Marek’s disease virus (MDV) RLORF4 has been shown to be directly involved in the attenuation of MDV upon serial passages in vitro; however, the exact function of this protein during viral infection was not well characterized. This study demonstrated that RLORF4 significantly inhibits cGAS-STING-mediated NF-κB activation by binding to the Rel homology domains of the NF-κB subunits p65 and p50, interrupting their translocation to the nuclei and thereby inhibiting IFN-β production. Furthermore, RLORF4 deficiency promoted the induction of IFN-β and downstream IFN-stimulated genes during MDV infection in chickens. Our results suggest that the contribution of RLORF4 to MDV virulence may stem from its inhibition of viral DNA-triggered IFN-β responses.
KEYWORDS: DNA sensing, Marek’s disease virus, NF-κB, RLORF4, innate immunity
ABSTRACT
Marek’s disease virus (MDV), which causes T cell lymphomas in chickens, is economically important and has contributed to knowledge of herpesvirus-associated oncogenicity. The DNA-sensing pathway induces innate immune responses against DNA virus infection, and nuclear factor κB (NF-κB) signaling is critical for the establishment of innate immunity. Here, we report that RLORF4, an MDV-specific protein directly involved in viral attenuation, is an inhibitor of the DNA-sensing pathway. The results showed that ectopically expressed RLORF4 blocked beta interferon (IFN-β) promoter activation induced by cyclic GMP-AMP synthase (cGAS) and stimulator of interferon genes (STING). RLORF4 selectively inhibited the activation of NF-κB but not IFN-regulatory factor 7. RLORF4 was found to bind the endogenous NF-κB subunits p65 and p50, and it also bound to the Rel homology domains of these subunits. Furthermore, RLORF4 suppressed the nuclear translocation of p65 and p50 mediated by tumor necrosis factor alpha and interferon-stimulatory DNA. Finally, deletion of RLORF4 from the MDV genome promoted IFN-β and interleukin-6 (IL-6) production in vitro and in vivo. In the absence of RLORF4, the host cellular immunity was significantly increased, and reduced viral titers were observed during infection of chickens. Our results suggest that the RLORF4-mediated suppression of the host antiviral innate immunity might play an important role in MDV pathogenesis.
IMPORTANCE Marek’s disease virus (MDV) RLORF4 has been shown to be directly involved in the attenuation of MDV upon serial passages in vitro; however, the exact function of this protein during viral infection was not well characterized. This study demonstrated that RLORF4 significantly inhibits cGAS-STING-mediated NF-κB activation by binding to the Rel homology domains of the NF-κB subunits p65 and p50, interrupting their translocation to the nuclei and thereby inhibiting IFN-β production. Furthermore, RLORF4 deficiency promoted the induction of IFN-β and downstream IFN-stimulated genes during MDV infection in chickens. Our results suggest that the contribution of RLORF4 to MDV virulence may stem from its inhibition of viral DNA-triggered IFN-β responses.
INTRODUCTION
The innate immune response to microbial invasion involves the activation of pattern recognition receptors (PRRs), which recognize the conserved pathogen-associated molecular patterns (PAMPs) of microorganisms to trigger the production of proinflammatory cytokines and type I interferons (IFNs) (1, 2). Viral components such as viral nucleic acids exposed in the cytoplasm are signs of foreign invasion and can be detected by many host cell PRRs. A number of cytosolic DNA sensors recognizing microbial pathogen DNA have been identified (3–6). The recognition of double-stranded DNA (dsDNA) by DNA sensors is a central host cellular defense against DNA virus infection. Among these DNA sensors, cyclic GMP-AMP synthase (cGAS) has been found to be the predominant cytosolic DNA sensor recognizing a variety of DNA ligands in different cell types (6–8). Upon binding to DNA fragments, cGAS utilizes ATP and GTP to synthesize the second messenger cyclic GMP-AMP (cGAMP) (9) to activate stimulator of interferon genes (STING). Activated STING then recruits TANK-binding kinase 1 (TBK1) to phosphorylate nuclear factor κB (NF-κB) and IFN-regulatory factor 3 (IRF3) (10, 11). Consequently, NF-κB and IRF3 translocate to the nuclei to induce IFN-β production (12, 13).
The NF-κB pathway is composed of the IκB kinase (IKK) complex and NF-κB essential modulators (14, 15). The IKK complex, which comprises IKKα and IKKβ, is activated by upstream signals, which results in the phosphorylation, K48-linked polyubiquitination, and ultimately proteasomal degradation of IκBs (16–18). Subsequently, NF-κB essential modulators, including p65/RelA, RelB, p50/NF-κB1, p52/NF-κB2, and c-Rel, which share a structurally conserved N-terminal Rel homology domain (RHD) that is important for protein dimerization, are released from cytoplasmic sequestration and translocate to the nuclei to initiate NF-κB-dependent gene transcription and regulate the innate immune responses (19, 20).
Herpesviruses are formidable pathogens that have evolved a variety of strategies to survive in a hostile cellular environment. One such strategy is to encode proteins that mimic the function of cellular proteins, interact with cellular pathways, and alter the host environment to facilitate their propagation (21). Such molecular mimicry can allow viruses to modulate cellular pathways, resulting in immunosuppression and oncogenic transformation (22). Marek’s disease virus (MDV), a member of the Alphaherpesvirinae subfamily of the Herpesviridae family (23), is one such virus. Infection with MDV results in the induction of T cell lymphomas in chickens as early as 3 to 4 weeks after infection (24). In addition to being an economically important disease affecting the health and welfare of poultry (25), Marek’s disease (MD) has great significance for the understanding of herpesvirus-associated oncogenicity. MD lymphoma neoplasia has many biological characteristics similar to those of lymphoid tissue infection by Epstein-Barr virus (EBV) (26). The genome of MDV shares close functional and structural similarities to those of the mammalian herpesviruses such as herpes simplex virus (HSV) (27). Whereas the innate immune response has been shown to be of paramount importance against human herpesviruses, little is known regarding the role of innate immunity in the control of MDV in chickens. Thus, it is necessary to investigate the host-virus interactions and host immunity against MDV.
RLORF4, as a single transcript encoding a 142-amino-acid protein in wild-type strains, was shown to be directly involved in the attenuation of MDV upon serial passages in vitro, and the deletion of RLORF4 from virulent MDV markedly reduced viral pathogenicity in vivo (28–30). However, the exact function of this protein during viral infection was not well characterized. We thus aimed to clarify the mechanism through which RLORF4 mediates host-virus interaction. In the present study, we found that overexpression of RLORF4 specifically inhibits the viral DNA-triggered IFN-β response. In contrast, RLORF4 deficiency resulted in enhanced induction of IFN-β and downstream IFN-stimulated genes (ISGs). Furthermore, we found that RLORF4 interacts with p65 and p50, interrupting their translocation to the nuclei and thereby inhibiting their transcriptional activity. Our findings suggest that MDV RLORF4 blocks dsDNA-triggered activation of the NF-κB promoter and dampens activation of the cellular DNA-sensing pathway and subsequent IFN-β production.
RESULTS
RLORF4 inhibits cGAS-STING-mediated IFN-β production.
