Chikungunya virus (CHIKV) is a medically important pathogen responsible for recent large-scale epidemics. The development of efficient therapies against CHIKV has been hampered by gaps in our understanding of how nonstructural proteins (nsPs) function to form the viral replicase and replicate virus RNA. Here we describe an extremely sensitive assay to analyze the effects of mutations on the virus RNA synthesis machinery in cells of both mammalian (host) and mosquito (vector) origin. Using this system, several lethal mutations in CHIKV nsP1 were shown to reduce but not completely block the ability of its replicase to synthesize viral RNAs. However, in contrast to related alphaviruses, CHIKV replicase was completely inactivated by mutations preventing palmitoylation of nsP1. These data can be used to develop novel, virus-specific antiviral treatments.
KEYWORDS: chikungunya virus, RNA polymerases, RNA replication, alphavirus, mosquito, replicase
ABSTRACT
Chikungunya virus (CHIKV) is a mosquito-borne alphavirus. It has a positive-sense RNA genome that also serves as the mRNA for four nonstructural proteins (nsPs) representing subunits of the viral replicase. Coupling of nsP and RNA synthesis complicates analysis of viral RNA replication. We developed trans-replication systems, where production of replication-competent RNA and expression of viral replicase are uncoupled. Mammalian and mosquito RNA polymerase I promoters were used to produce noncapped RNA templates, which are poorly translated relative to CHIKV replicase-generated capped RNAs. It was found that, in human cells, constructs driven by RNA polymerase I promoters of human and Chinese hamster origin performed equally well. In contrast, RNA polymerase I promoters from Aedes mosquitoes exhibited strong species specificity. In both mammalian and mosquito cells, novel trans-replicase assays had exceptional sensitivity, with up to 105-fold higher reporter expression in the presence of replicase relative to background. Using this highly sensitive assay to analyze CHIKV nsP1 functionality, several mutations that severely reduced, but did not completely block, CHIKV replicase activity were identified: (i) nsP1 tagged at its N terminus with enhanced green fluorescent protein; (ii) mutations D63A and Y248A, blocking the RNA capping; and (iii) mutation R252E, affecting nsP1 membrane anchoring. In contrast, a mutation in the nsP1 palmitoylation site completely inactivated CHIKV replicase in both human and mosquito cells and was lethal for the virus. Our data confirm that this novel system provides a valuable tool to study CHIKV replicase, RNA replication, and virus-host interactions.
IMPORTANCE Chikungunya virus (CHIKV) is a medically important pathogen responsible for recent large-scale epidemics. The development of efficient therapies against CHIKV has been hampered by gaps in our understanding of how nonstructural proteins (nsPs) function to form the viral replicase and replicate virus RNA. Here we describe an extremely sensitive assay to analyze the effects of mutations on the virus RNA synthesis machinery in cells of both mammalian (host) and mosquito (vector) origin. Using this system, several lethal mutations in CHIKV nsP1 were shown to reduce but not completely block the ability of its replicase to synthesize viral RNAs. However, in contrast to related alphaviruses, CHIKV replicase was completely inactivated by mutations preventing palmitoylation of nsP1. These data can be used to develop novel, virus-specific antiviral treatments.
INTRODUCTION
Genome replication of RNA viruses is carried out by an RNA-dependent RNA polymerase complex (replicase) that consists of one or more virus-encoded proteins and that may also incorporate host cell proteins. For positive-strand RNA viruses, the virus-encoded component(s) of replicase is directly translated from the RNA genome. Its expression depends on RNA replication and vice versa; RNA replication depends on the expression and functional activity of viral replicase. Thus, for these viruses, RNA synthesis and replicase expression are functionally coupled.
Alphaviruses (family Togaviridae) comprise a group of positive-strand RNA viruses that includes important human pathogens, such as Chikungunya virus (CHIKV), as well as several well-studied model viruses, including Semliki Forest virus (SFV) and Sindbis virus (SINV). Most alphaviruses are mosquito vectored (1) and can efficiently replicate in cells of vertebrate and arthropod origin. Alphavirus infection in vertebrate cells is highly cytotoxic. In contrast, infection of arthropod cells is noncytotoxic and results in a persistent low-level infection. Alphaviruses have RNA genomes that are approximately 12 kb in size with a 5′ cap and a 3′ poly(A) tail. The genome consists of two open reading frames (ORFs). The 3′ ORF encodes a structural polyprotein which is translated from a subgenomic (SG) RNA transcribed under the control of an SG promoter in infected cells (2). The virus-encoded replicase subunits, termed nonstructural proteins (nsPs), are expressed in the form of a nonstructural (ns) polyprotein precursor(s) (P123 and P1234) from the ORF located at the 5′ two-thirds of the virus genome (3). The ns polyprotein is proteolytically processed by nsP2, which has protease activity (4), initially resulting in a short-lived negative-strand RNA polymerase (P123 + nsP4) that is subsequently converted into a stable positive-strand RNA polymerase (nsP1 + nsP2 + nsP3 + nsP4) (5). All four nsPs, as well as most of their processing intermediates (P1234, P123, and P23), are strictly required for alphavirus RNA replication (3).
The replicative functions of alphavirus replicase proteins are relatively well studied. nsP1 is a virus-specific methyl- and guanylyltransferase and serves as a membrane anchor for virus replicase (6, 7). nsP2 is a papain-like cysteine protease (8), nucleoside triphosphatase (NTPase) (9), RNA triphosphatase (10), and RNA helicase (11). nsP3 of Old World alphaviruses binds cellular G3BP proteins that are required for RNA replication (12). It also contains an N-terminal macrodomain with the ability to bind ADP-ribose and remove it from mono-ADP-ribosylated substrates. This is crucial for viral RNA replication (13). nsP4 is the catalytic subunit of viral RNA polymerase (14, 15) and also has terminal adenylyltransferase activity (16). In addition, all of these proteins have a number of essential functions that are not directly linked to viral RNA replication. Thus, nsP1 has been reported to antagonize an antiviral protein, tetherin (17); nsP2 of Old World alphaviruses triggers the degradation of host cell RNA polymerase II (18) and antagonizes type I interferon signaling (19); and nsP3 modulates the phosphatidylinositol-3-kinase (PI3K)-Akt-mammalian target of rapamycin (mTOR) pathway (20) and is responsible for translational shutoff in infected cells (21). nsP4 is involved in suppression of the unfolded protein response (22). While the enzymatic functions of nsPs are well conserved for all alphaviruses, their nonreplicative functions exhibit a significant variation, depending on the virus and its host. For example, nsP2 is cytotoxic only in the case of Old World alphaviruses (23) and only in vertebrate cells (18). The multiple functions of nsPs, their variation between different alphaviruses, and the coupled nature of nsP expression and viral RNA replication complicate the analysis of the functional significance of nsPs and their different mutations, as well as investigation of the role of host cell interaction partners in alphavirus RNA replication.
Decoupling the viral replicase protein(s) expression from viral RNA synthesis represents a powerful approach to study viral RNA replication. This has been proven useful to study the replication of influenza virus (24, 25), the flavivirus Kunjin virus (26), and the nodavirus Flock House virus (27). In addition, the ability of viral replicase to amplify the RNA template provided in trans has also allowed construction of Saccharomyces cerevisiae yeast-based replication systems for bromo-, noda-, and tombusviruses (28–30). In the case of alphaviruses, decoupled replication systems are generally designed to amplify truncated RNA templates where the ns and structural ORFs of alphavirus genome are replaced by sequences encoding different reporter proteins; such systems are often referred to as trans-replication systems. trans-Replication systems have been developed for SINV, SFV, and CHIKV and used to study alphavirus replication complex biogenesis (31–33) and RNA template sequence requirements (34), for analysis of the impact of template length on the size of replication complexes (35), for tagging of different replicase proteins (36), and for analysis of the impact of different mutations introduced into replicase proteins on viral RNA replication in mammalian (8, 37, 38) and mosquito (39) cells. While these studies have universally found the alphavirus trans-replication systems to be efficient and robust tools, they have also revealed certain technical limitations. Namely, to produce replication-competent template RNA and mRNA for expression of replicase polyproteins, alphavirus trans-replication systems have traditionally used either bacteriophage T7 RNA polymerase and the corresponding promoters (33) or cellular RNA polymerase II promoters, such as the immediate early promoter of human cytomegalovirus (CMV) or the Aedes aegypti polyubiquitin promoter (Ubi) (36, 39). The major drawback of the use of bacteriophage T7 RNA polymerase promoters is that the use of such trans-replication systems is restricted to cell lines expressing T7 RNA polymerase, which are generally not available for cell types relevant for in vivo alphavirus infections. The use of promoters for cellular RNA polymerase II allows the use of a wider range of cell types. However, such systems suffer from reduced sensitivity, especially with regard to the replicase-mediated amplification of reporter activity used to replace the ns ORF of the virus genome and, thus, expressed from the full-length RNA template. The effect is due to the high background activity of the reporter resulting from its efficient translation using RNA polymerase II-generated capped transcripts. This activity is often comparable to or even higher than the reporter activity produced from much more abundant viral replicase-generated full-length positive-strand RNAs (36, 39).
In this study, several approaches were applied to overcome the above-mentioned limitations of alphavirus trans-replicase systems. It was found that inserting the sequence corresponding to the 5′ end of the CHIKV genome directly at the start site of the human, Chinese hamster, Aedes aegypti, or Aedes albopictus RNA polymerase I promoter allowed production of replication-competent transcripts. These RNAs are, presumably due to the lack of the 5′ cap structure, poorly translated, resulting in significantly reduced background levels of included reporters and enhanced sensitivity of the trans-replication assay. The template constructs with human or Chinese hamster RNA polymerase I promoters were active in various primate cell lines. Higher species specificity was observed for template constructs harboring either the Aedes aegypti or Aedes albopictus RNA polymerase I promoter.
The increased sensitivity of the system was used to reevaluate 13 mutant versions of CHIKV replicase previously reported to be inactive or to have activities close to the background level. A set of mutations in CHIKV nsP1, including both mutations in the methyl- and guanylyltransferase active site and mutations affecting membrane anchoring of nsP1, was also analyzed using CHIKV trans-replicase and infectious clone systems. Highly consistent results confirmed some findings previously reported for SFV; however, it was also found that, unlike with SFV, mutation of the palmitoylation site of nsP1 in CHIKV is lethal in the context of the viral genome and completely abolishes its replicase activity. These findings confirm the improved trans-replication system to be an extremely sensitive and robust system to study alphavirus RNA replication.
RESULTS
trans-Replicases of alphaviruses allow analysis of the synthesis of full-length negative- and positive-strand copies of suitable template RNAs as well as SG RNAs transcribed from an SG promoter(s) included in template RNA. However, directly analyzing such RNAs using Northern blotting and/or strand-specific reverse transcription-quantitative PCR is time- and resource-consuming and not convenient for large-scale experiments, such as screening libraries of antiviral compounds. Incorporation of one or more reporter-coding sequences into template RNA constructs allows the use of the much more convenient measurement of reporter activities as a proxy for the synthesis of corresponding positive-strand RNAs and is also applicable in a high-throughput format. Therefore, in the current study, as previously (8, 36, 39), firefly luciferase (Fluc) was used as a substitute for most of the ns ORF and Gaussia luciferase (Gluc) was used as a substitute for the structural ORF in template constructs (Fig. 1B and 2B). For simplicity, here the full-length RNA serving as the template for Fluc translation is termed “genomic RNA” (and its synthesis is referred to as “replication”), RNA synthesized from the SG promoter that serves as the template for Gluc translation is termed “SG RNA” (and its synthesis is referred to as “transcription”), and all RNAs synthesized by CHIKV trans-replicase are referred to as “viral RNAs.”
FIG 1.
