 1 to 2 post-ART. Lines indicate best-fit linear regressions. (B) Circle plot highlighting significant associations between bacterial families and either weight or intestinal T-cell IFN-γ expression at weeks 1 to 2 post-ART, considering all animals. Circles represent relative mean bacterial taxon size. Families of significance are filled in and identified to the right, with each family’s identifying phylum encircled in black and identified above the phylum ring. Significance was assessed by Spearman correlation with resultant rs and P values directly indicated (A) or represented as according to the color legend (B).
DISCUSSION
Antiretrovirals have significantly improved the life span and quality of life for people living with HIV and have proven greater than 75% effective at preventing HIV transmission between sero-discordant partners (38, 39). The introduction of these drugs has, without question, significantly impeded the HIV epidemic. However, prolonged treatment is not without complication for the greater than 21 million individuals accessing ARTs worldwide (40). Non-AIDS-related comorbidities are now a leading cause of mortality for people living with HIV and include cardiovascular events, neurocognitive disorders, gastrointestinal disease, and neoplasia, all of which have been linked to persistent inflammation in treated individuals (1). Although a causal relationship between these comorbidities and ART is confounded by post-HIV replicative damage, gastrointestinal complications are among reported side effects by uninfected individuals taking ARVs as PreP (38, 39). Here, we sought to determine whether ART alone might contribute to gastrointestinal inflammation and, given the intimate link between gastrointestinal health and the composition of the intestinal microbiome, whether ART might contribute to bacterial dysbiosis in a nonhuman primate model of AIDS. We observed a modest induction of inflammation independent of prolonged changes in specific bacterial taxa; however, these inflammatory perturbations coincided with significant differences in bacterial diversity, suggesting that ART alone may destabilize intestinal bacterial communities.
The modest perturbations in proinflammatory responses of intestinal lymphocytes following short-term (1 to 2 weeks) administration of the IN inhibitor L812 and long-term (5 to 6 weeks) administration of the PR inhibitor regimen darunavir-ritonavir are not sufficient to explain the degree of immune dysfunction noted in HIV-infected, ARV-treated individuals. However, these results are in line with previous observations that attribute gastrointestinal complications to PR inhibitor regimens in HIV-infected individuals (9, 10) and to RT inhibitor regimens in users of PreP (38, 39). HIV PR inhibitors target the HIV-1 PR catalytic domain, which maintains many of the same characteristics as nonviral aspartic proteases while nucleoside/nucleotide analog RT inhibitors (NRTIs; currently, all licensed PreP formulations) are incorporated indiscriminately into cellular DNA prior to editing and excision. In vitro, PR inhibitors have been shown to interfere with calcium-dependent chloride conductance (41) and epithelial proliferation and survival (42). Although the role of RT inhibitors in epithelial cell dysfunction is more nuanced, balanced by mitochondrial toxicity and oxidative stress on one hand (43) and inflammasome inhibition on the other (44), direct interference by ARVs with host cellular processes can undoubtedly contribute to gastrointestinal complications.
We along with others have previously reported a posttherapy association between bacterial dysbiosis and intestinal inflammation in HIV-infected individuals (12–14, 45) and SIV-infected macaques (18). Many HIV-1 ARVs target biochemical processes that are conserved across taxonomic kingdoms, with several ARVs showing bioactivity in bacterial, fungal, and protozoal culture systems (33–35). Thus, to our surprise, we did not observe persistent changes in individual taxa in our ARV-treated, uninfected macaques (Fig. 3B and C). There are several, nonexclusive, explanations for our observations. In lentivirus-infected, ARV-treated individuals and macaques, bacterial dysbiosis may be a feature of immune restoration. In SIV-infected macaques, dysbiosis does not appear to be a routine feature of disease progression (19, 46–48) but, rather, has been associated with the initiation of ART (18) and the onset of end-stage disease (46). Thus, in otherwise healthy macaques, ART may not be sufficient to induce dysbiosis. Second, ART-associated dysbiosis may be a feature of combination ART rather than individual ARVs. The combined effects of several ARVs may overwhelm the host cellular or bacterial capacity to mitigate independent off-target effects and, consequently, result in a confluence of abnormalities that promote the prolonged dysbiosis of specific taxa. As combination ART is not currently approved for PreP, such an effect cannot account for intestinal dysfunction in uninfected individuals. However, perhaps it is not a specific dysbiosis per se that is induced by ART during lentiviral infection but, rather, marked instability of the bacterial microbiome, as suggested by our data (Fig. 4).
Disruptions in intestinal bacterial diversity have been described pursuant to HIV infection (13, 14, 16), as well as with progressive (46) and natural SIV infections (49). Although several cross-sectional studies in people living with HIV have described specific alterations in intestinal bacterial distributions (12–15), studies in SIV-infected macaques more commonly describe only temporal disturbances (18, 46). To this end, although we did not observe persistent alterations in specific bacterial taxa after the introduction of ARVs, we observed evidence of distinct phylogenetic communities between cohorts posttreatment as well as sharp increases in the number of differentially abundant taxa compared to levels in control animals (Fig. 3). Our results are in keeping with a more generalized disruptive effect of ART. Indeed, a PCoA examination of the PR inhibitor-treated group alone suggested that continued PR inhibitor treatment in these animals did not promote phylogenetic clustering of intestinal communities posttreatment but, rather, was associated with community destabilization within each animal.
