Abstract
Autism spectrum disorder (ASD) is a neurodevelopmental condition characterized by core social impairments. ASD remains poorly understood because of the difficulty in studying disease biology directly in patients and the reliance on mouse models that lack clinically relevant, complex social cognition abilities. We use ethological observations in rhesus macaques to identify male monkeys with naturally occurring low sociality. These monkeys showed differences in specific neuropeptide and kinase signaling pathways compared to socially competent male monkeys. Using a discovery and replication design, we identified arginine vasopressin (AVP) in cerebrospinal fluid (CSF) as a key marker of group differences in monkey sociality; we replicated these findings in an independent monkey cohort. We also confirmed in an additional monkey cohort that AVP concentration in CSF is a stable traitlike measure. Next, we showed in a small pediatric cohort that CSF AVP concentrations were lower in male children with ASD compared to age-matched male children without ASD (but with other medical conditions). We demonstrated that CSF AVP concentration was sufficient to accurately distinguish ASD cases from medical controls. These data suggest that AVP and its signaling pathway warrant consideration in future research studies investigating new targets for diagnostics and drug development in ASD.
INTRODUCTION
Autism spectrum disorder (ASD) is a neurodevelopmental condition characterized by core social impairments (x1). Despite its prevalence (currently 1 in 68 U.S. children) and societal impact ($236 billion expended in the United States annually) (2, 3), ASD’s pathophysiology remains poorly understood. Consequently, there are no laboratory-based diagnostic tests to detect ASD or approved medications that can effectively treat this disorder. Scientific progress has been hindered by the difficulty of studying disease biology directly in patients and by reliance on mouse models, which as a species lack many of the clinically relevant social cognition abilities most relevant to understanding ASD (4, 5). These limitations underscore the value in developing an animal model of social impairments with more behavioral and biological homologies to the human disease.
Rhesus monkeys are an ideal model organism for advancing this objective. Like humans, rhesus monkeys are a highly social species capable of complex social cognition, and both species display stable and pronounced individual differences in social functioning (6). Previous research has shown that monkeys with low sociality (low-social) initiate fewer affiliative interactions, spend less time in physical contact and grooming with conspecifics, and display more inappropriate social behaviors compared to monkeys with high sociality (high-social), suggesting lower social motivation and impaired social skills (5, 7, 8). Because endophenotypic autistic traits are common and continuously distributed across the general human population (9, 10), the naturally occurring variations in rhesus monkey social behavior make them a useful model of the complex environmental and polygenic risk factors contributing to idiopathic ASD in human patients (11).
Studies of low-social monkeys stand to advance our understanding of the neurobiology of social deficits and allow us to hone hypothesis-driven questions that can be tested subsequently in valuable and volume-limited ASD patient samples. Here, we used a hypothesis-generating and cost-effective extreme group approach (12) to test, on the basis of three selection criteria, whether low-social versus high-social male monkeys exhibited differences in biological signaling pathways previously implicated in mammalian prosocial functioning (13–15), in idiopathic ASD (16–20), and in neurogenetic syndromes with high penetrance for ASD (21–23). These signaling molecules and pathways included arginine vasopressin (AVP), oxytocin (OXT), RAS–MAPK (mitogen-activated protein kinase), and PI3K (phosphatidylinositol 3-kinase)–AKT. We then tested the translational utility of our preclinical finding in a small patient cohort comprised of children with and without ASD.
RESULTS
Low- and high-social monkeys are identified and biological measurements differentiate these groups
Using legacy behavioral data, we identified n = 42 male rhesus monkeys (selected from a pool of n = 222 male monkeys) that were expected to show extremes in social functioning (see Materials and Methods). We then performed ethological observations on these individuals in their familiar social groups using focal-animal sampling methods and identified a subset of n = 15 low-social and n = 15 high-social monkeys based on their social interactions with others in their troop. To test the validity of classifying animals from ethological data, personality trait ratings were also collected, and a sociability score was calculated (24). As expected, sociability scores statistically predicted the ethological classification of animals into low- and high-social groups [likelihood ratio (LR) χ2 = 19.94; P < 0.0001].
Having validated the social groups, we then tested whether low-social versus high-social monkeys exhibited differences in a number of signaling molecules and pathways. We measured cerebrospinal fluid (CSF) concentrations of AVP and OXT; blood plasma concentrations of AVP and OXT; gene expression of oxytocin receptor (OXTR) and arginine vasopressin receptor V1a (AVPRV1a) in peripheral blood mononuclear cells (PBMCs); and the ratios of phosphorylated/total extracellular signal–regulated kinase (ERK), phosphatase and tensin homolog (PTEN), and AKT (components of the RAS-MAPK and PI3K-AKT signaling pathways) in PBMCs. Because false discovery is always a concern in these types of biological studies (25, 26), we adopted a statistical winnowing strategy, whereby at each stage of analysis, we excluded nonpredictive and collinear biological measures from further consideration (27).
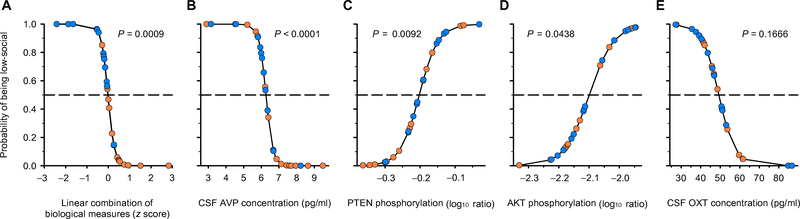
We first tested whether our biological data set, considered as a whole, could accurately distinguish low-social from high-social monkeys. Discriminant analysis yielded a 93% correct classification of monkeys into social groups (LR χ2 = 26.36; P < 0.0001; fig. S1 and table S1). We then used logistic regression to identify which biological measures were significantly predictive. Including all of the biological measures yielded an overspecified model, which is prone to false discovery (28). In the process of identifying a robust model, we excluded blood concentrations of AVP and OXT, OXTR gene expression in blood, the phosphorylated ERK/total ERK ratio in blood (all of which were nonpredictive), and AVPRV1a gene expression in blood (which was collinear with other variables in the model). Thus, the stable logistic regression model included CSF concentrations of AVP and OXT as well as the ratios of phosphorylated PTEN/total PTEN and phosphorylated AKT/total AKT (Fig. 1 and table S2.) These four biological measures when combined predicted social group classification (LR χ2 = 18.49; P = 0.0009; Fig. 1A). Analysis of the significance of each biological measure in this model revealed that CSF AVP concentration (LR χ2 = 16.55; P < 0.0001; Fig. 1B) and the ratios of phosphorylated PTEN/total PTEN (LR χ2 = 6.792; P = 0.0092; Fig. 1C) and phosphorylated AKT/total AKT (LR χ2 = 4.064; P = 0.0438; Fig. 1D) in blood predicted social group classification, whereas CSF OXT con centration did not (LR χ2 = 1.913; P = 0.1666; Fig. 1E).
Fig. 1. Biological measurements that differentiate low- and high-social monkeys in the discovery cohort.
(A) The logistic regression model [which, in this panel, includes the four combined biological measures that are plotted separately in (B) to (E)] correctly classified 24 of 27 monkeys (89%). (B to E) Each panel depicts a regression line that represents the effect of an individual biological measure. Each biological variable on the x axis is plotted against the probability of being low-social on the y axis and is corrected for the other variables in the analysis. Low-social monkeys (blue circles) plotted above, and high-social monkeys (orange circles) plotted beneath, the dashed lines (which represent 50% probability) are correctly classified. P values from LR χ2 tests are reported to indicate the strength of the plotted relationships, and the biological measures are presented in order of their contribution to the predictive power of the logistic regression model.
