Escherichia coli is one of the most prevalent facultative anaerobes of the human gut. E. coli normally exists as a harmless commensal but can also cause disease following the acquisition of genes that enhance its pathogenicity. Adhesion is an important first step in colonization of the host and is mediated by an array of cell surface components. In E. coli, these include a family of adhesins secreted by the type V secretion system. Here, we identified and characterized new proteins from an emerging subclass of the type V secretion system known as the inverse autotransporters (IATs). We found that IAT-encoding genes are present in a wide range of strains and showed that three novel IATs were localized on the E. coli cell surface and mediated biofilm formation. Overall, this study provides new insight into the prevalence, function, and regulation of IATs in E. coli.
KEYWORDS: Escherichia coli, autotransporter proteins, biofilms
ABSTRACT
Proteins secreted by the type V secretion system possess multiple functions, including the capacity to mediate adhesion, aggregation, and biolfilm formation. The type V secretion system can be divided into five subclasses, one of which is the type Ve system. Proteins of the type Ve secretion system are also referred to as inverse autotransporters (IATs). In this study, we performed an in silico analysis of 126 completely sequenced Escherichia coli genomes available in the NCBI database and identified several distinct IAT-encoding gene families whose distribution varied throughout the E. coli phylogeny. The genes included three characterized IATs (intimin, fdeC, and yeeJ) and four uncharacterized IATs (here named iatA, iatB, iatC, and iatD). The four iat genes were cloned from the completely sequenced environmental E. coli strain SMS-3-5 and characterized. Three of these IAT proteins (IatB, IatC, and IatD) were expressed at the cell surface and possessed the capacity to mediate biofilm formation in a recombinant E. coli K-12 strain. Further analysis of the iatB gene, which showed a unique association with extraintestinal E. coli strains, suggested that its regulation is controlled by the LeuO global regulator. Overall, this study provides new data describing the prevalence, sequence variation, domain structure, function, and regulation of IATs found in E. coli.
IMPORTANCE Escherichia coli is one of the most prevalent facultative anaerobes of the human gut. E. coli normally exists as a harmless commensal but can also cause disease following the acquisition of genes that enhance its pathogenicity. Adhesion is an important first step in colonization of the host and is mediated by an array of cell surface components. In E. coli, these include a family of adhesins secreted by the type V secretion system. Here, we identified and characterized new proteins from an emerging subclass of the type V secretion system known as the inverse autotransporters (IATs). We found that IAT-encoding genes are present in a wide range of strains and showed that three novel IATs were localized on the E. coli cell surface and mediated biofilm formation. Overall, this study provides new insight into the prevalence, function, and regulation of IATs in E. coli.
INTRODUCTION
Escherichia coli is one of the most prevalent facultative anaerobes of the human gut and harbors genes encoding a wide array of surface-expressed factors that promote the colonization of specific niches. One such factor includes the highly abundant group of proteins secreted by the type V secretion system (1, 2). All proteins secreted by this system share several common features: (i) an N-terminal signal sequence that targets the protein to the Sec machinery for transport across the inner membrane, (ii) a passenger domain that is either cell surface exposed or secreted, and (iii) a translocator (or β-barrel) domain that is embedded in the outer membrane and helps to facilitate the translocation of the passenger domain (2–5). The passenger domain of these proteins determines the unique functional characteristics of an individual protein. Overall, proteins secreted by the type V system possess a wide range of functions, including adhesion, cell-to-cell aggregation, and biofilm formation (6–8), as well as protease and cytotoxic activity (9, 10). For example, the well-characterized autotransporter (AT) protein antigen 43 (Ag43) of uropathogenic E. coli (UPEC) contributes to adhesion, cell-to-cell aggregation, biofilm formation, and long-term persistence in the urinary tract (11–14).
AT proteins can be classified into five subclasses, namely, types Va (monomeric AT), Vb (two-partner secretion system), Vc (trimeric), Vd (fused two-partner secretion system), and Ve (inverse ATs [IATs]) (1, 15, 16). The domain organization of IATs resembles that of the classical type Va AT proteins but with the passenger and translocation domains in opposite locations within the primary amino acid sequence. Two well-studied proteins from the type Ve subclass include intimin from E. coli and invasin from enteropathogenic Yersinia species (17–19). Intimin is an adhesin expressed by enteropathogenic E. coli (EPEC) and enterohemorrhagic E. coli (EHEC) and contributes to the formation of actin pedestals leading to attaching and effacing lesions in the gut (20, 21). Invasin-mediated adherence of enteropathogenic Yersinia to host cells triggers the envelopment of bacterial cells through host cell-mediated autophagy and plays an early role in the infection cycle by binding directly to host β1-integrins (22, 23).
The enormous volume of data available from genome sequencing has facilitated the identification of IATs from different phyla of bacteria (17, 24). However, many IATs still remain to be identified and characterized. In this study, we sought to identify the complement of IAT proteins found in E. coli and to characterize their phenotypic properties. To this end, we first probed 126 completely sequenced E. coli genomes available in the NCBI database for the presence of genes encoding IAT proteins. Next, we cloned, expressed, and characterized the function of three new IAT proteins, one of which was also examined at the regulatory level. Overall, this study has defined the set of IAT proteins found in E. coli.
RESULTS
E. coli possesses a diverse range of IAT genes.
The β-barrel domain represents the most conserved region of IAT proteins, and the presence of an intimin-like β-barrel domain defines the IAT family (18, 24). As such, we used the β-barrel amino acid sequences of intimin and invasin to probe a database of annotated protein sequences from 126 completely sequenced E. coli genomes (see Data Set S1 in the supplemental material). The subsequent list of hits represented proteins that contained the IAT β-barrel Pfam domain (PF11924). The amino acid sequences of these β-barrel domains were aligned, revealing seven distinct groups of IAT proteins (Fig. S1). Among these, intimin (encoded by the eaeA gene) has been very well characterized and thus was not examined further. The remaining IATs, the genes for which were all present in the environmental E. coli strain SMS-3-5, were selected for further study (Table 1). The six IAT genes from SMS-3-5 include the previously studied fdeC and yeeJ genes (25–27) and four uncharacterized IAT genes found at different locations on the chromosome and renamed as follows: EcSMS35_1920 (iatA), EcSMS35_2661 (iatB), EcSMS35_4024 (iatC), and EcSMS35_4876 (iatD).
TABLE 1.
E. coli SMS-3-5 type Ve AT proteins
| Locus tag | GenBank accession no. |
Gene | Gene size (bp) |
Protein size (aa)a |
|---|---|---|---|---|
| EcSMS35_0331 | ACB16013.1 | fdeC | 4,245 | 1,415 |
| EcSMS35_1146 | ACB16711.1 | yeeJ | 7,077 | 2,359 |
| EcSMS35_1920 | ACB19099.1 | iatA | 1,395 | 465 |
| EcSMS35_2661 | ACB17431.1 | iatB | 2,175 | 725 |
| EcSMS35_4024 | ACB17037.1 | iatC | 8,802 | 2,934 |
| EcSMS35_4876 | ACB20062.1 | iatD | 5,241 | 1,747 |
aa, amino acids.
Cladogram of the β-barrel domain from putative IAT proteins found in 126 complete E. coli genomic sequences. A database of annotated proteins from each strain was probed using fasta36 to identify putative β-barrel domain-containing IAT proteins. The context of the β-barrel domain was examined in each extracted protein sequence to ensure that it was located at the N-terminal end of the protein. The six prototype putative proteins chosen from SMS-3-5 are highlighted in red, and the branch representing intimin is in gray. Alignments and phylogenetic trees were drawn in MEGA7 and visualized with FigTree. Download FIG S1, PDF file, 0.2 MB (202.1KB, pdf) .
