Abstract
In response to strong selection, new mutations can arise quickly and sweep through populations, particularly, if survival and reproduction depend on certain allele copies for adaptation to rapidly changing environments, like resistance against deadly diseases or strong toxins. Since the 1950s, resistance to anticoagulant rodenticides in several rodents has emerged through single nucleotide mutations in the vitamin-K-epoxid-reductase-complex-subunit-1 (VKORC1) gene, often located in its exon 3. Detection of high prevalence and concentrations of anticoagulant rodenticides in non-target vertebrates, including carnivorous Mustelidae, let us assume that secondary exposure by feeding on poisoned prey may also cause selection along the food chain and we hypothesized that VKORC1-based resistance might also have evolved in rodents’ predators. Using newly-developed mustelid-specific primers for direct sequencing of genomic DNA, we studied VKORC1-DNA-polymorphisms in 115 mustelids of five species (Martes martes, M. foina, Mustela nivalis, M. erminea, M. putorius), obtained from northern Denmark, yielding six sites with nonsynonymous and several synonymous amino acid polymorphisms in exon 3. Comparison of these VKORC1-genotypes with hepatic rodenticide residues (obtained by HPLC combined with fluorescence or mass spectrometry) in 83 individuals (except M. martes), using generalized linear models, suggested that anticoagulant levels depended on species and specific polymorphisms. Although most VKORC-1 polymorphisms may present standing genetic variation, some are situated in resistance-mediating membrane parts of the VKORC1-encoded protein, and might be a result of selection due to exposure to anticoagulant poisons. Our new molecular markers might allow detecting indirect effects of anticoagulant rodenticides on rodent predator populations in the future.
Introduction
Under common selection scenarios of complex environments, evolutionary changes may gradually accumulate by subtle polygenic adaptation [e.g. 1,2]. However, in response to strong selection, new mutations can arise quickly and sweep through populations [1,2,3]. This is especially expected if survival and reproduction depend on the possession of certain allele copies, like for adaptation to rapidly changing environments [4], such as resistance against deadly diseases [5] or strong toxins [6], as for example, the application of strong rodenticides. While gene variants arising after such deadly impacts may rarely stem from de novo mutations, the majority of these polymorphisms is thought to rather arise from standing genetic variation due to allele frequency changes and may be accompanied by hard or soft selective sweeps [1,4,7].
Shortly after isolation of 4-hydroxycoumarins from spoiled sweet clover hay (Melilotus officinalis), identification and synthesizing (e.g. dicoumarol [8,9]), coumarin derivatives such as warfarin have been developed to lower blood coagulation and clogging for both therapeutic applications [10,11] and rodent control [12,13]. Metabolized coumarin derivatives affect blood coagulation in vertebrates by inhibition of the vitamin K epoxide reductase complex subunit 1 (VKORC1) [14,15,16], a small transmembrane protein of the endoplasmic reticulum of most eukaryotes [17,18]. It recycles vitamin K 2,3-epoxide to vitamin K hydroquinone, which is essential for the posttranslational γ-carboxylation of many blood coagulation factors [17]. In humans, VKORC1 gene consists of three exons with 6126 base pairs and codes for the VKORC1 protein, comprising 163 amino acids.
Since the late 1950s, resistance against warfarin-containing (later on so-called 1st generation ARs) rodenticides has emerged in several Rattus norvegicus and Mus musculus populations in Europe [19, 20, 21] but remained mechanistically unexplained. In 2004, when examining human disorders (e.g., VKCFD2), mutations in the membrane-imbedded part of the VKORC1 protein, were demonstrated to induce coumarin resistance by these haplotypes [14,18, 22]. Since about that time, missense mutations in exon 3 of VKORC1, have been shown to encode warfarin resistance in several Rattus species [23,24,25,26], in Mus musculus [27,28] as well as in Mastomys mice [29] (S1 Table). In rodents, the best-characterized mutations are the amino acid (AA) replacements Tyr139Cys, Tyr139Phe and Leu120Glu, known to mediate resistance to all first-generation and to some of the second-generation anticoagulant rodenticides (i.e. bromadiolone and difenacoum) [30].
The adaptive alleles in R. norvegicus have been shown to segregate in the ancestral environment [31] and formed the genetic basis for rapid resistance evolution against warfarin with several natural allelic VKORC1 variants conferring resistance. These alleles occurred in different populations of brown rats throughout Europe as part of their standing genetic variation [7,26,31,32]. Wild resistant rodents have been shown to bear higher residues in their livers, supposedly rather based on the prolonged survival time than on accumulation of anticoagulant rodenticides [33–34].
Resistance caused the application of the more toxic and persistent second-generation rodenticides (i.e. brodifacoum, flocoumafen and difethialon), which involve higher exposure risks for non-target mammals and birds [35,36,37]. Since decades, high anticoagulant rodenticide (AR) residues (used here and throughout the paper in the sense of “traces” or “concentrations”) have been detected in predators of rodents, particularly carnivores of the Mustelidae family (e.g., UK [38,39]; France [40]; USA [41]). In our focal research area, Denmark, rats, a potential prey of mustelids, are declared a notifiable species and systematically regulated by professional pest controllers, involving intense use of anticoagulants [42]. Furthermore, until 2012 anticoagulants were used to control voles in forests, and until 2014, private people could use AR s to kill mice in Denmark. Anticoagulant rodenticide resistance, facilitated by Tyr139Cys, has been frequently detected in R. norvegicus [42,43]. In rodents, resistance has not been reported from the anticoagulants brodifacoum and flocoumafen, used in Denmark, but are known for coumatetralyl, bromadiolone and difenacoum [27].
Residues have been detected in 97% of 61 stoats (Mustela erminea), 95% of 69 weasels (Mustela nivalis), 94% of 69 polecats (Mustela putorius) and 98% in 71 stone martens (Martes foina) [44,45]. In all four species, animals with hepatic concentrations of bromadiolone higher than 1000 ng/g ww (wet weight) were recorded, particularly in Martes foina. A subset of these animals was also used in our study (see Materials and Methods). Mustelids are highly susceptible to ARs [45,46, 47]. High concentrations might be due to recent ingestion of a high dosage of the rodenticides. However, since regulations command baits to be placed inaccessibly for non-target species, rodenticide residues in mustelids most likely originate from feeding on poisoned prey, even in food generalists as the genus Martes [44,45]. Therefore, the question arose whether resistance, similar to that evolved in their rodent prey may also be acquired through poison transfer along the food chain in the mustelid predators.
Thus, in the present study, we examine the prevalence of VKORC-1 non-synonymous substitutions in mustelids that are exposed to rodenticides. As previously shown (see Discussion), these mustelid species largely experience secondary pesticide exposure, most probably via predator-prey-interactions in food webs. To understand this, we have i) developed molecular markers with focus on exon 3 of VKORC1 and ii) identified polymorphisms in five free-living mustelid species. Based on great evolutionary conservation of VKORC1 among mammals, we assumed variation to occur potentially also as recessive heterozygotes and thus standing genetic variation that may counterbalance potentially negative mutational effects as documented in rodents. The co-measured residues from individual mustelids allowed us iii) examining whether VKORC1 polymorphisms may show relationships to AR-loads. We aimed at evaluating the potential of VKORC1 polymorphisms as indicators for ongoing or future evolution of resistance to coumarine-derivative ARs in mustelids.
