An emergent adenoviral human pathogen, HAdV-B76, associated with a fatality in 1965, shows a remarkable degree of genome identity with two recently isolated simian adenoviruses that contain cross-species genome recombination events from three hosts: human, chimpanzee, and bonobo. Zoonosis (nonhuman-to-human transmission) and anthroponosis (human to nonhuman transmission) may play significant roles in the emergence of human adenoviral pathogens.
KEYWORDS: adenovirus, anthroponosis, emerging pathogen, genomics, homologous recombination, virus evolution, zoonosis
ABSTRACT
Genomics analysis of a historically intriguing and predicted emergent human adenovirus (HAdV) pathogen, which caused pneumonia and death, provides insight into a novel molecular evolution pathway involving “ping-pong” zoonosis and anthroponosis. The genome of this promiscuous pathogen is embedded with evidence of unprecedented multiple, multidirectional, stable, and reciprocal cross-species infections of hosts from three species (human, chimpanzee, and bonobo). This recombinant genome, typed as HAdV-B76, is identical to two recently reported simian AdV (SAdV) genomes isolated from chimpanzees and bonobos. Additionally, the presence of a critical adenoviral replication element found in HAdV genomes, in addition to genes that are highly similar to counterparts in other HAdVs, reinforces its potential as a human pathogen. Reservoirs in nonhuman hosts may explain periods of apparent absence and then reemergence of human adenoviral pathogens, as well as present pathways for the genesis of those thought to be newly emergent. The nature of the HAdV-D76 genome has implications for the use of SAdVs as gene delivery vectors in human gene therapy and vaccines, selected to avoid preexisting and potentially fatal host immune responses to HAdV.
IMPORTANCE An emergent adenoviral human pathogen, HAdV-B76, associated with a fatality in 1965, shows a remarkable degree of genome identity with two recently isolated simian adenoviruses that contain cross-species genome recombination events from three hosts: human, chimpanzee, and bonobo. Zoonosis (nonhuman-to-human transmission) and anthroponosis (human to nonhuman transmission) may play significant roles in the emergence of human adenoviral pathogens.
INTRODUCTION
Infectious disease outbreaks caused by emergent and reemergent pathogens are seemingly more frequent, have global impact, and cause intense public concern. Zoonosis plays a major role in the genesis of some of these novel pathogens (1–6), as revealed using rapid, high-resolution, and cost-effective genomics technologies, coupled with computational tools and growing databases. Three recent examples of suspected zoonotic transmission exemplify this social and scientific convergence: the severe acute respiratory syndrome coronavirus (SARS-CoV) outbreak in Hong Kong and China (3), the Middle East respiratory syndrome coronavirus (MERS-CoV) outbreak in the Middle East (7), and the Ebola virus outbreak in Africa (4). A fourth example of a pathogen with zoonosis associated as a pathway for pandemics is influenza virus (1, 5). In assessing these emergent and, in some cases, reemergent pathogens, genomics and bioinformatics have provided critical tools for exact and rapid identification and for characterizing potential vaccine and drug targets.
Human adenoviruses (HAdVs) were among the first, well-studied human viral pathogens since their coisolations as “Respiratory Illness (RI)” viruses ca. 1953 (8, 9). There are currently 90 accepted genotypes characterized by whole-genome analysis (http://hadvwg.gmu.edu/) (10–12); these are parsed into seven species (A to G) based on biological, clinical, and genomic attributes (13). Ironically, they are used as gene and epitope delivery vectors as well as oncolytic agents (14), despite their roles as human pathogens. HAdV is implicated in a wide spectrum of diseases involving the respiratory, ocular, gastrointestinal, and genitourinary systems in humans (13). Infections may be latent and persistent or acute. The latter can cause diseases ranging from mild to severe, including death (13). Zoonotic transmission of simian adenoviruses (SAdV) and subsequent genome recombination with HAdV have led to the emergence and establishment of novel HAdV types, as evidenced by a human pathogen of current concern, HAdV-E4 (15, 16). HAdV-E4 was one of the first human adenoviruses cultured and characterized (9). With what is essentially a chimpanzee SAdV genome, HAdV-E4 remains a major human respiratory pathogen, evidenced by being only one of two HAdVs for which a vaccine has been developed, twice, and at considerable cost (17).
Genomic analysis provides unprecedented views on the role and extent of zoonosis in the genesis of emergent pathogens, including insights into the origins of historically important as well as currently circulating pathogens, e.g., 1918 H1N1 influenza virus, from pigs (5); human immunodeficiency virus (HIV), from chimpanzees and gorillas (18–23); and human herpes simplex virus 2, from an ancestor of modern chimpanzees (24). To date, zoonotic transmissions appear mainly unidirectional, from nonhumans to humans. Instances of human-to-nonhuman transfer, termed anthroponosis or “reverse zoonosis,” are also recognized but noted as “seldom-documented” (2, 25). In this study, a historically intriguing, then-emergent human adenovirus (HAdV) causing an upper respiratory infection and death in 1965 has been sequenced in a large-scale HAdV genomics project. Its genome is embedded with evidence of unprecedented “ping-pong” zoonosis and anthroponosis, indicating stable and recurring infections in hosts across three species (human, chimpanzee, and bonobo). Even more remarkable is the near identity of this human pathogen to two recently sequenced SAdV genomes. Analyses of these three genomes and comparisons with other HAdVs and SAdVs suggest an emergent human pathogen of concern.
RESULTS
Comparative genomics analysis.
The genome of HAdV-B76 was found to have nearly complete identity to two recently sequenced SAdV genomes that were isolated from stools of apparently healthy hosts: chimpanzee (SAdV-B35.1; University of Louisiana New Iberia Research Center, Lafayette, LA) and bonobo (SAdV-B35.2; San Diego Zoo, San Diego, CA) (26). The HAdV-B76 nucleotide sequence is 99.5% and 99.6% identical to counterparts from SAdV-B35.1 and SAdV-B35.2, respectively. For reference, the SAdV-B21 and HAdV-B21 (the closest HAdV) genomes present 94.9% and 89.7% nucleotide identities with HAdV-B76, respectively; the SAdV-B35.1 and SAdV-B35.2 genomes are 99.7% identical to each other. The genome near identity of HAdV-B76 to SAdV-B35.1 and SAdV-B35.2 is remarkable, as the human virus was isolated in 1965 (27) and the SAdV-B35.1 and SAdV-B35.2 viruses were isolated ca. 2008 (26).
Amino acid sequence percent identities of the three major capsid proteins are presented in Table 1. Several adenoviruses are included, based on BLAST results, with respiratory pathogens HAdV-B7 (subspecies B1) and HAdV-B14 (subspecies B2) noted for reference. The counterparts of HAdV-B76 are nearly identical with those in SAdV-B35.1 and SAdV-B35.2. For example, the penton base is 100% identical among the three. For reference, the penton base of the next closest counterpart is HAdV-B21 (99.3%) and SAdV-B21 (98.2%). A second chimpanzee AdV (SAdV-B27.1) shares 97.2% similarity, with this protein being the most similar of the three capsid proteins to that of HAdV-B76. The hexon protein, site of the epsilon epitope responsible for serum neutralization (SN) (28), shows the highest sequence similarity with HAdV-B21 (98.1%), accounting for the past SN observations (27, 29). HAdV-B16 is also reported to cross-react strongly by SN (27, 29) but shows a lower sequence similarity (84.7%). If the SN data are correct, then all viruses listed in Table 1 should also neutralize HAdV-B76. In fact, their cross-reaction should be stronger, as their sequence similarities are higher than that of HAdV-B16. This has implications in the use of SAdVs in place of HAdVs as vectors for vaccines and gene therapy.
TABLE 1.
Amino acid identities of HAdV-B76 proteins to homologsa
| Virus | Host | % amino acid identity |
||||
|---|---|---|---|---|---|---|
| Penton base | Hexon | Fiber | Fiber, proximal | Fiber, distal | ||
| SAdV-B35.1 | Chimpanzee | 100 | 99.8 | 100 | 100 | 100 |
| SAdV-B35.2 | Bonobo | 100 | 100 | 99.4 | 99.2 | 100 |
| SAdV-B21 | Chimpanzee | 98.2 | 92.3 | 81.6 | 72.6 | 100 |
| SAdV-B27.1 | Chimpanzee | 97.2 | 86.1 | 57.5 | 57.8 | 56.8 |
| HAdV-B16 | Human | 84.5 | 84.7 | 97.5 | 97.5 | 97.4 |
| HAdV-B21 | Human | 99.3 | 98.1 | 54.3 | 55.7 | 51.7 |
| HAdV-B7 | Human | 85.5 | 86 | 53.6 | 53.6 | 53.8 |
| HAdV-B14 | Human | 91.8 | 89.6 | 51.8 | 51.3 | 52.9 |
Comparison of representative HAdV-B76 proteins to their homologs in select adenoviruses. Amino acid identities of the three major capsid proteins were calculated using EMBOSS software (http://www.ebi.ac.uk/Tools/psa/emboss_needle), with default parameters. Specifically, global alignments were performed using the Needle program. The BLOSUM62 matrix was used for the sequence analysis. Reflecting the bipartite nature of the fiber protein, the shaft was analyzed separately from the gamma epitope and cell tropism determinant knob: proximal (shaft; 237 amino acids) and distal sequences (knob; 117 amino acids). The sequence identities of these three capsid proteins reflect the nearly identical genomes of HAdV-B76 and the two SAdV-B35 counterparts.