Copyright © 2019 Goh et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Prevalence of IAT proteins in the collection of 126 completely sequenced E. coli genomes. Download Data Set S1, XLSX file, 0.1 MB (21.6KB, xlsx) .
Copyright © 2019 Goh et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Analysis of the six IAT genes in E. coli.
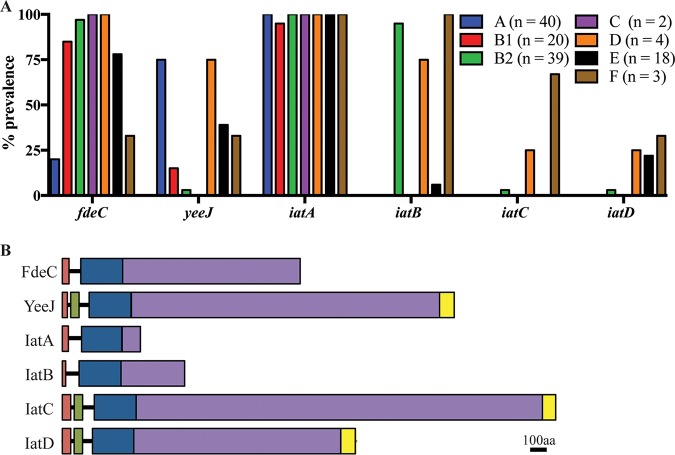
The prevalence of the six IAT genes was assessed in the 126 completely sequenced E. coli genomes. A complete gene was found in 67% of strains for fdeC (84/126), 35% for yeeJ (44/126), 99% for iatA (124/126), 35% for iatB (44/126), 3% for iatC (4/126), and 6% for iatD (7/126). A breakdown of the prevalence of the genes within each phylogroup is depicted in Fig. 1A. To extend this analysis, we examined the well-defined E. coli Reference (ECOR) collection of 72 strains using a PCR screening approach. The ECOR collection comprises strains isolated from a variety of hosts and locations and is representative of the ecological and phylogenetic diversity of the E. coli species. The correct-sized PCR product was found in 83% of strains for the fdeC gene, 36% for yeeJ, 90% for iatA, 36% for iatB, 19% for iatC, and 10% for iatD (Data Set S2).
FIG 1.
(A) Prevalence of IAT-encoding genes within phylogroups A (n = 40), B1 (n = 20), B2 (n = 39), C (n = 2), D (n = 4), E (n = 18), and F (n = 3). (B) Schematic diagram of E. coli SMS-3-5 IAT proteins. Predicted domains are shown as colored boxes (orange, signal sequence; green, LysM domain; blue, β-barrel domain; purple, passenger domain; yellow, C-type lectin domain). aa, amino acids.
Prevalence of iat genes in the ECOR collection. Download Data Set S2, XLSX file, 0.1 MB (11.8KB, xlsx) .
Copyright © 2019 Goh et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Next, we examined the genomic context of each of the E. coli IAT genes. The six IAT genes occur at different chromosomal locations; note that the position of each orthologue is conserved in all of the strains examined (Fig. S2). Both iatA and iatD were highly conserved (amino acid identity of >98%). In contrast, the iatB and iatC genes varied in size, with the predicted IatB protein ranging from 725 to 743 amino acids (amino acid identity of >89%) and the predicted IatC protein ranging from 2,459 to 2,965 amino acids (amino acid identity of >78%). IatB and IatC possessed a highly conserved signal sequence and translocation domain, respectively, with their variability attributed solely to sequence changes in the passenger domain.
Genomic context of the IAT genes (highlighted in red) in SMS-3-5. Depicted are two genes upstream and downstream of the IAT gene of interest and the location of the fragment on the genome of SMS-3-5. Genes with known functions are labeled with gene names, and genes of unknown function are labeled with locus tags. Download FIG S2, PDF file, 0.2 MB (218.5KB, pdf) .
Copyright © 2019 Goh et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The six IAT genes from SMS-3-5 encode proteins with similar domain organizations.
A schematic diagram representing the domain organizations of the six IAT proteins from SMS-3-5 is shown in Fig. 1B. The signal sequence of each protein is followed by an IAT β-barrel Pfam domain (PF11924). Modeling of the β-barrel domains from all six proteins revealed that they are very similar in size (243 to 246 amino acids) and are predicted to fold into a 12-stranded β-barrel structure. Overall, the β-barrel domains of these proteins share 34 to 88% amino acid identity. A comparative amino acid alignment against the β-barrel sequences from subtype Va AT proteins (Ag43, UpaH, Sat, and Vat) revealed that the sequences of each subgroup cluster independently and are highly variable between the different subgroups of AT proteins (Fig. S3).
Comparison of β-barrel amino acid sequences from IAT and type Va AT proteins. (Left) Cladogram generated using the β-barrel amino acid sequences from IAT and type Va AT proteins. The sequences from the different subgroups cluster separately. (Right) Pairwise comparison of each β-barrel sequence. Numbers represent the percent identity. Download FIG S3, PDF file, 0.2 MB (223.3KB, pdf) .
Copyright © 2019 Goh et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Analysis of the C-terminal passenger domain of each IAT on InterPro revealed that FdeC, YeeJ, IatC, and IatD contain various numbers of Big1 repeats, which are typical of IATs. Nonetheless, structural modeling of the passenger domains of IatA and IatB on Phyre2 suggests that they possess structural characteristics (e.g., a fibronectin III domain) similar to those of other bacterial immunoglobulin superfamily (IgSF) domains. The passenger domains of the three larger proteins (YeeJ, IatC, and IatD) are capped with a C-type lectin domain. Additionally, a LysM domain is present at the N-terminal end of the β-barrel domain of these three larger proteins.
Cloning and expression of IAT genes from SMS-3-5.
In order to examine the functional properties of the four uncharacterized IAT proteins, the iatA, iatB, iatC, and iatD genes were amplified from SMS-3-5 and cloned into the isopropyl-β-d-1-thiogalactopyranoside (IPTG)-inducible pSU2718 expression vector to generate plasmids pIatA, pIatB, pIatC, and pIatD, respectively. A list of strains and plasmids used is provided in Data Set S3. The plasmids were transformed into the E. coli K-12 flu deletion strain MS427, which is unable to mediate cell aggregation and biofilm formation normally associated with Ag43 expression (28). Specific antisera were generated against the C-terminal passenger domains of IatB, IatC, and IatD and used to confirm their expression by Western blot analysis of whole-cell lysates prepared from IPTG-induced cultures grown overnight (Fig. S4). These experiments resulted in the detection of bands corresponding to full-length IatB, IatC, and IatD proteins as well as lower-molecular-weight bands presumed to be breakdown products based on their specific antibody cross-reactivity. Despite our efforts, we were unable to generate an IatA antiserum of sufficient quality and could not reliably detect the expression of the IatA protein. Hence, only IatB, IatC, and IatD were further characterized.
Western blot analysis of the different IAT proteins. For each blot, − represents whole-cell lysates prepared from the MS427(pSU2718) vector control, and + represents whole-cell lysates prepared from MS427 expressing the different IAT proteins, IatB (81 kDa) (A), IatC (307 kDa) (B), and IatD (188 kDa) (C). Download FIG S4, PDF file, 0.1 MB (129.6KB, pdf) .
Copyright © 2019 Goh et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Strains and plasmids used in this study. Download Data Set S3, XLSX file, 0.1 MB (12.3KB, xlsx) .
Copyright © 2019 Goh et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
IatB, IatC, and IatD are located on the cell surface.
To examine if the IAT proteins were localized to the outer membrane, immunofluorescence microscopy was performed using antisera specific to each protein. A strong fluorescence signal was observed for MS427(pIatB), MS427(pIatC), and MS427(pIatD), suggesting that these three proteins were effectively translocated to the cell surface in our recombinant E. coli strain (Fig. 2).
FIG 2.
