Abstract
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is a core enzyme of the aerobic glycolytic pathway with versatile functions and is associated with cancer development. Recently, Kornberg et al. published the detailed correlation between GAPDH and di- or monomethyl fumarate (DMF or MMF), which are well-known GAPDH antagonists in the immune system. As an extension, herein, we report the crystal structure of MMF-bound human GAPDH at 2.29 Å. The MMF molecule is covalently linked to the catalytic Cys152 of human GAPDH, and inhibits the catalytic activity of the residue and dramatically reduces the enzymatic activity of GAPDH. Structural comparisons between NAD+-bound GAPDH and MMF-bound GAPDH revealed that the covalently linked MMF can block the binding of the NAD+ co-substrate due to steric hindrance of the nicotinamide portion of the NAD+ molecule, illuminating the specific mechanism by which MMF inhibits GAPDH. Our data provide insights into GAPDH antagonist development for GAPDH-mediated disease treatment.
Keywords: crystallography, glyceraldehyde-3-phosphate dehydrogenase, inhibitor, monomethyl fumarate
INTRODUCTION
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is a classic glycolytic enzyme that catalyses the conversion of glyceraldehyde-3-phosphate (G3P) to “1,3-bisphospho-D-glycerate” in the glycolytic pathway, using nicotinamide adenosine dinucleotide (NAD+) and an inorganic phosphate as the co-substrate and co-factor, respectively. Recently, researchers have revealed the function of human GAPDH (hGAPDH) in metabolic and cancer-related diseases, and its association with cell death mechanisms, such as apoptosis and autophagy (Colell et al., 2007; 2009; Sirover, 2005; 2011; 2012). For example, overexpression of GAPDH has been demonstrated to cause hepatocarcinogenesis, in which it interacts with colony-stimulating factor-1 (CSF-1), c-Jun, and mammalian target of rapamycin complex-1 (mTORC1) (Araki et al., 2007; Villanueva et al., 2008; Yamamoto et al., 1999; Zhou et al., 2008; Zhu et al., 2008). Thus, hGAPDH is considered as a molecular target for the treatment of various diseases, and several preclinical trials on hGAPDH targeting have been conducted over the years (Colell et al., 2009; Ganapathy-Kanniappan et al., 2012).
Koningic acid (KA, also known as heptelidic acid) is a selective natural hGAPDH antagonist that covalently links with a catalytic cysteine (Endo et al., 1985; Kato et al., 1992; Liberti et al., 2017; Sakai et al., 1991) have demonstrated that KA affects the metabolic network by decreasing the glycolytic rate and cytotoxicity due to the inhibition of hGAPDH. In addition to KA, there are other chemical-based hGAPDH antagonists, such as 3-bromopyruvate, methylglyoxal, and saframycin-A, which are derived from natural and/or synthetic sources (Ganapathy-Kanniappan et al., 2009; Ray et al., 1997; Xing et al., 2004). All of these chemicals including KA have been well-characterized as promising GAPDH inhibitors and are currently used in preclinical trials for cancer treatment (Ganapathy-Kanniappan et al., 2012).
Recent studies have described hGAPDH as a therapeutic target in autoimmunity where it inhibits aerobic glycolysis using dimethyl fumarate (DMF, trade name Tecifera) (Kornberg et al., 2018). DMF is a well-known oral immune-modulator for the treatment of relapsing-remitting multiple sclerosis (RRMS) and psoriasis (Blair, 2018; Bomprezzi, 2015). However, the detailed mechanism of its action is remained for an elucidation. Kornberg et al. (2018) have reported that DMF is an important regulator of immune cell activation. It functions by inhibiting the catalytic activity of hGAPDH and blocking hGAPDH-related aerobic glycolysis. Previous studies have demonstrated a decrease in lactate production and a lower glycolytic capacity in DMF-treated cells, supporting the recent observations of reduced glycolysis in T-cells from multiple sclerosis patients treated with DMF (Kornberg et al., 2018). Furthermore, DMF covalently modifies the catalytic cysteine (Cys152) of hGAPDH via succination. This modification leads to the irreversible inactivation of GAPDH enzymatic activity. The half-life of DMF is approximately 12 min and it is rapidly hydrolysed to monomethyl fumarate (MMF), which has a half-life of 36 h (Mrowietz et al., 1999). Hence, DMF is considered to function as a prodrug to MMF. Although the biological responses for DMF and MMF are different (Ahuja et al., 2016; Landeck et al., 2018), the inhibition-related responses to hGAPDH are similar (Kornberg et al., 2018).