The fiber protein was analyzed in two parts, corresponding to the shaft and the knob, which contains the gamma epitope (28). The corresponding fiber regions from HAdV-B76 are 97.5% and 97.4% similar to their counterparts in HAdV-B16, consistent with prior hemagglutination inhibition (HI) observations (27, 29). The next closest is SAdV-B21, with 72.6% and 100% sequence similarity. For reference, the counterparts from HAdV-B21 are 55.7% and 51.7% similar, respectively, consistent with no cross-reactions in HI assays (27, 29).
Sequence recombination analysis.
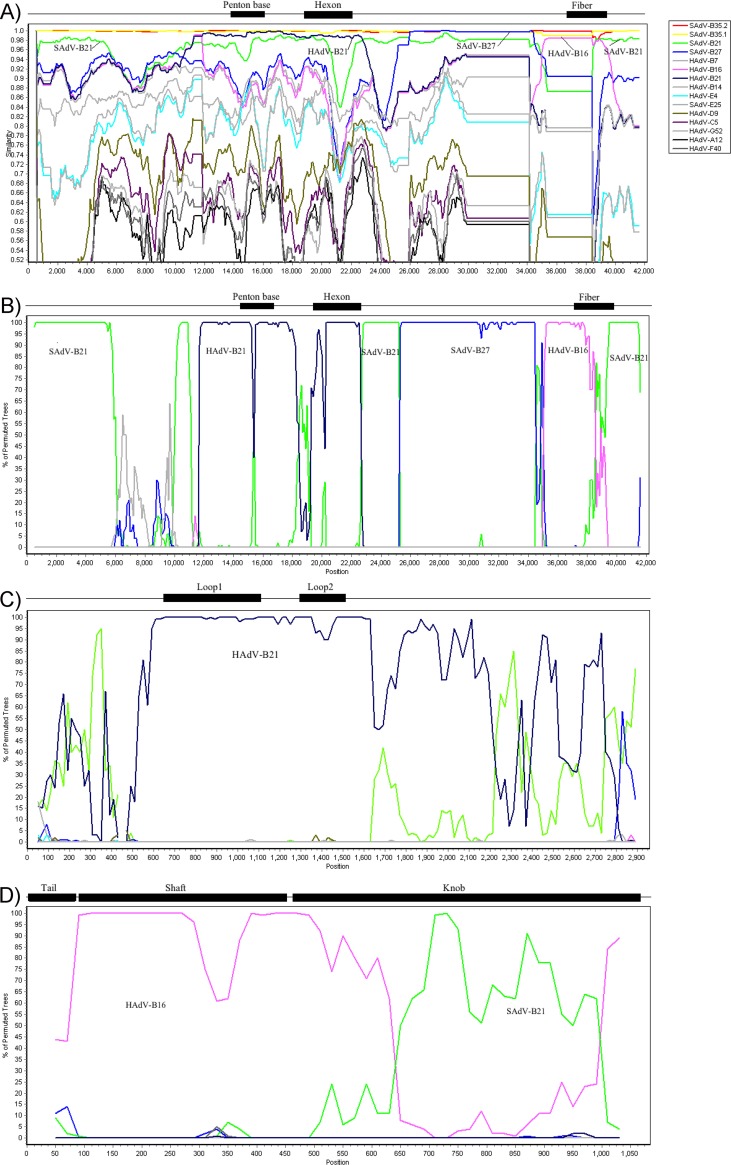
Comparative genome analysis, including BLAST and protein percent identities of the HAdV-B76 genes, indicated evidence of multiple recombination events embedded in the genome. A detailed query for lateral genomics transfer in the HAdV-B76 genome sequence was completed using Simplot, which provides the percent similarity of the genome sequences (Fig. 1A) and a bootscan plot that highlights regions of recombination using the percent similarities (Fig. 1B). Bootscan plots are presented for the hexon gene (Fig. 1C) and the fiber gene (Fig. 1D) for higher-resolution views. These analyses provide a schematic view of molecular evolution events involving zoonotic and anthroponotic infections, coinfections, and lateral genomics sequence transfers. Specifically, there are contributions from HAdV-B16 (fiber; human host), HAdV-B21 (penton base and hexon; human host), SAdV-B21 (3′-proximal genomics region and 5′-distal genomics region; chimpanzee host), and SAdV-B27 (genomics region between hexon and fiber, including E3 genes; chimpanzee host) in the genome of HAdV-B76 (Fig. 1A and B). SAdV-B21 was isolated from a chimpanzee with respiratory illness at a National Institutes of Health (NIH) facility in 1954 (30), and SAdV-B27 was recently isolated from an asymptomatic chimpanzee at the New Iberia Research Center (26). A region of low sequence similarity in the whole-genome profile (Fig. 1B) indicates that a 5′ genomic region between the inverted terminal repeat (ITR) and penton base contains sequences from unknown (yet to be reported and/or sequenced) adenovirus(es). As SAdV-B21 appeared to contribute sequences throughout the SAdV-B35 genome, including the 5′-end region, two internal regions, and the 3′-end region, it is likely the ancestral genome to which the other viruses contributed. The hexon bootscan profile shows HAdV-B21 providing the variable regions, loops 1 and 2, comprising the SN epitope (Fig. 1C). Bootscan analysis of the fiber indicates that it is recombinant, as the shaft is from HAdV-B16 while the knob domain is from SAdV-B21; it is a novel fiber protein that is identical to counterparts from SAdV-B35.1 and -B35.2 (chimpanzee and bonobo, respectively).
FIG 1.
Survey of recombination events embedded in HAdV-B76. HAdV-B76 was analyzed for recombination using software(http://sray.med.som.jhmi.edu/SCRoftware/simplot/) which provides a SimPlot analysis (A) as well as a Bootscan analysis (B) of the genome. The results are consistent with lateral transfers of genomics regions from SAdV-B27, HAdV-B21, and HAdV-B16 into the genome chassis of SAdV-B21 and resulting in the recombinant HAdV-B76. The box on the right shows the reference genomes and their colors. Additional inspection of the hexon gene (C) and the fiber gene (D) provides detailed views, confirming the original serotyping or virus neutralization of this virus as HAdV-B21 (hexon) and hemagglutination as HAdV-B16 (fiber). Hexon loops 1 and 2 contain the epitopes upon which serotyping is based. The fiber knob contains the epitope for hemagglutination inhibition. Initially, MAFFT software was used to align the sequences prior to analysis of recombination events (http://mafft.cbrc.jp/alignment/server/). Default parameter settings for the Simplot software applied to the gene sequences were as follows: window size, 200 nucleotides; step size, 20 nucleotides; replicates used, n = 100; gap stripping, on; distance model, Kimura; and tree model, neighbor joining. For the whole-genome analysis, only the window size and step size were altered (1,000 and 200, respectively), with the remaining default parameters unchanged. Genome nucleotide positions are noted along the x axis, and the percentages of nucleotide identities (Simplot) or permuted trees (bootscan) that supported grouping are marked along the y axis. For reference, genome-specific or gene-specific landmarks are noted above each graph; these are approximate in location and size.
Phylogenomics analysis.
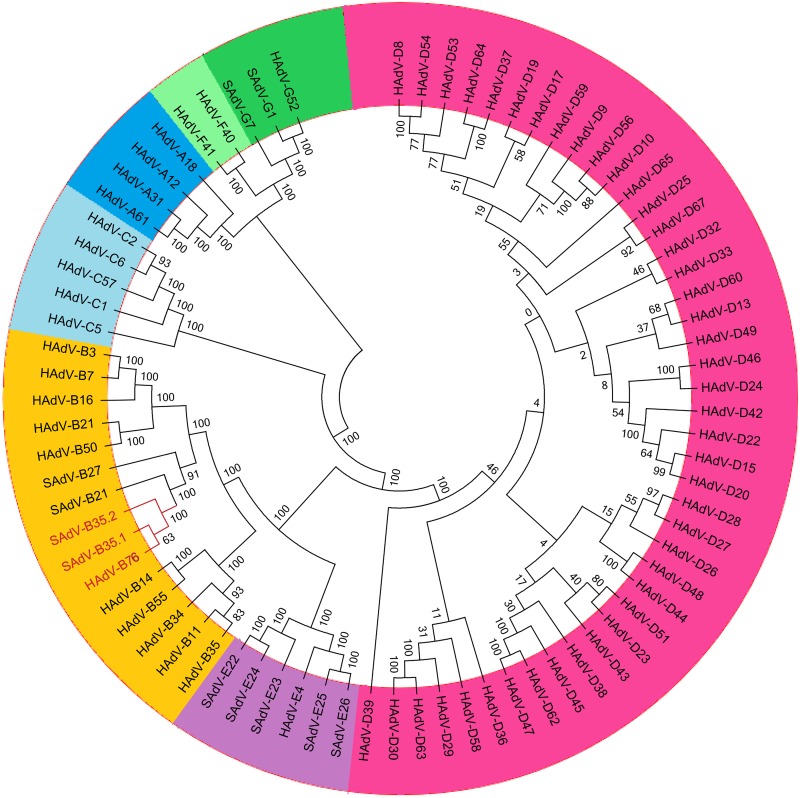
A whole-genome phylogeny analysis of HAdV and SAdV genomes is shown in Fig. 2. This group of genomes was chosen to place into context the relationships between HAdV-B76, SAdV-B35.1, and SAdV-B35.2 (red lettering) and other SAdVs and HAdVs. It is noted that within the context of the data, the simplest explanation of these sequence relationships is that the HAdV-B76, SAdV-B35.1, and SAdV-B35.2 genomes share significant sequence similarities to several SAdV and HAdV genomes, particularly ones within species B, and that the SAdVs are both intermingled with HAdVs, suggesting recombinants and common ancestry, and in a separate subclade. Phylogenetic analysis of the whole-genome data using neighbor-joining (Fig. 2), maximum likelihood, and maximum parsimony algorithms all reveal similar well-resolved phylogenetic relationships, with similar topologies (data not shown).