The potential causes for altered bacterial diversity and destabilization in in pre- and postexposure ARV prophylaxis are unclear but likely to be varied. The stability of discrete cross-kingdom networks relies on keystone members, the disruption of which can lead to network restructuring or collapse (50). Previous studies in SIV-infected macaques have revealed a prolonged disruption of the enteric virome (19, 46), a feature which we did not assess here. We also did not assess the influence of ART on systemic inflammation or diversity of microbial populations, which may influence HIV acquisition. A contribution of bacterial dysbiosis to HIV acquisition has been described in females, where vaginal Lactobacillus nondominance, here, increased diversity, is associated with increased genital inflammation (51, 52). Although a contribution of PreP to genital dysbiosis and diversity has not been described, Klatt et. Al. (53) recently demonstrated that vaginal Lactobacillus nondominance (Gardnerella vaginalis dominance) resulted in a 3-fold-reduced efficacy in the CAPRISA-004 topical microbicide (tenofovir) PreP trial and, furthermore, that Gardnerella vaginalis can actively metabolize tenofovir. A full metagenomic analysis of systemic ecosystems will be necessary to fully appreciate the effects of ART on immunity and immune dysfunction.
Specific immunological abnormalities within the GI tract are known to contribute directly to progressive HIV/SIV infections. T regulatory (Treg) CD4+ cells, Th17 cells, Tc17 cells, and innate lymphocyte type III (ILC3) cells are preferentially and persistently lost from the lamina propria of the GI tract in people living with HIV and in SIV-infected macaques and are not restored with the administration of ARVs (54–56). The loss of these cells and altered infiltration of neutrophils into GI tract tissues (57) contribute to epithelial barrier defects, microbial translocation, and inflammation. Our data suggest that ARVs themselves are unlikely to significantly contribute to immunological perturbations observed in ARV-treated, HIV-infected individuals (Fig. 2), and, thus, repairing these immunological perturbations might require adjuvant therapy. The administration of probiotics (live bacterial taxa which are thought to provide benefit) has been shown to impart immunological benefit to people living with HIV and in SIV-infected macaques on ART (58–62). This immunological benefit might be related to probiotics stabilizing the composition of the microbiome, which we observed to become disrupted after administration of ARVs both here in healthy macaques as well as after SIV infection (18).
In conclusion, we demonstrate that ARVs alone induce intestinal bacterial instability and a modestly proinflammatory milieu in a nonhuman primate model for AIDS. Importantly, given the significant alterations to the composition of the microbiome and to the GI tract immune system observed in people living with HIV and in SIV-infected macaques, ARV-induced instability to the microbiome may exacerbate these disruptions. Our work can inform an essential framework for understanding how ART itself contributes to immune reconstitution and non-AIDS-related comorbidities in people living with HIV. The identification of keystone organisms or biochemical processes altered by ART is necessary for informing the design of next-generation ARVs and palliative therapeutics.
MATERIALS AND METHODS
Study design.
Thirty-one rhesus macaques (Macaca mulatta) were assigned to a control (n = 6), PR inhibitor (n = 13), IN inhibitor (n = 6), or RT inhibitor (n = 6) group and treated daily for 1 to 6 weeks as detailed in Table 1. Groups were stratified by weight and sex, and animals were sampled as mixed populations. PR inhibitor-treated animals received darunavir and ritonavir (400 mg and 100 mg, respectively, orally twice a day [b.i.d.]), IN inhibitor-treated animals received L812 (120 mg orally b.i.d.), and RT inhibitor-treated animals received PMPA or FTC (30 mg/kg subcutaneously [s.c.] once daily [s.i.d.]).
The NIAID Division of Intramural Research Animal Care and Use Program, as part of the NIH Intramural Research Program, approved all experimental procedures (protocol LVD 26). The Program complies with all applicable provisions of the Animal Welfare Act and other federal statutes and regulations relating to animals.
Animals were housed and cared for at the NIH Animal Center, under the supervision of the Association for the Assessment and Accreditation of Laboratory Animal Care (AAALAC)-accredited Division of Veterinary Resources and as recommended by the Office of Animal Care and Use Nonhuman Primate Management Plan. Husbandry and care met the standards set forth by the Animal Welfare Act, Animal Welfare Regulations, as well as of The Guide for the Care and Use of Laboratory Animals (63). The physical conditions of the animals were monitored daily. Animals in this study were exempt from contact social housing due to scientific justification, per IACUC protocol, and were housed in noncontact, social housing where primary enclosures consisted of stainless steel primate caging. Animals were provided continuous access to water and offered commercial monkey biscuits twice daily as well as fresh produce, eggs, and bread products twice weekly and a foraging mix consisting of raisins, nuts, and rice thrice weekly. Enrichment to stimulate foraging and play activity was provided in the form of food puzzles, toys, cage furniture, and mirrors.
Sample collection.
Blood, stool, and intestinal biopsy specimens were collected longitudinally, with each measurement resulting from a single sample per time point. Biopsy specimens were maintained in RPMI medium prior to processing. Mononuclear cells were isolated from blood by Ficoll gradient centrifugation. Stool was collected fresh from each animal by inserting a sterile swab 2 cm into the rectum and swirling to collect available sample. For RB specimens, fecal material was removed from the rectum, and biopsy specimens were obtained with biopsy forceps. Jejunal biopsy specimens were obtained by video-guided endoscopy. Sampling occurred in random order. Neither the investigators nor the animal handlers were blinded to group allocation in order to ensure multilateral supervision of design and palliative treatment. Animals were sedated with tiletamine and zolazepam (Telazol) at 3 to 4 mg/kg intramuscularly (i.m.) and with isoflurane gas by intubation, to effect, for sample collection. Successful anesthetization was monitored by response to stimuli.
Immune phenotyping and functional assessment.