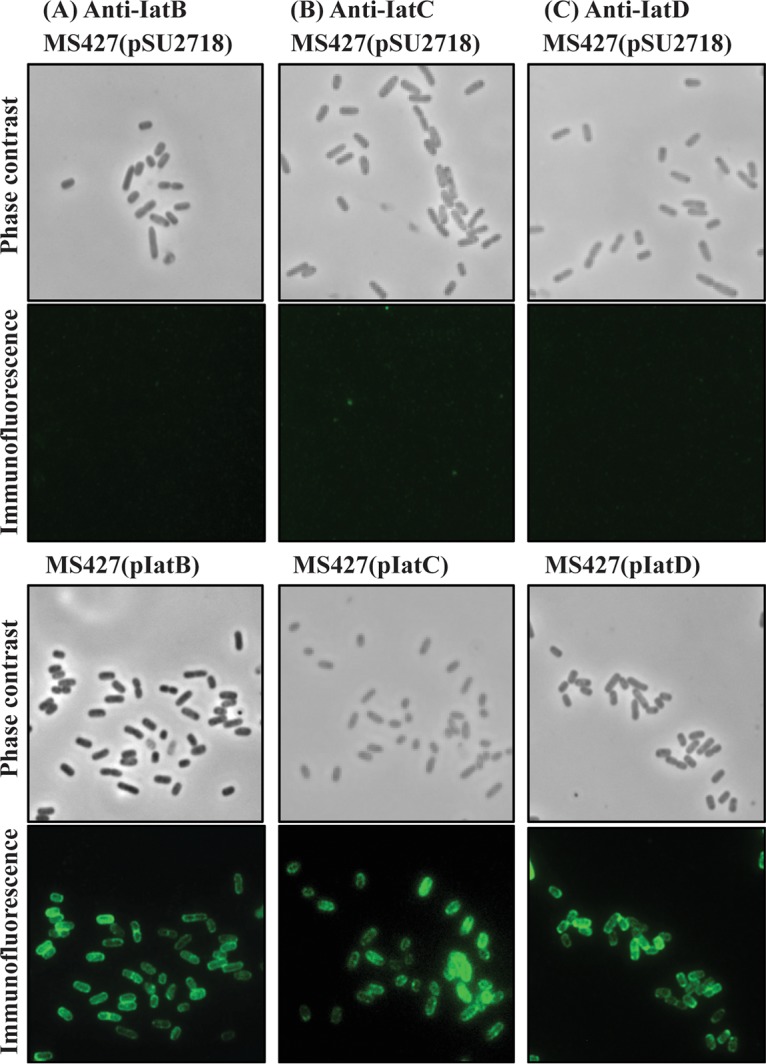
IatB, IatC, and IatD are localized at the cell surface when overexpressed in the MS427 background. Shown are images from phase-contrast and immunofluorescence microscopy using specific antisera against proteins IatB (A), IatC (B), and IatD (C). Positive reactions indicating the surface localization of IatB, IatC, and IatD were detected in MS427 (bottom) but not in the MS427(pSU2718) vector control (top).
Phenotypic properties of IatB, IatC, and IatD.
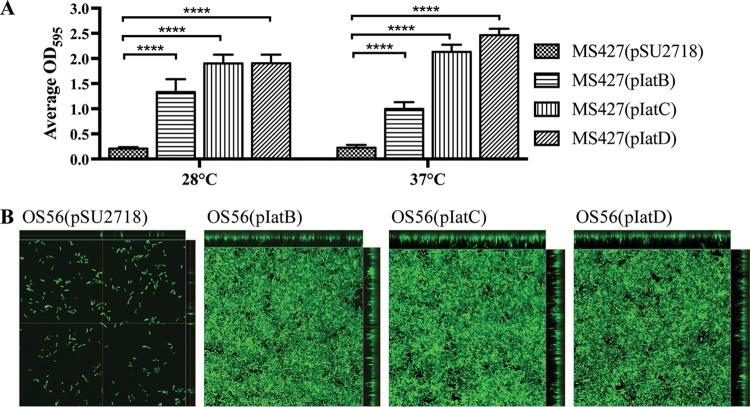
AT proteins are often associated with functions including cell aggregation, adhesion to extracellular matrix (ECM) proteins, and biofilm formation. While overexpression of the IatB, IatC, and IatD IAT proteins in MS427 did not lead to autoaggregation or adhesion to ECM proteins, all three IATs promoted strong biofilm formation. We first assessed this phenotype using a static microtiter plate biofilm assay, where MS427(pIatB), MS427(pIatC), and MS427(pIatD), but not the vector control strain MS427(pSU2718), were able to form a biofilm (Fig. 3A). The ability of the three IAT proteins to mediate biofilm formation was further explored using the gfp-tagged OS56 strain (a derivative of MS427) in a continuous-flow chamber setup, which permitted the distribution of cells within the biofilm to be monitored using confocal laser scanning microscopy. Consistent with the microtiter plate biofilm analyses, OS56(pIatB), OS56(pIatC), and OS56(pIatD) formed a significant biofilm with higher total biovolume, substratum coverage, and mean thickness (P < 0.0001) than for the vector control strain after 16 h of growth in M9 minimal medium supplemented with 1 mM IPTG (Fig. 3B). These results demonstrate that IatB, IatC, and IatD are able to mediate biofilm formation when overexpressed in a recombinant K-12 background.
FIG 3.
IatB, IatC, and IatD mediate biofilm formation. (A) Biofilm formation by E. coli strain MS427 in microtiter plates harboring plasmids pSU2718 (vector control), pIatB, pIatC, and pIatD. All strains were grown in M9 minimal medium in the presence of 1 mM IPTG to induce IAT protein expression, and plates were incubated at either 28°C or 37°C. Bar charts represent the average absorbance values at 595 nm, and error bars show the standard deviations calculated from three separate experiments (****, P < 0.0001). (B) Confocal laser scanning microscopy images of biofilms formed on plastic coverslips under continuous-flow conditions 16 h after inoculation with OS56(pSU2718) (vector control), OS56(pIatB), OS56(pIatC), and OS56(pIatD). The images represent horizontal sections within each biofilm. Displayed at the top and right of each image are vertical sections representing the xz and yz planes, at the positions indicated by the green and red lines, respectively.
Identification of genes involved in regulation of iatB.
Based on the high prevalence of iatB in E. coli phylogroup B2 (37/39 strains) (Fig. 1A), we selected this gene for further analysis and attempted to understand its regulation in SMS-3-5. The SMS-3-5 strain carries a large 130-kb plasmid (pSMS35_130; GenBank accession number CP000971.1) containing nine antibiotic resistance genes [aadA2, aph(3′)-la, strA, strB, blaTEM-1B, catA2, sul2, tet(A), and drfA14). To enable genetic manipulation of this strain, we first cured plasmid pSMS35_130 using a counterselectable vector strategy (29) to generate the strain SMS-3-5c.
To investigate the genetic basis of iatB regulation, we generated a chromosomal iatB promoter-lacZ reporter fusion strain (SMS-3-5clacIZ iatB::lacZ). SMS-3-5clacIZ iatB::lacZ was subjected to transposon mutagenesis using a mini-Tn5 cassette, generating approximately 30,000 transposon mutants that were screened for blue color development on lysogeny broth (LB) plates containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal). In this screen, we identified 12 colonies that were dark blue, indicating increased iatB promoter activity, and this was confirmed by their increased β-galactosidase activity compared to that of the SMS-3-5clacIZ iatB::lacZ parent strain (Fig. 4A). Further analysis revealed that the mutants all contained independent insertions within three regions of the leu operon: (i) three insertions in an intergenic region between the leuL and leuO genes, (ii) one insertion within the leuL gene, and (iii) eight insertions in an intergenic region between the leuL and leuA genes (Fig. 4B). Detailed analysis of the 12 Tn5 insertions revealed that they were all oriented in the same direction, with the chloramphenicol resistance gene placed in the same orientation as the downstream leuO gene. In previous studies, we have shown that the promoter of the chloramphenicol resistance gene in this Tn5 transposon can drive the transcription of a downstream gene if the insertion position is favorable (30, 31). Hence, we hypothesized that the Tn5 insertions caused increased transcription of the leuO gene, which in turn resulted in enhanced iatB promoter activity.