FIG 2.
Whole-genome phylogenomics analysis of HAdV-B76. A neighbor-joining bootstrap-confirmed phylogenetic tree was generated following multiple sequence alignment using the MAFFT software (http://mafft.cbrc.jp/alignment/server/) with default parameters. The tree is from 1,000 replicates, constructed using a maximum-composite-likelihood model in the MEGA 5.1 software (http://megasoftware.net). Bootstrap numbers shown at the nodes indicate the percentages of 1,000 replications producing the clade, with values above 80 considered robust. The seven human adenovirus species are contrasted by color, with the HAdV-B76 and nearly identical genomes of SAdV-B35.1 and SAdV-B35.2 in red lettering (species A, dark blue; species B, orange; species C, light blue; species D, magenta; species E, lavender; species F, light green; and species G, dark green).
A sampling of selected genomes from the phylogenetically closest respiratory pathogen subspecies B1 is presented in Fig. 3A, along with selected SAdVs. This analysis showed the genomes of HAdV-B76, SAdV-B35.1, and SAdV-B35.2 forming a subclade, with HAdV-B76 nearly indistinguishable from SAdV-B35.1 and -B35.2 at a bootstrap value of 69 (bootstrap values above 80 are considered robust). The next nearest genome is a subclade of SAdV-B21 (bootstrap value, 100), and these four are separated from the next nearest genome, SAdV-B27 (bootstrap value, 100). HAdVs comprising the respiratory pathogens of species B1, including HAdV-B21, are clustered in a separate subclade from these SAdVs and HAdV-B76 (bootstrap value, 100). Subspecies B2 virus HAdV-B14, also a respiratory pathogen, forms a subclade from this group.
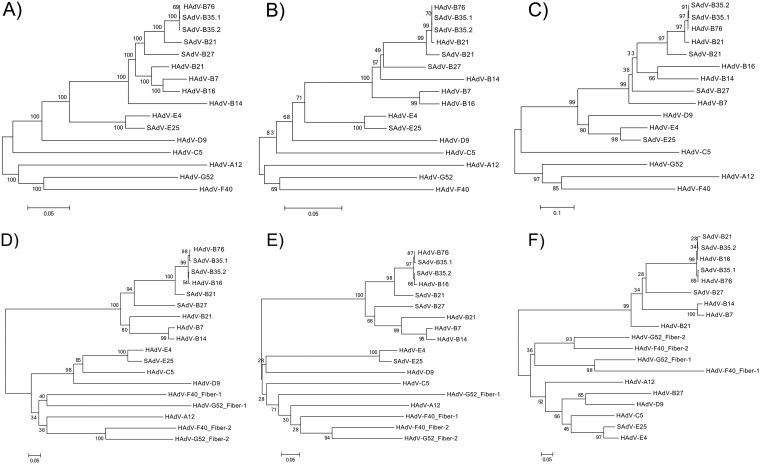
FIG 3.
Phylogenomics analysis of the HAdV-B76 genome and selected genes in the context of the recombinant genome. To provide a clear view of the recombination events, neighbor-joining bootstrap-confirmed phylogenetic trees of the whole-genome (A), penton base (B), hexon (C), and fiber (D) sequences are presented. The fiber gene (D) was further dissected for the detailed analyses of two domains: the shaft, encoded by the proximal sequences (716 bp) (E), and the knob (F), encoded by the distal sequences that include the variable region specifying the hemagglutination inhibition epitope (gamma) and dictating cell tropism (346 bp). The fiber gene appeared recombinant, so domains were extracted using the HAdV-B76 gene sequence and its bootscan profile as a reference, as the shaft and knob lengths varied per HAdV species. These phylogenetic trees were generated after multiple-sequence alignments using the MAFFT software (http://www.ebi.ac.uk/tools/mafft) and default parameters. Bootstrapped, neighbor-joining trees with 1,000 replicates were constructed using MEGA 5.1 software (http://megasoftware.net) with a maximum-composite-likelihood model. Bootstrap values depicted at the nodes indicate the percentages of 1,000 replications producing the clade, with values above 80 considered robust. The scale bar is in units of nucleotide substitutions per site.
To obtain higher-resolution views of lateral gene transfers, several genes were examined in greater detail. It is noted that within the context of the data, the direction of gene transfer is difficult to establish with precision, but the thesis that there are cross-infections and coinfections of human and simian hosts by adenoviruses, with lateral genomic transfers, is supported by these observations. The phylogenetic analysis of the penton base genes showed HAdV-B76 as indistinguishable from both SAdV-B35.1 and -B35.2 and HAdV-B21 (Fig. 3B). SAdV-B21 and SAdV-B27 form branches from this subclade and from each other; however, the bootstrap value (i.e., 49) indicated that this separation is not significant.
An inspection of the hexon also showed HAdV-B76 as indistinguishable from both SAdV-B35.1 and -B35.2, but HAdV-B21 forms a branch with a higher bootstrap value (97), as does SAdV-B21 (bootstrap value, 97) (Fig. 3C). SAdV-B27 appears more distant, but this separation may not be significant, with a bootstrap value of 36. The overall tree had a profile similar to that of the penton base tree (Fig. 3B), suggesting that both genes were participants in one recombination event.
The third gene examined was the fiber (Fig. 3D). As observed for the penton base and hexon, HAdV-B76 is in a subclade with both SAdV-B35.1 and -B35.2; however, HAdV-B16 is also contained within this subclade. The phylogenetic distances among them are small but the bootstrap values are suggestive, at 98 for HAdV-B76 and SAdV-B35.1 and 99 for this pair and the pair SAdV-B35.2 and HAdV-B16, the latter two indistinguishable (bootstrap value of 54). SAdV-B21 is the next closest, followed by SAdV-27. As the fiber appears to be recombinant via pairwise genome alignments, protein percent similarities, and recombination detection algorithms, it was reanalyzed as two domains: 716 bases were extracted for the shaft (Fig. 3E), and the abutting 346 bases were extracted for the knob (Fig. 3F). The shaft sequence has high similarity with HAdV-B16, and the knob has high similarity with SAdV-B21; this is reflected in the phylogenetic trees. Phylogenetic distribution of the shaft sequences showed HAdV-B76 forming a subclade with both SAdV-B35.1 and -B35.2 and with HAdV-B16, with a bootstrap value of 66 separating the latter. Again, both SAdV-B21 and -B27 form separate branches from this subclade. In contrast, the knob sequences presented a profile in which SAdV-B21 is also contained within the HAdV-B76 subclade, essentially indistinguishable, with a bootstrap value of 28. This is consistent with the recombination analysis indicating that the 3′ end of the genome is that of SAdV-B21.
Comparative sequence analysis of the hexon epsilon epitope.
HAdV-B76 is historically intriguing for two reasons. It was a human pathogen that emerged in association with a fatal infection and serotyped, unusually, as two distinct HAdVs: 16 and 21 (27, 29). Its nonstandard name (“21 + 16H16”) reflected the latter feature. To provide higher resolution, and using the molecular modeling and monoclonal antibody (MAb) neutralization data of Yuan et al. (31), based on earlier genetic mapping data from Crawford-Miksza and Schnurr (32), the five sequence motifs across two hypervariable loops that comprise the epsilon epitope (28) were compared (Fig. 4). Across these five motifs, HAdV-B76 shares identical amino acid sequences with SAdV-B35.2 and nearly identical amino acid sequences with SAdV-B35.1, with only one amino acid different (motif 2, E to K). HAdV-B21 shares identity or near identity across all five, consistent with its SN. However, the HAdV-B16 motifs are divergent and do not reconcile with HAdV-B76, also serotyping as HAdV-B16 (27, 29). This brings into question whether these are the sole SN epitopic regions and to what degree serotyping provides clarity in distinguishing HAdV types (29, 33).
FIG 4.
Multiple sequence alignment of the serum neutralization (SN) domain of the hexon gene. Five amino acid motifs that correspond to the hypervariable sequences encoding the putative serotyping (SN) or virus neutralization (VN) epitopes are highlighted in loops 1 and 2 of the hexon gene. HAdV-B76, SAdV-B35.1, and SAdV-B35.2 contain identical amino acid sequences in these regions, except at one amino acid (SAdV-B35.1; motif 2, E to K). These are nearly identical to HAdV-B21, accounting for the SN observations. However, divergent sequences contained in HAdV-B16 do not explain why HAdV-B76 is desribed in the literature to serotype as HAdV-B16.
Comparative sequence analysis of the fiber gamma epitope.
