Abstract
Resident microbiota activates regulatory cells that modulate intestinal inflammation and promote and maintain intestinal homeostasis. IL-10 is a key mediator of immune regulatory function. Our studies describe the functional importance and mechanisms by which gut microbiota and specific microbial components influence the development of intestinal IL-10–producing B cells. Using fecal transplant into germ-free (GF) Il10+/EGFP reporter and Il10–/– mice, we demonstrated that microbiota from specific pathogen–free mice primarily stimulated IL-10–producing colon-specific B cells and T regulatory 1 cells in ex-GF mice. IL-10 in turn downregulated microbiota-activated mucosal inflammatory cytokines. TLR2 and -9 ligands and enteric bacterial lysates preferentially induced IL-10 production and the regulatory capacity of intestinal B cells. Analysis of Il10+/EGFP mice crossed with additional gene-deficient strains and B cell cotransfer studies demonstrated that microbiota-induced IL-10–producing intestinal B cells ameliorated chronic T cell–mediated colitis in a TLR2-, MyD88-, and PI3K-dependent fashion. In vitro studies implicated downstream signaling of PI3Kp110δ and AKT. These studies demonstrated that resident enteric bacteria activated intestinal IL-10–producing B cells through TLR2, MyD88, and PI3K pathways. These B cells reduced colonic T cell activation and maintained mucosal homeostasis in response to intestinal microbiota.
Keywords: Gastroenterology, Immunology
Keywords: B cells, Inflammatory bowel disease, T cells
Introduction
Inflammatory bowel diseases (IBDs), including Crohn’s disease and ulcerative colitis, are T cell–mediated, chronic disorders whose development is influenced in part by dysregulated, aggressive intestinal mucosal immune reactions to resident intestinal bacteria (1–3). Enteric microbiota is essential for inducing and exacerbating chronic, immune-mediated intestinal inflammation in most rodent models of IBD. This has been most clearly demonstrated in gnotobiotic experiments in which most germ-free (GF), genetically susceptible mice and rats exhibit minimal intestinal inflammation and immune activation, but rapidly develop chronic colitis and pathogenic T cell–mediated immune responses after colonization with specific pathogen–free (SPF) or specific enteric bacteria (4–6). Recent studies have identified microbial factors that selectively stimulate pro- or antiinflammatory immune responses in the intestine. For example, segmented filamentous bacteria strongly activate largely proinflammatory Th17 responses in the small intestine (7). On the other hand, in normal hosts, some resident enteric bacterial species stimulate regulatory immune responses that help maintain intestinal mucosal homeostasis. Selected Clostridium species induce mucosal Foxp3+CD4+ T cells (8), and Bacteroides fragilis polysaccharide A stimulates IL-10–producing Tregs (9). However, these bacteria coexist in a consortium with many other species in the complex mammalian intestinal ecosystem, where a delicate balance of putative pro- and antiinflammatory species helps maintain mucosal homeostasis. In IBD, a characteristic expansion of putative aggressive bacterial groups and contraction of protective species (dysbiosis) is thought to contribute to IBD pathogenesis (1–3). Additional mechanistic studies are required in order to better understand how these resident bacterial communities interact with the host immune system to maintain intestinal homeostasis or cause IBD.
IL-10 inhibits proinflammatory, effector innate, and adaptive immune cells and helps maintain mucosal homeostasis. Mice with genetic deletion of IL-10 or the IL-10 receptor (IL-10R) spontaneously develop Th1/Th17-mediated colitis when normal enteric microbiota is present (4, 5, 10), while administration of exogenous recombinant IL-10 prevents the onset of experimental enterocolitis, improves epithelial barrier function, and inhibits microbiota-stimulated, proinflammatory cytokine production in the mucosa (4, 11–13). In addition, IL-10 or IL-10R gene polymorphisms are associated with ulcerative colitis and early onset of Crohn’s disease (14–16). IL-10 is produced by many types of immune cells, including CD4+ and CD8+ T cells, B cells, macrophages, DCs, neutrophils, NK cells, innate lymphoid cells, and eosinophils (17). Multiple studies implicate IL-10–producing regulatory CD4+ T cell subsets (Tregs, T regulatory 1 [Tr1] cells) in mucosal homeostasis (8, 18–20), while we and others have reported that IL-10–producing antigen-presenting cells (APCs) (21, 22) and regulatory B cells (23–28) are present in humans and prevent bacterial antigen-driven, T cell–mediated chronic experimental colitis.
Previous in vitro studies have demonstrated that TLR and Myd88 signaling contributes to the development of bacteria-induced, IL-10–secreting immune cells (24, 29, 30). However, our current understanding of how bacterial products stimulate IL-10 production is based mainly on in vitro studies using cell lines, bone marrow–derived cells, and narrowly defined stimulants, with conflicting results in different cell types (31). Therefore, further in vivo mechanistic studies in mucosal immune cells stimulated with physiologically relevant microbial stimuli are needed to more fully understand how IL-10 mediates intestinal homeostasis.
Given our recent work demonstrating a protective role of bacteria-stimulated IL-10–secreting B cells through activation of Tregs in chronic colitis and the ability of bacterial lysates to stimulate the regulatory capacity of B cells in vitro (23), we hypothesized that TLR-dependent IL-10 production by resident bacteria is critical for B cell–mediated protection against experimental colitis. In this study, we demonstrate that resident enteric bacteria promote the in vivo development of IL-10–producing colonic lamina propria (LP) immune cells, especially T cell subsets and B cells. Intestinal bacterial colonization transiently activated proinflammatory cytokines in the colon in conjunction with prolonged stimulation of IL-10 in ex-GF Il10+/+ mice, but led to sustained aggressive activation of inflammatory cytokines in the absence of IL-10. We also show that TLR2 and MyD88 signaling is essential for bacteria-inducible IL-10–producing B cells to regulate T cell–mediated colitis and that TLR2-dependent bacterial activation of IL-10 production by B cells is mediated through the PI3K pathway. These findings increase our understanding of the pathogenesis of IBD and regulation of mucosal homeostasis by resident microbiota.
Results
Resident enteric microbiota increases intestinal IL-10–producing T and B cells in vivo.
We used a combination of in vivo and in vitro strategies to elucidate how resident intestinal bacteria and their components stimulate protective IL-10 production by colonic LP immune cells. We employed the Il10+/EGFP reporter mice (32) that we derived GF (23). This model allowed us to explore the kinetics and types of IL-10–producing colonic LP cells activated by resident bacterial colonization without the ex vivo manipulation required for intracellular staining of IL-10. We first verified that GFP+ cells in our Il10+/EGFP reporter mice were suitable for investigating the kinetics of IL-10–producing cells by PCR, ELISA, and flow cytometry. We found that GFP+ B cells were functional, producing abundant IL-10 and lower levels of inflammatory cytokines compared with GFP– B cells following TLR ligation (Supplemental Figure 1, A–C; supplemental material available online with this article; https://doi.org/10.1172/JCI93820DS1). We developed flow cytometry methods and gating strategies to measure low-frequency GFP+ cells based on basal levels of GFP fluorescence intensity in WT SPF mice and stained WT cells with the antibodies used for target samples (Supplemental Figure 1D). Our results demonstrating that the number of GFP+ cells closely correlated with tissue IL-10 protein and gene expression (see below) validated the use of GFP+ cells to measure physiologic IL-10 activation in vivo.
Next, to investigate whether resident bacteria induce colonic IL-10 and the kinetics of this response, we colonized GF IL-10–sufficient Il10+/EGFP reporter mice and GF Il10−/− mice with feces from a WT isogenic C57BL/6J mouse housed in SPF conditions and measured various immune functions 3 and 7 days later. Age- and sex-matched adult mice raised in GF or SPF conditions served as controls. We confirmed by 16S rRNA assays that SPF bacteria stably colonized the intestines of ex-GF mice (Supplemental Figure 2). Spontaneous IL-10 secretion by ex vivo cultured colonic tissue explants, the frequency of GFP+ IL-10 reporter cells in the colonic LP, and Il10 mRNA expression in the distal colon significantly increased over time after SPF enteric bacterial colonization (Figure 1A). The frequency of GFP+ LP cells strongly correlated with tissue Il10 mRNA expression (Supplemental Figure 1E) and protein secretion (Supplemental Figure 1F). In addition, SPF bacterial colonization was associated with significantly greater colonic levels of phosphorylated STAT3, a key downstream molecule in both IL-10 and IL-6 signaling (Figure 1, B and C). These results suggest that the induction of IL-10 is associated with functional signaling, though we did not investigate the relative contributions of IL-10 versus IL-6 in activating STAT3.
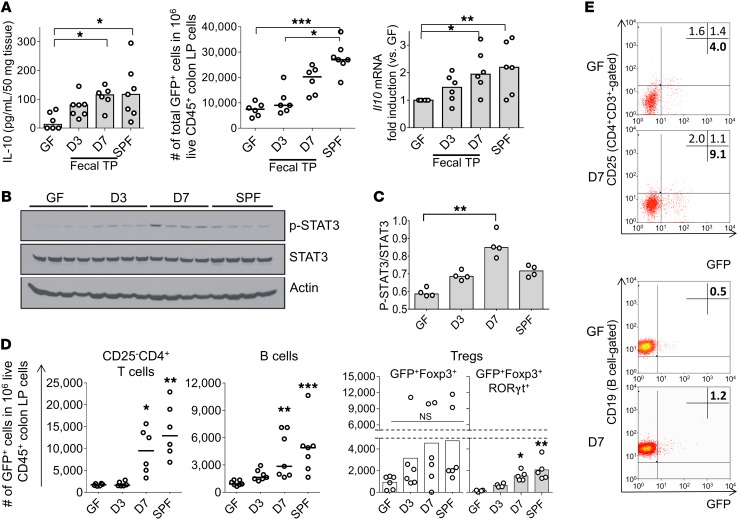
Figure 1. Resident intestinal microbiota increases the frequency of intestinal IL-10–producing immune cells and enhance IL-10 production.
(A) Left: Spontaneous IL-10 secretion by colonic tissue explants; middle: number of total IL-10–producing (GFP+) colon LP cells; right: Il10 mRNA expression in distal colon tissue normalized by expression in GF mice in GF, SPF-raised, or GF-conventionalized Il10+/EGFP reporter mice 3 days and 7 days (D3 and D7) after fecal transplantation (TP) with SPF feces. n = 6–9 mice/group, combined from 2 independent experiments. (B and C) Phosphorylation levels of STAT3 in the distal colon were evaluated by Western blot analysis and quantified by densitometry. n = 4 mice/group. (D) IL-10–producing (GFP+) colon LP cells were characterized by flow cytometry identifying cell subsets with antibodies to the indicated cell surface proteins. CD25–CD4+ T cells (CD25–CD4+CD3+), B cells (B220+CD19+), and Tregs, including GFP+Foxp3+RORγt+CD4+ T cells within the total GFP+Foxp3+CD4+ T cell population. n = 5–7 mice/group, combined from 2 independent experiments. (E) Representative dot plots for colon LP GFP+CD25–CD4+ T cells (CD4+ T cell–gated) and GFP+ B cells (B cell–gated) in GF and conventionalized day 7 Il10+/EGFP mice. For flow cytometry, the GFP+ population in live CD45+ colon LP cells was assessed using WT (GFP–) cells stained with the same antibodies as target samples as a control (see Flow cytometry in Methods). All data are presented as median values; *P < 0.05, **P < 0.01, ***P < 0.001, Kruskal-Wallis test with Dunn’s post hoc test.