Polychromatic flow cytometry was performed on stained mononuclear cells as previously described (61). Antibodies against the following antigens were used for staining and gating at predetermined concentrations: CCR5 (clone 3A9) conjugated to phycoerythrin (PE), CCR7 (3D12) PE-Cy7 or Alexa Fluor 647, CD3 (SP34-2) Alexa Fluor 700, CD8 (SK1) allophycocyanin (APC)-H7, CD20 (L27) PE-Cy7, CD28 (CD28.2) PE-Cy5, CD45 (D058-1283) BVio786 or PE-CF594, HLA-DR (G46-6) PE, or HLA-DR (L243) APC-H7 (all, BD Biosciences); CD4 (OKT4) eFluor450, CD8 (SK1) peridinin chlorophyll protein (PerCP)-eFluor710, IL-17 (eBio64Dec17) Alexa Fluor 488, IL-21 (3A3-N2) PE, IL-22 (IL22JOP) APC or PerCP-eFluor710, IFN-γ (4S.B3) eFluor450, TNF-α (Mab11) PE-Cy7 (all, Thermo Fisher Scientific); CD95 (DX2) PE-Cy5 or BVio650, HLAD-DR (L243) BVio711, IL-2 (MQ1-17H12) BVio650, TNF-α (Mab11) BVio605 (all, Biolegend); and CD28 (CD28.2) energy-coupled dye (ECD) (Beckman Coulter). Cell viability was assessed using Live/Dead Aqua Fixable Dead Cell Stain (Thermo Fisher Scientific). Gating was performed as previously described (17). In brief, CD4+ and CD8+ TM cells were defined as CD95+ singlet, clean, live, CD3+ lymphocytes. Positive/negative gating was based on clearly grouped populations, historically determined expression, and the use of internal controls. A minimum threshold of 100 collected events in the parent population was utilized for all subset expression analysis.
16S isolation and analysis.
A total of 250 mg of stool was transferred to soil-grinding Percellys tubes (Bertin Technologies) and homogenized at room temperature on a Precellys 24 homogenizer at 5,000 rpm in four successive 20-s intervals, with 750 μl of PowerBead solution (Qiagen) and 60 μl of solution C1 (Qiagen). Residual particulate was removed from the supernatant by centrifugation at 10,000 × g for 30 s at room temperature. Supernatant was transferred to a 96-well deep-well plate (Costar) and processed as follows with solution addition and supernatant transfer/removal facilitated using a Biomek NKP (Beckman Coulter) system: (step 1) 250 μl of solution C2 (Qiagen) was added to supernatant and mixed at 1,000 rpm for 10 min at room temperature on an orbital shaker; (step 2) the 96-well plate was sealed with an aluminum plate sealer and centrifuged at 4,500 × g for 10 min at 4°C; (step 3) 580 μl of supernatant was transferred to a new 96-well deep-well plate; (step 4) 200 μl of solution C3 (Qiagen) was added to the supernatant and mixed at 1,000 rpm for 10 min at room temperature; (step 5) 0.6 ng of an internal amplification control (GenBank accession number FJ357008) was added to each well as an internal amplification control; (step 6) the 96-well plate was sealed with an aluminum plate sealer and centrifuged at 4,500 × g for 10 min at 4°C; (step 7) 450 μl of supernatant was transferred to a new 96-well deep-well plate; (step 8) 450 μl of ClearMag (Qiagen) magnetic bead solution consisting of 19 μl of beads and 431 μl of binding solution was added to the supernatant and mixed at 1,000 rpm for 10 min at room temperature; (step 9) supernatant-containing plate was transferred to rest on a magnetic plate (Magnum FLX; Alpaqua), and bead-bound DNA was allowed to precipitate for 15 min at room temperature; (step 10) 820 μl of supernatant was removed following precipitation; (step 11) the deep-well plate was removed from the magnet, and bead-bound DNA washed three times with 500 μl of solution C5-D (Qiagen) on an orbital shaker, with between-wash magnetic pelleting and supernatant removal as in steps 9 and 10; (step 12) after final washing, pelleted DNA was allowed to dry overnight at room temperature; (step 13) 200 μl of sterile water was added to elute the DNA and mixed at 1,000 rpm for 15 min at room temperature; (step 14) the deep-well plate was transferred back to the magnetic plate where DNA-free beads were allowed to pellet for 10 min; (step 15) 200 μl of DNA-containing supernatant was transferred to a new deep-well plate. Eluted DNA was cleaned using a Qiagen DNeasy Blood and Tissue kit per the manufacturer’s protocol, beginning with the addition of buffer AL (64).
Total DNA (105 ng) was subjected to dual-index amplification and sequencing for nonhuman primate fecal pellets, using barcoded universal primers spanning base pairs 515(F) to 806(R) of the bacterial 16S rRNA V4 region. 16S amplicons were purified using Agencourt AMPure XP PCR purification (Beckman Coulter) both before and after index amplification. Indexed 16S amplicons were quantified by a KAPA quantitative PCR (qPCR) library quantification kit for Illumina Platforms (6-carboxy-X-rhodamine [ROX] low; KAPA Biosystems), normalized to 15 pM, spiked in with a 15% phiX control library (Illumina), and sequenced on a MiSeq platform (Illumina MiSeq, version 3, reagent cartridge kit; MiSeq, version 3, reagent kit; flow cell; PR2 buffer kit) utilizing a 300-paired-end reagent plate.