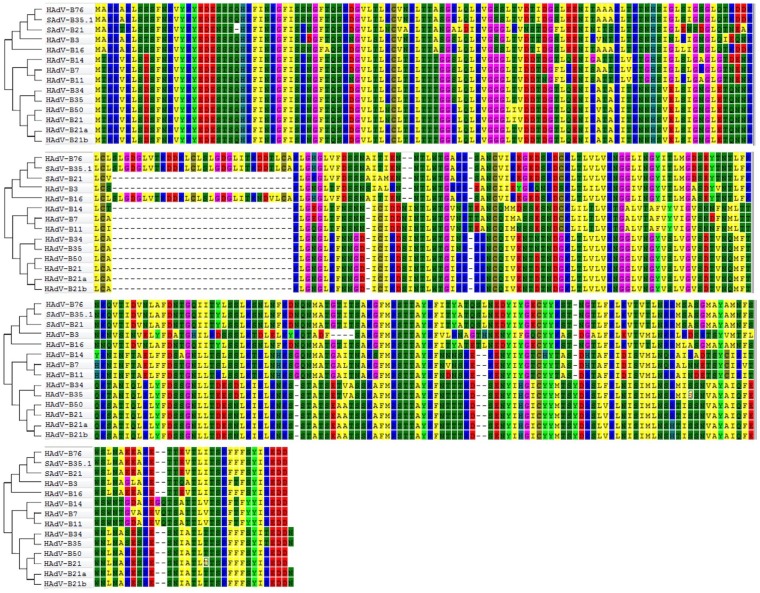
The fiber contains both the cell tropism determinant and the HI determinant. HI assays are reported to be difficult to perform and to repeat, particularly with regard to the quality and availability of reagents (29, 33, 34). Aside from the nucleotide differences, HAdV-B76, SAdV-B35.1, and HAdV-B16 share a large insertion and two small deletions (2 bases and 1 base), depicted in the second row of the phylogenetically noted amino acid alignments in Fig. 5. SAdV-B21 does not have this insertion but has the two small deletions. Consistent with other computational data presented, the third and fourth rows show that SAdV-B21 sequences are identical with those of HAdV-B76 and SAdV-B35.1. HAdV-B21 sequences show divergence, consistent with the HI observations that this virus is a recombinant with a HAdV-B16-like fiber (27, 29).
FIG 5.
Comparison of fiber protein sequences. HAdV-B76 fiber amino acid sequences were aligned against all species B homologs, including three variants of HAdV-B21 (prototype 21 and two recent isolates, 21a and 21b) and two simian adenoviruses, SAdV-B35.1 and SAdV-B21. The sequences support prior hemagglutination inhibition observations that the gamma epitope of HAdV-B76 is the same as for HAdV-B16. For this multiple-sequence alignment, MAFFT version 7 was used (http://mafft.cbrc.jp/alignment/server/), with default parameters. Each amino acid is color-coded to provide contrast, with dashes for alignment spacing. This alignment was also used to construct the phylogenetic relationships displayed at the left, with Mega 5.01 (http://megasoftware.net) and the following parameters: bootstrapped, neighbor-joining trees with 1,000 replicates and a maximum-composite-likelihood model.
ITR sequence analysis.
The ITR sequences at the ends of the linear genome contain the viral replication elements. For HAdVs, these include the core origin and the NF-I and NF-III motifs. The latter two elements reflect multiple observations that HAdVs utilize two host transcription factors, nuclear factors I and III, as part of the viral DNA replication complex for optimal replication (35–38). Except for variations in the genotypes of species D, all HAdVs have identical or highly similar NF-I and NF-III motifs. In contrast, SAdVs are missing the NF-I motif. It was reported that the prototype (1953) and a concurrently circulating strain of HAdV-E4, a human pathogen that is essentially a chimpanzee virus, is missing the NF-I motif, but recent isolates, e.g., HAdV-E4FS1, have acquired the motif (16, 39). As displayed in Fig. 6, the HAdV-B76 and SAdV-B35.1 genomes contain an NF-I motif (TGGAATGGTGCCAA) that is identical to those from HAdVs, particularly the respiratory pathogens within species B, including a deletion of nucleotide (nt) 8 of the SAdV NF-I sequences. This may represent an adaptation to the human host, as HAdV-E4 has apparently expanded beyond limited outbreaks in military venues to include larger civilian outbreaks (16, 39). In contrast, SAdV-B21 is missing this motif.
FIG 6.
Multiple sequence alignment of the inverted terminal repeat (ITR) sequences. Sequences of ca. 61 bases from the 5′ termini of the SAdV-E25 genome, as a reference, were aligned and compared with counterpart sequences from selected SAdVs and HAdVs using Clustal Omega (www.ebi.ac.uk/Tools/msa/clustalo/). Three adenovirus DNA replication sequence motifs are labeled and boxed: core origin, NF-I, and NF-III. This Clustal Omega-generated alignment was manually edited to position gaps to generate an optimal alignment. The HAdV-B76 sequence was identical to the SAdV-B35.1 counterpart and to HAdV-E4FS1, a currently circulating human respiratory pathogen, as well as to the respiratory pathogens of species B.
DISCUSSION
A genomics and bioinformatics approach to identifying, characterizing, and understanding emergent and reemergent infectious disease agents is critical to the rapid intervention and prevention of pandemics. A prime example is the 2009 H1N1 pandemic, in which a novel influenza A virus strain emerged in Mexico and the United States in mid-April (40). The genome was sequenced within weeks, if not days, with the exact identity initiating vaccine development, validation, and production, as well as genomics-based diagnostic protocol development and validation. In mid-September, both public health measures were available as the FDA approved the vaccine (40).
Several recent emergent and reemergent infectious disease agents causing considerable public concern have been shown through this genomics and bioinformatics approach to have zoonotic origins, including SARS-CoV (3), MERS-CoV (7), and Ebola virus (4). In addition to the initiation of public health measures, as noted for H1N1 (40), genomics provide high-resolution insights that are embedded in the genomes of these novel pathogens, including their histories and origins, that may allow predictions and genetic profiles of the next occurrence. As two examples, the genome sequence of a bat coronavirus contains a 190-nucleotide sequence that is identical to its counterpart in the human MERS-CoV genome (41), indicating a bat bioreactor and recombination pathway for an emergent MERS-CoV, and several genomes of MERS-CoV isolated from camels are identical to human MERS-CoV (7), indicating a second reservoir.
To explore pathways of pathogen emergence and the genesis of novel viruses, genomics analyses of archived, historically intriguing pathogens provide additional insights into current infectious disease agents, especially zoonotic ones. For example, retrospective genome analyses of archived “previously unidentified” samples provided the perspective that human immunodeficiency virus (HIV) was not uniquely or newly emergent in the 1980s but had a pedigree including multiple cross-species infections dating back to the 1920s (42). Similarly, genomics analyses of simian immunodeficiency viruses (SIVs) from both chimpanzee and gorilla provided insight that contributed to the origins of HIV, groups M and groups O and P, respectively (19, 23). Recently, Anoh et al. showed cross-species transmission of novel cytomegaloviruses in six nonhuman primates (43).
Revisiting previously well-studied HAdVs with a genomics approach has provided surprising and unexpected perspectives. For example, one of the first HAdV pathogens isolated and characterized in the 1950s was shown by whole-genome analysis to be a zoonotic chimpanzee AdV (15, 16). Applying whole-genome analysis has demonstrated recombination as another important molecular evolution mechanism by which novel HAdV pathogens arise, e.g., genotypes 53 and 64, recombinants that present epitopes of nonpathogens but are pathogens (44, 45), and genotype 55, a recombinant that presents the epitope of a kidney pathogen but is a respiratory pathogen (46). Taking advantage of a collaborative project between our Adenovirus Genome Sequencing Consortium and the J. Craig Venter Institute (JCVI) Genomic Sequencing Center for Infectious Diseases, historically intriguing HAdV pathogens were selected from three archives to ascertain additional recombinant pathogens that may have been misidentified with serological methods (12). One virus, originally named “Ad 21 + 16H16,” was an emergent pathogen that caused pneumonia and rapid progression to death in a 6-year-old boy (27). As supported by comparative genome analysis, including sequence alignments, sequence similarities, and phylogenomics, HAdV-B76 is a recombinant virus. Furthermore, the recombination events are multiple and include numerous adenoviruses, analogous to the genome of HAdV-D53 (44). Surprisingly, two recombination parents are chimpanzee AdVs (SAdV-B21 and -B27), indicating multiple, separate, and stable entries into this host. Unexpectedly, the HAdV-B76 genome is nearly identical to two recently isolated and sequenced genomes from a chimpanzee (SAdV-B35.1) and a bonobo (SAdV-B35.2), which are housed in different locations and have never interacted. Given that two HAdVs (types 16 and 21) are also parents of this recombinant, with multiple, separate, and stable entries, HAdV-B76 is the first example of a HAdV product of unprecedented “ping-pong” zoonotic and anthroponotic, cross-species, stable and recurring infections in hosts from three species (human, chimpanzee, and bonobo). This may represent a novel and significant molecular evolution pathway for the emergence of viruses with significant potential to cause human disease. Recent examples of SAdVs that appear to have infected humans include the novel Titi monkey adenovirus, which caused a remarkable outbreak of pneumonia in New World monkeys (47). An animal handler working closely with the monkeys developed flu-like symptoms, as did a member of his family; both were shown to have seroconverted when tested 4 months following their illnesses. According to another report, a baboon adenovirus caused an outbreak of pneumonia in captive infant baboons and was linked to flu-like symptoms in animal facility personnel, who also seroconverted (48). Neither of these examples led to a larger human outbreak. Even more convincing evidence for successful monkey-to-human-to-human transmission is the case of HAdV-E4, originally a chimpanzee virus. HAdV-E4 later host adapted to humans, enabling a wider circulation in the global population (15, 16); it has remained a significant human pathogen since 1953 and is now an even greater public health concern (49).
