The efficacy of influenza virus vaccines could be improved by enhancing the immunogenicity of the NA protein. One of the reasons for its poor immunogenicity is the immunodominance of the HA over the NA in many seasonal influenza virus vaccines. Here we demonstrate that, in the mouse model, extending the stalk domain of the NA protein can enhance its immunogenicity on virus particles and overcome the immunodominance of the HA without affecting antibody responses to the HA. The antibody repertoire is broadened by the extended NA and includes additional ADCC-active antibodies. Our findings may assist in the efforts toward more effective influenza virus vaccines.
KEYWORDS: immunodominance, influenza virus, neuraminidase, vaccine
ABSTRACT
Influenza viruses express two surface glycoproteins, the hemagglutinin (HA) and the neuraminidase (NA). Anti-NA antibodies protect from lethal influenza virus challenge in the mouse model and correlate inversely with virus shedding and symptoms in humans. Consequently, the NA is a promising target for influenza virus vaccine design. Current seasonal vaccines, however, poorly induce anti-NA antibodies, partly because of the immunodominance of the HA over the NA when the two glycoproteins are closely associated. To address this issue, here we investigated whether extending the stalk domain of the NA could render it more immunogenic on virus particles. Two recombinant influenza viruses based on the H1N1 strain A/Puerto Rico/8/1934 (PR8) were rescued with NA stalk domains extended by 15 or 30 amino acids. Formalin-inactivated viruses expressing wild-type NA or the stalk-extended NA variants were used to vaccinate mice. The virus with the 30-amino-acid stalk extension induced significantly higher anti-NA IgG responses (characterized by increased in vitro antibody-dependent cellular cytotoxicity [ADCC] activity) than the wild-type PR8 virus, while anti-HA IgG levels were unaffected. Similarly, extending the stalk domain of the NA of a recent H3N2 virus enhanced the induction of anti-NA IgGs in mice. On the basis of these results, we hypothesize that the subdominance of the NA can be modulated if the protein is modified such that its height surpasses that of the HA on the viral membrane. Extending the stalk domain of NA may help to enhance its immunogenicity in influenza virus vaccines without compromising antibody responses to HA.
IMPORTANCE The efficacy of influenza virus vaccines could be improved by enhancing the immunogenicity of the NA protein. One of the reasons for its poor immunogenicity is the immunodominance of the HA over the NA in many seasonal influenza virus vaccines. Here we demonstrate that, in the mouse model, extending the stalk domain of the NA protein can enhance its immunogenicity on virus particles and overcome the immunodominance of the HA without affecting antibody responses to the HA. The antibody repertoire is broadened by the extended NA and includes additional ADCC-active antibodies. Our findings may assist in the efforts toward more effective influenza virus vaccines.
INTRODUCTION
Influenza A and B viruses express two surface glycoproteins, hemagglutinin (HA) and neuraminidase (NA) (1). Nine subtypes of the NA (N1 to N9) are described for influenza A virus, two (N10 and N11) for bat influenza A-like viruses, and one for influenza B viruses (2). Although the NA is recognized as a promising target for vaccines (2, 3), current seasonal influenza virus vaccines do not reliably induce robust anti-NA immunity (2, 4–9). Anti-NA antibodies (Abs) have been shown to protect against lethal influenza virus challenge in mice (10–14), and human challenge studies revealed an inverse correlation between anti-NA titers and disease symptoms (15–18). In humans, neuraminidase-inhibiting (NI) antibody titers represent a correlate of protection that is independent of anti-HA antibody titers (16, 19, 20). The NA undergoes antigenic drift, indicating that the NA is under immune pressure as well (6, 21, 22). In contrast to anti-HA antibodies and despite the observed antigenic drift in the NA, anti-NA antibodies often cross-react within one subtype, such as N1 (11, 23–26) or N2 (11, 27, 28), or among influenza B virus NAs (11, 29, 30).
Many antibodies that target NA have been shown to inhibit its enzymatic activity, thereby preventing viral release from infected cells (2). These antibodies typically exert in vitro NI activity. More recently, it has been shown that broadly protective anti-NA antibodies can also act by activating Fc receptor-mediated effector functions, such as antibody-dependent cellular cytotoxicity (ADCC) (29). These antibodies potentially contribute to protection by mediating the killing of virus-infected cells. Future influenza virus vaccines designed to elicit strong anti-NA antibody responses, in addition to anti-HA antibodies, may be more effective than current vaccines, as immunity against the NA is often broad and viral escape is less likely if the two glycoproteins are potently targeted simultaneously (2, 3).
One reason for the poor induction of anti-NA antibodies upon infection or vaccination may be the immunodominance of the HA over the NA when both proteins are in close association, as is the case in virus particles and many seasonal vaccines (31–34). However, when mice are immunized with purified HA and NA proteins that are administered separately, the NA becomes more immunogenic (35). Consequently, supplementing seasonal vaccines with isolated NA proteins can enhance anti-NA responses in mice (36). More recently, NA vaccine candidates based on recombinant tetrameric NA proteins (11–13, 37), DNA plasmids (10), virus-like or replicon particles (23, 25), and modified vaccinia virus Ankara vectors (38) have been shown to induce heterologous anti-NA immunity in mice. In humans, purified N2 protein has been shown to induce heterologous antibody responses (28). In addition, the NA is a validated drug target, as FDA-approved drugs such as oseltamivir and zanamivir are NA inhibitors which are broadly active against influenza A and B viruses (1). Therefore, a promising approach to increase the breadth of protection afforded by seasonal influenza virus vaccines is to include an immunogenic NA component (11, 13, 36, 39). Another path would be to modify the NA protein such that it becomes more immunogenic when expressed on the virus particle. If the modified NA remains functional and supports viral replication in eggs, it could be used in existing manufacturing processes for seasonal vaccines.
The NA protein is composed of the globular head domain that contains the enzymatically active site and the stalk domain (1). Truncations of various lengths in the hypervariable stalk have been identified in the NAs of subtypes N1 to N3 and N5 to N7, mostly in those from avian influenza viruses (40). In contrast, there is currently no evidence for variation in the NA stalk length in influenza viruses circulating in humans (40). The stalk tolerates large artificially introduced insertions. For example, the NA stalk of the H1N1 A/WSN/1933 (WSN) virus has been shown to tolerate insertions of up to 28 amino acids without growth disadvantages in tissue culture or eggs (41). Another study found that up to 41 amino acids could be inserted into the NA stalk of the WSN virus without affecting viral growth in tissue culture (42), and the NA stalk of the H1N1 A/Puerto Rico/8/1934 (PR8) virus tolerated an insertion of 20 amino acids (43). It is presently unknown, however, whether extending the NA stalk influences the protein’s immunogenicity. To address this issue, we generated variants of the PR8 virus that have the NA stalk domain extended by 15 or 30 amino acids. Vaccination studies in mice revealed that the virus with a stalk domain extended by 30 amino acids induced significantly higher anti-NA IgG responses than the wild-type PR8 virus, while anti-HA IgGs were induced to similar levels. No differences were observed in the NI activity of the antibody responses, but antisera raised with the stalk extended by 30 amino acids exerted increased in vitro ADCC activity. To determine if this effect of enhanced immunogenicity would hold true for clinically relevant strains, we generated variants of the H3N2 A/Hong Kong/4801/2014 (HK14) virus that have a 15-amino-acid extension or a 25-amino-acid deletion in the N2 stalk. As with the N1 of PR8, we show that increasing the stalk length of N2 improves its immunogenicity. Our results show that extending the stalk domain of the NA is a promising approach to enhance its immunogenicity and overcome the immunodominance of the HA, which may help in the design of improved influenza virus vaccines.
RESULTS
Design, rescue, and characterization of influenza viruses expressing NA proteins with extended stalk domains.
We selected PR8 as a model influenza virus to study whether the length of the stalk domain of NA correlates with its immunogenicity. We hypothesized that an extended stalk domain would increase the visibility of the NA protein on the surface of virus particles to the humoral immune system, thereby enhancing its immunogenicity.