We then sought to determine which subsets of colonic LP immune cells produce IL-10 in response to bacterial colonization. Bacterial colonization was associated with greater proportions of GFP+ B cells and Tr1 cells (Foxp3negCD25–CD4+ T cells) (Figure 1, D and E, and Supplemental Figure 3A), while proportions of GFP+ macrophages, DCs, CD8+ T cells, and CD25+CD4+ T cells were not significantly altered (Supplemental Figure 3B). Although the frequency of total Foxp3+ Tregs was not significantly altered by SPF fecal colonization (Supplemental Figure 3C), a newly identified Treg population (Foxp3+RORγt+IL-10+CD4+ T cells) (33, 34) capable of greater immune suppression than the other Treg types significantly increased compared with GF levels (Figure 1D, right, gray bars). These results indicate that subsets of B cells and CD4+ T cells are important sources of microbiota-activated IL-10 production in the colonic LP. Further, the proportions of GFP+ cell populations in mesenteric lymph nodes (MLNs) and spleen were not significantly altered by bacteria colonization (Supplemental Figure 3D), indicating that the observed changes were selectively induced in the colon during the experiment. These results demonstrate that bacterial colonization of GF adult mice stimulates progressively increased IL-10 production, which after 7 days approaches levels seen in SPF mice colonized at birth with resident microbiota. This IL-10 induction is slow, with significant increases in IL-10 levels compared with baseline GF levels occurring only after 7 days.
Distribution and lineage of IL-10–producing immune cells in SPF mice.
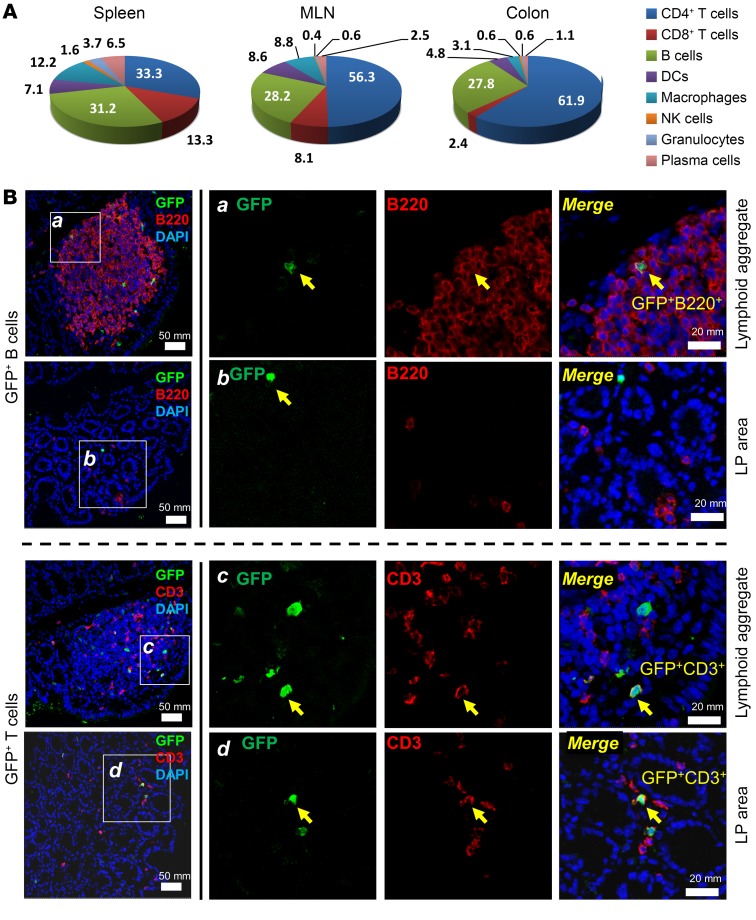
We next evaluated the distribution and lineage of physiologic IL-10–producing immune cells. In mice raised in SPF conditions from birth, CD4+ T cells were the most abundant type of IL-10+ cells in the colon LP (61.9%), followed by B cells (27.8%). B cell frequency among IL-10+ cells of the colon LP was similar to that observed in the spleen and MLNs (Figure 2A). Colonic GFP+B220+CD19+ cells expressed CD5+CD1dhiCD21hiCD23–CD24hiMHC-IIhiIgMloIgDlo, sharing certain phenotypic features with splenic marginal zone B cells and B10 cells (35) (Supplemental Figure 4). Compared with GFP– B cells, GFP+ LP B cells had lower IgM and IgD levels and higher CD24 intensity. In mice raised in SPF conditions, IL-10–producing B cells were located primarily within colonic mucosal lymphoid aggregates, while GFP+ T cells were more diffusely distributed in both the LP and lymphoid aggregates (Figure 2B). Similarly, chronic mucosal inflammation induced by repeated dextran sulfate sodium administration for 3 cycles in the presence of resident bacteria markedly increased total mucosal IL-10+ cells (Supplemental Figure 5A), with diffuse expansion of GFP+ T cells in the LP; in contrast, GFP+ B cells remained limited to the lymphoid aggregates (Supplemental Figure 5, B–D), as seen in noninflamed mice (Figure 2B). Together, these results indicate that accumulation of certain subsets of IL-10–producing T and B cells in the colonic LP depends on the presence of microbiota, and these cells’ prevalence increases with inflammation. This finding is consistent with previously reported induction of regulatory B cells by IL-1β and IL-6 (30).
Figure 2. Distribution and characteristics of SPF mucosal and splenic GFP+ T and B cells.
Physiological IL-10–producing immune cells were characterized in colonic LP, MLN, and spleen from SPF-raised Il10+/EGFP reporter mice. (A) Percentages of cell types among total GFP+ cells were assessed by flow cytometry using antibodies to cell surface markers as described in Methods, Flow cytometry. (B) Immunofluorescence was performed on distal colon tissue from SPF-reared Il10+/EGFP mice to characterize the distribution of GFP+ B cells and GFP+ T cells in lymphoid aggregates and the LP of the distal colon. GFP+ cells are shown in green. B cells (B220+) and T cells (CD3+) are shown in red. Merged images show B and T cells that are GFP+. Scale bars: far left column, 50 mm; right panels, 20 mm.
IL-10 regulates proinflammatory genes induced by bacterial colonization.
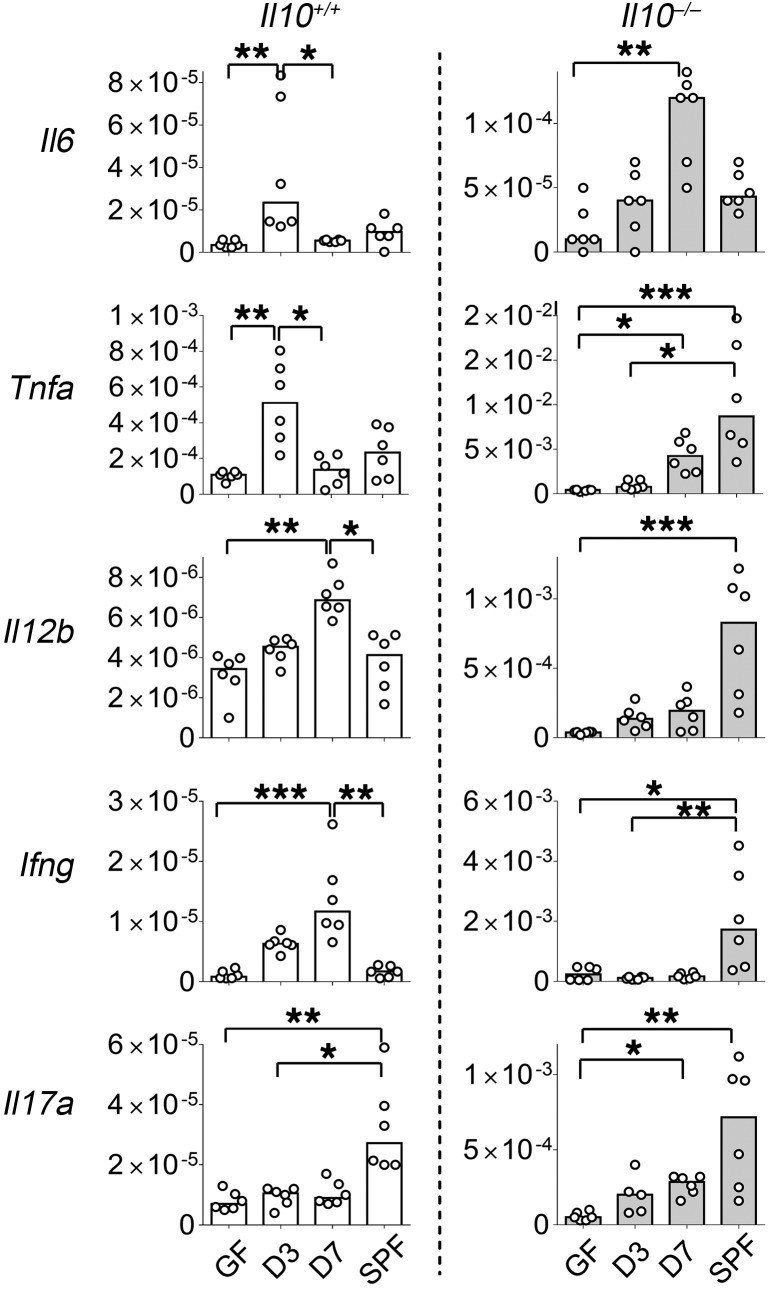
Because IL-10 regulates mucosal homeostasis, we investigated the kinetics of other inflammatory and regulatory intestinal immune responses in ex-GF mice colonized with resident enteric bacteria with and without endogenous IL-10. To do this, we examined real-time PCR expression of immune-related transcription factors and cytokines in whole tissue from the distal colons of Il10+/EGFP (Il10+/+) and Il10–/– mice reared GF, SPF-colonized ex-GF, or SPF, following the same time course used to study IL-10 induction (Figure 1A). Regulatory Il10 and Tgfb1 were persistently upregulated in Il10+/+ mice after colonization with bacteria (Figure 1A and Supplemental Figure 3E). In contrast, expression of innate proinflammatory cytokine genes, including Tnfa and Il6, was transiently upregulated 3 days after colonization (Figure 3), but downregulated by 7 days, when colonic IL-10 reached higher levels (Figure 1A). The primarily adaptive cytokine genes Ifng and Il12b and transcription factor Tbet were continuously expressed up to 7 days after colonization, but expression of these genes was not different between GF and SPF mice (Figure 3 and Supplemental Figure 3E), suggesting that either more prolonged stimulation after bacterial exposure or early-life exposure to microbiota is required in order to downregulate these cytokines. In contrast, Il17a expression did not increase following bacterial colonization of GF mice, but was higher in mice born SPF than in GF mice. Rorgt was persistently upregulated in Il10+/+ mice (Supplemental Figure 3E), which might be associated with induction of Foxp3+RORγt+IL-10+ Tregs (Figure 1D, right). Moreover, whole colonic tissue Foxp3 mRNA expression was not significantly altered by bacterial colonization, consistent with the lack of significant changes in the frequency of total LP Foxp3+ T cells in GF, ex-GF-colonized, and SPF mice (Figure 1D and Supplemental Figure 3, C and E). To determine the role of endogenous IL-10 in regulating these proinflammatory genes, we preformed similar studies in Il10–/– mice. In contrast to the transient increase in inflammatory genes in WT mice, colonization of Il10–/– mice with identical SPF fecal microbiota was associated with a far more robust and persistent increase in proinflammatory cytokine gene expression in the distal colon, with levels 1- to 2-log higher than in Il10+/+ mice (Figure 3). We had previously shown similar bacterial composition up to 1 week after fecal transplantation in WT and Il10–/– mice (36). Tgfb1 was also continuously upregulated in Il10–/– mice (data not shown), suggesting that transient upregulation of proinflammatory cytokine genes with subsequent suppression in Il10+/+ animals was either mainly IL-10 dependent or that an antiinflammatory effect of TGF-β1 was negated in the absence of IL-10. These results suggest that resident bacteria–induced IL-10 helps maintain intestinal homeostasis by downregulating aggressive proinflammatory responses induced by resident microbiota. This conclusion is consistent with our previous observation that in vivo neutralization of IL-10R increases IFN-γ responses to resident bacterial antigens (13).