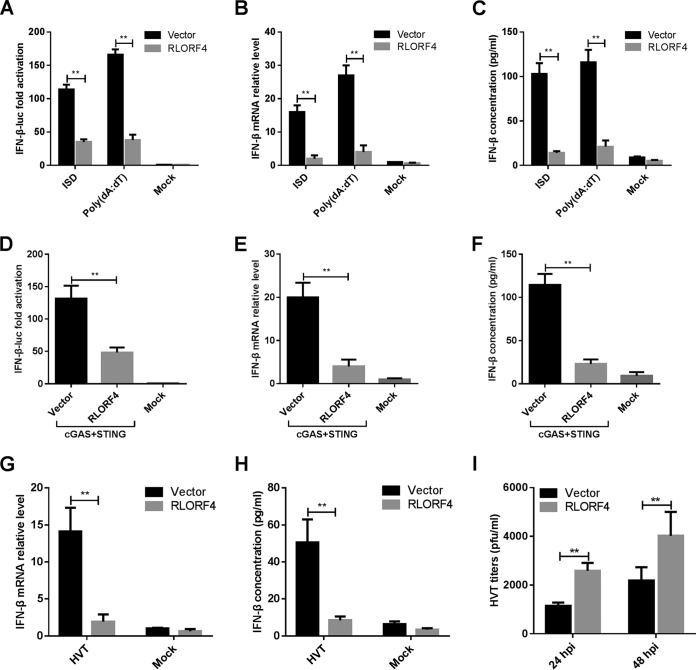
DF-1, a chicken fibroblast cell line, is known to respond to foreign DNA such as interferon-stimulatory DNA (ISD) and poly(dA·dT) (31). The IFN-β promoter was highly activated by cotransfecting the same amounts of cGAS and STING expression plasmids in DF-1 cells (32). With this model, we screened MDV proteins that could antagonize IFN-β production mediated by cGAS-STING and identified RLORF4 as one candidate. To verify the role of RLORF4 in the regulation of IFN-β production, we transfected DF-1 cells with an RLORF4-expressing plasmid or empty vector (EV) and different exogenous DNA-sensing stimuli, including ISD and poly(dA·dT), along with an IFN-β-luc promoter reporter plasmid and then used dual-luciferase reporter (DLR) assays to detect IFN-β promoter activity. As shown in Fig. 1A, the ectopic expression of RLORF4 markedly inhibited the DNA-triggered activation of the IFN-β promoter. Further, the mRNA and protein levels of IFN-β in cells transfected with these fragments were measured by real-time quantitative PCR (qPCR) and enzyme-linked immunosorbent assay (ELISA). IFN-β mRNA and protein levels were significantly increased in the EV-transfected cells in response to DNA stimuli, which was attenuated in RLORF4-expressing DF-1 cells (Fig. 1B and C), suggesting that RLORF4 inhibits the cytosolic DNA-induced IFN-β pathway.
FIG 1.
RLORF4 inhibits cGAS-STING-mediated IFN-β production and promotes herpesvirus of turkey (HVT) replication. (A) The IFN-β-luc reporter was cotransfected with ISD or poly(dA·dT), as well as an RLORF4 expression plasmid or empty vector, into DF-1 cells, and 24 h later, cells were harvested and subjected to a dual-luciferase reporter assay. (B) DF-1 cells were transfected with an empty vector or RLORF4 expression plasmid for 24 h and transfected with ISD or poly(dA·dT) for another 8 h, and IFN-β mRNA levels were measured by qPCR. (C) DF-1 cells were transfected as for panel B and stimulated with ISD and poly(dA·dT) for another 24 h, and IFN-β protein levels were measured by ELISA. (D) cGAS and STING expression plasmids were cotransfected with RLORF4-Flag plasmid or an empty vector, with the IFN-β-luc reporter, into DF-1 cells, and IFN-β promoter luciferase activity was measured at 24 h posttransfection. (E and F) The cGAS and STING expression plasmids were cotransfected with RLORF4-Flag or an empty vector into DF-1 cells, and IFN-β mRNA and protein levels were measured by qPCR and ELISA at 24 h posttransfection. (G and H) DF-1 cells were transduced with an empty vector or RLORF4-expressing lentivirus and then left uninfected or infected with HVT (multiplicity of infection [MOI] = 0.1). IFN-β mRNA levels in these cells were measured by qPCR at 12 h postinfection, and IFN-β protein was measured by ELISA at 24 h postinfection. (I) Transduced DF-1 cells were infected with HVT (MOI = 0.01). At 24 or 48 h postinfection (hpi), the HVT viral titer was tested by plaque assays. The relative amount of IFN-β mRNA was normalized to actin mRNA levels in each sample, and the fold differences between the treated samples and the mock controls were calculated. Data are presented as means ± SDs from at least three independent experiments. Statistical analysis was performed with Student’s t test (**, P < 0.01).
To determine whether RLORF4 could inhibit the cGAS-STING-mediated IFN-β production, the RLORF4 expression plasmid was transfected into DF-1 cells along with cGAS and STING expression plasmids and an IFN-β promoter reporter plasmid, and cells were subjected to DLR assays. As shown in Fig. 1D, the ectopic expression of RLORF4 significantly inhibited cGAS-STING-mediated activation of IFN-β. Furthermore, IFN-β transcription and protein levels were also significantly reduced in RLORF4-expressing DF-1 cells compared to those in the EV-transfected cells (Fig. 1E and F). These results indicated that RLORF4 inhibits the cGAS-STING-mediated DNA sensing pathway.
We then tested whether IFN-β induction, triggered by DNA virus infection, was blocked in RLORF4-expressing DF-1 cells. We infected RLORF4- and EV-expressing DF-1 cells with herpesvirus of turkey (HVT) and found that RLORF4 expression led to a diminished IFN-β response to this virus, compared to that observed in control cells, at both the mRNA (Fig. 1G) and protein (Fig. 1H) levels. Concordantly, the HVT titer in the infected RLORF4-expressing cells was much higher than that in EV-expressing cells (Fig. 1I). These data suggest that RLORF4 is an inhibitor of cGAS-STING-mediated signaling.
RLORF4 inhibits IFN-β production by targeting p65 and p50.
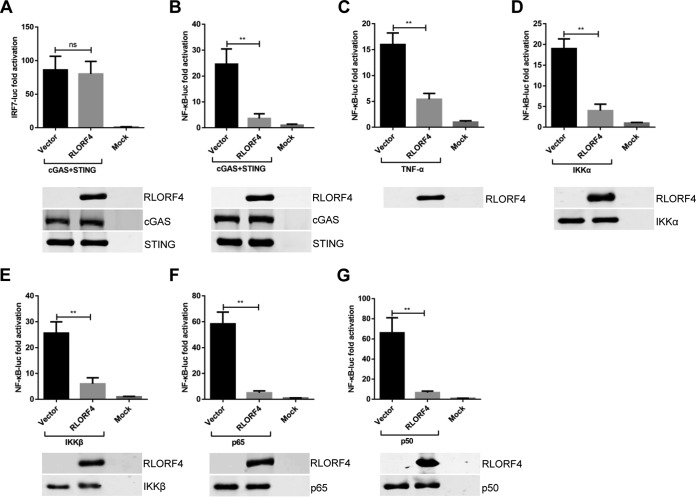
The innate immune system in chickens differs from that in mammals. Chickens are IRF3 deficient; however, the presence of functional IRF7 is considered to compensate for the IRF3 deficiency (33). IRF7 and NF-κB are the most important transcriptional activators that induce the expression of type I IFNs and other cytokines in chicken cells (33). To determine whether RLORF4 could suppress the cGAS-STING-mediated activation of IRF7 and NF-κB, we constructed the luciferase reporters pchIRF7-luc and pchNF-κB-luc, as previously described (31, 34), and then DLR assays were performed to analyze the effect of RLORF4 on IRF7 and NF-κB regulation. The results showed that RLORF4 negatively regulated the cGAS-STING-mediated IFN-β activation by targeting NF-κB but not IRF7 (Fig. 2A and B). Moreover, RLORF4 also inhibited the activation of NF-κB promoter activity mediated by tumor necrosis factor alpha (TNF-α), a key cytokine stimulating the NF-κB pathway (Fig. 2C). To further clarify the pathway components targeted by RLORF4 during NF-κB activation, DF-1 cells were cotransfected with an RLORF4 expression or vector plasmid, NF-κB-luc reporter plasmid, and expression plasmids encoding NF-κB signaling pathway components, including IKKα, IKKβ, p65, and p50. All NF-κB adaptor proteins resulted in an approximately 10- to 80-fold induction of NF-κB-luc reporter activity (Fig. 2D to G). However, NF-κB activation mediated by all NF-κB signaling pathway components was inhibited by RLORF4 (Fig. 2D to G). These findings indicate that RLORF4 probably suppresses cGAS-STING-mediated NF-κB signaling at or downstream of p65 and p50.
FIG 2.