From the perspective of other viral pathogens, HIVs have been shown to be recombinant among themselves to “a surprisingly large number” (50). SIVs have been reported as participating in cross-species zoonotic events from chimpanzee to gorilla (21) as well as from chimpanzee and gorilla to humans (19, 23). Zoonosis is also reported for SAdVs isolated from wild primates (51). In a comparison of a gorilla AdV and other SAdV sequences, Wevers et al. noted “the penton base gene of GgorAdV-B7 showed a striking similarity (99.7%) only to that of SAdV-29 (chimpanzee AdV)” (52). Additionally, a recombinant SIV genome was characterized in chimpanzees, containing distinct lineages of SIV from two different monkey species, from which all known strains of chimpanzee SIVs are derived (19). However, it has not previously been reported that these or other viral pathogens contain genomic elements across multiple host species, including humans.
The recognition of zoonosis, anthroponosis, and recombination as key components and contributors of HAdV and SAdV genome and viral diversity suggests caution in the use of SAdV genomes as alternative vectors for epitope and gene delivery in vaccine development and gene therapy protocols (53). The hexon and fiber proteins, which are principal sites of antigenic epitopes of HAdV-B76, SAdV-B35.1, and SAdV-B35.2, are derived from HAdV-B21 and HAdV-B16, respectively, or are nearly identical such that they cross-react with antisera for these viruses (27, 29). Detailed amino acid sequence comparisons of both genes and the putative epitope sequences confirm the serological data but also reveal inconsistencies, such as the cross-reaction serum neutralization of HAdV-B16, of which the epitope sequences are not similar. One explanation for inconsistent SN results may be that the nature of the neutralizing antibodies (NAbs) is not entirely clear. For example, HAdV-C5 “NAbs target both hexon and fiber following vaccination and natural infection” (54). This complemented an earlier report that HAdV-C5-specific NAbs induced by natural infection were observed to be directed primarily against the fiber, while vaccination produced HAdV-C5-specific NAbs that were directed primarily against nonfiber capsid proteins (55). The ambiguous recognition by SN antisera again illustrates a major pitfall of serology-based methods for HAdV typing and supports the use of unambiguous DNA sequence data for HAdV identification and typing (10, 11, 33).
HAdV-B76 is predicted to be a human pathogen of concern, given isolation of nearly identical viruses in a zoo and a primate colony. Additionally, analogous to a current major respiratory human pathogen arising from zoonosis, HAdV-E4, HAdV-B76 contains all three viral replication elements presumably optimized for human lung cells. The HAdV genes contained in its genome also are optimized for the human host, including the cell tropism determinant HAdV-B16-like fiber protein, as revealed by sequence analysis and previously reported serological profiling. A question arises as to the prevalence of HAdV-B76 in the current population. Due to the “pregenomics” era and current protocols of using only one or two epitopes for HAdV identification and typing (56), there is a strong possibility that HAdV-B76, with its putative serotypes of HAdV-B21 and -B16, were and are mistyped as HAdV-B21 or -B16 and therefore unrecognized as an emergent recombinant, HAdV-B76. More likely, as most respiratory pathogenic HAdVs are neither commonly screened nor reported due to their insignificant mortality rates, it may be that HAdV-B76 is simply underreported. Or perhaps, as in the epidemiology of other HAdV pathogens, there are long periods of absences. For example, HAdV-B21 infections have been sparsely and sporadically reported since its first isolation in 1956 (57) until recently (58–60). These recent reports include cases resulting in deaths: HAdV-B21a (GenBank accession no. KF577595 [2005] and KF577598 [2013]) (59) and an isolate typed as HAdV-B21 but named “Agent Y” (accession no. AJ012091 [1997]) (60). In the latter report, the virus was identified only with the hexon sequence (60). A third study of HAdV-B21 examined genomes of 20 currently circulating strains along with gene sequences of a selection of 173 archived HAdV-B21 isolates from U.S. military and civilian populations (1996 to 2014), finding similarities to the prototype genome (58). A 20-year period of absence between the first report of HAdV-B21 and the next was noted (1956 to 1978), followed by sporadic reports that include intervening periods of absence of ∼10 years (58). Therefore, these sporadically reported HAdV respiratory pathogens are still of concern even if they appear to be presently absent from the infectious disease public health landscape, for when they reemerge after these periods of absence, particularly in an immune-naive population, the effects may be devastating, as for HAdV-21 noted above and also recently for HAdV-B55 and HAdV-B14 (61–64). The latter two reemergent pathogens are currently reported globally, with HAdV-B55 (originally named HAdV-B11a) absent from 1974 to 2005 and HAdV-B14 absent from 1963 to 2006. Note that prior to its emergence in the United States in 2006, HAdV-B14 had never been reported in the Americas.
In summary, this report demonstrates that the emergent adenoviral human pathogen HAdV-B76, associated with a fatality in 1965, is the product of remarkable zoonotic and anthroponotic, cross-species transmission and genome recombinations, involving human, chimpanzee, and bonobo hosts. In light of the known penchant of HAdVs to undergo homologous recombination, cross-species transmission may play a previously underappreciated role in the emergence of human adenoviral pathogens.
MATERIALS AND METHODS
Sample processing and next-generation sequencing.
“Ad 21+ 16H16” was isolated in 1965 from “a disoriented, lethargic to stuporous 6-year-old” boy who died of pneumonia (Wilford Hall U.S. Air Force Hospital, San Antonio, TX) (27). This virus was among a collection of HAdVs gifted to A. Heim (Institut für Virologie, Medizinische Hochschule, Hannover, Germany) by T. Adrian and R. Wigand (Institut for Medizinische Mikrobiologie und Hygiene, Abteilung for Virologie, Universitätskliniken, Homburg, Germany) upon their retirement. Genomic DNA was prepared as described earlier (65), with the virus recovered from a freezer stock and propagated on A549 cells, and DNA was isolated and shipped to the J. Craig Venter Institute (JCVI) Genomics Sequencing Center for Infectious Diseases (Rockville, MD) and sequenced using Illumina HiSeq and Roche 454 GS-FLX platforms as described previously (12).
Genome assembly.
Following DNA sequencing, sequence reads for both sets were deconvoluted by bar code identity; sequences were trimmed for quality and for the removal of SISPA hexamer primer sequences. Both sets of reads were then assembled de novo using clc_novo_assemble command-line assembly software (CLC Bio, Aarhus, Denmark), and the resulting contigs were BLAST searched against a database of GenBank whole HAdV genome sequences to find the closest reference. Both sets of reads, from the two platforms, were then mapped to the selected reference genome using clc_ref_assemble_long command-line assembly algorithm. At loci where the GS-FLX and Illumina sequence data sets agreed on a variation (compared to the reference sequence), the reference sequence was updated to reflect the variation. Final mapping of all sequences to the updated reference sequences was performed using CLC Bio software.
Since genomes showed variation compared to selected reference sequences, manual reference extension and editing were performed based on the sequencing reads. This was followed by another round of mapping assembly, as noted. To improve the resulting genome consensus sequence and to bridge gaps in the consensus, custom primers were designed using automated primer design software (66) and applied in complementing targeted PCR-based DNA sequencing reactions. These PCR products were sequenced either using the Sanger dideoxy chemistry for short-range amplicons (<1 kb) or using an IonTorrent for long-range amplicons (2 to 4 kb). Finally, the CLC Bio command-line assembler was used to merge the initial data with the closure-generated sequences. These sequences were verified for functional completeness and adequate sequence coverage using in-house quality assurance (QA) software tools (JCVI, Rockville, MD). The resultant finished genome sequences from this pipeline have average genome coverages of 214.2×.
Genome annotations.
Genome sequences were annotated with viral annotation software, Viral Genome ORF Reader (VIGOR) (67). For identification and adenoviral typing of the penton base, hexon, and fiber genes, BLAST was used as well as a genotyping software tool for the hypervariable hexon regions, comprising loops 1 and 2. This genotyping tool was developed by Kalpana Dommaraju (unpublished data), following the criteria published by Madisch et al. (68).
To complement and extend the “first-pass” preliminary JCVI in-house, automated, pipeline annotation in the GenBank record, the genome was reexamined independently. Verification and reannotation of the coding sequences were performed using the software tool Genome Annotation Transfer Utility (GATU) (69) with SAdV-B35.1 as a reference. The genome viewer tool Artemis (http://www.sanger.ac.uk/resources/software/artemis/) was used to record and visualize the annotation (70).
Computational genome analyses.
Publicly accessible software tools were used to perform computational analyses on the genes and genomes, as described previously (44). MAFFT, multiple-sequence alignment software that utilizes a fast Fourier transform algorithm (http://www.ebi.ac.uk/Tools/mafft/), was used to perform sequence alignments (71) using the default gap parameters provided. zPicture (http://zpicture.dcode.org), a sequence alignment tool that analyzes pairs of genome sequences against each other for regions of similarity, provided pairwise alignments, comparisons, and visualization of genomes (72). The NIH version of BLAST was used to query for sequence similarities against the GenBank database (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Sequence percent identity analysis.
