Zika fever, an infectious disease caused by the Zika virus (ZIKV), normally results in mild symptoms. Severe infection can cause Guillain-Barré syndrome in adults and birth defects, including microcephaly, in newborns. Although ZIKV was first identified in Uganda in 1947 in rhesus monkeys, a widespread epidemic of ZIKV infection in South and Central America in 2015 and 2016 raised major concerns. To date, there is no vaccine or specific medicine for ZIKV. The significance of our research is the systematic discovery of small molecule candidates that modulate ZIKV infection, which will allow the development of antiviral therapeutics. In addition, we identified MLK3, a key mediator of host signaling pathways that can be activated during ZIKV infection and limits virus replication by inducing multiple inflammatory cytokines. These findings broaden our understanding of ZIKV pathogenesis.
KEYWORDS: cytokine, MLK3, URMC-099, ZIKV
ABSTRACT
The Zika virus (ZIKV) life cycle involves multiple steps and requires interactions with host factors. However, the inability to systematically identify host regulatory factors for ZIKV has hampered antiviral development and our understanding of pathogenicity. Here, using a bioactive compound library with 2,659 small molecules, we applied a high-throughput and imaging-based screen to identify host factors that modulate ZIKV infection. The screen yielded hundreds of hits that markedly inhibited or potentiated ZIKV infection in SNB-19 glioblastoma cells. Among the hits, URMC-099, a mixed-lineage kinase 3 (MLK3) inhibitor, significantly facilitated ZIKV replication in both SNB-19 cells and the neonatal mouse brain. Using gene silencing and overexpression, we further confirmed that MLK3 was a host restriction factor against ZIKV. Mechanistically, MLK3 negatively regulated ZIKV replication through induction of the inflammatory cytokines interleukin-6 (IL-6), IL-8, tumor necrosis factor alpha (TNF-α), and monocyte chemoattractant protein 1 (MCP-1) but did not modulate host interferon-related pathways. Importantly, ZIKV activated the MLK3/MKK7/Jun N-terminal protein kinase (JNK) pathway in both SNB-19 cells and neonatal mouse brain. Together, these findings reveal a critical role for MLK3 in regulating ZIKV infection and facilitate the development of anti-ZIKV therapeutics by providing a number of screening hits.
IMPORTANCE Zika fever, an infectious disease caused by the Zika virus (ZIKV), normally results in mild symptoms. Severe infection can cause Guillain-Barré syndrome in adults and birth defects, including microcephaly, in newborns. Although ZIKV was first identified in Uganda in 1947 in rhesus monkeys, a widespread epidemic of ZIKV infection in South and Central America in 2015 and 2016 raised major concerns. To date, there is no vaccine or specific medicine for ZIKV. The significance of our research is the systematic discovery of small molecule candidates that modulate ZIKV infection, which will allow the development of antiviral therapeutics. In addition, we identified MLK3, a key mediator of host signaling pathways that can be activated during ZIKV infection and limits virus replication by inducing multiple inflammatory cytokines. These findings broaden our understanding of ZIKV pathogenesis.
INTRODUCTION
Zika virus (ZIKV) is an emerging arthropod-borne virus in the genus Flavivirus and family Flaviviridae (1). Recently, a widespread epidemic of Zika fever was reported, first in Brazil and later across the Pacific, reaching approximately 60 countries since early 2015 (2). It is now clear that serious infection with ZIKV can cause neurological problems, such as Guillain-Barré syndrome in adults and severe brain anomalies, including microcephaly, in newborns (3). However, there is currently no vaccine or specific medicine for Zika fever.
ZIKV can be transmitted from an infected pregnant woman to her fetus and replicates efficiently in the fetal brain (4). ZIKV particles and viral RNA were found in congenitally infected fetal brain samples and were associated with agyria, hydrocephalus, and multifocal dystrophic calcifications (5). However, most infection and pathogenesis studies have been performed in mice and cultured cells. ZIKV frequently leads to increased death of neural progenitor cells (NPCs) (6). In addition, multiple cellular signaling pathways have been reported to regulate ZIKV infection, including type I interferon (IFN) (7), type II IFN (8), Akt-mTOR (9), and STING-dependent antiviral cellular defenses (10). Despite these findings, the mechanisms of ZIKV infection and pathogenicity are largely unknown. To better understand ZIKV and its host interactions, systems biology and high-throughput methods are urgently needed. Scaturro and colleagues recently reported an integrated proteomics approach for characterization of the cellular responses to ZIKV infection at the proteome and phosphoproteome levels, as well as affinity proteomics for the identification of ZIKV protein-associated cellular targets (11). Although a system-level network of ZIKV-associated proteins and cellular pathways was established, functional characterization is still needed.
We performed high-throughput screening (HTS) of a bioactive compound library in this study and found that host mixed-lineage kinase 3 (MLK3) was involved in ZIKV infection. MLK3, which is also named mitogen-activated protein kinase (MAPK) kinase kinase 11, belongs to the serine/threonine kinase family, which is activated in response to a wide variety of signals and mediates signal transduction cascades that regulate cell growth, differentiation, apoptosis, and innate immunity. MLK3 contains an SH3 domain and a leucine zipper basic motif (12). This kinase preferentially activates MAPK8/Jun N-terminal protein kinase (JNK) kinase and functions as a positive regulator of the JNK signaling pathway (13). MLK3 can directly phosphorylate and activate IκB kinases α and β and is involved in the transcriptional activity of NF-κB mediated by Rho family GTPases and CDC42 (14). Investigations of the involvement of MLK3 in the viral life cycle are limited. MLK3 can be activated by enterovirus 71 (15) and hepatitis B virus (16), and MLK3 deficiency delays influenza virus clearance in the lung (17). However, the role of MLK3 during ZIKV infection remains unknown.
Small molecule inhibitor screens are regularly used for the discovery of new candidate targets for therapies for cancer and viral disease (18, 19). Here, we used the human glioblastoma cell line SNB-19 as an in vitro infection model for HTS with bioactive compounds to systematically search for important host factors that have the potential to regulate ZIKV infection. The pathways targeted by effective compounds can yield insights into ZIKV biology. Importantly, by characterizing URMC-099, an MLK3 inhibitor, we revealed the protective role of MLK3 against ZIKV infection in human cells and the neonatal mouse brain. Our results suggest a host regulatory network and reveal a substantial role for MLK3 in ZIKV infection.
RESULTS
Systematic screening of bioactive compound libraries identifies potential host factors that regulate ZIKV infection.
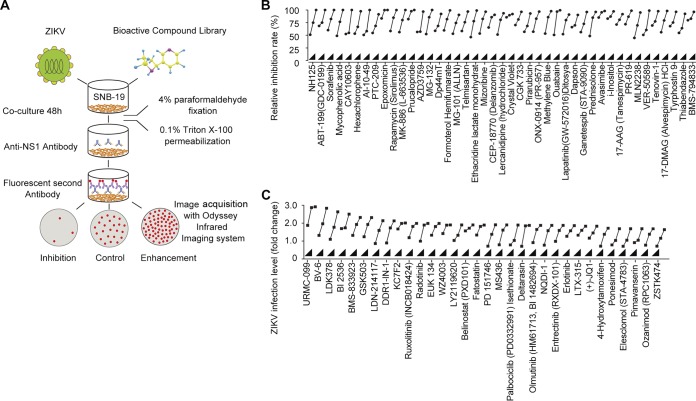
ZIKV infection and propagation require virus-host interaction, in which a large number of host factors may be involved. An in-depth understanding of the host factors and pathways involved in the regulation of the ZIKV life cycle has far-reaching significance for the prevention and treatment of the disease. An expeditious way to identify host candidates that regulate ZIKV infection is systematic screening using a bioactive compound library. The library used in this study is a unique collection of 2,659 bioactive chemical compounds with specific targets for HTS, including most Selleck inhibitors, natural products, chemotherapeutic agents, and compounds that have been approved by the FDA or are in clinical trials. The flowchart of the screening process is depicted in Fig. 1A. We established a cell culture model with ZIKV-infected SNB-19 glioblastoma cells. Glial cells are an important target cell type for ZIKV infection in humans (5). The Chinese ZIKV isolate SZ01 (20) can readily infect SNB-19 cells without reducing cell viability. Distinct from a previous HTS report (19), immunofluorescence staining of the ZIKV NS1 protein and a focus-forming unit (FFU) assay were used as the readouts in this study, which can directly reflect changes in virus replication. The advantage of this screening strategy is that it can identify not only inhibitory but also enhancing compounds for viral replication. The full data sets for the screen are summarized in Tables S1 and S2 in the supplemental material. Based on the primary screening results using 5 μM bioactive compounds, we selected the top-ranked candidate compounds for dose-dependent validation and found that 111 compounds inhibited ZIKV infection by more than 95%, whereas 12 compounds enhanced ZIKV infection by more than 2-fold. Some of these results are presented in Fig. 1B and C. These findings are partially consistent with previously reported observations for compounds such as AT7519, dinaciclib, flavopiridol (alvocidib), seliciclib, and SNS-032 (19). By applying direct virological evaluation strategies, this screening strategy has identified more new hits that will play a beneficial role in the study of ZIKV.
FIG 1.
Development of a bioactive small molecule high-throughput screen for the identification of host factors regulating ZIKV infection. (A) Schematic view of the screening workflow. Briefly, SNB-19 cells were plated in 96-well plates 36 h prior to treatment with 2,659 bioactive compounds and infection with ZIKV (SZ01) for 48 h, followed by fixation and staining. The final concentration of the compounds was 5 μM, and three duplicates were tested for each compound. The infection ratio was normalized and calculated with DMSO as a negative control across screening plates. Representative hits were further evaluated at concentrations of 200 nM, 1 μM, and 5 μM and categorized into two groups with inhibitory (B) or enhancing (C) effects on ZIKV infection.