Figure 3. Bacterial colonization influences the gene expression kinetics of the majority of proinflammatory cytokines in an IL-10–dependent fashion.
Kinetic expression of selected genes in distal colons from GF, SPF-raised (SPF), and ex-GF conventionalized (3 and 7 days after fecal transplantation) Il10+/EGFP reporter mice (Il10+/+) or Il10–/–mice was determined by real-time PCR of colonic tissues. All gene expression was normalized with Actb. n = 6 mice/group, combined from 2 independent experiments. Data are presented as median; *P < 0.05, **P < 0.01, ***P < 0.001, Kruskal-Wallis test with Dunn’s post hoc test.
Luminal bacteria increase the regulatory capacity of intestinal IL-10–secreting B cells ex vivo.
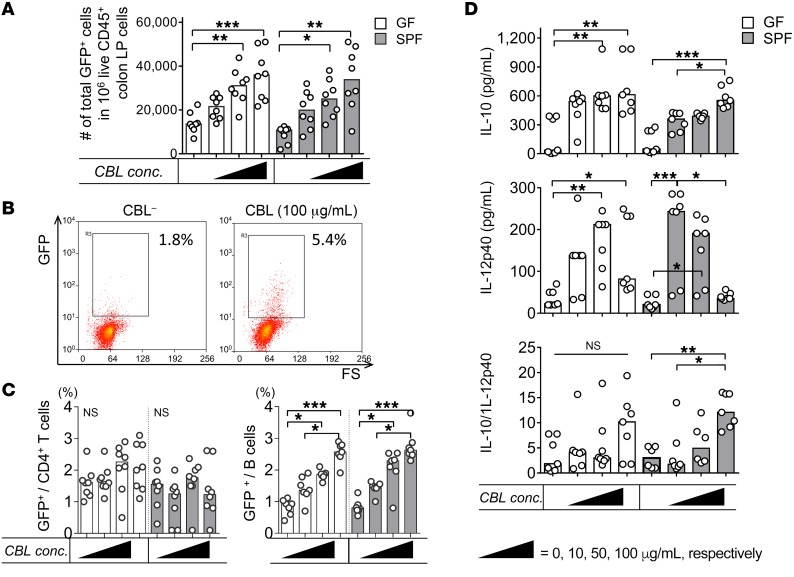
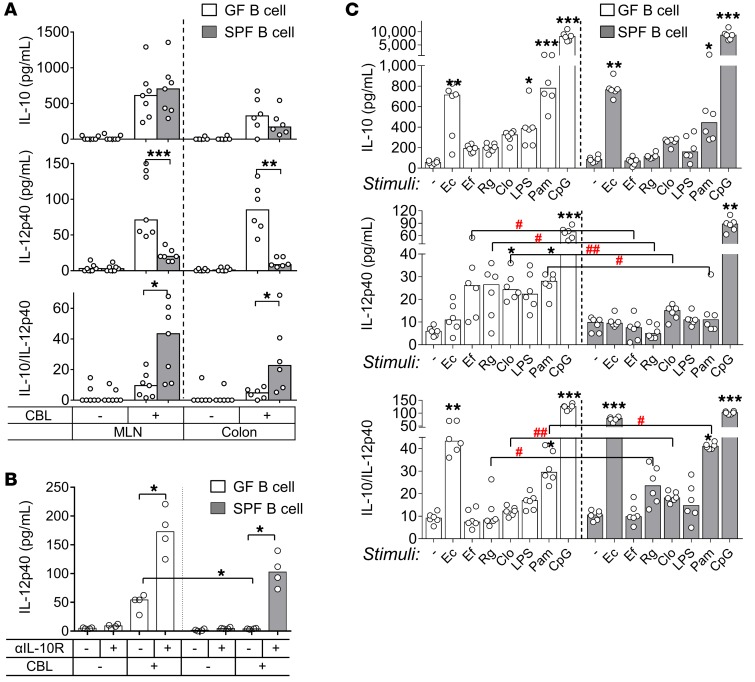
Having established that resident microbiota induces in vivo IL-10 expression in the colon and that endogenous IL-10 inhibits proinflammatory responses, we next characterized the IL-10–secreting immune cell types that were activated following ex vivo cecal bacterial lysate (CBL) treatment of cultured, unfractionated colon LP cells from GF versus SPF Il10+/EGFP mice. We have used CBL from SPF WT mice to replicate physiologic bacterial stimulation in previous ex vivo cellular activation studies (5, 21, 23). In the current experiments, ex vivo CBL stimulation significantly increased the percentage of GFP+ immune cells in a dose-dependent manner regardless of the source of cells (GF vs. SPF) (Figure 4, A and B). We observed a dose-dependent increase in numbers of GFP+ B cells, but not CD4+ T cells, following CBL stimulation (Figure 4C). IL-10 protein levels also increased in response to CBL stimulation, although the GF LP cells appeared to be more sensitive to low concentrations of CBL than did SPF cells, so that a dose-dependent response was not seen in GF cells. IL-10 protein secretion correlated strongly with the frequency of GFP+ B cells (r2 = 0.766, P < 0.0001). Levels of IL-12p40, a representative inflammatory cytokine that induces Th1/Th17 immune responses, in cells derived from SPF mice steadily decreased as CBL concentrations increased, but decreased only with the highest dose of CBL in GF cells. These results indicate a bacteria-mediated regulatory phenotype with elevated IL-10/IL-12p40 ratios in response to a high dose of CBL (Figure 4D).
Figure 4. CBL stimulates IL-10–producing colonic LP B cells ex vivo.
Unfractionated colonic LP cells from GF or SPF-reared Il10+/EGFP reporter mice were cultured with 0–100 μg/mL CBL from SPF mice for 2 days, as indicated. (A) The frequency of GFP+ cells was assessed by flow cytometry. conc., concentration. (B) Representative dot plots for GFP+ cells in GF colonic cells stimulated with 0 (CBL–), 10, 50, or 100 μg/mL CBL. (C) Using flow cytometry, we determined frequencies of GFP-expressing CD4+ T cells (CD4+CD3+) or B cells (CD19+B220+) as a percentage of total T or B cells using control WT (GFP–) cells that stained with the same antibodies as target samples. (D) Supernatant levels of IL-10 and IL-12p40 were measured by ELISA, and IL-10/IL12 p40 ratios were calculated. Data are presented as median of 8 separated cell cultures, with cells in each culture pooled from 2–4 mice, combined from 3 independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, Kruskal-Wallis test with Dunn’s post hoc test.
To examine the role of endogenous IL-10 in regulating in vitro induction of proinflammatory IL-12 by intestinal bacterial stimulation, we performed similar studies in cells from IL-10–deficient mice and in WT IL-10–sufficient mouse cells treated with neutralizing IL-10R to block IL-10 signaling. High-dose bacterial lysate–induced downregulation of IL-12p40 was not seen in LP cells from SPF Il10–/– mice (Supplemental Figure 6A), and levels of IL-12p40 in these mice were 2- to 3-fold greater than those found in WT cells stimulated by the lower doses of CBL (Figure 4D). These contrasting results in WT and Il10–/– cells suggest that IL-10 plays an important role in inducing the regulatory capacity of LP cells. This conclusion was confirmed by the observation that anti–IL-10R antibody administration significantly increased IL-12p40 production by WT LP cells stimulated ex vivo with high-dose CBL (Supplemental Figure 6B).
Bacterial TLR ligands selectively induce IL-10 production by LP innate immune cells.
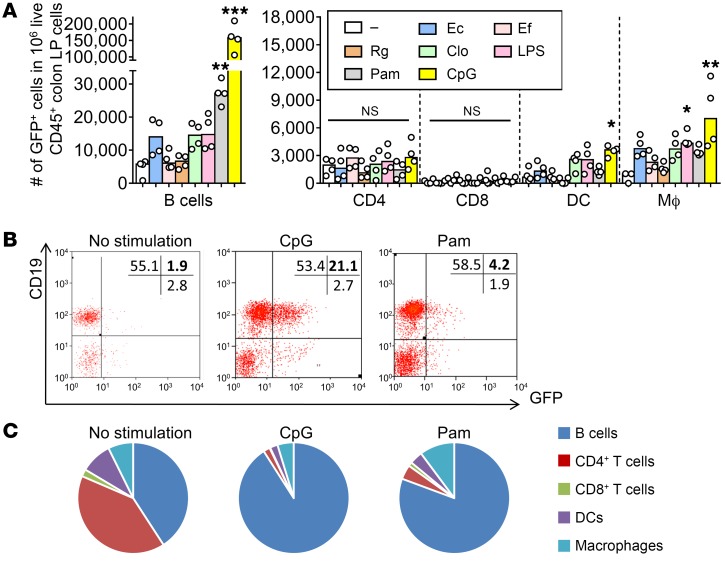
Having established that unfractionated bacterial microbiota induces IL-10 production by LP CD4+ T cells and B cells, we next sought to identify the bacterial components that induce IL-10 production by colonic LP cells. We performed in vitro stimulation using representative TLR ligands and lysates of representative classes of resident bacteria implicated in IBD pathogenesis and mucosal protection. We stimulated colonic LP cells from GF mice with (i) the TLR ligands Pam3csk4 (Pam3: TLR 1/2 agonist), LPS (TLR4 agonist), and CpG-DNA (TLR9 ligand); (ii) lysates from putative proinflammatory human bacterial species Escherichia coli LF82, Enterococcus faecalis, and Ruminococcus gnavus (more prevalent in patients with active IBD and inducers of experimental colitis in gnotobiotic IL-10–deficient mice; refs. 2, 36); and (iii) a mixture of 17 antiinflammatory strains of human Clostridium species that protect against experimental colitis (8) and are decreased in IBD patients (3, 37). We detected increased frequencies of GFP+ colon LP B cells in cultures that were treated with Pam3 or CpG-DNA (Figure 5, A and B). Frequencies of GFP+ macrophages and DCs also increased following ex vivo stimulation with CPG-DNA, but the number of these cells was lower than that of IL-10–producing B cells by 10- to 25-fold (Figure 5C). These results are consistent with a higher level of secreted IL-10 following ex vivo stimulation of colonic LP cells with Pam3 and CPG (Supplemental Figure 6C). The frequencies of GFP+ CD4+ T cells were not significantly altered by any of the in vitro stimuli tested (Figure 5A). These data indicate that ex vivo stimulation with bacterial products preferentially induces increased numbers of immunoregulatory IL-10–producing B cells compared with CD4+ T cells. CD4+ T cells may require additional signals, longer activation times, or specific antigenic stimulation of interacting APCs that are more efficient in vivo to produce IL-10.
Figure 5. Bacterial products predominantly expand GFP+ B cells and innate immune cells ex vivo.
Unfractionated colonic LP cells from GF Il10+/EGFP reporter mice were cultured without (–, media only) or with 200 ng/mL LPS, 50 ng/mL Pam3csk (Pam), 1 nM CpG-DNA (CpG), 10 μg/mL lysates of E. coli LF82 (Ec), E. faecalis (Ef), or R. gnavus (Rg) or a mixture of 17 strains of Clostridia species (Clo). (A) Frequencies of GFP-expressing cell types were determined by flow cytometry using antibodies to cell surface markers as described in Methods. Data are presented as median of 4 separate cell cultures, with cells in each culture pooled from 2–4 mice; *P < 0.05, **P < 0.01, ***P < 0.001 (vs. no stimulation), Kruskal-Wallis test with Dunn’s post hoc test. (B) Representative dot plots for GFP (IL-10)+ B cells stimulated with or without CpG or Pam3. (C) Summary pie charts for cell populations expressing GFP.