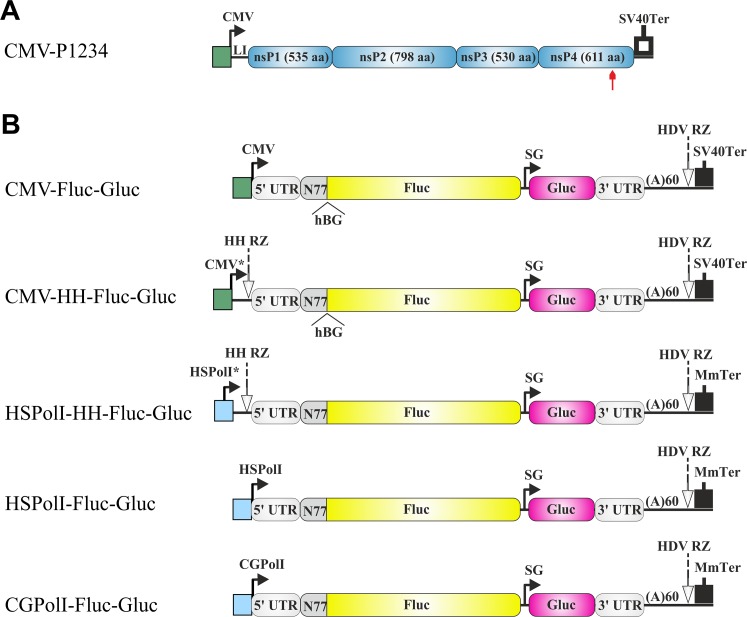
Schematic representation of the CMV-P1234 plasmid for CHIKV replicase expression and plasmids for expression of the RNA templates used in mammalian cells. (A) Expression construct for CHIKV ns proteins. CMV, CMV immediate early promoter; LI, leader region of the herpes simplex virus thymidine kinase gene with an artificial intron; SV40Ter, simian virus 40 late polyadenylation region. The red arrow highlights the position of the inactivating mutation in the nsP4 catalytic site. (B) Constructs expressing template RNAs. CMV*, CMV immediate early promoter followed by the TMV-derived leader; HSPolI*, full-length promoter (residues −211 to +20) for human RNA polymerase I (PolI); HSPolI, truncated promoter (residues −211 to −1) for human RNA polymerase I; CGPolI, truncated promoter (residues −227 to −1) for Chinese hamster RNA polymerase I; HH RZ, hammerhead ribozyme. The 5′ and 3′ UTRs are from CHIKV; N77, region encoding the 77 N-terminal amino acid residues of nsP1; SG, CHIKV SG promoter; HDV RZ, antisense-strand ribozyme of hepatitis delta virus; MmTer, terminator for RNA polymerase I from mouse (Mus musculus). The position of the second intron of the human beta-globin gene (hBG) in CMV-Fluc-Gluc and CMV-HH-Fluc-Gluc is marked. The vector backbone is not shown, and the drawings are not in scale.
FIG 2.
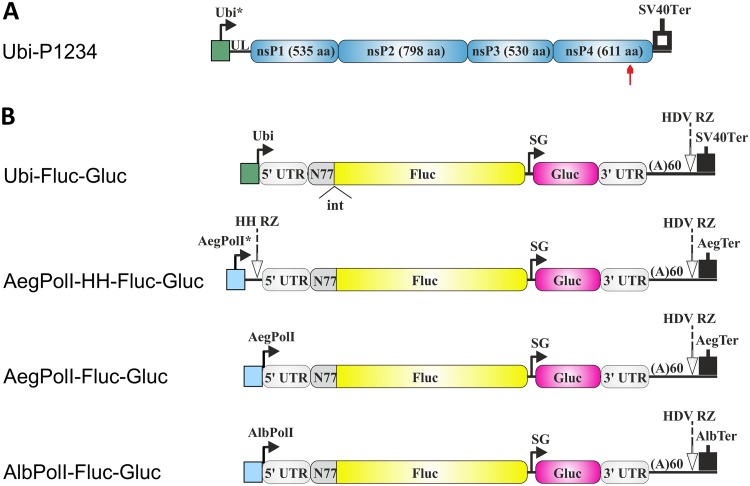
Schematic representation of the Ubi-P1234 plasmid for CHIKV replicase expression and plasmids for expression of the RNA templates used in mosquito cells. (A) Expression construct for CHIKV ns polyprotein. Ubi*, full-length Aedes aegypti polyubiquitin promoter; UL, transcribed leader of the polyubiquitin gene containing a naturally occurring intron; SV40Ter, SV40 late polyadenylation region. The red arrow highlights the position of the inactivating mutation in the nsP4 catalytic site. (B) Constructs expressing template RNAs. Ubi, truncated polyubiquitin promoter; AegPolI*, full-length promoter (residues −250 to +50) for Aedes aegypti RNA polymerase I; AegPolI, truncated promoter (residues −250 to −1) for Aedes aegypti RNA polymerase I; AlbPolI, truncated promoter (residues −250 to −1) for Aedes albopictus RNA polymerase I; HH RZ, hammerhead ribozyme; AegTer, tentative terminator for Aedes aegypti RNA polymerase I; AlbTer, tentative terminator for Aedes albopictus RNA polymerase I. The 5′ and 3′ UTRs are from CHIKV. N77, the region encoding the 77 N-terminal amino acid residues of nsP1; SG, CHIKV SG RNA promoter; HDV RZ, antisense-strand ribozyme of hepatitis delta virus. In Ubi-Fluc-Gluc, the position of the second intron of the Drosophila melanogaster alcohol dehydrogenase gene (int) is marked. The vector backbone is not shown, and the drawings are not in scale.
It has been observed that alphavirus trans-replication systems, where the initial template RNA subsequently used by virus replicase is produced by cellular RNA polymerase II, can be efficiently used for analysis of transcription but not of RNA replication (36, 39). The intrinsic problem is that although large amounts of genomic RNAs are synthesized from such templates by viral replicase (36), the activity of the Fluc reporter from such RNAs is masked by high background levels originating from the efficient translation of smaller amounts of initial capped template RNA transcripts. Therefore, the increase of Fluc activity is observed only for replicase mutants that boost viral RNA synthesis to levels considerably exceeding these achieved by wild-type (wt) replicase (39). Even the amplification of a signal for Gluc, which in commonly used cell lines is typically more than 1,000-fold for wt replicase (36), can be insufficient for the analysis of mutant replicases with severely reduced RNA synthesis activity. Therefore, systems with higher sensitivity and with a more prominent boost of Gluc and, especially, Fluc activities are required.
Design of plasmids for production of noncapped template RNAs.
It has been observed that the use of inefficiently translated template RNA transcripts synthesized by RNA polymerase of bacteriophage T7 increased the sensitivity of a trans-replication system (36). Importantly, these data also indicate that replicases of alphaviruses are capable of binding to and initiating the replication of template RNAs lacking a 5′ cap structure. This property was used in four new templates designed for use in mammalian cells. In the CMV-HH-Fluc-Gluc template, the leader sequence corresponding to residues 1 to 137 of tobacco mosaic virus (TMV), including the first 23 codons of its replicase ORF, followed by three terminator codons and a hammerhead ribozyme (HH RZ), was inserted between the start site of the CMV promoter and a nucleotide corresponding to the 5′ end of the CHIKV genome (Fig. 1B), so that the primary capped transcript has 193 nonviral nucleotides (Table 1) upstream of the CHIKV-specific sequence. Upon ribozyme cleavage, noncapped RNAs that have the authentic 5′ end of the CHIKV genome and that are therefore suitable for use by CHIKV replicase are generated. Similarly, in HSPolI-HH-Fluc-Gluc constructs, a full-length promoter for human RNA polymerase I was used to drive the transcription of initial template RNA, while HH RZ was used to generate the authentic 5′ end (Fig. 1B; Table 1). In addition, two constructs utilizing the design that has been successfully used to develop reverse genetics systems of negative-strand RNA viruses (40–44) were made. These constructs are HSPolI-Fluc-Gluc and CGPolI-Fluc-GLuc and use only the nontranscribed parts of RNA polymerase I promoters from humans and Chinese hamster, respectively (Fig. 1B; Table 1).
TABLE 1.
Sequences of promoter-CHIKV 5′ UTR junctions for plasmids expressing template RNAs
| Plasmid | Promoter residues −10 to −1 | Downstream part of promoter sequence | Leader sequence + HH RZa | CHIKV residues 1 to 10 |
|---|---|---|---|---|
| CMV-Fluc-Gluc | AGTGAACCGT | ATGGCTGCGT | ||
| CMV-HH-Fluc-Gluc | AGTGAACCGT | GTATTTTTACAACAATTACCAACAACAACAAACAACAAACAACATTAC AATTACTATTTACAATTACAATGGCATACACACAGACAGCTACCACAT CAGCTTTGCTGGACACTGTCCGAGGAAACAACTCCTTGGTCTAATAAT AAAGCCATCTGATGAGAGCGAAAGCTCGAAACTGGAGGAAACTCCAGTC |
ATGGCTGCGT | |
| HSPolI-HH-Fluc-Gluc | CCGGGTTATT | GCTGACACGCTGTCCTCTGG | AGCCATCTGATGAGAGCGAAAGCTCGAAACTGGAGGAAACTCCAGTC | ATGGCTGCGT |
| HSPolI-Fluc-Gluc | CCGGGTTATT | ATGGCTGCGT | ||
| CGPolI-Fluc-Gluc | TGACACGCTT | ATGGCTGCGT | ||
| Ubi-Fluc-Gluc | AAACCAGCTC | ATGGCTGCGT | ||
| AegPolI-HH-Fluc-Gluc | AAAACCCTTC | AGGGAGGAAGGCAGTGT GCGTGGACCGGCAGGAA AATGTTCCGAAAGCAA |
AGCCATCTGATGAGAGCGAAAGCTCGAAACTGGAGGAAACTCCAGTC | ATGGCTGCGT |
| AegPolI-Fluc-Gluc | AAAACCCTTC | ATGGCTGCGT | ||
| AlbPolI-Fluc-Gluc | AAAACCCTAT | ATGGCTGCGT |
The boldface sequence represents the HH RZ.
RNA polymerase I promoters and terminators have been mapped for both principal vectors of CHIKV, Aedes aegypti (45) and Aedes albopictus (46, 47) mosquitoes. Interestingly, in these promoters, the region with the highest sequence similarity is located immediately downstream of the transcription start site of rRNA (45). Three Aedes-specific template RNA-expressing constructs were designed. First, in the AegPolI-HH-Fluc-Gluc constructs, the full-length upstream promoter of Aedes aegypti RNA polymerase I and HH RZ were used (Table 1); the construct also contained the Aedes aegypti RNA polymerase I terminator (Fig. 2B). Next, in the template construct designated AegPolI-Fluc-Gluc, only the nontranscribed part of the promoter was used (Table 1). Finally, in the template construct designated AlbPolI-Fluc-Gluc, the Aedes aegypti RNA polymerase I promoter and terminator were replaced with their counterparts from Aedes albopictus (Table 1). The hepatitis delta virus negative-strand ribozyme (HDV RZ) sequence was added to ensure cleavage of all RNA templates immediately downstream of the poly(A) sequence (Fig. 1B and 2B).
Template RNA-encoding plasmids utilizing human and Chinese hamster RNA polymerase I promoters are efficient in primate cells.
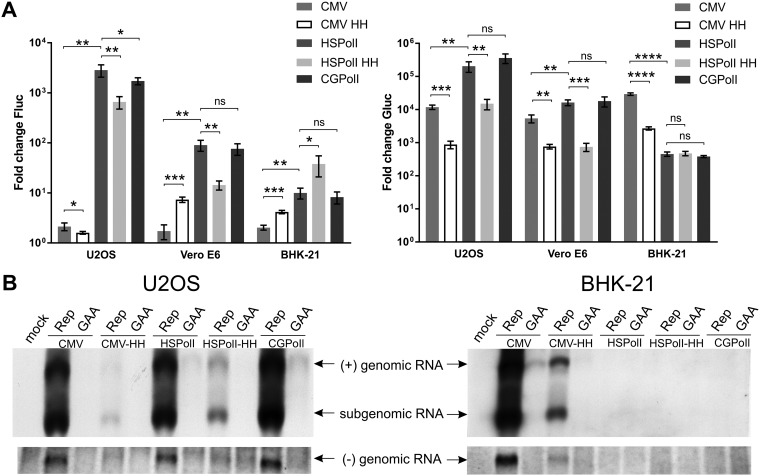
A panel of constructs consisting of CMV-Fluc-Gluc and four new templates (CMV-HH-Fluc-Gluc, HSPolI-Fluc-Gluc, HSPolI-HH-Fluc-Gluc, and CGPolI-Fluc-Gluc) was analyzed in two primate (U2OS [human], Vero E6 [African green monkey]) and two rodent (BHK-21 [Syrian golden hamster] and CHO [Chinese hamster]) cell lines. The Fluc and Gluc activities detected in CHO cells were always close to the background level, suggesting that CHIKV replicase works inefficiently, if at all, in CHO cells; for this reason, these cells were excluded from further analysis. Results obtained for the CMV-Fluc-Gluc template were consistent with those previously observed (36); the expression of the Fluc marker also occurred at a high level in cells cotransfected with CMV-Fluc-Gluc + CMV-P1234GAA, a negative control containing a polymerase-inactivating mutation in nsP4. The increase of Fluc activity in the presence of active (versus inactive) replicase was very low (∼2-fold) (Fig. 3A). In contrast, a strong boost of Gluc activity was observed in all cell lines, being the highest in U2OS and BHK-21 cells (∼20,000-fold) and slightly lower in Vero E6 cells (∼5,000-fold) (Fig. 3A).
FIG 3.