URMC-099 enhances ZIKV replication in SNB-19 cells and mice.
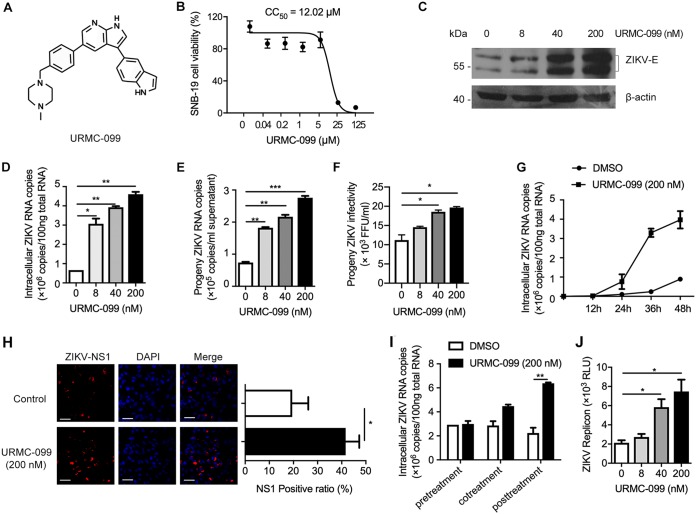
Among the screened hits, compounds that enhanced ZIKV infection were of interest because the targets of these compounds might critically restrict ZIKV. URMC-099 is an orally bioavailable and brain-penetrating inhibitor of MLK3 (21). The chemical structure of URMC-099 is shown in Fig. 2A. The cytotoxicity of URMC-099 was measured by an MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide] assay in SNB-19 cells, and the 50% cytotoxic concentration (CC50) value was estimated to be 12.02 μM (Fig. 2B). We observed cytotoxicity in some cell types when the drug concentration was higher than 1 or 5 μM. This might be due to a nonspecific effect of URMC-099. Therefore, we use 200 nM as the maximal dose, which is noncytotoxic in most cell types. We further analyzed the effects of URMC-099 on ZIKV infection in terms of viral protein and RNA levels and growth characteristics in SNB-19 cells. We used a multiplicity of infection (MOI) of 0.1 and infection duration of 48 h because URMC-099 is an enhancer but not an inhibitor for ZIKV. The dosage and time of ZIKV infection in the cell types used in this study were decided by optimization. As shown in Fig. 2C and D, URMC-099 treatment significantly enhanced ZIKV infection in a dose-dependent manner. URMC-099 treatments increased the copies of ZIKV RNA in culture supernatant (Fig. 2E) and the infectious level of progeny ZIKV (Fig. 2F). By measuring intracellular ZIKV RNA copies at 12-h intervals up to 48 h, the magnitude of the effect of URMC-099 on intracellular ZIKV RNA increased gradually and was dramatically larger at 36 and 48 h. (Fig. 2G). A similar enhancement was also observed by using indirect immunofluorescence staining with an antibody specific for viral NS1 in SNB-19 cells (Fig. 2H). To gain more insight into the mechanism of URMC-099 enhancement, a time-of-addition experiment was conducted to determine the effects of URMC-099 on viral attachment, cell entry, or the postentry step during ZIKV infection. As shown in Fig. 2I, URMC-099 significantly enhanced viral infection at postentry steps, but not entry steps. Because of the lack of a viral structural gene, the replicon can bypass viral entry and release. To further confirm the regulation of URMC-099 on ZIKV replication, a replicon system with a Renilla luciferase reporter was used for evaluation. As shown in Fig. 2J, URMC-099 enhanced the ZIKV replicon at a comparable level to live virus infection (Fig. 2D). These results suggest that the main step affected by URMC-099 is viral replication.
FIG 2.
URMC-099 promotes ZIKV infection in SNB-19 cells. (A) Chemical structure of URMC-099. (B) SNB-19 cell viability was determined by an MTT assay. (C and D) SNB-19 cells were inoculated with ZIKV (MOI = 0.1) and the indicated concentrations of URMC-099 for 48 h. The infection level of ZIKV was determined by either Western blotting (C) or qRT-PCR (D). ZIKV E protein shows double bands in Western blotting results. (E) SNB-19 cells were infected with ZIKV (MOI = 0.1) in the presence of various concentrations of URMC-099 for 36 h. Fresh media were changed, and the cells were allowed for another 12-h culture before collection of supernatants for viral RNA quantification by qRT-PCR. (F) The collected culture supernatants containing progeny virus were added into naive Vero cells in a 96-well plate, and the cells were cultured for 48 h before ZIKV FFU assay. (G) ZIKV growth curve in SNB-19 cells with or without URMC-099. Titers at time zero represent the initial inoculum. Titers at each successive time point represent intracellular ZIKV RNA copies. (H) Indirect immunofluorescence of ZIKV infection in SNB-19 cells. The cells were stained with an antibody specific for ZIKV NS1 to detect virus-positive cells and DAPI to visualize nuclei. The scale bar represents 50 μm. The ratio of NS1-positive cells is shown in the right panel. (I) Time-of-addition experiment for analysis of the effect of URMC-099 on the stages of ZIKV entry and postentry. (J) Analysis of the effect of URMC-099 on viral replication in SNB-19 cells using the ZIKV replicon that contains a Renilla luciferase reporter. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Data on infection are expressed as percentages relative to vehicle-treated cells. Means and standard deviations were recorded for treated cells as a percentage of the value for untreated cells.
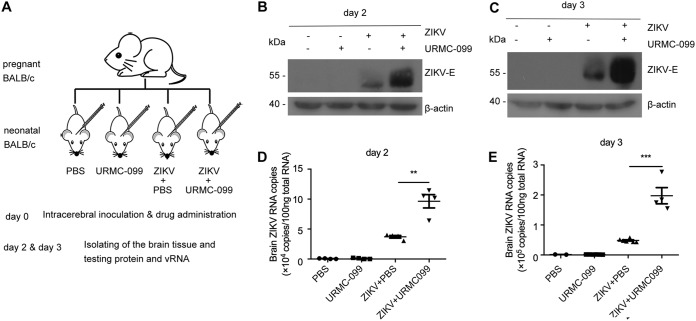
To further evaluate the effect of URMC-099 on ZIKV replication in vivo using the intracerebral inoculation of ZIKV into neonatal mice (Fig. 3A), we found that the levels of the ZIKV E protein and viral RNA in the brain were greatly increased by URMC-099 on days 2 and 3 (Fig. 3B to E). These results confirm that ZIKV replication can be enhanced by URMC-099 both in vitro and in vivo.
FIG 3.
URMC-099 promotes ZIKV infection in vivo. (A) Experimental scheme of the effect of URMC-099 in a neonatal mouse model of ZIKV infection. The ZIKV SZ01 strain (1 μl virus stock per animal) was intracerebrally injected into neonatal BALB/c mice on day 0 after birth. URMC-099 (5 mg/kg) was supplemented together with ZIKV on day 0. The same volume of PBS was used as a control. The infected brains were isolated for analysis on days 2 and 3. (B to E) Viral envelope protein levels and the viral RNA load in whole brains (n = 4) from ZIKV-infected neonatal mice were determined by Western blotting and qRT-PCR on day 2 (B and D) and day 3 (C and E). Experiments were performed twice, and the data are presented as the mean ± standard error of the mean (SEM) from one independent experiment. Statistical analysis was performed using an unpaired, two-tailed t test. **, P < 0.01; ***, P < 0.001.
Confirmation of MLK3 as a key regulator of ZIKV infection.
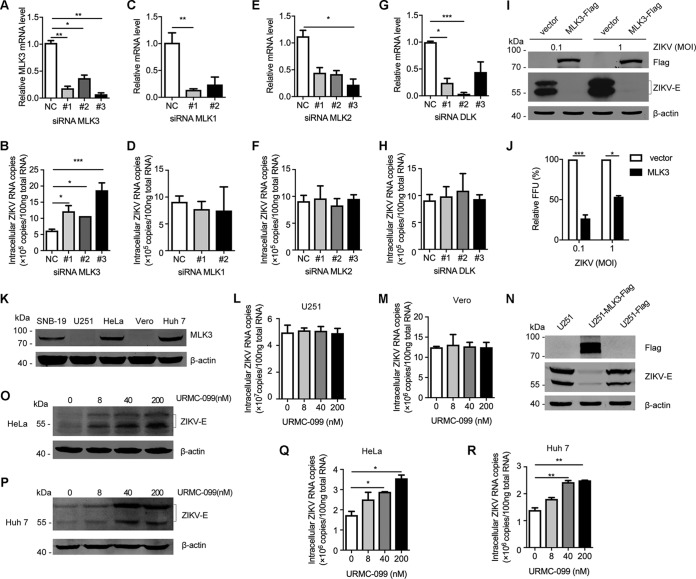
To confirm the functional relevance of MLK3, the target of URMC-099, in ZIKV infection, three small interfering RNAs (siRNAs) targeting MLK3 were designed and transfected into SNB-19 cells 36 h before infection with ZIKV. Quantitative reverse transcription-PCR (qRT-PCR) assays showed efficient knockdown of endogenous MLK3 (Fig. 4A), and MLK3 deficiency enhanced ZIKV infectivity (Fig. 4B). The knockdown results showed that other members of the MLK family (MLK1, MLK2, and DLK) were not involved in ZIKV infection (Fig. 4C to H). In addition to MLK3 knockdown, we also generated SNB-19 cells stably overexpressing MLK3. Compared to that in the vector control SNB-19 cells, ZIKV replication was significantly suppressed in cells overexpressing MLK3 by detection of viral E protein expression (Fig. 4I) and analysis of viral FFU (Fig. 4J). To further characterize the function of MLK3, a panel of cell lines was evaluated for endogenous expression of MLK3. SNB-19, HeLa, and Huh7 cells showed detectable MLK3 protein, whereas U251 and Vero cells lacked detectable MLK3 (Fig. 4K). Using these distinct cell lines, we were able to further confirm the regulatory role of MLK3 in ZIKV replication. ZIKV replicated robustly in MLK3-deficient U251 and Vero cells (107 viral RNA copies), and ZIKV replication in these cells was insensitive to URMC-099 (Fig. 4L and M). ZIKV replication was significantly suppressed in U251 cells stably overexpressing MLK3 (Fig. 4N). In contrast, ZIKV infection was enhanced by URMC-099 in MLK3-positive HeLa and Huh7 cells (Fig. 4O to Fig. 4R). These results demonstrate that MLK3 is an important host factor that restricts ZIKV replication in our system.
