Abstract
Purpose
Small cell carcinoma of the prostate (SCCP) is an aggressive disease that can arise de novo or by transdifferentiation from prostate adenocarcinoma. Alterations in anaplastic lymphoma kinase (ALK) gene are involved in neuroblastoma, lung cancer, and other malignancies but its role in SCCP has not been documented. We describe a patient with refractory de novo SCCP with ALK F1174C activating mutation who obtained clinical benefit from treatment with ALK inhibitor.
Experimental Design
Next-generation sequencing (NGS) was used to analyze primary and circulating tumor DNA (ctDNA). Prostate cancer databases were queried for alterations in ALK gene, mRNA and its impact in clinical outcomes. In vitro prostate cell line/organoid models were generated by lentiviral-mediated expression of ALK and ALK F1174C and assessed for response to ALK inhibitors crizotinib and alectinib.
Results
NGS analysis of the primary tumor and ctDNA of a 39-year old patient with refractory SSCP identified ALK F1174C mutation. Treatment with second-generation ALK inhibitor alectinib resulted in radiographic stable disease for over 6 months, symptomatic improvement, and significant molecular response as reflected by declining ctDNA allele fraction. Analysis of prostate cancer datasets showed that ALK amplification was associated with poor outcome. In prostate cancer cells and organoids, ALK F1174C expression enhanced growth and induced expression of the neuroendocrine marker neuron specific enolase. Alectinib was more effective than crizotinib in inhibiting ALK F1174C-expressing cell growth.
Conclusions
These findings implicate ALK activating mutations in SCCP pathogenesis and suggest the therapeutic potential of targeting ALK molecular alterations in some patients with SCCP.
Keywords: ALK F1174, Small cell carcinoma, Neuroendocrine prostate cancer
Introduction
Small cell carcinoma of the prostate (SCCP) accounts for 0.5–2% of prostate cancers and displays an aggressive clinical course (1). While many patients present with de novo disease, 40–50% of SCCP patients have a history of prostate adenocarcinoma and prior androgen-deprivation therapy (ADT) (2). Despite initial response to platinum-based chemotherapy, metastatic SCCP has a poor prognosis with median overall survival of 9–16 months related to molecular disruption of several pathways including amplification of Aurora Kinase A (AURKA) and MYCN genes (3,4). Advances in the understanding of molecular pathogenesis of SCCP have not translated into meaningful clinical benefit to patients. Hence, the important need to identify novel therapeutic targets.
Anaplastic lymphoma kinase (ALK) is a receptor tyrosine kinase involved in the pathogenesis of neuroblastoma, non-small cell lung cancer (NSCLC) as well as other malignancies but its importance in SCCP or adenocarcinoma of the prostate has not been documented (5). Aberrant ALK activation due to point mutations or gene fusions stimulates kinase activity and oncogenic transformation through downstream signaling pathways including RAS/ERK, JAK/STAT, and PI3K/AKT/MTOR. While ALK mutations have been described as oncogenic drivers in neuroblastoma (NB) and anaplastic thyroid cancer, ALK fusion proteins have been associated with NSCLC and non-Hodgkin’s lymphoma (NHL) (6). Specifically, ALK F1174L mutation induces auto phosphorylation and constitutive activation of ALK and is associated with resistance to ALK tyrosine kinase inhibitors (TKIs). Activated full length ALK regulates transcription of MYCN (gene encoding N-Myc), an important mediator of neuroendocrine differentiation (4,7).
While ALK TKIs are used to treat NSCLC positive for ALK rearrangements such as EML4-ALK, their efficacy in tumors harboring ALK point mutations has been described only in neuroblastoma and anaplastic large cell lymphoma (8). The F1174C mutation has been associated with resistance to crizotinib and ceritinib, but maintained sensitivity to alectinib in patients with NSCLC (9). Comprehensive genomic profiling of 114,200 clinical cases identified ALK rearrangements in a variety of tumor types beyond NSCLC and highlighted ALK as a therapeutic target in tumors with small cell and neuronal differentiation (10). We report the first case of a potentially oncogenic ALK point mutation in SCCP associated with molecular response to alectinib as reflected by declining cell-free circulating tumor DNA (ctDNA) allele fraction and stable radiologic disease. Database and functional analyses in vitro support the role of ALK in the pathogenesis of SCCP.
Materials and Methods
Patient Studies
Informed consent was obtained from the patient described in this study.
cBioportal and CCLE data analysis
To determine the spectrum of genomic alterations and expression pattern of ALK in prostate cancer patients, datasets available in cbioportal were used (www.cbioportal.org). Patients were stratified based on the presence ALK alterations and survival data was extracted for available datasets. Mutational frequencies of ALK were compared in COSMIC (www.cancer.sanger.ac.uk/cosmic). The gene expression of ALK was compared among prostate cancer cell lines in Comprehensive cell line encyclopedia cell line (CCLE) database (www.broadinstitute.org/ccle/home) by extracting gene-centric RMA-normalized mRNA expression data. ALK gene expression in prostate cancer PDX data was analysed by extracting mRNA expression data deposited in NCBI Geoprofile database with accession number GSE41193.