Comparison of template constructs in mammalian cells. (A) U2OS, Vero E6, and BHK-21 cells were all cotransfected with CMV-P1234 and one of CMV-Fluc-Gluc (CMV), CMV-HH-Fluc-Gluc (CMV HH), HSPolI-Fluc-Gluc (HSPolI), HSPolI-HH-Fluc-Gluc (HSPolI HH), or CGPolI-Fluc-Gluc (CGPolI). Control cells were all cotransfected with CMV-P1234GAA and the same template-expressing plasmids. Cells were lysed at 18 h posttransfection. The Fluc (replication, left) and Gluc (transcription, right) activities generated by the active replicase were normalized to those for the controls. Each column represents an average from three independent experiments; error bars represent standard deviations. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant (Student’s unpaired t test). (B) U2OS and BHK-21 cells were all cotransfected with CMV-P1234 and one of CMV-Fluc-Gluc, CMV-HH-Fluc-Gluc, HSPolI-HH-Fluc-Gluc, HSPolI-Fluc-Gluc, or CGPolI-Fluc-Gluc; control cells were all cotransfected with CMV-P1234GAA and the same template-expressing plasmids or were mock transfected. Samples were collected at 18 h posttransfection. RNA was analyzed by Northern blotting using a probe corresponding to the Fluc reporter gene to detect negative strands (bottom) or a probe complementary to the Gluc reporter gene to detect positive strands (top). “Genomic RNA” designates the full-length template RNA; note that an RNA of the same size is also synthesized by cellular RNA polymerases I and II and is therefore, for some promoters, also detectable in the presence of inactive replicases (GAA). “Subgenomic RNA” designates RNA synthesized by CHIKV replicase using the SG promoter. The experiment was repeated two times with similar results; data from one experiment are shown.
Adding a leader and HH RZ to the template reduced the Fluc background in control cells less than 10-fold. The high Fluc activity in the presence of inactive replicase indicates that additional TMV sequences and HH RZ cleavage were not sufficient to block Fluc translation. It is likely that CHIKV replicase cannot efficiently use templates with 193 additional residues upstream of the native 5′ end of the viral genome (Table 1). If so, boosting of expression of both reporters by active replicase (Fig. 3A) indicates that ribozyme-mediated cleavage did occur at some level. The low cleavage efficiency/speed apparently led to only small amounts of templates with correct 5′ ends in cells transfected with CMV-HH-Fluc-Gluc, which in turn resulted in reduced Fluc and, especially, Gluc activity in the presence of CHIKV-P1234. Consequently, the boost of Fluc expression remained low (Fig. 3A). Compared to those achieved for CMV-Fluc-Gluc, boosts of Gluc activities for CMV-HH-Fluc-Gluc were significantly lower in all three cell lines, typically less than 1,000-fold (Fig. 3A). Considering these findings together, it was concluded that while the approach used was not efficient enough to prevent Fluc translation from RNA polymerase II-generated transcripts, HH RZ was able to generate at least some amount of initial transcripts with proper 5′ ends that were subsequently utilized by CHIKV replicase.
The use of RNA polymerase I promoters greatly improved system performance. Although the absolute levels of Fluc and Gluc activities observed in cells transfected with HSPolI-Fluc-Gluc + CMV-P1234 or CMV-Fluc-Gluc + CMV-P1234 were similar in U2OS cells, the much lower background activity of both reporters expressed by HSPolI-Fluc-Gluc made the assay using the latter template generally much more sensitive. First and most importantly, the boost of Fluc activity was significantly increased in all cell lines used, being the highest (>2,000-fold) in U2OS cells (Fig. 3A). Second, the boost of Gluc activity also significantly increased in U2OS cells, where it exceeded 100,000-fold; the effect was less pronounced but still significant in Vero E6 cells. In contrast, in BHK-21 cells, CMV-Fluc-Gluc reached higher boost levels of Gluc expression than HSPolI-Fluc-Gluc (Fig. 3A). As CHIKV replicates well in both Vero E6 and BHK-21 cells, the observed trend most likely reflects a reduced activity of the human RNA polymerase I promoter in monkey cells and especially in rodent cells relative to that of the human-derived U2OS cells. The effect of incorporating HH RZ was consistent with the observation made above: expression of reporters in cells transfected with the HSPolI-HH-Fluc-Gluc template was reduced, which resulted in significantly diminished boost efficiencies of both markers in primate cells (Fig. 3A). Only in BHK-21 cells the boost of Gluc activity achieved through the use of HSPolI-HH-Fluc-Gluc was similar to that achieved through the use of HSPolI-Fluc-Gluc, and the boost of Fluc activity was even more prominent (Fig. 3A). Most likely, the presence of the downstream region of the human RNA polymerase I promoter allowed it to be used more efficiently in rodent cells.
Finally, construct CGPolI-Fluc-Gluc was tested in all three cell lines. Surprisingly, the construct performed poorly (∼8-fold boost of Fluc activity and ∼380-fold boost of Gluc activity) in BHK-21 cells. In contrast, it was highly active in U2OS and Vero E6 cells, showing boosts comparable to those observed for the HSPolI-Fluc-Gluc construct, suggesting that this rodent RNA polymerase I promoter is highly active in primate cells (Fig. 3A). Taken together, these experiments revealed U2OS cells and the HSPolI-Fluc-Gluc construct to be the most sensitive combination; therefore, it was selected for subsequent experiments.
Measurement of Fluc and Gluc activities is a convenient, but indirect, method for analysis of CHIKV replicase activity. To demonstrate that Fluc and Gluc activities indeed reflect the synthesis of viral positive-strand RNAs, Northern blot analysis was performed using transfected U2OS and BHK-21 cells. Negative-strand synthesis, which is not detectable through the measurement of reporter activities (34), was also assessed. The efficient synthesis of genomic and SG RNAs was observed in U2OS cells cotransfected with CMV-Fluc-Gluc, HSPolI-Fluc-Gluc, or CGPolI-Fluc-Gluc and CMV-P1234. The presence of HH RZ reduced positive-strand RNA synthesis for both CMV and HSPolI promoter-based vectors; again, most likely this indicates that the HH RZ is not efficient enough in generating RNAs with proper 5′ ends. The synthesis of negative-strand RNA was found to follow the same pattern (Fig. 3B, left). In BHK-21 cells, strong replication/transcription was observed only for CMV-Fluc-Gluc-derived template RNAs. The replication of template RNA generated from CMV-HH-Fluc-Gluc was diminished, and no replication products could be detected in cells transfected with CMV-P1234 and HSPolI-Fluc-Gluc, HSPolI-HH-Fluc-Gluc, or CGPolI-Fluc-Gluc (Fig. 3B, right). Taken together, these results demonstrate an excellent correlation between the levels of replicase-generated RNAs and the expression of reporters translated from viral positive-strand RNAs.
Template RNA-encoding plasmids utilizing RNA polymerase I promoters from Aedes mosquitoes display species specificity.
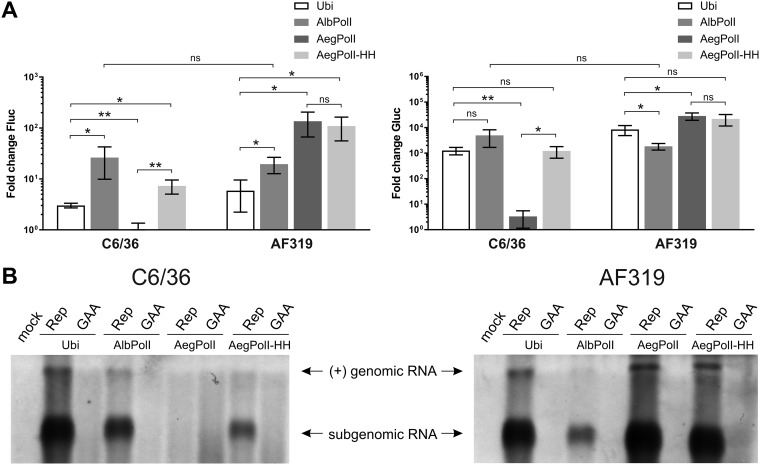
A panel consisting of Ubi-Fluc-Gluc and three new constructs (AegPolI-HH-Fluc-Gluc, AegPolI-Fluc-Gluc, and AlbPolI-Fluc-Gluc) was analyzed in Aedes albopictus mosquito-derived C6/36 cells and Aedes aegypti mosquito-derived AF319 cells. As previously observed (39), the background activity of the Fluc reporter in cells transfected with Ubi-Fluc-Gluc limited the boost of its expression by active CHIKV replicase; only an ∼3-fold or ∼6-fold increase of Fluc activity was observed in C6/36 and AF319 cells, respectively (Fig. 4A). In contrast, the boost of Gluc activity was ∼800-fold in C6/36 cells and ∼9,000-fold in AF319 cells (Fig. 4A).
FIG 4.
Comparison of template constructs in mosquito cells. (A) C6/36 and AF319 cells were all cotransfected with Ubi-P1234 and one of Ubi-Fluc-Gluc (Ubi), AegPolI-Fluc-Gluc (AegPolI), AegPolI-HH-Fluc-Gluc (AegPolI HH), or AlbPolI-Fluc-Gluc (AlbPolI). Control cells were all cotransfected with Ubi-P1234GAA and the same template-expressing plasmids. Cells were lysed at 48 h posttransfection. The Fluc (replication, left) and Gluc (transcription, right) activities generated by the active replicase were normalized to those for the controls. Each column represents an average from three independent experiments; error bars represent standard deviations. *, P < 0.05; **, P < 0.01; ns, not significant (Student’s unpaired t test). (B) C6/36 and AF319 cells were all cotransfected with Ubi-P1234 and one of Ubi-Fluc-Gluc, AlbPolI-Fluc-Gluc, AegPolI-Fluc-Gluc, or AegPolI-HH-Fluc-Gluc; control cells were all cotransfected with Ubi-P1234GAA and the same template-expressing plasmids or were mock transfected. Samples were collected at 48 h posttransfection. Positive-strand RNAs were revealed, and the data are presented as described in the legend to Fig. 3B, except that 5-fold more total RNA and a longer exposure were used to obtain the image. The experiment was repeated two times with similar results; data from one experiment are shown.
Both the AegPolI-HH-Fluc-Gluc and AegPolI-Fluc-Gluc constructs were highly active in AF319 cells. Again, the use of the RNA polymerase I promoter reduced the background levels of both reporters and, subsequently, increased the boost of both markers: a highly increased (∼100-fold) boost of Fluc activity and a more slightly, but still significantly, increased (∼20,000-fold) boost of Gluc activity were observed (Fig. 4A). Unlike in human cells, where the construct lacking hammerhead ribozyme significantly outperformed the HH RZ version (Fig. 3A), no significant difference between AegPolI-HH-Fluc-Gluc and AegPolI-Fluc-Gluc was observed in AF319 cells (Fig. 4A). This may indicate that in Aedes aegypti cells the presence of the downstream region of the RNA polymerase I promoter has a bigger impact on the activity of the RNA polymerase I promoter and, thus, compensates for the inefficient generation of initial transcripts with authentic 5′ ends by HH RZ cleavage. Alternatively, or in addition, it may indicate more efficient cleavage of primary transcripts by HH RZ, as a lower temperature (28°C instead of 37°C) may facilitate the formation of the ribozyme structure.
AlbPolI-Fluc-Gluc was also highly active and revealed little cell specificity: Fluc and Gluc boosts in homologous mosquito cells were only slightly higher (Fig. 4A), and the difference between mosquito cell lines was not statistically significant (P = 0.276 for Fluc and P = 0.090 for Gluc). In contrast, AegPolI-Fluc-Gluc was almost completely inactive in C6/36 cells. Surprisingly, the activity of AegPolI-HH-Fluc-Gluc in C6/36 cells was rather similar to that of Ubi-Fluc-Gluc (Fig. 4A). These data indicate that the upstream part of the RNA polymerase I promoter of Aedes aegypti is not sufficient to drive transcription in Aedes albopictus cells. However, the presence of highly conserved sequences located downstream of the transcription start site (45) compensates for this defect. Taken together, it was found that the most efficient template/mosquito cell combination was AegPolI-Fluc-Gluc and AF319 cells; for C6/36 cells, the best performance was observed for the AlbPolI-Fluc-Gluc template (Fig. 4A). These combinations were therefore selected for subsequent experiments.
To confirm that the observed differences in Fluc and Gluc reporter activities indeed reflect differences in the amount of viral RNAs, Northern blot analysis was performed. Again, an excellent correlation between the amount of replicase-generated RNAs and the expression levels of reporter proteins was observed. Thus, in C6/36 cells, the highest level of replicase-generated RNAs was observed for Ubi-Fluc-Gluc-derived templates, followed by AlbPolI-Fluc-Gluc-derived templates. Replicase-generated RNAs were also clearly detected in C6/36 cells transfected with AegPolI-HH-Fluc-Gluc but not with AegPolI-Fluc-Gluc (Fig. 4B, left). In AF319 cells, the highest RNA levels were observed when AegPolI-Fluc-Gluc or AegPolI-HH-Fluc-Gluc was used, with minimal differences between these two. High levels of replicase-generated RNAs were also detected in cells transfected with Ubi-Fluc-Gluc; in cells transfected with AlbPolI-Fluc-Gluc, their levels were much lower (Fig. 4B, right). It was also observed that, as with the reporter activities, the levels of replicase-made positive-strand transcripts in mosquito cells were lower than those in mammalian cells; in order to obtain comparable images, a 5-fold excess of mosquito cell-derived RNAs and a longer exposure of the Northern blot were required. Consistently, we were easily able to detect negative-strand RNAs in mammalian cells (Fig. 3B), while in mosquito cells, their levels were below the limit of detection of the assay.