FIG 4.
Confirmation of MLK3 as a key regulator of ZIKV infection. (A) MLK3-specific siRNAs (50 nM) were transiently transfected into SNB-19 cells. MLK3 mRNA levels were determined by qRT-PCR. (B) Viral RNA was quantified by qRT-PCR after 72 h of infection in SNB-19 cells that were transfected with MLK3 siRNAs 36 h before ZIKV (MOI = 0.1) infection. A scrambled siRNA was used as a negative control. (C to H) Knockdown of MLK1, MLK2, and DLK and its effects on ZIKV infection. (I and J) SNB-19 cells stably transfected with a plasmid overexpressing MLK3 or vector control cells were infected with ZIKV (MOI = 0.1 or 1) for 48 h, and virus propagation was analyzed by Western blotting (I) or FFU assay (J). (K) Endogenous MLK3 expression profiles were analyzed by Western blotting in SNB-19, U251, HeLa, Vero, and Huh7 cells. (L and M) The effects of URMC-099 on MLK3-defecient U251 and Vero cells. (N) ZIKV infection was inhibited in U251 cells stably expressing MLK3. (O to R) Characterization of the effects of URMC-099 on ZIKV infection (MOI = 0.1) in HeLa and Huh7 cells with endogenous MLK3 by Western blotting (O and P) and qRT-PCR (Q and R).
Effects of URMC-099 on other RNA viruses.
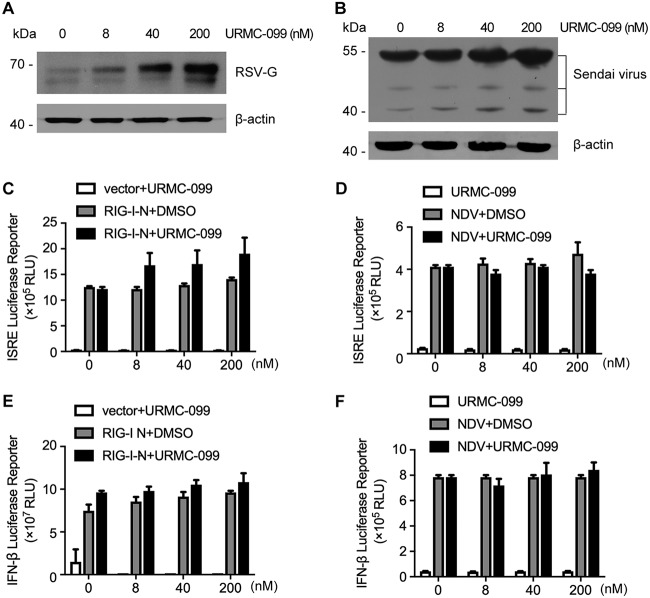
To investigate the specificity and possible regulatory mechanism of MLK3 for ZIKV, we selected two other RNA viruses, respiratory syncytial virus (RSV) and Sendai virus (SeV), for further assessment. The viral protein expression of RSV and SeV was increased in response to URMC-099 treatment (Fig. 5A and B). These results indicated a broad-spectrum regulatory role of URMC-099 and suggested that the host IFN antiviral mechanism might be modulated. RSV and SeV are both RNA viruses and sensitive to IFN-producing and effector pathways (22). Therefore, we used promoter-driven reporter genes, including those coding for the interferon-stimulated response element (ISRE) (Fig. 5C and D) and IFN-β (Fig. 5E and F), as readouts. Unexpectedly, after stimulation with the retinoic acid-inducible gene I N-terminal CARD domain (RIG-I-N) or Newcastle disease virus (NDV), these reporter genes remained unchanged after URMC-099 treatment. These results suggest the involvement of other regulatory mechanisms rather than host IFN pathways.
FIG 5.
URMC-099 treatment increases RSV and SeV infection. (A) HeLa cells were infected with the RSV A2 strain (MOI = 0.5) in the presence of various concentrations of URMC-099 for 48 h. Viral protein levels were determined by Western blotting. (B) 293T cells were infected with SeV (MOI = 0.5) in the presence of various concentrations of URMC-099 for 24 h. Viral protein levels were determined by Western blotting. (C and D) An ISRE promoter-driven reporter construct was transfected into SNB-19 cells, in combination with transfection of an RIG-I-N overexpression plasmid (C) or infection with NDV (D), and the cells were then treated with various concentrations of URMC-099 for 24 h. Luciferase activity was determined. The x axis indicates the concentrations of URMC-099 for the drug treatment group. For the non-drug-treatment group, the x axis indicates the corresponding volume of DMSO solvent. (E and F) An experimental design similar to that described in panels C and D was used, except for the substitution of the ISRE reporter with an IFN-β promoter-driven reporter.
URMC-099 inhibits ZIKV-induced inflammatory cytokine production.
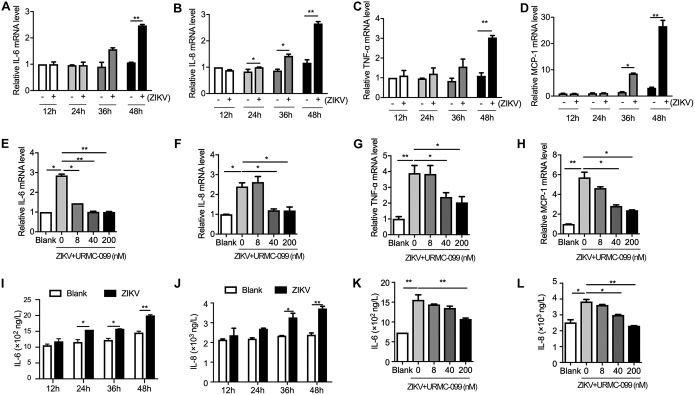
Since IFN antiviral pathways were not involved in MLK3-regulated ZIKV replication, other mechanisms may exist. Viral infection usually causes an acute inflammatory response that is required for the clearance of pathogens. MLK3 was reported to regulate cytokine production by activating the JNK and p38 pathways (23). In mouse models of sepsis, removal of MLK2/3 significantly reduced cytokine and chemokine production (24). Therefore, we examined the levels of major cytokines, including interleukin-6 (IL-6), IL-8, tumor necrosis factor alpha (TNF-α), and monocyte chemoattractant protein 1 (MCP-1). As shown in Fig. 6A to D, by using qRT-PCR, we found that mRNA expression of all these cytokines was significantly induced by ZIKV in SNB-19 cells at 36 and 48 h postinfection. When we compare Fig. 6A to D with Fig. 2G, a rough correlation between cytokine production and increased virus production could be concluded. Notably, treatment with URMC-099 suppressed ZIKV-induced inflammatory cytokine production in a dose-dependent manner (Fig. 6E to H), which was similar to the enhancement of viral replication by URMC-099. In addition, we measured the protein levels of secreted IL-6 and IL-8 in SNB-19 cells. Similarly, uninfected cells produced 732 ng/liter IL-6 and 2,300 ng/liter IL-8. When the cells were exposed to ZIKV, cytokine production increased over the basal level (Fig. 6I and J). URMC-099 treatment reversed ZIKV-induced IL-6 and IL-8 production (Fig. 6K and L). These results indicate that MLK3 is a crucial mediator of ZIKV-induced cytokine production.
FIG 6.
URMC-099 inhibits ZIKV-induced inflammatory cytokine production. (A to D) SNB-19 cells were inoculated with ZIKV (MOI = 0.1) for 12, 24, 36, or 48 h. The relative mRNA level of IL-6, IL-8, TNF-α, or MCP-1 was assayed by qRT-PCR. Uninfected SNB-19 cells in each time point were set as negative controls. (E to H) SNB-19 cells were inoculated with ZIKV (MOI = 0.1) in the absence or presence of URMC-099 (8, 40, or 200 nM) for 48 h. qRT-PCR was performed to measure IL-6, IL-8, TNF-α, or MCP-1 expression. Blank cells were used as negative controls. (I to L) Quantification of secreted IL-6 and IL-8 from SNB-19 cells by ELISA. Data are presented as the mean ± SEM. *, P < 0.05; **, P < 0.01.
Anti-ZIKV effect of inflammatory cytokines.
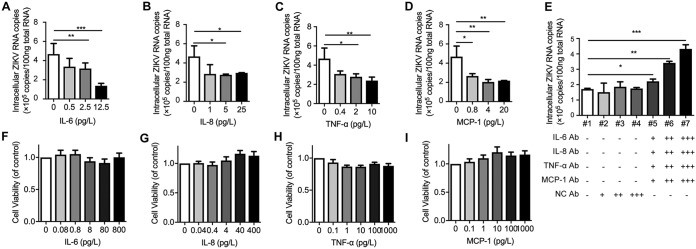
In addition to modulating immune function, we wondered whether these cytokines had direct antiviral effects against ZIKV in vitro. For this reason, a cell model of ZIKV-infected SNB-19 cells and recombinant cytokines was used. As shown in Fig. 7A to D, recombinant IL-6, IL-8, TNF-α, and MCP-1 inhibited ZIKV infection in a dose-dependent manner. Nevertheless, treatments with cytokines had no influence on cell viability (Fig. 7F to I). In addition to recombinant protein treatments, we also investigated the antiviral effect of ZIKV-induced endogenous cytokines. To this end, a combination of neutralizing antibodies was applied. The cytokine antibody combination but not the IgG isotype control significantly promoted intracellular ZIKV RNA replication (Fig. 7E).