CSF AVP concentration differs between low- and high-social monkeys and is positively associated with time spent in social grooming
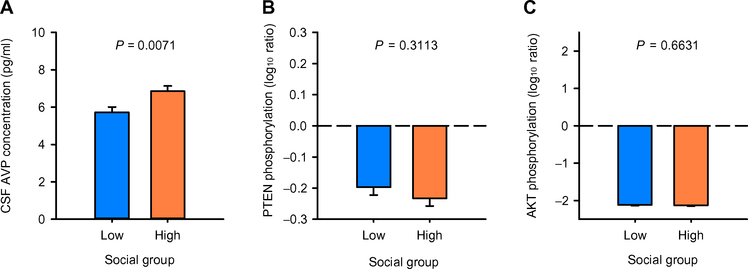
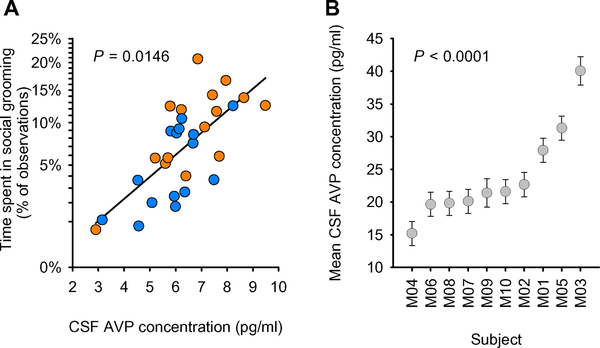
We then tested using a general linear model whether social group predicted differences in the three remaining significant biological measures independently (Fig. 2 and table S3). CSF AVP concentrations were significantly lower in low-social versus high-social monkeys (F1,18 = 9.236; P = 0.0071; Fig. 2A), whereas the ratios of phosphorylated PTEN/total PTEN (F1,17 = 1.089; P = 0.3113; Fig. 2B) and phosphorylated AKT/total AKT (F1,17 = 0.1966; P = 0.6631; Fig. 2C) in blood did not differ between groups. Thus, only CSF AVP concentration survived the full winnowing strategy. For an initial validation of this result, we then tested whether CSF AVP concentration could predict a continuously distributed and important measure of social competence in nonhuman primates, social grooming. We confirmed that CSF AVP concentration positively predicted time spent engaged in this social activity (F1,18 = 7.2914; P = 0.0146; partial r = +.54; Fig. 3A and table S4).
Fig. 2. Group comparisons for three biological measurements in the monkey discovery cohort.
General linear model F tests were used to evaluate whether the low- and high-social monkey groups differed in the three biological measurements (A to C) that were found to significantly predict monkey social group classification (as shown in Fig.1). Data are presented as least-squares means ± SEM. Only CSF AVP concentration differed significantly between the low-social (blue bars) and high-social (orange bars) monkey groups [n = 30 (CSF AVP); n = 28 (PTEN and AKT)].
Fig. 3. CSF AVP concentration is positively associated with social grooming in the monkey discovery cohort and is a stable, trait-like measure in an additional monkey cohort.
(A) The relationship between CSF AVP concentration and percentage of time spent in social grooming was tested with a general linear model F test (n = 30). The resulting regression line is plotted, corrected for the other variables in the analysis. Low-social monkeys are depicted in blue circles, and high-social monkeys are depicted in orange circles. (B) Within-individual consistency of CSF AVP concentrations across the four measurement time points was evaluated with intraclass correlation coefficients calculated from a mixed model. Least-squares means ± SEM CSF AVP concentrations are presented for each monkey (labeled M01 to M10; n = 10) and ordered on the x axis by ascending average CSF AVP concentration.
CSF AVP concentration is a stable trait-like measure
To test whether similar concentrations of CSF vasopressin were evident within an individual across multiple samplings, we evaluated banked CSF samples from a separate cohort of n = 10 adult male monkeys (see Materials and Methods). CSF samples were collected on four different occasions across a 4-month period, with each collection separated by an average of 40 days (intercollection interval range, 27 to 57 days). As expected, CSF AVP concentration showed stability within individuals across multiple time points (test-retest reliability intraclass correlation coefficient = 0.78; Fig. 3B and table S7). This intraclass correlation coefficient was significant (F9,25 = 12.88; P < 0.0001) and was considered a large effect size (29).
The specificity and validity of the statistical winnowing strategy is confirmed
We performed several post hoc quality control checks using general linear models. First, we tested for group differences in each of the original nine biological measures. Second, because blood involves less invasive collection procedures than CSF, we performed a separate analysis to test whether blood AVP concentration predicted CSF AVP concentration. Consistent with the rationale behind our statistical winnowing strategy, we observed no group differences in any biological measures except CSF AVP concentration (table S5). Blood AVP concentration was also unrelated to CSF AVP concentration (F1,18 = 0.001; P = 0.9963). These quality control checks thus further supported our rationale for selecting CSF AVP concentration as the focus of all subsequent analyses.
CSF AVP concentration is associated with group differences in monkey sociality in a replication cohort
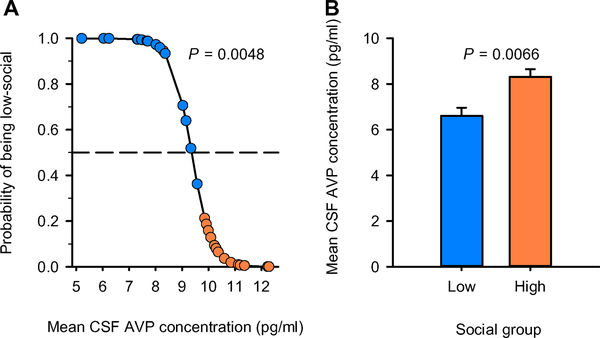
Having established that CSF AVP concentration statistically predicted social group classification in the discovery cohort and that our statistical winnowing strategy did not produce a false-negative result, we sought to replicate this CSF AVP finding in an independent monkey replication cohort. We observed n = 164 male monkeys and completed social behavior observations using a higher-throughput scan sampling-based method to identify a new cohort of n = 15 low-social and n = 15 high-social monkeys for CSF sample collection (see Materials and Methods). As before, CSF AVP concentration classified monkeys by social group (LR χ2 = 7.969; P < 0.0048; Fig. 4A), with low-social monkeys showing lower CSF AVP concentrations compared to high-social monkeys (F1,24 = 8.847; P = 0.0066; Fig. 4B and table S6).
Fig. 4. CSF AVP concentration distinguishes low- and high-social monkeys and differs between social groups in the monkey replication cohort.
(A) The logistic regression model correctly classified 28 of 30 monkeys (93%) and was evaluated for significance using LR χ2. CSF AVP concentration is plotted against the probability of being low-social; the resulting regression line is corrected for the other variables in the analysis. Low-social monkeys (blue circles) plotted above, and high-social monkeys (orange circles) plotted beneath, the dashed line (which represents 50% probability) are correctly classified. (B) A general linear model F test was used to test whether CSF AVP concentration was lower in low-social (blue bars) compared to high-social (orange bars) monkey groups in this replication cohort. Data are presented as least-squares means ± SEM.
CSF AVP concentrations are lower in children with ASD compared to children without ASD
To test the translational utility of our monkey model, we quantified AVP concentrations in CSF samples that had been previously collected as part of routine medical care from n = 14 male children [n = 7 children with ASD; n = 7 age-matched children who did not have ASD but who did have unrelated medical conditions (medical controls)].
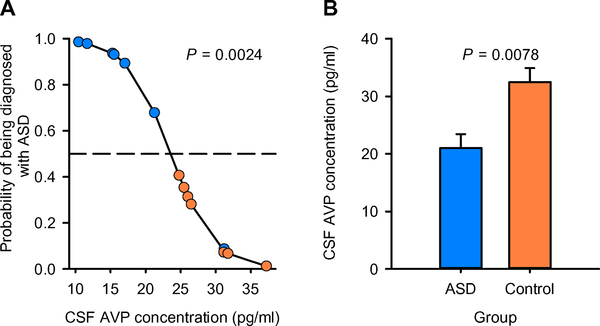
Participant demographic and medical characteristics are provided in Table 1. Using logistic regression, we found that CSF AVP concentration was sufficient to distinguish ASD cases from controls, whereby individuals with lower CSF AVP concentrations were more likely to have been previously diagnosed with ASD (LR χ2 = 9.233; P = 0.0024; Fig. 5A). Like low-social monkeys, ASD patients showed significantly lower CSF AVP concentrations compared to medical control children (F1,10 = 11.02; P = 0.0078; Fig. 5B). Because one pair of subjects (that is, an ASD patient and his matched control) was older than the other pairs of subjects in the analysis (see Table 1), we reran all analyses excluding this pair, and the results remained significant (Fig. 5 and table S8).