RLORF4 inhibits IFN-β activation via p65 and p50. (A and B) cGAS and STING expression plasmids was cotransfected with the IRF7-luc (A) or NF-κB-luc (B) reporters, as well as the RLORF4-Flag plasmid or an empty vector, into DF-1 cells; 24 h later, cells were harvested and subjected to the dual-luciferase reporter assay. (C) DF-1 cells were transfected with the NF-κB-luc reporter, together with the RLORF4-Flag plasmid or empty vector. At 24 h posttransfection, cells were treated with or without 10 ng/ml chicken TNF-α and incubated for an additional 6 h, followed by cell lysis and dual-luciferase reporter assay. (D to G) The NF-κB-luc reporter was transfected with plasmids expressing IKKα (D), IKKβ (E), p65 (F), or p50 (G) together with the RLORF4-Flag plasmid or an empty vector. Dual-luciferase reporter assays were performed at 24 h posttransfection. All cells were transfected with pRL-TK as an internal control to normalize transfection efficiency. Data are presented as means ± SDs from three independent experiments. Statistical analysis was performed with Student’s t test (**, P < 0.01; ns, no significant difference).
RLORF4 inhibits the nuclear translocation of p65 and p50.
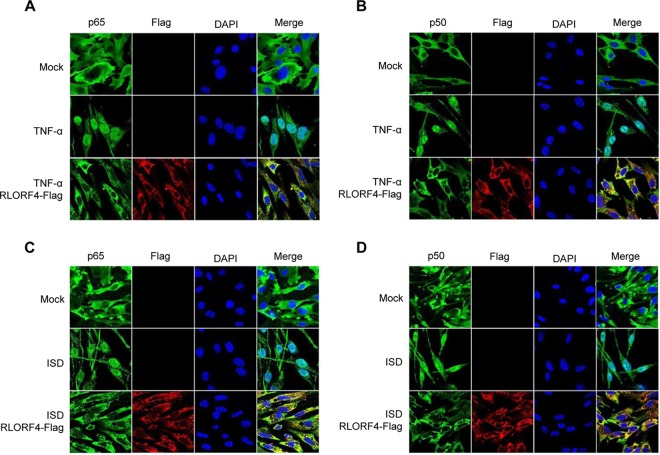
The initiation of IFN-β transcription requires the migration of NF-κB subunits to the nuclei and their binding to the IFN-β promoter region. To determine whether RLORF4 abrogates the trafficking of these NF-κB subunits, DF-1 cells were transfected with an RLORF4-Flag expression plasmid or an empty vector, and the TNF-α- or ISD-stimulated nuclear trafficking of endogenous p65 and p50 was monitored. As shown in Fig. 3A and B, stimulation with TNF-α led to increased levels of p65 and p50 in the nuclei. However, ectopic expression of RLORF4 reduced TNF-α-induced nuclear trafficking of p65 and p50. Furthermore, RLORF4 also inhibited ISD-induced nuclear trafficking of p65 and p50 (Fig. 3C and D). These results suggest that RLORF4 inhibited IFN-β response by targeting p65 and p50.
FIG 3.
RLORF4 blocks p65 and p50 nuclear translocation. DF-1 cells were transfected with the RLORF4-Flag plasmid or an empty vector. At 24 h posttransfection, cells were treated with TNF-α (10 ng/ml) for 30 min or with ISD (2 μg/ml) for 12 h as indicated. Cells were stained with mouse anti-Flag monoclonal antibody and rabbit anti-p65 (A and C) or anti-p50 (B and D) polyclonal antibody. Alexa 488-conjugated goat anti-rabbit (green) and Alexa 546-conjugated goat anti-mouse (red) were used as the secondary antibodies.
RLORF4 interacts with p65 and p50.
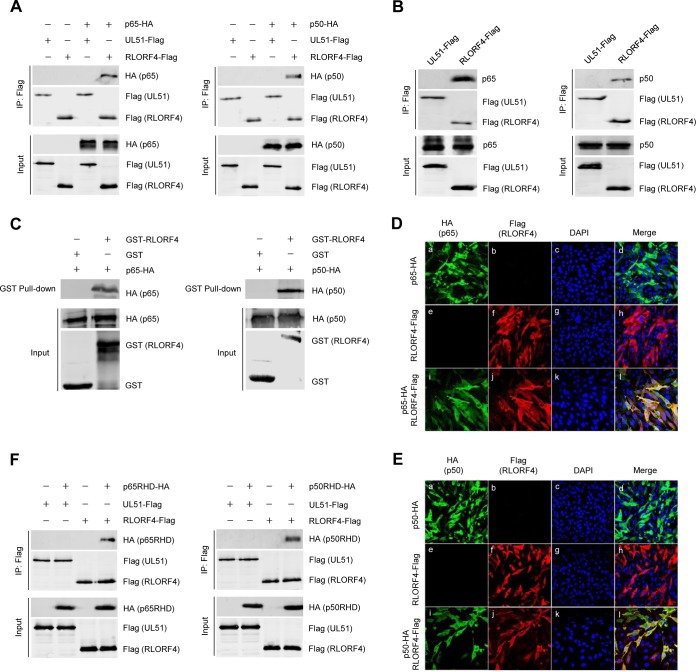
To elucidate the molecular mechanisms through which RLORF4 suppresses NF-κB activation, RLORF4-Flag or a nonrelevant viral protein UL51-Flag expression plasmid were cotransfected with either p65-hemagglutinin (p65-HA) or p50-HA into HEK293T cells, and coimmunoprecipitation (co-IP) analysis was performed with anti-Flag antibodies. As shown in Fig. 4A, both p65 and p50 were efficiently coimmunoprecipitated with RLORF4 but not with UL51. We further investigated the interactions between RLORF4 and endogenous p65 and p50. An RLORF4-Flag expression plasmid was transfected into DF-1 cells, and at 36 h posttransfection, both endogenous p65 and p50 proteins were efficiently coimmunoprecipitated with RLORF4 using anti-Flag antibodies (Fig. 4B). In vitro glutathione S-transferase (GST) pulldown assays further confirmed that RLORF4 interacted directly with p65 and p50 (Fig. 4C). In addition, confocal microscopy confirmed that RLORF4 colocalized with p65 (Fig. 4D) and p50 (Fig. 4E).
FIG 4.
RLORF4 interacts with p65 and p50. (A) HEK293T cells were cotransfected with RLORF4-Flag or a negative-control plasmid expressing Flag-tagged nonrelevant protein (UL51-Flag), as well as p65-HA or p50-HA plasmids, for 36 h, which was followed by a coimmunoprecipitation assay using an anti-Flag antibody. Western blotting was then performed with the indicated antibodies. (B) DF-1 cells were transfected with the RLORF4-Flag plasmid or a negative-control plasmid (UL51-Flag), and at 36 h posttransfection, coimmunoprecipitation assays were performed with an anti-Flag antibody. Western blotting was then performed with the indicated antibodies. (C) Purified GST or GST-RLORF4 was used to pull down transiently expressed p65-HA and p50-HA as indicated. (D and E) DF-1 cells were transfected with RLORF4-Flag and/or p65-HA (D) or p50-HA (E) expression plasmids for 24 h and then fixed and processed for dual labeling. Cell nuclei were counterstained with DAPI (blue). RLORF4 (red) and p65 (green) or p50 (green) proteins were visualized by immunostaining with anti-Flag and anti-HA antibodies, respectively. (F) HEK293T cells were cotransfected with RLORF4-Flag and p65RHD-HA or p50RHD-HA plasmids. At 36 h posttransfection, cells were harvested and lysed, which was followed by coimmunoprecipitation assays using anti-Flag antibody. Western blotting was performed with the indicated antibodies.
Both p65 and p50 contain an RHD, which is critical for protein dimerization, DNA binding, interactions with IκB, and nuclear translocation. To determine whether RLORF4 binds to the RHD of p65 or p50, HEK293T cells were cotransfected with RLORF4-Flag and either p65RHD-HA or p50RHD-HA expression plasmids. The results showed that p65RHD-HA and p50RHD-HA were efficiently coimmunoprecipitated by RLORF4 (Fig. 4F). These results suggest that RLORF4 inhibits the nuclear accumulation of p65 and p50 by binding to their RHDs.
RLORF4 plays an important role in MDV immune evasion.