Liquid Biopsy NGS
Circulating tumor DNA analysis was performed using a commercially available digital next-generation sequencing (NGS) assay (Guardant360, Guardant Health, Inc. Redwood City, CA). Guardant360 is a 73-gene ctDNA NGS panel from a CLIA-licensed, CAP-accredited laboratory. It provides complete exon sequencing of 19 cancer genes, critical exons of 54 genes and amplifications (18 genes), fusions (6 genes) and indels (23 genes) with high clinical sensitivity rates (85% in stage III/IV solid tumors) and ultra-high specificity as described previously (27).
Tissue Biopsy NGS
NGS of DNA from primary tumor site was performed by FoundationOne panel as described (28)
Cell Culture and reagents
LNCaP, DU145, 22Rv1 and PC-3 cells were obtained from the American Type Culture Collection (ATCC) and grown in RPMI medium (Gibco, Thermofisher Scientific) supplemented with 10% fetal bovine serum, 100 units/mL penicillin G, and 100 μg/mL streptomycin sulfate. Cells were maintained in a humidified incubator at 37°C and 5% CO2 atmosphere. For ALK inhibition, crizotinib and alectinib (Selleck Chemicals) were dissolved in DMSO and used at the indicated concentrations.
Lentiviral Vectors and Preparation
R777-E007 Hs.ALK was a gift from Dominic Esposito (Addgene plasmid # 70291). FM1 lentiviral vector was kindly provided by Dr. Jeffrey Milbrandt, which we modified to express CFP reporter. Human ALK cDNA was amplified from R777-E007, mutagenesis was performed at c.3521T and both WT and ALK mutant were subcloned into FM1 CFP at BstbI restriction site (Genscript). Lentiviruses were prepared as previously described. Briefly, lentiviral titers were calculated infecting HEK293T cell line with lentivirus expressing CFP (FM1-CFP, FM1-ALK WT-CFP and FM1-ALK F1174C-CFP). Titer is expressed as transducing units (TU)/ml calculated from CFP-positive cells (%) measured by flow cytometry and cells were infected with 50 and 10 and 1 Multiplicity of Infection (MOIs) for all lentiviruses. Lentiviral infection was performed with 6 ug/ml polybrene by centrifugation at 1,200 rpm for 3 hrs at room temperature (RT).
Organoid culture
LNCaP cells were plated at 2*105 /well density in 6 well plates in RPMI complete media and allowed to adhere overnight. Cells were then transduced with Control (FM1-CFP), ALK WT or ALK F1174C lentivirus particles. For matrigel basement growth, transduced cells were seeded at 5000 cells/well density in ultra-low attachment 96 well plates (Coring) and cultured in Hepatocyte growth media (Corning) containing primocin (Invitrogen) and matrigel (BD Biosciences) as described previously(29). At day 8 organoids were treated with drugs and at day 15, viability was assessed as per manufacturer’s protocol (Cell Titer Glo-3D viability assay, Promega). Organoids were imaged, and area was measured using ImageJ.
Cell Proliferation
Cell Proliferation was measured using the CellTiter 96 AQueous One Solution Cell Proliferation Assay assay (Promega). 5,000 cells/well were seeded in 96-well and incubated with DMSO control, crizotinib and alectinib and indicated concentrations. Absorbance was measured at 72 h at 490 nm in a microplate reader and cell viability was calculated as follows: cell viability = absorbance of test group / absorbance of control cell group ×100%.
Western Blotting
Cells transduced with ALK expressing lentivirus were harvested after lentivirus transduction for 7 days. Total protein was extracted with RIPA buffer (Invitrogen) and protein concentration was determined by BCA assay. Protein extracts were separated by 4–20 % SDS-PAGE and transferred to PVDF membranes. The membranes were blocked with 3% BSA in Tris-buffered saline with 0.1% Tween 20 (TBST) for 1 h at 37 °C, and then incubated overnight at 4 °C in blocking buffer with primary antibody including ALK (#3333, cell signaling), pALK (#3341, cell signaling), pMAPK (#9101, cell signaling), NSE (MAB324, Millipore) and GAPDH (V18, sc20357, Santa cruz). Following incubation with horseradish peroxidase-conjugated goat anti-rabbit/mouse secondary antibody (Biorad) for 1 h, the membranes were detected using Supersignal West Femto kit (Thermofisher).
Immunohistochemistry
FFPE sections were deparaffinized and stained using hematoxylin and eosin (H&E) or specific antibodies. Sections were incubated in citrate buffer (pH 6) for antigen retrieval, followed by 3% H2O2 to block peroxidase. Biotinylated secondary antibodies (Vector Labs) were used followed by incubation with ABC reagent (Vector Labs, PK-7100), and 3,3’-diaminobenzidine (Sigma, Cat#D4168–50SET). Sections were counterstained with hematoxylin, dehydrated, and mounted.
Statistical Analysis
All experiments were repeated at least three times and conducted in triplicates each time. The results are expressed as mean values of SEM. Groups were compared by one-way or two-way ANOVA followed by appropriate post-tests for multiple comparisons using GraphPad Prism 7.0 software. Differences between groups were considered statistically significant at p < 0.05.