Table 1.
Patient demographics and medical characteristics. ALL, acute lymphoblastic leukemia.
| Patient number | Group | CSF AVP (pg/ml) | Age (years) | Ethnicity | Sample collection time | Indications for CSF procedure | Diagnosis/ pathology report |
|---|---|---|---|---|---|---|---|
| 1 | ASD | 32.97 | 6.58 | Asian | 09:30 | Evaluation for metabolic disorder | No abnormality determined |
| 2 | ASD | 11.47 | 6.08 | Caucasian | 10:00 | Maintenance chemotherapy | ALL in remission |
| 3 | ASD | 24.09 | 6.75 | Asian | 07:50 | Evaluation for metabolic disorder | No abnormality determined |
| 4 | ASD | 30.12 | 18.83 | Caucasian | 11:40 | Maintenance chemotherapy | ALL in remission |
| 5 | ASD | 19.77 | 9.92 | Caucasian | 11:16 | Evaluation for autoimmune or degenerative encephalopathies | No abnormality determined |
| 6 | ASD | 12.07 | 5.30 | Asian | 09:40 | Evaluation for metabolic disorder or degenerative encephalopathies | No abnormality determined |
| 7 | ASD | 15.80 | 6.92 | Caucasian | 12:37 | Evaluation for autoimmune or degenerative encephalopathies | Elevated inflammatory markers |
| 8 | Control | 27.45 | 6.33 | Caucasian | 09:40 | Maintenance chemotherapy | ALL in remission |
| 9 | Control | 26.42 | 5.25 | Caucasian | 08:29 | Maintenance chemotherapy | ALL in remission |
| 10 | Control | 40.68 | 7.83 | Caucasian | 08:40 | Maintenance chemotherapy | ALL in remission |
| 11 | Control | 41.41 | 19.50 | Caucasian | 18:26 | Evaluation for meningitis | No abnormality determined |
| 12 | Control | 32.69 | 10.42 | Asian | 08:30 | Induction chemotherapy | Relapsed ALL |
| 13 | Control | 26.81 | 6.42 | Caucasian | 08:40 | Maintenance chemotherapy | ALL in remission |
| 14 | Control | 32.55 | 7.50 | Caucasian | 13:09 | Maintenance chemotherapy | ALL in remission |
Fig. 5. CSF AVP concentration distinguishes ASD cases from medical controls.
(A) The logistic regression model correctly classified 13 of 14 children (93%) in a small patient cohort comprising seven children with ASD and seven medical control children without ASD (but with other medical conditions) and was evaluated for significance using LR χ2. CSF AVP concentration is plotted against the probability of being diagnosed with ASD; the resulting regression line is corrected for the other variables in the analysis. Children with ASD (blue circles) plotted above, and medical control children (orange circles) plotted beneath, the dashed line (which represents 50% probability) are correctly classified. (B) A general linear model F test was used to test whether children with and without ASD differed in CSF AVP concentrations. Similar to low-social monkeys studied in our preclinical model (that is, as shown in Figs. 2A and 4B), CSF AVP concentration was found to be lower in children with ASD (blue bars) compared to medical control children without ASD (orange bars). Data are presented as least-squares means ± SEM.
DISCUSSION
This study demonstrated in a nonhuman primate model of naturally occurring low sociality that there were differences between low- and high-social monkeys regarding several signaling pathways previously implicated in idiopathic and syndromic ASD (16, 23). Further analysis revealed that CSF AVP concentration was a key marker of group differences in monkey sociality. We confirmed this finding in an independent validation cohort of male rhesus monkeys using different, higher-throughput behavioral methods. We also verified in a separate monkey cohort that CSF AVP concentration is a stable trait-like measure. As our monkey study investigated the biology of low-social functioning, a nonpathological entity, we next sought to translate these preclinical findings to a small pediatric cohort composed of seven age-matched male ASD patients and seven male medical controls. We found that CSF AVP concentrations were lower in children with ASD compared to medical control children, and that CSF AVP concentration differentiated individual ASD cases and controls accurately. These findings demonstrate the value of developing a primate model of social impairments for investigating the pathophysiology of ASD.
Although the patient CSF AVP concentration data are very preliminary, they are of interest because there are currently no laboratory-based diagnostic tests for detecting ASD. The lack of ASD diagnostic markers may be due to the fact that most ASD biological investigations have been largely restricted to peripheral tissues such as blood. Blood requires less invasive collection procedures, and although it may be related to measurements in brain tissue under some circumstances (20, 30, 31), it is also less representative of brain biochemistry than is CSF. Here, we observed no relationship between blood and CSF measurements of AVP concentration, and in a previous study, we observed no differences in blood AVP concentrations between children with and without ASD (20). Other brain disorders, such as dementia and multiple sclerosis, have benefited from CSF biomarker discovery efforts (32, 33). For example, one dementia study showed that antemortem CSF τ protein and β amyloid concentrations predicted neuropathological features (that is, amyloid load and neurofibrillary tangles) of Alzheimer’s disease in postmortem brain tissue (34). In our monkey model, although kinase measurements did not differ by social group, kinase signaling in PBMCs, particularly in the PI3KAKT pathway, improved the accuracy of CSF AVP to correctly classify low-social and high-social monkeys. This suggests that diagnostic marker discovery efforts for ASD should continue using both CSF and blood samples.
We currently have limited capacity to improve social outcomes in ASD patients, in part, because no robust “druggable” targets have been identified. However, there is long-standing evidence from multiple rodent species that AVP enhances, whereas selective AVPRV1a antagonists impair, social functioning (35–37). These pharmacological effects are particularly evident in male rodents, and given ASD’s male-biased prevalence in human patients (4:1 male/female ratio) (2), it has been previously hypothesized that brain AVP signaling deficits may be relevant to understanding ASD pathophysiology (15, 20, 38). Findings from our current study lend additional support to the notion that boosting brain AVP signaling could be a therapeutic strategy for ameliorating social impairments in ASD patients. Whether AVP administration could enhance social abilities in people with ASD has not yet been tested. However, intranasally administered AVP results in elevated CSF concentrations of AVP in healthy people, suggesting that AVP is able to access the brain via this delivery route (39). Because ASD is etiologically and phenotypically heterogeneous, CSF AVP concentrations could be measured before treatment to enable identification of ASD patients most likely to benefit from AVP pharmacotherapy.
There are several limitations to our study. First, our monkey and human cohorts were restricted to males because of ASD’s male-biased prevalence. Studies inclusive of females are now needed for both species. Second, we used an extreme group approach in our monkey studies. This design was selected because it is well suited for conducting hypothesis-generating research, increasing the power to detect an effect in a limited sample size, and improving cost efficiency (12). Nevertheless, this design is not without shortcomings; research is now required to test whether CSF AVP concentrations permit identification of low-social monkeys among conspecifics distributed across the social continuum. Third, the human sample was one of convenience, comprising children with and without ASD undergoing clinically indicated CSF collection. Although our control children did not have ASD, some of them had serious medical conditions (Table 1). This was inevitable given the challenges and ethical considerations involved in obtaining CSF samples from healthy children to serve as controls. Although it is possible that our CSF findings in ASD patients were somehow driven by medications or disease states not relevant to ASD, our monkey studies were conducted in medication and disease-free animals, and two independent monkey cohorts yielded similar findings to those obtained in the ASD patient cohort. Nevertheless, extension of these CSF AVP findings to adults with ASD would ethically permit recruitment of a healthy adult comparison group for CSF collection. Additionally, assessment of adults with ASD would allow us to test whether the CSF AVP finding persists into adulthood. Finally, owing to the very small size of our ASD sample, we consider our human findings to be preliminary and in need of replication in a much larger, well-phenotyped cohort. Especially important is the need to test whether low CSF AVP concentration is specific to ASD (versus other neurodevelopmental disorders) and whether CSF AVP concentration predicts ASD symptom severity.