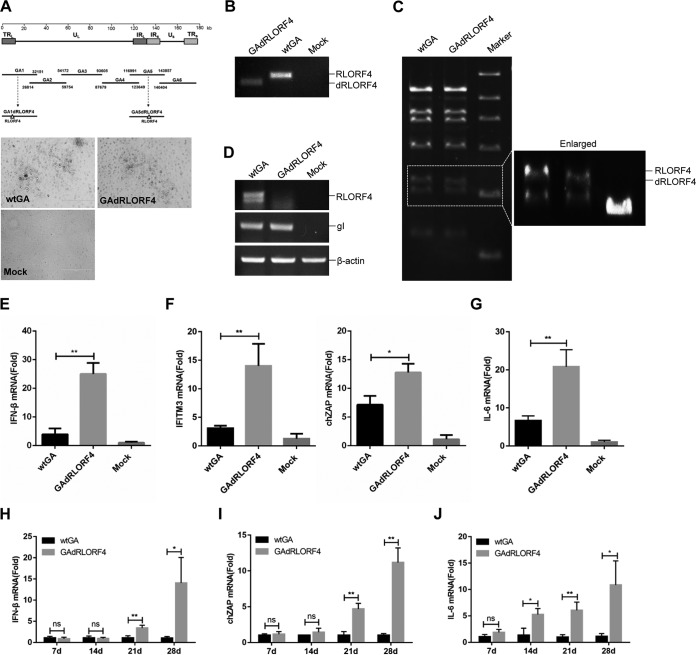
To further investigate whether RLORF4 acts as an IFN-β inhibitor in the context of MDV infection, we generated RLORF4-deficient MDV (GAdRLORF4) by using overlapping fosmid clones of the virulent MDV strain GA (Fig. 5A). The GAdRLORF4 virus yielded plaques similar to those resulting from the wild-type GA (wtGA) virus (Fig. 1A). The deletion of the RLORF4 gene from the MDV genome was confirmed by PCR analyses (Fig. 5B). Pulsed-field gel electrophoresis analysis of the GAdRLORF4 and wtGA virus DNA digested with BamHI further confirmed the successful deletion of RLORF4 from the MDV genome (Fig. 5C). The mRNA transcription of MDV RLORF4 and glycoprotein I (gI) at 4 days postinfection (p.i.) in infected chicken embryo fibroblast (CEF) cultures is shown in Fig. 5D. Deletion of the RLORF4 sequence completely abolished RLORF4 mRNA transcription, while gI transcription was not affected.
FIG 5.
RLORF4 deficiency enhances MDV-triggered IFN-β production in vitro and in vivo. (A) Schematic diagram of the recombinant fosmids for constructing recombinant MDVs (top) and the cytopathic effects induced by wtGA and GAdRLORF4 in DEFs (bottom). Bar length, 400 μm. (B) PCR analyses of the RLORF4-deficient MDV. (C) Pulsed-field gel electrophoresis analysis of the wtGA and GAdRLORF4 virus DNAs digested with BamHI. (D) Reverse transcription-PCR (RT-PCR) for RLORF4, gI, and β-actin mRNA expression was performed using total RNA collected from wtGA- or GAdRLORF4-infected cells. (E to G) CEFs were infected with wtGA or GAdRLORF4 (MOI = 0.1) for 12 h before the analysis of IFN-β (E), ZAP and IFITM3 (F), and IL-6 (G) mRNA levels. (H to J) Chickens were infected with 2,000 PFU of wtGA or GAdRLORF4. IFN-β (H), ZAP (I), and IL-6 (J) production in chickens was measured by qPCR at the indicated time points after MDV infection. Statistical analysis was performed with Student’s t test (*, P < 0.05, **, P < 0.01; ns, no significant difference).
We next examined the expression of IFN-β, the IFN-stimulated gene products, and the NF-κB-regulated cytokine interleukin-6 (IL-6) in cells infected with wild-type or RLORF4-deficient MDV. The results indicated that mRNA levels of IFN-β, IFN-stimulated gene products ZAP and IFN-inducible transmembrane protein 3 (IFITM3), and IL-6 induced by GAdRLORF4 were notably higher than those induced by wild-type MDV in CEFs (Fig. 5E to G). Consistently, GAdRLORF4 also induced significantly higher levels of IFN-β, ZAP, and IL-6 in chickens after 21 and 28 days of MDV infection (Fig. 5H to J).
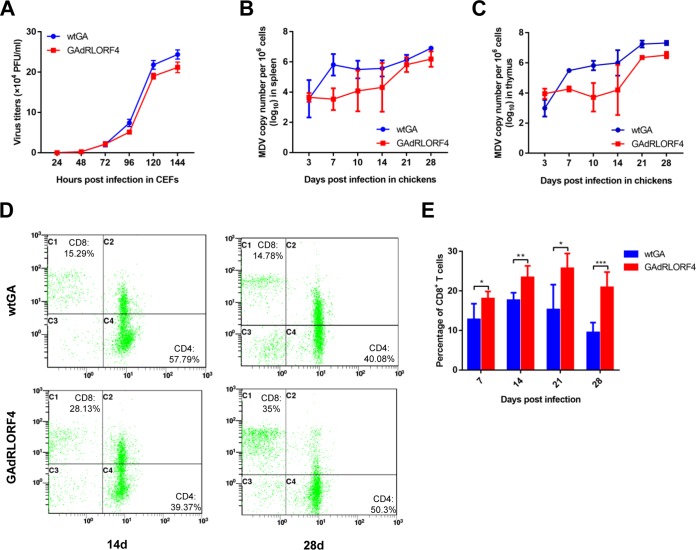
To determine the effects of RLORF4 on viral replication in vitro and in vivo, we first determined the replication curves of GAdRLORF4 and wtGA in cell cultures. As shown in Fig. 6A, the replication ability of GAdRLORF4 was decreased in comparison with that of wtGA in CEFs. The viral DNA loads in the spleen (Fig. 6B) and thymus (Fig. 6C) were then analyzed by qPCR during MDV infection in chickens. MDV infection in the lymphoid organs is known to switch from an early cytolytic phase to a latent phase at 7 to 8 days p.i. (25, 26). Thus, the viral titers detected beyond 7 to 8 days p.i. are likely due to the reactivation of virus from latency. As shown in Fig. 6B and C, the GAdRLORF4 virus replicated at the parental wtGA level during early cytolytic infection on day 3 p.i.; however, GAdRLORF4 replication measured at 7, 10, and 14 days after infection was reduced 119-, 26-, and 18-fold in spleen, and 14-, 128-, and 62-fold in thymus, respectively, compared with parental wtGA levels. These results indicate that RLORF4 is dispensable for early cytolytic infection; however, it might play a role in latency and reactivation. Therefore, we speculated that the effect of RLORF4 on viral replication may involve its ability to inhibit the host immune response, which resulted in enhanced viral replication in vivo.
FIG 6.
RLORF4 deficiency facilitates CD8+ T cell response and attenuates viral replication. (A) Growth properties of wtGA and GAdRLORF4 in CEF cell cultures. (B to E) Chickens were infected with 2,000 PFU of wtGA or GAdRLORF4. (B and C) Virus genome copy numbers in spleen (B) and thymus (C) were monitored at the indicated time points. (D and E) Chicken peripheral blood lymphocytes were obtained to analyze the percentage of CD8+ T cells at the indicated time points postinoculation. Statistical analysis was performed with Student’s t test (*, P < 0.05, **, P < 0.01, ***, P < 0.001).
Type I IFNs have multiple effects on infected cells and have the ability to link innate and adaptive immune responses (35). Recent studies revealed a link between type I IFN profiles and the CD8+ T cell response against tumor-associated antigens in vivo (36). Since RLORF4 deficiency significantly diminishes viral tumorigenic potential (28), we suspect that the inhibitory effect of RLORF4 on IFN-β induction might affect host antitumor immunity. To address this hypothesis, T cell subsets in the infected chickens were analyzed by flow cytometry. As shown in Fig. 6D and E, the percentage of CD8+ T cells was drastically reduced in wtGA-infected chickens in comparison to that in chickens infected with GAdRLORF4. These results suggested that RLORF4 affects the host CD8+ T cell response by inhibiting the innate antiviral response. Taken together, these data revealed that RLORF4 plays an important role in MDV immune evasion and contributes to the viral replication.