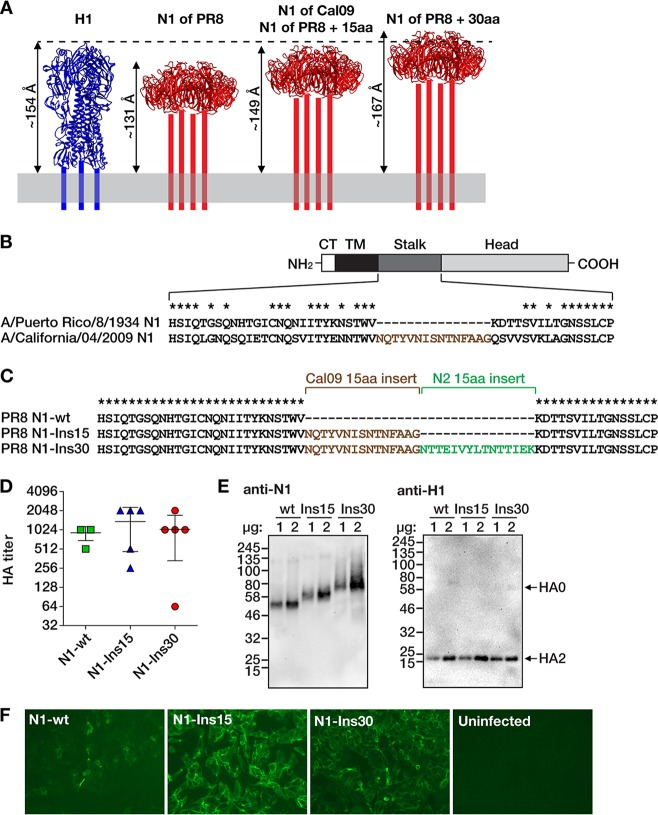
Compared to circulating H1N1 strains (pre- and postpandemic), the NA of the PR8 virus has a 15-amino-acid deletion in the stalk domain (44). It has been estimated by molecular dynamics calculations that the NA protein of the H1N1pdm09 A/California/04/2009 (Cal09) virus extends from the membrane by 149 Å, which is slightly less than the estimated height of the HA protein (154 Å) (45) (Fig. 1A). It was also calculated that each amino acid in the stalk domain contributes to ∼1.2 Å of the total height of the NA protein (45). Consequently, the NA of PR8 virus has an estimated height of 131 Å. Adding 15 amino acids to the PR8 NA would increase its height to that of the Cal09 NA (149 Å), and inserting 30 amino acids would raise the height to 167 Å, which would be 13 Å taller than that of the HA protein (Fig. 1A). Since unrelated sequences could perturb the structure of the PR8 NA protein, we chose to introduce stalk sequences of other NA proteins that, despite the variability of amino acid sequences, likely share structural features with those in the stalk of PR8 NA (41, 42).
FIG 1.
Design and rescue of influenza viruses with extended N1 neuraminidase stalk domains. (A) Estimated lengths of the ectodomains of N1 proteins with different stalk lengths (red) compared with the ectodomain of H1 hemagglutinin (blue). The structure of the NA stalk has not been determined and is indicated by four bars. The lengths of the ectodomains are estimates from molecular dynamics simulations, as reported before (45). The depiction of H1 is based on the crystal structure of the PR8 HA (PDB number 1RU7 [62]), and the depictions of N1 are based on the crystal structure of the NA of A/California/04/2009 (Cal09) virus (PDB number 3TI3 [63]). The structures are not to scale and were visualized with UCSF Chimera (64). aa, amino acids. (B) The four domains of the NA protein are indicated (CT, cytoplasmic tail; TM, transmembrane domain). The diagram is not to scale. The amino acid sequences comprising the stalk region are defined as previously described (42). Asterisks denote conserved amino acids. A 15-amino acid region of the Cal09 NA stalk that is not present in the PR8 NA is shown with brown letters. (C) Alignment of the three NA proteins with different stalk lengths. The 15-amino-acid N2 insertion is derived from the NA stalk domain of the A/New York/61/2012 (H3N2) virus. wt, wild type. (D) Hemagglutination (HA) titers of allantoic fluids from plaque-purified viruses. Data points represent individual plaques (n = 5 per virus). Horizontal bars show the mean values and whiskers the standard deviations. Phosphate-buffered saline (PBS) control wells showed no hemagglutination (not shown). (E) Western blots of proteins from concentrated viruses (left, anti-NA; right, anti-HA). One or two micrograms of total protein content of each virus preparation was analyzed, as indicated above the blots. Protein marker sizes (in kilodaltons) are indicated to the left of the blots. The bands corresponding to HA0 (uncleaved HA) and HA2 (cleavage product of HA0) are indicated with arrows (the antibody is specific to the C-terminal portion of the HA protein and therefore does not react with the HA1 polypeptide). (F) Immunofluorescence microscopy of MDCK cells infected with the indicated viruses and stained with anti-N1 monoclonal antibody 4A5 (11).
Alignment of the NA protein sequences of the PR8 and Cal09 viruses revealed the position of the 15-amino-acid deletion in the stalk of PR8 NA (Fig. 1B). At that position, the corresponding 15 amino acids of the Cal09 NA were inserted into the PR8 NA (Fig. 1C). This mutant was designated N1-Ins15. An additional sequence of 15 amino acids was derived from the NA stalk domain of the H3N2 A/New York/61/2012 (NY12) virus. A mutant of the PR8 NA that contained both the 15 amino acids of Cal09 NA and the 15 amino acids of NY12 NA was designated N1-Ins30 (Fig. 1C).
The nucleotide sequences of the NA gene segments from the Cal09 and NY12 viruses were used to create the recombinant RNAs encoding the N1-Ins15 and N1-Ins30 proteins. The modified segments were used to rescue viruses expressing these NAs in the PR8 backbone by reverse genetics. As a control, we also rescued the wild-type (wt) PR8 virus in parallel, whose NA was designated N1-wt. After growing for 48 h in embryonated chicken eggs, the plaque-purified and sequence-confirmed viruses grew to comparable hemagglutination titers (Fig. 1D). Thus, confirming previous reports (41–43), there was no evidence that the stalk insertions significantly affected viral growth. Western blots with proteins isolated from virus particles revealed distinct size shifts of the extended NA proteins compared to the wild-type NA (Fig. 1E). NA and HA expression levels were comparable in the different viruses (Fig. 1E). The three viruses were able to infect Madin-Darby canine kidney (MDCK), cells which resulted in expression of NA on the surface (Fig. 1F).
In summary, we successfully rescued two viruses in the PR8 backbone with NA stalk domains extended by 15 or 30 amino acids that replicated in eggs and MDCK cells. On virus particles, the mutated NA proteins were expressed at comparable levels, and the levels of HA appeared to be unaffected by the mutations in the NA.
Extending the stalk domain by 30 amino acids enhances the immunogenicity of the NA in mice.
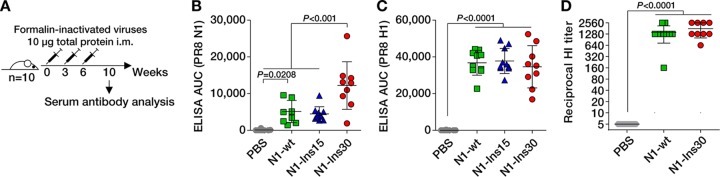
Next, we assessed whether the length of the stalk domain influences the immunogenicity of the NA protein in a mouse model. Three groups of 10 BALB/c mice were immunized intramuscularly with formalin-inactivated viruses expressing the N1-wt protein, N1-Ins15 protein, or N1-Ins30 protein three times at 3-week intervals with doses of 10 μg total protein (Fig. 2A). A fourth group of mice receiving phosphate-buffered saline (PBS) served as a control. At 4 weeks after the third immunization, the mice were sacrificed and serum IgG responses were determined by enzyme-linked immunosorbent assays (ELISAs). Compared to the PBS controls, immunization with all three viruses induced significant IgG responses against recombinant NA protein from PR8 (Fig. 2B). While the viruses with N1-wt and N1-Ins15 NAs induced comparable levels of anti-NA IgG, the virus carrying the N1-Ins30 NA elicited significantly stronger (∼2.5-fold) anti-NA IgG responses. In contrast, the three viruses induced comparable IgG responses against recombinant PR8 HA protein (Fig. 2C). In addition, the NA stalk length did not affect hemagglutination inhibition (HI) titers (Fig. 2D). Thus, extending the stalk domain by 30 amino acids significantly enhanced the IgG responses against NA, without compromising anti-HA antibody levels.