In conclusion, we developed a rhesus monkey model of naturally occurring social impairments to identify markers of primate social functioning using a discovery and replication cohort design. This approach identified CSF AVP concentration as a key marker of group differences in male monkey sociality and as a stable trait-like measure. We next translated these findings to a small cohort of ASD patients and found that CSF AVP concentration was lower in male children with ASD versus male medical control children without ASD and that CSF AVP concentration was sufficient to accurately distinguish ASD cases from controls. Collectively, these data suggest that a low CSF AVP concentration may be an important marker of low sociality in rhesus monkeys and that the AVP signaling pathway warrants consideration in future research endeavors that seek to develop new diagnostic tools and treatments for ASD.
MATERIALS AND METHODS
Study design
The goal of this research was to identify biological markers of sociality in low- and high-social rhesus monkeys. This research used a discovery and replication cohort design in monkeys, with subsequent translation to a small cohort of seven ASD patients and seven medical control children without ASD (but with other medical conditions). Experimenters were blinded to monkey (that is, low-social versus high-social) and patient (that is, ASD versus control) groups during behavioral observations in monkeys and biological quantification [e.g., enzyme immunoassay, quantitative polymerase chain reaction (qPCR), and Western blot procedures] in both species.
Monkey subjects
Subjects and study site
Subjects studied in the discovery and replication cohorts (that is, monkey cohorts 1 and 2, respectively) were n = 206 male rhesus monkeys (Macaca mulatta), ranging in age from 1 to 5 years. CSF samples previously collected and banked from an additional cohort of n = 10 male rhesus monkeys (that is, monkey cohort 3), 5 to 7 years of age, were also used in this research. All monkeys were born and reared at the California National Primate Research Center (CNPRC). Subjects lived in outdoor, half-acre (0.2 ha) field corrals, measuring 30.5 m wide × 61 m deep × 9 m high. Each corral contained up to 221 animals of all ages and both sexes. Subjects were tattooed as infants and dye-marked before behavioral observation for this study to facilitate easy identification. Monkeys had ad libitum access to Lixit-dispensed water, primate laboratory chow was provided twice daily, and fruit and vegetable supplements were provided twice weekly. Various toys, swinging perches, and other enrichments in each cage, along with outdoor and social housing, provided a stimulating environment. All procedures were approved by the relevant Institutional Animal Care and Use Committees and complied with National Institutes of Health policies on the care and use of animals.
Subject selection and behavioral data collection for monkey cohort 1
Subjects participated as infants in the colony-wide BioBehavioral Assessment (BBA) Program at CNPRC. The BBA comprises a set of highly standardized behavioral and physiological assessments focused on quantifying naturally occurring variation in temperament, behavioral responses, and pituitary-adrenal regulation as described elsewhere (40, 41). BBA-enrolled monkeys are each tested between 3 and 4 months of age. To select subjects for cohort 1, we developed an algorithm using an existing behavior data set previously collected from n = 80 adult male monkeys that were BBA “graduates.” We created a factor scale (α = 0.89) and identified animals with z scores of ≤−1.0 (low-social) or ≥+1.0 (high-social). Logistic regression revealed that BBA measures produced 88.9% classification. We applied this statistical model to a new set of BBA graduates (n = 222) and selected n = 42 of these monkeys for study in cohort 1.
Subjects were observed unobtrusively in their home field corrals. Interobserver reliabilities of >85% agreement were established on behavioral categories and age and sex classes of interaction partners before commencing experimental data collection. Each animal was then observed for two 10-min focal samples per day (08:00 to 10:30 and 10:30 to 13:00) for 8 days over a 2-week period (called a biweek). A maximum of eight subjects, residing in one or two corrals, were observed per biweek. Behavior was recorded at 30-s intervals using instantaneous sampling and time-ruled check sheets. Five social behaviors were recorded: nonsocial (subject is not within an arm’s reach of any other animal and is not engaged in play), proximity (subject is within arm’s reach of another animal), contact (subject is touching another animal in a nonaggressive manner), groom (subject is engaged in a dyadic interaction with one animal inspecting the fur of the other animal using its hands or mouth), and play [subject is involved in chasing, wrestling, slapping, shoving, grabbing, or biting accompanied by a play face (wide eyes and open mouth without bared teeth) or a loose, exaggerated posture and gait; the behavior must be deemed nonaggressive to be scored]. Only nonaggressive proximities and contacts were included for those two behaviors. Aggression was not analyzed here because data from a separate CNPRC cohort of n = 78 comparably aged male monkeys had shown that animals of this age engage in aggression rarely (on average 0.29 aggressive events per hour) (42), suggesting that aggression would have minimal impact on our data. The identities and age and sex classes of all social partners were recorded. At the end of a biweek, subjects were rated on 29 behavioral traits using a standardized instrument, with each trait evaluated on a seven-point scale (24).
After completion of behavioral data collection, subjects were rank-ordered on their total frequency of nonsocial behavior (summarized across the 320 focal behavior samples collected per subject). The n = 15 monkeys with the greatest frequency of nonsocial behavior were classified as low-social, and the n = 15 monkeys with the lowest frequency of nonsocial behavior (and therefore the highest frequency of all prosocial behaviors) were classified as high-social. CSF and blood samples were subsequently collected from these n = 30 subjects (see below).
Subject selection and behavioral data collection for monkey cohort 2
Data from cohort 1 demonstrated that the final social classification of the monkey subjects was essential. Thus, for the validation cohort, we adopted more high-throughput behavioral methods, drawing our sample from all available male subjects that were born into, and were living in, the field corrals. Instead of focal sampling, we adopted a scan sampling approach, allowing us to score multiple animals in the same group at the same time. Scan sampling, like the instantaneous sampling used for cohort 1, is a procedure that estimates durations of behavior (scan sampling is for groups, instantaneous sampling is for individuals) (43). Thus, the same five core behaviors were estimated in both cohorts using an appropriate sampling technique to estimate behavioral durations. Before commencing experimental data collection, interobserver reliabilities of >85% agreement were again established on behavioral categories, subject identities, and age and sex classes.
Subjects were observed unobtrusively in their home field corrals. Each observer conducted scan samples for a given corral during two observation periods per day (09:00 to 12:00 and 13:00 to 16:00). In each observation period, scan sampling was conducted at 20-min intervals, at a rate of 18 scans per day, for a total of 5 days per corral. Thus, about n = 90 scans were performed per corral. During each scan, the subjects in each corral were identified, and observers then recorded the occurrence of the following behaviors: nonsocial, proximity, contact, groom, and play as defined above. We also recorded proximity and contact for aggressive episodes to confirm the aforementioned findings generated by an independent CNPRC research team that aggression in this age class was minimal (42). Consistent with these previous findings, we observed no aggressive bouts by our subjects during data collection for this cohort. After completion of data collection, monkeys were rank-ordered on their total frequency of nonsocial behavior (summarized across the 90 scan samples). The n = 15 monkeys with the greatest frequency of nonsocial behavior were classified as low-social, and the n = 15 monkeys with the lowest frequency of nonsocial behavior (and therefore the highest frequency of prosocial behavior) were classified as high-social. To further improve biological precision for cohort 2, we collected two CSF samples from each subject and averaged the CSF AVP concentrations. Averaging the CSF AVP concentration also avoids collinearity between the two samples when statistically predicting social classification (see Statistical analyses below).
Subject selection for monkey cohort 3
Consistent with the 3R’s (replacement, reduction, and refinement) principle that guides ethical animal research practice (44), we used CSF samples that had been previously collected and banked from a separate cohort of n = 10 adult male monkeys (45) to evaluate whether CSF AVP concentration was a stable trait-like measure. Monkey cohort 3’s CSF samples were obtained on four different occasions across a 4-month period. CSF samples were collected and stored using standard CNPRC operating protocols (see below).