EMBOSS software, including the Needle tool, was used to perform global sequence alignments (73), utilizing a BLOSUM62 matrix for the amino acid sequence analysis and a DNA full matrix for DNA sequence analysis. Analysis of the hexon L1 and L2 regions, corresponding to the putative SN epsilon epitope (31, 32), and the flanking sequences were analyzed using the sequence coordinates as described by Madisch et al. (68). For a higher-resolution analysis, the fiber gene was divided into two segments, shaft and knob domains, using the zPicture analysis as an approximate guide to the boundary.
Genome and gene recombination analysis.
Simplot was used to query the genes and genomes for sequence recombination. This software includes a bootscan that was used to complete the sequence recombination assessment of the MAFFT-aligned genes (74). For genes, default parameter settings were used for the window size (200 nucleotides [nt]), step size (20 nt), replicates used (n = 100), gap stripping (on), distance model (Kimura), and tree model (neighbor joining). Similarly, whole genomes were analyzed in the same manner, starting with an initial alignment using MAFFT and ensuing with recombination analysis using Simplot and bootscan. For this much larger sequence, only the window size and step size were altered (1,000 and 200, respectively), with the remainder of the default parameters unchanged.
Phylogenomics analysis.
Whole-genome and gene sequence alignments were performed using MAFFT as a precursor for phylogenetic analysis. After sequence alignment, the results were ported to tree-generating software, MEGA 5.1 (http://megasoftware.net), which provided boot-strapped, neighbor-joining trees with 1,000 replicates, constructed using the maximum-composite-likelihood method (75). All other parameters used were default values. Detailed analysis of the fiber called for the gene sequence to be divided into two sections: a proximal domain comprising the shaft, 716 bp, and the distal domain comprising the knob, 346 bp. The knob contains the domain encoding the hemagglutination epitope (gamma) and cell tropism determinant (28).
To provide additional support for the phylogenetic results, in silico analyses were carried out using maximum parsimony and maximum likelihood algorithms (both available in the MEGA software) for the whole-genome sequences; the fiber gene and its domains were reexamined with the above-described algorithms as well as a Bayesian algorithm (TOPALi v.2; http://www.topali.org). The following settings and parameters were used for each method: (i) neighbor joining with a bootstrap of 1,000 and the maximum likelihood model (MEGA), (ii) maximum parsimony with a bootstrap of 1,000 and the MP search method with close-neighbor interchange (CNI) on random trees (MEGA; number of initial trees, 1,000, and MP search level), (iii) maximum likelihood with bootstrap of 1,000 and the Tamura-Nei model (MEGA; ML heuristic method, nearest-neighbor interchange [NNI]) and Bayesian analysis with TOPALi v2 (MrBayes; default setting used: substitution model, HKY; number of generation, 100,000; sample frequency, 10).
GenBank accession numbers.
Genomes used for these analyses are archived in GenBank: HAdV-A12 (AC_000005), HAdV-B7 (AY594255), HAdV-B14 (AY803294), HAdV-B16 (AY601636), HAdV-B21 (AY601633), HAdV-C5 (AC_000008), HAdV-D9 (AJ854486), HAdV-E4 prototype (AY594253), HAdV-E4FS1 (also known as NHRC 42606; AY599835), HAdV-F40 (NC_001454), HAdV-G52 (DQ923122), SAdV-B21 (AC_000010), SAdV-E22 (AY530876), SAdV-E25 (AC_000011), SAdV-B27.1 (FJ025909), SAdV-B35.1 (FJ025912.1), and SAdV-B35.2 (FJ025910.1). HAdV-B76 was assigned GenBank accession number KF633445 (12).
Data availability.
Following visual inspection and additional manual editing, genome sequence and annotation data were deposited into GenBank as part of Bioproject number PRJNA70469.
ACKNOWLEDGMENTS
This project was funded in part with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract number HHSN272200900007C, and was supported in part by U.S. Public Health Service grants EY013124 and EY014104 and a Research to Prevent Blindness Senior Scientific Investigator Award (to J.C.).
REFERENCES
- 1.Kalthoff D, Globig A, Beer M. 2010. (Highly pathogenic) avian influenza as a zoonotic agent. Vet Microbiol 140:237–245. doi: 10.1016/j.vetmic.2009.08.022. [DOI] [PubMed] [Google Scholar]
- 2.Messenger AM, Barnes AN, Gray GC. 2014. Reverse zoonotic disease transmission (zooanthroponosis): a systematic review of seldom-documented human biological threats to animals. PLoS One 9:e89055. doi: 10.1371/journal.pone.0089055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guan Y, Zheng BJ, He YQ, Liu XL, Zhuang ZX, Cheung CL, Luo SW, Li PH, Zhang LJ, Guan YJ, Butt KM, Wong KL, Chan KW, Lim W, Shortridge KF, Yuen KY, Peiris JS, Poon LL. 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 4.Gire SK, Goba A, Andersen KG, Sealfon RS, Park DJ, Kanneh L, Jalloh S, Momoh M, Fullah M, Dudas G, Wohl S, Moses LM, Yozwiak NL, Winnicki S, Matranga CB, Malboeuf CM, Qu J, Gladden AD, Schaffner SF, Yang X, Jiang PP, Nekoui M, Colubri A, Coomber MR, Fonnie M, Moigboi A, Gbakie M, Kamara FK, Tucker V, Konuwa E, Saffa S, Sellu J, Jalloh AA, Kovoma A, Koninga J, Mustapha I, Kargbo K, Foday M, Yillah M, Kanneh F, Robert W, Massally JL, Chapman SB, Bochicchio J, Murphy C, Nusbaum C, Young S, Birren BW, Grant DS, Scheiffelin JS, Lander ES, Happi C, Gevao SM, Gnirke A, Rambaut A, Garry RF, Khan SH, Sabeti PC. 2014. Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science 345:1369–1372. doi: 10.1126/science.1259657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taubenberger JK, Morens DM. 2006. 1918 influenza: the mother of all pandemics. Emerg Infect Dis 12:15–22. doi: 10.3201/eid1201.050979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carroll D, Watson B, Togami E, Daszak P, Mazet JA, Chrisman CJ, Rubin EM, Wolfe N, Morel CM, Gao GF, Burci GL, Fukuda K, Auewarakul P, Tomori O. 2018. Building a global atlas of zoonotic viruses. Bull World Health Organ 96:292–294. doi: 10.2471/BLT.17.205005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Briese T, Mishra N, Jain K, Zalmout IS, Jabado OJ, Karesh WB, Daszak P, Mohammed OB, Alagaili AN, Lipkin WI. 2014. Middle East respiratory syndrome coronavirus quasispecies that include homologues of human isolates revealed through whole-genome analysis and virus cultured from dromedary camels in Saudi Arabia. mBio 5:e01146-14. doi: 10.1128/mBio.01146-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rowe WP, Huebner RJ, Gilmore LK, Parrott RH, Ward TG. 1953. Isolation of a cytopathogenic agent from human adenoids undergoing spontaneous degeneration in tissue culture. Proc Soc Exp Biol Med 84:570–573. doi: 10.3181/00379727-84-20714. [DOI] [PubMed] [Google Scholar]
- 9.Hilleman MR, Werner JH. 1954. Recovery of new agent from patients with acute respiratory illness. Proc Soc Exp Biol Med 85:183–188. doi: 10.3181/00379727-85-20825. [DOI] [PubMed] [Google Scholar]
- 10.Seto D, Chodosh J, Brister JR, Jones MS, Members of the Adenovirus Research Community. 2011. Using the whole-genome sequence to characterize and name human adenoviruses. J Virol 85:5701–5702. doi: 10.1128/JVI.00354-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seto D, Jones MS, Dyer DW, Chodosh J. 2013. Characterizing, typing, and naming human adenovirus type 55 in the era of whole genome data. J Clin Virol 58:741–742. doi: 10.1016/j.jcv.2013.09.025. [DOI] [PubMed] [Google Scholar]