Materials and methods
Samples
We have studied five mustelid species based on initially 120 samples, comprising 20 Martes martes, 30 Martes foina, 20 Mustela nivalis, 20 Mustela erminea and 30 Mustela putorius. The mustelid carcasses were collected between 2000 and 2013 in Denmark (55°–57° N, 8°–10° E; Fig 1). Date, cause of death (mostly road kills, predation and culling), location and geographic coordinates (S2 Table) were recorded for each individual. Liver or muscle samples of all animals were stored frozen. For the DNA analyses subsamples were stored in 96% ethanol at room temperature. Due to insufficient DNA quality, VKORC1-genotypes could be only obtained from 115 samples (Results).
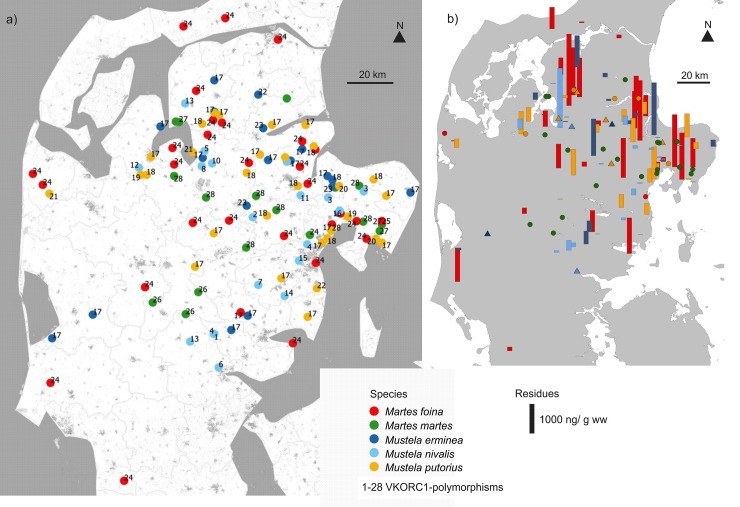
Fig 1.
a) Sampling sites and VKORC1-polymorphisms for the five mustelid carnivores in Denmark (map space approximately 55°–57° N, 8°–10° E). b) Individual hepatic AR residue amounts (sum of the residues of coumatetralyl, bromadiolone and difenacoum), indicated by bar heights; triangles indicate sampling sites for mustelids with no detectable AR residues (0 ng/g wet weight) in their liver; dots in (panel b) indicate sampling sites for individuals, which were not included in the AR-measurements [44,45].
Residues analyses
To examine potential relationships with VKORC1 genotypes, we included published hepatic AR-data of the same individual mustelids (S3 Table). In two previous studies [44,45], the concentration of five regionally relevant ARs (i.e. coumatetralyl, bromadiolone, difenacoum, brodifacoum and flocoumafen) was analyzed in 92 individuals from four species (except Martes martes). Martes martes was not included in these former studies and thus, residue data were unavailable. In Mustela nivalis and M. erminea, residues in liver tissue (sample size mean wet weight ± standard error: 0.59 g ± 0.28 g and 0.99 g ± 0.29 g) were determined by high pressure liquid chromatography (HPLC) coupled with a fluorescence and photodiode array detector [48]. For details on extraction, clean-up and quality assurance procedures, see [44]. For our study, AR-concentrations in Mustela erminea and M. nivalis were corrected for the average recovery rates per substance (coumatetralyl 51%, bromadiolone 81%, difenacoum 75%, brodifacoum 78% and flocoumafen 78%; [44]). Recovery rates were assessed from spiked control samples of chicken liver with known rodenticide concentrations.
In Mustela putorius and Martes foina, rodenticide residue levels were also determined in liver tissues (sample size mean wet weight ± standard error: 0.52 g ± 0.01 g and 0.53 g ± 0.03 g) by high pressure liquid chromatography (HPLC) coupled to tandem mass spectrometry (MS-MS) [49]. Concentrations were corrected for recovery rates, for each AR substance (coumatetralyl 77%, bromadiolone 36%, difenacoum 59%, brodifacoum 36% and flocoumafen 50%). Recovery rates were assessed from spiked controls of chicken liver with known rodenticide concentrations (details: [45]). The MS-MS method is more sensitive than the older fluorescence-photodiode array detection method [44,45]. To compare and analyse data from both studies, ARs concentrations in Martes foina, Mustela putorius and M. erminea were adjusted to the highest AR-limits as used to measure AR in Mustela nivalis (i.e. values in Martes foina, Mustela putorius and M. erminea that were below the detection limits in M. nivalis were set formally to 0 ng/g wet weight; S3 Table).
Genetic analyses
Subsamples of liver or muscle were taken and DNA extracted using Puregene Core Kit A (Qiagen) at the Julius Kuehn Institute. Subsequent molecular work (i.e. primer development, PCRs and cloning) was performed at the Leibniz-Institute of Freshwater Ecology and Inland Fisheries (IGB). Primers were developed by aligning DNA sequences of VKORC1, available from GenBank, for mice and rats and few available haplotypes of carnivores, namely panda, dog, cat and that of the only complete mustelid genome Mustela putorius, available on Ensemble. Two primer pairs were developed de novo (Table 1): DogEx3F and DogEx3R are both exon-based and amplify almost the entire exon 3 (ca. 200 bp) of VKORC1. Primers Must1F/Must1R stretch into the flanking intron 2 and amplify about 500 bp.
Table 1. Sequences of the newly developed mustelid-specific primers targeting VKORC1-exon 3 (MustF/R) and a flanking intron (DogEx3F/R).
| Primer name | Sequence (5' = >3') | PCR product size |
|---|---|---|
| Must1F | GRCCCGCTGGGCATCTAT | ca. 500 bp |
| Must1R | AGGGTCCCTCRCAGACAGA | |
| DogEx3F | CCGCTGGGCATCTATCCT | ca. 200 bp |
| DogEx3R | CAGTGCCCCTTGACCTTG |
PCR reactions comprised volumes of 25 μl with the following ingredients: 9.875 μl of ddH2O, 2.5 μl of TopTaq buffer including 1.5 mM MgCl2 supplemented with 1 μl of MgCl2 solution, 2.5 μl of solution Q, 2.5 μl of dNTP (2.5 mM each), 1.25 μl of each of two primers (in most cases MustF1/MustR1), 0.125 μl of TopTaq polymerase (all ingredients: Qiagen) and usually 4 ul of DNA (of 20–60 ng/μl; roughly quantified using Nanodrop). In case of apparent DNA degradation, we increased the DNA volume to up to 6 ul and reduced the water. Amplifications were performed on Mastercycler® ep gradient (Eppendorf) and comprised 5 min 95°C (denaturation), followed by 40 cycles of [1 min 95°C (denaturation), 1 min 58.1°C (annealing), 1 min 72°C (elongation)], and 5 min 72°C (final elongation). PCR products were visualized on 1.5% agarose gels in 1xTAE buffer electrophorese, running at 80 V for 40 min, using ethidium bromide and the GelDocTM XR+ system, and then directly Sanger-sequenced with the forward primers.