FIG 7.
Inflammatory factors inhibit ZIKV infection in a dose-dependent manner. (A to D) SNB-19 cells were inoculated with ZIKV (MOI = 0.1) with different concentrations of IL-6, IL-8, TNF-α, or MCP-1 for 48 h. Total cellular RNA was isolated, and ZIKV RNA copies were measured by qRT-PCR. (E) Antibody neutralization assay. SNB-19 cells were inoculated with ZIKV (MOI = 0.1) for 6 h, and the cells were then washed and treated with various concentrations of the antibody cocktail for another 48 h: +, 0.04 mg/liter; ++, 0.2 mg/liter; +++, 1 mg/liter. Intracellular viral RNA was measured by qRT-PCR. Isotype mouse IgG was used as a negative control. (F to I) SNB-19 cell viability was determined by MTT assay after 48 h of incubation with different concentrations of IL-6 (F), IL-8 (G), TNF-α (H), and MCP-1 (I). Values represent the mean ± SEM from independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
ZIKV activates the MLK3 and MLK3/MKK7/JNK pathways.
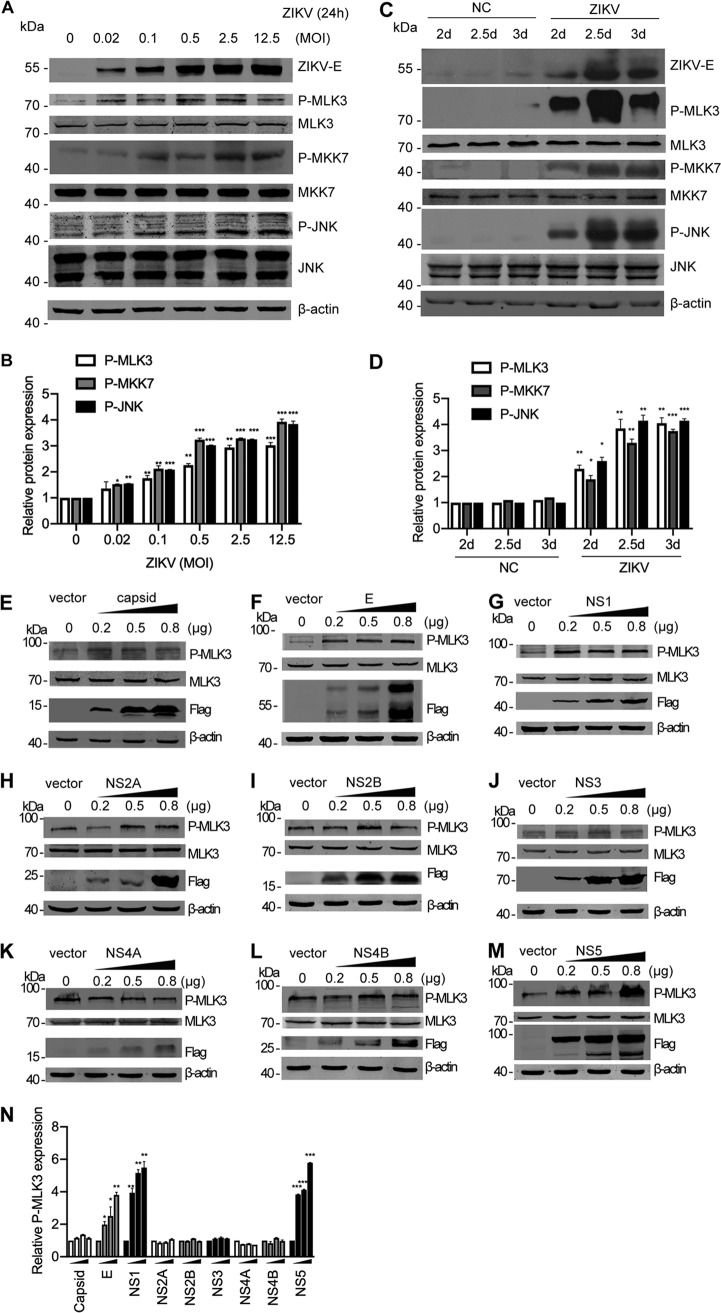
Since MLK3 plays an important antiviral role by inducing cytokine production, we next wanted to determine whether the activation status of the MLK3 signaling pathway changes during ZIKV infection. ZIKV-infected SNB-19 cells and neonatal mouse brains were used. In SNB-19 cells, the phosphorylation level of MLK3, MKK7, and JNK was increased at 24 h post-ZIKV infection in an MOI-dependent manner, whereas the total protein level remained unchanged (Fig. 8A). Band intensities of the phosphorylated proteins were normalized and quantified (Fig. 8B). A more significant activation of MLK3 signaling by ZIKV was observed in vivo. In brain tissues, ZIKV infection triggered an obvious increase in the phosphorylation of components of the MLK3/MKK7/JNK pathway compared to that of the mock-infected negative controls (Fig. 8C and D). ZIKV encodes 10 viral proteins, including the structural proteins capsid/preM/E and the nonstructural proteins NS1/2A/2B/3/4A/4B/5 (8). To determine which protein is responsible for MLK3 activation, we constructed individual protein overexpression plasmids and transfected them into SNB-19 cells at different concentrations (0.2, 0.5, and 0.8 μg). Interestingly, ectopic expression of ZIKV E, NS1, and NS5 but not capsid, NS2A, NS2B, NS3, NS4A, and NS4B increased the phosphorylation level of MLK3 (Fig. 8E to M). Western blotting bands in Fig. 8E to M were quantified with ImageJ software; fold change is shown in Fig. 8N.
FIG 8.
ZIKV infection activates the MLK3/MKK7/JNK pathway. (A) SNB-19 cells were infected with ZIKV (MOI = 0.02, 0.1, 0.5, 2.5, and 12.5) for 24 h. Cell lysates were subjected to Western blotting with the indicated antibodies. The same blots were reprobed with an anti-β-actin antibody to ensure equal loading. (B) Bands of the phosphorylated MLK3, MKK7, and JNK in panel A were quantified with ImageJ software; fold change is shown. The phosphorylated proteins were normalized to corresponding nonphosphorylated proteins to indicate their phosphorylation levels. (C) The ZIKV SZ01 strain was intracerebrally injected into neonatal BALB/c mice on day 0 after birth. “NC” represents PBS injection as a negative control. Brain tissues were collected at 2, 2.5, or 3 days postinfection (dpi), and Western blotting was performed with the indicated antibodies. (D) Bands of the phosphorylated MLK3, MKK7, and JNK in panel C were quantified. (E to M) SNB-19 cells in 24-well plates were transfected with individual Flag-tagged ZIKV protein constructs (0.2, 0.5, or 0.8 μg each well). Western blotting was performed to measure phosphorylated MLK3, MLK3 total protein, and Flag-tagged viral proteins. β-Actin was used as an equal loading control. (N) Western blotting bands in Fig. 8E to M were quantified as fold change.
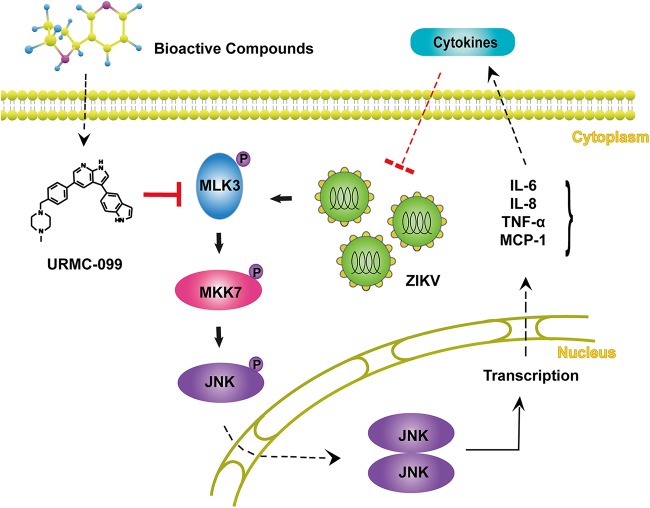
Taken together, our data reveal that MLK3 is a host regulator for ZIKV infection. A schematic regulation mechanism is illustrated in Fig. 9.
FIG 9.
Schematic representation of the ZIKV-MLK3 regulatory mechanism. ZIKV infection activates MLK3 and the MLK3/MKK7/JNK pathway and promotes production of some cytokines, such as IL-6, IL-8, TNF-α, and MCP-1. URMC-099 enhances ZIKV infection through inhibition of MLK3.