- 12.Ismail AM, Cui T, Dommaraju K, Singh G, Dehghan S, Seto J, Shrivastava S, Fedorova NB, Gupta N, Stockwell TB, Halpin R, Madupu R, Heim A, Kajon AE, Romanowski EG, Kowalski RP, Malathi J, Therese KL, Madhavan HN, Zhang Q, Ferreyra LJ, Jones MS, Rajaiya J, Dyer DW, Chodosh J, Seto D. 2018. Genomic analysis of a large set of currently—and historically—important human adenovirus pathogens. Emerg Microbes Infect 7:10. doi: 10.1038/s41426-017-0004-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lion T. 2014. Adenovirus infections in immunocompetent and immunocompromised patients. Clin Microbiol Rev 27:441–462. doi: 10.1128/CMR.00116-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang W, Fu J, Liu J, Wang H, Schiwon M, Janz S, Schaffarczyk L, von der Goltz L, Ehrke-Schulz E, Dorner J, Solanki M, Boehme P, Bergmann T, Lieber A, Lauber C, Dahl A, Petzold A, Zhang Y, Stewart AF, Ehrhardt A. 2017. An engineered virus library as a resource for the spectrum-wide exploration of virus and vector diversity. Cell Rep 19:1698–1709. doi: 10.1016/j.celrep.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 15.Purkayastha A, Ditty SE, Su J, McGraw J, Hadfield TL, Tibbetts C, Seto D. 2005. Genomic and bioinformatics analysis of HAdV-4, a human adenovirus causing acute respiratory disease: implications for gene therapy and vaccine vector development. J Virol 79:2559–2572. doi: 10.1128/JVI.79.4.2559-2572.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dehghan S, Seto J, Liu EB, Walsh MP, Dyer DW, Chodosh J, Seto D. 2013. Computational analysis of four human adenovirus type 4 genomes reveals molecular evolution through two interspecies recombination events. Virology 443:197–207. doi: 10.1016/j.virol.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lyons A, Longfield J, Kuschner R, Straight T, Binn L, Seriwatana J, Reitstetter R, Froh IB, Craft D, McNabb K, Russell K, Metzgar D, Liss A, Sun X, Towle A, Sun W. 2008. A double-blind, placebo-controlled study of the safety and immunogenicity of live, oral type 4 and type 7 adenovirus vaccines in adults. Vaccine 26:2890–2898. doi: 10.1016/j.vaccine.2008.03.037. [DOI] [PubMed] [Google Scholar]
- 18.Locatelli S, Peeters M. 2012. Cross-species transmission of simian retroviruses: how and why they could lead to the emergence of new diseases in the human population. AIDS 26:659–673. doi: 10.1097/QAD.0b013e328350fb68. [DOI] [PubMed] [Google Scholar]
- 19.Bailes E, Gao F, Bibollet-Ruche F, Courgnaud V, Peeters M, Marx PA, Hahn BH, Sharp PM. 2003. Hybrid origin of SIV in chimpanzees. Science 300:1713. doi: 10.1126/science.1080657. [DOI] [PubMed] [Google Scholar]
- 20.D'Arc M, Ayouba A, Esteban A, Learn GH, Boue V, Liegeois F, Etienne L, Tagg N, Leendertz FH, Boesch C, Madinda NF, Robbins MM, Gray M, Cournil A, Ooms M, Letko M, Simon VA, Sharp PM, Hahn BH, Delaporte E, Mpoudi Ngole E, Peeters M. 2015. Origin of the HIV-1 group O epidemic in western lowland gorillas. Proc Natl Acad Sci U S A 112:E1343–E1352. doi: 10.1073/pnas.1502022112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keele BF, Van Heuverswyn F, Li Y, Bailes E, Takehisa J, Santiago ML, Bibollet-Ruche F, Chen Y, Wain LV, Liegeois F, Loul S, Ngole EM, Bienvenue Y, Delaporte E, Brookfield JF, Sharp PM, Shaw GM, Peeters M, Hahn BH. 2006. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science 313:523–526. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao F, Bailes E, Robertson DL, Chen Y, Rodenburg CM, Michael SF, Cummins LB, Arthur LO, Peeters M, Shaw GM, Sharp PM, Hahn BH. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436–441. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- 23.Van Heuverswyn F, Li Y, Neel C, Bailes E, Keele BF, Liu W, Loul S, Butel C, Liegeois F, Bienvenue Y, Ngolle EM, Sharp PM, Shaw GM, Delaporte E, Hahn BH, Peeters M. 2006. Human immunodeficiency viruses: SIV infection in wild gorillas. Nature 444:164. doi: 10.1038/444164a. [DOI] [PubMed] [Google Scholar]
- 24.Wertheim JO, Smith MD, Smith DM, Scheffler K, Kosakovsky Pond SL. 2014. Evolutionary origins of human herpes simplex viruses 1 and 2. Mol Biol Evol 31:2356–2364. doi: 10.1093/molbev/msu185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bruhn CA, Nielsen SC, Samaniego JA, Wadsworth J, Knowles NJ, Gilbert MT. 2015. Viral meningitis epidemics and a single, recent, recombinant and anthroponotic origin of swine vesicular disease virus. Evol Med Public Health 2015:289–303. doi: 10.1093/emph/eov026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roy S, Vandenberghe LH, Kryazhimskiy S, Grant R, Calcedo R, Yuan X, Keough M, Sandhu A, Wang Q, Medina-Jaszek CA, Plotkin JB, Wilson JM. 2009. Isolation and characterization of adenoviruses persistently shed from the gastrointestinal tract of non-human primates. PLoS Pathog 5:e1000503. doi: 10.1371/journal.ppat.1000503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crandell RA, Dowdle WR, Holcomb TM, Dahl EV. 1968. A fatal illness associated with two viruses: an intermediate adenovirus type (21-16) and influenza A2. J Pediatr 72:467–473. doi: 10.1016/S0022-3476(68)80335-X. [DOI] [PubMed] [Google Scholar]
- 28.Norrby E. 1969. The structural and functional diversity of adenovirus capsid components. J Gen Virol 5:221–236. doi: 10.1099/0022-1317-5-2-221. [DOI] [PubMed] [Google Scholar]
- 29.Wigand R, Sehn N, Hierholzer JC, de Jong JC, Adrian T. 1985. Immunological and biochemical characterization of human adenoviruses from subgenus B. I. Antigenic relationships Arch Virol 84:63–78. doi: 10.1007/BF01310554. [DOI] [PubMed] [Google Scholar]
- 30.Rowe WP, Hartley JW, Huebner RJ. 1956. Additional serotypes of the APC virus group. Proc Soc Exp Biol Med 91:260–262. doi: 10.3181/00379727-91-22231. [DOI] [PubMed] [Google Scholar]
- 31.Yuan X, Qu Z, Wu X, Wang Y, Liu L, Wei F, Gao H, Shang L, Zhang H, Cui H, Zhao Y, Wu N, Tang Y, Qin L. 2009. Molecular modeling and epitopes mapping of human adenovirus type 3 hexon protein. Vaccine 27:5103–5110. doi: 10.1016/j.vaccine.2009.06.041. [DOI] [PubMed] [Google Scholar]
- 32.Crawford-Miksza L, Schnurr DP. 1996. Analysis of 15 adenovirus hexon proteins reveals the location and structure of seven hypervariable regions containing serotype-specific residues. J Virol 70:1836–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wigand R. 1987. Pitfalls in the identification of adenoviruses. J Virol Methods 16:161–169. doi: 10.1016/0166-0934(87)90001-2. [DOI] [PubMed] [Google Scholar]
- 34.Kajon AE, de Jong JC, Dickson LM, Arron G, Murtagh P, Viale D, Carballal G, Echavarria M. 2013. Molecular and serological characterization of species B2 adenovirus strains isolated from children hospitalized with acute respiratory disease in Buenos Aires, Argentina. J Clin Virol 58:4–10. doi: 10.1016/j.jcv.2013.06.030. [DOI] [PubMed] [Google Scholar]
- 35.Hatfield L, Hearing P. 1991. Redundant elements in the adenovirus type 5 inverted terminal repeat promote bidirectional transcription in vitro and are important for virus growth in vivo. Virology 184:265–276. doi: 10.1016/0042-6822(91)90843-Z. [DOI] [PubMed] [Google Scholar]
- 36.Hatfield L, Hearing P. 1993. The NFIII/OCT-1 binding site stimulates adenovirus DNA replication in vivo and is functionally redundant with adjacent sequences. J Virol 67:3931–3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mul YM, Verrijzer CP, van der Vliet PC. 1990. Transcription factors NFI and NFIII/oct-1 function independently, employing different mechanisms to enhance adenovirus DNA replication. J Virol 64:5510–5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pruijn GJ, van Miltenburg RT, Claessens JA, van der Vliet PC. 1988. Interaction between the octamer-binding protein nuclear factor III and the adenovirus origin of DNA replication. J Virol 62:3092–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Houng HS, Clavio S, Graham K, Kuschner R, Sun W, Russell KL, Binn LN. 2006. Emergence of a new human adenovirus type 4 (Ad4) genotype: identification of a novel inverted terminal repeated (ITR) sequence from majority of Ad4 isolates from US military recruits. J Clin Virol 35:381–387. doi: 10.1016/j.jcv.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 40.del Rio C, Guarner J. 2010. The 2009 influenza A (H1N1) pandemic: what have we learned in the past 6 months. Trans Am Clin Climatol Assoc 121:128–140. [PMC free article] [PubMed] [Google Scholar]