Raw Illumina FASTQ files were first demultiplexed using a custom Python script. Returned paired-end FASTQ reads were filtered and processed using QIIME (version 1.9.1) (65) through the NIAID/NIH Microbiome Analysis Platform, Nephele (Office of Cyber Infrastructure and Computational Biology/NIAID/NIH [http://nephele.niaid.nih.gov]) (66). Before quality trimming, approximately 5 million reads were included in the 51 samples with an average of 98,654 reads (median of 89,350 reads, maximum of 173,767 reads, and minimum of 12,324 reads) per sample. Sequences with any degenerate bases (e.g., N), a Phred quality score of less of than 25 per base, and more than three consecutive low-quality base calls were filtered out. Assessment of the read quality was performed using MultiQC (67). QIIME quality trimming resulted in approximately 3.8 million high-quality reads for all the samples, with an average of 75,099 reads (median of 43,534, maximum of 167,988, and minimum of 11,827 reads) per sample. On average, 76.13% of reads passed the quality filtering. All samples were included in the downstream quantitative analyses. Sequences were binned into OTUs and taxonomically assigned at 99% identity using the QIIME open reference OTU picking workflow with SortMeRNA (68). Sequences were taxonomically classified using the Silva 99% database (release 132). Frequencies of microbial taxa are calculated relative to all returned bacterial OTUs mapping to nonmitochondrial (Rickettsiales mitochondria) taxa.
Statistical analyses.
Unpaired, two-way t tests were used in statistical analyses of single-parameter lymphocyte phenotype and function (Prism, version 7.0; GraphPad Software, Inc.). Significance of polyfunctional cytokine expression was assessed using the Spice (version 6.0; NIAID) permutation test on relative expression values. The empirical identification of differentially abundant bacterial taxa between groups was determined using the LEfSe algorithm (37) one-against-all multiclass analysis, normalized to 106 reads. Microbial alpha diversity was assessed by Chao1 and Shannon indices from 10 random subsamplings of the QIIME OTU table at a sampling depth of 11,179 reads per sample, with statistical significance assessed by one-way ANOVA with Dunnett’s posttest at a 0.95 confidence level (Prism, version 7). Beta diversity was assessed by unweighted and weighted UniFrac (69) and Bray-Curtis dissimilarity indices, with statistical significance assessed by PERMANOVA (R-vegan function Adonis). Within-group analyses in Fig. 3 refer to comparisons run within a treatment group (control versus control animals, PR inhibitor-treated versus PR inhibitor-treated animals, and IN inhibitor-treated versus IN inhibitor-treated animals) and are derived from the specified beta diversity matrix. PERMANOVA analyses in Fig. 3 and 4 consider intergroup indices (control versus PR inhibitor-treated animals, control versus IN inhibitor-treated animals, and PR inhibitor-treated versus IN inhibitor-treated animals). PCoA plots generated by XLSTAT (version 20.7; Addinsoft). Instability indices were identified as beta-diversity indices of single animals across treatment-relative time points, with statistical significance assessed by one-way ANOVA or unpaired t test as required by number of comparisons. Spearman correlations were performed without multiple-comparison adjustments on OTUs identified as differentially abundant in both the PR and IN inhibitor LEfSe analyses or for bacterial families where greater than one-half of the samples had nonzero reads. No data that met minimum threshold requirements as outlined in Materials and Methods were excluded.
Data availability.
16S miSeq data were deposited in the NCBI Sequence Read Archive (SRA) under project number PRJNA526520.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Heather Kendall, JoAnne Swerczek, Richard Herbert, and all the veterinary staff at the NIH animal center for their excellent veterinary care. We thank Merck and Gilead for providing PMPA, FTC, and L812. We thank Miriam Quiñones and the NIAID Microbiome Program for technical and analytical assistance.
Funding for this study was provided in part by the Division of Intramural Research/NIAID/NIH.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00472-19.
REFERENCES
- 1.Hunt PW, Lee SA, Siedner MJ. 2016. Immunologic biomarkers, morbidity, and mortality in treated HIV infection. J Infect Dis 214(Suppl 2):S44–S50. doi: 10.1093/infdis/jiw275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zeng M, Smith AJ, Wietgrefe SW, Southern PJ, Schacker TW, Reilly CS, Estes JD, Burton GF, Silvestri G, Lifson JD, Carlis JV, Haase AT. 2011. Cumulative mechanisms of lymphoid tissue fibrosis and T cell depletion in HIV-1 and SIV infections. J Clin Invest 121:998–1008. doi: 10.1172/JCI45157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wherry EJ. 2011. T cell exhaustion. Nat Immunol 12:492–499. doi: 10.1038/ni.2035. [DOI] [PubMed] [Google Scholar]