In vivo exposure to resident microbiota programs protective B cell responses to ex vivo bacterial stimulation of IL-10 and inflammatory cytokine production.
Given that treatment with CBLs and selected TLR ligands increased the frequency of IL-10–producing B cells and IL-10 production in ex vivo cultures of LP cells isolated from GF mice, we next examined (i) whether in vivo exposure to resident microbiota influences innate immune responses to ex vivo bacterial products and (ii) whether these products differentially activate protective IL-10 and a key proinflammatory cytokine, IL-12p40. We stimulated colonic LP B cells from GF and SPF mice first with a physiologic colonic microbiota lysate, CBL, then with representative bacterial lysates and TLR ligands. We measured the relative induction of IL-10 and IL-12p40 secretion by ELISA. CBL-stimulated intestinal (MLN and colon LP) B cells produced abundant IL-10 ex vivo regardless of the source of cells (GF vs. SPF) (Figure 6A). However, IL-12p40 production by CBL-stimulated intestinal B cells from SPF mice was significantly lower than from B cells derived from GF mice, resulting in a more protective phenotype, as indicated by increased IL-10/IL-12p40 ratios in CBL-stimulated SPF B cells (Figure 6A). Blockade of IL-10 signaling with anti–IL-10R antibody increased IL-12p40 production in both SPF and GF colonic B cells, indicating that IL-10 signaling is essential for inducing a regulatory phenotype in mucosal B cells (Figure 6B). To assess the functional significance of physiologic bacterial lysate activation of regulatory B cells, we created B cell/T cell (CD4+) cocultures with and without CBL stimulation and measured T cell proliferation and effector cytokine production. Colonic B cells from SPF WT mice suppressed T cell proliferation and IFN-γ and IL-17a production in a B cell number–dependent manner with, but not without, CBL stimulation (Supplemental Figure 7, A–C). These results illustrate the importance of resident enteric microbiota and IL-10 signaling for the healthy development and function of intestinal regulatory B cells.
Figure 6. Enteric resident bacteria in mice increase the capacity of intestinal B cells to suppress ex vivo bacterial lysate–stimulated inflammatory cytokine production that is mediated by IL-10 signaling.
(A) Cytokine secretion by 10 μg/mL CBL-stimulated colonic LP B cells isolated from GF or SPF-raised WT mice in culture for 2 days. Cytokine concentrations were measured by ELISA. Data are presented as median of 6–7 separate cell cultures, with cells in each culture pooled from 3–4 mice. Mann-Whitney U test (GF vs. SPF). *P < 0.05, **P < 0.01, ***P < 0.001. (B) Colonic B cells from GF or SPF-reared WT mice were cultured with anti–IL-10R or isotype control antibodies in the presence or absence of 10 μg/mL CBL for 24 hours. Supernatant levels of IL-12p40 were measured by ELISA. Data are presented as median of 4 separate cell cultures, with cells in each culture pooled from 2–4 mice; *P < 0.05, Kruskal-Wallis test with Dunn’s post hoc test. (C) Cytokine secretion by MLN B cells from GF or SPF-raised WT mice cultured for 2 days with the indicated bacteria lysates or TLR ligands (see Figure 5 legend). IL-10 and IL-12p40 in culture supernatants were measured by ELISA. Data are presented as median of 4 separate cell cultures, with cells in each culture pooled from 3–6 mice. *P < 0.05, **P < 0.01, ***P < 0.001 (vs. no stimulation), Kruskal-Wallis test with Dunn’s post hoc test; #P < 0.05, ##P < 0.01 (GF vs. SPF), Mann-Whitney U test.
To identify the specific bacterial species and components that stimulate B cells to secrete IL-10, we treated MLN B cells from GF versus SPF mice with the spectrum of bacterial species lysates and TLR ligands described in the experiments shown in Figure 5. Both GF and SPF MLN B cells secreted abundant IL-10 in response to Pam3, CpG-DNA, and E. coli LF82 lysate (Figure 6C). However, while secretion of IL-10 by stimulated MLN B cells was similar in GF and SPF mice, MLN B cells from SPF-raised animals produced less IL-12p40 compared with GF mice when treated with lysates of E. faecalis, R. gnavus, and Clostridium species, and with Pam3 but not CpG-DNA. IL-10/IL-12p40 ratios were also higher in cultures of cells from SPF versus GF mice for R. gnavus, Clostridia, and Pam3 stimuli (Figure 6C). These observations are similar to those in CBL-stimulated MLN B cells (Figure 6A). In aggregate, these results demonstrate that intestinal B cells acquire a strong regulatory (tolerogenic) phenotype following in vivo exposure to complex resident bacteria that is maintained by continuous ex vivo stimulation with TLR2 and -9 ligands.
IL-10–secreting B cells are induced and inhibit colitis in a TLR2- and MyD88-dependent manner.
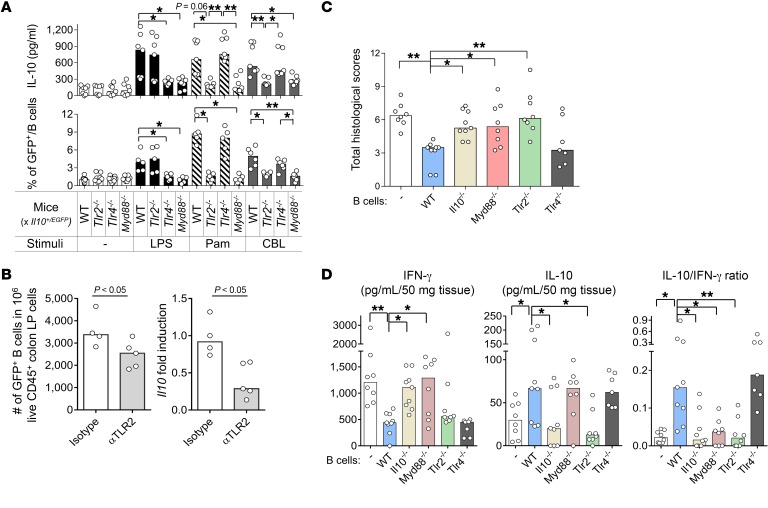
Based on our observations that in vitro exposure of resident microbiota and bacterial TLR ligands induces IL-10–secreting mucosal cells, including B cells with a regulatory phenotype, we next sought to determine (i) whether TLR signaling helps develop physiological or bacteria-induced IL-10–secreting mucosal cells; and (ii) whether this pathway mediates mucosal protection by regulatory B cells. Based on our observations that a TLR2 ligand consistently activated IL-10 production, we created SPF Il10+/EGFP Tlr2–/–, Il10+/EGFP Tlr4–/–, and Il10+/EGFP Myd88–/– double mutants to enable in vivo and ex vivo functional studies. We used TLR4 as a thoroughly investigated parallel pathway for LPS- and Gram-negative bacteria–induced activation and MyD88 as a common TLR2 and TLR4 downstream signaling molecule. We first quantified IL-10–secreting cells in the colon LP of these mice in vivo and after ex vivo stimulation with the TLR ligands LPS, Pam3, and CBL. We found that the absence of TLR2, TLR4, or MyD88 signaling did not significantly alter either the percentage of colonic LP GFP+ B cells or basal secretion of IL-10 by cultured LP B cells without ex vivo stimulation (Figure 7A and Supplemental Figure 8A). However, ex vivo CBL stimulation of cultured colonic LP cells significantly increased IL-10 secretion and the proportion of GFP+ B cells in a TLR2/MyD88-dependent, but TLR4-independent, manner (Figure 7A and Supplemental Figure 8A). As expected, LPS and Pam3 stimulation of cultured colonic LP cells increased IL-10 secretion and the proportion of GFP+ B cells in a TLR4/MyD88-dependent and TLR2/MyD88-dependent manner, respectively (Figure 7A). As in other experiments, the number of GFP+ B cells strongly correlated with IL-10 protein production (r2 = 0.724, P < 0.0001). We then verified that B cells are a primary source of IL-10 production following ex vivo intestinal bacterial stimulation in the absence of other cell populations (Supplemental Figure 9). These data suggest that in Tlr2–/– or Myd88–/– mice, TLR2/MyD88-dependent IL-10 production by B cells is not due to abnormal immune activation by other cell types, for example, aberrant T cell activation. On the other hand, CBL-stimulated IL-12p40 production by unfractionated colonic LP cells was not altered by lack of TLR signaling (Supplemental Figure 8B), likely due to predominant IL-12p40 production by non-B cell populations (i.e., macrophages and DCs) by other stimulatory pathways. In aggregate, these results demonstrate that global resident bacterial induction of IL-10–secreting B cells ex vivo is TLR2 and MyD88 dependent. In addition, in vivo blockade of TLR2 by anti-TLR2 antibody in GF Il10+/EGFP reporter mice during colonization with SPF bacteria resulted in diminished numbers of colonic GFP+ B cells and decreased colonic Il10 mRNA expression (Figure 7B). These results emphasize the importance of TLR2 signaling in enteric bacteria–mediated, intestinal IL-10–producing B cell development and activation.
Figure 7. TLR2/MyD88 signaling increases the frequency of IL-10–producing B cells upon bacterial product stimulation ex vivo and mediates the suppression of experimental colitis by IL-10–producing B cells in vivo.
(A) Colonic LP B cells from 8- to 10-week-old WT Il10+/EGFP, Tlr2−/− Il10+/EGFP, Tlr4−/− Il10+/EGFP, and Myd88−/− Il10+/EGFP mice were cultured without (–) or with 10 μg/mL CBL, 50 ng/mL Pam3, or 200 ng/mL LPS for 24 hours, after which IL-10 levels in culture supernatants were measured by ELISA (top panel). GFP+ populations in B cells (live CD45+CD19+B220+) were analyzed with flow cytometry in reference to WT (GFP–) control cells stained with the same antibodies as target samples. Data are presented as median of 7–8 separate cell cultures, with cells in each culture pooled from 2–3 mice, combined from 3 independent experiments (bottom panel). *P < 0.05, **P < 0.01. (B) Eight-week-old GF Il10+/EGFP reporter mice were conventionalized with SPF fecal bacterial transplantation. Mice were given anti-TLR2 antibody (αTLR2) or isotype control i.v. on days 0 and 4 after fecal bacterial transplantation and harvested on day 7 for IL-10 analysis. Colonic LP cells were isolated, and live CD45+GFP+ B cells (CD19+B220+) were analyzed by flow cytometry (left) as described above; Il10 mRNA levels were normalized with Actb (right). n = 4–5 mice. Data are presented as median, Mann-Whitney U test. (C and D) Splenic B cells (1 × 106) from SPF-raised WT, Tlr2−/−, Tlr4−/−, Myd88−/−, or Il10−/− mice were cotransferred with 5 × 105 WT naive CD4+ T cells isolated by a naive CD4+ T cell isolation kit (see Methods, Cell purification) into SPF Rag2−/− Il10−/− recipients. Six weeks after cell transfer, mice were evaluated for (C) severity of colitis by histology and (D) measurement of spontaneously secreted IL-10 and IFN-γ in colonic tissue explants by ELISA. n = 7–9 mice/group, combined from 2 independent experiments. Data are presented as median; *P < 0.05 and **P < 0.01 (vs. WT mice), Kruskal-Wallis test with Dunn’s post hoc test.