Sample collection and processing procedures
Samples were collected in the morning for all subjects (between 09:00 and 11:00 for cohorts 1 and 2; between 08:00 and 10:00 for cohort 3) to minimize any potential circadian effects on biological measurements. Each subject in cohorts 1 and 2 was captured from his home corral (or from his cage, because subjects in cohort 3 had been moved indoors), rapidly immobilized with Telazol (5 to 8 mg/kg), and moved to an indoor procedure room. Supplementary ketamine (5 to 8 mg/kg) was used as needed to facilitate complete immobilization. Collection of both CSF and blood samples was accomplished within 15 min of initial cage entry; only one monkey per day was sampled from the same corral. The latency from cage entry to subject capture (to control for possible variation in stress effects on the biological measures) and collection time (to account for possible circadian effects on the biological measures) were recorded and used as statistical covariates.
Immediately after capture, CSF (2 ml) was drawn from the cisterna magna using standard sterile procedure. CSF samples were immediately aliquoted into 1.5-ml siliconized polypropylene tubes and flash-frozen on dry ice. Next, whole-blood samples (up to 25 ml) were drawn from the femoral vein and collected into (i) EDTA-treated Vacutainer tubes and either placed on wet ice (for neuropeptide quantification) or left at room temperature (for kinase quantification) and (ii) PAXgene tubes and left at room temperature for 2 hours or longer (for neuropeptide receptor gene expression). Whole-blood samples for neuropeptide quantification were promptly centrifuged (1600g at 4°C for 15 min), the plasma fraction aliquoted into 1.5-ml polypropylene tubes and flash-frozen on dry ice. Whole-blood samples for kinase signaling quantification were spun over a Ficoll-h ypaque gradient, and mononuclear cells collected from the interface were washed in phosphate-buffered saline twice, pelleted, and solubilized [in 50 mM tris (pH 7.4), 10 mM EGTA, 0.5% NP-40, and protease and phosphatase inhibitor cocktails]. This mixed fraction was then centrifuged at 15,000 rpm for 30 min at 4°C. The soluble fraction was removed and kept frozen at −80°C until further processing. PAXgene tubes were subsequently transferred to −20°C for 24 hours and then transferred to −80°C as per manufacturer’s guidelines. All samples were stored at −80°C until quantification. After sample collection, each subject was administered replacement fluids and ketoprofen as needed. Subjects were placed in a standard laboratory cage located in a hospital/transition room for recovery overnight and then returned to their home corrals the next day.
Neuropeptide quantification
CSF and blood OXT and AVP concentrations were quantified at Stanford University using commercially available enzyme immunoassay kits (Enzo Life Sciences) (20, 30, 31, 46–49). These kits have been validated for use in rhesus monkeys, are highly specific, and exclusively recognize OXT and AVP, respectively, and not related peptides (that is, the OXT cross-reactivity with AVP is 0.6% and the minimum assay sensitivity is 11.7 pg/ml; and the AVP cross-reactivity with OXT is <0.001% and the minimum assay sensitivity is 3.39 pg/ml). A trained technician blinded to experimental conditions performed sample preparation and OXT and AVP quantification following established procedures recommended by the technical division of the assay manufacturer. Specifically, the CSF samples were directly assayed (without previous extraction) for OXT and AVP. The plasma samples were extracted for each hormone before assay to preclude known matrix interference effects of large blood-borne proteins in the accurate quantification of the neuropeptides (50), using the following methods.
Plasma samples for use in OXT assays were extracted as follows: Plasma samples (1000 μl per animal) were thawed in an ice bath, acidified with 0.1% trifluoroacetic acid (TFA), and centrifuged (17,000g at 4°C for 15 min). Phenomenex Strata-X columns (Phenomenex Inc.) were activated with 4 ml of high-performance liquid chromatography grade methanol followed by 4 ml of molecular biology grade water. Sample supernatants were applied, drawn through columns by vacuum after column activation, and eluted by sequentially applying 4 ml of wash buffer (89:10:1 water/acetonitrile/TFA) and 4 ml of elution buffer (20:80 water/acetonitrile).
Plasma samples for use in AVP assays were extracted as follows: Equal volumes of 40:60 butanol/diisopropyl ether were added to plasma samples (1000 μl per animal) before centrifugation at 8000g for 5 min at room temperature. The top organic layer was discarded, and the aqueous solution was transferred to a new microcentrifuge tube. A 2:1 volume of ice-cold acetone was then added to all samples before centrifugation at 12,000g for 20 min at 4°C. The supernatant was then transferred to 15-ml Falcon tubes, and a volume of 5:1 ice-cold petroleum ether was added. Samples were briefly vortexed and centrifuged at 3350g for 10 min at 1°C, and the top ether layer was discarded.
Plasma samples for each neuropeptide assay were then evaporated at room temperature using compressed nitrogen. Each evaporated plasma sample was reconstituted in 250 μl of assay buffer before OXT and AVP quantification to provide sufficient sample volume to run each sample in duplicate wells (100 μl per well). Given the sensitivity limitations of the commercial assays, plasma extraction ensured that the plated samples contained high enough quantities of OXT or AVP to be read above the limit of detection. The program used to calculate picogram per milliliter concentrations of OXT or AVP allows for extrapolation based on the sample concentration factor. That is, the program extrapolates the final OXT or AVP concentrations by dividing the results by the fold difference in original sample volume. This method increases the concentration of OXT or AVP in each well and ensures that each sample falls within the linear portion of the standard curve, above the assay’s limit of detection, when it is initially read. All CSF and plasma samples were assayed in duplicate (100 μl per well) with a tunable microplate reader for 96-well format as per manufacturer’s instructions.
Neuropeptide receptor quantification
Measurement of OXTR and AVPRV1a gene expression was conducted at Stanford University using protocols developed for rhesus monkeys. Total RNA was isolated and purified using a PAXgene blood RNA kit from blood stabilized in PAXgene RNA tubes (Qiagen). The first strand complementary DNA (cDNA) synthesis reaction was carried out with iScript Reverse Transcription Supermix (Bio-Rad) with a starting RNA quantity of 1 μg in a 20-μl final volume. qPCR was performed to determine OXTR and AVPRV1a gene expression using RT2 qPCR Primer Assays for Rhesus Macaque OXTR and AVPRV1a (Qiagen), and endogenous control [glyceraldehyde-3-phosphate dehydrogenase (GAPDH), Life Technologies] was used for normalization. qPCR was performed on the StepOnePlus Real-Time PCR System (Life Technologies) with SYBR Green (Qiagen). cDNA was PCR-amplified in triplicate, and cycle threshold (Ct) values from each sample were obtained using StepOnePlus software. Analyses were conducted using the comparative Ct method (2−ΔΔCt) (51).
Kinase signaling quantification
Blood kinase activation was determined at University of California, San Francisco (UCSF) using previously published protocols (16), which were optimized for use in rhesus monkeys. Denatured protein (20 μg per lane) from the previously generated soluble cytoplasmic fraction was electrophoresed through an 8 to 12% polyacrylamide gradient gel for 2 hours at 100 V at room temperature. The gel was electroblotted onto a polyvinylidene difluoride membrane for 1 hour at 50 V at 4°C. After blocking, the blot was incubated with primary antibody [Ras, MAPK kinase 1/2 (MEK1/2), phospho-MEK1/2, Erk1/2, phospho-Erk1/2, Pan AKT and phospho-AKT, PTEN and phospho-PTEN, and GAPDH; Cell Signaling] overnight at 4°C, washed, and followed by fluorescent secondary antibody (anti-rabbit immunoglobulin G, Cell Signaling) incubation for 1 hour at room temperature. The blots were visualized, and bands were quantified using the LiCoR Odyssey Imager.
Statistical analyses
Data were analyzed using JMP Pro 13 (SAS Institute Inc.). We began our analysis of monkey cohort 1 using a set of multidimensional biological measures. Our overall goal was to identify the biological measures most strongly associated with group differences in monkey sociality. As discussed above, because false discovery is a risk in this type of biological study, we adopted a statistical winnowing strategy, whereby at each stage of analysis, we excluded nonpredictive or collinear biological measures from further consideration, via a series of increasingly demanding analyses. Significance for all statistical tests described below was determined to be P < 0.05.