FIG 2.
The extended stalk domain enhances IgG responses to the N1 neuraminidase. (A) Immunization regime. Mice received three doses intramuscularly (i.m.) that contained 10 μg of formalin-inactivated viruses. Serum obtained 4 weeks after the third immunization was analyzed for antibodies against N1 neuraminidase and H1 hemagglutinin proteins. (B and C) Serum IgG levels corresponding to recombinant tetrameric N1 neuraminidase (B) and recombinant trimeric H1 hemagglutinin (C) from PR8 virus, as measured by ELISA. AUC, area under the curve. (D) Hemagglutination inhibition (HI) titers against wild-type PR8 virus. Statistical significance was inferred by one-way ANOVA with Bonferroni correction and P values are indicated in the graphs. Note that the Ins30 group comprised only 9 sera, as one animal in that group died from causes unrelated to the experiment.
Stalk extension enhances the induction of antibodies with in vitro effector functions.
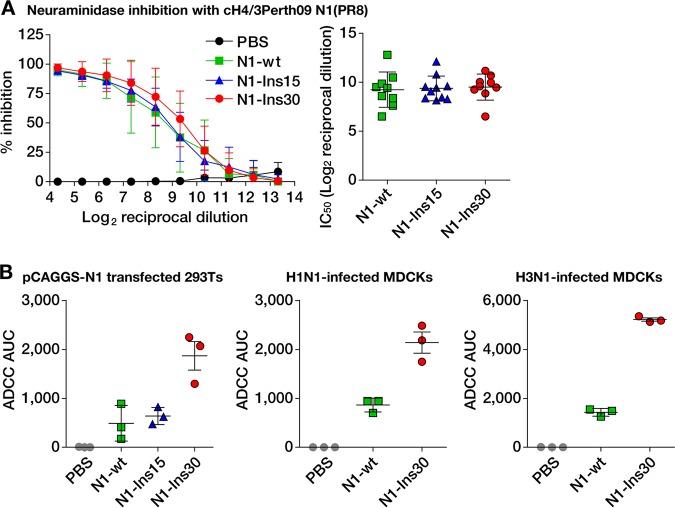
We next sought to assess the functional properties of the antibodies elicited by the different viruses. In general, the majority of anti-NA antibodies are thought to prevent binding of the enzymatically active site to its substrate sialic acid (2). As these types of antibodies typically exert in vitro NI activity, we performed NI assays with the sera obtained from the immunized mice. Although the N1-Ins30-expressing virus elicited higher total anti-NA IgG titers than the other two viruses, as measured by ELISA (Fig. 2B), levels of NI activities were similar between the groups of mice immunized with the three different viruses (Fig. 3A).
FIG 3.
The extended stalk domain enhances ADCC active antibody responses to the N1 neuraminidase. (A) Results of the neuraminidase inhibition assay. The two subpanels show the same data. The left subpanel shows percent inhibition relative to the serum dilution with data points representing mean values corresponding to 9 (Ins30 group) or 10 (all other groups) individual mice ± standard deviations. The right subpanel shows the 50% inhibitory concentrations (IC50 values) calculated from the curves in the left subpanel. (B) Results from antibody-dependent cellular cytotoxicity (ADCC) reporter assays. From left to right, the subpanels show assays performed with HEK 293T cells transfected with a pCAGGS expression plasmid for the N1 protein of the PR8 virus (left) and MDCK cells infected with either an H1N1 virus (PR8) (middle) or an H3N1 virus (right) (H3 was from A/Hong Kong/4801/2014; all other proteins were from PR8). Data points represent pooled sera measured in triplicate; horizontal bars show the mean values, and the whiskers indicate the standard deviations.
Another previously described function of anti-NA antibodies is the induction of Fc receptor-mediated effector functions, such as ADCC (29). To assess whether the induction of antibodies with effector functions was influenced by the stalk length of NA, we subjected the murine immune sera to an in vitro ADCC reporter assay (46). Using human embryonic kidney (HEK) 293T cells expressing the PR8 NA protein, sera raised with the N1-Ins30-expressing virus showed substantially higher ADCC activity than sera induced with viruses carrying N1-wt or N1-Ins15 (Fig. 3B). Similar results were obtained using MDCK cells that were infected with either wild-type PR8 virus or an H3N1 virus expressing the PR8 NA and the HA of the HK14 H3N2 virus (47) (Fig. 3B). In summary, extending the stalk domain of the NA enhanced antibody responses with in vitro ADCC activity but not the induction of NI active antibodies.
Stalk extension improves the immunogenicity of the NA of a recent clinically relevant H3N2 strain.
Next, we assessed if extending the stalk could improve NA immunogenicity in other influenza virus strains. To test this, we generated viruses containing HK14 H3N2 HA and NA with PR8 internal segments. Cryogenic electron micrographs of H3N2 virus show that both the NA and the HA extended from the membrane by ∼150 Å, with the NA being slightly taller than the HA (48). Thus, we generated viruses containing NAs with no stalk changes (N2-wt), NAs with a 25-amino-acid stalk deletion (N2-Del25), and NAs with a 15-amino-acid stalk insertion derived from part of the N1 stalk of Cal09 (N2-Ins15) (Fig. 4A).
FIG 4.
Design, rescue, and immunogenicity of influenza viruses with extended N2 neuraminidase stalk domains. (A) The four domains of the NA protein are indicated (CT, cytoplasmic tail; TM, transmembrane domain). The diagram is not to scale. The N2-Del25 protein has a deletion of 25 amino acids. The 15-amino-acid insertion of the N2-Ins15 protein is derived from the N1 protein of the Cal09 virus (see Fig. 1B). Asterisks denote conserved amino acids. (B) Hemagglutination (HA) titers of allantoic fluids from plaque-purified viruses measured in duplicate. Phosphate-buffered saline (PBS) control wells showed no hemagglutination (not shown). (C) Western blots of proteins from concentrated viruses. One microgram of total protein content (left blots) or amounts that were normalized to achieve equal intensities for the NP (right blots) of each virus were analyzed, as indicated above the blots. The normalization factors for Del25, wt, and Ins15 viruses were 0.802, 1.0, and 0.765, respectively. Approximate protein sizes (in kilodaltons) are indicated to the right of the blots. (D) Immunization regime. Mice received an amount of formalin-inactivated virus equivalent to 10 μg of N2-wt virus as determined by normalization to NP. Sera obtained 4 weeks after the immunization were analyzed for IgGs against N2 neuraminidase and H3 hemagglutinin by ELISA and for HI-reactive antibodies. (E and F) Serum IgG levels to recombinant tetrameric N2 neuraminidase (E) or recombinant trimeric H3 hemagglutinin (F) from HK14 virus, as measured by ELISA. AUC, area under the curve. Statistical significance was inferred by one-way ANOVA with Bonferroni correction, and P values are indicated in the graphs. n.s., not significant. (G) Hemagglutination inhibition (HI) titers against HK2014-wt virus. Pooled sera were analyzed in triplicate.
These viruses were rescued by reverse genetics as described above. After growing for 72 h in embryonated chicken eggs, the different plaque-purified and sequence-confirmed viruses achieved comparable hemagglutination titers (Fig. 4B). Western blot analyses revealed that the expression levels of the N2 and H3 glycoproteins differed between the N2-Del25-expressing, N2-wt-expressing, and N2-Ins15-expressing viruses (Fig. 4C). Therefore, we normalized vaccination doses to the expression levels of the nucleoprotein (NP). Three groups of five BALB/c mice were immunized intramuscularly once with formalin-inactivated viruses expressing the N2-wt protein, the N2-Del25 protein, or the N2-Ins15 NA protein. Mice received an amount of formalin-inactivated virus equivalent to 10 μg of wild-type virus as determined by normalization to NP content. A fourth group of three mice receiving PBS served as a control. Serum obtained 4 weeks postvaccination was subjected to antibody analysis by ELISA against recombinant tetrameric HK14 N2 protein (Fig. 4C and D). Immunization with N2-Ins15 virus elicited anti-NA IgG responses that were ∼3-fold and ∼4.5-fold stronger than those elicited by immunization with N2-wt and N2-Del25 viruses, respectively (Fig. 4E). Thus, extending the stalk domain improved the immunogenicity of N2 on virus particles. Similarly to the observations obtained with the H1N1 virus above, the stalk length of N2 did not significantly affect anti-H3 antibody titers (Fig. 4F) or the levels of HI-reactive antibodies (Fig. 4G).