- 4.Hellmuth J, Slike BM, Sacdalan C, Best J, Kroon E, Phanuphak N, Fletcher JLK, Prueksakaew P, Jagodzinski LL, Valcour V, Robb M, Ananworanich J, Allen IE, Krebs SJ, Spudich S, Search RV, SEARCH 010/RV254 and SEARCH013/RV304 Study Groups. 21 January 2019. Very early ART initiation during acute HIV infection is associated with normalization of cerebrospinal fluid but not plasma markers of immune activation. J Infect Dis. doi: 10.1093/infdis/jiz030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tauriainen J, Scharf L, Frederiksen J, Naji A, Ljunggren HG, Sonnerborg A, Lund O, Reyes-Teran G, Hecht FM, Deeks SG, Betts MR, Buggert M, Karlsson AC. 2017. Perturbed CD8+ T cell TIGIT/CD226/PVR axis despite early initiation of antiretroviral treatment in HIV infected individuals. Sci Rep 7:40354. doi: 10.1038/srep40354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sereti I, Krebs SJ, Phanuphak N, Fletcher JL, Slike B, Pinyakorn S, O'Connell RJ, Rupert A, Chomont N, Valcour V, Kim JH, Robb ML, Michael NL, Douek DC, Ananworanich J, Utay NS, RV254/SEARCH 010, RV304/SEARCH 013 and SEARCH 011 protocol teams. 2017. Persistent, albeit reduced, chronic inflammation in persons starting antiretroviral therapy in acute HIV infection. Clin Infect Dis 64:124–131. doi: 10.1093/cid/ciw683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deleage C, Schuetz A, Alvord WG, Johnston L, Hao XP, Morcock DR, Rerknimitr R, Fletcher JL, Puttamaswin S, Phanuphak N, Dewar R, McCune JM, Sereti I, Robb M, Kim JH, Schacker TW, Hunt P, Lifson JD, Ananworanich J, Estes JD. 2016. Impact of early cART in the gut during acute HIV infection. JCI Insight 1:e87065. doi: 10.1172/jci.insight.87065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tetteh RA, Yankey BA, Nartey ET, Lartey M, Leufkens HG, Dodoo AN. 2017. Pre-exposure prophylaxis for HIV prevention: safety concerns. Drug Saf 40:273–283. doi: 10.1007/s40264-017-0505-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Logan C, Beadsworth MB, Beeching NJ. 2016. HIV and diarrhoea: what is new? Curr Opin Infect Dis 29:486–494. doi: 10.1097/QCO.0000000000000305. [DOI] [PubMed] [Google Scholar]
- 10.Dikman AE, Schonfeld E, Srisarajivakul NC, Poles MA. 2015. Human immunodeficiency virus-associated diarrhea: still an issue in the era of antiretroviral therapy. Dig Dis Sci 60:2236–2245. doi: 10.1007/s10620-015-3615-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bares SH, Sandkovsky US, Talmon GA, Hutchins GF, Swindells S, Scarsi KK. 2016. Dolutegravir-induced colitis in an HIV-infected patient. J Antimicrob Chemother 71:281–282. doi: 10.1093/jac/dkv296. [DOI] [PubMed] [Google Scholar]
- 12.Vujkovic-Cvijin I, Dunham RM, Iwai S, Maher MC, Albright RG, Broadhurst MJ, Hernandez RD, Lederman MM, Huang Y, Somsouk M, Deeks SG, Hunt PW, Lynch SV, McCune JM. 2013. Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci Transl Med 5:193ra91. doi: 10.1126/scitranslmed.3006438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monaco CL, Gootenberg DB, Zhao G, Handley SA, Ghebremichael MS, Lim ES, Lankowski A, Baldridge MT, Wilen CB, Flagg M, Norman JM, Keller BC, Luevano JM, Wang D, Boum Y, Martin JN, Hunt PW, Bangsberg DR, Siedner MJ, Kwon DS, Virgin HW. 2016. Altered virome and bacterial microbiome in human immunodeficiency virus-associated acquired immunodeficiency syndrome. Cell Host Microbe 19:311–322. doi: 10.1016/j.chom.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lozupone CA, Li M, Campbell TB, Flores SC, Linderman D, Gebert MJ, Knight R, Fontenot AP, Palmer BE. 2013. Alterations in the gut microbiota associated with HIV-1 infection. Cell Host Microbe 14:329–339. doi: 10.1016/j.chom.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dillon SM, Lee EJ, Kotter CV, Austin GL, Dong Z, Hecht DK, Gianella S, Siewe B, Smith DM, Landay AL, Robertson CE, Frank DN, Wilson CC. 2014. An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol 7:983–994. doi: 10.1038/mi.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noguera-Julian M, Rocafort M, Guillen Y, Rivera J, Casadella M, Nowak P, Hildebrand F, Zeller G, Parera M, Bellido R, Rodriguez C, Carrillo J, Mothe B, Coll J, Bravo I, Estany C, Herrero C, Saz J, Sirera G, Torrela A, Navarro J, Crespo M, Brander C, Negredo E, Blanco J, Guarner F, Calle ML, Bork P, Sonnerborg A, Clotet B, Paredes R. 2016. Gut microbiota linked to sexual preference and HIV infection. EBioMedicine 5:135–146. doi: 10.1016/j.ebiom.2016.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ortiz AM, Flynn JK, DiNapoli SR, Vujkovic-Cvijin I, Starke CE, Lai SH, Long ME, Sortino O, Vinton CL, Mudd JC, Johnston L, Busman-Sahay K, Belkaid Y, Estes JD, Brenchley JM. 2018. Experimental microbial dysbiosis does not promote disease progression in SIV-infected macaques. Nat Med 24:1313–1316. doi: 10.1038/s41591-018-0132-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klase Z, Ortiz A, Deleage C, Mudd JC, Quinones M, Schwartzman E, Klatt NR, Canary L, Estes JD, Brenchley JM. 2015. Dysbiotic bacteria translocate in progressive SIV infection. Mucosal Immunol 8:1009–1020. doi: 10.1038/mi.2014.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Handley SA, Thackray LB, Zhao G, Presti R, Miller AD, Droit L, Abbink P, Maxfield LF, Kambal A, Duan E, Stanley K, Kramer J, Macri SC, Permar SR, Schmitz JE, Mansfield K, Brenchley JM, Veazey RS, Stappenbeck TS, Wang D, Barouch DH, Virgin HW. 2012. Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome. Cell 151:253–266. doi: 10.1016/j.cell.2012.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belkaid Y, Harrison OJ. 2017. Homeostatic immunity and the microbiota. Immunity 46:562–576. doi: 10.1016/j.immuni.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maier L, Pruteanu M, Kuhn M, Zeller G, Telzerow A, Anderson EE, Brochado AR, Fernandez KC, Dose H, Mori H, Patil KR, Bork P, Typas A. 2018. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 555:623–628. doi: 10.1038/nature25979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Falony G, Joossens M, Vieira-Silva S, Wang J, Darzi Y, Faust K, Kurilshikov A, Bonder MJ, Valles-Colomer M, Vandeputte D, Tito RY, Chaffron S, Rymenans L, Verspecht C, De Sutter L, Lima-Mendez G, D’hoe K, Jonckheere K, Homola D, Garcia R, Tigchelaar EF, Eeckhaudt L, Fu J, Henckaerts L, Zhernakova A, Wijmenga C, Raes J. 2016. Population-level analysis of gut microbiome variation. Science 352:560–564. doi: 10.1126/science.aad3503. [DOI] [PubMed] [Google Scholar]