We first used quadratic (unequal covariance) discriminant analysis to test whether the biological measures considered as a whole could predict monkey social group classification. This technique is a form of directed machine learning that seeks to classify monkeys by social group as a linear combination of the predictors (that is, the biological measures). Discriminant analysis answers the general question “Can the biological measures predict social group?” but is agnostic as to which biological measures are drivers, and which may be mediators or moderators, of the social group classification algorithm. CSF and blood concentrations of AVP and OXT, blood AVPRV1a and OXTR gene expression, and ratios of phosphorylated ERK/total ERK, phosphorylated PTEN/total PTEN, and phosphorylated AKT/total AKT in blood were all included as predictors. The resulting confusion matrix (the table of actual versus predicted social group) was then tested as a logistic regression to yield the LR (and associated P value) that the overall algorithm could predict social group given the biological measures. Two animals were excluded because of missing kinase data (that is, n = 28).
To test which biological measures were predictive, we next moved to a logistic regression model. The model containing the full biological measurement panel showed overspecification and quasi-complete separation, indicating collinearity between predictors, and an artifactually overprecise classification prone to false-positive results (28). Through a process of elimination of collinear variables, we identified a final stable model that maximized the number of biological measures included (that is, CSF AVP, CSF OXT, phosphorylated PTEN/total PTEN, and phosphorylated AKT/total AKT) and biologically relevant control variables (or “stratifiers”) (for example, field corral and Western blot). Two animals were excluded because of missing kinase data, and as a result, a third animal had to be excluded so that each field corral yielded at least one animal in each social group (that is, n = 27).
We next tested whether any of the three key biological measures identified in the logistic regression showed group differences. For CSF AVP concentration, we used a general linear model where field corral, capture latency, and sample collection time were included as blocking factors, and social group was the predictor of interest. There was no need to control for assay run because all animals’ samples were run on a single plate. Thirty monkeys yielded AVP data suitable for analysis. For the PTEN and AKT phosphorylation ratios, we used a general linear model, again controlling for field corral, as well as Western blot (to control for between-assay variance) and tested for the effect of social group. Twenty-eight monkeys yielded kinase data suitable for analysis. To test whether CSF AVP concentration predicted time spent in social grooming (a measure recorded during behavioral data collection), we used the same general linear model and blocking factors as described above, with CSF AVP concentration included in the model as the predictor of interest (n = 30). Time spent in social grooming was square root–transformed to meet the assumptions of general linear models (normality of error and linearity and homogeneity of variance). Finally, to confirm the specificity and validity of the statistical winnowing strategy, we tested for group differences in each of the biological measures. We used the same general linear models with the same blocking factors as detailed above for this cohort.
For the replication cohort (that is, monkey cohort 2), we focused on CSF AVP concentration because this was the only biological measure that showed group differences in cohort 1. Thus, we used logistic regression to test whether CSF AVP concentration could predict social group, controlling for capture latency and sample collection time, as well as behavioral observer. We also tested whether social group conversely predicted CSF AVP concentration using a general linear model that similarly controlled for capture latency, sample collection time, and behavioral observer. We used the same analyses as described above. All n = 30 monkeys yielded biological data suitable for analysis. Average values were used for the two sampling time points for CSF AVP concentration, capture latency, and sample collection time for all cohort 2 analyses. As before, appropriate quality-control checks were performed for each analysis.
For monkey cohort 3, we first examined the repeated CSF AVP sample data as a test-retest reliability estimate, for which we used a mixed model intraclass correlation coefficient. Following McGraw and Wong (52), we calculated a case 3A intraclass correlation coefficient (C,1) (that is, an intraclass correlation coefficient of consistency estimated from a Restricted Maximum Likelihood Mixed Model, in which monkey is the subject, and time point is treated as a fixed-effect repeated observation), equivalent to the mean of all possible correlation coefficients between time points. To assess the significance of this result, we also performed a repeated-measures general linear model and tested the significance of the random effect representing the subject. This analysis tested whether monkeys differed significantly from each other in a consistent manner from time point to time point. All n = 10 monkeys yielded biological data suitable for analysis, with n = 8 monkeys yielding data for four time points, and n = 2 monkeys yielding data for three time points.
Human participants
Participants and recruitment
This multisite research study and its associated consent and assent forms were reviewed and approved by the relevant institutional review boards (that is, Stanford University, Sutter Health, and UCSF) before study initiation. Participants were n = 14 male children (n = 7 boys with ASD and n = 7 medical control boys) who were undergoing clinically indicated lumbar punctures and were recruited to participate in this research study. Participants were between 5 and 19 years old.
Participants were recruited from Stanford Hospital and Clinics, the Lucile Packard Children’s Hospital, UCSF Benioff Children’s Hospital, and Sutter Medical Foundation. Clinical indications for CSF collection for the study participants included rule-out diagnoses (for example, clinical assessment to eliminate from consideration the possible presence of a condition or disease) and blood/tissue diseases such as leukemia that required CSF access in diagnosis or treatment. CSF aliquots for this study were either provided as an additional amount to the volume acquired for clinical purposes or reserved at the time of clinical procedure in lieu of disposal. Patients scheduled for these clinical procedures were identified by health care providers as potential study participants. All patients were research-consented before the sample collection (that is, in addition to the medical consent obtained by health care providers for the standard-of-care procedure). Parents or legal guardians of pediatric participants provided written consent. If the child was deemed intellectually capable of understanding the study (that is, those aged 7 years or older), then written assent was also obtained from the child participant.
Inclusion criteria for all participants consisted of a clinically indicated reason for CSF collection, English-speaking, any ethnicity, any gender, and between 6 months and 99 years of age. Children with ASD were required to meet diagnostic criteria for ASD [The Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition (Text Revision) (DSM-IV-TR) or The Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition (DSM-5)] (1, 53), on the basis of clinical evaluation, and be free of other severe or comorbid mental disorders (for example, schizophrenia and bipolar disorder). Medical control children were required to be diagnosed with a medical problem other than ASD. Exclusion criteria for all children included declining to participate in the study or having parents who declined to participate in the study. Children with ASD (all of whom were male) were matched with control children 1:1 on the basis of gender and within a 1-year band on age.
CSF sample collection and processing procedures
CSF was obtained using standard sterile procedures by clinical staff after administration of either local or general anesthetic. CSF was collected from the lumbar region by introduction of a 23-gauge spinal needle into the subarachnoid space at the L3–4 or L4–5 interspace below the conus medullaris. After collection of the clinical CSF samples, CSF samples for research were immediately aliquoted into siliconized polypropylene tubes and flash-frozen on dry ice. All samples were stored at −80°C until quantification.
CSF neuropeptide quantification
CSF AVP concentrations were quantified at Stanford University using the same commercially available enzyme immunoassay kits as used in the rhesus monkey experiments. [AVP is a highly conserved nonapeptide and is structurally identical in rhesus monkeys and humans (54).] Sample preparation and AVP quantification were performed following established procedures. As with the monkeys, the human CSF samples were directly assayed (without previous extraction) and run in duplicate as per the manufacturer’s technical guidelines.
Statistical analyses
Clinical data were managed using REDCap (55) and analyzed using JMP Pro 13 (SAS Institute Inc.). All n = 14 participants yielded biological data suitable for analysis. Patient data were analyzed using a parallel approach to that used in the monkey studies. First, we used logistic regression to test whether CSF AVP concentration could differentiate ASD cases and medical controls. Our first statistical model included CSF AVP concentration as a predictor, as well as standard control variables used in past clinical studies (that is, age, sample collection time, and ethnicity). There was no need to control for assay run because all patients’ samples were run on a single plate. The initial model showed quasi-complete separation (that is, particular combinations of predictors uniquely identified individuals, thereby bearing a high risk for false positives). We thus chose to remove ethnicity from the model because this factor was not significant (and we had no reason to a priori hypothesize that ethnicity would influence CSF AVP concentrations). We did retain age and sample collection time as factors in the model to control for potential developmental changes or circadian variation in CSF AVP concentration. Next, as done for the monkey cohorts, we tested whether patient group (ASD versus medical control) predicted CSF AVP concentration using a general linear model, with the same control factors used in the stable logistic regression model. Significance for all statistical tests was determined to be P < 0.05. The assumptions of general linear models (normality of error and linearity and homogeneity of variance) were confirmed post hoc for all analyses.