DISCUSSION
During viral infection, viral nucleic acids are released into the host nucleus or cytoplasm, depending on the cell type (3). In the cytoplasm, RNA viruses are detected by RIG-I-like receptors (37), whereas DNA viruses are recognized by various DNA sensors, including cGAS (7), absent in melanoma 2 (AIM2) (38), gamma interferon-inducible protein 16 (IFI16) (39), RNA polymerase III (40), and DNA-dependent activator of IFN regulatory factors (DAI) (41). Among these sensors, cGAS has been demonstrated to be a major cytosolic DNA sensor that is activated in response to DNA virus infection in various mammalian cell lines (7, 42). It has been reported that herpesvirus DNA activates the cGAS-STING DNA-sensing pathway in the cytoplasm, resulting in the production of IFNs and inflammatory chemokines and mediating the repression of viral replication (10). However, many anti-IFN response inhibitors encoded by herpesviruses have been identified, including HSV-1 VP16 (43), US3 (44), VP24 (45), ICP27 (46), VP22 (47), VP11/12 (48), UL24 (49), ICP0 (50), UL41 (51), UL42 (52), and UL36 (53), in addition to viral proteins encoded by human cytomegalovirus (54, 55), Kaposi’s sarcoma-associated herpesvirus (56–58), EBV (59), murine gammaherpesvirus 68 (60), and feline herpesvirus 1 (61). Compared to the case for mammalian viruses, strategies used by chicken DNA viruses to block the cGAS-STING pathway were not well understood. The strategies were suggested to be especially important for MDV, because periodic lytic replication and de novo infection occur naturally and are required for long-term persistence and pathogenesis in its hosts. We recently reported that the cGAS-STING pathway mediates the IFN-β responses to MDV in chicken fibroblasts and macrophages, and the VP23 protein encoded by MDV inhibited this pathway by suppressing IRF7 activation (32). In the present study, based on a screen of MDV open reading frames (ORFs), we identified MDV RLORF4 as a novel efficient inhibitor of IFN-β induction. By interacting with chicken p65 and p50 of the NF-κB pathway, RLORF4 suppresses their nuclear translocation, which eventually inhibits the IFN-β response during MDV infection.
NF-κB is a critical regulator of innate immunity and inflammation in response to pathogens (62). Manipulation of the NF-κB pathway is a common principle used by viruses to inhibit the host innate immune system. This intervention can occur at various levels, ranging from the interaction with cellular receptors to the modulation of downstream enzyme activities. For example, the HSV-1 kinase US3 interacts with p65 and blocks its nuclear translocation through hyperphosphorylation (63). Moreover, the HSV-1 immediate early protein ICP0 was previously shown to degrade p50 through its E3 ubiquitin ligase activity (50). In the present study, our results revealed for the first time that MDV RLORF4 markedly suppresses NF-κB activation induced by cGAS-STING. Mechanistically, RLORF4 interacts with the Rel homology domains of p65 and p50, inhibits their translocation to the nuclei, and consequently suppresses the production of IFN-β. RLORF4 might thus be one regulator that maintains a balance between viral infection and the antiviral responses.
RLORF4 is an MDV-specific gene that is located in the repeat long regions of the genome and is directly involved in viral attenuation (28, 64). In this study, deletion of RLORF4 from the MDV genome resulted in improved type I IFN responses in vitro and in vivo. Furthermore, the improved type I IFN responses in RLORF4-deficient MDV-infected chickens led to a marked reduction in viral growth, highlighting the indispensable role of RLORF4 in the relationship between the type I IFN response and viral growth in vivo. In addition, we observed that MDV lacking RLORF4 was severely attenuated with respect to lytic virus replication at 7 to 14 days postinfection. These results suggest that the involvement of RLORF4 in the viral pathogenesis might occur primarily during the latency and reactivation phases.
Some studies have shown that IFN-β induction is a pivotal component of the growing tumor recognition by the innate immune system and have determined a link between type I IFN activity and the CD8+ T cell response against tumor-associated antigens in vivo (65). Self-DNA from dying tumor cells has been suggested to be an important danger signal that triggers the cGAS-STING pathway to induce interferons, which in turn facilitates antigen presentation by major histocompatibility complex class I molecules and the activation of CD8+ T cells (66, 67). Therefore, the cGAS-STING pathway is important for the recognition of tumors by the innate immune system and has a critical role in intrinsic antitumor immunity. A previous analysis of T cell subsets in chickens showed that chickens infected with MDV exhibited downregulation of CD8+ T cell numbers; however, the deletion of MDV oncogenic gene meq restored the numbers of CD8+ T cells to the healthy-control level (68). In this study, the percentage of CD8+ T cells in GAdRLORF4-infected chickens was significantly higher than that in the wtGA group. Thus, it is reasonable to speculate that MDV oncogenes might not only directly interact with tumor suppressor proteins to induce tumors in the host but also affect host antitumor immunity by inhibiting the type I IFN response. Our findings could shed new light on the long-standing question of why MDV can rapidly cause cancer in chickens.
In summary, a new role of MDV RLORF4 in inhibition of IFN-β production through selectively targeting NF-κB signaling has been reported here. The identification of the MDV-encoded IFN antagonists will expand our knowledge on the pathogenesis of MDV, which may facilitate the development of more-effective vaccines against MDV infection.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
DF-1 and HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco-BRL) supplemented with 10% fetal bovine serum (FBS) and antibiotics (100 U/ml penicillin and 100 g/ml streptomycin). CEFs and duck embryo fibroblasts (DEFs) were prepared from 10-day-old specific-pathogen-free (SPF) embryos by standard methods and maintained in DMEM supplemented with 5% FBS and antibiotics. The virulent MDV strain GA (GenBank accession no. AF147806) and HVT strain FC126 (GenBank no. AF291866) were propagated in CEFs or DF-1 cells and titrated as described previously (69). Commercially available antibodies were used, including rabbit anti-HA, mouse anti-Flag, and mouse antiactin (Sigma-Aldrich, St. Louis, MO, USA). Anti-p65 antibodies were prepared in our laboratory, and anti-p50 antibodies were purchased from Abcam (Cambridge, UK). ISD and poly(dA·dT) were purchased from InvivoGen (San Diego, CA, USA), and chicken TNF-α was purchased from USCN Life Science (Wuhan, China).
Plasmid construction.
The chicken IFN-β promoter luciferase reporter plasmid (pchIFN-β-luc) was constructed by inserting the fragment from position −158 to +14 of the chicken IFN-β promoter into the pGL3-basic vector, as described previously (31, 34). The pIRF7-luc reporter contained four copies of the IRF7-binding positive regulatory domain (GCA AAT AGA AAGC), and the pNF-κB-luc reporter contained four copies of the NF-κB-binding positive regulatory domain (GGG AAT TCT C). A pRL-TK plasmid (Promega) expressing Renilla luciferase was used as a control. The RLORF4 gene was amplified from the MDV genome and cloned into the pCAGGS vector with a Flag tag fused to its 3′ end to yield RLORF4-Flag. Plasmids encoding chicken cGAS (GenBank no. XM_419881), STING (GenBank no. KP893157), IKKα (GenBank no. NM_001012904), IKKβ (GenBank no. NM_001031397), p65 (GenBank no. NM_205129), or p50 (GenBank no. D13719) were constructed by cloning the synthesized sequences into pCAGGS with Flag or HA tags fused to the 3′ ends.
RNA isolation and real-time qPCR.
Total RNA was extracted using the RNAiso Plus reagent (TaKaRa, Otsu, Japan). The cDNA was obtained using the ReverTra Ace qPCR RT kit (Toyobo, Osaka, Japan) and was used as a template for quantitative PCR (qPCR). The qPCR primers for IFN-β, IL-6, chicken ISGs ZAP, IFITM3, and β-actin were synthesized by Invitrogen (Shanghai, China). To determine the MDV viral DNA copy numbers, total DNA was extracted using the AxyPrep BodyFluid viral DNA/RNA miniprep kit (Corning Life Sciences, Shanghai, China) and tested by real-time qPCR by measuring copy numbers of the MDV meq gene as the MDV genome target and the chicken ovotransferrin gene as a reference, as described previously (70). All controls and treated samples were examined in triplicate in the same plate.