CHIKV infection triggers replication of plasmid-encoded template RNAs.
Template RNA-encoding constructs also have the potential to serve as sensors of virus infection in vitro and in vivo (48). For efficient detection, an advanced sensor must possess high sensitivity (high on/off ratio) and, in the induced (infected) state, express the reporter(s) at a level suitable for detection. In the CHIKV trans-replicase system, both of these requirements are clearly met: expression of Fluc and Gluc markers occurs at a high level, and the on/off ratios are extremely high, especially for Gluc (>100,000-fold; Fig. 3A). However, a trans-replication system is different from natural virus infection. Therefore, the response of selected reporters to virus was also analyzed. For these experiments, U2OS cells and the CMV-Fluc-Gluc or HSPolI-Fluc-Gluc template as well as C6/36 cells and the Ubi-Fluc-Gluc or AlbPolI-Fluc-Gluc template were used.
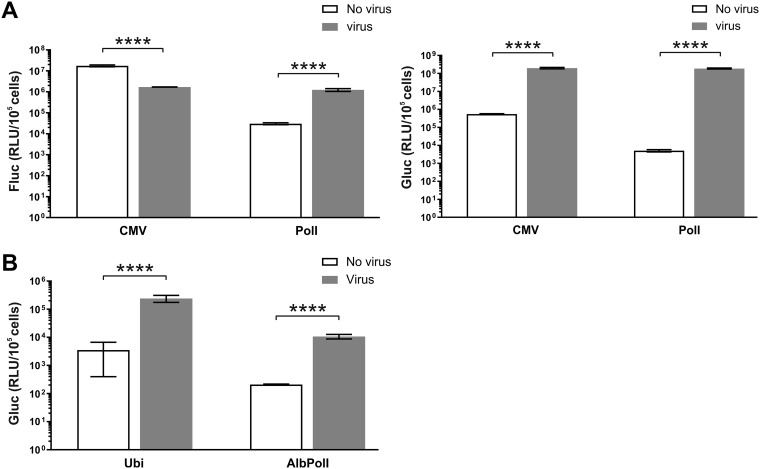
Infection of U2OS cells transfected with CMV-Fluc-Gluc resulted in a significant (∼10-fold) decrease of Fluc activity (Fig. 5A, left). Most likely, this effect was caused by inhibition of expression of Fluc from capped transcripts produced in the nucleus, either due to shutdown of their synthesis by RNA polymerase II or because of virus-induced translational shutdown. In contrast, expression of the Gluc marker increased ∼350-fold, indicating efficient synthesis of Gluc-expressing SG RNAs by incoming virus replicase (Fig. 5A, left). In U2OS cells transfected with HSPolI-Fluc-Gluc, CHIKV infection significantly increased expression of both markers. Expression of Fluc was boosted ∼50-fold. The effect, opposite that observed in cells transfected with CMV-Fluc-Gluc, presumably derives from the much lower background level of Fluc activity in noninfected cells. It is also possible that the RNA polymerase I is less susceptible to CHIKV infection-induced degradation, which might then contribute to the observed effect. The boost of the Gluc marker was, as expected, even more prominent (∼30,000-fold increase).
FIG 5.
CHIKV infection triggers replication and transcription of template RNAs produced from the CMV-Fluc-Gluc, HSPolI-Fluc-Gluc, Ubi-Fluc-Gluc, and AlbPolI-Fluc-Gluc plasmids. (A) U2OS cells were transfected with the CMV-Fluc-Gluc or HSPolI-Fluc-Gluc plasmid. At 18 h posttransfection, cells were either infected with CHIKV at an MOI of 10 or mock infected. Cells were collected at 24 hpi and lysed, and Fluc (replication, left) and Gluc (transcription, right) activities were measured and normalized to the number of transfected cells. (B) C6/36 cells were transfected with the Ubi-Fluc-Gluc or AlbPolI-Fluc-Gluc plasmid. At 36 h posttransfection, cells were either infected with CHIKV at an MOI of 10 or mock infected. Cells were collected at 72 hpi and lysed, and Gluc activities were measured and normalized to the number of transfected cells. Each column represents an average from at least three independent experiments; error bars represent standard deviations. ****, P < 0.0001 (Student’s unpaired t test). RLU, relative light units.
Infection of C6/36 cells transfected with Ubi-Fluc-Gluc or AlbPolI-Fluc-Gluc resulted in a minor increase of Fluc activity. Although the effect was statistically significant, the increase of Fluc activity upon infection never exceeded 2-fold. As in mammalian cells, the boost of Gluc activity was more prominent, ∼55-fold for both vector types, and highly significant (Fig. 5B). The absolute values of Gluc activity in cells transfected with Ubi-Fluc-Gluc were ∼15-fold higher than those in cells transfected with AlbPolI-Fluc-Gluc (Fig. 5B). This correlates with the observation that the SG RNA levels in the trans-replication system were also higher when Ubi-Fluc-Gluc was used as the source of template RNA for CHIKV replicase (Fig. 4B, left). It may indicate that in C6/36 cells the polyubiquitin promoter is stronger than the truncated Aedes albopictus RNA polymerase I promoter.
trans-Replication systems utilizing RNA polymerase I can be used to distinguish lethal mutations from ones having a strong negative impact on CHIKV RNA synthesis.
Many of the mutations introduced into the alphavirus ns proteins tend to reduce the infectivity of the mutant viruses by 1,000- to 10,000-fold (38, 49). For that reason, the sensitivity of the systems relying on the use of the CMV-Fluc-Gluc template in U2OS cells or the T7-Fluc-Gluc template in BSR cells may not be sufficient to distinguish between truly lethal (completely blocking RNA replication) and strongly attenuating mutations which often allow virus to be rescued and undergo reversion/pseudoreversion or compensation during subsequent replication. Based on this consideration, we reevaluated the phenotypes of 12 CHIKV replicase mutants previously reported to be inactive or possess strongly reduced replicase activity.
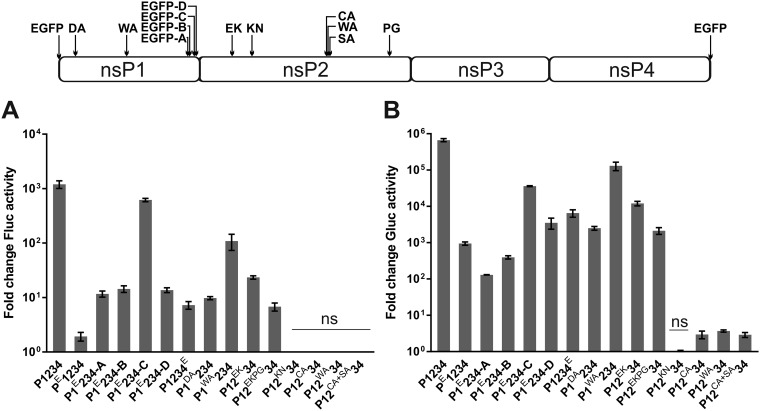
Analysis of mutants harboring enhanced green fluorescent protein (eGFP) at the C-terminal region of nsP1 confirmed previous findings. Insertion of eGFP after amino acid residue 516 of nsP1 (P1E234-C) was considerably better tolerated than similar insertions after residue 492, 497, or 525 (P1E234-A, P1E234-B, and P1E234-D, respectively) (Fig. 6A and B). It was also observed that from these three mutants the P1E234-D mutant displayed the highest transcription activity in U2OS cells (Fig. 6B). In contrast, in BSR cells, it had the lowest transcription activity (36). Most likely, this reflects a host cell-specific effect of the mutation. Indeed, we have previously observed that in rodent cells, CHIKV replicase mutations tend to have a more prominent effect on the trans-replicase activity than in human cells (39). The ability of a mutant replicase with eGFP attached to the C terminus of nsP4 (P1234E) to perform low-level (activity ∼1% of that of wt replicase) replication and transcription was also confirmed (Fig. 6A and B); this is consistent with the previous observation that virus harboring a similar insertion is viable but rapidly loses the inserted marker (36). A newly constructed replicase harboring eGFP at the N terminus of nsP1 displayed Fluc activity of only ∼2-fold over the background (activity in the presence of CMV-P1234GAA); however, this difference was statistically significant (P < 0.01). The transcription was better detectable (the boost of Gluc activity was ∼900-fold over the background) and highly significant (P < 0.0001). Thus, addition of eGFP to the N terminus of nsP1 strongly inhibits, but does not completely block, the activity of CHIKV replicase. A virus harboring the corresponding insertion might therefore be viable but likely also highly unstable.
FIG 6.
Reevaluation of activities of replicase mutants possessing no or strongly reduced RNA synthesis abilities. U2OS cells were all cotransfected with HSPolI-Fluc-Gluc and one of CMV-PE1234, CMV-P1E234-A, CMV-P1E234-B, CMV-P1E234-C, CMV-P1E234-D, CMV-P1234E, CMV-P1DA234, CMV-P1WA234, CMV-P12EK34, CMV-P12EKPG34, CMV-P12KN34, CMV-P12CA34, CMV-P12WA34, or CMV-P12CA+SA34. Control cells were cotransfected with HSPolI-Fluc-Gluc and CMV-P1234 or CMV-P1234GAA. Cells were lysed at 18 h posttransfection. Fluc (replication) (A) and Gluc (transcription) (B) activities generated by the active replicase were normalized to those for the controls. Each column represents an average from three independent experiments; error bars represent standard deviations. The names of the mutant polyproteins expressed by replicase expression plasmids are indicated below the graphs. With the exception of those marked ns (not significant), all other mutants showed activity significantly higher than that of the negative control (P1234GAA) (Student’s unpaired t test).
A mutation in the active site of the capping enzyme (P1DA234) allowed both Fluc (Fig. 6A) and Gluc (Fig. 6B) marker expression to occur at levels significantly above the background (P < 0.0001 and P < 0.001, respectively), clearly indicating the synthesis of CHIKV positive-strand RNAs. As for this mutant, the replicase-made RNAs are presumably noncapped and therefore poorly translated, and their relative amounts were probably even higher than those that could be deduced from the increase of reporter expression. This finding clearly supports the hypothesis that the synthesis of alphavirus positive-strand RNAs can occur in the absence of nsP1 capping activity (37, 39). The effects observed for the mutation in the membrane binding peptide of nsP1 (P1WA234) and for mutations known to reduce cytotoxicity (P12EK34, P12EKPG34) (Fig. 6A and B) were consistent with previous findings (36, 39).
A mutation in the active site of nsP2 helicase/NTPase/triphosphatase (P12KN34) was clearly lethal for CHIKV, as no activity above the background was displayed (Fig. 6A and B). Interestingly, a similar mutation in the context of SFV has been reported to allow reversions to occur (50). If so, the effect of this mutation is different among different alphaviruses. As the properties of the P1DA234 mutant suggest that RNA capping is not an absolute requirement for RNA synthesis, it can be concluded that it was a lack of the RNA helicase and/or NTPase activity of nsP2 that blocked the synthesis of CHIKV positive-strand RNAs. No Fluc expression above the background was observed for the P12CA34, P12WA34, and P12CA+SA34 mutants, which have mutations in the active site of nsP2 protease. This is in agreement with our previous findings that CHIKV harboring such mutations is not viable (8). All three mutants displayed very weak (∼3-fold over the background) but still significant (P values, <0.01, <0.0001, and <0.01, respectively) transcription activities. It is possible that this small activity may reflect some minor ability of unprocessed P1234 to perform viral RNA synthesis and/or that the bond between nsP3 and nsP4 in P1234 undergoes very inefficient spontaneous hydrolysis or cleavage by cellular enzymes. Considering the findings together, the trans-replication system used here allowed the clear separation of lethal replicase mutations from those that allow virus to be rescued. This conclusion is supported by data obtained using the RNA polymerase I-based trans-replication system for analysis of the effects of mutations in the helicase domain of CHIKV nsP2 (51). Similarly, in difficult-to-transfect murine neuronal cells, the differences between lethal and viable mutants of the nsP3 macrodomain were clearly observed (13).
Mutations in membrane binding regions of nsP1 have a strong negative effect on CHIKV RNA replication and infectivity.