Based on these results showing TLR2/MyD88-dependent induction of IL-10–producing B cells in vivo and in vitro, we examined the regulatory function of bacteria-induced IL-10–secreting B cells in vivo using our established CD4+ T and B cell cotransfer colitis model (23). We cotransferred splenic naive CD4+ T cells from WT mice with or without splenic B cells isolated from SPF-raised WT, Tlr2–/–, Tlr4–/–, Il10–/–, and Myd88–/– mice into Il10–/– Rag2–/– mice, and evaluated the severity of colitis 6 weeks after cell transfer. Transferred WT and Tlr4–/– B cells substantially suppressed histological evidence of colonic inflammation (Figure 7C and Supplemental Figure 10) and were associated with lower spontaneous IFN-γ secretion, higher spontaneous IL-10 secretion, and elevated IL-10/IFN-γ ratios by colonic explants (Figure 7D). On the other hand, B cells lacking IL-10, TLR2, or MyD88 exhibited no protective function (Figure 7, C and D, and Supplemental Figure 10), although transferred Tlr2–/– B cells were present in the colon (data not shown). Based on these results in the T and B cell cotransfer colitis model, we propose that mucosal IL-10–secreting B cells protect against chronic colitis in a microbiota-dependent manner through selective TLR signaling.
TLR2-specific CBL activation in B cells activates the PI3K/AKT/glycogen synthase kinase 3β signaling pathway.
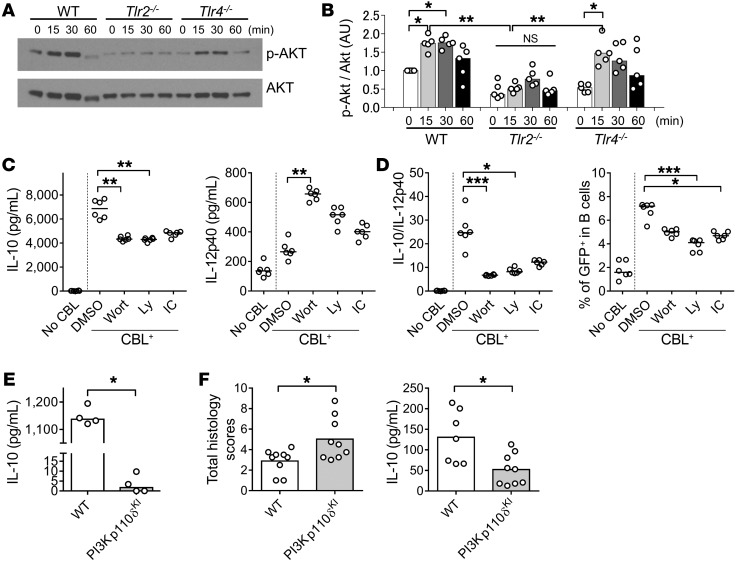
Since colonization with resident microbiota or ex vivo stimulation with CBL induces IL-10 production by colonic B cells with a regulatory phenotype in a TLR2-dependent fashion, we next explored downstream signaling pathways by measuring activation of signaling components by Western blotting and selective pharmacologic and genetic blockade of key signaling molecules. We first investigated the TLR2-specific activation of the PI3K/AKT pathway, which has been found to facilitate IL-10 production (31, 38, 39). We cultured colonic LP B cells from SPF WT, Tlr2–/–, and Tlr4–/– mice with CBL, and measured phosphorylation of AKT, a key downstream molecule in thePI3K pathway. We found that phosphorylation of AKT was activated to a significantly lesser extent by CBL in Tlr2–/– B cells compared with WT and Tlr4–/– B cells (Figure 8, A and B). We also observed that the PI3K pathway–related molecules PDK and glycogen synthase kinase 3β (GSK3β), but not p85, were activated in a TLR2-dependent manner, while the Myd88 signaling–associated molecules p38 and ERK were not dependent on TLR2 activation (Supplemental Figure 11, A and B). These results indicate that CBL stimulation preferentially activates the PI3K/AKT/GSK3β signaling pathway downstream of TLR2 in B cells. To confirm these findings functionally, we performed targeted inhibitor studies using CBL-stimulated B cell production of IL-10 and IL-12p40 (Figure 8C). We found that PI3K, including the PI3Kp110δ subunit, was involved in CBL-stimulated IL-10 production by B cells and regulatory function characterized by an increased IL-10/IL-12p40 ratio (Figure 8C). Increased CBL-stimulated GFP+ B cell frequency was also inhibited by pan-PI3K and PI3Kp110δ-specific inhibitors (Figure 8D and Supplemental Figure 12). PI3K110δKI B cells were neither able to produce abundant IL-10 following in vitro CBL stimulation (Figure 8E) nor ameliorate T cell–mediated colitis in the CD4+ T and B cell cotransfer model (Figure 8F). Together, our in vivo and in vitro studies indicate that protective IL-10 production by B cells is activated by CBL through TLR2 and downstream signaling through the PI3K (p110δ subunit) pathway.
Figure 8. The PI3K/AKT pathway is involved in TLR2-dependent IL-10 production by CBL-stimulated B cells and their regulatory effect on T cell–mediated colitis.
Splenic B cells (1 × 106/well) from 8- to 10-week-old SPF WT, Tlr2−/−, and Tlr4−/− mice were cultured without or with 10 μg/mL CBL. Cells were harvested 0, 15, 30, and 60 minutes after CBL stimulation, and phosphorylation of AKT was analyzed by Western blot. (A) Representative blots and (B) densitometric analysis of phospho-AKT/AKT are shown. n = 5 mice/group, combined from 2 independent experiments. *P < 0.05, **P < 0.01, Kruskal-Wallis test with Dunn’s post hoc test. (C) Splenic B cells (1 × 106/well) from 8- to 10-week-old SPF Il10+/EGFP mice were cultured with 1 μM pan-PI3K inhibitor wortmannin (Wort) or Ly294002 (Ly), 1 μM PI3Kp110δ-specific inhibitor IC-87114 (IC), or DMSO alone, with stimulation with 10 μg/mL CBL for 48 hours. Supernatant levels of IL-10 and IL-12p40 were measured by ELISA. (D) GFP+ population in B cells (live CD45+CD19+B220+) were analyzed by flow cytometry in reference to WT (GFP–) control cells stained with the same antibodies as target samples. Data are presented as median of 6 separate cell cultures, combined from 2 independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, Kruskal-Wallis test with Dunn’s post hoc test. (E) Splenic B cells (5 × 105/well) from 8- to 10-week-old SPF WT and PI3Kp110δKI mice were cultured with 10 μg/mL CBL for 48 hours. Supernatant levels of IL-10 were measured by ELISA. Data are presented as median. n = 4 mice, combined from 2 independent experiments. *P < 0.05, Mann-Whitney U test. (F) Splenic B cells (1 × 106) from SPF-raised WT or PI3Kp110δKI mice were cotransferred with WT naive CD4+ T cells (5 × 105) isolated by a naive CD4+ T cell isolation kit into SPF Rag2−/− Il10−/− recipients. Six weeks after cell transfer, mice were evaluated for severity of colitis by histology and measurement of spontaneously secreted IL-10 levels in colonic tissue explants by ELISA. n =7–9 mice/ group, combined from 2 independent experiments. Data are presented as median; *P < 0.05, Mann-Whitney U test.
Discussion
In this study, we describe the kinetics of in vivo development of IL-10–secreting intestinal B cells and subsets of CD4+ T cells in ex-GF Il10+/EGFP reporter mice colonized with enteric resident microbiota and the importance of TLR2 and PI3K signaling in the induction of protective resident bacteria–induced IL-10–producing B cells.
The consortium of resident enteric bacteria provides protective signals to host immune defenses (40–42). Colonization of GF WT mice with resident fecal microbiota rapidly stimulated mucosal proinflammatory cytokine production manifested by transient release of TNF-α, IL-12p40, and IFN-γ, but not IL-17a, within 3 days, while intestinal IL-10 levels gradually increased to maximum sustained levels in 7 days (43). The finding that transient in vivo stimulation of proinflammatory cytokine gene expression was followed by downregulation to basal levels in ex-GF Il10+/+ but not in Il10–/– mice advances the hypothesis that resident microbiota induces IL-10 production by LP immune cells that mediates mucosal homeostasis. That neutralization of IL-10 signaling in B cells by anti–IL-10R antibody potentiated physiologic bacterial stimulation of IL-12p40 further supports our hypothesis. These results are consistent with our previous work showing that in vivo administration of anti–IL-10R antibody prevented downregulation of ex vivo bacterial antigen–stimulated Th1 immune responses in ex-GF mice colonized with enteric bacteria (13).
Based on our current and previous observations, we propose that induction of IL-10 by enteric resident bacteria is essential for maintaining intestinal mucosal homeostasis. Our studies included several features that we believe to be novel: the use of GF Il10+/EGFP reporter (Vert-X) mice to study the in vivo kinetics of IL-10 activation after colonization with resident enteric bacteria; identification of the colonic LP cell types that produce IL-10 following microbial colonization; and the study of the signaling pathways of IL-10 induction by physiologic resident bacterial stimulation. Our use of GFP+ cells as indicators of cellular IL-10 production was validated by the strong correlations we found between the frequency of GFP+ cells and both IL-10 RNA transcription and IL-10 protein secretion following bacterial stimulation. Previous studies of systemic IL-10–secreting immune cells have relied on nonphysiologic, ex vivo stimulation of cultured immune cells (i.e., anti-CD40, anti-CD3/CD28, or concanavalin A [ConA]) or on mice with septic, infectious, or autoimmune disease (32). Cells isolated from normal mice typically require prestimulation with nonspecific agents such as PMA/ionomycin to detect and analyze cell-specific IL-10 secretion by flow cytometry (44). Our physiologic approach using GF Il10+EGFP reporter mice allowed us, without additional nonphysiologic stimulation, to more accurately detect and quantify the effects of complex intestinal microbiota lysates, specific bacterial species, and defined bacterial products on IL-10–producing cells in vitro; and the effects of in vivo physiologic colonization by complex resident microbiota on IL-10–producing cells in different mouse tissues over time.
Our somewhat unexpected observation that bacterial colonization of GF mice increased the frequency of colonic IL-10–producing B cells, in addition to subsets of CD4+ T cells, led us to investigate the roles of IL-10–producing B cells in maintaining mucosal homeostasis and the signaling pathways involved in IL-10 induction by resident microbiota (23–28). Our results and observations by others suggest that bacterial stimulation through the MyD88 pathway activates Il10 expression and causes B cells to produce IL-10, which helps maintain gut homeostasis. B cell–specific Myd88–/– mice are more susceptible to dextran sulfate sodium–induced mucosal damage compared with mice lacking MyD88 in other cell types, including T cells, macrophages, DCs, and epithelium (45). In the present study, we demonstrated that ex vivo activation of the TLR2/MyD88 pathway is essential for induction of bacteria-activated mucosal IL-10–producing B cells, but not T cells. However, the absence of TLR signaling through MyD88 did not alter the relative number of mucosal IL-10–producing B cells in SPF-raised animals in vivo, indicating that physiologic development of intestinal IL-10–producing B cells is TLR independent, although in vivo protection against colitis by B cells is IL-10, TLR2, and MyD88 dependent. We postulate that basal activation of in vivo IL-10–producing B cells and ex vivo bacteria-stimulated IL-10–secreting B cells likely requires different pathways in response to actively metabolizing resident intestinal bacteria, or that this activation depends on complex interactions with cell types not represented in our in vitro cultures. Similar disparities exist in the regulation of IL-10–producing CD4+ cells, which increased in frequency following colonization of GF mice with SPF microbiota but are not activated ex vivo by physiologic cecal microbial lysates. Resolving these disparities will require further mechanistic studies.