Due to low DNA quality, presumably caused by degradation in road kills, in several cases, bands of weak PCR-products were entirely cut from Low Melting Plaque Agarose (Biozyme) preparative gels and melted at 55°C in 1.5 μl Eppendorf tubes on a thermo-block. Four microliters of liquidized PCR-product + agarose then served as template in a 2nd PCR, otherwise identical to the 1st but comprising only 30 cycles.
PCR-products for all unique haplotypes inferred from direct sequencing with primers Must1F/Must1R, were frozen at -20°C, immediately after PCR, subsequently cloned using the TOPO®TA Cloning®Kit and pCRTMII-TOPO® vector (Invitrogen), following the manufacture’s protocols, and then re-amplified by colony-PCR using the universal vector-based primers M13F/M13R. At least twelve successfully amplified clones per sample were then Sanger-sequenced using the nested vector-based primers T7/SP6, aligned to each-other, edited by eye for very rare singletons (explicable by PCR-errors) and reduced to the two most frequently occurring allelic VKORC1-haplotypes, as expected for diploid organisms (edition made assuming that multiple clones of the same genotype present a true sequence allele).
Sequence evaluation and analyses
Chromatograms of Sanger sequences of mustelids were visualized using the programs Geneious (Biomatters) and/or Sequencher v.4.9, aligned to wildtype exon 3 sequences and adjacent gene regions of humans, brown rat (Rattus norvegicus), and house mouse (Mus musculus) as available from www.ensembl.org. For direct sequences, heterozygote single nucleotide polymorphisms (SNPs) were primarily detected by clear double peaks at a certain position.
Statistical analyses
Statistical analyses were performed using R v. 3.5.1 [50]. The amount of ARs hereafter called the sum of anticoagulant residues, was calculated as the sum of coumatetralyl, bromadiolone and difenacoum residues and analyzed using generalized linear models (GLM) with gamma family and log link for strictly positive continuous data. We excluded nine samples with zero amount of AR from the statistical analysis since our focus was to detect patterns in genotypes that point towards resistance to anticoagulant rodenticides. If an animal was not exposed to ARs the information on the genotype is statistically of minor importance. First, a global model was fitted with the nonsynonymous polymorphisms and species as explanatory variables. We could not test for multiplicative effects due to the incomplete factorial design (i.e. many combinations of the nonsynonymous polymorphisms factor levels were not found). A multi-model inference approach [51] was used to fit several candidate models with different sets of explanatory variables. Species was always included as explanatory variable to account for different detection and correction methods as described above. Candidate models were compared using the Bayesian Information Criterion (BIC) and the package MuMIn [52]. The Akaike weight (wi), the second order Akaike information criterion, corrected for small sample sizes (AICc), and the Pseudo R2 are also reported. Variables’ importance was calculated as the sum of Akaike weight (w+(j)) across all models [51]. It is determined for each predictor variable j by summing the Akaike weights wi across those models in the set, where the predictor variable j occurs. The models with the lowest BIC and those models within dBIC < 2, relative to the best model, were used for interpretation. Using the package emmeans [53], estimated marginal means, 95% confidence intervals and posthoc tests for pairwise comparisons at an alpha level of 0.05 with p-value adjustment by the Tukey method were performed. Model diagnostics was performed by plotting deviance residuals against fitted values and explanatory variables using the package ggfortify [54]. Residuals of models were not spatially autocorrelated (Moran’s I test, with p-values of the interpreted models > 0.81).
Ethics statements
None of the animals analyzed in the study were killed for research purposes. They were accidentally killed by cars, domestic cats or legally culled according to the hunting regulations of the Danish Environmental Agency. The carcasses were collected by Aarhus University for research purposes (BEK no. 330, 19/03/2013 and earlier statutory orders) via the general public and private taxidermists [44,45].
Results
VKORC1 polymorphisms
In total, we have obtained sequence information for VKORC1 from 115 tissue samples of the five mustelid species (S4 Table; S1 Data; in five mustelids, low DNA quality prevented successful amplification). Cloning was applied to a small subset of PCR products and confirmed sequences inferred from direct sequencing. Polymorphisms were detected in nine AA-positions, nonsynonymous polymorphisms in the six AA-positions: 123, 125, 127, 134, 146, 154 and synonymous polymorphisms in the same and further three positions: 137, 149 and 155 (S4 Table; S1 Data). In all five species, isoleucine and phenylalanine were most the frequent amino acids in positions 123 and 125, respectively (Table 2; Fig 2). Furthermore, at these positions we found homozygote states with two copies for tyrosine and heterozygote genotypes, where leucine co-occurred with another amino acid, respectively. In position 127, isoleucine was the most frequent amino acid in all Mustela but also occurred in Martes species. Additionally, valine was found in homo- and heterozygote genotypes. In AA-position 134, all examined mustelid species exhibited valine and in position 154 argenine, except for Mustela nivalis. In Mustela nivalis, however, we found individuals with homo- and heterozygote allele combinations involving isoleucine in position 134 as well as with homo- and heterozygote genotypes involving tryptophan. Three different amino acids (valine, leucine, methionine) occurred under homo- or heterozygote states in the AA-position 146. Mustela nivalis was characterized by the highest AA-variability in position 146, only homozygote leucine has not been found.
Table 2. Non-synonymous amino acid polymorphisms in exon 3 detected in the five mustelid species.
Four polymorphisms (#17, 18, 22, 24) were found in more than one species. For synonymous polymorphisms see S5 Table.