In order to perform a detailed analysis of CHIKV nsP1 functions, the D63A mutation was introduced into Ubi-P1234 and into CMV-ICRES1, an infectious cDNA (icDNA) clone of CHIKV. In addition, three additional mutations were introduced into CMV-ICRES1, CMV-P1234, and Ubi-P1234. First, Tyr248 was replaced with Ala (Y248A). This residue is located in the membrane binding peptide region of nsP1 of alphaviruses and is important for the membrane binding of nsP1 (52, 53) and/or for its activity as a capping enzyme (37, 54). Second, Arg252, located inside the membrane binding peptide, was replaced with Glu (R252E). Third, Cys residues 417 to 419, representing the palmitoylation site of CHIKV nsP1, were replaced with Ala residues (3C3A). All these mutations have been previously studied in the context of SFV trans-replicase and, with the exception of D64A (in SFV, numeration is based on the amino acid residue numbers in its nsP1), also in the context of SFV icDNA. Briefly, in the SFV trans-replicase system, none of the mutants allowed positive-strand RNA synthesis above the background level; however, mutations D64A and Y249A did allow negative-strand RNA synthesis and spherule formation (37). In the context of the SFV genome, the 3C3A mutation allowed virus rescue (55) and the generation of second-site adaptive mutations (56), while all three mutations in the membrane binding peptide were lethal (57). As we have already revealed, the phenotypes of SFVW259A and CHIKVW258A are clearly different (39). It was also very recently reported that mutation of C417-C419 to A417-A419 in the context of the CHIKV 181/25 genome results in a drastic reduction of virus RNA levels in cells transfected with transcripts from a corresponding mutant icDNA (58). However, the authors of this study did not report the presence or absence of infectious virus progeny. Therefore, it was of interest to compare the effects of nsP1 mutations on the rescue and RNA replication of SFV and CHIKV.
The infectious center assay (ICA) revealed that only the CMV-ICRES1DA clone was able to form plaques; however, its infectivity was ∼100,000-fold lower than that of wt CHIKV. The lack of infectivity for CMV-ICRES1YA, CMV-ICRES1RE, and CMV-ICRES13C3A was confirmed by Western blotting, which could not reveal the presence of capsid protein in cells transfected with any of these three plasmids. In contrast, synthesis of the capsid protein was observed for CHIKVD63A, consistent with the data on infectious virus rescue (Fig. 7A). Sequencing of the mutated region in the genome of rescued CHIKVDA revealed, however, that the introduced mutation had reverted. Nevertheless, these data clearly confirm that the lack of capping activity did not completely prevent positive-strand RNA synthesis, which, in turn, allowed reversion to take place. As no infectious progeny was found for CMV-ICRES13C3A, it was also concluded that, in contrast to SFV, the mutation of the nsP1 palmitoylation site is lethal for CHIKV.
FIG 7.
Effects of mutations in nsP1 of CHIKV for infectious virus rescue and trans-replicase activities in mammalian and mosquito cells. (A) BHK-21 cells were transfected with one of the following plasmids: CMV-ICRES1, CMV-ICRES1DA, CMV-ICRES1YA, CMV-ICRES1RE, or CMV-ICRES13C3A. (Left) Results of ICA. (Right) Western blot of lysates from transfected cells collected at 24 h posttransfection. The CHIKV capsid protein was revealed by the corresponding rabbit polyclonal antiserum; β-actin was used as a loading control. Data from one reproducible experiment out of two independent experiments are shown. (B) U2OS cells were all cotransfected with HSPolI-Fluc-Gluc and one of CMV-P1234, CMV-P1DA234, CMV-P1YA234, CMV-P1RE234, CMV-P1WA234, CMV-P13C3A234, or CMV-P1234GAA. Samples were collected at 18 h posttransfection. Production of positive-strand RNAs was estimated by measuring the activities of Fluc (left) and Gluc (right) as described in the legend to Fig. 3A. Each column represents an average from three independent experiments; error bars represent standard deviations. Viral protein expression was verified by Western blotting using anti-nsP1 antiserum. (C) AF319 cells were all cotransfected with AegPolI-Fluc-Gluc and one of Ubi-P1234, Ubi-P1DA234, Ubi-P1YA234, Ubi-P1RE234, Ubi-P1WA234, Ubi-P13C3A234, or Ubi-P1234GAA. Samples were collected at 48 h posttransfection. Production of positive-strand RNAs was estimated by measuring the activities of Fluc (left) and Gluc (right) as described in the legend to Fig. 3A. Each column represents an average from three independent experiments; error bars represent standard deviations. Viral protein expression was verified by Western blotting as described in the legend to panel B.
Next, the effects of these mutants were analyzed in U2OS cells using the HSPolI-Fluc-Gluc template and CMV-P1234 replicase plasmids. With the exception of nsP1RE, all nsP1 proteins were found to be expressed at similar levels (Fig. 7B). As the R253E mutation has a similar effect on SFV nsP1 (37), it can be hypothesized that nsP1RE fails to bind to membranes and this results in destabilization of the protein. The CMV-P1WA234 mutant was able to boost both Gluc and Fluc expression (∼60,000-fold and ∼150-fold, respectively). In both cases, the difference in the levels achieved using wt CMV-P1234 was ∼5-fold (Fig. 7B). At 39°C, however, only an ∼800-fold boost of Gluc expression and an ∼4-fold boost of Fluc expression were observed for CMV-P1WA234, a difference from wt CMV-P1234 of >300-fold and highly significant (P < 0.0001) for both replication and transcription markers. Thus, the temperature-sensitive phenotype resulting from the W258A mutation, formerly revealed only for transcription (39), is clearly applicable to replication as well. Consistent with data from ICA and capsid protein expression, no replicase activity above the background level was observed for CMV-P13C3A234 and CMV-P1RE234 (Fig. 7B). When the assay was performed at 28°C, the CMV-P13C3A234 mutant remained completely inactive. In contrast, at a reduced temperature, Fluc and Gluc activities were boosted by CMV-P1RE234 ∼3.3-fold and ∼350-fold, respectively, and were both significantly above the background level (P < 0.01 for Fluc and P < 0.05 for Gluc). Thus, in the context of CHIKV replicase, the R252E mutation results in a strong temperature-sensitive phenotype. The substitution introduced into CMV-ICRESRE (CGC codon to GAG) requires two nucleotide substitutions for pseudoreversion (GAG to CGG or AGG Arg codons). Based on our experience, such mutations do not revert when introduced into alphavirus icDNAs; therefore, rescue of CMV-ICRES1RE at a permissive temperature (28°C) was not attempted. CMV-P1DA234 was, again, able to boost expression of both Fluc and Gluc (Fig. 7A), indicating the presence of sufficient replicase activity for virus rescue to occur. These activities were, apparently, reduced too strongly (compared to wt CMV-P1234 activity, ∼130-fold for replication and ∼230-fold for transcription) to allow preservation of the introduced mutation that could be reverted by the change of a single nucleotide residue (GCT for Ala to GAT for Asp). CMV-P1Y248A234 displayed even stronger attenuation, yet both Fluc and Gluc activities were clearly and significantly (P < 0.05) above the background levels (Fig. 7B). The failure to rescue CMV-ICRES1YA (Fig. 7A) is most likely due to the nature of the introduced substitution (TAC codon to GCC), as two nucleotide substitutions are required for reversion. Considering these findings together, in U2OS cells, only the palmitoylation site mutation resulted in a replicase that was completely inactive under all conditions used. The D63A, Y248A, and R252E substitutions all displayed some, albeit strongly reduced, replicase activity, at least under some of the tested conditions.
Finally, the same set of mutations was analyzed in Aedes aegypti-derived AF319 cells using the AegPolI-Fluc-Gluc template and Ubi-P1234 replicase plasmids. As in mammalian cells, all nsP1 proteins except nsP1RE were found to be expressed at similar levels (Fig. 7C). Again, only the W258A mutant generated a replication signal similar to that generated by the wt. These data do not, however, exclude the synthesis of positive-strand genomic RNAs by the D63A mutant (and, possibly, the Y248A mutant), and as for this mutant, the noncapped replicase-made RNAs would not have a translation advantage over the initial RNA polymerase I-made transcripts. Indeed, the analysis of the transcription signal, which offers higher sensitivity, revealed Gluc activities significantly above the background for the D63A (P < 0.05) and Y248A (P < 0.05) mutants, indicating the synthesis and translation of SG RNAs (Fig. 7C). Ubi-P1RE234 also displayed an ability to increase Gluc levels; however, in this case the boost was minor (∼3.3-fold) and not statistically significant (P = 0.167). Finally, no replication or transcription was detected for replicase encoded by Ubi-P13C3A234. The correlation of data from mammalian and mosquito cells indicates that defects caused by nsP1 mutations were not host cell specific. Our data also confirm that the phenotypes caused by the D63A and Y248A mutations in CHIKV and their counterparts in SFV are similar. This might also be the case for the R252E mutation, but the tools used to study it in the context of SFV had inadequate sensitivity. In contrast, the phenotype caused by the 3C3A mutation in the context of SFV and CHIKV was clearly different: in the former it must allow replicase activity necessary for virus rescue, while in the latter no evidence of RNA replication could be revealed by any of the highly sensitive assays.
DISCUSSION
The use of promoters and terminators of cellular RNA polymerase I resulted in a trans-replicase system of unprecedented sensitivity. The increased sensitivity was mostly due to the fact that the background level of reporter expression was considerably reduced with little (mosquito cells) or no (mammalian cells) loss of marker expression in the presence of viral replicase. Similarly, it may also be advantageous that transcripts made by RNA polymerase I do not undergo splicing. This may be important for sequences derived from the genomes of positive-strand RNA viruses, which often contain cryptic splicing signals. Use of an RNA polymerase I-based system may also improve the stability of RNA transcripts. RNAs made from alphavirus template constructs (Fig. 1B and 2B) have a bicistronic structure. If made by RNA polymerase II, these RNAs behave as mRNAs with an abnormally long 3′ untranslated region (UTR) that contains a nontranslated Gluc ORF and may therefore be recognized by the cellular mRNA degradation machinery. Finally, compared to promoters of RNA polymerase II, the RNA polymerase I promoter, which is normally used for the synthesis of rRNAs, is less likely to have strongly differential expression or silencing in different tissues.
A caveat for the construction of efficient RNA polymerase I-mediated template RNA expression constructs is species specificity, which was observed for both mammal-derived (Fig. 3) and Aedes mosquito-derived (Fig. 4) promoters. Interestingly, no clear correlation between promoter origin and the efficiency of the corresponding system in cells from different organisms was observed. Thus, a human RNA polymerase I promoter had no obvious advantage over a Chinese hamster polymerase I promoter in primate cells (Fig. 3) and was also found to work in murine cells (13). In contrast, none of these promoters worked efficiently in BHK-21 cells. The reasons for this remain unknown, though it can be speculated that the RNA polymerase I of the Syrian golden hamster cannot recognize the promoter fragments used. This species specificity represents a significant problem for the design of template RNA-expressing constructs for nonmodel species where the RNA polymerase I promoters/terminators are not well characterized. Given the low conservation of the sequences of these elements, an analysis of the RNA polymerase I-based transcription of these organisms may be required before efficient template RNA-producing constructs can be designed. An obvious way to alleviate at least some of these problems would be the use of ribozymes to trim the 5′ and 3′ ends of RNA transcripts. In this regard, it should be noticed that while we experienced no difficulties with trimming of the 3′ ends of transcripts with an HDV RZ, the use of HH RZ to generate authentic 5′ ends essentially failed in mammalian cells. However, it worked reasonably well in the case of mosquito cells, alleviating problems related to the observed species specificity (Fig. 4). Therefore, it can be considered a potential approach for species where the information about the RNA polymerase I promoter sequence and function is limited or absent.
Consistent with our previous findings (36), relatively little transcript was generated from CMV-Fluc-Gluc by RNA polymerase II, and these RNAs were poorly detectable by Northern blot analysis (Fig. 3B). This was also the case for Ubi-Fluc-Gluc transcripts in mosquito cells (Fig. 4B). In contrast, genomic RNAs synthesized by wt CHIKV replicase were abundant (Fig. 3B and 4B). However, the increase of RNA copy number resulted in a disproportionate boost of Fluc expression (Fig. 3A and 4A). Most likely, this lack of correlation between RNA and Fluc reporter levels is a consequence of a basic property of alphavirus infection: ns proteins are synthesized only at the early stages of infection, while the synthesis of their mRNAs (virus genomes) remains active until cell death (59). Hence, only a small amount of RNA genomes is used as mRNAs and in late infection the ratio of ns proteins to RNA genomes drops dramatically. Our trans-replication system appears to capture this important property of alphavirus infection, but this effect limits the use of genomic (Fluc) reporter activity for the analysis of genomic RNA synthesis. For the RNA polymerase II-based system, the nucleus-made capped RNA transcripts are the first and most efficiently translated mRNAs for Fluc. The increase in replicase-generated genomic RNAs, observed by Northern blotting, occurs at a later stage, when they are poorly translated, if at all. Therefore, in the RNA polymerase II-based system, translation of the genomic reporter (Fluc) from the initial transcripts can mask that from replicase-generated RNAs. The use of noncapped transcripts made by RNA polymerase I avoids this effect.