Patient Studies
Informed consent was obtained from the patient described in this study. This research was approved by Quorum IRB for the generation of de-identified data sets for research purposes (Guardant protocol) and Northwestern University IRB (protocol STU00205723).
Results
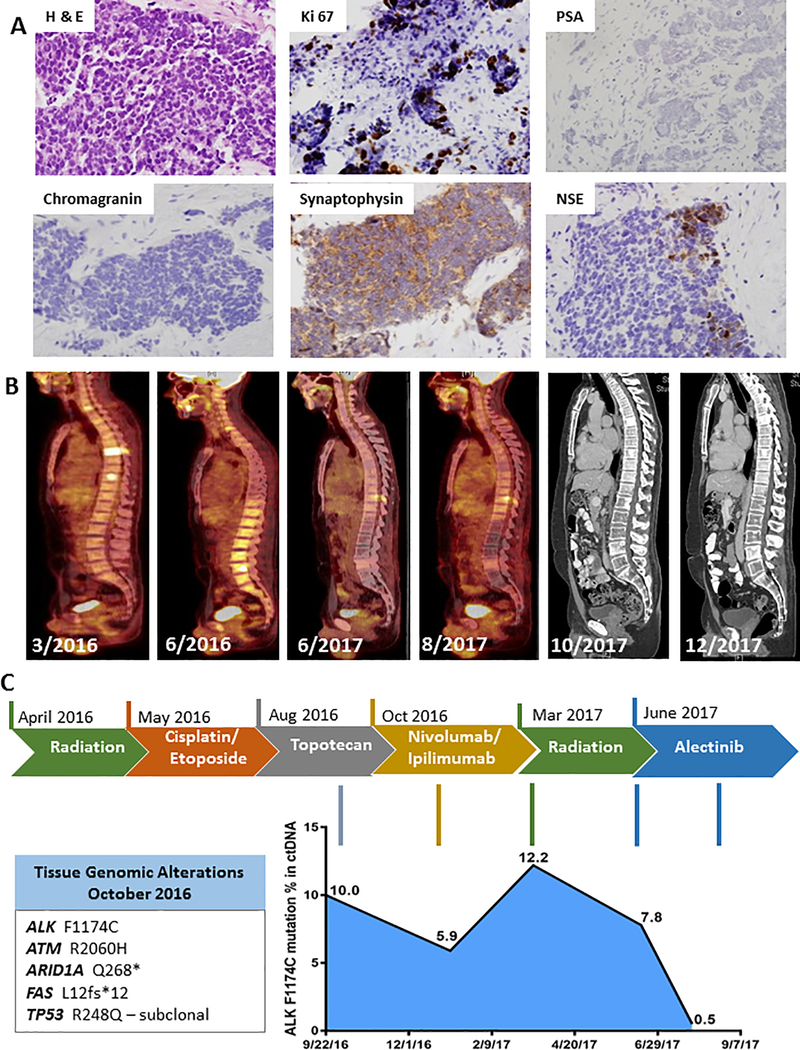
A 39-year-old man with no family history of malignancies presented with urinary urgency, and physical examination revealed nodularity of the prostate. Prostate-specific antigen (PSA) was 1.6 ng/mL. Transrectal ultrasound showed a right prostate mass (4.1 × 1.8 × 1.9 cm), and biopsy revealed small cell carcinoma with immunohistochemistry (IHC) panel positive for AE1/AE3 and synaptophysin, focally positive for neuron specific enolase (NSE), negative for chromogranin, with a Ki-67 index of 30% (Figure 1A). Positron Emission Tomography- Computed Tomograpy (PET-CT) showed hypermetabolic activity of the right prostate tumor and extensive bone metastases without visceral disease. The patient was treated with palliative radiation therapy to the thoracic spine, denosumab, and four cycles of cisplatin and etoposide. Repeat PET-CT showed progression of disease with new bone metastases. He received three cycles of topotecan, but the prostate tumor and bone metastases progressed further. Next, he received nivolumab/ipilumumab with imaging at 12 weeks showing stable disease. After 18 weeks of immunotherapy the patient developed symptoms of radiculopathy caused by progression of an epidural cervical lesion and right brachial plexus involvement. At the same time, a brain magnetic resonance imaging (MRI) brain showed 3 new sub-centimeter metastases. He was treated with gamma knife stereotactic radiosurgery and intensity modulated radiotherapy (IMRT) to the cervical lesion. Shortly thereafter he had progression of bone metastases in the pelvis and lumbar-sacral spine with compression of his cauda equina requiring radiation therapy (Figure 1B).
Figure 1. Clinical, histopathological and molecular features of the patient.
(A). Immunohistochemistry shows characteristics of small cell by H&E and staining of ki67, PSA, synaptophysin, chromogranin and NSE. Brown staining - DAB, counterstain - hematoxylin. (B) PET and CT scans during course of treatment. (C) Patient treatment timeline and corresponding tumor response map of ctDNA NGS findings is shown. Alterations detected in primary tumor tissue NGS are shown in box.