- 23.Ejim L, Farha MA, Falconer SB, Wildenhain J, Coombes BK, Tyers M, Brown ED, Wright GD. 2011. Combinations of antibiotics and nonantibiotic drugs enhance antimicrobial efficacy. Nat Chem Biol 7:348–350. doi: 10.1038/nchembio.559. [DOI] [PubMed] [Google Scholar]
- 24.Imhann F, Bonder MJ, Vich Vila A, Fu J, Mujagic Z, Vork L, Tigchelaar EF, Jankipersadsing SA, Cenit MC, Harmsen HJ, Dijkstra G, Franke L, Xavier RJ, Jonkers D, Wijmenga C, Weersma RK, Zhernakova A. 2016. Proton pump inhibitors affect the gut microbiome. Gut 65:740–748. doi: 10.1136/gutjnl-2015-310376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jackson MA, Goodrich JK, Maxan ME, Freedberg DE, Abrams JA, Poole AC, Sutter JL, Welter D, Ley RE, Bell JT, Spector TD, Steves CJ. 2016. Proton pump inhibitors alter the composition of the gut microbiota. Gut 65:749–756. doi: 10.1136/gutjnl-2015-310861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun L, Xie C, Wang G, Wu Y, Wu Q, Wang X, Liu J, Deng Y, Xia J, Chen B, Zhang S, Yun C, Lian G, Zhang X, Zhang H, Bisson WH, Shi J, Gao X, Ge P, Liu C, Krausz KW, Nichols RG, Cai J, Rimal B, Patterson AD, Wang X, Gonzalez FJ, Jiang C. 2018. Gut microbiota and intestinal FXR mediate the clinical benefits of metformin. Nat Med 24:1919–1929. doi: 10.1038/s41591-018-0222-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qiu X, Zhang F, Yang X, Wu N, Jiang W, Li X, Li X, Liu Y. 2015. Changes in the composition of intestinal fungi and their role in mice with dextran sulfate sodium-induced colitis. Sci Rep 5:10416. doi: 10.1038/srep10416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haaber J, Leisner JJ, Cohn MT, Catalan-Moreno A, Nielsen JB, Westh H, Penadés JR, Ingmer H. 2016. Bacterial viruses enable their host to acquire antibiotic resistance genes from neighbouring cells. Nat Commun 7:13333. doi: 10.1038/ncomms13333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arts EJ, Hazuda DJ. 2012. HIV-1 antiretroviral drug therapy. Cold Spring Harb Perspect Med 2:a007161. doi: 10.1101/cshperspect.a007161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.El Safadi Y, Vivet-Boudou V, Marquet R. 2007. HIV-1 reverse transcriptase inhibitors. Appl Microbiol Biotechnol 75:723–737. doi: 10.1007/s00253-007-0919-7. [DOI] [PubMed] [Google Scholar]
- 31.Ghosh AK, Osswald HL, Prato G. 2016. Recent progress in the development of HIV-1 protease inhibitors for the treatment of HIV/AIDS. J Med Chem 59:5172–5208. doi: 10.1021/acs.jmedchem.5b01697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Craigie R. 2014. The road to HIV-1 integrase inhibitors: the case for supporting basic research. Future Virol 9:899–903. doi: 10.2217/fvl.14.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andrews KT, Fairlie DP, Madala PK, Ray J, Wyatt DM, Hilton PM, Melville LA, Beattie L, Gardiner DL, Reid RC, Stoermer MJ, Skinner-Adams T, Berry C, McCarthy JS. 2006. Potencies of human immunodeficiency virus protease inhibitors in vitro against Plasmodium falciparum and in vivo against murine malaria. Antimicrob Agents Chemother 50:639–648. doi: 10.1128/AAC.50.2.639-648.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dunn LA, Andrews KT, McCarthy JS, Wright JM, Skinner-Adams TS, Upcroft P, Upcroft JA. 2007. The activity of protease inhibitors against Giardia duodenalis and metronidazole-resistant Trichomonas vaginalis. Int J Antimicrob Agents 29:98–102. doi: 10.1016/j.ijantimicag.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 35.Monari C, Pericolini E, Bistoni G, Cenci E, Bistoni F, Vecchiarelli A. 2005. Influence of indinavir on virulence and growth of Cryptococcus neoformans. J Infect Dis 191:307–311. doi: 10.1086/426828. [DOI] [PubMed] [Google Scholar]
- 36.Betts MR, Nason MC, West SM, De Rosa SC, Migueles SA, Abraham J, Lederman MM, Benito JM, Goepfert PA, Connors M, Roederer M, Koup RA. 2006. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood 107:4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. 2011. Metagenomic biomarker discovery and explanation. Genome Biol 12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grant RM, Lama JR, Anderson PL, McMahan V, Liu AY, Vargas L, Goicochea P, Casapia M, Guanira-Carranza JV, Ramirez-Cardich ME, Montoya-Herrera O, Fernandez T, Veloso VG, Buchbinder SP, Chariyalertsak S, Schechter M, Bekker LG, Mayer KH, Kallas EG, Amico KR, Mulligan K, Bushman LR, Hance RJ, Ganoza C, Defechereux P, Postle B, Wang F, McConnell JJ, Zheng JH, Lee J, Rooney JF, Jaffe HS, Martinez AI, Burns DN, Glidden DV, iPrEx Study Team. 2010. Preexposure chemoprophylaxis for HIV prevention in men who have sex with men. N Engl J Med 363:2587–2599. doi: 10.1056/NEJMoa1011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baeten JM, Donnell D, Ndase P, Mugo NR, Campbell JD, Wangisi J, Tappero JW, Bukusi EA, Cohen CR, Katabira E, Ronald A, Tumwesigye E, Were E, Fife KH, Kiarie J, Farquhar C, John-Stewart G, Kakia A, Odoyo J, Mucunguzi A, Nakku-Joloba E, Twesigye R, Ngure K, Apaka C, Tamooh H, Gabona F, Mujugira A, Panteleeff D, Thomas KK, Kidoguchi L, Krows M, Revall J, Morrison S, Haugen H, Emmanuel-Ogier M, Ondrejcek L, Coombs RW, Frenkel L, Hendrix C, Bumpus NN, Bangsberg D, Haberer JE, Stevens WS, Lingappa JR, Celum C, Partners PrEP Study Team. 2012. Antiretroviral prophylaxis for HIV prevention in heterosexual men and women. N Engl J Med 367:399–410. doi: 10.1056/NEJMoa1108524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.UNAIDS. 2017. Right to health. https://www.unaids.org/en/20171120_right_to_health_report.