DISCUSSION
Although other reasons for the poor immunogenicity of the NA of current seasonal vaccines are recognized, such as the differing and unreliable amounts of NA proteins (34) and their inconsistent stability (49), it has been established that anti-NA antibody responses are suppressed in current vaccines due to the immunodominance of the HA (31–34). Here, we demonstrated that a simple extension of the NA stalk can significantly enhance the anti-NA immune response without compromising the immunogenicity of the HA. Consistent with previous studies (41–43), mutant viruses in the PR8 backbone carrying NA proteins with NA stalk domains that were extended by 15 or 30 amino acids replicated in eggs and MDCK cells without any apparent growth disadvantages compared to wild-type PR8 virus. In mice, immunization studies performed with formalin-inactivated virus particles revealed that a 30-amino-acid extension to PR8 NA, but not a 15-amino-acid extension, significantly enhanced total anti-NA IgG responses without affecting IgG responses to the HA or the levels of HI-reactive antibodies. Based on published molecular dynamics simulations (45), a 30-amino-acid extension in PR8 (but not a 15-amino-acid extension) is predicted to increase the height of the NA such that it surpasses the height of the HA, suggesting that the improvement in immunogenicity is dependent on the visibility of the NA relative to that of the HA. Similarly, extending the stalk of the N2 of HK14 virus—the H3N2 strain used in the 2016–2017 and 2017–2018 seasonal vaccines (47)—significantly enhanced NA immunogenicity without affecting anti-HA IgG levels or HI titers, demonstrating that this approach is a viable strategy for improving immunogenicity.
We found that stalk extension of PR8 NA did not affect the induction of NI reactive antibodies but substantially increased the production of antibodies with in vitro ADCC activity. This suggests that the longer stalk makes novel epitopes accessible that are targeted by ADCC active antibodies but not by NI active antibodies, while the immunogenicity of regions recognized by NI reactive antibodies is preserved. Of note, it has been shown that ADCC active IgGs recognizing the stalk domain of the HA (50, 51) or the NA protein (52) can protect against lethal influenza virus infection in mice in an Fc gamma receptor-dependent manner. A recent study also showed that ADCC-active and NI inactive anti-NA monoclonal antibodies (MAbs) targeting the lateral surface of the head domain could confer protection in mice (53). We hypothesize that our approach of extending the stalk may enhance the exposure of epitopes below the head domain and/or on the lateral surface of the head domain, thereby increasing the repertoire of induced antibodies. Moreover, broadly reactive anti-NA antibodies that target conserved epitopes are often ADCC active (29). Therefore, extending the NA stalk domain may not only increase the immunogenicity of the NA on virus particles but also enhance the breadth of protection afforded by the induced anti-NA antibodies.
Unlike other pursued approaches designed to enhance anti-NA immunity that are based on isolated or recombinant NA proteins (11–13, 37), DNA plasmids (10), virus-like and replicon particles (23, 25), or virus-vectored vaccines (38), the NA stalk extension described here could be implemented in existing manufacturing processes for seasonal influenza virus vaccines, as the mutated NAs can be expressed on virus particles that efficiently replicate in eggs. Moreover, we provide evidence that the subdominance of the NA results in part from the height of the protein relative to the HA. We hypothesize that immunodominance is associated with the viral epitopes that are most distal from the surface of the virus or the infected cell and that immunodominance may simply be a consequence of being more easily recognized by B-cell receptors of the infected host.
MATERIALS AND METHODS
Recombinant neuraminidase genes and cloning.
The recombinant NA segments were based on the NA gene of the PR8 virus or the NA gene of the HK14 virus (47). The nucleotide sequences used for the 15-amino-acid insertions were retrieved from the Influenza Research Database (https://www.fludb.org). They were derived from the NA sequences of the Cal09 (H1N1pdm09) virus (GenBank accession number FJ966084) and the A/New York/61/2012 (H3N2) virus (accession number KF790392). Sequences were aligned with Clustal X 2.0 (54). DNA fragments encoding the NA gene segments that contained 15-bp cloning sites specific for the pDZ vector at the 5′ and 3′ ends were obtained as synthetic double-stranded DNAs from Integrated DNA Technologies, using the gBlocks Gene Fragments service. The NA DNAs were cloned into ambisense pDZ vector, which was digested with SapI restriction enzyme (New England Biolabs), by the use of an In-Fusion HD cloning kit (Clontech). Sequences were confirmed by Sanger sequencing (Macrogen). Sequencing primers pDZ_forward (TACAGCTCCTGGGCAACGTGCTGG) and pDZ_reverse (AGGTGTCCGTGTCGCGCGTCGCC) were obtained from Life Technologies.
Cell culture.
HEK 293T cells were cultured in Dulbecco’s modified Eagle medium (DMEM; Gibco) with 10% (vol/vol) fetal bovine serum (FBS) (HyClone) and 100 units/ml penicillin and 100 μg/ml streptomycin (Pen-Strep; Gibco). MDCK cells were maintained in minimum essential medium (MEM; Gibco) with 10% (vol/vol) FBS, Pen-Strep, 2 mM l-glutamine (Gibco), 0.15% (wt/vol) sodium bicarbonate (Corning), and 20 mM HEPES (Gibco). Both cell lines were maintained at 37°C with 5% CO2.
Rescue of recombinant influenza viruses.
Reassortant viruses were rescued by transfecting HEK 293T cells with 0.7 μg of NA-encoding pDZ plasmid, 0.7 μg of HA-encoding pDZ plasmid, and 2.1 μg of a pRS-6 segment plasmid that drives ambisense expression of the six segments of PR8 virus (except NA and HA) and that is described elsewhere (55) by the use of TransIT-LT1 transfection reagent (Mirus Bio). After 48 h, cells were treated for 30 min with 1 μg per ml tosyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin at 37°C. Supernatants were collected, clarified by low-speed centrifugation, and injected into 8-to-10-day-old specific-pathogen-free embryonated chicken eggs (Charles River Laboratories) that were incubated at 37°C. After 48 h, eggs were incubated at 4°C overnight and allantoic fluids were harvested and clarified by low-speed centrifugation. The presence of influenza virus in the allantoic fluids was determined by hemagglutination assays performed as described below. Virus cultures with positive results were plaque purified on confluent MDCK cell layers in the presence of TPCK-treated trypsin and expanded in embryonated chicken eggs. Sequences of the NA and HA genes were confirmed by isolating viral RNA from allantoic fluids with a High Pure viral RNA kit (Roche) followed by reverse transcription-PCR (RT-PCR) using a SuperScript III one-step RT-PCR system with Platinum Taq High Fidelity DNA polymerase (Thermo Fisher) and primers PR8_NA_for (CGAAAGCAGGGGTTTAAAATG) and PR8_NA_rev (TTTTTGAACAGACTACTTGTCAATG), PR8_HA_for (CCGAAGTTGGGGGGGAGCAAAAGCAGGGGAAAATAA) and PR8_HA_rev (GGCCGCCGGGTTATTAGTAGAAACAAGGGTGTTTTT), HK14_NA_for (GGGAGCAAAAGCAGGAGTAAAGATG) and HK14_NA_rev (TTATTAGTAGAAACAAGGAGTTTTTTCTAAAATTGCG), or HK14_HA_for (GGGAGCAAAAGCAGGGGATAATTC) and HK14_HA_rev (GGGTTATTAGTAGAAACAAGGGTGTTTTTAATTAATG) (obtained from Integrated DNA Technologies). The PCR products were purified from a 1% agarose gel by the use of a NucleoSpin Gel and PCR clean-up kit (Macherey-Nagel) and were submitted for Sanger sequencing (Genewiz) with the primers described above. No egg-adaptive mutations were observed for any of the sequenced viral genes.
Preparation of formalin-inactivated viruses for vaccination.