FIG 4.
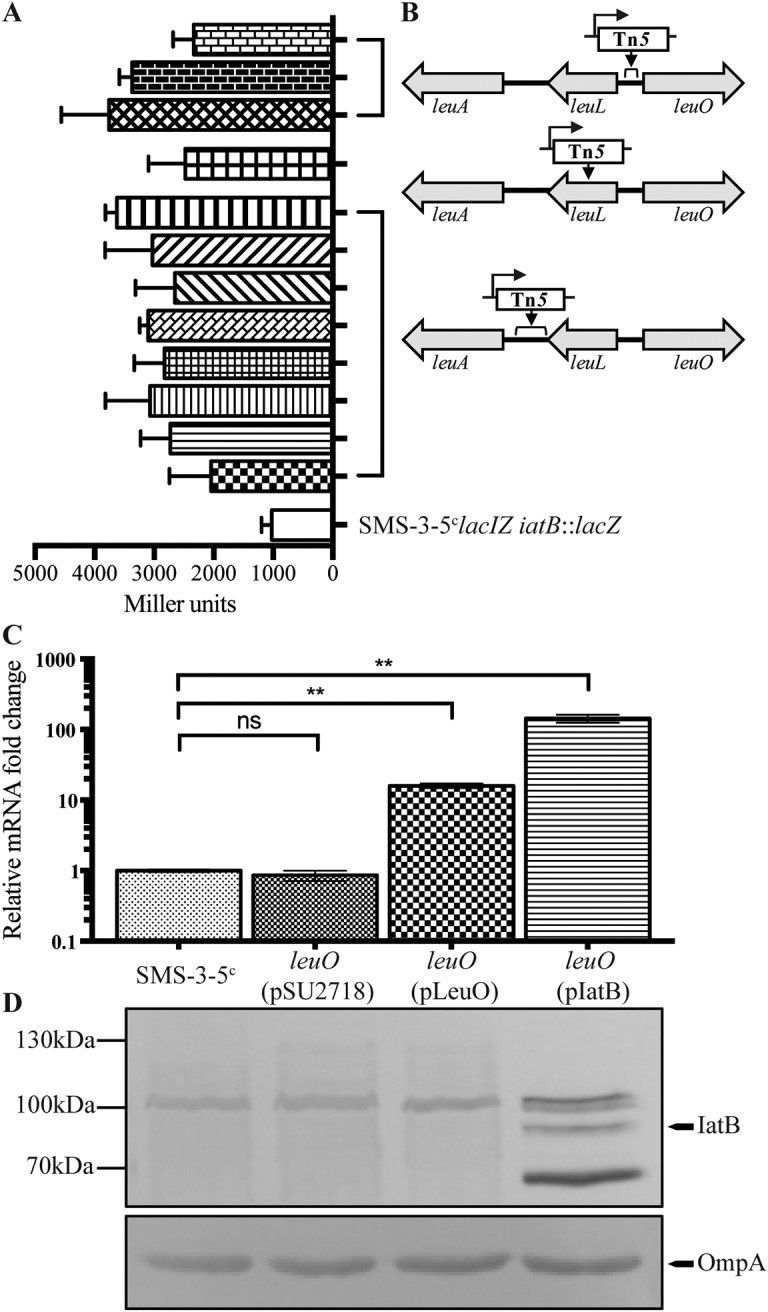
Effect of LeuO on iatB transcription in SMS-3-5c. (A) β-Galactosidase activity (in Miller units) of each mutant in comparison to the SMS35clacIZ iatB::lacZ control strain. (B) Schematic diagram depicting the location and orientation of the Tn5 insertions. (C) qRT-PCR showing the relative fold differences of iatB transcript levels in SMS-3-5c, SMS-3-5cleuO(pSU2718), SMS-3-5cleuO(pLeuO), and SMS-3-5cleuO(pIatB) (**, P = 0.0069 to 0.009; ns, not significant). (D) Western blot analysis of IatB in the respective strains. A band corresponding to IatB was detected only in the SMS-3-5cleuO(pIatB) strain.
iatB mRNA transcript levels are affected by the global regulator LeuO.
To directly examine the effect of LeuO on iatB expression, the leuO gene was mutated in SMS-3-5c to generate strain SMS-3-5cleuO. In addition, the leuO gene was also PCR amplified, cloned into the expression vector pSU2718 (to generate the plasmid pLeuO), and transformed into the SMS-3-5cleuO mutant. The expression of iatB was first examined in SMS-3-5c, SMS-3-5cleuO(pSU2718), SMS-3-5cleuO(pLeuO), and SMS-3-5cleuO(pIatB) by quantitative reverse transcription PCR (qRT-PCR) (Fig. 4C). In this assay, overexpression of leuO led to an ∼15-fold increase in relative iatB transcript levels, whereas iatB transcript levels in the SMS-3-5cleuO(pSU2718) vector control strain were similar to those in wild-type SMS-3-5c. In comparison, the level of the iatB transcript in SMS-3-5c(pIatB) was ∼144-fold higher than in SMS-3-5c. Next, the expression of IatB was examined by direct detection of the protein using Western blotting (Fig. 4D). Although the IatB protein was clearly detected in SMS-3-5c(pIatB), it was not detected in any of the other strains. Taken together, these results suggest that overexpression of leuO increases iatB transcript levels in SMS-3-5c, but this increase in transcription does not translate into detectable levels of the IatB protein under the experimental conditions employed in this study.
DISCUSSION
Proteins secreted by the type V secretion system exhibit extensive diversity, and we show here that this variation extends to IATs in E. coli. The three IAT proteins characterized in this study, IatB, IatC, and IatD, all promoted biofilm formation, suggesting that they may contribute to surface colonization under certain environmental conditions. By focusing on the regulation of iatB, which displayed a high prevalence in phylogroup B2 strains frequently associated with extraintestinal infection, we also identified LeuO as a putative regulator of IatB.
The presence of a short C-terminal passenger domain in IAT proteins, as observed for IatA, is not uncommon. IAT proteins are almost exclusively found among the Gammaproteobacteria, and many IATs possess short C-terminal extensions that share structural characteristics with bacterial immunoglobulin superfamily (IgSF) domains (e.g., a fibronectin III domain) (24, 32). These domains can participate in protein-protein interactions and have been identified in factors that mediate the translocation of protein substrates across the outer membrane (32). Modeling of the IatA passenger domain in Phyre2 suggests that it possesses structural characteristics similar to those of a fibronectin III domain (data not shown). IatA is also the most common IAT protein of E. coli, and by analogy to other proteins that contain IgSF domains, it is possible that IatA could assist in the translocation of other proteins across the outer membrane.
IatB is an orthologue of SinH from Salmonella and shares 74% amino acid identity. Analysis of this locus revealed that the coding sequences immediately downstream of iatB (i.e., EcSMS35_2660) and sinH (i.e., sinI) also share sequence similarity and encode predicted proteins that lack a β-barrel domain. These gene clusters are analogous to the well-characterized zirTSU operon found in Salmonella. The ZirT protein shares a similar domain organization with SinH and IatB: it contains an N-terminal IAT β-barrel domain and a C-terminal passenger containing IgSF domains. In addition, ZirT serves as a platform for the translocation of ZirS and ZirU (33, 34), and thus, it is possible that IatB (and SinH) could possess similar functional properties. Our results demonstrate that IatB is able to mediate biofilm formation independent of the downstream EcSMS35_2660 protein. However, whether IatB also acts as a transport platform for other proteins remains an intriguing subject for further investigation.