| Number of | 123 | 125 | 127 | 134 | 146 | 154 | Polymorphism number | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Species | individuals | Ile | Thr | Phe | Phe/Leu | Val | Val/Ile | Ile | Val | Val/Ile | Ile | Val | Val/Leu | Leu | Val/Met | Met | Trp | Trp/Arg | Arg | |
| M. nivalis | 1 | x | x | x | x | x | x | 1 | ||||||||||||
| 1 | x | x | x | x | x | x | 2 | |||||||||||||
| 2 | x | x | x | x | x | x | 3 | |||||||||||||
| 2 | x | x | x | x | x | x | 4 | |||||||||||||
| 1 | x | x | x | x | x | x | 5 | |||||||||||||
| 1 | x | x | x | x | x | x | 6 | |||||||||||||
| 1 | x | x | x | x | x | x | 7 | |||||||||||||
| 1 | x | x | x | x | x | x | 8 | |||||||||||||
| 1 | x | x | x | x | x | x | 9 | |||||||||||||
| 1 | x | x | x | x | x | x | 10 | |||||||||||||
| 1 | x | x | x | x | x | x | 11 | |||||||||||||
| 1 | x | x | x | x | x | x | 12 | |||||||||||||
| 2 | x | x | x | x | x | x | 13 | |||||||||||||
| 1 | x | x | x | x | x | x | 14 | |||||||||||||
| 1 | x | x | x | x | x | x | 15 | |||||||||||||
| 1 | x | x | x | x | x | x | 16 | |||||||||||||
| sum | 19 | 19 | 14 | 5 | 2 | 2 | 15 | 7 | 9 | 3 | 7 | 2 | 7 | 3 | 3 | 9 | 7 | |||
| M. putorius | 15 | x | x | x | x | x | x | 17 | ||||||||||||
| 8 | x | x | x | x | x | x | 18 | |||||||||||||
| 2 | x | x | x | x | x | x | 19 | |||||||||||||
| 2 | x | x | x | x | x | x | 20 | |||||||||||||
| 2 | x | x | x | x | x | x | 21 | |||||||||||||
| 1 | x | x | x | x | x | x | 22 | |||||||||||||
| sum | 30 | 25 | 5 | 30 | 2 | 4 | 24 | 30 | 2 | 13 | 15 | 30 | ||||||||
| M. erminea | 15 | x | x | x | x | x | x | 17 | ||||||||||||
| 1 | x | x | x | x | x | x | 18 | |||||||||||||
| 1 | x | x | x | x | x | x | 22 | |||||||||||||
| 3 | x | x | x | x | x | x | 23 | |||||||||||||
| sum | 20 | 16 | 4 | 20 | 20 | 20 | 2 | 18 | 20 | |||||||||||
| M. foina | 2 | x | x | x | x | x | x | 18 | ||||||||||||
| 27 | x | x | x | x | x | x | 24 | |||||||||||||
| 1 | x | x | x | x | x | x | 25 | |||||||||||||
| sum | 30 | 30 | 30 | 28 | 2 | 30 | 27 | 3 | 30 | |||||||||||
| M. martes | 2 | x | x | x | x | x | x | 24 | ||||||||||||
| 3 | x | x | x | x | x | x | 26 | |||||||||||||
| 3 | x | x | x | x | x | x | 27 | |||||||||||||
| 8 | x | x | x | x | x | x | 28 | |||||||||||||
| sum | 16 | 16 | 16 | 8 | 8 | 16 | 2 | 3 | 11 | 16 | ||||||||||
| 115 | 106 | 9 | 110 | 5 | 40 | 6 | 69 | 103 | 9 | 3 | 38 | 20 | 33 | 10 | 14 | 3 | 9 | 103 | ||
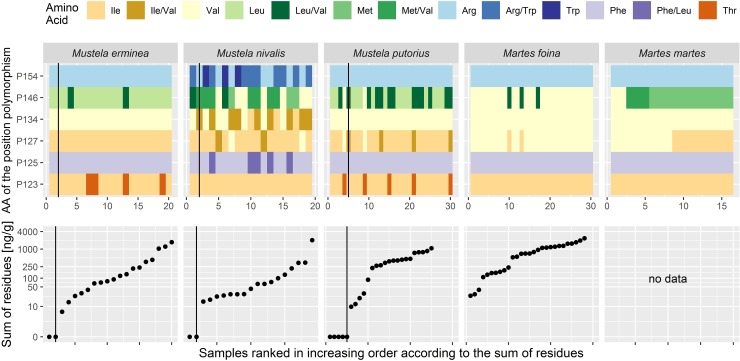
Fig 2. Amino acids at six selected polymorphic sites in VKORC1 (exon 3), detected in five mustelid species.
Individual Mustela erminea, M. nivalis, M. putorius and Martes foina are arranged in increasing order according to the sum of anticoagulant rodenticides (AR)-residues, which are displayed below. The vertical black lines separate individuals with zero sum from those with detected residues. No AR-data were available for Martes martes; amino acids are labelled according to the official three letter code.
Overall, the highest variability of amino acid polymorphisms was detected in Mustela nivalis, comprising 16 out of the 28 polymorphisms as found in all mustelid samples (Fig 2). Four polymorphisms occurred in more than one species, three other polymorphisms (Table 2: last column #17, 18, 22, 24) in two species and one (18) in the three species Mustela putorius, Mustela erminea and Martes foina.
Residues of anticoagulant rodenticides
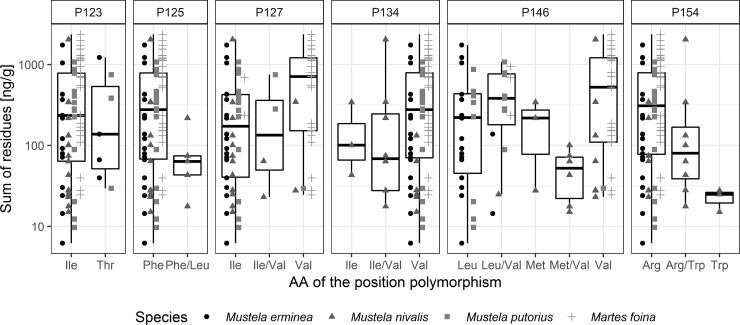
VKORC1-polymorphisms could be analysed in 115 samples (the missing five samples had to be excluded due insufficient DNA quality), for 92 samples AR residues were available, of which 83 showed detectable values (Fig 2). Their amounts varied between the four species and among the six inferred nonsynonymous polymorphisms (Fig 3). The nonsynonymous polymorphisms that best explained the sum of anticoagulant residues were (in decreasing order based on the sum of Akaike weight): P134 (w+(P134) = 0.61), P125 (w+(P125) = 0.55), P154 (w+(P154) = 0.30), P123 (w+(P123) = 0.09), P127 (w+(P127) = 0.02) and P146 (w+(P146) = 0.01). Comparisons yielded two models with considerable statistical support by a dBIC < 2 (Table 3). The best model indicated a higher sum of residues in Phe vs. Phe/Leu in P125 and a higher sum of residues in Ile/Val vs. Val in P134. Ile did not differ from Ile/Val and Val in P134. In the samples examined here, the lowest sum of residues was found in Mustela nivalis, i.e. lower than in M. erminea, M. putorius and Martes foina (Table 4). The second-best model indicated higher sum of residues in Arg/Trp vs. Trp in P154, which both did not differ from Arg. The effect of species was similar to the best model.
Fig 3. Sum of the anticoagulant rodenticide (AR-) residues of coumatetralyl, bromadiolone and difenacoum [ng/ g wet weight liver] in individuals vs. six nonsynonymous polymorphisms and four mustelid species (AA: Amino acid).
Table 3. Summary of the best fitting candidate models (dBIC < 2) and the null- and the global model to explain the sum of anticoagulant residues by non-synonymous VKORC1-mutations and species in 83 AR positive mustelids.
| Models | Explanatory variables | BIC | dBIC | Akaike weight | AICc | R2 | df |
|---|---|---|---|---|---|---|---|
| Top 1 candidate model | P125 + P134 + Species | 1188.6 | 0.0 | 0.35 | 1171.2 | 0.31 | 8 |
| Top 2 candidate model | P154 + Species | 1190.3 | 1.7 | 0.15 | 1174.8 | 0.26 | 7 |
| Candidate model with species only | Species | 1191.1 | 2.5 | 0.10 | 1179.8 | 0.17 | 5 |
| Null model | 1 | 1193.1 | 4.5 | 0.02 | 1188.4 | 0.00 | 2 |
| Global model | P123+ P125 + P127+ P134 + P146 + P154 + Species | 1221.2 | 32.6 | 0.00 | 1189.5 | 0.37 | 17 |
BIC = Bayesian information criterion, dBIC = delta BIC to the best model, AICc = second order Akaike information criterion corrected for small sample size, R2 = pseudo-R2 of the model, df = degrees of freedom.
