Host anti-RNA virus innate immunity relies mainly on the recognition by retinoic acid-inducible gene I and melanoma differentiation-associated protein 5 and subsequently initiates downstream signaling through interaction with MAVS. On the other hand, viruses have developed various strategies to counteract MAVS-mediated signaling. The mechanism for paramyxoviruses regulating MAVS to benefit their infection remains unknown. In this article, we demonstrate that the V proteins of NDV and several other paramyxoviruses target MAVS for ubiquitin-mediated degradation through E3 ubiquitin ligase RING-finger protein 5 (RNF5). MAVS degradation leads to the inhibition of the downstream IFN-β pathway and therefore benefits virus proliferation. Our study reveals a novel mechanism of NDV evading host innate immunity and provides insight into the therapeutic strategies for the control of paramyxovirus infection.
KEYWORDS: MAVS, NDV, RNF5, degradation, paramyxovirus
ABSTRACT
Paramyxovirus establishes an intimate and complex interaction with the host cell to counteract the antiviral responses elicited by the cell. Of the various pattern recognition receptors in the host, the cytosolic RNA helicases interact with viral RNA to activate the mitochondrial antiviral signaling protein (MAVS) and subsequent cellular interferon (IFN) response. On the other hand, viruses explore multiple strategies to resist host immunity. In this study, we found that Newcastle disease virus (NDV) infection induced MAVS degradation. Further analysis showed that NDV V protein degraded MAVS through the ubiquitin-proteasome pathway to inhibit IFN-β production. Moreover, NDV V protein led to proteasomal degradation of MAVS through Lys362 and Lys461 ubiquitin to prevent IFN production. Further studies showed that NDV V protein recruited E3 ubiquitin ligase RNF5 to polyubiquitinate and degrade MAVS. Compared with levels for wild-type NDV infection, V-deficient NDV induced attenuated MAVS degradation and enhanced IFN-β production at the late stage of infection. Several other paramyxovirus V proteins showed activities of degrading MAVS and blocking IFN production similar to those of NDV V protein. The present study revealed a novel role of NDV V protein in targeting MAVS to inhibit cellular IFN production, which reinforces the fact that the virus orchestrates the cellular antiviral response to its own benefit.
IMPORTANCE Host anti-RNA virus innate immunity relies mainly on the recognition by retinoic acid-inducible gene I and melanoma differentiation-associated protein 5 and subsequently initiates downstream signaling through interaction with MAVS. On the other hand, viruses have developed various strategies to counteract MAVS-mediated signaling. The mechanism for paramyxoviruses regulating MAVS to benefit their infection remains unknown. In this article, we demonstrate that the V proteins of NDV and several other paramyxoviruses target MAVS for ubiquitin-mediated degradation through E3 ubiquitin ligase RING-finger protein 5 (RNF5). MAVS degradation leads to the inhibition of the downstream IFN-β pathway and therefore benefits virus proliferation. Our study reveals a novel mechanism of NDV evading host innate immunity and provides insight into the therapeutic strategies for the control of paramyxovirus infection.
INTRODUCTION
As the first barrier of host defense against invading pathogens, innate immunity signaling pathways, including interferon (IFN) production and NK cell activation, play an important role in antiviral response. The most important cytoplasmic pathogen recognition receptors toward RNA virus are retinoic acid-inducible gene I (RIG-I) and melanoma differentiation-associated protein 5 (MDA5) (1, 2). Both contain a DExD/H-box RNA helicase domain that directly senses viral RNA (3). After RNA recognition, the two caspase recruitment domains (CARDs) at the N termini of RIG-I and MDA5 initiate downstream signaling through interaction with the CARD domain of the same adaptor protein as a mitochondrial antiviral signaling protein (MAVS; also known as Cardif, IPS-1, or VISA) (4–7). MAVS contains a CARD-like region, a proline-rich region (PRR), and a transmembrane (TM) domain, and the CARD and TM domains are essential for MAVS signaling (6). The interaction between RIG-I/MDA5 and MAVS signals IκB kinases (IKKs) and TANK-binding kinase 1 (TBK1) for the activation of interferon regulatory factor 3 (IRF3) and nuclear factor kappa B (NF-κB) pathways, which subsequently drives expression of IFN-β and other IFN-mediated antiviral immunities (8, 9).
MAVS plays a central role in the production of antiviral and other proinflammatory cytokines induced by virus infection (10, 11). However, in the long-term coexistence between virus and host, viruses have developed various strategies to counteract MAVS-mediated signaling. The cleavage of MAVS by virus protein is one of the most important antagonistic mechanisms. The function of MAVS relies on its mitochondrial localization. Several virus proteins possess the activity of protease and could cleave MAVS from mitochondrial membrane to inhibit downstream signaling. For example, hepatitis C virus (HCV) protease NS3/4A cleaves MAVS at position 508 (cysteine), resulting in the dislocation of the N-terminal fragment of MAVS from the mitochondria (12). The 3C protein of coxsackievirus B and porcine reproductive and respiratory syndrome virus cleaves MAVS at positions 148 (glutamine) and 268 (glutamic acid), respectively (13, 14). Except for cleavage of MAVS, several viruses inhibit MAVS-mediated signaling by degradation of MAVS. The hepatitis B virus X protein (HBX) interacts with MAVS, promoting the degradation of MAVS through Lys136 ubiquitin in MAVS protein, and disrupts innate immunity (15). The open reading frame 9b (ORF-9b) of severe acute respiratory syndrome coronaviruses (SARS-CoV) usurps poly(C)-binding protein 2 (PCBP2) and the homologous E6-AP carboxyl terminus (HECT) domain E3 ligase AIP4 to trigger the degradation of MAVS signalosome (16).
The Paramyxoviridae family includes various viruses, such as Newcastle disease virus (NDV), Sendai virus, measles virus, Hendra virus, mumps virus, Nipah virus, and parainfluenza virus (PIV). The protein products of the P/V/C genes of paramyxoviruses were known to antagonize the induction of IFN-β at several levels (17). The +1 frameshift of the P gene encodes the V gene, the product of which is the most well-known IFN antagonist among four P-gene-derived products (P/V/C/W). The +2 frameshift of the P gene encodes the W gene, the function of which is not well understood. Almost all paramyxovirus V proteins could bind the helicase domain of MDA-5 and inhibit its activation by blocking double-stranded RNA (dsRNA) binding and consequent self-association, subsequently inhibiting downstream IFN production (18, 19). Another example is that paramyxovirus V proteins hijack the DDB1-Cul4A ubiquitin ligase complex to degrade STATs, the transcription factors which are essential for IFN signaling, and thereby block IFN signaling (20–22). NDV, a member of the Paramyxoviridae family, causes respiratory disease and death in a wide variety of bird species (23). Oncolytic NDV can selectively replicate in tumor cells and serve as a potential oncolytic agent (24). Previous reports by us and others have shown that the NDV V protein could inhibit the pathway of RIG-I-like receptors (RLRs) by targeting MDA5, STAT1, or laboratory of genetics and physiology 2 (LGP2) (25–27). Here, we report that V proteins of NDV and several other paramyxoviruses target MAVS for ubiquitin-mediated degradation through E3 ubiquitin ligase RING-finger protein 5 (RNF5). Our study reveals a novel mechanism of NDV evading host innate immunity and provides insight into the therapeutic strategies for the control of paramyxovirus infection.
RESULTS
NDV infection induced MAVS degradation through the ubiquitin-proteasome pathway.
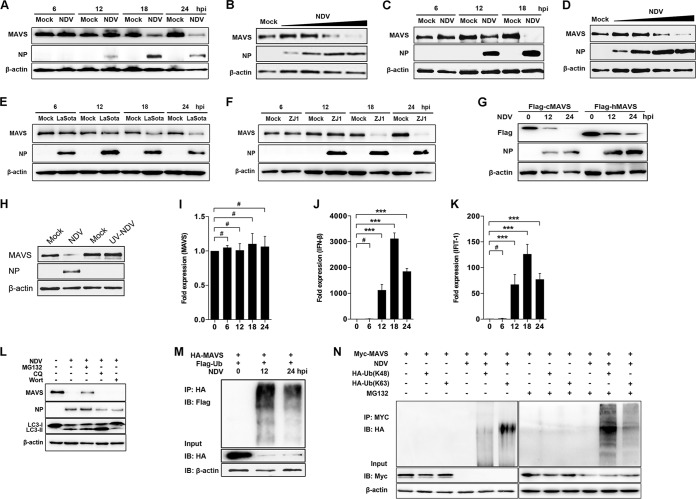
RLR signaling helps to defend the host against infection of many RNA viruses, including NDV (28, 29). On the other hand, paramyxoviruses exploit a series of strategies to antagonize RLR signaling by targeting MDA5, LGP2, etc. (25, 26). To explore whether NDV targets MAVS to inhibit RLR pathways, we investigated the expression of MAVS at both transcriptional and translational levels following NDV infection. The results showed that a significant degradation of endogenous MAVS was detected at 18 and 24 h after NDV Herts/33 infection (NDV Herts/33 strain is abbreviated as NDV here) (Fig. 1A). NDV degrades MAVS in a multiplicity of infection (MOI)-dependent manner (Fig. 1B). In addition to HeLa cells, A549 cells were utilized to test whether the NDV-triggered MAVS degradation was cell type dependent. The results showed that NDV also degraded MAVS in A549 cells in a time- and dose-dependent manner (Fig. 1C and D). To study whether the degradation is strain dependent, we used lentogenic LaSota and velogenic ZJ1 strains to evaluate their capability for MAVS degradation. As expected, both LaSota and ZJ1 strains apparently could degrade MAVS at 18 and 24 h postinfection (hpi) (Fig. 1E and F). Since NDV is an important avian virus, we next explored whether NDV could also degrade chicken MAVS. As expected, NDV triggered degradation of exogenous human and chicken MAVS (Fig. 1G). To investigate whether viral replication is required in NDV-mediated MAVS degradation, UV-inactivated NDV was used for the experiment. The results showed that UV-inactivated NDV did not induce the MAVS degradation and the NDV nucleoprotein (NP) was not detected either, suggesting that NDV replication is required for MAVS degradation (Fig. 1H). Since NDV infection degrades MAVS at the protein level, we next evaluated whether NDV infection affects MAVS expression at the transcriptional level. The results indicated that NDV infection did not affect the mRNA level of MAVS (Fig. 1I). Meanwhile, the mRNA levels of IFN-β and downstream interferon-stimulated gene (ISG) interferon-induced protein with tetratricopeptide repeats 1 (IFIT1) were investigated. As expected, NDV infection induced high mRNA levels of both IFN-β and IFIT1 at 12, 18, and 24 hpi and peaked at 18 hpi (Fig. 1J and K). To determine whether the proteasome or lysosome pathway plays a role in NDV-induced MAVS degradation, the proteasome inhibitor MG132, lysosome inhibitor chloroquine (CQ), or autophagy inhibitor wortmannin was used to evaluate the inhibitory effects. In the presence of MG132, NDV infection-induced MAVS degradation was partially inhibited, but in the presence of CQ or wortmannin treatment no inhibition was detectable, indicating the proteasome pathway plays a role in NDV-induced MAVS degradation (Fig. 1L). To further verify the role of the ubiquitin-proteasome pathway in NDV-induced MAVS degradation, HEK-293T cells were cotransfected with hemagglutinin (HA)-MAVS and Flag-ubiquitin followed by NDV infection. The immunoprecipitation results showed that the ubiquitination of MAVS was detected at 12 and 24 h after NDV infection (Fig. 1M). To illustrate which type of ubiquitin was involved in NDV-induced MAVS degradation, cells were transfected with either K48- or K63-linked ubiquitin plasmids, followed by NDV infection. The results showed that in the presence of MG132, NDV induced both K48 and K63 ubiquitination of MAVS and K48 ubiquitination was dominant (Fig. 1N). Collectively, our results demonstrate that replication-competent NDV triggers MAVS degradation through the ubiquitin-proteasome pathway.