The typical IAT passenger domain contains variable repeats of Big1 domains with a similar fold despite their low sequence similarity (17, 27). Of the four new IAT proteins identified in this study, IatB and IatC contain passenger domains of various lengths. The passenger domains of some IATs, such as the invasin-like molecule of Yersinia ruckeri, contain Ig domains that are almost identical (35). The presence of tandem sequence repeats can result in misassembly of a gene sequence due to erroneous sequencing analyses (36). However, we found no evidence to support the idea that there is variability in the number of tandem repeats in IatC or IatD. Other AT proteins, like UpaH and Ag43, exhibit sequence variation that results in altered levels of biofilm formation by different variants (13, 37). Thus, it is possible that different variants of IatB and IatC may possess different functional properties, such as various degrees of biofilm formation.
We showed that IatB, IatC, and IatD were able to mediate biofilm formation when expressed in a recombinant K-12 strain. Like intimin and invasin, bioinformatic analysis predicts that the passenger domains of IatC and IatD are capped with a C-type lectin domain. The C-type lectin domain of intimin mediates adhesion of enteropathogenic E. coli strains to the intestinal epithelium via interaction with its receptor Tir, whereas the domain found in invasin mediates binding to β1-integrins. Thus, it is possible that the C-type lectin domains of IatC and IatD could also recognize specific receptors on host surfaces that remain to be identified. The IatC and IatD proteins also contain a LysM domain, which has also been identified in other large IAT proteins (26, 38). The LysM domain is found in many peptidoglycan-binding proteins and may contribute to their localization and stability in the outer membrane (38, 39).
We also sought to understand the regulation of the iatB gene. The regulation of AT protein expression in E. coli is complex; for example, many AIDA-I AT proteins are not expressed during standard laboratory growth, and multiple control mechanisms have been described (7, 8, 40). Consistent with this, we were unable to detect expression of the IAT proteins in wild-type SMS-3-5 via Western blot analysis following static or shaking growth in either LB or M9 minimal medium at 28°C, 37°C, or 42°C (data not shown). Using a mutagenesis approach, we identified LeuO as a potential activator of iatB. However, while overexpression of LeuO led to an increase in the iatB transcript in SMS-3-5c, direct expression of the protein could not be detected via Western blotting. This may have been due to its low level of expression or instability or even the lack of additional regulatory factors that are required for its optimal expression. LeuO is a LysR-type transcriptional regulator (LTTR) that contains an N-terminal helix-turn-helix DNA-binding domain (41) and can be found in other members of the Enterobacteriaceae, including Salmonella, Shigella, and Yersinia spp. (41). LeuO is involved in coordination of the bacterial stress response, and expression of LeuO is increased upon entry into stationary phase (42–44). Additionally, LeuO activates several cryptic fimbriae of E. coli, and overexpression of LeuO in E. coli led to increased cell adhesion and biofilm formation (42). Our findings are consistent with these phenotypic properties, suggesting that further work is now required to understand the molecular mechanisms by which LeuO controls IatB expression and to determine if LeuO plays a role in the coordinated regulation of other IATs.
Overall, this study has identified the complement of IAT proteins present in E. coli and provides new insight into their diversity, function, and regulation. Four of these IAT proteins were new, and three were functionally characterized. We found that the IATs IatB, IatC, and IatD are surface exposed, mediate biofilm formation, and thus may comprise part of the arsenal of factors used by E. coli to colonize different surfaces. Further work is now needed to understand the molecular mechanisms that control their expression.
MATERIALS AND METHODS
Bioinformatics analysis.
The E. coli database was represented by 126 published complete genomes available in the NCBI database. Sequence comparisons were examined using the fasta36 software package (45). The database of annotated proteins from each strain was generated using the cds_extractor v0.7.1 tool (46) and probed using fasta36 to identify putative β-barrel domain-containing IAT proteins. The context of the β-barrel domain was examined in each extracted protein sequence to ensure that it was located at the N-terminal end of the protein. The prevalence of genes was determined using tfastx36 and a cutoff of >75% identity over an 80% amino acid sequence alignment. Any proteins lacking an N-terminal signal sequence or the β-barrel domain were discarded. Multilocus sequence typing (MLST) analysis was performed using the sequences of seven housekeeping genes as previously described (47). The E. coli strains were classified into major phylogroups (A, B1, B2, D, E, and F) as previously described (48). Briefly, strains were sorted into the different phylogroups based on an in silico analysis of the arpA, chuA, yjaA, and TSPE4.C2 loci using fasta36 with a cutoff of >90% identity over a 95% nucleotide sequence alignment. The genomic context of genes was analyzed and drawn with Easyfig (49). Alignments were constructed in MEGA7 (50) using the Muscle algorithm with default settings. Trees were produced with MEGA7 using the maximum likelihood method with default settings and supported with 100 bootstraps. The Conserved Domain Database (CDD) (51), InterPro (52), and Phyre2 (53) were used to analyze protein structures, and SignalP4.1 (54) was used to predict the presence of signal sequences.
Bacterial strains and culture conditions.
Strains were routinely cultured on solid or in liquid lysogeny broth (LB) or M9 minimal medium and supplemented with the following antibiotics where appropriate: gentamicin (Gent) (20 μg/ml), ampicillin (Amp) (100 μg/ml), kanamycin (Kan) (50 μg/ml), and chloramphenicol (Cm, 30 μg/ml). Expression of genes was induced with either 1 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG) or 0.2% l-arabinose when required. The bacterial strains and plasmids used in this study are outlined in Data Set S3 in the supplemental material.
Molecular methods.
Methods for DNA extraction, purification, sequencing, and PCR were performed as previously described (8, 55, 56). Deletion mutants were constructed using a modified λ red recombinase gene replacement system as described previously (57–59). For RNA extraction, exponentially growing cells grown in LB (500 μl) (optical density at 600 nm [OD600] = 0.6) were stabilized in 1 ml of RNAprotect bacterial reagent (Qiagen). Subsequent RNA extraction, DNase I treatment, first-strand cDNA synthesis, and qRT-PCR were performed as previously described (60). Gene expression levels were determined with the 2−ΔΔCT method (61), with relative fold differences expressed against wild-type SMS-3-5. All experiments were performed in three independent replicates. RT-PCRs were performed with primers specific for each gene. The full list of primers used is shown in Data Set S4.
Primers used in this study. Download Data Set S4, XLSX file, 0.1 MB (12.5KB, xlsx) .
Copyright © 2019 Goh et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Generation of polyclonal antibodies.
A fragment of the iatA-D genes corresponding to the C-terminal passenger domain was PCR amplified from SMS-3-5, cloned as an N-terminal 6×His fusion protein in plasmid pMCSG7 via ligation-independent cloning (LIC) (62), and transformed into E. coli BL21(DE3). The expression of each of the 6×His-tagged fusion proteins was induced with 1 mM IPTG and purified on a Ni-nitrilotriacetic acid (NTA) spin column (Qiagen). The purified proteins were quantified with a bicinchoninic acid protein assay kit (Sigma) and assessed for purity via SDS-PAGE. These purified proteins were used to generate polyclonal antisera in rabbits at the Walter and Eliza Hall Institute of Medical Research Antibody Facility.
Protein sample preparation and immunoblotting.
Whole-cell lysates were prepared by pelleting 1 ml of OD600-standardized cells and resuspending the cells in 50 μl water and 50 μl 2× SDS loading buffer (100 mM Tris-HCl, 4% [wt/vol] SDS, 20% [vol/vol] glycerol, 0.2% [wt/vol] bromophenol blue [pH 6.8]). SDS-PAGE and transfer of proteins onto polyvinylidene difluoride (PVDF) membranes for Western blot analysis were performed as previously described (63). Polyclonal antisera specific for each protein were used to probe for the respective proteins, and alkaline phosphatase-conjugated anti-rabbit antisera (Sigma-Aldrich) were used as the secondary antibodies.