Genomic profiling of prostate biopsy specimen by next-generation sequencing (NGS) revealed the following mutations: ALK F1174C, ATM R2060H, ARID1A Q268*, FAS L12FS*12, TP53 R248Q. Variants of unknown significance were identified on the genes AXIN1, FAT1, PIK3R2, SMARCA4, KDM5A and JAK2. Germline testing revealed an ATM p.R2060H germline variant of unknown significance (germline panel included the following genes ATM, BRCA1, BRCA2, EPCAM, MLH1, MSH2, MSH6, PMS2, CHEK2, HOXB13, NBN, TP53, and SMARCA4).
ctDNA was evaluated by NGS after initial progression on cisplatin and etoposide, revealing the same ALK F1174C mutation detected on primary tumor. This mutation had not been described in prostate cancer or small cell carcinoma despite being reported as the most frequent ALK mutation in neuroblastoma (44.5%) (5). The ALK F1174 variant allele frequency (VAF) in peripheral blood fell from 10.0% prior to immunotherapy to 6.4% after 12 weeks of immunotherapy. It rose to 12.2% at progression of epidural disease and dipped to 7.8% after completing radiation to several areas of disease (Figure 1C). After progression on combination immunotherapy and radiation, the patient was not eligible for available clinical trials. Based on the ALK F1147C mutation and its association with sensitivity to alectinib in NSCLC setting, the patient started treatment with alectinib 150mg daily. After 1 month of therapy the patient’s ALK F1174C VAF decreased to 0.5% and the patient described significant improvement of fatigue, appetite with 10lb weight gain. PET-CT (after 2 months of treatment) and CT scans of chest/abdomen/pelvis after 4 and 6 months of therapy demonstrated overall stable disease (Figure 1B). The patient remained on alectinib at the time of submission of manuscript (7 months of treatment) with continued clinical benefit. The somatic alteration frequencies of ALK F1174C and other mutations detected through serial liquid biopsy are listed in Supplementary Table 1.
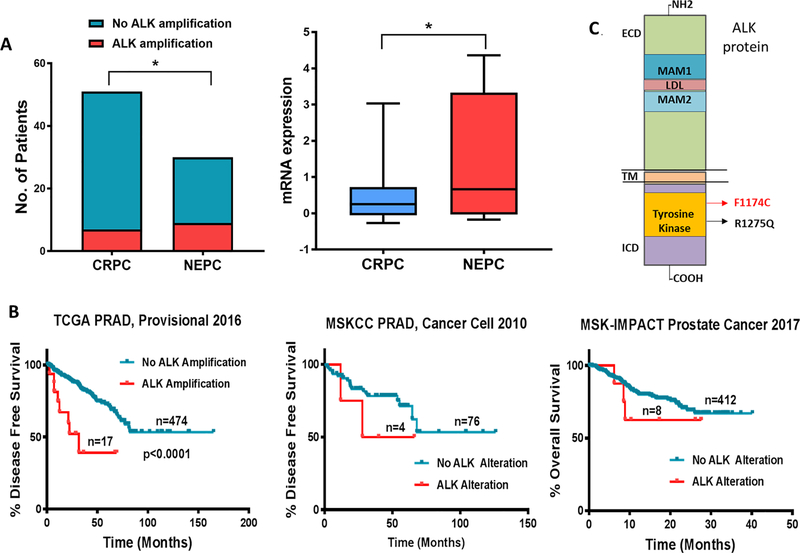
To explore the role of ALK in prostate cancer (PCa), we first analysed several pan-cancer datasets in cBioPortal. The MSK-IMPACT Clinical Sequencing Cohort consists of sequencing and Copy Number Alterations (CNA) data for 10336 patients representing 62 tumor types (11). Analyses of MSK-IMPACT dataset identified ALK alteration in 4% of cases, occurring most frequently in neuroblastoma (also known as embryonal tumour) (12.8% mutations and 3.5% amplifications) followed by small cell lung cancer (12.2% mutations) (Supplementary Figure 1A). Among 717 cases of PCa, ALK was altered in 20% of SCCP cases (1 deep deletion in 5 cases) and 1.1% in prostate adenocarcinoma (1 deep deletion, 1 truncating and 6 missense mutations among 705 cases). Further analysis of missense mutations revealed only one putative driver mutation (R1275Q) mapping to the tyrosine kinase domain of ALK (Figure 2B). No alterations were detected in ALK in samples of castration-resistant PCa with neuroendocrine differentiation (t-NEPC; 7 cases) and mixed prostate carcinoma patients (5 cases) in this dataset. However, analysis of ALK aberrations among PCa cases in cBioportal database that included cases of pure castration-resistant adenocarcinoma prostate cancer (CRPC) or NEPC revealed significantly higher ALK amplification: 27% in NEPC (9 out of 30 cases) as compared to 13% in CRPC (7 out of 51 cases). ALK mRNA expression was also higher in NEPC. Only one missense passenger mutation was identified in NEPC (Figure 2A) (12). The Kaplan–Meier survival curves of TCGA prostate adenocarcinoma (PRAD) dataset (NCI genomic commons) showed that patients with amplification in ALK gene exhibited significantly shorter disease-free survival (DFS) as compared to those with no alterations in ALK (P<0.0001). DFS and overall survival rates followed the same trend in MSKCC PRAD(13) and MSK-IMPACT prostate cancer (11) datasets (Figure 2B). The most frequent ALK mutations occur at residues F1174 and R1275 with a reported 95 and 83 occurrences in the Catalog of Somatic Mutations in Cancer (COSMIC) database respectively. Both mutations map to the tyrosine kinase domain of ALK (Figure 2C) and induce autophosphorylation leading to constitutive activation of ALK (6). Analysis of ALK F1174 mutation in databases from Guardant Health and Foundation Medicine identified the current report to be the only case of small cell carcinoma of the prostate with the ALK F1174 mutation. Among 2,182 prostate cancer cases in Guardant Health database, 54 had non-synonymous ALK mutations; 13 of these, including the current F1174 and a single case of R1275Q described below were in the TKI domain and 39 were outside of the TKI domain. The other 11 were variants of uncertain significance. The patient with the ALK R1275Q mutation had a diagnosis of prostate adenocarcinoma 8 years prior to his ctDNA analysis, but never had a re-biopsy to confirm small cell transformation. His ctDNA results did show two RB1 loss of function mutations which are suggestive of a small cell phenotype (14). Together, these results suggest the potential role of ALK point mutations as prognostic marker in SCCP.