DMF and MMF regulate the anti-inflammatory innate and adaptive immune cell populations (Kornberg et al., 2018). An in vitro study using mouse CD4+ T cells revealed that treatment with either DMF or MMF inhibited the differentiation of naïve CD4+ T cells into T helper 1 or T helper 17 cells through the inactivation of mouse GAPDH. However, the detailed mechanism by which DMF or MMF can inactivate GAPDH, especially hGAPDH, is yet to be determined. Therefore, to attain a better understanding of the mechanism, we determine the crystal structure of MMF-bound human hGAPDH at 2.29 Å. The structure shows the detailed location of MMF as well as the binding mode of MMF to hGAPDH. Subsequently, we compared the structure with an iodoacetate (IA)-bound complex structure; IA is a well-known antagonist of hGAPDH (McKee et al., 1965; Schmidt and Dringen, 2009). Finally, the results from our current study reveal a difference in the inhibitory mechanisms between the two inhibitors.
MATERIALS AND METHODS
Protein purification
hGAPDH was cloned into the modified pET28a (Novagen; Merck Biosciences, USA) that is fused with a tobacco etch virus (TEV) protease cleavage site after attaching an N-terminal 6×-His affinity tag. The pET28a-hGAPDH gene was transformed into BL21 (DE3) cells. Cells were grown in high salt LB medium at 37°C until optical density at 600 nm (OD600) reached about 0.8, the temperature was then decreased to 17°C and cells were treated with 0.1 mM isopropyl 1-thio-β-D-galactopyranoside (IPTG). After 16 h, cells were harvested by centrifugation and the pellet was frozen at −20°C. The pelleted cells were resuspended in lysis buffer (20 mM Tris-HCl pH 7.5, 300 mM NaCl, 30 mM imidazole) and then disrupted using a sonicator on chilled ice. The cell lysate was centrifuged and filtered through a cellulose acetate membrane (0.45 μm). The supernatants were loaded on a Ni-NTA resin column, washed using 3 column volumes of lysis buffer and eluted with elution buffer (20 mM Tris-HCl pH 8.0, 300 mM NaCl, 250 mM imidazole). The eluted proteins were digested with TEV protease for 16 h at 17°C. The proteins were loaded on the HiLoad 26/600 (GE Healthcare, USA) with gel filtration buffer (20 mM Tris pH 8.0, 100 mM NaCl). The purified GAPDH protein (5 mg) was incubated with 50 μM DMF in 50 ml of HEN buffer (250 mM HEPES-NaOH pH 7.7, 1 mM EDTA, 0.1 mM neocuproine) at 37°C for 3 h and filtered through cellulose acetate membrane (0.45 μm). The DMF-bound recombinant hGAPDH protein was concentrated to ~8.9 mg/ml and stored at −80°C after flash freezing using liquid nitrogen.
Crystallization, data collection, structure determination, and refinement
The protein crystal was formed in 0.1 M sodium acetate, 0.1 M HEPES-NaOH pH 7.5, 22% (w/v) PEG 4000, 4.4% (v/v) formamide at 290 K. After adding 20% (v/v) glycerol to the protein crystals as a cryo-protectant, crystals were flash frozen in liquid nitrogen. X-ray diffraction data were collected on beamline 7A at Photon Factory (Korea) and the indexing and scaling processes were carried out using the program HKL2000. The released human NAD+-bound GAPDH structure (Protein Data Bank [PDB] code: 4WNC) was used for molecular replacement using the program MOLREP from CCP4. REFMAC5 and COOT were used for structure refinement and further modelling, respectively. All figures were prepared using PyMOL.
Peripheral blood mononuclear cell (PBMC) isolation
Healthy participants were recruited following approval of the protocol by the Yonsei University and Severance Hospital Ethic Review Committee (approval No. 4-2010-0213). All study subjects gave informed consent for the use of the samples obtained. PBMCs were isolated from the whole blood of healthy participants using density gradient centrifugation with Percoll (Sigma Aldrich, USA) and stored in vapor phase of liquid nitrogen until use.
Treatment of PBMCs with DMF
Whole human PBMCs (200,000/well) labelled with CellTrace-Violet dye (CTV; Thermo Fisher Scientific, USA) were resuspended in complete RPMI medium (10% foetal bovine serum, 1% penicillin/streptomycin, 50 mM b-mercaptoethanol, and 1% L-Glu). CTV-labelled PBMCs were stimulated with 3 mg/ml soluble a-CD3 (BD Pharmingen, USA) and 3 mg/ml soluble a-CD28 antibody (BD Pharmingen) for four days at 37°C with 5% CO2. Cells were treated with the indicated doses of DMF at the onset of culture.
Flow cytometry and staining
For detection of cytokine production by T cells, human PBMCs were re-stimulated with PMA/ionomycin in presence of Golgi plug/Golgi stop (BD Biosciences, USA) for 5 h prior to staining. The cells were stained with fluorochrome-conjugated antibodies in phosphate-buffered saline containing 0.2% foetal bovine serum (Pan Biotech, Germany): antibodies against CD4 (OKT), CD8 (SK1), and interferon gamma (IFN-γ) (4S.B3) (BioLegend) and an antibody against CD25 (M-A251) (BD Biosciences). To remove the dead cell population, the Live/Dead Fixable Dead Cell Stain Kit (Invitrogen, USA) was used in the staining procedures. For intracellular staining of cytokines, cells were fixed and permeabilized using BD Cytofix/Cytoperm (BD Biosciences) in accordance with the manufacturer’s instructions. Flow cytometry was performed on the CytoFLEX system (Beckman Coulter, USA). Data were analysed using FlowJo 10.5.3 software (FlowJo, LLC, USA).