Immunofluorescence.
Cultures grown overnight and supplemented with the appropriate antibiotics and 1 mM IPTG were fixed to an OD600 of 0.4, spotted onto a glass slide, and allowed to dry. The cells were fixed with 4% paraformaldehyde (PFA), blocked with 0.5% bovine serum albumin (BSA), and incubated with a 1:100 dilution of the appropriate primary antibody in phosphate-buffered saline (PBS) for 30 min. The cells were washed and incubated with secondary goat anti-rabbit antiserum coupled to fluorescein isothiocyanate (FITC) diluted 1:500 in PBS. The slides were washed and air dried, mounted with Prolong gold antifade reagent (Life Technologies), and examined under a Zeiss Axioplan 2 epifluorescence light microscope.
Phenotypic assays.
Biofilm assays were performed in 96-well PVC plates (Corning) as previously described (64). Statistical analyses were performed using unpaired two-tailed Student’s t test. Flow cell experiments were performed as previously described (65, 66). Biofilm thickness, coverage, and total biomass measurements were collected from 10 z-stacks for each strain and analyzed with the COMSTAT program (67). The nonparametric Kruskal-Wallis test within GraphPad Prism 7 software was used for statistical analysis; P values of <0.05 were considered significant. β-Galactosidase assays were performed as previously described (68). Each strain was assessed in quadruplicate, and experiments were performed in two independent replicates.
Transposon mutagenesis.
Transposon mutagenesis of SMS-3-5lacIZ iatB::lacZ was performed using the Epicenter EZ::Tn5 transposome construction kit as previously described (60). The transposon insertion site of the mutants was identified via 2-step arbitrary PCR as previously described (69).
Curing of plasmid pSMS35_130.
Curing of the pSMS35_130 plasmid was performed as previously described (29). Briefly, the incompatibility regions (IncFIA and IncFII) and antitoxins (sok, vagC, and pemI) from pSMS35_130 were synthesized (Epoch Life Science Inc.) and incorporated directly into plasmid pMDP4, which contained a chloramphenicol resistance gene cassette, the gfp gene, and the sacB gene. This plasmid is referred to as pSMS35_130cure. Plasmid pSMS35_130cure was electroporated into SMS-3-5, and transformants were plated on LB agar containing chloramphenicol. Colonies were screened for fluorescence indicating the presence of the pSMS35_130cure plasmid. After one round of subculture, cells were plated onto LB agar containing 5% sucrose for subsequent selection of plasmid-free cells. Plasmid loss was confirmed by antibiotic sensitivity testing.
ACKNOWLEDGMENTS
This work was supported by grants from the National Health and Medical Research Council (NHMRC) of Australia (GNT1106590) and the Australian Research Council (DP180102987). M.A.S. is supported by an NHMRC senior research fellowship (GNT1106930).
REFERENCES
- 1.Grijpstra J, Arenas J, Rutten L, Tommassen J. 2013. Autotransporter secretion: varying on a theme. Res Microbiol 164:562–582. doi: 10.1016/j.resmic.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 2.Leyton DL, Rossiter AE, Henderson IR. 2012. From self sufficiency to dependence: mechanisms and factors important for autotransporter biogenesis. Nat Rev Microbiol 10:213–225. doi: 10.1038/nrmicro2733. [DOI] [PubMed] [Google Scholar]
- 3.Benz I, Schmidt MA. 2011. Structures and functions of autotransporter proteins in microbial pathogens. Int J Med Microbiol 301:461–468. doi: 10.1016/j.ijmm.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 4.Desvaux M, Parham NJ, Henderson IR. 2004. The autotransporter secretion system. Res Microbiol 155:53–60. doi: 10.1016/j.resmic.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Henderson IR, Navarro-Garcia F, Desvaux M, Fernandez RC, Ala’Aldeen D. 2004. Type V protein secretion pathway: the autotransporter story. Microbiol Mol Biol Rev 68:692–744. doi: 10.1128/MMBR.68.4.692-744.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valle J, Mabbett AN, Ulett GC, Toledo-Arana A, Wecker K, Totsika M, Schembri MA, Ghigo JM, Beloin C. 2008. UpaG, a new member of the trimeric autotransporter family of adhesins in uropathogenic Escherichia coli. J Bacteriol 190:4147–4161. doi: 10.1128/JB.00122-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Totsika M, Wells TJ, Beloin C, Valle J, Allsopp LP, King NP, Ghigo JM, Schembri MA. 2012. Molecular characterization of the EhaG and UpaG trimeric autotransporter proteins from pathogenic Escherichia coli. Appl Environ Microbiol 78:2179–2189. doi: 10.1128/AEM.06680-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allsopp LP, Beloin C, Ulett GC, Valle J, Totsika M, Sherlock O, Ghigo JM, Schembri MA. 2012. Molecular characterization of UpaB and UpaC, two new autotransporter proteins of uropathogenic Escherichia coli CFT073. Infect Immun 80:321–332. doi: 10.1128/IAI.05322-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drago-Serrano ME, Parra SG, Manjarrez-Hernández HA. 2006. EspC, an autotransporter protein secreted by enteropathogenic Escherichia coli (EPEC), displays protease activity on human hemoglobin. FEMS Microbiol Lett 265:35–40. doi: 10.1111/j.1574-6968.2006.00463.x. [DOI] [PubMed] [Google Scholar]
- 10.Guyer DM, Henderson IR, Nataro JP, Mobley HL. 2000. Identification of sat, an autotransporter toxin produced by uropathogenic Escherichia coli. Mol Microbiol 38:53–66. doi: 10.1046/j.1365-2958.2000.02110.x. [DOI] [PubMed] [Google Scholar]
- 11.Danese PN, Pratt LA, Dove SL, Kolter R. 2000. The outer membrane protein, antigen 43, mediates cell-to-cell interactions within Escherichia coli biofilms. Mol Microbiol 37:424–432. doi: 10.1046/j.1365-2958.2000.02008.x. [DOI] [PubMed] [Google Scholar]
- 12.Hasman H, Chakraborty T, Klemm P. 1999. Antigen-43-mediated autoaggregation of Escherichia coli is blocked by fimbriation. J Bacteriol 181:4834–4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ulett GC, Valle J, Beloin C, Sherlock O, Ghigo JM, Schembri MA. 2007. Functional analysis of antigen 43 in uropathogenic Escherichia coli reveals a role in long-term persistence in the urinary tract. Infect Immun 75:3233–3244. doi: 10.1128/IAI.01952-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heras B, Totsika M, Peters KM, Paxman JJ, Gee CL, Jarrott RJ, Perugini MA, Whitten AE, Schembri MA. 2014. The antigen 43 structure reveals a molecular Velcro-like mechanism of autotransporter-mediated bacterial clumping. Proc Natl Acad Sci U S A 111:457–462. doi: 10.1073/pnas.1311592111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leo JC, Grin I, Linke D. 2012. Type V secretion: mechanism(s) of autotransport through the bacterial outer membrane. Philos Trans R Soc Lond B Biol Sci 367:1088–1101. doi: 10.1098/rstb.2011.0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leo JC, Oberhettinger P, Schutz M, Linke D. 2015. The inverse autotransporter family: intimin, invasin and related proteins. Int J Med Microbiol 305:276–282. doi: 10.1016/j.ijmm.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 17.Tsai JC, Yen MR, Castillo R, Leyton DL, Henderson IR, Saier MH Jr.. 2010. The bacterial intimins and invasins: a large and novel family of secreted proteins. PLoS One 5:e14403. doi: 10.1371/journal.pone.0014403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fairman JW, Dautin N, Wojtowicz D, Liu W, Noinaj N, Barnard TJ, Udho E, Przytycka TM, Cherezov V, Buchanan SK. 2012. Crystal structures of the outer membrane domain of intimin and invasin from enterohemorrhagic E. coli and enteropathogenic Y. pseudotuberculosis. Structure 20:1233–1243. doi: 10.1016/j.str.