Table 4. Estimated marginal mean (EMM), lower (LCI) and upper confidence interval (UCI) of the sum of anticoagulant residues for each explanatory variable in the models.
Results from posthoc test: different letters indicate significant differences for pairwise comparisons at alpha 0.05 with p-value adjustment of the Tukey method.
| Explanatory variables | EMM | LCI | UCI | Posthoc test | |
|---|---|---|---|---|---|
| Top 1 candidate model | |||||
| P125 | |||||
| Phe | 1052 | 523 | 2119 | a | |
| Phe_Leu | 172 | 58 | 510 | b | |
| P134 | |||||
| Ile | 532 | 136 | 2078 | ab | |
| Ile_Val | 1334 | 504 | 3533 | a | |
| Val | 108 | 55 | 215 | b | |
| Species | |||||
| Mustela erminea | 523 | 210 | 1299 | a | |
| Martes foina | 1277 | 538 | 3030 | b | |
| Mustela nivalis | 76 | 42 | 139 | c | |
| Mustela putorius | 642 | 262 | 1576 | ab | |
| Top 2 candidate model | |||||
| P154 | |||||
| Arg | 341 | 250 | 465 | ab | |
| Arg_Trp | 939 | 323 | 2734 | a | |
| Trp | 61 | 14 | 269 | b | |
| Species | |||||
| Mustela erminea | 260 | 101 | 676 | a | |
| Martes foina | 637 | 260 | 1564 | b | |
| Mustela nivalis | 99 | 55 | 179 | a | |
| Mustela putorius | 320 | 125 | 818 | ab | |
Discussion
Potential of VKORC1 polymorphisms as indicator for ongoing or substrate for future rapid evolution of resistance to anticoagulant rodenticides
Although the genetic analysis is restricted to a single key gene, to our knowledge, this study is the first to co-examine the occurrence of VKORC1 polymorphisms and anticoagulant residues in carnivores, focusing on the standing genetic variation and its potential for rapid evolution of resistance. In total, across all five mustelid species, we have found synonymous polymorphisms in nine AA-positions and nonsynonymous ones in six positions. Our results suggest that some mutations may provide resistance given the large number of nonsynonymous amino acid replacements in a functional part of this gene. Others may comprise presumably recessive heterozygotes that may nevertheless counterbalance potentially negative mutational effects [55]. Despite relationships to AR load, possibly indicating some resistance to AR, some of this variation might represent standing (and thus possibly neutral) genetic variation.
Under ARs, mustelids might be more susceptible to become traffic victims and thus, measured AR-residues cannot differentiate between recent (high) and (low) long-term exposure to ARs. Survival also depends on ingested AR dosage and compound, some of which can persist several months in survivors’ livers (t½ >200 d) [56]. Thus, AR residues in mustelid tissues may have accumulated over many months or result from recent preying on heavily poisoned rodents.
Our statistical analyses revealed that differences in anticoagulant residues in mustelids (as measured in liver tissues) may depend on the possession of specific polymorphisms. For example, polymorphisms in positions 134, 125 and 154 were associated with the sum of anticoagulant residues. Given the exploratory nature of the data and the occurrence vs. absence of certain nonsynonymous polymorphisms in some species (Fig 2), our statistical results must be interpreted with caution. In addition, due to the incomplete factorial design (not all possible amino acid polymorphisms co-occurred in all positions), epistatic effects could not be assessed (see also Material and Methods). As stated, we excluded few (nine) samples with zero AR from the statistical analysis as we aimed at detecting genotypes that point towards resistance. Without detectable AR-exposure, genotype information is less meaningful, as we do not know whether animals may have survived previous exposure. While such case-only studies may be susceptible to bias, a meta-analysis [57] has shown it to be negligible compared to case-control studies and case-only designs may prove of considerable value. In our study, a case-control design would require more animals with different genetic background and data on exposure to ARs as well as survival, which is not feasible in wild animals.
The structure of the protein encoded by VKORC1 in the endoplasmic reticulum of mammalian cells [17,18] contains two important amino acid motifs, encoded by exon 3, that have been biochemically well characterized: i) the CIVC-motif, with the amino acids cysteine, isoleucine, valine and cysteine, and ii) the TYA-motif, with the amino acids threonine, tyrosine and alanine. The CIVC motif (amino acid positions 132–135), plays an important role in reducing vitamin K epoxide to vitamin K [58], which is then catalyzed into its active form, vitamin K hydroquinone [59]. The TYA-motif (positions 138–140) was identified as a binding site for warfarin/coumarin [60], where mutations lead to a resistance [61]. However, even in mammals including humans, the 3D-structure of the VKORC1 is still under discussion [18]. Therefore, it remains partly speculative whether the VKORC1-polymorphisms found in exon 3 of the mustelids may indeed facilitate resistance since we have currently no mustelid-specific proof for their functional effects. However, several of the mutations clearly sit in the protein within the membrane of the endoplasmic reticulum and in close vicinity of known resistance-inducing mutations. In brown rats and house mice, both homo- and heterozygote replacements of Tyr by Cys in position 139 are responsible for resistance to the substances warfarin, coumatetraly, chlorophacinon, bromadiolone and difenacoum [30,61].
Especially in Mustela nivalis, the SNPs in position 134 (valine/isoleucine) resides in the CIVC-motif, and there, mutations may result in lowering its function as a catalyst. At position 146 (valine/leucin or valine/methionine) another SNP was found within the membrane in all mustelids species. A SNP detected exclusively in Mustela nivalis at amino acid position 154 (arginine/tryptophan) seems to be located in the C-terminus of the protein and thus in the cytoplasm, i.e. at the site of action of the anticoagulants, which may have partial functional relevance. Blood clotting tests, as described for rats [62,63] would be one way of testing the functional relevance of VKORC1 polymorphisms in the future.
Known patterns of anticoagulant selection, susceptibility and resistance in mammals
In resistant rats, which exhibit increased vitamin K food requirements and reduced litter sizes as consequences of their resistance [64,65], there are indications for selection against such resistant individuals, when exposure to the toxin (bromadiolone) is removed. Under bromadiolone regime, however, their increased tolerance suggested that continuous anticoagulant selection would increase the proportion of highly resistant rats in the population [66]. In addition, resistance that allows survival of higher dosages of anticoagulants, can nevertheless be associated with intoxication effects. In non-target species, secondary exposure to ARs may lead to death from hemorrhaging. Despite the observed presence of VKORC1 polymorphisms that might indicate increased AR tolerance, there is an inverse relationship between AR-concentrations and general body condition in Mustela erminea and Mustela nivalis in the examined populations [41]. Persistent sublethal exposure in humans and rats include specific pathologies such as arterial calcification, severe skin irritation and both immune activation and suppression [67 and refs. therein). In conclusion, mutations in VKORC1 may be sometimes under balancing selection, favoring wild-type and resistance [55].