Statistical analysis
Data were analysed using the two-tailed unpaired Student’s t-test with Prism 5.0 software (GraphPad Software, USA). Results are presented as mean ± standard error of the mean (SEM). Differences with a P value less than 0.05 were considered to be statistically significant.
RESULTS
Effects of DMF on the proliferation and function of human T cells
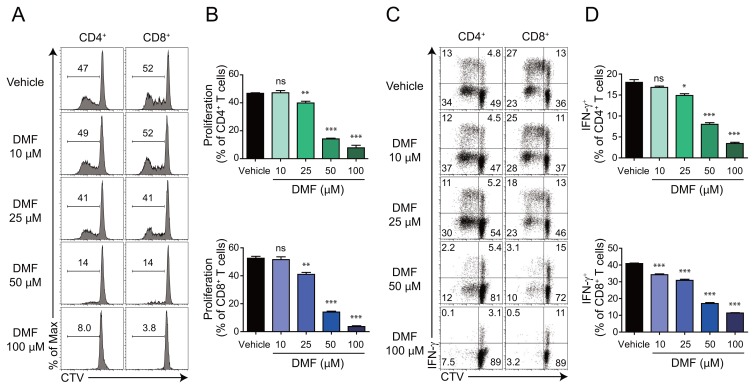
To investigate the effects of DMF on the proliferation and function of human CD4+ and CD8+ T cells, we activated human PBMCs with a-CD3 and a-CD28 antibodies in the presence of various doses of DMF. DMF impaired the viability of human lymphocytes in a dose-dependent manner (Supplementary Fig. S1). Although 10 mM DMF had no effect on the proliferation of both human CD4+ and CD8+ T cells, a dose-dependent inhibitory effect of DMF was clearly observed for doses > 25 mM (Figs. 1A and 1B). Consistent with previous reports on mouse CD4+ T cells (Kornberg et al., 2018), DMF inhibited IFN-γ production from human CD4+ T cells. Furthermore, a low-dose (10 μM) of DMF could inhibit IFN-γ production from human CD8+ T cells, indicating that CD8+ T cells are more sensitive to DMF than CD4+ T cells (Figs. 1C and 1D). Taken together, these results indicate that DMF suppresses both proliferation and cytokine production in human CD4+ and CD8+ T cells in a dose-dependent manner.
Fig. 1. DMF inhibits the proliferation and function of human CD4+ and CD8+ T cells.
CellTrace-Violet (CTV)-labelled human PBMCs were activated with 3 μg/ml soluble α-CD3 and 3 μg/ml soluble α-CD28 antibody in culture media alone or supplemented with the indicated doses of DMF for four days. (A) Representative flow cytometric plot of proliferating (CTVlow) cells that is gated on live CD4+ and CD8+ T cells. (B) Frequency of proliferating (CTVlow) CD4+ and CD8+ T cells as in A. (C) CTV profile showing proliferation versus IFN-γ gating on live CD4+ and CD8+ T cells. (D) Frequency of IFN-γ-secreting cells among live CD4+ and CD8+ T cells, as in C. Results are representative of three independent experiments. Data are shown as mean ± SEM and statistical significance was determined using a two-tailed unpaired Student’s t-test; *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant.
Overall structure and MMF binding mode of hGAPDH
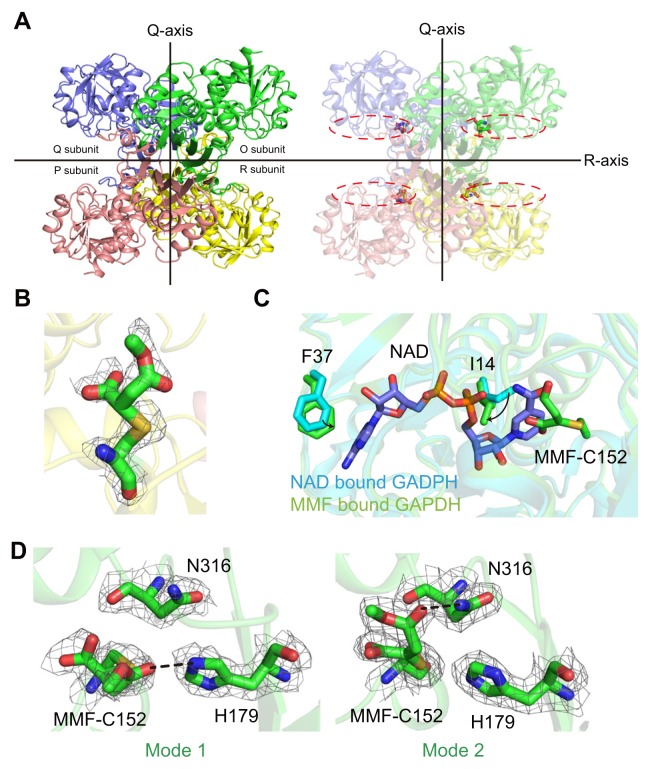
We determined the crystal structure of MMF-bound hGAPDH structure at 2.29 Å with a P21 space group (Table 1). A total of eight GAPDH molecules were found in an asymmetric unit and the refined structure indicated that two tetrameric GAPDH molecules were packed against each other. MMF-bound hGAPDH has a homo-tetramer structure based on the dimer of a dimer through the Q and R axis. These results indicate that MMF does not affect the tetrameric structure of hGAPDH (Fig. 2A, left panel). To identify the structural homologues of the MMF-bound GAPDH structure, we searched the DALI server and found that MMF-bound hGAPDH is highly similar to the NAD+-bound hGAPDH structure (PDB code: 4WNC, root mean square deviation [r.m.s.d.]: 0.2 Å). Amongst other species, GAPDH from Escherichia coli O157:H7 (PDB code: 5O0V) has the most similar structure (r.m.s.d: 0.7 Å) to MMF-bound hGAPDH (Table 2).