Supplementary Material
Fig. S1. Receiver operating characteristic for the discriminant analysis in the monkey discovery cohort.
Table S1. Raw data for fig. S1.
Table S2. SAS code for the data and analysis shown in Fig. 1.
Table S3. SAS code for the data and analysis shown in Fig. 2.
Table S4. SAS code for the data and analysis shown in Fig. 3A.
Table S5. Confirmation of the specificity and validity of the statistical winnowing strategy for the monkey discovery cohort.
Table S6. SAS code for the data and analysis shown in Fig. 4.
Table S7. SAS code for the data and analysis shown in Fig. 3B.
Table S8. SAS code for the data and analysis shown in Fig. 5.
Acknowledgments
We thank S. Seil, K. Bone, and J. Madrid for their help with monkey behavioral observations; J. Yee for monkey biological sample preparation; L. Calonder for help with the BBA testing; C. Parise, D. Carson, K. Hornbeak, S. Hannah, L. Jackson, R. Sumiyoshi, T. Trujillo, J. Moss, B. Benabides, N. Marlen, B. Fregeau, and many clinicians for their help with various aspects of patient identification/recruitment and sample collection/processing; S. Hyde, R. Sumiyoshi, and T. Niethamer for their help with aspects of biological quantification; and the patients and their families for participating.
Funding: The monkey research was supported by grants from the NIH (R21HD079095 and R01HD087048 to K.J.P.; P51OD011107, CNPRC base operating grant, and R24OD010962 to J.P.C.) and the Simons Foundation (274472 and 342873 to K.J.P.). The human research was supported by grants or gifts to K.J.P. from the Mosbacher Family Fund for Autism Research, Stanford University’s Bio-X NeuroVentures Program, the Weston Havens Foundation, Stanford University Child Health Research Institute, the Katherine D. McCormick Fund, and the Yani Calmidis Memorial Fund for Autism Research.
Competing interests: The Board of Trustees of the Leland Stanford Junior University and the Regents of UCSF have filed provisional patent applications related to biological measures studied herein (Stanford University: US 62/634,142 “Methods for diagnosing and for determining severity of an autism spectrum disorder”; UCSF: US 15/545,621 “Methods of diagnosing and treating autism spectrum disorders”). E.H.S. serves on the advisory boards for Invitae Corp and Retrophin Inc. M.G.C. is a paid speaker for Eisai Co. Ltd., Lundbeck Inc., and Sunovian Pharmaceuticals Inc. and has research funding from Eisai Co. Ltd., Greenwich Pharmaceuticals, and Marinus Pharmaceuticals Inc. A.Y.H. has research funding from BioElectron Technology Corp. and either serves as a consultant or is on the advisory board of Roche, Hoffmann Technologies, CheckOrphan, and Realiteer. All other authors declare that they have no competing interests.
Footnotes
Data and materials availability: All data are presented in the paper.
SUPPLEMENTARY MATERIALS
www.sciencetranslationalmedicine.org/cgi/content/full/10/439/eaam9100/DC1
REFERENCES AND NOTES
- 1.American Psychiatric Association, Diagnostic and Statistical Manual of Mental Disorders (American Psychiatric Association, ed. 5, 2013). [Google Scholar]
- 2.Christensen DL, Baio J, Van Naarden Braun K, Bilder D, Charles J, Constantino JN, Daniels J, Durkin MS, Fitzgerald RT, Kurzius-Spencer M, Lee L-C, Pettygrove S, Robinson C, Schulz E, Wells C, Wingate MS, Zahorodny W, Yeargin-Allsopp M, Prevalence and characteristics of autism spectrum disorder among children aged 8 years—Autism and developmental disabilities monitoring network, 11 sites, United States, 2012. MMWR Surveill. Summ 65, 1–23 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buescher AVS, Cidav Z, Knapp M, Mandell DS, Costs of autism spectrum disorders in the United Kingdom and the United States. JAMA Pediatr. 168, 721–728 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Garner JP, Gaskill BN, Weber EM, Ahloy-Dallaire J, Pritchett-Corning KR, Introducing therioepistemology: The study of how knowledge is gained from animal research. Lab Anim. 46, 103–113 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Sclafani V, Del Rosso LA, Seil SK, Calonder LA, Madrid JE, Bone KJ, Sherr EH, Garner JP, Capitanio JP, Parker KJ, Early predictors of impaired social functioning in male rhesus macaques (Macaca mulatta). PLOS ONE 11, e0165401 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Phillips KA, Bales KL, Capitanio JP, Conley A, Czoty PW, ‘t Hart BA, Hopkins WD, Hu SL, Miller LA, Nader MA, Nathanielsz PW, Rogers J, Shively CA, Voytko ML, Why primate models matter. Am. J. Primatol 76, 801–827 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Capitanio JP, Personality dimensions in adult male rhesus macaques: Prediction of behaviors across time and situation. Am. J. Primatol 47, 299–320 (1999). [DOI] [PubMed] [Google Scholar]
- 8.Capitanio JP, Sociability and responses to video playbacks in adult male rhesus monkeys (Macaca mulatta). Primates 43, 169–177 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Constantino JN, Todd RD, Autistic traits in the general population: A twin study. Arch. Gen. Psychiatry 60, 524–530 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Baron-Cohen S, Wheelwright S, Skinner R, Martin J, Clubley E, The autism-spectrum quotient (AQ): Evidence from Asperger syndrome/high-functioning autism, males and females, scientists and mathematicians. J. Autism Dev. Disord 31, 5–17 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Robinson EB, St Pourcain B, Anttila V, Kosmicki JA, Bulik-Sullivan B, Grove J, Maller J, Samocha KE, Sanders SJ, Ripke S, Martin J, Hollegaard MV, Werge T, Hougaard DM; iPSYCH-SSI-Broad Autism Group, Neale BM, Evans DM, Skuse D, Mortensen PB, Børglum AD, Ronald A, Smith GD, Daly MJ, Genetic risk for autism spectrum disorders and neuropsychiatric variation in the general population. Nat. Genet 48, 552–555 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Preacher KJ, Rucker DD, MacCallum RC, Nicewander WA, Use of the extreme groups approach: A critical reexamination and new recommendations. Psychol. Methods 10, 178–192 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Baribeau DA, Anagnostou E, Oxytocin and vasopressin: Linking pituitary neuropeptides and their receptors to social neurocircuits. Front. Neurosci 9, 335 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson ZV, Young LJ, Oxytocin and vasopressin neural networks: Implications for social behavioral diversity and translational neuroscience. Neurosci. Biobehav. Rev 76, 87–98 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ebstein RP, Knafo A, Mankuta D, Chew SH, Lai PS, The contributions of oxytocin and vasopressin pathway genes to human behavior. Horm. Behav 61, 359–379 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Faridar A, Jones-Davis D, Rider E, Li J, Gobius I, Morcom L, Richards LJ, Sen S, Sherr EH, Mapk/Erk activation in an animal model of social deficits shows a possible link to autism. Mol. Autism 5, 57 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu S, Jia M, Ruan Y, Liu J, Guo Y, Shuang M, Gong X, Zhang Y, Yang X, Zhang D, Positive association of the oxytocin receptor gene (OXTR) with autism in the Chinese Han population. Biol. Psychiatry 58, 74–77 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Yirmiya N, Rosenberg C, Levi S, Salomon S, Shulman C, Nemanov L, Dina C, Ebstein RP, Association between the arginine vasopressin 1a receptor (AVPR1a) gene and autism in a family-based study: Mediation by socialization skills. Mol. Psychiatry 11, 488–494 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Modahl C, Green L, Fein D, Morris M, Waterhouse L, Feinstein C, Levin H, Plasma oxytocin levels in autistic children. Biol. Psychiatry 43, 270–277 (1998). [DOI] [PubMed] [Google Scholar]