FIG 1.
NDV infection induced MAVS degradation. (A) HeLa cells were mock treated or infected with NDV at an MOI of 1. Cells were harvested at 6, 12, 18, and 24 hpi and detected using immunoblot (IB) analysis with anti-MAVS, anti-NP, or anti-β-actin antibody. (B) HeLa cells were mock treated or infected with NDV at an MOI of 0.01, 0.1, 1, or 5 (wedge). Cells were harvested at 18 hpi and detected using immunoblot analysis with anti-MAVS, anti-NP, or anti-β-actin antibody. (C) A549 cells were mock treated or infected with NDV Herts/33 strain at an MOI of 5. Cells were harvested at 6, 12, and 18 hpi and detected using immunoblot analysis with anti-MAVS, anti-NP, or anti-β-actin antibody. (D) A549 cells were mock treated or infected with NDV Herts/33 strain at an MOI of 0.01, 0.1, 1, or 5 (wedge). Cells were harvested at 18 hpi and detected using immunoblot analysis with anti-MAVS, anti-NP, or anti-β-actin antibody. (E and F) HeLa cells were mock treated or infected with NDV LaSota (E) or ZJ1 (F) strain at an MOI of 1. Cells were harvested at 6, 12, 18, and 24 hpi and detected using immunoblot analysis with anti-MAVS, anti-NP, or anti-β-actin antibody. (G) HEK-293T was transfected with Flag-tagged chicken MAVS (cMAV) and human MAVS (hMAVS). At 24 h after transfection, cells were infected with NDV at an MOI of 1. Cells were harvested at 12 and 24 hpi and detected using immunoblot analysis with anti-Flag, anti-NP, or anti-β-actin antibody. (H) HeLa cells were mock treated or infected with NDV (MOI of 1) or UV-treated NDV (MOI of 10). Cells were harvested at 18 hpi and detected with anti-MAVS, anti-NP, or anti-β-actin antibody. (I to K) HeLa cells were mock treated or infected with NDV at an MOI of 1. Cells were harvested at 6, 12, 18, and 24 hpi and detected using qRT-PCR with MAVS (I), IFN-β (J), or IFIT1 (K) primers. (L) HeLa cells were mock infected or infected with NDV at an MOI of 1 and maintained in the presence or absence of the lysosome inhibitor CQ (50 μM) or the autophagy inhibitor wortmannin (Wort; 100 nM) for 12 h. Cells were harvested and detected using immunoblot analysis with anti-MAVS, anti-NP, anti-LC3, or anti-β-actin antibody. For proteasome inhibition assay, the infected cells were treated with MG132 (20 μM) for 6 h prior to immunoblot analysis. (M) HEK-293T cells were cotransfected with HA-MAVS and Flag-ubiquitin. At 24 h after transfection, cells were infected with NDV at an MOI of 1. At 12 and 24 hpi, cells were harvested, immunoprecipitated (IP) with anti-HA antibody, and further detected using immunoblot analysis with anti-Flag antibody. Expression levels of the proteins were analyzed by immunoblot analysis of the lysates with anti-HA or anti-β-actin antibody. (N) HEK-293T cells were cotransfected with Myc-MAVS and either HA-ubiquitin (K48) or HA-ubiquitin (K63). At 24 h after transfection, cells were infected with NDV at an MOI of 1 and maintained in the presence or absence of the proteasome inhibitor MG132 (20 μM, 6 h prior to immunoprecipitation). At 18 hpi, cells were harvested, immunoprecipitated with anti-Myc antibody, and further detected using immunoblot analysis with anti-HA antibody. Expression levels of the proteins were analyzed by immunoblot analysis of the lysates with anti-Myc or anti-β-actin antibody. Data are presented as means from three independent experiments. #, P > 0.05; ***, P < 0.001.
NDV V protein targets MAVS for ubiquitination and degradation.
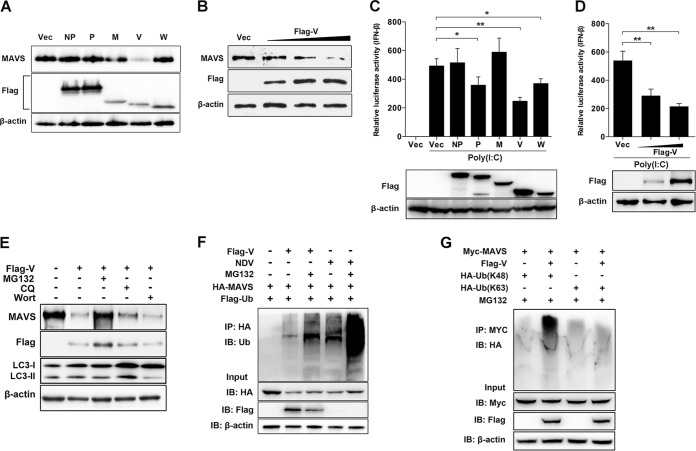
Since NDV infection induced MAVS degradation, we next evaluated which components of viruses were responsible for this degradation. Paramyxovirus V and W accessory proteins were previously proved to possess IFN antagonist activity (30, 31); therefore, they might be involved in MAVS degradation. In addition, NDV NP and P proteins have been reported to induce autophagy pathways (32), and they might be associated with MAVS degradation. The NDV M protein was used as an unrelated protein control, since no reports show its association with the degradation. The vector was used as a control. The results showed that MAVS was degraded by NDV V protein transfection but not by NDV NP, P, M, and W protein transfections (Fig. 2A). NDV V protein degrades endogenous MAVS in a dose-dependent manner (Fig. 2B). The role of virus proteins on downstream IFN-β activation was then evaluated. HEK-293T cells were cotransfected with poly(I·C) and each plasmid encoding NDV NP, P, M, V, or W protein or empty vector. As expected, the IFN-β promoter activity was inhibited by 50%, 29%, and 24% after cotransfection of Flag-V, -P, and -W, respectively, compared with that of the empty vector. In contrast, cotransfection of poly(I·C) and Flag-NP and -M did not affect IFN-β activation (Fig. 2C). Moreover, NDV V inhibited poly(I·C)-mediated IFN-β activation in a dose-dependent manner (Fig. 2D). These results demonstrated that NDV V protein inhibited IFN-β activation by degrading the adaptor MAVS. Pharmacological experiments showed that Flag-V transfection induced less MAVS degradation under treatment with MG132, while CQ or wortmannin treatment had no influence on MAVS degradation (Fig. 2E). By immunoprecipitation, we confirmed that Flag-V transfection increased the ubiquitination of MAVS (Fig. 2F). Further results demonstrated V-mediated MAVS ubiquitination with ubiquitin-K48 but not with ubiquitin-K63 (Fig. 2G). These results demonstrated the critical role of NDV V protein in MAVS degradation.
FIG 2.
NDV V protein triggers MAVS degradation through ubiquitin-proteasome pathway. (A) HeLa cells were transfected with either empty vector (Vec) or Flag-tagged NP, P, M, V, or W. At 12 h posttransfection (hpt), cells were transfected with poly(I·C) (20 μg/ml). Cells were harvested at 12 hpt with poly(I·C) and detected using immunoblot analysis with anti-MAVS, anti-Flag, or anti-β-actin antibody. (B) HeLa cells were transfected with either empty vector or Flag-V (0.5, 1, or 2 μg/ml; wedge). At 12 hpt, cells were transfected with poly(I·C) (20 μg/ml). Cells were harvested at 12 hpt with poly(I·C) and detected using immunoblot analysis with anti-MAVS, anti-Flag, or anti-β-actin antibody. (C) HEK-293T cells were cotransfected with p-125Luc, PRL-TK, and either empty vector or Flag-tagged NP, P, M, V, or W. At 12 hpt, cells were mock treated or transfected with poly(I·C) (20 μg/ml). Cells were harvested at 12 hpt and assessed for luciferase activity. The results are presented as relative luciferase activity. Expression levels of expressed proteins were analyzed by immunoblot analysis of the lysates with anti-Flag or anti-β-actin antibody. (D) HEK-293T cells were cotransfected with p-125Luc, PRL-TK, and either empty vector or Flag-V (0, 0.5, or 1 μg/ml; wedge). At 12 hpt, cells were transfected with poly(I·C) (20 μg/ml). Cells were harvested at 12 hpt and assessed for luciferase activity. The results are presented as relative luciferase activity. Expression levels of expressed proteins were analyzed by immunoblot analysis of the lysates with anti-Flag or anti-β-actin antibody. (E) HeLa cells were transfected with either empty vector or Flag-V and maintained in the presence or absence of the proteasome inhibitor MG132 (20 μM, 6 h prior to immunoblot analysis), the lysosome inhibitor CQ (50 μM), or the autophagy inhibitor wortmannin (100 nM) for 12 h. Cells were harvested and detected using immunoblot analysis with anti-MAVS, anti-NP, or anti-β-actin antibody. (F) HEK-293T cells were cotransfected with HA-MAVS, Flag-ubiquitin (Ub), and either empty vector or Flag-V and maintained in the presence or absence of the proteasome inhibitor MG132 (20 μM, 6 h prior to immunoprecipitation). At 24 hpt, cells were harvested, immunoprecipitated with anti-HA antibody, and further detected using immunoblot analysis with anti-ubiquitin MAbs. For the NDV infection group, HEK-293T cells were cotransfected with HA-MAVS and Flag-ubiquitin. At 24 hpt, cells were infected with NDV at an MOI of 1 and maintained in the presence or absence of MG132 (20 μM, 6 h prior to immunoprecipitation). At 12 hpi, cells were harvested for immunoprecipitation assay. Expression levels of the proteins were analyzed by immunoblot analysis of the lysates with anti-HA, anti-Flag, or anti-β-actin antibody. (G) HEK-293T cells were cotransfected with Myc-MAVS, HA-ubiquitin (K48), or HA-ubiquitin (K63) and either empty vector or Flag-V and maintained in the presence of the proteasome inhibitor MG132 (20 μM, 6 h prior to immunoprecipitation). At 24 hpt, cells were harvested, immunoprecipitated with anti-Myc antibody, and further detected using immunoblot analysis with anti-HA antibody. Expression levels of the proteins were analyzed by immunoblot analysis of the lysates with anti-Myc, anti-Flag, or anti-β-actin antibody. Data are presented as means from three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
NDV V protein negatively regulates MAVS-mediated signaling.