DISCUSSION
Increasing evidence has demonstrated a causal relationship between ZIKV infection in pregnant women and an unpredicted rise in the incidence of microcephaly. The global burden of Zika fever reflects the lack of a ZIKV prophylactic vaccine and specific antiviral drugs. An understanding of ZIKV-mediated pathogenic mechanisms and the determinant intracellular pathways for viral replication is urgently needed for the development of an efficient control strategy. In this study, we established an HTS strategy based on the direct evaluation of ZIKV antigen in infected SNB-19 cells. We selected a bioactive compound library with 2,659 small molecules. In addition to FDA-approved drugs, the library also includes the vast majority of known host protein inhibitors, agonists, active pharmaceutical ingredients (APIs), natural products, and chemotherapeutic agents, which are structurally diverse and medicinally active and exhibit various levels of cell permeability. This screening strategy ensures that more clues about host targets can be obtained. In total, we identified 111 compounds that inhibited ZIKV infection by more than 95% at 5 μM, suggesting that these compounds have potential implications for future clinical treatment of ZIKV. Many of these compounds have been used during pregnancy for the indications accepted in their safety profiles. Our findings are consistent with previously reported observations for some compounds (19, 25–33), such as SNS-032, flavopiridol (alvocidib), dinaciclib, tenovin-1, and mycophenolate mofetil. These findings are partially consistent with compounds such as mometasone furoate, lovastatin, roflumilast, methotrexate, and YM201636. In our system, treatment with the following chemicals induced partial cell desquamation and anti-ZIKV activity: dioscin, amuvatinib (MP470), flubendazole, mevastatin, and Ki8751. Some chemicals, including moxifloxacin HCl, celgosivir HCl, niclosamide, and NITD008, could not be found in the bioactive compound library. These results may establish target networks, reveal virus-host interactomes, and serve as an alternative approach for identification of anti-ZIKV molecules.
Interestingly, 12 inhibitors enhanced ZIKV infection by more than 2-fold, including URMC-099, BV-6, LDK378, BI2536, BMS-833923, GSK503, LDN-214117, DDR1-IN-1, KC7F2, ruxolitinib (INCB018424), radotinib, and EUK 134, suggesting that the targets of these inhibitors might exert restrictive effects against ZIKV.
URMC-099 was selected for further in-depth characterization because it showed a very robust enhancement of ZIKV replication in our assays. URMC-099 promoted viral replication by as much as 2.9-fold in the primary screen and was efficient both in vitro and in vivo. URMC-099 is an orally bioactive, low-cytotoxicity, brain-penetrating MLK3 inhibitor (21). In this study, we demonstrated that URMC-099 enhances ZIKV propagation at the replication step. We have used comprehensive approaches to illustrate the efficacy of URMC-099 in facilitating ZIKV infection. The involvement of MLK3 in antiviral responses was recently reported, and MLK3 deficiency delayed viral clearance in the lung and was associated with diminished influenza-induced cytopathic effects in infected cells (17). However, the detailed mechanism is still unclear.
We have demonstrated that MLK3 plays a critical role in host defense against ZIKV, as overexpression of MLK3 strongly suppresses infection in multiple MLK3-deficient cell types. Furthermore, we showed that the MLK3 inhibitor URMC-099 facilitates multiple viral infections, including ZIKV, RSV, and SeV. SeV is sensitive to the IFN signaling pathway. However, URMC-099 had no effect on the activity of the ISRE and IFN-β promoters. ZIKV infection activated MLK3, and the ZIKV E, NS1, and NS5 proteins played a major role in this effect. In addition, the MLK3/MKK7/JNK cascade promoted an increased level of some cytokines and chemokines, such as IL-6, IL-8, TNF-α, and MCP-1. The host innate response to viral infection is critical in controlling viral replication by accelerating the inflammatory process. These cytokines inhibited ZIKV replication in a dose-dependent manner and protected against ZIKV infection. Supplementation with URMC-099 inhibited the activity of MLK3 and reduced antiviral cytokine production through the MLK3/MKK7/JNK signaling pathway. Although ZIKV infection was associated with the induction of proinflammatory mediators that was evidenced by our and others’ data (34, 35), the influence of these elevated cytokines and chemokines on Zika virological events is completely unclear. The antiviral nature of some cytokines, such as TNF-α, has been generally well accepted. However, the contribution of other cytokines, such as MCP-1 and IL-8, to cell-intrinsic immunity is not defined. We assume that they might play a role in establishing the inflammatory microenvironment for both recruitment of leukocytes and initiation of intracellular pathways involved in multiple host essential and restriction factors for ZIKV infection. Despite the use of different technical setups, the observations obtained from this study and other studies revealed similar results (17), underscoring the biological relevance and general importance of MLK3 in controlling viral infections.
In summary, this study revealed a preliminary mechanism by which URMC-099 enhances ZIKV infection through the inhibition of the MLK3/MKK7/JNK signaling pathway, reducing ZIKV-induced production of inflammatory cytokines and weakening the host antiviral response. Individual compounds or compound combinations that enhance virus replication may be applied to the production of attenuated vaccines by increasing virus titers. Our work highlights the unique antiviral mechanism of MLK3 and suggests that protein agonists of the MLK3/MKK7/JNK signaling pathway may have clinical utility when repurposed as ZIKV treatments.
MATERIALS AND METHODS
Cells and viruses.
The human glioblastoma cell line SNB-19 (ATCC; CRL-2219) was maintained in Dulbecco’s modified Eagle’s medium (DMEM)–F-12 medium (1:1 vol/vol) (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS) (Thermo Fisher Scientific) and 1% penicillin and streptomycin (100 mg/liter). U251 (formerly known as U-373 MG; ECACC, 09063001), HEp2 (CCL-23; ATCC), HeLa (CCL-2; ATCC), Vero (CCL-81; ATCC), and Huh7 cells were provided by Apath, Inc. (Brooklyn, NY, USA) with permission from Charles Rice (Rockefeller University), LLC-MK2 (CCL-7; ATCC) and HEK293T (CRL-3216; ATCC) cells were cultured in DMEM as described above.
ZIKV Asian strain SZ01 (GenBank no. KU866423), isolated from a Chinese patient returning from Samoa in 2016 (20), was kindly provided by Cheng-Feng Qin (Beijing Institute of Microbiology and Epidemiology, Beijing, China). Viral stock (6.5 × 105 PFU/ml) was amplified in Vero cells. Briefly, Vero cells in a 10-cm dish were inoculated with 1 ml of viral stock and underwent overnight culture at 37°C. The cells were split into 3 new dishes on the second day and cultured for another 3 days before the supernatant was harvested. The titer of Vero-amplified ZIKV was determined in SNB-19 cells with serially diluted virus. The immunofluorescence FFU assay was performed at 48 h postinfection, and the titer was calculated as the multiplicity of infection (MOI) per ml.
Stock of the human respiratory syncytial virus (RSV) A2 strain (VR-1540; ATCC) was a gift from Jianwei Wang (Institute of Pathogen Biology, CAMS, China). The RSV A2 stock was amplified in HEp2 cells, and the titer of a 50% tissue culture infective dose (TCID50) of 1 × 107 per ml was determined (36). The titer of HEp2-amplified RSV A2 was determined in HeLa cells with serially diluted virus. The FFU assay was performed at 48 h postinfection, and the titer was calculated as MOI per ml. For RSV infection and drug treatment, 12-well plates were seeded with HeLa cells (3 × 105) 1 day before virus inoculation. The cells were infected with 1 ml 100× diluted RSV A2 stock in Opti-MEM for 2 h, and then the culture medium was replaced with DMEM containing 3% FBS in the presence of various concentrations of URMC-099. Cells were harvested at 48 h after infection for Western blotting.
Sendai virus (SeV) and Newcastle disease virus expressing green fluorescent protein (NDV-GFP) were kindly provided by Zhendong Zhao (Institute of Pathogen Biology, CAMS, China) (37). SeV was propagated in the allantoic fluid of embryonated chicken eggs. The titer was determined by plaque assay in LLC-MK2 cells and calculated as MOI per ml. For infections, 12-well plates were seeded with HEK293T cells 1 day before virus addition. The cells were infected with SeV for 2 h, and then various doses of URMC-099 were added. Virus infection was allowed for 24 h before the cells were harvested for Western blotting.
Reagents.
A bioactive compound library (L-1700) was purchased from Selleck (Houston, TX, USA). Mouse monoclonal antibodies (MAbs) specific for ZIKV E (BF-1176-56) and ZIKV NS1 (BF-1225-36) were purchased from BioFront Technologies (Tallahassee, FL, USA). Mouse monoclonal anti-β-actin (A5441) and anti-Flag (F3165) antibodies, MTT (M2128), DMSO (D5879), and bovine serum albumin (BSA [B2064]) were purchased from Sigma-Aldrich (St. Louis, MO, USA). A mouse polyclonal antibody specific for SeV was from MBL (Woburn, MA, Japan). A rabbit polyclonal anti-RSV-G antibody (11070-RP01) and a rabbit monoclonal antibody specific for RSV-F (11049-R009) were from SinoBiological (Beijing, China). The ViaFect transfection reagent (E4981), the luciferase assay system (E1501), and Renilla luciferase assay system (E2710) were from Promega (Madison, WI, USA). A rabbit polyclonal antibody specific for MLK3 (phospho-T277+S281; ab191530) was obtained from Abcam (Cambridge, MA, USA). The rabbit polyclonal anti-MLK3 antibody (11996-1-AP) was purchased from Proteintech (Rosemont, IL, USA). TRIzol reagent (15596-026) and prestained protein ladder (26616) were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Rabbit antibodies specific for phospho-SAPK/JNK (Thr183/Tyr185; 4668), SAPK/JNK (9252), phospho-MKK7 (Ser271Thr275; 4171), and MKK7 (4172) were obtained from Cell Signaling Technology (Danvers, MA, USA). Rabbit polyclonal antibodies specific for IL-6 (bs-4539R), IL-8 (bs-0780R), TNF-α (bs-2081R), and MCP-1 (bs-1101R) were purchased from Bioss Antibody (Boston, MA, USA). Horseradish peroxidase-conjugated secondary antibodies were purchased from Jackson ImmunoResearch (West Grove, PA, USA). The IRDye-conjugated secondary antibodies 800CW goat anti-rabbit IgG (926-32211), 800CW donkey anti-mouse IgG (925-32212), and 680RD goat anti-mouse IgG (926-680701) were from LI-COR Biosciences (Lincoln, NE, USA). The cytokines IL-6 (200-6), IL-8 (200-08M), TNF-α (300-01A), and MCP-1 (300-04) and enzyme-linked immunosorbent assay (ELISA) kits for IL-6 (BGK05231), IL-8 (BGK10145), TNF-α (BGK01375), and MCP-1 (BGK13500) were obtained from PeproTech (Rocky Hill, NJ, USA). Paraformaldehyde (P1110), Triton X-100 (T8200), skim milk (232100), protein loading buffer (P1015), and DAPI (4′,6-diamidino-2-phenylindole; C0060) were purchased from Solarbio (Beijing, China).