Figure 2. ALK alteration in prostate cancer is associated with poor survival.
(A) ALK gene amplification frequency and comparative mRNA expression in CRPC and NEPC cohorts of Trenton/Broad/Cornell dataset from cBioportal database. (B) Kaplan–Meier survival curves relative ALK alteration from prostate cancer datasets in cBioportal were generated. Total number of patients in the two categories are shown. (C) ALK protein domains and the location of the two identified activating mutations is shown. *P ≤ 0.10 for Fisher’s exact test at 90% C.I. for ALK amplification. *P ≤ 0.05 for t-test of ALK mRNA expression.
We extracted mRNA expression data from the Geoprofile database related to a recent study on patient derived xenografts (PDX) of prostate cancer (15) and identified higher expression of ALK in NEPC (Supplementary Figure 1B). NEPC PDX in the reported study were derived from prolonged exposure to androgen withdrawal leading to CRPC which eventually transdifferentiated into NEPC. A similar association between NEPC and high ALK levels was confirmed by analyzing gene expression data for all the prostate cancer cell lines within the Cancer Cell Line Encyclopedia (CCLE) database where NEPC cell line NCI H660 showed highest expression followed by PC-3 cells which also exhibit features of small cell carcinoma (Supplementary Figure 1D). To assess protein expression of ALK in prostate cancer, we performed immunoblotting of LNCaP (Hormone sensitive PCa), NE1.3 (LNCaP cell derivative isolated by chronic growth in androgen-deprived conditions to model NEPC), and PC3 cells (PCa with small cell-like features). As anticipated, PC-3 cells showed elevated expression of total and phosphorylated ALK (Supplementary Figure 1C).
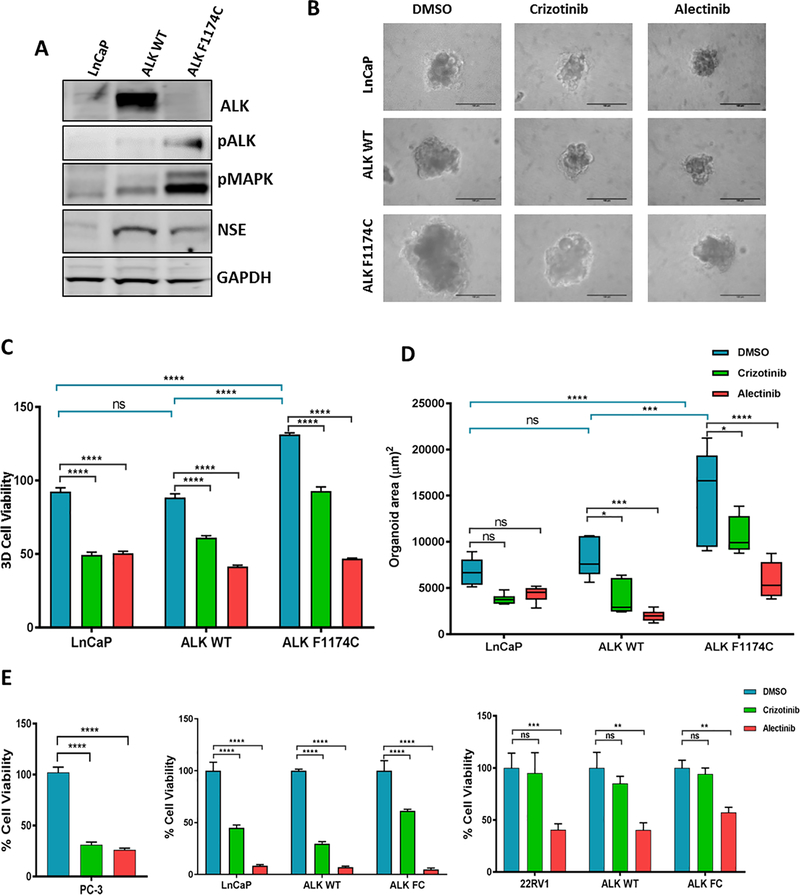
Next, to explore the effects of ALK in prostate tumorigenesis and response to therapy, we transduced LNCaP cells with lentivirus overexpressing ALK Wild Type (ALK WT) or ALK mutant (ALK F1174C). Consistent with previous reports, we observed increased activity of ALK shown by higher levels of phosphorylated ALK and phosphorylated MAP kinase, a key downstream mediator of ALK signaling pathway (6). We also observed an increase in NSE, a neuroendocrine marker in both ALK WT and ALK F1174C infected cells (Figure 3A). These results support the hypothesis that upregulation of ALK contributes to development of de novo and treatment emergent SCCP.