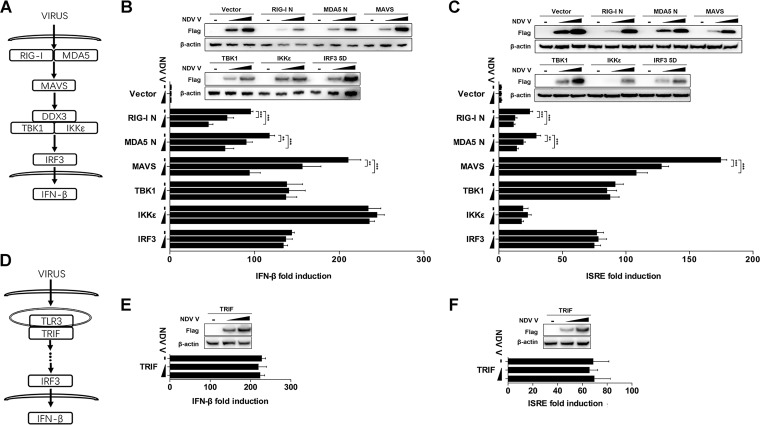
The molecules involved in the initiation and activation of the RLR pathway could be divided into four categories, receptors (RIG-I and MDA5), adaptors (MAVS), signaling transducers (TBK1 and IKKε), and transcript factors (IRF3) (Fig. 3A). To explore the effects of NDV V on the RLR pathway, HEK-293T cells were cotransfected with Flag-tagged V and each RLR pathway-related expression plasmid of RIG-I N, MDA5 N, MAVS, TBK1, IKKε, and IRF3-5D (Fig. 3A). The transfection of each RIG-I N, MDA5 N, MAVS, TBK1, IKKε, and IRF3-5D expression plasmid activated the transcription of IFN-β by over 100-fold compared with that of the empty vector transfection control. When Flag-V was cotransfected with RIG-I N, MDA5 N, or MAVS, the IFN-β promoter activity was inhibited in a dose-dependent manner. Cotransfection of Flag-V with TBK1, IKKε, or IRF3-5D had no inhibition of IFN-β promoter activity (Fig. 3B). Similarly, the V protein inhibited IFN sequence response element (ISRE) promoter activity induced by RIG-I N, MDA5 N, or MAVS but not TBK1, IKKε, or IRF3-5D (Fig. 3C). Toll-like receptor 3 (TLR3) is an endosomal TLR that also senses viral dsRNA and mediates IFN-β production via toll-like receptor (TLR) domain-containing adapter-inducing IFN-β (TRIF) (Fig. 3D) (33–35). Therefore, we next evaluated whether NDV V could impair TLR/TRIF-mediated IFN-β production. The results showed that NDV V did not affect the promoter activity of either IFN-β or ISRE driven by TRIF (Fig. 3E and F). These results demonstrated that NDV V specifically targets MAVS to inhibit the innate immunity response.
FIG 3.
NDV V protein suppresses MAVS-mediated IFN-β signaling transduction. (A) Schematic diagram of RIG-I/MDA5-mediated IFN-β signaling pathway. (B and C) HEK-293T cells were cotransfected with PRL-TK, expression plasmid RIG-I N, MDA5 N, MAVS, TBK1, IKKε, or IRF3-5D or empty plasmid (vector), and either empty vector (−) or Flag-V (1 or 2 μg/ml; wedge) as well as p-125Luc (B) or pISRE-luc (C). At 24 hpt, cells were harvested and assessed for luciferase activity. The results are presented as relative luciferase activity. Expression levels of expressed proteins were analyzed by immunoblot analysis of the lysates with anti-Flag or anti-β-actin antibody. (D) Schematic diagram of TLR3-mediated signaling pathway. (E and F) HEK-293T cells were cotransfected with PRL-TK, TRIF, and either empty vector or Flag-V (1 or 2 μg/ml; wedge) as well as p-125Luc (E) or pISRE-luc (F). At 24 hpt, cells were harvested and assessed for luciferase activity. The results are presented as relative luciferase activity. Expression levels of expressed proteins were analyzed by immunoblot analysis of the lysates with anti-Flag or anti-β-actin antibody. Data are presented as means from three independent experiments. **, P < 0.01; ***, P < 0.001.
Characterization of the interaction between NDV V protein and MAVS.
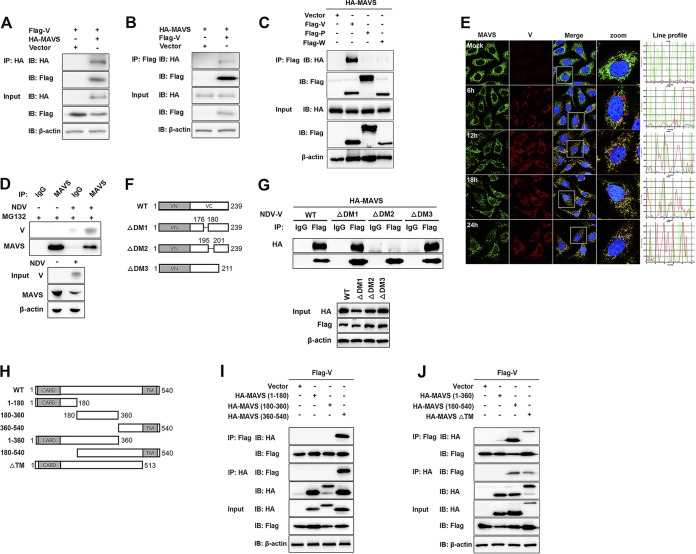
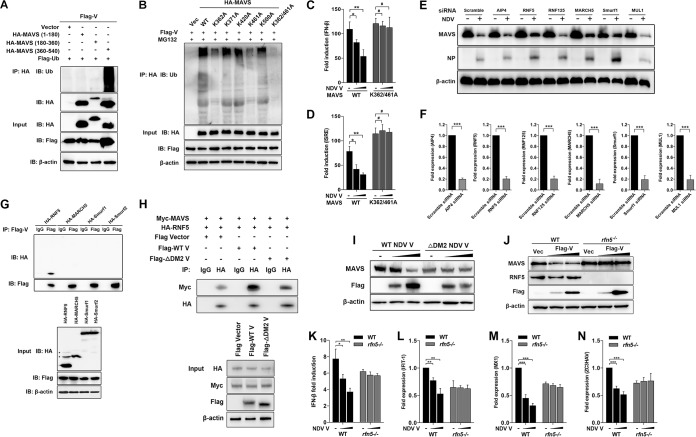
To investigate a possible interaction between NDV V protein and MAVS, HEK-293T cells were cotransfected with Flag-V and either HA-MAVS or pRK-HA empty vector. The lysates were immunoprecipitated with anti-HA antibody and analyzed by immunoblotting. As shown in Fig. 4A, the specific interaction between Flag-V and HA-MAVS was detected in the cotransfection group. A reverse immunoprecipitation experiment was performed with anti-Flag antibody and also confirmed the interaction between NDV V and MAVS (Fig. 4B). As shown above, V, but not P or W, specifically degraded MAVS, indicating the shared N domain was not responsible for MAVS degradation (Fig. 2A). Here, we further confirmed that V was the protein specifically interacting with MAVS (Fig. 4C). The endogenous coimmunoprecipitation results showed the interaction between MAVS and V in the course of NDV infection (Fig. 4D). To detect whether NDV V was associated with endogenous MAVS in the course of infection, HeLa cells seeded on coverslips were infected with NDV at an MOI of 1 and collected at 6, 12, 18, and 24 hpi. Endogenous MAVS was specifically located in the cytoplasm in uninfected or infected cells. The NDV V protein could be detected as the aggregated fluorescent plaque in the cytoplasm at 6 hpi. Interestingly, although MAVS and V were adjacent to each other, almost no colocalization was observed at 6 hpi. After that, the NDV V protein was detected as dispersed fluorescent granules, and some of them were observed to be colocalized with MAVS at 12, 18, and 24 hpi. (Fig. 4E). Paramyxovirus V proteins contain the conserved cysteine-rich C-terminal domains, which in this study we referred to as domain 1 (DM1) (amino acids [aa] 176 to 180), DM2 (aa 195 to 201), and DM3 (aa 201 to 211) (26) (Fig. 4F). To characterize the necessary domains involved in the interaction, HA-MAVS was cotransfected with wild-type (WT) Flag-V, each deletion mutant Flag-V (ΔDM1, ΔDM2, and ΔDM3), or empty vector and then subjected to immunoprecipitation. The results showed that deletion of DM2 completely abrogated the interaction between V and MAVS, indicating that DM2 of V is essential for MAVS-V interaction (Fig. 4G). MAVS contains an N-terminal CARD-like domain, a PRR domain, and a C-terminal TM domain (6) (Fig. 4H). To analyze the critical domain of MAVS responsible for MAVS-V interaction, Flag-V was cotransfected with either empty vector, WT HA-MAVS, truncated (aa 1 to 180, aa 180 to 360, aa 360 to 540, aa 1 to 360, or aa 180 to 540) HA-MAVS, or a TM domain deletion mutant (ΔTM) of HA-MAVS, followed by immunoprecipitation. The results showed that HA-MAVS (aa 360 to 540) had specifically interacted with V (Fig. 4I). Deletion of the TM domain did not abrogate MAVS-V interaction (Fig. 4J). Therefore, V interacted with aa 360 to 513 of MAVS. Collectively, these results strongly indicated that NDV V interacted with MAVS in both infection and transfection models.
FIG 4.
NDV V interacts with MAVS. (A) HEK-293T cells were cotransfected with Flag-V and either empty vector or HA-MAVS. At 24 hpt, cells were harvested, immunoprecipitated with anti-HA antibody, and further detected using immunoblot analysis with anti-HA or anti-Flag antibody. Expression levels of the proteins were analyzed by immunoblot analysis of the lysates with anti-HA, anti-Flag, or anti-β-actin antibody. (B) HEK-293T cells were cotransfected with HA-MAVS and either empty vector or Flag-V. At 24 hpt, cells were harvested, immunoprecipitated with anti-HA antibody, and further detected using immunoblot analysis with anti-HA or anti-Flag antibody. Expression levels of the proteins were analyzed by immunoblot analysis of the lysates with anti-HA, anti-Flag, or anti-β-actin antibody. (C) HEK-293T cells were cotransfected with HA-MAVS and either empty vector or Flag-V, -P, or -W. At 24 hpt, cells were harvested, immunoprecipitated with anti-Flag antibody, and further detected using immunoblot analysis with anti-HA or anti-Flag antibody. Expression levels of the proteins were analyzed by immunoblot analysis of the lysates with anti-HA, anti-Flag, or anti-β-actin antibody. (D) HeLa cells were mock treated or infected with NDV at an MOI of 1. At 24 hpt, cells were harvested, immunoprecipitated with anti-IgG or MAVS antibody, and further detected using immunoblot analysis with anti-V or anti-MAVS antibody. Expression levels of the proteins were analyzed by immunoblot analysis of the lysates with anti-V, anti-MAVS, or anti-β-actin antibody. (E) HeLa cells were mock treated or infected with NDV at an MOI of 1. Cells were harvested at 6, 12, 18, and 24 hpi and detected using IF assay with anti-MAVS or anti-V antibody. Shown is an intensity profile of the linear region of interest (ROI) across the HeLa cell costained with V and MAVS. (F) Schematics of a series of Flag-tagged truncated V constructs. (G) HEK-293T cells were cotransfected with HA-MAVS and either empty vector, WT, or truncated Flag-V (ΔDM1, ΔDM2, and ΔDM3). At 24 hpt, cells were harvested, immunoprecipitated with anti-IgG or anti-Flag antibody, and further detected using immunoblot analysis with anti-HA or anti-Flag antibody. Expression levels of the proteins were analyzed by immunoblot analysis of the lysates with anti-HA, anti-Flag, or anti-β-actin antibody. (H) Schematics of a series of HA-tagged truncated MAVS constructs. (I and J) HEK-293T cells were cotransfected with Flag-V and either empty vector or truncated HA-MAVS aa 1 to 180, aa 180 to 360, and aa 360 to 540 (I) or aa 1 to 360, aa 180 to 540, and ΔTM (J). At 24 hpt, cells were harvested, immunoprecipitated with anti-Flag or anti-HA antibody, and further detected using immunoblot analysis with anti-HA or anti-Flag antibody. Expression levels of the proteins were analyzed by immunoblot analysis of the lysates with anti-HA, anti-Flag, or anti-β-actin antibody.
RNF5 is the E3 ubiquitin ligase involved in NDV V protein-induced MAVS degradation.