HTS of the bioactive compound library.
Ninety-six-well plates were seeded with SNB-19 cells 36 h before ZIKV infection (MOI = 0.1). A 5 μM concentration of each compound in the library (total of 2,659 bioactive compounds) or the same volume of dimethyl sulfoxide (DMSO) as a negative control was added at the time of infection. After 48 h of coculture, the effects of compounds on ZIKV infection were determined by detecting FFU in each well using the In-Cell Western (ICW) assay.
The ICW and FFU assays.
ICW, created by LI-COR, is a quantitative immunofluorescence assay performed in microplates (96- or 384-well format) that combines the specificity of Western blotting with the reproducibility and throughput of ELISA. It is a kind of alternative form of the quantitative immunofluorescence staining or FFU assay, using a spectrally distinct infrared scanner instead of a fluorescence microscope. Briefly, at 48 h postinfection, the cells were fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100, followed by immunostaining with a ZIKV NS1 antibody and an IRDye-conjugated secondary antibody. Images were obtained on an Odyssey Infrared Imaging System (LI-COR) with the reader set to read a 96-well plate. Scanning was performed using the 800-nm channel to visualize fluorescent viral spots. The number of FFU was determined by counting the number of dots in each well using LI-COR Odyssey software, according to the manufacturer’s instructions.
ZIKV infection of cultured cells.
Cells were seeded in 12-well plates 24 h prior to virus addition. Virus stocks were thawed and diluted to the desired MOI according to the experimental purpose and added to the culture medium for 2 h at 37°C with 5% CO2. The inoculum was then removed, the cells were washed once with phosphate-buffered saline (PBS) to remove unbound viruses, and fresh culture medium was added to each well. The cells were harvested at the indicated time points. For mock-infected controls, the cells were incubated with an equivalent volume of uninfected culture supernatant.
Time-of-addition experiment.
The time-of-addition assay was conducted to determine the effects of URMC-099 on viral attachment, cell entry, or postentry step during ZIKV infection. For pretreatment, cells were pretreated with URMC-099 for 6 h, washed and infected with ZIKV for 2 h, and then washed and supplemented with fresh medium for 48 h. For cotreatment, cells were infected with ZIKV and treated with URMC-099 simultaneously for 2 h and then washed and supplemented with fresh medium for 48 h. For posttreatment, cells were infected with ZIKV for 2 h, and then washed and supplemented with fresh medium in the presence of URMC-099 for 48 h.
ZIKV replicon.
A ZIKV SZ01 strain-based replicon system was kindly provided by Bo Zhang (Wuhan Institute of Virology, CAS, China) and Cheng-Feng Qin (38). Briefly, replicon RNA was in vitro transcribed, purified, and transfected into SNB-19 cells (Promega). At 9 h posttransfection, the cells containing the ZIKV replicon were treated with URMC-099 or DMSO for 36 h. Cell lysates were used for measurement of Renilla luciferase activity in a GloMax-96 microplate luminometer (Promega).
ZIKV infection of mice.
Animal procedures were reviewed and approved by the institutional committees for animal ethics and biosafety. Pregnant BALB/c mice were purchased from Beijing HFK Bioscience (Beijing, China). Neonatal BALB/c mice were breast-fed by their own mothers and divided into different groups. As shown in Fig. 3A, neonatal BALB/c mice were injected in the ventricle of the postnatal brain with 1 μl (6.5 × 105 FFU/ml) of virus stock or URMC-099 (5 mg/kg) on day 0, the mice were sacrificed, and the brains were used for further analysis on days 2, 2.5, and 3. Subsequently, viral RNA copies and viral loads were determined. Total RNA was extracted using TRIzol reagent (Thermo Fisher Scientific) according to the manufacturer's instructions. RLT lysis buffer (Qiagen) was added to the EP tubes, and whole-brain tissue homogenates were prepared using an automated homogenizer. Tissues were ground with PBS and centrifuged at 12,000 × g at 4°C for 15 min. The supernatants were collected and stored at −80°C for later use. ZIKV copy analysis was performed using GraphPad Prism 5.0 software.
Constructs and stable cell line generation.
A codon-optimized version of MLK3 in the pcDNA3.1-C-Myc backbone was used as the original plasmid for subcloning of the coding region of MLK3 into the pMIR-cFlag lentiviral vector. The plasmids were transfected into HEK293T cells for lentivirus packaging. SNB-19 and U251 cells stably expressing MLK3 or control empty pMIR-cFlag vector were generated by lentiviral infection and puromycin addition. Genes encoding ZIKV capsid (C), envelope (E), NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5 were codon optimized and synthesized by GenScript (Nanjing, China) and cloned into the pcDNA3.1-N-Flag backbone. Plasmids were maxiprepped and transfected into SNB-19 cells with Lipofectamine 3000 transfection reagent (Thermo Fisher Scientific).
Gene silencing by siRNA.
All small interfering RNAs (siRNAs) were obtained from RiboBio (Guangzhou, China). The siRNA sequences are as follows: siMLK1#1, CGACCATCTTTCACGAATA; siMLK1#2, CAAACGAGATCCTAACCAA; siMLK2#1, CCTCTGGCTTTGAGCATAA; siMLK2#2, GCATGAACTACCTACACAA; siMLK2#3, CCTGGAAACTGGTCTCCTT; siMLK3#1, CCTATGGCGTAGCTGTTAA; siMLK3#2, GCCACATGGTACCTGGATT; siMLK3#3, GCATGCCACTCGACTTCAA; siDLK#1, GAAGTAGAGCTGACATCAA; siDLK#2, CCAACATGCTAATCACCTA; and siDLK#3, CCAACAACCTGTATATGGA. Transfection of siRNA was performed using Lipofectamine RNAi-MAX (Thermo Fisher Scientific) according to the manufacturer's protocol. A scrambled siRNA was used as a negative control.
Luciferase reporter assay.
Briefly, 24-well plates were seeded with SNB-19 cells at a cell density of 2 × 105 cells per well. The next day, the cells were transfected with the pGL3-IFN-β-Luc or pGL3-ISRE-Luc (generous gifts form Zhengdong Zhao) reporter plasmid, along with a plasmid for RIG-I-N or NDV stimulation. The culture medium was changed, and URMC-099 was added at 8 h posttransfection. The total amount of DNA was kept constant by adding empty control plasmid. At 48 h after transfection, the cells were harvested, and the cell lysates were used to determine luciferase activities according to the manufacturer's instructions (Promega).
Cytotoxicity assay.
To investigate cellular toxicity, 96-well plates were seeded with SNB-19 cells and incubated at 37°C in an atmosphere of 5% CO2 for 36 h. The next day, the cells were treated with URMC-099 at the indicated concentration for 48 h, and 20 μl of MTT solution (5 g/liter in PBS) was added to all wells. After incubation of the cells for 4 h at 37°C, the MTT was removed, and 150 μl of DMSO was added. The mixture was shaken, and the formazan crystals were fully dissolved for approximately 10 min. The value of each well was measured at a test wavelength of 490 nm.
Real-time qRT-PCR.
Total cellular RNA was extracted using TRIzol reagent (Thermo Fisher Scientific) according to the manufacturer's instructions. The absolute quantification of ZIKV RNA was described previously. The forward (F) and reverse (R) primer sequences for ZIKV, MLK3, IL-6, IL-8, TNF-α, MCP-1, and β-actin are as follows (6, 39–44): ZIKV, F, GGTCAGCGTCCTCTCTAATAAACG, and R, GCACCCTAGTGTCCACTTTTTCC; MLK3, F, GCAGCCCATTGAGAGTGAC, and R, CACTGCCCTTAGAGAAGGTGG; IL-6, F, ATGAACTCCTTCTCCACAAGCGC, and R, GAAGAGCCCTCAGGCTGGACTG; IL-8, F, ATGACTTCCAAGCTGGCCGTGGCT, and R, TCTCAGCCCTCTTCAAAAACTTCTC; TNF-α, F, CGGTGCTTGTTCCTCAGC, and R, GCCAGAGGGCTGATTAGAGA; MCP-1, F, CATTGTGGCCAAGGAGATCTG, and R, CTTCGGAGTTTGGGTTTGCTT; and β-actin, F, CCAACCGCGAGAAGATGA, and R, CCAGAGGCGTACAGGGATAG. Real-time quantification of mRNAs was performed by using a QuantiFast SYBR green RT-PCR kit (Qiagen, Düsseldorf, Germany). All quantifications were performed with the ABI Prism 7900 system (Thermo Fisher Scientific).
Immunofluorescence staining and confocal microscopy.