- 41.Rufo PA, Lin PW, Andrade A, Jiang L, Rameh L, Flexner C, Alper SL, Lencer WI. 2004. Diarrhea-associated HIV-1 APIs potentiate muscarinic activation of Cl− secretion by T84 cells via prolongation of cytosolic Ca2+ signaling. Am J Physiol Cell Physiol 286:C998–C1008. doi: 10.1152/ajpcell.00357.2003. [DOI] [PubMed] [Google Scholar]
- 42.Braga Neto MB, Aguiar CV, Maciel JG, Oliveira BM, Sevilleja JE, Oria RB, Brito GA, Warren CA, Guerrant RL, Lima AA. 2010. Evaluation of HIV protease and nucleoside reverse transcriptase inhibitors on proliferation, necrosis, apoptosis in intestinal epithelial cells and electrolyte and water transport and epithelial barrier function in mice. BMC Gastroenterol 10:90. doi: 10.1186/1471-230X-10-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hladik F, Burgener A, Ballweber L, Gottardo R, Vojtech L, Fourati S, Dai JY, Cameron MJ, Strobl J, Hughes SM, Hoesley C, Andrew P, Johnson S, Piper J, Friend DR, Ball TB, Cranston RD, Mayer KH, McElrath MJ, McGowan I. 2015. Mucosal effects of tenofovir 1% gel. Elife 4:e04525. doi: 10.7554/eLife.07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fowler BJ, Gelfand BD, Kim Y, Kerur N, Tarallo V, Hirano Y, Amarnath S, Fowler DH, Radwan M, Young MT, Pittman K, Kubes P, Agarwal HK, Parang K, Hinton DR, Bastos-Carvalho A, Li S, Yasuma T, Mizutani T, Yasuma R, Wright C, Ambati J. 2014. Nucleoside reverse transcriptase inhibitors possess intrinsic anti-inflammatory activity. Science 346:1000–1003. doi: 10.1126/science.1261754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pinto-Cardoso S, Lozupone C, Briceño O, Alva-Hernández S, Téllez N, Adriana A, Murakami-Ogasawara A, Reyes-Terán G. 2017. Fecal bacterial communities in treated HIV infected individuals on two antiretroviral regimens. Sci Rep 7:43741. doi: 10.1038/srep43741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Handley SA, Desai C, Zhao G, Droit L, Monaco CL, Schroeder AC, Nkolola JP, Norman ME, Miller AD, Wang D, Barouch DH, Virgin HW. 2016. SIV infection-mediated changes in gastrointestinal bacterial microbiome and virome are associated with immunodeficiency and prevented by vaccination. Cell Host Microbe 19:323–335. doi: 10.1016/j.chom.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McKenna P, Hoffmann C, Minkah N, Aye PP, Lackner A, Liu Z, Lozupone CA, Hamady M, Knight R, Bushman FD. 2008. The macaque gut microbiome in health, lentiviral infection, and chronic enterocolitis. PLoS Pathog 4:e20. doi: 10.1371/journal.ppat.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ortiz AM, Brenchley JM. 2018. Microbial translocation: translating simian immunodeficiency virus to HIV. Curr Opin HIV AIDS 13:15–21. doi: 10.1097/COH.0000000000000424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moeller AH, Shilts M, Li Y, Rudicell RS, Lonsdorf EV, Pusey AE, Wilson ML, Hahn BH, Ochman H. 2013. SIV-induced instability of the chimpanzee gut microbiome. Cell Host Microbe 14:340–345. doi: 10.1016/j.chom.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tipton L, Muller CL, Kurtz ZD, Huang L, Kleerup E, Morris A, Bonneau R, Ghedin E. 2018. Fungi stabilize connectivity in the lung and skin microbial ecosystems. Microbiome 6:12. doi: 10.1186/s40168-017-0393-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anahtar MN, Byrne EH, Doherty KE, Bowman BA, Yamamoto HS, Soumillon M, Padavattan N, Ismail N, Moodley A, Sabatini ME, Ghebremichael MS, Nusbaum C, Huttenhower C, Virgin HW, Ndung'u T, Dong KL, Walker BD, Fichorova RN, Kwon DS. 2015. Cervicovaginal bacteria are a major modulator of host inflammatory responses in the female genital tract. Immunity 42:965–976. doi: 10.1016/j.immuni.2015.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zevin AS, Xie IY, Birse K, Arnold K, Romas L, Westmacott G, Novak RM, McCorrister S, McKinnon LR, Cohen CR, Mackelprang R, Lingappa J, Lauffenburger DA, Klatt NR, Burgener AD. 2016. Microbiome composition and function drives wound-healing impairment in the female genital tract. PLoS Pathog 12:e1005889. doi: 10.1371/journal.ppat.1005889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Klatt NR, Cheu R, Birse K, Zevin AS, Perner M, Noel-Romas L, Grobler A, Westmacott G, Xie IY, Butler J, Mansoor L, McKinnon LR, Passmore JS, Abdool Karim Q, Abdool Karim SS, Burgener AD. 2017. Vaginal bacteria modify HIV tenofovir microbicide efficacy in African women. Science 356:938–945. doi: 10.1126/science.aai9383. [DOI] [PubMed] [Google Scholar]