Our coimmunoprecipitation experiments demonstrated that aa 360 to 513 of MAVS was responsible for V-MAVS interaction. We next proved that NDV V caused ubiquitination of wild-type MAVS and MAVS (aa 360 to 540) but not MAVS (aa 1 to 180) and MAVS (aa 180 to 360). These results together suggest that the ubiquitination sites of MAVS are located in the region of aa 360 to 513 of MAVS (Fig. 5A). Sequence analysis identified five lysine residues in this region, which are K362, K371, K420, K462, and K500. We then constructed MAVS mutants in which the lysine residues were mutated to alanines individually or simultaneously and investigated their ubiquitination by NDV V. As shown in Fig. 5B, K362A or K461A MAVS reduced V-mediated ubiquitination of MAVS, whereas simultaneous mutation of both K362 and K461 to alanines abolished V-catalyzed ubiquitination (Fig. 5B). Transfection of V specifically inhibited the promoter activities of both IFN-β (Fig. 5C) and ISRE (Fig. 5D) triggered by WT, but not K362/461A, MAVS.
FIG 5.
Characterization of RNF5 as the E3 ubiquitin ligase for V-mediated MAVS degradation. (A) HEK-293T cells were cotransfected with Flag-V, Flag-ubiquitin, and either empty vector or truncated HA-MAVS aa 1 to 180, 180 to 360, and 360 to 540. At 24 hpt, cells were harvested, immunoprecipitated with anti-HA antibody, and further detected using immunoblot analysis with anti-ubiquitin or anti-HA antibody. Expression levels of the proteins were analyzed by immunoblot analysis of the lysates with anti-HA, anti-Flag, or anti-β-actin antibody. (B) HEK-293T cells were cotransfected with Flag-V, Flag-ubiquitin, and either empty vector, WT, or mutated HA-MAVS K362A, K371A, K420A, K461A, K500A, and K362/461A and maintained in the presence of the proteasome inhibitor MG132 (20 μM, 6 h prior to immunoprecipitation). At 24 hpt, cells were harvested, immunoprecipitated with anti-HA antibody, and further detected using immunoblot analysis with anti-ubiquitin antibody. Expression levels of the proteins were analyzed by immunoblot analysis of the lysates with anti-HA, anti-Flag, or anti-β-actin antibody. (C and D) HEK-293T cells were cotransfected with PRL-TK, HA-MAVS, or K362/461A MAVS and either empty vector (−) or Flag-V (1 or 2 μg/ml; wedge) as well as p-125Luc (C) or pISRE-luc (D). At 24 hpt, cells were harvested and assessed for luciferase activity. The results are presented as relative luciferase activity. (E) HeLa cells were transfected with either scrambled siRNA or specific siRNA targeting AIP4, RNF5, RNF125, MARCH5, Smurf1, or MUL1. At 48 h after transfection, cells were infected with NDV at an MOI of 1. Cells were harvested at 18 hpi and detected using immunoblot analysis with anti-MAVS, anti-NP, or anti-β-actin antibody. (F) The transfection experiments were performed as described for panel A. Cells were harvested and detected using qRT-PCR with primers for AIP4, RNF5, RNF125, MARCH5, Smurf1, or MUL1. (G) HEK-293T cells were cotransfected with Flag-V and HA-tagged RNF5, MARCH5, Smurf1, or Smurf2. At 24 hpt, cells were harvested, immunoprecipitated with anti-IgG or anti-Flag antibody, and further detected using immunoblot analysis with anti-HA or anti-Flag antibody. Expression levels of the proteins were analyzed by immunoblot analysis of the lysates with anti-HA, anti-Flag, or anti-β-actin antibody. (H) HEK-293T cells were cotransfected with Myc-MAVS, HA-RNF5, and either WT or ΔDM2 Flag-V vector. At 24 hpt, cells were harvested, immunoprecipitated with anti-IgG or anti-HA antibody, and further detected using immunoblot analysis with anti-Myc or anti-HA antibody. Expression levels of the proteins were analyzed by immunoblot analysis of the lysates with anti-HA, anti-Myc, anti-Flag, or anti-β-actin antibody. (I) HeLa cells were transfected with either WT or ΔDM2 Flag-V vector (1 or 2 μg/ml; wedge). At 12 hpt, cells were transfected with poly(I·C) (20 μg/ml). Cells were harvested at 12 hpt with poly(I·C) and detected using immunoblot analysis with anti-MAVS, anti-Flag, or anti-β-actin antibody. (J) WT and rnf5−/− HeLa cells were transfected with either vector or Flag-V (1 or 2 μg/ml; wedge). At 12 hpt, cells were transfected with poly(I·C) (20 μg/ml). Cells were harvested at 12 hpt with poly(I·C) and detected using immunoblot analysis with anti-MAVS, anti-RNF5, anti-Flag, or anti-β-actin antibody. (K) WT and rnf5−/− HeLa cells were cotransfected with p-125Luc, PRL-TK, and either vector or Flag-V (1 or 2 μg/ml; wedge). At 12 hpt, cells were transfected with poly(I·C) (20 μg/ml). Cells were harvested at 12 hpt with poly(I·C) and assessed for luciferase activity. The results are presented as relative luciferase activity. (L to N) The transfection experiments were performed as described for panel D. Cells were harvested at 12 hpt with poly(I·C) and detected using qRT-PCR with IFIT1 (L), MX1 (M), or ZC3HAV (N) primers.
NDV V per se does not possess the ability to degrade the substrate. A previous report identified K362 and K461 as two ubiquitin sites for E3 ubiquitin ligase RNF5-mediated MAVS degradation (36). To explore which E3 ubiquitin ligase is involved in MAVS degradation, we designed the small interfering RNAs (siRNAs) targeting various E3 ligases and tested their roles in NDV-triggered MAVS degradation. Among these candidate E3 ligases, RNF5, MARCH5, and Smurf1 were proven to attenuate NDV-induced MAVS degradation (Fig. 5E and F). We next cloned the genes encoding RNF5, MARCH5, Smurf1, and Smurf2 and investigated their abilities to interact with NDV V. The results showed that only RNF5 was associated with V (Fig. 5G). Furthermore, WT V, but not ΔDM2 V, significantly increased the interaction between MAVS and RNF5 (Fig. 5H). To further confirm whether V recruits RNF5 to MAVS for degradation, HeLa cells were transfected with WT or ΔDM2 V independently, followed by MAVS detection. The results indicated the loss of V interaction with MAVS (ΔDM2 V) led to a loss of MAVS degradation (Fig. 5I). We next generated RNF5 knockout HeLa cells by using the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 system. The Western blotting confirmed the deletion of endogenous RNF5 in rnf5−/− cells (Fig. 5J, lanes 4 to 6). Transfection of NDV V led to the degradation of MAVS in WT but not rnf5−/− cells, indicating that RNF5 plays an essential role in V-mediated MAVS degradation (Fig. 5J). We next evaluated whether RNF5 knockout affects the levels of IFN-β and downstream ISGs after V transfection. As expected, in WT cells, transfection of V inhibits the promoter activity of IFN-β and mRNA levels of IFIT-1, MX1, and ZC3HAV driven by poly(I·C). In comparison, the levels of IFN-β and ISGs were not significantly affected by V transfection in rnf5−/− cells (Fig. 5K to N). These results demonstrated that E3 ligase RNF5 was involved in NDV V-mediated MAVS degradation and inhibition of the IFN signaling pathway.
V-deficient NDV induced less MAVS degradation.
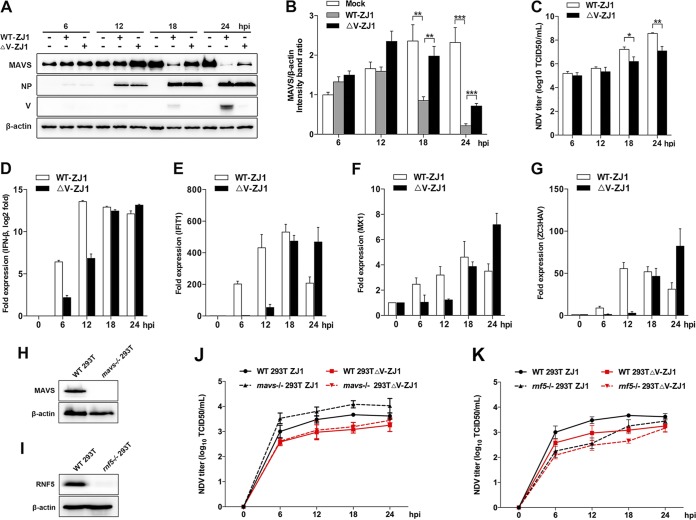
V-deficient NDV ZJ1 strains (ΔV ZJ1) that we previously constructed based on reverse genetics (27) were used to further verify the role of V in NDV-mediated MAVS degradation. HeLa cells were infected with WT ZJ1 or ΔV ZJ1, followed by Western blotting. At 18 and 24 hpi, the expression of V and the obvious degradation of MAVS were observed in WT ZJ1-infected cells. In comparison, ΔV ZJ1 infection induced less MAVS degradation than WT ZJ1 infection (Fig. 6A and B). Correspondingly, the NDV titers in the supernatant of WT ZJ1-infected cells were 10.5- and 30.4-fold higher than that of ΔV ZJ1-infected cells at 18 and 24 hpi, respectively (Fig. 6C), suggesting the V protein degrades MAVS to benefit NDV proliferation. Interestingly, depletion of V showed no observable effect on the intracellular virus replication, as evidenced by the similar expression of the NDV NP protein (Fig. 6A). To examine whether the depletion of V affects IFN-β signaling pathways, the mRNA levels of IFN-β and downstream ISGs were tested. The results showed that WT ZJ1 infection induced much higher mRNA levels of cellular IFN-β, as well as downstream ISGs (IFIT-1, MX1, and ZC3HAV), than the ΔV ZJ1 infection did at 6 hpi and 12 hpi. However, WT ZJ1 induced similar lower mRNA levels of IFN-β and ISGs at 18 and 24 hpi, respectively (Fig. 6D to G). These results demonstrated that with the expression of the V protein at 18 and 24 hpi, the cellular MAVS was accordingly degraded, and the IFN-β signaling pathway was also inhibited. These data further confirmed that the NDV V protein functions on MAVS degradation and subsequent IFN-β signaling inhibition to benefit virus late-stage proliferation. To evaluate the significance of V-mediated MAVS degradation, the replication curves of WT ZJ1 and ΔV ZJ1 in WT and mavs−/− cells were determined. Knockout of mavs and rnf5 genes was verified by Western blotting (Fig. 6H and I). In both WT (two solid lines) and mavs−/− (two dotted lines) cells, the NDV titers in the WT ZJ1-infected cell supernatants were higher than those in the ΔV ZJ1-infected cell supernatants. Interestingly, when comparing virus titers in WT and MAVS−/− 293T cells, we observed that MAVS knockout significant increased virus infection in the WT ZJ1 (two black lines) but not ΔV ZJ1 (two red lines) infection group, suggesting NDV V protein is necessary for specifically antagonizing MAVS-mediated innate immunity and virus infection (Fig. 6J). To further confirm the significance of RNF5 in V-mediated MAVS degradation and virus replication, we compared the replication curve of WT ZJ1 and ΔV ZJ1 in WT and rnf5−/− cells. The results showed that knockout of RNF5 significantly impaired virus infection in both the WT ZJ1 and ΔV ZJ1 infection group, indicating the complex role of RNF5 in NDV infection (Fig. 6K). Collectively, these results clearly demonstrated that V-mediated MAVS degradation benefited NDV replication.
FIG 6.