We showed that upstream of MyD88 signaling, TLR2, but not TLR4, signaling increased the numbers of bacterially activated GFP+ B cells and B cell IL-10 secretion, which contributed to B cell attenuation of T cell–mediated colitis. These TLR2 signaling findings represent what we believe to be a novel molecular mechanism of bacteria-stimulated, intestinal IL-10–producing B cells. Other investigators have shown that certain subsets of resident enteric bacteria induce regulatory immune responses, including IL-10 secretion, via TLR2 signaling (9, 46–49). Likewise, cecal luminal lysates and SPF bacteria appeared to preferentially activate mucosal IL-10–producing regulatory immune cells through TLR2. An important mechanistic observation from our study is that TLR2-mediated CBL stimulation of B cell production of IL-10 is mediated through activation of PI3K/AKT/GSK3β signaling. Furthermore, we demonstrated that WT but not PI3K signaling–deficient B cells attenuated colitis in a T and B cell cotransfer CD4+ colitis model, specifically through the p110δ subunit. Similarly, TLR2-dependent PI3K signaling has been reported to mediate phagocytosis of pathogens by macrophages, and PI3Kp110δ regulates APC activation of Th1/Th17 cells and colitis (39, 50).
We demonstrated that in vivo microbial activation of colonic B cells and continued exposure to bacterial components was important for their homeostatic function. We observed that with ex vivo bacterial activation, GF mucosal B cells, compared with SPF mucosal B cells, produced equal amounts of IL-10, but higher levels of the inflammatory cytokine IL-12p40. These results show that in vivo exposure to resident microbiota enhances the regulatory capacity of intestinal B cells by selectively downregulating production of bacteria-stimulated inflammatory cytokines, but not IL-10. We previously demonstrated that with in vivo bacterial exposure, transferred GF WT B cells acquire regulatory phenotypes that help ameliorate colitis, with effects similar to those of B cells derived from SPF WT mice (23). In the current study, we showed that SPF B cells were unable to suppress effector T cell function in vitro without continued CBL exposure. These observations indicate that continuous bacterial stimulation is required in order to maintain regulatory B cells. Others have shown that bacterial colonization reduces the IL-10–mediated inflammatory hyporesponsiveness present in GF animals and makes them less susceptible to infection (40). We further demonstrated that in vivo physiologic intestinal microbial colonization selectively activated IL-10 production by subsets of intestinal LP T and B cells, with no concomitant stimulation of IL-10–producing cells in MLN and splenic cells in the same mice. These findings are consistent with the lack of differences in the number or function of IL-10–producing peritoneal regulatory B cells in GF versus SPF mice (28). The results emphasize the important site-specific interactions between the microbiota and intestinal regulatory B cells in mediating mucosal homeostasis.
We also explored the ability of the microbiota to induce T cell production of IL-10. Bacterial colonization of GF mice markedly increases intestinal GFP+ Tr1 cells (Foxp3negCD25–CD4+ T cells) and RORγt+Foxp3+GFP+CD4+ T cells (33, 34, 51), which made up only a small portion of the total number of GFP+CD4+ T cells. In contrast, ex vivo intestinal bacterial stimulation of immune cells predominantly increased GFP+ IL-10–producing B cells but not CD4+ T cells. The lack of TLR/MyD88 signaling did not affect the frequency or activity of mucosal Tr1 cells either in vivo or ex vivo. These results suggest that resident enteric bacteria–stimulated Tr1 cell induction is TLR/MyD88 independent. Why different pathways are required for activation of IL-10–producing B and Tr1 cells is not entirely clear. CD4+ IL-10–producing T cells may require physiologic stimulation by live bacterial antigens or metabolites, such as butyrate (52), rather than bacterial lysates; longer incubation periods with other signals not present ex vivo, or interactions with epithelial or mesenchymal cells. Cong and colleagues demonstrated that IL-10–secreting Tr1 cells from SPF mice were expanded by CBL antigen-pulsed APCs in vitro and that a CBL-responsive Tr1 clone could inhibit colitis in a T cell cotransfer model (53). We recently showed that Tr1 cells were induced by IL-10–producing B cells ex vivo in the presence of CBL when equal numbers of APCs and naive T cells were cocultured, a ratio considerably higher than the ratio of APCs to T cells in the intestinal LP (23). In these ex vivo coculture studies, B cells were the primary source of secreted IL-10. Our results suggest that ex vivo activation of IL-10 by T cells requires boosted antigen presentation by greater than physiologic numbers of APCs.
In summary, complex resident enteric microbiota selectively induced colonic LP IL-10–producing B cells and subsets of CD4+ T cells, and increased the regulatory capacity of intestinal B cells. Fecal microbiota colonization of GF mice transiently activated inflammatory cytokines in the colon, which were subsequently downregulated by sustained IL-10 production. These inflammatory cytokines progressively increased from a higher baseline in IL-10–deficient mice, which develop colitis dependent on bacterial colonization. Resident bacteria stimulated IL-10 production through TLR2, MyD88, and PI3Kp110δ signaling, which mediated B cell antiinflammatory responses that, in turn, attenuated T cell–mediated colitis. We conclude that resident enteric bacteria activate IL-10–producing intestinal B cells in a MyD88/TLR2/PI3K-dependent manner that suppresses aggressive immune responses and maintains mucosal homeostasis.
Methods
Further information can be found in Supplemental Methods.
Mice.
C57BL/6 WT, Il10−/−, Rag2–/–, Tlr2−/−, Tlr4–/–, and Myd88–/– mice were purchased from the Jackson Laboratory. Il10+/EGFP reporter (Vert-X) mice have been described previously (32). Pi3kp110δD910A/D910A (PI3Kp110δKI) mice were previously obtained from B. Vanhaesebroeck (Queen Mary University of London, London, United Kingdom) (39). Il10−/−Rag2−/− double-knockout (DKO) mice were generated by crossing Il10−/− with Rag2–/– mice. Tlr2−/− Il10+/EGFP, Tlr4−/− Il10+/EGFP, and Myd88−/− Il10+/EGFP mice were generated by crossing Tlr2−/− with Il10+/EGFP mice, Tlr4−/− with Il10+/EGFP mice, and Myd88−/− with Il10+/EGFP mice, respectively. These mice were maintained in SPF conditions at UNC and were used at 8–12 weeks of age. Il10+/EGFP, WT, and Il10–/– mice were derived into GF conditions by embryo transfer and maintained in the UNC Center for Gastrointestinal Biology and Disease Gnotobiotic Core. In some experiments, GF mice were inoculated with feces from SPF WT mice.
Cell isolation.
Mononuclear cells were isolated from colonic LP, MLN, and spleen as described previously (24). MLNs were crushed through 70-μm filters into PBS with 2.5% FBS (Gibco, Invitrogen). Spleens were mechanically dissociated, and red blood cells were lysed with Red Blood Cell Lysing Buffer (Sigma-Aldrich). For isolation of colonic LP lymphocytes (LPLs), colons were washed with cold PBS, opened longitudinally, and cut into 10-mm pieces. Intestinal fragments were incubated in 1 mM DTT (Sigma-Aldrich) in Ca2+- and Mg2+-free HBSS (Gibco, Invitrogen) for 15 minutes at room temperature. Next, the tissues were incubated in 1 mM EDTA (Sigma-Aldrich) in HBSS for 20 minutes at 37°C with shaking, which was repeated after a thorough washing. The cell suspensions were removed, and remaining fragments were transferred to flasks containing HBSS with 1 mg/mL collagenase type III (Sigma-Aldrich) and 1% penicillin-streptomycin (Gibco, Invitrogen), then stirred gently for 60 minutes at 37°C. Cell suspensions containing LPLs were filtered through a nylon mesh and centrifuged; then the LPLs were purified using a 44%/70% discontinuous Percoll gradient (GE Healthcare). After centrifugation at 800 g for 20 minutes at 22°C, mononuclear cells were collected from the interface.
Cell purification.
MLN or colon LP B cells were purified magnetically by positive selection with anti-CD19 microbeads after negative selection using a mixture of anti-CD90.2, anti-CD11c, and anti-Ter119 microbeads. Final CD19+ cell fractions were confirmed to be greater than 99.5% pure by flow cytometry, while cell viability was shown to be greater than 88% by eosin Y exclusion. In some experiments, colon LP cells were separated into B cell and non-B cell populations with anti-CD19 MicroBeads. Naive CD4+ T cells were isolated by use of a Naive CD4+ T Cell Isolation Kit, Mouse, and confirmed to be greater than 94.7% pure and 95% viable. All microbeads including the naive CD4+ T cell isolation kit used in this study were purchased from Miltenyi Biotec.
Bacterial lysate.
CBL was prepared from the contents of ceca from SPF-raised C57BL/6 WT mice as described previously (21). Lysates of each bacteria strain were prepared from individual colonies of E. coli LF82, E. faecalis, and R. gnavus, as previously described (36). A lysate of Clostridia was prepared from the cecal contents of ex-GF mice selectively colonized with a mixture of 17 strains of Clostridium species (8).
Cell cultures.
Unfractionated MLN cells (1 × 106) or colon LP cells isolated from GF or SPF-raised Il10+/EGFP mice were cultured in 200 μl medium/well (96-well plates) for 24–72 hours at 37°C with 5% CO2. The culture medium was RPMI 1640 (Gibco, Invitrogen) containing 10% FBS, 1% penicillin-streptomycin-amphotericin B (Gibco, Invitrogen), and 5 × 10–5 mol/L 2-mercaptoethanol (Sigma-Aldrich) with or without the following bacterial products; 10, 50, or 100 μg/mL CBL; 10 μg/mL lysate of Clostridia, E. coli LF82, E. faecalis, or R. gnavus; 200 ng/mL LPS, and 50 ng/mL Pam3CSK4 (InvivoGen) or 1 nM CpG-DNA (Sigma-Aldrich). In some experiments, 1 μM of the pan-PI3K inhibitor wortmannin (Sigma-Aldrich) or Ly294002 (Sigma-Aldrich); 1 μM of the PI3Kp110δ-specific inhibitor IC-87114 (Sigma-Aldrich); or vehicle (DMSO) alone was added at various concentrations indicated in the Figure legends. Following cell culture, the supernatants were collected for measurements of cytokines by ELISA, while cells were analyzed by flow cytometry.
Cytokine measurements.
IFN-γ, IL-12/23p40, or IL-10 ELISAs were performed by the Immunoassay Core of the UNC Center for Gastrointestinal Biology and Disease using the following products: mouse anti–IL-10, –IL-12/23p40, and –IFN-γ (BD Biosciences) as previously described (21, 23). Concentrations of cytokines were established in triplicate culture supernatants by comparison with standard curves generated using the appropriate recombinant cytokine per the manufacturer’s instructions.
Real-time PCR.
Total RNA was isolated from distal colon using the RNeasy Micro Kit (QIAGEN). First-strand complementary DNA was synthesized from 1 μg total RNA using M-MLV Reverse Transcriptase (Gibco, Invitrogen) according to the manufacturer’s instructions. Quantitative reverse transcription PCR was performed on a Mastercycler ep realplex 2S (Eppendorf) using the SYBR Green quantitative PCR SuperMix (Invitrogen) to quantify gene expression. The PCR primers used in this study were reported previously (21, 23, 54). Each complementary DNA sample was analyzed in duplicate or triplicate for quantitative assessment of RNA amplification, and the results are expressed relative to the housekeeping gene β-actin.
Immunohistochemistry.
Distal colons were harvested, fixed with 4% paraformaldehyde (Sigma-Aldrich), and embedded in O.C.T. compound (Sakura). Tissue sections (5 μm) were blocked with TNB buffer (PerkinElmer) for 30 minutes at room temperature and incubated overnight at 4°C with rabbit anti-GFP antibody (Invitrogen) and rat anti–mouse B220 or Armenian hamster anti–mouse CD3 antibody (BD Biosciences). Rabbit IgG (Invitrogen) was used as a negative control for rabbit anti-GFP antibody. Primary antibodies are listed in Supplemental Table 1. After washing, tissue sections were treated with DyLight 488–conjugated donkey anti-rabbit IgG and HRP-conjugated donkey anti-rat IgG or HRP-conjugated donkey anti-hamster IgG (Jackson ImmunoResearch Laboratories Inc.). The sections were finally incubated with TSA Plus Cyanine 5 amplification reagent (PerkinElmer) for 10 minutes at room temperature and counterstained with DAPI (Sigma-Aldrich). All sections were analyzed by confocal microscopy (TCS SP2, Leica).