Secondary exposure of non-target species of rodenticides can occur by feeding on poisoned baits or along the food chain, i.e. by feeding on poisoned prey. Several studies provided experimental evidence that ARs can cause the death of mustelides by secondary exposure (Mustela nivalis, Warfarin: [46]; Mustela erminea, Bromadiolone: [47,68]) and have suggested bromadiolone to cause mustelid decline from abundances in field, however, without analyzing AR residues. Rodent control with brodifacoum in New Zealand resulted in the death of Mustela erminea and M. putorius (liver AR: 1000–2000 ng/g) [69]. In our study, brodifacoum levels in individuals were lower than 500 ng/g, and it is hard to know whether these present sublethal dosages. It seems likely that the exposure of the five examined predators in Denmark to these rodenticides is a result of secondary exposure through feeding on poisoned prey. For Mustela erminea, Mustela nivalis and Mustela putorius, this seems very probable, due to their high amount of rodent prey. The martens (Martes martes, Martes foina) though appear to have a broader food spectrum and may thus also occasionally feed on poisoned baits. Nevertheless, in Denmark, rodenticides baits should not be applied where martens and other non-target species have access; therefore, in practice, direct consumption appears negligible as a source of the detected anticoagulants [44,45].
Conclusions
Across all five mustelid species, we found nonsynonymous (= amino-acid-change-causing) exon-3-polymorphisms in six positions and several synonymous polymorphisms in all species. Statistical analyses suggested tolerance to AR-exposure depends on species and certain amino acid polymorphisms. While several polymorphisms may present standing (and thus potentially neutral) genetic variation, some are situated in membrane parts of the VKORC1-encoded protein and might have the potential for rapid evolution of resistance in the population if the advantage of AR resistance outweighs the costs of the resistance—as seen in rats. The new molecular markers may allow quantifying indirect effects of ARs on rodent predators.
Our results, however, can only be a first indication for the role of polymorphisms, anticoagulant residues and potential tolerance, but we are far from fully understanding the potential costs of these VKORC1 polymorphisms on individual mustelids’ fitness and on the species’ conservation status. Future research is required to fully understand the genomic, toxicological and conservation consequences of secondary AR-exposure.
Supporting information
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(DOCX)
(FA)
Acknowledgments
We thank several taxidermists and naturalists for collecting mustelid carcasses in Denmark. Rossana Bossi, Karin R. Petersen, Jeanette Rasmussen, Charlotte Dahl Schiødt and Ellen Christiansen are acknowledged for their assistance with rodenticide analyses. We thank Valeska Gajewski for DNA-extractions, Mareike Brehm for help in the laboratory, and Michael T. Monaghan and Hans-Joachim Pelz for comments on previous versions of the work.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by Leibniz-Institute of Freshwater Ecology and Inland Fisheries to MS, and Julius Kühn Institute to AE. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Pritchard JK, Pickrell JK, Coop G. The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Curr Biol 2010;20: R208–R215. 10.1016/j.cub.2009.11.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stephan W. Signatures of positive selection: from selective sweeps at individual loci to subtle allele frequency changes in polygenic adaptation. Mol Ecol 2016;25: 79–88. 10.1111/mec.13288 [DOI] [PubMed] [Google Scholar]
- 3.Messer PW, Ellner SP, Hairston NG. Can population genetics adapt to rapid evolution? Trends Genet 2016;32: 408–418. 10.1016/j.tig.2016.04.005 [DOI] [PubMed] [Google Scholar]
- 4.Hermisson J, Pennings PS. Soft sweeps: Molecular population genetics of adaptation from standing genetic variation. Genetics 2005;169: 2335–2352. 10.1534/genetics.104.036947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karlsson EK, Kwiatkowski DP, Sabeti PC. Natural selection and infectious desease in human populations. Nat Rev Genet 2014;15: 379–393. 10.1038/nrg3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reid NM, Proestou DA, Clark BW, Warren WC, Colbourne JK, Shaw JR, et al. The genomic landscape of rapid repeated evolutionary adaptation to toxic pollution in wild fish. Science 2016;354: 1305–1308. 10.1126/science.aah4993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barrett RDH, Schluter D. Adaptation from standing genetic variation. Trends Ecol Evol 2007;23: 38–44. 10.1016/j.tree.2007.09.008 [DOI] [PubMed] [Google Scholar]
- 8.Stahmann A, Huebner C, Link KP. Studies on the hemorrhagic sweet clover disease. V. Identification and synthesis of the hemorrhagic agent. Biol Chem194;138: 513–527. [Google Scholar]
- 9.Link KP. The anticoagulant from spoiled sweet clover hay. Harvey Lect 1942;39: 162–216. [Google Scholar]
- 10.Wright IS, Marple CD, Fahs Beck D. Report of the committee for elevation of anticoagulants in the treatment of coronary thrombosis with myocardial infarction. Am Heart J 1948;36: 801–815. 10.1016/0002-8703(48)90278-6 [DOI] [PubMed] [Google Scholar]
- 11.Link K. The discovery of dicumarol and its sequels. Circulation 1959;19: 97–107. 10.1161/01.cir.19.1.97 [DOI] [PubMed] [Google Scholar]
- 12.O'Connor JA. The use of blood anti-coagulants for rodent control. Research 1948;1: 334–336. [PubMed] [Google Scholar]
- 13.Seidman M, Robertson DM, Link KP. Studies on 4-Hydroxycoumarins. X. Acylation of 3-(a-Phenyl-p-acetylethyl)-4-hydroxycoumarin. J Am Chem Soc 1950;72: 5193–5195. [Google Scholar]
- 14.Rost S, Fregin A, Ivaskevicius V, Conzelmann E, Hörtnagel K, Pelz H-J, Lappegard K, et al. Mutations in VKORC1 cause warfarin resistance and multiple coagulation factor deficiency type 2. Nature 2004;427: 537–541. 10.1038/nature02214 [DOI] [PubMed] [Google Scholar]
- 15.Watanabe KP, Saengtienchai A, Tanaka KD, Ikenaka Y, Ishizuka M. Comparison of warfarin sensitivity between rat and bird species. Comp Biochem Physiol C 2010;152: 114–119. [DOI] [PubMed] [Google Scholar]