Depletion of V attenuated NDV-induced MAVS degradation. (A) HeLa cells were mock treated or infected with WT ZJ1 or ΔV ZJ1 at an MOI of 1. Cells were harvested at 6, 12, 18, and 24 hpi and detected using immunoblot analysis with anti-MAVS, anti-NP, anti-V, or anti-β-actin antibody. (B) The intensity band ratio of MAVS to β-actin. Representative results are shown with graphs representing the ratio of MAVS to β-actin normalized to the control condition. Data are presented as means from three independent experiments. (C) Extracellular virus yields in WT ZJ1 or ΔV ZJ1 infection group. (D to G) Virus infection experiments were performed as described for panel A. Cells were harvested and detected using qRT-PCR with IFN-β (D), IFIT-1(E), MX1 (F), or ZC3HAV (G) primers. (H and I) WT and either mavs−/− (H) or rnf5−/− (I) 293T cells were seeded in 6-well plates (1 × 106 cells/well) and collected for immunoblot analysis with anti-MAVS, anti-RNF5, or anti-β-actin antibody. (J) WT and mavs−/− HEK-293T cells were infected with WT ZJ1 or ΔV ZJ1, respectively, at an MOI of 1. Cells were harvested at 6, 12, 18, and 24 hpi. The extracellular virus yields in the WT ZJ1 or ΔV ZJ1 infection group were detected as TCID50 on DF-1 cells. (K) WT and rnf5−/− HEK-293T cells were infected with WT ZJ1 or ΔV ZJ1, respectively, at an MOI of 1. Cells were harvested at 6, 12, 18, and 24 hpi. The extracellular virus yields in the WT ZJ1 or ΔV ZJ1 infection group were detected as TCID50 on DF-1 cells. Data are presented as means from three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
V proteins from several other paramyxoviruses degrade MAVS and block downstream signaling.
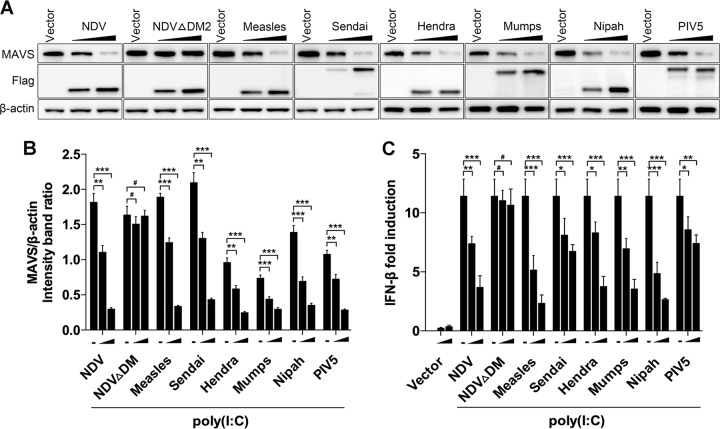
Paramyxovirus V proteins contain the conserved cysteine-rich C-terminal domains (26), which we have proven interact with MAVS. Therefore, we deduced that other paramyxovirus V proteins could degrade MAVS and subsequently inhibit downstream IFN-β production. Flag-tagged V proteins from several other paramyxoviruses, including Sendai virus, measles virus, Hendra virus, mumps virus, Nipah virus, and parainfluenza virus type 5 (PIV5), were transfected into HeLa cells to test their roles in MAVS degradation. As expected, all of the transfected V proteins worked on degrading endogenous MAVS (Fig. 7A and B). Accordingly, the IFN-β promoter activities in these paramyxovirus V protein-transfected cells were also downregulated compared with those in vector-transfected cells (Fig. 7C). In contrast, transfection of NDV ΔDM2 did not affect MAVS degradation and IFN-β promoter activity. These results indicated that other paramyxovirus V proteins also possess the capability for MAVS degradation.
FIG 7.
Paramyxovirus V proteins trigger MAVS degradation. (A) HeLa cells were transfected with either empty vector or Flag-tagged V of NDV, NDV ΔDM2, measles virus, Sendia virus, Hendra virus, mumps, Nipah virus, or PIV5. At 12 hpt, cells were transfected with poly(I·C) (20 μg/ml). Cells were harvested at 12 hpt with poly(I·C) and detected using immunoblot analysis with anti-MAVS, anti-Flag, or anti-β-actin antibody. (B) The intensity band ratio of MAVS to β-actin. Representative results are shown with graphs representing the ratio of MAVS to β-actin normalized to the control condition. Data are presented as means from three independent experiments. (C) HeLa cells were cotransfected with p-125Luc, PRL-TK, and either empty vector or Flag-tagged V of NDV, NDV ΔDM2, measles virus, Sendai virus, Hendra virus, mumps virus, Nipah virus, or PIV5. At 12 hpt, cells were mock treated or transfected with poly(I·C) (20 μg/ml). Cells were harvested at 12 hpt and assessed for luciferase activity. The results are presented as relative luciferase activity. Data are presented as means from three independent experiments. #, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
DISCUSSION
NDV and other paramyxoviruses, such as Sendai virus, were proven to be able to induce high levels of IFN-β and have been widely used as models to study IFN signaling pathways (3, 6, 37). However, growing evidence has shown that paramyxoviruses employ multiple strategies to counteract IFN signaling (17). We and others previously reported that NDV V protein can inhibit RLR-mediated interferon signaling by targeting MDA-5, LGP2, STAT1, etc. (22, 25–27). In this study, we provided several lines of evidence to demonstrate that NDV V protein targets MAVS degradation and subsequently inhibits RLR signaling to benefit virus proliferation at late stages. Both NDV infection and V protein transfection lead to the significant degradation of MAVS and disruption of IFN signaling. NDV V protein specifically interacted with MAVS, targeting its ubiquitination and degradation through E3 ligase RNF5. V-deficient NDV attenuated MAVS degradation, upregulated IFN-β production, and decreased virus late-stage proliferation, suggesting that NDV V protein functions on the degradation of MAVS and inhibition of IFN signaling, which benefit virus proliferation during infection. Finally, we proved that V proteins from other paramyxoviruses also were able to degrade MAVS and block downstream signaling.
We first described that NDV infection degraded MAVS at the protein level but had no effect on the mRNA level of MAVS. The V protein of NDV was then proven to be involved in MAVS degradation. It should be noted that V degrades MAVS only in the presence of poly(I·C). Our data also indicated that the basal level of IFN production is very low in steady-state cells (Fig. 2C and 3B). RIG-I and MAVS will be under conformational changes to initiate the downstream signaling pathway when cells are infected by RNA virus or treated with RNA ligands (38). Therefore, the activation of the IFN pathway is a prerequisite for MAVS degradation triggered by NDV V protein. The pharmacological experiments further demonstrated that the proteasome pathway is responsible for the MAVS degradation; however, only partial degradation of MAVS induced by NDV infection was recovered under MG132 treatment. By comparison, V transfection-induced MAVS degradation was almost totally recovered after treatment with the proteasome inhibitor MG132. Therefore, we deduced that except for V-mediated MAVS degradation, NDV infection might trigger MAVS degradation in other unknown mechanisms. In this study, we found that in addition to NDV V protein, NDV P and W proteins could also inhibit IFN-β promoter activity to some extent (Fig. 2C), which might be caused by the same amino-terminal sequence shared among the P, V, and W proteins (39). Previous reports also demonstrated the interferon-antagonist activity for paramyxovirus P and W proteins (31, 40, 41). Therefore, it is possible that NDV P and W proteins also possess the ability to impair the poly(I·C)-triggered IFN signaling pathway in a MAVS-independent manner. In comparison, the deletion of C-terminal DM2 abrogated the interaction between V and MAVS, indicating the C terminus of V protein was necessary for V-MAVS interaction and IFN signaling. NDV V inhibited IFN-β promoter activity triggered by transfection of MAVS and upstream RIG-I and MDA5. Paramyxovirus V was identified to directly or indirectly target MDA5 or RIG-I to inhibit interferon induction (19, 25). These results together indicated that V could specifically inhibit the IFN pathway at the level of the RLR receptor (RIG-I/MDA5) and adaptor (MAVS).
Both coimmunoprecipitation and immunofluorescence (IF) assays indicated that NDV V protein was associated with MAVS (Fig. 4A to C). The IF results showed that NDV V protein was colocalized with endogenous MAVS at 12 and 24 hpi and less colocalized at 6 hpi, which is in accord with the time course of the V protein expression and MAVS degradation (Fig. 1A and 4D). The fluorescence intensity of MAVS in NDV-infected cells at 18 or 24 hpi is much weaker than that in mock-infected cells. In the single cell, the MAVS fluorescence in mock-infected cells is stronger than that in cells at 18 or 24 hpi (Fig. 4D, line profile), indicating the degradation of MAVS accompanied by virus infection. The immunoprecipitation confirmed the interaction between MAVS and NDV V protein. Previous reports indicated that both the N-terminal CARD-like domain and the C-terminal TM domain are essential for MAVS signaling (6). Furthermore, the TM domain is essential for MAVS mitochondrial localization and is usually targeted by viruses for cleavage (12). As described previously, HCV NS3/4A cleaves MAVS off the mitochondria at the position of Cys508, which is located in the TM domain of MAVS, to evade innate immunity (12). Our results indicated that deletion of either the CARD or TM domain did not abolish the interaction between V and MAVS, indicating that these two functional domains were not involved in V-mediated MAVS degradation (Fig. 4H and I). Paramyxovirus V proteins contain 3 conserved cysteine-rich C-terminal domains, and we identified that DM2 (aa 195 to 201) of NDV V was important for V-MAVS interaction (Fig. 4F). The fact that the DM2 domain is conserved in almost all paramyxoviruses led us to explore the role of other paramyxovirus V proteins in MAVS degradation, which was then confirmed. These results demonstrated that MAVS, in addition to previously reported MDA5, LGP2, and STAT1 (22, 25, 26), could be considered another important target for V proteins of paramyxoviruses. We explored potential molecular mechanisms underlying MAVS degradation by NDV V protein. The pharmacological experiments showed that both NDV infection and V protein transfection degrade MAVS through the ubiquitin-proteasome pathway (Fig. 1H and 2E). Several E3 ligases were identified to be involved in MAVS degradation to avoid excessive activation of IFN. For example, both Smurf1 and Smurf2 E3 ubiquitin ligases interact with MAVS and target MAVS for K48-linked ubiquitination (42, 43). The mitochondrial ubiquitin ligase MARCH5 induces proteasome-mediated degradation of MAVS at Lys7 and Lys500 and modulates MAVS-mediated antiviral signaling (44). Until now, only the HECT domain E3 ligase AIP4 has been identified to be involved in virus-mediated MAVS degradation. As described above, SARS ORF-9b usurps AIP4 for the degradation of MAVS, TRAF3, and TRAF6 (16). In our experiment, Lys362 and Lys461 were characterized as two critical ubiquitination sites for V-mediated MAVS degradation. A previous report identified that RNF5 targeted MAVS at Lys362 and Lys461 for K48-linked ubiquitination and degradation (36). As expected, RNF5 was associated with V protein and involved in V-mediated MAVS degradation (Fig. 6A and B). Moreover, the results that the loss of V interaction with MAVS (ΔDM2 V) led to a loss of MAVS degradation and impaired MAVS-RNF5 demonstrated that V specifically recruits RNF5 to MAVS for degradation. It should be noted that knockdown of RNF5 only partially attenuated NDV-triggered MAVS degradation. Other E3 ligases, such as MARCH5 or Smurf1, also seemed to be involved in virus-induced MAVS degradation (Fig. 5E). Therefore, we speculated that NDV usurps E3 ligases, except RNF5, to degrade MAVS. In addition, V-deficient NDV still induced a considerable amount of MAVS degradation relative to that of WT virus (Fig. 6A), and MG132 treatment did not completely rescue NDV-induced MAVS degradation (Fig. 1L), indicating mechanisms other than the ubiquitin-proteosome pathway also play a role in the degradation, which should be investigated further.