Figure 3. Alectinib suppresses ALK F1174C induced proliferation in prostate cancer cells.
(A) Western blot analysis of ALK, pALK, pMAPK and NSE in LNCaP cell infected with ALK WT and ALK F1174C expressing lentivirus. GAPDH and Tubulin are internal loading controls. Data represent three independent experiments. (B) Representative images showing growth of LNCaP organoids Cells were infected with ALK WT or ALK F1174C and grown in matrigel supplemented media to form organoids for 7 days. At day 8, organoids were treated with 5μm crizotinib or alectinib for 7 days. Scale bars: 100μm. (C) Quantification of percent 3D cell viability assay (D) Quantification of organoid area (μm2). (E) Percent cell viability after 72 h treatment with 10μm crizotinib or alectinib in PC-3, LNCaP and 22Rv1 cells. Data represent mean ± standard error, n=6. ns=non significant, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ***P ≤ 0.001.
In in vitro organoid cultures, ALK F1174C overexpressing LNCaP cells displayed enhanced proliferation with significant increase in organoid area and percent cell viability. A similar trend was also observed with ALK WT overexpressing cells (Figure 3B). We then tested the capability of ALK inhibitors on ALK driven proliferation. Both ALK WT and ALK F1174C were significantly sensitive to treatment with the second-generation inhibitor alectinib in organoid culture. ALK F1174C was less sensitive to crizotinib when compared to ALK WT (Figure 3C, D) consistent with clinical experience in neuroblastoma (9). Western blotting of hormone sensitive LNCaP cells and CRPC 22Rv1 cells treated with crizotinib or alectinib further demonstrated the efficacy of alectinib in inhibiting ALK pathway as seen by decrease in pALK and pMAPK (Supplementary Figures 2A, B). We then tested various PCa cell lines for their sensitivity to ALK inhibitors. While both drugs induced dose dependent cytotoxicity in LNCaP cells, alectinib had lower IC50 in both ALK WT and ALK F1174C cells (Supplementary Figure 2C). PC-3 cells have higher endogenous ALK and respond to both drugs. Hormone sensitive LNCaP cells were the most sensitive to ALK inhibitors. CRPC cell line 22Rv1 was sensitive to alectinib but not crizotinib (Figure 3E). AR negative DU145 cells responded equally to both crizotinib and alectinib, attributed to the expression of c-Met in these cells, an additional target of crizotinib (Supplementary Figure 2D) (16). In summary, these data suggest that ALK expression induces cell growth and alectinib treatment results in favourable response in both ALK WT and ALK F1174C expressing PCa cells.
Discussion
The results describe for the first time the presence of ALK F1174C mutation in a patient with de novo small cell carcinoma of the prostate. They also demonstrate the higher frequency of alterations and elevated expression of ALK in NEPC. These findings led to analysis of a large plasma-based NGS database to identify a likely second case, highlighting the importance of publically searchable large NGS databases to identify novel targetable genomic alterations (17).
In NSCLC, ALK translocation leads to dimerization with fusion proteins and stimulates kinase activity and oncogenic transformation involving downstream signaling pathways including the RAS/ERK, JAK/STAT, and PI3K/AKT/MTOR pathways (5). Recent case reports discussed the unusual transformation of ALK positive NSCLC or lung adenocarcinoma to small cell lung cancer following treatment with alectinib illustrating the complex resistance mechanisms to ALK inhibitors while also highlighting the role of ALK in small cell transformation (18,19). Our data in LNCaP cells suggests that similar mechanisms involving ALK might regulate transdifferentiation to neuroendocrine phenotype in prostate cancer.
ALK mutation F1174L has been shown to induce constitutive activation, regulate MYCN and enhance its oncogenic activity in neuroblastoma (20). Although little is known about the molecular drivers of SCCP, it has been suggested that MYCN drives neuroendocrine differentiation of prostate (21) and that Aurora Kinase A (AURKA) and MYCN amplifications co-occur and co-operate to induce a small cell phenotype in prostate cells (22). In anaplastic large-cell lymphoma, AURKA is more highly expressed in ALK-positive than in ALK-negative tumors (23). These reports support the notion that ALK, AURKA and MYCN form a key signaling axis in SCCP and our findings highlight the need to further explore the role of ALK to delineate the molecular mechanisms underlying SCCP progression.