It was also observed that, in contrast to replication in mammalian cells, replication in mosquito cells resulted in an abnormally high SG RNA-to-genomic RNA ratio (Fig. 3B and 4B). This may relate to the use of a truncated 3′ UTR in the template RNAs. The native 3′ UTR of the CHIKV LR2006OPY1 strain comprises 498 nucleotide residues. In contrast, the template RNAs used in this study contained only the last 110 nucleotides (36). The missing part of the CHIKV 3′ UTR contains repeated sequence motifs which have only a minimal effect on CHIKV replication in mammalian cells (60). In contrast, however, a deletion of this region caused a prominent reduction of CHIKV replication in both Aedes aegypti and Aedes albopictus cells (61). To the best of our knowledge, the molecular basis of this defect has not been reported. Thus, it is plausible that the defect may include a shift of the SG RNA-to-genomic RNA ratio in favor of the former. This shift might reflect an altered ratio of synthesis or instability of bicistronic genomic RNA with a truncated 3′ UTR. If so, this trans-replication system may serve as a useful tool for studies on the role of the upstream part of the 3′ UTR in RNA synthesis and/or stability. Such an analysis was, however, outside the scope of the current study.
As shown here, alphavirus replicase can use RNA polymerase I-generated RNAs as templates; this emphasizes that the 5′ cap structure is not required for RNA replication. Even the ribozyme-generated 5′ ends that lack 5′ phosphate can still be used by alphavirus replicase. At the same time, it is well-known that in vitro-generated noncapped transcripts from alphavirus icDNAs are not infectious. Our data indicate that this is most likely a consequence of a low level of replicase protein synthesis rather than a lack of the ability of the replicase to use such transcripts as templates. If so, the role of the cap structure of viral RNA is to ensure the correct level of ns protein expression, stabilize the RNA in the cellular environment (62), and prevent its recognition by cellular pattern recognition receptors. It has recently been demonstrated that alphaviruses synthesize noncapped RNAs in a normal infection and that these RNAs are packed into virions (62). Furthermore, it was subsequently shown that an artificial increase of capping efficiency leads to the overproduction of viral nsPs and a decrease of virion formation (63). The authors of these studies also revealed that virions made in mammalian cells contain mostly noncapped RNAs, while virions made in mosquito cells contain mostly capped RNAs and therefore have lower particle-per-PFU ratios (62). These findings clearly correlate with the behavior of our trans-replication templates. In mammalian cells, the replicase of an incoming virus was able to use both capped and noncapped template RNAs equally well, resulting in similar final levels of marker expression (Fig. 5A). In C6/36 mosquito-derived cells, capped transcripts performed clearly better, reaching higher expression levels (Fig. 5B). This may indicate that in C6/36 cells the noncapped RNAs are used by viral replicase less efficiently than capped ones. The lack of a cap applies, however, only for the initial replication event, as CHIKV replicase facilitates the capping of the RNAs made by the formed replication complexes; hence, thereafter, the replication processes in the cell should proceed identically regardless of the capping status of the initial RNA transcripts. If so, the lower efficiency of replication (Fig. 5) could indicate that replication is initiated in relatively few C6/36 cells. A similar phenomenon has previously been observed in C6/36 cells transfected with a construct expressing the SINV template; upon infection with SINV, the replication of the RNA template was evident in only ∼5% of the infected cells (48). In line with these observations, we have identified several replicase mutations that boost replication of the CHIKV template RNA in mosquito cells between 10- and 30-fold (39). In contrast, a mutation in P1234 that is capable of boosting the replication of template RNA in mammalian cells considerably above the level achieved by wt P1234 has never been identified. All these findings indicate that initiation of template RNA replication in mosquito cells by virus-encoded replicase is relatively inefficient. Most likely, it is a consequence of the different environments in mammalian and mosquito cells. The initiation of template RNA replication is a complex process which includes RNA recognition by virus- and host-encoded replicase components, their interaction with host cell membranes, the formation and maturation of replication complexes, and so on; analysis of this multistep process is the topic of ongoing research in our laboratories, and different template RNA expression constructs represent valuable tools for such studies.
As shown in previous studies, alphavirus trans-replicase is an excellent tool for the analysis of nsP functions. Here we took advantage of the increased sensitivity of the RNA polymerase I-based system and analyzed the effects of different mutations on the ability of CHIKV nsP1 to support viral RNA synthesis. The analysis not only confirmed our previous findings that the C terminus of nsP1 tolerates insertion of the eGFP tag (36) but also revealed that such a tag can be incorporated into the N terminus of nsP1. Though the replicase harboring such a tag had severely diminished activity, the finding still clearly demonstrates that N-terminally tagged nsP1 is a functional protein. It is unclear why an insertion of eGFP into the N terminus of nsP1 has a higher negative impact on its function than a similar insertion in the C-terminal region. It may be a consequence of the N terminus proximity to important functional motifs, such as the catalytic His residue of guanylyltransferase and the catalytic Asp residue of methyltransferase, which are located only 37 and 63 amino acid residues downstream, respectively (64). At this point, it is not clear whether the similarity of the properties of PE1234 and P1DA234 (Fig. 5) is coincidental or reflects the similar nature of the defect (presumably, a lack of RNA capping) caused by these mutations. It was also found that mutations Y248A and R252E, counterparts of which have been shown to abolish the ability of SFV replicase to synthesize positive-strand RNAs (37), severely reduced but did not completely block CHIKV replicase activity. In these cases, the observed difference between SFV and CHIKV could most likely be explained by the more sensitive assay used in this study and the temperature-sensitive nature of the defect(s) caused by the R252E mutation. More likely than not, both of these mutations affect SFV and CHIKV in the same way. In contrast, the effect of a mutation in the palmitoylation site of CHIKV nsP1 was clearly different from that reported in SFV. No activity of this mutant CHIKV genome or its replicase was evident in our studies, indicating the lethal nature of the mutation. Interestingly, another defect of a similar type has also been described. The inability of nsP3 of SFV to bind cellular G3BP proteins results in an attenuated phenotype (65); in contrast, binding of G3BPs by nsP3 is crucial for CHIKV replication (12). nsP1 palmitoylation is required for correct interactions of alphavirus replicase proteins and their association with membranes (56). It has also been suggested that G3BPs participate in the formation of replicase complexes (12). Therefore, it can be speculated that both of these differences between CHIKV and SFV may have a common cause.
The replication and transcription of reporter-encoding template RNA have been used for the detection of SINV infection in mosquitoes. In vivo, up to a 10-fold activation of reporter expression upon infection by homologous virus was observed. Interestingly, when the same experiment was performed using transiently transfected C6/36 cells, the boost of marker expression was more modest, ∼2-fold (48). In a comparable setup, both Ubi-Fluc-Gluc- and AlbPolI-Fluc-Gluc-derived template RNAs showed an ∼55-fold boost of reporter expression (Fig. 5B), clearly outperforming the previously analyzed SINV template construct. Therefore, it is likely that the constructs reported in this study would have superior properties that would make them suitable for the generation of stable insect cell lines and transgenic mosquitoes. The same applies, likely to a much bigger extent, to mammalian cells and the HSPolI-Fluc-Gluc template expression construct. In a transient setup, a nearly 30,000-fold activation of Gluc expression was observed upon virus infection (Fig. 5A). As the background of the Gluc expression was negligible and the on/off ratio was as good as or superior to that of any inducible expression system reported to date, the design may have several potential uses. This includes in vitro applications, such as inducible expression of toxic proteins in cell culture or construction of cell lines for easy detection and quantification of alphavirus infection. In addition, a potential in vivo application might be the generation, through transgenesis, of animals carrying such an inducible reporter. This could, in principle, be used to monitor and trace virus infection with unprecedented sensitivity and accuracy.
MATERIALS AND METHODS
Cells.
All mammalian cell lines were maintained at 37°C in a humidified atmosphere with 5% CO2. U2OS human bone osteosarcoma cells (ATCC HTB-96) were maintained in Iscove’s modified Dulbecco’s medium (Gibco) containing 10% fetal bovine serum (FBS; GE Healthcare) and 2 mM l-glutamine. Vero E6 African green monkey kidney cells (ATCC CCL-81) were grown in Dulbecco’s modified Eagle’s medium (Gibco) containing 10% FBS and 2 mM l-glutamine. BHK-21 baby hamster kidney cells (ATCC CCL-10) were grown in Glasgow’s minimal essential medium (Gibco) containing 10% FBS, 2% tryptose phosphate broth (TPB), and 200 mM HEPES, pH 7.2. All mosquito-derived cell lines were maintained at 28°C with no additional CO2. Aedes albopictus C6/36 cells were maintained in Leibowitz’s L-15 medium (Corning) containing 10% FBS. The Aedes aegypti Aag2 cell-derived Dicer2 knockout cell line AF319 (66) was maintained in Leibowitz’s L-15 medium (Corning) containing 20% FBS, 10% TPB, and 1× nonessential amino acids. All media were supplemented with 100 U/ml penicillin and 0.1 mg/ml streptomycin.
Construction of plasmids for production of RNA templates in mammalian cells.
CMV-Fluc-Gluc, a vector designed for expression of replication-competent template RNA of CHIKV using cellular RNA polymerase II in mammalian cells, has been previously described (36). A 193-bp-long sequence, corresponding to 137 5′ residues of TMV including the first 23 codons of its replicase ORF, followed by 3 in-frame stop codons and an HH RZ designed to cleave RNA transcript immediately upstream of the residue corresponding to the 5′ end of CHIKV genome, was inserted between the start site of the CMV promoter and the beginning of a CHIKV-specific sequence in CMV-Fluc-Gluc using synthetic DNA fragments (GenScript, USA) and subcloning procedures; the generated plasmid was designated CMV-HH-Fluc-Gluc. Similarly, a synthetic DNA fragment containing sequences corresponding to the human RNA polymerase I promoter (from residues −211 to +20 with respect to the transcription start site) and HH RZ was used to replace the T7 RNA promoter in T7-Fluc-Gluc (36); a 100-bp-long sequence corresponding to the mouse RNA polymerase I terminator was inserted downstream of the sequence corresponding to HDV RZ. The generated plasmid was designated HSPolI-HH-Fluc-Gluc. The deletion of the sequence corresponding to the downstream region of the human RNA polymerase I promoter (residues +1 to +20) and HH RZ was performed using PCR-based mutagenesis, and the generated plasmid was designated HSPolI-Fluc-Gluc. Finally, the human RNA polymerase I promoter in HSPolI-Fluc-Gluc was replaced by the corresponding sequence (positions −227 to −1 with respect to the transcription start site) of the Chinese hamster (Cricetulus griseus), resulting in a plasmid designated CGPolI-Fluc-Gluc (Fig. 1B). The sequences of all plasmids were verified using Sanger sequencing. The sequences from residue −10 of the promoter to residue 10 of CHIKV are shown in Table 1; full sequences are available from the authors upon request.
Construction of plasmids for production of RNA templates in mosquito cells.
Ubi-Fluc-Gluc, a vector designed for the expression of replication-competent template RNA of CHIKV using cellular RNA polymerase II in mosquito cells, has been previously described (39). To obtain mosquito RNA polymerase I-based constructs, an intron, present in Ubi-Fluc-Gluc, was removed. As the RNA polymerase I promoters of Aedes aegypti and Aedes albopictus share little similarity (45, 46), separate vectors for template RNA production were constructed for cells derived from these two mosquito species. To obtain a vector for Aedes albopictus cells, the polyubiquitin promoter and simian virus 40 (SV40) terminators of Ubi-Fluc-Gluc were replaced with the Aedes albopictus RNA polymerase I promoter and putative terminator (100 bp), respectively. A 250-bp promoter fragment (residues −250 to −1) was used, such that the first nucleotide of the CHIKV genome corresponds to the transcription start. The generated plasmid was designated AlbPolI-Fluc-Gluc. Similar substitutions were made using the RNA polymerase I promoter (residues −250 to −1) and putative terminator (100 bp) of Aedes aegypti. This resulted in a plasmid designated AegPolI-Fluc-Gluc. Finally, the sequence corresponding to residues +1 to +50 of the RNA polymerase I promoter of Aedes aegypti followed by the sequence corresponding to an HH RZ was inserted between the start site of the promoter and the residue corresponding to the 5′ end of the CHIKV genome in AegPolI-Fluc-Gluc; the resulting plasmid was designated AegPolI-HH-Fluc-Gluc (Fig. 2B). The sequences of all plasmids were verified using Sanger sequencing. The sequences from residue −10 of the promoter to residue 10 of CHIKV are shown in Table 1; full sequences are available from the authors upon request.
Construction of plasmids for expression of mutant replicase and mutant icDNA constructs.