ELISA.
Secreted IFN-β in cell cultures was analyzed using the chicken IFN-β enzyme-linked immunosorbent assay (ELISA) kit (USCN Life Science, Wuhan, China) according to the manufacturer’s instructions.
Transfection and DLR assays.
DF-1 cells were cotransfected with a firefly luciferase reporter plasmid at 0.2 μg/well (IFN-β-luc, IRF7-luc, or NF-κB-luc) and the Renilla luciferase reporter pRL-TK at 0.02 μg/well, with or without expression plasmids as indicated, using the TransIT-X2 dynamic delivery system (Mirus, Madison, WI, USA). At 24 h posttransfection, cells were lysed, and samples were assayed for firefly and Renilla luciferase activities with the dual-luciferase reporter (DLR) assay system (Promega, Madison, WI, USA) as described previously (31). The reporter assays were repeated at least three times.
Co-IP assays and Western blot analysis.
The expression plasmids harboring Flag or HA tags were transfected into HEK293T cells with the TransIT-X2 dynamic delivery system (Mirus). At 36 h posttransfection, cells were lysed in ice-cold Pierce immunoprecipitation (IP) buffer (Thermo Fisher Scientific, Waltham, MA, USA) containing 1 mM phenylmethylsulfonyl fluoride (PMSF). The lysates were obtained by centrifugation and incubated with the indicated antibodies at 4°C overnight. Protein G-Sepharose beads (Roche) were added, and samples were incubated for another 6 h. The beads were washed six times with phosphate-buffered saline (PBS) and boiled in 5× SDS loading buffer before analysis by Western blotting with the indicated antibodies. The images were collected with the Odyssey infrared imaging system (Li-Cor Biosciences, USA).
GST pulldown assay.
GST-RLORF4 was bound to glutathione-agarose beads and incubated for 4 h with lysates from HEK293T cells transiently expressing p65-HA or p50-HA at 4°C. The beads were washed five times with NP-40 lysis buffer (Beyotime Biotechnology, Shanghai, China), mixed with 5× SDS loading buffer, and boiled for 10 min. The input/elutes were resolved by SDS-PAGE and analyzed by Coomassie blue staining and/or immunoblot analysis.
Subcellular localization.
DF-1 cells grown in 35-mm culture dishes were transfected with the plasmids using the TransIT-X2 dynamic delivery system, and 24 h later, they were treated with 10 ng/ml of TNF-α for 30 min or with 2 μg/ml of ISD for 12 h. For confocal imaging, cells were first fixed with 4% paraformaldehyde for 30 min and permeabilized with 0.1% Triton X-100 in PBS for 15 min, which was followed by blocking with 5% bovine serum albumin in PBS for 1 h. The cells were then incubated with rabbit anti-p65 and anti-p50 and mouse anti-Flag antibodies. The cells were washed five times with PBS and incubated with Alexa 488–anti-rabbit and Alexa 546–anti-mouse secondary antibodies (Abcam). Finally, nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich). After washing five times with PBS, the cells were examined using a confocal microscope system (LSM880; Zeiss, Oberkochen, Germany).
Generation of RLORF4-deleted recombinant MDV.
Six fosmid clones, GA1 to GA6, containing sequences encompassing the entire genome of the virulent MDV strain GA were constructed in our preliminary studies and were used here for the generation of recombinant GA viruses lacking the RLORF4 gene (Fig. 5A). Fosmids GA1 and GA5, containing a copy of the coding sequence of RLORF4, were used for the deletion of this gene using the Counter-Selection BAC modification kit (Gene Bridges). GA1 and GA5 fosmid clones in which the RLORF4 gene had been deleted, namely, GA1dRLORF4 and GA5dRLORF4, respectively, were identified by PCR analyses and sequencing. To rescue the RLORF4-deleted recombinant virus, GAdRLORF4, 2 μg of each NotI-digested and purified fosmid DNA (GA1dRLORF4, GA2, GA3, GA4, GA5dRLORF4, and GA6) was used to transfect primary DEFs in 60-mm dishes. Five days after transfection, cells were trypsinized, seeded onto a 100-mm dish, and monitored for cytopathic effects. The viral stocks were subsequently made in DEFs for further analysis.
Animal studies.
In total, 90 1-day-old SPF chickens were randomly divided into three groups, with 30 chickens in each group. Two groups were inoculated subcutaneously on the back of the neck with 2000 PFU of wtGA or GAdRLORF4, and the third group was mock injected with DMEM. On days 3, 7, 10, 14, 21, and 28, five birds from each group were humanely euthanized. The spleen and thymus were collected for the analysis of IFN-β and chicken ISG expression and viral DNA copy numbers, and the anticoagulant blood samples were collected to obtain peripheral blood lymphocytes using a chicken peripheral blood lymphocyte separation fluid kit (TBD, Tianjin, China). The cell suspensions were stained with a fluorescein isothiocyanate (FITC)-conjugated anti-chicken CD4 monoclonal antibody (MAb), an R-phycoerythrin (R-PE)-conjugated anti-chicken CD8a MAb, and an R-PE/cyanine 5 (SPRD)-conjugated anti-chicken CD3 MAb (Southern Biotechnology Associates, Birmingham, AL, USA) for 30 min at 4°C. After cells were washed with PBS, the relative immunofluorescence of cells was analyzed using a flow cytometer (Cytomics TM FC 500; Beckman Coulter, USA). The protocol of the animal study was approved by the Committee on the Ethics of Animal Experiments of the Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences (approval number SYXK [Heilongjiang] 2011-022).
Statistical analysis.
The data are presented as the means ± standard deviations (SDs). Statistical significance between groups was determined by performing a Student t test with GraphPad Prism 7.0 software. A P value of <0.05 was considered significant.
ACKNOWLEDGMENTS
This research was supported by grants from the National Natural Science Foundation of China (31600127) and the National Key Research and Development Program of China (2016YFE0203200 and 2017YFD0500704).