Several ALK inhibitors have been developed among which, crizotinib, ceritinib, brigatinib, alectinib and lorlatinib are the most extensively studied. Alectinib has emerged as a highly potent and selective ALK inhibitor capable of overcoming resistance to crizotinib treatment in neuroblastoma and NSCLC (24). In a recent clinical trial, alectinib showed superior efficacy and lower toxicity in primary treatment of ALK-fusion positive NSCLC (25). For ALK F1174C in particular, studies have shown that this variant is resistant to crizotinib and ceritinib but sensitive to alectinib (26). The remarkable molecular response we observed in the patient and our results from prostate cancer cell lines support the efficacy of alectinib over crizotinib in ALK wildtype as well as ALK activating mutant cancers. This report also highlights the clinical utility of ctDNA to identify novel therapeutic targets and monitor response to treatment by providing an indirect measure of tumor burden.
In conclusion, we identified for the first time ALK F1174C mutation in a de novo SCCP patient who responded to alectinib with stable radiographic findings after 6 months of therapy, molecular response as reflected by declining circulating tumor DNA (ctDNA) allele fraction and symptomatic improvement. Further studies are needed to refine the role of ALK in the pathogenesis of SCCP and confirm its role as a therapeutic target in this disease.
Supplementary Material
Translational relevance.
Small cell carcinoma of the prostate (SCCP) is an androgen signaling independent lethal subtype of prostate cancer with limited treatment options. The molecular drivers responsible for de novo small cell or treatment emergent neuroendocrine phenotype of prostate cancer are not well understood. We report the first case of an activating ALK F1174C mutation-driven de novo small cell carcinoma of prostate. We observed a remarkable molecular response to second generation ALK inhibitor alectinib and clinical benefit as reflected by declining circulating tumor DNA (ctDNA) allele fraction and stable disease. Database and functional analyses in vitro support the role of ALK in de novo and treatment-emergent neuroendocrine prostate cancer and the therapeutic potential of ALK tyrosine kinase inhibitors.
Acknowledgements
We extend our sincerest gratitude to the patient and his family for their contribution to this study. We thank Dr. Devalingam Mahalingam for helpful comments. We thank the Flow Cytometry Core Facility and the Pathology Core at Northwestern University for their technical help.
Grant Support:
This work was supported by NCI grants 1P50CA180995, R01CA123484 and R01 CA167966 (to S.A.A.
Footnotes
Disclosure of potential conflicts of interest:
Lanman RB has ownership interest (including patents) in Guardant Health, Inc. Ross J, Gay L, Elvin J and Ali S are employees of and shareholders in Foundation Medicine Inc. No potential conflicts of interest were disclosed by the other authors.
References
- 1.Nadal R, Schweizer M, Kryvenko ON, Epstein JI, Eisenberger MA. Small cell carcinoma of the prostate. Nature reviews Urology 2014;11(4):213–9 doi 10.1038/nrurol.2014.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aggarwal R, Zhang T, Small EJ, Armstrong AJ. Neuroendocrine Prostate Cancer: Subtypes, Biology, and Clinical Outcomes. Journal of the National Comprehensive Cancer Network 2014;12(5):719–26. [DOI] [PubMed] [Google Scholar]
- 3.Aparicio AM, Harzstark AL, Corn PG, Wen S, Araujo JC, Tu SM, et al. Platinum-based chemotherapy for variant castrate-resistant prostate cancer. Clin Cancer Res 2013;19(13):3621–30 doi 10.1158/1078-0432.CCR-12-3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beltran H, Rickman DS, Park K, Chae SS, Sboner A, MacDonald TY, et al. Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Discov 2011;1(6):487–95 doi 10.1158/2159-8290.CD-11-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hallberg B, Palmer RH. The role of the ALK receptor in cancer biology. Ann Oncol 2016;27 Suppl 3:iii4–iii15 doi 10.1093/annonc/mdw301. [DOI] [PubMed] [Google Scholar]
- 6.Roskoski R Jr. Anaplastic lymphoma kinase (ALK): structure, oncogenic activation, and pharmacological inhibition. Pharmacol Res 2013;68(1):68–94 doi 10.1016/j.phrs.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 7.Chen Y, Takita J, Choi YL, Kato M, Ohira M, Sanada M, et al. Oncogenic mutations of ALK kinase in neuroblastoma. Nature 2008;455(7215):971–4 doi http://www.nature.com/nature/journal/v455/n7215/suppinfo/nature07399_S1.html. [DOI] [PubMed] [Google Scholar]