It seems controversial that NDV infection induced high levels of IFN-β and ISGs while also triggering MAVS degradation (Fig. 1A, F, and G). Except for MAVS, NDV V protein could target several other RLR signaling pathway-related molecules, including MDA5 and LGP2, and inhibit IFN-β production (25, 26). It should also be noted that even though IFN and ISG levels are highly induced, NDV still replicates very well in the cells (Fig. 7C). Another example is that double-stranded RNA (dsRNA)-dependent protein kinase R (PKR) is obviously activated by dsRNA generated by NDV infection and leads to IFN-β production (45). An earlier report showed that paramyxovirus P and V proteins were involved in limiting PKR activation and shutoff of host and virus protein synthesis (40). Collectively, it is possible that both NDV-mediated IFN induction and V-mediated IFN inhibition are at play and that the virus has developed mechanisms to overcome the antiviral effects of IFN and downstream ISGs in tumor cells. We reported that NDV V antagonizes the innate immune response at the late stage of infection here and in previous literature. Our results also showed that at the early time points, ΔV ZJ1 generated less innate immune activation than the WT ZJ1, whose V protein is intact (Fig. 6D to G). The results could be addressed in the following ways. (i) MAVS was degraded at the late stage of infection, corresponding to the accumulation of V. At the early stage of the infection, NDV produced undetectable levels of V expression, and no degradation of MAVS was shown. Therefore, the difference of the innate immune activation between ΔV ZJ1 and WT ZJ1 should be caused by factors other than V protein. (ii) NDV V protein has dual functions, playing a direct role in virus replication as well as serving as a virulence factor (46). Our result also showed that replication of ΔV ZJ1 was weaker than that of WT ZJ1 at the early stage of infection (Fig. 6H and I). Therefore, compared with ΔV ZJ1, the faster replication rate of WT ZJ1 leads to stronger innate immune activation at the early stage of infection. Nevertheless, the increase of IFN in the WT ZJ1 infection group was decelerated and even decreased at the late stage of infection (18 to 24 hpi), accompanied by the expression of V, targeting MAVS for degradation, and finally impairing IFN signaling (Fig. 1F and G and 7A and D to G). In comparison, ΔV ZJ1 infection gradually upregulated the mRNA levels of IFN-β and ISGs. The comparison of these two trends, together with the trend of virus-induced MAVS degradation, leads us to conclude that V-mediated MAVS degradation and IFN antagonism mainly occur at the late stage of infection.
To our surprise, in both WT and MAVS−/− 293T cells, the virus titers in the supernatant of NDV-infected cells were higher than those in ΔV ZJ1-infected cells. Since paramyxovirus V protein could target several other RLR signaling pathway-related molecules, including MDA5, LGP2, and STAT1, and inhibit the IFN-β signaling pathway (25–27), knockout of MAVS alone is not sufficient to cover the effect of V deletion. However, interestingly, the finding that MAVS knockout significantly increased virus infection in NDV but not the ΔV ZJ1 infection group suggests NDV V protein is necessary for specifically antagonizing MAVS-mediated innate immunity and virus infection. In comparison, knockout of RNF5 significantly impaired virus infection in both the WT ZJ1 and ΔV ZJ1 infection group. Since the E3 ubiquitin ligase RNF5 could target several substrates, such as MITA/STING, JNK-associated membrane protein (JAMP), and autophagy-specific protein ATG4, for degradation (47–49), we propose that RNF5 knockout leads to the loss of degradation of several substrates, which could not all be antagonized by NDV V protein.
Overall, we showed in this study that the NDV-derived V triggers the degradation of the key adaptor molecule MAVS as a strategy to evade innate antiviral immunity. The mechanism behind V-mediated MAVS degradation was explored, and this mechanism is presumably shared by paramyxoviruses. Our findings therefore revealed a new mechanism of paramyxovirus V-mediated evasion of the host’s innate immunity.
MATERIALS AND METHODS
Reagents and antibodies.
Proteasome inhibitor MG-132 (S2619; Selleck Chemicals, Houston, TX, USA) was used at 25 μM. Autophagy inhibitors wortmannin (W1628; Sigma-Aldrich, St. Louis, MO, USA) and chloroquine (C6628; Sigma-Aldrich) were used at 300 nM and 25 μM, respectively. Mouse monoclonal anti-MAVS (sc-166583) was purchased from Santa Cruz Biotechnology (Dallas, TX, USA). Mouse monoclonal anti-Flag (F1804), anti-HA (H9658), and anti-β-actin antibody (A1978) were purchased from Sigma-Aldrich. Rabbit monoclonal anti-IgG (ab133470) and rabbit polyclonal anti-RNF5 (ab83466) were purchased from Abcam (Cambridge, MA, USA). Rabbit polyclonal anti-MAVS (A300-782A) was purchased from Bethyl Laboratories (Montgomery, TX, USA). Monoclonal antibody against NDV nucleoprotein (NP) and V protein was prepared in our laboratory. Horseradish peroxidase (HRP)-conjugated goat anti-rabbit or -mouse secondary antibodies were purchased from Jackson ImmunoResearch (West Grove, PA, USA). Alexa Fluor 488-labeled goat anti-mouse (A-11029) and Alexa Fluor 594-labeled goat anti-rabbit (A11012) were purchased from Thermo Fisher Scientific (Waltham, MA, USA).
Cell cultures and virus infection.
HeLa, A549, HEK-293T, and DF-1 cells were purchased from the American Type Culture Collection (ATCC). mavs−/− and rnf5−/− HEK-293T cells were obtained from Q. Zhu (Lanzhou Veterinary Research Institute, Lanzhou, China). These cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific). NDV strains Herts/33 and LaSota were obtained from the China Institute of Veterinary Drug Control (Beijing, China). NDV strain ZJ1 was provided by X. Liu (Yangzhou University, Yangzhou, China). V-deficient NDV strain ZJ1 (ΔV ZJ1) was constructed in our laboratory by introducing a stop codon in the ORF with a +1 frameshift (V) following the RNA edit motif of the full-length ZJ1 cDNA clone (27). The expression of P and W was not affected in ΔV ZJ1. The UV-inactivated NDV Herts/33 strain was prepared as described in Sun et al. (50). Cells were infected with NDV at a multiplicity of infection (MOI) of 1 at 37°C. Following a 1-h absorption period, unattached viruses were removed and the cells were then washed three times with serum DMEM and cultured in maintenance medium at 37°C. Viral titers on DF-1 cells were determined as median tissue culture infective doses (TCID50) as described previously (51).
Plasmids.
Flag-tagged MAVS, Flag-tagged TBK1, and HA-tagged wide-type and deletion constructs of MAVS (amino acids 1 to 180, 1 to 360, 180 to 360, 180 to 540, and 360 to 540) were provided by Z. Jiang (Peking University, Beijing, China). Flag-tagged ubiquitin and IRF3(5D) were provided by S. Zhang (Tsinghua University, Beijing, China). pRK5-HA-ubiquitin-K63 (17606), pRK5-HA-ubiquitin-K48 (17605), pcDNA-IKKε-FLAG (26201), and pcDNA-TRIF-CFP (13644) were obtained from Addgene. Flag-tagged RIG-I N and MDA5 N were constructed by inserting the sequence of the CARD domain of RIG-I (aa 1 to 186) and MDA5 (aa 1 to 196) into plasmid p3XFLAG-CMV-14 (Sigma). Flag-tagged chicken MAVS (Flag-cMAVS) and Flag-tagged virus proteins (NP, P, M, V, and W) were constructed in our laboratory (32, 52). RNF5, MARCH5, Smurf1, and Smurf2 gene fragments were amplified from cDNA made from A549 cells and then cloned into plasmid pCMV-HA (Clontech Laboratories, Inc., Palo Alto, CA) to generate HA-tagged proteins. The full-length sequences of V genes from several paramyxoviruses, including Sendai virus, measles virus, Hendra virus, mumps virus, Nipah virus, and PIV5 virus, were synthesized by Genewiz Biotechnology, Co., Ltd. (Suzhou, China), and cloned into plasmid p3XFLAG-CMV-14 (Sigma). Several C-terminally deleted mutations of V protein were constructed by deletion of aa 176 to 180, aa 195 to 201, and aa 201 to 211 from V proteins to generate ΔDM 1, ΔDM2, and ΔDM3 mutants, respectively. Point mutants of HA-MAVS (K462A, K371A, K420A, K461A, K500A, and K362A/K461A) were generated by site-directed mutagenesis as described previously (53, 54) (Table 1). Each mutation was confirmed by sequencing. For reporter assays, the IFN-β promoter luciferase reporter (p-125Luc) was provided by T. Fujita (Kyoto University, Japan). The IFN sequence response element (ISRE) luciferase reporters (pISRE-TA-luc) were purchased from Beyotime (Nantong, China).
TABLE 1.
Primers and siRNAs used in this study
| Primer and application | Sequence (5′–3′) |
|---|---|
| Site mutation | |
| K362A L | GGCATGGTGCCATCCGCAGTGCCTACTAGCATG |
| L362A R | CATGCTAGTAGGCACTGCGGATGGCACCATGCC |
| K371A L | AGCATGGTGCTCACCGCGGTGTCTGCCAGCACA |
| L371A R | TGTGCTGGCAGACACCGCGGTGAGCACCATGCT |
| K420A L | GGGTCGGAGCTGAGTGCGCCTGGCGTGCTGGCA |
| L420A R | TGCCAGCACGCCAGGCGCACTCAGCTCCGACCC |
| K461A L | GAGGAGAATGAGTATGCGTCCGAGGGCACCTTT |
| L461A R | AAAGGTGCCCTCGGACGCATACTCATTCTCCTC |
| K500A L | CCACAAGCCGACCGGGCGTTCCAGGAGAGGGAG |
| L500A R | CTCCCTCTCCTGGAACGCCCGGTCGGCTTGTGG |
| NDV VΔDM1L | CGTCCCTGGAAACCGCACAGCATATCATGG |
| NDV VΔDM1R | CCATGATATGCTGTGCGGTTTCCAGGGACG |
| NDV VΔDM2L | GTCACAACTATCAGCCCAGTCAGGGCAGAG |
| NDV VΔDM2R | CTCTGCCCTGACTGGGCTGATAGTTGTGAC |
| NDV VΔDM3L | CCGGAATTCCGGATGGCCACCTTTACAGATGCG |
| NDV VΔDM3R | CGCGGATCCGCGGGAGTATTGTCTTGGCTCTGC |
| RIG-I N L | CGGGGTACCCCGATGACCACCGAGCAGCGAC |
| RIG-I N R | CGCGGATCCGCGCCACAGTTCACTGAACTTGTTCCTTT |
| MDA5 N L | CGGGGTACCCCGATGTCGAATGGGTATTCCACAGAC |
| MDA5 N R | CGCGGATCCGCGCTCTTGGACAAGTTCATTGTTTCCT |
| Cloning | |
| RNF5 L | CCGCTCGAGCGATGGCAGCAGCGGAGGAG |
| RNF5 R | TGGCGGCCGCGTTCAAATACTGAGCAGCCAAAAAAAG |
| MARCH5 L | CCGCTCGAGCGATGCCGGACCAAGCCCTAC |
| MARCH5 R | TGGCGGCCGCGTTTATGCTTCTTCTTGTTCTGGATAATTC |
| Smurf 1 L | CCGCTCGAGCGATGTCGAACCCCGGGACAC |
| Smurf 1 R | TGGCGGCCGCGTTCACTCCACAGCAAACCCG |
| Smurf 2 L | CCGGAATTCCGATGTCTAACCCCGGAGGC |
| Smurf 2 R | TGGCGGCCGCGTCTCATTCCACAGCAAATCCAC |
| qRT-PCR | |
| qIFIT-1 F | GCCATTTTCTTTGCTTCCCCT |
| qIFIT-1 R | TGCCCTTTTGTAGCCTCCTTG |
| qβ-actin F | GATCTGGCACCACACCTTCT |
| qβ-actin R | GGGGTGTTGAAGGTCTCAAA |
| qIFN-β L | ACGACAGCTCTTTCCATGA |
| qIFN-β R | AGCCAGTGCTCGATGAATCT |
| qMX1 F | TGCGCCCCTGCATCGACCT |
| qMX1 R | GTTTCTTCAGTTTCAGCACCA |
| qMAVS F | CTTCAATACCCTTCAGCGGC |
| qMAVS R | GTAGACAGAGGCCACTTCGT |
| qAIP4-F | TGCAAGGACCAGCAAACAAA |
| qAIP4-R | ATCCTCCTACTGGCAATCGG |
| qRNF5-F | AACGGCAAGAGTGTCCAGTA |
| qRNF5-R | TTAATCTGGGATCCTGGGGC |
| qRNF125-F | ACTTGAGGAGACAGCAGCAA |
| qRNF125-R | CCTCCGTTCCGATCTGTGAT |
| qMarch5-F | CAGGAATAATGGTCGGCTCTATC |
| qMarch5-R | AGGATCAGCTCTCTCCATAACA |
| qSmuf1-F | TTGCTGTGCCATGAAATGCT |
| qSmuf1-R | TGGTCGGGGTTGATTGAAGA |
| qMUL1-F | AGTTCCACCCCTCGATTCAG |
| qMUL1-R | TTCAGCATCTCCTCGGTCTC |
| Gene knockdown | |
| siRNF125 | CCGUGUGCCUUGAGGUGUUTT |
| siRNF5 | GUGUCCAGUAUGUAAAGCUTT |
| siMARCH5 | GGGUGGAAUUGCGUUUGUUTT |
| siAIP4 | AGUUGGACUCAAGGAUUUATT |
| siMUL1 | UGUGCGGUCUGUUAAAGAATT |
| siSmurf1 | ACUUGUCUUAUUUCCACUUTT |
IF assay.