Glass coverslips in 24-well plates were seeded with SNB-19 cells before viral infection, and URMC-099 was administered. The treated cells were fixed with 4% paraformaldehyde, permeabilized with 0.1% (vol/vol) Triton X-100, blocked in 0.3% (wt/vol) bovine serum albumin (BSA), and incubated with primary antibodies specific for ZIKV NS1. Samples were washed with PBS and subsequently incubated with either fluorescein isothiocyanate (FITC)- or rhodamine-conjugated secondary antibodies according to the manufacturer’s instructions (Santa Cruz Biotechnology). The nucleus was stained with DAPI. Images were captured by a Leica TCS-SL confocal microscope, edited using Metamorph, and assembled using Photoshop CS (Adobe).
Western blot analysis.
For cell lysate preparation, the monolayer cells were lysed with lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% Nonidet P-40, 50 mM NaF, 1 mM Na3VO4, 5 mM β-glycerophosphate, 1 mM dithiothreitol, and 1 mM phenylmethylsulfonyl fluoride) supplemented with a protease inhibitor cocktail (Sigma-Aldrich) on ice. The lysate was cleared by centrifugation at 14,000 × g for 20 min. The samples were boiled in 2× SDS loading buffer and loaded onto a 10 to 12% polyacrylamide gel. After electrophoresis, the separated proteins were transferred onto a nitrocellulose membrane (Bio-Rad, Hercules, CA). To test protein phosphorylation, the resulting blots were blocked with 10% milk or 5% BSA for 1 h at room temperature and then incubated with a primary monoclonal antibody overnight at 4°C. Secondary antibodies conjugated to IRDye 680 RD or 800CW were used for detection by the Odyssey Infrared Imaging System (LI-COR Biosciences), with quantification by Odyssey software.
ELISA.
The secretion of IL-6, IL-8, TNF-α, and MCP-1 into the SNB-19 culture supernatant was confirmed by sandwich ELISA according to the manufacturer’s protocol (PeproTech).
Antibody neutralization.
Twenty-four-well plates were seeded with SNB-19 cells. After overnight incubation, the cells were simultaneously infected with ZIKV and incubated with an antibody mixture specific for IL-6, IL-8, TNF-α, MCP-1, and control IgG. At 48 h postinfection, the total RNA in the infected cells was extracted with TRIzol and quantified with qRT-PCR. The percentage of infected cells was obtained and normalized to those of the positive and negative controls.
Statistical analysis.
The values shown in graphs are presented as the mean ± standard deviation from at least three independent experiments, unless otherwise noted. Statistical analyses were performed using GraphPad Prism 8 software, and P values of <0.05 (obtained from Student's t test) were considered statistically significant: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jianwei Wang, Zhendong Zhao, Lili Ren (Institute of Pathogen Biology, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China), Lei Shi (Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China), Cheng-Feng Qin (Beijing Institute of Microbiology and Epidemiology, Beijing, China), and Bo Zhang (Wuhan Institute of Virology, Chinese Academy of Sciences) for providing viruses and constructive discussions.
This work was supported by the CAMS Initiative for Innovative Medicine (grant no. 2016-I2M-3-020), the National Basic Research Program of China (grant no. 2015CB554301), the National Natural Science Foundation of China (grant no. 81672030 and 81871667), and the National Science and Technology Major Project (grant no. 2017ZX10304402-001-013).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00758-19.
REFERENCES
- 1.Dick GW, Kitchen SF, Haddow AJ. 1952. Zika virus. I. Isolations and serological specificity. Trans R Soc Trop Med Hyg 46:509–520. doi: 10.1016/0035-9203(52)90042-4. [DOI] [PubMed] [Google Scholar]
- 2.Metsky HC, Matranga CB, Wohl S, Schaffner SF, Freije CA, Winnicki SM, West K, Qu J, Baniecki ML, Gladden-Young A, Lin AE, Tomkins-Tinch CH, Ye SH, Park DJ, Luo CY, Barnes KG, Shah RR, Chak B, Barbosa-Lima G, Delatorre E, Vieira YR, Paul LM, Tan AL, Barcellona CM, Porcelli MC, Vasquez C, Cannons AC, Cone MR, Hogan KN, Kopp EW, Anzinger JJ, Garcia KF, Parham LA, Ramírez RMG, Montoya MCM, Rojas DP, Brown CM, Hennigan S, Sabina B, Scotland S, Gangavarapu K, Grubaugh ND, Oliveira G, Robles-Sikisaka R, Rambaut A, Gehrke L, Smole S, Halloran ME, Villar L, Mattar S, et al. . 2017. Zika virus evolution and spread in the Americas. Nature 546:411–415. doi: 10.1038/nature22402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broutet N, Krauer F, Riesen M, Khalakdina A, Almiron M, Aldighieri S, Espinal M, Low N, Dye C. 2016. Zika virus as a cause of neurologic disorders. N Engl J Med 374:1506–1509. doi: 10.1056/NEJMp1602708. [DOI] [PubMed] [Google Scholar]
- 4.Rasmussen SA, Jamieson DJ, Honein MA, Petersen LR. 2016. Zika virus and birth defects—reviewing the evidence for causality. N Engl J Med 374:1981–1987. doi: 10.1056/NEJMsr1604338. [DOI] [PubMed] [Google Scholar]
- 5.Mlakar J, Korva M, Tul N, Popović M, Poljšak-Prijatelj M, Mraz J, Kolenc M, Resman Rus K, Vesnaver Vipotnik T, Fabjan Vodušek V, Vizjak A, Pižem J, Petrovec M, Avšič Županc T. 2016. Zika virus associated with microcephaly. N Engl J Med 374:951–958. doi: 10.1056/NEJMoa1600651. [DOI] [PubMed] [Google Scholar]
- 6.Li C, Xu D, Ye Q, Hong S, Jiang Y, Liu X, Zhang N, Shi L, Qin C, Xu Z. 2016. Zika virus disrupts neural progenitor development and leads to microcephaly in mice. Cell Stem Cell 19:120–126. doi: 10.1016/j.stem.2016.04.017. [DOI] [PubMed] [Google Scholar]
- 7.Bowen JR, Quicke KM, Maddur MS, O Neal JT, McDonald CE, Fedorova NB, Puri V, Shabman RS, Pulendran B, Suthar MS. 2017. Zika virus antagonizes type I interferon responses during infection of human dendritic cells. PLoS Pathog 13:e1006164. doi: 10.1371/journal.ppat.1006164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaudhary V, Yuen KS, Chan JF, Chan CP, Wang PH, Cai JP, Zhang S, Liang M, Kok KH, Chan CP, Yuen KY, Jin DY. 2017. Selective activation of type II interferon signaling by Zika virus NS5 protein. J Virol 91:e00163-17. doi: 10.1128/JVI.00163-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang Q, Luo Z, Zeng J, Chen W, Foo S, Lee S, Ge J, Wang S, Goldman SA, Zlokovic BV, Zhao Z, Jung JU. 2016. Zika virus NS4A and NS4B proteins deregulate Akt-mTOR signaling in human fetal neural stem cells to inhibit neurogenesis and induce autophagy. Cell Stem Cell 19:663–671. doi: 10.1016/j.stem.2016.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding Q, Gaska JM, Douam F, Wei L, Kim D, Balev M, Heller B, Ploss A. 2018. Species-specific disruption of STING-dependent antiviral cellular defenses by the Zika virus NS2B3 protease. Proc Natl Acad Sci U S A 115:E6310–E6318. doi: 10.1073/pnas.1803406115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scaturro P, Stukalov A, Haas DA, Cortese M, Draganova K, Płaszczyca A, Bartenschlager R, Götz M, Pichlmair A. 2018. An orthogonal proteomic survey uncovers novel Zika virus host factors. Nature 561:253–257. doi: 10.1038/s41586-018-0484-5. [DOI] [PubMed] [Google Scholar]
- 12.Ing YL, Leung IW, Heng HH, Tsui LC, Lassam NJ. 1994. MLK-3: identification of a widely-expressed protein kinase bearing an SH3 domain and a leucine zipper-basic region domain. Oncogene 9:1745–1750. [PubMed] [Google Scholar]
- 13.Hartkamp J, Troppmair J, Rapp UR. 1999. The JNK/SAPK activator mixed lineage kinase 3 (MLK3) transforms NIH 3T3 cells in a MEK-dependent fashion. Cancer Res 59:2195–2202. [PubMed] [Google Scholar]
- 14.Hehner SP, Hofmann TG, Ushmorov A, Dienz O, Wing-Lan LI, Lassam N, Scheidereit C, Droge W, Schmitz ML. 2000. Mixed-lineage kinase 3 delivers CD3/CD28-derived signals into the IkappaB kinase complex. Mol Cell Biol 20:2556–2568. doi: 10.1128/MCB.20.7.2556-2568.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi W, Hou X, Li X, Peng H, Shi M, Jiang Q, Liu X, Ji Y, Yao Y, He C, Lei X. 2013. Differential gene expressions of the MAPK signaling pathway in enterovirus 71-infected rhabdomyosarcoma cells. Braz J Infect Dis 17:410–417. doi: 10.1016/j.bjid.2012.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang RX, Kong FY, Fan BF, Liu XM, You HJ, Zhang P, Zheng KY. 2012. HBx activates FasL and mediates HepG2 cell apoptosis through MLK3-MKK7-JNKs signal module. World J Gastroenterol 18:1485–1495. doi: 10.3748/wjg.v18.i13.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desmet EA, Hollenbaugh JA, Sime PJ, Wright TW, Topham DJ, Sant AJ, Takimoto T, Dewhurst S, Maggirwar SB. 2010. Mixed lineage kinase 3 deficiency delays viral clearance in the lung and is associated with diminished influenza-induced cytopathic effect in infected cells. Virology 400:224–232. doi: 10.1016/j.virol.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKenzie BA, Zemp FJ, Pisklakova A, Narendran A, McFadden G, Lun X, Kenchappa RS, Kurz EU, Forsyth PA. 2015. In vitro screen of a small molecule inhibitor drug library identifies multiple compounds that synergize with oncolytic myxoma virus against human brain tumor-initiating cells. Neuro Oncol 17:1086–1094. doi: 10.1093/neuonc/nou359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu M, Lee EM, Wen Z, Cheng Y, Huang W, Qian X, Tcw J, Kouznetsova J, Ogden SC, Hammack C, Jacob F, Nguyen HN, Itkin M, Hanna C, Shinn P, Allen C, Michael SG, Simeonov A, Huang W, Christian KM, Goate A, Brennand KJ, Huang R, Xia M, Ming G, Zheng W, Song H, Tang H. 2016. Identification of small-molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat Med 22:1101–1107. doi: 10.1038/nm.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng Y, Zhao H, Li X, Zhang N, Liu Z, Jiang T, Gu D, Shi L, He J, Wang H, Sun Z, Ye Q, Xie D, Cao W, Qin C. 2016. Isolation, identification and genomic characterization of the Asian lineage Zika virus imported to China. Sci China Life Sci 59:428–430. doi: 10.1007/s11427-016-5043-4. [DOI] [PubMed] [Google Scholar]