The construction of CMV-P1234, CMV-P1234GAA, CMV-P1E234-A, CMV-P1E234-B, CMV-P1E234-C, CMV-P1E234-D, CMV-P1234E, CMV-P12EK34, CMV-P12EKPG34, CMV-P12KN34, CMV-P1DA234, and CMV-P1WA234 has been previously described (36, 39). In order to generate the constructs designated CMV-P12CA34, CMV-P12WA34, and CMV-P12CASA34, the region corresponding to mutated nsP2 protease was transferred from the T7-P12CA34, T7-P12WA34, and T7-P12CA+SA34 plasmids (8) to the CMV-P1234 plasmid. To fuse eGFP to the N terminus of nsP1, a flexible Gly-Gly-Ser-Gly-Gly-Ser linker was added to the C terminus of eGFP. Using site-directed PCR mutagenesis and subcloning, a plasmid designated CMV-PE1234 was generated. Additional point mutations were incorporated in CMV-P1234 using site-directed PCR mutagenesis: Y248A (CMV-P1YA234), R252E (CMV-P1RE234), and replacement of cysteine residues 417 to 419 of nsP1 with alanine residues (CMV-P13C3A234). The last three mutations as well as the D63A substitution were also incorporated into CMV-ICRES1 (also called DREP-ICRES1), an icDNA clone of the CHIKV LR2006OPY1 isolate (67), using site-directed PCR mutagenesis and subcloning procedures. The resulting clones were designated CMV-ICRES1DA, CMV-ICRES1YA, CMV-ICRES1RE, and CMV-ICRES13C3A. The sequences of all plasmids were verified using Sanger sequencing.
trans-Replication assay.
The trans-replication assay was performed as previously described (36). Briefly, U2OS, Vero E6, and BHK-21 cells grown in 12-well plates were cotransfected with 1 μg of a template-expressing vector (CMV-Fluc-Gluc, CMV-HH-Fluc-Gluc, HSPolI-HH-Fluc-Gluc, HSPolI-Fluc-Gluc, or CGPolI-Fluc-Gluc) and 1 μg of CMV-P1234 (or its mutant variants) using the Lipofectamine LTX (Thermo Fisher Scientific) reagent according to the manufacturer’s instructions. Transfected cells were incubated at 37°C for 18 h. C6/36 and AF319 cells grown in 12-well plates were cotransfected with 0.5 μg of a template-expressing vector (Ubi-Fluc-Gluc, AegPolI-HH-Fluc-Gluc, AegPolI-Fluc-Gluc, or AlbPolI-Fluc-Gluc) and 0.5 μg of Ubi-P1234 (or its mutant variants) using Lipofectamine LTX and incubated at 28°C for 48 h. After incubation, the cells were lysed and Fluc and Gluc activities were measured using a dual-luciferase reporter assay on a Glomax SIS luminometer (Promega). All Fluc and Gluc activities were normalized to those obtained for cells cotransfected with plasmids expressing the corresponding template RNA and CMV-P1234GAA or Ubi-P1234GAA (for mammalian and mosquito cell experiments, respectively) controls. All assays were repeated at least three times.
Northern blotting.
U2OS, BHK-21, C6/36, and AF319 cells were cotransfected with plasmids coding for CHIKV replicase and the RNA template as described above. At 18 h (U2OS, BHK-21) or 48 h (C6/36, AF319) posttransfection, total RNA was extracted using the TRIzol reagent (Life Technologies). Equal amounts of total RNA (for mammalian cells, 2 μg for positive-strand analysis and 10 μg for negative-strand analysis; for mosquito cells, 10 μg for positive-strand analysis and 10 μg for negative-strand analysis) were denatured for 10 min at 70°C in 2× RNA loading dye (Thermo Scientific), cooled on ice, and separated on a denaturing gel (1% agarose, 6% formaldehyde) using 1× MOPS (morpholinepropanesulfonic acid) buffer. RNA was transferred to a Hybond-N+ filter (GE Healthcare) and fixed using a UV Stratalinker 1800 cross-linker (Stratagene). A digoxigenin (DIG)-labeled RNA probe complementary to residues 42 to 390 of the sequence encoding the Gluc marker was used to detect positive-strand RNAs; a probe corresponding to residues 51 to 376 of the sequence encoding the Fluc marker was used to detect negative-strand RNAs. Filters were hybridized overnight; the blots were washed and developed according to the manufacturer's (Roche) protocols.
Activation of template replication by CHIKV infection.
U2OS cells grown in 12-well plates were transfected with the CMV-Fluc-Gluc or HSPolII-Fluc-Gluc plasmid. C6/36 cells grown in 12-well plates were transfected with the Ubi-Fluc-Gluc or AlbPolI-Fluc-Gluc plasmid. At 18 h (U2OS) or 36 h (C6/36) posttransfection, the cells were either infected with CHIKV at a multiplicity if infection (MOI) of 10 or mock infected. At 24 h postinfection (hpi) (U2OS cells) or 72 hpi (C6/36 cells), cells were collected and lysed and Fluc and Gluc activities were measured as described above.
Virus rescue and infectious center assay.
Virus rescue in BHK-21 cells was performed as previously described (68). The ICA was performed essentially as previously described (49), except that the cells were transfected with 5 μg of endotoxin-free plasmid CMV-ICRES1, CMV-ICRES1DA, CMV-ICRES1YA, CMV-ICRES1RE, or CMV-ICRES13C3A. Virus stocks were collected at 24 h (wt CHIKV) or at 48 h (mutant CHIKV variants) posttransfection. The obtained stocks were clarified by centrifugation at 3,000 × g for 10 min, and virus titers were determined using a standard plaque assay on BHK-21 cells.
The transfected cells were collected at the same time as the corresponding stocks. Cells were lysed by boiling in SDS gel-loading buffer (100 mM Tris-HCl, pH 6.8, 4% SDS, 20% glycerol, 200 mM dithiothreitol, 0.2% bromophenol blue). Lysate corresponding to 50,000 transfected cells was loaded on a 10% polyacrylamide gel. Proteins were separated by SDS-PAGE, transferred to polyvinylidene difluoride membranes, and detected using antibodies against CHIKV capsid protein (in-house); β-actin (catalog number sc-47778; Santa Cruz Biotechnology) was used as a loading control. The membranes were then incubated with appropriate secondary antibodies conjugated to fluorescent labels (LI-COR), and proteins were visualized using a LI-COR Odyssey Fc imaging system.
Statistical analysis.
Statistical analysis was done using GraphPad Prism software. Data were analyzed using Student’s unpaired one-tailed t test.
ACKNOWLEDGMENTS
We are grateful to Mark A. Kay for the pMC.BESPX vector and Alain Kohl for AF319 cells.
This work was supported by the European Regional Development Fund through the Centre of Excellence in Molecular Cell Engineering, Estonia (2014-2020.4.01.15-013 to A.M.), by institutional research funding (IUT20-27) from the Estonian Research Council (to A.M.), and by The Wellcome Trust (200171/Z/15/Z to A.M., R.F., and L.A.).
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We declare no conflict of interest.
REFERENCES
- 1.Chen R, Mukhopadhyay S, Merits A, Bolling B, Nasar F, Coffey LL, Powers A, Weaver SC, ICTV Report Consortium. 2018. ICTV virus taxonomy profile: Togaviridae. J Gen Virol 99:761–762. doi: 10.1099/jgv.0.001072. [DOI] [PubMed] [Google Scholar]
- 2.Strauss EG, Rice CM, Strauss JH. 1984. Complete nucleotide sequence of the genomic RNA of Sindbis virus. Virology 133:92–110. doi: 10.1016/0042-6822(84)90428-8. [DOI] [PubMed] [Google Scholar]
- 3.Rupp JC, Sokoloski KJ, Gebhart NN, Hardy RW. 2015. Alphavirus RNA synthesis and non-structural protein functions. J Gen Virol 96:2483–2500. doi: 10.1099/jgv.0.000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding MX, Schlesinger MJ. 1989. Evidence that Sindbis virus NSP2 is an autoprotease which processes the virus nonstructural polyprotein. Virology 171:280–284. doi: 10.1016/0042-6822(89)90539-4. [DOI] [PubMed] [Google Scholar]
- 5.Lemm JA, Rümenapf T, Strauss EG, Strauss JH, Rice CM. 1994. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J 13:2925–2934. doi: 10.1002/j.1460-2075.1994.tb06587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahola T, Kääriäinen L. 1995. Reaction in alphavirus mRNA capping: formation of a covalent complex of nonstructural protein nsP1 with 7-methyl-GMP. Proc Natl Acad Sci U S A 92:507–511. doi: 10.1073/pnas.92.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salonen A, Vasiljeva L, Merits A, Magden J, Jokitalo E, Kääriäinen L. 2003. Properly folded nonstructural polyprotein directs the Semliki Forest virus replication complex to the endosomal compartment. J Virol 77:1691–1702. doi: 10.1128/jvi.77.3.1691-1702.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rausalu K, Utt A, Quirin T, Varghese FS, Žusinaite E, Das PK, Ahola T, Merits A. 2016. Chikungunya virus infectivity, RNA replication and non-structural polyprotein processing depend on the nsP2 protease’s active site cysteine residue. Sci Rep 6:37124. doi: 10.1038/srep37124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rikkonen M, Peränen J, Kääriäinen L. 1994. ATPase and GTPase activities associated with Semliki Forest virus nonstructural protein nsP2. J Virol 68:5804–5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vasiljeva L, Merits A, Auvinen P, Kääriäinen L. 2000. Identification of a novel function of the alphavirus capping apparatus. RNA 5′-triphosphatase activity of Nsp2. J Biol Chem 275:17281–17287. doi: 10.1074/jbc.M910340199. [DOI] [PubMed] [Google Scholar]
- 11.Das PK, Merits A, Lulla A. 2014. Functional cross-talk between distant domains of chikungunya virus non-structural protein 2 is decisive for its RNA-modulating activity. J Biol Chem 289:5635–5653. doi: 10.1074/jbc.M113.503433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim DY, Reynaud JM, Rasalouskaya A, Akhrymuk I, Mobley JA, Frolov I, Frolova EI. 2016. New World and Old World alphaviruses have evolved to exploit different components of stress granules, FXR and G3BP proteins, for assembly of viral replication complexes. PLoS Pathog 12:e1005810. doi: 10.1371/journal.ppat.1005810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abraham R, Hauer D, McPherson RL, Utt A, Kirby IT, Cohen MS, Merits A, Leung AKL, Griffin DE. 2018. ADP-ribosyl-binding and hydrolase activities of the alphavirus nsP3 macrodomain are critical for initiation of virus replication. Proc Natl Acad Sci U S A 115:E10457–E10466. doi: 10.1073/pnas.1812130115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen MW, Tan YB, Zheng J, Zhao Y, Lim BT, Cornvik T, Lescar J, Ng LFP, Luo D. 2017. Chikungunya virus nsP4 RNA-dependent RNA polymerase core domain displays detergent-sensitive primer extension and terminal adenylyltransferase activities. Antiviral Res 143:38–47. doi: 10.1016/j.antiviral.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 15.Rubach JK, Wasik BR, Rupp JC, Kuhn RJ, Hardy RW, Smith JL. 2009. Characterization of purified Sindbis virus nsP4 RNA-dependent RNA polymerase activity in vitro. Virology 384:201–208. doi: 10.1016/j.virol.2008.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomar S, Hardy RW, Smith JL, Kuhn RJ. 2006. Catalytic core of alphavirus nonstructural protein nsP4 possesses terminal adenylyltransferase activity. J Virol 80:9962–9969. doi: 10.1128/JVI.01067-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones PH, Maric M, Madison MN, Maury W, Roller RJ, Okeoma CM. 2013. BST-2/tetherin-mediated restriction of chikungunya (CHIKV) VLP budding is counteracted by CHIKV non-structural protein 1 (nsP1). Virology 438:37–49. doi: 10.1016/j.virol.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akhrymuk I, Kulemzin SV, Frolova EI. 2012. Evasion of the innate immune response: the Old World alphavirus nsP2 protein induces rapid degradation of Rpb1, a catalytic subunit of RNA polymerase II. J Virol 86:7180–7191. doi: 10.1128/JVI.00541-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fros JJ, Liu WJ, Prow NA, Geertsema C, Ligtenberg M, Vanlandingham DL, Schnettler E, Vlak JM, Suhrbier A, Khromykh AA, Pijlman GP. 2010. Chikungunya virus nonstructural protein 2 inhibits type I/II interferon-stimulated JAK-STAT signaling. J Virol 84:10877–10887. doi: 10.1128/JVI.00949-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thaa B, Biasiotto R, Eng K, Neuvonen M, Götte B, Rheinemann L, Mutso M, Utt A, Varghese F, Balistreri G, Merits A, Ahola T, McInerney GM. 2015. Differential phosphatidylinositol-3-kinase-Akt-mTOR activation by Semliki Forest and chikungunya viruses is dependent on nsP3 and connected to replication complex internalization. J Virol 89:11420–11437. doi: 10.1128/JVI.01579-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akhrymuk I, Frolov I, Frolova EI. 2018. Sindbis virus infection causes cell death by nsP2-induced transcriptional shutoff or by nsP3-dependent translational shutoff. J Virol 92:e01388-18. doi: 10.1128/JVI.01388-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rathore APS, Ng M-L, Vasudevan SG. 2013. Differential unfolded protein response during Chikungunya and Sindbis virus infection: CHIKV nsP4 suppresses eIF2α phosphorylation. Virol J 10:36. doi: 10.1186/1743-422X-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garmashova N, Gorchakov R, Volkova E, Paessler S, Frolova E, Frolov I. 2007. The Old World and New World alphaviruses use different virus-specific proteins for induction of transcriptional shutoff. J Virol 81:2472–2484. doi: 10.1128/JVI.02073-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lutz A, Dyall J, Olivo PD, Pekosz A. 2005. Virus-inducible reporter genes as a tool for detecting and quantifying influenza A virus replication. J Virol Methods 126:13–20. doi: 10.1016/j.jviromet.2005.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moncorgé O, Mura M, Barclay WS. 2010. Evidence for avian and human host cell factors that affect the activity of influenza virus polymerase. J Virol 84:9978–9986. doi: 10.1128/JVI.01134-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu WJ, Sedlak PL, Kondratieva N, Khromykh AA. 2002. Complementation analysis of the flavivirus Kunjin NS3 and NS5 proteins defines the minimal regions essential for formation of a replication complex and shows a requirement of NS3 in cis for virus assembly. J Virol 76:10766–10775. doi: 10.1128/JVI.76.21.10766-10775.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quirin T, Chen Y, Pietilä MK, Guo D, Ahola T. 2018. The RNA capping enzyme domain in protein A is essential for Flock House virus replication. Viruses 10:E483. doi: 10.3390/v10090483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janda M, Ahlquist P. 1993. RNA-dependent replication, transcription, and persistence of brome mosaic virus RNA replicons in S. cerevisiae. Cell 72:961–970. doi: 10.1016/0092-8674(93)90584-D. [DOI] [PubMed] [Google Scholar]