REFERENCES
- 1.Ward CJ. 2010. Pathogen sensing in innate immunity. Expert Rev Vaccines 9:19–21. doi: 10.1586/erv.09.141. [DOI] [PubMed] [Google Scholar]
- 2.Cunha A. 2012. Innate immunity: pathogen and xenobiotic sensing—back to basics. Nat Rev Immunol 12:400. doi: 10.1038/nri3237. [DOI] [PubMed] [Google Scholar]
- 3.Wu J, Chen ZJ. 2014. Innate immune sensing and signaling of cytosolic nucleic acids. Annu Rev Immunol 32:461–488. doi: 10.1146/annurev-immunol-032713-120156. [DOI] [PubMed] [Google Scholar]
- 4.Nie Y, Wang YY. 2013. Innate immune responses to DNA viruses. Protein Cell 4:1–7. doi: 10.1007/s13238-012-2122-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Unterholzner L. 2013. The interferon response to intracellular DNA: why so many receptors? Immunobiology 218:1312–1321. doi: 10.1016/j.imbio.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 6.Sun L, Wu J, Du F, Chen X, Chen ZJ. 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai X, Chiu YH, Chen ZJ. 2014. The cGAS-cGAMP-STING pathway of cytosolic DNA sensing and signaling. Mol Cell 54:289–296. doi: 10.1016/j.molcel.2014.03.040. [DOI] [PubMed] [Google Scholar]
- 8.Xia P, Wang S, Gao P, Gao G, Fan Z. 2016. DNA sensor cGAS-mediated immune recognition. Protein Cell 7:777–791. doi: 10.1007/s13238-016-0320-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao P, Ascano M, Wu Y, Barchet W, Gaffney BL, Zillinger T, Serganov AA, Liu Y, Jones RA, Hartmann G, Tuschl T, Patel DJ. 2013. Cyclic [G(2',5')pA(3',5')p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell 153:1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Q, Sun L, Chen ZJ. 2016. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat Immunol 17:1142–1149. doi: 10.1038/ni.3558. [DOI] [PubMed] [Google Scholar]
- 11.Shu C, Li X, Li P. 2014. The mechanism of double-stranded DNA sensing through the cGAS-STING pathway. Cytokine Growth Factor Rev 25:641–648. doi: 10.1016/j.cytogfr.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanaka Y, Chen ZJ. 2012. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal 5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abe T, Barber GN. 2014. Cytosolic-DNA-mediated, STING-dependent proinflammatory gene induction necessitates canonical NF-kappaB activation through TBK1. J Virol 88:5328–5341. doi: 10.1128/JVI.00037-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiu YH, Zhao M, Chen ZJ. 2009. Ubiquitin in NF-kappaB signaling. Chem Rev 109:1549–1560. doi: 10.1021/cr800554j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonizzi G, Karin M. 2004. The two NF-kappaB activation pathways and their role in innate and adaptive immunity. Trends Immunol 25:280–288. doi: 10.1016/j.it.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 16.Wu CJ, Conze DB, Li T, Srinivasula SM, Ashwell JD. 2006. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-kappaB activation. Nat Cell Biol 8:398–406. doi: 10.1038/ncb1384. [DOI] [PubMed] [Google Scholar]
- 17.Woronicz JD, Gao X, Cao Z, Rothe M, Goeddel DV. 1997. IkappaB kinase-beta: NF-kappaB activation and complex formation with IkappaB kinase-alpha and NIK. Science 278:866–869. doi: 10.1126/science.278.5339.866. [DOI] [PubMed] [Google Scholar]
- 18.Iwai K. 2014. Diverse roles of the ubiquitin system in NF-kappaB activation. Biochim Biophys Acta 1843:129–136. doi: 10.1016/j.bbamcr.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 19.Hayden MS, Ghosh S. 2008. Shared principles in NF-kappaB signaling. Cell 132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 20.Gilmore TD. 2006. Introduction to NF-kappaB: players, pathways, perspectives. Oncogene 25:6680–6684. doi: 10.1038/sj.onc.1209954. [DOI] [PubMed] [Google Scholar]
- 21.Ma Z, Damania B. 2016. The cGAS-STING defense pathway and its counteraction by viruses. Cell Host Microbe 19:150–158. doi: 10.1016/j.chom.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia-Sastre A. 2017. Ten strategies of interferon evasion by viruses. Cell Host Microbe 22:176–184. doi: 10.1016/j.chom.2017.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davison AJ. 2002. Evolution of the herpesviruses. Vet Microbiol 86:69–88. doi: 10.1016/S0378-1135(01)00492-8. [DOI] [PubMed] [Google Scholar]
- 24.Schat KA, Markowski-Grimsrud CJ. 2001. Immune responses to Marek’s disease virus infection. Curr Top Microbiol Immunol 255:91–120. [DOI] [PubMed] [Google Scholar]
- 25.Boodhoo N, Gurung A, Sharif S, Behboudi S. 2016. Marek’s disease in chickens: a review with focus on immunology. Vet Res 47:119. doi: 10.1186/s13567-016-0404-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nair V. 2013. Latency and tumorigenesis in Marek’s disease. Avian Dis 57:360–365. doi: 10.1637/10470-121712-Reg.1. [DOI] [PubMed] [Google Scholar]
- 27.Lee LF, Wu P, Sui D, Ren D, Kamil J, Kung HJ, Witter RL. 2000. The complete unique long sequence and the overall genomic organization of the GA strain of Marek’s disease virus. Proc Natl Acad Sci U S A 97:6091–6096. doi: 10.1073/pnas.97.11.6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jarosinski KW, Osterrieder N, Nair VK, Schat KA. 2005. Attenuation of Marek’s disease virus by deletion of open reading frame RLORF4 but not RLORF5a. J Virol 79:11647–11659. doi: 10.1128/JVI.79.18.11647-11659.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jarosinski KW, Schat KA. 2007. Multiple alternative splicing to exons II and III of viral interleukin-8 (vIL-8) in the Marek’s disease virus genome: the importance of vIL-8 exon I. Virus Genes 34:9–22. doi: 10.1007/s11262-006-0004-9. [DOI] [PubMed] [Google Scholar]
- 30.Jarosinski KW, Donovan KM, Du G. 2015. Expression of fluorescent proteins within the repeat long region of the Marek’s disease virus genome allows direct identification of infected cells while retaining full pathogenicity. Virus Res 201:50–60. doi: 10.1016/j.virusres.2015.02.012. [DOI] [PubMed] [Google Scholar]
- 31.Cheng Y, Sun Y, Wang H, Yan Y, Ding C, Sun J. 2015. Chicken STING mediates activation of the IFN gene independently of the RIG-I gene. J Immunol 195:3922–3936. doi: 10.4049/jimmunol.1500638. [DOI] [PubMed] [Google Scholar]
- 32.Gao L, Li K, Zhang Y, Liu Y, Liu C, Zhang Y, Gao Y, Qi X, Cui H, Wang Y, Wang X. 2018. Inhibition of DNA-sensing pathway by Marek’s disease virus VP23 protein through suppression of interferon regulatory factor 7 activation. J Virol 93:e01934-18. doi: 10.1128/JVI.01934-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santhakumar D, Rubbenstroth D, Martinez-Sobrido L, Munir M. 2017. Avian interferons and their antiviral effectors. Front Immunol 8:49. doi: 10.3389/fimmu.2017.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sick C, Schultz U, Münster U, Meier J, Kaspers B, Staeheli P. 1998. Promoter structures and differential responses to viral and nonviral inducers of chicken type I interferon genes. J Biol Chem 273:9749–9754. doi: 10.1074/jbc.273.16.9749. [DOI] [PubMed] [Google Scholar]
- 35.Corrales L, Matson V, Flood B, Spranger S, Gajewski TF. 2017. Innate immune signaling and regulation in cancer immunotherapy. Cell Res 27:96–108. doi: 10.1038/cr.2016.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kolumam GA, Thomas S, Thompson LJ, Sprent J, Murali-Krishna K. 2005. Type I interferons act directly on CD8 T cells to allow clonal expansion and memory formation in response to viral infection. J Exp Med 202:637–650. doi: 10.1084/jem.20050821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoneyama M, Fujita T. 2009. RNA recognition and signal transduction by RIG-I-like receptors. Immunol Rev 227:54–65. doi: 10.1111/j.1600-065X.2008.00727.x. [DOI] [PubMed] [Google Scholar]