HeLa cells were washed in phosphate-buffered saline (PBS) and fixed in 4% neutral formaldehyde, permeabilized with 1× Tris-buffered saline with Tween 20 (TBST) with 0.5% Triton X-100 for 10 min, incubated in blocking buffer containing 1× TBST with 3% bovine serum albumin (BSA), and then incubated with primary antibody for 1 h at 37°C. Cells were washed three times with 1× TBST and incubated with secondary antibody. Cells were then washed and incubated with another primary antibody, followed by incubation of secondary antibody. Cells were washed and nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) (Thermo Fisher Scientific) for 10 min, and then the coverslips were mounted on slide glasses and visualized using an LSM 880 Zeiss confocal microscope (Carl Zeiss, Jena, Germany).
Immunoblot analysis.
Immunoblot analysis was performed as described previously (50). Briefly, cells were washed thoroughly and lysed in cell lysis buffer containing a protease inhibitor cocktail (Merck Millipore, Darmstadt, Germany). The lysates were denatured and then subjected to SDS-PAGE and transferred to nitrocellulose membranes (Whatman, Maidstone, UK). The membranes were then blocked and reacted with primary antibodies overnight at 4°C and horseradish peroxidase (HRP)-conjugated secondary antibodies for 1 h at room temperature. The antibody-antigen complex was visualized using enhanced chemiluminescence (Thermo Fisher Scientific) and quantified using Image J software.
Coimmunoprecipitation.
HEK-293T cells were lysed with a cell lysis buffer (150 mM NaCl, 50 mM Tris-HCl, pH 8.0, 5 mM EDTA, 0.5% NP-40) containing 1 mM phenylmethylsulfonyl fluoride and protease inhibitors (Merck-Millipore). Lysates were centrifuged at 12,000 × g for 10 min and precipitated with anti-Flag, anti-HA, or anti-IgG monoclonal antibodies, in conjunction with protein G-agarose beads (ThermoFisher Scientific), overnight at 4°C. The beads were washed with lysis buffer four times and eluted with SDS loading buffer by boiling for 10 min. Cell lysates (50 μl) were eluted with SDS loading buffer and boiled for 10 min. Proteins isolated from the beads and the cell lysates were subjected to immunoblot analysis.
qRT-PCR.
Quantitative real-time PCR (qRT-PCR) was performed as described previously (55). Briefly, total RNA was extracted and reverse transcribed to cDNA. Primers were designed using Primer3 software (http://www.broad.mit.edu/cgi-bin/primer/primer3_www.cgi) or derived from previous reports (45, 52) (Table 1). qRT-PCR was performed using Premix Ex Taq reagents (TaKaRa, Dalian, China). The comparative threshold cycle (ΔΔCT) method was used to calculate the relative abundance of mRNAs (56). All experiments were carried out in triplicate.
Cell transfection, luciferase assay, and RNA interference.
Cells were transfected by using FuGENE HD (Promega, Madison, WI, USA) or Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s instructions. For luciferase assay, HEK-293T cells (1.25 × 105) were cultured in 24-well plates and cotransfected with 100 ng of plasmid expressing proteins involved in the RLR pathway (RIG-I N, MDA5 N, MAVS, TKB1, etc.), 100 or 150 ng of Flag-V, 100 ng of firefly luciferase reporter (IFN-β-Luc and ISRE-luc), and 10 ng of the constitutive Renilla luciferase reporter pRL-TK (Promega). Luciferase activity was measured at 24 h posttransfection (hpt) using the Dual-Luciferase reporter assay system (Promega) according to the manufacturer’s protocol.
Small interfering RNA (siRNA) oligonucleotides for targeting endogenous RNF125, RNF5, MARCH5, AIP4, MUL1, and Smurf1 were purchased from Gene Pharma (Table 1). Cells were transfected with the indicated siRNA or scramble siRNA as described previously (50). At 6 hpt, cells were washed three times with PBS and incubated for an additional 48 h before virus infection.
Generation of RNF5 knockout cells.
A guide RNA (gRNA) specific to the RNF5 gene was designed using the online CRISPR design tool (http://crispr.mit.edu/), and the sequence was 5′-ACCTCCTGCCCACGATCGTT-3′. The gRNA oligonucleotides were annealed and cloned into the LentiCRISPR-V2 vector (plasmid 52961; Addgene) using the BsmBI enzyme site as previously described (57, 58). HEK-293T cells were seeded into 100-mm dishes and transfected with lentiCRISPR-single guide RNA (sgRNA) (15.4 μg) together with two package plasmids, psPAX2 (15.4 μg) and pMD2.G (10 μg). At 48 hpt, the supernatants were harvested. HeLa cells were infected with lentivirus supernatants supplemented with 8 μg/ml Polybrene (Sigma-Aldrich). Cells were horizontally centrifuged at 1,500 rpm at 32°C for 90 min and incubated at 37°C for 48 h. The supernatants were replaced with 10% fetal bovine serum (FBS; Thermo Fisher Scientific) supplemented with 1 μg/ml puromycin (Merck-Millipore) for 72 h prior to genomic DNA extraction. The genomic region surrounding the CRISPR target site was amplified by PCR using check primers (forward, 5′-GACTGGCAGGCATTCAGACC-3′; reverse, 5′-CTCACATTGTCCTGCCACTC-3′). Except for those used for PCR verification, cells were rechecked by immunoblot analysis using anti-RNF5 antibody. After confirmation of the activity of the designed sgRNA, HeLa cells were plated by limiting dilution, and the RNF-5 knockout HeLa cell line was established by the limiting dilution method in 96-well plates. The genomic DNA of the cells cultured from a single-cell clone was amplified using the check primers. The PCR products were purified and ligated into pEASY-T1 vector (TransGen, China). Six clones were sequenced to ensure the frameshifting mutation of both alleles of the established cell line. An immunoblot analysis was performed to confirm the knockout of RNF5.
Statistical analysis.
Data were expressed as means ± standard deviations. Significance was determined with the two-tailed independent Student's t test (P < 0.05) between two groups. One-way analysis of variance followed by Tukey’s test was used to compare multiple groups (>2).
ACKNOWLEDGMENTS
We thank X. Liu (Yangzhou University) for providing the NDV ZJ1 strain. We thank S. Zhang (Tsinghua University) for providing plasmid IRF3(5D) and technical assistance. We thank Takashi Fujita (Kyoto University) for providing plasmid p-125Luc. We thank Z. Jiang (Peking University) for providing truncated HA-MAVS plasmids. We thank Q. Zhu (Lanzhou Veterinary Research Institute) for providing mavs−/− and rnf5−/− HEK-293T cells.