- 29.Panavas T, Nagy PD. 2003. Yeast as a model host to study replication and recombination of defective interfering RNA of Tomato bushy stunt virus. Virology 314:315–325. doi: 10.1016/S0042-6822(03)00436-7. [DOI] [PubMed] [Google Scholar]
- 30.Price BD, Rueckert RR, Ahlquist P. 1996. Complete replication of an animal virus and maintenance of expression vectors derived from it in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 93:9465–9470. doi: 10.1073/pnas.93.18.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frolova EI, Gorchakov R, Pereboeva L, Atasheva S, Frolov I. 2010. Functional Sindbis virus replicative complexes are formed at the plasma membrane. J Virol 84:11679–11695. doi: 10.1128/JVI.01441-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hellström K, Kallio K, Utt A, Quirin T, Jokitalo E, Merits A, Ahola T. 2017. Partially uncleaved alphavirus replicase forms spherule structures in the presence and absence of RNA template. J Virol 91:e00787-17. doi: 10.1128/JVI.00787-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spuul P, Balistreri G, Hellström K, Golubtsov AV, Jokitalo E, Ahola T. 2011. Assembly of alphavirus replication complexes from RNA and protein components in a novel trans-replication system in mammalian cells. J Virol 85:4739–4751. doi: 10.1128/JVI.00085-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hellström K, Kallio K, Meriläinen H-M, Jokitalo E, Ahola T. 2016. Ability of minus strands and modified plus strands to act as templates in Semliki Forest virus RNA replication. J Gen Virol 97:1395–1407. doi: 10.1099/jgv.0.000448. [DOI] [PubMed] [Google Scholar]
- 35.Kallio K, Hellström K, Balistreri G, Spuul P, Jokitalo E, Ahola T. 2013. Template RNA length determines the size of replication complex spherules for Semliki Forest virus. J Virol 87:9125–9134. doi: 10.1128/JVI.00660-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Utt A, Quirin T, Saul S, Hellström K, Ahola T, Merits A. 2016. Versatile trans-replication systems for chikungunya virus allow functional analysis and tagging of every replicase protein. PLoS One 11:e0151616. doi: 10.1371/journal.pone.0151616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kallio K, Hellström K, Jokitalo E, Ahola T. 2015. RNA replication and membrane modification require the same functions of alphavirus nonstructural proteins. J Virol 90:1687–1692. doi: 10.1128/JVI.02484-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lulla V, Karo-Astover L, Rausalu K, Saul S, Merits A, Lulla A. 2018. Timeliness of proteolytic events is prerequisite for efficient functioning of the alphaviral replicase. J Virol 92:e00151-18. doi: 10.1128/JVI.00151-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bartholomeeusen K, Utt A, Coppens S, Rausalu K, Vereecken K, Ariën KK, Merits A. 2018. A chikungunya virus trans-replicase system reveals the importance of delayed nonstructural polyprotein processing for efficient replication complex formation in mosquito cells. J Virol 92:e00152-18. doi: 10.1128/JVI.00152-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Flick R, Flick K, Feldmann H, Elgh F. 2003. Reverse genetics for Crimean-Congo hemorrhagic fever virus. J Virol 77:5997–6006. doi: 10.1128/jvi.77.10.5997-6006.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flick R, Pettersson RF. 2001. Reverse genetics system for Uukuniemi virus (Bunyaviridae): RNA polymerase I-catalyzed expression of chimeric viral RNAs. J Virol 75:1643–1655. doi: 10.1128/JVI.75.4.1643-1655.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Freiberg A, Dolores LK, Enterlein S, Flick R. 2008. Establishment and characterization of plasmid-driven minigenome rescue systems for Nipah virus: RNA polymerase I- and T7-catalyzed generation of functional paramyxoviral RNA. Virology 370:33–44. doi: 10.1016/j.virol.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci U S A 97:6108–6113. doi: 10.1073/pnas.100133697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neumann G, Watanabe T, Ito H, Watanabe S, Goto H, Gao P, Hughes M, Perez DR, Donis R, Hoffmann E, Hobom G, Kawaoka Y. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc Natl Acad Sci U S A 96:9345–9350. doi: 10.1073/pnas.96.16.9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu CC, Fallon AM. 1998. Analysis of a ribosomal DNA intergenic spacer region from the yellow fever mosquito, Aedes aegypti. Insect Mol Biol 7:19–29. doi: 10.1046/j.1365-2583.1998.71194.x. [DOI] [PubMed] [Google Scholar]
- 46.Baldridge GD, Fallon AM. 1992. Primary structure of the ribosomal DNA intergenic spacer from the mosquito, Aedes albopictus. DNA Cell Biol 11:51–59. doi: 10.1089/dna.1992.11.51. [DOI] [PubMed] [Google Scholar]
- 47.Park YJ, Baldridge GD, Fallon AM. 1995. Promoter utilization in a mosquito ribosomal DNA cistron. Arch Insect Biochem Physiol 28:143–157. doi: 10.1002/arch.940280205. [DOI] [PubMed] [Google Scholar]
- 48.Steel JJ, Franz AWE, Sanchez-Vargas I, Olson KE, Geiss BJ. 2013. Subgenomic reporter RNA system for detection of alphavirus infection in mosquitoes. PLoS One 8:e84930. doi: 10.1371/journal.pone.0084930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Utt A, Das PK, Varjak M, Lulla V, Lulla A, Merits A. 2015. Mutations conferring a noncytotoxic phenotype on chikungunya virus replicons compromise enzymatic properties of nonstructural protein 2. J Virol 89:3145–3162. doi: 10.1128/JVI.03213-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rikkonen M. 1996. Functional significance of the nuclear-targeting and NTP-binding motifs of Semliki Forest virus nonstructural protein nsP2. Virology 218:352–361. doi: 10.1006/viro.1996.0204. [DOI] [PubMed] [Google Scholar]
- 51.Law Y-S, Utt A, Tan YB, Zheng J, Wang S, Chen MW, Griffin PR, Merits A, Luo D. 2019. Structural insights into RNA recognition by the Chikungunya virus nsP2 helicase. Proc Natl Acad Sci U S A 116:9558–9567. doi: 10.1073/pnas.1900656116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ahola T, Lampio A, Auvinen P, Kääriäinen L. 1999. Semliki Forest virus mRNA capping enzyme requires association with anionic membrane phospholipids for activity. EMBO J 18:3164–3172. doi: 10.1093/emboj/18.11.3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lampio A, Kilpeläinen I, Pesonen S, Karhi K, Auvinen P, Somerharju P, Kääriäinen L. 2000. Membrane binding mechanism of an RNA virus-capping enzyme. J Biol Chem 275:37853–37859. doi: 10.1074/jbc.M004865200. [DOI] [PubMed] [Google Scholar]
- 54.Ahola T, Karlin DG. 2015. Sequence analysis reveals a conserved extension in the capping enzyme of the alphavirus supergroup, and a homologous domain in nodaviruses. Biol Direct 10:16. doi: 10.1186/s13062-015-0050-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ahola T, Kujala P, Tuittila M, Blom T, Laakkonen P, Hinkkanen A, Auvinen P. 2000. Effects of palmitoylation of replicase protein nsP1 on alphavirus infection. J Virol 74:6725–6733. doi: 10.1128/jvi.74.15.6725-6733.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zusinaite E, Tints K, Kiiver K, Spuul P, Karo-Astover L, Merits A, Sarand I. 2007. Mutations at the palmitoylation site of non-structural protein nsP1 of Semliki Forest virus attenuate virus replication and cause accumulation of compensatory mutations. J Gen Virol 88:1977–1985. doi: 10.1099/vir.0.82865-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spuul P, Salonen A, Merits A, Jokitalo E, Kääriäinen L, Ahola T. 2007. Role of the amphipathic peptide of Semliki forest virus replicase protein nsP1 in membrane association and virus replication. J Virol 81:872–883. doi: 10.1128/JVI.01785-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang N, Zhao H, Zhang L. 2018. Fatty acid synthase promotes the palmitoylation of chikungunya virus nsP1. J Virol 93:e01747-18. doi: 10.1128/JVI.01747-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lachmi B, Kääriäinen L. 1977. Control of protein synthesis in Semliki forest virus-infected cells. J Virol 22:142–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen R, Wang E, Tsetsarkin KA, Weaver SC. 2013. Chikungunya virus 3′ untranslated region: adaptation to mosquitoes and a population bottleneck as major evolutionary forces. PLoS Pathog 9:e1003591. doi: 10.1371/journal.ppat.1003591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morley VJ, Noval MG, Chen R, Weaver SC, Vignuzzi M, Stapleford KA, Turner PE. 2018. Chikungunya virus evolution following a large 3′ UTR deletion results in host-specific molecular changes in protein-coding regions. Virus Evol 4:vey012. doi: 10.1093/ve/vey012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sokoloski KJ, Haist KC, Morrison TE, Mukhopadhyay S, Hardy RW. 2015. Noncapped alphavirus genomic RNAs and their role during infection. J Virol 89:6080–6092. doi: 10.1128/JVI.00553-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.LaPointe AT, Moreno-Contreras J, Sokoloski KJ. 2018. Increasing the capping efficiency of the Sindbis virus nsP1 protein negatively affects viral infection. mBio 9:e02342-18. doi: 10.1128/mBio.02342-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ahola T, Laakkonen P, Vihinen H, Kääriäinen L. 1997. Critical residues of Semliki Forest virus RNA capping enzyme involved in methyltransferase and guanylyltransferase-like activities. J Virol 71:392–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Varjak M, Zusinaite E, Merits A. 2010. Novel functions of the alphavirus nonstructural protein nsP3 C-terminal region. J Virol 84:2352–2364. doi: 10.1128/JVI.01540-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Varjak M, Maringer K, Watson M, Sreenu VB, Fredericks AC, Pondeville E, Donald CL, Sterk J, Kean J, Vazeille M, Failloux A-B, Kohl A, Schnettler E. 2017. Aedes aegypti Piwi4 is a noncanonical PIWI protein involved in antiviral responses. mSphere 2:e00144-17. doi: 10.1128/mSphere.00144-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hallengärd D, Kakoulidou M, Lulla A, Kümmerer BM, Johansson DX, Mutso M, Lulla V, Fazakerley JK, Roques P, Le Grand R, Merits A, Liljeström P. 2014. Novel attenuated chikungunya vaccine candidates elicit protective immunity in C57BL/6 mice. J Virol 88:2858–2866. doi: 10.1128/JVI.03453-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ulper L, Sarand I, Rausalu K, Merits A. 2008. Construction, properties, and potential application of infectious plasmids containing Semliki Forest virus full-length cDNA with an inserted intron. J Virol Methods 148:265–270. doi: 10.1016/j.jviromet.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]