Table 1.
Data collection and refinement statistics
| PDB code | 6IQ6 |
|---|---|
| Data collection | |
| Wavelength (Å) | 0.97932 |
| Space group | P21 |
| Cell dimensions | |
| a, b, c (Å) | 71.6, 108.5, 175.8 |
| α, β, σ (°) | 90.0, 96.4, 90.0 |
| Resolution (Å) | 40.5-2.3 (2.33–2.29)a |
| Total reflections | 630779 |
| Rmerge | 0.118 (0.576)a |
| I/σI | 9.0 (2.4)a |
| Completeness (%) | 99.8 (96.4)a |
| Redundancy | 5.2 (4.3)a |
| Refinement | |
| Resolution (Å) | 40.5-2.3 (2.33–2.29)a |
| Unique reflections | 120651 (6121)a |
| Rwork/Rfree | 0.2153/0.2551 |
| Total No. of atoms | 20298 |
| Protein | 20184 |
| Ligand/ion | 45 |
| Water | 69 |
| B factors | |
| Protein | 35.6 |
| Water | 23 |
| r.m.s.d. | |
| Bond lengths (Å) | 0.0083 |
| Bond angles (°) | 1.397 |
| Ramachandran statistics | |
| Most favoured (%) | 94.3 |
| Allowed (%) | 5.1 |
| Outlier (%) | 0.64 |
Values in parentheses are for the highest-resolution shell.
Fig. 2. Overall structure and MMF binding mode in the GAPDH active site.
(A) The crystal structure shows that MMF-bound hGAPDH has a tetrameric structure based on the Q and R axes (the P axis is not presented). The dotted line represents the NAD+ binding site of hGAPDH. (B) The Fo-Fc electron density map (contour 1.0 σ) for covalently linked MMF with Cys152 of hGAPDH. (C) Comparison of the NAD+ binding site between NAD+-bound hGAPDH and MMF-bound hGAPDH. Two molecules are superimposed based on the C-α carbon of the protein backbone. (D) Two different binding modes of MMF in each protomer of tetrameric hGAPDH. Hydrogen bonds are shown as black lines.
Table 2.
Structural comparison between MMF-bound hGAPDH and others based on the DALI server hits
| Protein | Species | Ligand | PDB code (chain) | r.m.s.d. (Å) | No. of aligned | Identity (%) | Z-score |
|---|---|---|---|---|---|---|---|
| GAPDH | Homo sapiens | NAD+ | 4wnc (A) | 0.2 | 333 | 100 | 57.3 |
| Escherichia coli O157:H7 | Apo | 5o0v (A) | 0.7 | 330 | 66 | 52.1 | |
| Trypanosoma cruzi | BRZ | 1k3t (A) | 1.1 | 359 | 53 | 49.1 | |
| Lactobacillus acidophilus | Apo | 5j9g (A) | 1.3 | 338 | 42 | 46.6 | |
| Streptococcus agalactiae | NAD+, G3H | 5jya (D) | 1.3 | 343 | 45 | 46.1 | |
| S. agalactiae | Apo | 5jyf (B) | 1.3 | 294 | 46 | 38.1 |
NAD+, nicotinamide-adenine-dinucleotide; BRZ, 6-(1,1-dimethylallyl)-2-(1-hydroxy-1-methylethyl)-2,3-dihydro-7h-furo[3,2-g]chromen-7-one; G3H, glyceraldehyde-3-phosphate.