- 38.Burckstummer T, Baumann C, Bluml S, Dixit E, Durnberger G, Jahn H, Planyavsky M, Bilban M, Colinge J, Bennett KL, Superti-Furga G. 2009. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol 10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 39.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG. 2010. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol 11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chiu YH, Macmillan JB, Chen ZJ. 2009. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, Ohba Y, Taniguchi T. 2007. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 42.Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ. 2013. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science 341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xing J, Ni L, Wang S, Wang K, Lin R, Zheng C. 2013. Herpes simplex virus 1-encoded tegument protein VP16 abrogates the production of beta interferon (IFN) by inhibiting NF-kappaB activation and blocking IFN regulatory factor 3 to recruit its coactivator CBP. J Virol 87:9788–9801. doi: 10.1128/JVI.01440-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang S, Wang K, Lin R, Zheng C. 2013. Herpes simplex virus 1 serine/threonine kinase US3 hyperphosphorylates IRF3 and inhibits beta interferon production. J Virol 87:12814–12827. doi: 10.1128/JVI.02355-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang D, Su C, Zheng C. 2016. Herpes simplex virus 1 serine protease VP24 blocks the DNA-sensing signal pathway by abrogating activation of interferon regulatory factor 3. J Virol 90:5824–5829. doi: 10.1128/JVI.00186-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Christensen MH, Jensen SB, Miettinen JJ, Luecke S, Prabakaran T, Reinert LS, Mettenleiter T, Chen ZJ, Knipe DM, Sandri-Goldin RM, Enquist LW, Hartmann R, Mogensen TH, Rice SA, Nyman TA, Matikainen S, Paludan SR. 2016. HSV-1 ICP27 targets the TBK1-activated STING signalsome to inhibit virus-induced type I IFN expression. EMBO J 35:1385–1399. doi: 10.15252/embj.201593458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang J, You H, Su C, Li Y, Chen S, Zheng C. 2018. Herpes simplex virus 1 tegument protein VP22 abrogates cGAS/STING-mediated antiviral innate immunity. J Virol 92:e00841-18. doi: 10.1128/JVI.00841-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deschamps T, Kalamvoki M. 2017. Evasion of the STING DNA-sensing pathway by VP11/12 of herpes simplex virus 1. J Virol 91:e00535-17. doi: 10.1128/JVI.00535-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu H, Su C, Pearson A, Mody CH, Zheng C. 2017. Herpes simplex virus 1 UL24 abrogates the DNA sensing signal pathway by inhibiting NF-kappaB activation. J Virol 91:e00025-17. doi: 10.1128/JVI.00025-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang J, Wang K, Wang S, Zheng C. 2013. Herpes simplex virus 1 E3 ubiquitin ligase ICP0 protein inhibits tumor necrosis factor alpha-induced NF-kappaB activation by interacting with p65/RelA and p50/NF-kappaB1. J Virol 87:12935–12948. doi: 10.1128/JVI.01952-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Su C, Zheng C. 2017. Herpes simplex virus 1 abrogates the cGAS/STING-mediated cytosolic DNA-sensing pathway via its virion host shutoff protein, UL41. J Virol 91:e02414-16. doi: 10.1128/JVI.02414-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang J, Wang S, Wang K, Zheng C. 2013. Herpes simplex virus 1 DNA polymerase processivity factor UL42 inhibits TNF-α-induced NF-κB activation by interacting with p65/RelA and p50/NF-κB1. Med Microbiol Immunol 202:313–325. doi: 10.1007/s00430-013-0295-0. [DOI] [PubMed] [Google Scholar]
- 53.Ye R, Su C, Xu H, Zheng C. 2017. Herpes simplex virus 1 ubiquitin-specific protease UL36 abrogates NF-κB activation in DNA sensing signal pathway. J Virol 91:e02417-16. doi: 10.1128/JVI.02417-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fu YZ, Su S, Gao YQ, Wang PP, Huang ZF, Hu MM, Luo WW, Li S, Luo MH, Wang YY, Shu HB. 2017. Human cytomegalovirus tegument protein UL82 inhibits STING-mediated signaling to evade antiviral immunity. Cell Host Microbe 21:231–243. doi: 10.1016/j.chom.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 55.Huang ZF, Zou HM, Liao BW, Zhang HY, Yang Y, Fu YZ, Wang SY, Luo MH, Wang YY. 2018. Human cytomegalovirus protein UL31 inhibits DNA sensing of cGAS to mediate immune evasion. Cell Host Microbe 24:69–80. doi: 10.1016/j.chom.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 56.Ma Z, Jacobs SR, West JA, Stopford C, Zhang Z, Davis Z, Barber GN, Glaunsinger BA, Dittmer DP, Damania B. 2015. Modulation of the cGAS-STING DNA sensing pathway by gammaherpesviruses. Proc Natl Acad Sci U S A 112:E4306–4315. doi: 10.1073/pnas.1503831112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu JJ, Li W, Shao Y, Avey D, Fu B, Gillen J, Hand T, Ma S, Liu X, Miley W, Konrad A, Neipel F, Sturzl M, Whitby D, Li H, Zhu F. 2015. Inhibition of cGAS DNA sensing by a herpesvirus virion protein. Cell Host Microbe 18:333–344. doi: 10.1016/j.chom.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang G, Chan B, Samarina N, Abere B, Weidner-Glunde M, Buch A, Pich A, Brinkmann MM, Schulz TF. 2016. Cytoplasmic isoforms of Kaposi sarcoma herpesvirus LANA recruit and antagonize the innate immune DNA sensor cGAS. Proc Natl Acad Sci U S A 113:E1034–E1043. doi: 10.1073/pnas.1516812113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu L, Fossum E, Joo CH, Inn KS, Shin YC, Johannsen E, Hutt-Fletcher LM, Hass J, Jung JU. 2009. Epstein-Barr virus LF2: an antagonist to type I interferon. J Virol 83:1140–1146. doi: 10.1128/JVI.00602-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun C, Schattgen SA, Pisitkun P, Jorgensen JP, Hilterbrand AT, Wang LJ, West JA, Hansen K, Horan KA, Jakobsen MR, O’Hare P, Adler H, Sun R, Ploegh HL, Damania B, Upton JW, Fitzgerald KA, Paludan SR. 2015. Evasion of innate cytosolic DNA sensing by a gammaherpesvirus facilitates establishment of latent infection. J Immunol 194:1819–1831. doi: 10.4049/jimmunol.1402495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tian J, Liu Y, Liu X, Sun X, Zhang J, Qu L. 2018. Feline herpesvirus 1 US3 blocks the type I interferon signal pathway by targeting interferon regulatory factor 3 dimerization in a kinase-independent manner. J Virol 92:e00047-18. doi: 10.1128/JVI.00047-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ghosh S, May MJ, Kopp EB. 1998. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol 16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 63.Wang K, Ni L, Wang S, Zheng C. 2014. Herpes simplex virus 1 protein kinase US3 hyperphosphorylates p65/RelA and dampens NF-kappaB activation. J Virol 88:7941–7951. doi: 10.1128/JVI.03394-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Spatz SJ, Silva RF. 2007. Polymorphisms in the repeat long regions of oncogenic and attenuated pathotypes of Marek’s disease virus 1. Virus Genes 35:41–53. doi: 10.1007/s11262-006-0024-5. [DOI] [PubMed] [Google Scholar]
- 65.Berraondo P, Minute L, Ajona D, Corrales L, Melero I, Pio R. 2016. Innate immune mediators in cancer: between defense and resistance. Immunol Rev 274:290–306. doi: 10.1111/imr.12464. [DOI] [PubMed] [Google Scholar]
- 66.Deng L, Liang H, Xu M, Yang X, Burnette B, Arina A, Li XD, Mauceri H, Beckett M, Darga T, Huang X, Gajewski TF, Chen ZJ, Fu YX, Weichselbaum RR. 2014. STING-dependent cytosolic DNA sensing promotes radiation-induced type I interferon-dependent antitumor immunity in immunogenic tumors. Immunity 41:843–852. doi: 10.1016/j.immuni.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ho SS, Zhang WY, Tan NY, Khatoo M, Suter MA, Tripathi S, Cheung FS, Lim WK, Tan PH, Ngeow J, Gasser S. 2016. The DNA structure-specific endonuclease MUS81 mediates DNA sensor STING-dependent host rejection of prostate cancer cells. Immunity 44:1177–1189. doi: 10.1016/j.immuni.2016.04.010. [DOI] [PubMed] [Google Scholar]
- 68.Li Y, Sun A, Su S, Zhao P, Cui Z, Zhu H. 2011. Deletion of the Meq gene significantly decreases immunosuppression in chickens caused by pathogenic Marek’s disease virus. Virol J 8:2. doi: 10.1186/1743-422X-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Geerligs H, Quanz S, Suurland B, Spijkers IE, Rodenberg J, Davelaar FG, Jongsma B, Kumar M. 2008. Efficacy and safety of cell associated vaccines against Marek’s disease virus grown in a continuous cell line from chickens. Vaccine 26:5595–5600. doi: 10.1016/j.vaccine.2008.07.080. [DOI] [PubMed] [Google Scholar]
- 70.Sun GR, Zhang YP, Zhou LY, Lv HC, Zhang F, Li K, Gao YL, Qi XL, Cui HY, Wang YQ, Gao L, Pan Q, Wang XM, Liu CJ. 2017. Co-infection with Marek’s disease virus and reticuloendotheliosis virus increases illness severity and reduces Marek’s disease vaccine efficacy. Viruses 9:158. doi: 10.3390/v9060158. [DOI] [PMC free article] [PubMed] [Google Scholar]