- 8.Mosse YP, Lim MS, Voss SD, Wilner K, Ruffner K, Laliberte J, et al. Safety and activity of crizotinib for paediatric patients with refractory solid tumours or anaplastic large-cell lymphoma: a Children’s Oncology Group phase 1 consortium study. Lancet Oncol 2013;14(6):472–80 doi 10.1016/s1470-2045(13)70095-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gainor JF, Dardaei L, Yoda S, Friboulet L, Leshchiner I, Katayama R, et al. Molecular Mechanisms of Resistance to First- and Second-Generation ALK Inhibitors in ALK-Rearranged Lung Cancer. Cancer Discov 2016;6(10):1118–33 doi 10.1158/2159-8290.CD-16-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ross JS, Ali SM, Fasan O, Block J, Pal S, Elvin JA, et al. ALK Fusions in a Wide Variety of Tumor Types Respond to Anti-ALK Targeted Therapy. Oncologist 2017. doi 10.1634/theoncologist.2016-0488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017;23(6):703–13 doi 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med 2016;22(3):298–305 doi 10.1038/nm.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010;18(1):11–22 doi 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan HL, Sood A, Rahimi HA, Wang W, Gupta N, Hicks J, et al. Rb loss is characteristic of prostatic small cell neuroendocrine carcinoma. Clin Cancer Res 2014;20(4):890–903 doi 10.1158/1078-0432.CCR-13-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin D, Wyatt AW, Xue H, Wang Y, Dong X, Haegert A, et al. High fidelity patient-derived xenografts for accelerating prostate cancer discovery and drug development. Cancer Res 2014;74(4):1272–83 doi 10.1158/0008-5472.CAN-13-2921-T. [DOI] [PubMed] [Google Scholar]
- 16.Dai Y, Siemann DW. Constitutively active c-Met kinase in PC-3 cells is autocrine-independent and can be blocked by the Met kinase inhibitor BMS-777607. BMC Cancer 2012;12:198 doi 10.1186/1471-2407-12-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grossman RL, Abel B, Angiuoli S, Barrett JC, Bassett D, Bramlett K, et al. Collaborating to Compete: Blood Profiling Atlas in Cancer (BloodPAC) Consortium. Clin Pharmacol Ther 2017;101(5):589–92 doi 10.1002/cpt.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujita S, Masago K, Katakami N, Yatabe Y. Transformation to SCLC after Treatment with the ALK Inhibitor Alectinib. J Thorac Oncol 2016;11(6):e67–72 doi 10.1016/j.jtho.2015.12.105. [DOI] [PubMed] [Google Scholar]
- 19.Takegawa N, Hayashi H, Iizuka N, Takahama T, Ueda H, Tanaka K, et al. Transformation of ALK rearrangement-positive adenocarcinoma to small-cell lung cancer in association with acquired resistance to alectinib. Ann Oncol 2016;27(5):953–5 doi 10.1093/annonc/mdw032. [DOI] [PubMed] [Google Scholar]
- 20.Berry T, Luther W, Bhatnagar N, Jamin Y, Poon E, Sanda T, et al. The ALK(F1174L) mutation potentiates the oncogenic activity of MYCN in neuroblastoma. Cancer Cell 2012;22(1):117–30 doi 10.1016/j.ccr.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee JK, Phillips JW, Smith BA, Park JW, Stoyanova T, McCaffrey EF, et al. N-Myc Drives Neuroendocrine Prostate Cancer Initiated from Human Prostate Epithelial Cells. Cancer Cell 2016;29(4):536–47 doi 10.1016/j.ccell.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mosquera JM, Beltran H, Park K, MacDonald TY, Robinson BD, Tagawa ST, et al. Concurrent AURKA and MYCN gene amplifications are harbingers of lethal treatment-related neuroendocrine prostate cancer. Neoplasia 2013;15(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanagal-Shamanna R, Lehman NL, O’Donnell JP, Lim MS, Schultz DS, Chitale DA, et al. Differential expression of aurora-A kinase in T-cell lymphomas. Mod Pathol 2013;26(5):640–7 doi 10.1038/modpathol.2012.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gadgeel SM, Gandhi L, Riely GJ, Chiappori AA, West HL, Azada MC, et al. Safety and activity of alectinib against systemic disease and brain metastases in patients with crizotinib-resistant ALK-rearranged non-small-cell lung cancer (AF-002JG): results from the dose-finding portion of a phase 1/2 study. Lancet Oncol 2014;15(10):1119–28 doi 10.1016/S1470-2045(14)70362-6. [DOI] [PubMed] [Google Scholar]
- 25.Peters S, Camidge DR, Shaw AT, Gadgeel S, Ahn JS, Kim DW, et al. Alectinib versus Crizotinib in Untreated ALK-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2017;377(9):829–38 doi 10.1056/NEJMoa1704795. [DOI] [PubMed] [Google Scholar]
- 26.Lin JJ, Riely GJ, Shaw AT. Targeting ALK: Precision Medicine Takes on Drug Resistance. Cancer Discov 2017;7(2):137–55 doi 10.1158/2159-8290.CD-16-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lanman RB, Mortimer SA, Zill OA, Sebisanovic D, Lopez R, Blau S, et al. Analytical and Clinical Validation of a Digital Sequencing Panel for Quantitative, Highly Accurate Evaluation of Cell-Free Circulating Tumor DNA. PLoS One 2015;10(10):e0140712 doi 10.1371/journal.pone.0140712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balko JM, Giltnane JM, Wang K, Schwarz LJ, Young CD, Cook RS, et al. Molecular profiling of the residual disease of triple-negative breast cancers after neoadjuvant chemotherapy identifies actionable therapeutic targets. Cancer Discov 2014;4(2):232–45 doi 10.1158/2159-8290.CD-13-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Unno K, Roh M, Yoo YA, Al-Shraideh Y, Wang L, Nonn L, et al. Modeling African American prostate adenocarcinoma by inducing defined genetic alterations in organoids. Oncotarget 2017;8(31):51264–76 doi 10.18632/oncotarget.17230. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.