MMF is located in the active site pocket (Fig. 2A, right panel). A previous study has demonstrated that the combination of dimethyl and monomethyl succination from DMF is covalently linked with catalytic Cys152 (Kornberg et al., 2018). Based on the result from model building, MMF is better well-fitted compared to DMF, even though DMF had been incubated with hGAPDH. The electron density map of MMF-bound hGAPDH instead of DMF indicates that MMF is covalently linked with Cys152 and this structure is in high agreement with the previous study (Kornberg et al., 2018) (Fig. 2B). In addition, the real-space correlation coefficient of the electron density map was measured and the score of the MMF-bound Cys was found to be 0.795 to 0.916 (overall 0.837), indicating that the MMF-bound cysteine residue is well-occupied in each hGAPDH protomer.
Moreover, a previous study has demonstrated that the modified cysteine residues by DMF or MMF are Cys152, Cys156, and Cys247. However, based on the crystal structure, only Cys152 was shown as an MMF-bound cysteine, whereas the electron density maps of MMF-linked Cys156 or Cys247 were not visible (Supplementary Fig. S2A). Structural analysis showed that only the thiol group of Cys152 is exposed to the surface of GAPDH, whereas the others are buried in the structure (Supplementary Fig. S2B), indicating that Cys152 is in an advantageous position to be modified by inhibitors. In addition, we suggest that without conformational changes, it is difficult for post-translational modifications of Cys156 and Cys247 to occur. Taken together, the structural data indicate that DMF is hydrolysed in solution to MMF as reported in a previous study (Kornberg et al., 2018) and that Cys152 is the main target of DMF or MMF.
Cys152 of hGAPDH is a catalytically important residue for its enzymatic activity, and covalent linkage of MMF inhibits the catalytic reaction of the cysteine (Kornberg et al., 2018; Nakajima et al., 2009). Our structure demonstrates that MMF also inhibits the binding of NAD+. Based on the superimposition between the NAD+-bound hGAPDH (White et al., 2015) (PDB code: 4WNC) and MMF-bound hGAPDH, the nicotinamide group of NAD+ poses as a steric hindrance with MMF (Fig. 2C). The sidechains of Ile14 and Phe37 fill the NAD+ binding site and this shows that the covalently linked MMF inhibits the function of Cys152 as well as NAD+ binding.
MMF is attached to Cys152 and is stabilized via hydrogen bonding. From a structural perspective, MMF interacts with His152 or Asn316 via hydrogen bonding, herein, termed mode 1 and mode 2, respectively. (Fig. 2D). The O, R and Q, P subunits undergo mode 1 and mode 2, respectively. In the NAD+-bound GAPDH structure, Asn316 interacts with the nicotinamide part of NAD+ via hydrogen bonding and His152 acts as a catalytic residue that participates in acid/base catalysis during the oxidoreduction stage of GAPDH (Reis et al., 2013). In the MMF-bound hGAPDH, His152 and Asn316 participate in the interaction with MMF to stabilize the ligand. Collectively, the MMF binding modes reveal that MMF occupies the NAD+ and G3P binding site at the same time because the interactions of Asn316-NAD+ and His152-G3P are well-defined. Moreover, MMF hijacks these two residues.
Furthermore, results from the B-factor analysis of the apo (F chain of 4WNC) and MMF-bound hGAPDH structures support modes 1 and 2, respectively. As compared to the apo form, all side chain B-factors of Cys152, His179, and Asn316 from the MMF-bound hGAPDH are reduced and this is in agreement with our structural analysis that three residues are involved in MMF stabilization (Collaborative Computational Project, Number 4, 1994) (Table 3). Put together, we suggest that MMF has two different inhibitory mechanisms that occur simultaneously—the first mechanism involves succination of the catalytic cysteine via a covalent linkage, whereas the second involves hijacking of the cofactor-binding site.
Table 3.
B-factor analysis results between apo- and MMF-bound hGAPDH
| Chain | B-factor (side chain) | |||
|---|---|---|---|---|
|
| ||||
| Cys152 | His179 | Asn316 | ||
| hGAPDH (Apo) | A | 0.1304 | 0.2934 | 0.1562 |
| B | 0.1166 | 0.2761 | 0.1776 | |
| C | 0.1701 | 0.2631 | 0.2196 | |
| D | 0.1257 | 0.3202 | 0.1309 | |
| hGAPDH (MMF-bound) | A | 0.0897 | 0.0681 | 0.0352 |
| B | 0.0682 | 0.0938 | 0.0633 | |
| C | 0.0856 | 0.0511 | 0.0727 | |
| D | 0.1037 | 0.0946 | 0.0391 | |
The Baverage program (Collaborative Computational Project, Number 4, 1994) was used for B-factor analysis.