- 16.Huang AC, Elliott JE, Hindmarch S, Lee SL, Maisonneuve F, Bowes V, Cheng KM, et al. Increased rodenticide exposure rate and risk of toxicosis in barn owls (Tyto alba) from southwestern Canada and linkage with demographic but not genetic factors. Ecotoxicol 2016;25: 1061–1071. [DOI] [PubMed] [Google Scholar]
- 17.Tie JK, Nicchitta C, Von Heijne G, Stafford DW. Membrane topology mapping of vitamin K epoxide reductase by in vitro translation/cotranslocation. J Biol Chem 280, 16410–16416 (2005). 10.1074/jbc.M500765200 [DOI] [PubMed] [Google Scholar]
- 18.Li T, Chang CY, Jin DY, Lin PJ, Khvorova A, Stafford DW. Identification of the gene for vitamin K epoxide reductase. Nature 2004;427: 541–544. 10.1038/nature02254 [DOI] [PubMed] [Google Scholar]
- 19.Boyle CM. Case of apparent resistance of Rattus norvegicus Berkenhout to anticoagulant poisons. Nature 2960;188: 517. [Google Scholar]
- 20.Rowe F, Redfern R. Toxicity tests on suspected warfarin resistant house mice (Mus musculus L.). J Hyg 1965;63: 417–425. 10.1017/s0022172400045307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greaves J. The present status of resistance to anticoagulants. Acta Zool Fenn 1985;173: 159–162. [Google Scholar]
- 22.McClain MR, Palomaki GE, Piper M, Haddow JE. A Rapid-ACCE review of CYP2C9 and VKORC1 alleles testing to inform warfarin dosing in adults at elevated risk for thrombotic events to avoid serious bleeding. Genet Med 2008;10: 89–98. 10.1097/GIM.0b013e31815bf924 [DOI] [PubMed] [Google Scholar]
- 23.Wang J, Feng Z, Yao D, Sui J, Zhong W, Li M, Dai J. Warfarin resistance in Rattus losea in Guangdong Province, China. Pest Biochem Physiol 2008;91: 90–95. [Google Scholar]
- 24.Huang BH, Feng ZY, Yue LF, Yao DD, Gao ZX, Wang DW, Liu XH. Warfarin resistance test and polymorphism screening in the VKORC1 gene in Rattus flavipectus. J Pest Sci 2011;84: 87–92. [Google Scholar]
- 25.Tanaka KD, Kawai YK, Ikenaka Y, Harunari T, Tanikawa T, Ando S, et al. The genetic mechanisms of warfarin resistance in Rattus rattus found in the wild in Japan. Pest Biochem Physiol 2012;103: 144–151. [Google Scholar]
- 26.Iacucci A, Colangelo P, Gamberi V, Mori E, Capizzi D, Baert K, et al. VKORC1 mutation in European populations of Rattus norvegicus with first data for Italy and the report of a new amino acid substitution. Hystrix It J Mamm 2018;29: 95–99. [Google Scholar]
- 27.Pelz H-J, Rost S, Müller E, Esther A, Ulrich RG, Müller CR. Distribution and frequency of VKORC1 sequence variants conferring resistance to anticoagulants in Mus musculus. Pest Manag Sci 2012;68: 254–259 10.1002/ps.2254 [DOI] [PubMed] [Google Scholar]
- 28.Šćepović T, Jokić G, Esther A, Kataranovski D, Vukša P, Vukša M. VKOR variant and sex are the main influencing factors on bromadiolone tolerance of the house mouse (Mus musculus L.). Pest Manag Sci 2016;72: 574–579. 10.1002/ps.4027 [DOI] [PubMed] [Google Scholar]
- 29.Gryseels S, Leirs H, Makundi R, Goüy de Bellocq J. Polymorphism in VKORC1 gene of natal multimammate mice, Mastomys natalensis, in Tanzania. J Hered 2015;106: 637–643. 10.1093/jhered/esv054 [DOI] [PubMed] [Google Scholar]
- 30.RRAC (2016) RRAC guidelines on anticoagulant rodenticide resistance management.30 pp; available (6 May 2019): http://about.rrac.info/fileadmin/downloads/RRAC_Guidelines_Resistance.pdf
- 31.Pelz H-J, Rost S, nerberg M, Fregin A, Heiberg A-C, Baert K, et al. The genetic basis of resistance to anticoagulants in rodents. Genetics 2005;170: 1839–1847. 10.1534/genetics.104.040360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berny P, Esther A, Jacob J, Prescott C. Risk mitigation measures for anticoagulant rodenticides as biological products. Final report. Luxembourg: Publications Office of the European Union; 2014. [Google Scholar]
- 33.Berny P, Fourel I, Lattard V Anticoagulant rodenticides: Resistance and residues in Norway rats in France, pp. 342–346. Proceedings of Vertebrate Pest Conference; 2014. [Google Scholar]
- 34.Desvars-Larrive A, Pascal M, Gasqui P, Cosson J-F, Benoît E, Lattard V, et al. Population genetics, community of parasites,and resistance to rodenticides in an urban brown rat (Rattus norvegicus) population. PLoS One 2017;12:e0184015 10.1371/journal.pone.0184015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Christensen TK, Lassen P, Elmeros M. High exposure rates of anticoagulant rodenticides in predatory bird species in intensively managed landscapes in Denmark. Arch Environ Contam Toxicol 2012;63: 437–444. 10.1007/s00244-012-9771-6 [DOI] [PubMed] [Google Scholar]
- 36.Geduhn A, Jacob J, Schenke D, Keller B, Kleinschmidt S, Esther A. Relation between intensity of biocide practice and residues of anticoagulant rodenticides in red foxes (Vulpes vulpes). PLoS ONE 2015;10: e0139191 10.1371/journal.pone.0139191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Geduhn A, Esther A, Schenke D, Gabriel D, Jacob J. Prey composition modulates exposure risk to anticoagulant rodenticides in a sentinel predator, the barn owl. Sci Total Environ 2016;544: 150–157. 10.1016/j.scitotenv.2015.11.117 [DOI] [PubMed] [Google Scholar]
- 38.McDonald RA, Harris S, Turnbull G, Brown P, Fletcher M. Anticoagulant rodenticides in stoats (Mustela erminea) and weasels (Mustela nivalis) in England. Environ Pollut 1998;103: 17–23. [Google Scholar]
- 39.Sainsbury KA, Shore RF, Schofield H, Croose E, Gloria Pereira M, Sleep D, et al. Long-term increase in secondary exposure to anticoagulant rodenticides in European polecats Mustela putorius in Great Britain. Environ Pollut 2018;236: 689–698. 10.1016/j.envpol.2018.02.004 [DOI] [PubMed] [Google Scholar]
- 40.Fournier-Chambrillon C, Berny PJ, Coiffier O, Barbedienne P, Dassé B, Delas G, et al. Evidence of secondary poisoning of free-ranging riparian mustelides by anticoagulant rodenticides in France: Implications for conservation of European mink (Mustela lutreola). J Wild Dis 2014;40: 688–695. [DOI] [PubMed] [Google Scholar]
- 41.Gabriel MW, Woods LW, Poppenga R, Sweitzer RA, Thompson C, Matthews SM, et al. Anticoagulant rodenticides on our public and community lands: spatial distribution of exposure and poisoning of a rare forest carnivore. PLoS ONE 2012;7: e40163 10.1371/journal.pone.0040163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.By- og Landskabsstyrelse. Plan for fokuseret forebyggelse og bekæmpelse af rotter i Danmark Copenhagen: Ministry of the Environment; 2010. [Google Scholar]