This work was funded by grants 31872453 (to Y.S.) and 31530074 (to C.D.) from the National Natural Science Foundation of China and by grant 2018YFD0500100 from the National Key Research and Development Program of China to C.D. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Akira S, Uematsu S, Takeuchi O. 2006. Pathogen recognition and innate immunity. Cell 124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Yoneyama M, Fujita T. 2009. RNA recognition and signal transduction by RIG-I-like receptors. Immunol Rev 227:54–65. doi: 10.1111/j.1600-065X.2008.00727.x. [DOI] [PubMed] [Google Scholar]
- 3.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Reis e Sousa C, Matsuura Y, Fujita T, Akira S. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 4.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol 6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 5.Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 6.Seth RB, Sun L, Ea CK, Chen ZJ. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 7.Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell 19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 8.Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ. 2011. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell 146:448–461. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun Q, Sun L, Liu HH, Chen X, Seth RB, Forman J, Chen ZJ. 2006. The specific and essential role of MAVS in antiviral innate immune responses. Immunity 24:633–642. doi: 10.1016/j.immuni.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 10.Bhoj VG, Sun Q, Bhoj EJ, Somers C, Chen X, Torres JP, Mejias A, Gomez AM, Jafri H, Ramilo O, Chen ZJ. 2008. MAVS and MyD88 are essential for innate immunity but not cytotoxic T lymphocyte response against respiratory syncytial virus. Proc Natl Acad Sci U S A 105:14046–14051. doi: 10.1073/pnas.0804717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng J, Chen Y, Liu G, Ren J, Go C, Ivanciuc T, Deepthi K, Casola A, Garofalo RP, Bao X. 2015. Mitochondrial antiviral-signalling protein plays an essential role in host immunity against human metapneumovirus. J Gen Virol 96:2104–2113. doi: 10.1099/vir.0.000178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li XD, Sun L, Seth RB, Pineda G, Chen ZJ. 2005. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci U S A 102:17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong J, Xu S, Wang J, Luo R, Wang D, Xiao S, Fang L, Chen H, Jiang Y. 2015. Porcine reproductive and respiratory syndrome virus 3C protease cleaves the mitochondrial antiviral signalling complex to antagonize IFN-beta expression. J Gen Virol 96:3049–3058. doi: 10.1099/jgv.0.000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mukherjee A, Morosky SA, Delorme-Axford E, Dybdahl-Sissoko N, Oberste MS, Wang T, Coyne CB. 2011. The coxsackievirus B 3C protease cleaves MAVS and TRIF to attenuate host type I interferon and apoptotic signaling. PLoS Pathog 7:e1001311. doi: 10.1371/journal.ppat.1001311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei C, Ni C, Song T, Liu Y, Yang X, Zheng Z, Jia Y, Yuan Y, Guan K, Xu Y, Cheng X, Zhang Y, Yang X, Wang Y, Wen C, Wu Q, Shi W, Zhong H. 2010. The hepatitis B virus X protein disrupts innate immunity by downregulating mitochondrial antiviral signaling protein. J Immunol 185:1158–1168. doi: 10.4049/jimmunol.0903874. [DOI] [PubMed] [Google Scholar]
- 16.Shi CS, Qi HY, Boularan C, Huang NN, Abu-Asab M, Shelhamer JH, Kehrl JH. 2014. SARS-coronavirus open reading frame-9b suppresses innate immunity by targeting mitochondria and the MAVS/TRAF3/TRAF6 signalosome. J Immunol 193:3080–3089. doi: 10.4049/jimmunol.1303196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Audsley MD, Moseley GW. 2013. Paramyxovirus evasion of innate immunity: diverse strategies for common targets. World J Virol 2:57–70. doi: 10.5501/wjv.v2.i2.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andrejeva J, Childs KS, Young DF, Carlos TS, Stock N, Goodbourn S, Randall RE. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc Natl Acad Sci U S A 101:17264–17269. doi: 10.1073/pnas.0407639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Childs KS, Andrejeva J, Randall RE, Goodbourn S. 2009. Mechanism of mda-5 inhibition by paramyxovirus V proteins. J Virol 83:1465–1473. doi: 10.1128/JVI.01768-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Didcock L, Young DF, Goodbourn S, Randall RE. 1999. The V protein of simian virus 5 inhibits interferon signalling by targeting STAT1 for proteasome-mediated degradation. J Virol 73:9928–9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li T, Chen X, Garbutt KC, Zhou P, Zheng N. 2006. Structure of DDB1 in complex with a paramyxovirus V protein: viral hijack of a propeller cluster in ubiquitin ligase. Cell 124:105–117. doi: 10.1016/j.cell.2005.10.033. [DOI] [PubMed] [Google Scholar]
- 22.Ulane CM, Kentsis A, Cruz CD, Parisien JP, Schneider KL, Horvath CM. 2005. Composition and assembly of STAT-targeting ubiquitin ligase complexes: paramyxovirus V protein carboxyl terminus is an oligomerization domain. J Virol 79:10180–10189. doi: 10.1128/JVI.79.16.10180-10189.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alexander DJ. 2000. Newcastle disease and other avian paramyxoviruses. Rev Sci Tech 19:443–462. doi: 10.20506/rst.19.2.1231. [DOI] [PubMed] [Google Scholar]
- 24.Fiola C, Peeters B, Fournier P, Arnold A, Bucur M, Schirrmacher V. 2006. Tumor selective replication of Newcastle disease virus: association with defects of tumor cells in antiviral defence. Int J Cancer 119:328–338. doi: 10.1002/ijc.21821. [DOI] [PubMed] [Google Scholar]
- 25.Childs K, Randall R, Goodbourn S. 2012. Paramyxovirus V proteins interact with the RNA Helicase LGP2 to inhibit RIG-I-dependent interferon induction. J Virol 86:3411–3421. doi: 10.1128/JVI.06405-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Childs K, Stock N, Ross C, Andrejeva J, Hilton L, Skinner M, Randall R, Goodbourn S. 2007. mda-5, but not RIG-I, is a common target for paramyxovirus V proteins. Virology 359:190–200. doi: 10.1016/j.virol.2006.09.023. [DOI] [PubMed] [Google Scholar]
- 27.Qiu X, Fu Q, Meng C, Yu S, Zhan Y, Dong L, Song C, Sun Y, Tan L, Hu S, Wang X, Liu X, Peng D, Liu X, Ding C. 2016. Newcastle disease virus V protein targets phosphorylated STAT1 to block IFN-I signaling. PLoS One 11:e0148560. doi: 10.1371/journal.pone.0148560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takeuchi O, Akira S. 2010. Pattern recognition receptors and inflammation. Cell 140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 29.Yoneyama M, Fujita T. 2010. Recognition of viral nucleic acids in innate immunity. Rev Med Virol 20:4–22. doi: 10.1002/rmv.633. [DOI] [PubMed] [Google Scholar]
- 30.Shaw ML, Cardenas WB, Zamarin D, Palese P, Basler CF. 2005. Nuclear localization of the Nipah virus W protein allows for inhibition of both virus- and toll-like receptor 3-triggered signaling pathways. J Virol 79:6078–6088. doi: 10.1128/JVI.79.10.6078-6088.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shaw ML, Garcia-Sastre A, Palese P, Basler CF. 2004. Nipah virus V and W proteins have a common STAT1-binding domain yet inhibit STAT1 activation from the cytoplasmic and nuclear compartments, respectively. J Virol 78:5633–5641. doi: 10.1128/JVI.78.11.5633-5641.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng JH, Sun YJ, Zhang FQ, Zhang XR, Qiu XS, Yu LP, Wu YT, Ding C. 2016. Newcastle disease virus NP and P proteins induce autophagy via the endoplasmic reticulum stress-related unfolded protein response. Sci Rep 6:24721. doi: 10.1038/srep24721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng J, Sun Y, Zhang X, Zhang F, Zhang S, Yu S, Qiu X, Tan L, Song C, Gao S, Wu Y, Ding C. 2014. Toll-like receptor 3 inhibits Newcastle disease virus replication through activation of pro-inflammatory cytokines and the type-1 interferon pathway. Arch Virol 159:2937–2948. doi: 10.1007/s00705-014-2148-6. [DOI] [PubMed] [Google Scholar]
- 34.Matsumoto M, Seya T. 2008. TLR3: interferon induction by double-stranded RNA including poly(I:C). Adv Drug Deliv Rev 60:805–812. doi: 10.1016/j.addr.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 35.Yamamoto M, Sato S, Hemmi H, Hoshino K, Kaisho T, Sanjo H, Takeuchi O, Sugiyama M, Okabe M, Takeda K, Akira S. 2003. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science 301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 36.Zhong B, Zhang Y, Tan B, Liu TT, Wang YY, Shu HB. 2010. The E3 ubiquitin ligase RNF5 targets virus-induced signaling adaptor for ubiquitination and degradation. J Immunol 184:6249–6255. doi: 10.4049/jimmunol.0903748. [DOI] [PubMed] [Google Scholar]
- 37.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol 5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 38.Yoneyama M, Fujita T. 2008. Structural mechanism of RNA recognition by the RIG-I-like receptors. Immunity 29:178–181. doi: 10.1016/j.immuni.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 39.Locke DP, Sellers HS, Crawford JM, Schultz-Cherry S, King DJ, Meinersmann RJ, Seal BS. 2000. Newcastle disease virus phosphoprotein gene analysis and transcriptional editing in avian cells. Virus Res 69:55–68. doi: 10.1016/S0168-1702(00)00175-1. [DOI] [PubMed] [Google Scholar]
- 40.Gainey MD, Dillon PJ, Clark KM, Manuse MJ, Parks GD. 2008. Paramyxovirus-induced shutoff of host and viral protein synthesis: role of the P and V proteins in limiting PKR activation. J Virol 82:828–839. doi: 10.1128/JVI.02023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park MS, Shaw ML, Munoz-Jordan J, Cros JF, Nakaya T, Bouvier N, Palese P, Garcia-Sastre A, Basler CF. 2003. Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W, and C proteins. J Virol 77:1501–1511. doi: 10.1128/JVI.77.2.1501-1511.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pan Y, Li R, Meng JL, Mao HT, Zhang Y, Zhang J. 2014. Smurf2 negatively modulates RIG-I-dependent antiviral response by targeting VISA/MAVS for ubiquitination and degradation. J Immunol 192:4758–4764. doi: 10.4049/jimmunol.1302632. [DOI] [PubMed] [Google Scholar]
- 43.Wang Y, Tong X, Ye X. 2012. Ndfip1 negatively regulates RIG-I-dependent immune signaling by enhancing E3 ligase Smurf1-mediated MAVS degradation. J Immunol 189:5304–5313. doi: 10.4049/jimmunol.1201445. [DOI] [PubMed] [Google Scholar]
- 44.Yoo YS, Park YY, Kim JH, Cho H, Kim SH, Lee HS, Kim TH, Sun Kim Y, Lee Y, Kim CJ, Jung JU, Lee JS, Cho H. 2015. The mitochondrial ubiquitin ligase MARCH5 resolves MAVS aggregates during antiviral signalling. Nat Commun 6:7910. doi: 10.1038/ncomms8910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang S, Sun Y, Chen H, Dai Y, Zhan Y, Yu S, Qiu X, Tan L, Song C, Ding C. 2014. Activation of the PKR/eIF2alpha signaling cascade inhibits replication of Newcastle disease virus. Virol J 11:62. doi: 10.1186/1743-422X-11-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mebatsion T, Verstegen S, De Vaan LT, Römer-Oberdörfer A, Schrier CC. 2001. A recombinant Newcastle disease virus with low-level V protein expression is immunogenic and lacks pathogenicity for chicken embryos. J Virol 75:420–428. doi: 10.1128/JVI.75.1.420-428.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuang E, Okumura CY, Sheffy-Levin S, Varsano T, Shu VC, Qi J, Niesman IR, Yang HJ, Lopez-Otin C, Yang WY, Reed JC, Broday L, Nizet V, Ronai ZA. 2012. Regulation of ATG4B stability by RNF5 limits basal levels of autophagy and influences susceptibility to bacterial infection. PLoS Genet 8:e1003007. doi: 10.1371/journal.pgen.1003007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhong B, Zhang L, Lei C, Li Y, Mao AP, Yang Y, Wang YY, Zhang XL, Shu HB. 2009. The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity 30:397–407. doi: 10.1016/j.immuni.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 49.Tcherpakov M, Delaunay A, Toth J, Kadoya T, Petroski MD, Ronai ZA. 2009. Regulation of endoplasmic reticulum-associated degradation by RNF5-dependent ubiquitination of JNK-associated membrane protein (JAMP). J Biol Chem 284:12099–12109. doi: 10.1074/jbc.M808222200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun Y, Yu S, Ding N, Meng C, Meng S, Zhang S, Zhan Y, Qiu X, Tan L, Chen H, Song C, Ding C. 2014. Autophagy benefits the replication of Newcastle disease virus in chicken cells and tissues. J Virol 88:525–537. doi: 10.1128/JVI.01849-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Romer-Oberdorfer A, Werner O, Veits J, Mebatsion T, Mettenleiter TC. 2003. Contribution of the length of the HN protein and the sequence of the F protein cleavage site to Newcastle disease virus pathogenicity. J Gen Virol 84:3121–3129. doi: 10.1099/vir.0.19416-0. [DOI] [PubMed] [Google Scholar]
- 52.Sun Y, Mao X, Zheng H, Wu W, Rehman ZU, Liao Y, Meng C, Qiu X, Tan L, Song C, Xu L, Yu S, Ding C. 2019. Goose MAVS functions in RIG-I-mediated IFN-beta signaling activation. Dev Comp Immunol 93:58–65. doi: 10.1016/j.dci.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 53.Li X, Qiu Y, Shen Y, Ding C, Liu P, Zhou J, Ma Z. 2008. Splicing together different regions of a gene by modified polymerase chain reaction-based site-directed mutagenesis. Anal Biochem 373:398–400. doi: 10.1016/j.ab.2007.10.021. [DOI] [PubMed] [Google Scholar]
- 54.Zheng L, Baumann U, Reymond JL. 2004. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res 32:e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qiu X, Yu Y, Yu S, Zhan Y, Wei N, Song C, Sun Y, Tan L, Ding C. 2014. Development of strand-specific real-time RT-PCR to distinguish viral RNAs during Newcastle disease virus infection. ScientificWorldJournal 2014:934851. doi: 10.1155/2014/934851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 57.Sanjana NE, Shalem O, Zhang F. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]