- 54.Shah SV, Manickam C, Ram DR, Reeves RK. 2017. Innate lymphoid cells in HIV/SIV infections. Front Immunol 8:1818. doi: 10.3389/fimmu.2017.01818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hartigan-O'Connor DJ, Hirao LA, McCune JM, Dandekar S. 2011. Th17 cells and regulatory T cells in elite control over HIV and SIV. Curr Opin HIV AIDS 6:221–227. doi: 10.1097/COH.0b013e32834577b3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mudd JC, Brenchley JM. 2016. Gut mucosal barrier dysfunction, microbial dysbiosis, and their role in HIV-1 disease progression. J Infect Dis 214(Suppl 2):S58–S66. doi: 10.1093/infdis/jiw258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hensley-McBain T, Wu MC, Manuzak JA, Cheu RK, Gustin A, Driscoll CB, Zevin AS, Miller CJ, Coronado E, Smith E, Chang J, Gale M Jr, Somsouk M, Burgener AD, Hunt PW, Hope TJ, Collier AC, Klatt NR. 2019. Increased mucosal neutrophil survival is associated with altered microbiota in HIV infection. PLoS Pathog 15:e1007672. doi: 10.1371/journal.ppat.1007672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kazemi A, Djafarian K, Speakman JR, Sabour P, Soltani S, Shab-Bidar S. 2018. Effect of Probiotic Supplementation on CD4 Cell Count in HIV-Infected Patients: A Systematic Review and Meta-analysis. J Diet Suppl 15:776–788. doi: 10.1080/19390211.2017.1380103. [DOI] [PubMed] [Google Scholar]
- 59.d'Ettorre G, Rossi G, Scagnolari C, Andreotti M, Giustini N, Serafino S, Schietroma I, Scheri GC, Fard SN, Trinchieri V, Mastromarino P, Selvaggi C, Scarpona S, Fanello G, Fiocca F, Ceccarelli G, Antonelli G, Brenchley JM, Vullo V. 2017. Probiotic supplementation promotes a reduction in T-cell activation, an increase in Th17 frequencies, and a recovery of intestinal epithelium integrity and mitochondrial morphology in ART-treated HIV-1-positive patients. Immun Inflamm Dis 5:244–260. doi: 10.1002/iid3.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.d'Ettorre G, Ceccarelli G, Giustini N, Serafino S, Calantone N, De Girolamo G, Bianchi L, Bellelli V, Ascoli-Bartoli T, Marcellini S, Turriziani O, Brenchley JM, Vullo V. 2015. Probiotics reduce inflammation in antiretroviral treated, HIV-infected individuals: results of the “Probio-HIV” clinical trial. PLoS One 10:e0137200. doi: 10.1371/journal.pone.0137200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ortiz AM, Klase ZA, DiNapoli SR, Vujkovic-Cvijin I, Carmack K, Perkins MR, Calantone N, Vinton CL, Riddick NE, Gallagher J, Klatt NR, McCune JM, Estes JD, Paiardini M, Brenchley JM. 2016. IL-21 and probiotic therapy improve Th17 frequencies, microbial translocation, and microbiome in ARV-treated, SIV-infected macaques. Mucosal Immunol 9:458–467. doi: 10.1038/mi.2015.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Klatt NR, Canary LA, Sun X, Vinton CL, Funderburg NT, Morcock DR, Quinones M, Deming CB, Perkins M, Hazuda DJ, Miller MD, Lederman MM, Segre JA, Lifson JD, Haddad EK, Estes JD, Brenchley JM. 2013. Probiotic/prebiotic supplementation of antiretrovirals improves gastrointestinal immunity in SIV-infected macaques. J Clin Invest 123:903–907. doi: 10.1172/JCI66227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academy Press, Washington, DC. [Google Scholar]
- 64.Qiagen. 2006. DNeasy blood & tissue handbook. Qiagen, Germantown, MD. [Google Scholar]
- 65.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weber N, Liou D, Dommer J, MacMenamin P, Quinones M, Misner I, Oler AJ, Wan J, Kim L, Coakley McCarthy M, Ezeji S, Noble K, Hurt DE. 2018. Nephele: a cloud platform for simplified, standardized and reproducible microbiome data analysis. Bioinformatics 34:1411–1413. doi: 10.1093/bioinformatics/btx617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ewels P, Magnusson M, Lundin S, Kaller M. 2016. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kopylova E, Noe L, Touzet H. 2012. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28:3211–3217. doi: 10.1093/bioinformatics/bts611. [DOI] [PubMed] [Google Scholar]
- 69.Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
16S miSeq data were deposited in the NCBI Sequence Read Archive (SRA) under project number PRJNA526520.