Comparison of the binding mode between IA and MMF
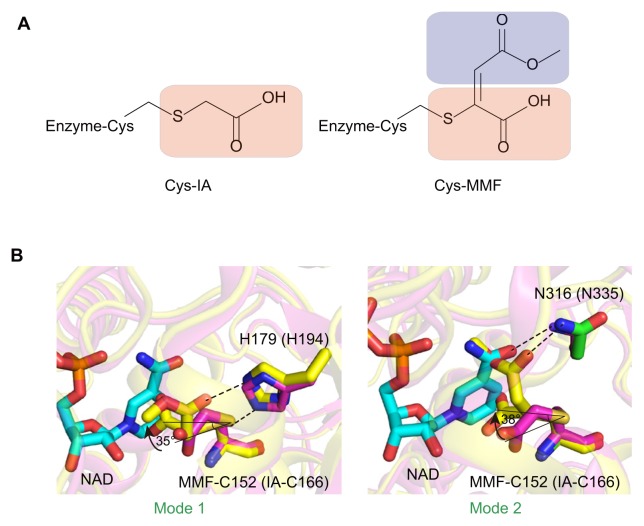
IA is a GAPDH inhibitor that covalently links to a catalytic cysteine. Previously, McKee et al. (1965) described the inhibitory role of IA against GAPDH in cancer cells at micromolar concentrations. The chemical structure of IA is highly similar to that of MMF (or DMF), with the exception of an additional extended methylated carboxyl group in MMF (Fig. 3A).
Fig. 3. Differences of binding modes to GAPDH between MMF and iodoacetate.
(A) Chemical structure of iodoacetate and MMF. The red and purple round-squares show common and different chemical structures for both inhibitors, respectively. (B) The difference in inhibitory modes between iodoacetate and MMF.
Interestingly, we demonstrate that the binding mode is different between the two inhibitors. In the IA-bound GAPDH structure from Trypanosoma cruzi (PDB code: 3DMT) (Guido et al., 2009), IA is covalently linked to Cys166, which corresponds to Cys152 in hGAPDH, whereas MMF is tilted towards the opposite direction of NAD+ at ~35° and ~38° in mode 1 and mode 2 as compared to IA, respectively (Fig. 3B). The IA inhibitor is stabilized via hydrophobic interaction with NAD+ as well as further hydrogen bonding with the nitrogen of the Cys166 backbone. The inhibitory mode of IA allowed the binding of NAD+, and thus a clear electron density map of the NAD+ was visible (Guido et al., 2009). On the other hand, in the case of MMF, due to its bulkier size (compared to IA) as well as the interaction of its methyl carboxylate group with His179 or Asn316, MMF inhibited the access of NAD+. In conclusion, NAD+ and catalytic cysteine are essential for the oxidoreduction catalysis mechanism catalysed by GAPDH. In addition, DMF can simultaneously inhibit the binding of NAD+ and the catalytic function of the Cys152 residue.
DISCUSSION
In recent years, the development of antagonists against GAPDH for the treatment of cancer or cancer-related diseases has become the centre of many structure-based drug design studies. In the current study, we report the crystal structure of MMF-bound hGAPDH and, based on the structure, we have elucidated the detailed mode of action by which MMF functions. Furthermore, we demonstrated that MMF has three types of inhibitory steps that occur simultaneously. First, MMF covalently links to catalytic Cys152, and the electron density map shows two other modes. Second, MMF is small enough to bind to the G3P binding site, which would inhibit the binding of substrate G3P. Third, it masks the nicotinamide part of NAD+. From a structural perspective, this binding step is different from that of IA, which lacks steric hindrance with NAD+. Put together, MMF is more efficient than IA from a structural perspective.
For the present study, we added DMF to recombinant GAPDH to obtain the DMF-bound GAPDH structure. However, the electron density map of MMF was visible, the map of DMF was not observed. In fact, the presence of an extra methyl group in DMF causes it to behave both chemically and structurally different from MMF. Hence, from a structural perspective, we suggest that the inhibitory effect of DMF would be significantly greater than MMF because the carboxyl group adjacent to the methyl group of MMF is close enough to Ile14 and/or Tyr320 (Supplementary Fig. S3). This indicates that Ile14 and/or Tyr320 is expected to interact with another methyl group that corresponds to the invisible part of DMF. This structural inference is in good agreement with previous experimental data showing that the inhibitory rate of GAPDH by DMF is more effective than MMF in a dose-dependent manner (Kornberg et al., 2018). However, without considering any off-target effects, the inhibitory efficacy of KA is greater than that of DMF at the same dose (Kornberg et al., 2018). The chemical structure is extensively different between KA and DMF, and a study of the detailed inhibitory mode of KA is necessary to explain the difference in inhibitory efficacy. Our structural data provide insights for the design and development of new antagonists of GAPDH.
Supplementary Material
ACKNOWLEDGMENTS
We thank the staff scientists at the beamline 5A and 1A of the Photon Factory and the beamline 11C of Pohang Light Source for their assistance. This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Ministry of Science and ICT (NRF-2016R1A2B2013305, 2016R1A5A1010764, and 2017M3A9F6029755) and by the Strategic Initiative for Microbiomes in Agriculture and Food funded by the Ministry of Agriculture, Food and Rural Affairs (918012-4).
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
Data availability
Coordinates and structure factors have been deposited at the Protein Data Bank under the accession code 6IQ6. Other data are available from the corresponding author upon reasonable request.