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oberhettinger P, Schutz M, Leo JC, Heinz N, Berger J, Autenrieth IB, Linke D. 2012. Intimin and invasin export their C-terminus to the bacterial cell surface using an inverse mechanism compared to classical autotransport. PLoS One 7:e47069. doi: 10.1371/journal.pone.0047069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Donnenberg MS, Tzipori S, McKee ML, O’Brien AD, Alroy J, Kaper JB. 1993. The role of the eae gene of enterohemorrhagic Escherichia coli in intimate attachment in vitro and in a porcine model. J Clin Invest 92:1418–1424. doi: 10.1172/JCI116718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jerse AE, Yu J, Tall BD, Kaper JB. 1990. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc Natl Acad Sci U S A 87:7839–7843. doi: 10.1073/pnas.87.20.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deuretzbacher A, Czymmeck N, Reimer R, Trulzsch K, Gaus K, Hohenberg H, Heesemann J, Aepfelbacher M, Ruckdeschel K. 2009. Beta1 integrin-dependent engulfment of Yersinia enterocolitica by macrophages is coupled to the activation of autophagy and suppressed by type III protein secretion. J Immunol 183:5847–5860. doi: 10.4049/jimmunol.0804242. [DOI] [PubMed] [Google Scholar]
- 23.Leo JC, Skurnik M. 2011. Adhesins of human pathogens from the genus Yersinia. Adv Exp Med Biol 715:1–15. doi: 10.1007/978-94-007-0940-9_1. [DOI] [PubMed] [Google Scholar]
- 24.Heinz E, Stubenrauch CJ, Grinter R, Croft NP, Purcell AW, Strugnell RA, Dougan G, Lithgow T. 2016. Conserved features in the structure, mechanism, and biogenesis of the inverse autotransporter protein family. Genome Biol Evol 8:1690–1705. doi: 10.1093/gbe/evw112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Easton DM, Allsopp LP, Phan MD, Moriel DG, Goh GK, Beatson SA, Mahony TJ, Cobbold RN, Schembri MA. 2014. The intimin-like protein FdeC is regulated by H-NS and temperature in enterohemorrhagic Escherichia coli. Appl Environ Microbiol 80:7337–7347. doi: 10.1128/AEM.02114-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez-Gil M, Goh KGK, Rackaityte E, Sakamoto C, Audrain B, Moriel DG, Totsika M, Ghigo JM, Schembri MA, Beloin C. 2017. YeeJ is an inverse autotransporter from Escherichia coli that binds to peptidoglycan and promotes biofilm formation. Sci Rep 7:11326. doi: 10.1038/s41598-017-10902-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nesta B, Spraggon G, Alteri C, Moriel DG, Rosini R, Veggi D, Smith S, Bertoldi I, Pastorello I, Ferlenghi I, Fontana MR, Frankel G, Mobley HL, Rappuoli R, Pizza M, Serino L, Soriani M. 2012. FdeC, a novel broadly conserved Escherichia coli adhesin eliciting protection against urinary tract infections. mBio 3:e00010-12. doi: 10.1128/mBio.00010-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reisner A, Haagensen JA, Schembri MA, Zechner EL, Molin S. 2003. Development and maturation of Escherichia coli K-12 biofilms. Mol Microbiol 48:933–946. doi: 10.1046/j.1365-2958.2003.03490.x. [DOI] [PubMed] [Google Scholar]
- 29.Hale L, Lazos O, Haines A, Thomas C. 2010. An efficient stress-free strategy to displace stable bacterial plasmids. Biotechniques 48:223–228. doi: 10.2144/000113366. [DOI] [PubMed] [Google Scholar]
- 30.Goh KGK, Phan MD, Forde BM, Chong TM, Yin WF, Chan KG, Ulett GC, Sweet MJ, Beatson SA, Schembri MA. 2017. Genome-wide discovery of genes required for capsule production by uropathogenic Escherichia coli. mBio 8:e01558-17. doi: 10.1128/mBio.01558-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phan M-D, Nhu NTK, Achard MES, Forde BM, Hong KW, Chong TM, Yin W-F, Chan K-G, West NP, Walker MJ, Paterson DL, Beatson SA, Schembri MA. 2017. Modifications in the pmrB gene are the primary mechanism for the development of chromosomally encoded resistance to polymyxins in uropathogenic Escherichia coli. J Antimicrob Chemother 72:2729–2736. doi: 10.1093/jac/dkx204. [DOI] [PubMed] [Google Scholar]
- 32.Bodelon G, Palomino C, Fernandez LA. 2013. Immunoglobulin domains in Escherichia coli and other enterobacteria: from pathogenesis to applications in antibody technologies. FEMS Microbiol Rev 37:204–250. doi: 10.1111/j.1574-6976.2012.00347.x. [DOI] [PubMed] [Google Scholar]
- 33.Gal-Mor O, Gibson DL, Baluta D, Vallance BA, Finlay BB. 2008. A novel secretion pathway of Salmonella enterica acts as an antivirulence modulator during salmonellosis. PLoS Pathog 4:e1000036. doi: 10.1371/journal.ppat.1000036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prehna G, Li Y, Stoynov N, Okon M, Vuckovic M, McIntosh LP, Foster LJ, Finlay BB, Strynadka NC. 2012. The zinc regulated antivirulence pathway of Salmonella is a multiprotein immunoglobulin adhesion system. J Biol Chem 287:32324–32337. doi: 10.1074/jbc.M112.357210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wrobel A, Ottoni C, Leo JC, Gulla S, Linke D. 2018. The repeat structure of two paralogous genes, Yersinia ruckeri invasin (yrInv) and a “Y. ruckeri invasin-like molecule”, (yrIlm) sheds light on the evolution of adhesive capacities of a fish pathogen. J Struct Biol 201:171–183. doi: 10.1016/j.jsb.2017.08.008. [DOI] [PubMed] [Google Scholar]
- 36.Allsopp LP, Totsika M, Tree JJ, Ulett GC, Mabbett AN, Wells TJ, Kobe B, Beatson SA, Schembri MA. 2010. UpaH is a newly identified autotransporter protein that contributes to biofilm formation and bladder colonization by uropathogenic Escherichia coli CFT073. Infect Immun 78:1659–1669. doi: 10.1128/IAI.01010-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Allsopp LP, Beloin C, Moriel DG, Totsika M, Ghigo JM, Schembri MA. 2012. Functional heterogeneity of the UpaH autotransporter protein from uropathogenic Escherichia coli. J Bacteriol 194:5769–5782. doi: 10.1128/JB.01264-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leo JC, Oberhettinger P, Chaubey M, Schütz M, Kühner D, Bertsche U, Schwarz H, Götz F, Autenrieth IB, Coles M, Linke D. 2015. The intimin periplasmic domain mediates dimerisation and binding to peptidoglycan. Mol Microbiol 95:80–100. doi: 10.1111/mmi.12840. [DOI] [PubMed] [Google Scholar]
- 39.Buist G, Steen A, Kok J, Kuipers OP. 2008. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol Microbiol 68:838–847. doi: 10.1111/j.1365-2958.2008.06211.x. [DOI] [PubMed] [Google Scholar]
- 40.Henderson IR, Owen P. 1999. The major phase-variable outer membrane protein of Escherichia coli structurally resembles the immunoglobulin A1 protease class of exported protein and is regulated by a novel mechanism involving Dam and oxyR. J Bacteriol 181:2132–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maddocks SE, Oyston PC. 2008. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 154:3609–3623. doi: 10.1099/mic.0.2008/022772-0. [DOI] [PubMed] [Google Scholar]
- 42.Shimada T, Bridier A, Briandet R, Ishihama A. 2011. Novel roles of LeuO in transcription regulation of E. coli genome: antagonistic interplay with the universal silencer H-NS. Mol Microbiol 82:378–397. doi: 10.1111/j.1365-2958.2011.07818.x. [DOI] [PubMed] [Google Scholar]
- 43.Fang M, Majumder A, Tsai KJ, Wu HY. 2000. ppGpp-dependent leuO expression in bacteria under stress. Biochem Biophys Res Commun 276:64–70. doi: 10.1006/bbrc.2000.3440. [DOI] [PubMed] [Google Scholar]