Plaque-purified and sequenced influenza viruses were expanded in 8-to-10-day-old embryonated chicken eggs. Pooled allantoic fluids from 10 to 20 eggs were added on top of 3 ml of a 20% (wt/vol) sucrose solution in 0.1 M NaCl–1 mM EDTA–10 mM Tris-HCl (pH 7.4) in 38.5-ml ultracentrifuge tubes (Denville). Following centrifugation at 25,000 rpm for 2 h at 4°C using an L7-65 ultracentrifuge (Beckman) equipped with an SW28 rotor, supernatants were carefully aspirated and pellets were recovered in 1 ml of PBS. After addition of 0.03% (vol/vol) formaldehyde, the virus suspensions were incubated for 48 h at 4°C with shaking. To remove the formaldehyde, virus suspensions were diluted with PBS and subjected to ultracentrifugation as described above. Pellets were resuspended in sterile PBS, and the total protein concentration was determined with a Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Fisher).
Western blots.
Purified virus particles were lysed in NP-40 lysis buffer (1% [vol/vol] NP-40, 150 mM NaCl, 50 mM Tris-HCl [pH 8.0], protease inhibitors [Halt protein and phosphatase inhibitor cocktail; Thermo Fisher], 1 mM dithiothreitol [DTT]). After incubation on ice for 30 min, samples were centrifuged for 10 min at 12,000 rpm in a table-top centrifuge. The supernatants were transferred to new microcentrifuge tubes, and the protein concentrations present after lysis were determined with a Pierce BCA protein assay kit (Thermo Fisher). Proteins (1 or 2 μg) were separated on 12.5% polyacrylamide gels under denaturing conditions in the presence of sodium dodecyl sulfate (SDS) and were then transferred onto polyvinylidene difluoride (PVDF) membranes. Color Prestained Protein Standard, Broad Range (New England Biolabs) (11 to 254 kDa), was used as a protein size marker. The membranes were blocked for 1 h using PBS–5% (wt/vol) skim milk powder and were washed three times with PBS containing 0.05% (vol/vol) Tween 20. The primary antibodies were as follows: mouse anti-N1 monoclonal antibody 4A5 (10) (1 μg per ml), rabbit anti-H1 (Thermo Fisher; catalog no. PA5-34929) (1:5,000 dilution), anti-N2 polyclonal serum raised in guinea pigs (generated in-house; 1:2,000 dilution), anti-H3 monoclonal antibody 12D1 (56), and anti-NP (Invitrogen; catalog no. PA5-32242) (1:3,000 dilution). The primary antibodies were diluted in PBS–1% (wt/vol) bovine serum albumin (BSA) and incubated on the membranes for 1 h. The membranes were washed three times with PBS containing 0.05% (vol/vol) Tween 20 and were incubated for 1 h with secondary horseradish peroxidase (HRP)-labeled antibodies (anti-mouse [catalog no. NXA931V] or anti-rabbit [catalog no. NA9340V]; both from GE Healthcare) diluted 1:3,000 in PBS–1% (wt/vol) BSA according to the manufacturer’s recommendations. After three washes with PBS containing 0.05% (vol/vol) Tween 20, developing solution (Pierce ECL Western blotting substrate; Thermo Scientific) was added to the membranes, which were subsequently developed in a ChemiDoc MP imaging system (Bio-Rad). NP band intensities were determined by the software provided on the ChemiDoc MP imaging system. Lanes were automatically detected with manual adjustment. Normalization factors were calculated by dividing the NP band intensity of one sample by the NP band intensity of N2-wt virus.
Immunization studies.
Animal experiments were performed with 6-to-8-week-old female BALB/c mice (Charles River) in accordance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) of the Icahn School of Medicine at Mount Sinai. Formalin-inactivated viruses were administered intramuscularly at a dose of 10 μg total protein per mouse after dilution in a total volume of 100 μl sterile PBS. At 4 weeks after the final immunization, mice were euthanized and blood was collected by cardiac puncture. Sera were prepared by removing red blood cells by centrifugation and were stored at −20°C until use.
Enzyme-linked immunosorbent assays (ELISAs).
The trimeric recombinant PR8 HA protein and the tetrameric recombinant PR8 and HK14 NA proteins were produced as described previously (57, 58). Proteins were coated onto Immulon 4 HBX 96-well microtiter plates (Thermo Scientific) at a concentration of 2 μg per ml in PBS (50 μl per well) for 16 h at 4°C. After one wash using PBS with 0.1% (vol/vol) Tween 20 (PBS-T), wells were blocked for 1 h with 5% (wt/vol) skim milk powder–PBS and washed once with PBS-T. Mouse sera diluted in PBS (50 μl per well) were added and incubated on the plates for 1 h. After three washes with PBS-T, wells were incubated with HRP-conjugated anti-mouse IgG antibody (GE Healthcare) diluted 1:5,000 in 5% (wt/vol) skim milk powder–PBS for 1 h, washed three times with PBS-T, and developed with 100 μl (per well) of SigmaFast OPD substrate (Sigma-Aldrich) for 20 min. Reactions were stopped by adding 100 μl (per well) of 3 M hydrochloric acid (HCl), and absorbance at 490 nm was determined on a Synergy 4 plate reader (BioTek). For each ELISA plate, averages plus 3 standard deviations of absorbance values of blank wells were used as a cutoff to calculate area under the curve (AUC) values in GraphPad Prism 5.03 (GraphPad Software).
Hemagglutination assays.
Using PBS, serial 2-fold dilutions of allantoic fluids were prepared in 96-well V-bottom microtiter plates to reach a final volume of 50 μl per well. To each well, 50 μl of a 0.5% suspension of turkey red blood cells (Lampire) in PBS was added. Plates were incubated at 4°C until the red blood cells in the PBS control samples settled to the bottom of the wells. The hemagglutination titer was defined as the reciprocal of the highest dilution of allantoic fluid that caused hemagglutination of red blood cells.
Hemagglutination inhibition (HI) assays.
One volume of mouse serum was treated with three volumes of receptor-destroying enzyme (RDE; Denka Seiken, Tokyo, Japan) at 37°C for 16 h (47). Then, a 2.5% sodium citrate solution (three volumes) was added. After incubation at 56°C for 30 min, PBS (three volumes) was added to reach a final dilution of 1:10. Two-fold dilutions (25 μl) of the RDE-treated serum–PBS solution were prepared in 96-well V-bottom microtiter plates and were combined with 25 μl per well of either PR8 wild-type virus or HK2014 wild-type virus (allantoic fluids), and the reaction mixtures were diluted in PBS to a final HA titer of 8 HA units per 50 μl. The samples were incubated for 30 min at room temperature to allow HA-specific antibodies to bind to the virus particles. Then, 50 μl of a 0.5% suspension of turkey red blood cells (Lampire) that was washed once with PBS was added to each well. The plates were incubated at 4°C until the red blood cells in the PBS control samples settled to the bottom of the wells. The HI titers were defined as the reciprocal of the highest serum dilution causing inhibition of hemagglutination of red blood cells.
Enzyme-linked lectin assay (ELLA) to determine neuraminidase inhibition (NI).
The ELLA was performed as previously described (59, 60). Microtiter 96-well plates (Immulon 4 HBX; Thermo Fisher Scientific) were coated with 50 μg per ml (150 μl per well) of fetuin (Sigma) diluted in coating solution (SeraCare Life Sciences Inc.) and incubated overnight at 4°C. The next day, heat-inactivated (56°C, 30 min) serum samples were serially diluted 1:2 in PBS in separate 96-well plates (leaving the first column as a virus-only control and the last column as the background), with a starting dilution of 1:20. The final volume of diluted serum samples was 75 μl per well. A recombinant influenza virus expressing a chimeric HA protein, cH4/3 [containing the H4 globular head domain from A/duck/Czech/1956 (H4N6) virus in combination with the H3 stalk domain from A/Perth/16/2009 (H3N2) virus (61)], and the remaining proteins of PR8 virus was diluted to a 90% effective concentration (EC90) in PBS containing 1% BSA, and 75 μl per well was added to the serially diluted serum samples and virus-only controls. PBS (75 μl) and 1% BSA were added to the background wells. The serum/virus plates were incubated for 2 h at room temperature to allow binding of antibodies to the virus particles. The fetuin-coated plates from the previous day were washed three times with PBS-T. The serum/virus mixtures (100 μl per well) were transferred to the washed fetuin-coated plates, which were then incubated for 2 h at 37°C. The plates were washed three times with PBS-T, and peanut agglutinin-horseradish peroxidase conjugate (PNA-HRP; Sigma-Aldrich) (100 μl per well) diluted to 5 μg per ml in PBS was added. The plates were incubated in the dark for 1 h at room temperature. After three washes with PBS-T, 100 μl per well of SigmaFast OPD substrate (Sigma-Aldrich) was added and the plates were incubated for 10 min. Reactions were stopped by adding 100 μl per well of 3 M HCl, and absorbance at 490 nm was determined on a Synergy 4 plate reader (BioTek). Serum sample reactivity was determined by subtracting background absorbance values (no virus, no serum) from the raw absorbance values of serum samples. The obtained values were divided by the average value determined from virus-only control wells and then multiplied by a factor of 100 to calculate the NA activity. Percent NI was determined by subtracting NA activity values from 100%. Using GraphPad Prism, percent NI data were fitted to a nonlinear regression to determine the 50% inhibitory concentration (IC50) of the serum samples.