Western blots.
Western blots were performed as previously reported (55). One centimeter of distal colons was homogenized in 1% RIPA lysis buffer (Boston Bio Products) containing cOmplete Protease Inhibitor Tablet (Roche) and PhosSTOP Phosphatase Inhibitor Tablet (Roche) using a Pro200 tissue homogenizer (PRO Scientific). Lysates were incubated on ice for an additional 10 minutes at 4°C with rotation. The lysates were cleared of cellular debris by centrifugation, and total protein concentration was determined by BCA assay (Thermo Fisher Scientific). A total of 30 μg protein from each sample was separated by SDS-PAGE using 4%–12% NuPAGE Bis-Tris gels (Invitrogen). The separated proteins were transferred onto 0.2-μm nitrocellulose membranes and were blocked for 1 hour at room temperature with TBS-T (10 mM Tris, 7.5 pH, 150 mM NaCl, and 0.1% Tween-20) containing 10% nonfat dry milk. The membranes were incubated overnight with anti–phospho-STAT3 (Tyr705; Cell Signaling Technology) and then washed 5 times with TBS-T and incubated for 2 hours at room temperature with HRP-conjugated goat anti-rabbit IgG secondary antibodies (Sigma-Aldrich) (1:40,000 dilution). The membranes were developed using SuperSignal West Chemiluminescent Substrate (Thermo Fisher Scientific) and were visualized by exposure to blue sensitive X-ray film (Genesee Scientific). To confirm even loading, the antibodies were removed from the membranes using Restore PLUS Western Blot Stripping buffer (Thermo Fisher Scientific), and membranes were blocked as previously described (55) and incubated with anti-STAT3 (Cell Signaling Technology) or anti-actin–HRP (Santa Cruz Biotechnology) (1:10,000 dilution), followed by HRP-conjugated goat anti-rabbit IgG secondary antibodies when appropriate.
Following in vitro stimulation of primary B cells with CBL, cells were washed with PBS at 4°C, pelleted by centrifugation at 400 g for 5 minutes at 4°C, and lysed in 1% RIPA buffer containing Complete Protease Inhibitor Tablet and PhosSTOP Phosphatase Inhibitor Tablet for 10 minutes on ice. Protein lysates were cleared of cellular debris by centrifugation, and the lysates were subjected to SDS-PAGE and Western blot as described above in Western blots using anti–phospho–Akt (Ser473) antibodies (Cell Signaling Technology; 1:1,000 dilution), followed by HRP-conjugated goat-anti-rabbit IgG secondary antibodies (Sigma-Aldrich) (1:40,000 dilution). Antibodies were removed from the membranes as described above, and even protein loading was confirmed using anti-Akt antibodies (Cell Signaling Technology; 1:1,000 dilution), followed by HRP-conjugated goat anti-rabbit IgG secondary antibodies (Sigma-Aldrich; 1:40,000 dilution).
Primary antibodies are listed in Supplemental Table 1. All antibodies were diluted in TBS-T containing 5% nonfat dry milk. Densitometric analysis was performed for all Western blots using ImageJ software (version 1.49; NIH).
Flow cytometry.
Spleen, MLN, and colonic LP cells were incubated for 15 minutes at 4°C with anti-CD16/CD32 (BD Biosciences) to block Fc receptors and then for 20 minutes at 4°C with 6–7 fluorochrome-conjugated antibodies (Supplemental Table 1) to evaluate the cell phenotype. All sample cells were stained with LIVE/DEAD Fixable Dead Cell Stain Kit (Near-IR, Thermo Fisher Scientific) and Pacific Orange–conjugated CD45 along with the following: PE, PE-Cy5, PE-Cy7, Pacific blue, allophycocyanin, or equivalent fluorescence-conjugated antibodies; B220 and CD19 for B cells; CD3 and CD4 for CD4+ T cells; CD3 and CD8 for CD8+ T cells; CD11b and CD11c for DCs; CD11b and F4/80 for macrophages; or CD3 and CD49b for NK cells. In addition, target antibodies or isotype control antibodies were simultaneously added. Enumeration of cells expressing Foxp3 or RORγt was performed by intracellular staining with the Fixation/Permeabilization Solution Kit according to the manufacturer’s instructions (BD Biosciences) or fixation buffer (PBS containing 4% paraformaldehyde; Electron Microscopy Sciences), 0.01% Tween-20 (Thermo Fisher Scientific), permeabilization buffer (PBS containing 0.1% Triton X-100; MP Biomedicals), 0.5% BSA (Sigma-Aldrich), and 2 mM EDTA. Cells were washed and then analyzed on a CyAn ADP flow cytometer (Beckman Coulter) or LSR II (BD Biosciences) at the UNC Flow Cytometry Core Facility. Cells from SPF-raised WT C57BL/6 (GFP–) mice were used to determine GFP basal intensity and set the GFP gate when Il10+/EGFP reporter cells were used. These WT cells were stained with the same antibodies as target cells except for GFP, as a fluorescence minus one (FMO) control to assure that spillover or spread from other fluorophores would not contribute to the signal in the GFP detector. Suitable positive control staining samples or IL-10 reporter cells were used for compensation. Seven parameters, including GFP channel, were used for running flow cytometry. Gated live CD45+ cells were analyzed with Summit 5.2 software (Beckman Coulter) or FlowJo version 10.3.
Transfer of CD4+ T and B cells.
DKO mice received i.p. injections of 5 × 105 splenic CD4+ cells with or without 1 × 106 splenic B cells from WT, Il10–/–, Myd88–/–, or PI3Kp110δKI donors. Cells were harvested 6 weeks after cell transfer, according to Association for Assessment of Laboratory Animal Care–approved (AALAC-approved) methods.
Evaluation of colitis.
Colitis severity was evaluated by histopathology scoring and cytokine ELISAs on culture media from colonic tissue explants. For histology, intestinal tissues were removed at necropsy and fixed in 10% buffered formalin. Paraffin sections were prepared and stained with H&E for assessment of colonic inflammation by the Histology Core of the UNC Center for Gastrointestinal Biology and Disease. Scoring of mucosal inflammation in cecum, proximal colon, and distal colon was performed in a blinded fashion, with each region being graded from 0 to 4 as described previously (56). Total histology scores (maximum score of 12) represent the summation of the scores from the 3 regions. For colonic tissue explant assays, cultures of colon fragments from the recipient mice were prepared as described previously (21).
Blockade of TLR2 signaling in vivo.
Anti-TLR2 antibody (BioLegend; 100 μg/mouse) or isotype control antibody (purified mouse IgG1, κ; BioLegend) was injected i.p. on days 0 and 4 after fecal transplantation with SPF feces. Mice were harvested on day 7, and we evaluated colonic GFP+ B cells by flow cytometry, as well as mRNA expression of Il10 in distal colon.
Statistics.
We used Prism 6 software (GraphPad) to compare medians between 2 groups with the Mann-Whitney U test; comparisons of means from multiple groups were analyzed with the Kruskal-Wallis test with Dunn’s post hoc test. P values less than 0.05 were considered significant.
Study approval.
These studies were reviewed and approved by the UNC IACUC, protocol 12-300.0.
Author contributions
YM and RBS conceptualized and designed the study; acquired, analyzed, and interpreted data; performed statistical analyses; and wrote the manuscript. AO, BL, JWH, CSE, TJF, EBS, IMC, LC, JEW, NCF, TN, AW, and JVG acquired, analyzed, and interpreted data. JJH and JPYT drafted the manuscript and provided administrative assistance. CLK provided reporter mice and technical/conceptual advice.
Supplementary Material
Acknowledgments
These studies were supported by NIH grants P01 DK094779, R01 DK53347, P40 OD010995, and P30 DK34987 (to RBS); R01 CA156330 (to JPYT); R01 AI123010 (to AW); and R01 AI111899, AI140799, and MH108179 (to JVG); and by Crohn’s and Colitis Foundation Microbiome Initiative and Gnotobiotic Facility grants (both to RBS) and Research Fellowship Award (to YM and AO). JEW was supported by a postdoctoral fellowship from the American Cancer Society (PF-13-401-01-TBE). The UNC Flow Cytometry Core is supported in part by Cancer Center Core Support Grant P30 CA016086 to the UNC Lineberger Comprehensive Cancer Center. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The views expressed herein do not necessarily represent the views of the Bill & Melinda Gates Foundation. The authors acknowledge the editorial assistance of Richard Youngblood of the UNC Translational and Clinical Sciences Institute.
Version 1. 06/18/2019
In-Press Preview
Version 2. 08/05/2019
Electronic publication
Version 3. 09/03/2019
Print issue publication
Funding Statement
The UNC Flow Cytometry core is supported in part by P30 CA016086 Cancer Center Core Support Grant to the UNC Lineberger Comprehensive Cancer Center.
Footnotes
CLK’s present address is: Global Health Discovery & Translational Sciences and Maternal, Neonatal and Child Health Discovery and Tools, The Bill & Melinda Gates Foundation, Seattle, Washington, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(9):3702–3716.https://doi.org/10.1172/JCI93820.
Contributor Information
Yoshiyuki Mishima, Email: mtmtyui@med.shimane-u.ac.jp.
Akihiko Oka, Email: akihiko.oka@gmail.com.
Bo Liu, Email: bo_liu@med.unc.edu.
Jeremy W. Herzog, Email: jeremy_herzog@med.unc.edu.
Chang Soo Eun, Email: cseun@hanyang.ac.kr.
Ting-Jia Fan, Email: tjfan@gustoglobal.com.
Emily Bulik-Sullivan, Email: ebsulliv@med.unc.edu.
Ian M. Carroll, Email: ian_carroll@med.unc.edu.
Jonathan J. Hansen, Email: jonathan_hansen@med.unc.edu.
Liang Chen, Email: chen.liang@abbvie.com.
Justin E. Wilson, Email: wilsonje@email.arizona.edu.
Nancy C. Fisher, Email: nancy_fisher@med.unc.edu.
Tomonori Nochi, Email: nochi@m.tohoku.ac.jp.
Angela Wahl, Email: angela_wahl@med.unc.edu.
J. Victor Garcia, Email: victor_garcia@med.unc.edu.
Christopher L. Karp, Email: Chris.Karp@gatesfoundation.org.