Disclosure
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Ahuja M., Ammal Kaidery N., Yang L., Calingasan N., Smirnova N., Gaisin A., Gaisina I.N., Gazaryan I., Hushpulian D.M., Kaddour-Djebbar I., et al. Distinct Nrf2 signaling mechanisms of fumaric acid esters and their role in neuroprotection against 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine-induced experimental Parkinson’s-like disease. J Neurosci. 2016;36:6332–6351. doi: 10.1523/JNEUROSCI.0426-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki K., Kishihara F., Takahashi K., Matsumata T., Shimura T., Suehiro T., Kuwano H. Hepatocellular carcinoma producing a granulocyte colony-stimulating factor: report of a resected case with a literature review. Liver Int. 2007;27:716–721. doi: 10.1111/j.1478-3231.2007.01468.x. [DOI] [PubMed] [Google Scholar]
- Blair H.A. Dimethyl fumarate: a review in moderate to severe plaque psoriasis. Drugs. 2018;78:123–130. doi: 10.1007/s40265-017-0854-6. [DOI] [PubMed] [Google Scholar]
- Bomprezzi R. Dimethyl fumarate in the treatment of relapsing-remitting multiple sclerosis: an overview. Ther Adv Neurol Disord. 2015;8:20–30. doi: 10.1177/1756285614564152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colell A., Green D.R., Ricci J.E. Novel roles for GAPDH in cell death and carcinogenesis. Cell Death Differ. 2009;16:1573–1581. doi: 10.1038/cdd.2009.137. [DOI] [PubMed] [Google Scholar]
- Colell A., Ricci J.E., Tait S., Milasta S., Maurer U., Bouchier-Hayes L., Fitzgerald P., Guio-Carrion A., Waterhouse N.J., Li C.W., et al. GAPDH and autophagy preserve survival after apoptotic cytochrome c release in the absence of caspase activation. Cell. 2007;130:385. doi: 10.1016/j.cell.2007.07.012. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Endo A., Hasumi K., Sakai K., Kanbe T. Specific-inhibition of glyceraldehyde-3-phosphate dehydrogenase by koningic acid (heptelidic acid) J Antibiot. 1985;38:920–925. doi: 10.7164/antibiotics.38.920. [DOI] [PubMed] [Google Scholar]
- Ganapathy-Kanniappan S., Geschwind J.F.H., Kunjithapatham R., Buijs M., Vossen J.A., Tchernyshyov I., Cole R.N., Syed L.H., Rao P.P., Ota S., et al. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is pyruvylated during 3-bromopyruvate mediated cancer cell death. Anticancer Res. 2009;29:4909–4918. [PMC free article] [PubMed] [Google Scholar]
- Ganapathy-Kanniappan S., Kunjithapatham R., Geschwind J.F. Glyceraldehyde-3-phosphate dehydrogenase: a promising target for molecular therapy in hepatocellular carcinoma. Oncotarget. 2012;3:940–953. doi: 10.18632/oncotarget.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guido R.V.C., Balliano T.L., Andricopulo A.D., Oliva G. Kinetic and crystallographic studies on glyceraldehyde-3-phosphate dehydrogenase from trypanosoma cruzi in complex with iodoacetate. Lett Drug Des Discov. 2009;6:210–214. doi: 10.2174/157018009787847774. [DOI] [Google Scholar]
- Kato M., Sakai K., Endo A. Koningic acid (heptelidic acid) inhibition of glyceraldehyde-3-phosphate dehydrogenases from various sources. Biochim Biophys Acta. 1992;1120:113–116. doi: 10.1016/0167-4838(92)90431-C. [DOI] [PubMed] [Google Scholar]
- Kornberg M.D., Bhargava P., Kim P.M., Putluri V., Snowman A.M., Putluri N., Calabresi P.A., Snyder S.H. Dimethyl fumarate targets GAPDH and aerobic glycolysis to modulate immunity. Science. 2018;360:449–453. doi: 10.1126/science.aan4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landeck L., Asadullah K., Amasuno A., Pau-Charles I., Mrowietz U. Dimethyl fumarate (DMF) vs. monoethyl fumarate (MEF) salts for the treatment of plaque psoriasis: a review of clinical data. Arch Dermatol Res. 2018;310:475–483. doi: 10.1007/s00403-018-1825-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberti M.V., Dai Z.W., Wardell S.E., Baccile J.A., Liu X.J., Gao X., Baldi R., Mehrmohamadi M., Johnson M.O., Madhukar N.S., et al. A predictive model for selective targeting of the Warburg effect through GAPDH inhibition with a natural product. Cell Metab. 2017;26:648–659.e8. doi: 10.1016/j.cmet.2017.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee R.W., Wong W., Landman M. Effects of iodoacetate on glycolysis and respiration in Ehrlich-Lettre ascites carcinoma cells. Biochim Biophys Acta. 1965;105:410–423. doi: 10.1016/S0926-6593(65)80227-2. [DOI] [PubMed] [Google Scholar]