Antibody-dependent cellular cytotoxicity (ADCC) reporter assays.
ADCC reporter assays were performed as described previously (46). Costar Corning 96-well white flat-bottom plates were seeded with 2 × 104 MDCK cells per well. After 18 h of incubation at 37°C, the MDCK cells were washed once with PBS and then infected with either wild-type PR8 virus or a 7:1 reassortant virus expressing the HA protein of A/Hong Kong/4801/2014 (H3N2) virus and the remaining proteins of PR8 virus (47) at a multiplicity of infection (MOI) of 5 for single-cycle replication. Alternatively, HEK 293T cells were plated in 96-well white flat-bottom plates treated with poly-d-lysine (Sigma-Aldrich) at a density of 2 × 104 cells per well and, after incubation for 4 h, were transfected with 100 ng per well of pCAGGS plasmid expressing the NA of PR8 virus using TransIT-LT1 transfection reagent (Mirus Bio). Infected MDCK cells or transfected HEK 293T cells were incubated for 16 h at 37°C. Then, the culture medium was aspirated and 25 μl of assay buffer (RPMI 1640 supplemented with 4% low-IgG FBS) was added to each well. Pooled sera were added in a volume of 25 μl at a starting dilution of 1:60, and serial 2-fold dilutions were prepared in assay buffer in triplicate. The sera were incubated with the cells for 30 min at 37°C. Genetically modified Jurkat cells expressing murine FcγRIV with a luciferase reporter gene under the control of the promoter nuclear factor-activated T cell (NFAT) (Promega) were added at 7.5 × 104 cells in 25 μl per well. After incubation for 6 h at 37°C, 75 μl per well of Bio-Glo Luciferase assay reagent (Promega) was added and luminescence was quantified using a Synergy 4 plate reader (BioTek). Fold induction was measured in relative light units and calculated by subtracting the background signal from wells without effector cells and then dividing signals of wells with antibody by those with no antibody added.
Immunofluorescence microscopy.
Tissue culture 96-well plates were seeded with 2 × 104 MDCK cells per well. After 24 h of incubation at 37°C, the MDCK cells were washed once with PBS and then infected with wild-type PR8 virus or with PR8 virus with N1-Ins15 NA or with PR8 virus with N1-Ins30 NA at an MOI of 5 for single-cycle replication. Infected MDCK cells were incubated for 16 h at 37°C. The culture medium was aspirated, and the cells were washed twice with PBS and then fixed with methanol-free 4% (vol/vol) paraformaldehyde–PBS for 15 min. After two washes with PBS, the wells were blocked with 5% (wt/vol) skim milk powder–PBS for 30 min. The cells were washed once with PBS and then incubated with the broadly N1-reactive 4A5 MAb (11) at 10 μg per ml diluted in 5% (wt/vol) skim milk powder–PBS for 2 h. After three washes with PBS, the cells were incubated with fluorescence-labeled anti-mouse IgG Alexa Fluor 488 antibody (Life Technologies) diluted 1:2,000 in 5% (wt/vol) skim milk powder–PBS for 1 h and then washed three times with PBS before pictures were taken on an EVOS fl inverted fluorescence microscope (AMG).
Statistics.
Statistical data were generated with GraphPad Prism. The statistical significance of results of comparisons between groups was determined by performing one-way analysis of variance (ANOVA) tests with Bonferroni correction for multiple comparisons. Levels of significance are indicated as follows: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
ACKNOWLEDGMENTS
We thank Shirin Strohmeier, Fatima Amanat, and Andres Javier for providing recombinant neuraminidase and hemagglutinin proteins.
This work was partially funded by the NIAID Centers of Excellence for Influenza Research and Surveillance (HHSN272201400008C to P.P. and F.K.), grants P01 AI097092 (P.P.) and U19 AI109946 (P.P. and F.K.). M.J.B. is supported by MSTP training grant NIH T32 GM007280 (P.P., principal investigator [PI]). N.S. thanks the Royal Golden Jubilee Ph.D. Program (PHD/0084/2558) for a scholarship.
For possible conflicts of interest, please see Peter Palese’s profile on the Icahn School of Medicine at Mount Sinai website (https://icahn.mssm.edu/profiles/peter-palese).
REFERENCES
- 1.Krammer F, Smith GJD, Fouchier RAM, Peiris M, Kedzierska K, Doherty PC, Palese P, Shaw ML, Treanor J, Webster RG, García-Sastre A. 2018. Influenza. Nat Rev Dis Primers 4:3. doi: 10.1038/s41572-018-0002-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krammer F, Fouchier RAM, Eichelberger MC, Webby RJ, Shaw-Saliba K, Wan H, Wilson PC, Compans RW, Skountzou I, Monto AS. 2018. NAction! How can neuraminidase-based immunity contribute to better influenza virus vaccines? mBio 9:e02332-17. doi: 10.1128/mBio.02332-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marcelin G, Sandbulte MR, Webby RJ. 2012. Contribution of antibody production against neuraminidase to the protection afforded by influenza vaccines. Rev Med Virol 22:267–279. doi: 10.1002/rmv.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kilbourne ED. 1976. Comparative efficacy of neuraminidase-specific and conventional influenza virus vaccines in induction of antibody to neuraminidase in humans. J Infect Dis 134:384–394. doi: 10.1093/infdis/134.4.384. [DOI] [PubMed] [Google Scholar]
- 5.Powers DC, Kilbourne ED, Johansson BE. 1996. Neuraminidase-specific antibody responses to inactivated influenza virus vaccine in young and elderly adults. Clin Diagn Lab Immunol 3:511–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sandbulte MR, Westgeest KB, Gao J, Xu X, Klimov AI, Russell CA, Burke DF, Smith DJ, Fouchier RA, Eichelberger MC. 2011. Discordant antigenic drift of neuraminidase and hemagglutinin in H1N1 and H3N2 influenza viruses. Proc Natl Acad Sci U S A 108:20748–20753. doi: 10.1073/pnas.1113801108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Couch RB, Atmar RL, Keitel WA, Quarles JM, Wells J, Arden N, Niño D. 2012. Randomized comparative study of the serum antihemagglutinin and antineuraminidase antibody responses to six licensed trivalent influenza vaccines. Vaccine 31:190–195. doi: 10.1016/j.vaccine.2012.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laguio-Vila MR, Thompson MG, Reynolds S, Spencer SM, Gaglani M, Naleway A, Ball S, Bozeman S, Baker S, Martínez-Sobrido L, Levine M, Katz J, Fry AM, Treanor JJ. 2015. Comparison of serum hemagglutinin and neuraminidase inhibition antibodies after 2010-2011 trivalent inactivated influenza vaccination in healthcare personnel. Open Forum Infect Dis 2:ofu115. doi: 10.1093/ofid/ofu115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krammer F. 2019. The human antibody response to influenza A virus infection and vaccination. Nat Rev Immunol 19:383–397. doi: 10.1038/s41577-019-0143-6. [DOI] [PubMed] [Google Scholar]