R. Balfour Sartor, Email: ryan_balfour_sartor@med.unc.edu.
References
- 1.Harris KG, Chang EB. The intestinal microbiota in the pathogenesis of inflammatory bowel diseases: new insights into complex disease. Clin Sci. 2018;132(18):2013–2028. doi: 10.1042/CS20171110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kostic AD, Xavier RJ, Gevers D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology. 2014;146(6):1489–1499. doi: 10.1053/j.gastro.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sartor RB, Wu GD. Roles for intestinal bacteria, viruses, and fungi in pathogenesis of inflammatory bowel diseases and therapeutic approaches. Gastroenterology. 2017;152(2):327–339.e4. doi: 10.1053/j.gastro.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berg DJ, et al. Enterocolitis and colon cancer in interleukin-10-deficient mice are associated with aberrant cytokine production and CD4(+) TH1-like responses. J Clin Invest. 1996;98(4):1010–1020. doi: 10.1172/JCI118861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sellon RK, et al. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect Immun. 1998;66(11):5224–5231. doi: 10.1128/iai.66.11.5224-5231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rath HC, et al. Normal luminal bacteria, especially Bacteroides species, mediate chronic colitis, gastritis, and arthritis in HLA-B27/human beta2 microglobulin transgenic rats. J Clin Invest. 1996;98(4):945–953. doi: 10.1172/JCI118878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ivanov, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139(3):485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Atarashi K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500(7461):232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 9.Round JL, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332(6032):974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yen D, et al. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. J Clin Invest. 2006;116(5):1310–1316. doi: 10.1172/JCI21404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steidler L, et al. Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science. 2000;289(5483):1352–1355. doi: 10.1126/science.289.5483.1352. [DOI] [PubMed] [Google Scholar]
- 12.Herfarth HH, Mohanty SP, Rath HC, Tonkonogy S, Sartor RB. Interleukin 10 suppresses experimental chronic, granulomatous inflammation induced by bacterial cell wall polymers. Gut. 1996;39(6):836–845. doi: 10.1136/gut.39.6.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu C, Sartor RB, Huang K, Tonkonogy SL. Transient activation of mucosal effector immune responses by resident intestinal bacteria in normal hosts is regulated by interleukin-10 signalling. Immunology. 2016;148(3):304–314. doi: 10.1111/imm.12612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franke A, et al. Sequence variants in IL10, ARPC2 and multiple other loci contribute to ulcerative colitis susceptibility. Nat Genet. 2008;40(11):1319–1323. doi: 10.1038/ng.221. [DOI] [PubMed] [Google Scholar]
- 15.Amre DK, et al. Interleukin 10 (IL-10) gene variants and susceptibility for paediatric onset Crohn’s disease. Aliment Pharmacol Ther. 2009;29(9):1025–1031. doi: 10.1111/j.1365-2036.2009.03953.x. [DOI] [PubMed] [Google Scholar]
- 16.Glocker EO, et al. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N Engl J Med. 2009;361(21):2033–2045. doi: 10.1056/NEJMoa0907206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sabat R, et al. Biology of interleukin-10. Cytokine Growth Factor Rev. 2010;21(5):331–344. doi: 10.1016/j.cytogfr.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 18.Roers A, et al. T cell-specific inactivation of the interleukin 10 gene in mice results in enhanced T cell responses but normal innate responses to lipopolysaccharide or skin irritation. J Exp Med. 2004;200(10):1289–1297. doi: 10.1084/jem.20041789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Groux H, Powrie F. Regulatory T cells and inflammatory bowel disease. Immunol Today. 1999;20(10):442–445. doi: 10.1016/S0167-5699(99)01510-8. [DOI] [PubMed] [Google Scholar]
- 20.Brockmann L, et al. Molecular and functional heterogeneity of IL-10-producing CD4. Nat Commun. 2018;9(1):5457. doi: 10.1038/s41467-018-07581-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu B, Tonkonogy SL, Sartor RB. Antigen-presenting cell production of IL-10 inhibits T-helper 1 and 17 cell responses and suppresses colitis in mice. Gastroenterology. 2011;141(2):653–662, 662.e1. doi: 10.1053/j.gastro.2011.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shouval DS, et al. Interleukin-10 receptor signaling in innate immune cells regulates mucosal immune tolerance and anti-inflammatory macrophage function. Immunity. 2014;40(5):706–719. doi: 10.1016/j.immuni.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mishima Y, Liu B, Hansen JJ, Sartor RB. Resident bacteria-stimulated IL-10-secreting B cells ameliorate T cell-mediated colitis by inducing Tr-1 cells that require IL-27-signaling. Cell Mol Gastroenterol Hepatol. 2015;1(3):295–310. doi: 10.1016/j.jcmgh.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mishima Y, et al. Decreased production of interleukin-10 and transforming growth factor-β in Toll-like receptor-activated intestinal B cells in SAMP1/Yit mice. Immunology. 2010;131(4):473–487. doi: 10.1111/j.1365-2567.2010.03318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El Fassi D, Nielsen CH, Kjeldsen J, Clemmensen O, Hegedüs L. Ulcerative colitis following B lymphocyte depletion with rituximab in a patient with Graves’ disease. Gut. 2008;57(5):714–715. doi: 10.1136/gut.2007.138305. [DOI] [PubMed] [Google Scholar]
- 26.Goetz M, Atreya R, Ghalibafian M, Galle PR, Neurath MF. Exacerbation of ulcerative colitis after rituximab salvage therapy. Inflamm Bowel Dis. 2007;13(11):1365–1368. doi: 10.1002/ibd.20215. [DOI] [PubMed] [Google Scholar]
- 27.Mizoguchi A, Mizoguchi E, Takedatsu H, Blumberg RS, Bhan AK. Chronic intestinal inflammatory condition generates IL-10-producing regulatory B cell subset characterized by CD1d upregulation. Immunity. 2002;16(2):219–230. doi: 10.1016/S1074-7613(02)00274-1. [DOI] [PubMed] [Google Scholar]
- 28.Maseda D, et al. Peritoneal cavity regulatory B cells (B10 cells) modulate IFN-γ+CD4+ T cell numbers during colitis development in mice. J Immunol. 2013;191(5):2780–2795. doi: 10.4049/jimmunol.1300649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yanaba K, Bouaziz JD, Matsushita T, Tsubata T, Tedder TF. The development and function of regulatory B cells expressing IL-10 (B10 cells) requires antigen receptor diversity and TLR signals. J Immunol. 2009;182(12):7459–7472. doi: 10.4049/jimmunol.0900270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosser EC, et al. Regulatory B cells are induced by gut microbiota-driven interleukin-1β and interleukin-6 production. Nat Med. 2014;20(11):1334–1339. doi: 10.1038/nm.3680. [DOI] [PubMed] [Google Scholar]
- 31.Saraiva M, O’Garra A. The regulation of IL-10 production by immune cells. Nat Rev Immunol. 2010;10(3):170–181. doi: 10.1038/nri2711. [DOI] [PubMed] [Google Scholar]
- 32.Madan R, et al. Nonredundant roles for B cell-derived IL-10 in immune counter-regulation. J Immunol. 2009;183(4):2312–2320. doi: 10.4049/jimmunol.0900185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lochner M, et al. In vivo equilibrium of proinflammatory IL-17+ and regulatory IL-10+ Foxp3+ RORgamma t+ T cells. J Exp Med. 2008;205(6):1381–1393. doi: 10.1084/jem.20080034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang BH, et al. Foxp3(+) T cells expressing RORγt represent a stable regulatory T-cell effector lineage with enhanced suppressive capacity during intestinal inflammation. Mucosal Immunol. 2016;9(2):444–457. doi: 10.1038/mi.2015.74. [DOI] [PubMed] [Google Scholar]
- 35.Mauri C, Menon M. The expanding family of regulatory B cells. Int Immunol. 2015;27(10):479–486. doi: 10.1093/intimm/dxv038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eun CS, et al. Induction of bacterial antigen-specific colitis by a simplified human microbiota consortium in gnotobiotic interleukin-10-/- mice. Infect Immun. 2014;82(6):2239–2246. doi: 10.1128/IAI.01513-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15(3):382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin M, Schifferle RE, Cuesta N, Vogel SN, Katz J, Michalek SM. Role of the phosphatidylinositol 3 kinase-Akt pathway in the regulation of IL-10 and IL-12 by Porphyromonas gingivalis lipopolysaccharide. J Immunol. 2003;171(2):717–725. doi: 10.4049/jimmunol.171.2.717. [DOI] [PubMed] [Google Scholar]
- 39.Steinbach EC, et al. Innate PI3K p110δ regulates Th1/Th17 development and microbiota-dependent colitis. J Immunol. 2014;192(8):3958–3968. doi: 10.4049/jimmunol.1301533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fagundes CT, et al. Transient TLR activation restores inflammatory response and ability to control pulmonary bacterial infection in germfree mice. J Immunol. 2012;188(3):1411–1420. doi: 10.4049/jimmunol.1101682. [DOI] [PubMed] [Google Scholar]
- 41.Kitajima S, Morimoto M, Sagara E, Shimizu C, Ikeda Y. Dextran sodium sulfate-induced colitis in germ-free IQI/Jic mice. Exp Anim. 2001;50(5):387–395. doi: 10.1538/expanim.50.387. [DOI] [PubMed] [Google Scholar]
- 42.Hegazy AN, et al. Circulating and tissue-resident CD4. Gastroenterology. 2017;153(5):1320–1337.e16. doi: 10.1053/j.gastro.2017.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sydora BC, MacFarlane SM, Lupicki M, Dmytrash AL, Dieleman LA, Fedorak RN. An imbalance in mucosal cytokine profile causes transient intestinal inflammation following an animal’s first exposure to faecal bacteria and antigens. Clin Exp Immunol. 2010;161(1):187–196. doi: 10.1111/j.1365-2249.2010.04140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muris AH, Damoiseaux J, Smolders J, Cohen Tervaert JW, Hupperts R, Thewissen M. Intracellular IL-10 detection in T cells by flowcytometry: the use of protein transport inhibitors revisited. J Immunol Methods. 2012;381(1-2):59–65. doi: 10.1016/j.jim.2012.04.011. [DOI] [PubMed] [Google Scholar]
- 45.Kirkland D, et al. B cell-intrinsic MyD88 signaling prevents the lethal dissemination of commensal bacteria during colonic damage. Immunity. 2012;36(2):228–238. doi: 10.1016/j.immuni.2011.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jeon SG, et al. Probiotic Bifidobacterium breve induces IL-10-producing Tr1 cells in the colon. PLoS Pathog. 2012;8(5):e1002714. doi: 10.1371/journal.ppat.1002714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sayi A, et al. TLR-2-activated B cells suppress Helicobacter-induced preneoplastic gastric immunopathology by inducing T regulatory-1 cells. J Immunol. 2011;186(2):878–890. doi: 10.4049/jimmunol.1002269. [DOI] [PubMed] [Google Scholar]
- 48.Liang Y, et al. Toll-like receptor 2 induces mucosal homing receptor expression and IgA production by human B cells. Clin Immunol. 2011;138(1):33–40. doi: 10.1016/j.clim.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 49.Mohamadzadeh M, et al. Regulation of induced colonic inflammation by Lactobacillus acidophilus deficient in lipoteichoic acid. Proc Natl Acad Sci U S A. 2011;108(suppl 1):4623–4630. doi: 10.1073/pnas.1005066107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shen Y, et al. Toll-like receptor 2- and MyD88-dependent phosphatidylinositol 3-kinase and Rac1 activation facilitates the phagocytosis of Listeria monocytogenes by murine macrophages. Infect Immun. 2010;78(6):2857–2867. doi: 10.1128/IAI.01138-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Britton GJ, et al. Microbiotas from humans with inflammatory bowel disease alter the balance of Gut Th17 and RORγt. Immunity. 2019;50(1):212–224.e4. doi: 10.1016/j.immuni.2018.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Furusawa Y, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504(7480):446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 53.Cong Y, Weaver CT, Lazenby A, Elson CO. Bacterial-reactive T regulatory cells inhibit pathogenic immune responses to the enteric flora. J Immunol. 2002;169(11):6112–6119. doi: 10.4049/jimmunol.169.11.6112. [DOI] [PubMed] [Google Scholar]
- 54.Liu B, et al. Irgm1-deficient mice exhibit Paneth cell abnormalities and increased susceptibility to acute intestinal inflammation. Am J Physiol Gastrointest Liver Physiol. 2013;305(8):G573–G584. doi: 10.1152/ajpgi.00071.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilson JE, et al. Inflammasome-independent role of AIM2 in suppressing colon tumorigenesis via DNA-PK and Akt. Nat Med. 2015;21(8):906–913. doi: 10.1038/nm.3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim SC, et al. Variable phenotypes of enterocolitis in interleukin 10-deficient mice monoassociated with two different commensal bacteria. Gastroenterology. 2005;128(4):891–906. doi: 10.1053/j.gastro.2005.02.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.