- Mrowietz U., Christophers E., Altmeyer P. Treatment of severe psoriasis with fumaric acid esters: scientific background and guidelines for therapeutic use. The German Fumaric Acid Ester Consensus Conference. Br J Dermatol. 1999;141:424–429. doi: 10.1046/j.1365-2133.1999.03034.x. [DOI] [PubMed] [Google Scholar]
- Nakajima H., Amano W., Kubo T., Fukuhara A., Ihara H., Azuma Y.T., Tajima H., Inui T., Sawa A., Takeuchi T. Glyceraldehyde-3-phosphate dehydrogenase aggregate formation participates in oxidative stress-induced cell death. J Biol Chem. 2009;284:34331–34341. doi: 10.1074/jbc.M109.027698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray M., Basu N., Ray S. Inactivation of glyceraldehyde-3-phosphate dehydrogenase of human malignant cells by methylglyoxal. Mol Cell Biochem. 1997;177:21–26. doi: 10.1023/A:1006831511527. [DOI] [PubMed] [Google Scholar]
- Reis M., Alves C.N., Lameira J., Tunon I., Marti S., Moliner V. The catalytic mechanism of glyceraldehyde 3-phosphate dehydrogenase from Trypanosoma cruzi elucidated via the QM/MM approach. Phys Chem Chem Phys. 2013;15:3772–3785. doi: 10.1039/c3cp43968b. [DOI] [PubMed] [Google Scholar]
- Sakai K., Hasumi K., Endo A. Identification of koningic acid (heptelidic acid)-modified site in rabbit muscle glyceraldehyde-3-phosphate dehydrogenase. Biochim Biophys Acta. 1991;1077:192–196. doi: 10.1016/0167-4838(91)90058-8. [DOI] [PubMed] [Google Scholar]
- Schmidt M.M., Dringen R. Differential effects of iodoacetamide and iodoacetate on glycolysis and glutathione metabolism of cultured astrocytes. Front Neuroenergetics. 2009;1:1. doi: 10.3389/neuro.14.001.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirover M.A. New nuclear functions of the glycolytic protein, glyceraldehyde-3-phosphate dehydrogenase, in mammalian cells. J Cell Biochem. 2005;95:45–52. doi: 10.1002/jcb.20399. [DOI] [PubMed] [Google Scholar]
- Sirover M.A. On the functional diversity of glyceraldehyde-3-phosphate dehydrogenase: biochemical mechanisms and regulatory control. Biochim Biophys Acta. 2011;1810:741–751. doi: 10.1016/j.bbagen.2011.05.010. [DOI] [PubMed] [Google Scholar]
- Sirover M.A. Subcellular dynamics of multifunctional protein regulation: mechanisms of GAPDH intracellular translocation. J Cell Biochem. 2012;113:2193–2200. doi: 10.1002/jcb.24113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva A., Chiang D.Y., Newell P., Peix J., Thung S., Alsinet C., Tovar V., Roayaie S., Minguez B., Sole M., et al. Pivotal role of mTOR signaling in hepatocellular carcinoma. Gastroenterology. 2008;135:1972–1983. doi: 10.1053/j.gastro.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M.R., Khan M.M., Deredge D., Ross C.R., Quintyn R., Zucconi B.E., Wysocki V.H., Wintrode P.L., Wilson G.M., Garcin E.D. A dimer interface mutation in glyceraldehyde 3-phosphate dehydrogenase regulates its binding to AU-rich RNA. J Biol Chem. 2015;290:4129. doi: 10.1074/jbc.A114.618165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing C., LaPorte J.R., Barbay J.K., Myers A.G. Identification of GAPDH as a protein target of the saframycin antiproliferative agents. Proc Natl Acad Sci U S A. 2004;101:5862–5866. doi: 10.1073/pnas.0307476101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S., Takashima S., Ogawa H., Kuroda T., Yamamoto M., Takeda A., Nakmaura H. Granulocyte-colony-stimulating-factor-producing hepatocellular carcinoma. J Gastroenterol. 1999;34:640–644. doi: 10.1007/s005350050387. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Yi X., Stoffer J.B., Bonafe N., Gilmore-Hebert M., McAlpine J., Chambers S.K. The multifunctional protein glyceraldehyde-3-phosphate dehydrogenase is both regulated and controls colony-stimulating factor-1 messenger RNA stability in ovarian cancer. Mol Cancer Res. 2008;6:1375–1384. doi: 10.1158/1541-7786.MCR-07-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X.D., Zhang J.B., Zhuang P.Y., Zhu H.G., Zhang W., Xiong Y.Q., Wu W.Z., Wang L., Tang Z.Y., Sun H.C. High expression of macrophage colony-stimulating factor in peritumoral liver tissue is associated with poor survival after curative resection of hepatocellular carcinoma. J Clin Oncol. 2008;26:2707–2716. doi: 10.1200/JCO.2007.15.6521. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.