- 10.Sandbulte MR, Jimenez GS, Boon AC, Smith LR, Treanor JJ, Webby RJ. 2007. Cross-reactive neuraminidase antibodies afford partial protection against H5N1 in mice and are present in unexposed humans. PLoS Med 4:e59. doi: 10.1371/journal.pmed.0040059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wohlbold TJ, Nachbagauer R, Xu H, Tan GS, Hirsh A, Brokstad KA, Cox RJ, Palese P, Krammer F. 2015. Vaccination with adjuvanted recombinant neuraminidase induces broad heterologous, but not heterosubtypic, cross-protection against influenza virus infection in mice. mBio 6:e02556. doi: 10.1128/mBio.02556-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu WC, Lin CY, Tsou YT, Jan JT, Wu SC. 2015. Cross-reactive neuraminidase-inhibiting antibodies elicited by immunization with recombinant neuraminidase proteins of H5N1 and pandemic H1N1 influenza A viruses. J Virol 89:7224–7234. doi: 10.1128/JVI.00585-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Job ER, Ysenbaert T, Smet A, Christopoulou I, Strugnell T, Oloo EO, Oomen RP, Kleanthous H, Vogel TU, Saelens X. 2018. Broadened immunity against influenza by vaccination with computationally designed influenza virus N1 neuraminidase constructs. NPJ Vaccines 3:55. doi: 10.1038/s41541-018-0093-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen YQ, Wohlbold TJ, Zheng NY, Huang M, Huang Y, Neu KE, Lee J, Wan H, Rojas KT, Kirkpatrick E, Henry C, Palm AE, Stamper CT, Lan LY, Topham DJ, Treanor J, Wrammert J, Ahmed R, Eichelberger MC, Georgiou G, Krammer F, Wilson PC. 2018. Influenza infection in humans induces broadly cross-reactive and protective neuraminidase-reactive antibodies. Cell 173:417–429.e10. doi: 10.1016/j.cell.2018.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murphy BR, Kasel JA, Chanock RM. 1972. Association of serum anti-neuraminidase antibody with resistance to influenza in man. N Engl J Med 286:1329–1332. doi: 10.1056/NEJM197206222862502. [DOI] [PubMed] [Google Scholar]
- 16.Couch RB, Kasel JA, Gerin JL, Schulman JL, Kilbourne ED. 1974. Induction of partial immunity to influenza by a neuraminidase-specific vaccine. J Infect Dis 129:411–420. doi: 10.1093/infdis/129.4.411. [DOI] [PubMed] [Google Scholar]
- 17.Clements ML, Betts RF, Tierney EL, Murphy BR. 1986. Serum and nasal wash antibodies associated with resistance to experimental challenge with influenza A wild-type virus. J Clin Microbiol 24:157–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Memoli MJ, Shaw PA, Han A, Czajkowski L, Reed S, Athota R, Bristol T, Fargis S, Risos K, Powers JH, Davey RT Jr, Taubenberger JK. 2016. Evaluation of antihemagglutinin and antineuraminidase antibodies as correlates of protection in an influenza A/H1N1 virus healthy human challenge model. mBio 7:e00417-16. doi: 10.1128/mBio.00417-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Couch RB, Atmar RL, Franco LM, Quarles JM, Wells J, Arden N, Niño D, Belmont JW. 2013. Antibody correlates and predictors of immunity to naturally occurring influenza in humans and the importance of antibody to neuraminidase. J Infect Dis 207:974–981. doi: 10.1093/infdis/jis935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monto AS, Petrie JG, Cross RT, Johnson E, Liu M, Zhong W, Levine M, Katz JM, Ohmit SE. 2015. Antibody to influenza virus neuraminidase: an independent correlate of protection. J Infect Dis 212:1191–1199. doi: 10.1093/infdis/jiv195. [DOI] [PubMed] [Google Scholar]
- 21.Kilbourne ED, Johansson BE, Grajower B. 1990. Independent and disparate evolution on nature of influenza A virus hemagglutinin and neuraminidase glycoproteins. Proc Natl Acad Sci U S A 87:786–790. doi: 10.1073/pnas.87.2.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao J, Couzens L, Burke DF, Wan H, Wilson P, Memoli MJ, Xu X, Harvey R, Wrammert J, Ahmed R, Taubenberger JK, Smith DJ, Fouchier RAM, Eichelberger MC. 2019. Antigenic drift of the influenza A(H1N1)pdm09 virus neuraminidase results in reduced effectiveness of A/California/7/2009 (H1N1pdm09)-specific antibodies. mBio 10:e00307-19. doi: 10.1128/mBio.00307-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Easterbrook JD, Schwartzman LM, Gao J, Kash JC, Morens DM, Couzens L, Wan H, Eichelberger MC, Taubenberger JK. 2012. Protection against a lethal H5N1 influenza challenge by intranasal immunization with virus-like particles containing 2009 pandemic H1N1 neuraminidase in mice. Virology 432:39–44. doi: 10.1016/j.virol.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wan H, Gao J, Xu K, Chen H, Couzens LK, Rivers KH, Easterbrook JD, Yang K, Zhong L, Rajabi M, Ye J, Sultana I, Wan XF, Liu X, Perez DR, Taubenberger JK, Eichelberger MC. 2013. Molecular basis for broad neuraminidase immunity: conserved epitopes in seasonal and pandemic H1N1 as well as H5N1 influenza viruses. J Virol 87:9290–9300. doi: 10.1128/JVI.01203-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Halbherr SJ, Ludersdorfer TH, Ricklin M, Locher S, Berger Rentsch M, Summerfield A, Zimmer G. 2015. Biological and protective properties of immune sera directed to the influenza virus neuraminidase. J Virol 89:1550–1563. doi: 10.1128/JVI.02949-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mendez-Legaza JM, Ortiz de Lejarazu R, Sanz I. 2019. Heterosubtypic neuraminidase antibodies against different A(H1N1) strains are elicited after seasonal influenza vaccination. Vaccines (Basel) 7:30. doi: 10.3390/vaccines7010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schulman JL. 1969. The role of antineuraminidase antibody in immunity to influenza virus infection. Bull World Health Organ 41:647–650. [PMC free article] [PubMed] [Google Scholar]
- 28.Kilbourne ED, Couch RB, Kasel JA, Keitel WA, Cate TR, Quarles JH, Grajower B, Pokorny BA, Johansson BE. 1995. Purified influenza A virus N2 neuraminidase vaccine is immunogenic and non-toxic in humans. Vaccine 13:1799–1803. doi: 10.1016/0264-410X(95)00127-M. [DOI] [PubMed] [Google Scholar]
- 29.Wohlbold TJ, Podolsky KA, Chromikova V, Kirkpatrick E, Falconieri V, Meade P, Amanat F, Tan J, tenOever BR, Tan GS, Subramaniam S, Palese P, Krammer F. 2017. Broadly protective murine monoclonal antibodies against influenza B virus target highly conserved neuraminidase epitopes. Nat Microbiol 2:1415–1424. doi: 10.1038/s41564-017-0011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piepenbrink MS, Nogales A, Basu M, Fucile CF, Liesveld JL, Keefer MC, Rosenberg AF, Martinez-Sobrido L, Kobie JJ. 2019. Broad and protective influenza B virus neuraminidase antibodies in humans after vaccination and their clonal persistence as plasma cells. mBio 10:e00066-19. doi: 10.1128/mBio.00066-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johansson BE, Moran TM, Kilbourne ED. 1987. Antigen-presenting B cells and helper T cells cooperatively mediate intravirionic antigenic competition between influenza A virus surface glycoproteins. Proc Natl Acad Sci U S A 84:6869–6873. doi: 10.1073/pnas.84.19.6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johansson BE, Moran TM, Bona CA, Popple SW, Kilbourne ED. 1987. Immunologic response to influenza virus neuraminidase is influenced by prior experience with the associated viral hemagglutinin. II. Sequential infection of mice simulates human experience. J Immunol 139:2010–2014. [PubMed] [Google Scholar]
- 33.Johansson BE, Kilbourne ED. 1993. Dissociation of influenza virus hemagglutinin and neuraminidase eliminates their intravirionic antigenic competition. J Virol 67:5721–5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wohlbold TJ, Krammer F. 2014. In the shadow of hemagglutinin: a growing interest in influenza viral neuraminidase and its role as a vaccine antigen. Viruses 6:2465–2494. doi: 10.3390/v6062465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johansson BE, Bucher DJ, Kilbourne ED. 1989. Purified influenza virus hemagglutinin and neuraminidase are equivalent in stimulation of antibody response but induce contrasting types of immunity to infection. J Virol 63:1239–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johansson BE, Matthews JT, Kilbourne ED. 1998. Supplementation of conventional influenza A vaccine with purified viral neuraminidase results in a balanced and broadened immune response. Vaccine 16:1009–1015. doi: 10.1016/S0264-410X(97)00279-X. [DOI] [PubMed] [Google Scholar]