- 41.Memish ZA, Mishra N, Olival KJ, Fagbo SF, Kapoor V, Epstein JH, Alhakeem R, Durosinloun A, Al Asmari M, Islam A, Kapoor A, Briese T, Daszak P, Al Rabeeah AA, Lipkin WI. 2013. Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerg Infect Dis 19:1819–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Faria NR, Rambaut A, Suchard MA, Baele G, Bedford T, Ward MJ, Tatem AJ, Sousa JD, Arinaminpathy N, Pepin J, Posada D, Peeters M, Pybus OG, Lemey P. 2014. HIV epidemiology. The early spread and epidemic ignition of HIV-1 in human populations. Science 346:56–61. doi: 10.1126/science.1256739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anoh AE, Murthy S, Akoua-Koffi C, Couacy-Hymann E, Leendertz FH, Calvignac-Spencer S, Ehlers B. 2017. Cytomegaloviruses in a community of wild nonhuman primates in Tai National Park, Côte D’Ivoire. Viruses 10:11. doi: 10.3390/v10010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walsh MP, Chintakuntlawar A, Robinson CM, Madisch I, Harrach B, Hudson NR, Schnurr D, Heim A, Chodosh J, Seto D, Jones MS. 2009. Evidence of molecular evolution driven by recombination events influencing tropism in a novel human adenovirus that causes epidemic keratoconjunctivitis. PLoS One 4:e5635. doi: 10.1371/journal.pone.0005635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou X, Robinson CM, Rajaiya J, Dehghan S, Seto D, Jones MS, Dyer DW, Chodosh J. 2012. Analysis of human adenovirus type 19 associated with epidemic keratoconjunctivitis and its reclassification as adenovirus type 64. Invest Ophthalmol Vis Sci 53:2804–2811. doi: 10.1167/iovs.12-9656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walsh MP, Seto J, Jones MS, Chodosh J, Xu W, Seto D. 2010. Computational analysis identifies human adenovirus type 55 as a re-emergent acute respiratory disease pathogen. J Clin Microbiol 48:991–993. doi: 10.1128/JCM.01694-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen EC, Yagi S, Kelly KR, Mendoza SP, Tarara RP, Canfield DR, Maninger N, Rosenthal A, Spinner A, Bales KL, Schnurr DP, Lerche NW, Chiu CY. 2011. Cross-species transmission of a novel adenovirus associated with a fulminant pneumonia outbreak in a new world monkey colony. PLoS Pathog 7:e1002155. doi: 10.1371/journal.ppat.1002155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chiu CY, Yagi S, Lu X, Yu G, Chen EC, Liu M, Dick EJ Jr, Carey KD, Erdman DD, Leland MM, Patterson JL. 2013. A novel adenovirus species associated with an acute respiratory outbreak in a baboon colony and evidence of coincident human infection. mBio 4:e00084-13. doi: 10.1128/mBio.00084-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang J, Kang J, Dehghan S, Sridhar S, Lau SKP, Ou J, Woo PCY, Zhang Q, Seto D. 2019. A survey of recent adenoviral respiratory pathogens in Hong Kong reveals emergent and recombinant human adenovirus type 4 (HAdV-E4) circulating in civilian populations. Viruses 11:129. doi: 10.3390/v11020129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robertson DL, Sharp PM, McCutchan FE, Hahn BH. 1995. Recombination in HIV-1. Nature 374:124–126. doi: 10.1038/374124b0. [DOI] [PubMed] [Google Scholar]
- 51.Wevers D, Metzger S, Babweteera F, Bieberbach M, Boesch C, Cameron K, Couacy-Hymann E, Cranfield M, Gray M, Harris LA, Head J, Jeffery K, Knauf S, Lankester F, Leendertz SA, Lonsdorf E, Mugisha L, Nitsche A, Reed P, Robbins M, Travis DA, Zommers Z, Leendertz FH, Ehlers B. 2011. Novel adenoviruses in wild primates: a high level of genetic diversity and evidence of zoonotic transmissions. J Virol 85:10774–10784. doi: 10.1128/JVI.00810-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wevers D, Leendertz FH, Scuda N, Boesch C, Robbins MM, Head J, Ludwig C, Kuhn J, Ehlers B. 2010. A novel adenovirus of Western lowland gorillas (Gorilla gorilla gorilla). Virol J 7:303. doi: 10.1186/1743-422X-7-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roy S, Gao G, Lu Y, Zhou X, Lock M, Calcedo R, Wilson JM. 2004. Characterization of a family of chimpanzee adenoviruses and development of molecular clones for gene transfer vectors. Hum Gene Ther 15:519–530. doi: 10.1089/10430340460745838. [DOI] [PubMed] [Google Scholar]
- 54.Bradley RR, Lynch DM, Iampietro MJ, Borducchi EN, Barouch DH. 2012. Adenovirus serotype 5 neutralizing antibodies target both hexon and fiber following vaccination and natural infection. J Virol 86:625–629. doi: 10.1128/JVI.06254-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cheng C, Gall JG, Nason M, King CR, Koup RA, Roederer M, McElrath MJ, Morgan CA, Churchyard G, Baden LR, Duerr AC, Keefer MC, Graham BS, Nabel GJ. 2010. Differential specificity and immunogenicity of adenovirus type 5 neutralizing antibodies elicited by natural infection or immunization. J Virol 84:630–638. doi: 10.1128/JVI.00866-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Salama M, Amitai Z, Amir N, Gottesman-Yekutieli T, Sherbany H, Drori Y, Mendelson E, Carmeli Y, Mandelboim M. 2016. Outbreak of adenovirus type 55 infection in Israel. J Clin Virol 78:31–35. doi: 10.1016/j.jcv.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 57.Bell SD Jr, Rota TR, McComb DE. 1960. Adenoviruses isolated from Saudi Arabia. III. Six new serotypes. Am J Trop Med Hyg 9:523–526. doi: 10.4269/ajtmh.1960.9.523. [DOI] [Google Scholar]
- 58.Kajon AE, Hang J, Hawksworth A, Metzgar D, Hage E, Hansen CJ, Kuschner RA, Blair P, Russell KL, Jarman RG. 2015. Molecular epidemiology of adenovirus type 21 respiratory strains isolated from US military trainees (1996–2014). J Infect Dis 212:871–880. doi: 10.1093/infdis/jiv141. [DOI] [PubMed] [Google Scholar]
- 59.Hage E, Huzly D, Ganzenmueller T, Beck R, Schulz TF, Heim A. 2014. A human adenovirus species B subtype 21a associated with severe pneumonia. J Infect 69:490–499. doi: 10.1016/j.jinf.2014.06.015. [DOI] [PubMed] [Google Scholar]
- 60.Cardosa MJ, Krishnan S, Tio PH, Perera D, Wong SC. 1999. Isolation of subgenus B adenovirus during a fatal outbreak of enterovirus 71-associated hand, foot, and mouth disease in Sibu, Sarawak. Lancet 354:987–991. doi: 10.1016/S0140-6736(98)11032-2. [DOI] [PubMed] [Google Scholar]
- 61.Zhu Z, Zhang Y, Xu S, Yu P, Tian X, Wang L, Liu Z, Tang L, Mao N, Ji Y, Li C, Yang Z, Wang S, Wang J, Li D, Xu W. 2009. Outbreak of acute respiratory disease in China caused by B2 species of adenovirus type 11. J Clin Microbiol 47:697–703. doi: 10.1128/JCM.01769-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kajon AE, Lu X, Erdman DD, Louie J, Schnurr D, George KS, Koopmans MP, Allibhai T, Metzgar D. 2010. Molecular epidemiology and brief history of emerging adenovirus 14-associated respiratory disease in the United States. J Infect Dis 202:93–103. doi: 10.1086/653083. [DOI] [PubMed] [Google Scholar]
- 63.Carr MJ, Kajon AE, Lu X, Dunford L, O’Reilly P, Holder P, De Gascun CF, Coughlan S, Connell J, Erdman DD, Hall WW. 2011. Deaths associated with human adenovirus-14p1 infections, Europe, 2009-2010. Emerg Infect Dis 17:1402–1408. doi: 10.3201/1708.101760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Q, Seto D, Zhao S, Zhu L, Zhao W, Wan C. 2012. Genome sequence of the first human adenovirus type 14 isolated in China. J Virol 86:7019–7020. doi: 10.1128/JVI.00814-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hage E, Gerd Liebert U, Bergs S, Ganzenmueller T, Heim A. 2015. Human mastadenovirus type 70: a novel, multiple recombinant species D mastadenovirus isolated from diarrhoeal faeces of a haematopoietic stem cell transplantation recipient. J Gen Virol 96:2734–2742. doi: 10.1099/vir.0.000196. [DOI] [PubMed] [Google Scholar]
- 66.Li K, Shrivastava S, Brownley A, Katzel D, Bera J, Nguyen AT, Thovarai V, Halpin R, Stockwell TB. 2012. Automated degenerate PCR primer design for high-throughput sequencing improves efficiency of viral sequencing. Virol J 9:261. doi: 10.1186/1743-422X-9-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang S, Sundaram JP, Stockwell TB. 2012. VIGOR extended to annotate genomes for additional 12 different viruses. Nucleic Acids Res 40:W186–W192. doi: 10.1093/nar/gks528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Madisch I, Harste G, Pommer H, Heim A. 2005. Phylogenetic analysis of the main neutralization and hemagglutination determinants of all human adenovirus prototypes as a basis for molecular classification and taxonomy. J Virol 79:15265–15276. doi: 10.1128/JVI.79.24.15265-15276.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tcherepanov V, Ehlers A, Upton C. 2006. Genome annotation transfer utility (GATU): rapid annotation of viral genomes using a closely related reference genome. BMC Genomics 7:150. doi: 10.1186/1471-2164-7-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Berriman M, Rutherford K. 2003. Viewing and annotating sequence data with Artemis. Brief Bioinform 4:124–132. doi: 10.1093/bib/4.2.124. [DOI] [PubMed] [Google Scholar]
- 71.Katoh K, Toh H. 2008. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform 9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 72.Ovcharenko I, Loots GG, Hardison RC, Miller W, Stubbs L. 2004. zPicture: dynamic alignment and visualization tool for analyzing conservation profiles. Genome Res 14:472–477. doi: 10.1101/gr.2129504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Olson SA. 2002. EMBOSS opens up sequence analysis. European Molecular Biology Open Software Suite. Brief Bioinform 3:87–91. doi: 10.1093/bib/3.1.87. [DOI] [PubMed] [Google Scholar]
- 74.Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol 73:152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Following visual inspection and additional manual editing, genome sequence and annotation data were deposited into GenBank as part of Bioproject number PRJNA70469.