- 20.Carson DS, Garner JP, Hyde SA, Libove RA, Berquist SW, Hornbeak KB, Jackson LP, Sumiyoshi RD, Howerton CL, Hannah SL, Partap S, Phillips JM, Hardan AY, Parker KJ, Arginine vasopressin is a blood-based biomarker of social functioning in children with autism. PLOS ONE 10, e0132224 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samuels IS, Saitta SC, Landreth GE, MAP’ing CNS development and cognition: An ERKsome process. Neuron 61, 160–167 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subramanian M, Timmerman CK, Schwartz JL, Pham DL, Meffert MK, Characterizing autism spectrum disorders by key biochemical pathways. Front. Neurosci 9, 313 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Francis SM, Sagar A, Levin-Decanini T, Liu W, Carter CS, Jacob S, Oxytocin and vasopressin systems in genetic syndromes and neurodevelopmental disorders. Brain Res. 1580, 199–218 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Capitanio JP, Widaman KF, Confirmatory factor analysis of personality structure in adult male rhesus monkeys (Macaca mulatta). Am. J. Primatol 65, 289–294 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Garner JP, The significance of meaning: Why do over 90% of behavioral neuroscience results fail to translate to humans, and what can we do to fix it? ILAR J. 55, 438–456 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ioannidis JPA, Why most published research findings are false. PLOS Med. 2, e124 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woodward M, Epidemiology: Study Design and Data Analysis (Chapman and Hall/CRC Press, 1999). [Google Scholar]
- 28.Allison PD, Logistic Regression Using the SAS System: Theory and Application (SAS Institute, 1999). [Google Scholar]
- 29.Cohen J, Statistical Power Analysis for the Behavioral Sciences (Lawrence Erlbaum Associates, ed. 2, 1988). [Google Scholar]
- 30.Carson DS, Berquist SW, Trujillo TH, Garner JP, Hannah SL, Hyde SA, Sumiyoshi RD, Jackson LP, Moss JK, Strehlow MC, Cheshier SH, Partap S, Hardan AY, Parker KJ, Cerebrospinal fluid and plasma oxytocin concentrations are positively correlated and negatively predict anxiety in children. Mol. Psychiatry 20, 1085–1090 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Carson DS, Howerton CL, Garner JP, Hyde SA, Clark CL, Hardan AY, Penn AA, Parker KJ, Plasma vasopressin concentrations positively predict cerebrospinal fluid vasopressin concentrations in human neonates. Peptides 61, 12–16 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Fitzner B, Hecker M, Zettl UK, Molecular biomarkers in cerebrospinal fluid of multiple sclerosis patients. Autoimmun. Rev 14, 903–913 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Blennow K, Hampel H, CSF markers for incipient Alzheimer’s disease. Lancet Neurol. 2, 605–613 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Tapiola T, Alafuzoff I, Herukka SK, Parkkinen L, Hartikainen P, Soininen H, Pirttilä T, Cerebrospinal fluid β-amyloid 42 and tau proteins as biomarkers of Alzheimer-type pathologic changes in the brain. Arch. Neurol 66, 382–389 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Donaldson ZR, Spiegel L, Young LJ, Central vasopressin V1a receptor activation is independently necessary for both partner preference formation and expression in socially monogamous male prairie voles. Behav. Neurosci 124, 159–163 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parker KJ, Lee TM, Central vasopressin administration regulates the onset of facultative paternal behavior in Microtus pennsylvanicus (meadow voles). Horm. Behav 39, 285–294 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Winslow JT, Hastings N, Carter CS, Harbaugh CR, Insel TR, A role for central vasopressin in pair bonding in monogamous prairie voles. Nature 365, 545–548 (1993). [DOI] [PubMed] [Google Scholar]
- 38.Insel TR, O’Brien DJ, Leckman JF, Oxytocin, vasopressin, and autism: Is there a connection? Biol. Psychiatry 45, 145–157 (1999). [DOI] [PubMed] [Google Scholar]
- 39.Born J, Lange T, Kern W, McGregor GP, Bickel U, Fehm HL, Sniffing neuropeptides: A transnasal approach to the human brain. Nat. Neurosci 5, 514–516 (2002). [DOI] [PubMed] [Google Scholar]
- 40.Golub MS, Hogrefe CE, Widaman KF, Capitanio JP, Iron deficiency anemia and affective response in rhesus monkey infants. Dev. Psychobiol 51, 47–59 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Capitanio JP, Mason WA, Mendoza SP, Del Rosso LA, Roberts JA, Nursery rearing and biobehavioral organization, in Nursery Rearing of Nonhuman Primates in the 21st Century, Sackett GP, Ruppenthal G, Elias K, Eds. (Springer, 2006), pp. 191–213. [Google Scholar]
- 42.Beisner BA, Jackson ME, Cameron A, McCowan B, Effects of natal male alliances on aggression and power dynamics in rhesus macaques. Am. J. Primatol 73, 790–801 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Altmann J, Observational study of behavior: Sampling methods. Behaviour 49, 227–267 (1974). [DOI] [PubMed] [Google Scholar]
- 44.Russell WMS, Burch RL, The Principles of Humane Experimental Technique (Methuen, 1959). [Google Scholar]
- 45.Capitanio JP, Cole SW, Social instability and immunity in rhesus monkeys: The role of the sympathetic nervous system. Phil. Trans. R. Soc. B 370, 20140104 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parker KJ, Hoffman CL, Hyde SA, Cummings CS, Maestripieri D, Effects of age on cerebrospinal fluid oxytocin levels in free-ranging adult female and infant rhesus macaques. Behav. Neurosci 124, 428–433 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clark CL, St John N, Pasca AM, Hyde SA, Hornbeak K, Abramova M, Feldman H, Parker KJ, Penn AA, Neonatal CSF oxytocin levels are associated with parent report of infant soothability and sociability. Psychoneuroendocrinology 38, 1208–1212 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuen KW, Garner JP, Carson DS, Keller J, Lembke A, Hyde SA, Kenna HA, Tennakoon L, Schatzberg AF, Parker KJ, Plasma oxytocin concentrations are lower in depressed vs. healthy control women and are independent of cortisol. J. Psychiatr. Res 51, 30–36 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parker KJ, Garner JP, Libove RA, Hyde SA, Hornbeak KB, Carson DS, Liao C-P, Phillips JM, Hallmayer JF, Hardan AY, Plasma oxytocin concentrations and OXTR gene variants predict social impairments in children with and without autism spectrum disorder. Proc. Natl. Acad. Sci. U.S.A 111, 12258–12263 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McCullough ME, Churchland PS, Mendez AJ, Problems with measuring peripheral oxytocin: Can the data on oxytocin and human behavior be trusted? Neurosci. Biobehav. Rev 37, 1485–1492 (2013). [DOI] [PubMed] [Google Scholar]
- 51.Livak KJ, Schmittgen TD, Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- 52.McGraw KO, Wong SP, Forming inferences about some intraclass correlation coefficients. Psychol. Methods 1, 30–46 (1996). [Google Scholar]
- 53.American Psychiatric Association, Diagnostic and Statistical Manual of Mental Disorders (American Psychiatric Association, ed. 4, Text Revision, 2000). [Google Scholar]
- 54.Wallis M, Molecular evolution of the neurohypophysial hormone precursors in mammals: Comparative genomics reveals novel mammalian oxytocin and vasopressin analogues. Gen. Comp. Endocrinol 179, 313–318 (2012). [DOI] [PubMed] [Google Scholar]
- 55.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG, Research electronic data capture (REDCap)—A metadata-driven methodology and workflow process for providing translational research informatics support. J. Biomed. Inform 42, 377–381 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Receiver operating characteristic for the discriminant analysis in the monkey discovery cohort.
Table S1. Raw data for fig. S1.
Table S2. SAS code for the data and analysis shown in Fig. 1.
Table S3. SAS code for the data and analysis shown in Fig. 2.
Table S4. SAS code for the data and analysis shown in Fig. 3A.
Table S5. Confirmation of the specificity and validity of the statistical winnowing strategy for the monkey discovery cohort.
Table S6. SAS code for the data and analysis shown in Fig. 4.
Table S7. SAS code for the data and analysis shown in Fig. 3B.
Table S8. SAS code for the data and analysis shown in Fig. 5.