- 43.Lodal J. Resistens hos brune rotter: monitering af resistens hos den brune rotte i Danmark 2008. Copenhagen: Danish Pest Infestation Laboratory & Danish Environmental Protection Agency; 2010. [Google Scholar]
- 44.Elmeros M, Christensen TK, Lassen P. Concentrations of anticoagulant rodenticides in stoats Mustela erminea and weasels Mustela nivalis from Denmark. Sci Total Environ 2011;409: 2373–2378. 10.1016/j.scitotenv.2011.03.006 [DOI] [PubMed] [Google Scholar]
- 45.Elmeros M, Lassen P, Bossi R, Topping CJ. Exposure of stone marten (Martes foina) and polecat (Mustela putorius) to anticoagulant rodenticides: Effects of regulatory restrictions of rodenticide use. Sci Total Environ 2018;612: 1358–1364. 10.1016/j.scitotenv.2017.09.034 [DOI] [PubMed] [Google Scholar]
- 46.Townsend MG, Bunyan PJ, Odam EM, Stanley PI, Wardall HP. Assessment of secondary poisoning hazard of warfarin to least weasel. J Wildl Manage 1984;48: 628–632. [Google Scholar]
- 47.Grolleau G, Lorgue G, Nahas K. Toxicité secondaire, en laboratoire, d'un rodenticide anticoagulant (bromadiolone) pour des prédateurs de rongeurs champêtres: buse variable (Buteo buteo) et hermine (Mustela erminea). EPPO Bulletin 1989;19: 633–648. [Google Scholar]
- 48.Jones A. HPLC determination of anticoagulant rodenticide residues in animal livers. Bull Environ Contam Toxicol 1996;56: 8–15. 10.1007/s001289900002 [DOI] [PubMed] [Google Scholar]
- 49.Albert CA, Wilson LK, Mineau P, Trudeau S, Elliott JE. Anticoagulant rodenticides in three owl species from Western Canada, 1988–2003. Arch Environ Contam Toxicol 2010;58: 451–459. 10.1007/s00244-009-9402-z [DOI] [PubMed] [Google Scholar]
- 50.R Core Team. R: A language and environment for statistical computing R Foundation for Statistical Computing, Vienna, Austria: 2018. Available from: https://www.R-project.org/ [Google Scholar]
- 51.Burnham KP, Anderson DR. Model selection and multimodel inference: a practical information-theoretic approach. New York, Springer, New York; 2002. [Google Scholar]
- 52.Barton K. MuMIn: Multi-model inference. R package version 1.42.1; 2018. [Google Scholar]
- 53.Lenth R. emmeans: Estimated Marginal Means, aka Least-Squares Means. R package version 1.2.3; 2018. [Google Scholar]
- 54.Tang Y, Horikoshi M, Li W. ggfortify: Unified interface to visualize statistical results of popular R packages. The R Journal 2016;8: 474–485. [Google Scholar]
- 55.Hedrick PW. What is the evidence for heterozygote advantage selection? Trends Ecol Evol 2012;27: 698–704. 10.1016/j.tree.2012.08.012 [DOI] [PubMed] [Google Scholar]
- 56.Vandenbroucke V, Bousquet-Melou A, De Backer P, Croubels S. Pharmacokinetics of eight anticoagulant rodenticides in mice after single oral administration. J Vet Pharmacol Ther 2008;31: 437–445. 10.1111/j.1365-2885.2008.00979.x [DOI] [PubMed] [Google Scholar]
- 57.Dennis J, Hawken S, Krewski D, Birkett N, Gheorghe M, Frei J, et al. Bias in the case-only design applied to studies of gene–environment and gene–gene interaction: a systematic review and meta-analysis. Int J Epidemiol, 2011:40:1329–1341. 10.1093/ije/dyr088 [DOI] [PubMed] [Google Scholar]
- 58.Litwack G. Vitamins and Hormones. vol. 78 Vitamin K, London: Academic Press; 2008. [Google Scholar]
- 59.Rost S, Fregin A, Hunerberg M, Bevans CG, Müller CR, Oldenburg J. Site-directed mutagenesis of coumarin-type anticoagulant-sensitive VKORC1: evidence that highly conserved amino acids define structural requirements for enzymatic activity and inhibition by warfarin. J Thromb Haemost 2005;94: 780–786. [DOI] [PubMed] [Google Scholar]
- 60.Myszka DG, Swenson RP. Synthesis of the photoaffinity probe 3-(pazidobenzyl)-4-hydroxycoumarin and identification of the dicoumarol binding site in rat liver NAD(P)H-quinone reductase. J Biol Chem 1991;266: 4789–4797. [PubMed] [Google Scholar]
- 61.Czogalla KJ, Biswas A, Wendeln A-C, Westhofen P, Mü CR, Watzka M, et al. Human VKORC1 mutation causes variable degrees of 4-hydroxycoumarin resistance and affect putative warfarin binding interfaces. BLOOD 2013;122: 2743–2750. 10.1182/blood-2013-05-501692 [DOI] [PubMed] [Google Scholar]
- 62.RRAC [Rodenticide Resistance Action Committee of CropLife International]. A reappraisal of blood clotting response tests for anticoagulant resistance and a proposal for a standardised BCR test methodology. Technical Monograph; 2003. [Google Scholar]
- 63.Prescott CV, Buckle AP, Hussain I, Endepols S. A standardised BCR resistance test for all anticoagulant rodenticides. Int J Pest Manage 2007;53: 265–272. [Google Scholar]
- 64.Markussen MDK, Heiberg A-C, Nielsen R, Leirs H. Vitamin K requirement in Danish anticoagulant-resistant Norway rats (Rattus norvegicus). Pest Manag Sci 2003;59: 913–920. 10.1002/ps.703 [DOI] [PubMed] [Google Scholar]
- 65.Jacob J, Endepols S, Pelz H-P, Kampling E, Cooper TG, Yeung C H, et al. Vitamin K requirement and reproduction in bromadiolone-resistant Norway rats. Pest Manag Sci 2012;68: 378–385s. 10.1002/ps.2273 [DOI] [PubMed] [Google Scholar]
- 66.Heiberg A-C, Leirs H, Siegismund HR. Reproductive success of bromadiolone‐resistant rats in absence of anticoagulant pressure. Pest Manag Sci 2006;62: 862–871. 10.1002/ps.1249 [DOI] [PubMed] [Google Scholar]
- 67.Fraser D, Mouton A, Serieys LEK, Cole S, Carver S, Vandewoude S, et al. Genome-wide expression reveals multiple systemic effects associated with detection of anticoagulant poisons in bobcats (Lynx rufus). Mol Ecol 2018;27: 1170–1187. 10.1111/mec.14531 [DOI] [PubMed] [Google Scholar]
- 68.Fernandez-de-Simon J, Coeurdassier M, Couval G, Fourel I, Giraudoux P. Do bromadiolone treatments to control grassland water voles (Arvicola scherman) affect small mustelid abundance? Pest Manag Sci 2019;75: 900–907. 10.1002/ps.5194 [DOI] [PubMed] [Google Scholar]
- 69.Alterio N. Secondary poisoning of stoats (Mustela erminea), feral ferrets (Mustela furo), and feral house cats (Felis catus) by the anticoagulant poison, brodifacoum. N Zeal J Zool 1996;23: 331–338. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(DOCX)
(FA)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.