- 37.Wohlbold TJ, Hirsh A, Krammer F. 2015. An H10N8 influenza virus vaccine strain and mouse challenge model based on the human isolate A/Jiangxi-Donghu/346/13. Vaccine 33:1102–1106. doi: 10.1016/j.vaccine.2015.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meseda CA, Atukorale V, Soto J, Eichelberger MC, Gao J, Wang W, Weiss CD, Weir JP. 2018. Immunogenicity and protection against influenza H7N3 in mice by modified vaccinia virus Ankara vectors expressing influenza virus hemagglutinin or neuraminidase. Sci Rep 8:5364. doi: 10.1038/s41598-018-23712-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krammer F, Palese P. 2015. Advances in the development of influenza virus vaccines. Nat Rev Drug Discov 14:167–182. doi: 10.1038/nrd4529. [DOI] [PubMed] [Google Scholar]
- 40.Li J, Zu Dohna H, Cardona CJ, Miller J, Carpenter TE. 2011. Emergence and genetic variation of neuraminidase stalk deletions in avian influenza viruses. PLoS One 6:e14722. doi: 10.1371/journal.pone.0014722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Castrucci MR, Kawaoka Y. 1993. Biologic importance of neuraminidase stalk length in influenza A virus. J Virol 67:759–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo G, Chung J, Palese P. 1993. Alterations of the stalk of the influenza virus neuraminidase: deletions and insertions. Virus Res 29:141–153. doi: 10.1016/0168-1702(93)90055-R. [DOI] [PubMed] [Google Scholar]
- 43.Kosik I, Angeletti D, Gibbs JS, Angel M, Takeda K, Kosikova M, Nair V, Hickman HD, Xie H, Brooke CB, Yewdell JW. 2019. Neuraminidase inhibition contributes to influenza A virus neutralization by anti-hemagglutinin stem antibodies. J Exp Med 216:304–316. doi: 10.1084/jem.20181624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steuler H, Rohde W, Scholtissek C. 1984. Sequence of the neuraminidase gene of an avian influenza A virus (A/parrot/ulster/73, H7N1). Virology 135:118–124. doi: 10.1016/0042-6822(84)90122-3. [DOI] [PubMed] [Google Scholar]
- 45.Durrant JD, Bush RM, Amaro RE. 2016. Microsecond molecular dynamics simulations of influenza neuraminidase suggest a mechanism for the increased virulence of stalk-deletion mutants. J Phys Chem B 120:8590–8599. doi: 10.1021/acs.jpcb.6b02655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bailey MJ, Broecker F, Leon PE, Tan GS. 2018. A method to assess Fc-mediated effector functions induced by influenza hemagglutinin specific antibodies. J Vis Exp 132:e56256. doi: 10.3791/56256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Broecker F, Liu STH, Sun W, Krammer F, Simon V, Palese P. 2018. Immunodominance of antigenic site B in the hemagglutinin of the current H3N2 influenza virus in humans and mice. J Virol 92:e01100-18. doi: 10.1128/JVI.01100-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harris A, Cardone G, Winkler DC, Heymann JB, Brecher M, White JM, Steven AC. 2006. Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc Natl Acad Sci U S A 103:19123–19127. doi: 10.1073/pnas.0607614103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sultana I, Yang K, Getie-Kebtie M, Couzens L, Markoff L, Alterman M, Eichelberger MC. 2014. Stability of neuraminidase in inactivated influenza vaccines. Vaccine 32:2225–2230. doi: 10.1016/j.vaccine.2014.01.078. [DOI] [PubMed] [Google Scholar]
- 50.DiLillo DJ, Tan GS, Palese P, Ravetch JV. 2014. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcγR interactions for protection against influenza virus in vivo. Nat Med 20:143–151. doi: 10.1038/nm.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DiLillo DJ, Palese P, Wilson PC, Ravetch JV. 2016. Broadly neutralizing anti-influenza antibodies require Fc receptor engagement for in vivo protection. J Clin Invest 126:605–610. doi: 10.1172/JCI84428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Henry-Dunand CJ, Leon PE, Huang M, Choi A, Chromikova V, Ho IY, Tan GS, Cruz J, Hirsh A, Zheng NY, Mullarkey CE, Ennis FA, Terajima M, Treanor JJ, Topham DJ, Subbarao K, Palese P, Krammer F, Wilson PC. 2016. Both neutralizing and non-neutralizing human H7N9 influenza vaccine-induced monoclonal antibodies confer protection. Cell Host Microbe 19:800–813. doi: 10.1016/j.chom.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yasuhara A, Yamayoshi S, Kiso M, Sakai-Tagawa Y, Koga M, Adachi E, Kikuchi T, Wang IH, Yamada S, Kawaoka Y. 2019. Antigenic drift originating from changes to the lateral surface of the neuraminidase head of influenza A virus. Nat Microbiol 4:1024–1034. doi: 10.1038/s41564-019-0401-1. [DOI] [PubMed] [Google Scholar]
- 54.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 55.Fulton BO, Sun W, Heaton NS, Palese P. 2018. The influenza B virus hemagglutinin head domain is less tolerant to transposon mutagenesis than that of the influenza A virus. J Virol 92:e00754-18. doi: 10.1128/JVI.00754-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang TT, Tan GS, Hai R, Pica N, Petersen E, Moran TM, Palese P. 2010. Broadly protective monoclonal antibodies against H3 influenza viruses following sequential immunization with different hemagglutinins. PLoS Pathog 6:e1000796. doi: 10.1371/journal.ppat.1000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krammer F, Margine I, Tan GS, Pica N, Krause JC, Palese P. 2012. A carboxy-terminal trimerization domain stabilizes conformational epitopes on the stalk domain of soluble recombinant hemagglutinin substrates. PLoS One 7:e43603. doi: 10.1371/journal.pone.0043603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Margine I, Palese P, Krammer F. 2013. Expression of functional recombinant hemagglutinin and neuraminidase proteins from the novel H7N9 influenza virus using the baculovirus expression system. J Vis Exp 81:e51112. doi: 10.3791/51112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gao J, Couzens L, Eichelberger MC. 6 September 2016, posting date. Measuring influenza virus neuraminidase inhibition antibody titers by enzyme-linked lectin assay. J Vis Exp. doi: 10.3791/54573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rajendran M, Nachbagauer R, Ermler ME, Bunduc P, Amanat F, Izikson R, Cox M, Palese P, Eichelberger M, Krammer F. 2017. Analysis of anti-influenza virus neuraminidase antibodies in children, adults, and the elderly by ELISA and enzyme inhibition: evidence for original antigenic sin. mBio 8:e02281-16. doi: 10.1128/mBio.02281-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Margine I, Krammer F, Hai R, Heaton NS, Tan GS, Andrews SA, Runstadler JA, Wilson PC, Albrecht RA, García-Sastre A, Palese P. 2013. Hemagglutinin stalk-based universal vaccine constructs protect against group 2 influenza A viruses. J Virol 87:10435–10446. doi: 10.1128/JVI.01715-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gamblin SJ, Haire LF, Russell RJ, Stevens DJ, Xiao B, Ha Y, Vasisht N, Steinhauer DA, Daniels RS, Elliot A, Wiley DC, Skehel JJ. 2004. The structure and receptor binding properties of the 1918 influenza hemagglutinin. Science 303:1838–1842. doi: 10.1126/science.1093155. [DOI] [PubMed] [Google Scholar]
- 63.Vavricka CJ, Li Q, Wu Y, Qi J, Wang M, Liu Y, Gao F, Liu J, Feng E, He J, Wang J, Liu H, Jiang H, Gao GF. 2011. Structural and functional analysis of laninamivir and its octanoate prodrug reveals group specific mechanisms for influenza NA inhibition. PLoS Pathog 7:e1002249. doi: 10.1371/journal.ppat.1002249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]