- 21.Goodfellow VS, Loweth CJ, Ravula SB, Wiemann T, Nguyen T, Xu Y, Todd DE, Sheppard D, Pollack S, Polesskaya O, Marker DF, Dewhurst S, Gelbard HA. 2013. Discovery, synthesis, and characterization of an orally bioavailable, brain penetrant inhibitor of mixed lineage kinase 3. J Med Chem 56:8032–8048. doi: 10.1021/jm401094t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komatsu T, Takeuchi K, Yokoo J, Tanaka Y, Gotoh B. 2000. Sendai virus blocks alpha interferon signaling to signal transducers and activators of transcription. J Virol 74:2477–2480. doi: 10.1128/jvi.74.5.2477-2480.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang G, Shi LZ, Chi H. 2009. Regulation of JNK and p38 MAPK in the immune system: signal integration, propagation and termination. Cytokine 48:161–169. doi: 10.1016/j.cyto.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kant S, Swat W, Zhang S, Zhang Z, Neel BG, Flavell RA, Davis RJ. 2011. TNF-stimulated MAP kinase activation mediated by a Rho family GTPase signaling pathway. Genes Dev 25:2069–2078. doi: 10.1101/gad.17224711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barrows NJ, Campos RK, Powell ST, Prasanth KR, Schott-Lerner G, Soto-Acosta R, Galarza-Muñoz G, McGrath EL, Urrabaz-Garza R, Gao J, Wu P, Menon R, Saade G, Fernandez-Salas I, Rossi SL, Vasilakis N, Routh A, Bradrick SS, Garcia-Blanco MA. 2016. A screen of FDA-approved drugs for inhibitors of Zika virus infection. Cell Host Microbe 20:259–270. doi: 10.1016/j.chom.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adcock RS, Chu Y, Golden JE, Chung D. 2017. Evaluation of anti-Zika virus activities of broad-spectrum antivirals and NIH clinical collection compounds using a cell-based, high-throughput screen assay. Antiviral Res 138:47–56. doi: 10.1016/j.antiviral.2016.11.018. [DOI] [PubMed] [Google Scholar]
- 27.Li Z, Brecher M, Deng YQ, Zhang J, Sakamuru S, Liu B, Huang R, Koetzner CA, Allen CA, Jones SA, Chen H, Zhang NN, Tian M, Gao F, Lin Q, Banavali N, Zhou J, Boles N, Xia M, Kramer LD, Qin CF, Li H. 2017. Existing drugs as broad-spectrum and potent inhibitors for Zika virus by targeting NS2B-NS3 interaction. Cell Res 27:1046–1064. doi: 10.1038/cr.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou T, Tan L, Cederquist GY, Fan Y, Hartley BJ, Mukherjee S, Tomishima M, Brennand KJ, Zhang Q, Schwartz RE, Evans T, Studer L, Chen S. 2017. High-content screening in hPSC-neural progenitors identifies drug candidates that inhibit Zika virus infection in fetal-like organoids and adult brain. Cell Stem Cell 21:274–283. doi: 10.1016/j.stem.2017.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Micewicz ED, Khachatoorian R, French SW, Ruchala P. 2018. Identification of novel small-molecule inhibitors of Zika virus infection. Bioorg Med Chem Lett 28:452–458. doi: 10.1016/j.bmcl.2017.12.019. [DOI] [PubMed] [Google Scholar]
- 30.Xu H, Hassounah SA, Colby-Germinario SP, Oliveira M, Fogarty C, Quan Y, Han Y, Golubkov O, Ibanescu I, Brenner B, Stranix BR, Wainberg MA. 2017. Purification of Zika virus RNA-dependent RNA polymerase and its use to identify small-molecule Zika inhibitors. J Antimicrob Chemother 72:727–734. doi: 10.1093/jac/dkw514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rausch K, Hackett BA, Weinbren NL, Reeder SM, Sadovsky Y, Hunter CA, Schultz DC, Coyne CB, Cherry S. 2017. Screening bioactives reveals nanchangmycin as a broad spectrum antiviral active against Zika virus. Cell Rep 18:804–815. doi: 10.1016/j.celrep.2016.12.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pascoalino BS, Courtemanche G, Cordeiro MT, Gil LH, Freitas-Junior L. 2016. Zika antiviral chemotherapy: identification of drugs and promising starting points for drug discovery from an FDA-approved library. F1000Res 5:2523. doi: 10.12688/f1000research.9648.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Retallack H, Di Lullo E, Arias C, Knopp KA, Laurie MT, Sandoval-Espinosa C, Mancia Leon WR, Krencik R, Ullian EM, Spatazza J, Pollen AA, Mandel-Brehm C, Nowakowski TJ, Kriegstein AR, DeRisi JL. 2016. Zika virus cell tropism in the developing human brain and inhibition by azithromycin. Proc Natl Acad Sci U S A 113:14408–14413. doi: 10.1073/pnas.1618029113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Connor MA, Tisoncik-Go J, Lewis TB, Miller CJ, Bratt D, Moats CR, Edlefsen PT, Smedley J, Klatt NR, Gale M, Fuller DH. 2018. Early cellular innate immune responses drive Zika viral persistence and tissue tropism in pigtail macaques. Nat Commun 9:3371. doi: 10.1038/s41467-018-05826-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ornelas AM, Pezzuto P, Silveira PP, Melo FO, Ferreira TA, Oliveira-Szejnfeld PS, Leal JI, Amorim MM, Hamilton S, Rawlinson WD, Cardoso CC, Nixon DF, Tanuri A, Melo AS, Aguiar RS. 2017. Immune activation in amniotic fluid from Zika virus-associated microcephaly. Ann Neurol 81:152–156. doi: 10.1002/ana.24839. [DOI] [PubMed] [Google Scholar]
- 36.Sun Y, López CB. 2016. Preparation of respiratory syncytial virus with high or low content of defective viral particles and their purification from viral stocks. Bio-Protoc 6:e1820. doi: 10.21769/BioProtoc.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang B, Xi X, Lei X, Zhang X, Cui S, Wang J, Jin Q, Zhao Z. 2013. Enterovirus 71 protease 2Apro targets MAVS to inhibit anti-viral type I interferon responses. PLoS Pathog 9:e1003231. doi: 10.1371/journal.ppat.1003231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J, Deng C, Gu D, Li X, Shi L, He J, Zhang Q, Zhang B, Ye H. 2018. Development of a replicon cell line-based high throughput antiviral assay for screening inhibitors of Zika virus. Antiviral Res 150:148–154. doi: 10.1016/j.antiviral.2017.12.017. [DOI] [PubMed] [Google Scholar]
- 39.Ibrahim SH, Hirsova P, Tomita K, Bronk SF, Werneburg NW, Harrison SA, Goodfellow VS, Malhi H, Gores GJ. 2016. Mixed lineage kinase 3 mediates release of C-X-C motif ligand 10-bearing chemotactic extracellular vesicles from lipotoxic hepatocytes. Hepatology 63:731–744. doi: 10.1002/hep.28252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mattoli S, Marini M, Fasoli A. 1992. Expression of the potent inflammatory cytokines, GM-CSF, IL6, and IL8, in bronchial epithelial cells of asthmatic patients. Chest 101(Suppl):27S–29S. doi: 10.1378/chest.101.3_Supplement.27S. [DOI] [PubMed] [Google Scholar]
- 41.Sahu U, Choudhury A, Parvez S, Biswas S, Kar S. 2017. Induction of intestinal stemness and tumorigenicity by aberrant internalization of commensal non-pathogenic E. coli. Cell Death Dis 8:e2667. doi: 10.1038/cddis.2017.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morimoto K, Horio J, Satoh H, Sue L, Beach T, Arita S, Tooyama I, Konishi Y. 2011. Expression profiles of cytokines in the brains of Alzheimer's disease (AD) patients compared to the brains of non-demented patients with and without increasing AD pathology. J Alzheimers Dis 25:59–76. doi: 10.3233/JAD-2011-101815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Locati M, Deuschle U, Massardi ML, Martinez FO, Sironi M, Sozzani S, Bartfai T, Mantovani A. 2002. Analysis of the gene expression profile activated by the CC chemokine ligand 5/RANTES and by lipopolysaccharide in human monocytes. J Immunol 168:3557–3562. doi: 10.4049/jimmunol.168.7.3557. [DOI] [PubMed] [Google Scholar]
- 44.Li X, Niu Y, Cheng M, Chi X, Liu X, Yang W. 2016. AP1S3 is required for hepatitis C virus infection by stabilizing E2 protein. Antiviral Res 131:26–34. doi: 10.1016/j.antiviral.2016.04.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.