- 44.Majumder A, Fang M, Tsai KJ, Ueguchi C, Mizuno T, Wu HY. 2001. LeuO expression in response to starvation for branched-chain amino acids. J Biol Chem 276:19046–19051. doi: 10.1074/jbc.M100945200. [DOI] [PubMed] [Google Scholar]
- 45.Pearson WR, Lipman DJ. 1988. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A 85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leimbach A. 21 December 2016. Bac-genomics-scripts: bovine E. coli mastitis comparative genomics edition. Zenodo doi: 10.5281/zenodo.215824. [DOI] [Google Scholar]
- 47.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clermont O, Christenson JK, Denamur E, Gordon DM. 2013. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep 5:58–65. doi: 10.1111/1758-2229.12019. [DOI] [PubMed] [Google Scholar]
- 49.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. 2011. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res 39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mitchell A, Chang HY, Daugherty L, Fraser M, Hunter S, Lopez R, McAnulla C, McMenamin C, Nuka G, Pesseat S, Sangrador-Vegas A, Scheremetjew M, Rato C, Yong SY, Bateman A, Punta M, Attwood TK, Sigrist CJ, Redaschi N, Rivoire C, Xenarios I, Kahn D, Guyot D, Bork P, Letunic I, Gough J, Oates M, Haft D, Huang H, Natale DA, Wu CH, Orengo C, Sillitoe I, Mi H, Thomas PD, Finn RD. 2015. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res 43:D213–D221. doi: 10.1093/nar/gku1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 54.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 55.Moriel DG, Tan L, Goh KG, Phan MD, Ipe DS, Lo AW, Peters KM, Ulett GC, Beatson SA, Schembri MA. 2016. A novel protective vaccine antigen from the core Escherichia coli genome. mSphere 1:e00326-16. doi: 10.1128/mSphere.00326-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nichols KB, Totsika M, Moriel DG, Lo AW, Yang J, Wurpel DJ, Rossiter AE, Strugnell RA, Henderson IR, Ulett GC, Beatson SA, Schembri MA. 2016. Molecular characterization of the vacuolating autotransporter toxin in uropathogenic Escherichia coli. J Bacteriol 198:1487–1498. doi: 10.1128/JB.00791-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kakkanat A, Phan MD, Lo AW, Beatson SA, Schembri MA. 2017. Novel genes associated with enhanced motility of Escherichia coli ST131. PLoS One 12:e0176290. doi: 10.1371/journal.pone.0176290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Totsika M, Beatson SA, Sarkar S, Phan MD, Petty NK, Bachmann N, Szubert M, Sidjabat HE, Paterson DL, Upton M, Schembri MA. 2011. Insights into a multidrug resistant Escherichia coli pathogen of the globally disseminated ST131 lineage: genome analysis and virulence mechanisms. PLoS One 6:e26578. doi: 10.1371/journal.pone.0026578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tan L, Moriel DG, Totsika M, Beatson SA, Schembri MA. 2016. Differential regulation of the surface-exposed and secreted SslE lipoprotein in extraintestinal pathogenic Escherichia coli. PLoS One 11:e0162391. doi: 10.1371/journal.pone.0162391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 62.Eschenfeldt WH, Lucy S, Millard CS, Joachimiak A, Mark ID. 2009. A family of LIC vectors for high-throughput cloning and purification of proteins. Methods Mol Biol 498:105–115. doi: 10.1007/978-1-59745-196-3_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ulett GC, Webb RI, Schembri MA. 2006. Antigen-43-mediated autoaggregation impairs motility in Escherichia coli. Microbiology 152:2101–2110. doi: 10.1099/mic.0.28607-0. [DOI] [PubMed] [Google Scholar]
- 64.Schembri MA, Klemm P. 2001. Biofilm formation in a hydrodynamic environment by novel fimH variants and ramifications for virulence. Infect Immun 69:1322–1328. doi: 10.1128/IAI.69.3.1322-1328.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schembri MA, Kjærgaard K, Klemm P. 2003. Global gene expression in Escherichia coli biofilms. Mol Microbiol 48:253–267. doi: 10.1046/j.1365-2958.2003.03432.x. [DOI] [PubMed] [Google Scholar]
- 66.Kjaergaard K, Schembri MA, Ramos C, Molin S, Klemm P. 2000. Antigen 43 facilitates formation of multispecies biofilms. Environ Microbiol 2:695–702. doi: 10.1046/j.1462-2920.2000.00152.x. [DOI] [PubMed] [Google Scholar]
- 67.Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersboll BK, Molin S. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146(Part 10):2395–2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- 68.Miller JH. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 69.O’Toole GA, Kolter R. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol 28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cladogram of the β-barrel domain from putative IAT proteins found in 126 complete E. coli genomic sequences. A database of annotated proteins from each strain was probed using fasta36 to identify putative β-barrel domain-containing IAT proteins. The context of the β-barrel domain was examined in each extracted protein sequence to ensure that it was located at the N-terminal end of the protein. The six prototype putative proteins chosen from SMS-3-5 are highlighted in red, and the branch representing intimin is in gray. Alignments and phylogenetic trees were drawn in MEGA7 and visualized with FigTree. Download FIG S1, PDF file, 0.2 MB (202.1KB, pdf) .
Copyright © 2019 Goh et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Prevalence of IAT proteins in the collection of 126 completely sequenced E. coli genomes. Download Data Set S1, XLSX file, 0.1 MB (21.6KB, xlsx) .
Copyright © 2019 Goh et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Prevalence of iat genes in the ECOR collection. Download Data Set S2, XLSX file, 0.1 MB (11.8KB, xlsx) .
Copyright © 2019 Goh et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genomic context of the IAT genes (highlighted in red) in SMS-3-5. Depicted are two genes upstream and downstream of the IAT gene of interest and the location of the fragment on the genome of SMS-3-5. Genes with known functions are labeled with gene names, and genes of unknown function are labeled with locus tags. Download FIG S2, PDF file, 0.2 MB (218.5KB, pdf) .
Copyright © 2019 Goh et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Comparison of β-barrel amino acid sequences from IAT and type Va AT proteins. (Left) Cladogram generated using the β-barrel amino acid sequences from IAT and type Va AT proteins. The sequences from the different subgroups cluster separately. (Right) Pairwise comparison of each β-barrel sequence. Numbers represent the percent identity. Download FIG S3, PDF file, 0.2 MB (223.3KB, pdf) .
Copyright © 2019 Goh et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Western blot analysis of the different IAT proteins. For each blot, − represents whole-cell lysates prepared from the MS427(pSU2718) vector control, and + represents whole-cell lysates prepared from MS427 expressing the different IAT proteins, IatB (81 kDa) (A), IatC (307 kDa) (B), and IatD (188 kDa) (C). Download FIG S4, PDF file, 0.1 MB (129.6KB, pdf) .
Copyright © 2019 Goh et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Strains and plasmids used in this study. Download Data Set S3, XLSX file, 0.1 MB (12.3KB, xlsx) .
Copyright © 2019 Goh et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in this study. Download Data Set S4, XLSX file, 0.1 MB (12.5KB, xlsx) .
Copyright © 2019 Goh et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.